Abstract
Background
Aloe vera supports a substantial global trade yet its wild origins, and explanations for its popularity over 500 related Aloe species in one of the world’s largest succulent groups, have remained uncertain. We developed an explicit phylogenetic framework to explore links between the rich traditions of medicinal use and leaf succulence in aloes.
Results
The phylogenetic hypothesis clarifies the origins of Aloe vera to the Arabian Peninsula at the northernmost limits of the range for aloes. The genus Aloe originated in southern Africa ~16 million years ago and underwent two major radiations driven by different speciation processes, giving rise to the extraordinary diversity known today. Large, succulent leaves typical of medicinal aloes arose during the most recent diversification ~10 million years ago and are strongly correlated to the phylogeny and to the likelihood of a species being used for medicine. A significant, albeit weak, phylogenetic signal is evident in the medicinal uses of aloes, suggesting that the properties for which they are valued do not occur randomly across the branches of the phylogenetic tree.
Conclusions
Phylogenetic investigation of plant use and leaf succulence among aloes has yielded new explanations for the extraordinary market dominance of Aloe vera. The industry preference for Aloe vera appears to be due to its proximity to important historic trade routes, and early introduction to trade and cultivation. Well-developed succulent leaf mesophyll tissue, an adaptive feature that likely contributed to the ecological success of the genus Aloe, is the main predictor for medicinal use among Aloe species, whereas evolutionary loss of succulence tends to be associated with losses of medicinal use. Phylogenetic analyses of plant use offer potential to understand patterns in the value of global plant diversity.
Electronic supplementary material
The online version of this article (doi:10.1186/s12862-015-0291-7) contains supplementary material, which is available to authorized users.
Keywords: Aloe vera, Evolution, Biogeography, Phylogeny, Medicinal use, Succulent plants
Background
The succulent leaf tissue of Aloe vera is a globally important commodity, with an estimated annual market of $13 billion [1]. The ‘gel’ tissue—polysaccharide-rich inner leaf mesophyll—provides a reservoir of water to sustain photosynthesis during droughts, and has been ascribed multiple bioactive properties associated with its use for skincare and digestive health [2]. Aloe vera has supported a thriving trade for thousands of years [3] and is arguably one of the most popular plants known in cultivation today, yet its origins in the wild have long been speculated. We have established that at least 25% of aloes (~120 species) are used for medicine yet fewer than 10 Aloe species are traded commercially, and these are used primarily for the purgative leaf exudate and on much lesser scales than Aloe vera (e.g. Aloe ferox in South Africa and Aloe arborescens in Asia) [4]. The immense market dominance of Aloe vera over other species of Aloe is not fully explained by available phytochemical evidence [5,6]. The extent to which the value of Aloe vera may be a consequence of evolutionary processes of selection and speciation, resulting in apparently unique properties and phylogenetic isolation, has not previously been considered.
Aloe (>500 species) is by far the most speciose of the six genera known collectively as aloes, which include Aloiampelos (7 species), Aloidendron (6 species), Aristaloe (1 species), Gonialoe (3 species) and Kumara (2 species). They are iconic in the African flora, and occur predominantly in eastern sub-Saharan Africa, and on the Arabian Peninsula, Madagascar and western Indian Ocean islands. Succulent plants are usually associated with arid environments; although numerous aloes occur in the drylands of Africa, they are also abundantly represented in tropical and subtropical vegetation infrequently impacted by drought. All aloes possess some degree of leaf succulence, as well as crassulacean acid metabolism (CAM) and a thick, waxy cuticle common in plants exhibiting a succulent syndrome [7]. Most are habitat specialists with narrow ranges and extraordinary rates of endemism, from an estimated 70% in southern Africa, 90% in Ethiopia, to 100% on Madagascar [8]. These centres of diversity coincide alarmingly with Africa’s biodiversity Hotspots, where a highly endemic biota is under substantial threat of extinction [9]. Risks posed by extensive habitat destruction and other threats to their survival are reflected by the inclusion of all aloes, except Aloe vera, in the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES). The species-level diversity, ecological importance and threats to aloes place them among the world’s most important succulent plant lineages, other examples of which are ice plants (Aizoaceae), cacti (Cactaceae) and Agave (Agavaceae) [10]. Phylogenetic studies of related groups have focussed on the South African endemic Haworthia (e.g. [11,12]), whereas aloes have received little attention (but see [13]), and the origins and diversification of Aloe have remained unclear. It has therefore not been possible to determine whether Aloe vera is phylogenetically distinct from its many relatives, nor whether such phylogenetic distance may account for any potentially unique properties underpinning the value of the succulent leaf tissue.
Phylogenetic prediction is emerging as a promising tool for exploring correlations between the phylogenetic diversity and useful attributes of medicinal plants [14-17]. Rich biocultural traditions surround the use of aloe leaves for medicine, cosmetics, digestive health and general wellbeing [4]. Two natural products are derived from the leaves: carbohydrate-rich succulent leaf mesophyll tissue, applied topically to the skin or taken internally for digestion; and exudate, a liquid matrix high in phenolic compounds and most often used as a potent purgative, or in veterinary medicine (see [18]). The literature describing these uses is an untapped resource for understanding plant use in an evolutionary context, and in particular the extraordinary case of Aloe vera, which is used almost exclusively for its succulent leaf tissue. One point of interest is whether leaf succulence in aloes, which ranges from barely succulent in some species to very fleshy in others, could influence their use.
We aimed to explore the Aloe vera ‘phenomenon’ [5] by combining the largest ever phylogenetic hypothesis for the aloes with predictive methods. We used this to infer a scenario for their evolution, addressing a persistent gap in the understanding of global succulent plant diversity and biogeography. Links between the medicinal usefulness of aloes, their phylogenetic history, and extent of leaf succulence were evaluated by identifying evolutionary correlations and phylogenetic signal in uses and habit. Our comprehensive sampling represents the full morphological and geographical diversity of the aloes, and enabled the origins, geographical range evolution and divergence times of Aloe and relatives to be inferred. We synthesized our findings to determine whether the global value of Aloe vera can be better explained by evolutionary distinctiveness or by historical anthropogenic factors.
Methods
Phylogenetic hypothesis
A dataset was assembled representing seven plastid and nuclear DNA regions in 239 taxa in Xanthorrhoeaceae, including 197 species in the genera Aloe, Aloidendron, Aloiampelos, Aristaloe, Gonialoe and Kumara. We generated 480 new sequences from leaf or floral specimens collected from natural populations or from curated living collections and DNA banks held primarily at the Royal Botanic Gardens, Kew. A further 279 sequences were obtained from GenBank (ncbi.nlm.nih.gov/genbank/), including 93 rbcL and 64 psbA sequences. Agapanthus africanus (Amaryllidaceae) was used as the outgroup taxon in all analyses.
Total genomic DNA was isolated from fresh plant material (ca. 1 g) or specimens dried in silica gel (ca. 0.3 g) using a modified CTAB protocol [19] or the Qiagen DNeasy kit (Qiagen, Copenhagen). Sequences of ITS, matK and trnL-F were amplified using methodology previously described by [6]. The trnQ-rps16 region was amplified with the primers trnQ(UUG)Aloe (5′-ATCTTRATACAATGTGATCCAC-3′; this study) and rps16x1 [20]. Sequences from the complementary strands were obtained for all taxa whenever possible, using the BigDye Terminator v3.1 on a 3730 DNA Analyzer (Applied Biosystems/Hitachi). Sequences were assembled in Sequencher 4.8 (Gene Codes, Ann Arbor) and submitted to GenBank (Additional file 1). Sequences were aligned automatically using MUSCLE [21] implemented with default settings in SeaView v4.2.12 [22], and adjusted manually in BioEdit v7.1.11 [23]. The DNA regions were aligned separately before the data were concatenated using an R [24] script to produce a final dataset comprising 240 taxa and 6732 nucleotides in seven DNA regions.
We used Bayesian inference, maximum likelihood and parsimony to produce a phylogenetic hypothesis for Aloe and allied genera, using single-partition (ITS, matK, rps16, psbA, rbcL, trnL-F intron and spacer) and combined datasets. We ran all analyses on the Cyber Infrastructure for Phylogenetic Research (CIPRES) portal [25]. Separate parsimony analyses of the ITS (175 taxa, 799 nucleotides) and plastid (231 taxa, 5933 nucleotides) datasets were undertaken with the parsimony ratchet implemented in PAUPRat [26], to check for strongly supported phylogenetic conflicts (bootstrap percentages >75), before proceeding with analyses based on a total evidence approach using all characters. A maximum likelihood analysis, comprising 1000 bootstrap replicates followed by a heuristic tree search, was executed in RAxML [27] with each partition assigned specific parameters under the recommended GTRCAT model. An additional 530 gaps and indels in the combined dataset of all DNA regions were coded using the algorithm described by [28] in the FastGap v1.2 interface [29]. Finally, we ran a Bayesian analysis of the combined dataset with gaps coded in MrBayes v3.1.2 [30]. Best-fitting models for each data partition for Bayesian inference were identified using the Akaike Information Criterion calculated in Modeltest v3.8 [31]. The Hasegawa, Kishino and Yano (HKY) model with gamma-shaped distribution of rate heterogeneity among sites (HKY + G) was selected for the ITS, matK, trnQ-rps16 and trnL-F data partitions, while the General Time Reversible (GTR) model with gamma distribution of rate heterogeneity among sites was selected for psbA (GTR + G), and with a proportion of invariable sites (GTR + I + G) for rbcL. For the Bayesian analysis, the parameters were unlinked between loci and four Metropolis Coupled Markov Chains with heating increments of 0.2 were run for 50 million generations and sampled every 1000th generation. The resulting parameters were summarised in Tracer 1.5.0 [32]. A quarter of the least likely trees were discarded, and a majority rule consensus tree with branch supports expressed as posterior probabilities (PP) was produced from the remaining trees.
Divergence time estimates and biogeographic scenario
Divergence times were estimated using a penalised likelihood (PL) approach previously applied in Hyacinthaceae, a related family in Asparagales, as described by [33]. In the absence of fossil data for aloes and related genera, analyses were constrained to the mean age of 34.2 Ma inferred for the crown node of Asphodeloideae in a recent study of all Asparagales families [34]. Due to the computational demands of analyses on the full Xanthorrhoeaceae dataset and our focus on the aloes (Aloe, Aloiampelos, Aloidendron, Aristaloe, Gonialoe and Kumara), we excluded subfamilies Xanthorrhoeoideae and Hemerocallidoideae from subsequent analyses and pruned the Bayesian consensus tree to 228 species in Asphodeloideae. The penalised likelihood method [35] was run on 1000 randomly selected trees from the Bayesian stationary distribution and summarised on the consensus tree [33]. The optimal rate smoothing value for this dataset was determined by cross validation on the pruned Bayesian consensus tree, using the Truncated Newton algorithm (S = 5) implemented in r8s v 1.8 [36]. The outgroup taxon was pruned prior to the estimation of divergence times, as required by r8s. Mean age values and 95% confidence intervals for the nodes on the Bayesian consensus tree were computed in TreeAnnotator [37].
A biogeographic scenario for the aloes was inferred using the dispersal-extinction-cladogenesis (DEC) likelihood model implemented in Lagrange v2.0.1 [38]. Species distribution data were compiled from authoritative checklists for Asphodeloideae [39,40] and standardised according to the Taxonomic Data Working Group (TDWG) guidelines [41]. We defined eight areas based on the statistically-delimited biogeographical regions of Africa, incorporating the faunal and floral diversity of the continent, described recently by [42]. For subfamily Asphodeloideae, Arabia, Madagascar and Eurasia were added to the Southern African, Zambezian and Congolian regions, together with expanded Ethiopian-Somalian and Saharan-Sudanian regions. Assigning species to areas was straightforward due to the typically narrow distribution of most Aloe species, and because neighbouring areas are separated by physical barriers or marked differences in climatic conditions. Ancestral area reconstructions in Lagrange [38] were performed on the dated consensus (allcompat) tree obtained from the penalised likelihood analysis. In brief, ancestral areas were computed at each node of the tree under the DEC likelihood model, following a method described in detail by [33]. Ancestral areas with a relative probability >1 were combined with the node age and lengths of the descendent branches on the tree to infer the frequency and nature of transition events between ancestral and descendant nodes [33]. The resulting biogeographic scenario was visualised on the dated Bayesian consensus tree using pie charts showing the likelihoods of all possible ancestral areas per node for subfamily Asphodeloideae.
Phylogenetic signal in utility and habit
We interrogated a dataset of over 1400 use records from the literature [18] to investigate phylogenetic signal in the uses of aloes. Data were coded according to the Economic Botany Data Standard [43] from which two categories of use were considered. In the first category, we combined all TDWG Level 1 data to yield a discrete binary character describing any documented use, while the second comprised data in the TDWG Level 2 Medicines category. General use (e.g. for food, materials, social purposes, etc.) and medicinal use specifically were scored as present (=1) or absent (=0) in each of the terminal taxa. Records describing a plant as not used are unusual in the ethnobotanical literature, and hence in all cases 0 indicated a lack of reported use, rather than definitive knowledge of no use. The consensus (allcompat) tree inferred by Bayesian analysis with gaps coded was pruned to 197 species representing Aloe, Aloiampelos, Aloidendron, Aristaloe, Gonialoe and Kumara.
We calculated phylogenetic signal using the D metric [44], a measure specifically developed for quantifying phylogenetic signal in binary characters, implemented in the R package caper [45]. D compares the number of observed changes in a trait over a phylogeny with the number that would be expected under two alternative simulated scenarios: one where there is strong phylogenetic dependence and the trait has evolved via a gradual Brownian motion model of evolution, and the second where there is no phylogenetic dependence and the trait is randomly scattered across the species, regardless of phylogeny. The D metric generates a value that usually lies between 0 and 1, where a value of 1 indicates that the trait has evolved in essentially a random manner (i.e. no phylogenetic signal), and 0 indicates that the trait is highly correlated with phylogeny, in a manner predicted by Brownian motion. Tests for significant differences from D = 1 (no phylogenetic signal) are derived by simulating the random distribution of the trait among species 1000 times to generate a null distribution for the D statistic. We conducted the analysis in two ways, one using just the consensus phylogeny, and the second using 1000 trees selected at random from the Bayesian posterior distribution calculating median values for D and associated P values.
The putative contribution of leaf succulence to the ‘usefulness’ of aloes was explored using a phylogenetic comparative approach. A character set describing the extent of water-storing mesophyll tissue in the leaves was assembled from species descriptions [46-48] and observations of leaf morphology in aloes. Species were broadly scored as ‘succulent’ or ‘barely succulent’ and additionally classified as barely succulent shrubs (the grass aloes, Aloe section Leptaloe), succulent shrubs (Aristaloe, Gonialoe and most of Aloe), branching trees (Aloidendron, Kumara) and scrambling shrubs with variably succulent leaves (Aloiampelos). These were visualised on the Bayesian consensus tree by reconstructing the ancestral states of three characters (succulence, habit and medicinal use), scored as binary traits, under the parsimony optimisation in Mesquite [49].
For calculation of phylogenetic signal using the D metric in these traits, they were coded as four separate dummy binary variables (e.g. succulence: 0 = no, 1 = yes). Pairwise comparison tests [50] were used to assess possible evolutionary correlations between habit and documented uses generally and medicinal uses specifically (dependent variables). This method takes phylogenetically independent pairs of species and observes any correlated differences in the states of two binary characters. For every gain or loss in one character (in this case, the measure of leaf succulence), it assesses whether there is an associated loss, gain or no change in the other (medicinal or general use), and compares any patterns with those expected if the second character were randomly distributed on the phylogeny. Pairwise comparison calculations were carried out using Mesquite [49]. As with our D metric calculations, to account for uncertainty in the phylogenetic topology and weak branch supports, we ran all the analyses on the Bayesian consensus (allcompat) topology (using 100 randomly selected sets of pairwise comparisons) and on a random sample of 1000 trees from the Bayesian posterior distribution, calculating median probability values associated with the correlation.
Results
Phylogenetic hypothesis
Our phylogenetic analyses of >7 kb plastid and nuclear characters (6732 nucleotides and 550 gaps) in ca. 40% of Aloe species substantiate current understanding of taxonomic relationships in Xanthorrhoeaceae subfamily Asphodeloideae [11,12,51] and divergence times within Asparagales [33] (Figure 1, Additional files 2 and 3). We sampled 26 genera and 240 species in Xanthorrhoeaceae, using a total evidence approach despite sequence data for some taxa being incomplete (Additional files 1 and 4). The effects of missing data on phylogenetic analyses have been widely debated, but there is convincing evidence for the accurate phylogenetic placement of taxa with considerable missing data (summarised by [52]). Model-based methods of phylogenetic inference perform better than parsimony in estimating trees from datasets with missing data [53,54], and we therefore based subsequent analyses on the Bayesian phylogenetic inference (Additional file 3). Low levels of genetic polymorphisms, taxonomic complexities, and the number of inaccessible, narrowly distributed species challenge the study of aloes; this is the first phylogeny to include >10% of Aloe species. Parsimony and maximum likelihood topologies (trees not shown) compared well to the Bayesian tree used in downstream analyses. Branching tree aloes (Aloidendron) are basal to the remainder of the alooids. A clade comprising the Cape endemic genus Kumara and Haworthia s.s. is sister to Aloiampelos, which is in turn sister to Aloe. Within the large Aloe clade (184 species), well-supported terminal branches highlight species-level relationships but the clades, which will ultimately underpin a taxonomic revision, are incompletely resolved. The placement of Aloiampelos juddii at the base of the alooid topology, on a branch sister to Kumara-Haworthia, warrants further investigation of reciprocal monophyly in Aloiampelos. We included four members of Astroloba, two Tulista, three Haworthiopsis and four Haworthia in our study and recovered these as paraphyletic with varying support. The haworthioid taxa were, until recently [12], phylogenetically problematic (e.g. [11]).
Figure 1.
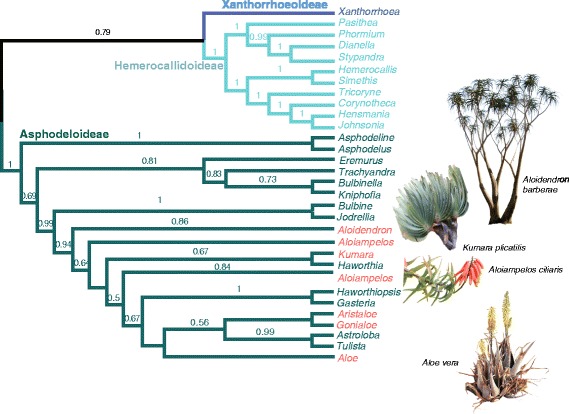
Subfamilies and genera of Xanthorrhoeaceae. Summary phylogram with Bayesian posterior probabilities (>0.5) above branches; red branches represent the six genera known collectively as aloes: Aloe, Aristaloe, Gonialoe, Kumara, Aloiampelos and Aloidendron.
Divergence time estimates and biogeographic scenario
Divergence times estimated using a penalised likelihood approach and ancestral area reconstructions revealed that aloes originated in southern Africa in the early Miocene, ~19 million years ago (Ma) (Additional file 3). Aloe vera was recovered in a strongly supported clade with eight other Arabian species, allowing us to infer its origins on the Arabian Peninsula within the last five million years. Two southern African species supporting commercial natural products industries, Aloe arborescens and A. ferox, were recovered together in a southern African clade. We estimate that the diversification of the genus Aloe began ~16 Ma in South Africa with a period of range expansion of ancestral taxa north-eastwards into the Zambezian and Ethiopian-Somalian regions ~10 Ma. Peripheral isolation and, to a lesser extent, vicariance were inferred to be the major speciation processes for the early diversification of aloes until around ~5 Ma, when a sharp increase in dispersal events occurred in several near-simultaneous radiations of the aloes at the extremities of their range, particularly in Madagascar. During this period, aloes reached West Africa, the Saharan-Sudanian region and the Arabian Peninsula via the Ethiopian-Somalian region, and arrived on Madagascar from the Zambezian region (Figure 2). This scenario identifies the Ethiopian-Somalian region as a cross-road for speciation processes in Aloe, as the majority of dispersal events (16 events) in our dataset were from here into each of the four adjacent regions. We identified multiple introductions to Madagascar (three dispersals). Similarly, diversification of aloes on the Arabian Peninsula resulted from one or more dispersals, as well as vicariance and peripheral isolation, with no evidence of dispersal back to continental Africa. A single southerly dispersal event was detected from the Zambezian to the Southern African regions.
Figure 2.
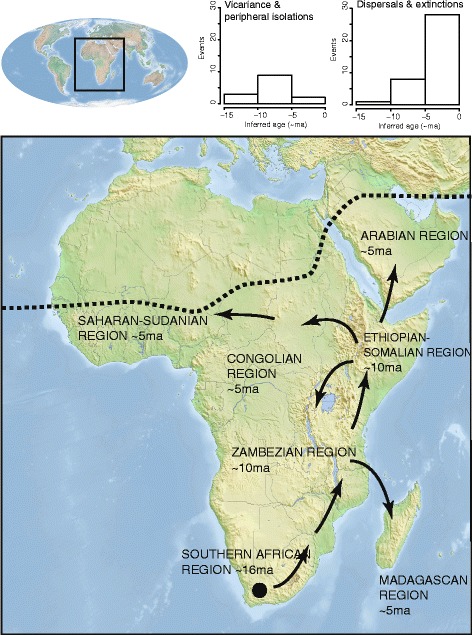
Biogeographic scenario for Aloe. Distribution and biogeographic scenario for Aloe inferred from nucleotide and plastid data for 228 taxa in Xanthorrhoeaceae subfamily Asphodeloideae. Enlarged map shows the natural distribution of Aloe, with northernmost limits indicated by dashed line. Direction and timing of diversification events inferred from ancestral state reconstruction and penalised likelihood dating are shown by arrows. Histograms show branch-based (dispersal and extinction) and node-based (vicariance and peripheral isolations) events in speciation processes since the divergence of the Aloe crown group ~16 Ma.
Phylogenetic signal in utility and habit
Leaf succulence increased steadily with the emergence of aloes in southern Africa, from the barely succulent tree aloes (Aloidendron and Kumara) and rambling aloes (Aloiampelos), to Aloe and neighbouring genera (Figures 1, 3 and 4). Though difficult to quantify, pronounced succulence is restricted to Aloe, and has been almost completely lost in several members of this genus, notably in the clade comprising southern African grass aloes, Aloe section Leptaloe, during the last ~10 Ma (Figure 3; Additional file 5 a-c). The habit of relatively large, succulent leaves borne in basal rosettes on an unbranched stem, typical of Aloe vera and other commercially valuable species, exhibits a strong phylogenetic signal. Using Fritz & Purvis’s D-metric [44] as our measure of phylogenetic signal, where D = 1 indicates no phylogenetic structure to the trait data and D = 0 indicates strong correlation between trait distribution and phylogeny (see methods for full description), we found the degree of phylogenetic signal in succulence per se was highly significant (D = 0.132, p < 0.001).
Figure 3.
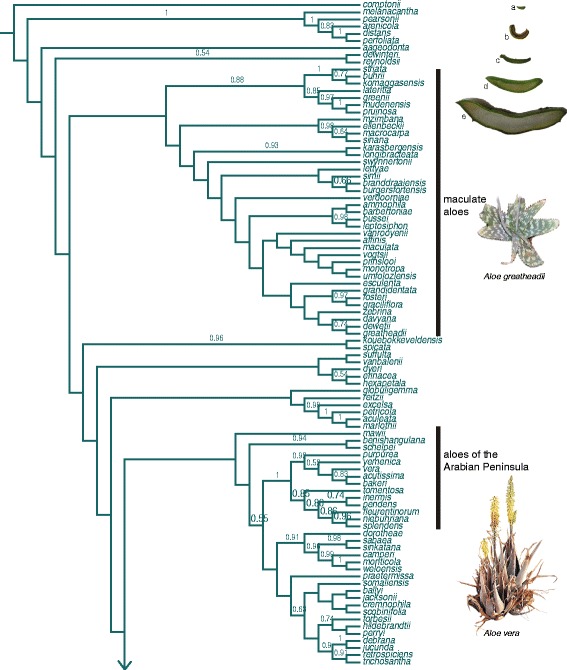
Bayesian consensus tree for Aloe. Core Aloe clade from a Bayesian analysis of Xanthorrhoeaceae highlighting relationships of interest in the biogeographical scenario. Inset shows representative variation in the extent of leaf succulence among aloes: a, Aloiampelos ciliaris; b, Aloidendron eminens; c, Kumara plicatilis; d, Aloe vera; e, Aloe marlothii.
Figure 4.
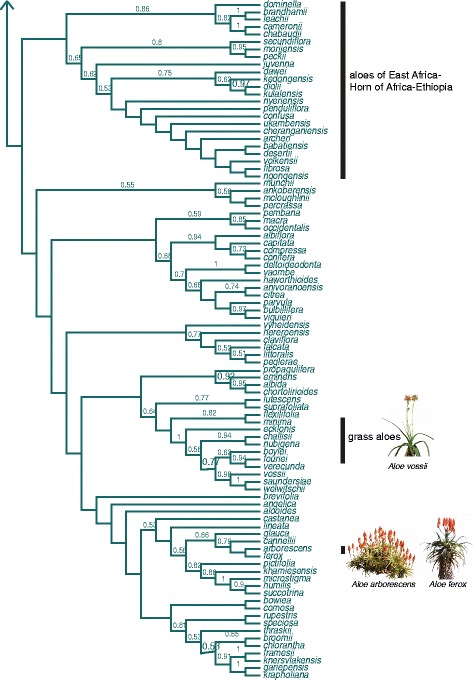
Bayesian consensus tree for Aloe (continued from Figure 3 ).
Uses are documented for 48% of the aloes sampled in this study. Of the 81 Aloe species in our analysis that have documented medicinal use, 98% have succulent leaves. By contrast, in 87% of the 15 species in which succulent leaf mesophyll has been almost entirely lost, there is negligible documented tradition of medicinal use, even in regions with thoroughly documented ethnoflora, such as South Africa. Whilst many succulent-leaved aloes do not have known medicinal uses, the likelihood of use is significantly higher in the succulent aloes (Fisher’s exact test comparing proportions of succulent vs. non-succulent species with medicinal utility, prior to considering phylogenetic effects: p = 0.014). Our pairwise comparison analyses indicated that there most likely have been six evolutionary losses of leaf succulence in aloes, as predicted from the Bayesian consensus phylogeny and 884 of the 1000 Bayesian posterior distribution trees. With the consensus tree, we tested the hypothesis that the use of an aloe for medicine diminishes or is lost entirely with a reduction in leaf succulence. Four of the six evolutionary transitions in aloes where succulence is severely reduced are associated with loss of medicinal use, providing weakly significant support for the hypothesis (pairwise comparison test: p = 0.065). The same analysis using 1000 trees sampled randomly from the Bayesian posterior distribution, was unable to resolve clearly whether four or three transitions in aloe leaf succulence were associated with loss of medicinal use (p = 0.125). We detected a weak phylogenetic signal in the general use of these genera (D = 0.828, p = 0.063). Focussing on the use of aloes for medicine, we also identified a weak, but significant, phylogenetic signal (D = 0.794, p = 0.029).
Species with documented medicinal use are not randomly distributed across the phylogeny. In a large clade comprising 29 species of maculate aloes, characterised by large, succulent leaves and a short stem, 55% are used for medicine. A clade of 18 closely related species native to East Africa, Ethiopia and the Horn of Africa (the Zambezian and Ethiopian-Somalian biogeographic regions) included 27% species with known medicinal uses. Aloe vera was among three medicinal species in a clade of eight species native to the Arabian Peninsula.
Discussion
The phylogenetic hypothesis for the aloes reveals a distinctive geographical pattern of major clades of Aloe with four biogeographical centres of diversity: Southern Africa (~170 species), Madagascar (~120 species), East Africa/Zambezian region (~100 species), and the Horn of Africa/Ethiopian-Somalian region (~90 species). Based on strongly supported close relationships with morphologically similar Arabian species, we clarify that Aloe vera is native to the Arabian Peninsula. Previous suggestions have included Sudan or the Arabian Peninsula, based on morphological affinities with Arabian species [55] and even further afield in the Canary Islands, Cape Verde Islands, Madeira or Spain [3], which could be explained by naturalised populations, introduced via ancient trade routes, being mistaken for indigenous elements of the flora. Our explicitly phylogenetic context places Aloe vera for the first time among related Arabian species at the northernmost natural range limit of aloes, in habitats at the extremes for aloes in terms of aridity and diurnal temperature fluctuations. Here, at the hot and dry edge of their natural range, aloes are characterised by leathery, glaucous leaves that likely protect the water-storing leaf mesophyll from diurnal temperature and radiation extremes. The evolutionary distinctiveness of aloes on the Arabian Peninsula which could account for atypical properties in Aloe vera, is thrown into question by their affinities with species in the Ethiopian-Somalian region [13,56]. We found evidence for at least one dispersal from the Ethiopian-Somalian region to the Arabian Peninsula within the last 5 Ma. The biogeographic scenario inferred here (Figure 2) elucidates the diversification of Aloe prior to its arrival in north-east Africa and the Arabian Peninsula, and reveals a southern African cradle for the genus ~16 Ma, in the early Miocene. Consequently, the longstanding hypothesis that aloes first appeared in southeast Africa considerably earlier, in the late Mesozoic-early Cenozoic [56] is contradicted by the molecular evidence in the present study.
The establishment of the Mediterranean climate in south-western Africa and the expansion of southern African deserts in the Miocene caused large-scale extinctions in the prevailing subtropical flora [57] and appear to have had a profound impact on the evolution of aloes. Habitat expansion has been proposed as the main driver for the simultaneous global diversification of plants with a succulent habit [10]. But on a local scale, loss of suitable habitat forced southern African aloes to migrate north-eastwards as these species struggled to adapt to bioclimatic changes at the southernmost tip of Africa. The establishment of the winter-rainfall region, in particular, appears to have largely excluded aloes from the semi-arid Succulent Karoo region, a celebrated global centre of succulent plant diversity with ~5,000 species and 40% endemism. The Succulent Karoo flora is characterised by short-lived, drought-sensitive dwarf and leaf-succulent shrubs [58] such as the ~1500 members of Aizoaceae subfamily Ruschioideae [59] and ~1000 species of Crassulaceae [60]. Aloes, in contrast, tend to be long-lived and drought tolerant, and are relatively poorly represented in the Succulent Karoo and winter-rainfall regions of southern Africa. Water-use efficiencies may have placed even the earliest, barely-succulent aloes at an ecological advantage over non-succulent lineages in the relictual subtropical vegetation.
The timing of two periods of diversification detected in aloes, in the late Miocene and more recently in the Pliocene, coincide remarkably with the simultaneous ‘burst’ of evolution in major succulent plant lineages globally, attributed to a rapid decline in atmospheric CO2 and increased aridity during the mid- to late Miocene [10]. Consistently low rates of extinction in our data agree with previous findings [56], suggesting continuous but irregular diversification of aloes. We identified a distinct shift from node-based speciation processes (namely, vicariance and peripheral isolations) to branch-based events (dispersals and extinctions) coincident with each of the radiations of the aloes. We interpret this as a period of range expansion and diversification of relatively widespread species until the Miocene-Pliocene boundary. The second rapid diversification was likely the result of species fragmentation and increased niche availability, when isolated taxa dispersed short distances into the rich habitat mosaics formed by geological processes during the Pliocene, giving rise to the present-day distribution of Aloe. This is evident in a five-fold increase in dispersal events in our dataset, while node-based processes and extinctions are low to negligible during the same period. The tempo of these pulsed radiations in Aloe is strikingly similar to that of Agave (Agavaceae), a New World group of ~200 species of leaf succulent rosette plants [61], adding new depth to this celebrated example of convergent evolution among succulents.
The evolution of leaf succulence followed the pattern of divergence in aloe and relatives, in tandem with the expansion of semi-arid habitats in Africa between ~ 10 and 5 Ma. Like the earliest cacti [62], ancestral aloes were barely succulent and tree-like. Large and markedly succulent leaves are restricted to the genus Aloe and, unlike other lineages in which succulence has arisen multiple times (e.g. Portulacineae [63] and Aizoaceae [59]), variation in the extent of leaf succulence among species of Aloe is due to loss of water-storing tissues (e.g. in the barely succulent grass aloes) (Figure 3). The idea that rich traditions of use in the aloes may be linked to the extent of leaf succulence has not been previously investigated, and our analyses suggest that a decrease in the proportion of water-storing leaf mesophyll reduces the possibility that a species is used for medicine, irrespective of whether the leaf mesophyll tissue and/or liquid exudate are used. Documented medicinal uses for barely succulent members of Aloidendron, Kumara and Aloiampelos focus on the roots or leaf exudate, and never the leaf mesophyll. Additionally, our phylogenetic reconstruction suggests that medicinal utility appears less likely in lineages where reduced succulence has evolved. For instance, we found very few documented medicinal uses for the barely-succulent grass aloes despite their relative abundance in regions with thoroughly documented ethnoflora, such as the fire-adapted grasslands of KwaZulu-Natal in South Africa. We detected weak, but significant, phylogenetic signals in the use of aloes generally, and for medicinal purposes specifically. A comparable study of the Amaryllidaceae, a family with well-characterised bioactive alkaloids, recovered a similar overall phylogenetic signal for medicinal use [15].
A link between leaf succulence and medicinal use suggests a traditionally pragmatic approach to the selection of aloes with large, succulent leaves for use in medicine [4]. Features such as firm leaf mesophyll, a short stem, small teeth on the leaf margins, and ease of propagation, are shared by Aloe vera and numerous other Aloe species used medicinally, including closely related species from the Arabian Peninsula and the Ethiopian-Somalian region. Our evolutionary hypothesis for Aloe locates Aloe vera in close phylogenetic proximity to seven other species native to the Arabian Peninsula, discounting a distinctive evolutionary history for Aloe vera which could imply unique leaf properties. Mounting anecdotal evidence for the beneficial properties of Aloe vera continues to stimulate research into the bioactivity of the succulent leaf mesophyll [2]. Recent studies of Aloe vera and a phylogenetically-representative sampling of nearly 30 Aloe species have shown very low levels of variation in the monosaccharide composition of leaf mesophyll carbohydrates [6,64], although differences in carbohydrate structure may yet be discovered pending systematic evaluation of these highly complex carbohydrates which are assumed to be responsible for the medicinal value of the leaf mesophyll [5]. On the other hand, documented traditions of use indicate that few of the closest relatives of Aloe vera are used medicinally. Records of the therapeutic uses of Aloe vera leaf mesophyll and exudate date to classical times [3,5,65]. Trade routes for Aloe vera were well established in the Red Sea and Mediterranean by the 4th century BCE [3] and, assuming the species occupied a narrow range typical of Arabian aloes, it may have been rapidly harvested to near-extinction to meet market demands. The remarkable contemporary market dominance of Aloe vera over other aloes therefore appears to be the consequence of its origins near important early trade routes, ancient selection for medicine and cultural history, which introduced the species into trade and cultivation thousands of years ago.
Conclusion
Phylogenetic investigation of plant use and leaf succulence among aloes has yielded new explanations for the extraordinary market dominance of Aloe vera. The evolutionary history inferred from our analyses of Aloe and related genera shows for the first time that Aloe vera is native to the Arabian Peninsula, and discounts phylogenetic distance as an explanation for its popularity over many other species of Aloe. The industry preference for Aloe vera appears to be due to its proximity to important historic trade routes, and early introduction to trade and cultivation. Well-developed succulent leaf mesophyll tissue, an adaptive feature that likely contributed to the ecological success of the genus Aloe, is the main predictor for medicinal use among Aloe species, whereas evolutionary losses of succulence tend to be associated with losses of medicinal use. Phylogenetic analyses of plant use offer potential to understand patterns in the value of global plant diversity.
Data accessibility
DNA sequences are deposited in GenBank and accession numbers are listed in Additional file 1.
The phylogenetic tree supporting the results of this article is available in the TreeBase repository, http://purl.org/phylo/treebase/phylows/study/TB2:S16954?format=html [66].
Acknowledgements
The authors thank Martin Årseth Hansen, Eshetu Fentaw, Amra Dzajic, Halima Amir, Livhuwani Nkuna, Erich van Wyk, Walter Mabatha, Neil Crouch, Arrie Klopper, Anthony Miller and Abdul Wali al Khulaidi for help with plant collecting, maintaining living collections, and laboratory work. This study was supported by grants awarded to OMG and NR in the Marie Curie Actions of the 7th European Community Framework Programme (grant ALOEDIVERSITY PIEF-GA-2009-251766) and Brødrene Hartmanns Fond, Denmark.
Additional files
GenBank sequence data for taxa studied. Accession/collectors’ numbers and international codes for herbaria where vouchers are deposited are given for sequences from this study: C, Copenhagen; E, Edinburgh; ETH, Addis Ababa; K, Kew; O, Oslo; NBG, Compton; PRE, Pretoria; DNA signifies DNA bank accession; — signifies no sequence.
Phylogenetic hypothesis for Xanthorrhoeaceae. Bayesian consensus tree for 240 species of Xanthorrhoeaceae subfamilies Xanthorrhoeoideae, Hemerocallidoideae and Asphodeloideae, with posterior probabilities >0.5 displayed above branches.
Ancestral area reconstructions for Xanthorrhoeaceae subfamily Asphodeloideae. a) Ancestral areas displayed on the penalised likelihood-dated Bayesian consensus tree; b) detail of the clade containing Aloe vera. Legend refers to regions modified from [57] for this analysis: A, Southern Africa; B, Zambezi; C, Congolian; D, Ethiopian-Somalian; E, Saharan-Sudanian; F, Arabian; G, Madagascan; H, Eurasian; Trash, sum of ancestral area probabilities <0.1.
Summary statistics for phylogenetic dataset. Taxon sampling, sequence length and model selection for data partitions.
Phylogenetic distribution of leaf succulence, habit and medicinal use in alooid taxa. Most parsimonious reconstructions of character states mapped to Bayesian consensus tree in a) leaf succulence, b) habit and c) medicinal uses.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
OMG and NRØN conceived and designed the study. OMG assembled sequence data, conducted the phylogenetic analyses and interpreted the results. SB conducted the node age estimate and ancestral area reconstructions, and interpreted these with OMG and FF. MRES conducted the phylogenetic signal analyses, and interpreted the results with OMG and NRØN. AEvW and GFS helped to interpret the results of phylogenetic and biogeographical analyses. RRK, CSB, SN and SD participated in obtaining essential plant material and/ or sequences for phylogenetic analysis. MSJS facilitated the utility analysis. OMG, SB, MRES and NRØN prepared the manuscript. All authors read and approved the final manuscript.
Contributor Information
Olwen M Grace, Email: o.grace@kew.org.
Sven Buerki, Email: s.buerki@nhm.ac.uk.
Matthew RE Symonds, Email: matthew.symonds@deakin.edu.au.
Félix Forest, Email: f.forest@kew.org.
Abraham E van Wyk, Email: braam.vanwyk@up.ac.za.
Gideon F Smith, Email: G.Smith@sanbi.org.za.
Ronell R Klopper, Email: R.Klopper@sanbi.org.za.
Charlotte S Bjorå, Email: charlotte.bjora@nhm.uio.no.
Sophie Neale, Email: s.neale@cmep.org.uk.
Sebsebe Demissew, Email: sebseb.demissew@gmail.com.
Monique SJ Simmonds, Email: m.simmonds@kew.org.
Nina Rønsted, Email: nronsted@snm.ku.dk.
References
- 1.Schulz H. Global aloe market estimated at $13 billion. Available at http://www.nutraingredients-usa.com/Markets/Global-aloe-market-estimated-at-13-billion.
- 2.Reynolds T, Dweck AC. Aloe vera leaf gel, a review updated. J Ethnopharmacol. 1999;68:3–37. doi: 10.1016/S0378-8741(99)00085-9. [DOI] [PubMed] [Google Scholar]
- 3.Hodge WH. The drug aloes of commerce, with special reference to the Cape species. Econ Bot. 1953;7:99–129. doi: 10.1007/BF02863057. [DOI] [Google Scholar]
- 4.Grace OM. Current perspectives on the economic botany of the genus Aloe Xanthorrhoeaceae. S Afr J Bot. 2011;98:980–987. doi: 10.1016/j.sajb.2011.07.002. [DOI] [Google Scholar]
- 5.Grindlay D, Reynolds T. The Aloe vera phenomenon – a review of the properties and modern uses of the leaf parenchyma gel. J Ethnopharmacol. 1986;16:117–151. doi: 10.1016/0378-8741(86)90085-1. [DOI] [PubMed] [Google Scholar]
- 6.Grace OM, Dzajic A, Jager AK, Nyberg NT, Onder A, Rønsted N. Monosaccharide analysis of succulent leaf tissue in Aloe. Phytochem. 2013;93:79–87. doi: 10.1016/j.phytochem.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 7.Ogburn RM, Edwards EJ. The ecological water-use strategies of succulent plants. In: Kader J-C, Delseny M, editors. Advances in Botanical Research Vol. 55. Burlington: Academic Press; 2010. pp. 179–225. [Google Scholar]
- 8.Grace OM. Systematics and biocultural value of Aloe L. (Asphodelaceae). PhD Thesis. University of Pretoria; 2009.
- 9.Mittermeier RA, Gill PR, Hoffman M, Pilgrim J. Hotspots Revisited, Earth’s biologically richest and most threatened terrestrial ecoregions. Washington: Cemex & Conservation International; 2004. [Google Scholar]
- 10.Arakaki M, Christian P-A, Nyffeler R, Lendel A, Eggli U, Ogurn RM, Spriggs E, Moore MJ, Edwards EJ, et al. Contemporaneous and recent radiations of the world’s major succulent plant lineages. Proc Natl Acad Sci U S A. 2011;108:8379–8384. doi: 10.1073/pnas.1100628108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daru BH, Manning JC, Boatwright JS, Maurin O, Maclean N, Schaefer H, Kuzmina M, Van der Bank M, et al. Molecular and morphological analysis of subfamily Alooideae (Asphodelaceae) and the inclusion of Chortolirion in Aloe. Taxon. 2013;62:62–76. [Google Scholar]
- 12.Manning JC, Boatwrights JS, Daru BH, Maurin O, Van der Bank M. A molecular phylogeny and generic classification of Asphodelaceae subfamily Alooideae: a final resolution of the prickly issue of polyphyly in the alooids? Syst Bot. 2014;39:55–74. doi: 10.1600/036364414X678044. [DOI] [Google Scholar]
- 13.Treutlein J, Smith GF, Van Wyk B-E, Wink M. Phylogenetic relationships in Asphodelaceae subfamily Alooideae inferred from chloroplast DNA sequences (rbcL, matK) and from genomic fingerprinting (ISSR) Taxon. 2003;52:193–207. doi: 10.2307/3647389. [DOI] [Google Scholar]
- 14.Zhu F, Qin C, Tao L, Liu X, Ma X, Jia J, Tan Y, Cui C, Link J, Tan C, Jiang Y, Chen Y, et al. Clustered patterns of species origins of nature-derived drugs and clues for future prospecting. Proc Natl Acad Sci U S A. 2011;108:12943–12948. doi: 10.1073/pnas.1107336108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rønsted N, Symonds MRE, Birkholm T, Christensen SB, Meerow AW, Molander M, Mølgaard P, Petersen G, Rasmussen N, Van Staden J, Stafford GI, Jäger AK. Can phylogeny predict chemical diversity and potential medicinal activity of plants? A case study of Amaryllidaceae. BMC Evol Biol. 2012;12:182. doi: 10.1186/1471-2148-12-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saslis-Lagoudakis CH, Savolainen V, Williamson EM, Forest F, Wagstaff SJ, Baral SR, Watson MR, Pendry CA, Hawkins JA, et al. Phylogenies reveal predictive power of traditional medicine in bioprospecting. Proc Natl Acad Sci U S A. 2012;109:15835–15840. doi: 10.1073/pnas.1202242109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guénard G, Von der Ohe PC, Walker SC, Lek S, Legendre P. Using phylogenetic information and chemical properties to predict species tolerances to pesticides. Proc R Soc B. 2014;281:20133239. doi: 10.1098/rspb.2013.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grace OM, Simmonds MSJ, Smith GF, Van Wyk AE. Documented utility and biocultural value of Aloe L. Ec Bot. 2009;63:167–178. doi: 10.1007/s12231-009-9082-7. [DOI] [Google Scholar]
- 19.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- 20.Shaw J, Lickey EB, Schilling WEE, Small RL. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in Angiosperms, the tortoise and the hare III. Am J Bot. 1994;94:275–288. doi: 10.3732/ajb.94.3.275. [DOI] [PubMed] [Google Scholar]
- 21.Edgar RC. MUSCLE, multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gouy M, Guindon S, Gascuel O. SeaView version 4, a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 23.Hall TA. BioEdit, a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1991;41:95–98. [Google Scholar]
- 24.R Core Team. R: A Language and Environment for Statistical Computing. Available at http://www.r-project.org.
- 25.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop GCE. 14 November 2010, New Orleans, LA.; 2010.
- 26.Nixon KC. The Parsimony Ratchet, a new method for rapid parsimony analysis. Cladistics. 1999;15:407–414. doi: 10.1111/j.1096-0031.1999.tb00277.x. [DOI] [PubMed] [Google Scholar]
- 27.Stamatakis A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics; 2014. doi.10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed]
- 28.Simmons MP, Ochoterena H. Gaps as characters in sequence-based phylogenetic analyses. Syst Biol. 2000;49:369–381. doi: 10.1093/sysbio/49.2.369. [DOI] [PubMed] [Google Scholar]
- 29.Borchsenius F. FastGap 1.2. Department of Biosciences, Aarhus University, Denmark. Available at http://www.aubot.dk/FastGap_home.htm.
- 30.Ronquist F, Hulsenbeck JP. MRBAYES 3, Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1752–1754. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 31.Posada D, Crandall KA. Modeltest, testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 32.Rambaut A, Drummond AJ. Tracer v1.5. Available at http://beast.bio.ed.ac.uk/Tracer.
- 33.Buerki S, Jose S, Yadav SR, Goldblatt P, Manning JC, Forest F. Contrasting biogeographic and diversification patterns in two Mediterranean-type ecosystems. PLoS One. 2012;7:e39377. doi: 10.1371/journal.pone.0039377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen S, Kim D-K, Chase MW, Kim J-H. Networks in a large-scale phylogenetic analysis, reconstructing evolutionary history of Asparagales (Lilianeae). based on four plastid genes. PLoS One. 2013;8:e59472. doi: 10.1371/journal.pone.0059472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times. A penalized likelihood approach. Mol Biol Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- 36.Sanderson MJ. r8s version 1.8. Available at http://loco.biosci.arizona.edu/r8s/.
- 37.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ree RH, Smith SA. Maximum-likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst Biol. 2008;57:4–14. doi: 10.1080/10635150701883881. [DOI] [PubMed] [Google Scholar]
- 39.Newton LE. Aloe. In: Eggli U, editor. CITES Aloe and Pachypodium Checklist. Kew: Royal Botanic Gardens; 2001. pp. 121–160. [Google Scholar]
- 40.Newton LE, Govaerts R. World Checklist of Xanthorrhoeaceae. Facilitated by the Royal Botanic Gardens, Kew. Available at http://apps.kew.org/wcsp/.
- 41.Brummitt RK. World Geographical Scheme for Recording Plant Distributions, 2nd ed. Pittsburg: Carnegie Mellon University; 2001. [Google Scholar]
- 42.Linder HP, De Klerk HM, Born J, Burgess NF, Fjelsda J, Rahbek C. The partitioning of Africa, statistically defined biogeographical regions in sub-Saharan Africa. J Biogeog. 2012;39:1189–1205. doi: 10.1111/j.1365-2699.2012.02728.x. [DOI] [Google Scholar]
- 43.Cook FEM. Economic Botany Data Collection Standard. Key: Royal Botanic Gardens; 1995. [Google Scholar]
- 44.Fritz SA, Purvis A. Selectivity in mammalian extinction risk and threat types, a new measure of phylogenetic signal in binary traits. Conserv Biol. 2010;24:1042–1051. doi: 10.1111/j.1523-1739.2010.01455.x. [DOI] [PubMed] [Google Scholar]
- 45.Orme D. Caper, Comparative Analysis of Phylogenetics and Evolution in R. Available at http://cran.r-project.org/package=caper.
- 46.Reynolds GW. The Aloes of South Africa. Johannesburg: Aloes of South Africa Book Fund; 1950. [Google Scholar]
- 47.Reynolds GW. The Aloes of Tropical Africa and Madagascar. Aloes Book Fund: Mbabane; 1966. [Google Scholar]
- 48.Carter S, Lavranos JJ, Newton LE, Walker CC. Aloes, The Definitive Guide. Kew: Royal Botanic Gardens; 2011. [Google Scholar]
- 49.Maddison WP, Maddison DR. Mesquite, a modular system for evolutionary analysis, v2.73. Available at http://mesquiteproject.wikispaces.com/.
- 50.Maddison WP. Testing character correlation using pairwise comparisons on a phylogeny. J Theor Biol. 2000;202:195–204. doi: 10.1006/jtbi.1999.1050. [DOI] [PubMed] [Google Scholar]
- 51.Grace OM, Klopper RR, Smith GF, Crouch NR, Figueiredo E, Rønsted N, Van Wyk AE. A revised generic classification of Aloe (Xanthorrhoeaceae subfam. Asphodeloideae) Phytotaxa. 2013;76:7–14. doi: 10.11646/phytotaxa.76.1.2. [DOI] [Google Scholar]
- 52.Buerki S, Forest F, Acevedo-Rodriguez P, Callmander MW, Nylander JAA, Harrington M, Sanmartin I, Kupfer P, Alvarez N, et al. Plastid and nuclear DNA markers reveal intricate relationships at subfamiliar and tribal levels in the soapberry family (Sapindaceae) Mol Phy Evol. 2009;51:238–258. doi: 10.1016/j.ympev.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 53.Wiens JJ. Can incomplete taxa rescue phylogenetic analyses from long-branch attractions? Syst Bio. 2005;54:731–742. doi: 10.1080/10635150500234583. [DOI] [PubMed] [Google Scholar]
- 54.Wiens JJ. Missing data and the design of phylogenetic analyses. J Biomed Inform. 2006;39:34–42. doi: 10.1016/j.jbi.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 55.Lavranos JJ. Notes on the aloes of Arabia with descriptions of six new species. J S Afr Bot. 1965;31:55–81. [Google Scholar]
- 56.Holland PG. An evolutionary biogeography of the genus Aloe. J Biogeog. 1978;5:213–226. doi: 10.2307/3038037. [DOI] [Google Scholar]
- 57.Linder HP. The radiation of the Cape Flora, southern Africa. Biol Rev. 2003;78:597–638. doi: 10.1017/S1464793103006171. [DOI] [PubMed] [Google Scholar]
- 58.Cowling RM, Rundel PW, Desmet PG, Esler KJ. Extraordinary high regional-scale plant diversity in southern African arid lands, subcontinental and global comparisons. Diversity Distrib. 1998;4:27–36. [Google Scholar]
- 59.Valente LM, Britton AW, Powell MP, Papadopulos AST, Burgoyne PM, Savolainen V. Correlates of hyperdiversity in southern African ice plants Aizoaceae. Bot J Linn Soc. 2013;174:110–129. doi: 10.1111/boj.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eggli U. Illustrated Handbook of Succulent Plants, Crassulaceae. Berlin: Springer-Verlag; 2003. [Google Scholar]
- 61.Good-Avila SV, Souza V, Gaut BS, Eguiarte LE. Timing and rate of speciation in Agave (Agavaceae) Proc Natl Acad Sci U S A. 2006;103:9124–9129. doi: 10.1073/pnas.0603312103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Edwards EJ, Donoghue MJ. Pereskia and the origin of the cactus life-form. Am Nat. 2006;167:777–793. doi: 10.1086/504605. [DOI] [PubMed] [Google Scholar]
- 63.Nyffeler R, Eggli U, Ogburn M, Edwards E. Variation on a theme, repeated evolution of succulent life forms in the Portulacineae (Caryophyllales) Haseltonia. 2008;14:26–36. doi: 10.2985/1070-0048-14.1.26. [DOI] [Google Scholar]
- 64.O’Brien C, Van Wyk B-E, Van Heerden FR. Physical and chemical characteristics of Aloe ferox leaf gel. S Afr J Bot. 2008;77:988–995. doi: 10.1016/j.sajb.2011.08.004. [DOI] [Google Scholar]
- 65.Morton JF. Folk uses and commercial exploitation of Aloe leaf pulp. Econ Bot. 1961;15:311–319. doi: 10.1007/BF02907852. [DOI] [Google Scholar]
- 66.Grace OM, Buerki S, Symonds MRE, Forest F, van Wyk AE, Smith GF, et al. Xanthorrhoeaceae Bayesian tree. 2015. http://purl.org/phylo/treebase/phylows/study/TB2:S16954?format=html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
DNA sequences are deposited in GenBank and accession numbers are listed in Additional file 1.
The phylogenetic tree supporting the results of this article is available in the TreeBase repository, http://purl.org/phylo/treebase/phylows/study/TB2:S16954?format=html [66].


