Summary
In C. elegans and Drosophila, removing germline stem cells increases lifespan. In C. elegans, this lifespan extension requires DAF-16, a FOXO transcription factor and DAF-12, a nuclear hormone receptor. To better understand the regulatory relationships between DAF-16 and DAF-12, we used microarray analysis to identify downstream genes. We found that these two transcription factors influence the expression of distinct but overlapping sets of genes in response to loss of the germline. In addition, we identified several new genes that are required for loss of the germline to increase lifespan. One, phi-62, encodes a conserved, predicted RNA binding protein. PHI-62 influences DAF-16-dependent transcription, possibly by collaborating with TCER-1, a putative transcription-elongation factor, and FTT-2, a 14-3-3 protein known to bind DAF-16. Three other genes encode proteins involved in lipid metabolism; one is a triacylglycerol lipase, and another is an acyl CoA reductase. These genes do not noticeably affect bulk fat storage levels; therefore, we propose a model in which they may influence production of a lifespan-extending signal or metabolite.
Keywords: lifespan, germline, steroid, DAF-16, DAF-12, aging
Introduction
Removing the germline of C. elegans, either by laser ablation of the germline precursor cells, or by mutation of genes required for germ cell proliferation, extends lifespan by approximately 60% (Hsin & Kenyon 1999). This lifespan extension appears to be caused specifically by loss of germline stem cells during adulthood (Arantes-Oliveira et al. 2002). Removing germline stem cells in Drosophila adults increases the fly's lifespan by up to 50% (Flatt et al. 2008), and in mice, too, signals from the reproductive system can extend lifespan (Cargill et al. 2003; Mason et al. 2009). How the germ cells, which give rise to the progeny, also control the rate of aging of the body in which they reside is a fascinating but unanswered question.
Two transcription factors, the FOXO-family transcription factor DAF-16 and the nuclear hormone receptor (NHR) DAF-12, are required for germline loss to extend lifespan in C. elegans (Hsin & Kenyon 1999). DAF-16/FOXO is best known for its ability to extend lifespan in response to reduced insulin/IGF-1 signaling (Kenyon 2010b). FOXO proteins have been linked to longevity in many animal species, and at least eight gene association studies suggest that they affect human longevity as well (Kenyon et al. 1993; Hwangbo et al. 2004; Giannakou et al. 2007; Taguchi et al. 2007; Willcox et al. 2008; Anselmi et al. 2009; Flachsbart et al. 2009; Li et al. 2009; Pawlikowska et al. 2009; Soerensen et al. 2010). In worms with reduced insulin/IGF-1 signaling, DAF-16 localizes to the nuclei of larval and adult tissues throughout the animal (Henderson & Johnson 2001; Lee et al. 2001; Lin et al. 2001). Loss of the germline has a different effect on DAF-16 nuclear localization, causing DAF-16 to accumulate primarily in the nuclei of one tissue, the intestine, during adulthood (Lin et al. 2001). The intestine appears to serve as C. elegans’ entire endoderm, carrying out functions associated with adipose tissue (fat storage), and the liver and pancreas (yolk production, and production of insulin and IGF-1-like hormones). DAF-16 functions in the intestine and other tissues to extend lifespan in response to inhibition of insulin/IGF-1 signaling, but it appears to function primarily in the intestine to extend lifespan in response to loss of the germ cells (Libina et al. 2003). DAF-16's function in the intestine/adipose tissue may potentially be conserved, as dFOXO expression exclusively in adipose tissue extends fly lifespan, and down-regulation of insulin signaling in mouse adipose tissue extends lifespan as well (Bluher et al. 2003; Giannakou et al. 2004; Hwangbo et al. 2004).
Loss of the germ cells also increases the levels of TCER-1, a putative transcription elongation factor, in the intestine (Ghazi et al. 2009). TCER-1 appears to have a rather focused activity in the worm, as its loss prevents germline ablation from extending lifespan but does not affect normal lifespan (Ghazi et al. 2009). A small set of genes up-regulated by DAF-16 in response to both germline loss and insulin/IGF-1 pathway inhibition has been identified, and TCER-1 is required for expression of some, but not all, of these genes in response to germline loss (Ghazi et al. 2009). TCER-1 is not up-regulated in insulin/IGF-1-pathway mutants, and its activity is not required for their DAF-16-dependent gene expression, or for lifespan extension. Likewise, an intestinal adaptor protein called KRI-1 is required for germline loss, but not insulin/IGF-1 pathway inhibition, to increase lifespan. Together these and other findings indicate that DAF-16's regulation and activity in the reproductive and insulin/IGF-1 pathways are distinct from one another (Kenyon 2010a).
The nuclear hormone receptor DAF-12 plays at least two distinct roles in germlineless animals. First, in animals that lack germ cells, DAF-12 is partially required for nuclear localization of DAF-16/FOXO (Berman & Kenyon 2006; Gerisch et al. 2007). However, DAF-12 must play an additional role in the germline pathway, as a mutant DAF-16 protein that is constitutively localized to the nucleus cannot extend lifespan in response to germline loss without DAF-12 activity (Berman & Kenyon 2006). DAF-12's second role could be linked to that of another, recently described, nuclear hormone receptor, NHR-80, as DAF-12, but not DAF-16, is required for overexpression of nhr-80 to extend lifespan in germline-deficient animals (Goudeau et al. 2011).
How DAF-12 ultimately influences lifespan is not known. In intact animals, a set of potential longevity genes regulated by DAF-12 has been identified using microarray analysis (Fisher & Lithgow 2006). Here, at low temperature, loss-of-function daf-12 alleles shorten lifespan, whereas gain-of-function alleles modestly lengthen lifespan (Gerisch et al. 2007). Interestingly, some of the down-regulated genes identified by Fisher and Lithgow are known to be functionally significant lifespan genes that are also down-regulated by DAF-16 in daf-2/insulin/IGF-1 receptor mutants. This finding suggests that DAF-12 and DAF-16 regulate at least some genes in common.
In this study, we have used microarray analysis to clarify the roles of DAF-16 and DAF-12 in the germline pathway. In particular, we were interested in learning to what extent the genes regulated by DAF-16 and DAF-12 in the germline pathway were likely to be the same as those regulated by these two transcription factors in other longevity pathways. In addition, we wanted to get a better idea of how DAF-16 and DAF-12 might interact to activate their target genes in the germline pathway. For example, some genes, such as the superoxide dismutase sod-3 (Yamawaki et al. 2010) and the lipase K04A8.5 (Wang et al. 2008), are known to be activated by DAF-16 independently of DAF-12 in germline-deficient animals, but the size and composition of this gene class is not known. Nor is it known whether DAF-16 and DAF-12 might co-regulate specific target genes in this pathway. Finally, we hoped that among the genes whose expression changed in response to germline loss we might identify some required for lifespan extension.
Results
Experimental design and initial validation
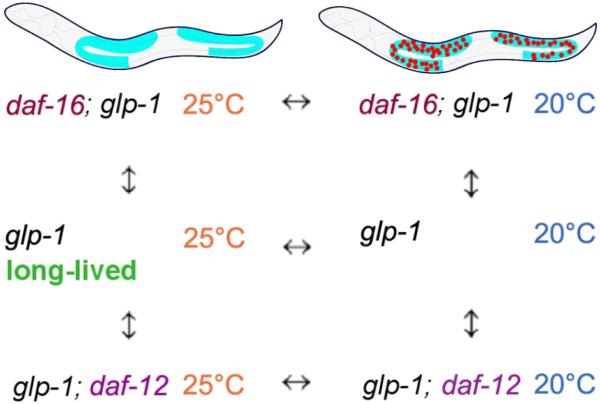
Because the germline comprises two thirds of the cells in C. elegans adults, a straightforward comparison of the differences in global transcription between intact and germline-deficient animals might identify mainly genes expressed in the germline, genes that are not necessarily involved in the germline's regulation of lifespan. Similarly, we might also expect that a comparison of daf-12 or daf-16 wild-type (+) versus mutant (-) animals would yield a broad range of these transcription factors' targets, not only those germane to the germline regulation of lifespan. In order to generate a comparison of gene expression under conditions that produce short and long lifespan that would not be overwhelmed by the many germline-expressed genes, we used ANOVA analysis of a simple block design (see Figure 1) (Kerr et al. 2000; Kerr & Churchill 2001). To remove the germ cells, we used a temperature-sensitive glp-1 mutation, which prevents germline proliferation at high temperature, and we used daf-12 and daf-16 null mutations to remove these two transcription-factor gene activities. Using the strategy outlined in Figure 1, we then compared the gene expression patterns of these animals to one another in various combinations. We hybridized 60 samples to arrays designed to probe 20,374 predicted C. elegans ORFs, as described previously (Cristina et al. 2009).
Figure 1.
Block design for microarray experiments. glp-1(e2141ts) animals lack a germ line and are long lived at 25°C, but are germline (+) and have a normal lifespan at 20°C.
To validate our method's ability to pick out specific functionally related classes of genes, we first compared the gene expression patterns of all the strains grown at high temperature (25°C, the germline-deficient condition) to the same strains grown at low temperature (the intact condition). The list of genes generated by this comparison was highly enriched for genes already known to have germline-specific functions related to early development and similar processes (Table S1). In addition, 1/3 of the genes we identified were also present in previous microarray experiments designed to identify germline-specific genes (p<1E-4) (Reinke et al. 2004).
Genes regulated by DAF-16
To identify genes that were regulated (directly or indirectly) by DAF-16/FOXO in response to germline loss, we looked for genes whose expression-change upon removal of the germline was most dependent on the presence of daf-16. (daf-12 mutants were excluded from this analysis.) Our list of germline-regulated DAF-16 targets contained 230 genes using a p<0.01 significance-level cutoff (Table S2). Of these, 38 were also present among 512 previously identified genes regulated by DAF-16 in a daf-2 mutant background (Murphy et al. 2003), a highly significant overlap (p<E-15). This striking correspondence suggests that DAF-16 may extend lifespan in germline-deficient animals and insulin/IGF-1 mutants via similar sets of downstream genes.
We performed a BiNGO analysis of this list of 230 genes for overrepresented biological processes (Shannon et al. 2003; Maere et al. 2005) (Table S3). This comparison yielded “aging”, “multicellular organismal aging”, and “determination of adult lifespan” as the three most significantly overrepresented processes. These three categories were all overrepresented on the basis of the same 13 genes (Table S4), of which 10 were annotated as being involved in lifespan in part because they were previously identified as targets of DAF-16 in a daf-2 mutant background, including mdt-15, which will be discussed further below. Other sets of closely related overrepresented categories contained genes involved in monocarboxylic acid metabolism, carboxylic acid metabolism, and organic acid metabolism. Genes in these overrepresented sets included fat-5, which encodes a fatty acid desaturase, and hacd-1, which encodes a hydroxyacyl-CoA dehydrogenase, both of which have been previously shown to be regulated by MDT-15 (Taubert et al. 2006), as well as gei-7, which encodes an isocitrate lyase / malate synthase that has been shown to be regulated both by DAF-16 in a daf-2(-) background (Murphy et al. 2003) and by MDT-15 (Taubert et al. 2006). In an independent study, Goudeau et al. recently identified fat-5 as a daf-16-regulated gene in glp-1 mutants as well (Goudeau et al. 2011).
Genes regulated by DAF-12 in response to germline loss
Our list of daf-12/NHR-dependent genes contained 130 genes using a p<0.01 significance-level cutoff (Table S5). Of these, 8 overlapped with 224 genes previously reported to be daf-12-regulated in intact animals grown at low temperature, again a highly significant overlap (p<1E-4) (Fisher & Lithgow 2006). As with DAF-16, this overlap suggests that many of the targets by which DAF-12 influences lifespan in response to germline loss may be shared in other contexts. A BiNGO analysis of this list of genes showed no overrepresented biological processes.
Genes regulated by both DAF-12 and DAF-16 in animals lacking germ cells
Having established that there was a conserved transcriptional output from each of these two transcription factors alone, in multiple lifespan-regulating contexts, we asked what target genes these two transcription factors might have in common in germline-deficient animals. Of the 230 predicted DAF-16 targets and the 130 predicted DAF-12 targets, 7 were common to both lists, which is statistically significant (p<1E-3), although less so than the overlap with previously reported targets of each transcription factor respectively (Murphy et al. 2003; Fisher & Lithgow 2006). Although the great majority of genes whose expression was most significantly affected by the loss of DAF-16 were distinct from those most significantly affected by the loss of DAF-12 (a finding that rules out the possibility of placing these two transcription factors in a simple linear pathway) we can say that they have a greater than random coincidence in transcriptional output. The seven genes regulated by both daf-16 and daf-12 in this strict analysis encoded a neuropeptide-like protein, a sodium neurotransmitter symporter family protein, a broad complex domain-containing protein, and several proteins of unknown function (Table S6). One of these proteins of unknown function, C46G7.2, was identified in an RNAi screen for increased paraquat resistance, and C46G7.2 RNAi was shown to increase wild-type lifespan by 44% in a daf-16 dependent manner (Kim & Sun 2007).
In any comparison of strict cutoff lists, the problem exists that genes with a less dramatic degree of regulation that is nonetheless shared may escape notice. In order to ask whether a more subtle enrichment of those genes might exist than that observed by the overlap of our two p<0.01 cutoff lists, we performed Gene Set Enrichment Analysis (Subramanian et al. 2005), testing the individual p<0.01 cutoff list of predicted DAF-16 targets against a list of all C. elegans genes present on our array ranked by likelihood of DAF-12 regulation, and vice-versa. In these more sensitive analyses, a more subtle enrichment was in fact detected: of 230 predicted DAF-16 targets, 103 were predicted to show DAF-12 regulation (Table S7), and of 130 predicted DAF-12 targets, 69 were predicted to show DAF-16 regulation (Table S8). In order to further investigate this overlapping regulation as well as validate our array results, we performed qPCR of lips-17, a gene identified in our long-vs.-short lived comparison; dod-8, a gene identified in this study as well as previous work (Murphy et al. 2003) as a DAF-16 target; and the previously identified DAF-16 targets gpd-2, nnt-1, and sod-3, (Murphy et al. 2003; Ghazi et al. 2009) in germline-deficient worms that were otherwise wild-type or null for either daf-12 or daf-16. Confirming our array results, we found that lips-17 expression was upregulated over 2.5-fold in germline-deficient animals, and that this increase was partially dependent on both daf-12 and daf-16 (Figure S1). Our array results were also confirmed by the finding that expression of dod-8 in germline-deficient worms was dependent on the presence of daf-16 (Figure S1). Interestingly, for dod-8 as well as the daf-16 reporters sod-3, nnt-1, and gpd-2, the decrease in expression upon removal of daf-16 was paralleled by a more modest although in some cases (gpd-2, nnt-1) statistically significant decrease upon removal of daf-12 (Figure S1). This again corresponds well with our array results, which show by gene set enrichment analysis that roughly half of those genes most strongly dependent on daf-16 show a more subtle dependence on daf-12, and vice-versa. We then subjected these predicted overlapping subsets of genes again to BiNGO analysis for overrepresented gene ontology categories. For the predicted daf-12 targets, as before, neither the daf-16 enriched nor daf-16 un-enriched subsets showed any gene ontology categories overrepresented. Among the daf-16 targets, those showing daf-12 enrichment no longer showed overrepresentation for aging and lifespan related biological processes, while those un-enriched still did. As discussed above, this is largely because the daf-16 targets that showed daf-12 enrichment from our array results in germline-deficient animals do not overlap significantly with previously published targets of daf-16 in the daf-2 pathway. Additionally, the daf-12 un-enriched daf-16 targets, that is, those daf-16 targets that we predict are not dependent on daf-12, also contained all of the genes responsible for overrepresentation of fatty acid metabolism in this group, and as a result fatty acid metabolism is reported as overrepresented in the daf-12 un-enriched subset but not the daf-12 enriched subset.
Regulatory analysis
One possible source of additional information about the regulation of these genes could come from an analysis of overrepresented sequences in their promoter regions. To identify such sequences, we used RSAT oligo analysis of 1 kb-upstream regions (van Helden et al. 1998). For our predicted DAF-16 targets, the two most significantly overrepresented sequences were, first, CTTATCAGT, and then TTGTTTAC. A portion of the first sequence, CTTATCA, was identified previously as being overrepresented in the promoter sequences of DAF-16 targets in the insulin/IGF-1 signaling pathway (Murphy et al. 2003), and was subsequently identified as a predicted ELT transcription factor binding site that was overrepresented in the promoters of several aging related gene sets (Budovskaya et al. 2008). The second sequence was originally identified as a consensus binding sequence for mouse DAF-16/FOXO proteins (Furuyama et al. 2000) and it was also found overrepresented in the promoters of DAF-16 targets in the C. elegans insulin/IGF-1 signaling pathway (Murphy et al. 2003).
For our predicted DAF-12 targets, we found the sequence TTGATAA, which has not been identified in previous aging or daf-12 studies, to be overrepresented. This sequence was not identified in the previous DAF-12 low-temperature studies (Fisher & Lithgow 2006). We analyzed this previously published list of predicted DAF-12 targets that might affect lifespan (Fisher & Lithgow 2006) using the same method and found a one base-pair variation of the same sequence (CTGATAA), further supporting the idea that this novel sequence may play a role in DAF-12's regulation of lifespan. Because our unseeded overrepresented oligo sequence results did not include either of two previously reported daf-12 binding sites (Ao et al. 2004; Shostak et al. 2004), we asked specifically whether either of these sequences was overrepresented in the promoters of our 130 predicted DAF-12 targets, and found that neither was. It is worth noting here that the first of these novel sequences, TTGATAA, has a reverse complement (TTATCAA) that differs by one base pair from the above-mentioned TTATCAC predicted ELT-binding sequence previously found to be overrepresented in the promoters of predicted daf-16 targets (Murphy et al. 2003), and in the promoters of several aging-related gene sets (Budovskaya et al. 2008). Finally, we note that, very recently, chromatin immunopreciptation was used to identify DAF-12 targets from intact larvae and adults (Hochbaum et al. 2011). Again their overrepresented DAF-12 binding sites did not coincide with ours.
In addition to these two analyses, we also looked for overrepresented sequences among genes whose change in expression correlated most significantly with lifespan; that is, whether the strain was long or short-lived. Among these “long-vs short-life” genes, we found a new sequence, AGTAACCC, and another portion of the putative ELT/DAF-16 binding site, TTATCAC, to be overrepresented (Budovskaya et al. 2008).
Functional Analysis
Next, we used RNAi to test the functional significance of genes identified in our microarray experiments. To do this, we fed intact and germline-deficient animals bacteria expressing the corresponding dsRNA sequences, and measured the worms’ lifespans. The most statistically-significant genes were tested for each of three categories. The first category contained genes whose expression differed most significantly between the long-lived germline-deficient condition and all of the remaining non-long-lived conditions. These non-long-lived conditions included, in a glp-1(ts) background, the following strains: daf-16(+); daf-12(+) animals grown at 20°C, and daf-12(-) and daf-16(-) worms mutants grown at either 20°C or 25°C. We began testing genes once we had carried out our first set of microarrays, which included only Day-1 adults. We tested a total of 19 genes from this set (see Table S10). Among the genes we identified in this screen (#12 in our list) was tcer-1, the putative transcription elongation factor discussed above. This finding too, validated our microarray strategy, and also indicated that the increase in the TCER-1::GFP signal observed in germline-deficient animals (Ghazi et al. 2009) likely results from changes in tcer-1 mRNA levels. We tested additional strains after completing all the arrays, which included mRNA samples from day 1-3 adults (Table S9). The second list we compiled and interrogated contained genes that were predicted to be (direct or indirect) DAF-16 targets in the germline pathway, and the third list contained genes that were predicted to be DAF-12 targets in the germline pathway.
From this analysis, we identified several genes that were required for the increased longevity of germline-defective animals but not for the longevity of wild type. These genes are discussed below.
phi-62 encodes a conserved RNA-binding protein required for germline-deficient animals to live long
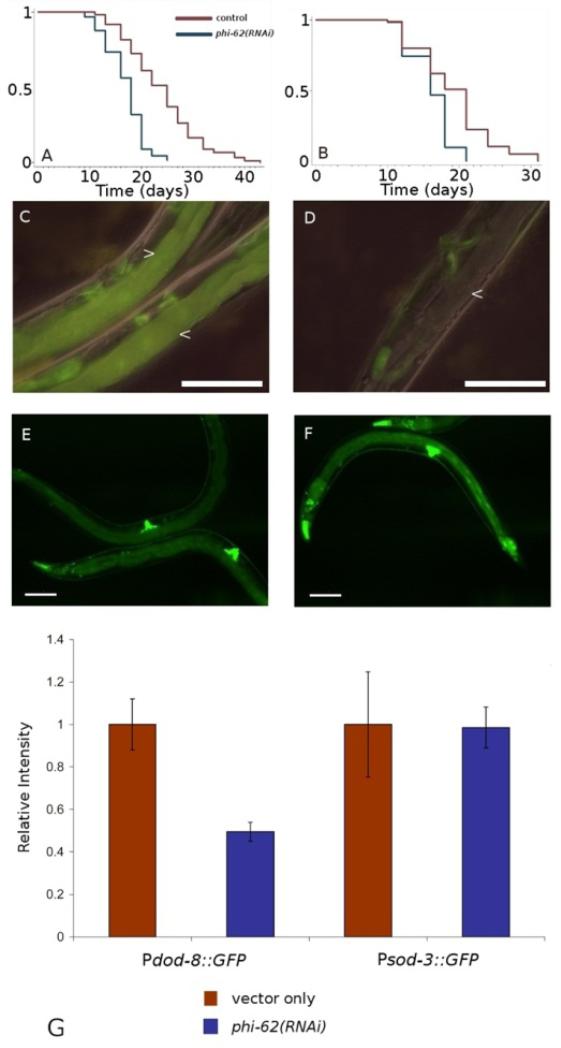
phi-62 was identified in our initial day-1 “long vs. short-lived” comparison (# 17 in our list), along with tcer-1. This gene encodes a small, 95-amino acid protein containing a predicted RNA-binding domain. phi-62 is C. elegans’ only member of a highly conserved protein family that has a single ortholog in many diverse animal species (Rampias et al. 2008). Although its biological function is unknown, the Ceratitis capitata ortholog has been shown to have ribonuclease activity in vitro (Rampias et al. 2003). RNAi knockdown of phi-62 has also been shown to accelerate protein aggregation in a worm model of polyglutamine aggregation diseases (Nollen et al. 2004). We found that RNAi knockdown of phi-62 reduced the lifespan of germline-deficient animals approximately to that of wild type, while reducing wild-type lifespan only slightly. This phenotype was similar to that caused by daf-16 RNAi (Figure 2A). With this in mind, we asked whether phi-62 was required for the expression of known daf-16-regulated genes using GFP reporters in transgenic animals. We examined dod-8 and sod-3, both of which are up-regulated in germline-deficient animals in a daf-16 dependent manner. Interestingly, dod-8 up-regulation was abolished by RNAi knockdown of phi-62, whereas sod-3 expression was unaffected (Figure 2B). This target specificity was reminiscent of that of tcer-1, which is also required for germline-deficient animals to live long (Ghazi et al. 2009). To investigate this apparent similarity more closely, we examined three more genes that were regulated by tcer-1. We found complete coincidence between the effects of phi-62 and tcer-1 on these genes (Figure S2). These findings raise the interesting possibility that this putative RNA binding protein functions in association (directly or indirectly) with TCER-1.
Figure 2.
phi-62 is required for removal of the germline to increase lifespan. It is also required for up-regulation of dod-8, a known daf-16-regulated gene, but not for sod-3, another daf-16-regulated gene, in animals that lack the germline. A) glp-1(e2141ts): vector-only control n = 96, m = 23.8 days, phi-62(RNAi) n = 72, m = 16.9 days, p < 0.0001. B) wild type (N2): vector-only control n = 70, m = 19.1 days, phi-62(RNAi) n = 62, m = 16.2 days, p < 0.0001. C,D) glp-1(e2141ts); Pdod-8::GFP, which is expressed in the intestine. C, vector-only control (two worms are shown), D, phi-62(RNAi), arrows indicate intestine. E,F) glp-1(e2141ts); Psod-3::GFP, E, vector, F, phi-62(RNAi). G) Relative GFP intensities. Pdod-8::GFP vector only vs. phi-62(RNAi), p < 0.01. Scale bar = 200μm.
TCER-1 activity is not required for all longevity pathways: for example, it is not required for the lifespan increased produced by daf-2 (insulin/IGF-1-receptor) mutation, eat-2 mutation (a condition that mimics caloric restriction) (Lakowski & Hekimi 1998) or clk-1 mutation (which affects ubiquinone biosynthesis) (Wong et al. 1995). Because of the similarity between the tcer-1(-) and phi-62(-) phenotypes, we tested whether loss of phi-62 activity might affect other lifespan pathways similarly to the way they are affected by loss of phi-62. Like knockdown of tcer-1, RNAi knockdown of phi-62 had no effect on the lifespans of daf-2 mutant animals in one trial, and caused only a slight reduction in lifespan in a second trial (Figure S3). phi-62 was partially required for lifespan extension caused by eat-2 and isp-1 mutations.
FTT-2 is predicted to bind to PHI-62 and is required for the increased lifespan of germline-deficient animals
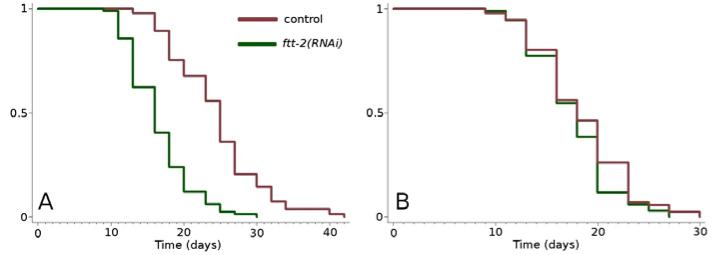
Using Gene Orienteer (www.geneorienteer.org) (Zhong & Sternberg 2006), we discovered that PHI-62 is predicted to bind FTT-2. This prediction is based on the interaction between the Drosophila orthologs of FTT-2 and PHI-62 in a yeast-two hybrid assay (Giot et al. 2003). FTT-2 is a 14-3-3 protein that has previously been shown to bind to DAF-16 (Berdichevsky et al. 2006; Wang et al. 2006; Li et al. 2007). We found that RNAi knockdown of ftt-2 reduced the long lifespan of germline-defective animals to that of wild type (Figure 3), while not shortening wild-type lifespan. Because FTT-2 has also been shown to interact with SIR-2.1 (Berdichevsky et al. 2006; Wang et al. 2006), we tested the lifespan of a glp-1(e2141ts); sir-2.1(ok434) double mutant. These animals were long lived, indicating that SIR-2.1 is not part of the germline longevity pathway (data not shown). These results suggest the possibility that PHI-62, TCER-1, FTT-2 and DAF-16 may act together in the regulation of DAF-16 target gene expression.
Figure 3.
ftt-2 RNAi prevents loss of the germline from increasing lifespan. A) glp- 1(e2141ts): vector-only control n = 90, m = 24.4, ftt-2(RNAi) n = 87, m = 16.4, p < 0.0001, B) wild type (N2): vector-only control n = 89, m = 18.4, ftt-2(RNAi) n = 85, m = 17.6, p < 0.1.
In an attempt to further characterize any phenotypic interactions between TCER-1, PHI-62, FTT-2, and DAF-16, we asked whether the previously reported lifespan extension of tcer-1 overexpressing worms was dependent on expression of the other three genes. As previously reported (Ghazi et al. 2009), we saw a modest but statistically significant increase in the lifespan of tcer-1 overexpressing worms relative to wild-type, and this increase did not occur upon RNAi knockdown of daf-16 (Table S10). We further saw that this increase of lifespan in tcer-1-overexpressing worms did not occur upon RNAi knockdown of ftt-2 or phi-62 (Table S10), suggesting that ftt-2 and phi-62 act downstream of, or in parallel to, tcer-1. To find out more about possible relationships between daf-16, ftt-2 and phi-62, we asked whether RNAi knockdown of ftt-2 or phi-62 could further shorten the already shortened lifespan of germline-deficient daf-16; glp-1 worms, and found that it could not in either case (Figure S4). This finding is consistent with the possibility that these three genes play related roles in the lifespan extension of germline-deficient animals.
We also asked whether the nuclear localization of DAF-16 in the intestines of germline-deficient animals was affected by RNAi knockdown of either phi-62 or ftt-2. We found that RNAi knockdown of phi-62 greatly reduced DAF-16 intestinal nuclear localization in germline-deficient animals, and knockdown of ftt-2 reduced it as well, though more modestly.
This latter finding was unexpected. Previous groups have shown an increase in DAF-16 nuclear localization upon RNAi knockdown of ftt-2, and no effect upon RNAi knockdown of its close homolog par-5 (Berdichevsky et al. 2006; Li et al. 2007). However, these studies examined intact rather than germline-deficient worms, and also employed an ftt-2 or par-5-specific RNAi construct, whereas the RNAi clone we used (from the Ahringer library) is likely to target both homologs. Therefore, we obtained the ftt-2 and par-5-specific constructs, and repeated these measurements in both glp-1(-) and wild-type worms. Again, we saw that DAF-16 intestinal nuclear localization was decreased in glp-1(-) mutants, and not increased in the wild type (Figure S5). Why do our findings differ? In our experiments, we initiated RNAi treatment at the time of hatching and examined the worms when they became adults, at 20°C, after shifting the animals at the L2-L4 stage to 25°C (as this removes the germline stem cells of glp-1(e2141ts) mutants). Li et al. initiated RNAi in L4 larvae at 16°C. They then examined L3 progeny, rather than same-generation adults. Berdichevsky et al. also initiated RNAi in L4 larvae, and observed nuclear DAF-16 localization in the L3s and adult progeny. Whether the timing of treatment, temperature, or another variable is responsible for the difference, the results reported here are relevant to regulation of DAF-16 in the germline context because (1) our treatment regime mimics exactly that used in the lifespan assays whose outcomes we seek to understand, and (2) work in our lab has shown that the up-regulation of DAF-16 reporters in germline-deficient animals is not noticeable until adulthood.
Finally, we asked whether RNAi knockdown of ftt-2 might affect the expression of genes that are normally switched on in a daf-16-dependent fashion in germline defective mutants. We assayed GFP reporters for sod-3, whose induction requires daf-16, and dod-8, whose induction not only requires daf-16but also tcer-1 and phi-62. Surprisingly, we did not see any change in the up-regulation of these two genes (Figure S6).
lips-17 and fard-1 are lipid-metabolism genes required for the longevity of germline-deficient animals
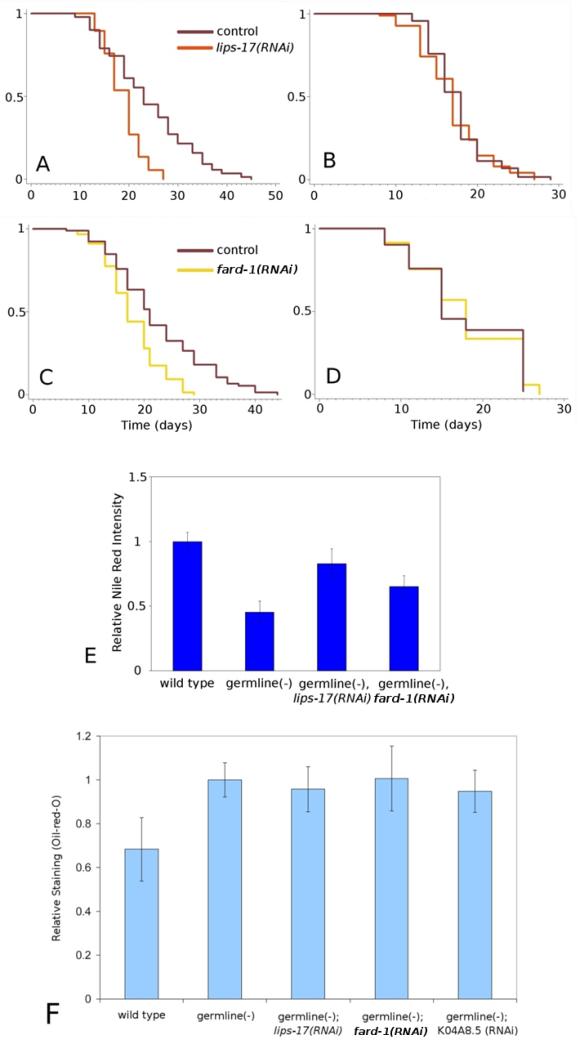
lips-17 encodes a predicted triacylglycerol lipase, and was present in our list of genes differentially regulated in long vs. short lived animals. Y71H10A.2, which we named fard-1, encodes a predicted fatty acyl reductase, and was identified as a gene upregulated in germline-deficient animals in a daf-12-dependent manner in our microarrays. RNAi knockdown of either lips-17 or fard-1 shortened the lifespan of germline-deficient animals roughly to that of wild type, while not shortening wild-type lifespan (Figure 4). Another triacylglycerol lipase, K04A8.5, was shown previously to be required for germline-deficient animals to live long (Wang et al. 2008), as were genes required for the synthesis of oleic acid (Goudeau et al. 2011).
Figure 4.
lips-17 and fard-1 are required for removal of the germline to increase lifespan. In addition, lips-17 and fard-1 RNAi alter Nile red lysosomal staining but not Oil-red-O lipid staining. A,B) lips-17: A, glp-1(e2141ts) vector-only control n = 89, m = 23.9; lips-17(RNAi) n = 77, m = 18.9, p < 0.0001. B, wild type (N2). vector-only control n = 90, m = 17.5; lips-17(RNAi) n = 92, m = 16.9, p > 0.4. C,D) fard-1. C, glp- 1(e2141ts). vector-only control n = 85, m = 22.3; fard-1(RNAi) n = 87, m = 17.8, p < 0.0001. D, wild type (N2): vector-only control n = 63, m = 17.8; fard-1(RNAi) n = 78, m = 17.9, p > 0.6. E) Nile red staining, p-values: wild type vs. germline-deficient p < 1E-6, germline-deficient vector-only control vs. lips-17(RNAi) p < 1E-5, germline-deficient vector only control vs. fard-1(RNAi) p < 0.01. F) Oil-red-O staining, p-values: wild type vs. germline(-), germline(-); lips-17(RNAi), and germline(-); K04A8.5(RNAi), all p < 0.01. wild type vs. germline(-); fard-1(RNAi) p = 0.02. germline(-) vs. all RNAi treatments, N.S.
Animals lacking a germline have been shown to have greatly reduced staining with the dye Nile red, and RNAi knockdown of K04A8.5 suppresses this abnormal Nile red phenotype (Wang et al. 2008). We asked what effect RNAi knockdown of lips-17 and fard-1 might have on Nile red staining, and found that knockdown of lips-17, and to a lesser extent knockdown of fard-1, increased Nile red staining in germline-deficient animals while not increasing staining in wild-type animals, a phenotype reminiscent of the K04A8.5-knockdown phenotype (Figure 5). Nile red has been thought to stain fat in C. elegans but it was recently shown to more specifically stain intestinal lysosomes (Schroeder et al. 2007; O'Rourke et al. 2009). Recently published work has suggested that oil-red-O staining is a better indicator of fat levels than is Nile red, as confirmed by solid-phase lipid extraction followed by GC/MS (O'Rourke et al. 2009; Soukas et al. 2009). Using oil-red-O staining, we found that overall fat storage was clearly increased in germline-defective animals relative to wild type, as first observed by O'Rourke et al. (O'Rourke et al. 2009). This increase was not affected by RNAi knockdown of lips-17, fard-1, or K04A8.5 (Figure 4). This finding suggests that these lipid-metabolizing genes are unlikely to influence lifespan by mechanisms that involve gross changes in total lipid stores.
Figure 5.
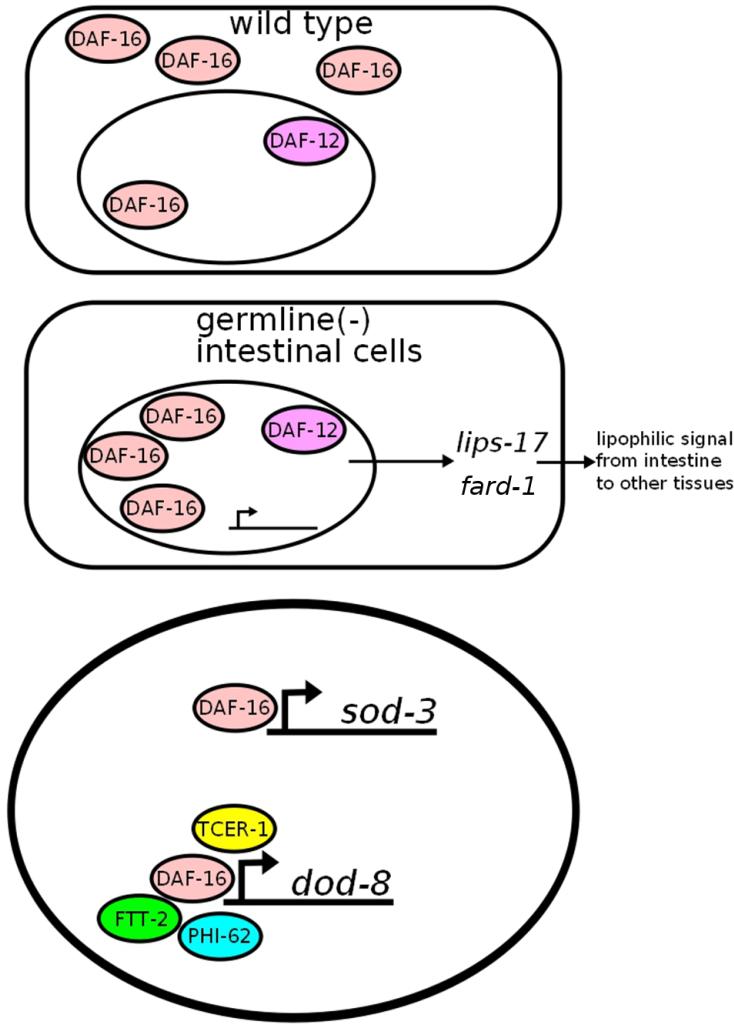
Summary and a hypothetical model. Upon removal of the germline, DAF-16 localizes to the nuclei of intestinal cells. Together with DAF-12, this causes upregulation of fard-1 and lips-17, which may be involved in the production of a hypothetical lipophilic signal from the intestine to other tissues. DAF-16, PHI-62, and FTT-2 (or their orthologs in other species) have been shown to interact physically, and are all required for lifespan extension by germline ablation, suggesting the possibility that they may act together in this process. PHI-62 and TCER-1 are required for changes in some but not all DAF-16 target genes (Mark I didn’t think FTT-2 affected gene expression, so I changed this, right?)
mdt-15(RNAi) worms are short-lived
One final RNAi knockdown, of mdt-15, produced a noteworthy phenotype. mdt-15 is a transcriptional mediator subunit orthologous to human MED15. In C. elegans, mdt-15 is required for expression of fatty-acid desaturase genes, genes induced during fasting, and genes involved in metabolic adaptation to ingested material (Taubert et al. 2006; Taubert et al. 2008). mdt-15 was identified as a gene upregulated in germline-deficient animals in a daf-16-dependent manner in our microarrays, and was also previously identified as a daf-16-regulated gene in the insulin signaling pathway (Murphy et al. 2003). In one trial, RNAi knockdown of mdt-15 was found to reduce the lifespan of germline-deficient animals. However, as reduction of mdt-15 function also shortens wild-type lifespan (Taubert et al. 2008), mdt-15 may not act in a germline-specific fashion. It is not clear whether mdt-15 plays a role in the expression or action of the lipid metabolism genes discussed above, but that is an interesting possibility.
Discussion
In this study, we used a block-design microarray approach to identify genes differentially expressed in long-lived germline-deficient animals. We further identified genes whose altered expression most strongly depended on either the DAF-16/FOXO transcription factor or the DAF-12 nuclear hormone receptor. We found that many of the genes dependent on DAF-16 in this germline-deficient context were the same genes previously identified in a daf-2(-) context, suggesting that there exists a transcriptional signature common to these two lifespan-extending pathways. This interpretation is strengthened by the finding that the same upstream-DNA sequences were overrepresented in DAF-16-regulated genes in these two longevity pathways. These overlapping targets may be of particular interest as genes central to DAF-16's regulation of lifespan. Because there were also many non-overlapping targets, it is likely that in addition to a shared signature of DAF-16 regulation, there are germline-specific and insulin/IGF-1-signaling specific targets as well.
As an independent assessment of this issue, in our laboratory, Ghazi et al. (2009) used GFP fusions to examine the expression of seventeen genes predicted from previous microarray and other experiments to be regulated by DAF-16. Several genes were regulated in the same way in daf-2 mutants and germline-defective animals, and others were up-regulated only in daf-2 mutants or only in germline-deficient animals. Interestingly, some were up-regulated in one set of tissues in animals lacking germ cells but in other tissues in daf-2 mutants. For example, a dod-8::GFP reporter was switched on in the intestine in response to germline loss, but in muscles and neurons in daf-2 mutants. Thus, while there does appear to be considerable overlap between the two sets of daf-16-dependent genes under these two conditions, there are distinct differences as well.
We also found a statistically very significant overlap between genes dependent on DAF-12 in the germline context and genes that are differentially expressed in daf-12 loss-of-function versus daf-12 gain-of-function mutants, which are short-lived and long-lived at low temperature, respectively (Fisher & Lithgow 2006). These findings suggest that DAF-12 can activate the same genes in more than one longevity pathway. As with DAF-16, this interpretation is strengthened by the finding that similar upstream DNA sequences were overrepresented in DAF-12-regulated genes in different longevity pathways.
One of the most interesting questions we wanted to address in this study was the question of the regulatory relationship between DAF-12 and DAF-16. Because DAF-12 is partially required for DAF-16 nuclear localization in germline-deficient animals, as is its known ligand dafachronic acid (Berman & Kenyon 2006; Gerisch et al. 2007), one could imagine that DAF-12 would be required for DAF-16 to affect the expression of any gene in response to loss of the germline. However, consistent with studies of a few specific daf-16-regulated genes (Wang et al. 2008; Yamawaki et al. 2010), we did not find this. A strict overlap comparison from our microarrays revealed a smaller overlap (7 genes, p<1E-3) between genes strongly influenced by both DAF-16 DAF-12 than one would expect if their target genes were all the same. We identified a much larger but not complete overlap when considering more subtle regulation of targets by both DAF-16 and DAF-12, which makes it clear that some, but not all, genes regulated by DAF-16 have a partial dependence on DAF-12, and vice versa, and in fact that is what we see using both qPCR (Figure S1), and GFP fusions to some of these genes in vivo (Yamawaki et al. 2010).
Some of the genes whose expression changed were known to have interesting effects on lifespan, including tcer-1 (Ghazi et al. 2009), and C46G7.2 (Kim & Sun 2007), one of the 7 genes co-regulated by daf-16 and daf-12. We tested additional candidate genes for RNAi lifespan phenotypes and identified several new genes that were completely required for loss of the germline to extend lifespan. One of these, phi-62, encodes a highly conserved putative RNA binding protein. One of our most intriguing findings was that the target specificity of PHI-62 seemed quite similar to the target specificity of TCER-1. This suggests the possibility that PHI-62 is part of the machinery that activates the expression of those DAF-16-regulated genes that utilize TCER-1 for transcription elongation. As such, this conserved protein could perform an essential function involving RNA metabolism during transcription. However, we did observe differences between these gene functions: knocking down tcer-1 changes DAF-16-dependent gene expression but not DAF-16 nuclear localization (Ghazi et al. 2009); whereas knocking down phi-62 reduces DAF-16 nuclear localization as well as DAF-16-dependent gene expression.
PHI-62 was predicted to bind to a known DAF-16-binding protein, the 14-3-3 protein FTT-2, so we tested FTT-2's role in the germline pathway. Like loss of PHI-62, loss of 14-3-3 activity does not shorten the lifespan of daf-2 mutants (Berdichevsky et al. 2006; Wang et al. 2006; Li et al. 2007), but it did completely abolish the longevity of germline-defective animals. (We note that because ftt-2 and its close homolog par-5 are predicted to cross-react with the ftt-2 RNAi clone we used to assay longevity, we do not know which is involved.) This finding suggests the possibility that DAF-16, PHI-62 and FTT-2 may act together to extend the lifespan of germline deficient animals (Figure 5). The role of 14-3-3 proteins in longevity is complex. In flies, loss of 14-3-3-protein function can be sufficient to trigger FOXO nuclear localization and extend wild-type lifespan (Nielsen et al. 2008). In contrast, in worms, loss of 14-3-3 proteins slightly shortens wild-type lifespan (Berdichevsky et al. 2006; Wang et al. 2006; Li et al. 2007). However, in our study we observed suppression of the glp-1-mutant longevity phenotype accompanied by a decrease in the level of DAF-16 nuclear localization, but no obvious decrease in the expression of the two DAF-16-regulated genes we examined. This makes the role of 14-3-3 proteins in this pathway enigmatic, suggesting that their further study could lead to new insights about their roles in FOXO-dependent signaling pathways.
Two of the genes that were required for the longevity of germline-deficient animals, lips-17 and fard-1, were involved in fat metabolism. Like K07A8.5, which was previously shown to be required for the longevity of germline-deficient animals, lips-17 encodes a triacylglycerol lipase (Wang et al. 2008). Thus, in germline-deficient animals, these two gene products presumably liberate long-chain fatty acids from triglycerides (forming fatty acyl CoA in the process). We considered the possibility that these two genes are both knocked down by the same RNAi clone, but this was not predicted to be the case when examined using the DEQOR software (Henschel et al. 2004). fard-1 encodes an acyl CoA reductase, which is a class of enzyme that reduces fatty acyl CoA molecules to aldehydes or alcohols. This enzyme could potentially act on the long-chain acyl-CoA molecules liberated by the triacylglycerol lipases. Inhibition of these three genes appeared to have little or no effect on overall fat levels (as measured by the fat indicator oil-red O). Because it is still not known how daf-16, which acts specifically in the intestine in this pathway, is able to affect the longevity of non-intestinal tissues, we propose that these three lipid metabolic genes may function in the synthesis or processing of a lipophilic longevity signal that allows intestinal DAF-16 to influence the lifespan of other tissues. Consistent with this model, KO4A8.5 is expressed in the intestine. These lipid-metabolic genes could be part of a pathway that also involves oleic acid, a monounsaturated fat that appears to be required for the longevity of germline-defective animals (Goudeau et al. 2011). The regulation of these fat metabolic genes is unexpectedly diverse. KO4A8.5 is regulated by daf-16 but not daf-12, whereas we identified fard-1 as a daf-12-regulated gene. The genes that synthesize oleic acid in response to germline loss are regulated by NHR-80 but not by DAF-12 or DAF-16. It will be interesting to learn whether lipid-metabolic genes regulated by different branches of the complex germline pathway might actually have distinct functions in lifespan determination.
Finally, we note that inhibition of a transcriptional mediator protein called MDT-15 shortens the lifespan of glp-1 mutants as well as wild type. This protein has previously been found to regulate many genes involved in fat metabolism, so it would be interesting to learn whether it regulates the expression of lips-17 and fard-1.
Experimental Procedures
Strains
All strains were maintained as described previously (Brenner 1974). CF1903: glp-1(e2141ts) III, CF1658: glp-1(e2141ts) III; daf-12(rh61rh411) X, CF1880: daf-16(mu86) I; glp-1(e2141) III, CF2032: muEx307 (Ptcer-1::tcer-1::gfp, Podr-1::rfp), CF2562: glp-1(e2141ts) III; sIs10314[Pdod-8::GFP + pCeh361], DA1116: eat-2(ad1116) II, CF1041: daf-2(e1370) III, CF1929: glp-1(e2141ts) II; muIs84[Psod-3::GFP], CF1935: daf-16(mu86)I; glp-1(e2141)III; muIs109 (Pdaf-16::daf-16::gfp, Podr-1::rfp),CF3201A: glp-1(e2141ts) III; sEx10466[Pnnt-1::GFP + pCeh361], CF3204: glp-1(e2141) III; sEx14516[pCes T21D12.9::GFP + pCeh361], CF3124: glp-1(e2141) III; sEx11128[Pgpd-2::GFP]
Microarray Analysis
Worms were grown synchronously at 20°C from eggs produced by bleaching mixed populations. They were transferred from 20° to 25°C between L2 and L4 to cause sterility in glp-1(-) strains, and harvested as early adults. Adults from these same harvested plates were set aside to confirm the expected lifespan phenotypes for all strains. 60 samples were hybridized, consisting of 20 wild-type, 12 daf-16, 9 daf-12, 9 glp-1, 3 glp-1; daf-12, and 7 daf-16; glp-1 samples. Data were analyzed using MAANOVA (Wu et al. 2003). For the long/short lived lists, a linear model with block-specific terms for daf-12, daf-16, germline(+)/germline(-), and long-lived was generated and the long-lived term was tested. For the specific target lists, a linear model with block-specific terms for daf-12 (or daf-16) and germline(+)/germline(-) was generated and the daf-12:germline(-) [or daf-16:germline(-)] interaction term was tested. For the daf-16 targets analysis, daf-12 mutant-containing arrays were excluded, and vice-versa. All lists used an Fs:Ptab 0.01 cutoff. Additional enrichment analysis was done using GSEA v2.0 (Subramanian et al. 2005).
Gene Ontology Analysis
Data were analyzed using the BiNGO 2.3 plugin (Maere et al. 2005) for Cytoscape 2.6 (Shannon et al. 2003), using a hypergeometric probability with Benjamini-Hochberg false discovery rate correction at a p<0.05 cutoff.
RNAi clone analysis
The identity of all RNAi clones was verified by sequencing the inserts using the M13-forward primer. The ftt-2 and par-5 specific RNAi constructs used to assay DAF-16 localization were kindly provided by the Lee lab (Li et al. 2007). pAD43 was used as the daf-16 RNAi clone (Dillin et al. 2002); all other clones were from Julie Ahringer's RNAi library (Kamath et al. 2003).
Lifespan analysis
Lifespan analysis was conducted at 20°C as described previously unless otherwise stated (Hsin & Kenyon 1999). For all germline(-) worms and parallel controls, animals were transferred from 20°C to 25°C after 24 hours at 20°C, and transferred back to 20°C as late L4/early adult. RNAi treatments were performed as whole-life treatments placed on plates seeded with the RNAi bacteria of interest. In the adult-only analysis, eggs were added to plates seeded with control RNAi bacteria, and adult animals were transferred to gene-specific RNAi-bacterial plates. The chemical 2’fluoro-5’deoxyuridine (FUDR, Sigma) was added to experimental and control adult worms (100 μM) in some experiments to prevent their progeny from developing. Strains were grown at 20°C under optimal growth conditions for at least two generations before use in lifespan analysis. STATA software was used for statistical analysis and to determine means and percentiles. In all cases, p-values were calculated using the Log-rank (Mantel-Cox) method.
Microscopy
All fluorescence images were captured using a Retiga EXi Fast1394 CCD digital camera (QImaging, Burnaby, BC, Canada) attached to a Zeiss Axioplan 2 compound microscope (Zeiss Corporation, Jena, Germany). Openlab 4.0.2 software (Improvision, Coventry, U.K.) was used for image acquisition. GFP assays were conducted on a Leica MZ16F (Wetzlar, Germany) stereomicroscope with fluorescence filter sets or the Zeiss Axioplan 2 compound microscope mentioned above.
Quantitative PCR
Total RNA was extracted using TRIzol reagent (Invitrogen) and purified using Qiagen RNAeasy Mini Kit. cDNA was generated using Protoscript First Strand cDNA Synthesis Kit (New England Biolabs). SybrGreen real-time Q-PCR reactions were performed on an Applied Biosystems 7300 Real Time PCR System. Oligos were used at a final concentration of 0.55 μM, with a 60°C annealing temperature for 30s. ama-1 was used as a reference gene. p-values were calculated using a two-tailed Student's t-test. Primers used were: gpd-2, 5’-AAG GCC AAC GCT CAC TTG AA-3’ and 5’-GGT TGA CTC CGA CGA CGA AC-3’, lips-17, 5’-ATC TGT TGC TGG AGC CAA TCG-3’ and 5’-TAT CCA ACT TTA TCG TCT CC-3’, nnt-1, 5’-CAG TAG AAA CTG CTG ACA TGC TTC-3’ and 5’-GAG CGA TGG GAT ATT GTG CCT GAG-3’, and sod-3, 5’-AAA GGA GCT GAT GGA CAC TAT TAA GC-3’ and 5’-AAG TTA TCC AGG GAA CCG AAG TC-3’. phi-62 has a very small ORF (95aa) and we were unable to amplify it for quantification successfully with the following four sets of primers: 5’-GCT TTC TGC ATG GTC ATG TCG G-3’ and 5’-CTC GAT GGA ACC TTT CCG TG-3’, 5’-CCA TCG AGT GTA ATC GAC GC-3’ and 5’-CGT GTT GTA TTT ATT CTG CC-3’, 5’-TCG GCT TTC TGC ATG GTC ATG TCG-3’ and 5’-ATG AAG ATC TGG GAA GAG AG -3’, and 5’-GGT TCC ATC GAG TGT AAT CGA CGC-3’ and 5’-AAG AGT AAC GCA TAG AGT CC-3’.
DAF-16::GFP Localization
Eggs were picked from parents raised on OP-50 bacteria at 20°C, and placed on IPTG-induced RNAi bacterial lawns on NG-carbenicillin plates to hatch. glp-1(e2141ts) worms were transferred to 25°C after 24hrs, then to 20°C at the late L4 stage. Adult worms were scored using a Zeiss SteREO Lumar.V12.
Supplementary Material
Acknowledgments
We thank members of the Kenyon Lab for valuable insights and discussions, and Elizabeth Tank for additional assistance with obtaining strains and reagents. We thank the Caenorhabditis Genetics Center (supported by the National Institutes of Health- National Center for Research Resources) for strains. M.M. was supported by a predoctoral fellowship in the biological sciences from the Howard Hughes Medical Institute. This work was funded by National Institute of Health grants RO1 AG020932 and RO1 AG032435 to C.K., who is an American Cancer Society Research Professor, and director of UCSF's Hillblom Center for the Biology of Aging.
References
- Anselmi CV, Malovini A, Roncarati R, Novelli V, Villa F, Condorelli G, Bellazzi R, Puca AA. Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 2009;12:95–104. doi: 10.1089/rej.2008.0827. [DOI] [PubMed] [Google Scholar]
- Ao W, Gaudet J, Kent WJ, Muttumu S, Mango SE. Environmentally induced foregut remodeling by PHA-4/FoxA and DAF-12/NHR. Science. 2004;305:1743–1746. doi: 10.1126/science.1102216. [DOI] [PubMed] [Google Scholar]
- Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science. 2002;295:502–505. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- Berdichevsky A, Viswanathan M, Horvitz HR, Guarente L. C. elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell. 2006;125:1165–1177. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- Berman JR, Kenyon C. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell. 2006;124:1055–1068. doi: 10.1016/j.cell.2006.01.039. [DOI] [PubMed] [Google Scholar]
- Bluher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, Johnson TE, Kim SK. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell. 2008;134:291–303. doi: 10.1016/j.cell.2008.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargill SL, Carey JR, Muller HG, Anderson G. Age of ovary determines remaining life expectancy in old ovariectomized mice. Aging Cell. 2003;2:185–190. doi: 10.1046/j.1474-9728.2003.00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristina D, Cary M, Lunceford A, Clarke C, Kenyon C. A regulated response to impaired respiration slows behavioral rates and increases lifespan in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000450. doi: 10.1371/journal.pgen.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Fisher AL, Lithgow GJ. The nuclear hormone receptor DAF-12 has opposing effects on Caenorhabditis elegans lifespan and regulates genes repressed in multiple long-lived worms. Aging Cell. 2006;5:127–138. doi: 10.1111/j.1474-9726.2006.00203.x. [DOI] [PubMed] [Google Scholar]
- Flachsbart F, Caliebe A, Kleindorp R, Blanche H, von Eller-Eberstein H, Nikolaus S, Schreiber S, Nebel A. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci U S A. 2009;106:2700–2705. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T, Min KJ, D'Alterio C, Villa-Cuesta E, Cumbers J, Lehmann R, Jones DL, Tatar M. Drosophila germ-line modulation of insulin signaling and lifespan. Proc Natl Acad Sci U S A. 2008;105:6368–6373. doi: 10.1073/pnas.0709128105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T, Nakazawa T, Nakano I, Mori N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J. 2000;349:629–634. doi: 10.1042/0264-6021:3490629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch B, Rottiers V, Li D, Motola DL, Cummins CL, Lehrach H, Mangelsdorf DJ, Antebi A. A bile acid-like steroid modulates Caenorhabditis elegans lifespan through nuclear receptor signaling. Proc Natl Acad Sci U S A. 2007;104:5014–5019. doi: 10.1073/pnas.0700847104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazi A, Henis-Korenblit S, Kenyon C. A transcription elongation factor that links signals from the reproductive system to lifespan extension in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000639. doi: 10.1371/journal.pgen.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakou ME, Goss M, Jacobson J, Vinti G, Leevers SJ, Partridge L. Dynamics of the action of dFOXO on adult mortality in Drosophila. Aging Cell. 2007;6:429–438. doi: 10.1111/j.1474-9726.2007.00290.x. [DOI] [PubMed] [Google Scholar]
- Giannakou ME, Goss M, Junger MA, Hafen E, Leevers SJ, Partridge L. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science. 2004;305:361. doi: 10.1126/science.1098219. [DOI] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL, Jr., White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Goudeau J, Bellemin S, Toselli-Mollereau E, Shamalnasab M, Chen Y, Aguilaniu H. Fatty Acid Desaturation Links Germ Cell Loss to Longevity Through NHR-80/HNF4 in C. elegans. PLoS Biol. 2011;9:e1000599. doi: 10.1371/journal.pbio.1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- Henschel A, Buchholz F, Habermann B. DEQOR: a web-based tool for the design and quality control of siRNAs. Nucleic Acids Res. 2004;32:W113–120. doi: 10.1093/nar/gkh408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochbaum D, Zhang Y, Stuckenholz C, Labhart P, Alexiadis V, Martin R, Knolker HJ, Fisher AL. DAF-12 Regulates a Connected Network of Genes to Ensure Robust Developmental Decisions. PLoS Genet. 2011;7:e1002179. doi: 10.1371/journal.pgen.1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kenyon C. A pathway that links reproductive status to lifespan in Caenorhabditis elegans. Ann N Y Acad Sci. 2010a;1204:156–162. doi: 10.1111/j.1749-6632.2010.05640.x. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010b;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kerr MK, Churchill GA. Experimental design for gene expression microarrays. Biostatistics. 2001;2:183–201. doi: 10.1093/biostatistics/2.2.183. [DOI] [PubMed] [Google Scholar]
- Kerr MK, Martin M, Churchill GA. Analysis of variance for gene expression microarray data. J Comput Biol. 2000;7:819–837. doi: 10.1089/10665270050514954. [DOI] [PubMed] [Google Scholar]
- Kim Y, Sun H. Functional genomic approach to identify novel genes involved in the regulation of oxidative stress resistance and animal lifespan. Aging Cell. 2007;6:489–503. doi: 10.1111/j.1474-9726.2007.00302.x. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RY, Hench J, Ruvkun G. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr Biol. 2001;11:1950–1957. doi: 10.1016/s0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- Li J, Tewari M, Vidal M, Lee SS. The 14-3-3 protein FTT-2 regulates DAF-16 in Caenorhabditis elegans. Dev Biol. 2007;301:82–91. doi: 10.1016/j.ydbio.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang WJ, Cao H, Lu J, Wu C, Hu FY, Guo J, Zhao L, Yang F, Zhang YX, Li W, Zheng GY, Cui H, Chen X, Zhu Z, He H, Dong B, Mo X, Zeng Y, Tian XL. Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Mason JB, Cargill SL, Anderson GB, Carey JR. Transplantation of Young Ovaries to Old Mice Increased Life Span in Transplant Recipients. J Gerontol A Biol Sci Med Sci. 2009 doi: 10.1093/gerona/glp134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Nielsen MD, Luo X, Biteau B, Syverson K, Jasper H. 14-3-3 Epsilon antagonizes FoxO to control growth, apoptosis and longevity in Drosophila. Aging Cell. 2008;7:688–699. doi: 10.1111/j.1474-9726.2008.00420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollen EA, Garcia SM, van Haaften G, Kim S, Chavez A, Morimoto RI, Plasterk RH. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci U S A. 2004;101:6403–6408. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke EJ, Soukas AA, Carr CE, Ruvkun G. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 2009;10:430–435. doi: 10.1016/j.cmet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlikowska L, Hu D, Huntsman S, Sung A, Chu C, Chen J, Joyner AH, Schork NJ, Hsueh WC, Reiner AP, Psaty BM, Atzmon G, Barzilai N, Cummings SR, Browner WS, Kwok PY, Ziv E. Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell. 2009;8:460–472. doi: 10.1111/j.1474-9726.2009.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampias TN, Fragoulis EG, Sideris DC. Genomic structure and expression analysis of the RNase kappa family ortholog gene in the insect Ceratitis capitata. FEBS J. 2008;275:6217–6227. doi: 10.1111/j.1742-4658.2008.06746.x. [DOI] [PubMed] [Google Scholar]
- Rampias TN, Sideris DC, Fragoulis EG. Cc RNase: the Ceratitis capitata ortholog of a novel highly conserved protein family in metazoans. Nucleic Acids Res. 2003;31:3092–3100. doi: 10.1093/nar/gkg414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V, Gil IS, Ward S, Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311–323. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- Schroeder LK, Kremer S, Kramer MJ, Currie E, Kwan E, Watts JL, Lawrenson AL, Hermann GJ. Function of the Caenorhabditis elegans ABC transporter PGP-2 in the biogenesis of a lysosome-related fat storage organelle. Mol Biol Cell. 2007;18:995–1008. doi: 10.1091/mbc.E06-08-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shostak Y, Van Gilst MR, Antebi A, Yamamoto KR. Identification of C. elegans DAF-12-binding sites, response elements, and target genes. Genes Dev. 2004;18:2529–2544. doi: 10.1101/gad.1218504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soerensen M, Dato S, Christensen K, McGue M, Stevnsner T, Bohr VA, Christiansen L. Replication of an association of variation in the FOXO3A gene with human longevity using both case-control and longitudinal data. Aging Cell. 2010;9:1010–1017. doi: 10.1111/j.1474-9726.2010.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 2009;23:496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi A, Wartschow LM, White MF. Brain IRS2 signaling coordinates life span and nutrient homeostasis. Science. 2007;317:369–372. doi: 10.1126/science.1142179. [DOI] [PubMed] [Google Scholar]
- Taubert S, Hansen M, Van Gilst MR, Cooper SB, Yamamoto KR. The Mediator subunit MDT-15 confers metabolic adaptation to ingested material. PLoS Genet. 2008;4:e1000021. doi: 10.1371/journal.pgen.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert S, Van Gilst MR, Hansen M, Yamamoto KR. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 2006;20:1137–1149. doi: 10.1101/gad.1395406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden J, Andre B, Collado-Vides J. Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J Mol Biol. 1998;281:827–842. doi: 10.1006/jmbi.1998.1947. [DOI] [PubMed] [Google Scholar]
- Wang MC, O'Rourke EJ, Ruvkun G. Fat metabolism links germline stem cells and longevity in C. elegans. Science. 2008;322:957–960. doi: 10.1126/science.1162011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Oh SW, Deplancke B, Luo J, Walhout AJ, Tissenbaum HA. C. elegans 14-3-3 proteins regulate life span and interact with SIR-2.1 and DAF-16/FOXO. Mech Ageing Dev. 2006;127:741–747. doi: 10.1016/j.mad.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A, Boutis P, Hekimi S. Mutations in the clk-1 gene of Caenorhabditis elegans affect developmental and behavioral timing. Genetics. 1995;139:1247–1259. doi: 10.1093/genetics/139.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Kerr MK, Cui X, Churchill GA. MAANOVA: a software package for the analysis of spotted cDNA microarray experiments. In: Parmigiani G, Garrett E, Irizarry R, Zeger S, editors. The Analysis of Gene Expression Data: Methods and Software. Springer-Verlag; New York: 2003. pp. 313–341. [Google Scholar]
- Yamawaki TM, Berman JR, Suchanek-Kavipurapu M, McCormick M, Maria Gaglia M, Lee SJ, Kenyon C. The somatic reproductive tissues of c. elegans promote longevity through steroid hormone signaling. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Sternberg PW. Genome-wide prediction of C. elegans genetic interactions. Science. 2006;311:1481–1484. doi: 10.1126/science.1123287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.