Abstract
A mutant(‘lab strain’) of the hyperthermophilicarchaeonPyrococcusfuriosusDSM3638 exhibited an extended exponential phase andatypical cell aggregation behavior. Genomic DNA from the mutant culturewas sequenced and compared to wild-type (WT) DSM3638, revealing145genes with one or more insertions, deletions, or substitutions (12 silent, 33amino acid substitutions, and 100 frame-shifts). Approximately half of the mutated genes weretransposases or hypothetical proteins.The WT transcriptomerevealednumerous changes in amino acid and pyrimidine biosynthesis pathways coincidental with growth phase transitions, unlike the mutant whose transcriptome reflected the observedprolonged exponential phase. Targeted gene deletions,based on frame-shifted ORFs in the mutant genome, in a genetically tractable strain of P. furiosus (COM1) could not generate the extended exponential phase behavior observed for the mutant. For example, a putative radical SAM family protein (PF2064) was the most highly up-regulated ORF (>25-fold) in the WT between exponential and stationary phase, although this ORF was unresponsive in the mutant; deletion of this gene in P. furiosus COM1 resulted in no apparent phenotype. On the other hand, frame-shifting mutations in the mutantgenome negatively impacted transcription ofa flagellar biosynthesis operon (PF0329-PF0338).Consequently, cells in the mutant culture lacked flagella and, unlike the WT, showed minimal evidence of exopolysaccharide-based cell aggregation in post-exponential phase. Electron microscopyofPF0331-PF0337 deletionsin P. furiosus COM1 showed that absence of flagella impacted normal cell aggregation behavior and, furthermore,indicatedthat flagella play a key role, beyond motility, in thegrowthphysiology ofP. furiosus.
Keywords: Pyrococcusfuriosus, hyperthermophile, archaeon, growth physiology, flagella
INTRODUCTION
Historically, the occurrence of unintended mutations in microorganisms(associated with ‘lab strains’) has been difficult to discern, unless the genetic change confers a readily observable loss or gain in function. Furthermore, even when a functional change is evident, it is usually not mapped back to a specific mutation that is responsible for the physiological alteration. However, as DNA sequencing capabilities have expanded and become more affordable, microbial genome integrity can be readily confirmed. Specific nucleotides associated with any mutationsgenome-wide can then be identified and examined in the context of possible effects on a microorganism’s growth physiology.
The susceptibility of microorganisms to mutations has often been considered from the impact that natural environments have on DNA integrity(Drake 1991a; Drake 1991b; Drake 1991c; Maki 2002). Of particular interest have been the extreme thermophiles that, by virtue of the high temperature biotopes that they inhabit, could be especially vulnerable to DNA damage (DiRuggiero et al. 1999; Drake 2009; Grogan 1998; Grogan et al. 2001; Mackwan et al. 2008; Majernik et al. 2004; Makarova et al. 2002; Martusewitsch et al. 2000; Omelchenko et al. 2005; Watrin and Prieur 1996; Williams et al. 2007). The relative rate of mutations in extreme thermophiles compared to mesophiles is not known in any comprehensive sense. Intuitively, high temperatures potentially present an intrinsic challenge to DNA integrity. However, extreme thermophiles may actually have lower mutation rates than less thermophilic microorganisms(Drake 2009).
The consequence of mutations has been investigated insome extremely thermophilic microorganisms. For example, spontaneous mutation characteristics in target genes in Thermusthermophilus (a bacterium) and Sulfolobusacidocaldarius (an archaeon) were found to be comparable, despite the phylogenetic and physiological differences that exist between the two microorganisms (Mackwan et al. 2008). Also, a chloramphenicol-resistant, mutant of the hyperthermophilic bacterium Thermotogamaritimacontainedseveral nucleotide changes in ribosomal genes, resulting in significant transcriptomic differences between the mutant and wild-type during growth in the presence and absence of the antibiotic (Montero et al. 2007).Mutations were identified in a laboratory strain of the hyperthermophilicarchaeon,PyrococcusfuriosusDSM 3638(Bridger et al. 2012). TermedCOM1, this strain was found to bea genetically tractable mutant with over 100 impacted genes through chromosomal insertions and deletions. COM1 has served as the basis for development of a genetic system for P. furiosus that has beenused to explore aspects of this archaeon’s physiology (Bridger et al. 2011; Lipscomb et al. 2011) and for metabolically engineeringpurposes to produce biofuels (Keller et al. 2013).
Here, a mutant of P. furiosusDSM3638arose over a period of time from efforts to improve growth media. P.furiosustypically is cultured on a variety of carbohydrates as primary carbon and energy sources, including maltose (Costantino et al. 1990), cellobiose(Voorhorst et al. 1999), β-glucans(Driskill et al. 1999), and starch (Lee et al. 2006), but also requires a complex component, such as yeast extract, tryptone or peptone for unspecified nutritional needs(Snowden et al. 1992). In all cases, CO2, acetate, alanine (Kengen et al. 1994), and H2 (or H2S, when grown on S°) are the primary metabolic products (Chou et al. 2007). While the use of pyruvate as the sole energy and carbon source has also been shown(Ward et al. 2000), defined media based on combinations of individual amino acids does not support significant growth of P. furiosus. However, successful attempts to grow other Thermococcales on mixtures of single amino acids have been reported (Hoaki et al. 1994; Hoaki et al. 1993; Watrin et al. 1995):Pyrococcusabyssi GE5 grew on a mixture of nine amino acids and vitamins (Watrin et al. 1995), andThermococcuslitoralis grew on a defined medium of 16 amino acids (Rinker and Kelly 1996; Rinker and Kelly 2000). Although single amino acids may not serve as sole carbon and energy sources for P. furiosus, they may improve growth as medium supplements.
In this study, maltose-based cultures containing yeast extractwere passed on media supplemented with L-tryptophan, L-glutamine, L-asparagine, and thiamine-HCl, resulting in a culture thathad an extended exponential phase and,consequently, achieved significantly higher final cell densities compared to the wild-type (WT). Furthermore, atypical cellaggregation patternswere observed for the mutant strain, especially at high cell densities. Sequencing of the genome of the mutant, in conjunction with transcriptomic analysis, revealed changes in severalgenesthatwere examined in light of the physiological differences between the mutant and WTcultures.
MATERIALS AND METHODS
Cultivation of Pyrococcusfuriosus DSM 3638 andmutant cultures
P. furiosus DSM3638 (WT) and acorresponding mutantwere cultured anaerobically at 80°C or 95°C on sea salts medium (containing per liter: 40 g of sea salts (Sigma-Aldrich, St. Louis, MO), 3.1 g of PIPES [piperazine-N,N-bis(2-ethanesulfonic acid)], 1 g of yeast extract, and 1 ml of a trace elements stock, which contained the following (per 100 ml):nitrilotriacetic acid, 1.50 g; FeCl2·6H2O, 0.50 g; Na2WO7·2H2O, 0.30 g; MnCl2·4 H2O, 0.40 g; NiCl2·6H2O, 0.2 g; ZnSO4·7H2O, 0.1 g; CoSO4·7H2O, 0.1 g; CuSO4·5H2O (10 mg/ml), 1.0 ml; and Na2MoO4·5H2O (10 mg/ml)) in an orbital shaking oil bath at 80 rpm. Cultures were grown in 1 L bottles with 370 mL sea salts medium containing 3.3 g/L maltose, using a 0.5% (v/v) inoculum already established on the growth substrate. Cell enumeration was performed throughout the growth by epifluorescence microscopy with acridine orange staining (Hobbie et al. 1977). Maltose consumption rates were determined for both WT andmutant cultures by measuring the amount of glucose present in the cell-free spent media, using anα-glucosidase (Sigma-Aldrich, St. Louis, MO) and a liquid glucose (oxidase) reagent set (Pointe Scientific Inc., Brussels, Belgium). A baseline was established by measuring glucose present in samples, with and without the addition of the α-glucosidase.
RNA isolation and purificationfor transcriptomics
WT and mutantcultures were harvested by rapid cooling, followed by centrifugation at 4,230 × g for 10 min; samples were taken at mid-exponential phase (10h, ~5 × 107 cells/mL), the bifurcation point in the growth curve of the two strains (14h, 9.7 × 107 cells/mL for WT and 1.2 × 108 cells/mL for the mutant), and stationary phase (24h, 2.7 × 108 cells/mL for WT and 1.6 × 109 cells/mL for mutant). After centrifugation, cells were washed in ice-cold TE buffer and stored at −80°C until further processing. Total RNA was isolated by re-suspending the cells in TRIzol reagent (Invitrogen, San Diego, CA). RNA was purified using an RNEasy kit (Qiagen, Valencia, CA). RNA was then reverse transcribed to cDNA using Superscript III (Invitrogen), random primers (Invitrogen), and the incorporation ofaminoallyl-DUTP, 5-(3-aminoallyl)-2’-deoxyuridine 5’-triphosphate (Ambion, Austin, TX), as described previously (Chou et al. 2007). cDNA was labeled with either Cy3 or Cy5 dye (GE Healthcare, Little Chalfont, United Kingdom), and hybridized to one of six microarray slides (Corning, Acton, MA) in the loop design (see Figure S1), following protocols previously described(Chou et al. 2007).
Transcriptional response analysis
Slides were scanned using an Axon scanner (Sunnyvale, CA) and quantified using GenePix Pro, version 6.0 software. Normalization of data and statistical analysis of a mixed effects ANOVA model for the microarray loop were performed using JMP Genomics (version 3.1) software (SAS, Cary, NC). Differential transcription was defined as a log2 value of ±1 with −log10 p-values greater than 4.7. Microarray data will be deposited to GEO upon acceptance of the manuscript.
Bioinformatics analysis
Sequence information for P. furiosus DSM3638 was obtained from NCBI database (http://www.ncbi.nlm.nih.gov/genome). KEGG ortholog annotation and pathway identification were performed on proteins encoded in ORFs meeting the log2 value of ±1 threshold and with −log10 p-values greater than 4.7, using the KO-Based Annotation System (http://kobas.cbi.pku.edu.cn/) (Wu et al. 2006). Protein alignments were performed using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2). Results from the Database of prOkaryoticOpeRons (DOOR) were used for operon prediction (Dam et al. 2007; Mao et al. 2009). The genome sequences were compared and analyzed with the help of ViroBLAST 2.2+ (Deng et al. 2007).
DNA isolation
The WT and SM cultureswere grown overnight on sea salts medium, supplemented with 1 g/L yeast extract and 3.3 g/L maltose. Cells were harvested at 4,230 × g for 10 min, and the cell pellet was washed with ice cold TE buffer. Genomic DNA was isolated using TE-saturated Phenol (pH 8.0) (Ambion, Austin, TX), with additional purification steps, including Proteinase K, SDS, and RNaseA (Invitrogen), followed by chloroform/isoamyl alcohol extraction and isopropanol precipitation. Genomic DNA was validated using 260/280 ratios, as well as visualization on 0.7% agarose gel. P. furiosusspecific primers for PF0073 were used to confirm the identity of both P. furiosusDSM3638 and the P. furiosusmutant.
Genome sequencing of theP.furiosusSM strain
Purified P. furiosus mutant genomic DNA was submitted to the Genomic Sciences Laboratory at North Carolina State University for sequencing. Sequencing was performed using a 454 GS FLX (Roche Diagnostics, Indianapolis, IN). Genome coverage was approximately 10x. The average read length was 378 nucleotides, with a median length of 430.
Electron Microscopy
Transmission Electron Microscopy (TEM) imaging of cells was carried out by performing a negative stain of cell sample using 1% (w/v) sodium phosphotungstate 5% (w/v) bacitracin solution (Hayat 1990). Scanning Electron Microscopy (SEM) imaging of cells was performed using 12-mm round glass cover slips coated with a 0.1% solution of poly-L-lysine in phosphate-buffered saline (pH 7.2) (Mazia et al. 1975).
Construction of a knock-out cassette by overlapping PCR
The knock-in cassette was created by overlapping PCR, where the primers used contained approximately 20-30 base pair overhangs (Table S1, Figures S2-S4).The selectable marker and flanking regions were amplified from pGLW021 (Lipscomb et al. 2011) and P. furiosusstrain COM1 genomic DNA, respectively. The diagrammatic representation of the knock-out(KO) cassette and sequence detail has been provided in FiguresS2-S4. PCR products corresponding to flanking region and marker cassette were purified using a commercial extraction protocol (Stratagene,Santa Clara, CA) and were used in overlapping PCR reactions using the Prime STARHS Premix (Clonetech). Final overlapping PCR products were gel purified and used for COM1 transformation, as previously described(Lipscomb et al. 2011).
Pyrococcusfuriosus transformation and construction of theΔPF0337,ΔPF0331-PF0337, and ΔPF1269, ΔPF2064 strains
Transformations were carried out using freshly grown COM1 strain (uracil auxotroph), a competent strain of P. furiosus(Bridger et al. 2012). For transformation, 500 ng DNA (knock-out cassette was mixed with 100 μl of an overnight culture of COM1 cells and grown on defined medium. After incubation at 90°C for 68 hours, plates were examined for colonies on defined medium plates for gain of the pyrF marker, after marker replacement, as transformed cells are able to grow without uracil. Three colonies were picked from defined medium plates, grown overnight in 5 ml defined media and 1.5 ml samples were used to isolate genomic DNA. PCR was used to confirm the correct insertion using the primers listed in Figures S2-S4, which were designed to bind outside of the homologous flanking regions (UFR) and marker cassette. PCR-positive colonies were further purified by three separate consecutive transfers in defined media lacking uracil in order to confirm the culture phenotype. PCR screening was done after each round to ensure proper incorporation of the knock-in cassette was maintained. All three knock-outs were confirmed by PCR using gene specific primers.
RNA isolation and qPCR for verification of knock-out
Total RNA was extracted from 10 ml of mid-log phase cultures grown in rich maltose medium using the Absolute RNA prep kit (Stratagene) and quantified by ThermoScientificNanodrop spectrophotometer. Before qPCR analyses, the RNA was treated with TURBODNase (Ambion; AppliedBiosystems, Bedford, CA, USA) for 30 min at 37°C and further purified using the DNase inactivation reagent (Ambion; Applied Biosystems, Bedford, CA). cDNA was prepared using the Affinity Script quantitative PCR (qPCR) cDNA synthesis kit (Agilent Technologies, Santa Clara, CA). All quantitative reverse transcription-PCR (RT-qPCR) experiments were carried out with an Mx3000P instrument (Stratagene) with the Brilliant SYBR Green qPCR master mix (Agilent Technologies). The genes encoding the pyruvate ferredoxinoxidoreductase gamma subunit (PF0971) were used as internal positive controls. Internal controls were taken into account in order to demonstrate that equal amounts of same cDNA were used for qPCR.
RESULTS
Growth dynamics of P. furiosusWT and mutant cultures
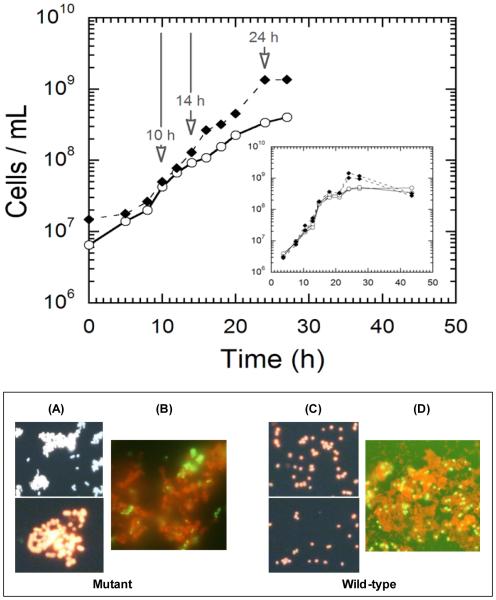
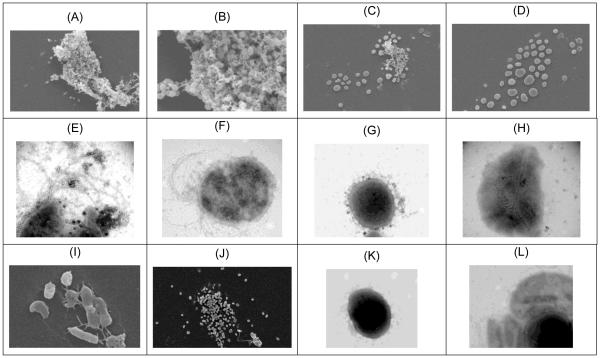
Growth ofP. furiosus on maltose-based media, in the absence of elemental sulfur,typically achieves cell densities of approximately 2-5 × 108 cells/mL, depending upon complex media components (e.g., yeast extract)(Blumentals et al. 1990). Previous analysis of the P. furiosusstationary phase transcriptome implicated certain amino acid and vitamin limitations in stationary phase onset (Chou and Kelly, unpublished data). In an effort to improve cell yields, P. furiosusWT was serially passed at 80°C on maltose (no sulfur), supplemented with L-tryptophan, L-glutamine, L-aspartate, and thiamine-HCl. Growth at 80°C, rather than 98°C (the Topt), was done to minimize thermally induced deamination of L-glutamine. Although the supplemented cultures grew to cell densities in excess of 109 cells/mL, it was subsequently determined that these cell density levels were also observed in some cultures lacking the additional amino acids and thiamine. As such, a mutant culturewas suspected. Growth rate comparisons were then done with a fresh stock culture of P. furiosus DSM3638 and the mutant at 80°C and 98°C. At 80°C, both cultures exhibited the same exponential growth rates (td= ~2 hours) (see Figure 1) and had similar maltose consumption profiles. Comparable results were observed at 98°C, although the growth rates were ~ 1.5-fold faster(data not shown). Growth of WTat 80°C leveled off at ~5 × 108 cells/mL. However, the mutant culture continued to grow to in excess of 109 cells/ml. Unlike the WT, the mutantcultures experienced a rapid loss in viability, coupled with cell lysis and little or no evidence of a longstationary phase (see Figure 1, top inset). Epifluorescence microscopy of culture samples taken post-24h after inoculation indicatedthatcells in the mutant culture were spatially closer compared to the WT. Calcofluor staining of cells post-24h revealed significant exopolysaccharide(EPS) formation for both WT and mutant (see Figure 1-B,D). However, SEM images of post-24h samples showed that while the WT was encased in EPS (Figure 2-A,B), very little EPS was associated with the mutant. SEM images also showed that cells in the mutant culture formed an array of spatially-proximate cells, but with no apparent physical connection between cells (Figure 2-C,D). Another distinctive difference noted through TEM was that, unlike the WT (Figure 2-E,F),cells in the mutant culture(Figure 2-G,H) were not flagellated. Taken together,results showed that the mutant culture had an extended exponential phase, rapidly lysed once the peak cell density was reached, produced EPS that, unlike the WT, was not directly associated with cell aggregates (by SEM imaging), and was likely non-motile given the absence of flagella.
Figure 1.
(Top) Growth of P. furiosusDSM 3638 (O) and the P. furiosusmutant (◆) at 80°C. Samples were taken at 10, 14 and 24 hours for transcriptomic analysis. Inset shows mutant cell density decreasing from cell lysis.
(Bottom) A and C: Epifluorescence micrographs (acridine orange stain) of mutant (left two panels) and wild-type (right two panels) P. furiosus cultures after 24 hours post-inoculation, showing significant cell aggregation in the mutant compared to wild-type strain, B and D: Epifluorescence micrographs (acridine orange and calcofluorstain) of the mutant (left panel) and WT (right panel) cultures at 14h showing production of EPS surrounding the cells from exponential phase.
Figure 2.
Electron micrographs of wild-type (WT) and mutantP. furiosus post-24h following inoculation. A,B. WT (SEM); C,D. mutant (SEM); E,F. WT (TEM); G,H. mutant (TEM); I. PF0337 KO(SEM); J. PF0331 KO (SEM); K. PF0337 KO(TEM); L. PF0331 KO (TEM).
Genome sequencing of the P. furiosusmutant strain
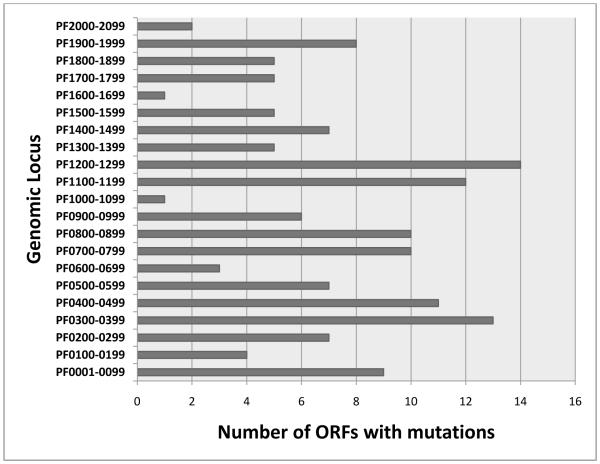
The basis for the differences betweenthe WT and the mutant was investigated by sequencing the genomic DNA of the mutant culture. Assembly of the ~157 K sequences yielded a draft genome for the mutant culture of 1.89 Mb (compared to 1.91 Mb for the wild-type), organized into 64 large contigs with an average size of ~29 Kb; the gaps between contigs likely accounted for most of the 20K difference between the two sequences.The contigs were compared to the P. furiosusDSM3638 genome sequence (NC_003413), revealingover 1,000 mutations, both within ORFs and within intergenic regions. A total of 145 ORFs in the mutant were not identical to the corresponding ORFs in DSM3638 (see TablesS2, S3).Of these, 12involved silent mutations, 33 had in-frame changes in amino acid residues, and 100 had frameshifts (see TablesS2, S3). Approximately half of the affected ORFs were annotated as either hypothetical proteins (57) or transposes (~18). It is interesting that many (~ 20) of the hypothetical proteins with mutations corresponded to ORFs of less than 400 nucleotides. The mutations within ORFs were distributed throughout the genome (see Figure 3), as were mutations in intergenic regions.
Figure 3.
Distribution of ORFs containing mutations in the P. furiosus mutant genome. Mutations in intergenic regions were also distributed throughout the genome (data not shown).
Genome-wide transcriptomic analysis of wild-type (WT)P. furiosusDSM3638 and the mutant
To further examine the nature and consequence of the substitutions, deletions and insertions in the mutant genome, a cDNA microarray for P. furiosus was used to compare the two strains. An experimental loop was designed (Figure S1) such that the growth phase trajectory of each strain could be tracked, as well as allowing for cross-strain contrasts at the three time points: exponentialphase (10 hours), a bifurcation point between the growth curves (14h), and at 24h when both cultures appeared to be in stationary phase.Transcriptomic changes corresponding to growth phase transitions were first analyzed for each strain, following which the two strains were compared at each sampling time.
Wild-type (WT) growthtranscriptome
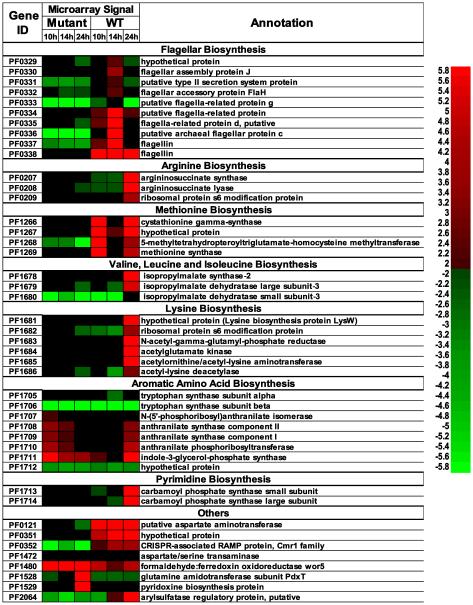
As microorganisms transition from exponential to stationary phase, a host of cellular responses are triggered in response to nutrient limitations, intercellular signaling,and inhibiting effect of metabolic products. For the WT, transition from exponential(10h) to post-exponentialphase (14h)was accompanied by the two-fold or more differential transcription of 92 ORFs (62 up/30 down), including up-regulation of genes involved in flagellarbiosynthesis (PF0330-0337) and down-regulation of genes involved in methionine/cysteine biosynthesis (PF1266-1269) (see TableS4,Figure 4).As the WT moved from post-exponential(14h) tostationary phase (24h), these patterns reversed such that flagellar protein synthesis was significantly reduced, concomitant with the up-regulation of many amino acid and purine/pyrimidine pathways. For this transition, 114 ORFs (69 up/45 down)were differentially transcribed. Of particular note in WT stationary phase were genes in the PF1678-1712 locus, which encodes proteins involved in amino acid biosynthesis. Thesegenes wereup-regulated as much as 14-fold.PF1713-1714, encoding carbamoyl phosphate synthase, which contributes to pyrimidine biosynthesis, was up-regulated 16- to 20-fold.From a transcriptomic perspective, it appears that the WT produces flagellar proteins, perhaps to promote cell-cell network formation (Nather et al. 2006), and minimizes biosynthetic operations upon entrance into post-exponential phase. However, in stationary phase, the situation reverses, suggesting efforts to synthesizenecessary metabolic building blocks and minimizing flagellar protein formation.
Figure 4.
Heat plot showing ORFs responding to growth phase transitions for the wild-type (WT) and mutant P. furiosus strains growing at 80°C. For the heat plot, red indicates higher transcript levels (log2(fold change)>1), green indicates lower (log2(fold change)<-1), and black denotes average transcript levels (The color scale for least squares mean from mixed effects ANOVA model shown on the right, where 0 is average transcript level).
Mutant growthtranscriptomes
The growthtranscriptome of the mutant was in stark contrast to the WT. For the period corresponding to 10hand 14h, only 27 ORFs in the mutant were differentially transcribed two-fold or more (21 up, 6 down), suggesting that the mutant culture remained in exponential phase. At around 24h, the results shown inFigure 1 suggest that the mutantgrowth slowed, presumably indicative of stationary phase onset, which was concomitant with the differential transcription of 96 genes (34 up/62 down). While there were a few genes in common between the phase transitions of the WT and mutant (e.g., PF0462, encoding a glycerol-3-phosphate 3-phosphatidyltransferase was down-regulated 10-fold or more in each strain), few discernible patterns were evident for the mutant. Of interest for the mutant culture in stationary phase was the up-regulation of PF1528-1529, which encodes pyridoxine biosynthesis proteins; this was not observed for the WT. Note also that PF2064, annotated as an arylsulfatase regulatory protein, was significantly up-regulated in the WT (~10-fold in post-exponential phase, and 3-fold further, or >25-fold, overall), while no differential transcription of this ORF was seen for the mutant throughout its growth.
Contrasting the WTand mutant growth transcriptomes
The experimental loop design allowed for direct comparisons between the WT and mutanttranscriptomes at the three time pointssampled (Figure 1). Table 1 lists those ORFs differentially transcribed 10-fold or more between the WT and mutant at 10, 14 and 24 h. For a complete list of all ORFs changing 2-fold or more for the WT vs. mutant contrast, see Table S5. In several cases, these large fold changes corresponded to genes that had disruptive mutations leading to frame shifts, apparently impacting transcription. In particular, the flagellar biosynthesis operon (PF0330-0337) was significantly down-regulated in the mutant at all three time points. Also, the locus PF0351-0352, implicated in CRISPR-related functions, was10- to 24-fold lower in the mutant than the WT(Hale et al. 2008; Hale et al.). Several ORFs associated with aspartate metabolism (PF0121, PF0207, PF1056, PF1472) were down-regulated in the mutantvs. WT. At all three time points, Met biosynthesis (PF1266-1269) was down-regulated in the mutant; note that PF1267 and PF1268 had mutations corresponding to frame shifts. It is interesting that PF1678-PF1684, which contains genes involved in Lys and Leu biosynthesis, were up-regulated 2 to 3-fold in the mutant at 14h, but down-regulated more than 10-fold at 24 h. PF2064, mentioned above as a putative arylsulfatase regulatory protein, was down 10- to 25-fold in the mutant at 14h and 24 h, respectively, compared to the WT.
Table 1.
ORFs differentially transcribed 10-fold or more for P. furiosus wild-type (WT) and mutant growth contrasts
| Gene ID |
Fold change | ||||
|---|---|---|---|---|---|
| Mutant vs. WT | |||||
| 10h | 14h | 24h | Annotation | Comment | |
| PF0121 | ↓9.2 | ↓17.1 | ↓18.4 | aspartate aminotransferase | PF0122 mutated (D) |
| PF0207 | 2.5 | 2.6 | ↓9.8 | argininosuccinate synthase | PF0209 mutated (S) |
| PF0333 | ↓4.0 | ↓14.9 | ↓2.1 | flagella-related protein g | Flagella biosynthesis; PF0332, PF0334, PF0337 mutated (FS) |
| PF0335 | ↓4.3 | ↓16.0 | ↓3.7 | flagella-related protein d | |
| PF0336 | ↓11.3 | ↓45.3 | ↓7.0 | hypothetical protein | |
| PF0351 | ↓7.0 | ↓13.0 | ↓12.1 | CRISPR-related protein | Both mutated (FS), CRISPR-related |
| PF0352 | ↓13.9 | ↓19.7 | ↓24.3 | RNA-binding protein (RAMP) | |
| PF1056 | 2.6 | ↓13.9 | Asp-semialdehyde dehydrogenase | Asp metabolism | |
| PF1133 | 5.7 | 8.6 | 13.0 | hypothetical protein (68 aa) | PF1132 mutated |
| PF1266 | ↓10.6 | ↓2.5 | ↓13.0 | cystathionine gamma-synthase | Met biosynthesis |
| PF1267 | ↓12.1 | ↓3.5 | ↓13.0 | hypothetical protein | Mutated |
| PF1268 | ↓24.3 | ↓3.5 | ↓24.3 | homocysteinemethyltransferase | Mutated |
| PF1269 | ↓13.0 | ↓2.0 | ↓7.5 | methionine synthase | Met biosynthesis |
| PF1472 | 2.5 | ↓9.8 | aspartate/serine transaminase | PF1474 mutated | |
| PF1480 | 3.0 | ↓11.3 | formaldehyde:ferredoxinoxidoreductase wor5 |
Mutated | |
| PF1528 | 14.9 | GInamidotransferase, PdxT | |||
| PF1529 | 2.3 | 13.0 | pyridoxine biosynthesis protein | ||
| PF1549 | ↓10.6 | ↓11.3 | ↓14.9 | RNA 3'-terminal-phosphate cyclase | PF1553 mutated |
| PF1664 | 2.8 | ↓13.9 | phosphoribosyl-AMP cyclohydrolase | ||
| PF1678 | 3.0 | ↓11.3 | 2-isopropylmalate synthase | Lys, Leu biosynthesis | |
| PF1679 | 2.8 | ↓14.9 | 3-isopropylmalate dehydratase | ||
| PF1680 | 2.6 | ↓13.9 | 3-isopropylmalate dehydratase | ||
| PF1682 | 2.1 | ↓10.6 | ribosomal protein s6 modification protein | ||
| PF1683 | 2.5 | ↓9.8 | N-acetyl-γ-glutamyl-P-reductase | ||
| PF1684 | 2.1 | ↓10.6 | acetylglutamate kinase | ||
| PF1713 | 2.3 | ↓10.6 | carbamoyl phosphate synthase | Pyrimidine metabolism | |
| PF2064 | ↓13.0 | ↓26.0 | arylsulfatase regulatory protein | SAM radical family | |
D = deletion; S = in-frame substitution; FS = Frame shift
Deletion of selected ORFs in the WT encoding mutated genes in the mutant
Certain genes that were the most transcriptionally dynamic in the WT as it progressed through phase transitions, and for which transcription in the mutantwas impacted by mutations, were selected for further analysis.
Methionine biosynthesis (PF1266-PF1269)
In the mutant, the methionine biosynthesis pathway (PF1266-69) was significantly down-regulated for all conditions tested. There were mutations in two of the ORFs in this operon. PF1267 encodes a putativemethylenetetrahydrofolatereductase (MTHFR) based on protein domain homology and presence in a methionine biosynthesis operon, which converts homocysteine to methionine, while PF1268 (metE) encodes a 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase, which catalyzes the transfer of a methyl group from 5-methyltetrahydrofolate to homocysteine to form methionine. This operon is not conserved in all Pyrococcus species. For example,Pyrococcusabyssilacks the MTHFR gene needed to convert homocysteine to methionine, making it a methionine auxotroph (Cohen et al. 2003). Here, in the WT, this operon was transcribed at high levels at 10h, down-regulated at 14h, and then up-regulated at 24h. These genes were transcribed at very low levels in the mutant, possibly because of polar effects related to mutations in PF1267 and PF1268. Deletion of PF1269(methionine synthase) resulted in no observable phenotype, although deletion of PF1267(methionine synthase) led to a non-viable phenotype such that the disruptant could not be isolated. Since the mutant was able to grow well even with this operon transcribed at very low levels and no effect was noted when additional methionine was added to the medium, it is likely that some compensatory mechanism was activated. Given the large number of hypothetical proteins responding during growth, one or more of these could be involved (see Table 1, Table S4, Table S5, and Figure 4).
Stationary phase response of PF2064
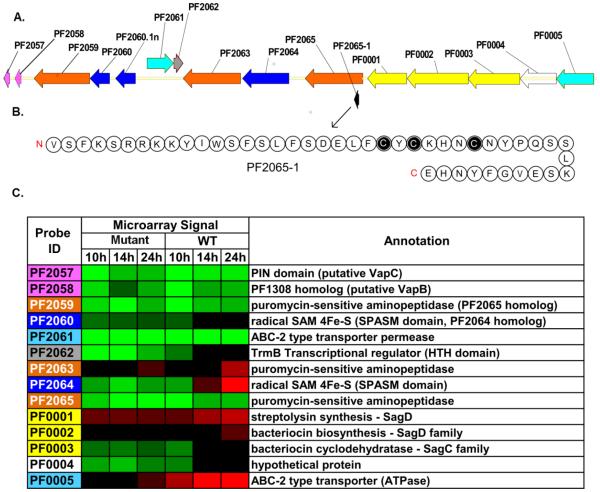
The most responsive (>25-fold) gene for the WT transition from exponential to stationary phase is for PF2064 and this is annotated as a putative arylsulfatase regulatory protein. This gene was unresponsive in the mutant over the time period examined (see Figure 4). Upon closer inspection, PF2064 contains a single [4Fe-4S] cluster domain and belongs to the radical SAM superfamily, with 38.4% identity to a bacteriocin maturation enzyme in Clostridium sp. BNL100. Furthermore, examination of this gene cluster (Figure 5) revealed an organization closely related to operons in Bacillus sp. that encode peptide bacteriocins that are post-translationally modified by radical SAM enzymes, such that the peptides contain Cys to α-carbon intrachain linkages, such as subtilosin A (Fluhe et al. 2012)and thuricin CD (Murphy et al. 2011). In the same reading frame as PF2064 is a putative 46-amino acid peptide, which has threeCys residues, similar to the two Bacillusbacteriocins. The fact that the locus containing PF2064 is induced in the WT in post-exponential and stationary phase could relate to competitive strategies that are triggered when nutrients are limited or other stationary phase drivers (quorum sensing) are activated. PF2064 transcript levels in the WT and SM were identical for exponential phase (Figure 4), but remain at low levels in the mutant, consistent with the observed extended exponential phase. Thus, transcription of PF2064 (and other ORFs in this gene cluster) could serve as an indicator of post-exponential phase onset in P. furiosus. Note that when the gene encoding PF2064 was deleted, no phenotypic differences were noted between the WT and the ΔPF2064 COM1 strain.The probe designed for PF2065 also included PF2065-1, which encodes the peptide shown in Figure 5B, although this ORF was not up-regulated during growth phase transitions in the WT or MLS.
Figure 5. PF2064 locus organization and gene response during growth phase transitions in WT and Mutant.
(A)The PF2064 locus includes small hypothetical ORFs, bacteriocin synthesis related genes (yellow), radical SAM proteins (dark blue), transporters (light blue), toxins/antitoxins (pink), and a transcriptional regulator containing HTH domain (grey). PF2065 is the last annotated gene in P. furiosusgenome and hence, PF0001-0005 genes are depicted to be a part of this locus. Gene organization was generated using Vector NTI (Invitrogen). (B) The amino acid sequence (46 residues) of a putative small peptide, presumably bacteriocin-like, discovered in the locus, its N and C termini indicated in red. Cysteines are highlighted in black. Amino acids are noted by their one letter code. (C) Transcriptional response of genes in the PF2064 locus. Red indicates higher transcript levels (log2(fold change)>1), green indicates lower (log2(fold change)<-1), and black denotes average transcript levels. The color scale used is the same as in Figure 4.
Flagellar biosynthesis
One striking difference between the transcriptomes of the mutant and WT involved genes related to flagellar protein biosynthesis(PF0329-PF0338). PF0332, PF0334 and PF0337 had frameshift mutations in the mutant, and this negatively impacted transcription of this operon to a significant extent, compared to the WT, perhaps because of polar effects. This is consistent with the images shown in Figure 2 that indicate that the mutant lacked monopolar flagella. PF0331-PF0337 and PF0337 (which was mutated in the mutant) were each deleted from the WT. Deletion of PF0337 (Figure 2-K), the last but one gene/ORF in the operon, did not completely eliminate formation of cell protrusions and possibly immature flagellaare present (Figure 2-I), but no EPS was observed,unlike the case for the WT (Figure 2-A,B). However, deletion of PF0331-PF0337 resulted in no observable flagella (Figure 2-J,L), minimal EPS formation, and cell patternsalong the lines seen with the mutant (Figure 2-C,D).
DISCUSSION
The accumulation of mutations over an extended period of time led to the creation of a ‘lab strain’ with distinguishing growth characteristics from the P. furiosus DSM3638 described above. Comparative genomics and transcriptomics were used in an attempt to determine the basis for the extended exponential phase and cell aggregation behavior.
Unlike the mutant, the WT transcriptome showed significant regulation of amino acid biosynthetic pathways during growth phase transitions (Figure 4). Failure to manage nutritional resources could have been responsible for the extended exponential growth and then rapid cell lysis at 24h noted for the mutant (see Figure 1). Following exponential phase, amino acid biosynthesis pathways in the WT, initially down-regulated, were triggered, in response to depletion of amino acid sources (Shaikh et al. 2010) (see Figure 4). In particular, operons involved in biosynthesis of branched chain amino acids (PF1678-80), lysine (PF1681-86), and aromatic amino acids (PF1705-12) were subsequently up-regulated as the WT culture entered stationary phase. Furthermore, components of the arginine (PF0207-PF1209) and methionine (PF1266-PF1269) biosynthesis pathways also followed this trend. As such, genes in the methionine pathway most significantly impacted by mutations in the mutant were examined. Although the mutation in the key gene involved in Met biosynthesis, PF1268 (metE),was silent, an insertion mutation in PF1267, annotated as a hypothetical protein in this operon, would lead to premature truncation. Low transcript levels for the PF1266-PF1269 were possibly due to translational polarity. Nonsense-codon-mediated polarity in operon expression has been demonstrated to occur in the closely-relatedhyperthermophileThermococcuskodakaraensis(French et al. 2007; Santangelo et al. 2008). Based on the transcription pattern observed in the PF1266-PF1269 operon and others (e.g. PF0329-38), it seems that translational polarity also occurs in P. furiosus. No clear effect of the mutated methionine biosynthesis pathway on cell growth could be identified. As mentioned above, addition of methionine to the mutant medium resulted in no differences with the WT. A knock-out strain lacking PF1269 (methionine synthase) was viable, but showed no significant differences in phenotype compared to the WT. Attempts were made to create a PF1267 knock-out mutant, but this strain was not viable, suggesting that PF1267 is an essential gene for the growth and sustainability of P. furiosus.
The extensive regulation of amino acid biosynthetic pathways in the WT, and not in the mutant, in post-exponential phase suggests certain global factors could be responsible. In E. coli, “feast” and “famine” modes (Calvo and Matthews 1994; Newman and Lin 1995) are associated with the Leucine-responsive regulatory protein (Lrp) (Chen and Calvo 2002). Despite limited information on archaeal transcription regulators, some are paralogs of the E. coli Lrp(Yokoyama et al. 2006), and have been referred to as Feast-Famine Regulatory Proteins (FFRPs) (Calvo and Matthews 1994). Interestingly, PyrococcusOT3 has one such FFRP, called FL11, that senses the levels of lysine to control growth mode by changing its oligomeric state, octamer or dimer, in the presence of high or low levels of this amino acid, respectively(Yokoyama et al. 2007). Also, there are indications of other similar FFRPs that respond to amino acid levels (FL4 with glutamic acid, FL9 and DM2 with glutamine) in the cell. At this point, it is not clear if methionine is involved in global cell metabolic regulation in P. furiosus through FFRPs, but merits further investigation given the results here.
A CRISPR-associated gene (PF0352) was also highly up-regulated in the WT compared with the mutant (over 20-fold at 24h). PF0352 has been proposed to be a cmr1-type RNA binding protein (RAMP); RAMP proteins are involved in a psiRNA-guided destruction of invading target RNA, thus providing phage-immunity (Hale et al. 2008; Hale et al. 2009). It is interesting to note that the only other cmr1-type RAMP protein identified in the P. furiosusgenome (Hale et al. 2008), PF1130, was also mutated, as was the neighboring CRISPR-cas6 family protein (PF1131); these were 2.5-fold up-regulated in the WT over the mutant, at 10h and 14h. Whether the low transcription levels of PF0352 in the mutant compared to the WT were related to the offset in phase transitions between the cultures is not known.
The largest fold changes observed for any of WT vs. mutant contrasts was PF2064, which was 13- and 26-fold up-regulated in the WT at 14h and 24h, respectively (Table 1, Figure 5). Whether PF2064 is involved in bacteriocin formation remains to be seen, but its up-regulation served as an indicator that P. furiosus had entered post-exponential phase. The master regulators that trigger post-exponential strategies by the extremely thermophilicarchaea are yet unknown, and whether these are unique to or similar to bacterial factors is just starting to be examined in detail (Orell et al. 2013b). The fact that P. furiosus and other extremely thermophilicarchaea form biofilms is clear (Frols 2013; Orell et al. 2013a), and likely involves quorum sensing and other ecological triggers. How the locus containing PF2064 is activated likely relates to regulation leading to post-exponential phase, and this issue is currently being examined.
Post-exponential microbial growth, triggered by sub-optimal nutrient concentrations, can be associated with initial stages of biofilm formation, driven by changes in regulatory factors and nucleotide second messengers (Serra et al. 2013). Recent studies focused on Escherichia coli revealed that flagella play an important architectural role in bacterial biofilm formation. Observations along these lines have been reported for P. furiosus, which produces monopolar flagella composed of up to 50 filaments of a glycoprotein that is similar to other archaealflagellins(Nather et al. 2006). Unlike bacteria, archaeal flagella are composedof flagellins that are more closely related to bacterial type IV pili; thesereportedly require 12 flagenes for synthesis, compared to 50 or more for bacteria (Friedrich et al. 2003; Thomas and Jarrell 2001; Zolghadr et al. 2010). Previous studies have shown that P. furiosusflagella are multifunctional, used for swimming, adhesion to surfaces, and forming cable-like, cell-cell connections, which could be involved in biofilm formation(Nather et al. 2006). Recently, it was proposed that, in E. coli, flagella tether cells together and, furthermore, through rotation, tighten up the linkages formed to create a robust macrocolony(Serra et al. 2013). This level of detail is starting to become available for the archaea (e.g., (Nather et al. 2006; Orell et al. 2013a; Orell et al. 2013b; Zolghadr et al. 2010) andit will interesting to see just how similar the trajectory of biofilm formation processes are between the two domains.
The results shown in Figure 2-G,H, in contrast to those ofFigure 2-E,Fdemonstrate that cells in themutant culture of P. furiosuslacked flagella. Furthermore, at 24h, while the WT showed significant formation of EPS-based cell aggregates (Figure 2-A,B), this was not the case for the mutant (Figure 2-C,D). PF0329-PF0338 encodes genes implicated in a flagellar biosynthesis operon, three of which (PF0332, PF0334, and PF0337) had frame-shifted mutations in the mutant. ORFs in this operon were transcribed at low levels in the mutant and, unlike the WT, were not responsive to growth phase transitions (see Figure 4). When PF0331-0337 was deleted, there was no flagella formation (Figure 2-L) and EPS-aggregates were not observed (Figure 2-J). Deletion of PF0337, which encodes a putative flagellin protein and is located towards the end of the operon, showed possible formation of immature flagella, but no EPS-aggregate formation (Figure 2-I). Taken together, these observations point to an essential role of flagella in P. furiosusin post-exponential cell aggregation and potentially biofilm formation.
The relationship between flagella and biofilm formation has been established in bacteria. In E. coli,the master biofilm regulator,CsgD, cross-regulates biofilm formation and flagellar synthesis (Ogasawara et al. 2011). Various factors, such as low temperature, starvation, low osmolarity and high cell density, are known to influence expression of CsgD. This regulator suppresses formation and function of flagella, leading to inhibition of planktonic growth and the switch to biofilm formation. Whether a similar regulatory process occurs in P. furiosus is not known. The closest homolog in P. furiosus to E. coliCsgD is a hypothetical protein encoded byPF1260 (28% identical and 48% similar over 55% of the 304 amino acid ORF in P. furiosus). This gene was essentially deleted in the mutant (theframeshiftwould have resulted in translation of 19 residues(Table S3),although no significant transcriptional differences between the mutantand WT were noted. The lack of a functional regulator could have been responsible for the unusual post-exponential behavior of the mutant. Since archaea lack σ-factorsand particularly those that regulate stationary phase and general stress response in bacteria, drivers for entrance to stationary phase are unknown, but any counterpart could interact with the CsgDortholog in P. furiosus. The identification of such factors remains to be seen.
Supplementary Material
ACKNOWLDEGMENTS
This work was supported by a grant from National Science Foundation (CBT-0730091) to RMK. Construction and characterization of the deletion mutants was fundedby a grant to MWA(FG05-95ER20175) from the Chemical Sciences, Geosciences and Biosciences Division of the Office of Basic Energy Sciences, Office of Science, and U.S. Department of Energy. The authors are grateful to Dr. JenniferSchaff, Genome Sciences Lab at NCSU, for her help with genome sequencing and Dr. Michael J. Dykstra, LAELOM, College of Veterinary Medicine, NCSU, for help with TEM and SEM imaging.
REFERENCES
- Blumentals, Brown SH, Schicho RN, Skaja AK, Costantino HR, Kelly RM. The hyperthermophilic archaebacterium, Pyrococcus furiosus. Development of culturing protocols, perspectives on scaleup, and potential applications. ANN NY Acad Sci. 1990;589:301–314. doi: 10.1111/j.1749-6632.1990.tb24254.x. [DOI] [PubMed] [Google Scholar]
- Bridger SL, et al. Deletion strains reveal metabolic roles for key elemental sulfur-responsive proteins in Pyrococcus furiosus. J Bacteriol. 2011;193:6498–6504. doi: 10.1128/JB.05445-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridger SL, Lancaster WA, Poole FL, 2nd, Schut GJ, Adams MW. Genome sequencing of a genetically tractable Pyrococcus furiosus strain reveals a highly dynamic genome. J Bacteriol. 2012;194:4097–4106. doi: 10.1128/JB.00439-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo JM, Matthews RG. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiological reviews. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Calvo JM. Leucine-induced dissociation of Escherichia coli Lrp hexadecamers to octamers. J Mol Biol. 2002;318:1031–1042. doi: 10.1016/S0022-2836(02)00187-0. [DOI] [PubMed] [Google Scholar]
- Chou CJ, Shockley KR, Conners SB, Lewis DL, Comfort DA, Adams MW, Kelly RM. Impact of substrate glycoside linkage and elemental sulfur on bioenergetics of and hydrogen production by the hyperthermophilic archaeon Pyrococcus furiosus. Appl Environ Microbiol. 2007;73:6842–6853. doi: 10.1128/AEM.00597-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen GN, et al. An integrated analysis of the genome of the hyperthermophilic archaeon Pyrococcus abyssi. Mol Microbiol. 2003;47:1495–1512. doi: 10.1046/j.1365-2958.2003.03381.x. [DOI] [PubMed] [Google Scholar]
- Costantino HR, Brown SH, Kelly RM. Purification and characterization of an alpha-glucosidase from a hyperthermophilic archaebacterium, Pyrococcus furiosus, exhibiting a temperature optimum of 105 to 115 degrees C. J Bacteriol. 1990;172:3654–3660. doi: 10.1128/jb.172.7.3654-3660.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dam P, Olman V, Harris K, Su Z, Xu Y. Operon prediction using both genome-specific and general genomic information. Nucleic Acids Res. 2007;35:288–298. doi: 10.1093/nar/gkl1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Nickle DC, Learn GH, Maust B, Mullins JI. ViroBLAST: a stand-alone BLAST web server for flexible queries of multiple databases and user's datasets. Bioinformatics. 2007;23:2334–2336. doi: 10.1093/bioinformatics/btm331. [DOI] [PubMed] [Google Scholar]
- DiRuggiero J, Brown JR, Bogert AP, Robb FT. DNA repair systems in archaea: mementos from the last universal common ancestor? J Mol Evol. 1999;49:474–484. doi: 10.1007/pl00006570. [DOI] [PubMed] [Google Scholar]
- Drake JW. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci U S A. 1991a;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW. Mutation: major evolutionary trends. Nucleic Acids Symp Ser. 1991b:159–160. [PubMed] [Google Scholar]
- Drake JW. Spontaneous mutation. Annu Rev Genet. 1991c;25:125–146. doi: 10.1146/annurev.ge.25.120191.001013. [DOI] [PubMed] [Google Scholar]
- Drake JW. Avoiding dangerous missense: thermophiles display especially low mutation rates. PLoS Genet. 2009;5:e1000520. doi: 10.1371/journal.pgen.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskill LE, Bauer MW, Kelly RM. Synergistic interactions among beta-laminarinase, beta-1,4-glucanase, and beta-glucosidase from the hyperthermophilic archaeon Pyrococcus furiosus during hydrolysis of beta-1,4-, beta-1,3-, and mixed-linked polysaccharides. Biotechnol Bioeng. 1999;66:51–60. [PubMed] [Google Scholar]
- Fluhe L, Knappe TA, Gattner MJ, Schafer A, Burghaus O, Linne U, Marahiel MA. The radical SAM enzyme AlbA catalyzes thioether bond formation in subtilosin. A Nature chemical biology. 2012;8:350–357. doi: 10.1038/nchembio.798. [DOI] [PubMed] [Google Scholar]
- French SL, Santangelo TJ, Beyer AL, Reeve JN. Transcription and translation are coupled in Archaea Molecular biology and evolution. 2007;24:893–895. doi: 10.1093/molbev/msm007. [DOI] [PubMed] [Google Scholar]
- Friedrich A, Rumszauer J, Henne A, Averhoff B. Pilin-like proteins in the extremely thermophilic bacterium Thermus thermophilus HB27: implication in competence for natural transformation and links to type IV pilus biogenesis. Appl Environ Microbiol. 2003;69:3695–3700. doi: 10.1128/AEM.69.7.3695-3700.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frols S. Archaeal biofilms: widespread and complex. Biochem Soc Trans. 2013;41:393–398. doi: 10.1042/BST20120304. [DOI] [PubMed] [Google Scholar]
- Grogan DW. Hyperthermophiles and the problem of DNA instability. Mol Microbiol. 1998;28:1043–1049. doi: 10.1046/j.1365-2958.1998.00853.x. [DOI] [PubMed] [Google Scholar]
- Grogan DW, Carver GT, Drake JW. Genetic fidelity under harsh conditions: analysis of spontaneous mutation in the thermoacidophilic archaeon Sulfolobus acidocaldarius. Proc Natl Acad Sci U S A. 2001;98:7928–7933. doi: 10.1073/pnas.141113098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C, Kleppe K, Terns RM, Terns MP. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008;14:2572–2579. doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayat MA, Miller SE. Negative staining. McGraw-Hill; New York, NY: 1990. [Google Scholar]
- Hoaki T, Nishijima M, Kato M, Adachi K, Mizobuchi S, Hanzawa N, Maruyama T. Growth requirements of hyperthermophilic sulfur-dependent heterotrophic archaea isolated from a shallow submarine geothermal system with reference to their essential amino acids. Appl Environ Microbiol. 1994;60:2898–2904. doi: 10.1128/aem.60.8.2898-2904.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoaki T, Wirsen CO, Hanzawa S, Maruyama T, Jannasch HW. Amino acid requirements of two hyperthermophilic archaeal isolates from deep-sea vents, Desulfurococcus strain SY and Pyrococcus strain GB-D. Appl Environ Microbiol. 1993;59:610–613. doi: 10.1128/aem.59.2.610-613.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie JE, Daley RJ, Jasper S. Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977;33:1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MW, et al. Exploiting microbial hyperthermophilicity to produce an industrial chemical, using hydrogen and carbon dioxide. Proc Natl Acad Sci U S A. 2013;110:5840–5845. doi: 10.1073/pnas.1222607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kengen SW, de Bok FA, van Loo ND, Dijkema C, Stams AJ, de Vos WM. Evidence for the operation of a novel Embden-Meyerhof pathway that involves ADP-dependent kinases during sugar fermentation by Pyrococcus furiosus. J Biol Chem. 1994;269:17537–17541. [PubMed] [Google Scholar]
- Lee HS, et al. Transcriptional and biochemical analysis of starch metabolism in the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 2006;188:2115–2125. doi: 10.1128/JB.188.6.2115-2125.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscomb GL, et al. Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl Environ Microbiol. 2011;77:2232–2238. doi: 10.1128/AEM.02624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackwan RR, Carver GT, Kissling GE, Drake JW, Grogan DW. The rate and character of spontaneous mutation in Thermus thermophilus. Genetics. 2008;180:17–25. doi: 10.1534/genetics.108.089086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majernik AI, Jenkinson ER, Chong JP. DNA replication in thermophiles. Biochem Soc Trans. 2004;32:236–239. doi: 10.1042/bst0320236. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki H. Origins of spontaneous mutations: specificity and directionality of base-substitution, frameshift, and sequence-substitution mutageneses. Annu Rev Genet. 2002;36:279–303. doi: 10.1146/annurev.genet.36.042602.094806. [DOI] [PubMed] [Google Scholar]
- Mao F, Dam P, Chou J, Olman V, Xu Y. DOOR: a database for prokaryotic operons. Nucleic Acids Res. 2009;37:D459–463. doi: 10.1093/nar/gkn757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martusewitsch E, Sensen CW, Schleper C. High spontaneous mutation rate in the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by transposable elements. J Bacteriol. 2000;182:2574–2581. doi: 10.1128/jb.182.9.2574-2581.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazia D, Schatten G, Sale W. Adhesion of cells to surfaces coated with polylysine. Applications to electron microscopy. The Journal of cell biology. 1975;66:198–200. doi: 10.1083/jcb.66.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero CI, et al. Responses of wild-type and resistant strains of the hyperthermophilic bacterium Thermotoga maritima to chloramphenicol challenge. Appl Environ Microbiol. 2007;73:5058–5065. doi: 10.1128/AEM.00453-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, O'Sullivan O, Rea MC, Cotter PD, Ross RP, Hill C. Genome mining for radical SAM protein determinants reveals multiple sactibiotic-like gene clusters. PloS one. 2011;6:e20852. doi: 10.1371/journal.pone.0020852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nather DJ, Rachel R, Wanner G, Wirth R. Flagella of Pyrococcus furiosus: multifunctional organelles, made for swimming, adhesion to various surfaces, and cell-cell contacts. J Bacteriol. 2006;188:6915–6923. doi: 10.1128/JB.00527-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman EB, Lin R. Leucine-responsive regulatory protein: a global regulator of gene expression in E. coli. Annual review of microbiology. 1995;49:747–775. doi: 10.1146/annurev.mi.49.100195.003531. [DOI] [PubMed] [Google Scholar]
- Ogasawara H, Yamamoto K, Ishihama A. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. J Bacteriol. 2011;193:2587–2597. doi: 10.1128/JB.01468-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omelchenko MV, et al. Comparative genomics of Thermus thermophilus and Deinococcus radiodurans: divergent routes of adaptation to thermophily and radiation resistance. BMC Evol Biol. 2005;5:57. doi: 10.1186/1471-2148-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orell A, Frols S, Albers SV. Archaeal biofilms: the great unexplored. Annual review of microbiology. 2013a;67:337–354. doi: 10.1146/annurev-micro-092412-155616. [DOI] [PubMed] [Google Scholar]
- Orell A, Peeters E, Vassen V, Jachlewski S, Schalles S, Siebers B, Albers SV. Lrs14 transcriptional regulators influence biofilm formation and cell motility of Crenarchaea. The ISME journal. 2013b;7:1886–1898. doi: 10.1038/ismej.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinker KD, Kelly RM. Growth physiology of the hyperthermophilic archaeon Thermococcus litoralis: Development of a sulfur-free defined medium, characterization of an exopolysaccharide, and evidence of biofilm formation. Appl Environ Microbiol. 1996;62:4478–4485. doi: 10.1128/aem.62.12.4478-4485.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinker KD, Kelly RM. Effect of carbon and nitrogen sources on growth dynamics and exopolysaccharide production for the hyperthermophilic archaeon Thermococcus litoralis and bacterium Thermotoga maritima. Biotechnol Bioeng. 2000;69:537–547. doi: 10.1002/1097-0290(20000905)69:5<537::aid-bit8>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Santangelo TJ, Cubonova L, Matsumi R, Atomi H, Imanaka T, Reeve JN. Polarity in archaeal operon transcription in Thermococcus kodakaraensis. J Bacteriol. 2008;190:2244–2248. doi: 10.1128/JB.01811-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra DO, Richter AM, Klauck G, Mika F, Hengge R. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. mBio. 2013;4:e00103–00113. doi: 10.1128/mBio.00103-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh AS, Tang YJ, Mukhopadhyay A, Martin HG, Gin J, Benke PI, Keasling JD. Study of stationary phase metabolism via isotopomer analysis of amino acids from an isolated protein. Biotechnol Prog. 2010;26:52–56. doi: 10.1002/btpr.325. [DOI] [PubMed] [Google Scholar]
- Snowden LJ, Blumentals, Kelly RM. Regulation of proteolytic activity in the hyperthermophile Pyrococcus furiosus. Appl Environ Microbiol. 1992;58:1134–1141. doi: 10.1128/aem.58.4.1134-1141.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas NA, Jarrell KF. Characterization of flagellum gene families of methanogenic archaea and localization of novel flagellum accessory proteins. J Bacteriol. 2001;183:7154–7164. doi: 10.1128/JB.183.24.7154-7164.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhorst WG, et al. Transcriptional regulation in the hyperthermophilic archaeon Pyrococcus furiosus: coordinated expression of divergently oriented genes in response to beta-linked glucose polymers. J Bacteriol. 1999;181:3777–3783. doi: 10.1128/jb.181.12.3777-3783.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward DE, Kengen SW, van Der Oost J, de Vos WM. Purification and characterization of the alanine aminotransferase from the hyperthermophilic Archaeon Pyrococcus furiosus and its role in alanine production. J Bacteriol. 2000;182:2559–2566. doi: 10.1128/jb.182.9.2559-2566.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrin L, Martin-Jezequel V, Prieur D. Minimal amino acid requirements of the hyperthermophilic archaeon Pyrococcus abyssi, isolated from deep-sea hydrothermal vents. Appl Environ Microbiol. 1995;61:1138–1140. doi: 10.1128/aem.61.3.1138-1140.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrin L, Prieur D. UV and ethyl methanesulfonate effects in hyperthermophilic archaea and isolation of auxotrophic mutants of Pyrococcus strains. Curr Microbiol. 1996;33:377–382. doi: 10.1007/s002849900131. [DOI] [PubMed] [Google Scholar]
- Williams E, Lowe TM, Savas J, DiRuggiero J. Microarray analysis of the hyperthermophilic archaeon Pyrococcus furiosus exposed to gamma irradiation. Extremophiles. 2007;11:19–29. doi: 10.1007/s00792-006-0002-9. [DOI] [PubMed] [Google Scholar]
- Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006;34:W720–724. doi: 10.1093/nar/gkl167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama K, et al. Feast/famine regulatory proteins (FFRPs): Escherichia coli Lrp, AsnC and related archaeal transcription factors. FEMS Microbiol Rev. 2006;30:89–108. doi: 10.1111/j.1574-6976.2005.00005.x. [DOI] [PubMed] [Google Scholar]
- Yokoyama K, et al. Feast/famine regulation by transcription factor FL11 for the survival of the hyperthermophilic archaeon Pyrococcus OT3. Structure. 2007;15:1542–1554. doi: 10.1016/j.str.2007.10.015. [DOI] [PubMed] [Google Scholar]
- Zolghadr B, Klingl A, Koerdt A, Driessen AJ, Rachel R, Albers SV. Appendage-mediated surface adherence of Sulfolobus solfataricus. J Bacteriol. 2010;192:104–110. doi: 10.1128/JB.01061-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.