Abstract
Non-receptor protein tyrosine kinases (NRTKs) are essential for cellular homeostasis, and thus are a major focus of current drug discovery efforts. Peptide substrates that can enhance lanthanide ion luminescence upon tyrosine phosphorylation enable rapid, sensitive screening of kinase activity, however design of suitable substrates that can distinguish between tyrosine kinase families is a huge challenge. Despite their different substrate preferences, many NRTKs are structurally similar even between families. Furthermore, the development of lanthanide-based kinase assays is hampered by incomplete understanding of how to integrate sequence selectivity with metal ion binding, necessitating laborious iterative substrate optimization. We used curated proteomic data from endogenous kinase substrates and known Tb3+-binding sequences to build a generalizable in silico pipeline with tools to generate, screen, align and select potential phosphorylation-dependent Tb3+-sensitizing substrates that are most likely to be kinase specific. We demonstrated the approach by developing several substrates that are selective within kinase families and amenable to HTS applications. Overall, this strategy represents a pipeline for developing efficient and specific assays for virtually any tyrosine kinase that use high throughput screening-compatible lanthanide-based detection. The tools provided in the pipeline also have the potential to be adapted to identify peptides for other purposes, including other enzyme assays or protein binding ligands.
Introduction
Protein kinases catalyze the reversible phosphorylation of proteins and play a ubiquitous role in the regulation of signal transduction pathways directing cellular processes including proliferation, survival and adhesion. Phosphorylation of a protein can result in changes in activity, conformation, and stability as well as facilitate protein-protein interactions through phospho-recognition domains. The human genome encodes more than 500 protein kinases, 32 of which are non-receptor tyrosine kinases (NRTKs). 1 This group of kinases has diverse roles in integrating signaling events initiated at the plasma membrane, including regulation of cell shape, motility, proliferation, and survival. NRTK deregulation occurs frequently in cancer through a variety of mechanisms including overexpression, gain-of-function mutation, or loss of negative regulators. 2-4 The association of many NRTKs with cancer and inflammatory disease has led to large drug discovery efforts, resulting in the development of 24 FDA-approved small molecule NRTK inhibitors since 2001. 5 However, despite their established clinical importance, approved inhibitors target only a small subset of NRTKs (5 out of 32). A major factor impeding development of kinase inhibitors is the difficulty in producing compounds that are highly specific, and several promising kinase inhibitors have failed clinical trials due to unanticipated off-target effects. Therefore, the development of broad-based tools that allow for sensitive detection of kinase activity has important applications in profiling kinase inhibitor specificity.
Typical strategies for monitoring kinase activity use radioactive ATP, antibodies, or proteomics to detect phosphorylation of native substrates. 6-8 While these methods have successfully generated a wealth of information about kinase activity, each suffers from several disadvantages. For example, redundancy among even otherwise disparate kinases can also confound the assignment of endogenous phosphorylation sites to a specific enzyme. Artificial peptide substrates offer an attractive strategy for examining kinase activity either in vitro or in intact cells, due to their diverse chemistries, compatibility with a wide variety of detection platforms, and their ability to directly report the function of a particular enzyme. A variety of detection methods have been utilized for assaying artificial substrates, including capillary electrophoresis, voltammetry, mass spectrometry, antibody-based detection (e.g. ELISA), light scattering based methods using SERS and RLS, and fluorescence-based methods such as chelation enhanced fluorescence (CHEF), FRET and fluorescence quenching. 9-19 In particular, CHEF methods that sensitize lanthanide ions such as terbium (Tb3+) in a phosphorylation-dependent manner 19-22 can enable high sensitivity and analytical reproducibility. Previously, we described the application of a kinase specific peptide substrate (SAStide) for the sensitive detection of spleen tyrosine kinase (Syk) activity in vitro through phosphorylation-dependent enhanced sensitization of Tb3+ luminescence. 22 The luminescence signal is generated when phosphorylation of the tyrosine residue results in exclusion of water and completion of the Tb3+ coordination sphere. Phosphorylation also alters the excitation wavelength of the aromatic side chain, increases the binding affinity for the peptide, and increases the luminescence lifetime,22 resulting in a large increase in signal to noise (16-fold in the case of SAStide). However, other than this example of a serendipitous case, most CHEF substrates are designed primarily to achieve optimal metal binding, which often comes at the expense of kinase selectivity and enzyme kinetics. Currently there is no general, streamlined method to identify and develop novel substrates that are simultaneously specific for an individual kinase and strong metal chelators. To develop such an approach, both elements (specificity and binding) must be taken into account.
In this report, we present a pipeline to develop peptide substrates for tyrosine kinases (using the NRTKs as a model system) that are compatible with phosphorylation dependent sensitization of Tb3+ (Fig. 1). We employed curated collections of known endogenous substrate sequences and data from positional scanning peptide library microarrays to develop an in silico positional scoring matrix model that enabled the rapid identification of selectivity determinants and assessed the relative importance of maintaining certain residues at each position. We used this information and Tb3+-binding motif alignment as sequence-space-filtering criteria to narrow down the potential substrate library generated from the motif for a given kinase. This yielded a manageable handful of sequences that could be empirically tested and thoroughly characterized. We applied this pipeline to generate biosensors for Abl, Jak2, and Src-Family tyrosine kinases and demonstrated HTS assays using the Abl substrate against a small molecule library to identify novel Abl inhibitors.
Figure 1. Design and development of phosphorylation-dependent enhanced Tb3+ luminescence tyrosine kinase peptide biosensors.
A) General biosensor design strategy for kinase biosensors capable of phosphorylation induced enhanced Tb3+ luminescence, where X is any amino acid, Φ is a hydrophobic antenna containing residue and [−] is an acidic amino acid. B) The detection strategy using the phosphorylation-dependent physical changes in the biosensors that result in enhanced Tb3+ luminescence. C) To develop a kinase specific peptide based biosensor, we first obtain all known phosphorylated substrates for a given kinase as the foreground as well as all unphosphorylated tyrosine centered sequences for the substrates and validated proteins that interact with the kinase as the background. Data from positional scanning peptide library screens from the Turk laboratory were also included23. (1) A positional scoring matrix, where values represent the preference for each amino acid at every position, and a site-selectivity matrix (SSM), representing the degree to which a given position “requires” a given amino acid, are generated from these data. SSM values are centered at one; values greater than one reflect a strong preference for a particular amino acid at that position and values less than one reflect a lack of preference. (2) A library of sequences were generated in silico based on substrate preferences at each positions using the site selectivity score (using the“Generator™” tool). (3) The library is scored against the kinase of interest as well as all other tyrosine kinases and clustered using bidirectional Euclidian distance and filtered to remove any nonspecific or nonsubstrate sequences for the kinases based on the PSM scores (using the “Screener™” tool). Scores are on a scale from 0 to 100, where binary classification (of “Substrate” or “Non-substrate”) was determined based on threshold values through cross-validation. (4) The remaining sequences are scored using a BLOSUM matrix to assess the similarity to the phosphorylation-dependent Tb3+-binding α-syn Y125 peptide24,25 (using the “Aligner™” tool), which enables filtering out of sequences that are predicted to be selective substrates but not to match the Tb3+ motif inherent in the target sequence (which in this case was the best-characterized model, the α-syn Y125 peptide, but could be another Tb3+-binding sequence of interest). (5) The remaining sequences are validated empirically for kinase specificity and photophysical properties associated with Tb3+ luminescence. For each relevant step, the score similarity for each kinase (columns) and sequence similarity to one another across kinases (rows) were clustered using bidirectional Euclidian distance.
Experimental Section
Positional Scoring Matrix (PSM) and Site Selectivity Matrix (SSM) generation
A blank “substrate informatics sheet” that can be used to perform the functions that yield the PSM and SSM, and workbooks for the Generator™, Screener™, and Aligner™ tools are provided as supplementary files. The calculations in the workbook were implemented as follows:
Positional Scanning Peptide Library
To combine the PSPL data from the Turk laboratory with the endogenous substrate information in the filtering algorithm, peptide phosphorylation signals for each array were quantified based on the median intensity for each spot. The median intensity values were then background corrected and signal intensity were then normalized by the following equation:
where Zij stands for the normalized score of amino acid j at position i having a signal score Sij and m stand for the total number of amino acids. Sci is the signal score of amino acid j at position i where i is defined in the summation of all the m amino acids.
Positional Probability (from endogenous substrates)
We computed the probability matrix, PM, as follows. It is experimentally known that kinase k phosphorylates n substrates (n1, n2, …, nn) consisting of nine amino acids, four on each side of the phosphorylation site. The frequency of each amino acid at each position in the collection of substrates was computed, fj,i, where j is amino acid (A, C, …, W, Y) at position i (−4, −3, …, 1). Due to the limitation of identified substrates for some kinases, when j = 0 for those amino acids the value of j = 1/n, where n is the number of substrate sequences for kinase k. The matrix values were computed by comparing the observed frequency, fi,j, within the substrates to the expected frequency (background frequency), bi,j, derived from the frequency of each amino acid in each protein containing a substrate sequence as well as non-phosphorylated interacting proteins (obtained from the Protein Information Resource (http://pir.georgetown.edu).26 This allowed for the background of amino acids to reflect the proteins with which the kinase naturally interacts. We constructed the probability matrix 20 × 9 for each amino acid and position defined as si,j = fi,j / bi,j.
Positional Scoring Matrix
The two individual matrices, PM and PSPLM, were then multiplied together to form the positional scoring matrix, PSM. The value for each amino acid can then be used to identify favorable and unfavorable residues at each position. Values greater than 0.9 were considered favorable or permissive for the kinase, while values less than 0.9 were consider unfavorable or impermissive.
For an nonapeptide of a given amino acid sequence the product of all si,j values yields the raw probability score, SR.
The raw score was normalized by probability of any nonapeptide being a substrate for kinase k, Ps. Ps was determined by the number kinase substrates collected n plus the number of significantly favorable amino acid from the PSPL compared to the total number tyrosine centered nonapeptides seen in substrate and interacting proteins and the 200 peptides from the PSPL for kinase k.
The site selectivity matrix was determined by the ratio of the number of significantly abundant residues found at the subsite, nsigi,j, to that the expected abundance from a random distribution, nsigaai,j, multiplied by the ratio of the number of the number of significantly abundant at the subsite to the total number of residues, naai,j.
An amino acid was defined as being significantly abundant if its frequency was found to be greater than two standard deviations above the mean.
Generation of kinase-focused virtual peptide libraries
Kinase focused virtual (i.e. in silico) peptide libraries were generated using the Generator™ tool based on the values of the PSM. All si,j > 0.9 were chosen as potential residues at each position. Combinatorial peptide sequences were generated from these residues and scored against each kinase using the Screener™ tool. Those peptides that scored positive for the kinase (or kinase family) of interest and negative for all other kinases (or kinase families) were then selected and added to virtual “focused libraries” in the Aligner™ Excel spreadsheet for further screening.
Terbium Binding in silico Screening
Following the generation of focused putative kinase substrate libraries, sequences were filtered for the potential to bind terbium in a phosphorylation-dependent manner using the Aligner tool™. A BLOSUM62 matrix was used to generate a sequence similarity score between the focused library of potential kinase substrates and the known terbium sensitizing sequence α-syn Y125 (DPDNEAYEMPSEEG). 24,25 The top several sequences (as desired) were chosen for further empirical evaluation.
Peptide synthesis and purification
Peptides were synthesized using a Protein Technologies Prelude parallel peptide synthesizer on Rink-amide resin (Peptides International, Louisville, KY). Coupling of standard Fmoc-protected amino acids (Peptides International, Louisville, KY) was achieved with HCTU (Peptides International, Louisville, KY)(100 mM) in the presence of NMM (Sigma-Aldrich, St. Louis, MO) (400 mM) in DMF (EMD Millipore, Billerica, MA) for two 10 min couplings. Fmoc deprotection was performed in 20% piperidine (Sigma-Aldrich, St. Louis, MO) in DMF for two 2.5 min cycles. Peptides were purified to >90% purity by preparative C18 reverse-phase HPLC (Agilent 1200 series) using a linear gradient 5%-38% acetonitrile/0.1%TFA and water/0.1%TFA and characterized using HPLC-MS (ThermoFinnegan Accela-LTQ).
In vitro kinase assays (Tb3+ luminescence)
Recombinant kinases; Abl, Src, Lyn, Csk, Jak2 and Hck (Millipore) and Syk, Btk, Fyn, Pyk2, and Fgr were expressed as described elsewhere.23 Recombinant kinases were incubated with the kinase reaction buffer (100 μM ATP, 10mM MgCl2, 125 ng/μL BSA and 25 mM HEPES pH 7.5, total volume 180 μL) containing 12.5 μM biosensor at 30°C. Aliquots (20 μL) were taken at designated time points (0.5, 5, 10, 15, 30, 45 and 60 min) and quenched in 6 M urea (20 μL). The quenched samples were then treated with the luminescence buffer (500 μM Tb3+ and 500 mM NaCl, 10 μL) for a total volume of 50 μL (final concentrations of sample components: 2.4 M urea, 40 μM ATP, 4 mM MgCl2, 50 ng/μL BSA and 10 mM HEPES pH 7.5). Time-resolved luminescence emission spectra were collected on a Biotek Synergy4 plate reader equipped with a monochromator at room temperature in black 384-well plates (Greiner Fluorotrac 200). Spectra were collected between 450 and 650 nm in 1 nm increments with 1 ms collection time and 10 reading per data point at a sensitivity of 180 after excitation at 266 nm with a Xenon flash lamp followed by a delay of 50 μs. area under each spectrum was integrated using GraphPad Prism. An additional aliquot (2 μL) of the kinase reaction mixture was taken at each time point for validation of phosphorylation using an ELISA-based chemifluorescent assay as previously described. 27
Chemifluorescent detection of phosphorylation
Each aliquot was quenched with 0.5 M EDTA and incubated in a 96-well Neutravidin coated plate (15 pmol biotin binding capacity per well, Thermo Scientific) in Tris-buffered saline (TBS, 25mM Tris-HCl and 150mM NaCl) containing 0.1% BSA and 0.05% Tween 20 for 1h. Following incubation, each well was washed with the TBS buffer and then incubated with mouse anti-phosphotyrosine monoclonal antibody 4G10 (Millipore, 1:10,000 dilution in TBS buffer) for 1h. Following incubation, each well was washed with TBS buffer and incubated with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) secondary antibody (Abcam) (1:1000 dilution) for 1h. Wells were then washed and treated with Amplex Red reaction buffer (Amplex Red reagent, Invitrogen, 20 mM H2O2 and sodium phosphate buffer) for 30 min. Fluorescence was measured using a Synergy4 multiwell plate reader (Biotek) with an excitation wavelength of 532 nm and emission wavelength of 590 nm.
Dose-response inhibition assay
Kinase (15 nM) was incubated with the kinase reaction buffer described above in the presence of DMSO (vehicle) or varying concentrations of kinase inhibitors (nilotinib, bosutinib, ruxolitinib) at 30°C for 10 min prior to the start of the reaction by adding the peptide substrate. The reaction was started with the addition of biosensor (37.5 μM, total reaction volume 20 μL). Each reaction was quenched after 30 min in 6 M urea (20 μL). The samples were then treated with the luminescence buffer (500 μM Tb3+ and 500 mM NaCl, 10 μL) for a total volume of 50 μL. Time-resolved luminescence spectra were collected as described above and the area under the emission curve determined. The IC50 value for each inhibitor was determined by fitting data to equation below where inhibitionmax is the bottom plateau of the curve, inhibitionmin is the top plateau of the curve, the Hill slope is the steepness of the curve and X is the concentration of the inhibitor.
High-throughput screening assay
Abl kinase (3 nM) was incubated with the kinase reaction buffer described above in the presence of DMSO (vehicle), imatinib (positive control) or a single compound from the GSK PKIS library (10 μM), at 30°C for 30 min prior to the start of the reaction by adding the peptide substrate. The reaction was started with the addition of the biosensor AbAStide (12.5 μM, total reaction volume 20 μL). Each reaction was quenched after 1 h in 6 M urea (20 μL). The samples were then treated with the luminescence buffer (500 μM Tb3+ and 500 mM NaCl, 10 μL) for a total volume of 50 μL. Time-resolved luminescence emission intensities were collected at the maxima of the four emission peaks and summed together to give total signaling for each well using the instrument settings described above. Percent inhibition was determined using the positive inhibition control, imatinib, and the negative inhibition control, DMSO.
Growth inhibition curves
K562 cells were seeded into 96-well plates at 10,000 cells per well in Iscove’s Modified Dulbecco’s Medium supplemented with 10% fetal bovine serum and pen/strep. The cells were dosed with the indicated inhibitor at the indicated concentrations (n = 4), and allowed to incubate for 3 days at 37° C. Following incubation, XTT reagent (ATCC) was added according to manufacturers protocol, and allowed to incubate at 37° C for 3 hours. Absorbance at 475 nm was measured on a Biotek Synergy4 plate reader. Values were calculated as percent of vehicle (0.1% DMSO), plotted in Graphpad Prism 6, and IC50 values generated by fitting a variable slope (four parameter) curve.
High-Throughput Screening Calculations
The Z’ factor was calculated according to Eq. 2.
| [2] |
The signal window was calculated according to Eq. 3
| [3] |
where n is the number of replicates, μpos and μneg are the average luminescence of the positive (phosphorylated peptide or uninhibited) and negative (unphosphorylated peptide or control inhibitor-treated) controls respectively; σpos and σneg are the standard deviation of the positive and negative controls.
Results
KINATEST-ID™: a substrate peptide sequence space filtering pipeline
Inspired by the general design rules of previous CHEF-based sensors for detection of kinase activity (i.e. Sox-Mg2+ and EF-hand-Tb3+ sensors), we aimed at developing a general approach to design biosensors for tyrosine kinase activity analysis using phosphorylation-dependent enhanced Tb3+ luminescence. Sensors were designed to combine nonreceptor tyrosine kinase substrate specificity with the excitation and chelation elements governing Tb3+ luminescence. To achieve this, each kinase biosensor was developed to contain an optimized substrate sequence with an embedded Tb3+ coordination motif, similar to that previously identified from the 14-residue fragment of α-synuclein surrounding Y125. (Figure 1A) Based on our previous work, we hypothesized that phosphorylation-dependent physical changes in the biosensor would enable enhanced Tb3+ luminescence of the phosphorylated biosensor compared to the unphosphorylated form (Figure 1B). 22
For each kinase, a focused virtual library of peptide biosensors was designed, optimized and selected in silico through a bioinformatic pipeline, KINATEST-ID™ (Kinase Terbium Emission Sensor Identification) comprised of three data processing tools: Generator™, Screener™, and Aligner™, implemented in Microsoft Excel workbooks that are available as supplementary files (Fig 1C). This method starts with the generation of a positional scoring matrix (PSM) (as described below) for a given kinase, which uses highly curated, biologically validated phosphorylation sites for individual kinases as well as empirically observed effects of amino acids from positional scanning peptide microarray data (unpublished, shared by Turk and co-workers) (Figure 1C Step 1). This matrix represents the relative preference the kinase has for each amino acid at each position within the sequence, yielding comparable preference motifs to those generated by state-of-the-art phosphosite prediction algorithms (e.g. NetPhorest and M3)28,29 (Tables S1-S7). A site selectivity matrix (SSM) (evaluating the importance of a particular site in the sequence to the preference of the kinase for that substrate) was also generated using the data (Table S8). These matrices were used to guide the generation of a focused in silico library of possible kinase-specific peptide substrates using the “Generator™” tool, where the motifs derived from the set of amino acids that were represented at >2 standard deviations from the mean were used to generate a list of all possible permutations of that set of amino acids at their respective positions (Figure 1C Step 2).
Each sequence in the focused library was given a score based on the PSM (which takes into account both the endogenous and positional scanning peptide library data) for the given kinase, as well as a score for all other kinases included in the analysis using their respective PSMs, using the “Screener™” tool, which effectively cross-references each sequence for its predicted selectivity amongst the kinases included in the analysis (Figure 1C Step 2). The focused library was then filtered using Screener™ based on classifying the sequences as predicted “substrates” or “nonsubstrates” for each kinase as well as “specific” or “nonspecific” for the given kinase. All nonsubstrate and nonspecific sequences were then filtered from the library. (Figure 1C Step 3) Cutoff scores for classifying the sequences as substrates or nonsubstrates for each kinase were selected based on the algorithm training parameters to give the lowest false discovery rate for the kinase of interest and the highest sensitivity for all off target kinases (Table S9). While not necessarily providing hard cutoffs, this at least ensured that all remaining sequences in the library would have a maximal likelihood to be substrates for the desired kinase and not for the other kinases. The remaining sequences were compared to the atypical Tb3+ sensitizing peptide derived from the α-synuclein Y125 center peptide using BLOSUM sequence alignment scoring using the “Aligner™” tool. (Figure 1C Step 4) Sequences with a BLOSUM score below the threshold of 25% similarity were considered “non-optimal” binders, however some were synthesized for testing to evaluate the predictive capabilities of the alignment score. Sequences could also be optimized for Tb3+ binding by changing amino acids at positions that are less important for substrate recognition (based on the site selectivity scores). This ultimately yielded a compressed library of potential kinase-specific peptide substrate sequences that were also likely to sensitize Tb3+ luminescence, from which a handful of the top ranked sequences were chosen for studies to demonstrate kinase specificity and Tb3+ sensitization. The site selectivity matrix was used when deciding the priority for sequences to empirically test, since it enabled more optimal balancing of both Tb3+ binding residues and residues the kinase preferred at specific sites (Figure 1C Step 5). Accordingly, particular sites that lack selectivity (thus having more flexibility for a given amino acid at that position) but are required for Tb3+ binding could be substituted with the appropriate Tb3+ binding residue, as opposed to a residue suggested by the catalytic preference motif.
Design of Abl, Jak2 and Src-family kinase substrate biosensors
To demonstrate its utility, KINATEST-ID™ was applied to generate Tb3+-sensitizing biosensors predicted to be specific for Abl, Jak2, and Src-family kinases. Initial potential substrate sequence libraries were generated by determining each kinase’s preference motif using the in silico model and listing all possible permutations of that motif in a virtual library using Generator™. These virtual libraries started with ~43,000, 92,000 and 5,500 sequences for Abl, Jak2 and Src-family kinases, respectively. These libraries were then filtered with Screener™ by PSM scores for each kinase in the analysis to remove sequences with favorable predictions for other kinases (i.e. nonspecific) and unfavorable predictions for the target kinase (i.e. nonsubstrate), which drastically reduced the library size by ~99% for each kinase. The Tb3+ binding alignment score filter was then applied using Aligner™, which reduced the size of the libraries by a further ~50%, leaving libraries ranging in size from 11-250 sequences. The remaining sequences for each of the kinases contained the identified kinase substrate motifs as well as the α-syn Y125 Tb3+ binding motif or slight shifts in that motif.
From these libraries, several sequences were selected to evaluate empirically for each kinase as kinase artificial substrate peptides (KAStides) for Abl (AbAStide), Jak2 (JAStide), and Src-family (SFAStide) kinases (Fig. 2A). Sequences from the pipeline were selected on the basis of highest predicted selectivity for the given kinases, and higher Tb3+ binding alignment scores (although a selection with a range of lower binding scores was also included, in order to test the relationship between alignment score and binding affinity). The specificity of these sensors was assessed by screening the peptides against a panel of kinases representing at least one member of each family of nonreceptor tyrosine kinases. The ability of the kinases to phosphorylate a given peptide was determined using an endpoint in vitro kinase assay. Phosphorylation of each peptide was determined quantitatively using chemifluorescent ELISA.30 Relative fluorescence units (representing the amount of phosphorylated peptide present) were measured and percent phosphorylation was interpolated from a calibration curve generated from synthetically phosphorylated peptide. (Figure 2B, Supporting information Figure S1.)
Figure 2. Identification, validation, and characterization of kinase specific biosensors using KINATEST-ID™.
A) The kinase substrate sequences selected for further evaluation and their prediction scores for the panel of kinases used in the assay, and Tb3+ alignment score. For kinase substrate prediction, scores >90 generally reflect “positive” substrates, whereas scores lower than 90 reflect some similarity with that kinase’s preferred motif but were below the thresholds defined by the Screener™ tool (i.e. lowest false discovery rate and highest sensitivity for the off-target kinases). Tb3+ alignment scores >25 were considered “positive” for Tb3+ binding, and generally, higher alignment scores correlated with longer luminescence lifetime of the phosphopeptide-Tb3+ complex (and thus higher occupancy of the chelated vs. hydrated form of Tb3+ in the equilibrium) (Fig. S4). B) Screening of kinase substrates against a panel of purified recombinant kinases (3 nM each, except 250 nM Csk which was the amount of recombinant Csk required to phosphorylate the positive control Src Y530-centered peptide in characterization experiments, data not shown) using ELISA–based chemifluorescence detection. Color-coded values represent the mean of experiments performed in triplicate (with individual graphs shown in Fig. S1). Substrate phosphorylation specificity per kinase (rows) and kinase specificity per substrate (columns) were clustered using bidirectional Euclidian distance. C-E) Recombinant, active Abl, Jak2 and Lyn (3 nM) were used to carry out the kinase reactions with 100 μM ATP and increasing concentrations of AbAStide, JAStide-E and SFAStide-A. Reaction progress was monitored using ELISA–based chemifluorescence detection. Initial rates of phosphorylation of the kinase specific biosensors (picomoles of phosphorylated product per minute) for AbAStide (C) JAStide-E (D) and SFAStide-A using Lyn (E) were calculated and fitted to the Michaelis-Menten equation. Values represent the mean ±SEM of experiments performed in triplicate.
All sequences were specific substrates of the intended kinases/kinase families (Fig. S1). AbAStide did display some nonspecific phosphorylation by Csk; however this was only observed at a very high concentration of Csk enzyme (250 nM, 83-fold greater than that used for the Abl assay). This suggested that, while not explicitly measured, the kcat and catalytic efficiency of AbAStide for Csk are most likely significantly lower than those for Abl. Analysis of Jak2 preference amongst the pool of substrates and kinases tested demonstrated that JAStide-E was the most efficient, with significantly more phosphorylation by JAK2 compared to the other potential JAK substrate sequences (P<0.0001 for JAStide-A and D and P<0.001 for JAStide-B and C). This was consistent with predicted preferences in Jak2 substrates, for which the −1 position demonstrated the greatest preference for acidic residues (JAStide-E) and reduced favorability for arginine (JAStide-A, B, C) and phenylalanine (JAStide-D). The SFAStides displayed comparable levels of phosphorylation across all Src family kinases, while maintaining selectivity against all other families. The variation in residue chemical properties between the sequences at the −3 and +2 positions demonstrated that SFKs tolerate substitutions at these positions with little effect on phosphorylation (which is in accordance with the positional selectivity matrix results). To our knowledge, these sequences are the first reported Jak2 specific substrates (JAStide-A-E), and the first demonstration of family-spanning specific substrates for Src-family kinases (SFAStide A and B). Notably, the core kinase recognition sequence (DEDIYEELD) in the substrate we term SFAStide-A has been previously identified as an optimal Lyn kinase motif,31 however it has not previously been analyzed in the context of the entire Src family. This gave us further confidence in the ability of our upstream informatic approach to identify appropriate substrate sequences, and also supported the importance of validating peptide substrates across a panel of kinases.
AbAStide, JAStide-E and SFAStide-A were selected for further characterization based on their specificities and efficiency of phosphorylation by their designated kinases. These biosensors were characterized using steady-state kinetics to model the kinase-substrate interaction and subsequent phosphorylation of the substrates. The initial in vitro reaction velocities for each kinase-substrate pair were measured and fitted to the Michaelis-Menten equation to derive Km, Vmax, turnover number (kcat) and catalytic efficiency (kcat/Km). (Figure 2C-E, Table S13, Supporting Information Figure S2) Overall, the kinetic parameters fell between those found in previously reported “optimal” substrate and those for endogenous substrates (which are often relatively low efficiency as short peptides when isolated from their protein context). The Km values for all the peptides were within ~50-200 μM, lower than is typically observed for endogenous substrates,32 but about ~2-5-fold greater than for relevant “optimal” substrates. Several reportedly Src specific peptide substrates have been developed using one-bead-one-peptide and oriented peptide libraries with Km values between 20-55 μM, similar to SFAStide-A (Km = 62 μM). AbAStide (Km = 99 μM) exhibited a substantially increased Km compared to the optimal substrate Abltide (Km = 4 μM) but comparable to the endogenous substrate CrkL Y207 (Km = 134 μM). Since JAStide-E (Km = 186 μM) represents the first report of an unnatural specific substrate for a JAK kinase, we could only compare it to the commonly used endogenous phosphorylation site STAT5 Y694 (Km = 306 μM), relative to which JAStide-E’s Km was 2-fold lower. The catalytic efficiencies for AbAStide, JAStide-E, and SFAStide-A sequences were excellent, comparable to those reported for the “optimal” kinase substrates. These results demonstrated that KINATEST-ID™ is capable of identifying sequences with a high likelihood of being selective substrates that have comparable kinetic parameters to the optimal substrates previously identified using traditional, fully empirical methods.
Tb3+ luminescence characterization of KINATEST-ID™ identified biosensors
The biosensors that displayed appropriate specificity in the screening panel were further evaluated for phosphorylation-dependent enhanced Tb3+ luminescence. Phosphorylated and unphosphorylated forms of the peptides were synthesized and Tb3+ luminescence emission was analyzed. Steady-state measurements of the biosensors revealed a modest range of enhancement (~1-2 fold) in Tb3+ luminescence upon phosphorylation. (Table S14) However, as we have previously observed for a Syk-specific peptide substrate (SAStide),22 time-resolved measurements significantly improved the enhancement of Tb3+ luminescence to the range of ~5-11 fold (approximately 3-5 fold improvement over steady-state measurements). As in that previous work, the enhancement of Tb3+ luminescence could be attributed to the differences in properties of the sensors including excitation wavelength (266 nm for the phosphorylated vs. 275 nm for the unphosphorylated), binding affinity, luminescence lifetime and hydration number (Table S14), which validated the phosphorylation-dependent design of the sensors. The unphosphorylated sequences exhibited binding constants (Kd) in the range of 9-80 μM, which were substantially weaker than the range of 1-12 μM observed for the phosphorylated forms (Table S14). The luminescence lifetimes of the all the biosensors were increased by an amount in the range of 100-200 μsec upon phosphorylation, enabling high signal to noise through time-resolved detection. Interestingly, these lifetimes appeared to be correlated with the Tb3+ binding sequence alignment score (Fig. S4), suggesting that the alignment parameter may be useful as a predictive measure for choosing sequences for further characterization as phosphorylation sensitive biosensors since longer lifetimes tended to result in better signal to noise. Overall, these results showed that this general design strategy can be applied to diverse tyrosine kinase substrates, and that these predicted substrates exhibit robust Tb3+ luminescence sensitization with photophysical properties consistent with the anticipated detection mechanism.
In vitro time-resolved Tb3+ luminescence-based detection of tyrosine kinase activity
AbAStide, JAStide-E, and SFAStide-A were further characterized for in vitro time-resolved Tb3+-luminescence-based detection of kinase activity. Conditions for optimal detection and calibration curves (using various ratios of phosphorylated and unphosphorylated forms of the sensors) were established in the kinase reaction buffer to account for potential interference from assay buffer components. All sensors displayed linear increases in Tb3+ luminescence with increasing percent phosphorylation allowing for quantitative determination of phosphorylation (Supporting Information Figure S3). High-throughput screening parameters were also derived from the calibration curves, including the Z’ factor and signal widow (SW), reflecting assay robustness. All sensors displayed appropriate parameters (Z’ factor < 0.5 and SW < 2) for application in HTS screening assays (Table S15)
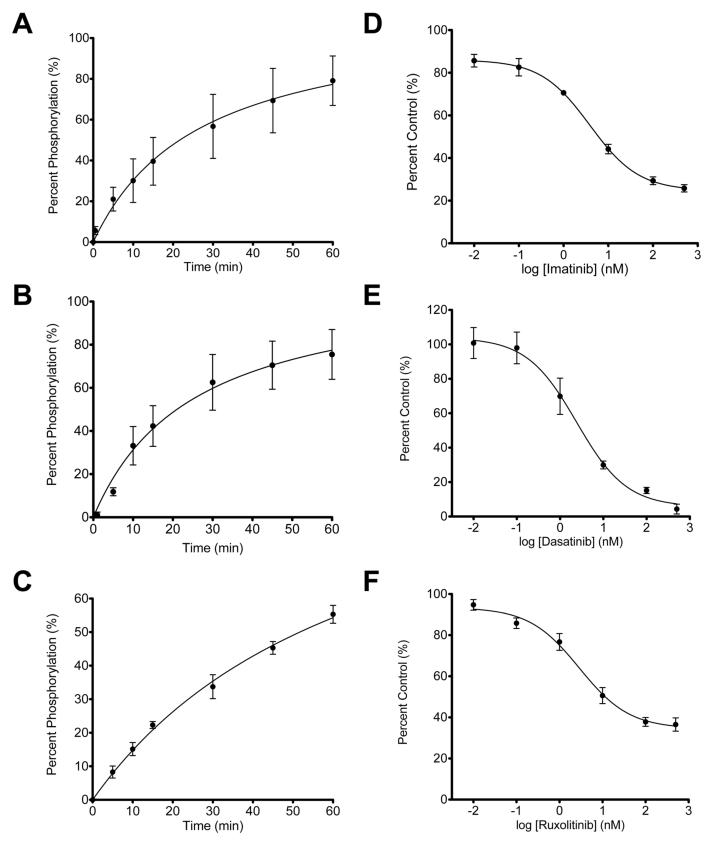
Quantitative in vitro kinase activity assays were performed using AbAStide, JAStide-E, and SFAStide-A and recombinant kinases over a 60-minute time course. Percent phosphorylation was interpolated from calibration curves, and followed the trends for those obtained using the quantitative ELISA-based read out (Fig. S5). (Figure 3 A-C) Dose-response inhibition of Abl, Jak2, and Hck kinase activity by the inhibitors imatinib, ruxolitinib, and dasatinib, respectively, was then assayed in an inhibitor dilution series from 10 pM to 500 μM. Luminescence emission spectra were collected and normalized to the vehicle (DMSO) control and reported as percent control. The observed IC50 values were 3.9 ± 1.3 nM, 2.9 ± 1.4 nM, and 2.3 ± 1.6 nM for imatinib/c-Abl, ruxolitinib/JAK2, and dasatinib/Hck, respectively. These values are in agreement with those reported in the literature for each drug/kinase combination. 33-35 The Z’ factor and SW for these assays were sufficient for HTS at some concentrations of inhibitor, indicating that characterization of the behavior at a given degree of inhibition will be necessary for optimizing screening assays (Table S16). The AbAStide biosensor was selected for further validation in an in vitro HTS for inhibitors of c-Abl.
Figure 3. Quantitative time-resolved phosphorylation-enhanced Tb3+ luminescence detection of nonreceptor tyrosine kinase activity and inhibition.
Kinase reaction progress curves for Abl (A) Hck (B) and Jak2 (C). Dose-response inhibition of Abl with imatinib, Hck with dasatinib, and Jak2 with ruxolitinib (D-F). Kinase reactions were performed with kinase reaction buffer containing ATP, MgCl2, HEPES, pH 7.5 and 15 nM recombinant kinase. IC50 values were determined values were generated by fitting the data to a variable slope (four parameter) curve. Data represent the average ± SEM of experiments performed in triplicate.
Application of AbAStide to High Throughput Screening for Small Molecule Inhibitors
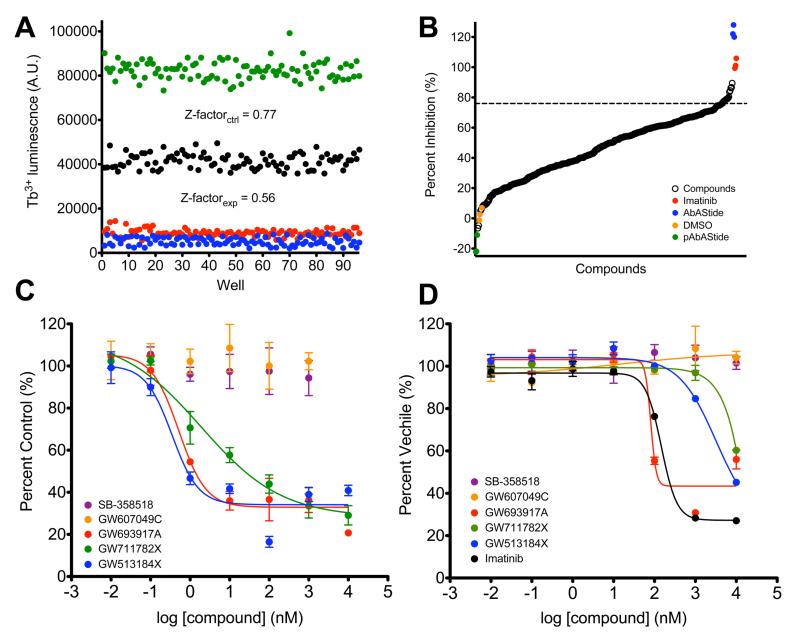
Replicate in vitro kinase assays were performed in a 384-well plate format in the presence (N=96) or absence (N=96) of imatinib to evaluate reproducible detection of Abl activity using AbAStide. To evaluate assay quality, positive and negative controls (containing the phosphorylated form and unphosphorylated form of the biosensor, respectively, N=96 for each) were also analyzed. Detection of AbAStide phosphorylation was robust and reproducible. (Fig. 4A) The Z’ factor and SW for the kinase reaction replicates were 0.56 and 84 respectively, comparable to those for control well readings, demonstrating sufficient performance for use in high-throughput screening. We leveraged this in a high-throughput screen using the GSK PKIS library, which consists of 364 compounds arrayed in 96-well plates as single compounds at 10 mM in DMSO (available to the research community upon request, see cited reference).36 The library was screened at a constant 1:1000 dilution, with 10 μM final concentration of compound in each well (1% DMSO). Compounds were incubated with the kinase for 30 minutes prior to start of the kinase reaction, which was initiated by the introduction of the biosensor substrate. The kinase reaction was allowed to proceed for one hour before being quenched with the Tb3+ luminescence buffer (containing urea and Tb3+). The time-resolved Tb3+ emission intensity was measured and the “percent inhibition” was determined compared to the biological positive and negative controls (known inhibitor imatinib and no inhibitor, respectively) (Fig. 4B). Primary hits were identified as compounds reducing Tb3+ luminescence by greater than 3-fold (the top 5% most potent inhibitors, which were the top 18 compounds). These top 5% primary hits were tested in a secondary screen using the same kinase reaction conditions, but employing a chemifluorescent ELISA-based detection instead of Tb3+-based detection. (Fig. S6) The secondary screen confirmed that all of the hits inhibited Abl kinase activity by at least 50% compared to vehicle (Table S17).
Figure 4. A high-throughput chemical screen using AbAStide biosensor identifies inhibitors of Abl tyrosine kinase.
A) The AbAStide in vitro kinase assay shows highly reproducible signal upon imatinib treatment. Green: synthetically phosphorylated peptide (positive control); Black: kinase reaction; Red: kinase reaction + imatinib; Blue: unphosphorylated peptide (negative control). B) Distribution of Abl inhibition identified in a high-throughput screen performed with the GSK PKIS library using AbAStide-sensitized Tb3+ luminescence. Green: synthetically phosphorylated peptide (positive control); Black: GSK PKIS compounds; Red: kinase reaction + imatinib (positive control); Blue: unphosphorylated peptide (negative control); Orange: DMSO (vehicle control). C) Dose-response inhibition of Abl kinase activity by selected compounds from the GSK PKIS, including the top three hits and two non-hits as negative controls. The extent of biosensor phosphorylation was interpolated from an externally generated calibration curve (not shown) and normalized to vehicle (DMSO) control. Data represent the average ± SEM of experiments performed in triplicate. D) XTT cytotoxicity assay for selected compounds (as in 4C), showing potencies in K562 cells. IC50 values were generated by fitting the data to a variable slope (four parameter) curve. Data represent the average ± SEM of experiments performed in triplicate.
The three most potent inhibitors from the HTS and validation screens were GW693917A, GW711782X and GW513184X, developed to target TIE2/VEGFR2, ALK5 and GSK3β respectively. These and two negative compounds (SB-358518 and GW607049C) were selected for further evaluation in vitro using a dose-response kinase assay with AbAStide to demonstrate the selectivity of the assay for identifying compounds correctly as inhibitors or noninhibitors. All three hits potently inhibited Abl kinase activity, with IC50 values of 0.52 nM, 1.91 nM, and 0.35 nM for GW693917A, GW711782X and GW513184X respectively, while the negative compounds gave no inhibition of Abl. (Fig. 4C) To determine whether the results of the in vitro inhibition studies translate to a CML model, the compounds were tested in cellular viability assays against the human CML cell line K562. Cellular IC50 for GW693917A was comparable to imatinib, at 81 nM compared to 147 nM. GW711782X and GW513184X were less potent in the cell viability assay, at 20 μM and 3.24 μM respectively. (Fig. 4D) Together these results demonstrate proof-of-concept that this strategy can produce an effective HTS assay for drug discovery applications.
Discussion
Synthetic peptide libraries are commonly used to identify determinants of kinase substrate specificity, these methods can be laborious to perform and require substantial quantities of purified kinase, which can limit widespread application. Here, we addressed these challenges by developing a straightforward computational strategy (KINATEST-ID™) which combines the identification of kinase specificity determinants with the prediction of kinase-substrate phosphorylation and peptide:Tb3+ complex formation, and used it to generate NRTK-specific biosensors for phosphorylation-dependent time-resolved Tb3+ luminescence detection. Traditionally, fluorescence-based kinase sensors have been generated through empirical design and iterative optimization, which slows down the pipeline for assay development. The design rules applied in KINATEST-ID™ facilitate substrate discovery by providing a set of in silico filters for sequence selection. The final sequences for AbAStide, SFAStide-A, and JAStide-E reported here demonstrate the utility of the design rules, yielding strong family-based selectivity and Tb3+ luminescence enhancement. Design of Tb3+ luminescence-based reporters of kinase activity has previously been difficult to streamline since Tb3+ binding motifs are not trivially compatible with all kinase preference motifs. Moreover, overall similarity in consensus sequences among NRTKs necessitates a tradeoff between optimal activity and specificity. By taking into account the importance of each given site in a substrate sequence to the recognition and selectivity of the cognate kinase, we successfully achieved a balance between the confounding factors involved. These substrates exhibited robust dynamic ranges and signal to noise, and their potential for high-throughput assay compatibility was demonstrated in an inhibitor screen. As efforts to expand the characterization of kinase-specific phosphoproteomes increase through the application of recently developed methods,37,38 the information available for generating the motifs, PSMs, and Screener™ selection for additional kinases will also expand. In next-generation applications of these sequences, incorporating docking motifs that target protein interaction domains could further increase the efficiency and potentially the selectivity of phosphorylation. Such modular substrates have previously been designed incorporating the D-domain and DEF-sites of Erk as well as the SH2 and SH3 domains of Abl and Hck. 39-42 We are currently pursing the application of these substrates in more complex mixtures of proteins, based on our previous work developing cell-deliverable substrates for Abl and Syk kinases,27,43 in order to exploit their selectivity to measure the activity and inhibition of specific kinases in a heterogeneous environment.
Overall, while we validated the method with well-known kinase drug targets (Abl, Src-family, and JAK2) as a model system, the generality of the approach suggests that the KINATEST-ID™ strategy should be able to be applied to develop new assays for other kinases that are currently underexplored in drug development. Even though we focused here on tyrosine kinases, the in silico focused library generation tools could be used to develop new artificial peptide substrates for serine/threonine (S/T) kinases as well. Such substrates could be employed in any type of phosphorylation read-out, however the BLOSUM alignment component could be used with any detection-related motif desired. For example, Tb3+-based S/T kinase detection requires a sensitizing chromophore such as tryptophan (W); accordingly, previously reported Tb-sensitizing, W- or unnatural amino acid-containing sequences (such as those reported by the Zondlo and Imperiali groups)20,44-47 could be used for the BLOSUM matrix to focus and filter the virtual library for empirical evaluation. This generality should make KINATEST-ID™ a useful approach to streamline the development of peptide-based kinase assays, as well as for broader applications towards other enzyme substrates or binding ligands for which sufficient training data are available.
Supplementary Material
Acknowledgements
We thank Dr. Benjamin Turk (Yale University) for sharing of positional scanning peptide library data.23 This work was supported by funding from the National Institutes of Health National Cancer Institute through grants R00CA127161, R21CA160129 and R01CA182543 (LLP), R25CA128770 (D. Teegarden) via the Cancer Prevention Internship Program (AML) administered by the Oncological Sciences Center and the Discovery Learning Research Center at Purdue University, and by an Innovative Pilot Project grant from the Purdue University Center for Cancer Research.
Footnotes
ASSOCIATED CONTENT
Supporting information. Additional characterization data for peptides, detailed motif characterization tables for the PSM, data from Michaelis-Menten kinetics characterization, and further information about HTS experiments is provided as supporting information. This material is available free of charge via the internet at http://pubs.acs.org.
References
- (1).Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298:1912. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- (2).Conde-Perez A, Larue L. Future Oncol. 2012;8:1109. doi: 10.2217/fon.12.106. [DOI] [PubMed] [Google Scholar]
- (3).Ren R. Nat Rev Cancer. 2005;5:172. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- (4).Tefferi A, Skoda R, Vardiman JW. Nat Rev Clin Oncol. 2009;6:627. doi: 10.1038/nrclinonc.2009.149. [DOI] [PubMed] [Google Scholar]
- (5).Uitdehaag JC, Verkaar F, Alwan H, de Man J, Buijsman RC, Zaman GJ. Br J Pharmacol. 2012;166:858. doi: 10.1111/j.1476-5381.2012.01859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Irish JM, Czerwinski DK, Nolan GP, Levy R. J Immunol. 2006;177:1581. doi: 10.4049/jimmunol.177.3.1581. [DOI] [PubMed] [Google Scholar]
- (7).Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, Balderas RS, Plevritis SK, Sachs K, Pe’er D, Tanner SD, Nolan GP. Science. 2011;332:687. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Monetti M, Nagaraj N, Sharma K, Mann M. Nat Methods. 2011;8:655. doi: 10.1038/nmeth.1647. [DOI] [PubMed] [Google Scholar]
- (9).Fernandes N, Allbritton NL. Biochem Biophys Res Commun. 2009 doi: 10.1016/j.bbrc.2009.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Zhang J, Ma Y, Taylor SS, Tsien RY. Proc Natl Acad Sci U S A. 2001;98:14997. doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Shults MD, Imperiali B. Biopolymers. 2005;80:515. [Google Scholar]
- (12).Stains CI, Tedford NC, Walkup TC, Lukovic E, Goguen BN, Griffith LG, Lauffenburger DA, Imperiali B. Chem Biol. 2012;19:210. doi: 10.1016/j.chembiol.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Yeh RH, Yan X, Cammer M, Bresnick AR, Lawrence DS. J Biol Chem. 2002;277:11527. doi: 10.1074/jbc.M111300200. [DOI] [PubMed] [Google Scholar]
- (14).Wang Q, Cahill SM, Blumenstein M, Lawrence DS. J Am Chem Soc. 2006;128:1808. doi: 10.1021/ja0577692. [DOI] [PubMed] [Google Scholar]
- (15).Kubota K, Anjum R, Yu Y, Kunz RC, Andersen JN, Kraus M, Keilhack H, Nagashima K, Krauss S, Paweletz C, Hendrickson RC, Feldman AS, Wu CL, Rush J, Villen J, Gygi SP. Nat Biotechnol. 2009;27:933. doi: 10.1038/nbt.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Parker LL, Brueggemeier SB, Rhee WJ, Wu D, Kent SB, Kron SJ, Palecek SP. Analyst. 2006;131:1097. doi: 10.1039/b607180e. [DOI] [PubMed] [Google Scholar]
- (17).Xu X, Liu X, Nie Z, Pan Y, Guo M, Yao S. Anal Chem. 2011;83:52. doi: 10.1021/ac102786c. [DOI] [PubMed] [Google Scholar]
- (18).Yue Z, Zhuang F, Kumar R, Wong I, Cronin SB, Liu YH. Spectrochim Acta A Mol Biomol Spectrosc. 2009;73:226. doi: 10.1016/j.saa.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zondlo SC, Gao F, Zondlo NJ. J Am Chem Soc. 2010;132:5619. doi: 10.1021/ja100862u. [DOI] [PubMed] [Google Scholar]
- (20).Balakrishnan S, Zondlo NJ. J Am Chem Soc. 2006;128:5590. doi: 10.1021/ja057692h. [DOI] [PubMed] [Google Scholar]
- (21).Zondlo NJ, Balakrishnan S, Gao F, Zondlo SC. Biopolymers. 2007;88:525. [Google Scholar]
- (22).Lipchik AM, Parker LL. Anal Chem. 2013;85:2582. doi: 10.1021/ac3023422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Deng Y, Alicea-Velazquez NL, Bannwarth L, Lehtonen SI, Boggon TJ, Cheng HC, Hytonen VP, Turk BE. J Proteome Res. 2014;13:4339. doi: 10.1021/pr500503q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Liu LL, Franz KJ. J Biol Inorg Chem. 2007;12:234. doi: 10.1007/s00775-006-0181-y. [DOI] [PubMed] [Google Scholar]
- (25).Liu LL, Franz KJ. J Am Chem Soc. 2005;127:9662. doi: 10.1021/ja043247v. [DOI] [PubMed] [Google Scholar]
- (26).Wu CH, Yeh LS, Huang H, Arminski L, Castro-Alvear J, Chen Y, Hu Z, Kourtesis P, Ledley RS, Suzek BE, Vinayaka CR, Zhang J, Barker WC. Nucleic Acids Res. 2003;31:345. doi: 10.1093/nar/gkg040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Lipchik AM, Killins RL, Geahlen RL, Parker LL. Biochemistry. 2012;51:7515. doi: 10.1021/bi300970h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Miller ML, Jensen LJ, Diella F, Jorgensen C, Tinti M, Li L, Hsiung M, Parker SA, Bordeaux J, Sicheritz-Ponten T, Olhovsky M, Pasculescu A, Alexander J, Knapp S, Blom N, Bork P, Li S, Cesareni G, Pawson T, Turk BE, Yaffe MB, Brunak S, Linding R. Sci Signal. 2008;1:ra2. doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Newman RH, Hu J, Rho HS, Xie Z, Woodard C, Neiswinger J, Cooper C, Shirley M, Clark HM, Hu S, Hwang W, Jeong JS, Wu G, Lin J, Gao X, Ni Q, Goel R, Xia S, Ji H, Dalby KN, Birnbaum MJ, Cole PA, Knapp S, Ryazanov AG, Zack DJ, Blackshaw S, Pawson T, Gingras AC, Desiderio S, Pandey A, Turk BE, Zhang J, Zhu H, Qian J. Mol Syst Biol. 2013;9:655. doi: 10.1038/msb.2013.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Lipchik AM, Killins RL, Geahlen RL, Parker LL. Biochemistry. 2012;51:7515. doi: 10.1021/bi300970h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Ruzzene M, Songyang Z, Marin O, Donella-Deana A, Brunati AM, Guerra B, Agostinis P, Cantley LC, Pinna LA. Eur J Biochem. 1997;246:433. doi: 10.1111/j.1432-1033.1997.t01-1-00433.x. [DOI] [PubMed] [Google Scholar]
- (32).Hantschel O, Warsch W, Eckelhart E, Kaupe I, Grebien F, Wagner KU, Superti-Furga G, Sexl V. Nat Chem Biol. 2012;8:285. doi: 10.1038/nchembio.775. [DOI] [PubMed] [Google Scholar]
- (33).Quintas-Cardama A, Vaddi K, Liu P, Manshouri T, Li J, Scherle PA, Caulder E, Wen X, Li Y, Waeltz P, Rupar M, Burn T, Lo Y, Kelley J, Covington M, Shepard S, Rodgers JD, Haley P, Kantarjian H, Fridman JS, Verstovsek S. Blood. 2010;115:3109. doi: 10.1182/blood-2009-04-214957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).O’Hare T, Walters DK, Stoffregen EP, Jia T, Manley PW, Mestan J, Cowan-Jacob SW, Lee FY, Heinrich MC, Deininger MW, Druker BJ. Cancer Res. 2005;65:4500. doi: 10.1158/0008-5472.CAN-05-0259. [DOI] [PubMed] [Google Scholar]
- (35).Manley PW, Drueckes P, Fendrich G, Furet P, Liebetanz J, Martiny-Baron G, Mestan J, Trappe J, Wartmann M, Fabbro D. Biochim Biophys Acta. 2010;1804:445. doi: 10.1016/j.bbapap.2009.11.008. [DOI] [PubMed] [Google Scholar]
- (36).Drewry DH, Willson TM, Zuercher WJ. Curr Top Med Chem. 2014;14:340. doi: 10.2174/1568026613666131127160819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Kettenbach AN, Wang T, Faherty BK, Madden DR, Knapp S, Bailey-Kellogg C, Gerber SA. Chem Biol. 2012;19:608. doi: 10.1016/j.chembiol.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Xue L, Wang WH, Iliuk A, Hu L, Galan JA, Yu S, Hans M, Geahlen RL, Tao WA. Proc Natl Acad Sci U S A. 2012;109:5615. doi: 10.1073/pnas.1119418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Sheridan DL, Kong Y, Parker SA, Dalby KN, Turk BE. J Biol Chem. 2008;283:19511. doi: 10.1074/jbc.M801074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Lee TR, Till JH, Lawrence DS, Miller WT. J Biol Chem. 1995;270:27022. doi: 10.1074/jbc.270.45.27022. [DOI] [PubMed] [Google Scholar]
- (41).Miller WT. Acc Chem Res. 2003;36:393. doi: 10.1021/ar020116v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Till JH, Chan PM, Miller WT. J Biol Chem. 1999;274:4995. doi: 10.1074/jbc.274.8.4995. [DOI] [PubMed] [Google Scholar]
- (43).Placzek EA, Plebanek MP, Lipchik AM, Kidd SR, Parker LL. Anal Biochem. 2010;397:73. doi: 10.1016/j.ab.2009.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Ende CW, Meng HY, Ye M, Pandey AK, Zondlo NJ. Chembiochem. 2010;11:1738. doi: 10.1002/cbic.201000056. [DOI] [PubMed] [Google Scholar]
- (45).Nitz M, Franz KJ, Maglathlin RL, Imperiali B. Chembiochem. 2003;4:272. doi: 10.1002/cbic.200390047. [DOI] [PubMed] [Google Scholar]
- (46).Nitz M, Sherawat M, Franz KJ, Peisach E, Allen KN, Imperiali B. Angew Chem Int Ed Engl. 2004;43:3682. doi: 10.1002/anie.200460028. [DOI] [PubMed] [Google Scholar]
- (47).Reynolds AM, Sculimbrene BR, Imperiali B. Bioconjug Chem. 2008;19:588. doi: 10.1021/bc700426c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.