Summary
• The structure and evolution of angiosperm mitochondrial genomes are driven by extremely high rates of recombination and rearrangement. An excellent experimental system for studying these events is offered by cybrid plants, in which parental mitochondria usually fuse and their genomes recombine. Little is known about the extent, nature, and consequences of mitochondrial recombination in these plants.
• We conducted the first study in which the organellar genomes of a cybrid – between Nicotiana tabacum and Hyoscyamus niger – were sequenced and compared to those of its parents.
• This cybrid mitochondrial genome is highly recombinant, reflecting at least 30 crossovers and five gene conversions between its parental genomes. It is also surprisingly large (41 and 64% larger than the parental genomes), yet contains single alleles for 90% of mitochondrial genes.
• Recombination produced a remarkably chimeric cybrid mitochondrial genome and occurred entirely via homologous mechanisms involving the double-strand break repair and/or break-induced replication pathways. Retention of a single form of most genes could be advantageous to minimize intracellular incompatibilities and/or reflect neutral forces that preferentially eliminate duplicated regions. We discuss the relevance of these findings to the surprisingly frequent occurrence of horizontal gene – and genome – transfer in angiosperm mitochondrial DNAs.
Keywords: chimeric, cybrid, homologous recombination, mitochondria, mtDNA, Solanaceae
Introduction
A hallmark of angiosperm mitochondrial DNAs (mtDNAs) is their high rate of recombination and rearrangement, which is reflected in a number of unusual properties. Most angiosperm mtDNAs exhibit extraordinarily high rates of reciprocal, intra- and intermolecular recombination between large direct and inverted repeats, along with less frequent recombination across smaller repeats. On an organismal time-scale, this leads to genomes consisting of a plethora of subgenomic and multimeric structures (Palmer & Shields, 1984; Backert et al., 1997; Marechal & Brisson, 2010), while on an evolutionary time-scale, it results in a highly scrambled gene order between closely related species and sometimes even within a species (Palmer & Herbon, 1988; Darracq et al., 2010; Sloan et al., 2012). Low-frequency recombination between short repeats is linked to the phenomenon of substoichiometric shifting of alternative configurations of the genome (Mackenzie, 2005; Arrieta-Montiel et al., 2009). Of great functional and economic importance are those rearrangements that create functionally novel, chimeric genes involved in cytoplasmic male sterility (Kubo et al., 2011). Finally, angiosperm mtDNAs incorporate foreign sequences remarkably often, from chloroplast and nuclear genomes of the same plant via intracellular gene transfer (Stern & Lonsdale, 1982; Knoop et al., 1996), and from other plants via horizontal gene transfer (Sanchez-Puerta et al., 2008; Rice et al., 2013; Xi et al., 2013).
Substantial progress has been made in understanding certain aspects of plant mitochondrial recombination, particularly through the use of Arabidopsis nuclear mutants that affect mitochondrial recombination and repair (Shedge et al., 2007; Arrieta-Montiel et al., 2009; Davila et al., 2011; Miller-Messmer et al., 2012). A major impediment to even greater understanding is the predominantly uniparental (usually maternal) inheritance of mitochondria and their genomes (Greiner & Bock, 2013), with the only known exception being two Pelargonium species for which predominantly biparental inheritance (in contrast to occasional paternal leakage; McCauley, 2013) has been shown (Weihe et al., 2009; Apitz et al., 2013). Fortunately, well-developed procedures are available in plants for creating parasexual hybrids (cybrids in particular) that overcome the sexual roadblock to studying mtDNA recombination at potentially the whole-genome level.
Somatic hybrids result from the fusion of protoplasts from two plant species (or varieties) followed by regeneration of hybrid plants containing genomes from both parents. Cybrids (cytoplasmic hybrids) are those somatic hybrids in which the nuclear genome is engineered to derive from one parent, whereas chloroplasts and mitochondria (and their genomes) follow entirely different, non-engineered pathways owing to fundamental biological differences: chloroplasts normally don't fuse with one another during plant growth and development, whereas mitochondria regularly do, sometimes massively (Arimura et al., 2004; Sheahan et al., 2005). Following protoplast fusion, plastids almost invariably sort out quickly (Morgan & Maliga, 1987; Earle et al., 1992), such that the chloroplast population of the cybrid plant is entirely of one parental type or the other (Belliard et al., 1979; Aviv et al., 1984a,b). By contrast, the mitochondrial genomes of somatic hybrid plants are usually recombinant (Belliard et al., 1979; Vedel et al., 1986; Temple et al., 1992;.
The evidence for this recombination is currently limited to detection of novel mitochondrial restriction fragments by electrophoresis of purified mtDNA, Southern blot hybridization, PCR amplification, or, only rarely, sequencing PCR fragments (Belliard et al., 1979; Galun et al., 1982; Nagy et al., 1983; Aviv et al., 1984b; Scotti et al., 2004; Morgan & Maliga, 1987). Novel fragments may be formed by interparental recombination (Vedel et al., 1986; Rothenberg & Hanson 1987; Temple et al., 1992; Akagi et al., 1995) or by selective amplification of pre-existing substoichiometric sequence arrangements (Bellaoui et al., 1998; Lossl et al., 1999; Rasmussen et al., 2000). In only a few cases have these novel fragments actually been shown, via cloning and DNA sequencing, to be the product of recombination between the fusion parents (Vedel et al., 1986; Temple et al., 1992; Akagi et al., 1995; Shikanai et al., 1998; Scotti et al., 2004). In only one case has the mitochondrial genome of a cybrid plant been sequenced (Wang et al., 2012), but the unfortunate lack of a genome sequence for one of the fusion parents precludes meaningful analysis of the recombination history of this cybrid genome.
In sum, little is known about the overall extent of mitochondrial recombination in cybrids, the mechanisms involved, and the constraints and consequences operating on mtDNA recombination in the context of nuclear-cytoplasmic interactions and incompatibilities. This knowledge gap makes cybrids an important subject for genome-level studies of mtDNA recombination and evolutionary dynamics, as well as nuclear-cytoplasmic coordination of gene expression and development (Levin, 2003; Greiner & Bock, 2013). To help fill this gap, we carried out a whole-genome sequencing study of a cybrid plant – the product of fusion between protoplasts from two distantly related members of the Solanaceae, Nicotiana tabacum (Nt; tobacco) and Hyoscyamus niger (Hn; henbane) (Fig. 1). These plants last shared common ancestry approximately 24 million years ago (Sarkinen et al., 2013). We carefully compared the cybrid mitochondrial genome to those of both parents (one of them also sequenced as part of this study) in order to address the following questions concerning plant mtDNA recombination: (1) How many recombination events (crossovers and gene conversions) occurred between the parental genomes to form the cybrid genome? (2) What is the balance between homologous and illegitimate recombination in generating these events? (3) Overall, what fraction of the cybrid genome is derived from one parent vs the other? (4) How much duplication – in the sense of retention of homologous regions (including genes) from both parents – is created by whole-genome recombination? (5) Does the cybrid contain chimeric and likely functional genes, created by homologous recombination between cognate parental genes? (6) Is there any bias towards retention of mitochondrial genes from Nt, whose nuclear genome predominates in the cybrid, presumably in order to preserve nuclear-cytoplasmic compatibilities?
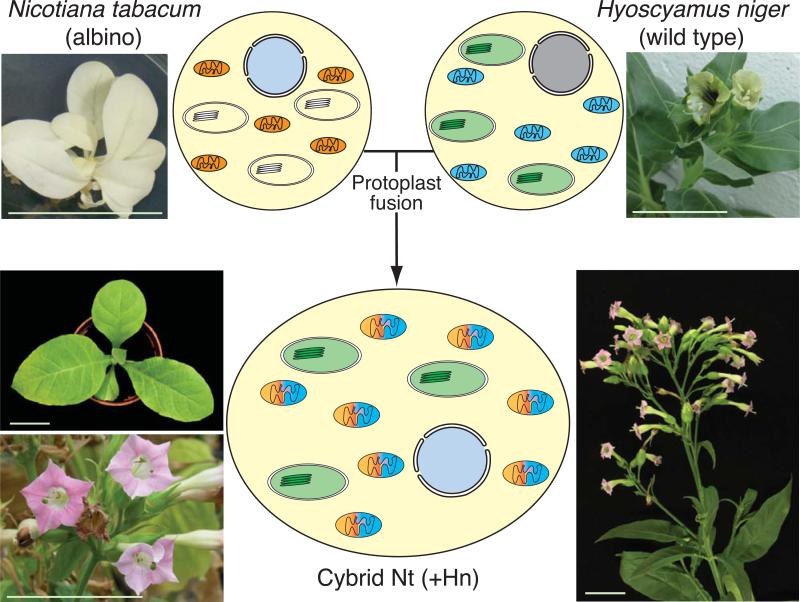
Fig. 1.
Production of a Solanaceae cybrid plant. Schematic production of a cybrid plant by chemical fusion of protoplasts from an albino mutant line of Nicotiana tabacum (Nt) and a wild type line of Hyoscyamus niger (Hn; Zubko et al., 1996). Green hybrid plants were backcrossed at least four times to wild type Nt (Zubko et al., 1996, 2001). The resulting cybrid plant has green chloroplasts derived from Hn, recombinant mitochondria derived from both parents, and a nuclear genome consisting predominantly of Nt sequences (Zubko et al., 1996). Bars: 5 cm, except top-left panel, 3 cm.
Materials and Methods
Genome sequencing, assembly and validation
Seeds of Hyoscyamus niger L. collection #A04750027 were obtained from the Nijmegen Botanical Garden (the Netherlands). Total DNA was extracted from leaves of individual plants of Hn and the cybrid Nt(+Hn) (Zubko et al., 1996) using a cetyl-trimethyl-ammonium-bromide DNA-extraction protocol (Doyle & Doyle, 1987) and sequenced at the Beijing Genomics Institute using Illumina Hiseq2000 sequencing technology (Illumina Inc.). This produced 70 million clean paired-end reads of 100 bp for each plant. Paired-end reads were assembled using the Velvet assembler 1.2.03 (Daniel Zerbino, European Bioinformatics Institute, Cambridge, UK) on the Mason large-memory computer cluster at Indiana University-Bloomington (IN, USA). To optimize parameters for the assembly (hash value, coverage cutoff, expected coverage), the program Velvet Optimiser 2.2 was used (Simon Gladman, CSIRO and Monash University, Australia). We conducted several independent Velvet assembly runs without scaffolding and using different assembly parameters each time. Taking advantage of the differences in read depths between nuclear (read depth <5) and organelle contigs, putative chloroplast (read depth >150) and mitochondrial (read depth between 20 and 150) contigs were separated. All chloroplast contigs from the set of Velvet runs of the cybrid were combined, as were all mitochondrial contigs of Hn and of the cybrid. The published cpDNA sequence of H. niger (Sanchez-Puerta & Abbona, 2014) is available in GenBank (KF248009). Genome assemblies and read-pair mapping patterns were visually inspected using Consed v26 (Gordon & Green, 2013). Mitochondrial contigs were manually connected into larger contigs using Sequencher 5.2 (Gene Codes Corporation, USA) based on consistent paired-end reads visualized in Consed v26. The DNA libraries had a mean size, estimated by Consed v26, of 768 bp (SD = 266 bp) for the cybrid mtDNA and 888 bp (SD = 200 bp) for Hn mtDNA. The resulting organellar genome assemblies were compared against all Illumina reads to identify errors until no errors remained, i.e., until the high-quality read depth and paired-end read depth were as expected at each base and the high quality mismatches were very low or zero.
To test whether the cybrid mitochondria maintained intact parental mtDNAs at low stoichiometries, all Illumina paired-end reads of the cybrid were uniquely mapped to the Nt or Hn mitochondrial genomes, in two separate analyses, using Consed v26 (Gordon & Green, 2013). This led to uneven depths across the parental mtDNAs of cybrid sequence reads and consistent paired-end reads, including regions not covered by any reads, as well as regions with inconsistent reads. Thus, the entire parental mitochondrial genomes are not present in the cybrid mitochondria and the highly chimeric nature of the cybrid mtDNA is not an artifact of the assembly.
All raw sequence data are available from the NCBI Sequence Read Archive sequence database as accession number SRP044148. The Annotated Hn and cybrid mitochondrial genomes were deposited in the GenBank data libraries under accession numbers KM207685 and KM207678-KM207684.
Genome annotation
Mitochondrial contigs were annotated using Mitofy (Alverson et al., 2010), the Blast tool (Altschul et al., 1990), and tRNAscan (Lowe & Eddy, 1997). Graphical genome maps were generated using OGDraw software (Lohse et al., 2007). The cybrid cpDNA was not annotated because it is identical to that of H. niger (Sanchez-Puerta & Abbona, 2014). Non-synonymous sites of RNA editing were predicted using PREP-Mt (Mower, 2005) with a cutoff value of 0.5.
Analysis of repeated sequences
Repeats larger than the DNA library size (~800 bp) were identified on the basis of the total read depth and consistent paired-end read depth calculated by Consed v26 (Gordon & Green, 2013). To determine repeat boundaries, we examined the reads flanking the region with an increased total read depth, in which we found reads that shared only part of their sequence, indicating the limits of a repeat region. Repeats shorter than 800 bp were detected in Consed by inspecting inconsistent and consistent reads and by pairwise Blastn analyses (using the default settings) of each genome with itself.
Comparisons between genomes
Chloroplast genomes were compared with Blastn using the default settings. Pairwise analyses of mitochondrial genomes were conducted using the Blast2seq tool and visualized with Genome Workbench. Putative interparental recombination events in the cybrid mtDNA were inferred by visual inspection of the Blast results and then tested by two computer programs: SBP (Single BreakPoint Recombination) and GeneConv. The statistical test SBP as implemented in the HyPhy software package (Kosakovsky Pond et al., 2005) was used to detect crossovers in the cybrid mtDNA by comparing it to the parental mitochondrial sequences. For those cases of putative recombination without crossing over, aligned mitochondrial regions of the cybrid and both parents were analyzed for evidence of gene conversion by the program GeneConv (Sawyer, 1989). Putative gene conversion events were identified visually using the criterion that three or more polymorphisms were shared by the cybrid and one parent within a region of the cybrid genome that was otherwise clearly derived from the other parent. Of the eight putative conversion events identified in this manner, GeneConv detected a gene conversion event in five cases with a p-value <0.01 in the global analyses with no mismatches. Genes were aligned and coding differences between Nt and Hn were scored as synonymous or non-synonymous substitutions using MacClade 4.07 (Maddison & Maddison, 2000).
cox1 transcript analysis
Total RNA was isolated using the RNeasy Plant Mini kit (QIAGEN). RNA was treated with DNase (Invitrogen) and then reverse-transcribed using the SuperScript III First Strand Synthesis System (Invitrogen) with random hexamers. cox1-specific cDNA products were amplified and sequenced using primers (5’-GGAGCAGTTGATTTAGC-3’ and 5’-GAGCAATGTCTAGCCC-3’) designed to not contain any predicted editing sites. RNA editing sites were determined by comparison of cDNA sequences to genomic sequences.
Results
The Hyoscyamus niger mitochondrial genome and comparison of parental organelle genomes
We sequenced and assembled the mitochondrial genome of Hn to enable genomic comparisons among both parents and the cybrid. The organellar genomes of Nt were sequenced previously (Shinozaki et al., 1986; Sugiyama et al., 2005), as was the chloroplast DNA (cpDNA) of Hn (Sanchez-Puerta & Abbona, 2014). The Hn cpDNA is highly similar to that of Nt in size and in sequence (98.4% identity excluding gaps), and the two genomes are identical in gene order and gene and intron content (Table 1).
Table 1.
Comparison of the mitochondrial and plastid genomes of the cybrid and its parents
| Genome features | Hyoscyamus niger | Nicotiana tabacum | Cybrid |
|---|---|---|---|
| Mitochondrial genome | |||
| Genome length in bp | 501,401 | 430,597 | 705,036 |
| Protein genesa | 37 (38) | 37 (40) | 37 (48) |
| rRNA genesa | 3 (4) | 3 (4) | 3 (3) |
| tRNA genesa | 22 (27)c | 22 (23)c | 23 (29) |
| Group II introns | 23 | 23 | 23 |
| cis-splicing | 17 | 17 | 17 |
| trans-splicing | 6 | 6 | 6 |
| Group I introns | 1 | 0 | 1 |
| Repeats in kb (% of genome)b | 97.4 (19.4%) | 68.6 (15.9%) | 229.5 (32.5%) |
| Short repeats in kb (100-250 bp) | 20.4 (4.1%) | 4.5 (1%) | 12.9 (1.8%) |
| GC content | 45.2% | 45.0% | 44.8% |
| Chloroplast-derived sequences | 2.3% | 2.2% | 1.8% |
| Chloroplast genome | |||
| Genome length in bp | 155,720 | 155,943 | 155,720 |
| Protein genesa | 80 (85) | 80 (85) | 80 (85) |
| rRNA + tRNA genesa | 34 (45) | 34 (45) | 34 (45) |
| Introns (all Group II) | 21 | 21 | 21 |
| GC content | 37.6% | 37.9% | 37.6% |
First value excludes duplicates; value in parentheses includes them.
Total length of repeats, including large, intermediate, and short repeats.
Hyoscyamus niger (Hn) and Nicotiana tabacum (Nt) share 21 of the 22 tRNA genes.
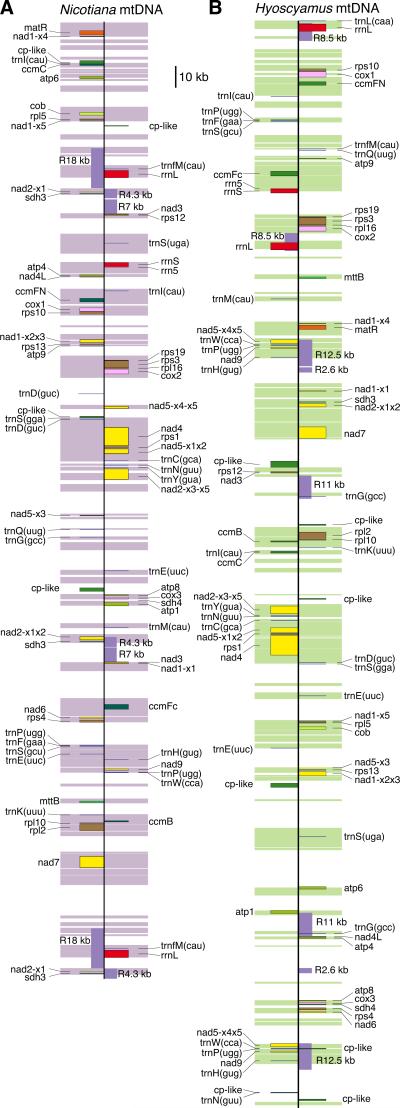
The mitochondrial genome of Hn assembled into a single linear molecule of 501,401 bp in length (Fig. 2b). This compares to a circular assembly of 430,597 bp (shown linearized in Fig. 2a) for the mtDNA of Nt, even though in vivo it forms a multipartite structure composed of subgenomic circular and branched molecules (Dale et al., 1983; Oldenburg & Bendich, 1996; Sugiyama et al., 2005). PCR across the ends of the Hn linear assembly did not yield any amplification product, but, based on paired-end reads, the ends recombine at a detectable frequency with other regions of the genome, forming alternative genome arrangements. Many additional arrangements of the Hn genome also occur in vivo, as the result of frequent recombination between the 15 largest (>250 bp) and best-matched (>90% identity) repeats present in the genome. This recombination was inferred for the four ‘large’ repeats (2.6–12.5 kb in length; Fig. 2b and Supporting Information S1A) by the presence in the assembly of alternative, recombination-diagnostic (Palmer & Shields, 1984) arrangements of unique sequences flanking the ends of these repeats and for the 11 ‘intermediate’ (250–800 bp) repeats by detecting inconsistent paired-end reads. The Hn genome contains >190 ‘small’ (100-250 bp), mostly imperfect repeats. Nt mtDNA contains a similar number and amount of large repeats (Fig. 2a), but only one intermediate repeat and 30 small repeats (Fig. S1B). Except for a few 100-bp repeats shared by both genomes, the Hn and Nt genomes do not contain any of the same repeats.
Fig. 2.
Parental mitochondrial genomes of the cybrid. Linearized, scale maps of the 430,597 and 501,401 bp mitochondrial genomes of Nicotiana tabacum (a) and Hyoscyamus niger (b), respectively. The wide violet (a) and green (b) shadings indicate regions present in the other mitochondrial genome, i.e., these represent the 203 kb of DNA shared between the two genomes. Shown are all full-length genes, repeats ≥1 kb in length (purple boxes labeled ‘R’, followed by the repeat lengths in kb), and chloroplast-derived regions >400 bp in length (green boxes labeled ‘cp-like’). Genes marked on different sides of the central black vertical lines are transcribed from opposite strands of the genome.
The Hn mtDNA contains the same 37 intact protein genes (not counting duplicates) and 23 group II introns (Table 1) as found in Nt mtDNA (Sugiyama et al., 2005). In addition, Hn contains a single group I intron (in cox1), whereas Nt contains no group I introns. Three of the 37 protein genes (ccmC, nad4L and sdh4) are identical in Nt and Hn, while the rest have an identity >97.9% (Table 2). The two genomes share 99% of RNA editing sites (449) with only six species-specific editing sites.
Table 2.
Features of protein and rRNA genes in parental (Nicotiana tabacum (Nt) and Hyoscyamus niger (Hn)) and cybrid (Cyb) mitochondrial genomes
| Gene namea | Length (bp)b | Hn/Nt differencesc | Hn/Nt differences in coding regions |
Hn/Nt differences in predicted editing sites | # intact genes |
Origin of cybrid genes | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Synonymous | Non-synonymous | Hn | Nt | Cyb | ||||||
| atp1 | 1530 | 25 | 19 | 6 | 0 | 1 | 1 | 2 | chimeric & Nt | |
| atp4 | 597 | 5 | 1 | 4 | 0 | 1 | 1 | 1 | Nt | |
| atp6 | 711 | 3 | 2 | 1 | 0 | 1 | 1 | 1 | Nt | |
| atp8 | 471 | 4 | 2 | 2 | 0 | 1 | 1 | 1 | Nt | |
| atp9 | 234 | 2 | 0 | 2 | 0 | 1 | 1 | 2 | Nt & Hn | |
| ccmB | 621 | 2 | 1 | 1 | 0 | 1 | 1 | 2 | Nt | |
| ccmC | 753 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | Hn | |
| ccmFc | 2271 | 14 | 2 | 0 | 0 | 1 | 1 | 1 | Nt | |
| ccmFn | 1740 | 37 | 7 | 30 | 2 | 1 | 1 | 2 | Hn | |
| cob | 1182 | 3 | 2 | 1 | 0 | 1 | 1 | 1 | Nt | |
| cox1 | 2551 | 11 | 9 | 2 | 2 | 1 | 1 | 2 | Hn | |
| cox2 | 2143 | 8 | 2 | 4 | 1 | 1 | 1 | 1 | chimeric | |
| cox3 | 798 | 2 | 1 | 1 | 0 | 1 | 1 | 1 | Nt | |
| matR | 1977 | 5 | 2 | 3 | 0 | 1 | 1 | 1 | chimeric | |
| mttB | 840 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | Nt | |
| nad1d | x1 | 387 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | Nt |
| x2-3 | 1485 | 12 | 1 | 0 | 0 | 1 | 1 | 1 | Nt | |
| x4 | 59 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | Hn | |
| x5 | 259 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | Nt | |
| nad2 | x1-2 | 1575 | 6 | 0 | 0 | 0 | 1 | 2 | 2 | Nt |
| x3-5 | 4796 | 24 | 1 | 0 | 0 | 1 | 1 | 1 | Nt | |
| nad3 | 357 | 4 | 2 | 2 | 0 | 1 | 2 | 2 | Nt | |
| nad4 | 8512 | 28 | 1 | 0 | 0 | 1 | 1 | 1 | chimeric | |
| nad4L | 303 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | Nt | |
| nad5 | x1-2 | 2290 | 8 | 1 | 1 | 0 | 1 | 1 | 1 | Nt |
| x3 | 22 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | Nt | |
| x4-5 | 1633 | 20 | 1 | 0 | 0 | 2 | 1 | 1 | chimeric | |
| nad6 | 687 | 3 | 2 | 1 | 0 | 1 | 1 | 1 | Nt | |
| nad7 | 5294 | 37 | 0 | 0 | 0 | 1 | 1 | 1 | chimeric | |
| nad9 | 573 | 2 | 0 | 2 | 0 | 2 | 1 | 1 | Nt | |
| rpl2 | 2898 | 36 | 4 | 4 | 0 | 1 | 1 | 1 | Nt | |
| rpl5 | 555 | 9 | 4 | 5 | 1 | 1 | 1 | 1 | Nt | |
| rpl10 | 480 | 4 | 2 | 2 | 0 | 1 | 1 | 2 | Hn | |
| rpl16 | 516 | 3 | 1 | 2 | 0 | 1 | 1 | 1 | Hn | |
| rps1 | 651 | 3 | 2 | 1 | 0 | 1 | 1 | 1 | Nt | |
| rps3 | 3469 | 27 | 4 | 6 | 0 | 1 | 1 | 1 | Hn | |
| rps4 | 1050 | 6 | 5 | 1 | 0 | 1 | 1 | 1 | Nt | |
| rps10 | 1138 | 4 | 1 | 2 | 0 | 1 | 1 | 2 | Hn | |
| rps12 | 378 | 2 | 1 | 1 | 0 | 1 | 1 | 1 | Nt | |
| rps13 | 351 | 3 | 1 | 2 | 0 | 1 | 1 | 1 | Nt | |
| rps19 | 285 | 3 | 1 | 2 | 0 | 1 | 1 | 2 | Hn | |
| rrn5 | 119 | 1 | NA | NA | ND | 1 | 1 | 1 | Nt | |
| rrn18 | 1902 | 4 | NA | NA | ND | 1 | 1 | 1 | Nt | |
| rrn26 | 3432 | 15 | NA | NA | ND | 2 | 2 | 1 | chimeric | |
| sdh3 | 327 | 3 | 1 | 2 | 0 | 1 | 3 | 3 | Nt | |
| sdh4 | 378 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | Nt | |
Intron-containing genes are in bold face.
Gene lengths are from the cybrid mtDNA.
These are nucleotide differences, excluding gaps.
nad1 is chimeric in the sense that the four separately-transcribed portions of this trans-spliced gene are derived from different parents.
NA, not available; ND, not determined. [Author, please check inserted text ‘NA, not available; ND, not determined.’ is correct. (or should it read ‘not applicable’?]
The amount and synteny of DNA sequences shared by the Hn and Nt mitochondrial genomes were assessed by pairwise Blast analysis. Despite their relatively similar sizes (501 and 431 kb), but in accord with observations made for other comparably related angiosperm pairs (Handa, 2003; Liu et al., 2013; Jo et al., 2014), many regions in both genomes are not present in the other genome, and those that are shared are highly rearranged (Figs 2, S1C). Excluding repeats, the two genomes have 203 kb of DNA in common; this represents 45% and 52% of the Hn and Nt genomes, respectively (Fig. 2). Unique and shared sequences are highly interspersed in these two genomes (Fig. 2). Shared regions include, but are by no means limited to, all mitochondrial genes and their flanking sequences and are highly similar, with an average sequence identity of 98.6% (excluding gaps).
A Blast search was performed to identify the sources of the 55% of the Hn genome that is not homologous to Nt mtDNA. Only c. 5% of the Hn mitochondrial genome is homologous to sequences in other plant mitochondrial genomes (but not in Nt), ~1.5% is homologous to plastid DNA not present in Nt mtDNA, <0.1% is homologous to nuclear DNA not present in Nt mtDNA, and the rest (~48%) did not show similarity to any sequence in the NCBI databases.
Cybrid Nt(+Hn) organellar genomes
The green cybrid line Nt(+Hn) FH-4 chosen for this study was produced by fusion of mesophyll protoplasts (with no chemical treatment or irradiation) from two Solanaceae species (Fig. 1): an albino plastid genome mutant of Nt and wild-type Hn (Zubko et al., 1996).This cybrid line is characterized by corolla-containing flowers, regular morphology, and phenotypic similarity to Nt (Zubko et al., 1996), accompanied by slight pigment deficiency and retarded growth during early development (Zubko et al., 2001). Its flowers (Fig. 1) are deficient in fertile pollen grains (a cytoplasmic-male-sterility phenotype) but are capable of seed production after backcrossing (Zubko et al., 1996). The line FH-4 was backcrossed four times with wild type Nt, which resulted in a nuclear genome containing the Nt chromosome number (2N = 48, compared to 2N = 34 in Hn) and consisting predominantly of Nt sequences (Zubko et al., 1996) (hence the ‘Nt(+Hn)’ designation of this cybrid). After recurrent backcrosses, sorting out of different mitochondrial types should be complete such that the mitochondrial genome is stable and largely homogeneous within each cybrid plant (Morgan & Maliga, 1987; Earle, 1995). Southern blot experiments showed that the FH-4 cybrid appears to have the chloroplast genome of Hn, but a recombinant mitochondrial genome (Zubko et al., 1996, 2001).
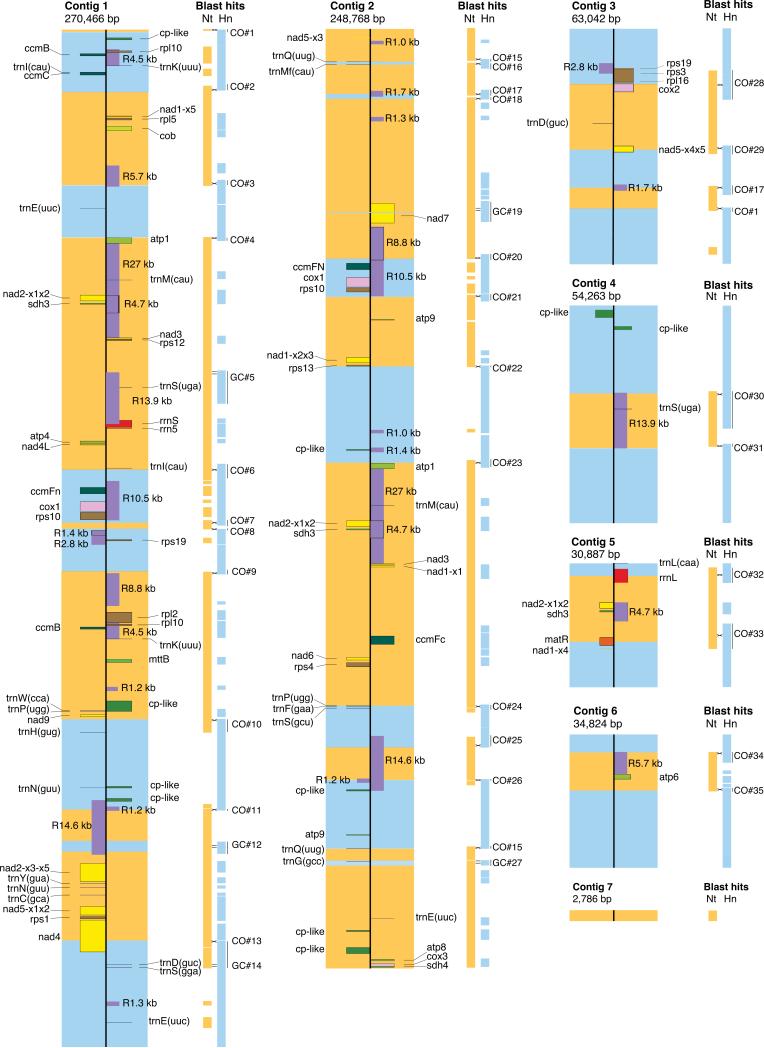
As expected, the cybrid chloroplast genome is derived from Hn; indeed, the two chloroplast genomes are identical with no substitutions, indels or rearrangements (Table 1). The cybrid mitochondrial genome assembled into seven contigs totaling 705,036 bp in size (Fig. 3). The size of the cybrid mtDNA is intermediate in comparison to the parental mtDNAs (501 and 431 kb) and the sum of the parental genomes (~932 kb). All mitochondrial contigs, except for the smallest one, are related to each other through large and intermediate repeats (Fig. 3). In addition, based on paired-end reads, the ends of all contigs appear to recombine with regions in other contigs, producing alternative genome structures. In light of previous observations (Oldenburg & Bendich, 1996; Grewe et al., 2009; Marechal & Brisson, 2010; Hecht et al., 2011; Mower et al., 2012b; Sloan, 2013; Gualberto et al., 2014;), the cybrid Nt(+Hn) mtDNA, like other angiosperm mitochondrial genomes, probably consists in vivo of a dynamically-changing population of alternative structures, such as overlapping linear and branched molecules.
Fig. 3.
Mitochondrial genome of the cybrid Nt(+Hn). Scale maps of the seven contigs that comprise the 705,036 bp mitochondrial genome of the cybrid. The wide orange and blue boxes located on the cybrid genome indicate regions inferred to derive from either the Nicotiana tabacum (Nt) or Hyoscyamus niger (Hn) parents, respectively. The thinner, orange and blue boxes to the right of the cybrid contig maps indicate regions shared (as defined by Blast hits) with the mitochondrial genomes of Nt or Hn, respectively. Numbers to the right of the blast hits designate the 35 intergenomic homologous recombination events detected in the cybrid (also see Table 3). Crossing and parallel lines mark inferred sites of crossover (CO) and gene conversion (GC) events, respectively. The cybrid contig maps show full-length genes, repeats ≥1 kb in length (purple boxes labeled ‘R’, followed by the repeat lengths in kb), and chloroplast-derived regions >400 bp in length (green boxes labeled ‘cp-like’). Genes marked on different sides of the central vertical lines are transcribed from opposite strands of the genome.
Repeats larger than 800 bp (library size) were identified by an increase in read depth in preliminary assemblies. The final assembly of the cybrid mitochondrial contigs shows a relatively even distribution of total reads (average read depth is 107; Fig. S2). Twenty-two repeats are larger than 500 bp and seven are 250–500 bp in length, while >80 are 100–250 bp in length (Fig. 4a). Repeats amount to almost 33% of the cybrid mtDNA (Table 1) and originated in three ways: (1) by duplication (the majority of repeats), i.e., by keeping two copies of a region present only once in a parental donor; (2) by retention of both copies of repeats already present in one or the other parental genome, and (3) by retention of homologous, single-copy regions from both parents, thereby creating ‘heterologous repeats.’
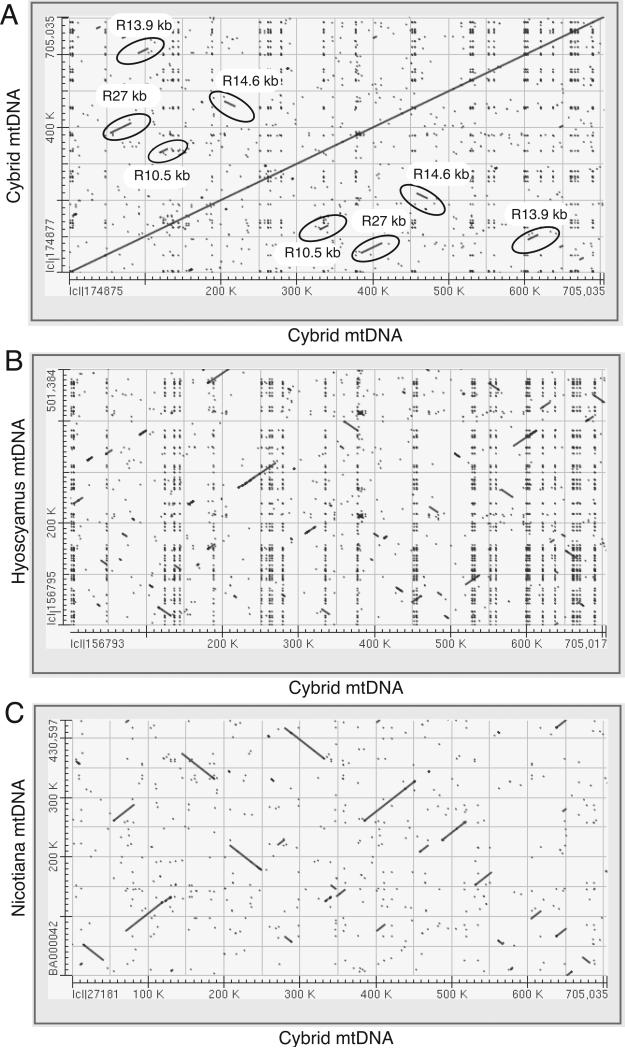
Fig. 4.
Dot-plot comparisons of mitochondrial genomes. (a) Self-self dot-plot analysis of the mitochondrial genome of the cybrid conducted to visualize repeats (ovals surround the largest repeats, which are labeled by ‘R’, followed by their lengths in kb). (b, c) Dot-plot comparisons of the mitochondrial genomes of the cybrid and Hyoscyamus niger (b), and the cybrid and Nicotiana tabacum (c), conducted to visualize regions of shared synteny. The cybrid mtDNA is represented by a concatenation of the seven contigs 1–7.
Recombination events and the origin of the cybrid mitochondrial genome
The cybrid mtDNA is highly chimeric (Fig. 3), as a result of at least 35 intergenomic recombination events between the parental mitochondrial genomes (Table 3). Consistent with its ‘intermediate’ size (see previous section), the cybrid mtDNA does not contain the entire parental mitochondrial genomes, only a subset of each of them. The cybrid mtDNA contains longer tracts of homology to Nt mtDNA than to Hn mtDNA (Fig. 3, see ‘Blast hits’, and Fig. 4b,c).
Table 3.
Recombination events between parental mitochondrial genomes (Nicotiana tabacum (Nt) and Hyoscyamus niger (Hn)) in the cybrid Nt(+Hn)
| Event # | Contig # | Recombining regions | Crossover (CO) or gene conversion (GC) event | Location of recombination | SBP test supporta | Length of gene conversion tract | # of recombination productsb | ||
|---|---|---|---|---|---|---|---|---|---|
| # bp | Identity | Genic | Breakpointa | ||||||
| 1b | 1 | 536 | 99% | CO | no | 417-587 | NS | 2b | |
| 2 | 1 | 1824 | 95% | CO | no | 16,493 | 100% | 1 | |
| 3 | 1 | 909 | 99% | CO | no | 41,000-41,150 | NS | 1 | |
| 4 | 1 | 398 | 99% | CO | atp1 | 55,160-55,330 | NS | 1 | |
| 5 | 1 | 8987 | 98% | GCc | no | 90,730-91,400 | NA | 18-371 | 1 |
| 6 | 1 | 4107 | 96% | CO | no | 117,050 | 100% | 1 | |
| 7 | 1 | 1597 | 98% | CO | no | 131,635 | 100% | 1 | |
| 8 | 1 | 226 | 99% | CO | no | 144,226 | 96% | 1 | |
| 9 | 1 | 862 | 98% | CO | no | 144,100-144,350 | NS | 1 | |
| 10 | 1 | 3815 | 99% | CO | no | 184,360 | 100% | 1 | |
| 11 | 1 | 199 | 99% | CO | no | 207,461 | 86% | 1 | |
| 12 | 1 | 3385 | 97% | GCc | no | 215,950-219,100 | NA | 2866-3150 | 1 |
| 13 | 1 | 7042 | 97% | CO | nad4 | 245,122 | 100% | 1 | |
| 14 | 1 | 7042 | 97% | GCc | no | 248,650-249,088 | NA | 151-344 | 1 |
| 15b | 2 | 634 | 99% | CO | trnQ | 8,592-8,794 | NS | 2b | |
| 16 | 2 | 1721 | 97% | CO | no | 9,245 | 100% | 1 | |
| 17b | 2 | 1691 | 98% | CO | no | 17,420 | 100% | 2b | |
| 18 | 2 | 678 | 99% | CO | no | 18,357-18,600 | NS | 1 | |
| 19 | 2 | 5716 | 99% | GCc | nad7 | 48,068-49,444 | NA | 168-1376 | 1 |
| 20 | 2 | 1669 | 96% | CO | no | 60,844 | 100% | 1 | |
| 21 | 2 | 1852 | 98% | CO | no | 72,300 | 100% | 1 | |
| 22 | 2 | 103 | 100% | CO | no | 89,332-89,435 | NS | 1 | |
| 23 | 2 | 2183 | 94% | CO | no | 114,942 | 100% | 1 | |
| 24 | 2 | 1003 | 99% | CO | no | 179,018-179,289 | NS | 1 | |
| 25 | 2 | 3385 | 99% | CO | no | 190,346 | 100% | 1 | |
| 26 | 2 | 199 | 99% | CO | no | 198,710-198,813 | NS | 1 | |
| 15b | 2 | 634 | 99% | CO | no | 216,921-216,928 | NS | 2b | |
| 27 | 2 | 1524 | 99% | GCc | no | 220,198-221,579 | NA | 1020-1381 | 1 |
| 28 | 3 | 3488 | 99% | CO | cox2 | 12,725 | 100% | 1 | |
| 29 | 3 | 1861 | 99% | CO | nad5 | 32,629 | 100% | 1 | |
| 17b | 3 | 1691 | 98% | CO | no | 42,718 | 100% | 2b | |
| 1b | 3 | 536 | 99% | CO | no | 48,170-48,240 | NS | 2b | |
| 30 | 4 | 9000 | 99% | CO | no | 22,140 | 100% | 1 | |
| 31 | 4 | 100 | 99% | CO | no | 35,525-35,624 | NS | 1 | |
| 32 | 5 | 3563 | 99% | CO | rrnL | 2,021 | 100% | 1 | |
| 33 | 5 | 6374 | 99% | CO | matR | 18,482 | 100% | 1 | |
| 34 | 6 | 2866 | 99% | CO | no | 4,726 | 95% | 1 | |
| 35 | 6 | 921 | 99% | CO | no | 14,413 | 99% | 1 | |
Breakpoint locations were calculated by the SBP (Single BreakPoint Recombination) program when they were significant as determined by this program. In those cases in which the SBP test was not significant (NS) but a crossover was nonetheless apparent because parental-unique sequences were found at each side of the putative crossover, the region of the breakpoint was estimated based on the length of the cybrid region that is identical to both parents and which is flanked by regions containing shared polymorphisms with one parent on one end and the other parent on the other end.
For only three of the 35 recombination events were two recombination products detected. These events (#'s 1, 15, and 17) therefore appear twice in this table.
The GeneConv program was significant (P<0.01) for these gene conversion events.
NA, not available. [Author, please check inserted text ‘NA, not available’ is correct (or should it read ‘not applicable’?]
No novel sequences were identified in the cybrid mtDNA, 405.9 kb (57.6%) of which came from Nt and 299.1 kb (42.4%) from Hn (Fig. 3). Sequence identity between each cybrid region and the corresponding parental region is extremely high (99.8-100%, with an average identity of 99.95%). The parental plants of the cybrid (N. tabacum R100a1 mutant and an ecotype of H. niger from Marburg Botanical Garden) are not the same as the Nt and Hn plants (H. niger A04750027 and N. tabacum cultivar Bright Yellow 4) whose sequenced organelle genomes were used for comparison to the cybrid genomes. However, the extremely high level of nucleotide identity between the cybrid mtDNA regions and the cognate region(s) in the sequenced Nt and/or Hn mtDNAs shows that the two different varieties of Nt and of Hn are virtually identical in mtDNA sequence. Furthermore, the chloroplast genomes of the cybrid and the sequenced strain of Hn are identical (see previous section).
Thirty cases of crossing-over were readily identified because parental-unique sequences were found at each side of the inferred crossover (Fig. 3). Five cases of gene conversion without crossing-over were detected at the P<0.01 level by GeneConv (Table 3). We did not find evidence for illegitimate recombination, i.e. non-homologous end joining (NHEJ). Eight of the 35 recombination events took place within gene sequences, with the rest occurring in intergenic regions (Table 3; Fig. 3). On average, there was one intergenomic recombination event every 20.1 kb of the resulting cybrid genome. The sites of recombination are not evenly distributed along the cybrid mtDNA: the closest pair of adjacent sites is only 0.6 kb apart, while the farthest is 64 kb apart (Fig. 3).
All detectable recombination events occurred in areas where parental mitochondrial sequences overlapped, i.e., were homologous (Fig. 3, see ‘Blast hits’). Overlapping homologous sequences from the fusion parents became repeats when the parental mitochondria fused, enabling homologous recombination to occur between their genomes. The homologous regions that engaged in the 35 intergenomic recombination events that were detected in the cybrid mitochondrial genome ranged between 100 bp and 9 kb in length, with an identity of at least >94% and usually much higher (Table 3). For 27 of the 30 crossover events, a single product was found at a detectable stoichiometry in the cybrid mtDNA, whereas in the other three cases two reciprocal recombination products were detected, and at similar stoichiometries (Table 3, events # 1, 15, and 17).
Gene content in the cybrid mitochondrial genome
Counting each gene only once, the cybrid mitochondrial genome contains 37 protein genes, three rRNA genes, and 23 tRNA genes (Table 1). Intron content is identical to Hn and includes 23 group II introns and one group I intron. The cybrid genome contains two copies of 15 genes and three copies of one gene. Six of the two-copy cases (atp1, atp9, trnK, trnN, trnQ, trnS) result from retention of either entire genes or partial genes in the form of intact chimeric genes (e.g. see atp1 in Table 2) from both parents. All other multi-copy cases result from duplication or triplication of genes inherited from only a single parent (Table 2; Fig. 3).
The cybrid genome possesses a total of 45 and 26 genes that were inherited from Nt and Hn, respectively. It also contains nine genes that are chimeric as a result of intergenic recombination (seven protein genes and one rRNA gene (Table 2), plus one tRNA gene (not tabulated)). Counting each gene only once, the cybrid mtDNA contains 21 protein, two rRNA, and 11 tRNA genes that were inherited exclusively from Nt, as compared to only eight protein and eight tRNA genes from Hn (Table 2). Of the 16 genes inherited exclusively from Hn, eight tRNA genes and one protein gene are identical to those in Nt and probably don't cause any nuclear-cytoplasmic incompatibility. By contrast, the other seven mitochondrial genes inherited only from Hn possess non-synonymous differences in coding sequences and, in two cases, differences in intron content and/or editing sites (Table 2).
Of special note, the cybrid cox1 gene, inherited from Hn, has a group I intron, which likely requires a nuclear factor for its splicing because its homolog in yeast mitochondria does not self-splice (Dujardin et al. 1982; Herbert et al. 1988). We showed that the cox1 gene is spliced and edited in the cybrid, which indicates that the cybrid possesses the necessary machinery for proper processing of the cox1 primary transcript (Fig. S3). Note that the Hn cox1 gene lacks any species-specific editing sites, whereas the Nt homolog contains two such sites.
Discussion
Generating new nuclear-cytoplasmic combinations in cybridization experiments is an important avenue for modeling evolution of the cytoplasm (Greiner & Bock 2013). It has been known for over three decades that somatic hybridization often results in novel mitochondrial genome arrangements (Belliard et al., 1979; Pelletier et al., 1985; Vedel et al., 1986). Yet this is the first study in which complete mitochondrial sequences from a cybrid and both of its parents are compared, allowing the first detailed examination of the extent and nature of recombination between two plant mitochondrial genomes.
The cybrid mitochondrial genome is unexpectedly large and highly chimeric
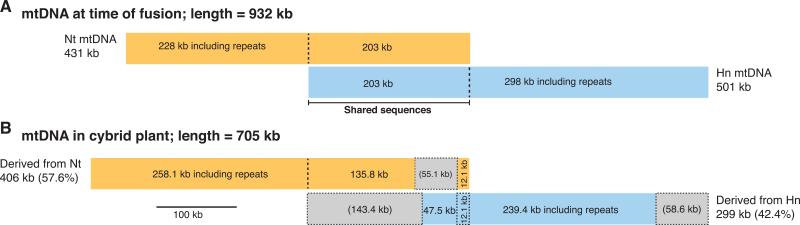
The cybrid mtDNA is ~705 kb in length, which is 227 kb less than the sum of its parental mtDNAs (932 kb) but considerably larger than each alone (431 and 501 kb). It is also remarkably chimeric as a result of at least 30 crossover and five gene conversion events between its parental genomes. Based on cytological studies of plant mitochondrial fusion (Arimura et al., 2004; Sheahan et al., 2005), we assume that during the formation of the cybrid, mitochondria from both parents fused massively, bringing both parental genomes into a single mitochondrion and thus enabling extensive recombination of the two genomes, accompanied (or followed) by loss of some parental sequences (Fig. 5). Some 57.6% and 42.4% of the cybrid mtDNA came from Nt and Hn, respectively; this corresponds to the loss of ~55 kb worth of Nt mitochondrial sequences and ~202 kb of the Hn mtDNA (Fig. 5).
Fig. 5.
Cybrid mitochondrial genome composition. Summary status of the cybrid mitochondrial genome with respect to its parental composition either (a) soon after protoplast fusion, at which point the single fusion cell probably possessed a mitochondrial network that contained the entirety of both parental mitochondrial genomes, or (b) as determined from the mtDNA sequence of the mature cybrid plant investigated in this study. In both parts, the cybrid genome comprises the sum of the orange Nicotiana tabacum (Nt)-derived, and blue, Hyoscyamus niger (Hn)-derived, sequences. The gray regions in (b) indicate the aggregate length of parental-shared or parental-unique regions that are absent from the genome of the cybrid plant.
The size of a hybrid mitochondrial genome relative to its parental genomes has been previously estimated in 12 Brassica somatic hybrids. In these cases, the hybrid mtDNA was either intermediate in size to its parental mtDNAs or at most 11% larger than the larger parental mtDNA (Temple et al., 1992; Wang et al., 2012). Strikingly, the Solanaceae cybrid mtDNA in this study is fully 41% and 64% larger than the Hn and Nt genomes, respectively. Mitochondrial genome size in somatic hybrids or cybrids is influenced by several factors, including: (1) cell type selected for protoplast isolation (particular cells may differ in the amount of mitochondria they contain and their ability to undergo massive mitochondrial fusion); (2) protoplast pre-treatments (chemical treatment or irradiation of protoplasts before fusion could inactivate or eliminate mitochondria from one parent); (3) nuclear genome composition (nuclear-mitochondrial interactions may impose a bias towards mitochondrial sequence retention or elimination); (4) recombination rate and efficiency between the parental mtDNAs depending on their overall synteny and similarity; and (5) culture, fusion and selection procedures (Earle et al., 1992; Sheahan et al., 2005; Preuten et al., 2010; Greiner & Bock, 2013;). Genome sequencing of many more cybrid mtDNAs, of diverse parentage and method of generation, is necessary to determine whether this Solanaceae cybrid genome is indeed unusually large, whether other cybrid genomes might be proportionately even larger, and whether any general patterns emerge with respect to cybrid genome size relative to parental genome sizes.
Mitochondrial rearrangements occurred only by homologous recombination Homologous recombination in plant mitochondria is restricted to sequences of low divergence. The mismatch repair system does not allow recombination between sequences that are highly divergent (Shedge et al., 2007; Arrieta-Montiel et al., 2009), acting as a safeguard to prevent deleterious recombination between non-homologous regions. Homologous recombination in plant mitochondria is mediated by nuclear-encoded recA homologs (Miller-Messmer et al., 2012). These events are classified as either crossovers, in which both strands of each homologous duplex are reciprocally broken and joined, so that alleles flanking the break site are exchanged; or non-crossovers, i.e., gene conversions, in which there is no reciprocal exchange and instead a non-reciprocal transfer of sequence information occurs from one homologous DNA segment to the other, with the donor sequence unchanged (Nieto Feliner & Rosello, 2012). The minimal pairing sequence of bacterial recA can be as small as 25 bp, but efficiency greatly increases with lengths larger than 200 bp (Gualberto et al., 2014). Knowing the minimum repeat length necessary for recombination in the mitochondrial genome is important for understanding how they rearrange over time and for identifying the mechanism(s) of these recombination events.
We posit that in the cybrid under study, homologous recombination took place across repeats that originated when homologous sequences from both parents were incorporated in the cybrid mitochondrial genome. The homologous regions that engaged in the 35 intergenomic recombination events that were detected in the cybrid mitochondrial genome ranged between 100 bp and 9 kb in length. Three of the five gene-conversion events detected extended for a minimum of 1kb (Table 3). These are longer than previous reports on plant mitochondria, which identified a converted region of no more than a few hundred nucleotides long (Hao & Palmer, 2009; Hao et al., 2010; Mower et al., 2010; Sloan et al., 2010). However, it is difficult to distinguish a long gene conversion from multiple crossover events because multiple crossovers mimic gene conversion (Santoyo et al., 2005).
Only a single recombinant product was detected for 27 of the observed crossover events, including all but one of the 19 crossovers that involve large (>1 kb) repeats (Table 3). This is surprising in the sense that in non-cybrid angiosperm mitochondrial genomes, repeats of this size are expected to mediate high frequency, reciprocal homologous recombination (Kuhn & Gualberto, 2012). This finding raises the possibility that recombination in the cybrid occurred in a predominantly non-reciprocal manner, presumably via the break-induced replication pathway (Marechal & Brisson 2010; Shedge et al., 2007). Alternatively, most or all crossovers may have occurred reciprocally, probably via the classic double-strand break repair pathway (Marechal & Brisson, 2010; Kuhn & Gualberto, 2012), but with only one of the recombination products retained at a detectable level in the cybrid.
Non-homologous end joining (NHEJ) refers to the ligation of two nonhomologous DNA ends, which may show regions of microhomology (Deriano & Roth, 2013). This process is suppressed by the nuclear gene MSH1 in Arabidopsis (Davila et al., 2011). No evidence for non-homologous (illegitimate) recombination was found in the cybrid Nt(+Hn) mitochondria. The absence of illegitimate recombination implies that homologous sequences were incorporated by recombination into this cybrid genome far more efficiently than non-homologous sequences.
Biased sequence loss in the cybrid mitochondrial genome
The cybrid mitochondrial genome shows two pronounced biases in sequence loss (Fig. 5). The first is related to parental origin: the cybrid genome has lost fully 40% (202 kb) of sequences from Hn but only 13% (55.1 kb) of Nt sequences. This could be the result of an unequal contribution of mitochondrial genome copies during protoplast fusion, i.e., the Nt protoplast may have contributed many more mtDNA copies than did Hn, such that the initial recombination events produced a cybrid genome that was biased toward Nt from the outset. Alternatively, the Nt-derived nuclear genome of the cybrid may have preferentially selected for retention of Nt mitochondrial sequences (especially genes) owing to nuclear-cytoplasmic incompatibilities with certain Hn sequences. This would be consistent with similar observations made in other cybrids (Wachocki et al., 1991; Yamaoka et al., 2000; Dey et al., 2000) and in introgressed populations (Burton et al., 2006).
The other bias in sequence loss involves duplicated vs unique sequences. The cybrid mtDNA has lost one copy of 94% (191 kb) of the 203 kb of sequences that are shared by the parental genomes and which presumably were initially present as ‘heterologous’ duplications in the cybrid. By contrast, only 11% (58.6 kb) of the 526 kb of the sequences that are unique to the parental genomes were lost in the cybrid (Fig. 5). This bias may reflect non-adaptive molecular mechanisms of sequence elimination that preferentially remove duplicated sequences. These mechanisms include reciprocal and non-reciprocal homologous crossovers between repeated sequences, gene conversion between homeoalleles, and illegitimate recombination (Shaked et al., 2001; Bennetzen et al., 2005; Shedge et al., 2007; Grover & Wendel, 2010; Davila et al., 2011; Leitch & Leitch, 2013). Alternatively, adaptive forces may be at work, involving deleterious effects of keeping potentially conflicting copies of coding regions derived from each parent. Relevant here is the striking pattern of allele retention in the cybrid mtDNA, for which 57 of the 63 mitochondrial genes (including 35 of 37 protein-coding genes) were found in a single form, derived from either a single parent or chimeric. In other words, 57 alleles of 63 genes were lost by the cybrid mitochondria.
Previous studies on somatic hybrids and cybrids showed a similar pattern of single-allele retention, although only one or a few genes per hybrid were analyzed in most of these studies (Temple et al., 1992; Bonnema et al., 1995; Motomura et al., 1996; Kanno et al., 1997; Moriguchi et al., 1997; Cardi et al., 1999; Lossl et al., 1999; Rasmussen et al. 2000; Bastia et al., 2001; Scotti et al. 2004). Also, a genomic study (Wang et al., 2012) on a Brassica cybrid showed that its mitochondrial genome contained only 59 genes, very similar to the gene content in each parental mtDNA (Tanaka et al., 2012), although the parental origin of the cybrid genes was not determined.
We propose that the co-existence of different alleles in the mtDNA of somatic hybrids and cybrids may be deleterious, as they might impair genetic homeostasis or global gene expression, resulting in an improved fitness of those hybrids that more efficiently eliminate these extra alleles. During this process of gene elimination, non-coding duplicated regions flanking the genes may also be deleted, resulting in an overall loss of duplicated regions.
This selective process probably also affects plant mtDNAs that acquire foreign mitochondrial genes by horizontal gene transfer (HGT). In the majority of known cases, a single allele of each mitochondrial gene - either the native, the foreign, or a chimeric form - has been retained (Bergthorsson et al., 2003; Barkman et al., 2007; Hao et al., 2010; Mower et al., 2010; Xi et al., 2013). The few angiosperms that maintain both foreign and native alleles appear to overcome the putative selective disadvantage of keeping two distinct alleles by silencing and/or rapid pseudogenization of one allele or the other, almost always the foreign one (Mower et al., 2004, 2010; Rice et al. 2013).
Further studies are necessary to establish whether the biased elimination of repeated sequences and genes seen in the Nt(+Hn) cybrid is due primarily to a selective advantage of having single alleles or to largely neutral molecular mechanisms that remove duplicated sequences from cybrid mitochondrial genomes.
Nuclear-cytoplasmic incompatibility
Nuclear-cytoplasmic incompatibilities are expected when the nuclear and cytoplasmic genomes have a different origin (Greiner & Bock, 2013). Some organelle genes in angiosperms are subjected to C-to-U RNA editing, which restores the conserved amino acid sequence (Freyer et al., 1997). This editing is controlled by the nuclear genome, opening the door to nuclear-cytoplasmic incompatibilities (Schmitz-Linneweber et al., 2005). The Nt-derived nuclear-encoded editing machinery in the cybrid Nt(+Hn) faces the challenge of editing three Hn-specific sites of chloroplast RNA editing, one in each of three genes (ndhA, ndhD, and rpoB) in which the cybrid (this study) and Hn (Sanchez-Puerta & Abbona, 2014) cpDNAs are predicted to undergo RNA editing at sites at which Nt cpDNA encodes a T and does not require editing. Those edits restore what would otherwise be non-synonymous changes at highly conserved amino acids. The Hn-specific editing sites in the cybrid cpDNA are presumably recognized and edited by the Nt nuclear-encoded machinery, although perhaps with reduced efficiency, given that the cybrid Nt(+Hn) is pale green in early development but photosynthetically functional (Zubko et al., 2001). Indeed, a study on N. tabacum (+Atropa belladonna) cybrids showed experimentally that the same three editing sites that are specific to Hn-derived sequences present in our cybrid were successfully edited by the Nt nuclear–encoded machinery in Atropa chloroplast transcripts (Schmitz-Linneweber et al., 2005). It is believed that the Nt nuclear genome has additional editing factors and thus is capable of heterologous editing (Schmitz-Linneweber et al., 2005). In an extreme case of nuclear-cytoplasmic incompatibility, cybrids Atropa belladonna (+Nt) with the Nt plastid genome manifested an albino phenotype (Kushnir et al., 1991). These cybrids failed to edit all or some of the five Nt-specific editing sites in four chloroplast genes (Schmitz-Linneweber et al., 2005).
Nuclear-mitochondrial incompatibilities are also expected in the cybrid under study given the observed differences in intron content, editing sites, and non-synonymous sites in those mitochondrial genes inherited exclusively from Hn. That the group I intron acquired from Hn was successfully spliced in the cybrid is surprising in the context of the best-studied cognate of this intron, from yeast, whose splicing requires nuclear factors (Dujardin et al., 1982; Herbert et al., 1988). On the other hand, this intron is well-established to have been horizontally transferred hundreds if not thousands of times among angiosperm mitochondrial genomes (Cho et al., 1998; Sanchez-Puerta et al., 2008, 2011), which suggests that such factor(s) were widely present in plants before the evolutionary spread of this intron (presumably because it/they play some other, important role). Other differences in Hn-derived mitochondrial genes and/or newly-formed ORFs could be related to the cytoplasmic male sterility and other phenotypic characteristics observed in the cybrid Nt(+Hn) (Zubko et al., 2001). The results from this study set the stage for in-depth investigation of the expression of these genes and ORFs.
Cybrids as a model system for studying HGT in plant mitochondrial genomes
Whereas mitochondrial-to-mitochondrial HGT occurs relatively often in angiosperms, HGT is unknown in their chloroplast genomes (Mower et al., 2012a; Rice et al., 2013; Xi et al., 2013). These evolutionary findings are consistent with evidence from both cell biology (Arimura et al., 2004; Sheahan et al., 2005) and genetics (Belliard et al., 1979; Earle et al., 1992; Morgan & Maliga, 1987; Temple et al. 1992) that mitochondria readily fuse with one another but not chloroplasts. These observations, plus those specific to the case of extraordinarily massive HGT in the Amborella mitochondrial genome (Rice et al., 2013), led Rice et al. (2013) to propose that the entry of foreign mtDNA into angiosperm mitochondrial genomes is driven principally, if not entirely, by the ease of mitochondrial fusion. Therefore, the fusion-driven creation of a novel, chimeric mitochondrial genome in cybrids is probably quite analogous to the early stage of the creation of a novel mitochondrial genome when HGT occurs in plant mitochondria. That is, soon after the fixation of an HGT event, the mitochondrial genome probably resembles those of cybrids in containing substantial portion of both ‘parental’ genomes (i.e., the HGT donor and recipient genomes). Over evolutionary time, selection for nuclear-mitochondrial compatibilities, in conjunction with neutral deletional forces that in most plant mtDNAs result in high rates of DNA loss, likely leads to loss of most of the foreign DNA and especially (see previous section) the foreign genes.
Nuclear vs mitochondrial allopolyploidy
The potential to combine the entirety of two distinct plant mitochondrial genomes through cybrid formation makes this process somewhat analogous to the well-studied and evolutionarily important process of allopolyploidization, in which sexually hybrid plants contain two copies of each parental nuclear genome (Ozkan et al., 2001; Adams et al., 2003). As in cybrid mitochondrial genomes, the doubled nuclear genome in allopolyploids is rapidly reduced by elimination of significant portions of both parental nuclear genomes. In particular, allopolyploids undergo rapid loss of non-coding regions that are shared by the parental species, presumably to suppress homeologous pairing at meiosis (Shaked et al., 2001; Leitch & Bennet, 2004; Doyle et al., 2008). However, in contrast to the single-copy fate of most genes in cybrid mitochondria, nuclear allopolyploids generally retain both parental alleles, although often silencing one of them (D'Arcy & Zhang, 1992; Ozkan et al., 2001; Adams et al., 2003). Another key difference is that recombination between homeologous chromosomes in nuclear allopolyploids is usually quite limited, whereas, as shown in this study, recombination between parental mitochondrial genomes in cybrids can be extensive, creating a highly novel and intensely chimeric genome.
Supplementary Material
Acknowledgements
We thank C. Abbona for laboratory assistant and J. Mower for critical reading of the manuscript. This work was supported by NIH (FIRCA-BB-R03-TW008353-01 to J.D.P. and M.V.S-P.), by ANPCyT (PICT-193 to M.V.S-P.), by CONICET/NSF (RD#138/13 to M.V.S-P), and by NSF (#1062432 to Indiana University), which supports the computer cluster on which many of the analyses were performed.
Footnotes
Supporting Information
Additional supporting information may be found in the online version of this article.
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
References
- Adams KL, Cronn R, Percifield R, Wendel JF. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Nat. Acad. Sci. 2003;100:4649–54. doi: 10.1073/pnas.0630618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi H, Shimada H, Fujimura T. High-frequency inter-parental recombination between mitochondrial genomes of rice cybrids. Curr. Genet. 1995;29:58–65. doi: 10.1007/BF00313194. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Alverson AJ, Wei X, Rice DW, Stern DB, Barry K, Palmer JD. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol. Biol. Evol. 2010;27:1436–48. doi: 10.1093/molbev/msq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apitz J, Weihe A, Pohlheim F, Börner T. Biparental inheritance of organelles in Pelargonium: evidence for intergenomic recombination of mitochondrial DNA. Planta. 2013;237:509–15. doi: 10.1007/s00425-012-1768-x. [DOI] [PubMed] [Google Scholar]
- Arimura S, Yamamoto J, Aida GP, Nakazono M, Tsutsumi N. Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proc. Nat. Acad. Sci. USA. 2004;101:7805–08. doi: 10.1073/pnas.0401077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta-Montiel MP, Shedge V, Davila J, Christensen AC, Mackenzie SA. Diversity of the Arabidopsis mitochondrial genome occurs via nuclear-controlled recombination activity. Genetics. 2009;183:1261–68. doi: 10.1534/genetics.109.108514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv D, Bleichman S, Arzee-Gonen P, Galun E. Intersectional cytoplasmic hybrids in Nicotiana. Theor. Appl. Genet. 1984a;67:499–504. doi: 10.1007/BF00264893. [DOI] [PubMed] [Google Scholar]
- Aviv D, Arzee-Gonen P, Bleichman S, Galun E. Novel alloplasmic Nicotiana plants by “donor-recipient” protoplast fusion: cybrids having N. tabacum or N. sylvestris nuclear genomes and either or both plastomes and chondriomes from alien species. Mol. Gen. Genet. 1984b;196:244–53. [Google Scholar]
- Backert S, Nielsen BL, Börner T. The mystery of the rings: structure and replication of mitochondrial genomes from higher plants. Trends Plant Sci. 1997;2:477–83. [Google Scholar]
- Barkman TJ, McNeal JR, Lim SH, Coat G, Croom HB, Young ND, dePamphilis CW. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evol. Biol. 2007;7:248. doi: 10.1186/1471-2148-7-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastia T, Scotti N, Cardi T. Organelle DNA analysis of Solanum and Brassica somatic hybrids by PCR with “universal primers”. Theor. Appl. Genet. 2001;102:1265–72. [Google Scholar]
- Bellaoui M, Martin-Canadell A, Pelletier G, Budar F. Low-copy-number molecules are produced by recombination, actively maintained and can be amplified in the mitochondrial genome of Brassicaceae: relationship to reversion of the male sterile phenotype in some cybrids. Mol. Gen. Genet. 1998;257:177–85. doi: 10.1007/s004380050637. [DOI] [PubMed] [Google Scholar]
- Belliard G, Vedel F, Pelletier G. Mitochondrial recombination in cytoplasmic hybrids of Nicotiana tabacum by protoplast fusion. Nature. 1979;281:401–03. [Google Scholar]
- Bennetzen JL, Ma JX, Devos TM. Mechanisms of recent genome size variation in flowering plants. Ann. Bot. 2005;95:127–32. doi: 10.1093/aob/mci008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U, Adams KL, Thomason B, Palmer JD. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature. 2003;424:197–201. doi: 10.1038/nature01743. [DOI] [PubMed] [Google Scholar]
- Bonnema AB, Castillo C, Reiter N, Cunningham M, Adams HP, O'Connell MA. Molecular and ultrastructural analysis of a nonchromosomal variegated mutant. Plant Physiol. 1995;109:385–92. doi: 10.1104/pp.109.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RS, Ellison CK, Harrison JS. The sorry state of F2 hybrids: consequences of rapid mitochondrial DNA evolution in allopatric populations. Am. Nat. 2006;168:S14–24. doi: 10.1086/509046. [DOI] [PubMed] [Google Scholar]
- Cardi T, Bastia T, Monti L, Earle E. Organelle DNA and male fertility variation in Solanum spp. and interspecific somatic hybrids. Theor. Appl. Genet. 1999;99:819–28. [Google Scholar]
- Cho Y, Qiu YL, Kuhlman P, Palmer JD. Explosive invasion of plant mitochondria by a group I intron. Proc. Natl. Acad. Sci. USA. 1998;95:14244–49. doi: 10.1073/pnas.95.24.14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Arcy W, Zhang Z-Y. Notes on the Solanaceae of China and neighbouring areas. Novon. 1992;2:124–28. [Google Scholar]
- Dale R, Wu M, Kiernan M. Analysis of four tobacco mitochondrial DNA size classes. Nucleic Acids Res. 1983;11:1673–85. doi: 10.1093/nar/11.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darracq A, Varre J, Touzet P. A scenario of mitochondrial genome evolution in maize based on rearrangement events. BMC Genomics. 2010;11:233. doi: 10.1186/1471-2164-11-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila J, Arrieta-Montiel M, Wamboldt Y, Cao J, Hagmann J, Shedge V, Xu Y, Weigel D, Mackenzie S. Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol. 2011;9:64. doi: 10.1186/1741-7007-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriano L, Roth D. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu. Rev. Genet. 2013;47:433–55. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- Dey R, Barrientos A, Moraes C. Functional constraints of nuclear-mitochondrial DNA interactions in xenomitochondrial rodent cell lines. J. Biol. Chem. 2000;275:31520–27. doi: 10.1074/jbc.M004053200. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid isolation procedure for small quantities of fresh leaf tissues. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- Doyle JJ, Flagel L, Paterson AH, Rapp RA, Soltis DE, Soltis PS, Wendel JF. Evolutionary genetics of genome merger and doubling in plants. Ann. Rev. Genet. 2008;42:443–61. doi: 10.1146/annurev.genet.42.110807.091524. [DOI] [PubMed] [Google Scholar]
- Dujardin G, Jacq C, Slonimski P. Single base substitution in an intron of oxidase gene compensates splicing defects of the cytochrome b gene. Nature. 1982;298:628–32. doi: 10.1038/298628a0. [DOI] [PubMed] [Google Scholar]
- Earle ED. Mitochondrial DNA in somatic hybrids and cybrids. In: Levings C, Vasil IK, editors. The molecular biology of plant mitochondria. Kluwer Academic Publishers; Dordrecht, the Netherlands: 1995. pp. 557–84. [Google Scholar]
- Earle ED, Temple M, Walters TW. Organelle assortment and mitochondrial DNA rearrangements in Brassica somatic hybrids and cybrids. Physiol. Plant. 1992;85:325–33. [Google Scholar]
- Freyer R, Kiefer-Meyer M, Kossel H. Occurrence of plastid RNA editing in all major lineages of land plants. Proc. Natl. Acad. Sci. USA. 1997;94:6285–90. doi: 10.1073/pnas.94.12.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galun E, Arzee-Gonen P, Fluhr R, Edelman M, Aviv D. Cytoplasmic hybridization in Nicotiana: mitochondrial DNA analysis in progenies resulting from fusion between protoplasts having different organelle constitutions. Mol. Gen. Genet. 1982;186:50–56. doi: 10.1007/BF00422911. [DOI] [PubMed] [Google Scholar]
- Gordon D, Green P. Consed: a graphical editor for next-generation sequencing. Bioinformatics. 2013;29:2936–37. doi: 10.1093/bioinformatics/btt515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner S, Bock R. Tuning a menage a trois: co-evolution and co-adaptation of nuclear and organellar genomes in plants. Bioessays. 2013;35:354–65. doi: 10.1002/bies.201200137. [DOI] [PubMed] [Google Scholar]
- Grewe F, Viehoever P, Weisshaar B, Knoop V. A trans-splicing group I intron and tRNA-hyperediting in the mitochondrial genome of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 2009;37:5093–104. doi: 10.1093/nar/gkp532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover C, Wendel JF. Recent insights into mechanisms of genome size change in plants. J. Bot. 2010;2010:1–8. [Google Scholar]
- Gualberto JM, Mileshina D, Wallet C, Niazi A, Weber-Lotfi F, Dietrich A. The plant mitochondrial genome: dynamics and maintenance. Biochimie. 2014;100:107–20. doi: 10.1016/j.biochi.2013.09.016. [DOI] [PubMed] [Google Scholar]
- Handa H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 2003;31:5907–16. doi: 10.1093/nar/gkg795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W, Palmer JD. Fine-scale mergers of chloroplast and mitochondrial genes create functional, transcompartmentally chimeric mitochondrial genes. Proc. Natl. Acad. Sci. USA. 2009;106:16728–33. doi: 10.1073/pnas.0908766106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W, Richardson AO, Zheng Y, Palmer JD. Gorgeous mosaic of mitochondrial genes created by horizontal transfer and gene conversion. Proc. Natl. Acad. Sci. USA. 2010;14:21576–81. doi: 10.1073/pnas.1016295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht J, Grewe F, Knoop V. Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: the root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol. Evol. 2011;3:344–58. doi: 10.1093/gbe/evr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert CJ, Labouesse M, Dujardin G, Slonimski P. The NAM2 proteins from S. cerevisiae and S. douglasii are mitochondrial leucyl-tRNA synthetases, and are involved in mRNA splicing. EMBO J. 1988;7:473–783. doi: 10.1002/j.1460-2075.1988.tb02835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo YD, Choi Y, Kim DH, Kim BD, Kang B-C. Extensive structural variations between mitochondrial genomes of CMS and normal peppers (Capsicum annuum L.) revealed by complete nucleotide sequencing. BMC Genomics. 2014;15:561. doi: 10.1186/1471-2164-15-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno A, Kanzaki H, Kameya T. Detailed analyses of chloroplast and mitochondrial DNAs from the hybrid plant generated by asymmetric protoplast fusion between radish and cabbage. Plant Cell Rep. 1997;16:479–84. doi: 10.1007/BF01092770. [DOI] [PubMed] [Google Scholar]
- Knoop V, Unseld M, Marienfeld JR, Brandt P, Sunkel S, Ullrich H, Brennicke A. copia-, gypsy- and LINE-like retrotransposon fragments in the mitochondrial genome of Arabidopsis thaliana. Genetics. 1996;142:579–85. doi: 10.1093/genetics/142.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond S, Frost S, Muse S. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–79. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Kubo T, Kitazaki K, Matsunaga M, Kagami H, Mikami T. Male sterility-inducing mitochondrial genomes: how do they differ? Crit. Rev. Plant Sci. 2011;30:378–400. [Google Scholar]
- Kuhn K, Gualberto JM. Recombination in the stability, repair and evolution of the mitochondrial genome. In: Marechal-Drouard L, editor. Mitochondrial genome evolution. Elsevier Ltd: Academic Press; 2012. pp. 215–252. [Author, please insert the place of publication (city, state (if US) and country] [Google Scholar]
- Kushnir S, Babiychuk E, Bannikova M, Momot V, Komarnitsky I, Cherep N, Gleba Y. Nucleo-cytoplasmic incompatibility in cybrid plants possessing an Atropa genome and a Nicotiana plastome. Mol. Gen. Genet. 1991;225:225–30. doi: 10.1007/BF00269852. [DOI] [PubMed] [Google Scholar]
- Leitch I, Bennet M. Genome downsizing in polyploid plants. Biol. J. Linnean Soc. 2004;82:651–63. [Google Scholar]
- Leitch I, Leitch A. Genome size diversity and evolution in land plants. In: Leitch I, editor. Plant genome diversity 2. Springer-Verlag; Wien, Austria: 2013. pp. 307–322. [Google Scholar]
- Levin DA. The cytoplasmic factor in plant speciation. Syst. Biol. 2003;28:5–11. [Google Scholar]
- Liu G, Cao D, Li S, Su A, Geng JN, Grover C, Hu S, Hua J. The complete mitochondrial genome of Gossypium hirsutum and evolutionary analysis of higher plant mitochondrial genomes. PLoS One. 2013;8:e69476. doi: 10.1371/journal.pone.0069476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007;52:267–74. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- Lossl A, Adler N, Horn R, Frei U, Wenzel G. Chondriome-type characterization of potato: mt alpha, beta, delta, epsilon and novel plastidmitochondrial configurations in somatic hybrids. Theor. Appl. Genet. 1999;98:1–10. [Google Scholar]
- Lowe T, Eddy S. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–64. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie SA. The influence of mitochondrial genetics on crop breeding strategies. Plant Breed. Rev. 2005;25:115–38. [Google Scholar]
- Maddison W, Maddison P. MacClade version 4: analysis of phylogeny and character evolution. Sinauer Associates; Sunderland, MA, USA: 2000. [Google Scholar]
- Marechal A, Brisson N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010;186:299–317. doi: 10.1111/j.1469-8137.2010.03195.x. [DOI] [PubMed] [Google Scholar]
- McCauley DE. Paternal leakage, heteroplasmy, and the evolution of plant mitochondrial genomes. New Phytol. 2013;200:966–77. doi: 10.1111/nph.12431. [DOI] [PubMed] [Google Scholar]
- Miller-Messmer M, Kuhn K, Bichara M, Le Ret M, Imbault P, Gualberto JM. RecA-dependent DNA repair results in increased heteroplasmy of the Arabidopsis mitochondrial genome. Plant Physiol. 2012;159:211–26. doi: 10.1104/pp.112.194720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A, Maliga P. Rapid chloroplast segregation and recombination of mitochondrial DNA in Brassica cybrids. Mol. Gen. Genet. 1987;209:240–46. doi: 10.1007/BF00329649. [DOI] [PubMed] [Google Scholar]
- Moriguchi T, Motomura T, Hidaka T, Akihama T, Omura M. Analysis of mitochondrial genomes among Citrus plants produced by the interspecific somatic fusion of ‘Seminole’ tangelo with rough lemon. Plant Cell Rep. 1997;16:397–400. doi: 10.1007/BF01146781. [DOI] [PubMed] [Google Scholar]
- Motomura T, Moriguchi T, Akihama T, Hidaka T, Omura M. Analysis of cytoplasmic genomes in somatic hybrids between ‘Hazzara (Abohar)’ (Citrus reticulata Blanco) and Microcitrus australis (Planch.) Swingle. J. Japan. Soc. Hort. Sci. 1996;65:497–503. [Google Scholar]
- Mower J, Stefanovic S, Young GJ, Palmer JD. Plant genetics: gene transfer from parasitic to host plants. Nature. 2004;432:165–66. doi: 10.1038/432165b. [DOI] [PubMed] [Google Scholar]
- Mower JP. PREP-Mt: predictive RNA editor for plant mitochondrial genes. BMC Bioinformatics. 2005;6:96. doi: 10.1186/1471-2105-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower JP, Sloan D, Alverson AJ. Plant mitochondrial genome diversity: The genomics revolution. In: Wendel JF, et al., editors. Plant genome diversity I. Springer-Verlag; Wien, Austria: 123-144: 2012a. [Google Scholar]
- Mower JP, Case A, Floro E, Willis JH. Evidence against equimolarity of large repeat arrangements and a predominant master circle structure of the mitochondrial genome from a monkeyflower (Mimulus guttatus) lineage with cryptic CMS. Genome Biol. Evol. 2012b;4:670–86. doi: 10.1093/gbe/evs042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower JP, Stefanovic S, Hao W, Gummow JS, Jain K, Ahmed D, Palmer JD. Horizontal acquisition of multiple mitochondrial genes from a parasitic plant followed by gene conversion with host mitochondrial genes. BMC Biol. 2010;8:150. doi: 10.1186/1741-7007-8-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy F, Lazar G, Menczel L, Maliga P. A heteroplasmic state induced by protoplast fusion is a necessary condition for detecting rearrangements in Nicotiana mitochondrial DNA. Theor. Appl. Genet. 1983;66:203–207. doi: 10.1007/BF00251143. [DOI] [PubMed] [Google Scholar]
- Nieto Feliner G, Rosello J. Concerted evolution of multigene families and homoeologous recombination. In: Wendel JF, et al., editors. Plant genome diversity I. Springer-Verlag; Wien, Austria: 2012. pp. 171–194. [Google Scholar]
- Oldenburg D, Bendich A. Size and structure of replicating mitochondrial DNA in cultured tobacco cells. Plant Cell. 1996;8:447–461. doi: 10.1105/tpc.8.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan H, Levy A, Feldman M. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell. 2001;13:1735–1747. doi: 10.1105/TPC.010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD, Shields C. Tripartite structure of the Brassica campestris mitochondrial genome. Nature. 1984;307:437–440. [Google Scholar]
- Palmer JD, Herbon LA. Plant mitochondrial-DNA evolves rapidly in structure, but slowly in sequence. J. Mol. Evol. 1988;28:87–97. doi: 10.1007/BF02143500. [DOI] [PubMed] [Google Scholar]
- Pelletier G, Vedel F, Belliard G. Cybrids in genetics and breeding. Hereditas. 1985;103:49–56. [Google Scholar]
- Preuten T, Cincu E, Fuchs J, Zoschke R, Liere K, Borner T. Fewer genes than organelles: extremely low and variable gene copy numbers in mitochondria of somatic plant cells. Plant J. 2010;64:948–959. doi: 10.1111/j.1365-313X.2010.04389.x. [DOI] [PubMed] [Google Scholar]
- Rasmussen J, Lossl A, Rasmussen O. Analysis of the plastome and chondriome origin in plants regenerated after asymmetric Solanum ssp. protoplast fusions. Theor. Appl. Genet. 2000;101:336–343. [Google Scholar]
- Rice DW, Alverson AJ, Richardson AO, Young GJ, Sanchez-Puerta MV, Munzinger J, Barry K, Boore JL, Zhang Y, dePamphilis CW, et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science. 2013;342:1468–1473. doi: 10.1126/science.1246275. [DOI] [PubMed] [Google Scholar]
- Rothenberg M, Hanson MR. Recombination between parental mitochondrial DNA following protoplast fusion can occur in a region which normally does not undergo intragenomic recombination in parental plants. Curr. Genet. 1987;12:235–40. [Google Scholar]
- Sanchez-Puerta MV, Abbona C. The chloroplast genome of Hyoscyamus niger and a phylogenetic study of the tribe Hyoscyameae (Solanaceae). PLoS One. 2014;9:e98353. doi: 10.1371/journal.pone.0098353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Puerta MV, Cho Y, Mower JP, Alverson AJ, Palmer JD. Frequent, phylogenetically local horizontal transfer of the cox1 group I intron in flowering plant mitochondria. Mol. Biol. Evol. 2008;25:1762–1777. doi: 10.1093/molbev/msn129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Puerta MV, Abbona C, Zhuo S, Tepe E, Bohs L, Olmstead RG, Palmer JD. Multiple recent horizontal transfers of the cox1 intron in Solanaceae and extended coconversion of flanking exons. BMC Evol. Biol. 2011;11:277. doi: 10.1186/1471-2148-11-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoyo G, Martínez-Salazar J, Rodriguez C, Romero D. Gene conversion tracts associated with crossovers in Rhizobium etli. J. Bacteriol. 2005;187:4116–4126. doi: 10.1128/JB.187.12.4116-4126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkinen T, Bohs L, Olmstead RG, Knapp S. A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. BMC Evol. Biol. 2013;13:214. doi: 10.1186/1471-2148-13-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 1989;6:526–538. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Kushnir S, Babiychuk E, Poltnigg P, Herrmann RG, Maier RM. Pigment deficiency in nightshade/tobacco cybrids is caused by the failure to edit the plastid ATPase alpha-subunit mRNA. Plant Cell. 2005;17:1815–1828. doi: 10.1105/tpc.105.032474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti N, Marechal-Drouard L, Cardi T. The rpl5-rps14 mitochondrial region: a hot spot for DNA rearrangements in Solanum spp. somatic hybrids. Curr. Genet. 2004;45:378–382. doi: 10.1007/s00294-004-0496-6. [DOI] [PubMed] [Google Scholar]
- Shaked H, Kashkush K, Ozkan H, Feldman M, Levy A. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell. 2001;13:1749–1759. doi: 10.1105/TPC.010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan MB, McCurdy DW, Rose RJ. Mitochondria as a connected population: ensuring continuity of the mitochondrial genome during plant cell dedifferentiation through massive mitochondrial fusion. Plant J. 2005;44:744–755. doi: 10.1111/j.1365-313X.2005.02561.x. [DOI] [PubMed] [Google Scholar]
- Shedge V, Arrieta-Montiel M, Christensen A, Mackenzie S. Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell. 2007;19:1251–1264. doi: 10.1105/tpc.106.048355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T, Kaneko A, Nakata S, Harada K, Watanabe KI. Mitochondrial genome structure of a cytoplasmic hybrid between tomato and wild potato. Plant Cell Rep. 1998;17:832–836. doi: 10.1007/s002990050493. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, et al. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986;5:2043–49. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan D. One ring to rule them all? Genome sequencing provides new insights into the ‘master circle’ model of plant mitochondrial DNA structure. New Phytol. 2013;200:978–985. doi: 10.1111/nph.12395. [DOI] [PubMed] [Google Scholar]
- Sloan D, MacQueen A, Alverson AJ, Palmer JD, Taylor DR. Extensive loss of RNA editing sites in rapidly evolving Silene mitochondrial genomes: selection vs. retroprocessing as the driving force. Genetics. 2010;185:1369–1380. doi: 10.1534/genetics.110.118000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan D, Muller KM, McCauley D, Taylor DR, Storchova H. Intraspecific variation in mitochondrial genome sequence, structure, and gene content in Silene vulgaris, an angiosperm with pervasive cytoplasmic male steritility. New Phytol. 2012;196:1228–1239. doi: 10.1111/j.1469-8137.2012.04340.x. [DOI] [PubMed] [Google Scholar]
- Stern DB, Lonsdale D. Mitochondrial and chloroplast genomes of maize have a 12-kilobase DNA sequence in common. Nature. 1982;299:698–702. doi: 10.1038/299698a0. [DOI] [PubMed] [Google Scholar]
- Sugiyama Y, Watase Y, Nagase M, Makita N, Yagura S, Hirai A, Sugiura M. The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Mol. Gen. Genomics. 2005;272:603–615. doi: 10.1007/s00438-004-1075-8. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Tsuda M, Yasumoto K, Yamagishi H, Terachi T. A complete mitochondrial genome sequence of Ogura-type male sterile cytoplasm and its comparative analysis with that of normal cytoplasm in radish (Raphanus satitus L.). BMC Genomics. 2012;13:352. doi: 10.1186/1471-2164-13-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple M, Makaroff C, Mutschler M, Earle E. Novel mitochondrial genomes in Brassica napus somatic hybrids. Curr. Genet. 1992;22:243–249. doi: 10.1007/BF00351732. [DOI] [PubMed] [Google Scholar]
- Vedel F, Chetrit P, Mathieu C, Pelletier G, Primard C. Several different mitochondrial DNA regions are involved in intergenomic recombination in Brassica napus cybrid plants. Curr. Genet. 1986;11:17–24. [Google Scholar]
- Wachocki S, Bonnema A, O'Connell M. Comparison of the organization of the mitochondrial genome in tomato somatic hybrids and cybrids. Theor. Appl. Genet. 1991;81:420–427. doi: 10.1007/BF00228686. [DOI] [PubMed] [Google Scholar]
- Wang J, Jiang J, Li X, Li A, Zhang Y, Guan R, Wang Y. Complete sequence of heterogenous-composition mitochondrial genome (Brassica napus) and its exogenous source. BMC Genomics. 2012;13:675. doi: 10.1186/1471-2164-13-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihe A, Apitz J, Salinas A, Pohlheim F, Borner T. Biparental inheritance of plastidial and mitochondrial DNA and hybrid variegation in Pelargonium. Mol. Genet. Genomics. 2009;282:587–593. doi: 10.1007/s00438-009-0488-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Wang Y, Bradley R, Sugumaran M, Marx C, Rest J, Davis CC. Massive mitochondrial gene transfer in a parasitic plant clade. PloS Genet. 2013;9:e1003265. doi: 10.1371/journal.pgen.1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka M, Isobe K, Shitara H, Yonekawa H, Miyabayashi S, Hayashi J. Complete repopulation of mouse mitochondrial DNA-less cells with rat mitochondrial DNA restores mitochondrial translation but not mitochondrial respiratory function. Genetics. 2000;155:301–307. doi: 10.1093/genetics/155.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko MK, Zubko EI, Patskovsky YV, Khvedynich OA, Fisahn J, Gleba YY, Schieder O. Novel ‘homeotic’ CMS patterns generated in Nicotiana via cybridization with Hyoscyamus and Scopolia. J. Exp. Bot. 1996;47:1101–1110. [Google Scholar]
- Zubko MK, Zubko EI, Ruban AV, Adler K, Mock HP, Misera S, Gleba YY, Grimm B. Extensive developmental and metabolic alterations in cybrids Nicotiana tabacum (+ Hyoscyamus niger) are caused by complex nucleocytoplasmic incompatibility. Plant J. 2001;25:627–639. doi: 10.1046/j.1365-313x.2001.00997.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.