Abstract
Human DNA mismatch repair (MMR) proteins correct DNA errors and regulate cellular response to DNA damage by signaling apoptosis. Mutations of MMR genes result in genomic instability and cancer development. Nonetheless, how MMR proteins are regulated has not yet been determined. While hMLH1, hPMS2, and hMLH3 are known to participate in MMR, the function of another member of MutL-related proteins, hPMS1, remains unclear. Here we show that DNA damage induces the accumulation of hPMS1, hPMS2, and hMLH1 through ataxia-telangiectasia-mutated (ATM)-mediated protein stabilization. The subcellular localization of PMS proteins is also regulated during DNA damage, which induces nuclear localization of hPMS1 and hPMS2 in an hMLH1-dependent manner. The induced levels of hMLH1 and hPMS1 are important for the augmentation of p53 phosphorylation by ATM in response to DNA damage. These observations identify hMutL proteins as regulators of p53 response and demonstrate for the first time a function of hMLH1-hPMS1 complex in controlling the DNA damage response.
The DNA mismatch repair (MMR) system plays a critical role in maintaining genomic integrity in both prokaryotes and eukaryotes (27). It recognizes and repairs the base-base mismatches and small insertion-deletion mispairs generated during DNA synthesis. The MMR system is composed of several protein complexes, including MutS, MutL, and MutH. There are six MutS homologues (MSH1 to MSH6) and four MutL homologues (MLH1, PMS1, PMS2, and MLH3) in eukaryotes. MSH2 heterodimerizes with MSH3 or MSH6 to form MutS complexes. These two complexes have different mispair recognition properties. MSH2-MSH3 heterodimer (MutSα) recognizes insertion-deletion mispairs, whereas MSH2-MSH6 heterodimer (MutSβ) interacts with both single-base-pair mismatches and insertion-deletion mispairs. There are two MutL-related heterodimeric complexes known to be involved in MMR: MLH1-PMS2 (mammalian PMS2, closely related to Saccharomyces cerevisiae PMS1) and MLH1-MLH3. MLH1-PMS2 heterodimer (MutLα) interacts with both MSH2-MSH3 and MSH2-MSH6 complexes and plays a major role in MMR. MLH1-MLH3 also plays a role in repairing mispairs (19) and promoting meiotic crossing-over (39).
Another MutL complex, MutLβ (hMLH1-hPMS1), was identified in human cells (28, 35). However, the hMLH1-hPMS1 complex does not have significant MMR activity in an in vitro MMR assay (35). Pms1-deficient mice also do not develop the tumors or manifest microsatellite instability seen in Mlh1- or Pms2-deficient mice (34, 39). Although a mutation of hPMS1 was found in a patient with hereditary nonpolyposis colon cancer, the colon cancer susceptibility in this family was later attributed to the hMSH2 mutation (31). Moreover, mutations in yeast gene mlh2 (human PMS1 homologue) do not lead to an increase in the mutation rate (22, 39). Thus, the biological function of hPMS1 remains unknown. Interestingly, the yeast mlh2 mutants are more resistant to cisplatin, carboplatin, and doxorubicin (17). Thus, the function of hPMS1 deserves investigation.
Recent evidence supports an additional role of MMR in signaling the DNA damage response. It has been reported that MMR is required for p53 phosphorylation in response to DNA methylator damage (15). UVB-induced apoptosis and p53 phosphorylation at serine 15 are significantly diminished in MSH2-deficient cells (33). MMR is also involved in the induction of the p53-related transcription factor p73. Cisplatin-induced accumulation of p73 depends on functional hMLH1 (20). An interaction between hPMS2 and p73 is induced upon cisplatin treatment, which leads to stabilization and activation of p73 (37). Moreover, the MMR system was shown to be required for activation of S-phase checkpoint in response to ionizing radiation (6). Therefore, the MMR system appears to play a dual role in response to abnormal DNA structures: MMR and DNA damage signaling. Although MMR repairs only DNA mismatches or mispairs, it is involved in apoptosis and checkpoint activation in response to various forms of DNA damage. Moreover, while the repair can function efficiently at subnormal levels of hMLH1, the checkpoint activation requires a full level of hMLH1 (12). Thus, the functions of MMR proteins in MMR and signaling may involve different molecular processes. How MMR proteins participate in the DNA damage signaling is not well understood. hMSH2-hMSH6 forms a sliding clamp on mismatched DNA (21). This sliding clamp was proposed to transduce a mismatch signal through interaction with other signaling molecules to activate apoptosis (18). In fact, MMR proteins, including hMSH2, hMSH6, and hMLH1, are part of a large complex containing other signaling molecules, such as breast cancer associated-1 (BRCA1) and ataxia-telangiectasia (AT)-mutated (ATM) molecules (40). It was also suggested that an MMR complex formed at the sites of DNA damage could serve as a scaffold to recruit both ATM and Chk2 and facilitate the activation of Chk2 by ATM (6).
How the function of MMR proteins is activated to induce apoptosis is not well understood. Cells lacking hMLH1 have very low levels of hPMS1 and hPMS2 proteins in spite of normal levels of RNA, suggesting that hPMS proteins are unstable in the absence of hMLH1 (8, 13, 28, 35). Thus, the stability of hPMS proteins may be regulated. It was also proposed that dimerization of hMLH1 and hPMS2 may regulate the nuclear import of hMLH1-hPMS2 (41). We have now examined the expression and subcellular localization of hMutL proteins in response to DNA damage. We find that DNA damage results in stabilization and accumulation of hMutL proteins. The induction depends on ATM, a kinase responsible for induction of other proteins such as p53 (4, 10, 26), E2F1 (30), and BLM (1). Furthermore, hMLH1 mediates DNA-damage-induced nuclear localization of hPMS1 and hPMS2. Consequently, the nuclear accumulation of hMLH1 and hPMS1 proteins augments the activation of p53 by ATM. Thus, the regulation of hMutL proteins could function as a sensor mechanism and fine tune the amplitude of DNA damage response according to the severity of DNA damage. A function of hMLH1-hPMS1 complex in the DNA damage response is demonstrated for the first time.
MATERIALS AND METHODS
Cell culture and transfection.
Human foreskin fibroblasts (HFF) and HEK293T, HEK293, MDA-MB468, and MCF7 cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS). H460, GM2184, GM3382, GM1526, GM3332, and GM719 cell lines were maintained in RPMI 1640 supplemented with 10% FBS. HCT116 (MLH1-deficient colorectal tumor line) cells were purchased from the American Type Culture Collection (ATCC) and maintained in McCoy's 5A medium supplemented with sodium bicarbonate (1.5 g/liter) and 10% FBS. 293T and 293 cells were transfected using a standard calcium phosphate method. HCT116 cells were transfected with Lipofectamine 2000 (Invitrogen) or a Gene Pulser Xcell electroporation system (Bio-Rad) according to the manufacturer's instructions. For electroporation, HCT116 cells were resuspended with 200 μl of McCoy's 5A medium in 2-mm-long cuvettes and electroporated with the following settings: 155 V, 1,000 μF, ∞ ohm. To obtain high-level transfection efficiency, HCT116 cells were transfected twice by electroporation on two consecutive days and were harvested 24 h after the second electroporation. To establish MCF7 cell lines stably expressing MLH1 short interfering RNAs (siRNA), MCF7 cells were transfected with pSUPER vector or pSUPER-siMLH1 along with pSV2Neo by use of a Gene Pulser Xcell electroporation system according to the manufacturer's instructions. The stable clones were then selected with G418.
Plasmid construction.
pCMV-SPORT6-hMLH1 was purchased from ATCC. To construct HcRed1-hMLH1, the XbaI fragment of hMLH1 cDNA from pCMV-SPORT6-hMLH1 was digested and inserted into the XbaI site of pHcRed1-C1. The hemagglutinin (HA)-tagged hPMS1 expression vector, pcDNA3-HA-hPMS1, was constructed by removing hPMS1 cDNA from pBS-hPMS1 (a gift from Bert Vogelstein) and inserting it into a pcDNA3 vector expressing HA-tagged fusion protein. pEGFP-hPMS1 was constructed by inserting the BamHI/ApaI fragment of pcDNA3 HA-hPMS1 into the pEGFP-C1 vector (Clontech). pEGFP-hPMS1 was digested with PflmI and religated to construct pEGFP-hPMSΔC, which contains amino acids 1 to 643 of hPMS1. pEGFP-hPMS2 was constructed by inserting the SalI/ApaI fragment of pCMV-SPORT6-hPMS2 (ATCC) into the pEGFP-C2 vector (Clontech). The pSUPER vector and its derivative pSUPER-siMLH1, expressing siRNA of hMLH1, were constructed according to the instructions of the manufacturer (7). The 19-nucleotide target sequence for siMLH1 is 5′-GGTTCACTACTAGTAAACT-3′; the target sequence for siPMS1 is 5′-GGAATCTACTCGTTTGTAT-3′.
Western blot analysis.
Equal amounts of protein lysates were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrotransferred to an Immobilon-P membrane (Millipore). Equal protein loading was confirmed with Ponceau-S staining. MLH1 antibody (G168-728) was purchased from BD Biosciences. The antibodies against hPMS1 (C-20), p53 (FL393), E2F1 (C20), PCNA (PC10), and green fluorescent protein (GFP) were obtained from Santa Cruz. The antibodies for PMS2 (Ab-1) and ATM (Ab-3) were purchased from Oncogene Research Products. The antibodies against phospho-Ser15 p53 and phospho-Ser20 p53 were obtained from Cell Signaling Technology.
Northern blot analysis.
Total RNA was extracted with TRIzol reagent (Gibco BRL) from H460 cells. Poly(A)+ mRNA was prepared with PolyTtract mRNA Isolation Systems III (Promega) following the manufacturer's instructions. A total of 3 μg of poly(A)+ mRNA was resolved on a 1% agarose-2% formaldehyde gel and transferred to membrane with a Nytran SuPerCharge kit (Schleicher and Schuell) following the manufacturer's instructions. The cDNAs of hMLH1, hPMS1, hPMS2, and GAPDH were labeled with [α-32P]dCTP by use of a Prime-It II Random primer labeling kit (Promega) and purified by Sephadex G-50 (Amersham) as the probes for Northern blotting.
Immunofluorescence studies.
HEK293, HCT116, or HFF cells were plated on collagen-coated coverslips in six-well plates. To visualize enhanced green fluorescent protein (EGFP)-PMS1 and HcRed1-MLH1, transfected cells were fixed in 3% paraformaldehyde for 20 min and the nuclei were stained with Hoechst 33258. For immunostaining, cells were fixed in 3% paraformaldehyde for 20 min followed by permeabilization in 0.5% Triton X-100 for 5 min. Cells were then blocked in 2% bovine serum albumin-phosphate-buffered saline at 4°C overnight and incubated with primary antibody in blocking solution for 1 h and fluorescein-conjugated AffiniPure donkey anti-mouse immunoglobulin G (IgG) (Jackson ImmunoResearch) or Texas Red-X goat anti-rabbit IgG (Molecular Probes) (1:500 dilution) for 1 h. MLH1 antibody G168-728 (1:100), PMS1 antibody (H-300; Santa Cruz) (1:100), and PMS2 antibody (AB-1) (1:100) were used for immunostaining. Images were captured on a Zeiss fluorescent microscope (Axioplan 2 imaging system).
RESULTS
DNA damage induces hPMS1, hPMS2, and hMLH1.
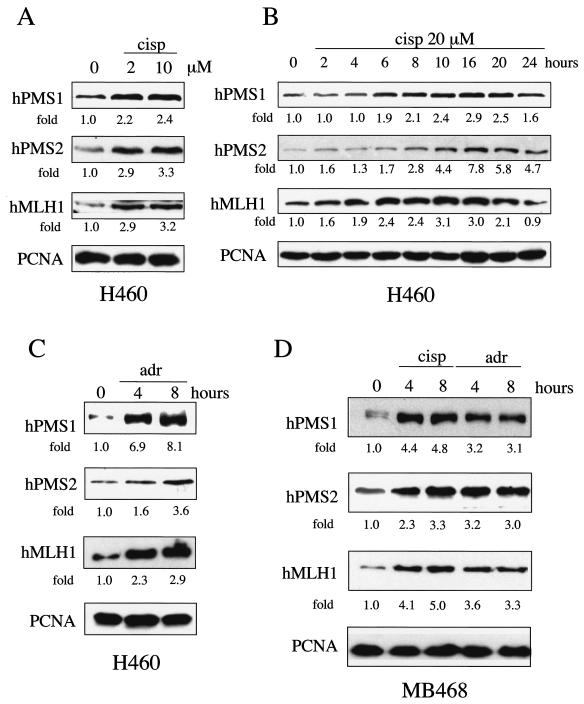
Several cell lines were treated with cisplatin or adriamycin, and the expression of hPMS1, hPMS2, and hMLH1 proteins was examined by Western blot analysis. We found that hPMS1, hPMS2, and hMLH1 proteins were induced in the H460 lung cancer cell line by cisplatin treatment in a dose-dependent (Fig. 1A) and time-dependent (Fig. 1B) manner. We also observed that not only cisplatin but also adriamycin induced the accumulation of hPMS1, hPMS2, and hMLH1 in H460 cells (Fig. 1C) and MDA-MB468 breast cancer cells (Fig. 1D). The hMutL proteins were also induced in other cell types such as human lymphoblastoid cell lines and fibroblasts upon treatment of DNA damage agents (see Fig. 3). Together, these data demonstrate that the induction of hMLH1, hPMS1, and hPMS2 proteins occurs in response to DNA damage and that the induction appears to be a general response to DNA damage.
FIG. 1.
Induction of hMLH1, hPMS1, and hPMS2 following DNA damage. (A) Western blot analysis of hMLH1, hPMS1, and hPMS2 in the H460 human lung cancer cell line following treatment with cisplatin (cisp) at 2 to 10 μM for 24 h. PCNA immunoblot analysis was performed to ensure equal levels of protein loading. (B) Western blot analysis of hMLH1, hPMS1, and hPMS2 in H460 cells following treatment with cisplatin (20 μM) for the indicated time periods. (C) Western blot analysis of hPMS1, hPMS2, and hMLH1 in H460 cells, which were treated with 500 nM adriamycin (adr) for 4 or 8 h. (D) Western blot analysis of hPMS1, hPMS2, and hMLH1 in the MDA-MB468 breast cancer cell line, following treatment with 50 μM cisplatin or 500 nM adriamycin for 4 or 8 h. The intensity of each immunoblot signal was quantified by densitometry. The severalfold change in induction relative to the results seen with untreated samples (0 μM or 0 h) is shown below each panel.
FIG. 3.
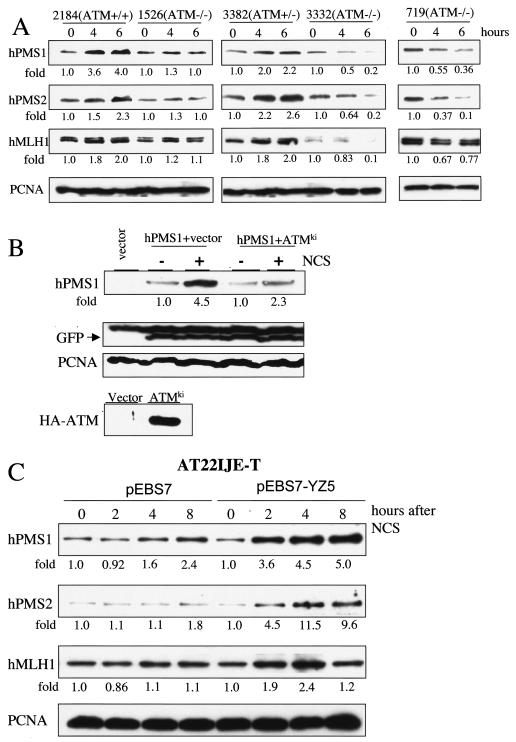
ATM mediates the induction of hMLH1, hPMS1, and hPMS2 accumulation in response to DNA damage. (A) Several human lymphoblastoid cell lines with the indicated ATM genotypes were treated with adriamycin (1 μM). hMLH1, hPMS1, and hPMS2 proteins were assayed by Western blot analysis. (B) Induction of hPMS1 in response to DNA damage requires ATM. 293T cells were transfected with pcDNA3- HA-hPMS1 and pGFP along with pBJ-1-HA-ATMki or an empty vector. The cells were then treated with NCS (300 ng/ml) for 5 h. Transfected hPMS1 was detected with an hPMS1-specific antibody. The expression of transfected GFP serves as a control of transfection efficiency. PCNA immunoblotting was performed to ensure equal levels of protein loading. Expression of transfected HA tagged-ATMki protein was confirmed by immunoprecipitation with HA beads followed by immunoblotting with ATM antibody (bottom panel). (C) ATM-deficient human AT22IJE-T fibroblasts, stably transfected with pEBS7 (an empty vector) or pEBS7-YZ5 (carrying FLAG-tagged wild-type ATM cDNA) (46), were treated with NCS (300 ng/ml). The expression of MutL proteins was detected by Western blot analysis. The intensity of each immunoblot signal was quantified by densitometry. The relative intensity compared to that of the untreated sample is shown below each panel.
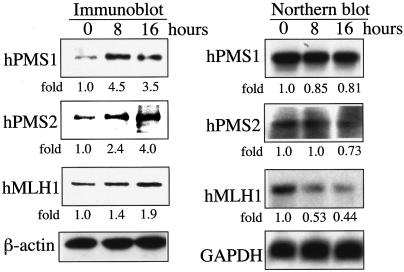
To further examine the mechanism of the induction, the levels of proteins and mRNA of hPMS1, hPMS2, and hMLH1 were measured in H460 cells after cisplatin treatment for 8 and 16 h. As shown in Fig. 2, while the hMutL proteins were induced by cisplatin treatment the level of hPMS1 mRNA did not significantly change. The levels of hPMS2 and hMLH1 mRNA indeed gradually decreased. Thus, cisplatin-induced accumulation of hMutL proteins is mediated by a posttranscriptional mechanism.
FIG. 2.
DNA damage-induced accumulation of hMLH1, hPMS1, and hPMS2 proteins is mediated through a posttranscriptional mechanism. Expression of the hMutL proteins (Immunoblot) and RNA (Northern blot) in H460 cells following treatment with cisplatin (20 μM) for 8 or 16 h is indicated. The intensity of each immunoblot signal was quantified by densitometry. The Northern blot was quantified by PhosphorImager (Typhoon 8600). The relative intensity value compared to that of the untreated sample is shown below each panel.
ATM mediates the accumulation of hPMS1, hPMS2, and hMLH1 in response to DNA damage.
Many lines of evidence have established the role of ATM/ATR in mediating the DNA damage response (see reviews in references 2, 25, and 36). ATM is directly and/or indirectly responsible for the stabilization of p53 (4, 10, 14, 23, 26), E2F1 (30), and BLM (1) during DNA damage. To explore the role of ATM in the induction of hMutL proteins, we examined the induction of hMutL proteins in a panel of lymphoid cell lines established from AT patients or normal subjects. As shown in Fig. 3A, adriamycin treatment induced the accumulation of hPMS1, hPMS2, and hMLH1 in cells harboring wild-type ATM (GM2184) as well as in cells that are heterozygous for the ATM mutation (GM3382). In contrast, the protein induction was not observed in the cell lines bearing an ATM homozygous mutation (GM1526, GM3332, and GM719). We also utilized a kinase-inactive ATM mutant (ATMki) as a dominant-negative inhibitor to block endogenous ATM activity in 293T cells (29, 30). Cells were transfected with HA-hPMS1 expressing plasmid alone or together with the ATMki mutant, treated with radiomimetic chemical neocarzinostatin (NCS), and then assayed for the hPMS1 expression. GFP was coexpressed and detected by immunoblotting to ensure equal levels of transfection efficiency. As shown in Fig. 3B, NCS induced the accumulation of hPMS1 and the induction was partially blocked (from 4.5- to 2.3-fold) by the ATMki mutant.
To further test the role of ATM in another cell type, the induction of hMutL proteins was examined in human AT fibroblasts and AT fibroblasts reconstituted with wild-type ATM (46). Whereas there was no induction of hMLH1and hPMS2 or minimal induction of hPMS1 in the AT fibroblasts upon NCS treatment, reconstitution of ATM significantly restored the induction of these proteins (Fig. 3C).
Taken together, these findings lead us to conclude that ATM mediates the induction of hMutL proteins in response to DNA damage.
Induction of hPMS1, hPMS2, and hMLH1 is mediated through protein stabilization.
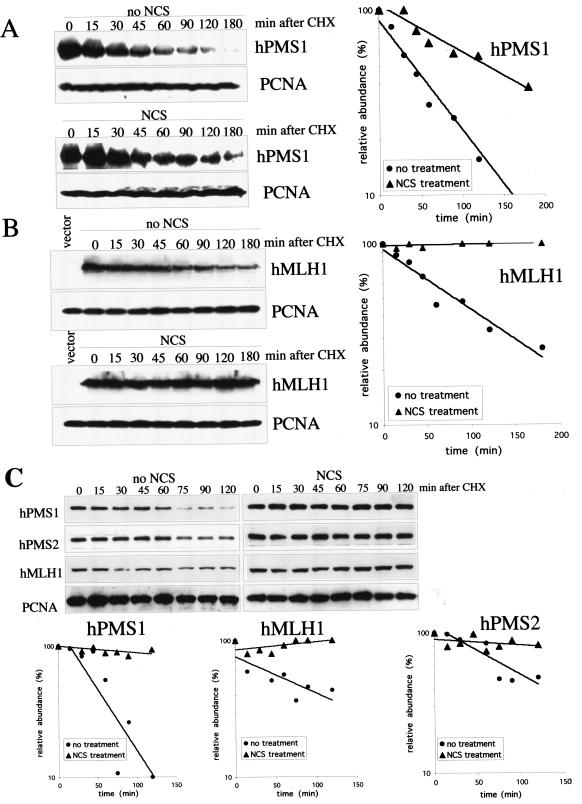
To test whether the ATM-dependent induction of hMutL proteins is due to protein stabilization, we measured the half-life of hPMS1 and hMLH1 proteins in 293T cells upon treatment with radiomimetic chemical NCS. Treatment of cells with NCS leads to double-strand DNA breaks and induction of ATM kinase activity (4). Indeed, many experiments suggest that this agent is a selective inducer of ATM. We expressed HA-tagged hPMS1 in 293T cells and examined the hPMS1 protein levels in the cells treated with cycloheximide in the absence or presence of NCS. The half-life of HA-hPMS1 protein was about 40 min in the absence of DNA damage and was extended to about 130 min upon NCS treatment (Fig. 4A). To measure the half-life of hMLH1 protein, HcRed1-tagged hMLH1 was expressed in 293T cells and its half-life was measured. The half-life of HcRed1-hMLH1 was about 80 min in the absence of NCS and was greatly prolonged by NCS treatment (Fig. 4B). We also measured the half-lives of endogenous proteins in HEK293 cells. The half-lives of endogenous hPMS1, hPMS2, and hMLH1 are about 45, 100, and 70 min, respectively (Fig. 4C). Upon NCS treatment, their half-lives were significantly prolonged (Fig. 4C). It is worth noting that the half-lives of endogenous proteins and overexpressed tagged proteins are very similar and that hPMS1 appears to have the shortest half-life among them. Taken together, these findings lead us to conclude that hPMS1, hPMS2, and hMLH1 proteins are stabilized by NCS treatment.
FIG. 4.
The half-lives of hMLH1, hPMS1, and hPMS2 are prolonged by NCS treatment. (A) HEK293T cells were transfected with pcDNA3- HA-hPMS1. Two days later, cells were either left untreated or treated with NCS (300 ng/ml) for 3 h followed by treatment with cycloheximide (CHX) at 20 μg/ml for the time periods as indicated. hPMS1 protein was detected by Western blot analysis using an antibody specific to hPMS1 (left panel) and quantified by densitometry (right panel). The relative abundance of hPMS1 following CHX treatment relative to that seen with no CHX treatment control was graphed (right panel). The PCNA immunoblot serves as a protein loading control. (B) HEK293T cells were transfected with pHcRed-hMLH1. The half-life of hMLH1 protein following CHX treatment was measured as described above. hMLH1 protein was detected by Western blot analysis with an hMLH1-specific antibody (left panels) and quantified by densitometry (right panel). (C) Growing HEK293 cells were either left untreated or treated with NCS (300 ng/ml) for 3 h followed by treatment with CHX (20 μg/ml) for the time periods as indicated. The endogenous hPMS1, hPMS2, and hMLH1 were detected by Western blot analysis (upper panels). The signals were quantified by densitometry, and the relative abundance of each protein following CHX treatment relative to that seen with the 0-min control was plotted (lower panels).
hMLH1 is required for full induction of hPMS1 and hPMS2 in response to DNA damage.
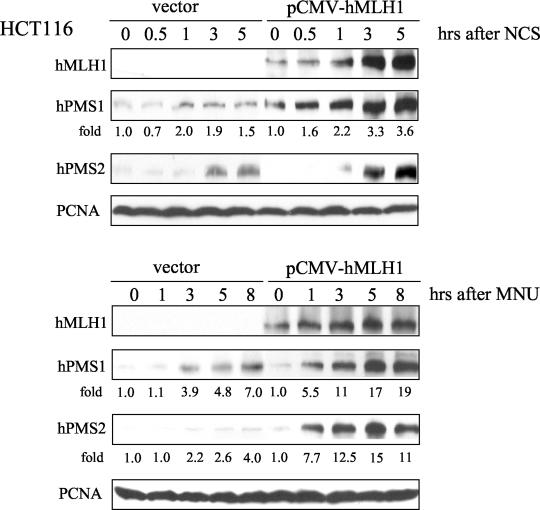
Previous studies have demonstrated a role of hMLH1 in maintaining normal levels of hPMS1 and hPMS2 proteins (8, 13, 28, 35). The hPMS1 and hPMS2 protein levels are significantly decreased in spite of normal levels of their RNA in the cells lacking hMLH1, suggesting that PMS proteins are unstable in the absence of hMLH1. Thus, we postulated that efficient induction of hPMS1 and hPMS2 might also require hMLH1. To test the role of hMLH1 in the stabilization of hPMS1 and hPMS2, we examined the induction of hPMS1 and hPMS2 in the hMLH1-deficient HCT116 colorectal tumor line and HCT116 cells reconstituted with hMLH1 following DNA damage. While there was induction of both hPMS1 and hPMS2 protein accumulation in HCT116 cells following treatment with NCS or N-methyl-N-nitrosourea (MNU), the induction was greatly enhanced by reconstitution with MLH1 (Fig. 5). This result is consistent with the notion that forming a complex with hMLH1 facilitates the protein stability of hPMS1 and hPMS2. While hMLH1 promoted efficient accumulation of hPMS proteins, induction of hPMS proteins still occurred to a lesser extent in the absence of hMLH1. Thus, it is likely that each component of the hMutL complexes is an independent target of ATM.
FIG. 5.
hMLH1 is required for full induction of hPMS1 and hPMS2 in response to DNA damage. hMLH1-deficient HCT 116 cells were transfected with pCMV-SPORT6-hMLH1 or an empty vector by a double-electroporation protocol (see Materials and Methods) and treated with NCS (80 ng/ml) (upper panels) or MNU (2 mM) (lower panels) for the indicated periods of time. Cell lysates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the immunoblots were probed with antibodies specific to hMLH1, hPMS1, and hPMS2. PCNA immunoblotting was performed to ensure equal levels of protein loading. The intensity of hPMS signals was quantified by densitometry. The relative intensity compared to that of the 0-h sample is shown below each panel.
hMLH1 mediates nuclear localization of hPMS1 and hPMS2.
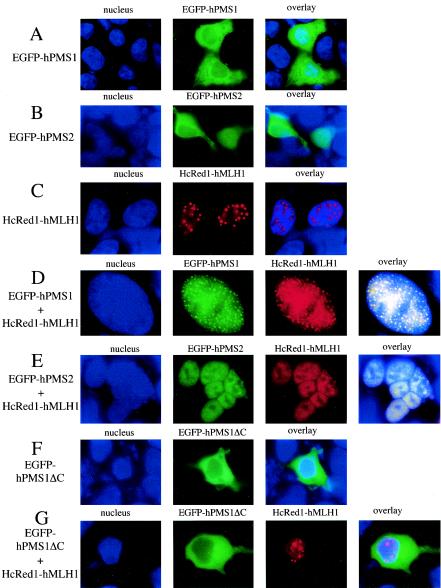
Nuclear compartmentalization is known to regulate the function of a number of nuclear factors such as p53, Cdc25, p21Cip1/WAF1, and adenomatous polyposis coli (11). For example, nuclear export of p53 is blocked in response to DNA damage (44). Both hMLH1 and hPMS2 possess nuclear localization signals and nuclear export sequences (41). To investigate whether the subcellular localization of MutL proteins is regulated during DNA damage, we examined the localization of EGFP-hPMS1, EGFP-hPMS2, and HcRed1-hMLH1 in HEK293 cells. First, we examined the subcellular localization of these proteins in growing HEK293 cells. EGFP-hPMS1 and hPMS2 proteins were predominantly localized in cytoplasm or distributed in the whole cells (Fig. 6A and B), and nuclear hPMS1 or hPMS2 was seen in only one-third of the population. In contrast, HcRed1-hMLH1 was localized exclusively in the nucleus and formed discrete nuclear punctates (Fig. 6C). When EGFP-hPMS1 was coexpressed with HcRed1-hMLH1, it was relocalized to nucleus and colocalized with HcRed1-hMLH1 in the nuclear foci (Fig. 6D). Coexpression with hMLH1 also resulted in the relocalization of hPMS2 to nucleus and the formation of a few nuclear foci (Fig. 6E). Interestingly, although both hPMS1 and hPMS2 were relocalized to nucleus by HcRed1-hMLH1 in HEK293 cells, the distribution of hPMS1 foci and hPMS2 foci was different. Coexpression of hPMS1 with hMLH1 greatly increased the number of hMLH1 foci, and nearly 50% of the cells contained numerous hPMS1-hMLH1 foci throughout the whole nucleus (Fig. 6D). In contrast, there were only a few hPMS2-hMLH1 foci seen in each nucleus of hMLH1- and hPMS2-coexpressed cells (Fig. 6E). The activity of hMLH1 to relocalize hPMS1 is specific, since HcRed1-MLH1 failed to relocalize a mutant hPMS1 (hPMS1ΔC) which lacks an hMLH1 interaction domain (Fig. 6F and G).
FIG. 6.
hMLH1 mediates nuclear localization of hPMS1 and hPMS2. HEK293 cells were transfected with expression plasmids as indicated on the left side of the figure. Cells were fixed, and nuclei were stained with Hoechst 33258. (A) When expressed alone, EGFP-hPMS1 was exclusively localized to the cytoplasm in 196 (49%) cells, distributed within the whole cells in 84 (21%) cells, and localized to the nucleus in 120 (30%) cells out of 400 transfected cells examined. (B) HEK293 cells were transfected with pEGFP-hPMS2. The distribution pattern showed that hPMS2 was located throughout the cells in 45%, was exclusively cytoplasmic in 23%, and was nuclear in 32% out of 400 GFP+ cells examined. (C) HcRed-hMLH1 was localized in the nucleus and formed discrete punctuates. The images reflect 396 (99%) of 400 transfected cell examined. Four cells exhibited diffuse nuclear distribution without the formation of foci. (D) Upon coexpression with hMLH1, EGFP-hPMS1 was translocated to nucleus and colocalized with HcRed1-hMLH1 in the nuclear foci in 400 (100%) out of 400 transfected cells examined. A total of 50% of cells contained numerous foci distributed throughout the nucleus as shown. The rest of the population exhibited a few nuclear foci (fewer than 20) in each nucleus. (E) HEK293 cells were cotransfected with pEGFP-hPMS2 and HcRed1-hMLH1. hPMS2 was exclusively localized to nucleus when it was coexpressed with HcRed1-hMLH1 in 400 (100%) out of 400 transfected cells examined. The images are representative of 99% of transfected cells. Only 1% of the cells contained more than 10 foci in the nucleus. (F) EGFP-hPMS1ΔC was exclusively localized in the cytoplasm. The images reflect 99% of 400 transfected cells examined. A total of 1% of the cells exhibited a whole-cell distribution pattern. (G) When coexpressed with hMLH1, EGFP-hPMS1ΔC remained in the cytoplasm. The images are representative of 99% of 400 transfected cells examined.
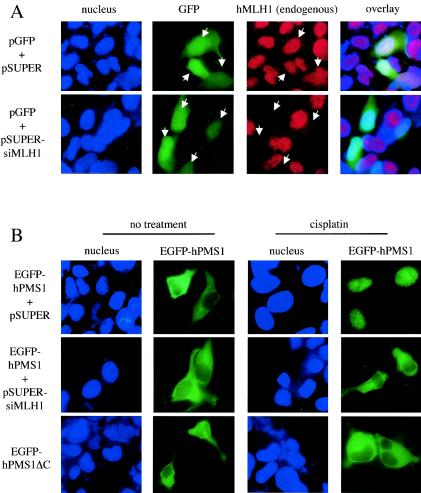
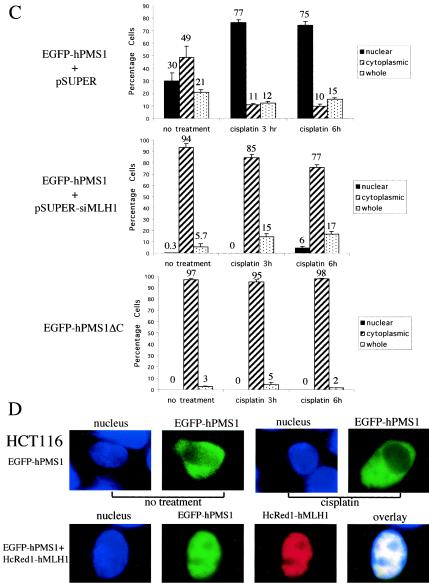
To address whether hMLH1 is required for the nuclear localization of hPMS1, we expressed hMLH1 siRNA in HEK293 cells by use of pSUPER vector (7) to inhibit the expression of hMLH1. HEK293 cells were transfected with pSUPER constructs and pGFP plasmid, and the transfected cells were identified using a fluorescent microscope to assess the expression of GFP. hMLH1 siRNA significantly knocked down the expression of endogenous hMLH1, as shown by dramatically reduced MLH1 immunofluorescent staining in the hMLH1 siRNA-transfected cells (Fig. 7A). We then coexpressed EGFP-hPMS1 with hMLH1 siRNA in HEK293 cells and treated the cells with cisplatin to examine the localization of hPMS1 during DNA damage (Fig. 7B and C). In growing HEK293 cells, the nuclear EGFP-hPMS1 pattern was seen in 30% of cells (no treatment; Fig. 7C, upper panel) but it was seen in only 0.3% of hMLH1 siRNA-transfected cells (no treatment; Fig. 7C, middle panel) and was not seen in the mutant hPMS1 lacking an hMLH1-interacting domain (hPMS1ΔC) (no treatment; Fig. 7C, lower panel). This result demonstrates that hMLH1 is required for the nuclear localization of hPMS1. Upon cisplatin treatment, EGFP-hPMS1 was shifted from being predominantly cytoplasmic or located throughout the whole cells (exclusively nuclear in only 30% of the cells) to being predominantly nuclear (in more than 75% of the cells) (Fig. 7C, upper panel) and formed a nuclear punctate pattern (Fig. 7B, upper panel). In sharp contrast, EGFP-hPMS1 remained predominantly cytoplasmic in the hMLH1 siRNA-transfected cells (in more than 75% of the cells) in spite of cisplatin treatment (Fig. 7B and C, middle panels). The mutant hPMS1 lacking an hMLH1-interacting domain (hPMS1ΔC) also remained in the cytoplasm (in 97% of the cells) during DNA damage (Fig. 7B and C, lower panels). To further verify the physiological role of hMLH1 in the translocation of hPMS1, we examined the localization of hPMS1 in hMLH1-deficient HCT116 cells. In contrast to the nuclear compartmentalization seen in HEK293 cells, EGFP-hPMS1 remained in the cytoplasm of HCT116 cells upon cisplatin treatment (Fig. 7D, upper panel). Reconstitution of hMLH1 relocalized EGFP-hPMS1 to the nucleus in HCT116 cells (Fig. 7D, lower panel). Thus, we conclude that hMLH1 is required for cisplatin-induced relocalization of hPMS1 to the nucleus.
FIG. 7.
hMLH1 is required for nuclear localization of hPMS1 during DNA damage. (A) HEK293 cells were transfected with pGFP and either pSUPER (upper panels) or pSUPER-siMLH1 (lower panels). The cells were fixed, and hMLH1 was immunostained with its specific antibody followed by Texas Red X-conjugated anti-mouse secondary antibody. Nuclei were stained with Hoechst 33258. The transfected cells (arrows) were identified by the expression of GFP. Transfection of pSUPER empty vector did not alter the level of hMLH1 expression, but transfection of pSUPER-siMLH1 knocked down the expression of endogenous hMLH1. (B) HEK293 cells transfected with pEGFP-hPMS1 and either pSUPER (upper panels) or pSUPER-siMLH1 (middle panels) or transfected with pEGFP-hPMS1ΔC (lower panels) were treated with cisplatin (50 μM) for 3 h. Cells were fixed, and nuclei were stained with Hoechst 33258. Most EGFP-hPMS1 translocated from cytoplasm to nucleus and formed a nuclear punctate pattern upon cisplatin treatment (upper panels). MLH1 siRNA prevented the cisplatin-induced relocalization of EGFP-hPMS1 (middle panels). The hPMS1 mutant (hPMS1ΔC), lacking an hMLH1-interacting domain, remained in the cytoplasm following cisplatin treatment (lower panels). (C) GFP-positive cells of each sample in the experiments described for panel B were scored for the localization of EGFP-hPMS1 or EGFP-hPMS1ΔC, and the percentages of cells displaying exclusively nuclear, exclusively cytoplasmic, or both nuclear and cytoplasmic (whole) staining were determined. At least 300 GFP-positive cells were scored for each experiment. The data shown represent the means ± standard errors of three independent experiments. (D) HCT116 cells were transfected with the expression plasmids as indicated on the left. The cells were either left untreated or treated with cisplatin (50 μM) for 6 h. The cells were then fixed, and nuclei were stained with Hoechst 33258. EGFP-hPMS1 remained in the cytoplasm of hMLH1-deficient cells following cisplatin treatment but was relocalized to nucleus in hMLH1-reconstituted HCT116 cells. The images reflect 100% (upper panels) or 90% (lower panels) of 200 transfected cells examined. In hMLH1-reconstituted HCT116 cells, the distribution of hPMS1 was as follows: 90% exclusively nuclear, 4% exclusively cytoplasmic, and 6% in the whole cell.
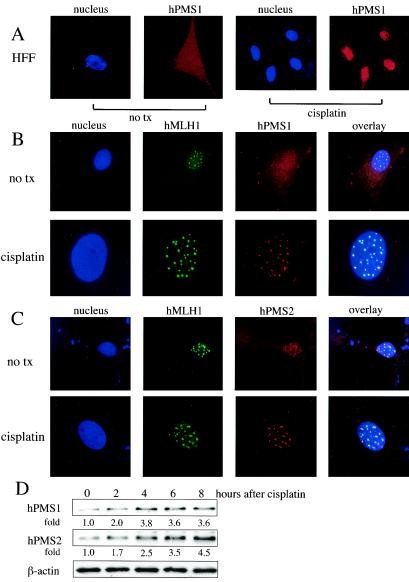
To examine the localization of endogenous hMutL proteins in primary nontransformed human cells, we performed immunofluorescent studies on primary HFFs. hPMS1 and hPMS2 were distributed throughout the whole cells in growing HFFs, whereas hMLH1 was localized in the nucleus and formed a nuclear punctate pattern (Fig. 8). The localization of hMLH1 in the nuclear foci was also reported in WI-38 cells (32), a human lung fibroblast cell line, and is consistent with the observation of HcRed1-hMLH1 foci in HEK293 cells (Fig. 6). Note the presence of hPMS2 in the hMLH1 foci in the HFF without cisplatin treatment (Fig. 8C). In contrast, there is much less hPMS1 (although it is still detectable) in the hMLH1 foci in cells without cisplatin treatment (Fig. 8B). However, both hPMS1 and hPMS2 were predominantly localized to nucleus and colocalized with hMLH1 foci upon cisplatin treatment (Fig. 8B and C). The disappearance of cytoplasmic hPMS proteins could in theory be due to either nuclear sequestration or degradation of cytoplasmic hPMS1/2. To rule out the possibility of protein degradation, we measured the hPMS protein levels in HFF following cisplatin treatment. The protein levels of both hPMS1 and hPMS2 increased upon cisplatin treatment (Fig. 8D). Thus, we conclude that hPMS1 and hPMS2 are compartmentalized within nucleus upon cisplatin treatment.
FIG. 8.
Localization of endogenous hPMS1, hPMS2, and hMLH1 proteins in primary HFFs. (A) Endogenous hPMS1 was distributed throughout the cells but was relocalized to nucleus by cisplatin treatment in primary HFFs. HFFs were either left untreated or treated with cisplatin (50 μM) for 6 h. hPMS1 was immunostained with its specific antibody followed by Texas Red X-conjugated anti-rabbit IgG. Nuclei were stained with Hoechst 33258. Images are representative of 99% of 300 cells examined. (B) Endogenous hMLH1 formed discrete nuclear foci and colocalized with nuclear hPMS1 in these foci. HFF were treated with cisplatin as described for panel A. The immunostaining was performed with specific antibodies against hMLH1 (monoclonal) and hPMS1 (polyclonal) followed by fluorescein isothiocyanate-conjugated anti-mouse IgG and Texas Red X-conjugated anti-rabbit IgG. Nuclei were stained with Hoechst 33258. Images are representative of at least 95% of 300 cells examined. no tx, no cisplatin treatment. (C) Endogenous hPMS2 is distributed throughout the cells but is relocalized to nuclei by cisplatin treatment and colocalized with hMLH1. HFF were treated and the immunostaining was performed as described for panel B except that an antibody for hPMS2 (polyclonal) was used to detect hPMS2. The images in the upper panels represent 89% of 300 cells examined. The other 11% of cells showed a whole-cell staining pattern of hPMS2 without nuclear foci. The images in the lower panels are representative of 93% of 300 cells examined (7% of cells had a nuclear hPMS2 pattern without foci). (D) HFFs were treated with cisplatin (50 μM) for the indicated periods of time. Total cellular lysates were subjected to Western blot analysis to detect endogenous hPMS1 and hPMS2. The relative intensity of the signals was quantified by densitometry and compared to that seen with the 0-h sample.
Together, these data indicate that hPMS1 and hPMS2 are relocalized to nucleus upon DNA damage and that their nuclear localization depends on hMLH1.
A role of hMLH1 and hPMS1 for p53 activation by DNA damage.
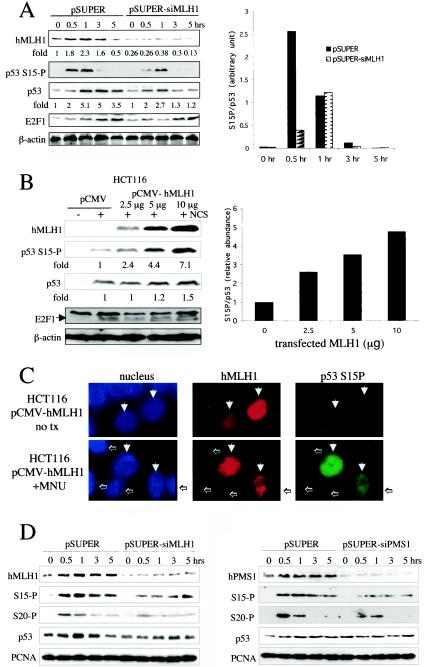
A role for hMLH1 for the p53 phosphorylation in response to DNA alkylation (15, 43) or UV (33) has been reported. The ATM-dependent induction of MutL proteins suggests that the increased MutL proteins may augment the p53 response, and the levels of MutL proteins may be important for deciphering the DNA damage response. In fact, activation of p53 by treatment of DNA-methylating agents requires a full complement of hMLH1 protein whereas MMR efficiency is fairly normal even at low levels of hMLH1 (12). To test that, we knocked down the levels of hMLH1 in MCF7 cells by use of hMLH1 siRNA. The DNA damage response in these cells was then evaluated. hMLH1 siRNA effectively decreased the expression of hMLH1 to about 25% of the endogenous level (Fig. 9A). The induction of p53 protein levels by NCS was affected by the presence of hMLH1 siRNA (levels of induction changed from 5.1-fold induction in pSUPER-transfected MCF7 cells to 2.7-fold induction in MLH1 siRNA-transfected cells at 1 h after NCS treatment). The Ser 15 phosphorylation of p53 was affected by hMLH1 at a much earlier time point (0.5 h) and more significantly. The ratio between Ser 15 phosphorylation and total p53 levels was measured, and the result clearly indicates that hMLH1 siRNA inhibits the phosphorylation of p53 at the Ser 15 residue during NCS treatment (Fig. 9A, right panel). The induction of E2F1, another DNA damage response protein, was not affected by the presence of hMLH1 siRNA. These results demonstrate that a full level of hMLH1 is required for effective activation of p53 during DNA damage.
FIG.9.
A role of hMLH1 and hPMS1 for p53 activation by DNA damage. (A) Inhibition of hMLH1 expression by siRNA blocked the induction of p53 but not of E2F1 in response to DNA damage. MCF-7 cells, stably transfected with hMLH1 siRNA or an empty vector, were treated with NCS (300 ng/ml) for 0.5 to 5 h. Western blot analysis was performed using antibodies as indicated (left panels). The intensity of the signals was quantified by densitometry. The relative levels of hMLH1 in pSUPER-siMLH1-transfected cells were compared to those in the untreated pSUPER-transfected cells. The ratio between the intensity reflecting Ser 15 phosphorylation and that of total p53 is shown in the right panel. (B) The extent of DNA damage-induced accumulation of p53, but not of E2F1, depends on the levels of hMLH1 protein. HCT116 cells were transfected with pCMV or increasing amounts of pCMV-SPORT6-hMLH1 as indicated on the top of the panels and treated with NCS (80 ng/ml) for 3 h (left panel). Cell lysates were subjected to Western blot analysis, and the intensity of the signals was quantified by densitometry. The ratio between Ser 15 phosphorylation and total p53 levels is shown in the right panels. The same results were also seen in the cells treated with NCS for 0.5 and 1 h. (C) HCT116 cells were transiently transfected with pCMV-SPORT6-hMLH1 and then left untreated (upper panels) or treated with MNU (lower panels) as described for panel B. Cells were fixed, and the immunostaining was performed with antibodies against hMLH1 (monoclonal) and phospho-Ser 15 p53 (polyclonal) followed by Texas Red X-conjugated anti-mouse IgG and fluorescein isothiocyanate-conjugated anti-rabbit IgG. Nuclei were stained with Hoechst 33258. The transfected cells expressing various levels of hMLH1 are indicated (↓). Cells which did not express hMLH1 are indicated (←). no tx, no MNU treatment. (D) HEK293 cells were transiently transfected with pSUPER, pSUPER-siMLH1, or pSUPER-siPMS1 and then treated with NCS (300 ng/ml) for 0.5 to 5 h. Western blot analysis was performed using antibodies as indicated.
Next, we examined DNA damage response in hMLH1-deficient HCT116 cells and HCT116 cells reconstituted with hMLH1. Although accumulation and phosphorylation of p53 in response to NCS (Fig. 9B) and MNU (data not shown) were seen in empty vector-transfected HCT116 cells (pCMV lanes), reexpression of hMLH1 significantly augmented the p53 phosphorylation and, to a lesser degree, p53 protein accumulation. The ratio of Ser 15 phosphorylation to total p53 correlated with the amount of hMLH1 reconstituted (Fig. 9B, right panel). Consistent with the results obtained in the siRNA experiments, the induction of E2F1 was not altered by hMLH1 expression. The effect of hMLH1 on the accumulation of Ser 15-phosphorylated p53 was also assessed by immunofluorescent microscopy. Reexpression of hMLH1 in HCT116 cells did not induce phosphorylation of p53 in the absence of DNA damage treatment (Fig. 9C, upper panel). Upon MNU treatment, the cells expressing hMLH1 (Fig. 9C, lower panels) contained a significant accumulation of Ser 15-phosphorylated p53 whereas the cells not expressing hMLH1 (Fig. 9C, lower panel) failed to accumulate Ser 15-phosphorylated p53. Similar experiments were also carried out using NCS, and hMLH1 also enhanced NCS-induced accumulation of Ser 15-phosphorylated p53, as observed in MNU treatment (data not shown). Taken together, these findings show that hMLH1 augments p53 phosphorylation during DNA damage.
To test whether hPMS1 is involved in the p53 phosphorylation, we examined the p53 response in hPMS1 siRNA-transfected or hMLH1 siRNA-transfected HEK293 cells. Both hPMS1 siRNA and hMLH1 siRNA knocked down the expression of their endogenous proteins very effectively (Fig. 9D). The NCS-induced phosphorylation of p53 at Ser 15 and Ser 20 was significantly inhibited or delayed by hMLH1 siRNA or by hPMS1 siRNA (Fig. 9D). Compared with the results seen with empty vector-transfected cells, hMLH1 siRNA reduced the Ser 15 phosphorylation (measured by the ratio between Ser 15 phosphorylation and total p53 levels) to 0.35-fold or 0.39-fold in the cells treated with NCS for 0.5 or 1 h, respectively. hPMS1 siRNA reduced the Ser 15 phosphorylation to 0.24-fold or 0.49-fold in the cells treated with NCS for 0.5 or 1 h. hMLH1 and hPMS1 siRNA inhibited the Ser 20 phosphorylation to 0.5-fold and 0.25-fold, respectively, in the cells treated with NCS for 0.5 h. Consistent with the results obtained in MCF7 cells, the induction of E2F1 was not affected by these siRNAs in HEK293 cells (data not shown). This result provides the first evidence for a role of the hMLH1-hPMS1 heterodimer in the activation of p53 during NCS treatment.
DISCUSSION
ATM mediates the protein stabilization of hMLH1, hPMS1, and hPMS2.
The induction of hMutL proteins was observed in multiple cell lines in response to several DNA-damaging agents, including cisplatin, adriamycin, MNU, and radiomimetic chemicals, indicating that stabilization of hMutL proteins is a general response to DNA damage. By employing AT cell lines and ATM dominant-negative mutant or ATM-reconstituted cells, we demonstrated that ATM is required for the induction of MutL proteins. ATM is a serine/threonine protein kinase that mediates the activation of multiple signaling transduction pathways following the induction of DNA double-strand breaks (2, 25, 36) and DNA-alkylating agents (3). ATM has been previously shown to be involved in the stabilization of several DNA damage-responsive proteins, including p53 (4, 10, 26), E2F1 (30), and BLM (1). Thus, the ATM-mediated protein stabilization appears to be a general mechanism by which certain proteins are induced by DNA damage.
The stabilization of hPMS1 and hPMS2 proteins also requires hMLH1. Nevertheless, induction of hPMS1 and hPMS2 can still be seen in the absence of hMLH1 but to a much less extent compared to the induction in the presence of hMLH1. This result is consistent with previous observations that the complex formation with hMLH1 promotes the stability of hPMS1 and hPMS2 (8, 13, 28, 35).
Subcellular localization of hPMS1 and hPMS2.
Besides protein stabilization, our studies also demonstrate a regulation at the localization of PMS proteins and a critical role of hMLH1 for their nuclear compartmentalization in response to DNA damage. Regulation of nuclear factors by both protein stability and nuclear localization has been shown in other proteins. For example, protein degradation and nuclear export of p53 (44) are both blocked by DNA damage. Through these two mechanisms, the intranuclear levels of hMLH1-hPMS1 and hMLH1-hPMS2 are induced by DNA damage. Interestingly, these two complexes form nuclear foci with different distribution characteristics. In HEK293 cells, coexpression of hMLH1 and hPMS1 relocalized hPMS1 to the nucleus and formed numerous hMLH1-hPMS1 foci throughout the nucleus; in contrast, only a few hMLH1-hPMS2 foci are apparent in each nucleus upon coexpression of hMLH1 and hPMS2. In HFFs, the endogenous proteins of both hPMS1 and hPMS2 colocalized with hMLH1 foci; however, hPMS2 foci were more apparent than hPMS1 foci in the growing HFFs. The significance of the difference between hPMS1 and hPMS2 foci in HEK293 cells and HFFs would require future investigation. It does raise a possibility of distinct functions for hMLH1-hPMS1 and hMLH1-hPMS2 complexes.
A role of hMLH1 and hPMS1 in the p53 phosphorylation.
ATM phosphorylates p53 at Ser 15 (4, 10), and phosphorylation of Ser 15 stimulates p53 transactivation and its apoptotic activity (16). Prior studies have demonstrated a role of hMLH1 for the p53 phosphorylation in response to DNA alkylation (15, 43) or UV (33). However, another study showed that MMR-deficient HCT116 cells still displayed rapid phosphorylation of p53 at Ser 15 after DNA alkylation (3). Thus, the exact role of hMLH1 for the p53 activation has not been completely settled. Our results indicate that although hMLH1 is not essential for p53 phosphorylation, it is required to augment the phosphorylation of p53 by ATM at Ser 15 residue. This conclusion is based on the following observations: (i) phosphorylation of p53 can be seen in hMLH1-deficient HCT116 cells in response to ionizing radiation (9, 24), alkylation agents (3), or NCS (Fig. 9B); (ii) reexpression of hMLH1 alone without exposure of DNA damage agents in HCT116 cells does not lead to phosphorylation of p53 (Fig. 9C) (15, 43); (iii) complementation of hMLH1 augments the p53 phosphorylation during MNU treatment (Fig. 9C) (43) and NCS treatment (Fig. 9B); and (iv) hMLH1 siRNA inhibits and delays p53 phosphorylation induced by DNA damage.
Thus, MMR proteins do not appear to be the “on-off switch” for controlling DNA damage signaling but rather act like a “volume control knob” to adjust the amplitude of p53 response. The up-regulation of the hMutL complexes would be important to control the cellular response to genotoxic stress. hMutL proteins, which are induced through ATM, in turn facilitate the activation of p53. Thus, ATM and MutL complexes may form a positive feedback loop once DNA is significantly injured to amplify the DNA damage-signaling response. Such positive-feedback loops often lead to irreversible biochemical responses (e.g., cell fate decision) due to positive-feedback-based bistable kinetics (42). Thus, a positive-feedback loop between ATM and MMR could be ignited by prolonged treatment of DNA damage agents and therefore lead to accentuated p53 induction and, eventually, apoptosis. hMLH1 interacts with ATM in a large complex (6, 40). MSH2 and p53 were found to colocalize in the nuclear foci containing recombinative repair complexes and coimmunoprecipitate within the same DNA-protein complexes (45). Thus, the MMR complex may bring ATM and p53 within the proximity of DNA damage sites and augment phosphorylation of p53 by ATM. This scaffold model has been proposed for the MMR-dependent mechanism controlling Chk2 activation (6).
The hMLH1-hPMS1 heterodimer has no MMR activity in an in vitro MMR assay (35). The function of this complex has not been delineated. Mutations in the yeast gene mlh2 (human PMS1 homologue) lead to resistance to cisplatin, carboplatin, and doxorubicin (17), suggesting a role of mlh2 in the DNA damage response. Our data indicate that hPMS1 is a DNA damage-responsive protein and demonstrate its role in facilitating p53 phosphorylation during NCS treatment. Interestingly, hPMS2 is required for the stimulation of p73 apoptosis function by cisplatin (37). Future study would be focused on defining the role of each individual hMutL complex regarding its function in the DNA damage apoptosis response.
Intriguingly, the induction of E2F1 by DNA damage is not regulated by hMLH1. The accumulation of E2F1 and p53 was often concurrently induced upon treatment with various DNA damage agents; therefore, it was suggested that the same mechanism is responsible for stabilization of both p53 and E2F1 (5). Although both p53 and E2F1 are induced through ATM/Chk2 (4, 10, 14, 30, 38), our data clearly demonstrate that p53 and E2F1 are induced by distinct mechanisms; one is regulated by the levels of MMR proteins, whereas the other is not. This observation suggests the presence of multiple ATM-containing signaling complexes for different targets.
Acknowledgments
We thank Bert Vogelstein for providing pBS-hPMS1 and William A. May for pSUPER plasmid. We thank Yosef Shiloh for ATM-reconstituted AT22IJE-T cells.
The work was supported by a General Motors Cancer Research Scholar Award (W.-C.L.), National Institutes of Health-National Cancer Institute grant K12 CA 7693705 (W.-C.L.), a UAB Avon/Breast Cancer SPORE Career Development Award (W.-C.L. and F.-T.L.), an HHMI Faculty Development Award (W.-C.L. and F.-T.L.), and grant CA100848 (F.-T.L.) from the National Institutes of Health.
All authors disclose that they have no financial interests that will pose a conflict of interest regarding this article.
REFERENCES
- 1.Ababou, M., S. Dutertre, Y. Lecluse, R. Onclercq, B. Chatton, and M. Amor-Gueret. 2000. ATM-dependent phosphorylation and accumulation of endogenous BLM protein in response to ionizing radiation. Oncogene 19:5955-5963. [DOI] [PubMed] [Google Scholar]
- 2.Abraham, R. T. 2003. Checkpoint signaling: epigenetic events sound the DNA strand-breaks alarm to the ATM protein kinase. BioEssays 25:627-630. [DOI] [PubMed] [Google Scholar]
- 3.Adamson, A. W., W. J. Kim, S. Shangary, R. Baskaran, and K. D. Brown. 2002. ATM is activated in response to N-methyl-N′-nitro-N-nitrosoguanidine-induced DNA alkylation. J. Biol. Chem. 277:38222-38229. [DOI] [PubMed] [Google Scholar]
- 4.Banin, S., L. Moyal, S. Shieh, Y. Taya, C. W. Anderson, L. Chessa, N. I. Smorodinsky, C. Prives, Y. Reiss, Y. Shiloh, and Y. Ziv. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281:1674-1677. [DOI] [PubMed] [Google Scholar]
- 5.Blattner, C., A. Sparks, and D. Lane. 1999. Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Mol. Cell. Biol. 19:3704-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, K. D., A. Rathi, R. Kamath, D. I. Beardsley, Q. Zhan, J. L. Mannino, and R. Baskaran. 2003. The mismatch repair system is required for S-phase checkpoint activation. Nat. Genet. 33:80-84. [DOI] [PubMed] [Google Scholar]
- 7.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 8.Buermeyer, A. B., C. Wilson-Van Patten, S. M. Baker, and R. M. Liskay. 1999. The human MLH1 cDNA complements DNA mismatch repair defects in Mlh1-deficient mouse embryonic fibroblasts. Cancer Res. 59:538-541. [PubMed] [Google Scholar]
- 9.Bunz, F., A. Dutriaux, C. Lengauer, T. Waldman, S. Zhou, J. P. Brown, J. M. Sedivy, K. W. Kinzler, and B. Vogelstein. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282:1497-1501. [DOI] [PubMed] [Google Scholar]
- 10.Canman, C. E., D. S. Lim, K. A. Cimprich, Y. Taya, K. Tamai, K. Sakaguchi, E. Appella, M. B. Kastan, and J. D. Siliciano. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281:1677-1679. [DOI] [PubMed] [Google Scholar]
- 11.Carmo-Fonseca, M. 2002. The contribution of nuclear compartmentalization to gene regulation. Cell 108:513-521. [DOI] [PubMed] [Google Scholar]
- 12.Cejka, P., L. Stojic, N. Mojas, A. M. Russell, K. Heinimann, E. Cannavo, M. di Pietro, G. Marra, and J. Jiricny. 2003. Methylation-induced G2/M arrest requires a full complement of the mismatch repair protein hMLH1. EMBO J. 22:2245-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang, D. K., L. Ricciardiello, A. Goel, C. L. Chang, and C. R. Boland. 2000. Steady-state regulation of the human DNA mismatch repair system. J. Biol. Chem. 275:29178. [PubMed] [Google Scholar]
- 14.Chehab, N. H., A. Malikzay, M. Appel, and T. D. Halazonetis. 2000. Chk2/hCds1 functions as a DNA damage checkpoint in G1 by stabilizing p53. Genes Dev. 14:278-288. [PMC free article] [PubMed] [Google Scholar]
- 15.Duckett, D. R., S. M. Bronstein, Y. Taya, and P. Modrich. 1999. hMutSα- and hMutLα-dependent phosphorylation of p53 in response to DNA methylator damage. Proc. Natl. Acad. Sci. USA 96:12384-12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dumaz, N., and D. W. Meek. 1999. Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J. 18:7002-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durant, S. T., M. M. Morris, M. Illand, H. J. McKay, C. McCormick, G. L. Hirst, R. H. Borts, and R. Brown. 1999. Dependence on RAD52 and RAD1 for anticancer drug resistance mediated by inactivation of mismatch repair genes. Curr. Biol. 9:51-54. [DOI] [PubMed] [Google Scholar]
- 18.Fishel, R. 1999. Signaling mismatch repair in cancer. Nat. Med. 5:1239-1241. [DOI] [PubMed] [Google Scholar]
- 19.Flores-Rozas, H., and R. D. Kolodner. 1998. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc. Natl. Acad. Sci. USA 95:12404-12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong, J. G., A. Costanzo, H. Q. Yang, G. Melino, W. G. Kaelin, Jr., M. Levrero, and J. Y. Wang. 1999. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 399:806-809. [DOI] [PubMed] [Google Scholar]
- 21.Gradia, S., D. Subramanian, T. Wilson, S. Acharya, A. Makhov, J. Griffith, and R. Fishel. 1999. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol. Cell 3:255-261. [DOI] [PubMed] [Google Scholar]
- 22.Harfe, B. D., B. K. Minesinger, and S. Jinks-Robertson. 2000. Discrete in vivo roles for the MutL homologs Mlh2p and Mlh3p in the removal of frameshift intermediates in budding yeast. Curr. Biol. 10:145-148. [DOI] [PubMed] [Google Scholar]
- 23.Hirao, A., Y. Y. Kong, S. Matsuoka, A. Wakeham, J. Ruland, H. Yoshida, D. Liu, S. J. Elledge, and T. W. Mak. 2000. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 287:1824-1827. [DOI] [PubMed] [Google Scholar]
- 24.Jallepalli, P. V., C. Lengauer, B. Vogelstein, and F. Bunz. 2003. The Chk2 tumor suppressor is not required for p53 responses in human cancer cells. J. Biol. Chem. 278:20475-20479. [DOI] [PubMed] [Google Scholar]
- 25.Kastan, M. B., D. S. Lim, S. T. Kim, and D. Yang. 2001. ATM—a key determinant of multiple cellular responses to irradiation. Acta Oncol. 40:686-688. [DOI] [PubMed] [Google Scholar]
- 26.Keramaris, E., A. Hirao, R. S. Slack, T. W. Mak, and D. S. Park. 2003. Ataxia telangiectasia-mutated protein can regulate p53 and neuronal death independent of Chk2 in response to DNA damage. J. Biol. Chem. 278:37782-37789. [Online.] [DOI] [PubMed] [Google Scholar]
- 27.Kolodner, R. D., and G. T. Marsischky. 1999. Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev. 9:89-96. [DOI] [PubMed] [Google Scholar]
- 28.Leung, W. K., J. J. Kim, L. Wu, J. L. Sepulveda, and A. R. Sepulveda. 2000. Identification of a second MutL DNA mismatch repair complex (hPMS1 and hMLH1) in human epithelial cells. J. Biol. Chem. 275:15728-15732. [DOI] [PubMed] [Google Scholar]
- 29.Lim, D. S., S. T. Kim, B. Xu, R. S. Maser, J. Lin, J. H. Petrini, and M. B. Kastan. 2000. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 404:613-617. [DOI] [PubMed] [Google Scholar]
- 30.Lin, W. C., F. T. Lin, and J. R. Nevins. 2001. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 15:1833-1844. [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, T., H. Yan, S. Kuismanen, A. Percesepe, M. L. Bisgaard, M. Pedroni, P. Benatti, K. W. Kinzler, B. Vogelstein, M. Ponz de Leon, P. Peltomaki, and A. Lindblom. 2001. The role of hPMS1 and hPMS2 in predisposing to colorectal cancer. Cancer Res. 61:7798-7802. [PubMed] [Google Scholar]
- 32.Pedrazzi, G., C. Perrera, H. Blaser, P. Kuster, G. Marra, S. L. Davies, G. H. Ryu, R. Freire, I. D. Hickson, J. Jiricny, and I. Stagljar. 2001. Direct association of Bloom's syndrome gene product with the human mismatch repair protein MLH1. Nucleic Acids Res. 29:4378-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peters, A. C., L. C. Young, T. Maeda, V. A. Tron, and S. E. Andrew. 2003. Mammalian DNA mismatch repair protects cells from UVB-induced DNA damage by facilitating apoptosis and p53 activation. DNA Repair (Amsterdam) 2:427-435. [DOI] [PubMed] [Google Scholar]
- 34.Prolla, T. A., S. M. Baker, A. C. Harris, J. L. Tsao, X. Yao, C. E. Bronner, B. Zheng, M. Gordon, J. Reneker, N. Arnheim, D. Shibata, A. Bradley, and R. M. Liskay. 1998. Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nat. Genet. 18:276-279. [DOI] [PubMed] [Google Scholar]
- 35.Raschle, M., G. Marra, M. Nystrom-Lahti, P. Schar, and J. Jiricny. 1999. Identification of hMutLβ, a heterodimer of hMLH1 and hPMS1. J. Biol. Chem. 274:32368-32375. [DOI] [PubMed] [Google Scholar]
- 36.Shiloh, Y. 2003. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer 3:155-168. [DOI] [PubMed] [Google Scholar]
- 37.Shimodaira, H., A. Yoshioka-Yamashita, R. D. Kolodner, and J. Y. Wang. 2003. Interaction of mismatch repair protein PMS2 and the p53-related transcription factor p73 in apoptosis response to cisplatin. Proc. Natl. Acad. Sci. USA 100:2420-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevens, C., L. Smith, and N. B. La Thangue. 2003. Chk2 activates E2F-1 in response to DNA damage. Nat. Cell Biol. 5:401-409. [DOI] [PubMed] [Google Scholar]
- 39.Wang, T. F., N. Kleckner, and N. Hunter. 1999. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc. Natl. Acad. Sci. USA 96:13914-13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, Y., D. Cortez, P. Yazdi, N. Neff, S. J. Elledge, and J. Qin. 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14:927-939. [PMC free article] [PubMed] [Google Scholar]
- 41.Wu, X., J. L. Platt, and M. Cascalho. 2003. Dimerization of MLH1 and PMS2 limits nuclear localization of MutLα. Mol. Cell. Biol. 23:3320-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiong, W., and J. E. Ferrell, Jr. 2003. A positive-feedback-based bistable ′memory module' that governs a cell fate decision. Nature 426:460-465. [DOI] [PubMed] [Google Scholar]
- 43.Yanamadala, S., and M. Ljungman. 2003. Potential role of MLH1 in the induction of p53 and apoptosis by blocking transcription on damaged DNA templates. Mol. Cancer Res. 1:747-754. [PubMed] [Google Scholar]
- 44.Zhang, Y., and Y. Xiong. 2001. A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science 292:1910-1915. [DOI] [PubMed] [Google Scholar]
- 45.Zink, D., C. Mayr, C. Janz, and L. Wiesmuller. 2002. Association of p53 and MSH2 with recombinative repair complexes during S phase. Oncogene 21:4788-4800. [DOI] [PubMed] [Google Scholar]
- 46.Ziv, Y., A. Bar-Shira, I. Pecker, P. Russell, T. J. Jorgensen, I. Tsarfati, and Y. Shiloh. 1997. Recombinant ATM protein complements the cellular A-T phenotype. Oncogene 15:159-167. [DOI] [PubMed] [Google Scholar]