Abstract
Cotinine is a proxy for secondhand smoke (SHS) exposure. Genetic variation along nicotine and cotinine metabolic pathways may alter the internal cotinine dose, leading to misinterpretations of exposure-health outcome associations. Caucasian children with available SHS exposure and hair cotinine data were genotyped for metabolism-related genes and. SHS-exposed children had 2.4-fold higher hair cotinine (0.14ng/mg±0.22) than unexposed children (0.06ng/mg±0.05, p<0.001). SHS-exposed children carrying the NAT1 minor allele had 2-fold higher hair cotinine (0.18ng/mg for heterozygotes and 0.17ng/mg for homozygotes) compared to major allele homozygotes (0.09ng/mg, p=0.0009), even after adjustment for SHS dose. These findings support that NAT1 has a role in the metabolic pathway of nicotine/cotinine and/or their metabolites. The increased cotinine levels observed for those carrying the minor allele may lead to SHS exposure misclassification in studies utilizing cotinine as a biomarker. Additional studies are required to identify functional SNP(s) in NAT1 and elucidate the biological consequences of the mutation(s).
Introduction
Second hand smoke (SHS) exposures have been estimated to cause between 150,000 and 300,000 lower respiratory tract infections and exacerbate 400,000 to 1,000,000 cases of childhood asthma annually1, 2. Exposures also cause acute and recurring middle ear disease, cough, phlegm, wheeze and breathlessness, and may be responsible for induction of new cases of asthma in children and adolescents3. Despite these health risks, 35% of children in the United States live in homes where residents or visitors smoke on a regular basis4 and 50–75% of children have detectable levels of serum cotinine, indicating systemic exposure to SHS1, 5.
Nicotine and cotinine measures serve as a proxy for systemic exposure to over 7000 chemicals present in cigarette smoke6. Although biomarkers of SHS are routinely used to determine systemic exposures, there is great deal of inter-individual variability in these measurements, even in pharmacokinetic studies utilizing labeled nicotine and cotinine where exposure levels are identical7–10. Some of this variation is due to known factors, such as race, gender, age and genetic variation11, 12.
Nicotine is mainly metabolized through hepatic oxidation, and 70 and 80% is converted to cotinine through a two-step process8, 13 mediated by a cytochrome p450 (CYP) system and catalyzed by a cytoplasmic aldehyde oxidase (AOX1)13–15. The CYP system consists of genes including CYP2A6, CYP2B6, CYP2D616–19 and CYP2A1312. CYP2A6 is responsible for approximately 90% of the oxidation of nicotine to cotinine8, 20, 21. Other liver enzymes, including UDP-glucuronosyltransfeases (UGTs), and flavin-containing monooxygenase (FMO) are involved in the metabolism of nicotine and cotinine to additional metabolites8, 11, 12. The genes that regulate the metabolism of nicotine are rich in polymorphisms, and many alter the conversion of nicotine to cotinine and their metabolites. Genetic variation in the CYP2A6 gene is well-documented to cause decreased or absent nicotine metabolism in adults. Smokers and adults dosed with labeled nicotine that carry variant CYP2A6 alleles have decreased nicotine metabolism and urinary cotinine levels22–24. No studies, however, have evaluated the effect of polymorphisms in these genes and other metabolism-related genes with cotinine levels in SHS exposed children.
The objective of this study is to determine how polymorphisms in genes along the nicotine and xenobiotic metabolism pathways contribute to cotinine level variability, with respect to parental reported SHS exposures. We genotyped Caucasian children participating in the Cincinnati Childhood Allergy and Air Pollution Study (CCAAPS) on a custom single nucleotide polymorphism (SNP) chip for 176 tagging and functional variants in 9 genes known to be involved in the metabolism of nicotine, cotinine or other constituents present in SHS.
Methods
Study Population and SHS Exposure Determination
Caucasian children were participants in CCAAPS, a birth cohort of 762 infants born to atopic parent(s)25. The study was limited to Caucasians because of the known differences in nicotine metabolism in Caucasians compared to African-Americans9. Infants were identified by birth records and eligible parents had at least one allergy symptom and were skin prick test (SPT) positive to at least one aeroallergen25. Children were examined annually at ages 1, 2, 3, 4 and 7 years of age for allergic disease development. SHS exposure histories were obtained by utilizing a panel of four questions26. Briefly, parents were asked about the smoking habits of the mother, the number of smokers living in the child's home, the number of hours per day the child is around SHS at any location and SHS exposures in the car. Children whose parents answered positively to any of these four questions at the age 2 or 4 clinical exam were defined as exposed to SHS while those answering no to all were considered unexposed.
Hair Cotinine Quantification
Hair samples were obtained by cutting two samples of approximately 20 strands from the root end of the hair in the occipital region in the scalp. Hair samples from the 2 and 4 years of age visit were adjusted for weight and analyzed for cotinine level by radioimmunoassay at the Hospital for Sick Children in Toronto, Ontario, Canada; the limit of detection was 0.02 ng/mg27. The average hair cotinine level from the age 2 and 4 samples was used. If a child only had one sample available from either of those ages, that value was used.
Gene and SNP Selection, DNA Isolation and Genotyping
Genes were selected for inclusion on a custom Illumina GoldenGate Assay (http://www.illumina.com) based on function and relevance to nicotine metabolism. Genes implicated in nicotine metabolism (AOX1, CYP1A1, CYP1B1, CYP2A6, CYP2A13, CYP2B6, CYP2D6, FMO3, UGT2B10) as well as those having functions in xenobiotic metabolism (AHR, CYP1A1, NAT1, NAT2) were included. Tagging SNPs in the CEU and YRI populations with a R-square cutoff of 0.8 and minor allele frequency (MAF) ≥10% that captured the common genetic variation were selected using HapMap (http://hapmap.ncbi.nlm.nih.gov/). Functional SNPs in the CYP2A6 gene were also included regardless of MAF. Table 1 describes the number of SNPs selected in each gene as well as the gene name, function and associated disorders.
Table 1.
Selected genes, functions and associated disorders.
| Total SNPs | SNPs after exclusions | Gene Name | Full Gene Name | Chr. | Reported Processes and Functiona | Reported Associated Disordersb |
|---|---|---|---|---|---|---|
| 18 | 12 | FMO3 | flavin containing monooxygenase 3 | 1q24.3 | intrinsic to ER membrane, flavin adenine dinucleotide binding, monooxygenase activity, NADP binding | trimethylaminuria, metabolic disorder, familial adenomatous polyposis, polyposis, adenoma |
| 10 | 7 | CYP1B1 | cytochrome P450, family 1, subfamily B, polypeptide 1 | 2p22.2 | angiogenesis, arachidonic acid and cellular aromatic compound metabolic process, endothelial cell migration and adhesion, epoxygenase and omega-hydroxylase activity, redox process, response to toxins, steroid and xenobiotic metabolism, visual perception, ER membrane, aromatase, monooxygenase, oxidoreductase and electron carrier activity, heme, oxygen and iron binding | breast cancer, congenital glaucoma tumors, cancer, glaucoma, prostate cancer, endometrial cancer, endometriosis, colorectal cancer, glaucoma open-angle |
| 46 | 37 | AOX1 | aldehyde oxidase 1 | 2p33.1 | aldehyde oxidase and electron carrier activity, metal ion and NAD binding | molybdenum cofactor deficiency, xanthinuria type I and II, tumors, genetic disorder, neurological disorders, renal disease, amyotrophic lateral sclerosis, influenza |
| 7 | 3 | UGT2B10 | UDP glucuronosyltransferase 2 family, polypeptide B10 | 4q13.2 | lipid metabolism, ER membrane, glucuronosyltransferase activity | none |
| 17 | 15 | AHR | aryl hydrocarbon receptor | 7p21.1 | apoptosis, blood vessel development, cell cycle, intracellular receptor signaling pathway, regulation of transcription from RNA polymerase II, prostate gland development, regulation of B-cell proliferation and transcription, response to stress and xenobiotics, xenobiotic metabolism, cytosolic AHR complex, DNA, transcription factor and protein binding | breast cancer, tumors, cancer, lung cancer, shock, adenocarcinoma lung, mammary tumor, retinoblastoma, inflammation, endometriosis |
| 19 | 10 | NAT1 | N-acetyltransferase 1 | 8p22 | small molecule and xenobiotic metabolism, cytosol, acetyltransferase activity | cancer, breast cancer, bladder cancer, colorectal cancer, polyps, tumors, lung cancer, prostate cancer, pancreatic cancer, colon cancer |
| 15 | 14 | NAT2 | N-acetyltransferase 2 | 8p22 | small molecule and xenobiotic metabolism, cytosol, acetyltransferase activity | bladder cancer, cancer, breast cancer, colorectal cancer, lung cancer, tumors, prostate cancer, polyps, colon cancer, endometriosis |
| 1 | 0 | CYP1A1 | cytochrome P450, family 1, subfamily A, polypeptide 1 | 15q24.1 | 9-cis-retinoic acid biosynthetic process, aging, amine, coumarin, insecticide, porphyrin and flavenoid metabolism, camera-type eye development, cell proliferation, dibenzo-p-dioxin catabolism, digestive tract development, embryo development, hepatocyte differentiation, hydrogen peroxide biosynthesis, parturition, antibiotic, food, herbicide, hyperoxia liposaccharide, nematode, organic cyclic compound, virus, vitamin A, wounding and arsenic response, demethylase, electron carrier, flavonoid 3'-monooxygenase and oxidoreductase activity, enzyme, heme and iron ion binding | lung cancer, breast cancer, cancer, tumors, prostate cancer, ahh inducibility, squamous cell carcinoma, oral cancer, colorectal cancer, breast carcinoma |
| 7 | 2 | CYP2A6 | cytochrome P450, family 2, subfamily A, polypeptide 6 | 19q13.2 | coumarin metabolism and catabolism, exogenous drug, steroid, xenobiotic and small molecule metabolism, cytoplasm microtubule, ER membrane, coumarin 7-hydroxylase, oxidoreductase and electron carrier activity, enzyme, heme and iron ion binding | lung cancer, cancer, tumors, adenocarcinoma, smoking habit, liver diseases, colorectal cancer, adenoma, renal disease, carcinoma |
| 30 | 13 | CYP2B6 | cytochrome P450, family 2, subfamily B, polypeptide 6 | 19q13.2 | electron carrier, monooxygenase and oxidoreductase activity, heme and iron binding, incorporation or reduction of molecular oxygen | tumors, prostate cancer, cancer, inflammation, breast cancer, immunodeficiency, hypertrophy, virus infection, metastasis, necrosis |
| 5 | 3 | CYP2A13 | cytochrome P450, family 2, subfamily A, polypeptide 13 | 19q13.2 | xenobiotic and small molecule metabolism, ER membrane, aromatase and electron carrier activity, heme and iron binding | cancer, lung cancer |
| 1 | 1 | CYP2D6 | cytochrome P450, family 2, subfamily D, polypeptide 6 | 22q13.2 | electron carrier and oxidoreductase activity, heme and iron binding, incorporation or reduction of molecular oxygen | breast cancer, Parkinson disease, schizophrenia, lung cancer, autoimmune hepatitis, cancer, hepatitis c, Alzheimer's disease, bladder cancer, tumors |
Obtained from Gene Ontoloqv website (www.qeneontoloqv.orq).
Reported disease associations were obtained from the too 10 Novoseek disease relationships hits (number of articles in which both the qene's symbol or description and the disease appear) from GeneCards® (www.qenecards.orq).
DNA was isolated from buccal cells collected via cytology brush or saliva and using the Zymo Research Genomic DNA II Kit™ (Orange, CA) or Oragene DNA kit (DNA Genotek Inc.). Genotypes were assigned using BeadStudio's genotyping module (BeadStudio v3.2, San Diego, CA). A total of 176 SNPs were considered for analyses.
Statistical Analyses
Hair cotinine values were log-transformed. SNPs were excluded (n=59) that failed Hardy Weinberg Equilibrium in the non-allergic control group (p<0.0001), had minor allele frequencies below 10% (except two SNPs in CYP2A6); or had missing call rates greater than 10%. Principal component analyses were performed using the 100 included ancestry-informative markers (AIMs) in EIGENSTRAT28, 29 to account for potential population stratification; λ=1 indication no adjustment needed. Using PLINK30, associations with hair cotinine were tested adjusting for sex using the additive linear regression model. To address multiple testing, we determined the average pairwise LD (as measured by r2) for all SNP combinations and calculated the Bonferroni correction using Simple Interactive Statistical Analyses Software (http://www.quantitativeskills.com/sisa/). Associations were therefore considered significant at 0.000677. Linkage disequilibrium between SNPs of interest was calculated using JMP Genomics 6.0 (SAS, Cary, NC).
Results
Demographics of Subjects
Of the 431 Caucasian children genotyped on the Illumina array, 51 who did not have available hair cotinine levels and/or had >20% of their total SNP calls missing were excluded, yielding 380 children for these analyses (Table 2). Children with parental reported SHS exposure had 2.4-fold higher hair cotinine levels (0.139 ng/mg ± 0.216) compared to children with no parental reported exposures (0.058 ng/mg ± 0.051, p<0.0001; Table 2). The proportion of males did not differ between groups (Table 2).
Table 2.
Sample size and covariates for the CCAAPS Caucasian population.
| Variable | Parental Report of SHS | No Parental Report of SHS |
|---|---|---|
|
| ||
| Total children (n) | 160 | 271 |
| Exclusions1 (n) | 14 | 37 |
| Children after exclusions (n) | 146 | 234 |
| Percent male | 54.8% | 54.3% |
| Mean (SD) ng/mg hair cotinineb | 0.139 (0.216) | 0.058 (0.051) |
Individuals without an available hair cotinine level or more than 20% of their total SNPs missing were excluded.
Hair cotinine was significantly higher in children whose parent(s) reported SHS exposure (p < 0.0001).
Genetic Associations with Hair Cotinine Level
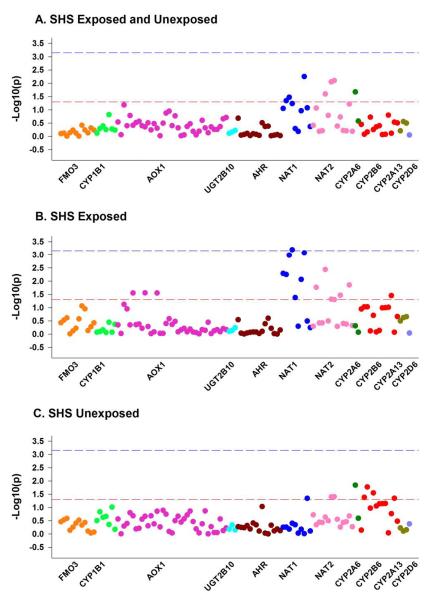
Genetic associations of the FMO3, CYP1B1, AOX1, UGT2B10, AHR, NAT1, NAT2, CYP2A6, CYP2B6, CYP2A13 AND CYP2D6 SNPs were evaluated adjusted for sex (Figure 1A). After consideration for multiple comparisons and stratification for reported SHS exposures, one tagging SNP in NAT1 (rs13253389) was significantly associated with hair cotinine level in children with parental-reported SHS exposures (p=0.0006, Figure 1B, Table 3). Other SNPs in the NAT1 gene were associated with hair cotinine level at a nominal level, but were not significant after multiple testing considerations (Table 3). Children without reported SHS exposures exhibited no significant genetic associations (Figure 1C).
Figure 1.
Associations of selected genes with early hair cotinine in CCAAPS Caucasian children.
Table 3.
Beta of Genetic Associations of SNPs in the NAT1 Gene with Hair Cotinine Level in Caucasian CCAAPS Children
| All Subjects | SHS Exposed | SHS Unexposed | ||||||
|---|---|---|---|---|---|---|---|---|
| n=380 | n=146 | n=234 | ||||||
|
| ||||||||
| SNP | Major/Minor Alleles | MAF | Beta | P-value | Beta | P-value | Beta | P-value |
| rs7017402 | G/A | 0.123 | 0.172 | 0.089 | 0.466 | 0.0052 | −0.067 | 0.5550 |
| rs4921580 | C/G | 0.126 | 0.204 | 0.046 | 0.492 | 0.0056 | −0.066 | 0.5481 |
| rs4921581 | A/G | 0.333 | 0.147 | 0.034 | 0.403 | 0.0010 | −0.033 | 0.6536 |
| rs13253389 | A/G | 0.344 | 0.132 | 0.058 | 0.419 | 0.0006 | −0.063 | 0.3933 |
| rs17693103 | G/T | 0.179 | 0.059 | 0.512 | 0.325 | 0.0417 | −0.074 | 0.4466 |
| rs6586714 | G/A | 0.086 | 0.054 | 0.659 | 0.152 | 0.5067 | 0.012 | 0.9266 |
| rs4921880 | A/T | 0.227 | 0.128 | 0.109 | 0.366 | 0.0086 | −0.037 | 0.6618 |
| rs11777998 | G/C | 0.123 | 0.286 | 0.006 | 0.547 | 0.0008 | 0.005 | 0.9680 |
| rs7003890 | T/C | 0.442 | 0.120 | 0.085 | 0.123 | 0.3240 | 0.149 | 0.0451 |
| rs8190845 | G/A | 0.126 | −0.081 | 0.431 | −0.115 | 0.5728 | −0.030 | 0.7693 |
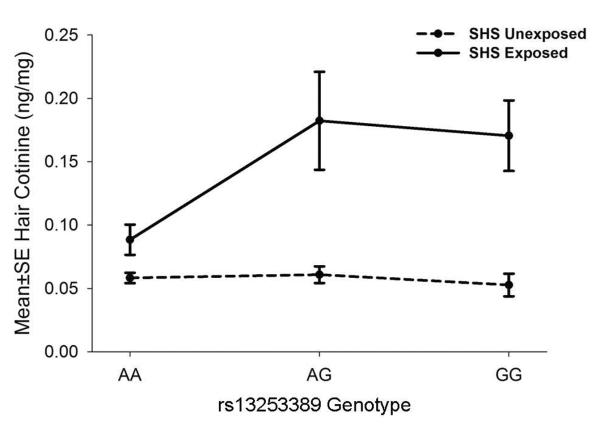
The significant tagging SNP in NAT1 is an A➔G polymorphism with a minor allele frequency of 34%. Children whose parents reported SHS exposures and were heterozygous or homozygous for the minor `G' allele had 2-fold higher hair cotinine levels (0.178 and 0.175ng/mg, respectively) than subjects that were homozygous for the major `A' allele (0.089ng/mg, p=0.0009) (Figure 3). Common genetic variation at this locus did not alter hair cotinine levels of children with no reported SHS exposures.
Figure 3.
Association of NAT1 SNPs with early hair cotinine level in CCAAPS Caucasian children.
To ensure that subjects carrying the AG or GG genotypes did not have higher hair cotinine levels due to higher SHS exposures, we evaluated the proportion of subjects whose parents reported SHS exposure by NAT1 rs13253389 genotype. There were no significant differences in the proportion of children with reported SHS exposures for subjects that were AA (35.6%), AG (41.8%), or GG (35.6%), p=0.45. In addition, we evaluated the association between the NAT1 SNP and hair cotinine level in children exposed to SHS adjusted for sex and the SHS dose, defined by the average number of cigarettes reportedly smoked by all household members at ages 2 and 4. Even after adjustment, we found that the NAT1 rs13253389 SNP still independently and significantly contributed to hair cotinine level (p=0.012, data not shown).
Discussion
Our data support that the NAT1 gene plays a role in the metabolic pathway of nicotine to its metabolites. Children whose parents indicate they are exposed to SHS that carry the minor `G' allele of the rs13253389 SNP had 2-fold higher hair cotinine levels than those that were homozygous for the major `A' allele. This association remained significant after adjustment for SHS exposure level, indicating that this NAT1 SNP independently contributes to hair cotinine level. In contrast, children whose parents indicated no SHS exposures had no differences in hair cotinine levels by genotype. These results collectively suggest that NAT1, either directly or indirectly, regulates the metabolism of nicotine or cotinine to its downstream metabolites. To our knowledge, this is the first report of an association between the NAT1 gene and hair cotinine level.
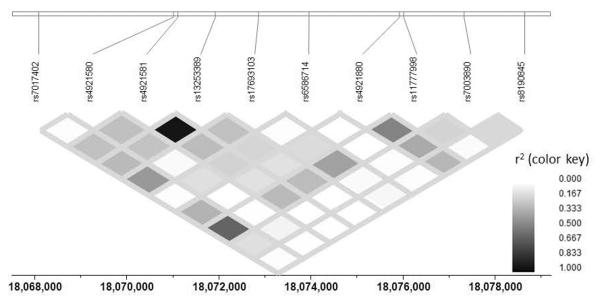
NAT1 rs13253389 is an intronic SNP located 4305 base pairs downstream of the transcription start site. While it is possible that this tagging SNP plays a direct role in the phenotype observed, it is likely that this SNP is in linkage disequilibrium with the causal variant. The NAT1 gene is part of the family of ayrlamine N-acetyltransferases that are xenobiotic conjugating enzymes. These enzymes are responsible for the acetylation of arylamines and arylhydrazines, O-acetylation of N-arylhydroxylamines, and N,O acetyl transfer of N-hydroxamic acid31. Genetic polymorphisms of both the NAT1 and NAT2 genes have been associated with slow and fast acetylator phenotypes31. While the the metabolic pathways of nicotine and cotinine have been extensively characterized, NAT1 has not yet been implicated in nicotine or cotinine metabolism.
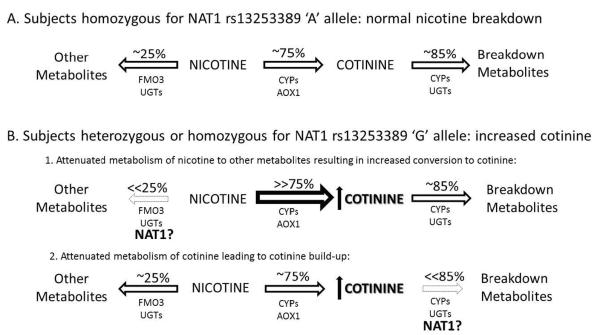
Nicotine is metabolized to six primary metabolites, and cotinine is quantitatively the most important11. Cotinine is also further broken down to six primary metabolites8, 11, 32–34. The majority of variation in the clearance of nicotine via the cotinine pathway is genetically influenced (heritability estimate 60.8%)35. After taking into account the effect of variation in CYP2A6, the heritability estimates are reduced but remain substantial at 51.8%35. Thus, a large proportion of the genetic variance in nicotine metabolism remains suggesting the involvement of additional genetic factors that have yet to be identified35, 36. There are many potential mechanisms by which NAT1 could, in-part, modulate cotinine levels (Figure 4). NAT1 might promote the breakdown of nicotine to an as yet unidentified metabolite and the SNP (or a causal SNP in linkage disequilibrium with this SNP) might reduce the breakdown of nicotine, resulting in a compensatory increase in cotinine formation (Figure 4B (1)). Alternatively, NAT1 may regulate cotinine metabolism with the SNP preventing cotinine breakdown resulting in cotinine accumulation (Figure 4B (2)). Since nicotine metabolism is also affected by intake of certain foods such as menthol and grapefruit juice11, NAT1 could play a role in regulating the breakdown of one of these foods and thereby impact cotinine levels. Finally, NAT1 might regulate pathways upstream of the CYP system or other known enzymes that metabolize nicotine, indirectly affecting cotinine levels.
Figure 4.
Potential mechanisms by which NAT1 modulates cotinine levels. (A) In subjects homozygous for the major NAT1 `A' allele, 75% of nicotine is broken down to cotinine and 25% to other metabolites. (B) The minor NAT1 `G' allele might affect cotinine levels by (1) regulating acetylation of an unknown nicotine metabolite or intermediate decreasing the breakdown of nicotine, resulting in a compensatory increase in cotinine formation; or (2) There might be an unknown intermediate that requires acetylation by NAT1 to further metabolize cotinine, and mutations in NAT1 could block this pathway, resulting in an accumulation of cotinine.
Cotinine is regarded as the biologic gold standard of SHS assessment5. Clinically, these observations are important because they highlight the NAT1 gene as a source of variability in cotinine measurement among Caucasian children. In our study, Caucasian children exposed to SHS that carry the NAT1 minor `G' allele had increased cotinine levels that were not due to increased exposures, potentially leading to exposure misclassification. This is important because exposure misclassification can produce spurious differences between groups or mask true associations, and decrease power to detect true effects37. Because biomarker variability leading to exposure misclassification has a considerable impact on all smoke exposure-related studies, sources of variability and limitations in biomarker measurement, including genetics, needs consideration.
These findings may also have an impact on the study of nicotine dependence since it is possible that the increased cotinine levels are due to an increased metabolism of nicotine. People who metabolize nicotine more quickly might smoke more cigarettes per day compared with slower metabolizers11. Genetic variations in other genes, such as CYP2A6, have also been reported to cause increased or decreased cotinine levels22–24 and associate with smoking behavior38, 39. However, the allele frequencies of these polymorphisms are low (0–8%, with a combined frequency of 9.1% in whites23), so it is not surprising that we did not find any associations with functional the CYP2A6 SNPs in our population given we were not powered to detect associations with minor allele frequencies < 10%.
Since the objective of this study was to examine how genetic variability in genes involved in nicotine and xenobiotic metabolism affects children's hair cotinine levels, we utilized parental report of SHS to stratify our exposure groups. Thus, it is possible that the carriers of the NAT1 minor allele(s) had greater SHS exposures due to parental underreporting which contributed to the observed increased cotinine levels. To minimize this likelihood, parental reported SHS exposures were defined by a four-question panel designed to provide the parent(s) an opportunity to report exposures without divulging personal habits26. Consequently, there were no significant differences in the proportion of children with parental reported SHS exposures by genotype. In addition, NAT1 rs13253389 remained significant in the multivariate model even after adjustment for the total number of cigarettes smoked per day by all smokers living in the child's home, indicating that this SNP independently contributes to children's hair cotinine level.
In conclusion, our data support that NAT1 has a role in the metabolic pathways from nicotine to cotinine and/or their associated metabolites, leading to increased cotinine levels. These findings are clinically relevant because genetic variability in NAT1 can affect cotinine biomarker levels independent of SHS exposure levels. Additional studies are required to identify functional SNP(s) in NAT1 and elucidate the biological consequences of the mutation(s).
Figure 2.
LD Plot of SNPs in the NAT1 Gene.
Acknowledgments
The authors thank the clinic staff for their efforts in study coordination, recruitment, data management, and data collection. We also thank all of the CCAAPS families for their time and commitment. This work was supported by the National Institute for Environmental Health Sciences grants R01-ES011170 and P30-ES006096.
Footnotes
Conflict of Interest The authors declare no conflict of interest.
References
- 1.ACS . Secondhand Smoke and Children Fact Sheet. American Cancer Society; Atlanta, GA: 2006. [Google Scholar]
- 2.Health Effects of Exposure to Environmental Tobacco Smoke. California Environmental Protection Agency; Jun, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Department of Health and Human Services, editor. The Health Consequences of Involuntary Exposure to Tobacco Smoke: A Report of the Surgeon General. Department of Health and Human Services, Centers for Disease Control and Prevention, Coordinating Center for Health Promotion, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health; Atlanta, GA: 2006. [Google Scholar]
- 4.Schuster MA, Franke T, Pham CB. Smoking patterns of household members and visitors in homes with children in the United States. Arch Pediatr Adolesc Med. 2002;156(11):1094–1100. doi: 10.1001/archpedi.156.11.1094. [DOI] [PubMed] [Google Scholar]
- 5.Benowitz NL. Cotinine as a biomarker of environmental tobacco smoke exposure. Epidemiol Rev. 1996;18(2):188–204. doi: 10.1093/oxfordjournals.epirev.a017925. [DOI] [PubMed] [Google Scholar]
- 6.Department of Health and Human Services PHS; Office of the Surgeon General, editor. How Tobacco Smoke Causes Disease: The Biology and Behavioral Basis for Smoking-Attributable Disease: A Report of the Surgeon General. Rockville, MD: 2010. p. 17. [PubMed] [Google Scholar]
- 7.Benowitz NL, Jacob P., 3rd Individual differences in nicotine kinetics and metabolism in humans. NIDA Res Monogr. 1997;173:48–64. [PubMed] [Google Scholar]
- 8.Benowitz NL, Jacob P., 3rd Metabolism of nicotine to cotinine studied by a dual stable isotope method. Clin Pharmacol Ther. 1994;56(5):483–493. doi: 10.1038/clpt.1994.169. [DOI] [PubMed] [Google Scholar]
- 9.Perez-Stable EJ, Herrera B, Jacob P., 3rd Benowitz NL. Nicotine metabolism and intake in black and white smokers. Jama. 1998;280(2):152–156. doi: 10.1001/jama.280.2.152. [DOI] [PubMed] [Google Scholar]
- 10.Benowitz NL, Perez-Stable EJ, Herrera B, Jacob P., 3rd Slower metabolism and reduced intake of nicotine from cigarette smoking in Chinese-Americans. Journal of the National Cancer Institute. 2002;94(2):108–115. doi: 10.1093/jnci/94.2.108. [DOI] [PubMed] [Google Scholar]
- 11.Benowitz NL, Hukkanen J, Jacob P., 3rd Nicotine chemistry, metabolism, kinetics and biomarkers. Handb Exp Pharmacol. 2009;(192):29–60. doi: 10.1007/978-3-540-69248-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mwenifumbo JC, Tyndale RF. Molecular genetics of nicotine metabolism. Handb Exp Pharmacol. 2009;(192):235–259. doi: 10.1007/978-3-540-69248-5_9. [DOI] [PubMed] [Google Scholar]
- 13.Hukkanen J, Jacob P., 3rd Benowitz NL. Metabolism and disposition kinetics of nicotine. Pharmacol Rev. 2005;57(1):79–115. doi: 10.1124/pr.57.1.3. [DOI] [PubMed] [Google Scholar]
- 14.Brandange S, Lindblom L. The enzyme “aldehyde oxidase” is an iminium oxidase. Reaction with nicotine delta 1'(5') iminium ion. Biochem Biophys Res Commun. 1979;91(3):991–996. doi: 10.1016/0006-291x(79)91977-6. [DOI] [PubMed] [Google Scholar]
- 15.Brandange S, Lindblom L. Synthesis, structure and stability of nicotine delta1'(5') iminium ion, an intermediary metabolite of nicotine. Acta Chem Scand B. 1979;33:187–191. [Google Scholar]
- 16.Yamazaki H, Inoue K, Hashimoto M, Shimada T. Roles of CYP2A6 and CYP2B6 in nicotine C-oxidation by human liver microsomes. Arch Toxicol. 1999;73(2):65–70. doi: 10.1007/s002040050588. [DOI] [PubMed] [Google Scholar]
- 17.Lang T, Klein K, Fischer J, Nussler AK, Neuhaus P, Hofmann U, et al. Extensive genetic polymorphism in the human CYP2B6 gene with impact on expression and function in human liver. Pharmacogenetics. 2001;11(5):399–415. doi: 10.1097/00008571-200107000-00004. [DOI] [PubMed] [Google Scholar]
- 18.Caporaso NE, Lerman C, Audrain J, Boyd NR, Main D, Issaq HJ, et al. Nicotine metabolism and CYP2D6 phenotype in smokers. Cancer Epidemiol Biomarkers Prev. 2001;10(3):261–263. [PubMed] [Google Scholar]
- 19.van Vleet T, Bombick D, Coulombe J, RA Inhinition of human cytochrome p450 2E1 by nicotine, cotinine and aqueous cigarette tar extract in vivo. Toxicol Sci. 2001;64(2):185–191. doi: 10.1093/toxsci/64.2.185. [DOI] [PubMed] [Google Scholar]
- 20.Messina ES, Tyndale RF, Sellers EM. A major role for CYP2A6 in nicotine C-oxidation by human liver microsomes. J Pharmacol Exp Ther. 1997;282(3):1608–1614. [PubMed] [Google Scholar]
- 21.Nakajima M, Yamamoto T, Nunoya K, Yokoi T, Nagashima K, Inoue K, et al. Characterization of CYP2A6 involved in 3'-hydroxylation of cotinine in human liver microsomes. J Pharmacol Exp Ther. 1996;277(2):1010–1015. [PubMed] [Google Scholar]
- 22.Malaiyandi V, Goodz SD, Sellers EM, Tyndale RF. CYP2A6 genotype, phenotype, and the use of nicotine metabolites as biomarkers during ad libitum smoking. Cancer Epidemiol Biomarkers Prev. 2006;15(10):1812–1819. doi: 10.1158/1055-9965.EPI-05-0723. [DOI] [PubMed] [Google Scholar]
- 23.Nakajima M, Fukami T, Yamanaka H, Higashi E, Sakai H, Yoshida R, et al. Comprehensive evaluation of variability in nicotine metabolism and CYP2A6 polymorphic alleles in four ethnic populations. Clin Pharmacol Ther. 2006;80(3):282–297. doi: 10.1016/j.clpt.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 24.Yang M, Kunugita N, Kitagawa K, Kang SH, Coles B, Kadlubar FF, et al. Individual differences in urinary cotinine levels in Japanese smokers: relation to genetic polymorphism of drug-metabolizing enzymes. Cancer Epidemiol Biomarkers Prev. 2001;10(6):589–593. [PubMed] [Google Scholar]
- 25.LeMasters GK, Wilson K, Levin L, Biagini J, Ryan P, Lockey JE, et al. High prevalence of aeroallergen sensitization among infants of atopic parents. J Pediatr. 2006;149(4):505–511. doi: 10.1016/j.jpeds.2006.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biagini Myers JM, Khurana Hershey GK, Deka R, Burkle JW, Levin LS, Bernstein DI, et al. Asking the right questions to ascertain early childhood secondhand smoke exposures. J Pediatr. 2012;160(6):1050–1051. doi: 10.1016/j.jpeds.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eliopoulos C, Klein J, Phan MK, Knie B, Greenwald M, Chitayat D, et al. Hair concentrations of nicotine and cotinine in women and their newborn infants. Jama. 1994;271(8):621–623. [PubMed] [Google Scholar]
- 28.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 29.Narayanaswamy CR, Raghavarao D. Principal Component Analysis of Large Dispersion Matrices. Appl Stat-J Roy St C. 1991;40(2):309–316. [Google Scholar]
- 30.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sim E, Walters K, Boukouvala S. Arylamine N-acetyltransferases: from structure to function. Drug Metab Rev. 2008;40(3):479–510. doi: 10.1080/03602530802186603. [DOI] [PubMed] [Google Scholar]
- 32.McKennis H, Jr., Turnbull LB, Bowman ER. N-Methylation of nicotine ad cotinine in vivo. J Biol Chem. 1963;238:719–723. [PubMed] [Google Scholar]
- 33.Neurath GB. Aspects of the oxidative metabolism of nicotine. Clin Investig. 1994;72(3):190–195. doi: 10.1007/BF00189309. [DOI] [PubMed] [Google Scholar]
- 34.Neurath GB, Pein FG. Gas chromatographic determination of trans-3'-hydroxycotinine, a major metabolite of nicotine in smokers. J Chromatogr. 1987;415(2):400–406. doi: 10.1016/s0378-4347(00)83234-x. [DOI] [PubMed] [Google Scholar]
- 35.Swan GE, Benowitz NL, Lessov CN, Jacob P, 3rd, Tyndale RF, Wilhelmsen K. Nicotine metabolism: the impact of CYP2A6 on estimates of additive genetic influence. Pharmacogenet Genomics. 2005;15(2):115–125. doi: 10.1097/01213011-200502000-00007. [DOI] [PubMed] [Google Scholar]
- 36.Keskitalo K, Broms U, Heliovaara M, Ripatti S, Surakka I, Perola M, et al. Association of serum cotinine level with a cluster of three nicotinic acetylcholine receptor genes (CHRNA3/CHRNA5/CHRNB4) on chromosome 15. Hum Mol Genet. 2009;18(20):4007–4012. doi: 10.1093/hmg/ddp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flegal KM, Brownie C, Haas JD. The effects of exposure misclassification on estimates of relative risk. Am J Epidemiol. 1986;123(4):736–751. doi: 10.1093/oxfordjournals.aje.a114294. [DOI] [PubMed] [Google Scholar]
- 38.Schoedel KA, Hoffmann EB, Rao Y, Sellers EM, Tyndale RF. Ethnic variation in CYP2A6 and association of genetically slow nicotine metabolism and smoking in adult Caucasians. Pharmacogenetics. 2004;14(9):615–626. doi: 10.1097/00008571-200409000-00006. [DOI] [PubMed] [Google Scholar]
- 39.Kubota T, Nakajima-Taniguchi C, Fukuda T, Funamoto M, Maeda M, Tange E, et al. CYP2A6 polymorphisms are associated with nicotine dependence and influence withdrawal symptoms in smoking cessation. The pharmacogenomics journal. 2006;6(2):115–119. doi: 10.1038/sj.tpj.6500348. [DOI] [PubMed] [Google Scholar]