Abstract
Diazaborine treatment of yeast cells was shown previously to cause accumulation of aberrant, 3′-elongated mRNAs. Here we demonstrate that the drug inhibits maturation of rRNAs for the large ribosomal subunit. Pulse-chase analyses showed that the processing of the 27S pre-rRNA to consecutive species was blocked in the drug-treated wild-type strain. The steady-state level of the 7S pre-rRNA was clearly reduced after short-term treatment with the inhibitor. At the same time an increase of the 35S pre-rRNA was observed. Longer incubation with the inhibitor resulted in a decrease of the 27S precursor. Primer extension assays showed that an early step in 27S pre-rRNA processing is inhibited, which results in an accumulation of the 27SA2 pre-rRNA and a strong decrease of the 27SA3, 27SB1L, and 27SB1S precursors. The rRNA processing pattern observed after diazaborine treatment resembles that reported after depletion of the RNA binding protein Nop4p/Nop77p. This protein is essential for correct pre-27S rRNA processing. Using a green fluorescent protein-Nop4 fusion, we found that diazaborine treatment causes, within minutes, a rapid redistribution of the protein from the nucleolus to the periphery of the nucleus, which provides a possible explanation for the effect of diazaborine on rRNA processing.
Translation of mRNA in the cytoplasm relies on the ribosome. While the principal steps in initiation and elongation of protein synthesis were uncovered in the middle of the last century, details of the biogenesis of the ribosome itself remained elusive. However, the development of novel techniques in affinity purification and refined techniques of protein identification by mass spectrometry within the last several years provided novel insights into the steps of assembly of the ribosomal subunits and how they are coordinated with the processing of the rRNA precursors (for recent reviews see references 15 and 47). Much of this detailed knowledge was worked out in the baker's yeast, Saccharomyces cerevisiae. This organism constitutes a simple model for the reaction sequence occurring in more complex eukaryotic cells.
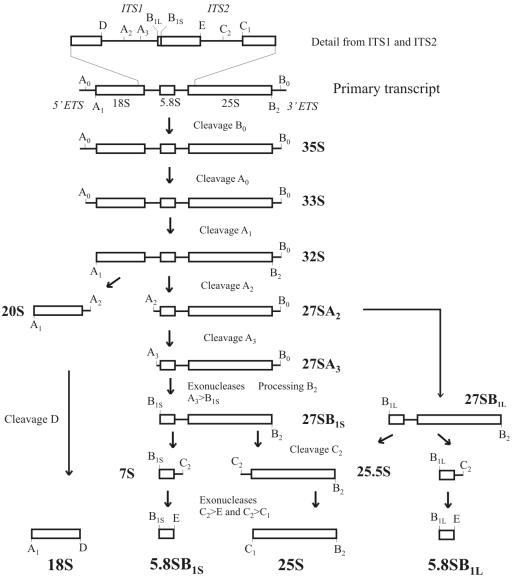
While the 5S rRNA is transcribed by RNA polymerase III, the 18S, 25S, and 5.8S rRNAs are transcribed as a single precursor molecule by RNA polymerase I. The primary transcript is cotranscriptionally processed at its 3′ external transcribed spacer (ETS) end into the 35S pre-rRNA (26). The rRNAs for both the large and the small subunits are then generated from the 35S pre-rRNA by several subsequent processing events. In the first two processing steps for the 35S rRNA, the 5′ ETS is successively removed by endonucleolytic cleavages at positions A0 and A1. The following step, which separates the rRNA precursors for the two ribosomal subunits, is a cleavage reaction at position A2 in internal transcribed spacer 1 (ITS1) located between the 18S and 5.8S rRNAs (23, 27). This cleavage reaction results in the release of the 20S pre-rRNA, which matures in the cytoplasm to the 18S rRNA present in the small subunit of the ribosome (49). The second product of the A2 cleavage is the 27SA2 pre-rRNA, which contains the 5.8S and the 25S rRNAs. The 27S pre-rRNA is covered with 60S subunit components and proteins required for 60S maturation and export. The subsequent endonucleolytic cleavage, which is performed by the RNP nuclease MRP, occurs at A3 (30, 39). Further trimming and cleavage reactions yield 5.8SB1S and 25S rRNAs (10, 17, 21, 33). There is also an alternative pathway, starting at the 27SA2 pre-rRNA, which produces mature 5.8SB1L and 25S rRNA, albeit in smaller amounts. The 5.8S rRNAs generated by the major and minor pathways differ slightly at their 5′ ends (21). The details of the rRNA processing pathway are outlined in Fig. 1.
FIG. 1.
Schematic diagram of the pre-rRNA processing pathway in S. cerevisiae. The processing of the primary transcript in the 3′ ETS results in formation of the 35S pre-rRNA. Two subsequent cleavages at positions A0 and A1 in the 5′ ETS generate the 5′ end of the 18S rRNA. The precursors of the rRNAs for the small and large subunits are separated by cleavage at A2. Cleavage of the 20S pre-rRNA at site D generates the mature 18S rRNA. Processing of 27SA2 occurs by two alternative pathways. Whereas in the major pathway cleavage at A3 is followed by exonucleolytic trimming to position B1S, the minor pathway creates a different 5′ end, resulting in the 27SB1L precursor, which is longer by 7 nucleotides. Cleavage at C2 splits the 27SB pre-rRNAs into a 7S pre-rRNA and a 25.5S pre-rRNA. 3′-5′ and 5′-3′ exonucleolytic trimming of these precursors produces the mature 5.8S and 25S rRNAs, respectively.
The processing reactions are coordinated by many different factors, including ribosomal and nonribosomal proteins and snoRNPs. The assembly of these factors occurs in a distinct order, and subpopulations of preribosomal particles with different protein patterns and rRNA processing intermediates were isolated on their way from the nucleolus through the nucleoplasm into the cytoplasm (3, 14, 20, 32, 34, 38). Processing and export of the two subunits mostly rely on different factors. The processing and maturation of the small ribosomal subunit involve fewer steps and are accomplished faster than those of the large subunit. This probably reflects the higher degree of complexity in the maturation of the large subunit (47). Successful processing of the pre-rRNA within the nucleus is a prerequisite for the pre-60S particle to gain export competence. The export of both subunits to the cytoplasm involves the exportin Xpo1p/Crm1p and RanGTP (16, 22).
Diazaborine is an antibacterial drug that inhibits fatty acid biosynthesis in Escherichia coli (5, 48). We found that diazaborine also inhibits growth of yeast cells, although in this organism fatty acid biosynthesis is not affected. Mutations in genes YAP1, PDR1, PDR3, and DRG1/AFG2 confer resistance to diazaborine in yeast (24, 51). In contrast to resistance mediated by YAP1, PDR1, and PDR3, where cross-resistance to other drugs is observed, resistance mediated by DRG1/AFG2 is specific for diazaborine (51). The function of gene DRG1/AFG2 is unknown, but it is essential and encodes an ATPase of the AAA family of proteins (46, 51, 52). AAA proteins are present in all classes of organisms, where they act as specific chaperones which catalyze the structural remodeling, unfolding, and disassembly of proteins and protein complexes (29).
We have demonstrated recently that treatment of S. cerevisiae with diazaborine results in accumulation of aberrant mRNA species of various genes. The aberrant mRNAs are extended at their 3′ end and terminate at alternative termination signals (25). No aberrant mRNAs were observed when the diazaborine-resistant DRG1-1 mutant ESY212 was treated with the inhibitor. Here we report that diazaborine treatment of yeast cells results in the inhibition of processing of the 27S pre-rRNA and causes a reduction of free 60S subunits in the cytoplasm. The drug-induced processing defects resemble the defects observed upon depletion of Nop4p/Nop77p, a nucleolar RNA binding protein involved in 60S subunit maturation (4, 44, 45). We further show that diazaborine treatment results in redistribution of a green fluorescent protein (GFP)-Nop4 fusion protein to the periphery of the nucleus, which explains the observed effects of the drug on rRNA processing.
MATERIALS AND METHODS
Strains and growth conditions.
The strains used in this study were W303 and the diazaborine-resistant isogenic DRG1-1 mutant ESY212, which were described previously (25). Strains were grown either in yeast extract-peptone-dextrose (YPD) complex medium or, for metabolic labeling, in synthetic dextrose (SD) medium supplemented with the appropriate amino acids. Plasmid-containing strains were grown in SD complete medium lacking uracil.
Construction of GFP fusions.
GFP fusions were constructed by PCR amplification of the respective genes with Expand High-Fidelity DNA polymerase with specific primers that introduce suitable restriction sites. The PCR products were cloned in frame into the appropriately cleaved pUG35 or pUG36 plasmid DNA to construct fusions with enhanced GFP (eGFP). pUG35 and pUG36 are vectors for the construction of C-terminal and N-terminal eGFP fusions, respectively (J. H. Hegemann, personal communication). The fusion constructs are expressed under the control of the MET35 promoter. The yellow fluorescent protein (YFP)-Nop7 and YFP-Drs1 fusions were constructed by exchanging the coding region of eGFP in pUG36 for that of YFP, which was derived from plasmid pDH5. Thereafter, the genes were cloned by PCR amplification with the use of specific primers as described above. The Rpl7A-YFP fusion was generated by chromosomal tagging of the gene in strains W303 and ESY212.
Fluorescence microscopy.
The strain to be investigated was grown at 25°C in synthetic complete medium lacking uracil and methionine to the early log phase (A600, 0.3 to 0.6). The culture was split, and one aliquot was treated with 5 μg of diazaborine/ml. After different periods of incubation in the presence of the inhibitor, cells were viewed by fluorescence microscopy with the use of small-band eGFP or YFP filters (Zeiss) on a Zeiss Axioskop microscope. For 4′,6′-diamidino-2-phenylindole (DAPI) staining, cells were treated with 70% ethanol for 2 min and stained for 2 min with DAPI (1 μg/ml) in phosphate-buffered saline, pH 7.0. Cells were harvested by centrifugation, washed two times with 1 ml of SD complete medium, and viewed with a DAPI filter and a fluorescein isothiocyanate (FITC) filter (Zeiss).
rRNA labeling and pulse-chase analyses.
For the pulse-chase analyses, strains were grown to an A600 of 1.2 in SD medium. Cells corresponding to 3.5 A600 units were harvested by centrifugation and resuspended in 1 ml of SD medium lacking uracil. Thereafter 5 μCi of [14C]uracil/ml (54 mCi/mmol) was added. After 2 min, a 50-fold excess of nonradioactive uracil and 5 μg of diazaborine/ml were added to the culture, and 0.5-ml samples were removed after different periods of incubation. Cells were broken after suspending them in 5 M guanidinium-HCl buffer by shaking vigorously with glass beads. RNA was extracted by the hot phenol method, separated on formaldehyde-1.2% agarose gels, and transferred onto nylon membranes by the capillary transfer method. After UV cross-linking the radioactivity on the membrane was detected by phosphoimaging with a tritium screen (Kodak).
Polysome analyses.
Strains were grown in 100 ml of YPD medium to the early log phase (A600 of 0.6), and subsequently cycloheximide (final concentration, 100 μg/ml) was added. In the case of diazaborine treatment the inhibitor (final concentration, 100 μg/ml) was added 30 min before the cycloheximide treatment. After addition of cycloheximide the culture flasks were incubated for 10 min in an ice-water bath. Cells were harvested by centrifugation at 4°C and 3,000 × g and washed twice in buffer A (100 mM Tris-HCl [pH 7.5], 100 mM KCl, 10 mM MgCl2) containing 50 μg of cycloheximide/ml, 200 μg of heparin/ml, and 0.02% diethyl pyrocarbonate. The cell pellet was resuspended in 1 ml of buffer A containing cycloheximide, heparin, and diethyl pyrocarbonate. Cells were opened by vigorous shaking eight times for 20 s with glass beads. The extract was centrifuged for 7 min at 6,000 × g and again for 10 min at 10,000 × g. A quantity of 6.5 A260 units from the supernatant was loaded at the top of a linear 15 to 50% sucrose gradient in buffer A. The gradients were centrifuged at 200,000 × g for 2.5 h and harvested from the top after the bottom of the centrifuge tube was pierced and a solution containing 70% sucrose, 10% glycerol, and 0.01% bromphenol blue was pumped in. Polysome profiles were recorded using an ISCO gradient harvester device.
Northern blotting.
Northern blotting was performed as described previously (25). RNA was separated on formaldehyde-1.2% agarose gels and blotted onto nylon membranes by capillary transfer. Hybridization was performed overnight with the radiolabeled oligonucleotides 001 (GGCCAGCAATTTCAAGTTA), 002 (GCGTTCTTCATCGATGC), 003 (CTCTCACCGTTTGGAATAGC), NME1 (GAAAGAGCAATCGTCATAACTATGGT), and SNR30 (GCTCGTAGTCTGACGACTCAAAACTCTG) at 50°C. After washing, radioactivity on the filters was detected by exposing X-ray films or by phosphoimaging.
Primer extension analyses.
Primer extension was performed with total RNA extracted from treated or untreated cells by using oligonucleotide 001. The experiment was performed using 2.5 μg of total RNA and 1 pmol of 32P-labeled oligonucleotides as described in reference 2.
Metabolic labeling of yeast cells with [35S]methionine.
Metabolic labeling of yeast cells with [35S]methionine was performed in SD medium lacking methionine. Cells were grown in 5 ml to an A600 of 0.6; thereafter 13 μCi of [35S]methionine (specific activity, 1,175 Ci/mmol) was added. After different periods of incubation, aliquots were removed and the cells were collected by filtration on 0.45-μm-pore-size filters (Millipore). Incorporated radioactivity was measured by liquid scintillation counting.
RESULTS
Diazaborine treatment results in a decrease of free 60S ribosomal subunits.
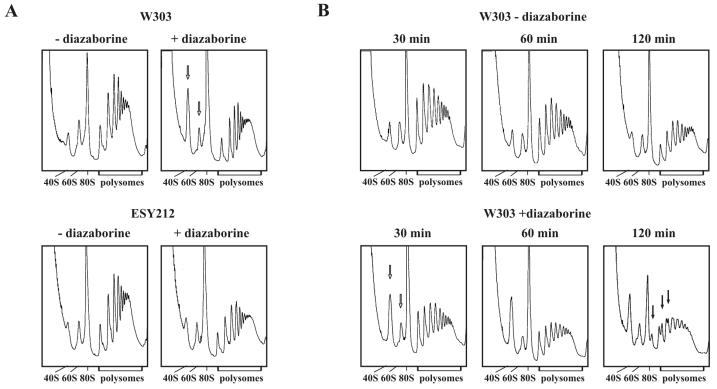
In the course of our studies on the effects of diazaborine on RNA metabolism of S. cerevisiae, we observed that diazaborine treatment resulted in marked alterations of polysome profiles. The wild-type strain W303 and the diazaborine-resistant DRG1-1 mutant ESY212 were grown in YPD medium to the early log phase and treated for 30 min with the inhibitor. Polysome profiles were obtained from the drug-treated strains and the untreated controls with the use of 15 to 50% sucrose gradients. As demonstrated in Fig. 2A, treatment of the wild-type strain with diazaborine resulted in a massive decrease of the free 60S subunits compared to the free 40S subunits. To estimate the ratio of the 40S and 60S subunits more precisely, we also analyzed the extracts on 10 to 25% sucrose gradients (data not shown). While the ratio of 60S to 40S subunits in the untreated W303 wild-type strain was estimated to be 1.5 ± 0.1 (mean of three independent experiments), it was shifted to 0.5 ± 0.1 in the diazaborine-treated strain after 30 min of incubation. The decrease of the 60S/40S ratio was not accompanied by the appearance of half-mers during this time period. The polysome profiles of the diazaborine-resistant DRG1-1 strain ESY212 showed a ratio of free 60S to 40S subunits of 1.4. The ratio of free 60S to 40S subunits in the ESY212 strain did not show such a pronounced change upon diazaborine treatment and was in the range of 1.0.
FIG. 2.
Treatment with diazaborine leads to a reduced level of 60S subunits. After incubation of the wild-type strain W303 and the diazaborine-resistant mutant ESY212 in the presence of the inhibitor for different periods of time, extracts were prepared and 6.5 A260 units each was loaded on 15 to 50% sucrose gradients as described in Materials and Methods. After ultracentrifugation for 2.5 h at 200,000 × g polysome profiles were collected. (A) W303 and ESY212 incubated for 30 min in the presence and absence of diazaborine. Arrows point to the free 40S and 60S subunit peaks to highlight the massive changes in the treated wild-type strain. (B) Time course of W303 incubated for 30, 60, and 120 min in the presence and absence of diazaborine. Free 40S and 60S subunits and half-mer polysomes observed after 120 min of treatment are indicated by open and filled arrows, respectively.
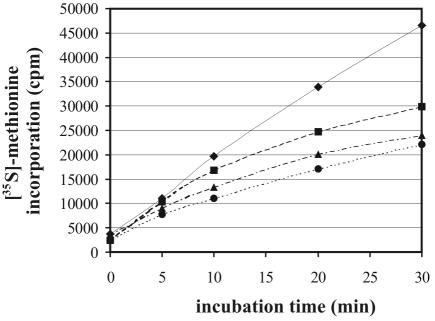
To investigate whether the diazaborine-induced shift in the ratio of 60S to 40S affects protein synthesis, we performed metabolic labeling studies with [35S]methionine in the presence of the inhibitor. Yeast cells are much more sensitive to diazaborine in minimal medium than in YPD medium. A diazaborine concentration of 5 μg/ml in minimal medium leads to a growth inhibition that is similar to that observed with 100 μg/ml in YPD medium. The concentration of the inhibitor was therefore reduced in experiments where cells were grown in minimal medium. For the [35S]methionine incorporation studies 15 μg of diazaborine/ml was used, which is three times above the MIC in SD complete medium. When the [35S]methionine incorporation was measured under these conditions, only a slight decrease compared to the untreated control was observed within the first 10 min of this experiment. This concentration already significantly affects growth of the diazaborine-resistant DRG1-1 mutant ESY212 (data not shown). In cultures pretreated for 20 min with the inhibitor the decrease was stronger, but still the cells synthesized proteins at a level of 50% of that of the untreated control (Fig. 3).
FIG. 3.
Metabolic labeling with [35S]methionine. The wild-type strain W303 was grown in SD complete medium lacking methionine to early log phase (A600 of 0.6), and 13 μCi of the radioactive tracer was added. Diazaborine was added immediately (squares), 10 min (triangles), or 20 min (circles) before addition of [35S]methionine. The untreated control is indicated by diamonds. Incorporated radioactivity was measured by the filter binding assay after different incubation periods. One representative experiment of three independent repeats is shown.
The findings that, in spite of the strong shift in the 60S to 40S ratio, diazaborine treatment of yeast cells resulted neither in an immediate block of protein synthesis nor in half-mer formation could be explained by the large amount of ribosomes present in the cytosol of wild-type cells before addition of the inhibitor. These ribosomes might allow the cell to carry out protein synthesis for some time even if the constant supply of 60S ribosomal subunits were inhibited. If this hypothesis were true, longer incubation in the presence of the inhibitor should result in a stronger depletion of 60S subunits and in formation of half-mers. We therefore obtained polysome profiles from the wild-type strain which was treated with the inhibitor for a longer period (Fig. 2B). The 60S/40S ratio in the untreated control was estimated to lie between 1.4 and 1.5 within the interval of 2 h. A decrease in the polysome fraction was observed for the untreated strain within 2 h. We attribute this decrease in polysomes to the depletion of nutrients at the end of the logarithmic growth phase. In the presence of the inhibitor the ratio of 60S to 40S was 0.6 after 30 min of incubation, similar to that found after 1 h (0.42) and 2 h (0.63) of treatment. However, after 1 h of incubation with the inhibitor a tailing of the polysome peaks was detected. After 2 h of incubation with the inhibitor a decrease of the 80S peak was observed and the polysomes exhibited a marked formation of half-mers. The polysome profiles of the diazaborine-resistant DRG1-1 mutant ESY212 did not show any half-mers after 2 h of incubation with the inhibitor (data not shown).
The reduced 60S subunit level is caused by a block in 27S pre-rRNA processing.
To uncover the reason for the strong decrease in free 60S subunits after 30 min of diazaborine treatment, we tested the effect of the drug on steady-state levels of pre-rRNA precursors. This was done by Northern blotting experiments using radiolabeled oligonucleotides as probes that hybridize to specific regions on the rRNA precursors for the large subunit (Fig. 4). In the RNA extracted from the treated wild-type cells, 7S pre-RNA stopped being detectable as soon as after 15 min of incubation with diazaborine. The 27S pre-rRNA was detected in an amount similar to that in the untreated control after 15 min, but an accumulation of the 35S precursor was observed. Longer incubation, e.g., for 60 min in the presence of diazaborine, resulted in a pronounced decrease of the 27S and 35S precursors. Treatment of the resistant strain ESY212 for 60 min caused only a slight reduction of the steady-state level of the 27S pre-rRNA and did not affect the level of 7S pre-rRNA. To investigate whether drug treatment results in the production of aberrant rRNA processing intermediates, we used an oligonucleotide which is complementary to a region between the 3′ end of the 18S rRNA and the A2 processing site. This probe allowed us to detect the 20S pre-rRNA precursor and the 23S aberrant processing product. The aberrant 23S RNA is generated from the 35S pre-rRNA by an endonucleolytic cleavage reaction at position A3 before cleavage at A0, A1, and A2 occurs and contains the 18S rRNA and flanking regions. As shown in Fig. 4 the amounts of the 23S RNA and the 20S pre-rRNA decreased after 15 min of treatment, and the signal from the 35S pre-rRNA strongly increased. No accumulation of the 23S aberrant processing product was observed. The effect of the drug on the formation of 20S pre-rRNA was not as pronounced as that observed for the 7S pre-rRNA. We interpret this result to mean that the inhibition occurs at a step after the conversion of the 32S intermediate to the two cleavage products and acts predominantly on the pathway leading to the 25S rRNA. We could also detect the small processing product ranging from site D to A2, which is removed from the 20S pre-rRNA in the course of 18S rRNA formation. The steady-state level of this fragment showed behavior similar to that of the 20S pre-rRNA. In the resistant mutant ESY212, the level of the 20S pre-rRNA remained constant and no increase in 35S pre-rRNA level was observed after 15 min of treatment. Longer incubation resulted in a slight increase of the 35S pre-rRNA.
FIG. 4.
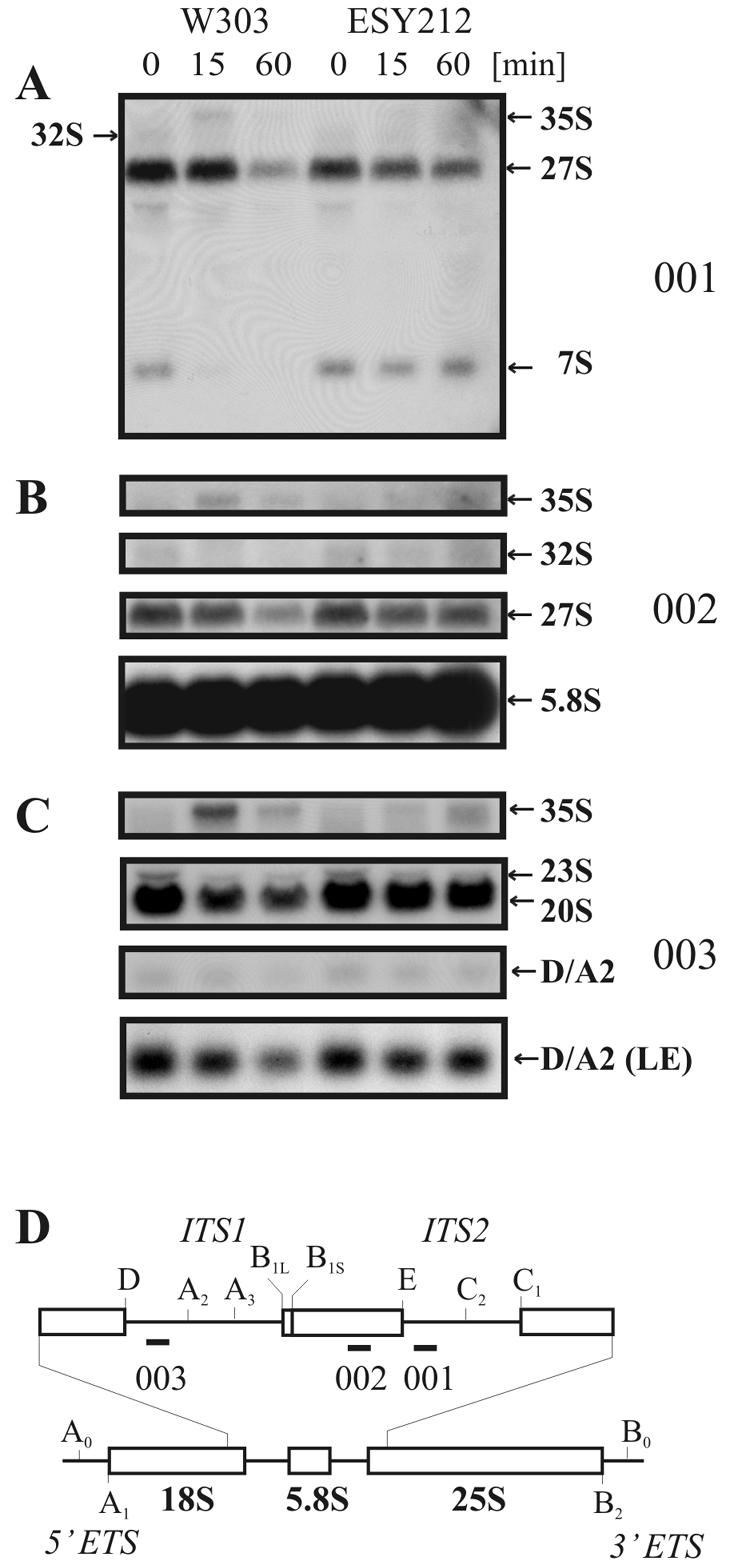
Diazaborine blocks processing of the 27S pre-rRNA to the 7S pre-rRNA. Strains W303 and ESY212 were incubated in the presence of diazaborine for 0, 15, and 60 min. RNA was extracted, and sequences complementary to oligonucleotides 001, 002, and 003 were detected by Northern hybridization. (A) Hybridization with probe 001, complementary to the 5′ region of ITS2. (B) Hybridization with probe 002, which binds to the 5.8S rRNA.(C) Hybridization with probe 003, which hybridizes to the 5′ region of ITS1. “(LE)” denotes the panel with a section of a longer exposure showing the D/A2 signal. (D) Schematic diagram of the 35S precursor rRNA with an enlarged view of the ITS1 and ITS2 region. Oligonucleotide probes (001, 002, and 003) used for hybridization are indicated as short bars.
We also performed pulse-chase experiments after incorporation of [14C]uracil. In the case of diazaborine treatment, the inhibitor was added at a concentration of 5 μg/ml to the chase solution. As shown in Fig. 5, processing of the 27S pre-rRNA to the mature 25S rRNA was inhibited in the wild-type strain by addition of diazaborine to the chase solution. Even after 30 min of chase significant amounts of the 27S pre-rRNA were detected. The 27S precursor was generated normally in the untreated and treated wild-type strains, demonstrating that earlier steps in pre-rRNA processing are not affected by diazaborine. Processing of the 20S pre-rRNA to the 18S rRNA in the treated wild-type strain occurred as fast as in the untreated strain and was complete in both instances after 15 min. This result shows that the formation of 18S rRNA is not inhibited by diazaborine. When the pulse-chase analysis was performed with the diazaborine-resistant mutant ESY212, no inhibition of 27S processing in the presence of the inhibitor was observed. The formation of the 25S rRNA occurred with the same kinetics as in the untreated wild-type strain.
FIG. 5.
Treatment with diazaborine inhibits formation of the 25S rRNA. Cells from the strains W303 and ESY212 were pulse-labeled with [14C]uracil for 2 min and chased with a 50-fold excess of nonradioactive uracil for 0, 3, 15, and 30 min in the presence or absence of 5 μg of diazaborine/ml. RNA was extracted by the hot phenol method. The RNA was separated on 1.2% formaldehyde agarose gels to monitor the processing of high-molecular-weight rRNA precursors. The gel was blotted, and the radioactivity was detected by phosphoimaging.
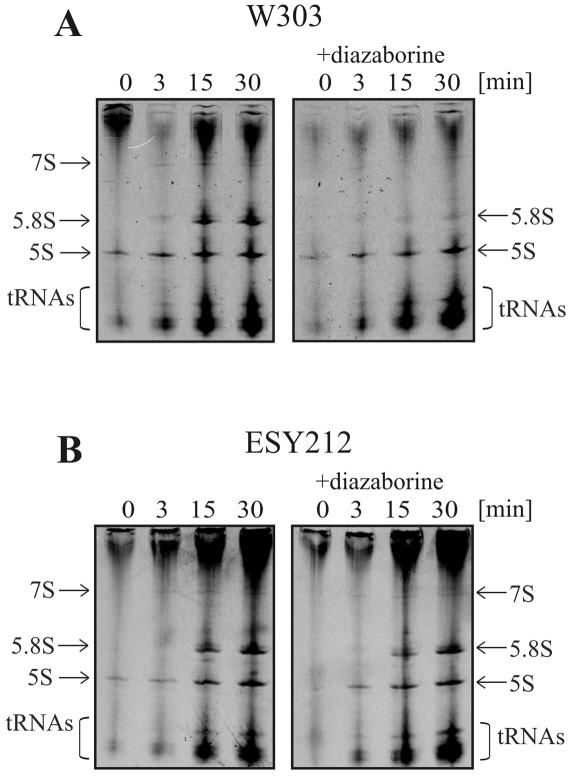
The effect of diazaborine on the formation of small RNAs was analyzed on 8% acrylamide-8 M urea gels (Fig. 6). In the untreated wild-type strain, we detected the 7S pre-rRNA and the mature 5.8SB1L and 5.8SB1S rRNAs. Moreover, the 5S rRNA and several tRNA species were also identified. Diazaborine treatment almost completely blocked the formation of the 7S and 5.8S pre-rRNAs. The formation of 5S rRNA and tRNA was not affected. The formation of 7S pre-rRNA and 5.8S rRNA was not affected by diazaborine in the resistant mutant. Taken together, these results show that diazaborine treatment specifically prevents the processing of the 27S pre-rRNA into its products, i.e., 7S pre-rRNA and 25.5S pre-rRNA.
FIG. 6.
Treatment with diazaborine inhibits formation of the 7S pre-rRNA and the 5.8S rRNA. Cells from the strains W303 and ESY212 were pulse-labeled with [14C]uracil for 2 min and chased with a 50-fold excess of nonradioactive uracil for 0, 3, 15, and 30 min in the presence or absence of 5 μg of diazaborine/ml. RNA was extracted by the hot phenol method and separated on 8% acrylamide-8 M urea gels to detect the formation of low-molecular-weight RNAs. The gel was dried, and the radioactivity was detected by exposure of an X-ray film.
The processing of the 27SA2 pre-rRNA is inhibited by diazaborine.
The 27S pre-rRNA detected by the Northern blotting and pulse-chase studies in fact represents a pool of at least four different processing intermediates with similar sizes that were not separated due to the low resolution of agarose gel electrophoresis. The first 27S RNA is generated by processing of the 32S pre-rRNA at A2, resulting in the 27SA2 intermediate. The 27SA2 pre-rRNA is subsequently processed near the 5′ end at position A3. This processing step is accomplished by the MRP endonuclease (39). The product of this cleavage reaction, the 27SA3 pre-rRNA, is further trimmed at the 5′ end by the exonuclease Rat1p or Xrn1p to the 27SB1S precursor (21). An alternative pathway that is used by approximately 15% of the precursors skips cleavage at A3 and ends up with the 27SB1L intermediate, which is a few nucleotides longer at its 5′ end than the 27SB1S intermediate is (21).
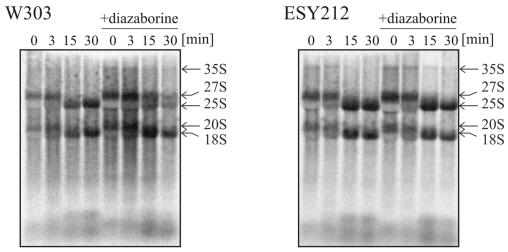
To identify the exact step affected by diazaborine treatment, we performed primer extension experiments with oligonucleotide 001, which hybridizes to ITS2 immediately beyond the 3′ end of the 5.8S rRNA. As shown in Fig. 7, B1S represents the most prominent band in the primer extension analysis of the untreated wild-type strain. The second prominent band is A2, while the A3 band is of faint appearance. When we looked at the RNA from the drug-treated strain, marked alterations of the precursor pattern were observed. After 15 min of incubation in the presence of diazaborine the A3 band disappeared, while the amount of A2 was increased. Concomitantly with the increase of A2 a pronounced decrease of the B1L and B1S precursors was observed. The ratio of 27SB1S to 27SA2 was 4.1 in the untreated strain and was shifted to 0.8 after 15 min of treatment. Longer incubation resulted in a further decrease of the B1L and B1S species, while the level of 27SA2 was only slightly reduced. These results show that diazaborine treatment inhibits processing of the 27SA2 pre-rRNA to the 27SA3 intermediate. The ratio of the 27SB1S to 27SB1L species, which are generated by different pathways originating at the 27SA2 precursor, remained constant and was estimated to be 5.3 in the untreated strain and 5.5 after 1 h of treatment. The same experiment was performed with the diazaborine-resistant DRG1-1 mutant ESY212. In the drug-treated resistant mutant, the ratio of A2 to B1S was similar to that in the untreated control and no decrease of the A3 precursor was observed.
FIG. 7.
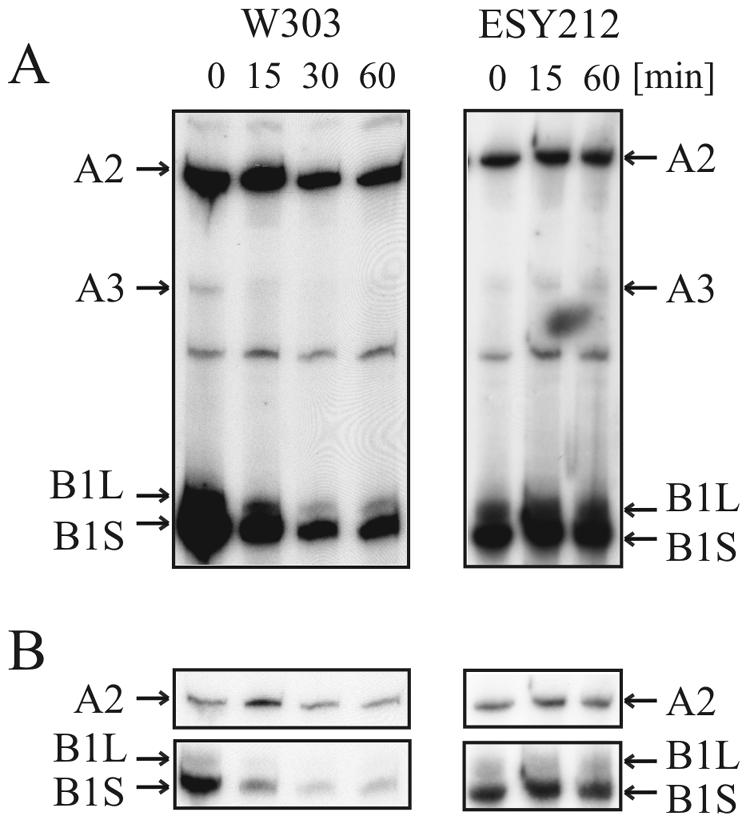
Diazaborine inhibits cleavage at position A3 of the 27S pre-rRNA. Strains W303 and ESY212 were treated with diazaborine, and RNA was extracted and subjected to primer extension analysis with oligonucleotide 001. (A) The positions of the primer extension stops A2, A3, B1L, and B1S are indicated by arrows. W303 was incubated for 0, 15, 30, and 60 min and ESY212 was incubated for 0, 15, and 60 min with diazaborine. (B) Sections of an autoradiogram containing the A2 and the B1L and B1S stops that was exposed only briefly to demonstrate the actual extent of the decrease of B1 relative to A2.
Formation of NME1 RNA and snR30 snoRNA is not affected by diazaborine.
It was reported previously that diazaborine treatment of yeast cells results in the accumulation of 3′-elongated mRNAs (25). Many small RNA species are important components of the rRNA processing machinery. Therefore, we tested whether the effects of the drug on rRNA processing are the consequence of aberrantly 3′-processed small RNAs. Since we found that diazaborine treatment specifically interferes with processing of the 27SA2 precursor at site A3, we were interested in whether the drug affects formation of NME1 RNA. This is the RNA component of the RNase P-like enzyme MRP, which is responsible for cleavage at A3 (9, 30). We performed Northern blot studies with total RNA extracted from yeast cells that were treated with diazaborine for different periods and used a radiolabeled oligonucleotide complementary to the NME1 RNA as a probe. As shown in Fig. 8 the NME1 RNA showed the same electrophoretic mobility in the treated and untreated samples, demonstrating that the drug does not affect 3′ processing of the NME1 RNA. Short-time diazaborine treatment did not affect the steady-state level of NME1 RNA. Although a reduction of the NME1 signal by 40% was observed after 60 min of treatment, the decrease in 7S pre-rRNA, as described above, was stronger and occurred much faster.
FIG. 8.
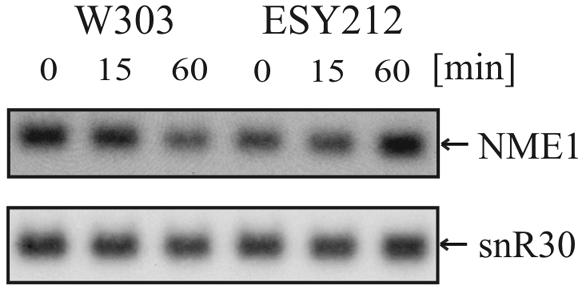
Short-term treatment with diazaborine does not affect electrophoretic mobility or steady-state levels of NME1 RNA and snR30 snoRNA. Cells from the strains W303 and ESY212 were incubated with diazaborine for 0, 15, and 60 min. The same Northern blot as that shown in Fig. 4 was hybridized with probes specific for NME1 RNA and snR30 snoRNA.
We also investigated whether the drug affects the steady-state level or the electrophoretic mobility of another important small RNA involved in rRNA processing. The snoRNA snR30 is involved in rRNA processing at sites A1 and A2. As shown in Fig. 8, no alteration in the electrophoretic mobility of snR30 RNA was observed in RNA extracted from the treated cells. This result shows that diazaborine does not exert a general effect on 3′ processing of snoRNAs. Moreover, the steady-state level of snR30 snoRNA was not affected by the inhibitor.
Diazaborine treatment results in relocalization of Nop4p/Nop77p to the nuclear periphery.
As mentioned above, our primer extension studies showed that diazaborine interferes with a very early step in the formation of the large ribosomal subunit. However, a survey of the literature shows that inactivation of most proteins involved in the early pre-27S rRNA processing pathway results in different processing patterns than that which we observed after diazaborine treatment. These mutants show either a strong decrease of the 27SA2 precursor or an accumulation of 27SA3 pre-rRNA which is accompanied by a shift in the ratio of 27SB1L to 27SB1S. Only depletion of Nop4p/Nop77p did not cause a decrease of 27SA2 pre-rRNA but resulted in a reduction of the 27SA3 and 27SB precursors without a shift in the ratio of B1L to B1S (4). This pattern of processing intermediates closely resembles that which we observed in the diazaborine-treated wild-type strain. Nop4p/Nop77p is an essential RNA binding protein which interacts with the fibrillarin Nop1p. Very recently it was shown that NOP4/NOP77 gene dosage reduction results in enhanced sensitivity to the anticancer drug 5-fluorouracil (18, 28).
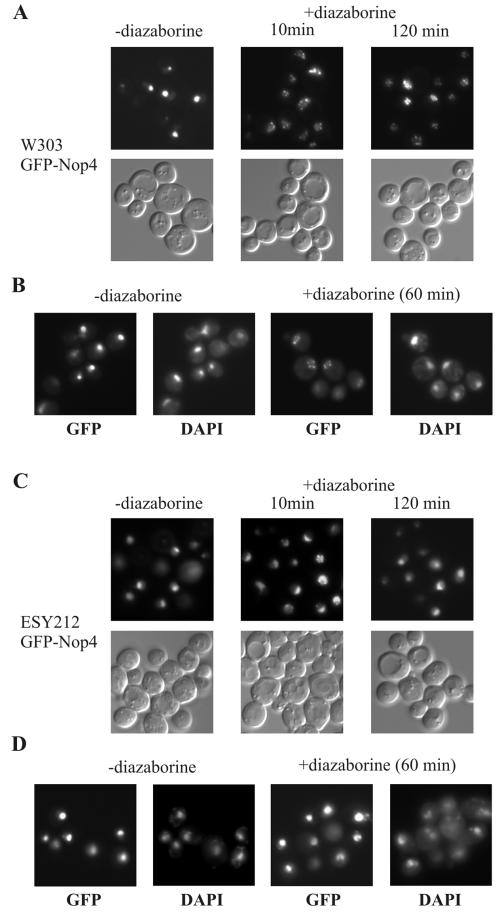
The correlation in the processing patterns of Nop4p/Nop77p-depleted and diazaborine-treated strains prompted us to investigate whether the inhibitor affects the localization of Nop4p/Nop77p. This protein is essential for 27SA2 pre-rRNA processing and was shown recently to be part of the pre-60S E1 complex (14). In line with these results we detected the GFP-Nop4 fusion mainly in the nucleolus of the untreated strain, although traces of the fusion protein also occurred in the nucleoplasm. The GFP-Nop4 fusion was fully functional and complemented the chromosomal nop4 deletion (data not shown). Incubation of the wild-type strain expressing the GFP-Nop4 fusion with 5 μg of diazaborine/ml resulted within a few minutes in a complete redistribution of the fusion protein into several small punctate structures. In contrast, treatment with 0.5 μg of cycloheximide/ml, which is also fully inhibitory under the tested conditions, did not result in a relocalization of the GFP-Nop4 fusion to the nuclear periphery (data not shown). The altered localization of the GFP-Nop4 fusion observed after diazaborine treatment is therefore not a general response to inhibition of protein synthesis or stress. Figure 9 shows the localization of the GFP-Nop4 fusion in the wild-type strain after 10 min of treatment with diazaborine. More than 96% of the cells showed a complete redistribution of the GFP-Nop4 fusion after this period. The punctate structures persisted after longer incubation in the presence of diazaborine when tested for up to 2 h. DAPI staining of cells that were treated with diazaborine for 1 h showed that these structures were located at the nuclear periphery. Thus, diazaborine treatment results in mislocalization of Nop4p/Nop77p from the nucleolus to the nuclear periphery. This finding provides an explanation for the similar rRNA processing patterns of strains depleted of Nop4p/Nop77p and diazaborine-treated wild-type strains.
FIG. 9.
Diazaborine leads to mislocalization of GFP-Nop4. The wild-type strain W303 or the diazaborine-resistant DRG1-1 mutant ESY212 expressing the GFP-Nop4 fusion was grown to the early log phase and treated with diazaborine. (A) W303 treated for 10 and 120 min with 5 μg of diazaborine/ml. Each field is represented by a fluorescence view along with that of Nomarski optics. (B) W303 cells treated with 5 μg of diazaborine/ml for 60 min were fixed as described in Materials and Methods, stained with DAPI, and viewed with the FITC and DAPI filters. (C) ESY212 was treated for 10 and 120 min with 5 μg of diazaborine/ml. (D) ESY212 cells treated with 5 μg of diazaborine/ml for 60 min were fixed, stained with DAPI, and viewed with the FITC and DAPI filters.
When the GFP-Nop4 fusion was expressed in the diazaborine-resistant strain ESY212, the fusion protein was also mainly located in the nucleolus (Fig. 9). After treatment of the resistant mutant for 2 h with diazaborine, the GFP-Nop4 fusion was still localized in the nucleolus in more than 80% of the cells. The rest of the cells showed punctate structures similar to those of the treated wild-type strain, but generally some fluorescence remained in the nucleolus.
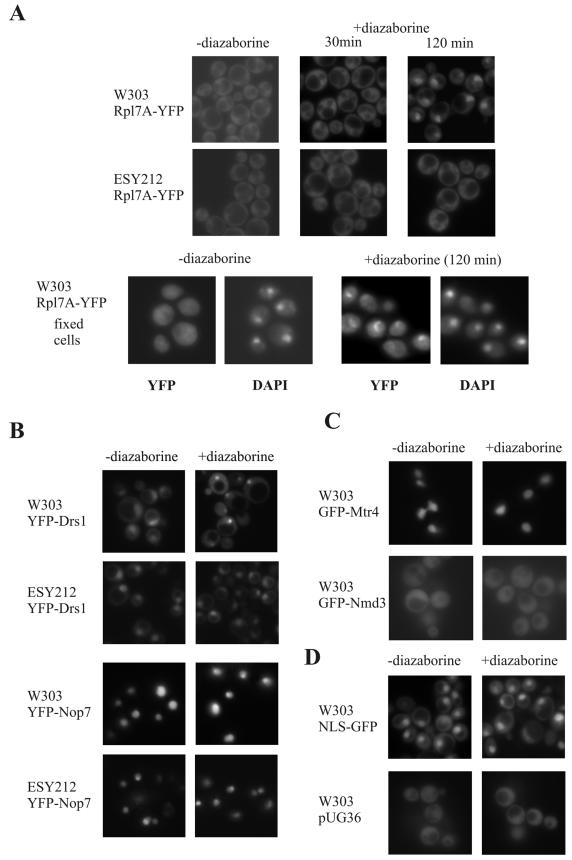
To investigate whether the punctate structures that we observed after diazaborine treatment in the nuclear periphery represent preribosome particles on their way from the nucleolus to the cytoplasm, we tested the effect of the drug on the localization of the ribosomal protein Rpl7A. This protein assembles early with the pre-rRNA and was detected in very early pre-60S particles (14, 37). W303 and ESY212 expressing a YFP-tagged version of Rpl7A exhibited fluorescence predominantly in the cytoplasm. After 30 min of treatment with diazaborine traces of the fusion protein were also detected in the nucleolus. The fluorescence in the nucleolus increased with longer incubation, resulting in a strong nucleolar accumulation of the fusion protein after 2 h (Fig. 10). This result indicates that the preribosome particles are retained in the nucleolus after diazaborine treatment. From the appearance of the Rpl7A-YFP and GFP-Nop4 fusions in different nuclear compartments after drug treatment, we conclude that the punctate structures do not represent preribosome particles on their way into the cytoplasm.
FIG. 10.
Long-term diazaborine treatment results in accumulation of Rpl7A-YFP, YFP-Drs1, and YFP-Nop7 in the nucleolus. The localization of GFP and YFP fusion proteins in untreated and diazaborine-treated wild-type and mutant cells was detected by fluorescence microscopy. Cells were treated with 5 μg of the inhibitor/ml. (A) W303 and ESY212 cells expressing Rpl7A-YFP were treated for 30 min or 2 h with diazaborine. DAPI staining of fixed cells is also shown. (B) YFP-Drs1 and YFP-Nop7 fusions expressed in W303 and ESY212. (C) For the Mtr4-GFP and Nmd3-GFP fusions only examples of the wild-type cells are shown, but the fusion proteins exhibited the same localization in the resistant mutant. (D) Cells with the empty plasmid pUG36 expressing GFP alone and cells expressing a nuclear localization signal-GFP fusion (pNLS) treated with diazaborine served as controls. Cells shown in panels B to D were treated for 120 min with the inhibitor.
The results from the fluorescence microscopy raise the question whether diazaborine treatment exerts a general effect on the localization of proteins involved in ribosome biogenesis. We therefore constructed GFP or YFP fusions with other proteins known to be involved in different steps in 60S subunit formation. Drs1p and Nop7p are known to act early in the 60S biogenesis pathway. Nop7p is the yeast pescadillo homologue and was shown to tandem affinity purification purify with several preribosomal particles, including the pre-60S E1, E2, and M complexes (20, 34). An allelic form of the NOP7 gene was found to be synthetically lethal with drs1 mutants (1). Drs1p is an RNA helicase that is involved in very early steps of 60S subunit formation (36). This protein is part of the pre-60S E1 and E2 complexes but not of the M complex (14). Drs1p therefore has to leave the preribosome particle at an earlier step than Nop7p does. Mtr4p and Nmd3p are proteins involved in late stages of processing and export. Mtr4p, also designated Dob1p, is a putative RNA helicase important for the activity of the exosome and is involved in generating the mature 3′ end of the 5.8S rRNA (12). Nmd3p is the export adaptor for the 60S subunit and binds to Rpl10p prior to export (16, 22). The W303 and ESY212 strains expressing GFP or YFP fusions of these proteins were investigated. As shown in Fig. 10 the YFP-Nop7p and the Mtr4p-GFP fusions were located exclusively in the nucleus. YFP-Nop7p clearly showed an increased level in the nucleolus, while the Mtr4-GFP fusion was uniformly distributed throughout the nucleoplasm. The YFP-Drs1p fusion was found in the nucleus, where it was enriched in the nucleolus, and it appeared in traces also in the cytoplasm. As described previously, the Nmd3-GFP fusion was located both in the nucleus and in the cytoplasm (22). When we explored the localization of the YFP and GFP fusion proteins after diazaborine treatment for short periods, no alterations were observed (data not shown). However, 2 h of drug treatment of the strains expressing the YFP-Drs1p and YFP-Nop7p fusions resulted in an accumulation of the fusion proteins in the nucleolus. The accumulation of the YFP-Drs1p fusion in the nucleolus after diazaborine treatment was accompanied by a strong decrease of the fluorescence in the nucleoplasm (Fig. 10). The accumulation of the YFP-Nop7p fusion in the nucleolus was less pronounced, and large amounts of the fusion protein were still present in the nucleoplasm. The reason for the nucleolar accumulation of these proteins after long-term diazaborine treatment is unknown. The localization of Nmd3-GFP and Mtr4-GFP was not affected by 2 h of treatment with the inhibitor.
When we expressed the YFP-Drs1p, YFP-Nop7p, Mtr4p-GFP, and Nmd3-GFP fusions in the diazaborine-resistant DRG1-1 mutant ESY212, the localizations of the fusion proteins were the same as for the wild-type strain. Moreover, no altered localization of the fusion proteins was observed after diazaborine treatment of ESY212 (Fig. 10 and data not shown).
DISCUSSION
By this work we demonstrate that diazaborine treatment of yeast cells causes an inhibition of processing of the rRNA precursors for the large ribosomal subunit. The defect in pre-60S rRNA processing leads to a depletion of free 60S subunits in the cytoplasm. Longer incubation in the presence of the inhibitor results in half-mer formation. Ribosome half-mers represent polysome populations containing a 43S preinitiation complex in addition to 80S ribosomes. Half-mers are often observed in mutants defective in 60S biogenesis and in pre-rRNA processing. Indeed, while the pulse-chase analyses demonstrated that the 27S pre-rRNA was formed normally in the treated strain, processing of the 27S precursor to the mature 25S rRNA, which is integrated into the large subunit, was inhibited by diazaborine treatment. Processing of the 20S pre-rRNA to the 18S rRNA, which is finally incorporated into the small ribosomal subunit, was only slightly affected by the inhibitor. The different effects of diazaborine on the formation of the pre-rRNAs for the large and small ribosomal subunits are consistent with reports in the literature which show that different factors are necessary for maturation of the two ribosomal subunits (15, 19, 47). The results from our rRNA processing studies are in line with the results from Northern blotting, which showed that the steady-state level of the 7S precursor is reduced in the treated cells. Northern blotting also showed an accumulation of 35S pre-rRNA in the presence of the inhibitor. Because our pulse-chase experiments demonstrated that the 27S precursor is formed with similar kinetics in treated and untreated strains, we conclude that the accumulation of the 35S precursor observed in our Northern blots is a consequence of the block in 27S processing. Increased levels of the 35S pre-rRNA are often observed in mutants in the 60S subunit biogenesis pathway (reviewed in reference 50). Longer incubation with diazaborine resulted in a drop of the 27S pre-rRNA level. This decrease was accompanied by a slight reduction in 20S pre-RNA. The decline of the 20S pre-rRNA is to be expected as a consequence of the buildup of the 35S precursor. Moreover, no significant increase in formation of the 23S aberrant processing product was observed upon drug treatment, which would be indicative of a block in 5′-ETS processing.
A detailed exploration of the processing pathway by primer extension analyses showed that diazaborine inhibits the conversion of 27SA2 to 27SA3. The cleavage of the 27SA2 precursor at site A3 is known to be accomplished by MRP RNase. This RNase is related to RNase P and is composed of an RNA component encoded by the NME1 locus and several proteins (7). It was demonstrated previously that mutations in genes coding for the protein components of MRP affect the stability and the steady-state level of the NME1 RNA (8, 13, 31, 39, 42, 43). We tested whether the amount of NME1 RNA is affected by diazaborine. No significant reduction in the RNA level was observed after brief incubation with the inhibitor. We therefore conclude that the MRP endonuclease is not affected by the drug. Moreover, the pre-rRNA processing pattern described for mutants in NME1 RNA or for the protein components of the MRP nuclease shows an increase of the B1L species, which is different from the pattern that we describe here (6, 8, 13, 31, 39, 40, 42, 43).
It was observed previously that diazaborine treatment of yeast strains results in the accumulation of aberrant 3′-elongated mRNAs (25). Such aberrant mRNAs were also observed in the untreated Δxrn1 strain. Since Xrn1p is also involved in rRNA processing, the possibility exists that the drug interferes with the activity of this enzyme. Xrn1p and its homologue Rat1p perform the exonucleolytic trimming of the 27SA3 precursor to the 27SB1S pre-rRNA (21). However, the rRNA processing pattern observed in xrn1 and rat1 mutants shows a typical ladder of processing intermediates characteristic for exonucleases and is clearly different from that of the diazaborine-treated strain (21). Moreover, we did not observe an accumulation of the D-A2 spacer fragment in drug-treated strains. This fragment is known to be degraded by Xrn1p, and consequently, it should be increased if the protein were inhibited (35, 41). The effect of the drug on pre-rRNA processing can therefore not be attributed to inhibition of Xrn1p.
The pattern of processing intermediates which we found after diazaborine treatment, i.e., a strong decrease of the 27SA3, 27SB1L, 27SB1S, and 7S intermediates and an increase of the 27SA2 pre-rRNA, is indicative of a block at a very early step in the 60S processing pathway. The earliest known preribosome particle for the large ribosomal subunit is the pre-60S E1 complex. This E1 complex contains the 27SA2 intermediate and several known 60S processing factors, like Ssf1p, Dbp9p, and Nop4p/Nop77p, which were not detected in the later E2 complex (14). For strains lacking these proteins the processing of the rRNA precursors was monitored (4, 11, 14). Depletion of both Ssf1p and Dbp9p resulted in a decrease in the 27SA2 rRNA precursor and subsequent processing intermediates. The processing pattern in these strains is therefore different from what we observed after diazaborine treatment.
The processing pattern reported upon Nop4p/Nop77p depletion closely resembles that which we observed after diazaborine treatment. Shutoff of the NOP4/NOP77 gene resulted in disappearance of the 27SA3 precursor, while the 27SA2 pre-rRNA level remained constant (4). Also a reduction of the 27SB1L and 27SB1S intermediates was observed, and the ratio between these two precursors did not change (4). To our knowledge this behavior is unique for the Nop4p/Nop77p-depleted strain, because most other mutants that are defective in 27S pre-rRNA processing exhibit a pronounced shift in the ratio of 27SB1L and 27SB1S when transferred to the restrictive temperature.
Nop4p/Nop77p is an RNA binding protein containing four RNA binding domains which is essential for rRNA processing (4, 44, 45). This protein is thought to mediate a specific interaction between Nop1p and pre-rRNA (4). As with the depletion of Nop4p/Nop77p, diazaborine treatment resulted in a reduction in the levels of B1L and B1S, keeping the ratio between the two precursors constant. As mentioned above, B1L and B1S are generated by alternative pathways diverging at the 27SA2 pre-rRNA. B1S results from cleavage at A3 and an exonucleolytic trimming reaction. In contrast, the alternative trimming pathway leading to B1L, which accounts for approximately 15% of the pre-27SB intermediates, skips the cleavage at A3. The similar processing patterns of yeast depleted of Nop4p/Nop77p and of diazaborine-treated cells suggest that a processing or assembly step of the pre-60S subunit which depends on Nop4p/Nop77p is affected by the drug.
We found that diazaborine treatment resulted within minutes in a redistribution of a GFP-Nop4 fusion from the nucleolus into several punctate structures at the periphery of the nucleus. The nature of these structures is unknown, but they do not represent a fragmented nucleolus, since other nucleolar proteins, like Drs1p or Nop7p, did not exhibit such behavior. The location at the periphery of the nucleus indicates that these structures might be connected with the nuclear membrane. We therefore tested whether the punctate structures represent preribosomal particles on the way from the nucleus to the cytoplasm. A fusion with the ribosomal protein Rpl7A, which assembles early with the pre-rRNA, did not show punctate structures in the nuclear periphery but instead accumulated in the nucleolus after diazaborine treatment. This finding indicates that the preribosome particles are retained in the nucleolus after drug treatment. Since the nucleolus is devoid of Nop4p/Nop77p under the same conditions, as our microscopy data show, the protein cannot interact with newly synthesized preribosomal particles to perform its physiological function in 27S pre-rRNA maturation. This might explain the similar processing patterns of drug-treated cells and cells depleted of Nop4p/Nop77p. The block in 27S pre-rRNA maturation leads to the retention of the pre-60S ribosomal particles in the nucleolus, which finally results in the imbalance of ribosomal subunits that we observed in our polysome profiles.
Acknowledgments
We thank Roger Schneiter, Ernst Müllner, and Günther Koraimann for helpful discussions; Dale Hailey and David S. Goldfarb for the plasmids pDH5 and pNLS-GFP; and Johannes H. Hegemann for the plasmids pUG35 and pUG36. We thank the Novartis Research Institute, Vienna, Austria, for a gift of diazaborine and Friederike Turnowsky for critically reading the manuscript.
This work was supported by grant no. P15458 of the Austrian Science Foundation (FWF).
REFERENCES
- 1.Adams, C. C., J. Jakovljevic, J. Roman, P. Harnpicharnchai, and J. L. Woolford, Jr. 2002. Saccharomyces cerevisiae nucleolar protein Nop7p is necessary for biogenesis of 60S ribosomal subunits. RNA 8:150-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M. (ed.). 1988. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Bassler, J., P. Grandi, O. Gadal, T. Lessmann, E. Petfalski, D. Tollervey, J. Lechner, and E. Hurt. 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell 8:517-529. [DOI] [PubMed] [Google Scholar]
- 4.Berges, T., E. Petfalski, D. Tollervey, and E. C. Hurt. 1994. Synthetic lethality with fibrillarin identifies NOP77p, a nucleolar protein required for pre-rRNA processing and modification. EMBO J. 13:3136-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergler, H., P. Wallner, A. Ebeling, B. Leitinger, S. Fuchsbichler, H. Aschauer, G. Kollenz, G. Högenauer, and F. Turnowsky. 1994. Protein EnvM is the NADH-dependent enoyl-ACP reductase (FabI) of Escherichia coli. J. Biol. Chem. 269:5493-5496. [PubMed] [Google Scholar]
- 6.Cai, T., T. R. Reilly, M. Cerio, and M. E. Schmitt. 1999. Mutagenesis of SNM1, which encodes a protein component of the yeast RNase MRP, reveals a role for this ribonucleoprotein endoribonuclease in plasmid segregation. Mol. Cell. Biol. 19:7857-7869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chamberlain, J. R., Y. Lee, W. S. Lane, and D. R. Engelke. 1998. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 12:1678-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu, S., J. M. Zengel, and L. Lindahl. 1997. A novel protein shared by RNase MRP and RNase P. RNA 3:382-391. [PMC free article] [PubMed] [Google Scholar]
- 9.Chu, S., R. H. Archer, J. M. Zengel, and L. Lindahl. 1994. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc. Natl. Acad. Sci. USA 91:659-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cote, C. A., and B. A. Peculis. 2001. Role of the ITS2-proximal stem and evidence for indirect recognition of processing sites in pre-rRNA processing in yeast. Nucleic Acids Res. 29:2106-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daugeron, M. C., D. Kressler, and P. Linder. 2001. Dbp9p, a putative ATP-dependent RNA helicase involved in 60S-ribosomal-subunit biogenesis, functionally interacts with Dbp6p. RNA 7:1317-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Cruz, J., D. Kressler, D. Tollervey, and P. Linder. 1998. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 17:1128-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dichtl, B., and D. Tollervey. 1997. Pop3p is essential for the activity of the RNase MRP and RNase P ribonucleoproteins in vivo. EMBO J. 16:417-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fatica, A., A. D. Cronshaw, M. Dlakic, and D. Tollervey. 2002. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell 9:341-351. [DOI] [PubMed] [Google Scholar]
- 15.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 16.Gadal, O., D. Strauss, J. Kessl, B. Trumpower, D. Tollervey, and E. Hurt. 2001. Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21:3405-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geerlings, T. H., J. C. Vos, and H. A. Raue. 2000. The final step in the formation of 25S rRNA in Saccharomyces cerevisiae is performed by 5′→3′ exonucleases. RNA 6:1698-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giaever, G., P. Flaherty, J. Kumm, M. Proctor, C. Nislow, D. F. Jaramillo, A. M. Chu, M. I. Jordan, A. P. Arkin, and R. W. Davis. 2004. Chemogenomic profiling: identifying the functional interactions of small molecules in yeast. Proc. Natl. Acad. Sci. USA 101:793-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grandi, P., V. Rybin, J. Bassler, E. Petfalski, D. Strauss, M. Marzioch, T. Schafer, B. Kuster, H. Tschochner, D. Tollervey, A. C. Gavin, and E. Hurt. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10:105-115. [DOI] [PubMed] [Google Scholar]
- 20.Harnpicharnchai, P., J. Jakovljevic, E. Horsey, T. Miles, J. Roman, M. Rout, D. Meagher, B. Imai, Y. Guo, C. J. Brame, J. Shabanowitz, D. F. Hunt, and J. L. Woolford, Jr. 2001. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell 8:505-515. [DOI] [PubMed] [Google Scholar]
- 21.Henry, Y., H. Wood, J. P. Morrissey, E. Petfalski, S. Kearsey, and D. Tollervey. 1994. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 13:2452-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho, J. H., G. Kallstrom, and A. W. Johnson. 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 151:1057-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes, J. M., and M. Ares, Jr. 1991. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 10:4231-4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jungwirth, H., F. Wendler, B. Platzer, H. Bergler, and G. Högenauer. 2000. Diazaborine resistance in yeast involves the efflux pumps Ycf1p and Flr1p and is enhanced by a gain-of-function allele of gene YAP1. Eur. J. Biochem. 267:4809-4816. [DOI] [PubMed] [Google Scholar]
- 25.Jungwirth, H., H. Bergler, and G. Högenauer. 2001. Diazaborine treatment of baker's yeast results in stabilization of aberrant mRNAs. J. Biol. Chem. 276:36419-36424. [DOI] [PubMed] [Google Scholar]
- 26.Kufel, J., B. Dichtl, and D. Tollervey. 1999. Yeast Rnt1 is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA 5:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, S. J., and S. J. Baserga. 1999. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol. Cell. Biol. 19:5441-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lum, P. Y., C. D. Armour, S. B. Stepaniants, G. Cavet, M. K. Wolf, J. S. Butler, J. C. Hinshaw, P. Garnier, G. D. Prestwich, A. Leonardson, P. Garrett-Engele, C. M. Rush, M. Bard, G. Schimmack, J. W. Phillips, C. J. Roberts, and D. D. Shoemaker. 2004. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell 116:121-137. [DOI] [PubMed] [Google Scholar]
- 29.Lupas, A. N., and J. Martin. 2002. AAA proteins. Curr. Opin. Struct. Biol. 12:746-753. [DOI] [PubMed] [Google Scholar]
- 30.Lygerou, Z., C. Allmang, D. Tollervey, and B. Seraphin. 1996. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science 272:268-270. [DOI] [PubMed] [Google Scholar]
- 31.Lygerou, Z., P. Mitchell, E. Petfalski, B. Seraphin, and D. Tollervey. 1994. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev. 8:1423-1433. [DOI] [PubMed] [Google Scholar]
- 32.Milkereit, P., O. Gadal, A. Podtelejnikov, S. Trumtel, N. Gas, E. Petfalski, D. Tollervey, M. Mann, E. Hurt, and H. Tschochner. 2001. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell 105:499-509. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell, P., E. Petfalski, and D. Tollervey. 1996. The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev. 10:502-513. [DOI] [PubMed] [Google Scholar]
- 34.Nissan, T. A., J. Bassler, E. Petfalski, D. Tollervey, and E. Hurt. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21:5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petfalski, E., T. Dandekar, Y. Henry, and D. Tollervey. 1998. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 18:1181-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ripmaster, T. L., G. P. Vaughn, and J. L. Woolford, Jr. 1993. DRS1 to DRS7, novel genes required for ribosome assembly and function in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:7901-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saveanu, C., A. Namane, P. E. Gleizes, A. Lebreton, J. C. Rousselle, J. Noaillac-Depeyre, N. Gas, A. Jacquier, and M. Fromont-Racine. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 23:4449-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schäfer, T., D. Strauss, E. Petfalski, D. Tollervey, and E. Hurt. 2003. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 22:1370-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitt, M. E., and D. A. Clayton. 1993. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:7935-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shuai, K., and J. R. Warner. 1991. A temperature sensitive mutant of Saccharomyces cerevisiae defective in pre-rRNA processing. Nucleic Acids Res. 19:5059-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevens, A., C. L. Hsu, K. R. Isham, and F. W. Larimer. 1991. Fragments of the internal transcribed spacer 1 of pre-rRNA accumulate in Saccharomyces cerevisiae lacking 5′→3′ exoribonuclease 1. J. Bacteriol. 173:7024-7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stolc, V., and S. Altman. 1997. Rpp1, an essential protein subunit of nuclear RNase P required for processing of precursor tRNA and 35S precursor rRNA in Saccharomyces cerevisiae. Genes Dev. 11:2926-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stolc, V., A. Katz, and S. Altman. 1998. Rpp2, an essential protein subunit of nuclear RNase P, is required for processing of precursor tRNAs and 35S precursor rRNA in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95:6716-6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun, C., and J. L. Woolford, Jr. 1994. The yeast NOP4 gene product is an essential nucleolar protein required for pre-rRNA processing and accumulation of 60S ribosomal subunits. EMBO J. 13:3127-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun, C., and J. L. Woolford, Jr. 1997. The yeast nucleolar protein Nop4p contains four RNA recognition motifs necessary for ribosome biogenesis. J. Biol. Chem. 272:25345-25352. [DOI] [PubMed] [Google Scholar]
- 46.Thorsness, P. E., K. H. White, and W. C. Ong. 1993. AFG2, an essential gene in yeast, encodes a new member of the Sec18p, Pas1p, Cdc48p, TBP-1 family of putative ATPases. Yeast 9:1267-1271. [DOI] [PubMed] [Google Scholar]
- 47.Tschochner, H., and E. Hurt. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13:255-263. [DOI] [PubMed] [Google Scholar]
- 48.Turnowsky, F., K. Fuchs, C. Jeschek, and G. Högenauer. 1989. envM genes of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 171:6555-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Udem, S. A., and J. R. Warner. 1973. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J. Biol. Chem. 248:1412-1416. [PubMed] [Google Scholar]
- 50.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 51.Wendler, F., H. Bergler, K. Prutej, H. Jungwirth, G. Zisser, K. Kuchler, and G. Högenauer. 1997. Diazaborine resistance in the yeast Saccharomyces cerevisiae reveals a link between YAP1 and the pleiotropic drug resistance genes PDR1 and PDR3. J. Biol. Chem. 272:27091-27098. [DOI] [PubMed] [Google Scholar]
- 52.Zakalskiy, A., G. Högenauer, T. Ishikawa, E. Wehrschütz-Sigl, F. Wendler, D. Teis, G. Zisser, A. C. Steven, and H. Bergler. 2002. Structural and enzymatic properties of the AAA protein Drg1p from Saccharomyces cerevisiae. Decoupling of intracellular function from ATPase activity and hexamerization. J. Biol. Chem. 277:26788-26795. [DOI] [PubMed] [Google Scholar]