Abstract
The members of the plakin family of proteins serve as epidermal cytolinkers and components of cell-cell and cell-matrix adhesion complexes, i.e., desmosomes and hemidesmosomes, respectively. Periplakin is a recently characterized member of this family. Human and mouse periplakin genomic loci are conserved, and the proteins are highly homologous, suggesting a role for periplakin in vertebrate physiology. In order to evaluate the functional role of periplakin, we generated periplakin null mice through targeted homologous recombination of mouse embryonic stem cells, followed by development of Ppl−/− mice. Mice homozygous for the targeted allele were born in the expected Mendelian frequency, developed normally, possessed grossly normal epidermis and hair, and were healthy and fertile. The epidermal barrier appeared to develop normally during fetal days E15.5 to E16.5, and the cornified envelope and desmosomes in the newborn mice were ultrastructurally normal. No compensatory increase in the expression of other epithelial proteins was detected in the neonatal mouse epidermis lacking periplakin. Consequently, the primary role of periplakin may not relate to the physiology of the cornified cell envelope in epidermal keratinocytes but may reside in the challenges, which normal laboratory mice do not encounter.
Periplakin is one of the five plakin proteins which share a common structure with a central rod domain flanked by N- and C-terminal globular domains; the other members of this family are desmoplakin, plectin, envoplakin, and the 230-kDa bullous pemphigoid antigen (BPAG1) (for reviews, see references 16, 21, 31, 32, and 43). In spite of the structural homology, each plakin protein displays a characteristic tissue-specific expression pattern and subcellular distribution, suggesting a distinct functional role for each individual protein.
Plectin, a versatile cytoskeletal linker protein, has an important role in maintaining the structural integrity of diverse cells and tissues, and it has been detected in desmosomal, hemidesmosomal, and perinuclear localizations in epidermal keratinocytes (7, 45, 55). Plectin deficiency in mice causes a blistering skin phenotype, abnormalities in skeletal muscle, and disintegration of intercalated disks in the heart, resulting in neonatal death at the age of 2 to 3 days (8). Hemidesmosomes have been found to be significantly reduced in number in plectin-deficient mice, indicating the role of plectin in the stability of hemidesmosomes. Human autosomal recessive disorder epidermolysis bullosa with muscular dystrophy results from defective or absent plectin due to the mutations in the plectin gene (PLEC1) (52).
BPAG1 was characterized as a major target for autoantibodies in the autoimmune blistering disease, bullous pemphigoid (38), but no human mutations have been identified as yet. The epithelial isoform of this protein is the intracellular component of hemidesmosomes, while the neuronal isoform functions as an essential cytoskeletal linker protein connecting actin microfilaments to intermediate filaments (57). Gene targeting of BPAG1 in mice results in hemidesmosomes which lack the inner plate and have no cytoskeleton attached (19). Mice also develop severe dystonia and sensory nerve degeneration due to the lack of the neural isoform binding and stabilizing microtubules (11, 56).
While plectin decorates intermediate filament (IF) networks and is a component of hemidesmosomes in stratified epithelia, desmoplakin connects IF to desmosomes. Desmoplakin, an integral component of inner desmosomal plague in a wide variety of tissues and cells, is essential for embryonic development (54). Mouse embryos deficient in desmoplakin proceed through implantation but do not survive beyond E6.5 due to major defects in the heart muscle and neuroepithelium (17, 18). In humans, a recessive mutation affecting desmoplakin tail and disrupting desmoplakin-IF interactions is not embryonic lethal but causes dilated cardiomyopathy, woolly hair, and keratoderma (40).
Envoplakin and periplakin, together with involucrin, were first characterized as precursors of the cornified cell envelope (CE) (44). Before the final assembly of the CE, envoplakin and periplakin have been detected in a soluble cytoplasmic pool, although these proteins mainly localize to desmosomes and interdesmosomal plasma membrane (47). No genetic disease has been found in humans due to the mutations in the genes encoding involucrin, envoplakin, or periplakin. However, all five plakin proteins, but most consistently envoplakin and periplakin, are recognized by circulating autoantibodies in patients with the autoimmune disease paraneoplastic pemphigus (9, 10, 35).
Although loricrin is a major protein of the epidermal CE, loricrin-deficient mice develop normally after a short delay in epithelial barrier development (28). As a surprise, involucrin knockout mice did not show any discernible phenotype (13), and the generation of envoplakin knockout mice revealed that this protein also is dispensable for normal viability (34).
The CE has been thought to play a crucial role in the epidermal barrier function, since it provides a rigid but flexible support to terminally differentiated keratinocytes in the cornified cell layer (23). Loricrin, involucrin, and envoplakin, as well as periplakin, become cross-linked to ceramides, the constituents of lipid layers, which provide the water barrier for the epidermis (36). Mice deficient for the sphingolipid activator proteins have revealed the importance of the extracellular lipid membranes for normal epidermal barrier formation and function (14). Transglutaminase 1 (TGase 1)-dependent cross-linking of proteins and lipids (39) is indispensable for normal epidermal development (27). TGase 1 is one of the genes implicated in autosomal recessive congenital ichthyosis in humans. Skin from TGase 1−/− mice, which die as neonates, lacks the normal insoluble cornified envelope and has impaired barrier function (30). At least 20 proteins become assembled into the CEs, and accordingly, it has been speculated that the cornified envelope assembly is guaranteed through compensatory mechanisms and the redundancy of its components (46).
Here, we report the generation of mice deficient in periplakin. Surprisingly, periplakin deficiency did not result in a discernible phenotype.
MATERIALS AND METHODS
Cloning of mouse periplakin gene and construction of the targeting vector.
Overlapping lambda clones spanning the mouse periplakin gene were sequentially isolated from a 129/Sv Lambda FIX II genomic DNA library (Stratagene) using a human periplakin 3′-cDNA fragment as an initial probe. The overlapping regions were identified through restriction enzyme digests, and the most 5′ fragments were used as probes for further library screenings. The entire periplakin gene was covered by five overlapping lambda clones. Exons and the immediate intervening sequences, as well as the 5′- and 3′-flanking regions, were subjected to direct DNA sequencing. A 4.5-kb insert in lambda clone 7, containing periplakin exons 13 to 16 with flanking intronic sequences, was released by digestion with NotI and XhoI. The insert was ligated to NotI-XhoI-digested pPNT vector (51). The resulting plasmid with the 5′ arm was digested with XbaI-EcoRI, and an internal 1.5-kb XbaI-EcoRI fragment of lambda clone 5 containing periplakin exon 22 was inserted there to serve as a 3′ arm. Prior to electroporation into the embryonic stem cells, the targeting plasmid was linearized by utilizing the NotI site located in the cloning vector at the 5′ side of the 5′ arm.
Generation of knockout mouse.
The NotI-linearized targeting vector (25 μg) was electroporated into the mouse embryonic stem (ES) cells (5 × 107 cells) cultured under standard conditions (29). Clones were selected with G418 (250 μg/ml) and with 1 μM ganciclovir (Sigma, St. Louis, Mo.). This strategy selects those cells which have undergone homologous recombination events by simultaneously selecting for a neo gene and against a herpes simplex virus thymidine kinase gene placed at the end of the targeting vector (51). The selected ES clones were screened by PCR and Southern blotting for correct homologous recombination.
ES cells were microinjected into blastocysts obtained from C57BL/6J mice (Jackson Laboratories, Bar Harbor, Maine) and transferred to pseudopregnant B6CBA F1 mice (Jackson Laboratories). The chimeric animals were mated with wild-type C57BL/6J mice to determine the germ line transmission of the targeted Ppl allele. Heterozygotes were crossbred to produce Ppl−/− homozygous animals. The use of mice was approved by the Institutional Animal Care and Use Committee of Thomas Jefferson University.
Genotyping of the ES cells and mice.
Both the ES cell clones and the animals were genotyped by using Southern blotting and/or PCR. For this purpose DNA from cells or mouse tissues was extracted by using standard protocols. Southern blotting was performed with genomic DNA digested with SacI restriction endonuclease and using a [32P]dCTP-labeled 1.3-kb PCR product as a probe. Primers for the amplification of the probe were 5′CTA TAA TTC CAA GTC ATC CAA GG-3′ (forward) and 5′-GCC ATA AAG TTC CCT ATC ACT-3′ (reverse). This probe recognizes a 7.5-kb SacI-fragment from the wild-type mouse DNA and a 4-kb SacI-fragment from the targeted allele.
In addition to Southern blotting, PCR was applied for genotyping of littermates. For this purpose, three primers were designed for the amplification of the mutant, wild type, or both alleles in the same reaction. Mouse periplakin exon 16-specific forward primer 5′-CTC ATA CGA GAA CAG GCT G-3′ and exon 17-specific reverse primer 5′-CTG CTT GGC CAC CTG TAG-3′ produce a 982-bp PCR product from the wild-type allele, while the exon 16-specific forward primer together with a PGK promoter-specific reverse primer, 5′-CCA GAG GCC ACT TGT GTA G-3′, produce a 602-bp PCR product from the targeted allele.
Western analysis.
A piece of newborn mouse dorsal skin was mixed with 150 μl of urea buffer (50 mM Tris-HCl [pH 7.4], 8 M urea, 1% Triton X-100), sonicated on ice, and cleared through centrifugation for 15 min at 10,000 × g in a microcentrifuge. An aliquot of the supernatant was mixed with an equal volume of Laemmli sample buffer (Bio-Rad) supplemented with β-ercaptoethanol. For Western analysis, proteins were separated either by sodium dodecyl sulfate (SDS)-5% polyacrylamide gel electrophoresis or by 4 to 20% gradient polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Perkin-Elmer Life Sciences). The membranes were blocked for 1 h at room temperature (RT) with 1% bovine serum albumin (BSA) and 5% nonfat dried milk in phosphate-buffered saline (PBS). After the primary antibody incubation in 1% BSA in PBS overnight at 8°C, the membranes were washed four times, for 10 min each wash, in 0.5% Tween 20 in Tris-buffered saline, followed by incubations for 1 h at RT with the secondary antibody, and then washed again. The signal was developed using Western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer Life Sciences).
Preparation of cornified envelopes.
Pieces of newborn mouse dorsal skin (5 mm by 5 mm) were heated to 100°C for 20 min in CE isolation buffer containing 20 mM Tris-HCl (pH 7.5), 5 mM EDTA, 10 mM dithiotreitol, and 2% SDS. Cornified envelopes were pelleted by a 10-min centrifugation at 6,000 × g, washed in CE isolation buffer with 0.2% SDS, repelleted, and resuspended in the washing buffer. Envelopes were examined in a hemacytometer under phase microscopy.
Electron microscopy.
Samples of neonatal mouse skin were fixed in half-strength Karnovsky fixative, and after secondary fixation in 1% osmium tetroxide, they were dehydrated in ethanol, stained en bloc using 1% uranyl acetate in 50% ethanol, and embedded in Epon resin (TAAB, Aldermaton, Berks, United Kingdom). Ultra-thin sections were stained with 1.5% uranyl acetate in methanol and with Reynold's lead citrate.
Dye exclusion assay.
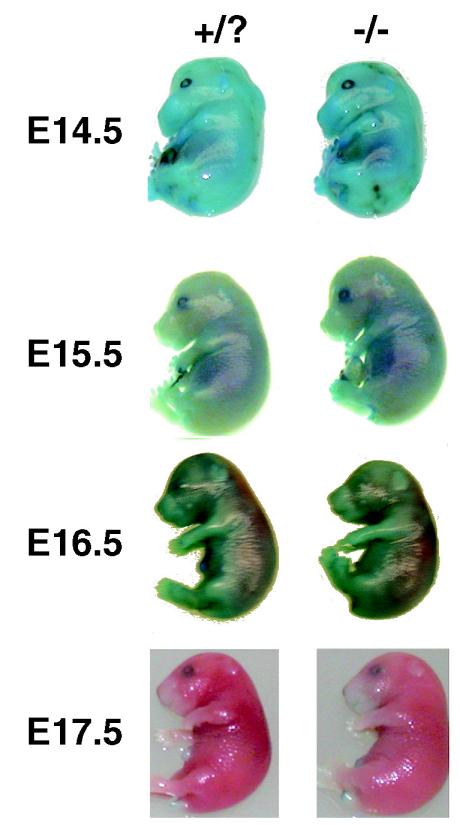
Periplakin heterozygous breeder mice were used to produce embryos to analyze skin water barrier formation by the dye exclusion assay. Embryos were harvested from pregnant females at the embryonic stages indicated in Fig. 5, the tail was removed for genotyping, and the dye exclusion assay was performed as described previously (20, 28, 34), using 0.0125% toluidine blue O (Fisher Scientific Corp, Pittsburgh, Pa.).
FIG. 5.
Dye exclusion analysis of skin barrier formation in mouse embryos from periplakin heterozygous mouse matings. The column of numbers on the left indicates the age (E14.5, E15.5, E16.5, or E17.5) of the embryos, shown to the right, where the day of the vaginal plug is considered to be E0.5. The left column of embryos depicts dye exclusion results from wild-type or heterozygous embryos (+/?; only the E16.6 embryo is heterozygous, while all others were wild type), and the right column depicts results from periplakin null embryos (−/−). Embryo pictures shown depict typical results for the pups from the indicated litters.
Immunofluorescence microscopy.
Tissue from 1- to 2-day-old mice were embedded in OCT medium and cut into 7-μm cryosections. For indirect immunofluorescence, slides were fixed with methanol for 5 to 10 min and, after 2 washes with PBS, were further permeabilized with 0.1% Triton X-100 in PBS for 5 min at RT, followed by three washes with PBS. Blocking was done with 1% BSA in PBS for 1 h at RT, followed by the primary antibody incubation at 8°C overnight. Slides were washed three times with PBS, each time for 5 min, followed by the secondary antibody incubation at RT for 1 h. After three washes with PBS, slides were mounted and studied under a fluorescence microscope (Axioskop, Carl Zeiss, Inc). The images were stored with ImagePro Plus 4.0 imaging software (Media Cybernetics) and processed with Photoshop 7.0 (Adobe Systems Inc.) and Canvas 7 (Deneba Software).
Antibodies.
A hybridoma culture supernatant containing mouse monoclonal antibody against the C-terminal region of the periplakin rod domain (PPL-488), recognizing an epitope within periplakin amino acids (aa) 1548 to 1583, was used in 1:10 dilution (1, 25). Affinity-purified rabbit polyclonal antienvoplakin antibody was a kind gift from L. Marekov and A. Kalinin and has been previously described (47). The following commercially available primary antibodies were used: goat antiplectin antibody against the C terminus of plectin (1:500; clone C20; Santa Cruz Biotechnology); rabbit antidesmoplakin antibody against the C-terminal nucleotides 4247 to 5228 (1:10,000; Serotech Ltd., Raleigh, N.C.); anti-involucrin monoclonal antibody (MAb) (1:2,000; NeoMarkers); antiactin MAb (1:10,000; Boehringer/Roche). Secondary antibodies were purchased from Jackson Laboratories. The horseradish peroxidase-conjugated anti-mouse, anti-goat, and anti-rabbit antibodies, as appropriate, were diluted 1:25,000 for the Western blotting. Species-specific secondary antibodies, conjugated with Texas Red and fluorescein isothiocyanate, were also purchased from Jackson Laboratories and used for indirect immunofluorescence in 1:500 dilution in PBS-1% BSA.
Nucleotide sequence accession numbers.
Mouse periplakin gene sequences determined in this study were deposited in GenBank under accession numbers AF116519 to AF116523.
RESULTS
Cloning and characterization of the mouse periplakin gene.
The gene encoding mouse periplakin was isolated in five overlapping lambda clones. Restriction enzyme mapping combined with the DNA sequencing of the exons, exon-intron junctions, and intron sequences immediately flanking the exons, revealed that the overall organization of human (4, 5) and mouse periplakin gene loci is similar. Both genes are composed of 22 exons, and the exon-intron distribution follows the same pattern for the length of exons and the split codons, but mouse introns are generally shorter, resulting in a more compact gene. Specifically, mouse periplakin gene was estimated to span more than 50 kb of genomic sequences, while human periplakin gene spans about 110 kb. Similar to the human promoter (5), mouse periplakin promoter does not contain CAT or TATA boxes but is specific for epithelial cells and is not functional in fibroblasts. The 3′ untranslated region in both genes ends in two AATAAA polyadenylation consensus sequences. Both human (2) and mouse periplakin genes are located in tail-to-tail orientation with the ubinuclein gene, separated only by about 200 bp of genomic sequences.
Gene targeting of periplakin locus.
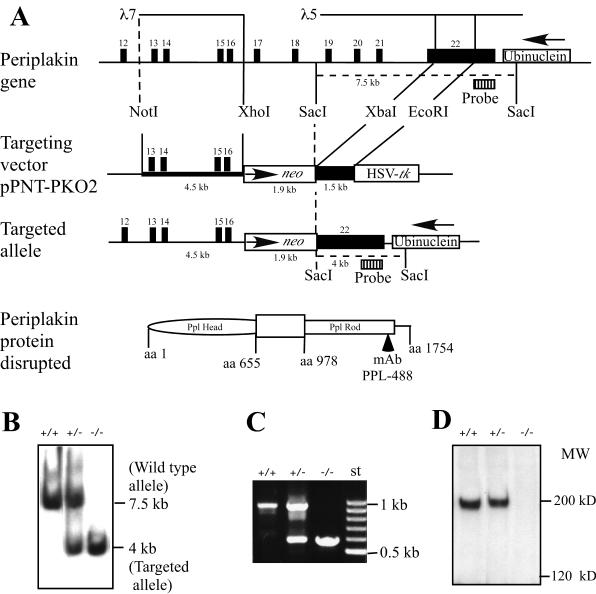
To generate periplakin knockout mice, we constructed a targeting vector from a 4.5-kb lambda clone insert, encoding exons 13 to 16, serving as a 5′ arm (Fig. 1A). To avoid the possibility of disturbing the ubinuclein gene, an internal fragment from periplakin exon 22 was used as the 3′ arm. The homologous recombination of the targeting construct results in deletion of an 8-kb XhoI-XbaI fragment, removing periplakin exons 17 to 21 and a part of exon 22, replacing those sequences with a 1.9-kb neo gene under the control of PGK1 promoter (Fig. 1A). Out of 112 clones obtained through the positive-negative selection protocol, Southern blotting revealed that 5 were correctly targeted. All five independent clones were injected into C57BL/6 blastocycts. The chimeric animals were crossed to C57BL/6J mice, and the litters obtained from heterozygous crossings were genotyped through Southern blotting and PCR (Fig. 1B and C). Crossing of the heterozygous animals produced all genotypes in the expected Mendelian ratio. Out of 59 mice born in 7 litters, 16 homozygotes (27%), 28 heterozygotes (48%), and 15 wild-type (25%) mice were obtained.
FIG. 1.
Gene targeting of periplakin locus. (A) Schematic diagram of the mouse periplakin gene (exons 12 to 22) and the position of the lambda clones used for the construction of the targeting vector. The homologous recombination results in the replacement of mouse periplakin exons 17 to 21 and part of exon 22 with the neomycin gene are shown. Black boxes represent periplakin exons, and the open boxes represent ubinuclein last exon (top) and the neo casette and herpes simplex virus thymidine kinase gene (middle line), as indicated. Thus, the 7.5-kb SacI fragment in the wild-type periplakin gene is replaced by a 4-kb SacI fragment in the targeted allele. Gene disruption alters periplakin reading frame, abolishing periplakin protein detected through MAb PPL-488. (B) Southern blotting revealed the expected wild-type and targeted SacI fragments from the DNA of the offspring of heterozygous crosses. (C) Oligonucleotide primers, exon 16-specific forward primer with the exon 17-specific reverse primer, produce a 982-bp PCR product from the wild-type allele, while exon 16-specific forward primer with PGK promoter-specific reverse primer produce a 602-bp PCR product from the mutant allele. (D) Periplakin protein, detected as a 195-kDa band by MAb PPL-488 from the skin of wild-type and heterozygous mice, is not present in the knockout littermate.
In the targeted allele, the sequences encoding mouse periplakin amino acid residues 656 to 977, including the C-terminal end of the head domain and the N-terminal end of the rod domain, are replaced with the neo gene (Fig. 1A). Periplakin MAb PPL-488, recognizing an epitope within human periplakin aa 1548 to 1583, detected a 195-kDa protein both from the Ppl+/+ and Ppl+/− mouse skin extract, but the Ppl−/− mouse skin was completely devoid of periplakin (Fig. 1D), indicating that the insertion of the neo gene is not bypassed by an in-frame cryptic splicing event.
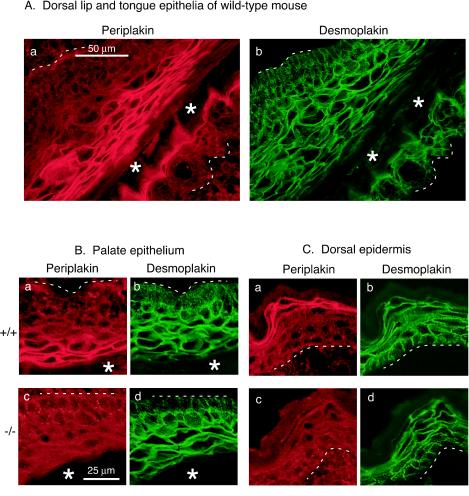
The epithelia of mice lacking periplakin appear normal.
In wild-type neonatal mouse mouth epithelium, as represented by the dorsal lip and dorsal tongue epithelia, periplakin is distinctly present throughout the upper granular cell layer, at the cell-cell junctions, and along the plasma membrane of the flattened keratinocytes (Fig. 2A). For comparison, desmoplakin is present at all cell-cell junctions throughout these epithelia. The morphology of palate epithelium (Fig. 2B) and dorsal skin (Fig. 2C) of a wild-type mouse and a periplakin knockout littermate was studied through double labeling for periplakin and desmoplakin. The upper granular cell layer stained positive for periplakin in the wild-type mouse, but the absence of periplakin was obvious in the Ppl−/− littermate. Double-labeling for desmoplakin confirmed the normal appearance of epithelia lacking periplakin (panels d in Fig. 2B and C).
FIG. 2.
Periplakin is a constituent of the cell-cell junctions in the upper granular cell layer of dorsal lip and tongue (Aa), palate (Ba), and dorsal epidermis (Ca) of normal mouse (+/+). The sections were double labeled for desmoplakin, a desmosomal protein which is present at the cell-cell junctions throughout the epithelia (Ab, Bb, and Cb, respectively). The epithelia of periplakin knockout mouse (−/−) are devoid of periplakin protein (Bc and Cc). Double labeling for desmoplakin confirms the normal structural architecture and appearance of periplakin-deficient epithelia (Bd and Cd). The oral cavity in panels A and B is indicated by an asterisk, and the basement membrane zone separating the epithelia from the underlying tissue is denoted by a broken line.
Mice lacking periplakin develop and reproduce normally.
The amino acid sequences of human and mouse periplakin proteins, deduced from the cDNA sequences, showed 88.1% identity and 94.4% similarity. The differences are evenly distributed along the periplakin polypeptide, as the 942-aa N-terminal head domain contained 87.2% of identical amino acid residues and the 701-aa rod domain contained 88.3%. However, the 111-aa C-terminal tail domain revealed 96.4% identity between human and mouse. Mouse exon 5, encoding the N-terminal plakin repeat Z, is six nucleotides shorter, resulting in a protein of 1,754 aa residues, compared to human periplakin with 1,756 aa residues. Thus, mouse periplakin is lacking residues corresponding to human aa G-186 and D-187. The high level of sequence homology suggests that periplakin protein has remained conserved during evolution and implies a common functional role in these species. However, the appearance of periplakin knockout mice did not provide clues to the functional role for periplakin protein. Mice lacking periplakin appeared grossly normal and showed no obvious phenotypic differences in comparison to their heterozygous or wild-type littermates. Ppl−/− females crossed with Ppl−/− males produced litters of normal size, excluding embryonic or fetal lethality and defects in fertility.
Normal CEs develop in the absence of periplakin.
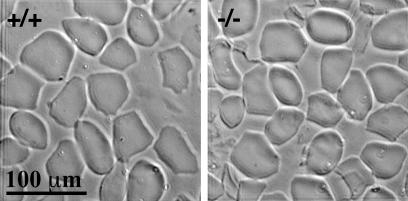
CEs are stable structures of differentiated keratinocytes (41, 50). Envelopes were prepared from the dorsal skin of neonatal mice. From four litters obtained from heterozygous crossing, a pair of wild-type and knockout mice was randomly selected, and the CEs were prepared and examined by phase-contrast microscopy (Fig. 3). The CEs prepared from ppl−/− mice were indistinguishable from those of wild-type mice. Both strains of mice possessed a mixture of balloon-shaped and angular envelopes. Similarly, it has been reported that CEs prepared from involucrin−/− mice as well as envoplakin−/− mice are ultrastructurally indistinguishable from those isolated from normal mice (13, 34), but the lack of loricrin, although compensated by other proteins, results in quasinormal CEs (22).
FIG. 3.
CEs extracted from the dorsal skin of wild-type (+/+) and periplakin-deficient (−/−) mice. There is no obvious difference between the appearance of the populations of CEs. Rounded and angular CEs are observed in both type of mice.
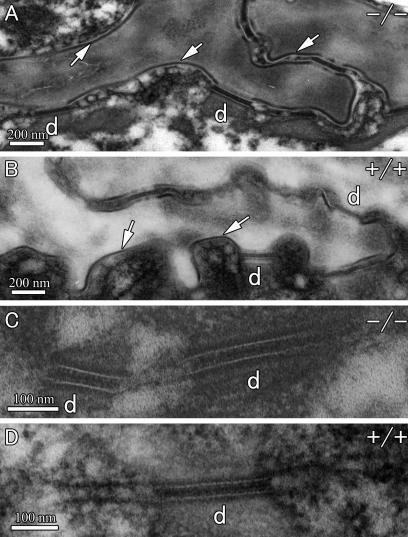
Electron microscopy imaging of epidermis from neonatal wild-type and periplakin knockout mice did not reveal apparent structural differences (Fig. 4). Specifically, hemidesmosomes, desmosomes, keratin filaments, lamellar granules, keratohyalin granules, and the CEs within the epidermis had normal appearance in both cases.
FIG. 4.
Transmission electron microscopy of the wild-type (+/+) and knockout (−/−) mouse epidermis, presenting the cornified cell layer-granular layer interface (A and B) and spinous cells (C and D). There is no apparent structural difference in the CEs (arrows) and desmosomes (d) between the wild-type and knockout mice.
Dye exclusion assay.
Mouse lines deficient in loricrin (28) or envoplakin (34) demonstrated a delay in skin barrier formation during embryonic development that resolved after birth. Involucrin null mice examined for skin barrier formation after birth displayed no defect in the CE (13). To determine if periplakin null mouse embryos also demonstrated a delay in skin barrier formation, the dye exclusion assay (20, 28, 34) was performed using embryos from 13.5 to 17.5 days after coitus (day of vaginal plug equals 0.5 or E0.5), using periplakin heterozygous breeders. As can be seen in Fig. 5, E14.5 and E 15.5 embryos have not yet developed the skin barrier and were stained uniformly blue with the staining in the earlier age being more intense. At E16.5, the skin barrier is partially formed with the dye being excluded first from the dorsal surface. By E17.5, the barrier has almost completely formed, with some embryos exhibiting only slight staining on the extreme ventral surface and tips of appendages. Our observations are consistent with the dorsal-to-ventral progression of skin barrier formation in normal mouse embryos as characterized by Hardman et al. (20), where the skin barrier was first detected by dye exclusion in E16/5 (estimated gestational age) embryos. As seen in Fig. 5, we could detect no consistent difference in the developmental sequence of barrier formation between normal (periplakin +/+ or +/−) or periplain null embryos. Thus, periplakin does not appear to be essential for the development of a normal skin barrier during mouse development.
Absence of periplakin is not compensated through the altered level of other epidermal proteins.
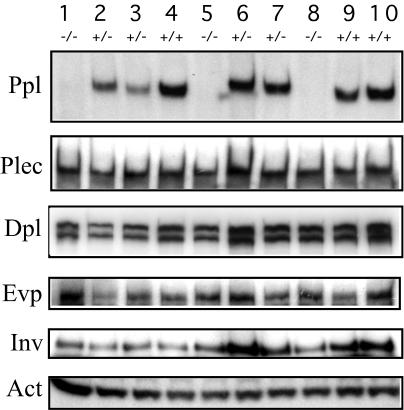
Since periplakin has been detected as a component of the CE, at the cell membranes associated with various adhesion structures, and in the cytoplasmic pool, it was of interest to determine how its absence will affect other epidermal proteins. We analyzed a litter of 10 mice obtained from a heterozygous crossing with three wild-type (+/+), four heterozygous (+/−), and three knockout (−/−) offspring, through the Western blotting analysis (Fig. 6). The complete absence of periplakin in knockout mice was confirmed. However, the level of plectin, desmoplakin, envoplakin, involucrin, and actin in each skin extract did not change appreciably with the genotype of the offspring, thus providing no evidence for a compensatory mechanism through the level of these proteins.
FIG. 6.
Absence of periplakin does not alter the level of other epidermal proteins. Western analysis of skin samples from a litter, obtained from a heterozygous crossing, confirms the absence of periplakin (Ppl) from the knockout mice (−/−). No clear quantitative difference between wild-type (+/+) and heterozygous (+/−) mice was observed. Furthermore, the levels of plectin (Plec), desmoplakin (Dpl), envoplakin (Evp), involucrin (Inv), and actin (Act) did not reflect the presence or absence of periplakin.
Lack of periplakin does not alter the distribution of other epidermal proteins.
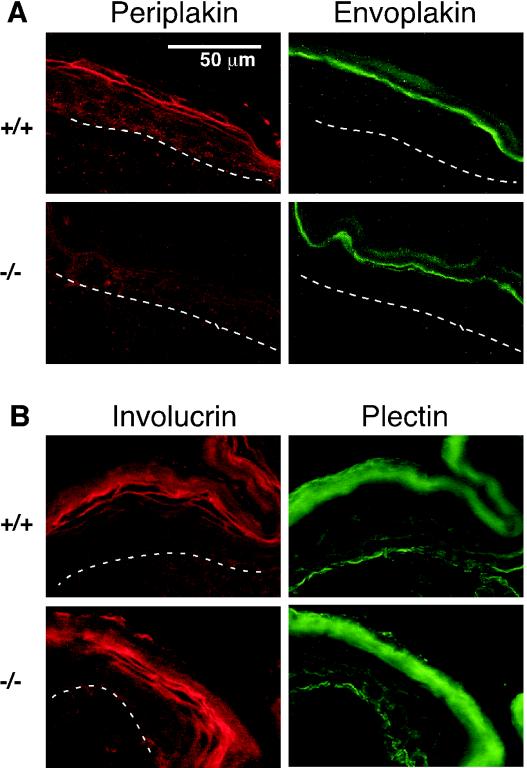
Cryosections of dorsal skin from wild-type and periplakin knockout mice were studied through double labeling and subsequent immunofluorescence analysis (Fig. 7). Periplakin, envoplakin, and involucrin are known as early constituents of the CE (47). In neonatal mouse epidermis, envoplakin was detected within a narrow zone between the granular and cornified cell layers, only partially overlapping with periplakin staining. Involucrin was present as a lamellar pattern, colocalizing with the periplakin staining pattern. The absence of periplakin did not result in changes in the appearance of envoplakin or involucrin staining patterns. Plectin staining within the basal cell layer along the basement membrane zone was not altered when the wild-type and periplakin-deficient epithelia were compared. Furthermore, the nonspecific signal obtained with the goat secondary antibody along the cornified cell layer also demonstrated similar structure for the wild-type and periplakin-deficient mouse epidermis.
FIG. 7.
Distribution of epidermal proteins in the neonatal wild-type (+/+) and knockout (−/−) littermates. (A) Double labeling for periplakin and envoplakin revealed envoplakin staining in the narrow area above the granular cell layer, barely overlapping with periplakin signal. The appearance of envoplakin is not altered due to the absence of periplakin. (B) Double labeling for involucrin and plectin revealed the presence of involucrin along the lamellar structures in the outer layers of epidermis and a distinct signal for plectin at the basement membrane zone in both types of mice. The nonspecific cornified layer staining with the combination of plectin antibody and the goat-specific secondary antibody also demonstrates that the absence of periplakin did not result in visible differences in appearance between knockout mice and the wild-type littermate. The dermal-epidermal junction is denoted by a broken line.
DISCUSSION
It has been shown previously that the members of the plakin family of proteins, desmoplakin, plectin, and BPAG1, which are integral components of cell-cell or cell-matrix adhesion structures, i.e., desmosomes and hemidesmosomes, respectively, are indispensable for the function and maintenance of the normal stratified epithelium. In contrast, elimination of the components of the CE results in no discernible phenotype or at most in a very subtle changes.
Clearly, as reported in this study or previously (13, 34), periplakin, envoplakin, and involucrin, when deleted through targeted inactivation of the corresponding genes in mice, are not crucial for CE formation and epithelial barrier function. The CE formation may be achieved through compensatory mechanisms. This has been demonstrated in loricrin-deficient mice, where an increased expression of members of the small proline-rich protein family and repetin are thought to compensate for the loricrin deficiency (28). Thus, not a single molecular constituent has been found thus far to be indispensable for CE integrity. Abundant but highly insoluble molecules, similar to periphilin, which we recently characterized through the yeast two-hybrid interaction, cDNA cloning, and overexpression in host cells (25), hold a potential to be indispensable for epithelial barrier function. In fact, we can speculate that periplakin, and perhaps also envoplakin and involucrin, may become cross-linked to the CE by default as a mechanism for disposing of these proteins during epithelial differentiation. Such a “dustbin” or “trashcan” hypothesis has been proposed (37), although a more organized model developed for the assembly of the CE does not support it (23).
Although periplakin was originally identified as a precursor of the CE in terminally differentiated epidermal keratinocytes (33, 42, 44), its expression has also been demonstrated in other tissues with a prominent epithelial component (4, 26). The yeast two-hybrid screening for periplakin interaction partners confirmed a specific binding of periplakin to keratin 8 and vimentin (26, 53). Transfection studies have shown that the overexpression of envoplakin resulted in the aggregate formation, but the overexpression of periplakin was able to rescue envoplakin from forming such aggregates and, instead, allowed it to resume its intermediate filament-associated localization (12). However, periplakin has been seen prominently aligned with the intermediate filaments only as a result of overexpression (24, 26). In cultured keratinocytes periplakin appears in a punctate cytoplasmic staining, and upon induction of differentiation, periplakin localizes to the cell-cell junctions and to the apical plasma membrane (33, 42). Periplakin was recently found in lens fiber cells, forming a cortical complex with ezrin, periaxin, and desmoyokin (49).
In epidermis, plectin is mainly localized to hemidesmosomes and displays staining along the basement membrane zone. Using a novel monoclonal antibody and carefully controlling the protocol for sample treatment, we recently showed that periplakin is also present in the basal cell layer in human neonatal foreskin epidermis, localizing along the basement membrane zone (1). The normal appearance of plectin in mouse epithelia in the absence of periplakin suggests independent interactions of these proteins with the hemidesmosomal transmembrane protein type XVII collagen/BP180 (1, 4, 6). Also, other novel protein-protein interactions, identified through yeast two-hybrid screening, suggest that periplakin serves other functions besides being a component of the CE (3). Unconventional, unexpected, and unexplained periplakin interaction partners include the intracellular domain of the hemidesmosomal transmembrane protein type XVII collagen/BPAG2/BP180 (1, 4), a G-protein receptor in brain (15), a tyrosine kinase, PKB/c-Akt, which is involved in regulation of apoptosis (53), and the 300-kDa mannose-6-phosphate receptor, involved in intracellular protein trafficking (48). In these reports, the overexpression of periplakin has been found to selectively inhibit μ-opioid receptor activation of G protein (15), modulate PKB signaling by binding and sequestration (53), and inhibit MPR-300-dependent endocytosis (48). Thus, although functional consequences for overexpression of periplakin have been suggested, the role of the endogenous periplakin in epithelial cells remains undisclosed.
In conclusion, the lack of a major phenotype for periplakin knockout mice suggests that the primary function of periplakin is not related to its physiological role as a component of the CE, but its role may be in pathological challenges which are not encountered by laboratory mice during their normal life span.
Acknowledgments
This work was supported by The National Institutes of Health grants R01 AR-33488 (S.A.), P01 AR-38923 (J.U.), and DAMD 17-99-2-9040 (J.F.K.) and grants from the Ministry of Education, Culture, Sports, Science and Technology and the Ministry of Health, Labor and Welfare of Japan (A.I.Y.).
The Animal Models Core Facility in the Department of Dermatology and Cutaneous Biology, Thomas Jefferson University (Philadelphia, Pa.), is acknowledged for the production of mice. Specifically, the technical assistance of David Manion and Dominic Leperi in the generation of ES cell lines and maintenance of the animals is acknowledged, and Minna Mannikko is acknowledged for preliminary protein and genetic analysis. The skillful assistance of Sue Gotta and the Morphology Core in obtaining the microscopic images is also gratefully acknowledged. We thank John McGrath for preparing tissue samples for EM studies and Lyuben Marekov and Andrey Kalinin for providing the envoplakin antibody. We acknowledge discussions and the expert opinions of Fiona Watt, to whom periplakin knockout mice have been forwarded for generation of double and triple knockouts with envoplakin and involucrin null mice.
REFERENCES
- 1.Aho, S. 2004. Many faces of periplakin: domain-specific antibodies detect the protein throughout the epidermis, explaining the multiple protein-protein interactions. Cell Tissue Res. 316:87-97. [DOI] [PubMed] [Google Scholar]
- 2.Aho, S., M. Buisson, T. Pajunen, Y. W. Ryoo, J.-F. Giot, H. Gruffat, A. Sergeant, and J. Uitto. 2000. Ubinuclein, a novel nuclear protein interacting with cellular and viral transcription factors. J. Cell Biol. 148:1165-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aho, S., and S. Kazerounian. 2003. Molecular interactions of periplakin in epithelial cells. Recent Res. Dev. Cell. Biochem. 1:131-141. [Google Scholar]
- 4.Aho, S., W. H. I. McLean, K. Li, and J. Uitto. 1998. cDNA cloning, mRNA expression, and chromosomal mapping of human and mouse periplakin gene. Genomics 48:242-247. [DOI] [PubMed] [Google Scholar]
- 5.Aho, S., K. Rothenberger, E. M. L. Tan, Y. W. Ryoo, B. H. Cho, W. H. I. McLean, and J. Uitto. 1999. Human periplakin: genomic organization in a clonally unstable region of chromosome 16p with an abundance of repetitive sequence elements. Genomics 56:160-168. [DOI] [PubMed] [Google Scholar]
- 6.Aho, S., and J. Uitto. 1997. Basement membrane zone protein-protein interactions disclosed by yeast two-hybrid system. J. Investig. Dermatol. 108:546. [Google Scholar]
- 7.Allen, P. G., and J. V. Shah. 1999. Brains and brawn: plectin as regulator and reinforcer of the cytoskeleton. BioEssays 21:451-454. [DOI] [PubMed] [Google Scholar]
- 8.Andrä, K., H. Lassmann, R. Bittner, S. Shorny, R. Fässler, F. Propst, and G. Wiche. 1997. Targeted inactivation of plectin reveals essential function in maintaining the integrity of skin, muscle, and heart cytoarchitecture. Genes Dev. 11:3143-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anhalt, G. J. 1997. Paraneoplastic pemphigus. Adv. Dermatol. 12:77-96. [PubMed] [Google Scholar]
- 10.Anhalt, G. J., S. Kim, J. R. Stanley, N. J. Korman, D. A. Jabs, M. Kory, H. Izumi, H. Ratrie, D. Mutasim, L. Ariss-Abdo, and R. S. Labib. 1990. Paraneoplastic pemphigus. An autoimmune mucocutaneous disease associated with neoplasia. N. Engl. J. Med. 323:1729-1735. [DOI] [PubMed] [Google Scholar]
- 11.Brown, A., G. Bernier, M. Mathieu, J. Rossant, and R. Kothary. 1995. The mouse dystonia musculorum gene is a neural isoform of bullous pemphigoid antigen 1. Nat. Genet. 10:301-306. [DOI] [PubMed] [Google Scholar]
- 12.DiColandrea, T., T. Karashima, A. Määttä, and F. M. Watt. 2000. Subcellular distribution of envoplakin and periplakin: insights into their role as precursors of the epidermal cornified envelope. J. Cell Biol. 151:573-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Djian, P., K. Easley, and H. Green. 2000. Targeted ablation of the murine involucrin gene. J. Cell Biol. 151:381-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doering, T., W. Holleran, A. Potratz, G. Vielhaber, P. Elias, K. Suzuki, and K. Dandhoff. 1999. Sphingolipid activator proteins are required for epidermal permeability barrier formation. J. Biol. Chem. 274:11038-11045. [DOI] [PubMed] [Google Scholar]
- 15.Feng, G. J., E. Kellett, C. A. Scorer, J. Wilde, J. H. White, and G. Milligan. 2003. Selective interactions between helix VIII of the human mu opioid receptors and the C-terminus of periplakin disrupt G protein activation. J. Biol. Chem. 278:33400-33407. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs, E., and Y. Yang. 1999. Crossroads on cytoskeletal highways. Cell 98:547-550. [DOI] [PubMed] [Google Scholar]
- 17.Gallicano, G. I., C. Bauer, and E. Fuchs. 2001. Rescuing desmoplakin function in extra-embryonic ectoderm reveals the importance of this protein in embryonic heart, neuroepithelium, skin and vasculature. Development 128:929-941. [DOI] [PubMed] [Google Scholar]
- 18.Gallicano, G. I., P. Kouklis, C. Bauer, M. Yin, V. Vasioukhin, L. Degenstein, and E. Fuchs. 1998. Desmoplakin is required early in development for assembly of desmosomes and cytoskeletal linkage. J. Cell Biol. 143:2009-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo, L., L. Degenstein, J. Dowling, Q.-C. Yu, R. Wollman, B. Perman, and E. Fuchs. 1995. Gene targeting of BPAG1: abnormalities in mechanical strength and cell migration in stratified epithelia and neurologic degeneration. Cell 81:233-243. [DOI] [PubMed] [Google Scholar]
- 20.Hardman, M., P. Sisi, D. Banbury, and C. Byrne. 1998. Patterned acquisition of skin barrier function during development. Development 125:1541-1552. [DOI] [PubMed] [Google Scholar]
- 21.Huber, O. 2003. Structure and function of desmosomal proteins and their role in development and disease. Cell. Mol. Life Sci. 60:1872-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarnik, M., P. A. de Viragh, E. Scharer, D. Bundman, M. N. Simon, D. Roop, and A. Steven. 2002. Quasi-normal cornified cell envelopes in loricrin knockout mice imply the existence of a loricrin backup system. J. Investig. Dermatol. 118:102-109. [DOI] [PubMed] [Google Scholar]
- 23.Kalinin, A. E., A. V. Kajava, and P. M. Steinert. 2002. Epithelial barrier function: assembly and structural features of the cornified cell envelope. BioEssays 24:789-800. [DOI] [PubMed] [Google Scholar]
- 24.Karashima, T., and F. M. Watt. 2002. Interaction of periplakin and envoplakin with intermediate filaments. J. Cell Sci. 115:5027-5037. [DOI] [PubMed] [Google Scholar]
- 25.Kazerounian, S., and S. Aho. 2003. Characterization of periphilin, a widespread, highly insoluble nuclear protein and potential constituent of the keratinocyte cornified envelope. J. Biol. Chem. 278:36707-36717. [DOI] [PubMed] [Google Scholar]
- 26.Kazerounian, S., J. Uitto, and S. Aho. 2002. Unique role for the periplakin tail in intermediate filament association: specific binding to keratin 8 and vimentin. Exp. Dermatol. 11:428-438. [DOI] [PubMed] [Google Scholar]
- 27.Kim, S.-Y., T. Jeitner, and P. Steinert. 2002. Transglutaminases in disease. Neurochem. Int. 40:85-103. [DOI] [PubMed] [Google Scholar]
- 28.Koch, P., P. de Viragh, E. Scharer, D. Bundman, M. Longley, J. Bickenbach, Y. Kawachi, Y. Suga, Z. Zhou, M. Huber, D. Hohl, T. Kartasova, M. Jarnik, A. Steven, and D. Roop. 2000. Lessons from loricrin-deficient mice: compensatory mechanisms maintaining skin barrier function in the absence of a major cornified envelope protein. J. Cell Biol. 151:389-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Köntgen, F., and C. L. Stewart. 1993. Simple screening procedure to detect gene targeting events in embryonic stem cells. Methods Enzymol. 225:878-889. [DOI] [PubMed] [Google Scholar]
- 30.Kuramoto, N., T. Takizawa, T. Takizawa, M. Matsuki, H. Morioka, J. M. Robinson, and K. Yamanishi. 2002. Development of ichthyosiform skin compensates for defective permeability barrier function in mice lacking transglutaminase 1. J. Clin. Investig. 109:243-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leung, C. L., K. J. Green, and R. K. H. Liem. 2002. Plakins: a family of versatile cytolinker proteins. Trends Cell Biol. 12:37-45. [DOI] [PubMed] [Google Scholar]
- 32.Leung, C. L., R. K. H. Liem, D. A. D. Parry, and K. J. Green. 2001. The plakin family. J. Cell Sci. 114:3409-3410. [DOI] [PubMed] [Google Scholar]
- 33.Ma, A. S.-P., and T.-T. Sun. 1986. Differentiation-dependent changes in the solubility of a 195-kD protein in human epidermal keratinocytes. J. Cell Biol. 103:41-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Määttä, A., T. DiColandrea, K. Groot, and F. M. Watt. 2001. Gene targeting of envoplakin, a cytoskeletal linker protein and precursor of the epidermal cornified envelope. Mol. Cell. Biol. 21:7047-7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahoney, M. G., S. Aho, J. Uitto, and J. R. Stanley. 1998. The members of the plakin family of proteins recognized by paraneoplastic pemphigus antibodies include periplakin. J. Investig. Dermatol. 111:308-313. [DOI] [PubMed] [Google Scholar]
- 36.Marekov, L. N., and P. M. Steinert. 1998. Ceramides are bound to structural proteins of the human foreskin epidermal cornified cell envelope. J. Biol. Chem. 273:17763-17770. [DOI] [PubMed] [Google Scholar]
- 37.Michel, S., R. Schmidt, S. M. Robinson, B. Shroot, and U. Reichert. 1987. Identification and subcellular distribution of cornified envelope precursor proteins in the transformed human keratinocyte line SV-K14. J. Investig. Dermatol. 88:301-305. [DOI] [PubMed] [Google Scholar]
- 38.Mueller, S., V. Klaus-Kovtum, and J. R. Stanley. 1989. A 230-kD basic protein is the major bullous pemphigoid antigen. J. Investig. Dermatol. 92:33-38. [DOI] [PubMed] [Google Scholar]
- 39.Nemes, Z., L. N. Marekov, L. Fesus, and P. Steinert. 1999. A novel function for transglutaminase 1: attachment of long-chain ω-hydroxyceramides to involucrin by ester bond formation. Proc. Natl. Acad. Sci. USA 96:8402-8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Norgett, E. E., S. J. Hatsell, L. Carvajal-Huerta, J.-C. R. Caberas, J. Common, P. E. Purkis, N. Whittock, I. M. Leigh, H. P. Stevens, and D. P. Kelsell. 2000. Recessive mutation in desmoplakin disrupts desmoplakin-intermediate filament interactions and causes dilated cardiomyopathy, woolly hair and keratoderma. Hum. Mol. Genet. 9:2761-2766. [DOI] [PubMed] [Google Scholar]
- 41.Rice, R. H., and H. Green. 1977. The cornified envelope of terminally differentiated human epidermal keratinocytes consists of cross-linked protein. Cell 11:417-422. [DOI] [PubMed] [Google Scholar]
- 42.Ruhrberg, C., M. A. N. Hajibagheri, D. A. D. Parry, and F. M. Watt. 1997. Periplakin, a novel component of cornified envelopes and desmosomes that belongs to the plakin family and forms complexes with envoplakin. J. Cell Biol. 139:1835-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruhrberg, C., and F. M. Watt. 1997. The plakin family: versatile organizers of cytoskeletal architecture. Curr. Opin. Genet. Dev. 7:392-397. [DOI] [PubMed] [Google Scholar]
- 44.Simon, M., and H. Green. 1984. Participation of membrane-associated proteins in the formation of the cross-linked envelope of the keratinocyte. Cell 36:827-834. [DOI] [PubMed] [Google Scholar]
- 45.Steinböck, F. A., and G. Wiche. 1999. Plectin: a cytolinker by design. Biol. Chem. 380:151-158. [DOI] [PubMed] [Google Scholar]
- 46.Steinert, P. 2000. The complexity and redundancy of epithelial barrier function. J. Cell Biol. 151:F5-F7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steinert, P. M., and L. N. Marekov. 1999. Initiation of assembly of the cell envelope barrier structure of stratified squamous epithelia. Mol. Biol. Cell 10:4247-4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Storch, S. 1999. Wechselwirkungen von Proteinen mit den zytoplasmatischen Domänen der Mannose-6-Phosphat-Rezeptoren. Doctoral thesis. Georg-August-Universität, Göttingen, Germany.
- 49.Straub, B., J. Boda, C. Kuhn, M. Schnoelzer, U. Korf, T. Kempf, H. Spring, M. Hatzfeld, and W. Franke. 2003. A novel cell-cell junction system: the cortex adherens mosaic of lens fiber cells. J. Cell Sci. 116:4985-4995. [DOI] [PubMed] [Google Scholar]
- 50.Sun, T.-T., and H. Green. 1976. Differentiation of epidermal keratinocyte in cell culture: formation of the cornified envelope. Cell 9:511-521. [DOI] [PubMed] [Google Scholar]
- 51.Tybulewicz, V., C. Crawford, P. Jackson, R. Bronson, and R. Mulligan. 1991. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell 65:1153-1163. [DOI] [PubMed] [Google Scholar]
- 52.Uitto, J., L. Pulkkinen, F. J. D. Smith, and W. H. I. McLean. 1996. Plectin and human genetic disorders of the skin and muscle. The paradigm of epidermolysis bullosa with muscular dystrophy. Exp. Dermatol. 5:237-246. [DOI] [PubMed] [Google Scholar]
- 53.van den Heuvel, A. P. J., A. M. M. de Vries-Smits, P. C. van Weeren, P. F. Dijkers, K. M. T. de Bruyn, J. A. Riedl, and B. M. T. Burgering. 2002. Binding of protein kinase B to the plakin family member periplakin. J. Cell Sci. 115:3957-3966. [DOI] [PubMed] [Google Scholar]
- 54.Vasioukhin, V., E. Bowers, C. Bauer, L. Degenstein, and E. Fuchs. 2001. Desmoplakin is essential in epidermal sheet formation. Nat. Cell Biol. 3:1076-1085. [DOI] [PubMed] [Google Scholar]
- 55.Wiche, G. 1998. Role of plectin in cytoskeleton organization and dynamics. J. Cell Sci. 111:2477-2486. [DOI] [PubMed] [Google Scholar]
- 56.Yang, Y., C. Bauer, G. Strasser, R. Wollman, J.-P. Julien, and E. Fuchs. 1999. Integrators of the cytoskeleton that stabilize microtubules. Cell 98:229-238. [DOI] [PubMed] [Google Scholar]
- 57.Yang, Y., J. Dowling, Q. C. Yu, P. Kouklis, D. W. Cleveland, and E. Fuchs. 1996. An essential cytoskeletal linker protein connecting actin microfilaments to intermediate filaments. Cell 86:655-665. [DOI] [PubMed] [Google Scholar]