Abstract
Minichromosome maintenance protein 1 (Mcm1) is required for efficient replication of autonomously replicating sequence (ARS)-containing plasmids in yeast cells. Reduced DNA binding activity in the Mcm1-1 mutant protein (P97L) results in selective initiation of a subset of replication origins and causes instability of ARS-containing plasmids. This plasmid instability in the mcm1-1 mutant can be overcome for a subset of ARSs by the inclusion of flanking sequences. Previous work showed that Mcm1 binds sequences flanking the minimal functional domains of ARSs. Here, we dissected two conserved telomeric X ARSs, ARS120 (XARS6L) and ARS131a (XARS7R), that replicate with different efficiencies in the mcm1-1 mutant. We found that additional Mcm1 binding sites in the C domain of ARS120 that are missing in ARS131a are responsible for efficient replication of ARS120 in the mcm1-1 mutant. Mutating a conserved Mcm1 binding site in the C domain diminished replication efficiency in ARS120 in wild-type cells, and increasing the number of Mcm1 binding sites stimulated replication efficiency. Our results suggest that threshold occupancy of Mcm1 in the C domain of telomeric ARSs is required for efficient initiation. We propose that origin usage in Saccharomyces cerevisiae may be regulated by the occupancy of Mcm1 at replication origins.
Replication of DNA must be inherently accurate and precisely regulated. It is therefore not surprising that the mechanism for the initiation of DNA replication is both complex and conserved. Initiation of DNA synthesis involves the assembly of a multicomponent complex at designated sites known as replication origins. While the protein components of the prereplication complex (pre-RC) used in this initiation process are conserved in all eukaryotes (5), there is little in common between the nucleotide sequences of replication origins within each eukaryote and between different eukaryotes (29).
In Saccharomyces cerevisiae, replication origins (ORI) consist of defined sequences of about 200 bp that can replicate autonomously independently of their native chromosomal environment. These autonomously replicating sequences (ARSs) are modular in structure. They contain a ubiquitous 11-bp (5′-WTTTAYRTTTW) ARS consensus sequence (ACS) (11, 22, 63) where the origin recognition complex (ORC) binds (4). In certain ARSs, a 10-of-11 match of the ACS is sufficient for ORC recognition (64). The ACS is essential but not sufficient for autonomous replication. cis elements on the 5′ (C domain) or the 3′ (B domain) flanking sequences of the ACS are also required (12). Elements in the B domain of one particular ARS, ARS1, have been characterized in great detail. Three elements, known as B1, B2, and B3, have been identified (44). B1 is protected by the ORC (55), whereas B3 is protected by Abf1 (41) or Mcm1 (17) in in vitro footprinting analyses. Initiation of DNA synthesis at ARS1 has been mapped to a region between the B1 and B2 elements (6). Two of the three B elements in combination with the ACS are sufficient to promote autonomous replication. Although B elements of different ARSs are not conserved in nucleotide sequence, they are interchangeable between certain ARSs (31, 54, 61). The differences in size and modular composition of replication origins suggest that there are many ways to assemble a functional replication origin from a finite set of modules (60-62, 65). The plasticity in the organization of replication origins is further illustrated by the dispensability of the B elements altogether in the presence of the C domain in several telomeric ARSs (13). However, because the C domain is generally larger, elements in the C domain have not been characterized.
The redundant functions of the B and C domains in promoting replication initiation suggest that although the process of pre-RC assembly may be conserved, there are many ways to create an environment conducive to this assembly process. The concept of alternative pathways for creating an environment for pre-RC assembly is especially appealing in higher eukaryotes, where there appears not to be a unifying mechanism for site selection for pre-RC assembly. In Xenopus oocytes, replication initiation occurs at random sequences (38). In mammalian cells, initiation occurs at multiple sites within replication zones that are defined by their chromosomal contexts rather than nucleotide sequences (29, 46). Indeed, the activity of replication origins is responsive to their contexts (9, 36). It has been shown that replication origins taken out of their native environment are no longer temporally regulated (35, 52) and that silent replication origins are no longer repressed (23). To better understand the principle of site selection in replication initiation and the regulation of origin activity, it is important to learn more about the extended sequences that provide the contexts for replication initiation.
Mcm1 is a combinatorial transcription factor that binds with exquisite specificity to diverse recognition sequences in combination with a cofactor (37, 58). By itself, Mcm1 binds to the degenerate sequence CCYWWWWNGN (17, 50, 58, 68). Like other MADS domain transcription factors (8), Mcm1 is a master regulator that specifies cell identity (33, 34, 51) and coordinates the expression of genes required for cell growth and proliferation (57).
Mcm1 is required for initiation of DNA replication. A P97L mutation (mcm1-1) that compromises the DNA binding activity of Mcm1 causes an increase in plasmid loss rate, as well as a decrease in the initiation frequency of chromosomal replication origins (17). However, Mcm1 appears to regulate the initiation of DNA synthesis at two levels. It modulates the transcriptional expression of several components of the pre-RC including Cdc6, Mcm3, Mcm5, Mcm6, and Mcm7 (25, 45, 58), which are preassembled at replication origins before the onset of S phase. It also acts directly at replication origins to stimulate DNA replication initiation. In vivo cross-linking studies show that Mcm1 occupies sites near replication origins. In vitro DNA binding studies confirm that Mcm1 binds multiple sites flanking the minimal functional domains (MFDs) of ARSs (17). Footprints of extended regions of ARSs indicate that Mcm1 binds a variable number of sites at different ARSs, suggesting that if Mcm1 plays a direct role at replication origins, its influences on different origins may be dissimilar. To elucidate the function of Mcm1 at replication origins independently of its indirect effects on the pre-RC, it is important to analyze Mcm1 binding site mutations. In this study, we investigated the function of Mcm1 at replication origins by exploiting natural variants of two conserved telomeric X ARSs (15) that respond differently to reduced Mcm1 binding activity and by varying the number of Mcm1 binding sites at one of the ARSs. Both approaches led to the conclusion that the number of Mcm1 binding sites at ARSs is critical for the efficient initiation of DNA replication especially when Mcm1 activity is limiting.
MATERIALS AND METHODS
Strains and plasmids.
Parent strain 8534-8C (MATα leu2-3,112 ura3-52 his4-Δ34) and mutant strain RM9-3A (MATa leu2-3,112 ura3-52 his3-11,15 mcm1-1) were used for minichromosome maintenance assays. The minichromosomes used were YCp1, YCp121, YCp131a, YCp120 (14, 43), and YCp121AB (64, 65). The vectors used for cloning were pLC5 (LEU2 CEN5), pC5L (CEN5 LEU2), and YCp56 (URA3 CEN4).
Minichromosome maintenance assay.
Yeast cells containing minichromosomes were grown in selective medium to saturation and then plated on complete or selective medium (complete-Leu or Cm-Ura) to determine the initial percentage of plasmid-containing cells. Cells were used to inoculate yeast extract-peptone-dextrose YEPD and allowed to grow for approximately 10 generations before plating onto YEPD plates and selective plates to determine the final percentage of plasmid-containing cells. Loss rate per generation was determined with the equation X = 1 − (F/I)1/n, where F and I represent the final and initial percentages of plasmid-containing cells and n is the number of generations. Each value is the average of at least three independent experiments, except for the values shown in Fig. 5C, which are averages of five or six independent experiments.
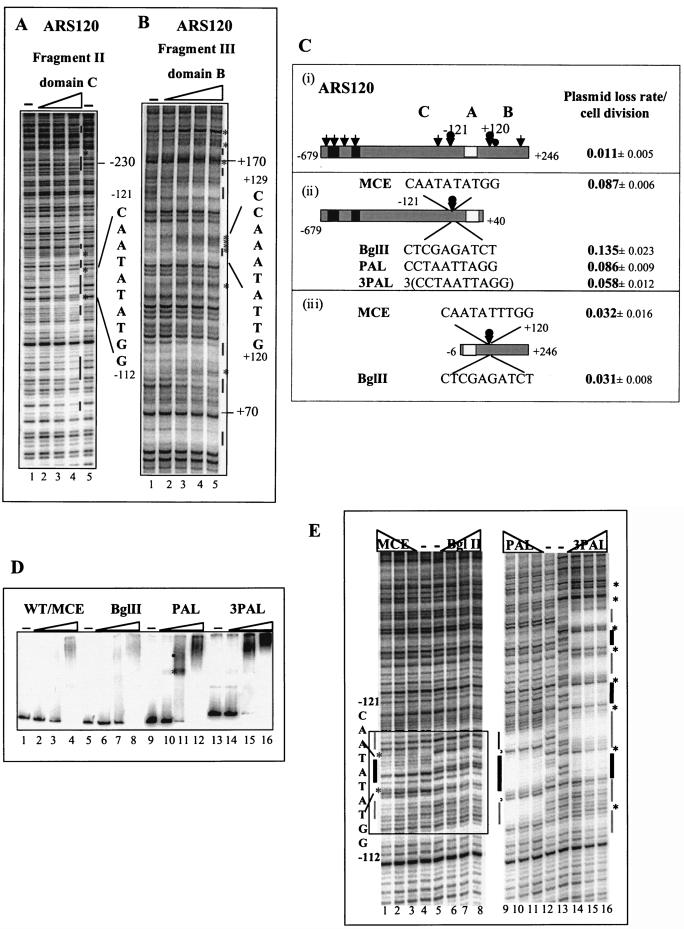
FIG. 5.
Footprinting analysis of the conserved region of ARS120. (A) Footprints of Mcm1 on fragment II of domain C. Protection at the −121 MCE is indicated. Amounts of Mcm1 added: lanes 1 and 5, 0 pmol; lane 2, 5 pmol; lane 3, 10 pmol; lane 4, 20 pmol. (B) Footprints of Mcm1 on fragment III of domain B. Protection at the +120 MCE is indicated. Amounts of Mcm1 added: lane 5, 0 pmol; lane 4, 5 pmol; lane 3, 10 pmol; lane 2, 20 pmol; lane 1, 40 pmol. A bar indicates protection. An asterisk indicates hypersensitivity. (C) Mutagenesis of MCEs at −121 and +120 of ARS120 and their effects on plasmid stability. (i) Full-length ARS120. Arrows indicate Mcm1 binding sites, a bobbed arrow highlights the site to be mutagenized, and a filled circle represents the Abf1 binding site. A black box represents nonconserved sequences. (ii) ARS120 without B domain. The MCE at −121 was replaced with BglII, PAL, or 3PAL as indicated. (iii) ARS120 without the C domain. The MCE at +120 was replaced with BglII as indicated. (D) Binding of Mcm1 to fragment II containing the native MCE or replacement of BglII, PAL, or 3PAL by EMSA. Fifty femtomoles of fragment II containing the native MCE at −121 (lanes 1 to 4), BglII (lanes 5 to 8), PAL (lanes 9 to 12), or 3PAL (lanes 13 to 16) was added to each lane. Amounts of Mcm1 added: lanes 1, 5, 9, and 13, 0 pmol; lanes 2, 6, 10, and 14, 2.2 pmol; lanes 3, 7, 11, and 15, 6.7 pmol; lanes 4, 8, 12, and 16, 20 pmol. WT, wild type. (E) DNase I protection of fragment II by Mcm1. Lanes: 1 to 4, MCE; 5 to 8, BglII; 9 to 12, PAL; 13 to 16, 3PAL. Amounts of Mcm1 added: lanes 4, 5, 12, and 13, 0 pmol; lanes 3, 6, 11, and 14, 5 pmol; lanes 2, 7, 10, and 15, 10 pmol; lanes 1, 8, 9, and 16, 20 pmol. The substituted sequence is boxed in lanes 1 to 8. Thick lines indicate protection of MCE or PAL. Thin lines indicate protection flanking MCE or PAL. An asterisk indicates hypersensitivity.
2-D gel electrophoresis.
Procedures for two-dimensional (2-D) gel electrophoresis analysis of replicating DNA have been described by Brewer and Fangman (10). For each strain, DNA from a single DNA preparation was used for the study of both ORI1 and ORI121. Probes for detecting ORI1 and ORI121 were prepared as follows. Plasmid DNA containing ARS1 or ARS121 was digested with NcoI or BamHI and EcoRI, respectively. Restriction fragments were gel purified and 32P labeled by random oligonucleotide priming.
DNA sequencing and sequence analysis.
The strategy used to determine the 1.0- to 1.5-kb DNA sequences of ARS120, ARS131n, ARS131A, ARS131j, and ARS131s is described in reference 16. To search for Mcm1 consensus elements (MCEs), we used the weighted matrix program of Mulligan et al. (47) and Goodrich et al. (30). The consensus matrix used in this search was compiled from 20 known Mcm1 binding sequences from promoters (16).
Deletions of ARS120.
The 5′ and 3′ deletions of ARS120 were constructed by two separate methods: exonuclease Bal31 treatment at a unique restriction site and subcloning of smaller restriction fragments by partial or full digestion at sites that produce a desired endpoint. The BamHI/SphI fragment of each deletion was then recloned between the BamHI/H3 sites of CEN vector pLC5. Other 5′ and 3′ deletions were made by subcloning smaller restriction fragments from the larger fragments (e.g., −679/+40 and −6/+834) into the BamHI/ScaI or H3/ScaI sites of CEN5 vector pLC5 (16).
Site-specific mutagenesis of MCEs in ARS120.
Plasmids pC−679/+40ΔBgl2, pC−679/+40PAL, pC−6/+246ΔBgl2, and pC−6/+246PAL were constructed by a three-step cloning-mutagenesis-cloning process. The first step is cloning the wild-type ARS fragment into a Bluescript KS− or SK+ vector, the second step is mutagenizing a specific 10-bp region containing an MCE, and the third step is transferring the mutated fragment into CEN5 vector YCp56. pC−679/+40+3PAL and pC−6/+246+3PAL were constructed by inserting a 75-bp sequence (TTCCTAATTAGGAATAAGATCCATT)3 containing three repeats of PAL (underlined). All enzyme manipulations and oligonucleotide mutagenesis were performed with the Bluescript KS− and SK+ vectors. All mitotic stability assays were carried out with the ARS fragments cloned into YCp56 (16).
DNA binding assays and DNase I footprinting analysis.
Electrophoretic mobility shift assay (EMSA) and DNase I footprinting analyses were carried out as previously described (17). The primers used for EMSA and footprinting of ARS120 as shown in Fig. 3A were as follows: fragment I, no. 7 (CATTTCATTTCCGGTTTTCTATCT) and 8 (AGCACCACCGTACCTCTAAGTTT); fragment II, no. 9 (AGAGGTACGGTGGTGCTA) and 10 (ATCCTTCAACAATAATACATAAAC); fragment III, no. 11 (TGACGGTATTAAGGAACATTT) and 12 (ATCTCAACTTACCCTACTTTCAC). The primers used for EMSA and footprinting of ARS131a were as follows: fragment I, no. 13 (CCATTTCATTTCCGATTTTC) and 14 (TGCTTTGATCCGGTTTACAGT); fragment II, no. 15 (TACCATTTTCCCTCCTTAT) and 16 (CACACTCAATTGGGTATCT); fragment III, no. 17 (TGACGATATTAAGGAACATTT) and 18 (CTCAAGTTACCCTACTCTCAC).
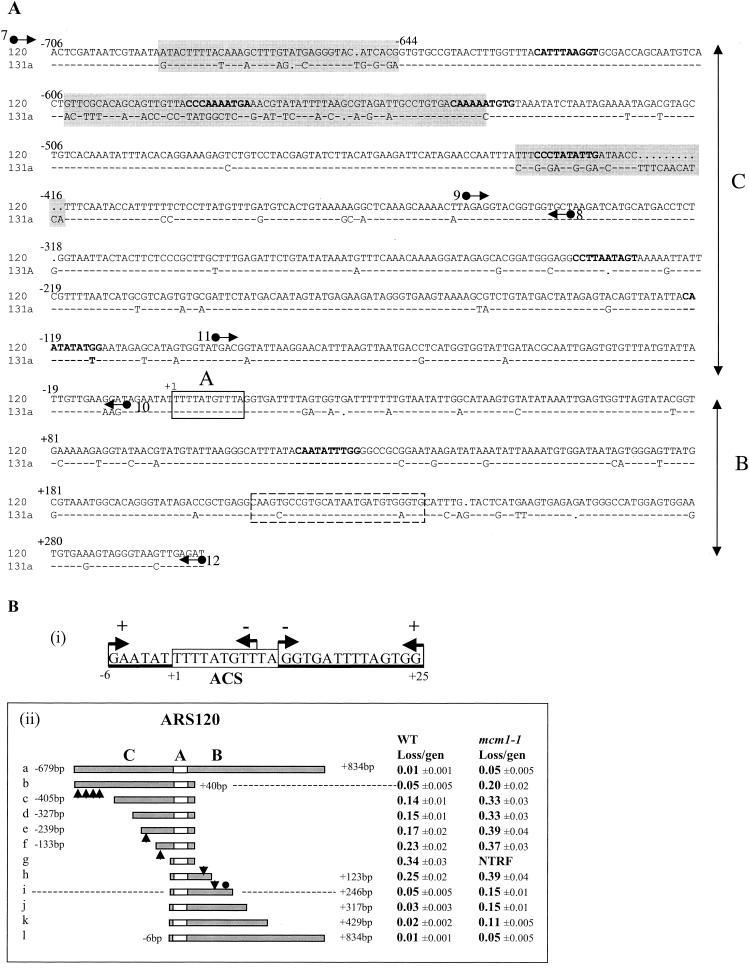
FIG. 3.
Dissection of ARS120. (A) Alignment of ARS120 and ARS131a DNA sequences. Domains C, A, and B are indicated. Nonconserved sequences clustered in the C domain are highlighted. ACS is boxed in solid lines. The Abf1 binding site is boxed in dashed lines. MCEs determined by the weighted matrix score (30, 47) are in bold type. Arrows represent primers used for sequence analysis in Fig. 4 and to generate DNA fragments by PCR for EMSA. Primers 7 and 8 were used to amplify fragment I by PCR. Primers 9 and 10 were used to amplify fragment II. Primers 11 and 12 were used to amplify fragment III. (B) Identification of the essential and important elements of ARS120. (i) Summary of HFT results of 5′- or 3′-end deletions generated by Bal31 digestion of the SacII/StuI fragment of ARS120. Endpoints of 5′ and 3′ deletions are represented by arrows pointing right and left, respectively. A plus sign indicates HFT activity, and a minus sign indicates no HFT activity. The boxed sequence is the ACS. (ii) Stability of minichromosomes containing the deletion fragments of ARS120 in the wild-type (WT) and mcm1-1 mutant strains. Endpoints of each deletion are indicated. Arrowheads represent locations of MCEs identified by the weighted matrix program. The filled circle represents the Abf1 binding site. The empty box represents the ACS. Domains C, B, and A are indicated. gen, generation; NTRF, no transformation.
Other methods.
Purification of glutathione S-transferase-Mcm1 has been described previously (17).
RESULTS
MREs in ARSs.
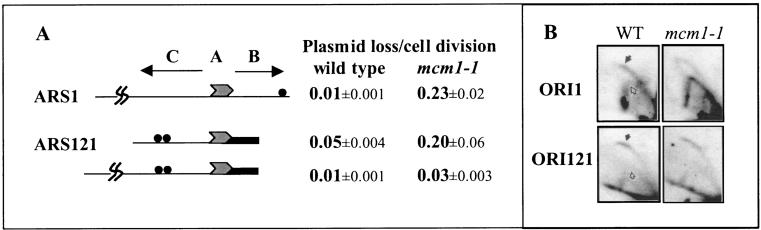
We have previously shown that plasmids containing ARS fragments of about 300 bp are destabilized by the mcm1-1 mutation (17). However, this destabilizing effect of mcm1-1 can be overcome in certain ARSs by additional flanking sequences. Figure 1A illustrates this ARS-specific destabilizing effect of mcm1-1 on minichromosomes containing large fragments of ARS1 and ARS121. In the mcm1-1 mutant, plasmids containing ARS1, which has few Mcm1 binding sites, are lost at a rate of about 0.2 per cell division, independently of the size of the ARS fragment. In contrast, plasmids containing ARS121, which has many Mcm1 binding sites, show dramatic differences in the loss rates, depending on the size of the ARS fragment. If we define the MFD of ARSs as the smallest ARS fragment that replicates efficiently in the wild-type strain, then sequences in the C domain outside of the MFD of ARS121 stabilize the minichromosome in the mcm1-1 mutant strain. These results suggest that Mcm1-responsive elements (MREs) located outside of the MFD in ARS121 are responsible for compensating for the reduced binding activity of Mcm1-1. These MREs are distinct from the MFD, where the pre-RC is assembled.
FIG. 1.
The C domain of ARS121 compensates for the destabilizing effects of the mcm1-1 mutation on minichromosome maintenance. (A) Plasmid loss rates of YCp1, YCpAB121, and YCp121 containing ARS1(853 bp), ARS121 (489 bp), and ARS121(877 bp), respectively. ACS is symbolized by shaded arrowheads. Abf1 binding sites are symbolized by filled circles. Thick bar represents AT-rich sequences containing near matches to the ACS found in the B domain of ARS121. (B) Autoradiograms of replicating DNA prepared from isogenic wild-type (WT) and mcm1-1 mutant strains separated by 2-D gel electrophoresis. Filled arrows point to initiation bubbles or “bubble” arcs. Unfilled arrows point to elongation forks or “Y” arcs. Portions of the results shown in panels A and B are reproduced from reference 17.
To investigate if reduced Mcm1 binding activity also affects replication initiation at chromosomal origins with an origin-specific effect, we examined the activity of ORI1 and ORI121 in the mcm1-1 mutant by 2-D gel analysis (Fig. 1B). Judging from the relative intensities of the “bubble” arc versus the “Y” arc in the wild-type and mutant strains, initiation of DNA synthesis appears to be significantly reduced in ORI1 but unaffected in ORI121 in the mcm1-1 mutant. This result indicates that larger ARS fragments reflect more faithfully the activity of the native replication origins. This result also indicates that further increases in the length of the ARS1 fragment are unlikely to compensate for the destabilizing effects of mcm1-1 since ORI1 inherently contains all of the flanking sequences on either side of ARS1. Because ORI1 and ORI121 are both early replicating origins (53) (S. Hunt, personal communication), this selectivity of Mcm1 does not appear to affect the temporal regulation of origin activation.
Two conserved telomeric X ARSs, ARS120 and ARS131A, respond differently to the mcm1-1 mutation.
We have previously shown that Mcm1 binds multiple sites outside of the MFD of ARSs (17). In Fig. 1A, we showed that MREs that alleviate the crippling effects of mcm1-1 on ARS activity are located outside of the MFD of ARS121 (65). Since Mcm1-1 has compromised DNA binding activity (17), it is likely that MREs and Mcm1 binding sites are identical. To examine whether Mcm1 binding sites are MREs, we need to show that the presence of Mcm1 binding sites directly correlates with the efficiency of replication.
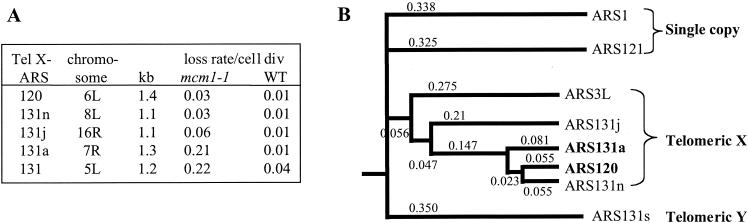
We took advantage of a family of highly conserved replication origins localized at the subtelomeric regions of chromosomes known as the X ARSs (14-16). Despite their sequence similarities, these ARSs respond differently to the mcm1-1 mutation (Fig. 2A). Plasmids containing ARS120 (XARS6L), ARS131n (XARS8L), ARS131j (XARS16R) (Fig. 2A), and ARS131c (43) are stable in the mcm1-1 mutant. In contrast, plasmids containing ARS131a (XARS7R) (Fig. 2A) and ARS131 (XARS5L) (43) are unstable. Therefore, variations in the sequences of these ARSs serve as a ready source of natural mutations for our study.
FIG. 2.
(A) Stability of minichromosomes containing telomeric (Tel) X ARSs in the wild-type (WT) strain and the mcm1-1 mutant. Sizes of ARS fragments and their chromosomal locations are indicated. div, division. (B) Pairwise similarities between telomeric X ARSs presented in a phylogenetic tree (26). Each number between one branch point and the next represents a match value (evolutionary divergence) for pair types (48).
A phylogenetic tree of a number of known ARSs is shown in Fig. 2B to illustrate the sequence relatedness or evolutionary distances between them. We focused our analysis on ARS120 and ARS131a, two of the most conserved telomeric X ARSs that show differential responses to the mcm1-1 mutation. Comparing these two sequences indicates that they are 89% identical within a 1-kb sequence with three stretches of nonconserved sequences clustered within a 300-bp region (−706 to −415) at the 5′ end of the C domains (Fig. 3A). We hypothesize that the nonconserved sequences of ARS120 contain the MREs that allow efficient replication of ARS120 in the mcm1-1 mutant. Inspection of sequences identified four potential MCEs (bold type) within this 300-bp region. Only one of them is conserved between ARS120 and ARS131a.
Deletion analysis and DNA binding assays localized Mcm1 binding sites in the nonconserved region of ARS120.
To identify the essential ARS consensus sequence of ARS120, we generated a series of end deletions by Bal31 digestion from either the 5′ or the 3′ side of a 350-bp SacII/StuI fragment of ARS120 (13). These deletion fragments were subcloned and tested for ARS function with a high-frequency transformation (HFT) assay (59). A sequence of 31 bp flanked by the leftmost or rightmost deletion endpoints that still preserved ARS function was identified (Fig. 3B, part i). The leftmost and rightmost deletion endpoints that destroyed HFT activity removed either part or all of an 11-bp sequence that is a perfect match to the ACS, confirming that this ACS is essential for ARS120 function. The first T of this ACS is designated position +1.
To identify the locations of elements important for efficient replication of ARS120, we constructed deletions along a 1.5-kb DNA sequence of ARS120 (Fig. 3A and B) and analyzed the ARS activity by assaying plasmid stability in the wild-type strain. The MFD, which retains almost the full ARS activity, is a 263-bp DNA fragment containing the A element and 246 bp of sequence in the B domain (Fig. 3B, part ii, i). Most of the B domain is dispensable in the presence of 679 bp of DNA in the C domain (Fig. 3B, part ii, b) confirming that elements in the B and C domains perform redundant functions. On the basis of the step increases in the loss rate of plasmids containing the deletion fragments, approximate locations of elements important for efficient replication of ARS120 can be determined. With the A element as a reference (position +1), important elements appear to be located at intervals between −679 and −405, between −239 and −133, and between −133 and −6 in the C domain and between +123 and +246 in the B domain, which also contains an Abf1 binding site (indicated by the filled circle). Interestingly, these intervals also contain MCEs (indicated by arrowheads) identified by the weighted matrix search.
The stability of the ARS120 deletion plasmids in the mcm1-1 mutant strain was also examined (Fig. 3B, part ii). The B domain is indispensable in the mcm1-1 mutant for full ARS activity (Fig. 3B, part ii, b). Removal of the interval containing four MCEs further destabilized ARS120 (Fig. 3B, part ii, c). Similarly, the C domain is indispensable for full ARS activity in the mcm1-1 mutant (Fig. 3B, part ii, k). The requirement for the C domain was alleviated by a DNA sequence between +429 and +834 3′ to the B domain (Fig. 3B, part ii, l). Removal of the sequence between +246 and +123 significantly increased the plasmid loss rate. No transformation was observed when both the B and C domains were removed (Fig. 3B, part ii, g).
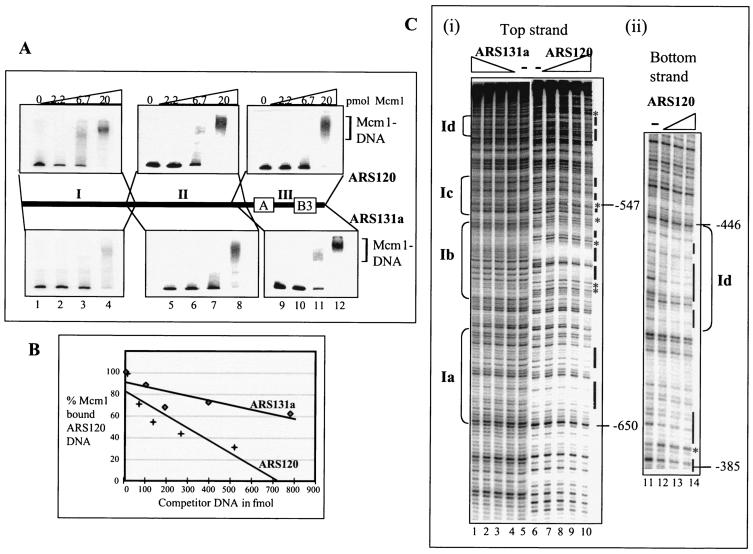
To investigate which of these intervals contain Mcm1 binding sites, we divided the 1-kb ARS120 sequence (Fig. 3A) into three fragments (I [−737 to −337], II [−354 to −7], and III [−94 to +304]) and assayed them for affinity for Mcm1 by EMSA (Fig. 4A). Mcm1 binds fragments I, II, and III of ARS120 with comparable efficiencies (top panel), consistent with the presence of Mcm1 binding sites in all three fragments. We also included the corresponding DNA fragments from ARS131a as a control. Mcm1 appears to bind fragments I, II, and III of ARS131a with an efficiency similar to that observed for ARS120 (bottom panel). Differences in the affinity of Mcm1 for fragments I of ARS120 and ARS131a were not immediately discernible by EMSA. To directly compare the affinities of Mcm1 for fragments I of ARS120 and ARS131a, we carried out competitive DNA binding assays. In this assay, Mcm1 bound to the ARS120 fragment I probe was competed with unlabeled fragment I DNA from ARS120 or ARS131a. The percentage of probe bound to Mcm1 was plotted against increasing concentrations of the unlabeled competitor DNA (Fig. 4B). On the basis of the slopes of the curves, ARS120 competed 3.5 times more efficiently than ARS131a for the Mcm1-bound DNA probe, suggesting that Mcm1 has a higher affinity for fragment I of ARS120.
FIG. 4.
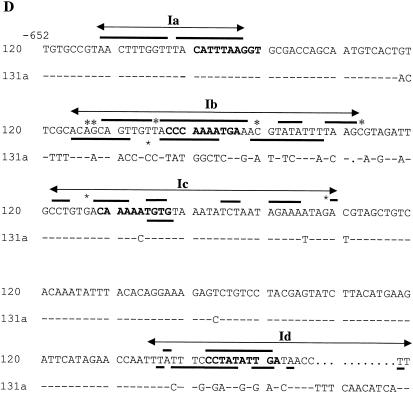
Binding of Mcm1 to ARS120 and ARS131a. (A) Binding of Mcm1 to fragments I, II (domain C), and III (domains A and B) of ARS120 and ARS131a by EMSA. Endpoints of fragments are indicated on the bar diagram. Fifty femtomoles of DNA was added in each lane. Amounts of Mcm1 added: lanes 1, 5, and 9, 0 pmol; lanes 2, 6, and 10, 2.2 pmol; lanes 3, 7, and 11, 6.7 pmol; lanes 4, 8, and 12, 20 pmol. (B) Competition for binding of Mcm1 by fragment I of ARS120 and ARS131a. Labeled fragment I of ARS120 bound to Mcm1 (4 pmol) was competed with increasing concentrations of unlabeled competitor DNA. (C) DNase protection in the nonconserved region of fragment I by Mcm1. (i) Top strand of ARS131a and ARS120. (ii) Bottom strand of ARS120. Amounts of Mcm1 added: lanes 5, 6, and 11, 0 pmol; lanes 2, 7, and 12, 5 pmol; lanes 3, 8, and 13, 10 pmol; lanes 4, 9, and 14, 20 pmol; lanes 5 and 10, 40 pmol. A bar indicates protection. An asterisk indicates hypersensitivity. Ia, Ib, Ic, and Id are footprints of Mcm1. (D) Sequence of DNase footprints of Mcm1 on fragment I. The protected sequence is underscored, and hypersensitivity is indicated by an asterisk. Protection patterns are deduced from at least two independent gels for both the top and bottom strands of ARS120 and ARS131a.
Nonconserved region of ARS120 is protected by Mcm1 from DNase I and required for efficient replication initiation.
Deletion analysis (Fig. 3B, part ii) and competitive DNA binding studies suggest that Mcm1 binding in the nonconserved region of ARS120 may be responsible for alleviating the high plasmid loss rate of ARS120 in the mcm1-1 mutant. To identify the exact locations of these putative Mcm1 binding sites, we carried out DNase I footprinting analysis of Mcm1-bound complexes on all three fragments of ARS120 and ARS131a. DNase I protection by Mcm1 was observed at four sites in both strands of fragment I of ARS120 (Fig. 4C, parts i and ii [only site Id is shown]). Sites Ia, Ib, Ic, and Id each cover about 20 to 40 bp, spanning a region of about 230 bp from −644 to −416 (Fig. 4D). The spacing of sites Ia, Ib, Ic, and Id is such that they each contain an MCE. Protection is observed at sites Ib, Ic, and Id, which are nonconserved sequences that include matches to the MCE. Protection is also observed at site Ia, which is conserved in both ARSs (Fig. 4D). However, Mcm1 does not show any protection in fragment I of ARS131a, not even in the conserved Ia site (Fig. 4C, part i). These results suggest that site Ia alone will not bind Mcm1 and that protection of site Ia in ARS120 requires the cooperation of sites Ib, Ic, and Id.
Mcm1 also binds to the C and B domains immediately flanking the ARS consensus sequence.
The immediate flanking sequences of the ACS in ARS120 and ARS131a are conserved. We observed DNase I protection by Mcm1 in fragments II and III of both ARS120 and ARS131a. Fragment II, which corresponds to the C domain proximal to the A element, is protected at multiple sites, but of particular interest is a site that contains a conserved MCE at positions −121 to −112 (Fig. 5A). Also protected in fragment II is the region between −239 and −230 that contains a match to the MCE (Fig. 5A). Fragment III, which corresponds to the B domain, is also protected at multiple sites, including the conserved MCE at positions +120 to +129 and half MCEs at positions +166 to +189 (Fig. 5B) about 20 bp from the Abf1 binding site (Fig. 3A). The locations of these Mcm1 binding sites correspond to the important elements identified in the deletion analysis (Fig. 3B, part ii). Although these DNase I protection and hypersensitive sites are subtle, they are reproducible.
Binding of Mcm1 to the C domain immediately flanking the ACS facilitates replication initiation.
To evaluate whether the subtle but reproducible footprinting patterns of Mcm1 at −121 and +120 are physiologically relevant, we mutagenized the MCEs at position −121 in the C domain and at position +120 in the B domain by site-directed mutagenesis and verified these mutations by DNA sequencing (16). Activity of ARS120 containing the ACS together with either the C domain (Fig. 5C, part ii) or the B domain (Fig. 5C, part iii) was assayed with the minichromosome maintenance assay. Replacing the 10-bp MCE (CAATATATGG) with a BglII restriction sequence (CTCGAGATCT [restriction sequence underlined]) at −121 in domain C of ARS120 reduced its activity from a plasmid loss rate of 0.087 to 0.135 (ii). Replacing the BglII site with the synthetic high-affinity Mcm1 binding site PAL (CCTAATTAGG) (50) restored stability (loss rate of 0.086). Inserting 75 bp containing 3PAL into the BglII site further stabilized the minichromosome (loss rate of 0.058). These results suggest that the Mcm1 binding site located at −121 bp from the ACS is important for the activity of ARS120 without a B domain and that the number of Mcm1 binding sites seems to make a difference. In contrast, replacement of the MCE with a BglII site at +120 in domain B had no significant effect on the activity of ARS120, which contains domain B but lacks domain C (Fig. 5C, part iii). Since domain B also contains the Abf1 binding site, this result suggests that Mcm1 may perform a redundant function in domain B or its function at a replication origin cannot be measured out of context in an autonomous plasmid replication assay.
In the absence of domain B, the number of Mcm1 binding sites in domain C correlates with the activity of ARS120.
To verify whether the substitution mutations actually destroyed or enhanced Mcm1 binding at position −121, we analyzed the binding of Mcm1 to these sequences first by EMSA (Fig. 5D) and then by DNase I footprinting assay (Fig. 5E). Fragment II containing either the wild-type MCE at position −121 or the mutant BglII, PAL, or 3PAL sequence was used in these experiments. Mcm1 formed heterogeneous complexes with fragment II containing either the MCE or BglII sequence at low Mcm1 concentrations (Fig. 5D, lanes 3 and 7). However, at higher concentrations, Mcm1 formed more discrete complexes with both templates, consistent with the presence of multiple Mcm1 binding sites in fragment II (Fig. 5A) and the fact that a higher concentration leads to full occupancy. The PAL sequence, which has a high affinity for Mcm1, formed discrete complexes with Mcm1, even at a lower concentration (6.7 pmol) (Fig. 5D, lane 11, asterisk). PAL also seemed to facilitate the binding of Mcm1 to other sites on fragment II to form discrete high-molecular-mass complexes (lane 11, dot). At a high concentration (20 pmol), all of the PAL templates were engaged in slow-migrating complexes with Mcm1 (lane 12). Cooperative binding of Mcm1 to the 3PAL template was evident (cf. lanes 11 and 15). At 2.2 pmol, Mcm1 bound poorly or not at all (lane 14). At 6.7 pmol, all of the 3PAL templates were engaged in discrete high-molecular-mass complexes (lane 15). This result further illustrates the propensity of Mcm1 to bind cooperatively to clustered sites.
Results from the footprinting analysis are consistent with the EMSA results and provide further insight into the importance of the MCE at −121 (Fig. 5E). Mcm1 protects the MCE at −121 (lanes 1 to 3, thick line) and an additional 3 to 4 bp on either side (thin line) separated by hypersensitive sites (*) but does not protect the BglII-substituted site (lanes 6 to 8). Although this MCE has only a modest effect on the binding of Mcm1 to fragment II (350 bp) (Fig. 5D), it has a significant effect on the activity of the larger (719-bp) ARS120 fragment (Fig. 5C, part ii) on a plasmid. Enhanced affinity of Mcm1 to the PAL sequence is evident (Fig. 5E, lanes 9 to 11). Mcm1 protects the PAL site (thick line) and an additional 4 or 5 bp on either side (thin line), as well as induces hypersensitive sites flanking the PAL sequence (*). Protection of the three PAL sites by Mcm1 is strong (lanes 14 to 16) and more extensive, covering the entire 75 bp including the 15-bp intervening sequences between the PAL sites. This protection pattern is consistent with the enhanced cooperativity of Mcm1 in binding clustered sites (Fig. 5D, lanes 14 to 15) to form a complex that excludes DNase I access, and it correlates with the improved ARS activity observed in the 3PAL insertion mutant. Together, these results indicate that the MCE at −121 is important for ARS120 activity and that increasing the number of MCEs in that region promotes ARS activity.
DISCUSSION
In the past, analyses of replication origins in S. cerevisiae have focused mainly on the MFD that includes only the A and B domains. Minichromosomes containing only the MFD of ARSs are unstable in the mcm1-1 mutant (17). In this study, we focused our attention on the C domain of two telomeric ARSs. The C domain contains extended DNA sequences that are required for autonomous replication of minichromosomes in the absence of the B domain in wild-type cells. In the mcm1-1 mutant, when Mcm1 DNA binding activity is limiting, the C domain is required even in the presence of the B domain. These observations led us to investigate the relationship between Mcm1 and the C domain.
Conserved telomeric replication origins that respond differently to limiting Mcm1 activity provide a source of natural mutations for dissecting domain C. The telomeric X ARSs ARS120 and ARS131a are conserved throughout the length of more than 1 kb, except for stretches of nonconserved sequences clustering within a 300-bp region at the 5′ end of the C domain. Deletion analyses coupled with DNA binding studies and Mcm1 footprinting analyses indicate that the binding of Mcm1 to this nonconserved region is responsible for overcoming limiting Mcm1 activity to allow for efficient replication of ARS120. Conversely, the lack of these Mcm1 binding sites is responsible for the crippled activity of ARS131a in limiting Mcm1 activity. We have also mutated conserved Mcm1 binding sites in the immediate vicinity of element A. We found that the MCE at −121 in domain C contributed significantly to the activity of ARS120 without domain B. In contrast, the MCE at +120 in domain B did not contribute significantly to the activity of ARS120 without domain C. These results together suggest that the activity of telomeric X ARSs is modulated by the occupancy of Mcm1 in domain C. The analyses of ARS120 (Fig. 3B) and ARS121 (Fig. 1) suggest that yeast replication origins may include regulatory elements located in the C domain hundreds of base pairs away from the ORC binding site—a novel concept for budding yeast but a common phenomenon for other eukaryotes. Although the B domains of replication origins are believed to be critical for assembling the pre-RC at the ACS, elements of the B domain are dispensable in the presence of C domains on plasmids. It is unclear what contributions each of these domains makes toward initiating DNA synthesis in native chromosomal replication origins. We have not explored the role of Mcm1 binding in the extended sequence 3′ of domain B in this study. Deletion analysis of ARS120 detects Mcm1-dependent elements located more than 400 bp 3′ of the ACS (Fig. 3B, part ii, l). Such Mcm1-dependent elements may result from Mcm1 binding, binding of Mcm1-interacting factors, or the indirect effect of Mcm1 on the regulation of these factors.
A cartoon that interprets the results presented in this study is shown in Fig. 6A. Mcm1 binds as a dimer to its cognate sequence to produce a 66° bend in the DNA at multiple sites in ARS120 (1, 66). We have shown that Mcm1 binds cooperatively to four MCEs in the C domain of ARS120 to protect four clustered sequences spanning a 250-bp region. The DNase I-hypersensitive sites interspersed between protected sequences are consistent with the distortion of DNA as a result of DNA bending. Our results showed that the MCEs are the actual binding sites of Mcm1. The larger protected areas presumably result from the inaccessibility of these sequences to DNase I owing to structural hindrance of the tertiary complex. This interpretation is also consistent with the protection pattern of Mcm1 in domain C of ARS121 reported in another study (17). Although one of the four MCEs is conserved in fragment I of ARS131a (Fig. 3A), Mcm1 binds nonspecifically to fragment I (Fig. 4A) without a discrete protection pattern (Fig. 4C, part i), underscoring the importance of a multiplicity of weak binding sites in promoting specific interactions. Footprinting analysis (Fig. 5A), supported by deletion analysis (Fig. 3B), indicates that two MCEs at −239 and −121 are bona fide Mcm1 binding sites. Mutating the MCE at −121 abolished Mcm1 binding at that site and diminished the activity of ARS120 (Fig. 5C, part ii). Reintroducing a synthetic high-affinity (PAL) sequence restored binding and ARS activity (Fig. 5C, part ii). We did not see a direct correlation between an increased affinity of Mcm1 for PAL and an increase in ARS activity (Fig. 5C, part ii, and E). It is important to bear in mind that Mcm1 is a combinatorial DNA binding factor and that in vitro binding studies of Mcm1 may not fully reflect the binding activity of Mcm1 in the presence of cofactors in vivo. Mutating the MCE at +120 had no significant effect on ARS120 activity, possibly because it has a redundant function in domain B. Alternatively, this site only functions in cooperation with binding sites in domain C and such long-range interactions only occur in intact replication origins or large ARS DNA fragments that include both the B and C domains.
FIG. 6.
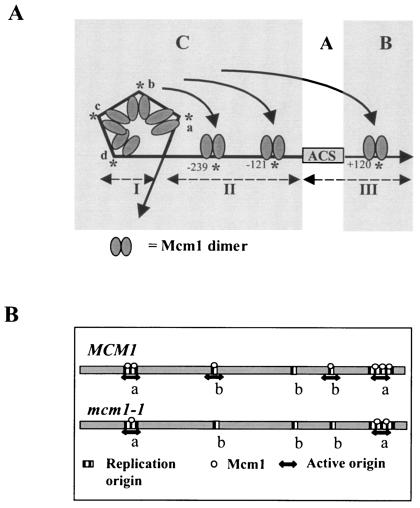
Models that incorporate the DNA binding and footprinting studies of ARS120. (A) The Mcm1 dimers bind cooperatively to the four MCEs (a, b, c, and d) in fragment I to form a large complex that protects an extensive region including the MCEs. Bending of DNA by Mcm1 results in a tertiary structure that occludes DNase I from accessing DNA. This structure is required for promoting ARS activity in the mcm1-1 mutant. Mcm1 also binds fragment II and protects the MCEs at −239 and −121 in domain C, as well as the MCE at +120 in domain B, but only the MCEs in domain C are shown to be important for ARS activity on the basis of plasmid stability assays. Long-range interactions depicted as arrows between MCEs in fragments I, II, and III cannot be assessed from DNA binding studies but may be important for creating a nucleosome-free zone for pre-RC assembly at the ACS. (B) Model for origin usage on the basis of occupancy of Mcm1. Origins are classified into two types. Type a origins contain many Mcm1 binding sites, whereas type b origins contain few Mcm1 binding sites. In wild-type cells, Mcm1 binds to most replication origins efficiently to promote replication initiation. In the mcm1-1 mutant or under conditions in which Mcm1 activity is low, Mcm1 only binds type a origins to promote their usage.
What might be the function of an array of bound Mcm1 that forms a tertiary complex spanning a region of about 250 bp? One possible function for this large complex is to exclude nucleosomes from occupying domain C to provide an environment for pre-RC assembly. Mcm1 has been shown previously to regulate donor preference in mating type switching by binding to the recombination enhancer and by altering the nucleosome display in a mating type-specific manner (67). Another possible function for this tertiary complex of Mcm1 in domain C is to limit the region where ORC can bind to enhance its specificity for the ACS because ORC has a propensity to bind AT-rich sequences (4, 19, 39). A third possible function for a tertiary complex is to bring Mcm1 closer to the pre-RC for direct physical interaction. These models are not mutually exclusive and testable. The binding of Mcm1 to the vicinity of a replication origin to promote replication initiation by altering local structure may be analogous to the binding of EBNA1 to adjacent regions of OriP in Epstein-Barr virus (27, 56) or dnaA at OriC in prokaryotes (32).
The number of Mcm1 binding sites appears to be important in altering the local structures of replication origins. DNA competition experiments suggest that the number can make up for the weak affinity of individual sites. Fragment II of ARS121, which contains eight or nine weak Mcm1 binding sites, competes well with the promoter sequence of MCM7, which contains two strong Mcm1 binding sites (17). Differential affinities of binding substrates specifying different roles have been described for other multifunctional regulators, such as ORC (20). This mode of action is also consistent with the known properties of Mcm1. Mcm1 is a degenerate DNA binding chromatin protein (25, 50), a property shared by other cell proliferation factors, such as E2F-RB (7, 49). It exerts its specific regulation of diverse genes through interactions with different complexes. The relatively weak and degenerate binding of Mcm1 reported in this study is the property of Mcm1 binding alone without a cofactor. We cannot rule out the possibility that these weak interactions are enhanced during replication initiation by as yet unknown cell cycle-specific cofactors. Candidates for such a cofactor include proteins Mcm2 to Mcm7, which are known to stimulate Mcm1 binding at early cell cycle gene promoters (25).
Our current model suggests that origin usage is dependent on the occupancy of Mcm1 (Fig. 6B). For simplicity, one can classify all replication origins in the yeast genome into two types, a and b. Type a origins, such as ARS121 and ARS120, contain many Mcm1 binding sites, while type b origins, such as ARS1 and ARS131a, contain few Mcm1 binding sites. Under conditions in which the cellular concentration of Mcm1 is high, most replication origins are occupied by Mcm1 and are competent to initiate DNA synthesis. When Mcm1 activity is limiting, as in the case of the mcm1-1 mutant, Mcm1 occupies few origins and few origins are competent to initiate DNA synthesis. The prediction is that activity of Mcm1 determines origin usage. Previous studies suggest that the activity of Mcm1 is regulated by the flux of glycolysis (18), carbon source (21), and osmotic shock (24, 69), as well as a variety of environmental stresses and growth phases (28). It would be interesting to investigate if under these different conditions, a set of replication origins different from that in cells growing under optimal conditions may be used. Although many MADS domain transcription factors play an important role in coordinating gene expression and cell proliferation in differentiating tissues, a direct role for these regulators in DNA replication has not been explored. Generalization of this model for Mcm1 may apply to other MADS domain transcription factors such as Agamous (8, 42), SRF (2), and MEF2 (40), as well as cell proliferation factors such as E2F-RB (7) and Myb (3), in plants and animals. Future studies to test this model may provide insight into the role of multifunctional regulators in coordinating cell division and gene expression during morphogenesis and development in metazoans.
Acknowledgments
We thank Maggie Merchant for performing the 2-D gel analysis and Ivan Liachko for technical assistance with figures. We thank Carol Newlon, Jim Theis, and Eric Alani for discussion and critical reading of our manuscript.
This work was supported by funding from the NIH (GM34190) and NSF (CHE-024 2328). J.J.D. is a GAANN fellow and is supported by NIH training grant GM07273.
REFERENCES
- 1.Acton, T. B., H. Zhong, and A. K. Vershon. 1997. DNA-binding specificity of Mcm1: operator mutations that alter DNA-bending and transcriptional activities by a MADS box protein. Mol. Cell. Biol. 17:1881-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arsenian, S., B. Weinhold, M. Oelgeschlager, U. Ruther, and A. Nordheim. 1998. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 17:6289-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beall, E. L., J. R. Manak, S. Zhou, M. Bell, J. S. Lipsick, and M. R. Botchan. 2002. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 420:833-837. [DOI] [PubMed] [Google Scholar]
- 4.Bell, S., and B. Stillman. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357:128-134. [DOI] [PubMed] [Google Scholar]
- 5.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 6.Bielinsky, A.-K., and S. A. Gerbi. 1999. Chromosomal ARS1 has a single leading strand start site. Mol. Cell 3:477-486. [DOI] [PubMed] [Google Scholar]
- 7.Bosco, G., W. Du, and T. L. Orr-Weaver. 2001. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat. Cell Biol. 3:289-295. [DOI] [PubMed] [Google Scholar]
- 8.Bowman, J. L., D. Smyth, and E. M. Meyerowitz. 1989. Genes directing flower development in Arabidopsis. Plant Cell 1:37-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brewer, B. J., and W. L. Fangman. 1994. Initiation preference at a yeast origin of replication. Proc. Natl. Acad. Sci. USA 91:3418-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brewer, B. J., and W. L. Fangman. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51:463-471. [DOI] [PubMed] [Google Scholar]
- 11.Broach, J., Y. Li, J. Feldman, M. Jayaram, J. Abraham, K. Nasmyth, and J. Hicks. 1983. Localization and sequence analysis of yeast origins of DNA replication. Cold Spring Harbor Symp. Quant. Biol. 47:1165-1173. [DOI] [PubMed] [Google Scholar]
- 12.Celniker, S. E., K. Sweder, F. Srienc, J. E. Bailey, and J. L. Campbell. 1984. Deletion mutations affecting autonomously replicating sequence ARS1 of Saccharomyces cerevisiae. Mol. Cell. Biol. 4:2455-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan, C. S. M. 1985. Ph.D. thesis. Cornell University, Ithaca, N.Y.
- 14.Chan, C. S. M., and B.-K. Tye. 1983. A family of Saccharomyces cerevisiae repetitive autonomously replicating sequences that have very similar genomic environments. J. Mol. Biol. 168:505-523. [DOI] [PubMed] [Google Scholar]
- 15.Chan, C. S. M., and B.-K. Tye. 1983. Organization of DNA sequences and replication origins at yeast telomeres. Cell 33:563-573. [DOI] [PubMed] [Google Scholar]
- 16.Chang, V. K. 1993. Ph.D. thesis. Cornell University, Ithaca, N.Y.
- 17.Chang, V. K., M. J. Fitch, J. J. Donato, T. W. Christensen, A. M. Merchant, and B.-K. Tye. 2003. Mcm1 binds replication origins. J. Biol. Chem. 278:6093-6100. [DOI] [PubMed] [Google Scholar]
- 18.Chen, Y., and B. K. Tye. 1995. The yeast MCM1 protein is regulated posttranscriptionally by the flux of glycolysis. Mol. Cell. Biol. 15:4631-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuang, R. Y., L. Chretien, J. Dai, and T. J. Kelly. 2002. Purification and characterization of the Schizosaccharomyces pombe origin recognition complex: interaction with origin DNA and Cdc18 protein. J. Biol. Chem. 277:16920-16927. [DOI] [PubMed] [Google Scholar]
- 20.DeBeer, M. A. P., U. Müller, and C. A. Fox. 2003. Differential DNA affinity specifies roles for the origin recognition complex in budding yeast heterochromatin. Genes Dev. 17:1817-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 22.Deshpande, A. M., and C. S. Newlon. 1992. The ARS consensus sequence is required for chromosomal origin function in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:4305-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubey, D. D., L. R. Davis, S. A. Greenfeder, L. Y. Ong, J. Zhu, J. Broach, C. Newlon, and J. A. Huberman. 1991. Evidence suggesting that the ARS elements associated with silencers of the yeast mating-type locus HML do not function as chromosomal DNA replication origins. Mol. Cell. Biol. 11:5346-5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fassler, J. S., W. M. Gray, C. L. Malone, W. Tao, H. Lin, and R. J. Deschenes. 1997. Activated alleles of yeast SLN1 increase Mcm1-dependent reporter gene expression and diminish signaling through the Hog1 osmosensing pathway. J. Biol. Chem. 272:13365-13371. [DOI] [PubMed] [Google Scholar]
- 25.Fitch, M. J., J. J. Donato, and B. K. Tye. 2003. Mcm7, a subunit of the presumptive MCM helicase, modulates its own expression in conjunction with Mcm1. J. Biol. Chem. 278:25408-25416. [DOI] [PubMed] [Google Scholar]
- 26.Fitch, W. M., and E. Margoliash. 1967. Construction of phylogenetic trees. Science 155:279-284. [DOI] [PubMed] [Google Scholar]
- 27.Frappier, L., and M. O'Donnell. 1992. EBNA1 distorts oriP, the Epstein-Barr virus latent replication origin. J. Virol. 66:1786-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilbert, D. M. 2001. Making sense of eukaryotic DNA replication origins. Science 294:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodrich, J. A., M. L. Schwartz, and W. McClure. 1990. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF). Nucleic Acids Res. 18:4993-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang, R. Y., and D. Kowalski. 1993. A DNA unwinding element and an ARS consensus comprise a replication origin within a yeast chromosome. EMBO J. 12:4521-4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jakimowicz, D., J. Majkadagger, G. Konopa, G. Wegrzyn, W. Messer, H. Schrempf, and J. Zakrzewska-Czerwinska. 2000. Architecture of the Streptomyces lividans DnaA protein-replication origin complexes. J. Mol. Biol. 298:351-364. [DOI] [PubMed] [Google Scholar]
- 33.Jarvis, E., D. C. Hagen, and G. F. Sprague, Jr. 1988. Identification of a DNA segment that is necessary and sufficient for α-specific and a-specific genes: implications for regulation of α-specific and a-specific genes. Mol. Cell. Biol. 8:309-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keleher, C. A., C. Goutte, and A. D. Johnson. 1988. The yeast cell-type-specific repressor α2 acts cooperatively with a non-cell-type-specific protein. Cell 53:927-936. [DOI] [PubMed] [Google Scholar]
- 35.Kim, S. M., and J. A. Huberman. 2002. Regulation of replication timing in fission yeast. EMBO J. 20:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohzaki, H., Y. Ito, and Y. Murakami. 1999. Context-dependent modulation of replication activity of Saccharomyces cerevisiae autonomously replicating sequences by transcription factors. Mol. Cell. Biol. 19:7428-7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar, R., D. M. Reynolds, A. Shevchenko, A. Shevchenko, S. D. Goldstone, and S. Dalton. 2000. Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr. Biol. 10:896-906. [DOI] [PubMed] [Google Scholar]
- 38.Laskey, R. A., and R. M. Harland. 1982. Replication origins in the Xenopus egg. Cell 31:503. [DOI] [PubMed] [Google Scholar]
- 39.Lee, J. K., K. Y. Moon, Y. Jiang, and J. Hurwitz. 2001. The Schizosaccharomyces pombe origin recognition complex interacts with multiple AT-rich regions of the replication origin DNA by means of the AT-hook domains of the spOrc4 protein. Proc. Natl. Acad. Sci. USA 98:13589-13594. [DOI] [PMC free article] [PubMed]
- 40.Lilly, B., B. Zhao, G. Ranganayakulu, B. Paterson, R. Schulz, and E. Olson. 1995. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science 267:688-693. [DOI] [PubMed] [Google Scholar]
- 41.Lipford, J. R., and S. P. Bell. 2001. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol. Cell 7:21-30. [DOI] [PubMed] [Google Scholar]
- 42.Lohmann, J. U., R. L. Hong, M. Hobe, M. A. Busch, F. Parcy, R. Simon, and D. Weigel. 2001. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105:793-803. [DOI] [PubMed] [Google Scholar]
- 43.Maine, G. T., P. Sinha, and B. K. Tye. 1984. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics 106:365-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marahrens, Y., and B. Stillman. 1992. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255:817-822. [DOI] [PubMed] [Google Scholar]
- 45.McInerny, C. J., J. F. Partridge, G. E. Mikesell, D. P. Greemer, and L. L. Breeden. 1997. A novel Mcm1-dependent element in the SWI4, CLN3, CDC6, and CDC47 promoters activates M/G1-specific transcription. Genes Dev. 11:1277-1288. [DOI] [PubMed] [Google Scholar]
- 46.Mesner, L. D., X. Li, P. A. Dijkwel, and J. L. Hamlin. 2003. The dihydrofolate reductase origin of replication does not contain any nonredundant genetic elements required for origin activity. Mol. Cell. Biol. 23:804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mulligan, J. E., D. K. Hawley, R. Entriken, and W. R. McClure. 1984. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 12:789-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Needleman, S. B., and C. D. Wunsch. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48:443-453. [DOI] [PubMed] [Google Scholar]
- 49.Ohtani, K. 1999. Implication of transcription factor e2f in regulation of DNA replication. Front. Biosci. 4d:793-804. [DOI] [PubMed] [Google Scholar]
- 50.Passmore, S., R. Elble, and B. K. Tye. 1989. A protein involved in minichromosome maintenance in yeast binds a transcriptional enhancer conserved in eukaryotes. Genes Dev. 3:921-935. [DOI] [PubMed] [Google Scholar]
- 51.Passmore, S., G. T. Maine, R. Elble, C. Christ, and B. K. Tye. 1988. Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MATα cells. J. Mol. Biol. 204:593-606. [DOI] [PubMed] [Google Scholar]
- 52.Raghuraman, M., B. J. Brewer, and W. L. Fangman. 1997. Cell cycle-dependent establishment of a late replication program. Science 276:806-809. [DOI] [PubMed] [Google Scholar]
- 53.Raghuraman, M. K., E. A. Winzeler, D. Collingwood, S. Hunt, L. Wodicka, A. Conway, D. J. Lockhart, R. W. Davis, B. J. Brewer, and W. L. Fangman. 2001. Replication dynamics of the yeast genome. Science 294:115-121. [DOI] [PubMed] [Google Scholar]
- 54.Rao, H., Y. Marahrens, and B. Stillman. 1994. Functional conservation of multiple elements in yeast chromosomal replicators. Mol. Cell. Biol. 14:7643-7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rao, H., and B. Stillman. 1995. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc. Natl. Acad. Sci. USA 92:2224-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, J. Harwood, J. F. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 20:4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simon, I., J. Barnett, N. Hannett, C. T. Harbison, N. J. Rinaldi, T. L. Volkert, J. J. Wyrick, J. Zeitlinger, D. K. Gifford, T. S. Jaakkola, and R. A. Young. 2001. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106:697-708. [DOI] [PubMed] [Google Scholar]
- 58.Spellman, P., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stinchcomb, D. T., K. Struhl, and R. W. Davis. 1979. Isolation and characterisation of a yeast chromosomal replicator. Nature 282:39-43. [DOI] [PubMed] [Google Scholar]
- 60.Theis, J. F., and C. S. Newlon. 1997. The ARS309 chromosomal replicator of Saccharomyces cerevisiae depends on an exceptional ARS consensus sequence. Proc. Natl. Acad. Sci. USA 94:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Theis, J. F., and C. S. Newlon. 1994. Domain B of ARS307 contains two functional elements and contributes to chromosomal replication origin function. Mol. Cell. Biol. 14:7652-7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Theis, J. F., and C. S. Newlon. 2001. Two compound replication origins in Saccharomyces cerevisiae contain redundant origin recognition complex binding sites. Mol. Cell. Biol. 21:2790-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Houten, J. V., and C. S. Newlon. 1990. Mutational analysis of the consensus sequence of a replication origin from yeast chromosome III. Mol. Cell. Biol. 10:3917-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walker, S. S., S. C. Francesconi, and S. Eisenberg. 1990. A DNA replication enhancer in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 87:4665-4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walker, S. S., A. K. Malik, and S. Eisenberg. 1991. Analysis of the interactions of functional domains of a nuclear origin of replication from Saccharomyces cerevisiae. Nucleic Acids Res. 19:6255-6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.West, A. G., and A. D. Sharrocks. 1999. MADS-box transcription factors adopt alternative mechanisms for bending DNA. J. Mol. Biol. 286:1311-1323. [DOI] [PubMed] [Google Scholar]
- 67.Wu, C., K. Weiss, C. Yang, M. Harris, B.-K. Tye, C. Newlon, R. Simpson, and J. Haber. 1998. Mcm1 regulates the recombination enhancer controlling donor preference in Saccharomyces mating-type control. Genes Dev. 12:1726-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wynne, J., and R. Treisman. 1992. SRF and MCM1 have related but distinct DNA binding specificities. Nucleic Acids Res. 20:3297-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu, G., R. J. Deschenes, and J. S. Fassler. 1995. The essential transcription factor, Mcm1, is a downstream target of Sln1, a yeast “two-component” regulator. J. Biol. Chem. 270:8739-8743. [DOI] [PubMed] [Google Scholar]