Abstract
RNA polymerase II (Pol II) termination is triggered by sequences present in the nascent transcript. Termination of pre-mRNA transcription is coupled to recognition of cis-acting sequences that direct cleavage and polyadenylation of the pre-mRNA. Termination of nonpolyadenylated [non-poly(A)] Pol II transcripts in Saccharomyces cerevisiae requires the RNA-binding proteins Nrd1 and Nab3. We have used a mutational strategy to characterize non-poly(A) termination elements downstream of the SNR13 and SNR47 snoRNA genes. This approach detected two common RNA sequence motifs, GUA[AG] and UCUU. The first motif corresponds to the known Nrd1-binding site, which we have verified here by gel mobility shift assays. We also show that Nab3 protein binds specifically to RNA containing the UCUU motif. Taken together, our data suggest that Nrd1 and Nab3 binding sites play a significant role in defining non-poly(A) terminators. As is the case with poly(A) terminators, there is no strong consensus for non-poly(A) terminators, and the arrangement of Nrd1p and Nab3p binding sites varies considerably. In addition, the organization of these sequences is not strongly conserved among even closely related yeasts. This indicates a large degree of genetic variability. Despite this variability, we were able to use a computational model to show that the binding sites for Nrd1 and Nab3 can identify genes for which transcription termination is mediated by these proteins.
RNA polymerases synthesize discrete transcripts by initiating and terminating transcription in response to specific sequence elements. To initiate transcription, DNA sequences direct the binding of transcription initiation factors and polymerase to appropriate sites upstream of genes. Termination signals, providing for the creation of functional 3′ ends, may reside either in the DNA template or, alternatively, are found in the nascent RNA transcript.
Eucaryotic RNA polymerases have evolved distinct mechanisms for termination. RNA polymerase III (Pol III) requires no protein factors but terminates efficiently after transcribing four to six consecutive U residues, presumably due to instability of the RNA-DNA hybrid in the enzyme active site (1, 7). RNA Pol I terminates in response to a protein factor, Reb1, which blocks further elongation by binding to a DNA sequence downstream of the termination site (28). The Reb1 site is situated in such a way that the paused polymerase contains an inherently unstable U-rich RNA-DNA hybrid in the active site.
The RNA Pol II termination mechanism is more complex than those employed by the other eucaryotic RNA polymerases, requiring a large multiprotein complex that recognizes the poly(A) signal in the nascent transcript (4, 17, 20, 21, 27, 43). Deletion or mutation of the poly(A) signal results in a failure to terminate messages at the appropriate site (9, 19). This observation suggests that processing of the nascent message is coupled to termination, but the mechanism of this coupling remains uncertain. In Saccharomyces cerevisiae, mutations in several yeast genes encoding components of the pre-mRNA 3′-end processing machinery involved in cleavage of the nascent transcript (RNA14, RNA15, and PCF11) result in readthrough transcripts, suggesting that cleavage of the nascent transcript is necessary for termination (5, 27). However, another mutant allele of PCF11 with impaired processing activity terminates properly, indicating that transcript cleavage is not an obligatory step in termination (30). Cleavage at the poly(A) site in mammalian transcripts is not required for termination in Xenopus laevis oocytes (25) or in HeLa extracts (38). Together, these results support an antitermination model in which the cleavage and polyadenylation machinery induces a termination-competent state in the polymerase in response to the emergence of the poly(A) signal in the nascent transcript (6, 9, 21). How this signal is imparted to the polymerase is not known but has been shown to require the carboxyl-terminal domain (CTD) of the largest subunit (21).
Not all Pol II transcripts, however, are processed by the cleavage-polyadenylation machinery. Pol II also transcribes a subset of noncoding small nuclear and nucleolar RNAs (snRNAs and snoRNAs). The 3′ ends of these transcripts are processed by a complex of 3′-to-5′ exonucleases, the exosome, which acts on nonpolyadenylated [non-poly(A)] 3′ ends (2, 13, 39). Previous studies have described a yeast pathway for termination of non-poly(A) Pol II transcripts which requires the RNA-binding proteins Nrd1 and Nab3, the helicase Sen1, a CTD kinase Ctk1, and the Pol II CTD (10, 36). More recent studies have implicated several components of a subcomplex of the cleavage and polyadenylation machinery in this pathway (12, 24, 35). Mutation in any of these genes leads to readthrough transcription of certain snRNA and snoRNA genes.
While proteins required for termination of non-poly(A) transcripts have been identified, the sequences that direct this process are poorly understood. Nrd1 has been shown to bind a sequence fortuitously present in an inverted U6 RNA gene (33, 34). Mutations that alter an internal GUAA in this sequence lead to readthrough transcription, indicating that this sequence is necessary for recognition by Nrd1p. Similar sequences are present in the regions downstream of some snoRNA and snRNA genes, together with a repeat of the sequence CUU (36). In this paper we present a detailed analysis of Nrd1 and Nab3 response elements in the downstream regions of two snoRNA genes, SNR13 and SNR47, and show that they are required for correct termination. This analysis confirms the presence of the Nrd1-responsive GUA[AG] motif and provides the first evidence that Nab3p recognizes the sequence UCUU. RNAs with mutations in either of these sequence motifs bind Nrd1p or Nab3p poorly and cause inefficient termination. We have applied a recently developed computational approach to demonstrate that together these elements provide a signal to Pol II that directs termination of yeast non-poly(A) transcripts.
MATERIALS AND METHODS
Yeast strains.
Yeast strains with integrated epitope-tagged nrd1-102 (YJC1166) and NRD1 (YJC1163) as well as the nab3-11 (YJC1099) strain have been described previously (10).
Plasmids.
To test the function of non-poly(A) terminators, we inserted snoRNA 3′ sequences upstream of a green fluorescent protein (GFP) gene driven either by an ADH promoter or by the natural snoRNA promoter and coding region. The GFP reporter vectors were based on the pRS shuttle vectors (32) and contained a GFP coding region followed by a CYC1 terminator. The GFP-416 vector expressing GFP driven by an ADH promoter has been described previously (36). We cloned 54-bp sequences comprising the upstream and downstream halves of the 108-bp fragment our group had previously shown to direct termination of the SNR13 gene (36) into a SpeI site located between the ADH promoter and the GFP coding sequences in GFP-416 and transformed this plasmid into YJC1163 and YJC1166. In the control plasmid snR13-Δ4-GFP (see Fig. 4), all of the sequences downstream of the mature snR13 RNA have been deleted.
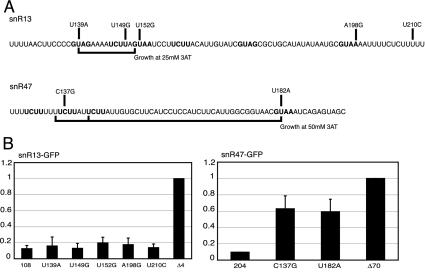
FIG. 4.
Site-directed mutagenesis of snoRNA-GFP constructs. (A) Sequences of a portion of snR13 or snR47 3′ downstream RNAs. Mutations are identified above the sequences. GUA[AG] and UCUU motifs are highlighted. Bars below the sequences represent mutants identified in the selection experiment (Fig. 3) that greatly impair the elements and result in growth at high concentrations of 3AT. (B) Northern blot analysis of total RNA derived from NRD1-HA WT yeast transformed with single mutant plasmids. The blots were probed for GFP as well as an SCR1 loading control. The snR47-Δ70 and snR13-Δ4 constructs were set at a value of 1, and all other deletions were expressed as a fraction of these maxima. snR13-108 and snR47-204 constructs represent basal levels of GFP expression, and all mutations were made from these plasmids.
To localize sequences important for non-poly(A) termination at the SNR47 locus, we constructed a set of plasmids containing the SNR47 promoter and snoRNA coding region together with various lengths of downstream sequence (Fig. 1B). DNA fragments were amplified by PCR from yeast genomic DNA, and the amplified DNA products were cloned as SacI/SpeI fragments into a SacI/SpeI-digested GFP-416 vector (snR47-398-GFP-416, snR47-116-GFP-416, and snR47-12-GFP-416) and transformed into YJC1163. We also cloned a 116-bp fragment derived from sequences immediately downstream of the mature SNR47 gene into the SpeI site of the ADH-driven GFP-416 vector (ADH-116-GFP-416). Plasmids were verified by sequencing and transformed into YJC1166 and YJC1099.
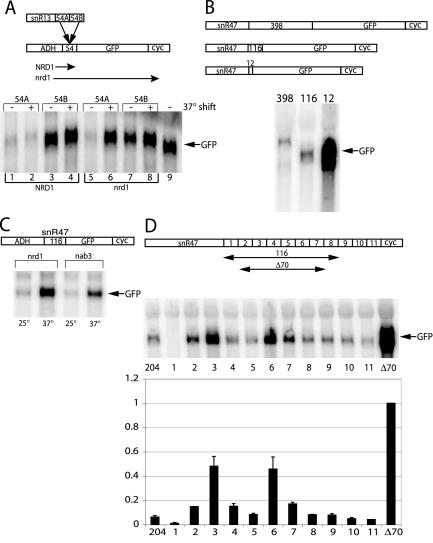
FIG. 1.
Definition of snoRNA 3′ downstream sequences that confer termination function. (A) Northern blot analysis of total RNA derived from a NRD1 WT or a ts nrd1-102 mutant strain at permissive (30°C, 120 min) or restrictive (37°C, 120 min) temperatures. Yeast were transformed with the ADH-snR13(54)-GFP plasmid constructs containing either the 54A or 54B fragment. (B) Northern blot analysis of total RNA derived from NRD1 WT yeast transformed with snR47-GFP plasmids containing various lengths of 3′ downstream sequence. (C) Northern blot analysis of total RNA derived from ts nrd1 or nab3 mutant strains at permissive (30°C, 120 min) or restrictive (37°C, 120 min) temperatures. Yeast were transformed with an ADH-snR47-GFP plasmid containing 116 bp of snR47 3′ downstream sequence. (D) Northern blot analysis of total RNA derived from NRD1 WT yeast transformed with the snR47 15-bp deletion series plasmids. The blot was probed for GFP as well as an SCR1 loading control. The Δ70 construct was set at a value of 1, and all other deletions were expressed as a fraction of this maximum. The snR47(204) plasmid contained an intact snR47 3′ downstream sequence from which all the deletions were derived.
To more clearly define the SNR47 termination region, we constructed a series of sequential, nonoverlapping 15-bp deletions spanning 166 bp immediately downstream of the SNR47 snoRNA coding region. The 15-bp deletions were constructed by PCR by using pairs of primers to amplify sequences upstream and downstream of the deletion. Upstream and downstream fragments were amplified separately and combined by a second PCR using only the outer primers. The inner primers were complementary to each other, comprising 20 bases on each side of the deletion. Amplified DNA products were cloned as SacI/SpeI fragments into a GFP-415 vector and transformed into YJC1163. The Δ70 construct (with sequences from +126 to +195 removed) was amplified by PCR using a similar strategy. All plasmids were verified by sequencing. The sequences of all primers and details of the PCR deletion protocol are available upon request.
Single base changes were introduced into the SNR13 and SNR47 downstream regions using the QuikChange protocol (Stratagene). The DNA fragments were amplified by PCR from the snR47-204-GFP plasmid or from the SNR13-108-GFP plasmid. Plasmids were then transformed into YJC1163.
Selection of mutant snR13 and snR47 termination elements.
To select for mutations in snoRNA termination elements, we developed a selectable marker vector based on the previously described GFP vectors. In this series, we exchanged the GFP for HIS3 to allow selection on media lacking histidine and containing 3-aminotriazol (3AT). Insertion of the 66-bp snR13 or the 70-bp snR47 terminator upstream of the HIS3 coding region resulted in a partially histidine-negative phenotype and a marked sensitivity to 3AT (Fig. 2). We selected mutant snoRNA termination elements from libraries of mutated DNA fragments in which the snoRNA downstream region was mutated at a frequency of less than one mutation per molecule. These fragments were generated by PCR amplification using DNA oligonucleotides containing a region spanning the snR47 or snR13 critical termination sequence defined by the deletion studies (Fig. 2B). These oligonucleotides contained a sequence at the 3′ end complementary to the 5′ end of the HIS3 gene. Using a second oligonucleotide complementary to a sequence internal to the HIS3 gene allowed amplification of a DNA fragment containing a snoRNA termination region immediately upstream of the HIS3 coding region. The 5′ end of the upstream oligonucleotide contained sequences complementary to the ADH promoter present in the vector. The snR47 and snR13 regions of these oligonucleotides were doped with a 1% change to the other three bases at each position. In addition, we changed the A residue 71 bases from the end of the mature snR47 snoRNA to a T (Fig. 3) to eliminate an ATG codon that might have led to nonsense-mediated decay of the nascent message. This change had no effect on the function of the snR47 terminator (data not shown). PCR products with mutated snoRNA sequences upstream of a HIS3 coding sequence were then transformed into YJC1163 along with gapped vector ADH-HIS3 digested with XbaI/MscI. Transformations were plated on synthetic complete (SC) medium lacking leucine (−leu) or on SC −leu−his medium containing either 10 mM 3AT (snR47) or 2.5 mM 3AT (snR13). Mutated bases that knock out the element allow for growth on 3AT. Single colonies from control or 3AT plates were used in a yeast colony PCR to amplify the snoRNA insert, and these PCR products were sequenced.
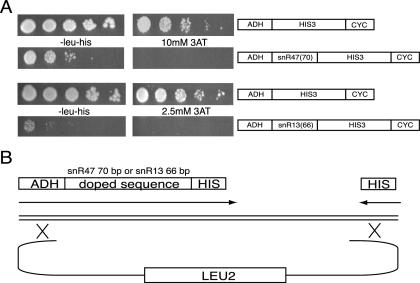
FIG. 2.
Assay for mutant snoRNA termination elements. (A) Growth of yeast harboring the plasmids shown at right. Note the lack of growth on 10 mM 3AT (snR47) and 2.5 mM 3AT (snR13) when snoRNA sequences are present in front of the HIS3 gene. (B) Design of strategy to generate a library of mutant snoRNA termination elements. snR47 70 bp or snR13 66 bp were doped at 1% per base. The doped oligonucleotide was used as an upstream primer to generate a PCR product with ends homologous to a gapped vector. The PCR product and gapped vector were transformed together into yeast. See Materials and Methods for specific protocols.
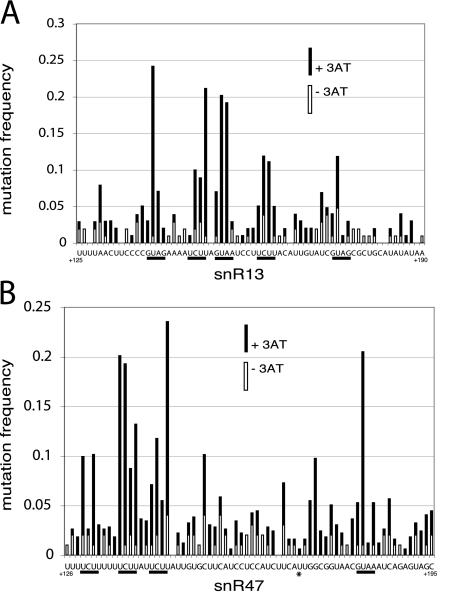
FIG. 3.
Distribution of mutations in snR13 (A) or snR47 (B) snoRNA termination elements. The mutation frequency at each base is plotted for selected (+3AT) and control (−3AT) populations. GUA[AG] and UCUU motifs are underlined. An asterisk underneath the snR47 sequence denotes an A nucleotide that was changed to a T nucleotide in all clones (see Materials and Methods). The positions of the sequences with respect to the 5′ end of the mature snoRNA are indicated at the beginnings and ends of the sequences.
Preparation and analysis of yeast RNA.
Total RNA was extracted from yeast as previously described (35). Northern blot analysis was performed using 20 μg of RNA per sample. The GFP riboprobe was made with T3 RNA polymerase, transcribing a linearized GFP-containing plasmid in the presence of [α-32P]CTP. The SCR1 control probe was made by random priming of Klenow DNA polymerase on an SCR1 PCR product in the presence of [α-32P]dCTP. Blots were scanned on a phosphorimager, and bands were quantified using ImageQuant software.
Protein expression and purification.
PCR primers were used to amplify a region of the Nrd1 gene encoding residues 308 to 560. This fragment was cloned into a derivative of pMAL-c2E (New England BioLabs) in which we placed a TEV protease cleavage site. The resulting plasmid, pMAL-Tev-Nrd1308-560, was used to express Nrd1 fused to the C terminus of the Escherichia coli maltose-binding protein (MBP) such that the Nrd1308-560 fragment could be released by TEV cleavage. In a similar manner, a Nab3 fragment encoding residues 277 to 565 was cloned into the same vector.
Expression of recombinant Nrd1308-560 protein in E. coli strain BL21(DE3) was induced in low-salt Luria broth with 0.3 mM isopropyl-d-thiogalactoside for 3 h at 25°C. Cell pellets from induced cultures were resuspended in column buffer (200 mM NaCl, 20 mM Tris · HCl [pH 7.4], 10 mM 2-mercaptoethanol, 1 mM EDTA, and protease inhibitors [Sigma P8465]). Cells were lysed by sonication, and cell debris was removed by centrifugation at 10,000 × g for 20 min.
The extract was passed over amylose resin and eluted with 10 mM maltose, and the maltose eluate was dialyzed into 20 mM Tris (pH 7.4), 20 mM NaCl, and 10% glycerol and run on a Q Sepharose column. The MBP-Nrd1 fusion protein which eluted at 0.5 M NaCl was then cleaved with TEV protease for 4 h at 30°C. The digested protein was again passed over a Q Sepharose column, and the cleaved Nrd1 polypeptide was recovered in the flowthrough, dialyzed into 20 mM Tris and 20 mM NaCl, and applied to a heparin agarose column. The Nrd1308-560 protein eluting at 0.7 M NaCl was dialyzed into RNA-binding buffer and frozen at −80°C. The Nab3277-565 protein was purified by the same strategy with the following changes. First, the protein was expressed in BL21(DE3)pLysS cells. Second, the protein eluted from Q Sepharose at 0.4 M NaCl and from heparin at 0.8 M NaCl.
RNA-binding assays.
RNA binding reactions were carried out in a buffer containing 100 mM NaCl, 20 mM Tris (pH 7.4), 10% glycerol, 1 mM dithiothreitol, 0.1 mM EDTA, 4 mM MgCl2, 40 μg of bovine serum albumin/ml, and 200 μg of tRNAPhe/ml. Labeled RNA probes were present at a final concentration of 2 nM, and protein concentrations determined using the Bradford method were as described in the figure legends. Binding was initiated by mixing 6 μl of RNA and 6 μl of protein and incubating for 20 min at room temperature. For competitions, 500 nM Nab3-specific or 50 μM Nrd1-specific unlabeled RNA oligonucleotide was used. The binding reaction was stopped by adding 4 μl of 40% sucrose and loading 12 μl of the binding reaction mixture onto the gel. Binding reaction mixtures were separated on 5% polyacrylamide gels (19:1 acrylamide-bisacrylamide, 50 mM Tris, 50 mM borate, 1 mM EDTA [pH 8.3], 5% glycerol) that had been prerun for 1 h at 160 V in a 4°C cold room and run for 3 h at 160 V. Gels were scanned on a LI-COR Odyssey infrared imaging system.
Labeled and unlabeled RNA probes were obtained from Dharmacon and corresponded to frequently mutated regions 3′ of the snR47 gene. The upstream 23-base fragment had the sequence UUUUCUUUUUUCUUAUUCUUAUU, while the downstream fragment had the sequence UUGGCGGUAACGUAAAUCAGA. Each of the oligonucleotides was synthesized with and without a 5′ Alexa Fluor 680 tag. In addition, we used mutant untagged RNA oligonucleotides with the sequences shown below in Fig. 6, in which the indicated residues correspond to frequently observed mutations that inactivate the snR47 Nrd1/Nab3 response element.
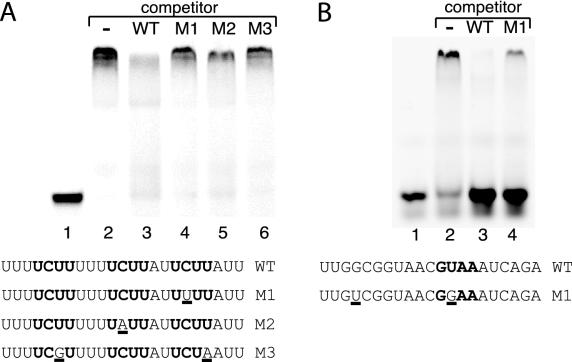
FIG. 6.
Gel mobility shift competition assays. Binding conditions were as described in Materials and Methods. Each reaction mixture contained 2 nM labeled probe RNA and 1.25 μM Nab3p (A) or Nrd1p (B) protein. The indicated competitor RNAs were at a final concentration of 500 nM (Nab3) or 50 μM (Nrd1).
Computational methods.
We used a computational model named GOMER (generalizable occupancy model of expression regulation) to determine how well the putative binding sites for Nrd1p and Nab3p identify genes for which transcription termination is mediated by these proteins. GOMER is based on a simple thermodynamic model for binding site occupancy used to rationalize coregulated genes (18), but we have generalized the model to include cooperative and competitive interactions (J. Granek and N. Clarke, unpublished data). The model was parameterized using a training set of snoRNAs for which transcription termination is affected by loss of Nrd1 (36). In addition to optimizing parameters for a cooperative Nrd1 and Nab3 model, we optimized parameters for models of binding by Nrd1 alone and by Nab3 alone. The initial values for the parameters, including the position weight matrices for Nrd1 and Nab3 binding, were estimated subjectively from a consideration of the mutational analysis (Fig. 3) and from inspection of the 3′ regions of the training set snoRNAs. The model parameters were then varied systematically. The varied parameters were (i) the region of sequence considered to be bound by the proteins, (ii) the “stringency” of the position weight matrices that describe the binding specificity of Nrd1p and Nab3p, and (iii) for the cooperative model, the interaction distance limit between binding sites for the two proteins. The stringency parameter has the effect of increasing or decreasing the weight of the favored bases over the unfavored and was applied equally to all positions; individual position weights were not varied independently. Sequence logos (31) for the position weight matrices obtained for each model showed that the cooperative model used binding sites of lower stringency than the single-protein models (see Fig. 8C).
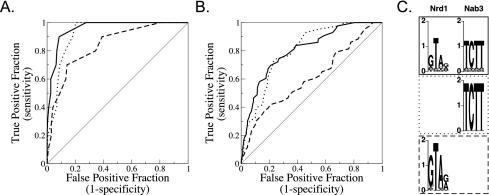
FIG. 8.
Tests of models predicting non-poly(A) terminators in the S. cerevisiae genome. (A) Receiver operator characteristic (ROC) curves (15) for the training set compared to those for all other open reading frames, snRNAs, and snoRNAs, using the optimized binding models for Nrd1 alone (dashed), Nab3 alone (dotted), and cooperative Nrd1-Nab3 binding (solid). (B) ROC curves for test set RNA genes compared to those for the nontest set. The models and parameters are the same for the training set curves and the test set curves. The ROC curve for a random regulatory model that has no predictive value at all would run along the diagonal, while a ROC curve for a perfectly predictive regulatory model would run up the y axis and then along the top of the plot. (C) Sequence logo representations (31) of the binding specificities of Nrd1 and Nab3 optimized for the cooperative model, Nrd1 alone, and Nab3 alone. The y axis shows bits of information. The line patterns of the boxes surrounding the sequence logos correspond to the lines in the ROC curves.
For a given set of parameters, sequence features annotated in the yeast genome as being open reading frames, snRNAs, or snoRNAs were scored according to the model and then rank ordered according to their score. The ranks of the training set genes were then evaluated using two rank-order metrics (8), which were used to guide the optimization of the model parameters. Each of the final optimized models was analyzed to determine its ability to distinguish a test set of snRNA and snoRNA genes from all other features. The test set consists of snRNAs and snoRNAs that are expected to be transcriptionally terminated by a mechanism independent of polyadenylation. Excluded from this test set list are two groups of snoRNAs which are not themselves transcriptionally terminated: one group consists of snoRNAs processed from introns, and the other group consists of snoRNAs that are cotranscriptionally expressed with other snoRNAs and are not at the 3′ end of the transcript. These two groups of snoRNAs were, however, included in the set of “non-test” sequence features to which the test set was compared. Additionally, the 10 snoRNAs used for training were excluded from both the test set and the non-test set, so as not to bias the analysis. In total there are 43 genes in the test set and 6,286 genes in the non-test set. It is important to note that there is no direct experimental evidence demonstrating a role for Nrd1 or Nab3 in termination of the genes in the test set. Complete lists of training and test set genes will be provided upon request.
RESULTS
Boundaries of SNR47 and SNR13 termination sequences.
Previous experiments have indicated that sequences downstream of snoRNA genes contain cis elements that are important for directing termination and/or 3′ processing of the primary RNA transcript (22, 35, 36). To further define the boundaries of these elements, we made deletions in two snoRNA downstream regions.
Previously, we showed that a Nrd1 response element was contained within 108 bp downstream of the end of the mature snR13 transcript (36). To test this region further, we divided the 108-nucleotide segment into two 54-bp halves, A and B. snR13 54A and 54B were placed upstream of a GFP reporter gene under the control of an ADH promoter. The constructs were expressed in either a wild-type (WT) or temperature-sensitive nrd1 mutant yeast strain, and readthrough of the plasmid-borne reporter gene was analyzed by Northern blotting by probing for GFP. Figure 1A shows that the 54A fragment contained snR13 sequences sufficient for termination. Readthrough into GFP resulted only in the nrd1 mutant strain shifting to the nonpermissive temperature. In contrast, the strains with a plasmid containing the 54B fragment expressed GFP mRNA in all cases. This demonstrates that the 54 bp adjacent to the 3′ end of the mature snR13 transcript contains elements that direct termination and that sequences downstream of this 54-base fragment are not, on their own, sufficient for termination.
The SNR47 gene was first studied using a plasmid containing both the snoRNA promoter and coding region. Deletions from the 3′ end of the downstream region revealed that elements conferring a termination function are contained within the 116 bp adjacent to the 3′ end of the mature snR47 transcript (Fig. 1B). Extending the sequence to the start codon of the downstream gene (398 bp) did not result in a greater repression of GFP expression. The 116-bp snR47 fragment was then placed in the ADH-GFP construct. Figure 1C shows that in either nrd1 or nab3 strains at the permissive temperature this sequence still repressed expression of GFP. However, upon shifting the nrd1 and nab3 mutant strains to the nonpermissive temperature, readthrough transcription was evident. SnR47, which is both Nrd1p and Nab3p responsive, stands in contrast to snR13, which seems to be primarily Nrd1p responsive (36).
Bipartite arrangement of SNR47 termination elements.
To further delineate specific snoRNA downstream elements, consecutive 15-bp deletions of the SNR47 downstream region were generated. SnR47 was utilized because it is both Nrd1p and Nab3p responsive (Fig. 1C). Constructs contained ∼550 bp upstream of and including the SNR47 gene and 204 bp downstream of the mature end of the transcript. A set of constructs was made that contained a series of 11 sequential 15-bp deletions of the downstream region (Fig. 1D). This region was followed by a GFP reporter and the CYC1 cleavage and polyadenylation signal. Readthrough of the plasmid-borne SNR47 gene was analyzed by Northern blotting by probing for GFP. Figure 1D shows that when regions 3 and 6 were deleted, significant readthrough into GFP resulted. However, these deletions independently did not show the maximum readthrough seen in the Δ70 construct (Fig. 1D), suggesting that important elements must synergize to obtain maximum termination function.
Genetic selection of mutant SNR13 and SNR47 termination elements.
Deletion analysis of the 3′ downstream regions of SNR13 and SNR47 snoRNA genes identified regions important in directing termination. However, we wished to determine the specific nucleotides within those regions that contribute to function. We designed an assay wherein we could select for variant sequences that knocked out the function of the snoRNA termination elements, thus highlighting those bases that were critical within the minimal sequence stretch we had identified.
Placing 66 bp of the snR13 sequence (extending from the 3′ end of the mature end of the snoRNA and continuing slightly beyond the 54A fragment), which contains sufficient termination function (Fig. 1A), in front of the HIS3 gene resulted in lack of growth on 2.5 mM 3AT for yeast harboring this plasmid (Fig. 2A). We exploited this fact to select for point mutants showing increased readthrough into the HIS3 gene. Oligonucleotide-directed mutagenesis was used to mutagenize the snoRNA terminator region at a rate of 1% per base, and PCR fragments containing these mutant terminators were tested for readthrough transcription by transformation and selection. Yeast colonies from both the 3AT-selected and unselected plates were picked (99 and 105 colonies, respectively), and the snR13 region was amplified by PCR and sequenced. The frequency of mutations in the unselected pool was ∼0.67, as expected from the percentage of bases in the doped nucleotide pools. The selected sequences contained an average of 2.09 mutations per element. Figure 3A shows a graph of the control (−3AT) and selected (+3AT) populations, in which the frequency of mutation at each base was plotted. It is evident that specific bases within the sequence were more critical than others in maintaining the function of the element. Specifically, a GUA[AG] and several UCUU repeats were important motifs along the 66-bp element.
The same strategy was used to probe the snR47 element. Placing a 70-bp stretch of snR47 sequence, extending from the middle of deletion region 2 to the middle of deletion region 7 (Fig. 1D) and encompassing the most critical stretches of the snR47 3′ downstream region, in front of the HIS3 gene resulted in reduced growth on SC −leu−his medium and a complete lack of growth on SC −leu−his medium containing 10 mM 3AT (Fig. 2A). Oligonucleotide design and transformation were done as for snR13 (Fig. 2B). Transformants were picked from SC −leu control plates (99 colonies) and SC −leu−his 10 mM 3AT plates (164 colonies) to identify faulty mutagenized snR47 elements. As with the snR13 experiment, the background frequency of mutation (0.77) was as expected for the percentage of doped nucleotides. The selected sequences contained an average of 2.47 mutations per element. Figure 3B shows a graph of the control (−3AT) and selected (+3AT) populations. The mutation frequency at each base is plotted. As was seen with snR13, the unselected mutations were randomly distributed, while the mutations in the selected pool identified certain bases within the 70-bp stretch that are more critical than others. Again, UCUU and GUAA motifs appeared to be important for termination function.
To determine which mutant elements contained the most severe mutations, all of the strains containing mutant elements were tested on higher concentrations of 3AT. The most deleterious mutations in each termination region contained multiple base changes in the short motifs (Fig. 4A). In the case of snR13, mutations that altered the two upstream GUA[AG] motifs were resistant to 25 mM 3AT. A triple mutation in the snR47 termination region affecting both a UCUU and a GUAA motif was resistant to 50 mM 3AT. This is consistent with the apparently synergistic effect of the Δ70 deletion compared to effects of individual 15-bp deletions in the snR47 downstream region (Fig. 1D).
Readthrough analysis of mutant SNR terminators in their natural context.
Genetic selection experiments defined specific bases within the snR13 and snR47 elements that were critical to their termination function. This selection was done in a heterologous context, however, and we wished to look at the effect of the most prevalent base changes in the context of the snoRNA promoter and gene itself.
A construct containing the entire SNR13 gene, including 108 bp of the 3′ downstream sequence followed by GFP, was mutated by site-directed mutagenesis to target specific bases within the snR13 sequence (Fig. 4A). The specific mutants constructed are shown above the RNA sequence in the figure and correspond to the most frequently observed base substitutions in the mutant population, with the exception of the two most-3′ mutations, which correspond to mutations identified by Steinmetz and Brow (35) in an independent analysis of snR13 termination elements. Plasmids were transformed into yeast, and GFP RNA was analyzed by Northern blotting. Figure 4B shows that making single base changes in the sequence did not cripple snR13 termination function to any great extent. Readthrough of the single mutations was about 10 to 20% of the maximum. Altering more than one base, in a number of the short sequence motifs, seems to have been necessary for knocking out termination function.
SnR47 was examined in the same way. A construct containing 204 bp of snR47 downstream sequence followed by GFP was mutagenized at specific bases within the snR47 sequence (Fig. 4A). Figure 4B shows that making single changes in the snR47 3′ downstream region had a significant effect on termination, with the level of readthrough reaching ∼60% of the maximum. This is a very different effect than that observed for snR13. Compromising the termination function of the snR47 element requires fewer changes than it does to knock out the snR13 element.
Nrd1 and Nab3 bind to snoRNA termination motifs.
We tested, using electrophoretic gel mobility shift assays, the binding of Nrd1 and Nab3 to labeled synthetic RNAs corresponding to elements in the snR47 downstream region. Both Nrd1 and Nab3 RNA-binding domains were expressed as MBP fusions in E. coli. The MBP domain was removed by TEV protease cleavage, and the Nrd1308-560 and Nab3277-565 proteins were purified to homogeneity by ion exchange chromatography (see Materials and Methods).
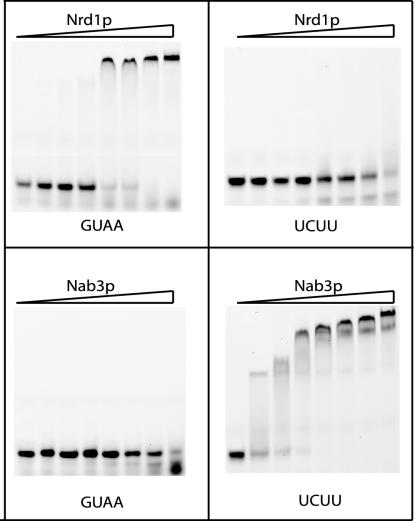
The probe labeled UCUU corresponds to sequences from 27 to 49 bases downstream of the 3′ end of the mature snR47 snoRNA. This RNA oligonucleotide contains three UCUU sequences that we have shown to be frequently mutated to yield readthrough transcription beyond the snR47 downstream region (Fig. 3). The 21-nucleotide GUAA oligonucleotide corresponds to sequences from 71 to 91 bases downstream of the mature snR47 3′ end and contains the single GUAA sequence that is frequently mutated (Fig. 3).
In Fig. 5 we show that Nrd1308-560 was able to form a retarded complex with discrete electrophoretic mobility with the GUAA RNA but not with the UCUU RNA. The appearance of the shifted band over a narrow concentration range indicates that Nrd1 binds cooperatively. Since mutagenesis revealed only a single important GUAA element in the labeled probe, we conclude that Nrd1 self-association leads to more efficient binding.
FIG. 5.
Gel mobility shift assays of purified Nrd1 and Nab3 proteins. Details of the protein purification, probe sequences, and binding reaction conditions are described in Materials and Methods. Each reaction mixture contained a 2 nM concentration of the labeled RNA probe and, from left to right in each set, 0, 0.156, 0.312, 0.625, 1.25, 2.5, 5.0, or 10 μM purified protein.
The Nab3277-565 protein was able to shift the mobility of the UCUU RNA but not that of the GUAA RNA. The complex formed by Nab3277-565 and UCUU was not discrete but, rather, we observed multiple bands as the concentration of protein was increased. This could have been due to the presence of multiple UCUU motifs in the RNA probe and/or to self-association of the Nab3 protein.
The sequence specificities of Nrd1308-560 and Nab3277-565 RNA interactions were tested in the competition experiments shown in Fig. 6. In these experiments an excess of unlabeled RNA was added to identical binding reaction mixtures (see Materials and Methods) which, in the absence of competitor RNA, resulted in a complete shift. In panel A an unlabeled RNA oligonucleotide corresponding to the wild-type 23-nucleotide UCUU probe was able to compete away (lane 3) the shifted Nab3277-565 complex observed in lane 2. Three mutant oligonucleotides in which one or two of the UCUU sequences were changed to sequences selected in the readthrough experiment were unable to compete to the same extent as the wild-type RNA. In panel B we observed a similar result with the Nrd1308-560 protein, which preferentially bound the wild type compared to the mutant (M1) oligonucleotide.
Conservation of non-poly(A) terminator motifs.
To further characterize important snoRNA termination elements, we searched for conserved sequences among the snoRNA downstream regions of different closely related yeast species and among different snoRNA genes in each yeast strain. Figure 7 shows that some, but not all, of the UCUU and GUA[AG] motifs are conserved in Saccharomyces paradoxus, Saccharomyces mikatae, and Saccharomyces bayanus. It seems clear that even in these closely related yeasts there are significant differences in the presence and spacing of motifs corresponding to those that are functional in S. cerevisiae. Figure 7 also shows the alignment of different snoRNA downstream regions derived from S. cerevisiae snoRNA genes that have previously been shown to terminate through the Nrd1 pathway. As we have observed for snR13 and snR47, these snoRNA downstream sequences contained multiple UCUU and GUA[AG] motifs, but the number and spacing were variable.
FIG. 7.
Alignment of snoRNA downstream sequences. In the top panel, sequences downstream of the SNR13 genes of several yeast species were aligned by using CLUSTAL (37). Indicated species are S. cerevisiae (cer), S. mikitae (mik), and S. bayanus (bay). The second panel shows a similar alignment of the SNR47 downstream sequences. In the bottom panel, the downstream regions of several S. cerevisiae snoRNA genes are aligned by their first downstream Nrd1 or Nab3 element to show the presence of sequence elements similar to those present in SNR13 and SNR47.
Prediction of snoRNA gene Nrd1/Nab3 response elements.
Are Nrd1, Nab3, or both implicated in termination of all snoRNAs? For some snoRNAs (those located in introns or upstream in polycistronic arrays), there is no expectation of finding Nrd1 or Nab3 sites 3′ to the coding region. For all others, we have asked whether the presence of Nrd1 and/or Nab3 binding sites in the 3′ region can be used to predict the presence of non-poly(A) terminators. We used a computer program named GOMER to determine how well the putative binding sites for Nrd1 and Nab3 can identify genes for which transcription termination is mediated by these proteins (see Materials and Methods). GOMER allows for the exploration of models of gene regulation that involve one or more proteins.
We examined three variants of a model for transcription termination, one that uses Nrd1p alone, one that uses Nab3p alone, and a third that includes cooperative interactions between the two proteins. Scores were obtained for a training set of snoRNAs known to be terminated by the Nrd1/Nab3 pathway, for a test set of snoRNAs that are candidates for Nrd1/Nab3 termination, and for the 3′ regions of more than 6,000 other genomic features that act as controls against which the test and training sets were compared. Table 1 shows the ranks of all 10 training set genes according to the cooperative model, as well as the ranks of the top 10 snoRNA genes from the test set. In support of this computational analysis, we subsequently observed readthrough transcripts from both SNR46 and SNR62 in both nrd1 and nab3 strains (data not shown).
TABLE 1.
SNR genes ranked by score in the Nrd1-Nab3 cooperative model
| Training set | Ranka | Test set | Ranka |
|---|---|---|---|
| SNR45 | 1 | SNR65 | 21 |
| SNR13 | 3 | SNR46 | 49 |
| SNR50 | 25 | SNR189 | 70 |
| SNR33 | 76 | SNR62 | 78 |
| SNR3 | 169 | SNR68 | 82 |
| SNR128 | 170 | SNR48 | 114 |
| SNR47 | 410 | SNR51 | 125 |
| SNR17A | 445 | SNR58 | 159 |
| SNR39B | 549 | SNR63 | 245 |
| SNR71 | 1756 | SNR32 | 252 |
Rank among 6,329 yeast 3′ noncoding regions.
Figure 8A presents the results of a computational analysis, showing that the cooperative binding model outperforms either the Nrd1-alone or Nab3-alone model in identifying the training set of snoRNAs known to be regulated by Nrd1 (see Materials and Methods). The cooperative model also performed substantially better than the Nrd1-alone model for the test set, which consisted of snoRNA genes that were candidates for termination by the Nrd1/Nab3 pathway (Fig. 8B). The superiority of the cooperative over the Nab3-alone model was less dramatic than for Nrd1 alone, though it did perform better for about 70% of the test set. The optimal cooperative model was found by decreasing the stringency of the binding site descriptions from those in the initial model but, surprisingly, no improvement in the fit to the training set was obtained by changing the initial values that defined the regulatory region (100 bp 3′ to the end of the annotated genome feature) or the interaction distance between putative Nrd1 and Nab3 sites (70 bp). This might reflect a good choice of initial parameter values, but it is also possible that the optimization of the binding site stringency (the first step in our optimization) trapped the parameterization in a local minimum. As further data accumulate, it should be possible to refine the algorithm to allow more-robust predictions of non-poly(A) terminators.
DISCUSSION
The ability of RNA sequences in the nascent transcript to signal the elongating RNA polymerase to terminate is a common feature of transcription regulation. The RNA signal can be primary sequence, as in the case of eucaryotic Pol III (1, 7), or secondary structures, as in intrinsic termination by procaryotic RNA polymerase (16, 42). Alternatively, the RNA signal can be recognized by protein factors like Rho in bacteria (29) or the cleavage-polyadenylation machinery in Pol II transcription (26, 43). In this paper we describe a set of cis elements that direct termination of yeast Pol II transcription in a mechanism that is not coupled to polyadenylation. This mechanism ensures that small, non-poly(A) transcripts are correctly terminated.
Efficient termination of non-poly(A) yeast Pol II transcripts requires the RNA-binding proteins Nrd1 and Nab3, the RNA helicase Sen1, the Pol II CTD, the CTD kinase catalytic subunit Ctk1 (10, 36), and several components of the cleavage-polyadenylation pathway, including Ssu72, Rna15, Pti1, and Ref2 (12, 24, 35). Together, these factors recognize the RNA signal that directs termination without coupling to the polyadenylation machinery. Whether this complex of proteins couples termination to processing of the snoRNA 3′ end is not known, but we have observed a weak two-hybrid interaction between the Nrd1 and the Rnt1 exonuclease (N. Conrad and J. Corden, unpublished observation). The requirement for some factors involved in cleavage and polyadenylation suggests that there are parallels between the poly(A) and the non-poly(A) termination mechanisms.
One clear distinction between the poly(A) and non-poly(A) pathways is the RNA signal that triggers the process. Using a mutational analysis, we have identified multiple short sequence motifs in the region downstream of two yeast snoRNA genes that are required for efficient termination. The most frequently mutated bases occur either in the sequence GUA[AG] or the sequence UCUU. The first sequence corresponds to the core sequence of the artificial U6R* sequence that was originally used to identify the yeast NRD1 gene. Steinmetz and Brow (33) showed that the Nrd1 protein interacts with RNA fragments containing this artificial sequence, and we have confirmed this observation using a naturally occurring sequence from the SNR47 downstream region. In addition, we show for the first time that the other frequently mutated sequence motif, UCUU, is specifically bound by Nab3. Precisely how these short signal sequences are recognized by the non-poly(A) termination machinery is not known, but the nature of the Nrd1 and Nab3 RNA-binding domains offers a substantial clue.
Both Nrd1 and Nab3 contain a single conserved RNA recognition motif (RRM), a domain common to proteins involved in RNA processing. RRM proteins contain one or more 80- to-100-amino-acid domains, each folded into a four-stranded antiparallel β-sheet (40). One side of the β-sheet is stabilized by a pair of α-helices that connect the β-strands, while the other side of the sheet contacts RNA (23). Each RRM makes contact with only five to six bases of single-stranded RNA and specifically recognizes only one or two bases through two conserved amino acid motifs, RNP1 and RNP2 (11, 14, 41). Most RRM proteins bind selectively because they contain multiple (two to four) RRM domains (3). The short sequences that comprise the non-poly(A) terminator are consistent with recognition by the RRM domains of Nrd1p and Nab3p but, with only a single RRM each, these proteins differ from the common class of RNA-binding proteins.
In the two cases we have examined, both Nrd1 and Nab3 binding elements are present, but the number, spacing, and relative orientation of the GUA[AG] and UCUU sequences is different in the two terminators. In the snR13 terminator, multiple Nrd1p binding sites are present and seem to be the most frequently mutated. In the snR47 case, there is only one Nrd1p binding site but multiple potential Nab3p binding sites. In the snR13 element, UCUU sequences are located downstream of GUA[AG], while in snR47 the opposite orientation occurs. In addition, the spacing of the elements is different. In snR13, sequence motifs are more clustered, whereas in snR47 the sequence motifs are more dispersed, with the UCUU sequences located more than 30 bases from the GUAA sequence. This spacing is not critical, however, because two 15-bp deletions between the Nrd1 and Nab3 binding sites do not substantially alter the efficiency of termination. How these composite terminators are recognized by the non-poly(A) termination machinery is not clear, but several possible mechanisms can be envisaged.
Our group's previous observation that Nrd1 and Nab3 directly interact both genetically and physically (10) suggests that a heteromeric complex of these RNA-binding proteins recognizes sequences in the nascent snoRNA transcript and directs Pol II to terminate without polyadenylation of the nascent transcript. We propose several possible mechanisms for the interaction of the Nrd1/Nab3 complex with the non-poly(A) signal RNA. One possibility is that the Nrd1-Nab3 RRMs are attached to their dimerization domains by flexible linkers. This would allow the spacing of recognition sites to vary and, if the linkers are long enough, to juxtapose. A second possibility is that Nrd1 and Nab3 proteins form higher-order multimers. In this case, the terminator RNA may simultaneously bind several Nrd1 or Nab3 RRMs and, depending on the arrangement of the protein subunits and the path of the RNA, the complex may accommodate many orientations of the GUA[AG] and UCUU sequences. Finally, the presence of multiple binding sites could be important simply to increase the local concentration of targets for Nrd1 and Nab3 during the window of opportunity as the elongating polymerase passes through the termination region. Further experiments will allow us to discriminate among these possibilities.
Do we have enough information to predict the presence of Nrd1-Nab3-dependent termination signals? Using a simple algorithm, we have been able to show that GUA[AG] and UCUU sequences are a predictor of non-poly(A) termination and that the explicit inclusion of cooperative binding terms into the model improves its ability to distinguish likely Nrd1/Nab3 targets. Cooperative binding by Nrd1 was suggested by the results of the gel shift experiments shown in Fig. 5. In addition, the observation of multiple shifted species with Nab3 suggests the possibility of cooperative binding. More detailed binding studies will be required to determine the degree to which binding of a Nrd1p/Nab3p complex is cooperative.
As is observed in yeast cleavage and polyadenylation signals, critical sequence elements are redundant. Single base changes in elements of the snR13 terminator have relatively little effect on the ability to terminate. This is likely due to the redundancy of both the GUA[AG] and UCUU motifs. Single base changes in the snR47 element are more deleterious but fail to reach the level of a complete deletion.
Another factor that may influence the binding of the non-poly(A) termination machinery is the presence of additional RNA-binding proteins. In addition to the CPSF and CstF, there are several other RNA-binding proteins present in the cleavage and polyadenylation complex, including Hrp1, Ydh1, Yth1, Yhh1, and Rna15 (26, 43). In the case of non-poly(A) termination, the presence of binding sites for additional proteins like Rna15, Pti1, or Ref2 could provide sequence recognition in addition to that provided by Nrd1 and Nab3. Sequences downstream of the snR13 termination fragment that we have used in these studies have been implicated in snoRNA termination in some studies (35) but not others (22). Perhaps the efficiency of termination is increased by such elements, but our results indicate that these sequences are not sufficient, on their own, to trigger termination.
The simplicity of the non-poly(A) terminator motifs we have described suggests that such terminators may be present in other locations in the genome. Indeed, we have previously observed that Nrd1 is able to autoregulate production of its own message (36). Sequences similar to those described here are located throughout the Nrd1 gene and its flanks. It will be interesting to see if the same signals that direct termination of non-poly(A) transcripts are also involved in an attenuation-type mechanism that may regulate the expression of Nrd1 and perhaps other mRNA-encoding genes.
Acknowledgments
We thank B. Cormack and J. Lorsch for helpful comments on the manuscript.
This work was supported by National Institutes of Health grants GM66108 to J.L.C. and GM065179 to N.D.C.
REFERENCES
- 1.Allison, D. S., and B. D. Hall. 1985. Effects of alterations in the 3′ flanking sequence on in vivo and in vitro expression of the yeast SUP4-o tRNATyr gene. EMBO J. 4:2657-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allmang, C., J. Kufel, G. Chanfreau, P. Mitchell, E. Petfalski, and D. Tollervey. 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18:5399-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antson, A. A. 2000. Single-stranded-RNA binding proteins. Curr. Opin. Struct. Biol. 10:87-94. [DOI] [PubMed] [Google Scholar]
- 4.Bentley, D. 2002. The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14:336-342. [DOI] [PubMed] [Google Scholar]
- 5.Birse, C. E., L. Minvielle-Sebastia, B. A. Lee, W. Keller, and N. J. Proudfoot. 1998. Coupling termination of transcription to messenger RNA maturation in yeast. Science 280:298-301. [DOI] [PubMed] [Google Scholar]
- 6.Calvo, O., and J. L. Manley. 2001. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol. Cell 7:1013-1023. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, F. E., Jr., and D. R. Setzer. 1992. Transcription termination by RNA polymerase III: uncoupling of polymerase release from termination signal recognition. Mol. Cell. Biol. 12:2260-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke, N. D., and J. A. Granek. 2003. Rank order metrics for quantifying the association of sequence features with gene regulation. Bioinformatics 19:212-218. [DOI] [PubMed] [Google Scholar]
- 9.Connelly, S., and J. L. Manley. 1988. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 2:440-452. [DOI] [PubMed] [Google Scholar]
- 10.Conrad, N. K., S. M. Wilson, E. J. Steinmetz, M. Patturajan, D. A. Brow, M. S. Swanson, and J. L. Corden. 2000. A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics 154:557-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deo, R. C., J. B. Bonanno, N. Sonenberg, and S. K. Burley. 1999. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell 98:835-845. [DOI] [PubMed] [Google Scholar]
- 12.Dheur, S., T. A. Vo le, F. Voisinet-Hakil, M. Minet, J. M. Schmitter, F. Lacroute, F. Wyers, and L. Minvielle-Sebastia. 2003. Pti1p and Ref2p found in association with the mRNA 3′ end formation complex direct snoRNA maturation. EMBO J. 22:2831-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fatica, A., M. Morlando, and I. Bozzoni. 2000. Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3′-processing apparatus. EMBO J. 19:6218-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handa, N., O. Nureki, K. Kurimoto, I. Kim, H. Sakamoto, Y. Shimura, Y. Muto, and S. Yokoyama. 1999. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature 398:579-585. [DOI] [PubMed] [Google Scholar]
- 15.Hanley, J. A., and B. J. McNeil. 1982. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143:29-36. [DOI] [PubMed] [Google Scholar]
- 16.Henkin, T. M., and C. Yanofsky. 2002. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. Bioessays 24:700-707. [DOI] [PubMed] [Google Scholar]
- 17.Hirose, Y., and J. L. Manley. 2000. RNA polymerase II and the integration of nuclear events. Genes Dev. 14:1415-1429. [PubMed] [Google Scholar]
- 18.Liu, X., and N. D. Clarke. 2002. Rationalization of gene regulation by a eukaryotic transcription factor: calculation of regulatory region occupancy from predicted binding affinities. J. Mol. Biol. 323:1-8. [DOI] [PubMed] [Google Scholar]
- 19.Logan, J., E. Falck-Pedersen, J. E. Darnell, Jr., and T. Shenk. 1987. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse beta maj-globin gene. Proc. Natl. Acad. Sci. USA 84:8306-8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 21.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S. D. Patterson, M. Wickens, and D. L. Bentley. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357-361. [DOI] [PubMed] [Google Scholar]
- 22.Morlando, M., P. Greco, B. Dichtl, A. Fatica, W. Keller, and I. Bozzoni. 2002. Functional analysis of yeast snoRNA and snRNA 3′-end formation mediated by uncoupling of cleavage and polyadenylation. Mol. Cell. Biol. 22:1379-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagai, K., C. Oubridge, T. H. Jessen, J. Li, and P. R. Evans. 1990. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature 348:515-520. [DOI] [PubMed] [Google Scholar]
- 24.Nedea, E., X. He, M. Kim, J. Pootoolal, G. Zhong, V. Canadien, T. Hughes, S. Buratowski, C. L. Moore, and J. Greenblatt. 2003. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J. Biol. Chem. 278:33000-33010. [DOI] [PubMed] [Google Scholar]
- 25.Osheim, Y. N., N. J. Proudfoot, and A. L. Beyer. 1999. EM visualization of transcription by RNA polymerase II: downstream termination requires a poly(A) signal but not transcript cleavage. Mol. Cell 3:379-387. [DOI] [PubMed] [Google Scholar]
- 26.Proudfoot, N., and J. O'Sullivan. 2002. Polyadenylation: a tail of two complexes. Curr. Biol. 12:R855-R857. [DOI] [PubMed] [Google Scholar]
- 27.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 28.Reeder, R. H., P. Guevara, and J. G. Roan. 1999. Saccharomyces cerevisiae RNA polymerase I terminates transcription at the Reb1 terminator in vivo. Mol. Cell. Biol. 19:7369-7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richardson, J. P. 2002. Rho-dependent termination and ATPases in transcript termination. Biochim. Biophys. Acta 1577:251-260. [DOI] [PubMed] [Google Scholar]
- 30.Sadowski, M., B. Dichtl, W. Hubner, and W. Keller. 2003. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 22:2167-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider, T. D., and R. M. Stephens. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steinmetz, E. J., and D. A. Brow. 1998. Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc. Natl. Acad. Sci. USA 95:6699-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steinmetz, E. J., and D. A. Brow. 1996. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol. Cell. Biol. 16:6993-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steinmetz, E. J., and D. A. Brow. 2003. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol. Cell. Biol. 23:6339-6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinmetz, E. J., N. K. Conrad, D. A. Brow, and J. L. Corden. 2001. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413:327-331. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tran, D. P., S. J. Kim, N. J. Park, T. M. Jew, and H. G. Martinson. 2001. Mechanism of poly(A) signal transduction to RNA polymerase II in vitro. Mol. Cell. Biol. 21:7495-7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Hoof, A., P. Lennertz, and R. Parker. 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varani, G., and K. Nagai. 1998. RNA recognition by RNP proteins during RNA processing. Annu. Rev. Biophys. Biomol. Struct. 27:407-445. [DOI] [PubMed] [Google Scholar]
- 41.Wang, X., and T. M. Tanaka Hall. 2001. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat. Struct. Biol. 8:141-145. [DOI] [PubMed] [Google Scholar]
- 42.Yarnell, W. S., and J. W. Roberts. 1999. Mechanism of intrinsic transcription termination and antitermination. Science 284:611-615. [DOI] [PubMed] [Google Scholar]
- 43.Zhao, J., L. Hyman, and C. Moore. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63:405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]