Abstract
The human T-cell leukemia virus type 1 (HTLV-1) is a retrovirus that integrates randomly into the T-cell genome. Two long terminal repeats (LTRs) flank the integrated provirus. The upstream and downstream LTRs carry identical promoter sequences. Studies with other retroviruses suggest that the downstream promoter is silent and that RNA polymerases initiating at the upstream promoter proceed through the 3′ LTR. In this study, we used the chromatin immunoprecipitation assay to compare the binding of transcription regulatory proteins at both the upstream and downstream promoters in HTLV-1-infected cell lines and adult T-cell leukemia-lymphoma cells. Unexpectedly, we detected a nearly equal distribution of activator (Tax, CREB, ATF-1, ATF-2, c-Fos, and c-Jun) and regulatory protein (CBP, p300, TAFII250, and polymerase II) binding at both the upstream and downstream promoters. Consistent with this observation, we found that the downstream promoter was transcriptionally active, suggesting that the two promoters are functionally equivalent. We also detected asymmetrical binding of histone deacetylases (HDAC-1, -2, and -3) at both promoters. All three HDACs strongly repressed Tax transactivation, and this repression correlated with displacement of Tax from the HTLV-1 promoter. These effects were reciprocal, as Tax expression reversed HDAC repression and displaced HDACs from the HTLV-1 promoter. These data suggest that HTLV-1 transcriptional regulation at both the 5′ and 3′ LTRs is mediated, in part, through the mutually exclusive binding of Tax and HDACs at the proviral promoters.
The human T-cell leukemia virus type 1 (HTLV-1) is a retrovirus responsible for an aggressive and fatal malignancy called adult T-cell leukemia-lymphoma (ATLL) (37, 46). Following infection, HTLV-1 integrates randomly into the host cell chromosome (40). During this event, the 5′ and 3′ ends of the retrovirus are duplicated, forming long terminal repeats (LTRs). The LTRs of the integrated provirus carry two identical U3 regions containing two identical promoters. The 5′ promoter directs synthesis of the genomic RNA, whereas the 3′ promoter, if active, synthesizes RNAs that extend into the adjacent host cell genome. Previous studies with other retroviruses have suggested that RNA polymerase II (Pol II) initiates at the upstream U3 promoter and transcription proceeds through the provirus and downstream U3 promoter, as the polyadenylation signal is located adjacent to the TATA sequence and the polyadenylation site is downstream of the U3 region (13).
The U3 promoter regions of HTLV-1 each carry three highly conserved 21-bp enhancer elements that are critical for Tax-activated transcription. These 21-bp elements are referred to as viral CREs. The viral CREs carry a central binding site for the cellular transcription factor CREB and/or other ATF/CREB family members (reviewed in reference 11). The CREs are flanked by GC-rich DNA sequences that are conserved in all HTLV family members. The virally encoded Tax oncoprotein is the regulatory protein responsible for strong transcriptional activation of HTLV-1. Tax interacts with both CREB and the GC-rich DNA sequences that immediately flank the CREB binding site in the viral CRE (18, 25, 26, 30). These protein-protein and protein-DNA interactions promote the assembly of very stable ternary complexes on the HTLV-1 promoter. The formation of the Tax/CREB promoter-bound complexes is critical for the recruitment of the multifunctional cellular coactivators CBP/p300 (10, 21) and subsequent strong transcriptional activation of the virus (8, 9, 29).
Until recently, very little was known about which activators, and/or other ancillary factors, interact with the viral promoter in vivo. The chromatin immunoprecipitation (ChIP) assay has been used to identify chromatin modifications, transcription factors, and coactivators assembled at the chromosomally integrated proviral promoter in HTLV-1-infected cells. Not surprisingly, these studies revealed the presence of Tax and a variety of ATF/CREB and AP-1 family members (CREB, CREB-2, ATF-1, ATF-2, c-Fos, and c-Jun) at the integrated HTLV-1 promoter in vivo (23). The coactivators p300 and CBP were also detected at the promoter (23, 29). Interestingly, we detected histone H3 and H4 acetylation throughout the proviral genome, with greater acetylation located in the gag gene (23). Unexpectedly, the repressive histone deacetylase (HDAC) complexes were also present at the viral promoter and, following their inhibition, histone H4 acetylation and viral RNA synthesis increased.
These studies revealed that a variety of transcriptional regulators bind the HTLV-1 promoter, but they did not discriminate between factor occupancy at the 5′ and 3′ LTRs. Therefore, the data showed the average of factor binding at the two LTRs. Although the 3′ U3 region carries promoter sequences identical to those in the 5′ U3 region, including the three viral CREs, it is currently not known whether this promoter is transcriptionally active. Work with other retroviruses suggests that transcription from the 3′ LTR is blocked when the 5′ LTR is active (2). The mechanism of this silencing is not well understood, but it may result from RNA Pol II elongation through the 3′ U3 region (transcriptional overlap interference [2]) or by unfavorable chromatin topology at a 3′ LTR linked to a transcriptionally active 5′ LTR (6). We were interested in examining whether the HTLV-1 upstream and downstream LTR promoters followed this same paradigm.
In this study, we used the ChIP assay to examine the binding of transcription regulatory proteins at the 5′ versus the 3′ promoter of the integrated provirus in both an HTLV-1-transformed cell line and ATLL patient cells. We found that the binding of most activators and coactivators (Tax, CREB, ATF-1, ATF-2, c-Fos, c-Jun, CBP, p300, TAFII250, and Pol II) were generally evenly distributed between the 5′ and 3′ U3 regions. Consistent with this observation, we detected RNA that accurately initiated from the downstream U3 region. We found that the binding of the HDACs 1 and 2 was higher at the 5′ promoter, whereas the binding of HDAC-3 was higher at the 3′ promoter. HDAC overexpression strongly inhibited Tax transactivation. Reciprocally, Tax reversed HDAC repression of HTLV-1 transcription. These effects appear to be mediated through the mutually exclusive binding of Tax and the HDAC proteins at the HTLV-1 promoter, as indicated by ChIP analysis. These data reveal a complex interplay between transcriptional activators and repressors in the regulated expression of the integrated HTLV-1 provirus.
MATERIALS AND METHODS
Cell culture and transient-transfection assays.
HTLV-1-transformed T cells (SLB-1 and MT2) and Jurkat T cells were cultured in Iscove's modified Dulbecco's medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and penicillin-streptomycin. ATLL cells, mononuclear cells obtained from the blood of patients with ATLL (acute-type leukemia) and separated on Ficoll, were a gift from R. Harrod (Southern Methodist University, Dallas, Tex.) and Shigeki Takemoto and Hirokuni Taguchi (Department of Hematology and Respiratory Medicine, Kochi Medical School Hospital, Kochi, Japan). The ATLL patient materials were obtained from HTLV-1-infected individuals after informed consent. The cells were maintained in culture as described above, except the medium also contained 50 U of interleukin-2 (Roche)/ml. CHOK1-Luc hamster ovary cells (34) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, penicillin-streptomycin, and 500 μg of G418 (Geneticin; Invitrogen)/ml. For transient-cotransfection assays, cells were transfected with Lipofectamine Plus reagent for adherent cells (Invitrogen) and a constant amount of DNA. After 24 h, the cells were harvested and lysed, and luciferase activity was measured using the dual-luciferase reporter assay system (Promega) with a Turner Designs model TD 20-e luminometer. Luciferase activity was normalized to Renilla luciferase activity from the herpes simplex virus thymidine kinase (TK) promoter (pRL-TK; Promega). For the ChIP experiments the cells were electroporated as described elsewhere (41). Briefly, 2 × 107 cells were electroporated with the GenePulser Xcell from Bio-Rad in the presence of 20 μg of total DNA. The cells were then harvested at 24 h for luciferase and ChIP analyses. The electroporation transfection protocol used in this study produced transfection efficiencies approaching 40%. Expression plasmids for Tax (pSG-Tax [39]), Flag-HDAC-1, Flag-HDAC-2, and Flag-HDAC-3 (16), and pminLuc-viral CRE (10) have been previously described.
Antibodies.
Antibodies against ATF-1 (C41-5.1), ATF-2 (N-96), c-Jun (H-79), c-Fos (H-125), p300 (N-15), CBP (A-22), TAFII250 (6B3), HDAC-2 (H-54), HDAC-3 (H-99), mSin3A (AK-11), N-CoR (H-303), and RNA Pol II (H-224) were purchased from Santa Cruz Biotechnology. Antibodies against acetylated lysines 9/14 of H3 (06-599), acetylated lysines 5/8/12/16 of H4 (06-866 or 06-598), dimethylated lysine 4 of H3 (07-030), and HDAC-1 (06-720) were purchased from Upstate Biotechnology. The CREB (CREB-1/p43) antibody (Ab 7540) and trimethylated lysine 4 of H3 (Ab 8580) were purchased from Abcam. Tax monoclonal antibody (hybridoma 168B17-46-92) was obtained from the NIH AIDS Research and Reference Reagent Program.
ChIP assay.
The ChIP assay was performed essentially as described elsewhere (20, 23).
Reverse transcriptase PCR.
Cytoplasmic RNA was extracted from MT2 cells that were mock treated or treated with trichostatin A (TSA; 400 nM) using the cytoplasmic RNA reagent (Invitrogen) to avoid DNA contaminants. mRNA (2 μg) was reverse transcribed using the I-Script cDNA synthesis kit provided by Bio-Rad.
PCRs and primers.
Real-time PCRs were performed as described elsewhere (23), using iQ SYBR Green Supermix (Bio-Rad) and a 100 to 200 nM concentration of primer per reaction mixture. PCR primers were as follows: primer 1, AATGACCATGAGCCCCA; primer 2, GTGAGGGGTTGTCGTCA; primer 3, AACGAAAAAGAGGCAGATGA; env, 5′-GTCTCTGTTCCATCCTCTTCTT and 5′-GGGGTCAAAGCAGTGGGT; +255 primer, GCCGCTACAGATCGAAAGTT; +462 primer, AAGATTTGGCCCATTGCCT; MT2 (clone 1), GAACAAGCTGGGCATCAGAA; MT2 (clone 2), GAGCAGCTTCTGTTCAGACT; B-myb forward, CGACGCGCTTGGCGGGAGATAGA; B-myb reverse, GCTCCTCGCTCGCAGGAACTG; elongation factor 1α (EF-1α) forward, GCCTCTCCAGGATGTCTACAAA; EF-1α reverse, GTTTGAGAACACCAGTCTCCACTC. Standard curves were generated for all primer sets using 10-fold serial dilutions of pHTLV-1563 (23) plasmid and/or genomic DNA from infected cells. PCR efficiencies ranged from 94 to 110%, with correlation coefficients of 0.990 or greater. Threshold cycle data were quantified relative to the input, as described previously (7). The value for an immunoglobulin G (IgG) sample in each experiment was used for background subtraction 2ΔCT(sample) − 2ΔCT(IgG). For 3′ versus 5′ LTR comparisons of a given immunoprecipitation, the values obtained using primers 1 and 2 were set at 100%, as these primers amplify both LTRs. The percent occupancy of the 5′ LTR was then determined by the difference between the total and the values obtained using primers 2 and 3, which specifically amplify the 3′ LTR.
Real-time PCR for quantification of mRNA transcripts was done using 2 μl of a 1:10 dilution of cDNA per 25-μl reaction mixture. Data were analyzed using the 2−ΔΔCT method as described elsewhere (28), taking into account any differences in PCR efficiencies between the housekeeping gene EF-1α and the 3′-LTR-generated transcripts as described elsewhere (36).
LM-PCR.
Cellular DNA sequences directly flanking the 3′ LTRs in MT2 cells were determined using ligation-mediated PCR (LM-PCR) as described elsewhere (43), with the exception that Pfu was used in place of Taq to limit mutations arising from PCR amplification. Two clones were identified, corroborating Southern blotting and real-time PCR experiments, showing that the MT2 cells carry two provirus copies. The sequences flanking the 3′ LTRs were as follows: MT2 clone 1, AGTAGAAGTCTTGATTTTCTGATGCCCAGCTTGTTCAT; MT2 clone 2, TATACCTATGTAACAAACCTGCACGTTGTGCACATG.
Primer extension.
Total RNA was extracted from MT2 cells with RNA-Bee reagent as described by the manufacturer (ISO-TEX Diagnostics). Primer extensions were done with avian myeloblastosis virus reverse transcriptase essentially as described by the manufacturer (Promega). To ensure reverse transcription of >400 nucleotides, three successive hybridization-extension steps were done for each reaction with 50 μg of RNA and 0.4 pmol of 32P-end-labeled primer added only at the first step. For both the second and third hybridization-extension steps, the reaction volumes were doubled but maintained the same concentrations of reaction buffer, deoxynucleoside triphosphates, and avian myeloblastosis virus reverse transcriptase as in the first step. Following reverse transcription, reactions were processed as described for in vitro transcription reactions (25).
Western blot analysis.
SLB-1 and ATLL cells were lysed in radioimmunoprecipitation assay buffer and resuspended in sodium dodecyl sulfate sample dyes. Proteins were separated on a sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis and analyzed by Western blotting with the indicated antibodies.
RESULTS
Transcriptional regulators and histone modifications are found at both the 5′ and 3′ LTRs in HTLV-1-transformed cell lines.
We have previously used the ChIP assay to show that a wide variety of transcriptional activators, coactivators, and repressors bind to the HTLV-1 promoter in living SLB-1 and MT2 cells (23). However, it is important to note that the PCR primers used in these assays amplified promoter sequences within both the 5′ and 3′ LTRs. Therefore, these studies revealed the average of factor binding at both the upstream and downstream promoter regions of the integrated provirus. Since it is currently unknown whether transcription regulatory proteins bind at this region or whether the 3′ U3 promoter of HTLV-1 is transcriptionally active, we performed a series of experiments to specifically address these questions.
We designed a PCR primer that anneals at position +7910 of the proviral genome located in the pX region, immediately upstream of (and slightly overlapping) the 3′ LTR (Fig. 1, primer 3). This primer was paired with primer 2, which anneals at −97 relative to the putative 3′ start site (Fig. 1). PCRs using this primer pair specifically amplify the 3′ U3 promoter region, including the three viral CREs. To assess the relative binding of factors at each promoter simultaneously, we performed parallel reactions using primers 1 and 2 to amplify both upstream and downstream promoters and primers 2 and 3 to specifically amplify the 3′ promoter region. We then deduced the binding at each promoter by subtracting the signal obtained from the 3′-specific PCRs from the 5′ plus 3′ PCR signal.
FIG. 1.
Schematic representation of the HTLV-1 provirus. The U3, R, and U5 regions of the 5′ and 3′ LTRs and the structural organization of the HTLV-1 genome are shown. The TATA sequence, transcription start site (U3-R junction; +1), and the three viral CREs (boxes), are also shown. The viral CREs are located at approximately −100, −200, and −250 relative to the transcriptional start site. The real-time PCR primers are indicated by arrows and are denoted according to the nucleotide position at the 5′ end of the primer. Positions are relative to the transcription start site. The primer 3 position is mapped relative to the 5′ LTR start site. Primers 1 and 2 amplify the viral CREs from the 5′ and 3′ LTRs. Primers 2 and 3 amplify only the 3′ LTR.
Using this approach, we examined the binding of the transcriptional activators Tax, ATF-1, ATF-2, CREB, c-Jun, and c-Fos, the coactivators CBP and p300, the TBP-associated factor TAFII250, and RNA Pol II in SLB-1 cells (Fig. 2A and B). Unexpectedly, we found that all of these proteins were present at both the 5′ and 3′ promoters. In general, transcription factor binding was nearly evenly distributed. However, we detected slightly higher binding of Tax and significantly higher binding of CREB at the 5′ promoter region. Similarly, we also observed modestly higher binding of CBP, p300, TAFII250, and Pol II at the 5′ LTR, although these factors were also clearly present at the 3′ promoter. Comparable results were obtained in another HTLV-1-infected cell line, MT2 (data not shown). Both SLB-1 and MT2 cells express significant amounts of Tax protein (14, 32). The association of activators and coactivators with the upstream and downstream promoters is consistent with both promoters existing in a transcriptionally active state in these cell lines.
FIG. 2.
Distribution of transcription factors, coactivators, and histone modifications between the 5′ and 3′ HTLV-1 LTRs in SLB-1 cells. (A) Quantification of the relative binding of Tax, ATF/CREB members, c-Jun, and c-Fos at the 5′ and 3′ LTRs. Real-time PCR was used to quantify the DNA in each immunoprecipitation. The graph shows data averaged from two to three independent ChIP experiments. (B) Quantification of relative binding of CBP, p300, TAFII250, and Pol II at the 5′ and 3′ LTRs. Experiments were performed as described for panel A. (C) Quantification of relative histone modification levels at the 5′ and 3′ LTRs. The graph shows data representative of two independent ChIP experiments.
TAFII250 and the cellular coactivators CBP and p300 harbor histone acetyltransferase activity, and we have previously shown that acetylated nucleosomes are present at the HTLV-1 promoter (23). Consistent with the observed enrichment of these factors at the 5′ LTR relative to the 3′ LTR, we detected enhanced acetylation of the histone H3 and H4 amino-terminal tails in the same region (Fig. 2C). However, dimethylation and trimethylation of lysine 4 on the H3 tail were evenly distributed between the upstream and downstream promoters. Both histone tail acetylation and methylation have previously been shown to be associated with transcriptionally active genes (17).
A similar pattern of transcription factor-coactivator binding is observed at the 5′ and 3′ LTRs in patient-derived ATLL cells.
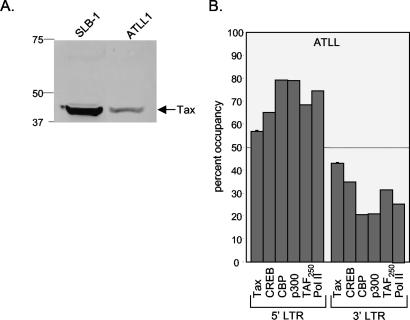
To verify the physiological significance of the factor binding profile detected in cultured HTLV-1-transformed cell lines, we tested activator-coactivator binding in ex vivo ATLL cells. These HTLV-1-infected T cells were recently isolated from the Ficoll-separated peripheral blood mononuclear cells from a patient with ATLL. ATLL cells generally carry a single integrated HTLV-1 provirus and express Tax, albeit at levels below that observed in SLB-1 cells (Fig. 3A). We focused on the binding of factors that are known to play an essential role in transcription from the provirus. In these cells, we detected the binding of Tax, CREB, CBP, p300, TAFII250, and Pol II, each with modestly higher levels at the 5′ LTR relative to that at the 3′ LTR (Fig. 3B). This pattern of factor binding at the 5′ and 3′ promoters was similar to the pattern observed in SLB-1 cells (Fig. 2A and B).
FIG. 3.
Distribution of transcription factors and coactivators between 5′ and 3′ HTLV-1 LTRs in patient-derived ATLL cells. (A) Comparison of Tax expression in SLB-1 and ATLL cells. A Western blot of SLB-1 and ATLL cell extracts (50 μg) was probed with a monoclonal antibody directed against Tax. Protein standards (in kilodaltons) are indicated. (B) Quantification of the relative binding of Tax, CREB, CBP, p300, TAFII250, and Pol II at the 5′ and 3′ LTRs. Real-time PCR was used to quantify the DNA in each immunoprecipitation. The graph shows data representative of two independent ChIP experiments.
The 3′ LTR of HTLV-1 is transcriptionally active.
Our observation that transcription regulatory proteins associate with the HTLV-1 3′ LTR suggests that this promoter is transcriptionally active. To directly address this question, we cloned and sequenced the cellular DNA that immediately flanks the 3′ LTRs in MT2 cells. We chose this cell line because it harbors only two copies of the provirus, while SLB-1 cells carry five copies of the provirus (data not shown) (4). We obtained two clones representative of the two proviruses in the MT2 cell line (see “LM-PCR”' of Materials and Methods for sequences) and designed reverse PCR primers from these sequences (clone 1 and clone 2) (Fig. 4A). Reverse transcriptase PCR revealed the presence of RNA generated from the 3′ LTR (data not shown). This level of amplification was significantly greater than background expression observed for the p56lck gene, which is transcriptionally silent in HTLV-1-infected T cells (24). We performed primer extension analysis (Fig. 4B) to confirm that the 3′ RNA accurately initiated from the downstream promoter and not from read-through transcription originating from the 5′ LTR. Furthermore, we wanted to quantitatively compare the levels of RNA that initiated from the 3′ LTR relative to those from the 5′ LTR. For these assays, we used the MT2 clone 1 and clone 2 primers for detection of 3′-initiated RNA (Fig. 4A). A primer that annealed at +462 was used to detect 5′-initiated RNA. As expected, the 5′ LTR-specific primer generated a product that mapped to the upstream U3-R junction (Fig. 4B, lane 2). Similarly, the extension products detected with primers specific to both clone 1 and clone 2 produced products that mapped to the downstream U3-R junction (Fig. 4B, lanes 3 and 4). Quantitation of the extension products revealed that the 3′ LTR was three- to fourfold less transcriptionally active than the 5′ LTR. These values are likely an underestimate of the amount of 3′ LTR transcription, as the 3′ RNA is potentially unstable due to improper processing. These results indicate that the 3′ LTR of the integrated HTLV-1 provirus is transcriptionally active, consistent with the binding of transcription factors and coactivators within this region.
FIG. 4.
The 3′ LTR is transcriptionally active. (A) Schematic representation of HTLV-1 provirus showing the 5′ and 3′ LTRs. The annealing positions of the primers used for primer extension are indicated by short arrows, and the sizes of the cDNA products obtained from primer extension are indicated by long arrows. (B) Primer extension analysis of transcripts initiated from the 5′ and 3′ LTRs. The gag-specific primer (+462) was used to measure 5′-initiated RNA, and clone 1 and clone 2 primers were each used to measure 3′-initiated RNA. Size markers and transcripts are indicated.
Distinct HDAC complexes bind the 5′ and 3′ HTLV-1 promoters in vivo.
The regulation of transcriptional activity through histone tail acetylation is strongly influenced by HDAC complexes that are generally associated with transcriptional repression (3, 15). Previously, we detected HDAC-1, -2, and -3 binding at the HTLV-1 promoters in SLB-1 cells (23). At the time, we postulated that a portion of the observed HDAC binding arose from the putatively inactive 3′ promoter region of the provirus. However, as discussed above, our assays did not distinguish between the two promoters. Furthermore, the data presented in Fig. 4 indicate that the 3′ LTR is transcriptionally active. For these reasons, we measured HDAC-1, -2, and -3 binding specifically at the upstream versus the downstream promoter in SLB-1 cells. Comparison of HDAC binding at the 5′ and 3′ LTRs revealed enrichment of both HDAC-1 and -2 at the 5′ LTR and enrichment of HDAC-3 at the 3′ LTR (Fig. 5A). We corroborated specific HDAC binding by measuring the association of two corepressors that have previously been shown to form a complex with the different deacetylases in vivo. The mSin3A corepressor associates with HDAC-1 and -2, whereas the N-CoR corepressor associates with HDAC-3 (12, 27, 44, 47). The pattern of corepressor binding at the 5′ and 3′ LTRs correlated with HDAC binding in SLB-1 cells (Fig. 5B), ATLL cells (Fig. 5C), and MT2 cells (data not shown). To assess the relevance of HDAC binding at the two LTRs, we also measured the binding of all three HDACs within the coding region of the virus. Figure 5D shows that the binding of HDACs 1 and 2 was similar to that observed at the 3′ LTR, whereas HDAC-3 binding was intermediate between the 5′ and 3′ LTRs. These data corroborate the observation of enhanced binding of HDAC-3 at the 3′ LTR.
FIG. 5.
Distribution of HDACs between 5′ and 3′ LTRs. (A) Quantification of HDAC-1, -2, and -3 binding at the 5′ and 3′ LTRs in SLB-1 cells. Real-time PCR was used to quantify the DNA in each immunoprecipitation. The graph shows data averaged from three independent ChIP experiments. (B) Quantification of mSin3A and NCoR binding at the 5′ and 3′ LTRs in SLB-1 cells. Real-time PCR was used to quantify each immunoprecipitation. The graph shows data representative of two independent ChIP experiments. (C) Quantification of HDAC-1, -2, and -3, mSin3A, and NcoR binding at the 5′ and 3′ LTRs in ATLL cells. Real-time PCR was used to quantify the DNA in each immunoprecipitation. The graph shows data representative of two independent ChIP experiments. (D) HDAC binding within the HTLV-1 provirus in SLB-1 cells. Quantification is shown of relative HDAC binding at the 5′ LTR, env region, and 3′ LTR of the provirus. Real-time PCR was used to quantify HDAC-1, -2, and -3 immunoprecipitations from SLB-1 cells. The HDAC-1 and -2 signals were normalized to the 3′ LTR, and HDAC-3 was normalized to the 5′ LTR. The graph is representative of two independent ChIP experiments.
The differential association of the mSin3A subunit to HDAC-1 and -2 was confirmed by coimmunoprecipitation. Transient transfection of either Flag-tagged HDAC-1 or HDAC-2 into CHOK1 cells led to enrichment of mSin3A in the anti-Flag antibody immunoprecipitates. In contrast, mSin3A was not present in the HDAC-3 complex (data not shown).
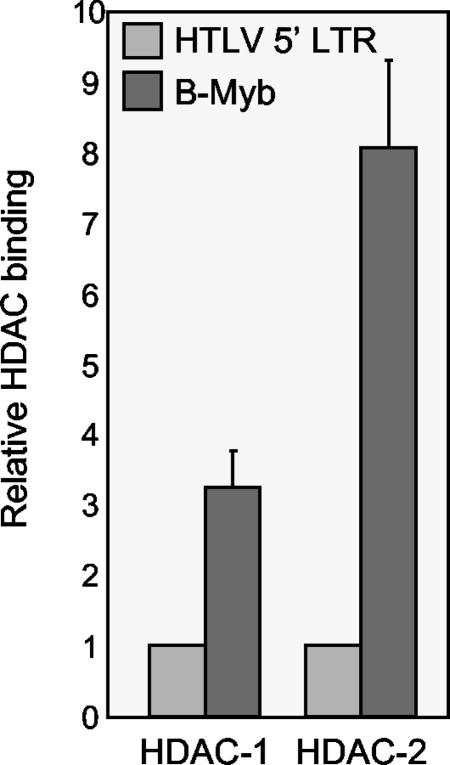
Finally, we were interested in assessing the relative level of HDAC protein binding to the transcriptionally active HTLV-1 promoter relative to that with a transcriptionally repressed promoter. We compared the binding of HDACs at the 5′ HTLV-1 promoter and the B-Myb promoter in SLB-1 cells. B-Myb has previously been shown to be transcriptionally repressed by Tax (33) and to bind HDAC-1 (38). Quantitation of ChIPs using antibodies against HDAC-1 and -2 revealed that HDAC binding was significantly greater at the transcriptionally repressed B-Myb promoter relative to that at the 5′ HTLV-1 promoter (Fig. 6). We did not detect HDAC-3 binding at the B-Myb promoter (data not shown). These data clearly support a role for HDAC proteins in transcriptional repression. However, the data also suggest that the HDAC proteins participate in dynamic interactions at transcriptionally active promoters.
FIG. 6.
Comparison of HDAC-1 and -2 binding at the B-Myb promoter and at the 5′ LTR in SLB-1 cells. Real-time PCR was used to quantify the DNA in each immunoprecipitation. The graph shows data averaged from two independent ChIP experiments. HDAC binding at the 5′ HTLV-1 LTR was set to 1.
Inhibition of HDACs leads to an increase in transcription from the 3′ LTR.
Our observation that HDAC-3 and, to a lesser extent, HDAC-1 and -2 bind to the 3′ HTLV-1 promoter prompted us to test whether the HDAC proteins are involved in transcriptional regulation from the downstream promoter. To test this hypothesis, we treated MT2 cells with the HDAC inhibitor TSA and measured the level of RNA generated from the 3′ LTR. RNA purified from TSA-treated or untreated cells was reverse transcribed, and real-time PCR was performed with a primer that anneals at +255 (U5 region) paired with the reverse primer from clone 1 or clone 2 (Fig. 4A). Figure 7 shows that TSA treatment produced a two- to threefold increase in transcript levels initiated from the 3′ LTR. This increase is similar to the TSA-induced increase in transcription from the 5′ LTR (23).
FIG. 7.
Inhibition of HDAC activity increases transcription initiated from the 3′ LTR, as shown by real-time PCR of reverse-transcribed cytoplasmic RNA from MT2 cells untreated or treated with TSA (400 nM) for 24 h. The amplification was normalized to that with the housekeeping gene EF-1α. The level of RNA expression for untreated cells was set to 1.
The Tax-responsive viral CREs are targets of HDAC repression in vivo.
The unexpected association of HDAC proteins and Tax with both the HTLV-1 upstream and downstream promoters led us to test the functional role of these proteins in Tax transactivation. A previous study showed that HDAC-1 specifically functions as a negative regulator of Tax transactivation (5). We were interested in extending these studies to test whether HDAC-2 and -3 were also involved in the repression of Tax transcriptional activity at the HTLV-1 promoter. To examine this possibility, we cotransfected expression plasmids for Tax and each of the three HDAC proteins into HTLV-1-negative CHOK1-Luc cells. These cells carry two to four copies of the integrated HTLV-1 promoter (U3 region) driving expression of the luciferase gene (34). A previous study showed that transfection of Tax into these cells produced strong transcriptional activation from the integrated HTLV-1 promoter (34). Figure 8A shows that all three HDAC proteins similarly repressed Tax transactivation in a dose-dependent fashion. Comparable results were obtained using Jurkat T cells transiently transfected with the HTLV-1 promoter driving luciferase (data not shown). To show that the deacetylase activities of the HDACs are essential for their repression of HTLV-1 transcription, we used HDAC mutants defective for deacetylase activity. A specific point mutation in the catalytic domain of HDAC-1, and carboxy-terminal deletions in HDACs 2 and 3, partially or fully relieved the HDAC repression of Tax transcription function (data not shown).
FIG. 8.
Reciprocal transcriptional repression between Tax and HDACs. (A) HDAC-1, -2, and -3 repress Tax transactivation of HTLV-1. CHOK1-Luc cells were transfected with pSG-Tax (100 ng) in the absence or presence of increasing amounts of HDAC-1, -2, and -3 (75, 150, or 300 ng) expression plasmids. Reported values for all transient-transfection experiments are the average luminescence from one experiment performed in duplicate. The standard error is indicated. Experiments were repeated at least three times. (B) HDAC-1 represses Tax transactivation from a minimal promoter carrying only the viral CREs. The pminLuc-viral CRE reporter plasmid (100 ng) (10) was cotransfected into Jurkat T cells with pSG-Tax (200 ng) and increasing amounts of HDAC-1 (100, 200, and 400 ng), as indicated. (C) Tax reverses HDAC repression of HTLV-1. CHOK1-Luc cells were transfected with an expression plasmid for Tax alone (100 ng) or with HDAC-1, -2, or -3 (300 ng), in the presence of increasing amounts of pSG-Tax (50 and 100 ng).
Since the HTLV-1 promoter contains multiple transcription factor binding sites, we were interested in determining whether the viral CREs, which are the primary cis regulatory elements involved in Tax transactivation, are the targets of HDAC repression. To address this question, we utilized a reporter plasmid carrying three tandem copies of the third viral CRE cloned upstream of a core promoter (pminLuc-viral CRE [10]). All of the other known cis regulatory elements in the HTLV-1 promoter are absent from this reporter construct. The pminLuc-viral CRE reporter plasmid was cotransfected with expression plasmids for Tax and HDAC-1 into Jurkat T cells. As with the full promoter, a dose-dependent decrease in Tax transactivation was observed (Fig. 8B). These data suggest that the viral CREs are sufficient for mediating HDAC-1 repression and support a role for Tax in this process.
We were next interested in testing whether the effects of Tax and HDACs on HTLV-1 transcription were reciprocal. To perform this experiment, we measured HTLV-1 promoter activity in CHOK1-Luc cells in the presence of a constant amount of an expression plasmid for HDAC-1, -2, or -3. We added increasing amounts of the Tax expression plasmid to determine whether Tax would oppose the HDAC-induced transcriptional repression. Figure 8C shows that in all cases, Tax partially recovered HTLV-1 transcription in the presence of the transfected HDACs.
Tax and HDAC binding at the HTLV-1 promoter are mutually exclusive.
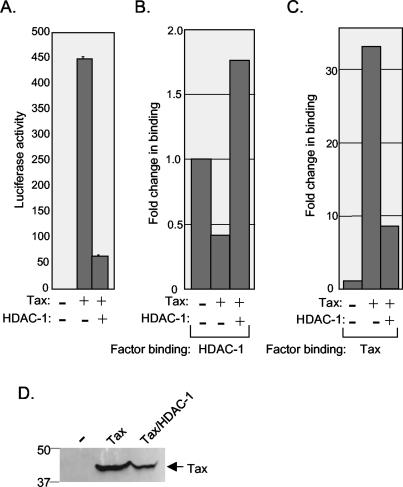
The opposing effects of Tax and the HDAC proteins on transcription from the HTLV-1 promoter raised the question as to whether these proteins displace one another from the promoter. To test this hypothesis, we used the ChIP assay to measure the binding of endogenous HDAC proteins at the chromosomally integrated HTLV-1 promoter in CHOK1-Luc cells in the absence and presence of Tax. Transfection of Tax into the CHOK1-Luc cells produced strong transcriptional activation from the integrated HTLV-1 promoter (Fig. 9A). As expected, this transcriptional activation correlated with the binding of Tax at the HTLV-1 promoter (Fig. 9B). Interestingly, transfection of the Tax expression plasmid significantly reduced the association of all three endogenous HDAC proteins at the promoter (Fig. 9C). In all cases, at least a twofold reduction in HDAC binding was observed in the presence, compared with the absence, of Tax.
FIG. 9.
Tax displaces HDAC-1, -2, and -3 from the HTLV-1 promoter. (A) Tax transactivation of HTLV-1 in CHOK1-Luc cells. Luciferase activity was determined in the same sample used for the ChIP experiments shown in panels B and C. (B) Quantification of relative Tax binding at the integrated HTLV-1 promoter. CHOK1-Luc cells were transfected with pSG-Tax, or with pUC19 (20 μg each) as a control. ChIP analysis was performed 24 h following transfection. Real-time PCR was used to quantify the DNA in each immunoprecipitation. The graph is representative of four independent ChIP experiments, and the background signal was normalized to 1. (C) Quantification of endogenous HDAC-1, -2, and -3 binding at the HTLV-1 promoter. CHOK1-Luc cells were transfected with pSG-Tax, or with pUC19 (20 μg each) as a control. ChIP analysis was performed 24 h following transfection. Real-time PCR was used to quantify the DNA in each immunoprecipitation. The graph is representative of four independent ChIP experiments, and HDAC binding in the absence of Tax was set to 1.
Since endogenous HDACs were displaced from the HTLV-1 promoter by Tax, we were interested in testing whether, conversely, Tax is displaced by the HDAC proteins. To test this hypothesis, we examined HTLV-1 promoter occupancy of Tax in the presence of transfected HDAC-1 in CHOK1 cells. As shown above, transfection of Tax increased transcription from the integrated HTLV-1 promoter, and cotransfection of HDAC-1 significantly repressed the effect of Tax (Fig. 10A). Factor occupancy mirrored this observation, as transfection of the HDAC-1 expression plasmid increased binding of HDAC-1 and decreased binding of Tax at the HTLV-1 promoter (Fig. 10B and C). Since the HDACs may also reduce expression of proteins from transfected plasmids, we measured Tax levels in the presence of HDAC-1. Western blot analysis showed that Tax was modestly lower in the presence of transfected HDAC-1 (Fig. 10D). Together, these data indicate that the opposing effects of Tax and the HDAC complexes may be carried out via a dynamic mechanism of factor displacement at the HTLV-1 promoter. Furthermore, the data suggest that the net effect of activators or repressors at a given promoter may be determined by their relative local concentrations.
FIG. 10.
HDAC-1 displaces Tax from the HTLV-1 promoter. (A) HDAC-1 repression of Tax transactivation of HTLV-1. Luciferase activity was determined in the same sample used for the ChIP experiments shown in panels B and C. (B) Quantification of relative HDAC-1 binding at the integrated HTLV-1 promoter. CHOK1-Luc cells were transfected with pSG-Tax (10 μg), pUC19 (20 μg), or pSG-Tax and HDAC-1 together (10 μg each). ChIP analysis was performed 24 h following transfection. Real-time PCR was used to quantify the DNA in the HDAC-1 immunoprecipitations, and HDAC binding in the absence of Tax was set to 1. (C) Quantification of Tax binding at the HTLV-1 promoter. CHOK1-Luc cells were transfected and processed as described for panel B. Real-time PCR was used to quantify the DNA in the anti-Tax immunoprecipitations, and the background signal was normalized to 1. (D) Western blot analysis of Tax expression in CHOK1-Luc cells following transfection.
DISCUSSION
In this study we compared transcription factor occupancy at the 5′ versus the 3′ LTR promoter of the integrated HTLV-1 provirus. ChIP analyses using cultured HTLV-1-transformed cell lines and ATLL cells revealed that Tax, ATF-1, ATF-2, CREB, c-Jun, c-Fos, CBP, p300, TAFII250, and Pol II were found at both the 5′ and 3′ LTRs. Although we found modest enrichment of most factors at the 5′ promoter, we also detected significant binding of transcription regulatory proteins at the 3′ promoter. This result was somewhat unexpected, as the 3′ LTRs of most mammalian retroviruses are generally believed to be transcriptionally silent (13). This pattern of factor binding was also observed in patient-derived ATLL cells, indicating that the results are not specific to long-term cell culture of productively infected T cells. It is intriguing to consider that transcription regulatory factors can associate with the HTLV-1 3′ promoter despite Pol II read-through from the highly active 5′ promoter.
Consistent with the binding of activators and coactivators at the 3′ U3 region, we also found that this promoter is transcriptionally active. Primer extension analysis revealed that the 3′ LTR-initiated transcripts represent 25 to 33% of the total viral RNA, demonstrating that the HTLV-1 3′ promoter has significant activity compared with other mammalian retroviruses. For example, transcripts initiated from the 3′ LTR of human immunodeficiency virus have not been detected and are estimated to account for less than 10% of LTR-generated transcription (19). Furthermore, a comparative analysis of the chromatin structure of the 5′ and 3′ LTRs of human immunodeficiency virus suggests that the 3′ promoter is inactive (42). It is possible that the transcriptional activity of the HTLV-1 3′ LTR is actually higher, since it is likely that these transcripts are not processed or packaged into virions and, therefore, are less stable. The functional significance of an active 3′ promoter in HTLV-1-infected T cells is currently unknown. It is formally possible that transcription from an active 3′ promoter of HTLV-1 contributes to oncogenic effects of this virus, as 3′ promoter activity has previously been shown to mediate malignant transformation with other retroviruses (2). However, HTLV-1 integration into T-cell DNA is essentially random, and only about 6% of the integration events occur in transcribed regions of the genome (22).
The HDACs are believed to play a major role in maintaining the balance between the acetylated and deacetylated states of lysine residues in the histone tails (3, 15). Our studies surprisingly revealed enrichment in the binding of HDAC-1 and -2 at the 5′ LTR and of HDAC-3 at the 3′ LTR. These data indicate that at least two HDAC complexes preferentially associate with the LTR sequences at the integrated proviral DNA. We also showed that RNA initiated from the 3′ LTR is increased by treatment with the HDAC inhibitor TSA, suggesting that the HDAC complex is involved in transcriptional regulation at the 3′ LTR. The mechanism that promotes differential HDAC distribution at the 5′ versus the 3′ LTR is unknown. Whether this distribution directly impacts relative transcription levels is also unknown. In both SLB-1 and MT2 cells we observed significant enrichment of CREB at the 5′ LTR. Since both HDAC-1 and HDAC-2, but not HDAC-3, have previously been coimmunoprecipitated with CREB (1), it is possible that CREB binding at the upstream promoter facilitates the selective binding of HDAC-1 and -2 to this region. None of the factors tested in the ChIP analysis was significantly enriched at the 3′ LTR. Therefore, the DNA binding factors that selectively recruit HDAC-3 to the 3′ LTR are unknown. Regulatory elements identified in the R and U5 regions of the LTR may bind a repressor protein necessary for HDAC-3 recruitment, and this repressor may be enriched within the 3′ R and U5 regions due to the lower level of polymerase read-through from the 3′ transcription unit (31, 35, 45).
All of the cell lines examined expressed the potent transcriptional activator protein Tax and, thus, the integrated proviruses are generally believed to be transcriptionally active. Although HDAC binding to the HTLV-1 LTRs is less than that observed at a transcriptionally repressed promoter, it is clear that the HDAC repressor complexes participate in regulation of HTLV-1 transcription in the presence of Tax. Therefore, we examined the interplay between Tax and the HDAC proteins on the HTLV-1 promoter. We found that endogenous HDAC proteins are associated with the integrated HTLV-1 promoter, and transfection of Tax led to their physical displacement with concomitant transcriptional activation. This relationship was reciprocal, as we also found that overexpressed HDAC proteins displaced Tax from the HTLV-1 promoter. These observations suggest that a dynamic exchange occurs between Tax and the HDAC proteins and that increased local concentrations of activator versus repressor complexes dictate what class of regulator is bound to the promoter. This exchange then influences the acetylation state of promoter histones, the recruitment of other regulators, and thus the transcriptional output of the virus. The mechanism of this regulation is unclear, as coimmunoprecipitation assays indicate that Tax physically interacts with HDAC-1 (5) as well as HDAC-2 and HDAC-3 (data not shown). These observations suggest that the physical interaction between Tax and the HDAC proteins facilitates the exchange process. It therefore appears that this interaction must be transient, enabling the dissociation of HDAC proteins concomitant with the stable binding of Tax to the viral CREs.
ADDENDUM IN PROOF
During the review of the manuscript, two reports showing that >50% of HLTV-1 integration sites occur within transcription units in cells derived from ATL patients were published (S. Hanai, T. Nitta, M. Shoda, M. Tanaka, N. Iso, I. Mizoguchi, S. Yashiki, S. Sonoda, Y. Hasegawa, T. Nagasawa, and M. Miwa, Cancer Sci., 95:306-310, 2004; T. Ozawa, T. Itoyama, N. Sadamori, Y. Yamada, T. Hata, M. Tomonaga, and M. Isobe, J. Hum. Genet. 49:154-165, 2004).
Acknowledgments
We are especially grateful to Robert Harrod, Shigeki Takemoto, and Hirokuni Taguchi for the ATLL cells, K.-T. Jeang for the CHOK1-Luc cells, and Ed Seto for the Flag-HDAC plasmids. We also thank Jill Livengood for critical reading of the manuscript.
This study was supported by grants from the National Institutes of Health, National Cancer Institute: CA55035 to J.K.N. and CA87540 to J.K.N. and P.J.L.
REFERENCES
- 1.Canettieri, G., I. Morantte, E. Guzman, H. Asahara, S. Herzig, S. D. Anderson, J. R. Yates III, and M. Montminy. 2003. Attenuation of a phosphorylation-dependent activator by an HDAC-PP1 complex. Nat. Struct. Biol. 10:175-181. [DOI] [PubMed] [Google Scholar]
- 2.Cullen, B. R., P. T. Lomedico, and G. Ju. 1984. Transcriptional interference in avian retroviruses—implications for the promoter insertion model of leukaemogenesis. Nature 307:241-245. [DOI] [PubMed] [Google Scholar]
- 3.Dehee, A., R. Cesaire, N. Desire, A. Lezin, O. Bourdonne, O. Bera, Y. Plumelle, D. Smadja, and J. C. Nicolas. 2002. Quantitation of HTLV-1 proviral load by a TaqMan real-time PCR assay. J. Virol. Methods 102:37-51. [DOI] [PubMed] [Google Scholar]
- 4.de Ruijter, A. J., A. H. van Gennip, H. N. Caron, S. Kemp, and A. B. van Kuilenburg. 2003. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370:737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ego, T., Y. Ariumi, and K. Shimotohno. 2002. The interaction of HTLV-1 Tax with HDAC1 negatively regulates the viral gene expression. Oncogene 21:7241-7246. [DOI] [PubMed] [Google Scholar]
- 6.Emerman, M., and H. M. Temin. 1984. Genes with promoters in retrovirus vectors can be independently suppressed by an epigenetic mechanism. Cell 39:449-467. [PubMed] [Google Scholar]
- 7.Frank, S. R., M. Schroeder, P. Fernandez, S. Taubert, and B. Amati. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15:2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Georges, S. A., H. A. Giebler, P. A. Cole, K. Luger, P. J. Laybourn, and J. K. Nyborg. 2003. Tax recruitment of CBP/p300, via the KIX domain, reveals a potent requirement for acetyltransferase activity that is chromatin dependent and histone tail independent. Mol. Cell. Biol. 23:3392-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Georges, S. A., W. L. Kraus, K. Luger, J. K. Nyborg, and P. J. Laybourn. 2002. p300-Mediated Tax transactivation from recombinant chromatin: histone tail deletion mimics coactivator function. Mol. Cell. Biol. 22:127-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giebler, H. A., J. E. Loring, K. Van Orden, M. A. Colgin, J. E. Garrus, K. W. Escudero, A. Brauweiler, and J. K. Nyborg. 1997. Anchoring of CREB binding protein to the human T-cell leukemia virus type 1 promoter: a molecular mechanism of Tax transactivation. Mol. Cell. Biol. 17:5156-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grant, C., K. Barmak, T. Alefantis, J. Yao, S. Jacobson, and B. Wigdahl. 2002. Human T cell leukemia virus type I and neurologic disease: events in bone marrow, peripheral blood, and central nervous system during normal immune surveillance and neuroinflammation. J. Cell. Physiol. 190:133-159. [DOI] [PubMed] [Google Scholar]
- 12.Guenther, M. G., W. S. Lane, W. Fischle, E. Verdin, M. A. Lazar, and R. Shiekhattar. 2000. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 14:1048-1057. [PMC free article] [PubMed] [Google Scholar]
- 13.Guntaka, R. V. 1993. Transcription termination and polyadenylation in retroviruses. Microbiol Rev. 57:511-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeang, K. T., D. Derse, M. Matocha, and O. Sharma. 1997. Expression status of Tax protein in human T-cell leukemia virus type 1-transformed MT4 cells: recall of MT4 cells distributed by the NIH AIDS Research and Reference Reagent Program. J. Virol. 71:6277-6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jepsen, K., and M. G. Rosenfeld. 2002. Biological roles and mechanistic actions of co-repressor complexes. J. Cell Sci. 115:689-698. [DOI] [PubMed] [Google Scholar]
- 16.Juan, L. J., W. J. Shia, M. H. Chen, W. M. Yang, E. Seto, Y. S. Lin, and C. W. Wu. 2000. Histone deacetylases specifically down-regulate p53-dependent gene activation. J. Biol. Chem. 275:20436-20443. [DOI] [PubMed] [Google Scholar]
- 17.Khorasanizadeh, S. 2004. The nucleosome: from genomic organization to genomic regulation. Cell 116:259-272. [DOI] [PubMed] [Google Scholar]
- 18.Kimzey, A. L., and W. S. Dynan. 1998. Specific regions of contact between human T-cell leukemia virus type I Tax protein and DNA identified by photocross-linking. J. Biol. Chem. 273:13768-13775. [DOI] [PubMed] [Google Scholar]
- 19.Klaver, B., and B. Berkhout. 1994. Comparison of 5′ and 3′ long terminal repeat promoter function in human immunodeficiency virus. J. Virol. 68:3830-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuo, M. H., and C. D. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19:425-433. [DOI] [PubMed] [Google Scholar]
- 21.Kwok, R. P., M. E. Laurance, J. R. Lundblad, P. S. Goldman, H. Shih, L. M. Connor, S. J. Marriott, and R. H. Goodman. 1996. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature 380:642-646. [DOI] [PubMed] [Google Scholar]
- 22.Leclercq, I., F. Mortreux, M. Cavrois, A. Leroy, A. Gessain, S. Wain-Hobson, and E. Wattel. 2000. Host sequences flanking the human T-cell leukemia virus type 1 provirus in vivo. J. Virol. 74:2305-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemasson, I., N. Polakowski, P. J. Laybourn, and J. K. Nyborg. 2002. Transcription factor binding and histone modifications on the integrated proviral promoter in HTLV-I-infected T-cells. J. Biol. Chem. 277:49459-49465. [DOI] [PubMed] [Google Scholar]
- 24.Lemasson, I., V. Robert-Hebmann, S. Hamaia, M. Duc Dodon, L. Gazzolo, and C. Devaux. 1997. Transrepression of lck gene expression by human T-cell leukemia virus type 1-encoded p40Tax. J. Virol. 71:1975-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lenzmeier, B. A., E. E. Baird, P. B. Dervan, and J. K. Nyborg. 1999. The tax protein-DNA interaction is essential for HTLV-I transactivation in vitro. J. Mol. Biol. 291:731-744. [DOI] [PubMed] [Google Scholar]
- 26.Lenzmeier, B. A., H. A. Giebler, and J. K. Nyborg. 1998. Human T-cell leukemia virus type 1 Tax requires direct access to DNA for recruitment of CREB binding protein to the viral promoter. Mol. Cell. Biol. 18:721-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, J., J. Wang, Z. Nawaz, J. M. Liu, J. Qin, and J. Wong. 2000. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 19:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔ CT) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 29.Lu, H., C. A. Pise-Masison, T. M. Fletcher, R. L. Schiltz, A. K. Nagaich, M. Radonovich, G. Hager, P. A. Cole, and J. N. Brady. 2002. Acetylation of nucleosomal histones by p300 facilitates transcription from tax-responsive human T-cell leukemia virus type 1 chromatin template. Mol. Cell. Biol. 22:4450-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundblad, J. R., R. P. Kwok, M. E. Laurance, M. S. Huang, J. P. Richards, R. G. Brennan, and R. H. Goodman. 1998. The human T-cell leukemia virus-1 transcriptional activator Tax enhances cAMP-responsive element-binding protein (CREB) binding activity through interactions with the DNA minor groove. J. Biol. Chem. 273:19251-19259. [DOI] [PubMed] [Google Scholar]
- 31.Montagne, J., and P. Jalinot. 1995. Characterization of a transcriptional attenuator within the 5′ R region of the human T cell leukemia virus type 1. AIDS Res. Hum. Retrovir. 11:1123-1129. [DOI] [PubMed] [Google Scholar]
- 32.Mori, N., P. S. Gill, T. Mougdil, S. Murakami, S. Eto, and D. Prager. 1996. Interleukin-10 gene expression in adult T-cell leukemia. Blood 88:1035-1045. [PubMed] [Google Scholar]
- 33.Nicot, C., R. Opavsky, R. Mahieux, J. M. Johnson, J. N. Brady, L. Wolff, and G. Franchini. 2000. Tax oncoprotein trans-represses endogenous B-myb promoter activity in human T cells. AIDS Res. Hum. Retrovir. 16:1629-1632. [DOI] [PubMed] [Google Scholar]
- 34.Okada, M., and K. T. Jeang. 2002. Differential requirements for activation of integrated and transiently transfected human T-cell leukemia virus type 1 long terminal repeat. J. Virol. 76:12564-12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okumura, K., G. Sakaguchi, S. Takagi, K. Naito, T. Mimori, and H. Igarashi. 1996. Sp1 family proteins recognize the U5 repressive element of the long terminal repeat of human T cell leukemia virus type I through binding to the CACCC core motif. J. Biol. Chem. 271:12944-12950. [DOI] [PubMed] [Google Scholar]
- 36.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poiesz, B. J., F. W. Ruscetti, A. F. Gazdar, P. A. Bunn, J. D. Minna, and R. C. Gallo. 1980. Detection and isolation of type C retrovirus particle from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA 77:7415-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rayman, J. B., Y. Takahashi, V. B. Indjeian, J. H. Dannenberg, S. Catchpole, R. J. Watson, H. te Riele, and B. D. Dynlacht. 2002. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev. 16:933-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rousset, R., C. Desbois, F. Bantignies, and P. Jalinot. 1996. Effects on NF-kappa B1/p105 processing of the interaction between the HTLV-1 transactivator Tax and the proteasome. Nature 381:328-331. [DOI] [PubMed] [Google Scholar]
- 40.Seiki, M., R. Eddy, T. B. Shows, and M. Yoshida. 1984. Nonspecific integration of the HTLV provirus genome into adult T-cell leukaemia cells. Nature 309:640-642. [DOI] [PubMed] [Google Scholar]
- 41.van den Hoff, M. J., V. M. Christoffels, W. T. Labruyere, A. F. Moorman, and W. H. Lamers. 1995. Electrotransfection with “intracellular” buffer. Methods Mol. Biol. 48:185-197. [DOI] [PubMed] [Google Scholar]
- 42.Verdin, E. 1991. DNase I-hypersensitive sites are associated with both long terminal repeats and with the intragenic enhancer of integrated human immunodeficiency virus type 1. J. Virol. 65:6790-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wattel, E., J. P. Vartanian, C. Pannetier, and S. Wain-Hobson. 1995. Clonal expansion of human T-cell leukemia virus type I-infected cells in asymptomatic and symptomatic carriers without malignancy. J. Virol. 69:2863-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wen, Y. D., V. Perissi, L. M. Staszewski, W. M. Yang, A. Krones, C. K. Glass, M. G. Rosenfeld, and E. Seto. 2000. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc. Natl. Acad. Sci. USA 97:7202-7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu, X., D. A. Brown, I. Kitajima, J. Bilakovics, L. W. Fey, and M. I. Nerenberg. 1994. Transcriptional suppression of the human T-cell leukemia virus type I long terminal repeat occurs by an unconventional interaction of a CREB factor with the R region. Mol. Cell. Biol. 14:5371-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshida, M., I. Miyoshi, and Y. Hinuma. 1982. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc. Natl. Acad. Sci. USA 79:2031-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, Y., R. Iratni, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 1997. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89:357-364. [DOI] [PubMed] [Google Scholar]