Abstract
Toll-like receptors (TLRs) recognize conserved products of microbial pathogens to initiate the innate immune response. TLR4 signaling is triggered upon binding of lipopolysaccharides (LPS) from gram-negative bacteria. Using comparative gene expression profiling, we demonstrate a master regulatory role of IκB kinase (IKK)/NF-κB signaling for immediate-early gene induction after LPS engagement in precursor B cells. IKK/NF-κB signaling controls a large panel of gene products associated with signaling and transcriptional activation and repression. Intriguingly, the induction of AP-1 activity by LPS in precursor B cells and primary dendritic cells fully depends on the IKK/NF-κB pathway, which promotes expression of several AP-1 family members, including JunB, JunD, and B-ATF. In pre-B cells, AP-1 augments induction of a subset of primary NF-κB targets, as shown for chemokine receptor 7 (CCR7) and immunoglobulin κ light chain. Thus, our data illustrate that NF-κB orchestrates immediate-early effects of LPS signaling and controls secondary AP-1 activation to mount an appropriate biological response.
Activation of transcription factor NF-κB plays a pivotal role in the regulation of diverse cellular processes such as inflammation, immune response, differentiation, proliferation, and apoptosis. The mammalian NF-κB family consists of five members, p50, p52, p65/RelA, c-Rel, and RelB, which are tightly controlled by a family of inhibitory molecules (IκBs) comprising IκBα, IκBβ, IκBɛ, and the precursor molecules for p50 and p52, p105 and p100, respectively. IκB proteins prevent nuclear entry and DNA binding of NF-κB. NF-κB activation is initiated through cytokine signaling, innate or adaptive immune responses, morphological signals, or environmental stress (14, 18, 28). All signaling pathways known to date converge at the IκB kinase (IKK) complex that consists of two catalytic subunits (IKKα and IKKβ) and one regulatory subunit (IKKγ/NEMO). The “canonical” IKKβ- and IKKγ-dependent signaling pathway involves stimulus-dependent phosphorylation of the IκB molecule IκBα, IκBβ, IκBɛ, or p105 at conserved serine residues. Phosphorylation triggers the recognition and polyubiquitination of IκBs by SCFβTrCP ubiquitin ligases and their subsequent destruction by the 26S proteasome. Released NF-κB enters the nucleus, where it activates transcription of target genes (14, 22). Aside from this canonical IκB degradation pathway, a “novel” pathway, which is activated by a subset of NF-κB stimuli, e.g., B-cell activating factor (BAFF), lymphotoxin β (LTβ), CD40 ligand, and lipopolysaccharide (LPS) and enhances the processing of p100 and the generation of p52 containing DNA binding complexes, has been described (6, 7, 10, 23, 36, 50). The p100 processing pathway requires NF-κB-inducing kinase, as shown for BAFF, LTβ, CD40, and IKKα, as demonstrated for LTβ. Furthermore, a requirement for ongoing protein synthesis was revealed for LTβ, CD40, and LPS.
The biological responses to IKK and NF-κB signaling largely depend on transcriptional up-regulation of a network of direct target genes that contain NF-κB consensus sites in their promoter or enhancer regions. About 150 target genes have been identified, mainly by studies with transfected promoters and by in vitro DNA binding assays (reviewed in reference 40). The gene expression profiles in response to well-established NF-κB stimuli, such as tumor necrosis factor alpha (TNF-α), interleukin-1β (IL-1β), CD40 ligand, and LPS, have been determined by microarray analyses (8, 31, 35, 51-53). Yet many stimuli known to trigger NF-κB activation also induce other signaling pathways, especially mitogen-activated protein kinase (MAPK) cascades, which ultimately lead to phosphorylation and enhanced transcriptional activity of transcription factors, e.g., c-Jun, JunB, JunD, ATF-2, and Elk-1/TCF (25). Previous reports have analyzed the contribution of IKK components and NF-κB activation for global gene expression by LPS, IL-1β, or TNF-α in pre-B cells and embryonal fibroblasts (27, 29) and by CD40 ligand in B cells (8). However, these microarray studies did not allow a differentiation between direct and indirect effects of NF-κB on target gene expression. By using gene profiling and chromatin immunoprecipitation, a network of direct NF-κB target genes has been identified in Hodgkin's disease-derived tumor cells, which indicated a central role of NF-κB in the pathogenesis of Hodgkin's lymphoma (19).
We have investigated the requirement of the IKK complex and NF-κB for gene expression by performing gene profiling in murine pre-B-cell lines after short-term LPS stimulation. We have taken advantage of the well-characterized 70Z/3 cell system, where the availability of an IKK-deficient mutant (1.3E2) and its revertant and efficient inhibition of “classical” NF-κB by IκBαΔN allows for an investigation of differential gene induction by LPS. About 70 genes were reproducibly up-regulated, the vast majority of which required both a functional IKK complex and NF-κB for full gene induction, indicating that the classical IκB degradation pathway is the central mediator of gene induction in the innate immune response. In addition, our data suggest that the IKK complex can mediate induction of a few genes by an alternative, at least partially NF-κB-independent, pathway. Primary IKK/NF-κB-triggered gene induction enhances the expression of a network of signaling components as well as positive and negative transcriptional regulators. LPS-induced AP-1 activity in 70Z/3 pre-B and primary human dendritic cells fully depends on IKK/NF-κB signaling. Subordinately activated AP-1 (JunD and JunB) complexes cooperate with NF-κB and are required for sustained, high-level chemokine receptor 7 (CCR7) and immunoglobulin κ (Igκ) light chain expression and production of surface IgM. Our data indicate that the IKK/NF-κB cascade plays a master role in the immediate-early gene response to LPS and that it mounts secondary transcriptional responses involving AP-1 and a network of other regulators.
MATERIALS AND METHODS
Cell culture and treatment.
Cell culture conditions for 70Z/3, 1.3E2, and 1.3E2 IKKγ/protein kinase C θ (PKCθ) have been described previously (24). Retroviral infection of 70Z/3 with pFBneoIκBαΔN, pLXSNA-Fos, or empty vector (pFBneo or pLXSN) and preparation of human primary dendritic cells were done as described previously (36). Except when stated otherwise, cells were treated with 10 μg of LPS (Sigma)/ml, 200 ng of phorbol myristate acetate (PMA) (Sigma)/ml, 20 μg of cycloheximide (CHX) (Calbiochem)/ml, or 10 ng of gamma interferon (IFN-γ) (Endogen)/ml. For UV light irradiation, cells were exposed to 120 J/m2 of UV light at 254 nm (UV-C) in a Stratagene UV cross-linker.
Plasmids and antibodies.
Flag-IκBαΔN was cloned into pFBneo (Stratagene) or pcDNA3 (Invitrogen), and Flag-A-Fos was cloned into pLXSN (Clontech). Antibodies used were as follows: anti-Flag M5 (Sigma), anti-JunD (sc-74), anti-cFos (sc-52; all from Santa Cruz), anti-JunB (33), anti-p65 (both from Biomol), and anti-mouse IgM (R6-60.2; both from Pharmingen).
DNA microarray analysis.
Total RNA was prepared from 70Z/3 and 1.3E2 cells, clones or pools of 70Z/3 cells infected with control virus (CV) (pFBneo; Stratagene), or 70Z/3 cells infected with IκBαΔN virus (ΔNV) (pFBneoIκBαΔN) left untreated or treated with LPS (90 min) by using an RNeasy kit (QIAGEN). Samples were prepared from 7.5 μg of total RNA according to manufacturer's instructions. Murine Genome U74Av2 GeneChip (Affymetrix) containing about 12,500 unique probe sets for genes and expressed sequence tags was hybridized with biotinylated cRNA for 16 h at 45°C, washed, and stained according to the manual. cRNA labeling and quality was confirmed by using GeneChip Test3 array (Affymetrix). DNA chips were scanned with a GeneChip scanner, and microarray image files were processed with Affymetrix Microarray Suite 5. For comparison, all chip files were scaled to a uniform intensity value of 1,000 U. Comparative analyses were done as follows. The values of LPS-stimulated 70Z/3, 1.3E2, 70Z/3CV, or 70Z/3ΔNV cells were taken as the experimental file and compared to the corresponding unstimulated cells as the baseline file. Probe sets that fit the following criteria were considered increased in 70Z/3 cells after 90 min of LPS stimulation (Table 1): the change call reflecting the variations of mRNA levels in the samples as calculated by the software was increased in both independent experiments, a twofold difference or more was obtained in at least one experiment, a presence call was given after LPS stimulation, and the signal value after LPS induction was 500 or more. Independent verifications by Northern blotting of genes that were included or excluded due to these criteria were performed (Fig. 1 and data not shown) to confirm that a reproducible increase was obtained even for genes that had increase calls of around twofold. For the comparison of 70Z/3CV and 70Z/3ΔNV cells, only probe sets that had an increase call in LPS-stimulated 70Z/3CV cells were chosen.
TABLE 1.
LPS and IKKγ target gene profiling in murine pre-B cellsa
| Category and protein | GenBank accession no. | Result for expt no.
|
Verified by Northern blotg | Reference(s)f | |||||
|---|---|---|---|---|---|---|---|---|---|
| 1
|
2
|
||||||||
| Change (fold) for:
|
Call for 1.3E2 cells | Change (fold) for:
|
Call for 1.3E2 cells | ||||||
| 70Z/3 cells | 1.3E2 cells | 70Z/3 cells | 1.3E2 cells | ||||||
| Extracellular ligands | |||||||||
| TNF-βc | M16819 | 16.0 | 1.3 | NC | 8.0 | 2.5 | NC | 19, 40 | |
| TNF-αc | D84196 | 10.6 | 9.8 | I | 7.5 | 1.6 | I | 19, 40 | |
| MIP-1α/Scya3c,d | J04491 | 9.2 | 2.3 | I | 24.3 | 2.6 | I | + | 19, 40 |
| 4-1BB ligandb | L15435 | 8.0 | 0.9 | NC | 4.0 | 1.0 | NC | + | |
| IP-10/ScyB10 | M33266 | 5.3 | 0.2 | NC | 4.9 | 0.6 | NC | 40 | |
| LT-β | U16985 | 3.7 | 0.8 | NC | 9.2 | 0.8 | NC | 40 | |
| WNT10A | U61969 | 3.2 | 0.9 | NC | 3.5 | 1.1 | NC | 27 | |
| EBI-3 | AF013114 | 2.6 | 0.9 | NC | 2.8 | 2.5 | NC | 27 | |
| RANK ligandb | AF019048 | 2.5 | 0.8 | NC | 5.3 | 1.1 | NC | ||
| IL-12 (p35) | M86672 | 2.0 | 1.1 | NC | 13.0 | 0.8 | NC | 15 | |
| Cell surface receptors | |||||||||
| ICAM1c | M90551 | 22.6 | 1.1 | NC | 16.0 | 1.1 | NC | 19, 40 | |
| CD40c | M83312 | 10.6 | 1.4 | NC | 13.9 | 0.4 | NC | + | 19 |
| Notch1 | Z11886 | 7.5 | 0.3 | NC | 4.3 | 6.1 | NC | 27 | |
| Igκ lighte | M80423 | 2.6 | 1.1 | NC | 3.5 | 1.0 | NC | 27, 40 | |
| Eck/Eph A2b | U07634 | 2.6 | 0.9 | NC | 2.1 | 1.1 | NC | + | |
| IL-10Rβb | U53696 | 2.0 | 0.8 | NC | 2.3 | 0.9 | NC | + | |
| CCR7/EBI1c | L31580 | 1.9 | 1.2 | NC | 2.3 | 0.6 | NC | + | 19, 27 |
| IFNαR2βb | Y09864 | 1.5 | 1.1 | NC | 2.5 | 1.1 | NC | ||
| CD5/Ly1b | M15177 | 1.5 | 0.9 | NC | 2.5 | 1.4 | NC | ||
| Intracelluar signaling | |||||||||
| A20 | U19463 | 48.5 | 1.1 | NC | 21.1 | 0.9 | NC | 40 | |
| COT/TPL2b | D13759 | 19.7 | 0.8 | NC | 3.7 | 0.2 | NC | + | |
| MyD118/GADD45Be | X54149 | 11.3 | 0.7 | NC | 6.5 | 1.1 | NC | 27 | |
| RGS16 | U94828 | 7.0 | 0.8 | NC | 9.2 | 1.2 | NC | 27 | |
| PIM1e | AA764261 | 5.7 | 0.9 | NC | 3.5 | 0.9 | NC | + | 54 |
| Pea-15/Mat-1 | L31958 | 5.3 | 0.9 | NC | 2.6 | 0.9 | NC | 27 | |
| PAC-1 | U09268 | 4.3 | 2.0 | I | 4.0 | 1.3 | I | 27 | |
| MKP7/MKPMb,c | AI642662 | 4.3 | 2.5 | I | 3.0 | 2.1 | I | + | |
| p21/CDKN1Ab,e | AW048937 | 2.6 | 1.5 | I | 2.8 | 1.3 | I | + | |
| CASPASE11 | Y13089 | 2.6 | 0.9 | NC | 3.0 | 0.8 | NC | 27 | |
| TRAF3b | U21050 | 2.5 | 1.0 | NC | 2.1 | 1.2 | NC | + | |
| BCL-10b | AJ006289 | 2.3 | 1.2 | NC | 1.9 | 1.1 | NC | ||
| F52/MLP1b | X61399 | 2.3 | 1.0 | NC | 1.9 | 1.1 | NC | ||
| PKCδ | X60304 | 2.0 | 1.1 | NC | 2.1 | 0.9 | NC | + | 19 |
| RNF14/TRIAD2b | AW123517 | 2.0 | 1.0 | NC | 1.7 | 1.0 | NC | + | |
| ABIN-1 | AJ242778 | 1.4 | 0.9 | NC | 2.0 | 1.0 | NC | + | 19, 27 |
| IAP-1 | U88908 | ND | ND | ND | 2.5 | 1.1 | NC | 40 | |
| A1/BFL1c,d | U23778 | 1.5 | 1.1 | NC | 2.6 | 1.1 | NC | 19, 40 | |
| Transcriptional activation/ repression | |||||||||
| JunBc,e | U20735 | 32.0 | 2.0 | NC | 7.5 | 0.9 | NC | + | 19, 40 |
| MAILb,d | AA614971 | 13.9 | 6.5 | I | 24.3 | 3.5 | I | + | |
| IκBαe | U57524 | 12.1 | 1.3 | I | 9.8 | 1.3 | I | 40 | |
| ReIBe | M83380 | 5.3 | 2.8 | NC | 8.0 | 0.8 | NC | 27 | |
| p100c | AW047899 | 4.0 | 0.9 | NC | 6.5 | 1.2 | NC | 40 | |
| TIS11/ZFP36b | M58566 | 3.2 | 0.9 | NC | 3.7 | 1.5 | I | + | |
| MPC2/CBX4b | U63387 | 3.2 | 1.0 | NC | 2.6 | 0.9 | NC | + | |
| NAB2b | U47543 | 3.0 | 1.6 | NC | 2.0 | 1.3 | NC | + | |
| IκBɛb | AF030896 | 2.8 | 0.9 | NC | 2.8 | 1.4 | NC | + | |
| OCT2 | X53654 | 2.8 | 0.2 | NC | 2.5 | 0.5 | NC | 4 | |
| BTG1b | Z16410 | 2.6 | 1.1 | NC | 2.0 | 1.1 | NC | + | |
| B-MYBb | X70472 | 2.5 | 0.9 | NC | 2.1 | 1.1 | NC | + | |
| IRF-8/ICSBP1b | M32489 | 2.5 | 1.0 | NC | 1.9 | 1.1 | NC | + | |
| Stra13/Dec1b | Y07836 | 2.5 | 0.9 | NC | 1.6 | 1.7 | I | + | |
| CHOP10/DDIT3b | X67083 | 2.5 | 1.2 | MI | 3.0 | 1.1 | I | + | |
| ZNF40/αACRYBP1b | L36829 | 2.5 | 2.3 | NC | 3.7 | 0.8 | NC | + | |
| B-ATF | AF017021 | 2.3 | 0.8 | NC | 2.8 | 0.3 | NC | + | 27 |
| IRF-5 | AF028725 | 2.3 | 1.2 | NC | 2.8 | 0.8 | NC | 27 | |
| BTG2b | M64292 | 2.1 | 1.1 | NC | 1.7 | 1.1 | NC | + | |
| LITAF/PIG7c | AI852632 | 2.0 | 1.4 | I | 2.0 | 1.1 | NC | + | 19 |
| p105 | M57999 | 2.0 | 0.9 | NC | 1.7 | 0.9 | NC | 40 | |
| NUR77/NR4A1b | X16995 | 2.0 | 1.3 | NC | 1.6 | 1.1 | NC | + | |
| NF-ATcb | AF087434 | 1.7 | 1.4 | I | 2.0 | 1.3 | NC | ||
| TIS7b | V00756 | ND | ND | ND | 2.0 | 1.4 | NC | + | |
| Miscellaneous | |||||||||
| IL4 induced 1 | U70430 | 18.4 | 0.7 | NC | 17.1 | 0.9 | NC | 27 | |
| ISG15 | X56602 | 7.0 | 0.5 | NC | 6.1 | 6.1 | NC | 27 | |
| ZNF151b | U14556 | 3.7 | 0.8 | NC | 4.0 | 1.4 | NC | + | |
| mKIAA0964b | AI840413 | 3.7 | 1.4 | NC | 2.0 | 0.9 | NC | + | |
| CD74 antigen (DHLAG)b | X00496 | 3.2 | 2.1 | NC | 3.2 | 0.5 | NC | ||
| HERPUD1b | AI846938 | 2.3 | 1.3 | I | 2.5 | 1.1 | NC | +/− | |
| SWAP-70b | AF053974 | 1.7 | 0.7 | NC | 2.5 | 1.4 | NC | + | |
Wild-type 70Z/3 cells and their IKKγ-lacking derivative 1.3E2 cells were left untreated or treated for 90 min with LPS before total RNA was prepared and subjected to DNA microarray analysis. LPS-inducible genes were grouped according to their potential cellular function. Shown are the results of two independent experiments (1 and 2). The indicated values represent the increases (fold) of RNA after LPS stimulation compared to the untreated controls as a baseline (70Z/3 LPS versus without 70Z/3 and 1.3E2 LPS versus without vs. 1.3E2; left and central columns, respectively). According to the sort criteria, all genes had an increased call after LPS stimulation in 70Z/3 cells. The change call for the genes in 1.3E2 cells upon LPS treatment is shown in the right column for each experiment (NC, not changed; I, increased; MI, mildly increased; ND, not determined).
Novel IKK and/or NF-κB target genes.
Genes activated by constitutive NF-κB in Hodgkin's disease tumor cells (19).
Genes induced by at least twofold in IKKγ-negative 1.3E2 cells in both experiments.
Multiple hits on the microarray.
Previously described NF-κB/IKK induced genes.
LPS-induced expression for the majority of novel IKK/NF-κB target genes have been verified by Northern blotting, as indicated (+) (compare to Figure 1).
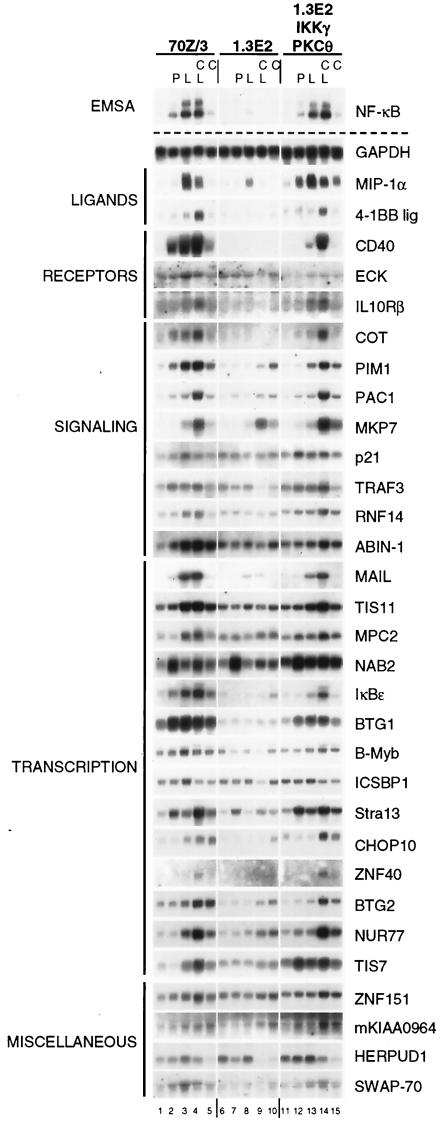
FIG. 1.
Analysis of LPS and IKKγ-dependent gene regulation by Northern blotting. 70Z/3, 1.3E2, or 1.3E2 IKKγ/PKCθ-expressing cells were stimulated for 2 h with PMA (P), LPS (L), LPS in the presence of CHX (C), or CHX alone. As a control, whole-cell extracts were prepared and analyzed by EMSA for NF-κB DNA binding activity (upper left panel). In parallel, total RNA was extracted and subjected to Northern blotting with cDNA probes for the indicated genes. Membranes were repetitively used after stripping, and equal loading was verified by using a GAPDH (glyceraldehyde-3-phosphate dehydrogenase) probe.
DNA binding assays and Western and Northern blotting.
Preparation of whole-cell extracts for Western blotting and electrophoretic mobility shift assay (EMSA) were essentially performed as described previously (24, 33). For Northern blotting, total RNA was prepared, separated by using gel electrophoresis, blotted, cross-linked, and hybridized as described previously (19). Probes were generated from IMAGE cDNA clones obtained from the Resource Center of the German Human Genome Project, Berlin, Germany (http://www.rzpd.de). A detailed description of probe generation is available upon request.
Flow cytometry.
Surface IgM expression was analyzed with fluorescein isothiocyanate-labeled anti-mouse IgM antibody. Cytometric analysis was done with a FACSCalibur cytometer (Becton Dickinson).
RESULTS
The IKK complex mediates the primary LPS response in pre-B cells.
To characterize the requirement of the IKK complex for the repertoire of genes induced by LPS, we performed gene expression profiling in pre-B cells. 70Z/3 and IKKγ-deficient 1.3E2 cells were left untreated or were stimulated with LPS for 90 min. This relatively short stimulation time was chosen to restrict the analysis to primary LPS/IKK-responsive genes. Total RNA was prepared and hybridized to high-density DNA microarrays comprising approximately 12,500 genes and expressed sequence tags. Table 1 shows all genes that had an increased call after LPS stimulation of 70Z/3 cells in two independent experiments and that were up-regulated by twofold or more in at least one experiment. Untreated 70Z/3 served as a baseline. The results were compared to 1.3E2 cells treated accordingly.
LPS stimulation for 90 min in 70Z/3 cells reproducibly up-regulated approximately 70 genes, which were grouped according to their assigned cellular functions (Table 1). Expression of none of these LPS response genes was increased in IKKγ-negative 1.3E2 cells to the same extent, and only three genes, namely, MIP-1α, MKP-7, and MAIL, were reproducibly induced in 1.3E2 cells (Table 1 and Fig. 1), indicating that an IKKγ-independent pathway(s) allows at least partial induction of these genes after LPS treatment. Moreover, we did not observe an increase in the expression of any gene after LPS treatment in 1.3E2 cells that was not increased in 70Z/3 cells (Table 1 and data not shown). These data indicate that the IKK complex is the central mediator for LPS-stimulated gene expression. In contrast, the baseline signal for most LPS-inducible genes in 70Z/3 cells was not significantly altered in 1.3E2 cells (data not shown), suggesting that the function of the IKK pathway is largely restricted to inducible gene activation rather than to basal expression. Furthermore, the expression of only very few genes decreased in 70Z/3 cells after 90 min of LPS treatment (data not shown), emphasizing that the IKK signaling pathway predominantly causes gene induction as a primary response.
A number of LPS response genes have been described previously as IKK/NF-κB-regulated genes (Table 1), but more then 50% of the genes were putative novel IKK/NF-κB-regulated genes (Table 1). We investigated the mRNA up-regulation for most of these putative IKK/NF-κB target genes (Table 1 and Fig. 1). Northern blotting was performed with mRNA from 70Z/3 and 1.3E2 cells and from a 1.3E2 clone expressing IKKγ and PKCθ to rescue LPS- and PMA-mediated NF-κB activation (24) (Fig. 1). Induction of nearly all LPS target genes was verified, indicating that the microarray data are highly reliable. Furthermore, most genes were not induced in 1.3E2 cells while induction was restored in cells complemented with IKKγ and PKCθ, proving that the induction of these genes by LPS requires intact IKK signaling. As expected from the microarray data (Table 1), MIP-1α and MAIL were partially induced in 1.3E2 cells, while induction of MKP-7 was completely independent of IKKγ.
LPS stimulation was also performed in the presence of the translation inhibitor CHX to discriminate between a primary immediate-early gene induction and secondary gene expression. As expected, CHX either did not affect or even enhanced the expression of most LPS target genes, underscoring that these genes actually constitute an immediate-early transcriptional response and do not require induced synthesis of other factors. For many genes, CHX treatment alone caused a slight enhancement of mRNA expression, and CHX-mediated up-regulation was seen in 1.3E2 cells as well (e.g., PIM1, IκBɛ, BTG2, and NUR77), perhaps due to a depletion of IκBs or other inhibitors by constitutive turnover. In contrast, up-regulation of MIP-1α was significantly inhibited by CHX treatment, indicating that either the blocked synthesis of labile costimulatory factors or a secondary NF-κB-dependent event might be required for full gene induction.
We also determined the mRNA amounts induced by PMA as a further IKK-activating agent. PMA directly activates PKC and mimics B-cell receptor signaling in 70Z/3 pre-B cells (24). For most genes, LPS was a stronger stimulus than PMA, which could be explained by the weaker and more transient NF-κB activation by PMA compared to the sustained NF-κB activation in response to LPS (36). Alternatively, LPS, but not PKC, activation might trigger costimulatory pathways that are required for full NF-κB-driven transcriptional activation. Only two genes, NAB2 and Stra13, were activated more strongly by PMA. Remarkably, PMA induced both genes in 1.3E2 cells as well, although LPS induction required the presence of IKKγ. These data indicate that in pre-B cells, PMA induces expression of a subset of genes by distinct pathways, independent of the IKK complex.
NF-κB activation is required for the induction of most LPS response genes.
To determine the contribution of NF-κB for LPS/IKK-mediated gene expression, we performed a genechip analysis using 70Z/3 cells retrovirally infected either with a CV or with an IκBα superrepressor-expressing virus (IκBαΔNV) to inhibit NF-κB activation. An IκBαΔN-expressing clone with strong expression of the transgene was used for the microarray analysis. In all IκBαΔN-expressing 70Z/3 cells, LPS-induced NF-κB activation and postinductive IκBα resynthesis were efficiently inhibited (see Fig. 6A and data not shown). Cells were left untreated or stimulated for 90 min with LPS before RNA extraction. Microarray analysis was performed, and the results are shown in Fig. 2.
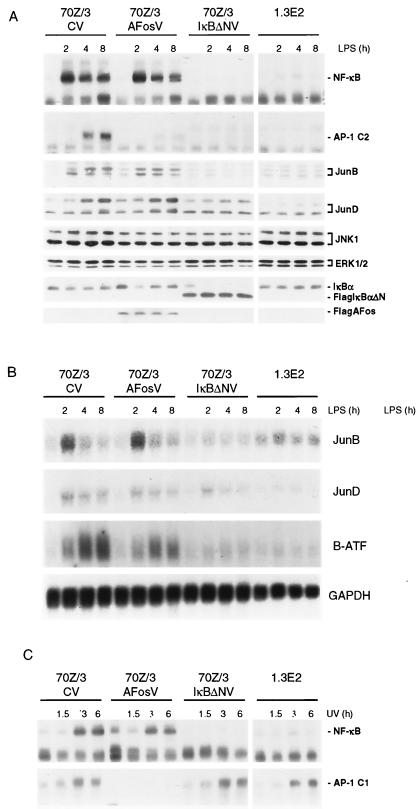
FIG. 6.
AP-1 activation in response to LPS correlates with NF-κB-mediated up-regulation of JunB, JunD, and B-ATF. (A) 70Z/3 cells infected with CV, Flag-A-Fos virus (AFosV), or Flag-IκBαΔNV(IκBΔNV) and 1.3E2 cells were stimulated for 0, 2, 4, and 8 h with LPS, and extracts were analyzed for NF-κB and AP-1 DNA binding activity by EMSA and for JunB, JunD, JNK1, ERK1/2, IκBα, Flag-IκBαΔN, and FlagA-Fos expression by Western blotting. (B) Cells were treated as described above (A), and total RNA was extracted and analyzed for JunB, JunD, B-ATF, and GAPDH expression by Northern blotting. (C) The different cell clones were stimulated with UV light, and extracts were prepared after 0, 1.5, 3, and 6 h and analyzed for NF-κB and AP-1 activity by EMSA.
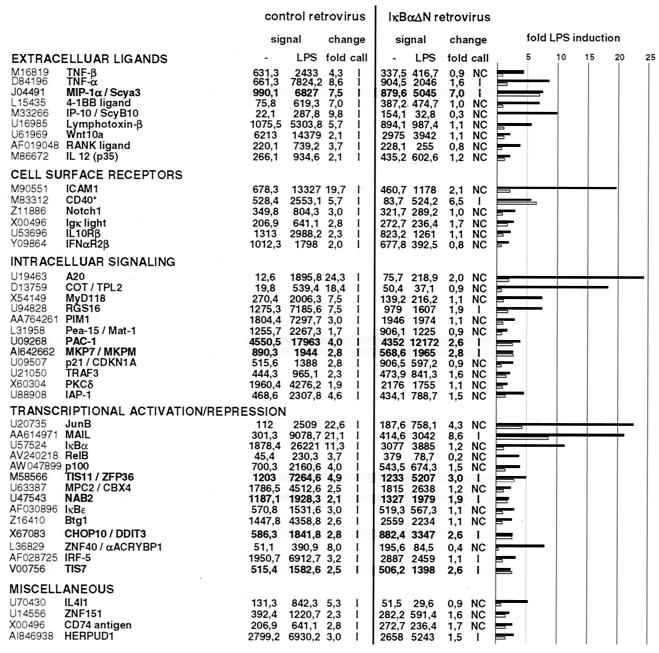
FIG. 2.
LPS and NF-κB-dependent target gene profiling in murine pre-B cells. Either mock-infected (control retrovirus) cells or a clone of 70Z/3 cells infected with IκBαΔN-expressing retrovirus (IκBαΔN retrovirus) were left untreated or stimulated with LPS for 90 min before total RNA was extracted and subjected to microarray analysis. Shown are the baseline signals (first column), the signal intensity after LPS stimulation (second column), the induction (fold) (third column), and the change call comparing LPS to baseline signals (fourth column) for 70Z/3 control (left) or IκBαΔN (right). A graphical representation of the severalfold inductions for each gene in 70Z/3CV cells (black) and 70Z/3 IκBΔN cells (gray) is shown. Genes were sorted as shown in Table 1. Genes in gray boxes are refractory to IκBαΔN-mediated LPS induction. Note that basal CD40 expression is highly reduced in IκBαΔN-expressing cells. The deceptive increase in 70Z/3 IκBαΔN cells after LPS treatment results from the very low expression level and could not be verified by Northern blotting (data not shown).
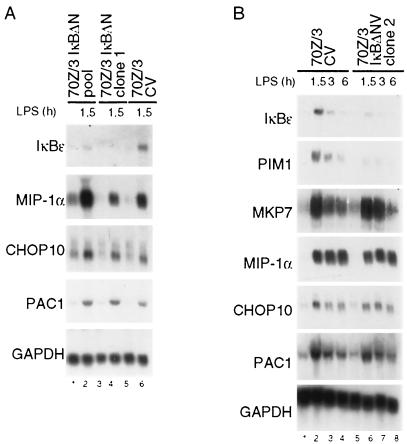
Retroviral infection led to an overall decrease in the induction of many target genes compared to uninfected cells (data not shown). Comparable results were also obtained with a pool of IκBαΔN-expressing cells, even though the block of induction of many target genes was less pronounced due to the slightly weaker expression of the superrepressor (data not shown). Expression of only 44 genes was increased in 70Z/3CV cells compared to the initial screen with 68 induced genes, suggesting that viral infection might render the cells less susceptible to LPS. Most genes whose expression was stimulated in 70Z/3 cells showed a considerable reduction in LPS-mediated gene activation upon IκBαΔN expression (Fig. 2). Nevertheless, some genes, namely, MIP-1α, PAC1, MKP-7, TIS11, NAB2, CHOP10, and TIS7, were hardly repressed in IκBαΔN-expressing cells, suggesting that their induction was partially or completely independent of NF-κB. Therefore, the induction of some of these genes was further analyzed by Northern blotting in different IκBαΔN-expressing 70Z/3 cell lots (Fig. 3). Whereas the induction of the bona fide IKK/NF-κB targets IκBɛ and PIM1 was strongly impaired upon NF-κB inhibition, MKP-7, MIP-1α, CHOP10, and PAC1 were completely unaffected.
FIG. 3.
Analysis of LPS-mediated NF-κB-independent gene regulation in 70Z/3 cells. (A and B) CV- or IκBΔNV-infected 70Z/3 cells (a pool or two independent clones, as indicated were treated with LPS for the times indicated, and RNA was prepared and subjected to Northern blotting as indicated. LPS-mediated induction of IκBɛ, PIM1, MKP-7, MIP-1α, CHOP10, PAC1, and GAPDH was analyzed.
The microarray analysis highlights the central role of the IKK complex and NF-κB for primary LPS-mediated gene activation. Since induction of a minority of IKKγ-dependent genes was not influenced by NF-κB inhibition, IKKγ appears to directly or indirectly control the activity of other transcription factors. Interestingly, genes whose induced expression was partially (MIP1-α) or completely (MKP-7) independent of IKKγ (compare Table 1 and Fig. 1) were also found in the group of genes that was insensitive to NF-κB inhibition. Thus, full LPS-mediated induction of these genes may require autonomous, IKK/NF-κB-independent signaling pathways.
Primary IKK/NF-κB activation is required for LPS-induced AP-1 activity in pre-B cells and dendritic cells.
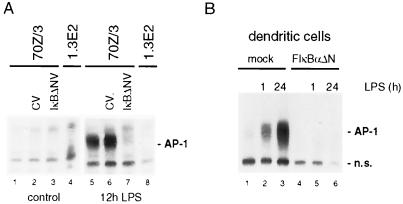
Besides the well-established autoregulation of NF-κB/IκB family members, LPS-triggered IKK signaling induced a large panel of further transcriptional activators (e.g., Oct2 and B-Myb) or repressors (e.g., MPC2, NAB2, Stra13, CHOP10, and TIS7). Furthermore, LPS strongly enhanced the expression of JunB and B-ATF, two potential AP-1 complex components (Table 1). We asked whether IKK/NF-κB signaling is necessary for LPS-mediated AP-1 activation in 70Z/3 pre-B cells (Fig. 4A). AP-1 DNA binding was induced by LPS, and activation was completely blocked in IKKγ-deficient 1.3E2 cells and in 70Z/3 cells infected with IκBαΔN retrovirus. Similarly, LPS-induced AP-1 activity in human primary dendritic cells was inhibited upon IκBαΔN expression (Fig. 4B). These observations provide the first evidence that NF-κB regulates an AP-1 activity in LPS-initiated responses.
FIG. 4.
NF-κB is required for LPS-induced AP-1 activation in pre-B cells and dendritic cells. (A) Untreated 70Z/3, CV- and IκBΔN-infected 70Z/3, or 1.3E2 cells were stimulated with LPS for 12 h, and AP-1 DNA binding activity was determined by EMSA. (B) Human primary dendritic cells were mock transfected or transfected with IκBαΔN expression vector and afterwards stimulated with LPS for 1 and 24 h. AP-1 DNA binding activity was analyzed by EMSA. IκBαΔN expression resulted in the complete loss of NF-κB DNA binding activity in dendritic cells (compare results to reference 36; data not shown). n.s., nonspecific.
To address the mechanism of NF-κB-mediated gene expression, we considered the expression patterns for all Jun/Fos/ATF/Maf family members represented on the microarray (Table 2). Besides JunB and B-ATF, expression levels of JunD and ATF-4 had an increase call after 90 min of LPS stimulation, which was below the threshold to be included in Table 1. In addition, MafF expression was enhanced, and detection call changed from absent to present even though, due to the low signal values, no increase call could be obtained. We confirmed the induction of JunB, JunD, B-ATF, and MafF by Northern blotting (Fig. 5A). In contrast, c-Jun and all Fos/Fra family members, components of the classical mitogen-induced AP-1 heterodimer, were absent in pre-B cells before and after induction (Table 2 and Fig. 5A). c-Jun, c-Fos, and FosB were also not induced after 4 h of LPS stimulation (data not shown). Other ATFs were expressed but displayed no change after stimulation (e.g., ATF-1 and ATF-2). These data suggest that the LPS-initiated IKK/NF-κB pathway could control AP-1 activity by up-regulation of a subset of AP-1 family members.
TABLE 2.
Expression of potential AP-1 complex components in 70Z/3 and 1.3E2 cellsa
| Family and protein | GenBank accession no. | Expression pattern for:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 70Z/3 cells
|
1.3E2 cells
|
||||||||||
| Control
|
LPS
|
Change call of LPS vs control | Control
|
LPS
|
Change call of LPS vs control | ||||||
| Signal | Call | Signal | Call | Signal | Call | Signal | Call | ||||
| Jun | |||||||||||
| JunB | U20735 | 60 | A | 1,766 | P | I | 180 | A | 370 | A | NC |
| JunD | J04509 | 20,892 | P | 27,285 | P | I | 15,413 | P | 27,694 | P | I |
| c-Jun | X12761 | 74 | A | 297 | A | NC | 214 | A | 197 | A | NC |
| Fos/Fra | |||||||||||
| c-Fos | V00727 | 249 | A | 52 | A | NC | 276 | A | 213 | A | NC |
| FosB | AF093624 | 75 | A | 66 | A | NC | 136 | A | 47 | A | NC |
| Fra1 | AF017128 | 36 | A | 49 | A | NC | 246 | A | 45 | A | NC |
| Fra2 | X83971 | 203 | A | 89 | A | NC | 671 | A | 251 | A | NC |
| ATF | |||||||||||
| B-ATF | AF017021 | 739 | A | 1,656 | P | I | 769 | A | 627 | A | NC |
| ATF4 | M94087 | 19,697 | P | 29,801 | P | I | 16,648 | P | 15,294 | P | NC |
| ATF1 | M63725 | 1,498 | P | 1,511 | P | NC | 1,774 | P | 2,281 | P | NC |
| ATF2 | U46026 | 1,300 | P | 767 | P | NC | 1,152 | P | 1,245 | P | NC |
| ATF3 | U19118 | 196 | A | 284 | P | NC | 281 | A | 165 | A | NC |
| ATF5/ATFx | AB012276 | 729 | P | 862 | P | NC | 425 | A | 817 | P | NC |
| Maf | |||||||||||
| MafF | AB009694 | 166 | A | 482 | P | NC | 80 | A | 252 | M | NC |
| MafG | AB009693 | 800 | A | 738 | P | NC | 867 | P | 669 | P | NC |
The complete sets of Jun/Fos/ATF/Maf family members present on the microarray chip from experiment 1 (Table 1) are shown. Signal values (arbitrary units) and detection call (A, absent; P, present; M, mildly expressed) are given for each sample (control and 90-min LPS). The change calls (I, induced; NC, no change) for LPS-treated cells versus control cells are indicated for each pair.
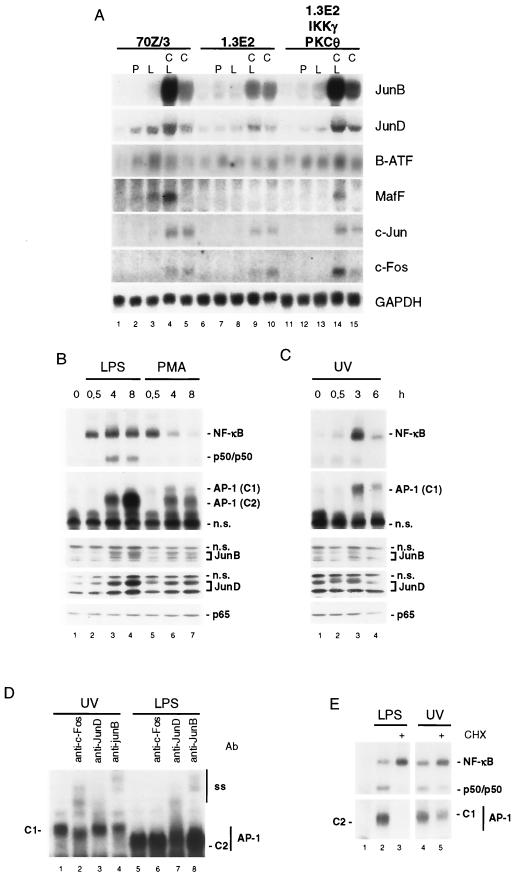
FIG. 5.
IKK/NF-κB activation regulates LPS induction of AP-1 family members in pre-B cells. (A) 70Z/3 or 1.3E2 cells or 1.3E2IKKγ/PKCθ-expressing cells were stimulated as described in the Fig. 1 legend and analyzed for expression of Jun/Fos/ATF/MAF family members as indicated. (B and C) 70Z/3 cells were stimulated with LPS or PMA (B) or UV light (C) for the times indicated, and whole-cell extracts were analyzed for NF-κB and AP-1 DNA binding and JunB, JunD, and p65 expression levels by Western blotting. The migrations of two AP-1 DNA complexes that have a differential mobility are indicated (C1 and C2). (D) AP-1 DNA binding activity after UV light (3 h) or LPS (8 h) stimulation was subjected to supershifting analysis with anti-JunB, anti-JunD, or anti-cFos antibody. Mobilities of AP-1 C1, AP-1 C2, and supershifted complexes (ss) are indicated. (E) 70Z/3 cells were stimulated with LPS or UV light for 4 h in the absence or presence of CHX. NF-κB and AP-1 DNA binding activity were analyzed by EMSA. n.s., nonspecific.
We compared the induction of AP-1 proteins and DNA binding activity by LPS, PMA, and UV light in 70Z/3 cells (Fig. 5B and C). The expression of JunB and JunD proteins was induced by LPS and, although weaker, by PMA (Fig. 5B). In contrast, UV light did not significantly enhance JunB and JunD expression levels but induced JunD phosphorylation (Fig. 5C). Whereas LPS or PMA activated NF-κB rapidly, the appearance of an AP-1 DNA binding complex was delayed and detectable at 4 h of LPS or PMA stimulation and strongest after 8 h of LPS incubation (complex C2) (Fig. 5B). The kinetics and intensity of AP-1 activation correlated with the up-regulation of JunB and JunD proteins (Fig. 5B). In contrast, UV light-induced NF-κB and AP-1 DNA binding activities peaked after 3 h, and this AP-1 complex displayed a slower migration (complex C1) (Fig. 5C). Supershift analysis (Fig. 5D) revealed that the UV-induced C1 complex, but not the LPS-induced C2 complex, contained c-Fos. While both complexes were reacting with JunB antibody, the LPS complex was strongly inhibited and supershifted by JunD antibody, demonstrating that UV light and LPS induce distinct AP-1 complexes. Moreover, the formation of the LPS-induced AP-1 complex was completely inhibited by CHX, while the induction of the C1 AP-1 complex by UV light was largely independent of protein de novo synthesis (Fig. 4).
To investigate the requirements for LPS-mediated AP-1 activation and to determine the functional consequences for AP-1 downstream of NF-κB, we infected 70Z/3 cells with the AP-1 inhibitor A-Fos, which specifically forms inactive heterodimers with Jun partners (39). We performed a kinetic analysis of NF-κB and AP-1 activation as well as induction of JunB and JunD protein amounts in 70Z/3 cells expressing either A-Fos or IκBαΔN and in IKKγ defective 1.3E2 cells (Fig. 6A). LPS-induced up-regulation of JunB and JunD and of AP-1 DNA binding activity was completely lost in IKKγ-deficient 1.3E2 cells and in 70Z/3 cells infected with IκBαΔN retrovirus but not in cells infected with CV (Fig. 6A). The AP-1 inhibitor A-Fos blocked AP-1, but not NF-κB, activity. However, A-Fos did not abolish induction of the JunB and JunD proteins by LPS (Fig. 6A). Likewise, LPS induction of JunB, JunD, or B-ATF mRNAs was reduced in the absence of IKKγ or when IκBαΔN was expressed, but not upon A-Fos expression (Fig. 6B). Interestingly, mRNA levels for JunD were initially increased in an NF-κB-independent manner, but the increase in IκBαΔN-expressing cells was much more transient than mock- or A-Fos-infected cells. Expression of c-Jun N-terminal kinase 1 (JNK1) and extracellular signal-regulated kinase 1 and 2 (ERK1/2), potential upstream regulators of AP-1, was not changed in response to LPS, even though ERK levels were slightly reduced in A-Fos virus-infected cells. Using phosphorylation-specificantibodies, we were not able to detect phosphorylated JNK or ERK after prolonged exposure to LPS (data not shown), indicating that delayed AP-1 activity does not correlate with the activation of MAPK signaling.
In contrast to the LPS-induced AP-1 complexes, the induction of AP-1 activity by UV light was blocked by A-Fos, but it was insensitive to NF-κB inhibition by IκBαΔN and not affected by the lack of IKKγ in 1.3E2 cells (compare Fig. 6C to A, top panels). We conclude from these experiments that UV light induces the formation of Fos/Jun containing C1-type AP-1 complexes by a classical pathway likely involving MAPK signaling, while induction of C2-type AP-1 by LPS requires IKK- and NF-κB-dependent induction of JunD and JunB and possibly of other AP-1 family members.
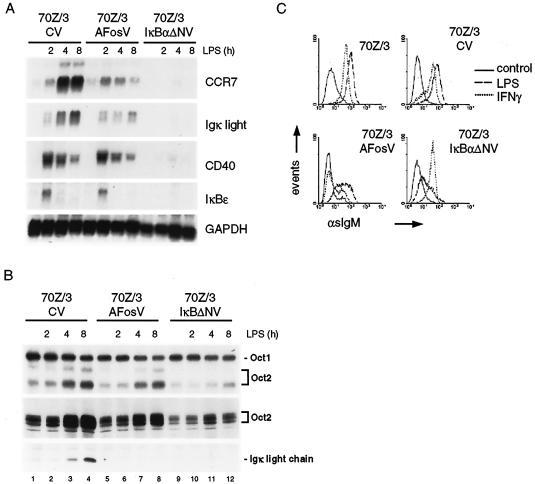
The LPS-IKK/NF-κB cascade utilizes the secondary AP-1 response for full induction of CCR7 and expression of surface IgM.
The IKK/NF-κB-dependent delayed AP-1 activation suggests that both transcription factors might cooperate in a second phase of gene induction in pre-B cells. For a target gene analysis, we selected the CCR7 gene (Table 1), whose LPS induction in 70Z/3 cells required de novo protein synthesis (19) to assess the functional requirement for IKK/NF-κB-dependent activation of AP-1. Initial LPS induction of CCR7 mRNA levels was seen at 2 h but was strongly enhanced thereafter (Fig. 7A). While AP-1 inhibition by A-Fos did not affect the immediate-early induction of CCR7, it completely inhibited the secondary increase of CCR7 mRNA after 4 and 8 h. Similarly, AP-1 inhibition diminished a second phase of induction of the Igκ light chain gene (Fig. 7A), which resulted in a drastic reduction of Ig light chain protein expression (Fig. 7B). In contrast, the induction of two other genes, CD40 and IκBɛ, whose expression peaked at early times of stimulation and decreased thereafter, was not affected by A-Fos (Fig. 7A). Likewise, LPS-induced Oct-2 expression and DNA binding activity remained largely unaffected upon AP-1 inhibition (Fig. 7B). However, as expected, expression of the NF-κB superrepressor IκBΔN completely eliminated LPS responsiveness of CCR7, Igκ light chain, CD40, IκBɛ, and Oct-2 (Fig. 7A and B).
FIG. 7.
NF-κB and AP-1 cooperate in transcriptional and biological responses to LPS. (A) 70Z/3CV-, Flag-A-FosV-, and IκBαΔNV-infected cells were stimulated with LPS for 0, 2, 4, and 8 h, and total RNA was prepared and analyzed for mRNA expression of the indicated genes by Northern blotting. (B) Cells were treated as described above (A), and DNA binding activity of Oct-1 and Oct-2 was determined by EMSA. OCT-2 and Igκ light chain expression was analyzed by Western blotting (lower panels). (C) Either parental 70Z/3 cells or control (pLXSN) infected and A-Fos- and IκBαΔN-infected 70Z/3 cells were treated for 20 h with LPS (1 μg/ml) or IFN-γ, and sIgM expression was examined by flow cytometry.
A biologically important consequence of LPS signaling at the transition from pre-B cell to B cell is the induction of transcription and V(D)J recombination of the Igκ light chain locus and the concomitant expression of surface IgM (sIgM) (49). In line with the IKK/NF-κB dependency of Igκ light chain mRNA induction by LPS, IκBαΔN strongly impaired LPS-stimulated sIgM expression (Fig. 7C). Concurrent with the effects on Igκ light chain mRNA induction, A-Fos expression strongly reduced the amount of LPS-induced sIgM as well. Interestingly, sIgM induction by IFN-γ was almost completely inhibited by A-Fos but not by IκBαΔN, underscoring the differential downstream requirements of LPS and IFN-γ to induce Igκ transcription (5). The observations that IFN-γ induces AP-1 (38) together with our findings indicate an essential role for AP-1 in IFN-γ signaling.
Taken together, our results demonstrate that in LPS signaling, IKK/NF-κB-induced AP-1 complexes are required to synergize with NF-κB in a secondary transcriptional response program.
DISCUSSION
A master function of IKK/NF-κB signaling in global LPS induced gene expression in pre-B cells.
The immediate-early induction of gene expression by LPS is known to critically depend on Toll-like receptor 4 (TLR4)-mediated parallel activation of MAPK and IKK signaling pathways. Whereas MAPK signaling involves phosphorylation of ERKs, JNKs, and p38 members and the subsequent activation of transcription factor Elk1, AP-1, SRF, or CREB, activation of IKK induces nuclear translocation and DNA binding of NF-κB (17). Given the range of activated signaling cascades, it could be expected that LPS-induced gene profiles display a heterogeneity that reflects the activation of a diverse spectrum of transcription factors. By use of gene chip analysis, we now provide evidence that the IKK complex is the central player for LPS-induced gene expression in 70Z/3 pre-B cells. With a few exceptions, no gene induction was seen upon LPS stimulation of IKK-defective 1.3E2 cells, but induction could be rescued by reintroduction of IKKγ. Thus, the LPS-initiated innate immune response in pre-B cells essentially depends on the IKK complex. However, MAPK pathways, although not sufficient to stimulate gene expression in the absence of IKK/NF-κB signaling, may have accessory functions. For instance, in dendritic cells, p38 activity is required for LPS-induced recruitment of NF-κB/p65 to a subset of target promoters (IL-6, IL-12p40, and MCP-1) (41). Since LPS activates none of these p38-dependent genes in 70Z/3 cells, we speculate that the relatively weak LPS-triggered p38 activity (data not shown) is below the threshold to promote histone H3 phosphorylation and subsequent NF-κB recruitment to these promoters (41).
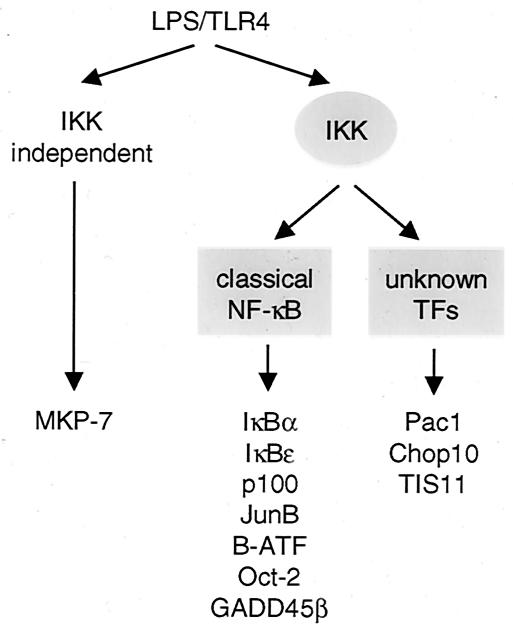
Obviously, the classical IKKγ/NF-κB signaling cassette induces expression of the vast majority of LPS response genes (Fig. 8). In contrast, the induction of very few genes is partially (MIP-1α or MAIL) or completely (MKP-7) independent of IKK (Fig. 1), indicating that other LPS-regulated factors contribute to various extents in target gene induction. LPS induction of a third group of genes, namely, PAC1, CHOP10, TIS11, NAB2, and TIS7, was not significantly inhibited by IκBαΔN but was completely lost in 1.3E2 cells, suggesting the interesting possibility that the IKK complex is involved in the regulation of other transcription factors. Here, it cannot be completely excluded that a weak, residual NF-κB activity in IκBαΔN-expressing cells may be sufficient for the activation of this set of genes. In line with the complete lack of IκBαΔN inhibition of CHOP10 and PAC1, the regulatory regions of both genes reveal no apparent NF-κB sites (16, 37; data not shown). It remains an open question which transcription factors other than NF-κB are activated by IKKγ-containing complexes and whether the catalytic subunits of the IKK complex are also required for the LPS induction of these genes. In contrast, the lack of IκBαΔN-mediated repression of the MIP1α gene, which contains NF-κB sites in its promoter region to which p65 is recruited upon LPS stimulation (40, 41), is most likely explained by an alternative IKK/NF-κB-independent pathway that also operates in 1.3E2 cells (Fig. 1).
FIG. 8.
Schematic representation of primary LPS response genes in 70Z/3 pre-B cells. Primary transcriptional response genes can be grouped according to the requirement of IKK and/or NF-κB for gene induction. Whereas the largest group of genes requires classical IKK and NF-κB signaling (center), induction of a single gene is completely independent of IKK and NF-κB activation (left). Induction of a few genes depends on IKK activation but unknown transcription factors, indicating that the IKK complex may regulate an alternative NF-κB independent pathway(s). Depicted are some representative genes.
Primary LPS-IKK/NF-κB target genes orchestrate a secondary response program: complete control of AP-1 activity by NF-κB.
Activation of NF-κB induces the expression of a set of transcriptional repressors. TIS7 is a corepressor that engages Sin3B, histone deacetylase, and NCoR to repress transcription (47). Nab2 represses Egr-1 and Krox20 (44). The polycomb protein Mpc2/CBX4 is a repressor for a number of genes (1). IRF-8/ICSBP has repressor and activator functions (26). IRF-8 functions together with IRF-4 as a genetic switch to down-regulate surrogate light chain expression and to induce conventional light chain transcription and coordinate the transition from pre-B cell to B cell (30). Stra13 and CHOP10 are repressors for E-box-binding basic helix-loop-helix members and for C/EBPs, respectively (3, 42). Thus, it appears that the LPS pathway reprograms the cellular gene expression repertoire and switches off genes whose induction is inappropriate under infection and stress conditions.
Perhaps the most provocative result is that we could show that LPS induction of AP-1 in pre-B cells and primary dendritic cells is entirely mediated by the IKK/NF-κB pathway. LPS-induced AP-1 activation was delayed compared to that of NF-κB and contained JunD and JunB as main components, and induction required protein de novo synthesis. Whereas the JunB gene contains NF-κB sites (13) and was identified as a primary IKK/NF-κB target gene, immediate-early induction of JunD mRNA was weaker and was apparently independent of NF-κB activation. Nevertheless, full up-regulation of JunD mRNA at later time points or of JunD protein amounts required IKK/NF-κB signaling (Fig. 6). The JunD promoter lacks bona fide NF-κB sites but contains a functional octamer motif (9), and the gene can be activated by CHOP10 (46), suggesting that IKK signaling may promote JunD expression indirectly via induction of CHOP10 and/or Oct-2 (Table 1). Induction of JunD was not inhibited by A-Fos, excluding an involvement of an autoregulatory process. Other mechanisms, e.g., regulation of mRNA stability, might also play a role. Besides JunB and JunD, other inducible factors, like B-ATF or MafF, or constitutively expressed AP-1 family members, e.g., ATF-1 or ATF-2, could be components of the AP-1 complex.
We demonstrated that IKK/NF-κB-induced AP-1 activity is needed to evoke, in cooperation with NF-κB, a secondary strong transcriptional response, as shown for the LPS induction of CCR7 and Igκ light chain. It can be envisioned that activation of classical NF-κB p50/p65 is sufficient for transient gene induction, as in the case of CD40 and IκBɛ, but that subordinate transcription factors like AP-1, NF-κB p52/RelB, or Oct-2 are required to maintain high expression levels of persistently activated genes, e.g., CCR7 and Igκ light chain.
TLRs activate proximal events that culminate in the activation of IKK/NF-κB as well as MAPKs ERK, p38, and JNK (2). Whereas the MAPKs JNK and ERK are only poorly activated by LPS in 70Z/3 pre-B cells, p38 was induced (24; data not shown). An intriguing aspect is the apparent disengagement of primary TLR4-mediated JNK signaling from secondary activation of functional AP-1. Moreover, LPS activates the expression of a panel of proteins which inhibit MAPK signaling. The MAPK phosphatase PAC-1 negatively interferes with MAPK activation in general (48), while Pea-15/Mat1 specifically inhibits ERK (12), and MYD118/GADD45β and MKP-7 are negative regulators of JNK (11, 32, 34). Furthermore, p21 can suppress JNK activity (20). This negative cross talk by IKK/NF-κB targets may be important primarily in limiting the duration of MAPK induction by LPS. However, the cause for the lack of any NF-κB-independent, initial AP-1 activation by LPS in pre-B and dendritic cells is not yet clear. Similar to LPS-stimulated AP-1 in pre-B cells, constitutive AP-1 activity in Hodgkin cells occurs in the absence of MAPK signaling (33). In Hodgkin's disease tumor cells, AP-1 activity contains c-Jun as a major component, which discriminates the AP-1 complex in LPS-stimulated pre-B cells and Hodgkin cells. In each case, the Jun proteins most likely heterodimerize with an unknown partner(s), and the different subunit composition will likely affect the biological outcome of the response. Future studies must determine the exact contribution of individual AP-1 factors for mounting an appropriate innate immune response in lymphoid cells.
The control of AP-1 activity by IKK/NF-κB could explain some of the observations made in previous studies. Cross-linking of surface Ig antigen receptor or CD40 ligand in primary B lymphocytes induced expression of JunB and JunD (21), and the concomitant activation of AP-1 was dependent on JunB de novo synthesis (45), suggesting an involvement of NF-κB. Furthermore, PKCθ deficiency in peripheral T cells causes a complete block of T-cell-receptor-initiated NF-κB and AP-1 activation, even though MAPK signaling was unaffected (43). Thus, we speculate that MAPK-independent AP-1 activation in T lymphocytes may involve NF-κB-mediated transcriptional up-regulation of AP-1 family members.
In conclusion, this study defines a novel type of cross talk between the transcription factors of the NF-κB and AP-1 families in the innate immune reaction elicited by LPS. Through NF-κB-dependent AP-1 activation, NF-κB can indirectly control the expression of AP-1 target genes. The IKK/NF-κB module acts as a central early switch. Apart from integrating AP-1 and NF-κB, its primary response genes are apt to significantly program the secondary transcriptional response to LPS.
Acknowledgments
We thank Erika Scharschmidt and Sabine Jungmann for expert technical assistance and Charles Vinson for providing A-Fos.
This work was supported in part by grants from Deutsche Forschungsgemeinschaft and National Genome Research Network NGFN to C.S.
REFERENCES
- 1.Alkema, M. J., J. Jacobs, J. W. Voncken, N. A. Jenkins, N. G. Copeland, D. P. Satijn, A. P. Otte, A. Berns, and M. van Lohuizen. 1997. MPc2, a new murine homolog of the Drosophila polycomb protein is a member of the mouse polycomb transcriptional repressor complex. J. Mol. Biol. 273:993-1003. [DOI] [PubMed] [Google Scholar]
- 2.Barton, G. M., and R. Medzhitov. 2003. Toll-like receptor signaling pathways. Science 300:1524-1525. [DOI] [PubMed] [Google Scholar]
- 3.Batchvarova, N., X. Z. Wang, and D. Ron. 1995. Inhibition of adipogenesis by the stress-induced protein CHOP (Gadd153). EMBO J. 14:4654-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bendall, H. H., D. C. Scherer, C. R. Edson, D. W. Ballard, and E. M. Oltz. 1997. Transcription factor NF-kappa B regulates inducible Oct-2 gene expression in precursor B lymphocytes J. Biol. Chem. 272:28826-28828. [DOI] [PubMed] [Google Scholar]
- 5.Briskin, M., M. D. Kuwabara, D. S. Sigman, and R. Wall. 1988. Induction of kappa transcription by interferon-gamma without activation of NF-kappa B. Science 242:1036-1037. [DOI] [PubMed] [Google Scholar]
- 6.Claudio, E., K. Brown, S. Park, H. Wang, and U. Siebenlist. 2002. BAFF-induced NEMO-independent processing of NF-kappa B2 in maturing B cells. Nat. Immunol. 3:958-965. [DOI] [PubMed] [Google Scholar]
- 7.Coope, H. J., P. G. Atkinson, B. Huhse, M. Belich, J. Janzen, M. J. Holman, G. G. Klaus, L. H. Johnston, and S. C. Ley. 2002. CD40 regulates the processing of NF-kappaB2 p100 to p52. EMBO J. 21:5375-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dadgostar, H., B. Zarnegar, A. Hoffmann, X. F. Qin, U. Truong, G. Rao, D. Baltimore, and G. Cheng. 2002. Cooperation of multiple signaling pathways in CD40-regulated gene expression in B lymphocytes. Proc. Natl. Acad. Sci. USA 99:1497-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Groot, R. P., M. Karperien, C. Pals, and W. Kruijer. 1991. Characterization of the mouse junD promoter—high basal level activity due to an octamer motif. EMBO J. 10:2523-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dejardin, E., N. M. Droin, M. Delhase, E. Haas, Y. Cao, C. Makris, Z. W. Li, M. Karin, C. F. Ware, and D. R. Green. 2002. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity 17:525-535. [DOI] [PubMed] [Google Scholar]
- 11.De Smaele, E., F. Zazzeroni, S. Papa, D. U. Nguyen, R. Jin, J. Jones, R. Cong, and G. Franzoso. 2001. Induction of gadd45β by NF-κB downregulates pro-apoptotic JNK signalling. Nature 414:308-313. [DOI] [PubMed] [Google Scholar]
- 12.Formstecher, E., J. W. Ramos, M. Fauquet, D. A. Calderwood, J. C. Hsieh, B. Canton, X. T. Nguyen, J. V. Barnier, J. Camonis, M. H. Ginsberg, and H. Chneiweiss. 2001. PEA-15 mediates cytoplasmic sequestration of ERK MAP kinase. Dev. Cell 1:239-250. [DOI] [PubMed] [Google Scholar]
- 13.Frazier-Jessen, M. R., C. D. Thompson, R. Brown, R. Rawat, R. P. Nordan, and G. M. Feldman. 2002. NF-κB elements contribute to junB inducibility by lipopolysaccharide in the murine macrophage cell line RAW264.7. FEBS Lett. 513:203-207. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-kappaB puzzle. Cell 109:S81-S96. [DOI] [PubMed] [Google Scholar]
- 15.Grumont, R., H. Hochrein, M. O'Keeffe, R. Gugasyan, C. White, I. Caminschi, W. Cook, and S. Gerondakis. 2001. c-Rel regulates interleukin 12 p70 expression in CD8+ dendritic cells by specifically inducing p35 gene transcription. J. Exp. Med. 194:1021-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grumont, R. J., J. E. Rasko, A. Strasser, and S. Gerondakis. 1996. Activation of the mitogen-activated protein kinase pathway induces transcription of the PAC-1 phosphatase gene. Mol. Cell. Biol. 16:2913-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guha, M., and N. Mackman. 2001. LPS induction of gene expression in human monocytes. Cell. Signal. 13:85-94. [DOI] [PubMed] [Google Scholar]
- 18.Hatada, E. N., D. Krappmann, and C. Scheidereit. 2000. NF-kappaB and the innate immune response. Curr. Opin. Immunol. 12:52-58. [DOI] [PubMed] [Google Scholar]
- 19.Hinz, M., P. Lemke, I. Anagnostopoulos, C. Hacker, D. Krappmann, S. Mathas, B. Dörken, M. Zenke, H. Stein, and C. Scheidereit. 2002. Nuclear factor κB-dependent gene expression profiling of Hodgkin's disease tumor cells, pathogenetic significance, and link to constitutive signal transducer and activator of transcription 5a activity. J. Exp. Med. 196:605-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, S., L. Shu, M. B. Dilling, J. Easton, F. C. Harwood, H. Ichijo, and P. J. Houghton. 2003. Sustained activation of the JNK cascade and rapamycin-induced apoptosis are suppressed by p53/p21(Cip1). Mol. Cell 11:1491-1501. [DOI] [PubMed] [Google Scholar]
- 21.Huo, L., and T. L. Rothstein. 1995. Receptor-specific induction of individual AP-1 components in B lymphocytes. J. Immunol. 154:3300-3309. [PubMed] [Google Scholar]
- 22.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 23.Kayagaki, N., M. Yan, D. Seshasayee, H. Wang, W. Lee, D. M. French, I. S. Grewal, A. G. Cochran, N. C. Gordon, J. Yin, M. A. Starovasnik, and V. M. Dixit. 2002. BAFF/BLyS receptor 3 binds the B cell survival factor BAFF ligand through a discrete surface loop and promotes processing of NF-κB2. Immunity 17:515-524. [DOI] [PubMed] [Google Scholar]
- 24.Krappmann, D., A. Patke, V. Heissmeyer, and C. Scheidereit. 2001. B-cell receptor- and phorbol ester-induced NF-κB and c-Jun N-terminal kinase activation in B cells requires novel protein kinase C's. Mol. Cell. Biol. 21:6640-6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kyriakis, J. M., and J. Avruch. 2001. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 81:807-869. [DOI] [PubMed] [Google Scholar]
- 26.Levi, B. Z., S. Hashmueli, M. Gleit-Kielmanowicz, A. Azriel, and D. Meraro. 2002. ICSBP/IRF-8 transactivation: a tale of protein-protein interaction. J. Interferon Cytokine Res. 22:153-160. [DOI] [PubMed] [Google Scholar]
- 27.Li, J., G. W. Peet, D. Balzarano, X. Li, P. Massa, R. W. Barton, and K. B. Marcu. 2001. Novel NEMO/IkappaB kinase and NF-kappa B target genes at the pre-B to immature B cell transition. J. Biol. Chem. 276:18579-18590. [DOI] [PubMed] [Google Scholar]
- 28.Li, Q., and I. M. Verma. 2002. NF-kappaB regulation in the immune system. Nat. Rev. Immunol. 2:725-734. [DOI] [PubMed] [Google Scholar]
- 29.Li, X., P. E. Massa, A. Hanidu, G. W. Peet, P. Aro, A. Savitt, S. Mische, J. Li, and K. B. Marcu. 2002. IKKalpha, IKKbeta, and NEMO/IKKgamma are each required for the NF-kappa B-mediated inflammatory response program. J. Biol. Chem. 277:45129-45140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu, R., K. L. Medina, D. W. Lancki, and H. Singh. 2003. IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Genes Dev. 17:1703-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malcolm, K. C., P. G. Arndt, E. J. Manos, D. A. Jones, and G. S. Worthen. 2003. Microarray analysis of lipopolysaccharide-treated human neutrophils. Am. J. Physiol. Lung Cell Mol. Physiol. 284:L663-L670. [DOI] [PubMed] [Google Scholar]
- 32.Masuda, K., H. Shima, M. Watanabe, and K. Kikuchi. 2001. MKP-7, a novel mitogen-activated protein kinase phosphatase, functions as a shuttle protein. J. Biol. Chem. 276:39002-39011. [DOI] [PubMed] [Google Scholar]
- 33.Mathas, S., M. Hinz, I. Anagnostopoulos, D. Krappmann, A. Lietz, F. Jundt, K. Bommert, F. Mechta-Grigoriou, H. Stein, B. Dorken, and C. Scheidereit. 2002. Aberrantly expressed c-Jun and JunB are a hallmark of Hodgkin lymphoma cells, stimulate proliferation and synergize with NF-kappa B. EMBO J. 21:4104-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuguchi, T., T. Musikacharoen, T. R. Johnson, A. S. Kraft, and Y. Yoshikai. 2001. A novel mitogen-activated protein kinase phosphatase is an important negative regulator of lipopolysaccharide-mediated c-Jun N-terminal kinase activation in mouse macrophage cell lines. Mol. Cell. Biol. 21:6999-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Messmer, D., B. Messmer, and N. Chiorazzi. 2003. The global transcriptional maturation program and stimuli-specific gene expression profiles of human myeloid dendritic cells. Int. Immunol. 15:491-503. [DOI] [PubMed] [Google Scholar]
- 36.Mordmuller, B., D. Krappmann, M. Esen, E. Wegener, and C. Scheidereit. 2003. Lymphotoxin and lipopolysaccharide induce NF-kappaB-p52 generation by a co-translational mechanism. EMBO Rep. 4:82-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nozaki, S., G. W. Sledge, Jr., and H. Nakshatri. 2001. Repression of GADD153/CHOP by NF-kappaB: a possible cellular defense against endoplasmic reticulum stress-induced cell death. Oncogene 20:2178-2185. [DOI] [PubMed] [Google Scholar]
- 38.O'Brien, D. P., E. M. Oltz, and B. G. Van Ness. 1997. Coordinate transcription and V(D)J recombination of the kappa immunoglobulin light-chain locus: NF-κB-dependent and -independent pathways of activation. Mol. Cell. Biol. 17:3477-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olive, M., D. Krylov, D. R. Echlin, K. Gardner, E. Taparowsky, and C. Vinson. 1997. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. J. Biol. Chem. 272:18586-18594. [DOI] [PubMed] [Google Scholar]
- 40.Pahl, H. L. 1999. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene 18:6853-6866. [DOI] [PubMed] [Google Scholar]
- 41.Saccani, S., S. Pantano, and G. Natoli. 2002. p38-dependent marking of inflammatory genes for increased NF-kappa B recruitment. Nat. Immunol. 3:69-75. [DOI] [PubMed] [Google Scholar]
- 42.St.-Pierre, B., G. Flock, E. Zacksenhaus, and S. E. Egan. 2002. Stra13 homodimers repress transcription through class B E-box elements. J. Biol. Chem. 277:46544-46551. [DOI] [PubMed] [Google Scholar]
- 43.Sun, Z., C. W. Arendt, W. Ellmeier, E. M. Schaeffer, M. J. Sunshine, L. Gandhi, J. Annes, D. Petrzilka, A. Kupfer, P. L. Schwartzberg, and D. R. Littman. 2000. PKC-theta is required for TCR-induced NF-kappaB activation in mature but not immature T lymphocytes. Nature 404:402-407. [DOI] [PubMed] [Google Scholar]
- 44.Svaren, J., B. R. Sevetson, E. D. Apel, D. B. Zimonjic, N. C. Popescu, and J. Milbrandt. 1996. NAB2, a corepressor of NGFI-A (Egr-1) and Krox20, is induced by proliferative and differentiative stimuli. Mol. Cell. Biol. 16:3545-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanguay, D. A., J. A. Dickinson, B. J. McManus, L. Huo, T. L. Rothstein, and T. C. Chiles. 1994. Activation of AP-1 in primary B lymphocytes by surface immunoglobulin requires de novo Jun-B synthesis. Cell Immunol. 158:281-291. [DOI] [PubMed] [Google Scholar]
- 46.Ubeda, M., M. Vallejo, and J. F. Habener. 1999. CHOP enhancement of gene transcription by interactions with Jun/Fos AP-1 complex proteins. Mol. Cell. Biol. 19:7589-7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vietor, I., S. K. Vadivelu, N. Wick, R. Hoffman, M. Cotten, C. Seiser, I. Fialka, W. Wunderlich, A. Haase, G. Korinkova, G. Brosch, and L. A. Huber. 2002. TIS7 interacts with the mammalian SIN3 histone deacetylase complex in epithelial cells. EMBO J. 21:4621-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ward, Y., S. Gupta, P. Jensen, M. Wartmann, R. J. Davis, and K. Kelly. 1994. Control of MAP kinase activation by the mitogen-induced threonine/tyrosine phosphatase PAC1. Nature 367:651-654. [DOI] [PubMed] [Google Scholar]
- 49.Weeks, R. S., and C. H. Sibley. 1987. Molecular analysis of immunoglobulin expression in variants of murine B lymphoma, 70Z/3 Somat. Cell Mol. Genet. 13:205-219. [DOI] [PubMed] [Google Scholar]
- 50.Yilmaz, Z. B., D. S. Weih, V. Sivakumar, and F. Weih. 2003. RelB is required for Peyer's patch development: differential regulation of p52-RelB by lymphotoxin and TNF. EMBO J. 22:121-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao, B., R. A. Bowden, S. A. Stavchansky, and P. D. Bowman. 2001. Human endothelial cell response to gram-negative lipopolysaccharide assessed with cDNA microarrays. Am. J. Physiol. Cell Physiol. 281:C1587-C1595. [DOI] [PubMed] [Google Scholar]
- 52.Zhao, B., S. A. Stavchansky, R. A. Bowden, and P. D. Bowman. 2003. Effect of interleukin-1beta and tumor necrosis factor-alpha on gene expression in human endothelial cells. Am. J. Physiol. Cell Physiol. 284:C1577-C1583. [DOI] [PubMed] [Google Scholar]
- 53.Zhou, A., S. Scoggin, R. B. Gaynor, and N. S. Williams. 2003. Identification of NF-kappa B-regulated genes induced by TNFalpha utilizing expression profiling and RNA interference. Oncogene 22:2054-2064. [DOI] [PubMed] [Google Scholar]
- 54.Zhu, N., L. M. Ramirez, R. L. Lee, N. S. Magnuson, G. A. Bishop, and M. R. Gold. 2002. CD40 signaling in B cells regulates the expression of the Pim-1 kinase via the NF-kappaB pathway. J. Immunol. 168:744-754. [DOI] [PubMed] [Google Scholar]