Background: Ubiquitination of histone H2B regulates gene expression.
Results: Bre1 RING domain recognizes nucleosome and directs Rad6 to specifically monoubiquitinate H2B Lys-123. Separate Bre1 domain augments ubiquitin transfer to nucleosome by Rad6 backside binding.
Conclusion: Bre1 RING and non-RING domains cooperate in coupling substrate targeting and E2 activation.
Significance: We provide detailed insights into the catalytic mechanism of histone H2B ubiquitination.
Keywords: Chromatin, E3 Ubiquitin Ligase, Histone, Ubiquitin, Ubiquitin-conjugating Enzyme (E2 Enzyme), Bre1, Rad6
Abstract
Ubiquitin signaling on chromatin is linked to diverse aspects of genome regulation, including gene expression and DNA repair. The yeast RING E3 ligase Bre1 combines with the E2 Rad6 to monoubiquitinate histone H2B during transcription. Little is known about how Bre1 directs Rad6 toward transferring only a single ubiquitin to a specific lysine residue. Using a defined in vitro system, we show that the Bre1 RING domain interaction with Rad6 is minimally sufficient to monoubiquitinate nucleosomes at histone H2B Lys-123. In addition, we reveal a cluster of charged residues on the Bre1 RING domain that is critical for recognizing the nucleosome surface. Notably, a second Rad6 binding domain of Bre1 interacts with the E2 backside and potentiates ubiquitin transfer to the substrate. Taken together, our study establishes a molecular framework for how distinct RING and non-RING E3 elements cooperate to regulate E2 reactivity and substrate selection during gene expression.
Introduction
The yeast E3 ligase Bre1 (human RNF20/40) associates with the RNA polymerase II machinery and plays a key role in coupling nucleosome modification with transcription elongation in eukaryotic cells (1–7). Bre1 specifically monoubiquitinates histone H2B at Lys-123 in yeast (equivalent to mammalian H2B Lys-120), a modification linked to gene regulation and cross-talk with histone methylation (8). The nucleosome core particle (NCP)3 is the basic unit of the eukaryotic genome formed by 147 bp of DNA wrapped around an octameric assembly of histone proteins (two copies each of H2A, H2B, H3, and H4) (9). Ubiquitin signaling on nucleosomes involves several more histone ubiquitin ligases and can collectively impart structural changes to the chromatin fiber, serve as a docking site for recruiting other factors during transcription and DNA repair, or mediate histone degradation (10). Other prominent examples include the monoubiquitination of metazoan H2A at Lys-119 by E3 ligases of the Polycomb repressive complex (PRC1) (11–13) or the ubiquitination at H2A Lys-13/Lys-15 by the E3 RNF168 after DNA damage (14). A hallmark of the above reactions is the highly selective choice of a substrate lysine residue, which is not seen in many other ubiquitination events (e.g. polyubiquitinated substrates for proteasomal degradation) (15). How lysine-targeting specificity on nucleosomes is achieved is still poorly understood, although recent studies suggest that recognition of multiple nucleosome surfaces and a direct role of the substrate may contribute (16, 17).
The ubiquitination reaction is catalyzed by an enzymatic cascade involving ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin-ligating (E3) enzymes (18). Most E3 ligases use a RING domain to activate an E2∼ubiquitin thioester and mediate ubiquitin transfer to a substrate (“∼” denotes a covalent and “-” denotes a noncovalent interaction). Ubiquitination occurs when the E3 binds to both the substrate and the E2 and brings them in proximity. Current models suggest that the E3 RING domain, the E2, and ubiquitin interact with each other to stabilize a stereochemically constrained (“closed”) E2∼Ub conformation that immobilizes and primes the thioester bond for nucleophilic attack by the substrate lysine residue (19–21). Besides the canonical RING domain-E2 interaction, several E3s can also bind E2s via regions separate from their RING domain. These noncanonical interactions can involve the E2 “backside,” a region opposite to the E2 active site (22).
Notably, E2 backside binding was suggested to play a role in restricting an E2 toward the transfer of a single ubiquitin molecule and to prevent the generation of polyubiquitin chains (23). The RING E3s Bre1, Rad18, and Ubr1 can all interact with the E2 Rad6 via regions separate from their RING domain. Although Ubr1 directs Rad6 to polyubiquitinate N-end rule substrates (24), both Bre1 and Rad18 adjust Rad6 to specifically monoubiquitinate the target. Rad18 monoubiquitinates proliferating cell nuclear antigen (PCNA) and interacts with Rad6 both canonically through its RING domain and via a distinct α-helix that binds the Rad6 backside (25–27). Interestingly, the Rad18-Rad6 backside interaction overlaps with a low affinity, noncovalent ubiquitin-binding site on Rad6, which was implicated in Rad6's intrinsic ability to form free ubiquitin chains in the absence of an E3. This lead to the hypothesis that Rad18 binding to the Rad6 backside displaces ubiquitin from this site and thereby directs the E2 toward mono- rather than polyubiqitination of PCNA (23). However, it is unclear whether these findings can be extrapolated to infer a similar mechanism for histone monoubiquitination by Bre1. A previous study showed that yeast Rad6 alone has an in vitro ability to nonspecifically ubiquitinate all core histones independently of an E3 (28). Bre1 is required to direct Rad6 toward the physiological H2B ubiquitination site in the nucleosomal context. This study further suggested that the C-terminal Bre1 RING domain is essential for H2B ubiquitination but dispensable for the interaction of Bre1 with Rad6, which appeared to proceed entirely through a separate N-terminal domain of Bre1.
To fully appreciate the role of Bre1 and Rad6 in gene regulation, it is important to understand their enzymatic mechanism and specificity toward a structurally complex substrate. Additionally, these enzymes may offer more general insights into how E3 ligases interact with their cognate E2s to target a specific acceptor lysine for mono- rather than polyubiquitination. Here, we show that Bre1 interacts with Rad6 both through the E2 backside and canonically via the RING. Backside binding, although dispensable for histone monoubiquitination at Lys-123, strongly enhances Rad6 activation and thereby accelerates the overall reaction. We define a direct role for the RING in nucleosome recognition and provide a detailed model of how RING and non-RING domains of Bre1 cooperate with Rad6 to achieve site-specific H2B ubiquitination.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Coding regions for Rad6, Bre1, ubiquitin, and histones were amplified by PCR from yeast genomic DNA and cloned into the respective expression vectors. Mutations and deletions were generated by PCR-based methods and confirmed by sequencing. All recombinant proteins used in this study were expressed in Escherichia coli BL21 CodonPlus (DE3) RIL cells. Expression was induced by addition of 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at 23 °C for 3 h. Strep-Bre1 constructs were affinity-purified on a 5-ml Strep-Tactin® Superflow column in buffer containing 100 mm NaCl, 50 mm CHES, pH 9.5. Proteins were eluted with 2.5 mm desthiobiotin. GST and GST-Rad6 were purified on glutathione-Sepharose 4B beads (GE Healthcare) in the following buffer: 100 mm NaCl, 50 mm Tris-HCl, pH 7.5, 1.5 mm MgCl2, 0.5 mm DTT, 0.15% Nonidet P-40. For GST pulldown assays, GST-Rad6 variants were co-expressed with Strep-Bre1 constructs. His6-TEV-Rad6 affinity purification was performed using a HisTrap HP 5-ml column (GE Healthcare) in a buffer containing 300 mm NaCl, 50 mm Tris-HCl, pH 7.5, 1.5 mm MgCl2, 50 mm imidazole, and eluted by 100 mm imidazole. Rad6 was further purified by size exclusion chromatography (SEC) on a Superdex 200 16/60 column (GE Healthcare) equilibrated with 50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 50 mm KCl, 10 mm MgCl2. His6-TEV-ubiquitin was obtained by Ni2+ affinity purification on nickel-Sepharose 6 Fast Flow beads (GE Healthcare) and imidazole elution followed by TEV-protease cleavage of the tag and SEC on a Superose 6 10/300 column (GE Healthcare).
Analytical Size Exclusion Chromatography
Rad6, Strep-RBD, and Strep-RBD K31D were run on Superdex 200 Increase 3.2/200 column (GE Healthcare) equilibrated with 50 mm Tris-HCl, pH 7.5, 50 mm NaCl. Protein mixtures of Rad6/Strep-RBD or Rad6/Strep-RBD K31D were preincubated in a 1:6 molar ratio for 5 min prior to chromatography. All fractions were analyzed by SDS-PAGE (14% gel) and Coomassie staining.
Histone Octamer Purification and NCP Reconstitution
A single step purification strategy for the histone octamer was developed. Yeast histones were cloned into the pST44 polycistronic expression vector (29) with an N-terminal His6-TEV-FLAG tag on histone H2B for co-expression in E. coli BL21 CodonPLus (DE3) RIL cells. Histone octamers were obtained by affinity purification on a HisTrap HP 5-ml column (GE Healthcare) equilibrated with 50 mm Tris-HCl, pH 7.5, 1 m NaCl, 1.5 mm MgCl2, 50 mm imidazole. Elution was performed with 500 mm imidazole. NCPs were reconstituted from histone octamers and the 167-bp 601 WIDOM positioning sequence using a KCl gradient (2–0.25 m) in the following buffer: 10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 1 mm DTT (30, 31). This was followed by Mono Q ion exchange chromatography (elution gradient: 0.25–1 m KCl in 20 mm Tris-HCl, pH 7.5) and SEC on a Superdex 200 HiLoad 16/60 column (GE Healthcare) equilibrated with 20 mm Tris-HCl, pH 7.5.
NCP Ubiquitination Assay
For NCP ubiquitination reactions, 100 nm E1, 3 μm Rad6, 36 μm ubiquitin, 18 μm Bre1 (unless otherwise indicated in figure legend), 3 mm ATP, and 0.1 mm DTT were mixed in a reaction volume of 20 μl (50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 50 mm KCl, 10 mm MgCl2) and incubated for 90 min at 30 °C. Reactions were stopped by addition of 4× SDS loading buffer and analyzed by SDS-PAGE (NuPAGE 4–12% gels, Invitrogen, MOPS buffer) followed by immunoblotting with anti-FLAG M2 (Sigma, F1804) or anti-ubiquitin P4D1 antibody (Santa Cruz Biotechnology). The concentration of Rad6-Bre1 chimeras used in the NCP ubiquitination assay was 3 μm.
In Vitro Binding Assays
Rad6-Bre1 eRING
Purified GST-Rad6 or GST was mixed with purified Strep-tagged Bre1 eRING in 1:2 and 1:5 molar ratios in B buffer (50 mm Tris HCl, pH 7.5, 100 mm NaCl, 20 μg of BSA), followed by incubation with glutathione-Sepharose 4B beads (GE Healthcare) for 90 min at 4 °C. Beads were washed with B buffer (without BSA, 0.5 mm DTT), and bound proteins were eluted with glutathione (GSH). After trichloroacetic acid (TCA) precipitation, the samples were analyzed by SDS-PAGE (NuPAGE 4–12% gel, MES buffer) and Coomassie staining or by Western blotting with an anti-Strep antibody (Qiagen, 34850).
NCP-Bre1
Reconstituted NCP was mixed with Bre1 constructs at a 1:5 molar ratio in binding buffer (50 mm Tris, pH 7.5, 100 mm NaCl, 1.5 mm MgCl2) and incubated with 30 μl of anti-FLAG M2 beads (Sigma). After washing, proteins were eluted by incubation with FLAG peptide (0.1 mg/ml in binding buffer) for 45 min at 4 °C, TCA-precipitated, and analyzed by SDS-PAGE (NuPAGE 4–12% gel, MOPS buffer) and immunoblotting.
E2 Discharging Assay
Rad6 pre-charging was performed with 250 nm E1 (Uba1; Boston Biochem), 20 μm ubiquitin, 5 μm Rad6, 0.2 mm DTT, and 3 mm ATP in R buffer (50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 50 mm KCl, 10 mm MgCl2), for 1 h at 30 °C. Charging reactions were stopped by adding EDTA, pH 8.0, at 50 mm final concentration and placed on ice for 2 min. Charged E2 was added to samples containing different concentrations of Bre1 constructs in a final reaction volume of 80 μl. Samples were incubated at 30 °C, and aliquots were collected after 0, 5, 10, and 15 min and analyzed by nonreducing SDS-PAGE (12% NuPAGE gels, MES buffer). Protein bands were visualized by Coomassie staining, and quantification of band intensity was performed by ImageJ software (rsb.info.nih.gov). The discharging assay for the Bre1 eRING was performed in 50 mm CHES, pH 8.0, 100 mm NaCl.
Measurement of Global H2B Ubiquitin Levels
The FLAG-H2B yeast strain used for the assay was described previously (32). BRE1 was deleted by a standard one-step PCR-based technique using clonNAT as a selectable marker. Immunoprecipitation of FLAG-tagged histone H2B was performed as described previously (33).
RESULTS
Non-RING Bre1 Motif Mediates a Stable Interaction with Rad6
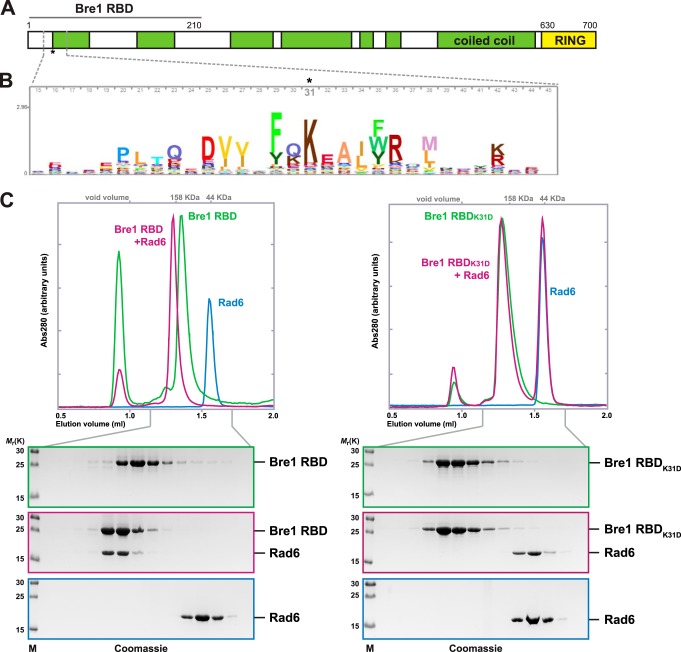
The yeast E3 ligase Bre1 is predicted to have a secondary structure comprising a series of interrupted coiled-coil segments and a C-terminal RING finger domain (Fig. 1A). RING domains typically coordinate two zinc ions and serve as a rigid, globular platform for E2 binding and activation (18). A previous study determined that Bre1 binds to Rad6 via an N-terminal domain (amino acids 1–210), which is both necessary and sufficient for the Rad6 interaction when assessed by pulldown experiments and immunoblotting. In contrast, the Bre1 RING domain was found to be dispensable for the interaction with Rad6 but essential for nucleosomal H2B ubiquitination (28). This suggested a potentially noncanonical mode of E2 activation. The Rad6-interacting site, which we term RBD (Rad6 binding domain), includes two predicted coiled-coil segments (Fig. 1A). Compared with the highly conserved RING domain, putative functional motifs were not readily apparent in the Bre1 RBD. However, using a Hidden Markov model-based algorithm applied to metazoan, plant, and fungal sequences, we identified a cluster of conserved residues centered around Bre1 Lys-31 at the beginning of the first coiled coil (Fig. 1B). To test the importance of this motif for Rad6 binding, we reconstituted the interaction between recombinant Rad6 and the Bre1 RBD (amino acids 1–210) proteins in vitro. When monitored by analytical SEC, Rad6 and the Bre1 RBD alone migrated as well defined protein species (Fig. 1C, left panel). However, upon co-incubation Rad6 was shifted completely into a higher molecular weight fraction together with the Bre1 RBD indicating the formation of a stable complex. Rad6 was present in smaller amounts in this complex possibly suggesting that one Rad6 molecule binds to a Bre1 dimer that may be stabilized by its coiled-coil portion. We also observed that the Bre1 RBD-Rad6 complex migrated with a higher molecular weight than expected (>158 kDa), possibly indicating the formation of higher order Bre1-Rad6 oligomers in vitro. Next, we tested the effects of inverting the charge of Bre1 Lys-31, the most conserved RBD residue. Strikingly, the K31D single-site mutation fully abolished the interaction of Bre1 with Rad6 as both proteins no longer co-migrated in the SEC assay (Fig. 1C, right panel). These results reveal a critical role of Lys-31 in mediating a direct interaction between Rad6 and the Bre1 RBD.
FIGURE 1.
Non-RING Bre1 motif mediates a stable interaction with Rad6. A, cartoon of the Bre1 domain organization. RBD denotes the Rad6 binding domain as determined previously (28). Boundaries are drawn to scale. The coiled-coil prediction was performed with COILS using default settings. B, HMM profile was generated using jackhammer from HMMER 3.1 with default parameters as follows: database UniProtKB; iteration 2; 13,186 total hits in which the most representative kingdoms among eukaryotes sum up to 57% metazoan, 19.1% fungi, and 14% viridiplantae. The logo includes Bre1 residues 15–45 with the Bre1 Lys-31 position highlighted and was generated using Skylign with default settings. C, analytical SEC reveals complex formation of the Bre1 RBD with Rad6 (left panel) and loss of the interaction with Rad6 in the RBD K31D mutant (right panel). Fractions were analyzed by SDS-PAGE and Coomassie staining. SEC elution profiles are shown on the top.
Bre1 RBD Binds to the Rad6 Backside
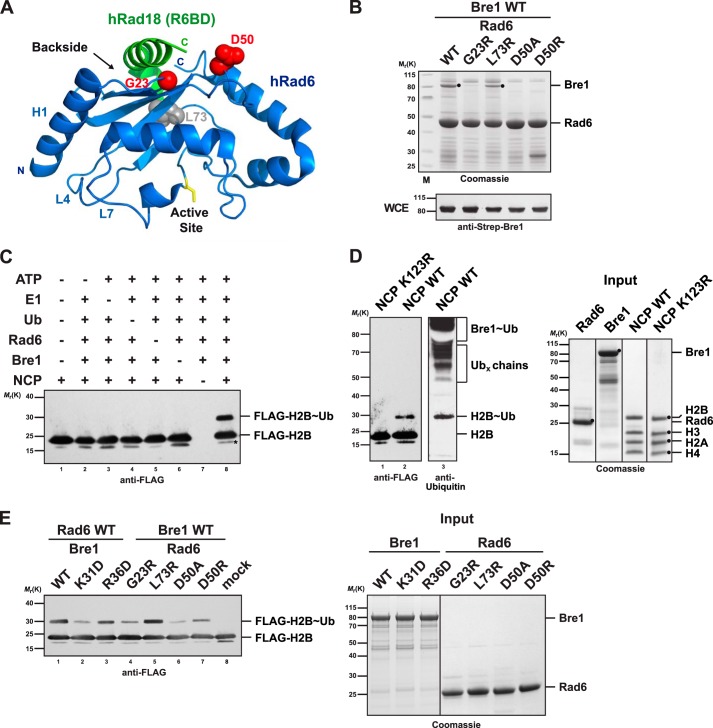
E2 enzymes have an evolutionarily conserved ∼150-residue catalytic Ubc domain, which includes four α-helices, a short helix near the catalytic site, and a four-stranded antiparallel β-sheet that forms a backside surface (Fig. 2A) (22). The canonical RING-E2 interaction site, generalized from different E2 structures bound to various E3s, involves residues on the N-terminal helix (or helix 1, H1) and loops 4 and 7 (L4 and L7) and is distinct from the Ubc domain backside. Because Rad6 binds Bre1 in a noncanonical way, we hypothesized that the Rad6 backside could be involved. To test this idea, we mutated specific Rad6 backside residues and tested whether binding to the RBD was impaired. The analysis was guided by a crystal structure of human Rad6 in complex with a helical (non-RING) part of Rad18 that is bound to the E2 backside (Fig. 2A) (23). We generated Rad6 D50A and D50R mutations as this residue directly interacts with the Rad18 C terminus in the structure. Rad6 Gly-23 was mutated because a G23R/T52A double mutant is specifically impaired in ubiquitin but not Rad18 backside binding (23). Rad6 Leu-73 is a conserved hydrophobic backside residue that is also surface-exposed but has not been implicated in binding. We performed GST pulldown assays after co-expressing GST-Rad6 and full-length Strep-tagged Bre1 in E. coli cells. Interestingly, the Rad6 G23A, D50A, and D50R mutants failed to interact with Bre1, whereas the L73A mutation had no major effect (Fig. 2B). None of the mutations affected Rad6 stability. We conclude that Bre1 binds via its RBD motif to the backside of Rad6. The interaction mode is similar to Rad18 in that the Rad6 Asp-50 residue is involved, yet it is different because Gly-23 also plays a role.
FIGURE 2.
Bre1 RBD binds to the Rad6 backside. A, ribbon structure of the human Rad6-Rad18 complex (Protein Data Bank 2YBF). Human and S. cerevisiae Rad6 are 85% identical and strictly conserved in the residues shown as spheres. Residues highlighted in red are involved in the Rad18 interaction (Asp-50) or are relevant for ubiquitin binding (Gly-23). The Rad6 backside and active site (yellow stick representation) are highlighted. The N-terminal helix1 (H1) and loops L4 and L7 comprise the canonical E1/E3-binding site. B, point mutations at the backside of Rad6 interfere with Bre1 binding. Strep-Bre1 and GST-Rad6 wild type (WT) or the indicated point mutants were co-expressed in E. coli and subjected to GST affinity purification. Proteins were eluted with GSH and analyzed by SDS-PAGE (4–12% gel, MES buffer) and Coomassie staining. Bre1 expression levels in E. coli whole cell extracts (WCE) were confirmed by immunoblotting with an anti-Strep antibody. C, requirements for the in vitro NCP ubiquitination reaction. The indicated combination of E1 (100 nm), Rad6 (3 μm), Bre1 (18 μm), ubiquitin (36 μm), and NCP (2 μm) was used. Reaction mixtures were analyzed by SDS-PAGE and immunoblotting against the FLAG-tagged histone H2B. Asterisk indicates a histone H2B degradation band. D, ubiquitination assay using the H2B K123R mutant NCP demonstrates site specificity of the reaction. Lanes 1 and 2 were probed with an anti-FLAG antibody and lane 3 with anti-ubiquitin antibody. Putative free Ub chains formed by Rad6 and bands corresponding to ubiquitinated Bre1 are indicated in the upper part of the gel in agreement with earlier reports (23, 28) Recombinant proteins used in the assays in C and D are shown in the Coomassie-stained gel on the right. E, Bre1 and Rad6 point mutants relevant for the E2 backside interaction show impaired NCP ubiquitination activity in vitro. NCP ubiquitination assay was performed with the indicated mutants. Input of the recombinant proteins is shown on the right.
To test the functional importance of the Rad6 backside-Bre1 interaction for H2B ubiquitination, we sought to fully reconstitute the reaction from defined components. Previous work had suggested that Bre1 can monoubiquitinate histone H2B in vitro when tested with natural oligonucleosomes derived from HeLa cells. In contrast, recombinant NCPs from Xenopus laevis lacking all post-translational modifications were poor substrates suggesting that some other histone modifications not present on recombinant NCPs may stimulate the ubiquitination reaction (28). We developed a rapid method for reconstituting recombinant yeast NCPs, which involves polycistronic expression of the His-FLAG-H2B, H2A, H3, and H4 proteins, nickel-affinity chromatography, and assembly with the Widom 601 DNA sequence by salt dialysis. We show that yeast Bre1 can efficiently monoubiquitinate recombinant yeast NCPs in an E1-, ATP-, and Rad6-dependent reaction (Fig. 2C). Using NCPs with a K123R mutation, we demonstrate that the reaction is highly site-specific on H2B (Fig. 2D). Immunoblotting with a ubiquitin antibody further revealed that monoubiquitinated H2B is the predominant ubiquitinated NCP species. This differs from the in vitro activity of free Rad6, which can mono- and polyubiquitinate free core histones when provided separately or as a histone octamer in the absence of Bre1 (28, 34). Rad6 also generates free ubiquitin chains and ubiquitinates Bre1 (23). We find that this in vitro side reaction rapidly consumes the available ubiquitin and possibly limits the extent of H2B ubiquitination in the assay (Fig. 2D).
Using Bre1-mediated ubiquitination of recombinant yeast NCPs, we could test the effects of disrupting the Rad6 backside-Bre1 RBD interface. Notably, the Bre1 K31D mutation, which significantly lowered the affinity to Rad6, was defective in histone H2B monoubiquitination. Mutating Bre1 Arg-36, another charged residue in the RBD motif, had a lesser effect (Figs. 1B and 2E). When analyzing the Rad6 backside mutants, we found that the G23R and D50A had the strongest impairment in histone H2B monoubiquitination activity. The Rad6 L73R mutant behaved like wild type consistent with its unperturbed affinity to the RBD. The Bre1 K31D mutant, which completely disrupted the interaction with Rad6 in the stringent SEC assay, still exhibited residual histone ubiquitination activity. This suggests that the Rad6 backside-Bre1 RBD interface is functionally important but not strictly required for H2B monoubiquitination in vitro.
Monoubiquitination of Histone H2B Lys-123 Relies on the Bre1 RING Domain-Rad6 Interaction with the Nucleosome
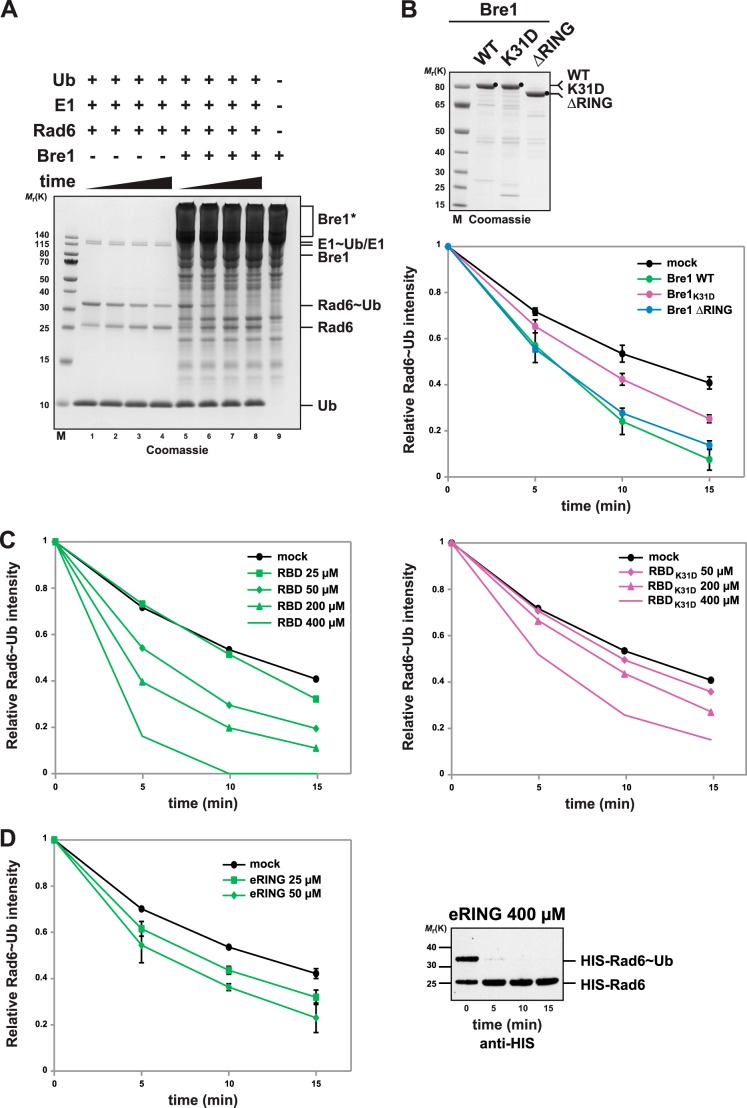
To define the minimal parts of Bre1 required for NCP monoubiquitination at Lys-123, we engineered a series of Bre1 constructs carrying internal deletions in the coiled-coil region (Fig. 3A), in which the remaining N- and C-terminal parts of Bre1 are connected by a flexible Gly-Ser-Gly-Ser linker. Purification of the N-terminally Strep-tagged Bre1 constructs from E. coli identified two Bre1 versions (Bre1Δ211–493 and Bre1Δ211–307), which were stably expressed, whereas the other three constructs, including one with a shortened RBD (Bre1Δ91–493), were susceptible to degradation (Fig. 3B). We proceeded with the two stable constructs and tested their ability to monoubiquitinate H2B. Notably, both constructs carried out the reaction as efficiently as wild-type Bre1 (Fig. 3C). The smaller construct (Bre1Δ211–493) comprises only the RBD and the RING domain with its adjacent coiled coil, which was included to promote RING domain stability. This led us to analyze the effects of removing the RBD entirely. Notably, this RING domain construct, termed eRING (extended RING; Bre1 amino acids 494–700), was sufficient to monoubiquitinate H2B in the NCP thus representing the minimal E3 core (Fig. 3D). However, the Bre1 eRING was less efficient than the construct with an appended RBD (Bre1Δ211–493). Titration of the eRING into the reaction could restore the H2B ubiquitinating activity to the level of the Bre1Δ211–493 construct when a 5-fold higher concentration of eRING was used (Fig. 3D, compare lanes 2 and 5). Moreover, the Bre1 eRING retains the lysine-targeting specificity of the wild-type protein as shown in a reaction with the K123R mutant NPCs (Fig. 3E). This suggests that the Bre1 RBD, which mediates the high affinity interaction with Rad6, is not essential for NCP ubiquitination at Lys-123 in vitro. Instead, the RBD imparts an ∼5-fold stimulation to the reaction when physically linked to the eRING domain. The ability to specifically monoubiquitinate H2B Lys-123 is provided by the eRING in conjunction with Rad6 and implies the existence of a canonical E2-RING interaction besides E2-RBD binding. To test this possibility, we reconstituted the interaction by immobilizing GST-Rad6 on beads and adding different concentrations of recombinant Bre1 eRING. This allowed us to demonstrate a specific Coomassie-stainable interaction, which was confirmed by immunoblotting (Fig. 3F). This finding contrasts with an earlier report that concluded that the Bre1 RING is dispensable for interaction with Rad6 (28). The different outcome is likely explained by the transient nature of RING-E2 interactions, which often escape detection in standard pulldown assays.
FIGURE 3.
Monoubiquitination of H2B Lys-123 is intrinsic to the Bre1 RING-Rad6 interaction with the nucleosome. A, schematic representation of Bre1 deletion constructs. Boundaries are drawn to scale. GSGS denotes a Gly-Ser-Gly-Ser linker. B, strep-affinity purification of recombinant Bre1 constructs. The internal deletions differentially affect protein stability. Black dots indicate Bre1 proteins, which were assigned according to their calculated molecular weight. Proteins were analyzed by SDS-PAGE (4–12% gel, MOPS buffer) and Coomassie staining. C, NCP ubiquitination assay with the indicated Bre1 deletion constructs that were used at equimolar concentrations (18 μm). D, Bre1 eRING is intrinsically capable of monoubiquitinating H2B together with Rad6. In vitro NCP ubiquitination assay was performed by adding incremental amounts of Bre1 eRING (18, 36, 90, and 180 μm final concentration). The Bre1 protein used for the assay is shown on the right. E, lysine targeting specificity of the eRING construct was assessed with the K123R mutant NCP. Bre1 eRING is used at 90 μm final concentration. Reactions were probed by anti-FLAG immunoblotting. F, Rad6 directly binds to the Bre1 eRING in vitro. Recombinant GST-Rad6 was immobilized on GSH beads and incubated with Strep-Bre1 eRING in 1:2 and 1:5 molar ratios. Recombinant GST was used as negative control. Proteins were eluted with GSH and analyzed by SDS-PAGE (12% gel, MOPS buffer) and Coomassie staining. Asterisks indicate Strep-Bre1 eRING. The same eluates were immunoblotted with an anti-Strep antibody.
Bre1 RBD Is Necessary and Sufficient to Promote Rad6 Activation
Transfer of ubiquitin to a substrate comprises multiple subreactions. To gain insight into which parts of Bre1 activate Rad6, we sought to directly analyze the discharge of a ubiquitin thioester (∼Ub) from the active site of Rad6 (35). By first charging Rad6 with ubiquitin in the presence of E1 and ATP and then treating the reactions with EDTA, subsequent Rad6 recharging was blocked. Rad6∼Ub was then incubated with different Bre1 fragments, and we quantified the decrease of Rad6∼Ub by SDS-PAGE analysis under nonreducing conditions. This assay measures a single turnover because only one ubiquitin is discharged per Rad6. Notably, Rad6 underwent rapid ubiquitin discharge independently of Bre1 such that ∼50% of the Rad6∼Ub conjugate was turned over after 15 min (Fig. 4, A, lanes 1–4, and B). This observation parallels previous reports about some E2s (e.g. Ubc13) that have an intrinsic ability to transfer ubiquitin independently of an E3, possibly by adopting closed E2 conformations that are more prone to ubiquitin transfer (36).
FIGURE 4.
Bre1 RBD is necessary and sufficient to promote Rad6 activation. A, Bre1 activates ubiquitin discharge from Rad6. Representative gel of a single turnover ubiquitin discharge experiment, in which Rad6, pre-charged with ubiquitin (Rad6∼Ub), was incubated without Bre1 (lanes 1–4) or with 25 μm Bre1 (lanes 5–8). Aliquots were collected after 0, 5, 10, and 15 min, and the reaction was stopped by denaturation. Samples were separated by nonreducing SDS-PAGE (12% gel, MES buffer) followed by Coomassie staining. Rad6 discharging is apparent by the time-dependent decrease of the Rad6∼Ub band intensity with a concomitant increase of the Rad6 band. Note that Rad6 can partially discharge without the E3. Lane 9 shows the Bre1 migration behavior under nonreducing conditions and formation of high molecular weight Bre1 aggregates marked by an asterisk. B, quantification of single turnover experiments as shown in A. Intensity of the Rad6∼Ub band was quantified from Coomassie-stained gels using ImageJ software, and the 0 time point was used for normalization. Three independent experiments per condition were used for quantification. Mean and standard deviation are indicated. Bre1 wild-type and mutant proteins used in the assay are shown on the Coomassie gel above. Mock indicates an assay without Bre1. C, single turnover assay showing Bre1-RBD stimulated Rad6 discharge and dose dependence of the reaction (left panel). Recombinant Bre1 RBD was added at the indicated concentrations. The same assay performed with the K31D mutant RBD (right panel) reveals critical role of RBD-Rad6 interaction for E2 activation. D, Bre1 eRING can activate Rad6. Rad6 discharging assay was performed with the indicated concentration of Bre1 eRING. Three independent experiments per condition were used for quantification. Mean and standard deviation are indicated. The eRING protein formed aggregates at high concentration (>100 μm) under nonreducing conditions, which increased the gel background and interfered with direct quantification of Rad6. To test Rad6 discharge at 400 μm eRING (right panel) His-Rad6 was detected by anti-His immunoblotting.
Based on these findings, we asked whether Bre1 accelerates the intrinsic catalytic activity of Rad6. Notably, adding wild-type Bre1 at 25 μm concentration significantly enhanced the discharge rate of Rad6 (Fig. 4, A, lanes 5–8, and B). Bre1 may stimulate Rad6 also at lower concentrations; however, we could not measure more subtle effects against the high background of intrinsic Rad6 ubiquitin discharge. To test a potential role of the Bre1 RBD in Rad6 activation, we examined the effects of the Bre1 K31D mutant, which has a reduced affinity for Rad6. Notably, the Rad6∼Ub thioester was significantly more stable when Rad6 binding to the Bre1 RBD is impaired (Fig. 4B). This is a key result as it suggested that the RBD of Bre1 could play a role in activating Rad6 independently of the RING. To assess the contribution of the Bre1 RING to Ub discharge from Rad6, we examined the activity of a RING-deleted Bre1 construct (Bre1 ΔRING). Surprisingly, the Rad6∼Ub thioester decayed only at a slightly lesser rate than wild-type Bre1 under the conditions of the assay (Fig. 4B). This result does not rule out a contribution of the RING or other parts of Bre1 to Rad6 discharge; however, it suggests that a predominant factor for Rad6 activation could be the RBD of Bre1. To confirm the influence of the Bre1 RBD on Rad6 activity and to eliminate the potential contribution of other parts of Bre1, we performed the assay with the Bre1 RBD fragment alone. This revealed that the Bre1 RBD was capable of activating Rad6∼Ub discharge in a dose-dependent manner, although it was less efficient than equimolar amounts of full-length Bre1 (Fig. 4C, left panel). In contrast, an RBD carrying the K31D mutation was significantly inhibited in Rad6 activation, again displaying dose dependence (Fig. 4C, right panel). Because we saw a slightly impaired ubiquitin discharge in the Bre1 ΔRING mutant, we directly assessed the effects of an eRING construct. Notably, the free eRING could also promote ubiquitin discharge from Rad6, even though this effect was largely masked in the context of full-length Bre1 (Fig. 4D). This might suggest that the canonical RING-E2 activation mechanism is attenuated in the full-length protein such that the RBD effect predominates under these conditions.
Regardless, these data establish that the Bre1 RBD-Rad6 interaction is both necessary and minimally sufficient for Rad6 activation. They further suggest a canonical, potentially less efficient, RING-E2 activation in the context of the full-length Bre1 protein. The latter finding is consistent with the ability of the Bre1 eRING to promote a low level of NCP ubiquitination independently of the RBD (Fig. 3D).
Tethering Rad6 to the Bre1 RING Can Partially Bypass the Role of the RBD
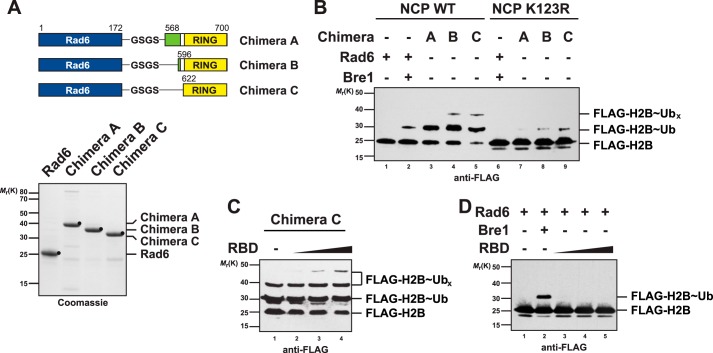
A stable interaction of Rad6 with the N-terminal RBD of Bre1 (Fig. 1C) raises the question as to how Rad6 engages in the canonical low affinity interaction with the C-terminal RING (Fig. 3F). A simple possibility is that the Bre1 RBD folds back onto the Bre1 C terminus and thereby positions Rad6 in proximity of the RING and substrate. We reasoned that the role of the RBD in tethering Rad6 to Bre1 might be bypassed experimentally by fusing Rad6 to the RING. To test this hypothesis, we designed three Rad6∼Bre1 RING chimeras (A, B, and C), which differed in the length of the N-terminal coiled-coil extensions that emanate from the RING (Fig. 5A). All constructs were biochemically stable indicating that the fused domains are correctly folded. Intriguingly, when tested in the NCP ubiquitination assay, all chimeras monoubiquitinated H2B at least as efficient as full-length Bre1. However, instead of a single monoubiqutinated H2B species, we observed an additional slower migrating histone H2B band in chimeras B and C (Fig. 5B, lanes 4 and 5). This could either indicate formation of a di-ubiquitin chain or two mono-ubiquitin marks (or both). To assess the site specificity of the chimeras, we analyzed the mutant K123R NCP. Interestingly, the slowest migrating H2B band disappeared, although a weaker H2B∼Ub species was still detected (Fig. 5B, lanes 7–9). This suggests that the chimeras mainly target Lys-123 on H2B; however, other lysine residues on H2B (and likely on other histones) can be modified, altogether indicating a partial loss of lysine specificity.
FIGURE 5.
Tethering Rad6 to the Bre1 RING can partially bypass the role of the RBD. A, cartoon depicting the design of the Rad6/Bre1 chimeras. Full-length Rad6 is fused to different fragments of Bre1. Boundaries are drawn to scale. Chimeras were subject to Strep-affinity purification and analyzed by SDS-PAGE. B, Rad6/Bre1 chimeras ubiquitinate H2B in vitro. Lane 2 shows the standard NCP ubiquitination assay performed with 3 μm Rad6 and 18 μm Bre1. All chimeras were used at 3 μm concentration. Lysine specificity was assessed by performing the assay with the K123R mutant NCP. UbX denotes additional ubiquitinated sites on H2B. C, Bre1 RBD provided in trans to a Rad6/Bre1 chimera can stimulate NCP ubiquitination. Increasing amounts of RBD (3, 6, and 18 μm) were titrated into the reactions. D, same reaction as in C except that the RBD (3, 6, and 18 μm) was added to free Rad6 (3 μm). Lane 2 shows a positive control with full-length Bre1 (18 μm).
Given that the Bre1 RBD promotes ubiquitin discharge from Rad6, we tested whether the RBD would enhance the chimeras' histone ubiquitinating activity when provided in trans. Notably, histone ubiquitination was further increased when titrating increasing amounts of RBD to the reaction (Fig. 5C). In contrast, the RBD could not activate free Rad6 to ubiquitinate the substrate (Fig. 5D). The appearance of another slowly migrating H2B∼Ub species may suggest that the ubiquitination zone covered by Rad6 is further enlarged such that more peripheral lysines are modified. Alternatively, the increased potency of the reaction could lead to ubiquitination of lysine residues that are not preferred under physiological conditions. We conclude that tethering Rad6 to the Bre1 RING can partially bypass the role of the RBD, however, at the expense of lysine-targeting specificity.
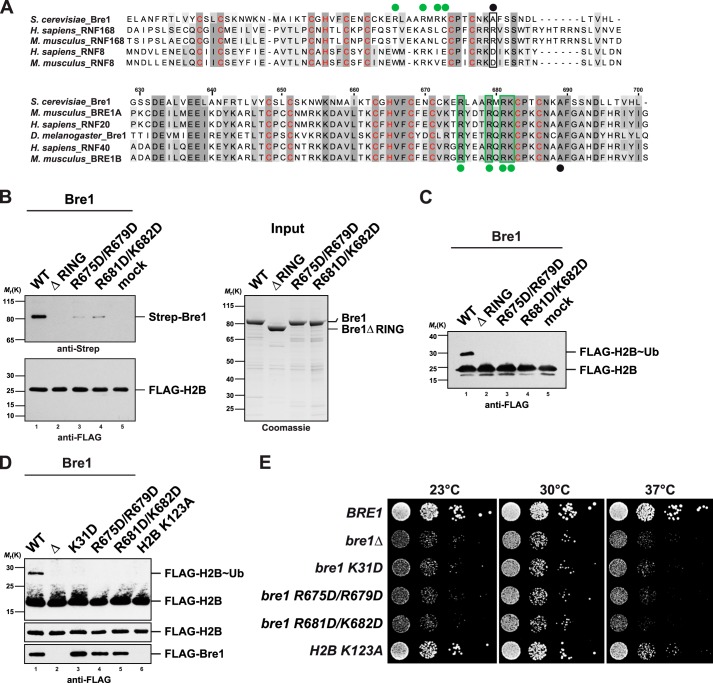
Bre1 RING Uses a Basic Region for Nucleosome Recognition
Given that the eRING of Bre1 is minimally required for H2B Lys-123 ubiquitination, we reasoned that substrate recognition might proceed through the highly conserved RING domain. Notably, the histone H2A ubiquitin ligase RNF168 specifically recognizes nucleosomes through a critical basic residue (Arg-57) (Fig. 6A, upper panel) (14). In contrast, RNF8, a second RING E3 ligase involved in DNA repair, has a negative charge in this position and is inactive toward nucleosomal H2A (14). The Bre1 residue corresponding to RNF168 Arg-57 is an alanine; however, we noticed a nearby stretch of four basic residues that were identical between yeast and human Bre1 (Fig. 6A, lower panel). We therefore created two double mutants with an inverted charge (i.e. R675D/R679D; R681D/K682D). These proteins were stably expressed suggesting that the overall fold of the RING domain was not affected. To test whether the Bre1 RING in general and the basic residues in particular were involved in a direct interaction with the nucleosome, we attempted to reconstitute the NCP-Bre1 interaction. This assay revealed that the RING domain is essential for the NCP interaction, and a charge inversion of the conserved basic residues resulted in a substantial loss of affinity (Fig. 6B). In agreement with these findings, NCP ubiquitination was abolished in all RING mutants (Fig. 6C) underscoring their essential contribution to the overall ubiquitination reaction.
FIGURE 6.
Bre1 RING domain uses a basic region for nucleosome recognition. A, multiple sequence alignment of the RING domains of S. cerevisiae Bre1 compared with the RNF168 and RNF8 E3 ligases. Zinc-coordinating residues are labeled in red. Upper alignment, black dot indicates position of a basic residue in human RNF168 that is critical for histone interaction (Arg-57). Note that RNF8 is inactive toward H2A and has a negatively charged residue at this position. Lower alignment, comparison of Bre1 orthologs shows a stretch of highly conserved basic residues labeled with green dots. Black dot corresponds to the position of Arg-57 in RNF168. The basic Bre1 residues are not conserved in the H2A ubiquitin ligase RNF168 (green dots in upper alignment). Alignments were generated with ClustalW and colored in Jalview. Numbers indicate residues of S. cerevisiae Bre1. B, Bre1 RING domain directly interacts with the nucleosome. In vitro binding assay with NCPs immobilized on anti-FLAG beads and recombinant Bre1 constructs added to it in a 1:5 molar ratio. Proteins bound to the beads were eluted with FLAG peptide and analyzed by SDS-PAGE and immunoblotting with an anti-Strep antibody to detect Strep-Bre1 and an anti-FLAG antibody to confirm equal loading of H2B. Mock indicates omission of Bre1 protein. Gel on the right shows input of Bre1 constructs. C, standard NCP ubiquitination assay was performed with the indicated Bre1 mutants (18 μm) and analyzed by anti-FLAG immunoblotting. Bre1 was omitted from mock sample. D, analysis of global histone H2B ubiquitin levels. BRE1 was deleted in a strain carrying FLAG-tagged histone H2B and then transformed with plasmid-based versions of wild-type FLAG-BRE1, empty vector, and the indicated mutants (lanes 1–5). A FLAG-H2B K123A strain containing genomic wild-type BRE1 (untagged) serves as a control (lane 6). Cell lysates were subject to anti-FLAG immunoprecipitation. Recovered proteins were analyzed by SDS-PAGE and Western blotting using an anti-FLAG antibody. Lower FLAG-H2B panel shows a shorter exposure of the blot membrane. As FLAG-Bre1 was poorly detectable in whole cell extracts, FLAG-Bre1 that co-precipitated together with FLAG-H2B on the beads served as an approximate measure of protein stability/expression. E, growth analysis of wild-type BRE1 and mutant strains used in D. Growth was followed on SDC-HIS-URA plates (−HIS to maintain the FLAG-H2B plasmid, −URA to maintain FLAG-BRE1). Cell density was normalized, and 10-fold serial dilutions were prepared and then plated. Plates were incubated for 2–3 days at 23, 30, and 37 °C.
Bre1 Interactions with Rad6 and the Substrate Are Functionally Important in Vivo
Having defined critical interfaces between Bre1, Rad6, and the nucleosome, we sought to test their in vivo relevance. To this end, we measured histone H2B ubiquitin levels in Saccharomyces cerevisiae cells. Perturbing the RBD-Rad6 and RING-nucleosome interaction by mutations resulted in a loss of H2B ubiquitination just like the deletion of the entire BRE1 gene or mutation of the H2B Lys-123 residue (Fig. 6D). To globally assess Bre1 function, we further analyzed the mutant yeast phenotypes at different temperatures. Deletion of BRE1 confers a growth defect that is most pronounced at 23 and 37 °C (Fig. 6E). Significantly, all bre1 RBD and RING mutants exhibited growth defects that were comparable with the loss of full-length BRE1. This supports the notion that the regulatory protein interfaces, which we have identified in this study, are physiologically important. Interestingly, all bre1 mutants displayed a more severe growth phenotype than the histone H2B K123A mutant. This may suggest that Bre1 has additional, yet unidentified, targets beyond histone H2B.
DISCUSSION
Important questions for monoubiquitin signaling on chromatin are as follows. How is the substrate recognized? How is a high lysine-targeting specificity achieved? How is an E2 restricted toward transferring only a single ubiquitin moiety? Our study sheds light on the molecular details of H2B ubiquitination by reconstituting the reaction from its minimal components. We propose the following framework for the reaction cycle (Fig. 7). The Bre1 RING domain directly recognizes the nucleosome. The RING also interacts with Rad6 and promotes Ub discharge. This is the minimal although inefficient part of the machinery capable of monoubiquitinating H2B at Lys-123. Bre1 further binds to the Rad6 backside through a spatially distinct RBD. The RBD potentiates ubiquitin transfer from the E2 to the substrate and may play a role in tethering Rad6 in proximity of the RING.
FIGURE 7.

Cooperation between Bre1 RING and RBD in coupling substrate targeting and E2 activation. Model of the Bre1/Rad6-catalyzed histone H2B monoubiquitination reaction. Critical steps are highlighted.
Our findings suggest an intricate division of labor between the different parts of the Bre1-Rad6 machinery, in which substrate recognition, positioning of the E2 relative to the substrate, and the timely activation of the reactive E2∼Ub are mechanistically coupled. We assume that the basic patch on the Bre1 RING domain is essential for positioning the E3 on a defined surface of the nucleosome, which might depend on features present on both histones and DNA. The interaction of Rad6∼Ub with the RING would then spatially orient the E2 and direct Ub transfer toward H2B Lys-123. Rad6 binding to the RING may further promote an increased occupancy of the closed E2∼Ub conformation in line with current structural models of E2 activation. Superimposed on this reaction is the noncanonical Rad6 backside activation by the RBD, which may serve to constitutively tether Rad6 to Bre1 and to add another layer of E2 regulation, as discussed further below.
Similarities and Differences between Rad6-Bre1 and Other Rad6-E3 Interactions
Rad6 is a versatile E2 that is intrinsically capable of synthesizing free polyubiquitin chains and also promotes the unspecific ubiquitination of free H2B independent of an E3 in vitro. It is interesting to consider how different E3s manipulate Rad6 for their purpose, in particular how Rad6 is adjusted to add only a single ubiquitin to a specific lysine. The Rad6-interacting RING E3s Bre1 and Rad18 both utilize regions outside of the canonical RING domain to target the Rad6 Ubc domain. Rad18 monoubiquitinates PCNA and interacts with Rad6 both canonically through its RING domain and via a separate α-helical segment that binds the Rad6 backside (Fig. 2A). Rad18 backside binding does not alter the rate of formation of the Rad6∼Ub thioester. Instead, Rad18 was shown to limit the intrinsic Ub chain-forming activity of Rad6. This led to the hypothesis that Rad18 backside binding to Rad6 displaces ubiquitin from this site and thereby directs the E2 toward mono- rather than polyubiquitination of PCNA (23). Our data, however, suggest that Rad6 backside binding by the Bre1 RBD does not explain why histone H2B receives only a single ubiquitin, because the RING domain alone was already sufficient for monoubiquitination. The Bre1 RBD-Rad6 interaction may, however, resemble aspects of how the human RING E3 gp78 stimulates its cognate E2 Ube2g2. In this case, the E2 is also contacted on its backside through a non-RING α-helix (37, 38), which leads to conformational changes in the E2 catalytic center. Interestingly, the gp78 α-helix is oriented roughly perpendicular to the axis of the Rad18 α-helix (Fig. 2A). Elucidating the structural details of how the Bre1 coiled-coil domain interacts with the Rad6 backside will be essential for understanding the precise mechanism of an expected allosteric regulation.
H2B Monoubiquitination May Be Dictated by the Intrinsic Structural Properties of the E3-E2-Nucleosome Interface
Our data indicate that monoubiquitination at H2B Lys-123 is an intrinsic property of the Bre1 RING-E2 interaction. A recent study on the RNF168 E3 ligase suggested that an acidic patch on the nucleosome can enhance E2 discharge and specificity toward H2A Lys-13/15 (16). Taking this into account, we consider the possibility that the Bre1 RING and the nucleosome surface may also locally activate Rad6. This would prevent other lysines than Lys-123 from carrying out a nucleophilic attack on the Rad6∼Ub thioester in the E2's catalytic center. A recent seminal study describing the PRC1 histone ubiquitin ligase module bound to a nucleosome revealed an intriguing multivalent interaction where different nucleosome surface residues interact with both the RING domain and the E2 UbcH5 to position the E2's catalytic center in the correct orientation toward H2A Lys-119 (17). The Bre1-Rad6 complex is clearly different from PRC1-UbcH5 and the latter structure did not include ubiquitin; however, both machineries may constrain the E2 in close proximity to the nucleosome surface and form a structural “cage” that prevents the addition of a second ubiquitin.
Although the Bre1 RING domain is expected to be structurally analogous to other RINGs, the overall fold of the entire Bre1 molecule poses interesting questions. Given that Rad6 stably interacts with the Bre1 N terminus and at least transiently with the C-terminal RING, this implies that the Bre1 N and C termini may approach each other at some point of the reaction cycle. This model is further supported by the finding that the RBD function can be partially bypassed by a direct fusion of Rad6 to the RING. Formation of a coiled-coil structure further predicts a potential for RING domain homo-dimerization. Structural data on the full-length Bre1 will be required to elucidate how its coiled-coil segments fold into a conformation that supports a dynamic interaction of the Bre1 RING with RBD-bound Rad6 and adaptation to the disk-like nucleosome surface.
Other Potential Functions of the Bre1 RBD-E2 Backside Interaction
Besides activating ubiquitin discharge from Rad6, the Bre1 RBD may play additional roles. An E2 needs to sequentially interact with both an E1 and an E3, and these interactions involve overlapping E2 surfaces (Fig. 2A). This implies that an E2 cannot be recharged with ubiquitin when bound to the RING (22). The Bre1 RBD could offer an interesting solution to this dilemma. First, the RBD interacts with the Rad6 backside, which is distinct from the canonical RING- and E1-interacting surface of E2s. Second, the RBD-Rad6 interaction is surprisingly robust and may allow Rad6 to remain bound to Bre1 after ubiquitin discharge. According to this idea, the Bre1 RBD could support a direct recharging of Rad6 by the E1 without dissociation from the Bre1 RING and be particularly relevant in the context of gene expression. Bre1-Rad6 are rather unique in that they “travel” with the elongating RNA polymerase II using the Paf complex (polymerase II-associated factor) as an adaptor (39). The polymerase II movement itself and the structural complexity of the chromatin fiber may complicate nucleosome ubiquitination and allow only a limited time window to successfully ubiquitinate a nucleosome en route. On-site E2 recharging with ubiquitin afforded by the Bre1 RBD may contribute to the efficiency and fidelity of the reaction. Further studies will be required to understand how the specific mechanistic features of Bre1 and Rad6 are integrated into their broader role of gene regulation.
Acknowledgments
We thank S. Schitter-Sollner for excellent advice and technical support and T. Clausen for comments on the manuscript; T. Richmond (ETH Zürich) for the 601 Widom plasmid; S. Tan (Pennsylvania State University) for the pST44 vector; Ali Shilatifard (Stowers Institute) for the FLAG-tagged H2B strains; and S. Westermann (IMP Vienna) for antibodies.
This work was supported by ERC Grant 281354, NPC GENEXPRESS (to the Köhler laboratory), and START Grant Y557-B11 from the Austrian Science Fund.
- NCP
- nucleosome core particle
- RBD
- Rad6 binding domain
- SEC
- size exclusion chromatography
- TEV
- tobacco etch virus
- CHES
- 2-(cyclohexylamino)ethanesulfonic acid
- PCNA
- proliferating cell nuclear antigen
- Ub
- ubiquitin.
REFERENCES
- 1. Fleming A. B., Kao C. F., Hillyer C., Pikaart M., Osley M. A. (2008) H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell 31, 57–66 [DOI] [PubMed] [Google Scholar]
- 2. Hwang W. W., Venkatasubrahmanyam S., Ianculescu A. G., Tong A., Boone C., Madhani H. D. (2003) A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 11, 261–266 [DOI] [PubMed] [Google Scholar]
- 3. Kim J., Guermah M., McGinty R. K., Lee J. S., Tang Z., Milne T. A., Shilatifard A., Muir T. W., Roeder R. G. (2009) RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 137, 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Robzyk K., Recht J., Osley M. A. (2000) Rad6-dependent ubiquitination of histone H2B in yeast. Science 287, 501–504 [DOI] [PubMed] [Google Scholar]
- 5. Wood A., Krogan N. J., Dover J., Schneider J., Heidt J., Boateng M. A., Dean K., Golshani A., Zhang Y., Greenblatt J. F., Johnston M., Shilatifard A. (2003) Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11, 267–274 [DOI] [PubMed] [Google Scholar]
- 6. Xiao T., Kao C. F., Krogan N. J., Sun Z. W., Greenblatt J. F., Osley M. A., Strahl B. D. (2005) Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 25, 637–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhu B., Zheng Y., Pham A. D., Mandal S. S., Erdjument-Bromage H., Tempst P., Reinberg D. (2005) Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol. Cell 20, 601–611 [DOI] [PubMed] [Google Scholar]
- 8. Weake V. M., Workman J. L. (2008) Histone ubiquitination: triggering gene activity. Mol. Cell 29, 653–663 [DOI] [PubMed] [Google Scholar]
- 9. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 10. Braun S., Madhani H. D. (2012) Shaping the landscape: mechanistic consequences of ubiquitin modification of chromatin. EMBO Rep. 13, 619–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bentley M. L., Corn J. E., Dong K. C., Phung Q., Cheung T. K., Cochran A. G. (2011) Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. EMBO J. 30, 3285–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Buchwald G., van der Stoop P., Weichenrieder O., Perrakis A., van Lohuizen M., Sixma T. K. (2006) Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 25, 2465–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R. S., Zhang Y. (2004) Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 [DOI] [PubMed] [Google Scholar]
- 14. Mattiroli F., Vissers J. H., van Dijk W. J., Ikpa P., Citterio E., Vermeulen W., Marteijn J. A., Sixma T. K. (2012) RNF168 ubiquitinates Lys-13–15 on H2A/H2AX to drive DNA damage signaling. Cell 150, 1182–1195 [DOI] [PubMed] [Google Scholar]
- 15. Mattiroli F., Sixma T. K. (2014) Lysine-targeting specificity in ubiquitin and ubiquitin-like modification pathways. Nat. Struct. Mol. Biol. 21, 308–316 [DOI] [PubMed] [Google Scholar]
- 16. Mattiroli F., Uckelmann M., Sahtoe D. D., van Dijk W. J., Sixma T. K. (2014) The nucleosome acidic patch plays a critical role in RNF168-dependent ubiquitination of histone H2A. Nat. Commun. 5, 3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McGinty R. K., Henrici R. C., Tan S. (2014) Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 514, 591–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deshaies R. J., Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 19. Dou H., Buetow L., Sibbet G. J., Cameron K., Huang D. T. (2013) Essentiality of a non-RING element in priming donor ubiquitin for catalysis by a monomeric E3. Nat. Struct. Mol. Biol. 20, 982–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Plechanovová A., Jaffray E. G., Tatham M. H., Naismith J. H., Hay R. T. (2012) Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489, 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pruneda J. N., Littlefield P. J., Soss S. E., Nordquist K. A., Chazin W. J., Brzovic P. S., Klevit R. E. (2012) Structure of an E3:E2∼Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol. Cell 47, 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wenzel D. M., Stoll K. E., Klevit R. E. (2011) E2s: structurally economical and functionally replete. Biochem. J. 433, 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hibbert R. G., Huang A., Boelens R., Sixma T. K. (2011) E3 ligase Rad18 promotes monoubiquitination rather than ubiquitin chain formation by E2 enzyme Rad6. Proc. the Natl. Acad. Sci. U.S.A. 108, 5590–5595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xie Y., Varshavsky A. (1999) The E2-E3 interaction in the N-end rule pathway: the RING-H2 finger of E3 is required for the synthesis of multiubiquitin chain. EMBO J. 18, 6832–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ulrich H. D., Jentsch S. (2000) Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 19, 3388–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bailly V., Prakash S., Prakash L. (1997) Domains required for dimerization of yeast Rad6 ubiquitin-conjugating enzyme and Rad18 DNA binding protein. Mol. Cell. Biol. 17, 4536–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Notenboom V., Hibbert R. G., van Rossum-Fikkert S. E., Olsen J. V., Mann M., Sixma T. K. (2007) Functional characterization of Rad18 domains for Rad6, ubiquitin, DNA binding and PCNA modification. Nucleic Acids Res. 35, 5819–5830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim J., Roeder R. G. (2009) Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast. J. Biol. Chem. 284, 20582–20592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tan S., Kern R. C., Selleck W. (2005) The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr. Purif. 40, 385–395 [DOI] [PubMed] [Google Scholar]
- 30. Luger K., Rechsteiner T. J., Flaus A. J., Waye M. M., Richmond T. J. (1997) Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 272, 301–311 [DOI] [PubMed] [Google Scholar]
- 31. Lowary P. T., Widom J. (1998) New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 [DOI] [PubMed] [Google Scholar]
- 32. Nakanishi S., Sanderson B. W., Delventhal K. M., Bradford W. D., Staehling-Hampton K., Shilatifard A. (2008) A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat. Struct. Mol. Biol. 15, 881–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Köhler A., Zimmerman E., Schneider M., Hurt E., Zheng N. (2010) Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell 141, 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jentsch S., McGrath J. P., Varshavsky A. (1987) The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature 329, 131–134 [DOI] [PubMed] [Google Scholar]
- 35. Petroski M. D., Deshaies R. J. (2005) Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell 123, 1107–1120 [DOI] [PubMed] [Google Scholar]
- 36. Pruneda J. N., Stoll K. E., Bolton L. J., Brzovic P. S., Klevit R. E. (2011) Ubiquitin in motion: structural studies of the ubiquitin-conjugating enzyme approximately ubiquitin conjugate. Biochemistry 50, 1624–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen B., Mariano J., Tsai Y. C., Chan A. H., Cohen M., Weissman A. M. (2006) The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc. Natl. Acad. Sci. U.S.A. 103, 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Das R., Mariano J., Tsai Y. C., Kalathur R. C., Kostova Z., Li J., Tarasov S. G., McFeeters R. L., Altieri A. S., Ji X., Byrd R. A., Weissman A. M. (2009) Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Mol. Cell 34, 674–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tomson B. N., Arndt K. M. (2013) The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states. Biochim. Biophys. Acta 1829, 116–126 [DOI] [PMC free article] [PubMed] [Google Scholar]