Background: Macrophages express Smyd2 during development and differentiation.
Results: Smyd2 inhibits macrophage IL-6 and TNF-α production.
Conclusion: Smyd2 negatively regulates M1 macrophage polarization.
Significance: These findings are important for understanding the regulation of macrophage polarization and provide new insights for autoimmune disease therapy.
Keywords: Autoimmunity, ChiP, Immunology, Macrophage, Signaling
Abstract
SET and MYND domain-containing 2 (Smyd2), a histone 3 lysine 4- and histone 3 lysine 36 (H3K36)-specific methyltransferase, plays critical roles in cardiac development and tumorigenesis. However, the role of Smyd2 in immunity and inflammation remains poorly understood. In this study, we report that Smyd2 is a novel negative regulator for macrophage activation and M1 polarization. Elevated Smyd2 expression suppresses the production of proinflammatory cytokines, including IL-6 and TNF, and inhibits the expression of important cell surface molecules, including major MHC-II and costimulatory molecules. Furthermore, macrophages with high Smyd2 expression inhibit Th-17 cell differentiation but promote regulatory T cell differentiation as a result of increased TGF-β production and decreased IL-6 secretion. In macrophages, Smyd2 specifically facilitates H3K36 dimethylation at Tnf and Il6 promoters to suppress their transcription and inhibits NF-κB and ERK signaling. Therefore, our data demonstrate that epigenetic modification by Smyd2-mediated H3K36 dimethylation at Tnf and Il6 promoters plays an important role in the regulation of macrophage activation during inflammation.
Introduction
Innate immunity is the first line of defense against infection by bacteria, viruses, parasites, and fungi. When infected by foreign pathogens, innate immunity is first activated by pattern recognition receptors expressed on the immune cells to recognize pathogen-associated molecular patterns associated with the pathogens (1–3). Toll-like receptors (TLRs)4 are one of the best-characterized pattern recognition receptors to help macrophages and other phagocytic cells recognize invading pathogens and induce immune responses, leading to the production of proinflammatory cytokines and chemokines (4–7). TLRs trigger multiple signaling pathways, including NF-κB and MAPK pathways, inducing activation of a set of transcription factors and kinases such as NF-κB, ERK, p38, and c-Jun. These transcription factors cooperatively promote the expression of multiple downstream genes such as TNF and IL-6, leading to macrophage activation (8–10).
Macrophages can be divided into subpopulations on the basis of their anatomical locations and functional phenotypes. Specialized tissue-resident macrophages include osteoclasts, alveolar macrophages, histiocytes, and Kupffer cells. According to their functional phenotypes, macrophages could be polarized into classically activated (M1) and alternatively activated (M2) macrophages. M1 macrophages produce a large amount of proinflammatory cytokines such as TNF-α and IL-6, are critical for clearing various pathogens, and also play a role in antitumor immunity (11, 12). M2 macrophages, characterized by their anti-inflammatory property, have essential roles in wound healing, tissue repair, angiogenesis, and tumor progression and also in immune response to parasite infection (13–15). Marked changes in the activity and gene expression profile of macrophages can occur when stimulated by pathogens or related cytokines, and these changes are tightly regulated by genetic, epigenetic, and transcriptional modification (16, 17). Disorders of macrophage identity and functional balance may cause chronic inflammation or autoimmune diseases such as Crohn disease, rheumatoid arthritis, multiple sclerosis, and autoimmune hepatitis (12).
Accumulating studies suggest a critical involvement of epigenetic modification in macrophage activation and functional differentiation (16). The basic structural unit of eukaryotic chromatin is the nucleosome, consisting of 146 base pairs of DNA wrapped by four core histones (H2A, H2B, H3, and H4). Gene expression can be regulated tightly by histone posttranslational modifications such as acetylation, methylation, and phosphorylation (18). Aberrant histone modifications are associated with multiple human diseases and are potential diagnostic biomarkers or therapeutic targets for some inflammatory or autoimmune diseases (19). Histone lysine methylation is one of the most characterized posttranslational modifications, such as histone 3 lysine 4 (H3K4), histone 3 lysine 27 (H3K27), and histone 3 lysine 36 (H3K36) methylation. Tri- or dimethylation of H3K4 and trimethylation of H3K36 are implicated with actively transcribed genes, whereas H3K36 dimethylation and H3K27 trimethylation corresponds with gene silencing (20, 21). It has been reported that histone lysine methylation plays a critical role in macrophage polarization and activation. One report showed that H3K27 demethyltransferase Jumonji domain containing 3 (Jmjd3) plays a critical role in M2 macrophage activation (22), whereas another report suggested that H3K4 histone methyltransferase Ash1l with a conserved SET domain regulated IL-6 and TNF-α production in LPS-induced macrophage activation (23).
The SMYD family consists of five methyltransferases, defined as Smyd1–5, that contain a SET domain that is split into two segments by an MYND domain (24). Smyd1–4 have been reported to play crucial roles in cardiomyocyte maturation and tumor cell proliferation (25). In the absence of HSP90α, Smyd2, a lysine methyltransferase, dimethylates H3K36, leading to repressed gene transcription (24, 26). However, in the presence of HSP90α, Smyd2 transfers the methyl to H3K4, leading to gene transcription (26). In addition, Smyd2 not only methylates histones but also methylates non-histone proteins. For example, tumor suppressor p53 methylation at lysine 370 leads to its functional impairment, and retinoblastoma protein methylation at lysine 860 results in repression of its target genes (27, 28). A recent study showed that Smyd2 was expressed in cells of the immune system (26). However, the functional role of Smyd2 in regulating immune response and inflammation is still elusive.
This study is prompted by our initial observation that Smyd2 expression is decreased dramatically when macrophages are activated by LPS stimulation, indicating that Smyd2 could be a negative regulator for macrophage activation. We found that overexpression of Smyd2 in primary macrophages could suppress TNF-α and IL-6 expression. Our data show that Smyd2 regulates the production of TNF-α and IL-6 by inhibiting the di- or trimethylation of H3K4 and increasing the dimethylation of H3K36. In addition, Smyd2 overexpression impairs NF-κB activation and reduces binding of RelA in the promoter region of targeting genes. Importantly, macrophages with altered Smyd2 expression exhibit significant modulatory function in the differentiation of regulatory T cells (Tregs) and Th-17 cells. Therefore, our study is important for understanding the regulation of macrophage polarization and provides new insights for autoimmune disease therapy.
EXPERIMENTAL PROCEDURES
Animals
C57BL/6 mice were purchased from the Shanghai Laboratory Animal Center, Chinese Academy of Sciences (Shanghai, China). Mice were kept in a specific pathogen-free facility in microisolator cages, and all experiments were performed with mice 6–8 weeks old. All animal protocols were approved by the Institutional Laboratory Animal Care and Use Committee of Soochow University.
Macrophage Isolation and Culture
Femur and tibia bone marrow cells were collected and cultured in macrophage differentiation medium consisting of RPMI 1640 medium (Invitrogen), 10% FBS, 100 units/ml penicillin, and 100 units/ml streptomycin supplemented with 100 ng/ml macrophage colony-stimulating factor (eBiosicence) at a cell density of 2 × 106/ml (6-wells plate) for 6 days, and the culture medium was refreshed after 3 days. After 6 days of culture, the adherent cells were collected and used for transfection. After 24 h, cells were collected and stimulated with 100 ng/ml LPS for macrophage activation.
T Cell Purification
CD4+ T cells were purified with a CD4 isolation kit (Miltenyi Biotech). CD4+CD25− T cells were further prepared by FACS, and the purity of these cells was approximately 95%.
T Cell Differentiation and Coculture with Macrophages
Purified CD4+CD25− cells were stimulated with antibodies to CD3 (3 μg/ml) and CD28 (2 μg/ml) under iTreg cell differentiation conditions (rhTGF-β1, 3 ng/ml) or Th-17 cell differentiation conditions (IL-6, 10 ng/ml; rhTGF-β1, 3 ng/ml; anti-IL-4, 10 μg/ml; and anti-IFNγ, 10 μg/ml) and cocultured with macrophages transfected with Smyd2 plasmid or control mutant plasmid at a ratio of 1:1 for 3 or 4 days, respectively.
Plasmid Transfection
The Smyd2 plasmid (catalog no. MC204022) and control vector (catalog no. PS100001) were purchased from OriGene. For Smyd2 catalytic mutant Y240F point mutation construction, primers 5′ CGAGGTGTTCACCAG CTTCATCGACCTGCTATATCC; 3′ GGATATAGCAGGTCGATGAAGCTGGTGAACACCTCG were used. The Il6 and Tnf luciferase reporter plasmids were constructed as described previously (23, 29, 30). The pGL3 luciferase vector and internal control TK plasmid were purchased from Promega. Primers for Il6 (forward, 5′-CCTCTAGATAGTGCGTTATGCCTAAGCA-3′; reverse, 5′-CCTCTAGAGTTTGAAGACAGTCTAAACAT-3′) and Tnf (forward, 5′-CCATCTGTGAAACCCAATAAACCTC-3′; reverse, 5′-GGGAGATATGGCGCCTTGG-3′) were used for reporter plasmid construction. All constructs were confirmed by DNA sequencing. Plasmids were transfected into macrophages with a mouse macrophage nucleofector kit (Lonza) according to the instructions of the manufacturer.
RNA Interference
The mouse Smyd2-specific siRNA1, ON-TARGET plus SMARTpool mouse Smyd2 (catalog no. 226830), and scrambled control RNA were purchased from Dharmacon. Another Smyd2-specific siRNA2 (catalog no. SC-76530) was purchased from Santa Cruz Biotechnology. siRNA duplexes were transfected into macrophages using a mouse macrophage nucleofector kit (Lonza) according to the instructions of the manufacturer.
Real-time PCR
Total RNA was extracted from cells with an RNeasy mini kit (Qiagen) according to the instructions of the manufacturer, and then cDNA was synthesized with an iScript cDNA synthesis RT kit (Bio-Rad). The gene expression profile was analyzed by quantitative real-time PCR in an ABI 7900HT (Applied Biosystems) with the following customized primer sets: mouse Smyd1, 5′-TCAGTGACCAGAGAGGGCTAC-3′ and 5′-AGCTCAATCTTGCCATTGTTGAA-3′; Smyd2, 5′-ACTGCGACGTGGAATGTCAG-3′ and 5′-CGCACAGTCTCCGAAGGAT-3′; Smyd3, 5′-CCGACCCCTTGGCTTACAC-3′ and 5′-CGGCATTGAGAACAACGCATC-3′; Smyd4, 5′-GGTGGATGAATGGAAGTCCTACC-3′ and 5′-CCTCAGGTTGAAGAAGGGAAGAA-3′; Smyd5, 5′-GGCACCCCCTCAATAAGCTG-3′ and 5′-ACCCAGTGGTCCTTGTCCTT-3′; IL-6, 5′-TAGTCCTTCCTACCCCAATTTCC-3′ and 5′-TTGGTCCTTAGCCACTCCTTC-3′; TNF-α, 5′-CCCTCACACTCAGATCATCTTCT-3′ and 5′-GCTACGACGTGGGCTACAG-3′; and β-actin, 5′-TGTCCACCTTCCAGCAGATGT-3′ and 5′-AGCTCAGTAACAGTCCGCCTAGA-3′.
Western Blot Analyis
Cells were lysed with radioimmune precipitation assay buffer containing PMSF and protease inhibitor mixture. The lysates were fractionated by SDS-PAGE and analyzed by immunoblotting. Specific antibodies to Smyd2, phospho-IKKα/β, IKKα/β, phospho-IκBα, IκBα, phospho-p65, p65, phospho-ERK1/2, ERK1/2, phospho-p38, p38, phospho-c-Jun, and c-Jun were purchased from Cell Signaling Technology. Anti-β-actin antibody was purchased from Sigma-Aldrich. Blots were exposed using ECL substrate with a Fujifilm LAS4000 luminescence imager (GE) and graphed with ImageJ software (National Institutes of Health).
ELISA
Levels of cytokines production in culture supernatants were measured using an ELISA kit purchased from R&D Systems for mouse TNF-α, IL-6, and IL-17 according to the instructions of the manufacturer.
Assay of Luciferase Reporter Gene Expression
RAW264.7 macrophages were cotransfected with a mixture of the indicated pGL3-luciferase reporter plasmid, the Prl-tk-Renilla-luciferase plasmid (internal control), and indicated amounts of Smyd2 or a control expression construct. After 24 h, the cells were stimulated with LPS. Luciferase activities were measured using a Dual-Luciferase reporter assay system (Promega) according to the instructions of the manufacturer. Data were normalized for transfection efficiency by dividing firefly luciferase activity with that of Renilla luciferase. The relative values are presented as -fold increase over the indicated control.
ChIP Assay
ChIP assays were conducted with a ChIP assay kit (Millipore) according to the protocol of the manufacturer. Immunoprecipitated DNA and input DNA were analyzed by quantitative real-time PCR, and results were presented of normalization to input DNA. ChIP antibody, Smyd2 (catalog no. ab108217), H3K4me2 (catalog no. ab7766), H3K4me3 (catalog no. ab12209), H3K27me3 (catalog no. ab6002), and H3K36me2 (catalog no. ab9048) were purchased from Abcam. The ChIP kit EZ-ChIPTM (catalog no. 17-371) was purchased from Millipore. The following promoter locus primers were used for amplification of the ChIP analysis: Il6, 5′-GCAGTGGGATCAGCACTAAC-3′ (forward) and 5′-GGTGGGTAAAGTGGGTGAAG-3′ (reverse); Tnf, 5′-CAGCCACTGCTTGGCTAGAC-3′ (forward) and 5′-CGGATCCCATGGACCAACTG-3′ (reverse); Tafaip3, 5′-TTGAATGGTGGTGGTCTTCA-3′ (forward) and 5′-TGAGGAGGAGGGGAATAACC-3′ (reverse); Il12b, 5′-CCCTGGATACAGACAACA-3′ (forward) and 5′-GTGAATAGAGGCGGCAAT-3′ (reverse); and Jmjd3, 5′-TAAGGATTAGGAGGGAAGAG-3′ (forward) and 5′-CTGGTGTAGGCAGGTTCT-3′ (reverse).
Flow Cytometry
For cell surface or intracellular staining, cells were stained with antibodies to murine CD4, CD11b, CD80, CD86, MHC-II, CD40, IL-17, Foxp3, and IFN-γ according to the instructions of the manufacturer. FACS antibodies were purchased from eBioscience. Before IL-17 and IFN-γ intracellular staining, cells were stimulated with phorbol 12-myristate 13-acetate (100 ng/ml) and ionomycin (100 ng/ml) in the presence of Golgi-stop (BD Biosciences, 1:100) for 5 h.
Statistical Analysis
Data were analyzed using two-tailed Student's t test, and p < 0.05 was considered statistically significant (*, p < 0.05; **, p < 0.01).
RESULTS
Smyd2 Expression Is Reduced following Macrophage Activation
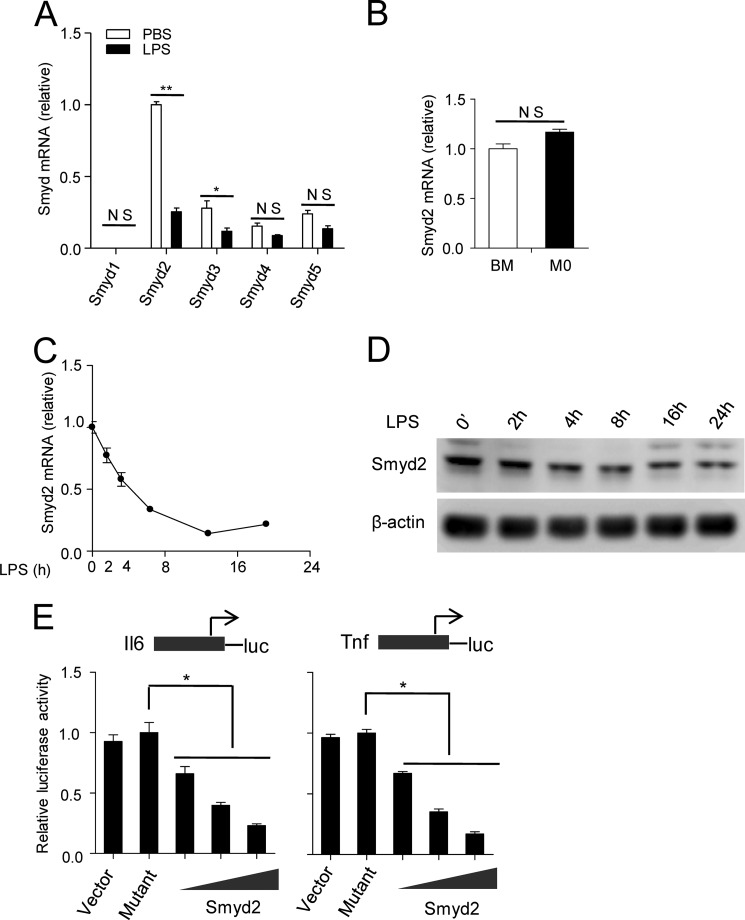
In this study, mouse bone marrow cells were differentiated into mature macrophages (i.e. M0 macrophages) by macrophage colony stimulation factor (MCSF). The M0 macrophages were polarized into activated macrophages through LPS stimulation. We first measured Smyd family member mRNA expression by LPS-induced macrophage activation. We found that only Smyd2 and Smyd3 had a significant decrease of mRNA expression in this procedure. However, Smyd2 mRNA decreased more significantly and had more abundant mRNA expression (Fig. 1A). Therefore, we selected Smyd2 to further investigate its role in LPS-induced macrophage activation. When Smyd2 expression during macrophage differentiation was analyzed, we found that there was no significant difference in Smyd2 transcription between bone marrow cells and differentiated M0 cells (Fig. 1B). To further examine Smyd2 expression during the course of LPS-induced M1 macrophage polarization, we analyzed its transcription and protein level at multiple time points following LPS stimulation. Smyd2 expression was decreased progressively during macrophage activation (Fig. 1, C and D). Smyd2 has been reported to have methyltransferase activity, so we constructed a Smyd2 full-length expression plasmid and Y240F point mutation catalytic mutant plasmid to investigate whether its methyltransferase activity is involved in LPS-induced macrophage activation. Using a luciferase reporter assay, we found that overexpression of Smyd2 markedly and dose-dependently repressed LPS-induced production of the TNF-α and IL-6 luciferase reporter genes (Fig. 1E) compared with a mock vector or the Smyd2 mutant control. These data indicate that Smyd2 may be a negative regulator during LPS-induced M1 macrophage polarization on the basis of its methyltransferase activity.
FIGURE 1.
Expression of Smyd2 during macrophage differentiation and activation. Mouse bone marrow (BM) cells were differentiated into M0 macrophages with MCSF for 6 days. A, M0 macrophages were stimulated with 100 ng/ml LPS or PBS for 6 h and collected later for detection of Smyd family mRNA expression by Q-PCR. B, the mRNA levels of Smyd2 in bone marrow cells and M0 cells detected by Q-PCR. C and D, the quantities of mRNA and protein of Smyd2 in bone marrow-derived macrophages stimulated with LPS for 0, 2, 4, 8, 16, or 24 h were measured by Q-PCR and Western blot analysis. E, luciferase (luc) assay of Raw 264.7 cells transfected with IL-6 or the TNF promoter reporter plasmid with the Smyd2 plasmid or its catalytic mutant control. *, p < 0.05; **, p < 0.01; NS, no significance. Data are mean ± S.D. of technical replicates from one of three independent experiments. In A–C and E, controls were normalized to one. In Q-PCR experiments, β-actin served as an internal control. The experiments were performed at least three times with similar results obtained.
Smyd2 Overexpression Suppresses IL-6 and TNF-α Production Depending on Its Methyltransferase Activity in Activated Macrophages
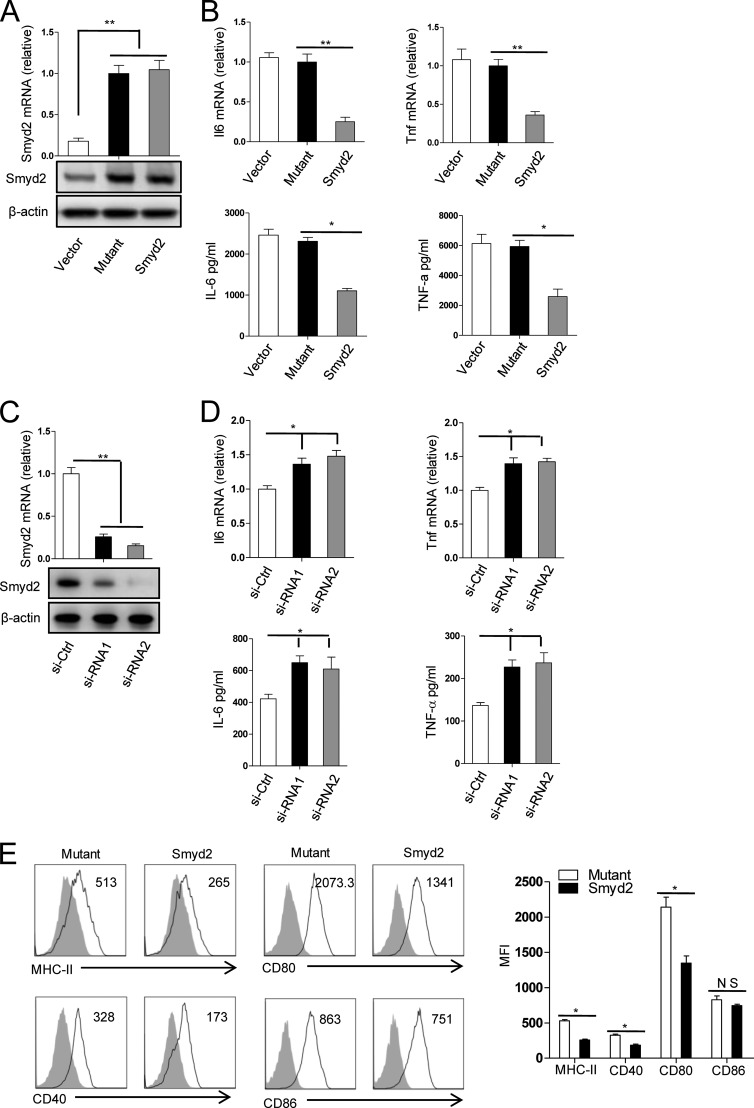
To determine the functional role of Smyd2 in macrophage polarization, we overexpressed Smyd2 and the control mutant plasmid in M0 macrophages and subsequently polarized the resulting macrophages by LPS stimulation. Following LPS stimulation, the production of major proinflammatory cytokines, such as TNF-α and IL-6, was determined by real-time PCR and ELISA. As illustrated in Fig. 2, A and B, overexpression of Smyd2 in macrophages significantly suppressed TNF-α and IL-6 expression at both mRNA and protein levels compared with the mutant control. Consistently, when Smyd2 expression was knocked down by Smyd2-specific siRNA, TNF-α and IL-6 expression was increased significantly and consistently at both mRNA and protein levels (Fig. 2, C and D). These results suggest that Smyd2 inhibits LPS-induced M1 macrophage activation by suppressing TNF-α and IL-6 production, depending on its methyltransferase activity.
FIGURE 2.
Smyd2 inhibits the production of proinflammatory cytokines and the expression of costimulatory molecules. A, the quantities of mRNA (top panel) and protein (bottom panel) of Smyd2 in MCSF-driven bone marrow-derived macrophages 24 h after transfection with the Smyd2 mutant (Mutant) or Smyd2 plasmid (Smyd2) were measured by Q-PCR and Western blot analysis. B, the expression of IL-6 and TNF mRNA (top panel) and protein (bottom panel) in macrophages transfected as described in A and subsequently stimulated with 100 ng/ml LPS for 4 and 24 h, respectively, were determined by Q-PCR and ELISA. C, the quantities of mRNA (top panel) and protein (bottom panel) of Smyd2 in MCSF-driven bone marrow-derived macrophages 24 h after transfection with control siRNA (si-Ctrl) or two kinds of Smyd2 siRNA (si-RNA1 and si-RNA2) were measured by Q-PCR and Western blot analysis. D, expression of IL-6 and TNF mRNA (top panel) and protein (bottom panel) in macrophages transfected as described in C and then stimulated with 100 ng/ml LPS for 4 and 24 h, respectively, were determined by Q-PCR and ELISA. E, macrophages transfected as described in A and subsequently stimulated for 24 h with 100 ng/ml LPS were analyzed for the expressions of the indicated surface markers by flow cytometry. The shaded area and solid line refer to the isotype control and the indicated markers, respectively. MFI, mean fluorescence intensity. *, p < 0.05; **, p < 0.01; NS, no significance. Data are mean ± S.D. of technical replicates from one of three independent experiments. In Q-PCR experiments, controls were normalized to one, and β-actin served as an internal control. The experiments were performed at least three times with similar results obtained.
Furthermore, we evaluated the expression of some important surface markers associated with macrophage function in Smyd2 overexpressed macrophages. Overexpression of Smyd2 significantly suppressed the expression of MHC-II and the costimulatory molecules CD80 and CD40 (Fig. 2E). However, CD86 expression was not changed significantly compared with the mutant control. These data indicate that Smyd2 may negatively regulate the macrophage antigen-presenting function and reduce macrophage-induced inflammatory immune responses through its methyltransferase action.
Smyd2 Negatively Regulates the NF-κB and MAPK Signaling Pathways in Macrophages
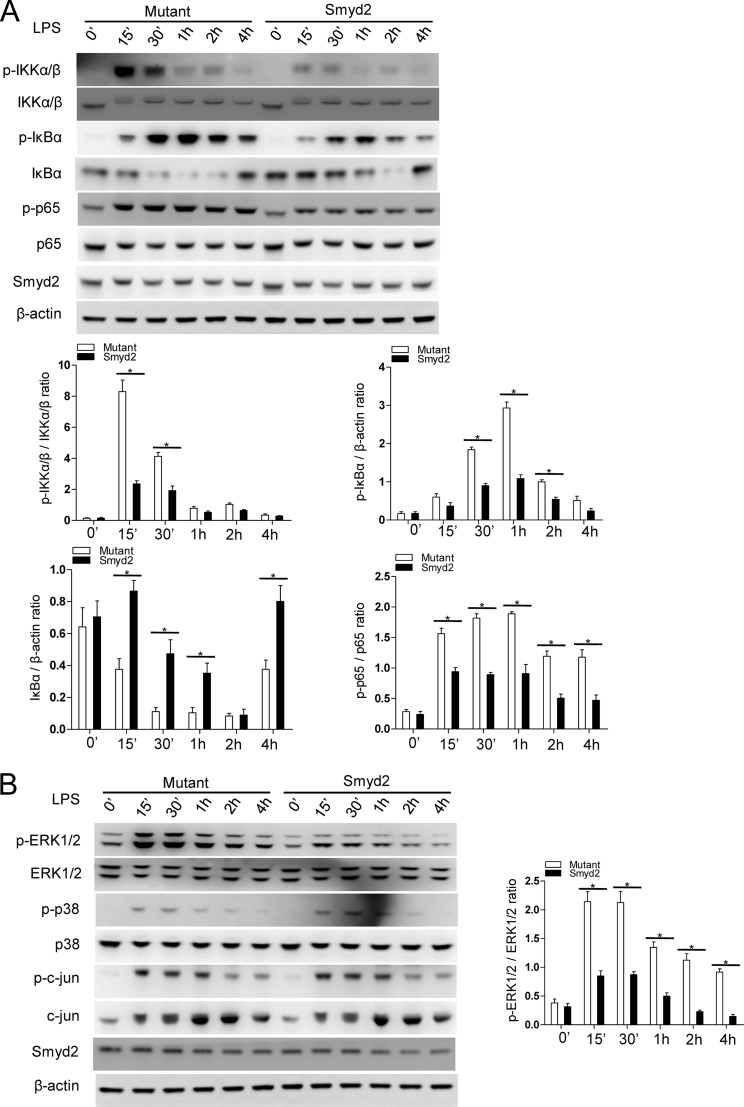
It has been reported that NF-κB- and MAPK-mediated signal transduction downstream of TLR4 is critical for TNF-α and IL-6 production in response to LPS stimulation (9, 31, 32). To investigate the molecular mechanism by which Smyd2 regulates TLR-triggered IL-6 and TNF-α production, we investigated the activation of key signaling molecules of the NF-κB and MAPK pathways in TLR-triggered macrophages overexpressing Smyd2. We found that the phosphorylation of IKKα, IKKβ, and IκBα was decreased significantly by Smyd2 overexpression. Additionally, the degradation of IκBα is ameliorated and delayed correlatively, and the phosphorylation of p65 was also decreased significantly compared with that of the mutant control (Fig. 3A). These data indicate that overexpression of Smyd2 in a time-dependent manner represses the activation of the NF-κB pathway in M1 macrophages. Furthermore, we evaluated the activation of ERK, p38, and c-Jun MAPK kinases during LPS-induced M1 polarization. As shown in Fig. 3B, we observed that overexpression of Smyd2 results in a dramatic decrease of ERK phosphorylation. However, only a minimal difference was observed in the phosphorylation of c-Jun and p38 between the experimental and control groups. These data indicate that Smyd2 only regulates the ERK- but not the JNK- and p38- mediated signal pathways. Therefore, our study suggests that Smyd2, via its methyltransferase activity, acts as a negative regulator in TLR-triggered macrophage activation by suppressing NF-κB and ERK signal transduction, resulting in decreased production of TNF-α and IL-6.
FIGURE 3.
Smyd2 suppresses TLR-mediated activation of the NF-κB and MAPK signaling pathways. A and B, MCSF-driven bone marrow-derived macrophages were transfected with the Smyd2 mutant (Mutant) control or the Smyd2 plasmid (Smyd2) for 24 h. After stimulation for 0–4 h with LPS, the phosphorylated (p-) or total protein in lysates of mutant control and macrophages overexpressing Smyd2 were analyzed by Western blot analysis. Total protein and β-actin served as loading controls. Graphed data were derived from the Western blot analysis by ImageJ software (National Institutes of Health). *, p < 0.05. Western blot data are representative of one of three independent experiments. Data are mean ± S.D. of technical replicates. The experiments were performed at least three times with similar results obtained.
Smyd2 Suppresses IL-6 and TNF-α Production through H3K36 Dimethylation
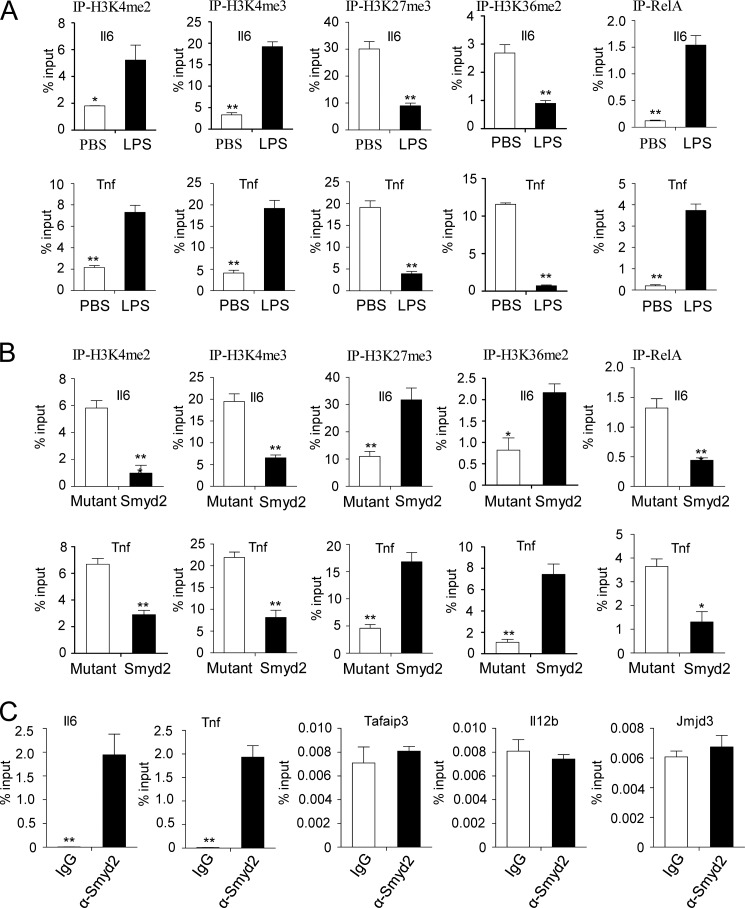
It has been reported that, as a histone methyltransferase, Smyd2 can methylate H3K4 or H3K36, which are associated with activation or repression of target gene transcription, respectively (24, 26). Our findings indicate that Smyd2 regulates H3K36 dimethylation and represses target gene expression, such as IL-6 and TNF-α, in TLR4-triggered macrophage activation. To investigate the exact role of Smyd2 in macrophage polarization, we measure the methylation levels of H3K4, H3K27, and H3K36 in the promoter regions of the TNF-α and IL-6 genes. As shown in Fig. 4A, when M0 macrophages are stimulated with LPS, significantly increased H3K4 tri- and dimethylation, decreased H3K27 trimethylation, and H3K36 dimethylation are observed in the TNF-α and IL-6 promoter regions, which is correlated with increased binding of the NF-κB transcription factor RelA in the same promoter regions (33). Importantly, when we overexpress smyd2 in M0 macrophages, a reversed pattern of methylation of H3K4, H3K27, and H3K36 as well as a decreased binding level of RelA is observed in the TNF-α and IL-6 promoter regions in these macrophages compare with the mutant control (Fig. 4B). These data indicate that smyd2 participates in dimethylation of H3K36, but not H3K4, in macrophages activation. Furthermore, to explore the direct or indirect role of Smyd2 in the modification of histone H3K36 dimethylation, we measured the binding activity of Smyd2 in the TNF-α, IL-6, A20, IL-12, and JMJD3 promoter regions. As shown in Fig. 4C, we observed that the Smyd2 is capable of specifically binding to the IL-6 and TNF-α promoter region supported by the low background observed at other loci (Tafaip3, Il12b, and Jmjd3 promoters). Taken together, our results indicate that Smyd2 suppresses IL-6 and TNF-α production through H3K36 dimethylation.
FIGURE 4.
Suppression of IL-6 and TNF-α production by Smyd2 through its H3K36 methyltransferase activity. A, the dimethylation of histone 3 lysine 4 (H3K4me2), trimethylation of histone 3 lysine 4 (H3K4me3), trimethylation of histone 3 lysine 27 (H3K27me3), dimethylation of histone 3 lysine 36 (H3K36me2), and RelA at the Il6 and Tnf promoter regions in MCSF-driven bone marrow-derived macrophages stimulated for 2 h with LPS or PBS were analyzed by ChIP assay. The promoter sequences were detected by quantitative PCR. B, the promoter sequences were detected in mutant control- or Smyd2 plasmid-transfected macrophages as in A. Macrophages were transfected for 24 h before stimulation with 100 ng/ml LPS for another 2 h. C, the recruitment of Smyd2 to the Il6, Tnf, Tafaip3, Il12b, or Jmjd3 promoter locus with Smyd2 antibody in MCSF-driven bone marrow-derived macrophages without stimulation were analyzed by ChIP assay. Promoter sequences in input DNA and DNA recovered from antibody-bound chromatin segments were detected by quantitative PCR. IgG served as a ChIP control and input as a normalized control. *, p < 0.05; **, p < 0.01. Data are mean ± S.D. of technical replicates from one of three independent experiments. The experiments were performed at least three times with similar results obtained.
Macrophages Overexpressing Smyd2 Suppress Th-17 Cell but Enhance iTreg Cell Differentiation
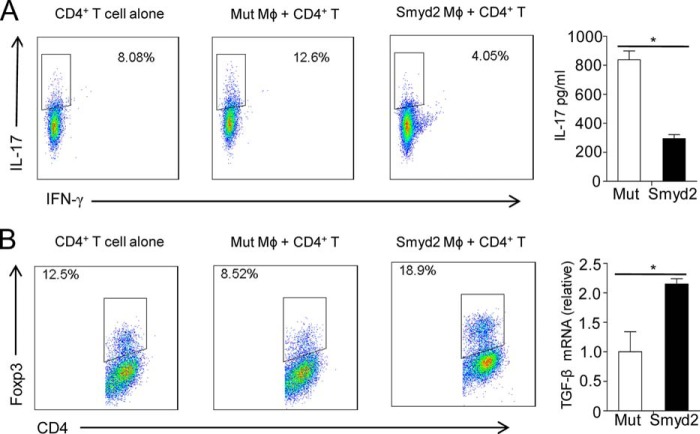
As potent functional antigen-presenting cells, macrophages initiate the adaptive immune responses by regulating CD4+ helper T cell differentiation (34, 35). Our data above indicate that Smyd2 suppresses IL-6 and TNF-α production by altering the histone modification pattern and signal transduction. Because IL-6 is a key cytokine for Th-17 differentiation and plays critical roles in the reciprocal differentiation of Treg and Th-17 cells (36), we wanted to examine whether macrophages overexpressing Smyd2 could regulate Treg and Th-17 cell differentiation. We stimulated mutant control and Smyd2 overexpressing macrophages with LPS and then cocultured them with purified CD4+CD25− T cells under either Treg cell differentiation conditions or Th-17 cell differentiation conditions. As shown in Fig. 5A, macrophages overexpressing Smyd2 dramatically decreased the percentage of Th-17 cells in CD4+ T cells and suppressed IL-17 secretion compared with the mutant control. Smyd2 overexpressing macrophages consistently and significantly promoted iTreg cell differentiation compared with the control macrophages (Fig. 5B, left panel). Because it is known that TGF-β is critical for Treg cell differentiation, we detected the TGF-β mRNA level in macrophages overexpressing Smyd2. We found that Smyd2 overexpression increases TGF-β mRNA transcription (Fig. 5B, right panel), suggesting a regulatory mechanism for a higher percentage of Treg cells in the Smyd2 overexpression group.
FIGURE 5.
Macrophages overexpressing Smyd2 suppress Th-17 but promote Treg cell differentiation. MCSF-driven bone marrow-derived macrophages were transfected with mutant control (Mut) or Smyd2 plasmid (Smyd2) and then stimulated for 4 h with LPS. A, the macrophages were cocultured with purified CD4+CD25− T cells (1:1) under suboptimal Th-17 differentiation conditions. After 4 days of culture, Th-17 cell differentiation was evaluated by flow cytometry gated on CD4+ T cells (left panel) or ELISA detection of IL-17 (right panel). B, the macrophages were cocultured under suboptimal Treg differentiation conditions. Three days later, iTreg cell differentiation was evaluated by flow cytometry detection of Foxp3 (left panel). The expression of TGF-β1 mRNA in cocultured macrophages transfected with the mutant or Smyd2 plasmid (right panel) was determined by Q-PCR. *, p < 0.05. Data are mean ± S.D. of technical replicates from one of three independent experiments. The experiments were performed at least three times with similar results obtained.
Collectively, our results show that macrophages with enhanced Smyd2 expression promote Treg cell and suppress Th-17 cell differentiation through inhibition of IL-6 and up-regulation of TGF-β. These data suggest that Smyd2 participates in the negative regulation of adaptive immune responses induced by regulating macrophage activation.
DISCUSSION
Recent studies have shown that epigenetic modification is very important to macrophage activation (17, 37, 38). In this study, we found that Smyd2, a lysine methyltransferase, plays critical roles in TLR4-induced macrophage activation and proinflammatory cytokine secretion. Our data show that H3K36 dimethylation regulated by Smyd2 suppresses TNF-α and IL-6 expression by directly interacting with the promoter regions of chromatin-remodeling events. Furthermore, overexpressed Smyd2 in macrophages impairs the activation of the NF-κB and ERK signal pathways. These data indicate that Smyd2 participates in macrophage activation, especially in proinflammatory classical M1 macrophages, as a negative regulator. Finally, we found that macrophages with altered Smyd2 expression exhibit an immune-regulatory property that suppresses Th-17 and promotes regulatory T cell differentiation. It has been reported that multiple mechanisms participate in macrophage activation and functional modulation, such as signal transduction, transcription factor activation, and epigenetic modification. Significantly, here we found that the dimethylation of H3K36 by Smyd2 only alters the production of TNF-α and IL-6. However, this epigenetic modification does not change other typical marker genes of M1 macrophage, such as IL-1β and IL-12, nor does it have an impact on the expression of M2 genes, including IL-10, Arg1, and Ym1 (data not shown). These data demonstrate a novel regulatory mechanism by which epigenetic modification operates at specific modules of gene expressing programs, fine-tuning macrophage functional differentiation and polarization.
Smyd2 methylates H3K4 and H3K36 sites in histone under different conditions (24, 26). When interacting with HSP90α, Smyd2 prefers to modify H3K4 and promotes target gene transcription. On the contrary, in the absence of HSP90α, Smyd2 targets dimethylation of H3K36 and suppresses gene expression. In the process of TLR4-triggered macrophage activation, our data show that enhanced expression of Smyd2 inhibits TNF-α and IL-6 transcription, indicating that it targets H3K36 dimethylation. Furthermore, we find that overexpression of Smyd2 reverses the pattern of H3K4 and H3K36 methylation in the promoters of TNF-α and IL-6, suggesting that, in macrophage activation induced by TLR4, Smyd2 targets H3K36 dimethylation independently of HSP90α. On the other hand, the NF-κB and MAPK signaling pathways play critical roles in TLR4-triggered macrophage activation. By dissecting the signaling pathways in macrophage polarization, we found that overexpression of Smyd2 suppresses the signal transduction of both the NF-κB and ERK pathways. The data indicate that Smyd2 not only has enzymatic activity on histone modification but that it also influences signal transduction. This can be explained because, similar to the role of Smyd2 in p53 and retinoblastoma activity (27, 39), Smyd2 may target some key transcription factors or coactivators through protein-protein interaction. This could be the next interesting question to answer: how does Smyd2 participate in the regulation of NF-κB and ERK signal transduction?
Macrophages are important members of phagocytes and antigen-presenting cells that integrate innate and adaptive immunity by different molecular mechanisms with remarkable functional complexity and plasticity (12, 34, 40, 41). Our findings demonstrate that enhanced expression of Smyd2 suppresses the expression of costimulatory molecules on macrophages, such as CD80, CD86, CD40, and MHC II, leading to impaired T cell activation. As antigen-presenting cells, an important function of macrophages is to help CD4 T cells differentiate into specific helper T cell subsets under specific conditions. We found that enhanced Smyd2 expression changes the properties of macrophages by suppression of IL-6 and TNF-α production and promotion of TGF-β secretion, consequently resulting in inhibited Th-17 cell and promoted Treg cell differentiation. In summary, this study demonstrates that Smyd2 acts as a negative regulator in proinflammatory macrophage activation through epigenetic modification and results in a rebalance of Th-17 and Treg cells, which provides mechanistic insights into the epigenetic modulation of immune responses and inflammation, providing new insights, strategies, or targets for understanding autoimmune disease and their therapy.
Footnotes
- TLR
- Toll-like receptor
- H3K4
- histone 3 lysine 4
- H3K36
- histone 3 lysine 36
- H3K4me3
- histone 3 lysine 36 methyltransferase 2
- H3K4me3
- histone 3 lysine 4 methyltransferase 3
- H3K27
- histone 3 lysine 27
- IKK
- IκB kinase
- Treg
- regulatory T cell
- iTreg
- induced regulatory T cell
- MCSF
- macrophage colony stimulation factor
- Q-PCR
- quantitative PCR.
REFERENCES
- 1. Kugelberg E. (2014) Innate immunity: a wee protection. Nat. Rev. Immunol. 14, 359. [DOI] [PubMed] [Google Scholar]
- 2. Bordon Y. (2012) Innate immunity: bitter enemies. Nat. Rev. Immunol. 12, 746. [DOI] [PubMed] [Google Scholar]
- 3. Papatriantafyllou M. (2012) Innate immunity: inflammasome triggered by cell swelling. Nat. Rev. Immunol. 12, 742. [DOI] [PubMed] [Google Scholar]
- 4. Takeda K., Kaisho T., Akira S. (2003) Toll-like receptors. Annu. Rev. Immunol. 21, 335–376 [DOI] [PubMed] [Google Scholar]
- 5. O'Neill L. A., Golenbock D., Bowie A. G. (2013) The history of Toll-like receptors: redefining innate immunity. Nat. Rev. Immunol. 13, 453–460 [DOI] [PubMed] [Google Scholar]
- 6. Kawai T., Akira S. (2010) The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11, 373–384 [DOI] [PubMed] [Google Scholar]
- 7. Kumar H., Kawai T., Akira S. (2009) Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun. 388, 621–625 [DOI] [PubMed] [Google Scholar]
- 8. Kawai T., Akira S. (2011) Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34, 637–650 [DOI] [PubMed] [Google Scholar]
- 9. Kawai T., Akira S. (2007) Signaling to NF-κB by Toll-like receptors. Trends Mol. Med. 13, 460–469 [DOI] [PubMed] [Google Scholar]
- 10. Arthur J. S., Ley S. C. (2013) Mitogen-activated protein kinases in innate immunity. Nat. Rev. Immunol. 13, 679–692 [DOI] [PubMed] [Google Scholar]
- 11. Alderton G. K. (2014) Tumour immunology: turning macrophages on, off and on again. Nat. Rev. Immunol. 14, 136–137 [DOI] [PubMed] [Google Scholar]
- 12. Murray P. J., Wynn T. A. (2011) Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 11, 723–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gordon S. (2003) Alternative activation of macrophages. Nat. Rev. Immunol. 3, 23–35 [DOI] [PubMed] [Google Scholar]
- 14. Gordon S., Martinez F. O. (2010) Alternative activation of macrophages: mechanism and functions. Immunity 32, 593–604 [DOI] [PubMed] [Google Scholar]
- 15. Martinez F. O., Helming L., Gordon S. (2009) Alternative activation of macrophages: an immunologic functional perspective. Annu. Rev. Immunol. 27, 451–483 [DOI] [PubMed] [Google Scholar]
- 16. Lawrence T., Natoli G. (2011) Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat. Rev. Immunol. 11, 750–761 [DOI] [PubMed] [Google Scholar]
- 17. Takeuch O., Akira S. (2011) Epigenetic control of macrophage polarization. Eur. J. Immunol. 41, 2490–2493 [DOI] [PubMed] [Google Scholar]
- 18. Goldberg A. D., Allis C. D., Bernstein E. (2007) Epigenetics: a landscape takes shape. Cell 128, 635–638 [DOI] [PubMed] [Google Scholar]
- 19. Koch M. W., Metz L. M., Kovalchuk O. (2013) Epigenetic changes in patients with multiple sclerosis. Nat. Rev. Neurol. 9, 35–43 [DOI] [PubMed] [Google Scholar]
- 20. Kubicek S., Jenuwein T. (2004) A crack in histone lysine methylation. Cell 119, 903–906 [DOI] [PubMed] [Google Scholar]
- 21. Wei G., Wei L., Zhu J., Zang C., Hu-Li J., Yao Z., Cui K., Kanno Y., Roh T. Y., Watford W. T., Schones D. E., Peng W., Sun H. W., Paul W. E., O'Shea J. J., Zhao K. (2009) Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30, 155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Satoh T., Takeuchi O., Vandenbon A., Yasuda K., Tanaka Y., Kumagai Y., Miyake T., Matsushita K., Okazaki T., Saitoh T., Honma K., Matsuyama T., Yui K., Tsujimura T., Standley D. M., Nakanishi K., Nakai K., Akira S. (2010) The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 11, 936–944 [DOI] [PubMed] [Google Scholar]
- 23. Xia M., Liu J., Wu X., Liu S., Li G., Han C., Song L., Li Z., Wang Q., Wang J., Xu T., Cao X. (2013) Histone methyltransferase Ash1l suppresses interleukin-6 production and inflammatory autoimmune diseases by inducing the ubiquitin-editing enzyme A20. Immunity 39, 470–481 [DOI] [PubMed] [Google Scholar]
- 24. Brown M. A., Sims R. J., 3rd, Gottlieb P. D., Tucker P. W. (2006) Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol. Cancer 5, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abu-Farha M., Lanouette S., Elisma F., Tremblay V., Butson J., Figeys D., Couture J. F. (2011) Proteomic analyses of the SMYD family interactomes identify HSP90 as a novel target for SMYD2. J. Mol. Cell Biol. 3, 301–308 [DOI] [PubMed] [Google Scholar]
- 26. Abu-Farha M., Lambert J. P., Al-Madhoun A. S., Elisma F., Skerjanc I. S., Figeys D. (2008) The tale of two domains: proteomics and genomics analysis of SMYD2, a new histone methyltransferase. Mol. Cell. Proteomics 7, 560–572 [DOI] [PubMed] [Google Scholar]
- 27. Huang J., Perez-Burgos L., Placek B. J., Sengupta R., Richter M., Dorsey J. A., Kubicek S., Opravil S., Jenuwein T., Berger S. L. (2006) Repression of p53 activity by Smyd2-mediated methylation. Nature 444, 629–632 [DOI] [PubMed] [Google Scholar]
- 28. Cho H. S., Hayami S., Toyokawa G., Maejima K., Yamane Y., Suzuki T., Dohmae N., Kogure M., Kang D., Neal D. E., Ponder B. A., Yamaue H., Nakamura Y., Hamamoto R. (2012) RB1 methylation by SMYD2 enhances cell cycle progression through an increase of RB1 phosphorylation. Neoplasia 14, 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. An H., Hou J., Zhou J., Zhao W., Xu H., Zheng Y., Yu Y., Liu S., Cao X. (2008) Phosphatase SHP-1 promotes TLR- and RIG-I-activated production of type I interferon by inhibiting the kinase IRAK1. Nat. Immunol. 9, 542–550 [DOI] [PubMed] [Google Scholar]
- 30. Lin L., Hou J., Ma F., Wang P., Liu X., Li N., Wang J., Wang Q., Cao X. (2013) Type I IFN inhibits innate IL-10 production in macrophages through histone deacetylase 11 by downregulating microRNA-145. J. Immunol. 191, 3896–3904 [DOI] [PubMed] [Google Scholar]
- 31. Chen Z. J. (2012) Ubiquitination in signaling to and activation of IKK. Immunol. Rev. 246, 95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kawai T., Akira S. (2007) TLR signaling. Semin. Immunol. 19, 24–32 [DOI] [PubMed] [Google Scholar]
- 33. Gürtler C., Carty M., Kearney J., Schattgen S. A., Ding A., Fitzgerald K. A., Bowie A. G. (2014) SARM regulates CCL5 production in macrophages by promoting the recruitment of transcription factors and RNA polymerase II to the Ccl5 promoter. J. Immunol. 192, 4821–4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chawla A., Nguyen K. D., Goh Y. P. (2011) Macrophage-mediated inflammation in metabolic disease. Nat. Rev. Immunol. 11, 738–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gause W. C., Wynn T. A., Allen J. E. (2013) Type 2 immunity and wound healing: evolutionary refinement of adaptive immunity by helminths. Nat. Rev. Immunol. 13, 607–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Littman D. R., Rudensky A. Y. (2010) Th17 and regulatory T cells in mediating and restraining inflammation. Cell 140, 845–858 [DOI] [PubMed] [Google Scholar]
- 37. Yang X., Wang X., Liu D., Yu L., Xue B., Shi H. (2014) Epigenetic regulation of macrophage polarization by DNA methyltransferase 3b. Mol. Endocrinol. 28, 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ivashkiv L. B. (2013) Epigenetic regulation of macrophage polarization and function. Trends Immunol. 34, 216–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang J., Sengupta R., Espejo A. B., Lee M. G., Dorsey J. A., Richter M., Opravil S., Shiekhattar R., Bedford M. T., Jenuwein T., Berger S. L. (2007) p53 is regulated by the lysine demethylase LSD1. Nature 449, 105–108 [DOI] [PubMed] [Google Scholar]
- 40. Bordon Y. (2013) Macrophages: metabolic master prompts a change of tack. Nat. Rev. Immunol. 13, 706. [DOI] [PubMed] [Google Scholar]
- 41. Moore K. J., Sheedy F. J., Fisher E. A. (2013) Macrophages in atherosclerosis: a dynamic balance. Nat. Rev. Immunol. 13, 709–721 [DOI] [PMC free article] [PubMed] [Google Scholar]