Abstract
TACC2 is a member of the transforming acidic coiled-coil-containing protein family and is associated with the centrosome-spindle apparatus during cell cycling. In vivo, the TACC2 gene is expressed in various splice forms predominantly in postmitotic tissues, including heart, muscle, kidney, and brain. Studies of human breast cancer samples and cell lines suggest a putative role of TACC2 as a tumor suppressor protein. To analyze the physiological role of TACC2, we generated mice lacking TACC2. TACC2-deficient mice are viable, develop normally, are fertile, and lack phenotypic changes compared to wild-type mice. Furthermore, TACC2 deficiency does not lead to an increased incidence of tumor development. Finally, in TACC2-deficient embryonic fibroblasts, proliferation and cell cycle progression as well as centrosome numbers are comparable to those in wild-type cells. Therefore, TACC2 is not required, nonredundantly, for mouse development and normal cell proliferation and is not a tumor suppressor protein.
The mitotic cell cycle is a precisely ordered series of processes that includes the formation, function, and dissolution of the mitotic spindle. Members of the transforming acidic coiled-coil-containing (TACC) family of proteins have been shown to be components of the centrosome-spindle apparatus (reviewed in reference 6). The family is characterized by a highly conserved coiled-coil domain that is essential for localization to the centrosome-spindle apparatus. There is a single TACC gene in Drosophila melanogaster, dTACC, that is hypothesized to play an essential role in stabilizing the centrosome-spindle structure (7, 15). Localization to the spindle requires the Drosophila Aurora A kinase (9), a kinase required for centrosome maturation (11). Mutants lacking dTACC have abnormally short and weak spindle and astral microtubules, a condition leading to chromosomal missegregation. A Xenopus laevis TACC family member, termed Maskin, has also been extensively studied (2, 10, 23). In this case, it is proposed that Maskin is involved in sequestering of mRNA complexes to the spindle and suppression of their translation. This process is mediated through the association of Maskin with a cytoplasmic polyadenylation element binding protein and with eIF4E. Cells injected with antibodies against Maskin have mitotic defects including disruption of the centrosome-spindle apparatus. More recently, a distantly related TACC protein was identified in Caenorhabditis elegans, TAC-1 (14, 22), which has been shown to regulate the growth and assembly of microtubules.
The mammalian TACC proteins (TACC1, TACC2, and TACC3) have only recently been identified. TACC1, the first family member identified, was discovered as the product of a gene that is amplified in breast cancer (24) and more recently was also shown to be overexpressed in prostate cancer (4). TACC3 was identified independently as an Ah receptor nuclear translocator protein-interacting protein and a putative player in hypoxic responses (21), the product of an erythropoietin-induced gene in hematopoietic progenitors (17), and a Stat5-interacting protein in a yeast two-hybrid screen (19). Initially, an amino-terminal splice variant of TACC2 was identified as the product of a gene with reduced expression in metastatic breast cancer cells (3) and subsequently as the product of a gene that is induced in human microvascular endothelial cells by erythropoietin (20). In the first situation, overexpression studies suggested that TACC2 was a tumor suppressor protein, while in the later case, a role in blood vessel formation was proposed.
The physiological roles of TACC3 have been recently addressed through the derivation of mice in which the TACC3 gene is disrupted (19). Deletion of TACC3 results in embryonic lethality at midgestation, with a number of cell types being affected. Among the lineages, the hematopoietic ones are some of the most profoundly affected, and this most likely results in failure to produce sufficient red cells to support embryo growth. Although the ability of hematopoietic stem cells to differentiate was largely unaltered, there was a very high rate of apoptosis, resulting in the lack of expansion of essential progenitors or lineage-committed cells. The inability to expand sufficiently for hematopoietic lineage functions was rescued by introduction of the TACC3 deficiency onto a p53 deficiency. These data demonstrated that TACC3 is involved in controlling the activity of p53 during the mitotic phase of the cell cycle, although this control does not appear to be dependent upon the direct sequestration of p53 by TACC3. Importantly, studies of TACC3- and p53-deficient hematopoietic cells failed to identify significant chromosome missegregation or altered centrosome replication.
In the studies described here, we have further pursued the roles of the mammalian TACC proteins by disrupting the gene for TACC2. Unlike TACC3 deficiency, TACC2 deficiency does not create any phenotypic alterations that would provide insight into the protein's potential functions, if any, in mammals.
MATERIALS AND METHODS
Northern blot analysis.
Northern blots with mRNAs from murine tissues were obtained from Ambion (Austin, Tex.). Total RNA from various tissues was prepared using RNAzol B reagent by following the protocol of the manufacturer (Tel Test, Friendswood, Tex.). Twenty micrograms of total RNA per lane was separated on 1% formaldehyde agarose gels. cDNA fragments of murine TACC1, TACC2, and TACC3 genes and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as a loading control were used as probes after being labeled with [α-32P]dCTP by using a random priming labeling kit (Amersham, Arlington Heights, Ill.). Membranes were hybridized using a rapid hybridization solution (Amersham) followed by final stringent washes in 0.2× SSC (1× SSC is 0.15 M sodium chloride plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate at 65°C.
Construction of the TACC2 targeting vector, ES cell screening using Southern blotting, and generation of TACC2 mutant mice.
The TACC2 gene was isolated from a 129 mouse/RW4 embryonic stem (ES) cell genomic library (Incyte Genomics, Palo Alto, Calif.) by using a mouse TACC2 cDNA probe. A 10-kb EcoRI fragment identified by Southern blotting was subcloned into pBluescript II SK(+). For the TACC2 targeting vector, a 7.5-kb AflII fragment containing part of exon 4 and exons 5 to 7 was replaced with a neomycin resistance cassette previously described (27). A diphtheria toxin A cassette mediating negative selection (1) was inserted into the 5′ end of the TACC3-neomycin gene construct. Twenty micrograms of the NotI-linearized targeting vector was electroporated into embryonic day 14 ES cells, and cells were grown under selection conditions with 350 μg of Geneticin (G418; Gibco, Rockville, Md.)/ml. Clones were picked and expanded 7 to 9 days after electroporation. Correctly targeted ES clones were identified by Southern analysis using a genomic probe including exons 11 and 12 (see Fig. 2B), and karyotypically normal clones were injected into blastocysts. Mice derived from two ES clones were analyzed in detail, and identical phenotypes were observed. Conditions for injection of ES cells into blastocysts and breeding to generate mice homozygous for the mutated TACC3 gene were as described previously (28).
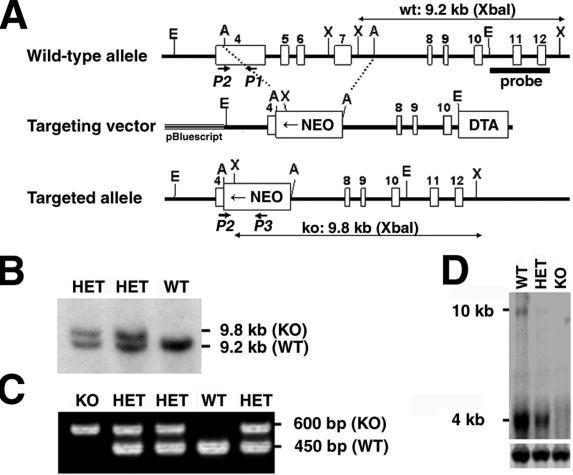
FIG. 2.
Targeted disruption of the TACC2 gene. (A) Schematic structure of and targeting strategy for the murine TACC2 locus. Empty boxes represent exons 4 to 12 of the murine TACC2 gene. The locations of the 5′ external probe and PCR primers for genotyping are indicated. Restriction enzyme sites are as follows: E, EcoRI; A, AflII; X, XbaI. wt, wild type; NEO, neomycin resistance cassette; DTA, diphtheria toxin A cassette; ko, knockout. Confirmation of successful targeting of ES cells was obtained by Southern blot analysis (B) and PCR screening of F1 mice for the presence of the disrupted allele (C). WT, wild type; KO, knockout; HET, heterozygote. (D) Northern blot analysis of kidney tissue with a TACC2-specific probe shows no hybridization signal in knockout tissue. Note that disruption of the TACC2 gene led to the complete absence of both the 4- and 10-kb transcripts, therefore generating a null mutation for all transcript variants. Hybridization for actin (lower panel) was used to control RNA loading. The positions of transcript sizes are indicated.
Genotyping of TACC2-deficient mice by genomic PCR.
Genomic DNA was amplified in 50-μl reaction mixtures using 2.5 U of AmpliTaq Gold (Perkin-Elmer, Branchburg, N.J.) in PCR buffer with a final concentration of deoxynucleotide triphosphates of 0.2 mM and a final concentration of MgCl2 of 2 mM. The PCR primers consisted of P1 primer (5′-CCTCAGAATAGTCAAACTCCAGC-3′) at 0.2 μM, P2 primer (5′-GGATCCAACAGTGCTTCCAGC-3′) at 0.3 μM, and the neomycin primer P3 (5′-ATCTCCTGTCATCTCACCTTGCT-3′) at 0.3 μM. The PCR cycle profile was as follows: 1 cycle at 94°C for 10 min followed by 35 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min with a 5-s autoextension in every cycle and finally 1 cycle at 72°C for 10 min. A 450-bp fragment indicated the presence of the wild-type allele, whereas a 600-bp fragment was amplified from the mutated allele.
Histology.
Tissues from 4-week-old wild-type and TACC2-deficient mice were collected and fixed in 10% neutral buffered formalin. Tissue samples were processed routinely and embedded in paraffin, sectioned at 4 μm, stained with hematoxylin and eosin, and analyzed by light microscopy.
Irradiation and checkpoint activation in neurons.
Pups 5.5 days old were irradiated with 18 Gy from a cesium irradiator at a rate of 0.9 Gy/min. After 4 or 6 h, the animals were killed. Tissues were collected after transcardial perfusion with 4% paraformaldehyde in phosphate-buffered saline (PBS). Fixed tissues were cryoprotected in 25% buffered sucrose solution and cryosectioned at 12 μm with a HM500 M cryostat (MICROM, Walldorf, Germany). Staining was performed with 1% neutral red (Aldrich Chemical, Milwaukee, Wis.) in 0.1 M acetic acid, pH 4.8, for 1 min, followed by dehydration in ethanol.
Cell culture and proliferation of MEFs.
Mouse embryonic fibroblasts (MEFs) were derived from 13.5-day-old embryos by using a 3T9 protocol based on a strategy of Todaro and Green (26). Following removal of the head and organs, embryos were rinsed with PBS, minced, and digested with trypsin (0.05% solution containing 0.53 mM EDTA) for 10 min at 37°C. Trypsin was inactivated by addition of Dulbecco modified Eagle medium containing 10% fetal bovine serum and 2 mM glutamine, 0.1 mM minimal essential medium nonessential amino acids, 55 μM 2-mercaptoethanol, and 10 mg of gentamicin/ml. Cells from single embryos were plated into two 60-mm-diameter in-culture dishes (Becton Dickinson, Bedford, Mass.) and incubated at 37°C in a 10% CO2 humidified chamber. Cells were maintained on a defined schedule (9 × 105 cells per 60-mm-diameter dish passaged every 3 days). Plating after disaggregation of embryos was considered to be passage 1, and the first replating 3 days later was considered to be passage 2. Growth curves at passages 2 and 6 were initiated with replicate cultures of 3 × 105 cells per T-25 tissue culture flask (Falcon, Lincoln Park, N.J.). The cell numbers from triplicate cultures were determined daily by automated counting after trypsinization (Beckman, Fullerton, Calif.).
Cell cycle analysis.
Primary MEF cells of passage 3 were used for cell cycle analysis assays. The cells were seeded onto 6-cm-diameter plates at a density of 3 × 105 cells/plate. All cells were cultured in complete medium over a period of 16 h and then starved for 24 h (in 0.1% fetal calf serum [FCS]). Starved cells were analyzed at this point. The growing fraction was analyzed after addition of 10% FCS-containing medium for another 24 h. In parallel, one fraction of these cells underwent treatment with 250 ng of nocodazole/ml for the same period of time and was analyzed afterwards. For determination of cell cycle distribution, cells were stained with propidium iodide (50 μg/ml in 0.1% sodium citrate-0.1% Triton X-100) and treated with 2 μg of RNase/ml for 30 min. The DNA content was assayed by flow cytometry (Becton Dickinson, Bedford, Mass.), and the percentages of cells within the different phases of the cell cycle were determined using ModFit software (Verify Software).
Determination of centrosome numbers.
Exponentially growing MEFs on four-well culture slides (Becton Dickinson) were fixed with a 1:1 mixture of methanol and acetone for 20 min at −20°C. The slides were blocked in PBS containing 10% FCS for 1 h, and cells were stained with mouse anti-γ-tubulin (1:500; Sigma, St. Louis, Mo.) for 1 h at room temperature. A cyanine-coupled (cy-3) anti-mouse antibody (Jackson Immunoresearch, West Grove, Pa.) was used as a secondary antibody (1:100). DNA was visualized in parallel by staining with DAPI (4′,6′-diamidino-2-phenylindole; Sigma Aldrich, St. Louis, Mo.). Slides were mounted with Permouth (Fisher Scientific, Pittsburgh, Pa.). Analysis was performed by confocal microscopy using a DM-IRBE microscope (Leica, Exton, Pa.) together with Leica TCS-NT software.
RESULTS
Tissue distribution of TACC family members.
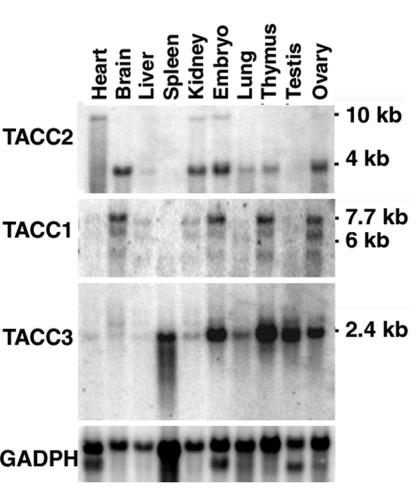
To begin to explore the possible biological functions of TACC2, its expression pattern was analyzed in adult tissues and compared with those of other family members. As illustrated in Fig. 1, a major 4-kb transcript was detected in brain, kidney, lung, thymus, and ovary as well as in whole embryo RNA. In addition, there was a minor 10-kb transcript detected in heart, kidney, and embryos. Previous studies indicated that a moderate level of expression of both TACC2 mRNAs can be detected throughout mouse embryogenesis (19) and in human fetal tissues (12). In general, the levels of transcripts for TACC2 were considerably less than those for TACC3, which is expressed in hematopoietic tissues, testis, and ovary and during embryonic development. Like TACC2, TACC1 is generally expressed at lower levels than TACC3. Expression was seen predominantly in brain, kidney, thymus, and ovary and during embryonic development. In summary, the TACC family members show both unique and overlapping patterns of mRNA expression. We have also attempted to examine TACC2 protein levels in various tissues. However, although all available antibodies readily detect overexpressed protein in 293T cells, immunoprecipitation and Western blotting with these reagents have failed to detect TACC2 protein in normal tissues (data not shown). We conclude that the protein is expressed at very low levels and/or is unstable.
FIG. 1.
Tissue distribution of TACC transcripts in the mouse. Northern blot analysis was performed with poly(A)+ RNA samples from different murine tissues. The blot was subsequently hybridized with probes specific for TACC2 (top panel), TACC1 (second panel), and TACC3 (third panel). Hybridization for GAPDH (fourth panel) was used to control RNA loading. Positions of standard markers and TACC transcript sizes in kilobases are indicated.
Generation of TACC2-deficient mice.
To assess the physiologic role of TACC2, we generated mice lacking TACC2. Most of exon 4 and all of exons 5 to 7 of the TACC2 gene were replaced by a neomycin resistance cassette in the targeting construct (Fig. 2A). The targeting vector was introduced into embryonic day 14 ES cells, and correctly targeted ES clones were identified by Southern blot analysis (Fig. 2B). Four independent heterozygous ES clones were used to generate chimeric mice that transmitted the mutated gene through the germ line. TACC2 heterozygous mice were interbred to produce homozygous mutant mice. All offspring were genotyped by PCR (Fig. 2C). Northern blot analysis of mutant animals confirmed the presence of lower levels of expression in heterozygous mice and the absence of any TACC2 transcripts in the homozygous TACC2-deficient mice, establishing this mouse strain as a TACC2 null mutant (Fig. 2D).
Normal development and lack of tumors in TACC2-deficient mice.
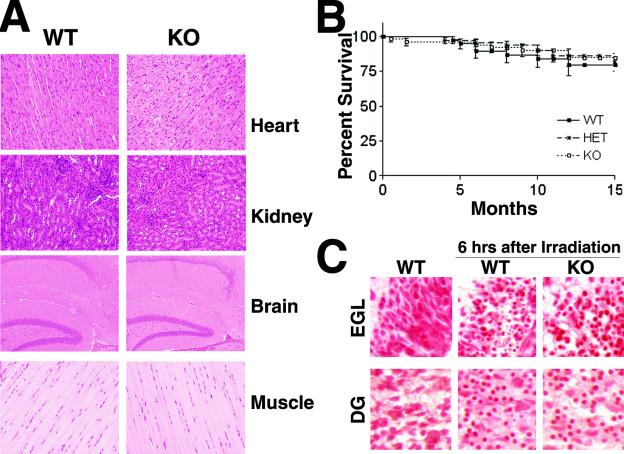
Interbreeding of heterozygous mice produced offspring with an expected Mendelian ratio that were indistinguishable from littermate controls in terms of growth and development. In addition, no abnormalities were observed in mutant mice with regard to their reproductive abilities and fosterage. Evaluation of histological sections of various tissues expressing the highest levels of TACC2 mRNA (muscle, kidney, heart, and brain) failed to reveal any pathological abnormalities (Fig. 3A). TACC2-deficient mice showed normal survival over a period of at least 15 months compared to wild-type and heterozygous control groups (Fig. 3B). None of the aging mutant mice developed tumors as judged by macroscopic and microscopic evaluations of various tissues (data not shown). These observations do not support suggestions that TACC2 is a tumor suppressor for breast cancer (3, 12).
FIG. 3.
Normal development and lack of pathology in TACC2-deficient mice. (A) Histological appearance of tissue sections from 8-week-old TACC2 wild-type (WT) and knockout (KO) mice. Representative paraffin-embedded and hematoxylin- and eosin-stained sections of heart, muscle, kidney, and brain are shown (magnification, ×20). (B) Survival curve of TACC2-deficient mice. The Kaplan-Meyer plot represents observed TACC2-deficient animals (n = 54) compared to their heterozygous (HET; n = 64) and wild-type (n = 37) littermates. The interval of observation was up to 15 months. Percentages of surviving mice are shown as a function of time. (C) Normal irradiation-induced checkpoint activation in neural cells from TACC2-deficient mice. Pups 5.5 days old were irradiated with 18 Gy and analyzed for induction of apoptosis by staining with neutral red. Stainings from nonirradiated wild-type brains are shown in the left panels. The middle and right panels show wild-type and TACC2-deficient tissues, respectively, 6 h after γ-irradiation. The external granular layer of the cerebellum (EGL) and the dentate gyrus (DG) are shown.
Lack of influence of TACC2 deficiency on cell proliferation and checkpoint function.
TACC2 is a protein which constitutively localizes at the centrosome and interacts with microtubules (8), similar to TACC1 and TACC3. Since TACC3 is a critical regulator in cell cycle progression and its absence is associated with the induction of a p53-dependent apoptotic response (19), we further explored the possibility that TACC2 may play a comparable role in cells in which it is expressed. Since TACC2 is predominately expressed in the brain, we determined whether TACC2-deficient neuronal cells would be more susceptible to DNA damage. In particular, brains of day 5.5 neonatal mice have areas containing both mitotic and postmitotic cell populations (16). Irradiation of these areas leads to a characteristic induction of rapid apoptotic cell death as visualized by the occurrence of cells with pycnotic nuclei. As shown in Fig. 3C, this response was unaltered in TACC2 mutant animals. In particular, no differences could be detected in the level of apoptosis in either mitotic or postmitotic parts of the brain (the dentate gyrus and the extragranular layer) compared to the levels in brain sections from wild-type mice. Therefore, TACC2-deficient cells showed no change in the frequency of induction of apoptosis in either the proliferating or postmitotic stage.
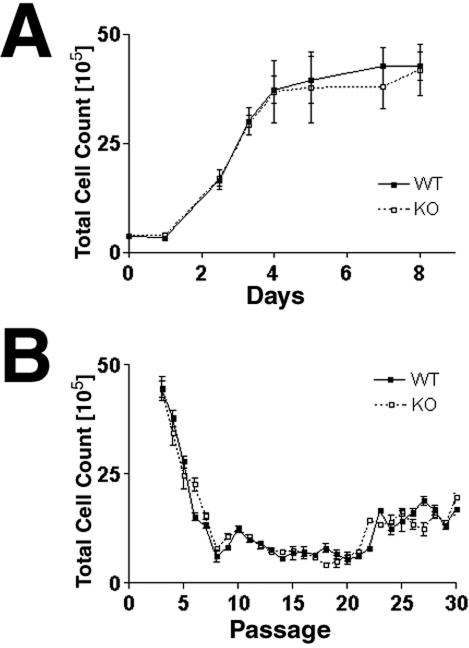
MEFs express TACC2 (data not shown), and therefore we examined the properties of MEFs derived from TACC2-deficient embryos. TACC2-deficient MEFs could be easily established ex vivo with proliferation kinetics indistinguishable from those of control cells (Fig. 4A). Furthermore, mutant MEFs derived on a 3T9 protocol (26) progressively lost proliferation capacity, underwent a senescence crisis comparable to that of wild-type control cells (Fig. 4B), and eventually gave rise to transformed cell lines at a frequency comparable to that observed with wild-type cells. Lastly, although TACC2 was expressed only at low levels in hematopoietic tissues, T- and B-cell development was unaltered and TACC2-deficient cells proliferated in response to antigen receptor engagement and cytokines comparably to wild-type cells (data not shown).
FIG. 4.
Normal proliferation and senescence of TACC2-deficient MEFs. (A) Proliferation assays using passage 2 MEFs. Cells from wild-type (WT) and TACC2 knockout (KO) embryos were seeded at 3 × 105 cells per T-25 flask. Duplicate dishes were harvested at daily intervals, and the total number of cells per culture was determined. The graph shows representative results from one of three experiments. (B) Cell proliferation on a 3T9 protocol. At 3-day intervals, the total numbers of wild-type and TACC2-deficient cells per culture dish were determined prior to redilution of the cells to a density of 9 × 105 cells per 60-mm-diameter dish for repassaging. Bars indicate standard errors of the mean. Representative results from one of three experiments are shown.
Normal centrosome numbers and lack of aneuploidy in TACC2-deficient cells.
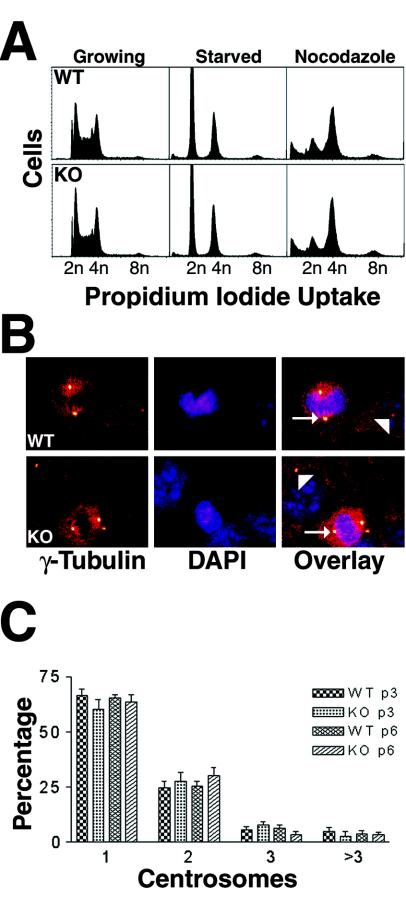
In Drosophila, dTACC plays an essential role in normal mitotic spindle function during embryogenesis (7) and its absence results in chromosomal segregation defects. We therefore determined whether the absence of TACC2 would be associated with aneuploidy, as assessed by DNA content, in late-passage MEFs (Fig. 5A) or in expanded T-cell populations (data not shown). In neither case was there evidence of aneuploidy. In addition, we treated MEFs with nocodazole for 24 h to destabilize the mitotic spindle and induce cells to arrest at the G2/M phase of the cell cycle. Primary TACC2-deficient MEFs arrested comparably to wild-type fibroblasts in response to nocodazole inhibition.
FIG. 5.
(A) DNA content analysis of proliferating and starved MEF cells at passage 3. Cells were starved for a 24-h period (in 0.1% serum) and thereafter restimulated with 10% serum (growing) or treated with nocodazole for 20 h. After staining with propidium iodide, the DNA content was measured. Representative results from one analysis of cells of each genotype are shown. WT, wild type; KO, knockout. (B) Normal centrosome numbers in TACC2-deficient MEF cells. Fixed wild-type and knockout MEFs were stained with anti-γ-tubulin antibodies (left), and DNA was visualized using DAPI (middle). Normal bipolar centrosomes are visible as yellow spots (arrowheads). The panel on the right shows an overlay of both stainings. The arrows indicate cells in metaphase. (C) Centrosome numbers in wild-type and knockout MEFs from passages 3 (p3) and 6 (p6). Each column represents two different cell lines. For each cell line, at least 1,800 cells were counted at a magnification of ×20.
Lastly, we examined the number of centrosomes in interphase and mitotic cells by immunostaining (Fig. 5B) and quantification (Fig. 5C) of γ-tubulin, a marker for centrosomes (18), and we also examined the mitotic spindle architecture by α-tubulin immunostaining (data not shown). TACC2 mutant cells displayed no significant differences in either dividing cells (>20 metaphases analyzed) or interphase cells compared to wild-type control cells. Furthermore, there were no signs of malfunctions in the spindle apparatus when cells were stained with anti-α-tubulin antibodies (data not shown). Collectively, these findings indicate that TACC2 is dispensable for centrosome duplication and mitotic spindle function.
DISCUSSION
The mammalian family of TACC proteins consists of TACC1, TACC2, and TACC3. Interestingly, each of the TACC family member genes chromosomally colocalizes with one of the fibroblast growth factor receptor genes (25) and, while there are four fibroblast growth factor receptor genes, a fourth TACC gene has yet to be identified. As illustrated in our work and in previous studies, each of the TACC family members has a unique pattern of expression that only partially overlaps with those of other family members. TACC2, in contrast to TACC3, is primarily expressed in postmitotic tissues, with the highest levels of expression in the brain. As noted above, numerous attempts have failed to find detectable levels of TACC2 protein, in contrast to TACC3 protein, which is readily detectable. This is consistent with the considerably low levels of TACC2 RNA relative to those of TACC3 RNA but may also indicate that the TACC2 protein is more unstable and/or alternatively expressed at low levels in vivo.
TACC2 has been identified independently in several laboratories and has been hypothesized to have several different biological functions. In one laboratory (3), a splice variant of the TACC2 gene was proposed to be a breast cancer tumor suppressor gene based on the observations that it was downregulated in more malignant cell clones and that, when overexpressed in cell lines, it could suppress cell growth and metastatic properties (3). The TACC2 gene was also identified as a gene that is induced in cultured microvascular endothelial cells in response to erythropoietin (20) and proposed to be an important component in the control of blood vessel growth and development. The detection of interaction of TACC2 with the nuclear histone acetyltransferase hGCN5L2 has led to the suggestion that it may function in the regulation of gene transcription (5), although TACC2 has been suggested to be in other complexes (13). The lack of any phenotypic changes when the TACC2 gene is deleted in mice demonstrates that TACC2 lacks any critical, nonredundant functions and fails to provide support for any of the proposed functions.
The lack of any phenotypic consequences of deleting TACC2 may indicate that it has functions that are redundantly provided by other family members. We consider this possibility somewhat unlikely since each of the family members has a number of specific sites of expression and, in the tissues we have examined, the deletion of TACC2 does not result in the activation or increased expression of the other family members. We also consider the possibility unlikely since there is little sequence similarity among the TACC family members outside of the coiled-coil domain at the C terminus. Nevertheless, to begin to address the issue of redundancy more definitively, we have introduced the TACC2 deficiency onto the TACC3 deficiency. Since TACC3 deficiency causes embryonic lethality at midgestation and affects a variety of cell types, we wished to assess whether the combined deficiency would result in earlier or more profound embryonic lethality. Characterization of a number of embryos from midgestation that were homozygously deficient for both TACC2 and TACC3 has failed to detect any differences from embryos with just the TACC3 deficiency (data not shown). Also, preliminary experiments indicate that MEFs lacking both TACC2 and TACC3 proliferate normally ex vivo (data not shown). Since a TACC1 deficiency is not yet available, we are unable to assess the possibly redundant roles of TACC2 and TACC1.
Evolutionarily, Drosophila has a single TACC gene whose product, like all the mammalian TACC proteins, interacts with the centrosome and the mitotic spindle (7, 15). This property of the TACC proteins is dependent upon the only conserved domain within the proteins, namely the coiled-coil domain. In Drosophila, reduction of dTACC expression leads to mitotic defects and death during embryogenesis and female sterility in adults. In Xenopus, however, Maskin, the TACC family member, is proposed to form complexes with certain mRNAs, localize them to the mitotic apparatus, and thereby regulate their expression (2). Previous studies with TACC3 demonstrated that its presence in hematopoietic progenitors is required to suppress p53-mediated apoptosis and that, in the absence of p53, cell replication and chromosomal segregation are not detectably affected. Based on these observations, it would appear that TACC proteins have acquired quite distinct functions with evolution and it is possible either that some of the family members that have evolved by duplication in mammals have not acquired functions or that evolution has eliminated their requirement.
Acknowledgments
The expert technical assistance of Mahnaz Paktinat, Kristen Rothammer, Neena Carpino, Sandy Moore, and Juliana Nunez is gratefully acknowledged. Special thanks go to Kirsteen Maclean and members of the department for experimental advice, suggestions, and fruitful discussions. Thanks go to Janet Partridge for revision of the manuscript. We also thank the Transgenic Core Unit and the personnel of the Animal Resources Center for assistance in generating TACC2-deficient mice and for care and monitoring of the animals.
This work was supported by the Cancer Center CORE grant CA21765, by the grant RO1 DK42932 to J.N.I., by grant PO1 HL53749, and by the American Lebanese Syrian Associated Charities (ALSAC).
REFERENCES
- 1.Adachi, M., S. Suematsu, T. Kondo, J. Ogasawara, T. Tanaka, N. Yoshida, and S. Nagata. 1995. Targeted mutation in the Fas gene causes hyperplasia in peripheral lymphoid organs. Nat. Genet. 11:294-300. [DOI] [PubMed] [Google Scholar]
- 2.Cao, Q., and J. D. Richter. 2002. Dissolution of the maskin-eIF4E complex by cytoplasmic polyadenylation and poly(A)-binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J. 21:3852-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, H. M., K. L. Schmeichel, I. S. Mian, S. Lelievre, O. W. Petersen, and M. J. Bissell. 2000. AZU-1: a candidate breast tumor suppressor and biomarker for tumor progression. Mol. Biol. Cell 11:1357-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhanasekaran, S. M., T. R. Barrette, D. Ghosh, R. Shah, S. Varambally, K. Kurachi, K. J. Pienta, M. A. Rubin, and A. M. Chinnaiyan. 2001. Delineation of prognostic biomarkers in prostate cancer. Nature 412:822-826. [DOI] [PubMed] [Google Scholar]
- 5.Gangisetty, O., B. Lauffart, G. V. Sondarva, D. M. Chelsea, and I. H. Still. 2004. The transforming acidic coiled coil proteins interact with nuclear histone acetyltransferases. Oncogene 23:2559-2563. [DOI] [PubMed] [Google Scholar]
- 6.Gergely, F. 2002. Centrosomal TACCtics. Bioessays 24:915-925. [DOI] [PubMed] [Google Scholar]
- 7.Gergely, F., D. Kidd, K. Jeffers, J. G. Wakefield, and J. W. Raff. 2000. D-TACC: a novel centrosomal protein required for normal spindle function in the early Drosophila embryo. EMBO J. 19:241-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gergely, F., C. Karlsson, I. Still, J. Cowell, J. Kilmartin, and J. W. Raff. 2000b. The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. Proc. Natl. Acad. Sci. USA 97:14352-14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giet, R., D. McLean, S. Descamps, M. J. Lee, J. W. Raff, C. Prigent, and D. M. Glover. 2002. Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J. Cell Biol. 156:437-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groisman, I., Y. S. Huang, R. Mendez, Q. Cao, W. Theurkauf, and J. D. Richter. 2000. CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell 103:435-447. [DOI] [PubMed] [Google Scholar]
- 11.Hannak, E., M. Kirkham, A. A. Hyman, and K. Oegema. 2001. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. J. Cell Biol. 155:1109-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lauffart, B., O. Gangisetty, and I. H. Still. 2003. Molecular cloning, genomic structure and interactions of the putative breast tumor suppressor TACC2. Genomics 81:192-201. [DOI] [PubMed] [Google Scholar]
- 13.Lauffart, B., S. J. Howell, J. E. Tasch, J. K. Cowell, and I. H. Still. 2002. Interaction of the transforming acidic coiled-coil 1 (TACC1) protein with ch-TOG and GAS41/NuBI1 suggests multiple TACC1-containing protein complexes in human cells. Biochem. J. 363:195-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Bot, N., M. C. Tsai, R. K. Andrews, and J. Ahringer. 2003. TAC-1, a regulator of microtubule length in the C. elegans embryo. Curr. Biol. 13:1499-1505. [DOI] [PubMed] [Google Scholar]
- 15.Lee, M. J., F. Gergely, K. Jeffers, S. Y. Peak-Chew, and J. W. Raff. 2001. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat. Cell Biol. 3:643-649. [DOI] [PubMed] [Google Scholar]
- 16.Lee, Y., M. J. Chong, and P. J. McKinnon. 2001. Ataxia telangiectasia mutated-dependent apoptosis after genotoxic stress in the developing nervous system is determined by cellular differentiation status. J. Neurosci. 17:6687-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKeveney, P. J., V. M. Hodges, R. N. Mullan, P. Maxwell, D. Simpson, A. Thompson, P. C. Winter, T. R. Lappin, and A. P. Maxwell. 2001. Characterization and localization of expression of an erythropoietin-induced gene, ERIC-1/TACC3, identified in erythroid precursor cells. Br. J. Haematol. 112:1016-1024. [DOI] [PubMed] [Google Scholar]
- 18.Moritz, M., and D. A. Agard. 2001. Gamma-tubulin complexes and microtubule nucleation. Curr. Opin. Struct. Biol. 11:174-181. [DOI] [PubMed] [Google Scholar]
- 19.Piekorz, R. P., A. Hoffmeyer, C. D. Duntsch, C. McKay, H. Nakajima, V. Sexl, L. Snyder, J. Rehg, and J. N. Ihle. 2002. The centrosomal protein TACC3 is essential for hematopoietic stem cell function and genetically interfaces with p53-regulated apoptosis. EMBO J. 21:653-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pu, J. J., C. Li, M. Rodriguez, and D. Banerjee. 2001. Cloning and structural characterization of ECTACC, a new member of the transforming acidic coiled coil (TACC) gene family: cDNA sequence and expression analysis in human microvascular endothelial cells. Cytokine 13:129-137. [DOI] [PubMed] [Google Scholar]
- 21.Sadek, C. M., S. Jalaguier, E. P. Feeney, M. Aitola, A. E. Damdimopoulos, M. Pelto-Huikko, and J. A. Gustafsson. 2000. Isolation and characterization of AINT: a novel ARNT interacting protein expressed during murine embryonic development. Mech. Dev. 97:13-26. [DOI] [PubMed] [Google Scholar]
- 22.Srayko, M., S. Quintin, A. Schwager, and A. A. Hyman. 2003. Caenorhabditis elegans TAC-1 and ZYG-9 form a complex that is essential for long astral and spindle microtubules. Curr. Biol. 13:1506-1511. [DOI] [PubMed] [Google Scholar]
- 23.Stebbins-Boaz, B., Q. Cao, C. H. de Moor, R. Mendez, and J. D. Richter. 1999. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol. Cell 4:1017-1027. [DOI] [PubMed] [Google Scholar]
- 24.Still, I. H., M. Hamilton, P. Vince, A. Wolfman, and J. K. Cowell. 1999. Cloning of TACC1, an embryonically expressed, potentially transforming coiled coil containing gene, from the 8p11 breast cancer amplicon. Oncogene 18:4032-4038. [DOI] [PubMed] [Google Scholar]
- 25.Still, I. H., P. Vance, and J. K. Cowell. 1999. The third member of the transforming acidic coiled-coil-containing gene family, TACC3, maps to 4p16, close to translocation breakpoints in multiple myeloma, and is upregulated in various cancer cell lines. Genomics 58:165-170. [DOI] [PubMed] [Google Scholar]
- 26.Todaro, G. J., and H. Green. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17:299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Deursen, J., R. Lovell-Badge, F. Oerlemans, J. Schepens, and B. Wieringa. 1991. Modulation of gene activity by consecutive gene targeting of one creatine kinase M allele in mouse embryonic stem cells. Nucleic Acids Res. 19:2637-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Deursen, J., A. Heerschap, R. Oerlemans, W. Ruitenbeek, P. Jap, H. ter Laak, and B. Wieringa. 1993. Skeletal muscles in mice deficient in muscle creatine kinase lack burst activity. Cell 74:621-631. [DOI] [PubMed] [Google Scholar]