Background: The relationship between chondroitin N-acetylgalactosaminyltransferase-1 (ChGn-1) and 2-phosphoxylose phosphatase (XYLP) in controlling the number of chondroitin sulfate chains is unclear.
Results: GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate) was detected in ChGn-1−/− but not in wild-type cartilage. ChGn-1-mediated addition of N-acetylgalactosamine was accompanied by rapid XYLP-dependent dephosphorylation.
Conclusion: GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate) is the preferred substrate for ChGn-1.
Significance: ChGn-1 and XYLP cooperatively regulate the number of CS chains.
Keywords: Chondroitin Sulfate, Glycosaminoglycan, Glycosyltransferase, Phosphatase, Proteoglycan Synthesis
Abstract
A deficiency in chondroitin N-acetylgalactosaminyltransferase-1 (ChGn-1) was previously shown to reduce the number of chondroitin sulfate (CS) chains, leading to skeletal dysplasias in mice, suggesting that ChGn-1 regulates the number of CS chains for normal cartilage development. Recently, we demonstrated that 2-phosphoxylose phosphatase (XYLP) regulates the number of CS chains by dephosphorylating the Xyl residue in the glycosaminoglycan-protein linkage region of proteoglycans. However, the relationship between ChGn-1 and XYLP in controlling the number of CS chains is not clear. In this study, we for the first time detected a phosphorylated tetrasaccharide linkage structure, GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate), in ChGn-1−/− growth plate cartilage but not in ChGn-2−/− or wild-type growth plate cartilage. In contrast, the truncated linkage tetrasaccharide GlcUAβ1–3Galβ1–3Galβ1–4Xyl was detected in wild-type, ChGn-1−/−, and ChGn-2−/− growth plate cartilage. Consistent with the findings, ChGn-1 preferentially transferred N-acetylgalactosamine to the phosphorylated tetrasaccharide linkage in vitro. Moreover, ChGn-1 and XYLP interacted with each other, and ChGn-1-mediated addition of N-acetylgalactosamine was accompanied by rapid XYLP-dependent dephosphorylation during formation of the CS linkage region. Taken together, we conclude that the phosphorylated tetrasaccharide linkage is the preferred substrate for ChGn-1 and that ChGn-1 and XYLP cooperatively regulate the number of CS chains in growth plate cartilage.
Introduction
Chondroitin sulfate (CS),2 a class of glycosaminoglycan (GAG), consists of linear polysaccharide chains comprising repeating disaccharide units ((-4GlcUAβ1–3GalNAcβ1-)n). Assembly of CS chains is initiated by synthesis of the GAG-protein linkage region, which is covalently linked to specific serine residues of specific core proteins. The linkage region tetrasaccharide is formed by sequential, stepwise addition of monosaccharide residues by four specific glycosyltransferases: xylosyltransferase, galactosyltransferase-I, galactosyltransferase-II, and glucuronyltransferase-I (GlcAT-I) (1). During maturation of the GAG-protein linkage region, the Xyl is transiently phosphorylated and dephosphorylated by FAM20B (a kinase) (2) and 2-phosphoxylose phosphatase (XYLP) (3), respectively. Transfer of the first N-acetylgalactosamine (GalNAc) to the non-reducing terminal GlcUA residue in the tetrasaccharide linkage region by N-acetylgalactosaminyltransferase-I (GalNAcT-I) activity triggers the synthesis of the chondroitin backbone (1, 4, 5). The repetitive disaccharide that is characteristic of CS is synthesized via alternate addition of GlcUA and GalNAc residues by GlcAT-II and GalNAcT-II activities, respectively (1, 6–8). During CS synthesis, numerous modifications, including phosphorylation, dephosphorylation, and sulfation, occur under tight spatiotemporal regulation and produce mature, functional CS chains that exert specific biological functions, which are dependent on their size, number, position, and degree of sulfation. Notably, CS is a major component of the cartilaginous extracellular matrix. Characteristic CS chains have specific functions during cartilage development, suggesting that the phosphorylation, dephosphorylation, sulfation, and number of CS chains are strictly regulated by these biosynthetic enzymes (1).
To date, six homologous glycosyltransferases, chondroitin synthase-1 (ChSy-1), ChSy-2, ChSy-3, chondroitin polymerizing factor (ChPF), and chondroitin N-acetylgalactosaminyltransferases 1 and 2 (ChGn-1 and ChGn-2), all of which are likely involved in CS biosynthesis, have been cloned by us and others (1, 4–9). We previously demonstrated chondroitin polymerization with alternating GalNAc and GlcUA residues when any two of the four enzymes ChSy-1, ChSy-2, ChSy-3, and ChPF were co-expressed (5–9). ChGn-1 and -2 are thought to catalyze chain initiation and elongation, exhibiting activities of GalNAcT-I and -II (4, 5). In addition, seven sulfotransferases involved in the sulfation of CS have been cloned to date (1). Four sulfotransferases that catalyze sulfation of position 4 of the GalNAc residue have been cloned, and chondroitin 4-O-sulfotransferases-1, -2, and -3 (C4ST-1, -2, and -3) sulfate position 4 of the GalNAc residues in CS (10–14).
Recently, we revealed that a deficiency in ChGn-1 reduced the number of CS chains, leading to skeletal dysplasias in mice (15). In addition, we found two missense mutations in the ChGn-1 gene that were associated with a profound decrease in enzyme activity in two patients with neuropathy (16). Thus, it is suggested that ChGn-1 regulates the number of CS chains and the total amount of CS in these patients and in growth plate cartilage. More recently, we demonstrated that XYLP regulates the number of CS chains by dephosphorylating the Xyl residue in the GAG-protein linkage region of proteoglycans (PGs) (3). However, the relationship between ChGn-1 and XYLP in the biosynthesis of CS was not clear. In the present study, we report that ChGn-1 and XYLP interact with each other and that ChGn-1-mediated addition of N-acetylgalactosamine was accompanied by rapid XYLP-dependent dephosphorylation during formation of the CS linkage region.
EXPERIMENTAL PROCEDURES
Animals
Mice (C57BL/6 background) were kept under pathogen-free conditions in an environmentally controlled, clean room at the Institute of Laboratory Animals, Kobe Pharmaceutical University; animals were maintained on standard rodent food and on a 12-h light/12-h dark cycle. All animal procedures were approved by the Kobe Pharmaceutical University Committee on Animal Research and Ethics. All experiments were conducted in accordance with the institutional ethical guidelines for animal experiments and safety guidelines for gene manipulation experiments.
Isolation of Linkage Region Oligosaccharides from Mouse Growth Plate Cartilage
Growth plate cartilage CSPG was extracted from E18.5 ChGn-1−/−, ChGn-2−/−, and wild-type mouse embryos with 4 m guanidinium chloride and 0.05 m Tris-HCl, pH 8.0 containing proteinase inhibitors as described (15, 17). The extract was centrifuged at 15,000 × g for 10 min to remove insoluble material. The protein concentration of each sample was determined using a BCA protein assay kit according to the manufacturer's instructions. The CSPG fractions were precipitated with 70% ethanol containing 5% sodium acetate. The partially purified CSPG fractions were dissolved in 1 m LiOH and incubated on a rotator at 4 °C for 16 h to release the O-linked saccharides from the core proteins (18, 19). After neutralization, the sample was applied to an AG 50W-X2 column (2.5-ml bed volume, H+ form; Bio-Rad). The flow-through fractions containing the O-linked oligosaccharide components were pooled and neutralized with 10% NH4HCO3.
Derivatization of the Isolated Oligosaccharide with 2-Aminobenzamide (2AB)
Derivatization of the oligosaccharides with 2AB was performed as described (18, 20). The labeled oligosaccharides were analyzed by high performance liquid chromatography (HPLC) on an amine-bound PA-03 column as described previously (3).
Enzyme Digestion
Enzyme digestion with chondroitinase ABC (EC 4.2.2.20) from Arthrobacter aurescens (10 mIU), chondroitinase AC-II (chondroitinase AC lyase; EC 4.2.2.5) from A. aurescens (10 mIU), or alkaline phosphatase (1 unit) (Roche Applied Science) was carried out in a total volume of 20 μl of appropriate buffer at 37 °C overnight (3).
Expression of Soluble Forms of ChGn-1, XYLP, FAM20B, or C4ST-2
The expression plasmids (6.0 μg) for ChGn-1 (4), XYLP (3), FAM20B (2), or C4ST-2 (10) were individually transfected into COS-1 cells on 100-mm plates using FuGENETM 6 (Roche Applied Science) according to the manufacturer's instructions. For co-transfection experiments, the ChGn-1 and XYLP expression plasmids (3.0 μg each) were co-transfected into COS-1 cells on 100-mm plates using FuGENE 6 as above. Two days after transfection, 1 ml of the culture medium was collected and incubated with 10 μl of IgG-Sepharose (GE Healthcare) for 12 h at 4 °C. The beads were recovered by centrifugation and washed with the assay buffer. The beads were then resuspended in the same buffer and tested for GalNAcT-I, phosphatase, and sulfotransferase activities as described previously (4, 5, 10, 21). To quantify the protein absorbed onto IgG-Sepharose beads, the bound protein was eluted with 1 m acetic acid and quantified using the BCA protein assay reagent (enhanced protocol; Pierce).
GalNAcT-I and Phosphatase Assays and Identification of Reaction Products
A phosphate transfer reaction was conducted as follows. α-Thrombomodulin (TM) containing the linkage region tetrasaccharide GlcUAβ1–3Galβ1–3Galβ1–4Xyl (1 nmol) (18) or the chemically synthesized tetrasaccharide peptide GlcUAβ1–3Galβ1–3Galβ1–4Xylβ1-O-Ser-Gly-Trp-Pro-Asp-Gly (1 nmol) (22) was used as an acceptor in each 20-μl incubation mixture, which contained 10 μl of beads bearing the soluble form of FAM20B as the enzyme source and 10 μm [γ-32P]ATP (1.11 × 105 dpm), 50 mm Tris buffer, pH 7.0, 10 mm MnCl2, 10 mm CaCl2, and 0.1% BSA as described (2, 3). The products of each reaction were then separated by gel filtration chromatography on a Superdex peptide column that had been equilibrated with elution buffer (0.25 m NH4HCO3 and 7% 1-propanol). The fractions containing the enzyme reaction products were pooled and dehydrated. The isolated reaction products were used as substrates for the GalNAcT-I and phosphatase reactions. GalNAcT-I reactions were simultaneously incubated in parallel in 20-μl reaction mixtures containing 5 pmol of GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-[32P]phosphate)β1-O-TM or GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-[32P]phosphate)β1-O-Ser-Gly-Trp-Pro-Asp-Gly, 0.25 mm UDP-[3H]GalNAc (5.28 × 105 dpm), 100 mm MES buffer, pH 5.8, 10 mm MnCl2, and 10 μl of the soluble form of ChGn-1- or ChGn-1/XYLP-bound beads as described (4, 5). Each such mixture was incubated at 37 °C for 4 h, and gel filtration chromatography was used to separate the radiolabeled products using a Superdex peptide column equilibrated with elution buffer. Fractions (0.4 ml each) were collected at a flow rate of 0.4 ml/min, and the radioactivity was measured using a liquid scintillation counter.
In addition, phosphatase reactions were simultaneously incubated in parallel in 20-μl reaction mixtures containing 5 pmol of GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-[32P]phosphate)β1-O-TM, 0.25 mm UDP-GalNAc, 50 mm Tris buffer, pH 5.8, 10 mm MnCl2, and 10 μl of the soluble form of XYLP- or ChGn-1/XYLP-bound beads (3) as the enzyme source. Each of these mixtures was incubated at 37 °C for 4 h, and the products of each reaction were then separated by gel filtration chromatography on a Superdex peptide column equilibrated with elution buffer (3). Fractions (0.4 ml each) were collected at a flow rate of 0.4 ml/min, and a liquid scintillation counter was used to measure the radioactivity.
Polymerization Assay and Identification of Polymerization Reaction Products
First, a phosphate transfer reaction was conducted as follows. α-TM (1 nmol) was used as an acceptor in each 20-μl incubation mixture, which contained 10 μl of beads bearing the soluble form of FAM20B as the enzyme source and 10 μm [γ-32P]ATP (1.11 × 105 dpm), 50 mm Tris buffer, pH 7.0, 10 mm MnCl2, 10 mm CaCl2, and 0.1% BSA as described (2). Next, a GalNAc transfer reaction was conducted using GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-[32P]phosphate)β1-O-TM as an acceptor in the 30-μl incubation mixture, which contained 10 μl of the soluble form of ChGn-1-bound beads as the enzyme source, 0.25 mm UDP-GalNAc, 100 mm MES buffer, pH 6.5, and 10 mm MnCl2. Subsequent polymerization reactions were simultaneously incubated in parallel in 20-μl reaction mixtures containing 1 nmol of GalNAcβ1–4GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-[32P]phosphate)β1-O-TM, 0.25 mm UDP-GalNAc, 0.25 mm UDP-GlcUA, 100 mm MES buffer, pH 6.5, 10 mm MnCl2, and 10 μl of the soluble form of ChSy-1/ChPF-, ChSy-1/ChSy-2-, ChSy-1/ChSy-3-, ChSy-2/ChPF-, ChSy-2/ChSy-3-, or ChSy-3/ChPF-bound beads as described (6–9). Each such mixture was incubated at 37 °C overnight, and a Superdex peptide column equilibrated with elution buffer and gel filtration chromatography were used to separate the radiolabeled products. The radioactive fractions containing the enzyme reaction products were pooled, and the mixtures were dehydrated. The dried reaction products were subjected to reductive β-elimination with NaBH4/NaOH, and Superdex 200 columns and eluent containing 0.25 m NH4HCO3 and 7% 1-propanol were then used to analyze the products.
Pulldown Assays
Pulldown assays were performed as described previously (3). The His-tagged and protein A-tagged expression vectors (3, 4) were co-transfected into COS-1 cells grown on 100-mm plates. FuGENE 6 was used according to the manufacturer's instructions. Two days after transfection, 1 ml of the culture medium was collected and incubated with 10 μl of Ni-NTA-agarose (Qiagen) overnight at 4 °C. The beads were recovered by centrifugation, washed with TBS buffer containing Tween 20 three times, and subjected to SDS-PAGE (7% gel); the separated proteins were transferred to a polyvinylidene difluoride (PVDF) membrane. The membrane was incubated in PBS containing 2% skim milk and 0.1% Tween 20, then incubated with IgG antibody, and then treated with anti-mouse IgG conjugated with horseradish peroxidase (GE Healthcare). An enhanced chemiluminescence (ECL) Select Detection Reagent (GE Healthcare) was used to visualize antibody-labeled protein bands.
Preparation of Embryonic Fibroblasts
Wild-type, ChGn-1−/−, and ChGn-2−/− mouse embryonic fibroblasts (MEFs) were generated from homozygous intercrosses (wild type × wild type, ChGn-1−/− × ChGn-1−/−, and ChGn-2−/− × ChGn-2−/−, respectively). Primary MEFs were harvested from embryonic day 14 embryos. Pregnant female mice were anesthetized using pentobarbital, the uteruses were isolated, and the embryos were extracted and placed into a 10-cm Petri dish. The head, limbs, and liver were then removed, and the embryos were subsequently minced and incubated at 37 °C in the presence of 6 ml of 0.05% trypsin and 0.02% EDTA for 20 min in a humidified incubator. Trypsin-treated embryos were homogenized by trituration until a viscous fluid was obtained with only a few tissue clumps remaining. The homogenized embryos were again incubated in the presence of 6 ml of 0.05% trypsin and 0.02% EDTA for 20 min. After the addition of 2 ml of fetal bovine serum, the homogenized embryos were centrifuged at 100 × g for 5 min. Cell pellets were suspended in fresh DMEM (Wako, Osaka, Japan) containing 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin, and each cell suspension was then transferred to a 10-cm dish.
Chondrocyte Cultures
Immature chondrocytes were isolated from long bone cartilages of newborn (5-day-old) wild-type and ChGn-1−/− mice as described (23) and maintained in DMEM containing 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. The passage 2 cultures were used for subsequent analyses including gene delivery as described below and cytokine treatment. To induce anabolic processes that are characteristic of chondrocytes, the subconfluent cultures were stimulated with 200 ng/ml recombinant human insulin-like growth factor-1 (IGF-1; R&D Systems) for 48 h. The cell harvests were then utilized either to extract total RNA or to isolate the linkage region oligosaccharides as described above. For assessment of the amounts of CS chains, GAGs from chondrocytes were prepared as described previously (7). The purified GAG fraction containing CS was digested with chondroitinase ABC at 37 °C for 2 h. The digests were derivatized with the fluorophore 2AB and then analyzed via anion exchange HPLC as described above. Identification and quantification of the resulting disaccharides were achieved by comparison with authentic unsaturated CS disaccharides (Seikagaku, Tokyo, Japan).
Subcellular Localization
pEGFP-N1-XYLP was constructed previously (3), and FuGENE 6 was used to transfect EGFP-tagged expression vectors (3.0 μg each) into wild-type, ChGn-1−/−, or ChGn-2−/− MEFs and wild-type or ChGn-1−/− immature chondrocytes that were grown on coverslips (Matsunami Glass, Osaka, Japan) according to the manufacturer's instructions. After 24 h of culture, cells were fixed in 4% paraformaldehyde for 30 min, permeabilized with 0.2% Triton X-100 for 10 min, and then blocked with 3% BSA for 30 min with PBS washes between each step. The primary antibodies were GM130 (1:100; BD Pharmingen) and anti-GFP (1:1,000; Invitrogen). The secondary antibodies were obtained from Molecular Probes. A laser-scanning confocal microscope (LSM700, Carl Zeiss, Tokyo, Japan) was used to obtain fluorescence images.
Quantitative Real Time RT-PCR
TRIzol reagents were used to extract total RNA from intact or IGF-1-treated chondrocytes. An aliquot of each total RNA was pretreated with RNase-free DNase to serve as template for cDNA synthesis. FastStart DNA Master plus SYBR Green I and a LightCycler ST300 (Roche Applied Science) were used to perform quantitative real time RT-PCR. The following primer sets were used: glyceraldehyde-3-phosphate dehydrogenase (Gapdh), forward primer, 5′-CATCTGAGGGCCCACTG-3′ and reverse primer, 5′-GAGGCCATGTAGGCCATGA-3′; ChGn-1, forward primer, 5′-TAAACAGCCCTGTGGAGAG-3′ and reverse primer, 5′-GTCGAAATAGGACAAGTCGC-3′; XYLP, forward primer, 5′-AGCAAGAGTCGAAAGAGGAT-3′ and reverse primer, 5′-CATGAACGGAGACCAGCTTA-3′; FAM20B, forward primer, 5′-TTGTCTTTAAGCCTAAGCGGT-3′ and reverse primer, 5′-GGCTTAACCTTCTGTCCGCA-3′; C4ST-2, forward primer, 5′-ATCAGCATCACCAGCAACA-3′ and reverse primer, 5′-TTGGTCATGCTGCCCTG-3′; aggrecan core protein (Acan), forward primer, 5′-TGGGAAGAGCCTCGAATC-3′ and reverse primer, 5′-GCGACAAGAAGACACCA-3′; and type II collagen (Col2a1), forward primer, 5′-ATGTAGAGATGAGGGCCG-3′ and reverse primer, 5′-CATGGGTGCGATGTCAATAAT-3′. The expression level of each gene was normalized to that of Gapdh.
Statistical Analysis
Data are expressed as mean ± S.D. The Student's t test was used to assess statistical significance. Differences were considered to be significant with a p value less than 0.05.
RESULTS
Accumulation of Phosphorylated Pentasaccharide and Tetrasaccharide Linkages in ChGn-1−/− Growth Plate Cartilage
It was shown previously that a deficiency in ChGn-1 reduced the number of CS chains, leading to skeletal dysplasias in mice (15, 24). Although these results indicated that ChGn-1 regulates the number of CS chains by transferring the first GalNAc to the tetrasaccharide in the protein linkage region of CS, ChGn-1−/− mice still produced more than half the amount of CS present in wild-type growth plate cartilage. However, the mechanism underlying this regulation is not clear. To further examine whether linkage region maturation is influenced by ChGn-1, the linkage oligosaccharides obtained from the growth plate cartilage CSPGs of E18.5 ChGn-1−/− mice were compared with those obtained from ChGn-2−/− and wild-type mice. The linkage oligosaccharides in each growth plate cartilage sample were isolated after mild alkaline treatment with LiOH as described previously (3, 25). The isolated oligosaccharides were derivatized with the fluorophore 2AB and analyzed using HPLC. Surprisingly, the truncated linkage tetrasaccharide GlcUAβ1–3Galβ1–3Galβ1–4Xyl-2AB was detected in all growth plate cartilages examined. Notably, GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate)-2AB and GlcNAcα1-4GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate)-2AB were isolated from ChGn-1−/− growth plate cartilage but not from ChGn-2−/− and wild-type growth plate cartilage (Table 1). GlcNAcα1–4GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate) was recently demonstrated to be formed by EXTL2 and considered to be a biosynthetic intermediate of an immature GAG chain (25). In addition, the truncated linkage pentasaccharide GalNAcβ1–4GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate)-2AB was not detected in any of the growth plate cartilage tissues examined.
TABLE 1.
Proportion of linkage region saccharides from wild-type, ChGn-1−/−, or ChGn-2−/− cartilage
| Structure | Wild type | ChGn-1−/− | ChGn-2−/− |
|---|---|---|---|
| pmol/mg protein (%) | pmol/mg protein (%) | pmol/mg protein (%) | |
| ΔHexUA-Gal-Gal-Xyl-2AB | 2682 ± 32 (88) | 1042 ± 22 (39) | 2387 ± 188 (85) |
| GlcUA-Gal-Gal-Xyl-2AB | 373 ± 15 (12) | 1150 ± 83 (42) | 437 ± 62 (15) |
| GlcUA-Gal-Gal-Xyl(2P)-2ABa | NDb | 312 ± 50 (12) | ND |
| GalNAc-GlcUA-Gal-Gal-Xyl(2P)-2AB | ND | ND | ND |
| GlcNAc-GlcUA-Gal-Gal-Xyl(2P)-2AB | ND | 200 ± 44 (7) | ND |
| Total | 3055 ± 137 | 2704 ± 182 | 2824 ± 255 |
a 2P represents 2-O-phosphate.
b ND, not detected (<0.1 pmol/mg of protein).
GlcUAβ1–3Galβ1–3Galβ1–4Xyl-2AB, GlcUAβ1–3Galβ1-3Galβ1–4Xyl(2-O-phosphate)-2AB, and GlcNAcα1–4GlcUAβ1-3Galβ1–3Galβ1–4Xyl(2-O-phosphate)-2AB were digested with alkaline phosphatase; β-glucuronidase, which catalyzes hydrolysis of β-GlcUA residues from the non-reducing termini of sugar chains; heparitinase, which cleaves the α1–4 linkage of GlcNAcα1–4GlcUA (3, 25); and chondroitinase AC-II, which cleaves the β1–4 linkage of GalNAcβ1–4GlcUA, resulting in co-elution with each authentic standard (data not shown). These results indicate that ChGn-1 might preferentially transfer GalNAc to the phosphorylated linkage tetrasaccharide in the protein linkage region of CS.
A Phosphorylated Tetrasaccharide Structure Facilitates ChGn-1-transferase Activity
We next examined whether transfer of a GalNAc residue to the phosphorylated linkage tetrasaccharide structure GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate) was preferentially catalyzed by ChGn-1. We used α-TM bearing a tetrasaccharide (GlcUA-Gal-Gal-Xyl) as a primer and recombinant FAM20B as an enzyme source to generate a phosphorylated linkage structure, GlcUA-Gal-Gal-Xyl(2-O-phosphate), attached to α-TM. This phosphorylated structure (GlcUA-Gal-Gal-Xyl(2-O-phosphate)-TM) was incubated with ChGn-1 and UDP-[3H]GalNAc as a donor substrate. As shown in Table 2, the GalNAcT-I activity of ChGn-1 for GlcUA-Gal-Gal-Xyl-(2-O-phosphate)-TM was more than 100-fold higher than for GlcUA-Gal-Gal-Xyl-TM. These results indicate that ChGn-1 preferentially transfers a GalNAc residue to the phosphorylated tetrasaccharide in vitro.
TABLE 2.
Comparison of the acceptor specificity of ChGn-1 or co-transfection of ChGn-1 and XYLP secreted into culture medium by transfected COS-1 cells
| Acceptor substrate | GalNAc-transferase activitya |
|
|---|---|---|
| ChGn-1 | ChGn-1/XYLP | |
| pmol/mg/h | ||
| GlcUA-Gal-Gal-Xyl-thrombomodulin | 0.05 ± 0.01 | 0.06 ± 0.01 |
| GlcUA-Gal-Gal-Xyl(2P)-thrombomodulinb | 1.34 ± 0.8 | 1.8 ± 0.2 |
| GlcUA-Gal-Gal-Xyl-O-Ser-Gly-Trp-Pro-Asp-Gly | NDc | ND |
| GlcUA-Gal-Gal-Xyl(2P)-O-Ser-Gly-Trp-Pro-Asp-Gly | ND | 62.6 ± 5.7 |
a The values are the mean ± S.D. of three measurements.
b 2P represents 2-O-phosphate.
c ND, not detected (<0.01 pmol/mg/h).
Interactions between ChGn-1 and XYLP
We showed previously that GalNAc-GlcUA-Gal-Gal-Xyl(2-O-phosphate) was not detected in cells (3). Moreover, as shown in Table 1, GalNAc-GlcUA-Gal-Gal-Xyl(2-O-phosphate)-2AB was not detected in ChGn-1−/−, ChGn-2−/−, and wild-type growth plate cartilage. This suggested that ChGn-1-mediated addition of GalNAc can be accompanied by XYLP-dependent dephosphorylation during completion of the linkage pentasaccharide formation. To evaluate the interactions between ChGn-1 and XLYP, ChGn-1 and XLYP were co-expressed. We first examined whether the co-expression of these two proteins augments GalNAcT-I or dephosphorylation activity. As shown in Table 2, when GlcUA-Gal-Gal-Xyl(2-O-phosphate)-TM was used as an acceptor, co-expressed ChGn-1 and XLYP showed higher GalNAcT-I activity than when GlcUA-Gal-Gal-Xyl-TM was used as an acceptor. Notably, when GlcUA-Gal-Gal-Xyl(2-O-phosphate)-Ser-Gly-Trp-Pro-Asp-Gly was used as an acceptor, only co-expression of ChGn-1 and XLYP showed markedly elevated GalNAcT-I activity. Moreover, dephosphorylation activity was evident with enzymes from cells co-expressing ChGn-1 and XYLP when GlcUA-Gal-Gal-Xyl(2-O-[32P]phosphate)-TM was used as a substrate in the presence of UDP-GalNAc (Table 3), whereas dephosphorylation activity was not observed when only XYLP was present as an enzyme source. These results suggest that addition of the GalNAc residue by ChGn-1 was accompanied by rapid dephosphorylation by XYLP.
TABLE 3.
Comparison of phosphatase activities of XYLP and co-transfected XYLP and ChGn-1
| Substrate | XYLP | XYLP/ChGn-1a |
|---|---|---|
| nmol/mg/h | nmol/mg/h | |
| GlcUA-Gal-Gal-Xyl(2P)-TM + UDP-GalNAcb | NDc | 4.5 ± 0.3 |
a The value is the mean ± S.D. of two measurements.
b 2P represents 2-O-phosphate.
c Not detected (<0.01 nmol/mg/h).
Next, we used pulldown assays to determine whether ChGn-1 and XYLP interact. For this analysis, a soluble protein A-tagged XYLP fusion protein (XYLP-ProA) and soluble His6-tagged ChGn-1 and ChGn-2 fusion proteins (ChGn-1-His and ChGn-2-His, respectively) were generated. In addition, to test the specificity of the interaction, we also performed these assays with ChGn-2. Ni-NTA-agarose was added to the culture medium to pull down the His-tagged proteins, and the proteins were separated by SDS-PAGE and blotted. No band was detected in samples from co-transfectants expressing XYLP-ProA and ChGn-2-His (Fig. 1A). However, a protein band with a molecular mass of ∼90 kDa, corresponding to the predicted size of XYLP-ProA, was detected in samples from co-transfectants expressing XYLP-ProA and ChGn-1-His (Fig. 1A). These results indicated that XYLP and ChGn-1 interact with each other and that ChGn-1-mediated addition of GalNAc can be accompanied by rapid, XYLP-dependent dephosphorylation during the completion of linkage pentasaccharide formation in CS.
FIGURE 1.
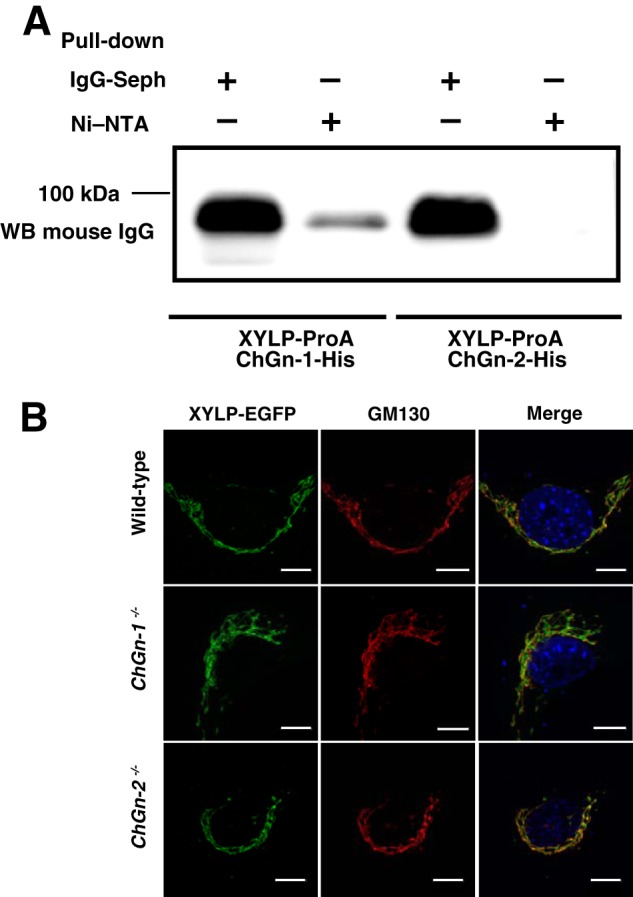
Interactions between ChGn-1 and XYLP. A, culture medium from cells co-expressing XYLP-ProA and ChGn-1-His or XYLP-ProA and ChGn-2-His was incubated with Ni-NTA-agarose to purify the His6-tagged ChGn and any associated proteins. The purified proteins were separated by SDS-PAGE and transferred to PVDF membranes, which were incubated with an IgG primary antibody with ECL Select Detection Reagent used to visualize immunoreactive proteins. B, XYLP-EGFP (green) was co-localized with cis-Golgi (GM130; red) in wild-type, ChGn-1−/−, and ChGn-2−/− MEFs. Scale bars, 10 μm. Seph, Sepharose; WB, Western blot.
Subcellular Localization of ChGn-1 and XYLP
To examine the effect of ChGn-1 on the intracellular localization of XYLP, XYLP-EGFP was expressed in wild-type, ChGn-1−/−, and ChGn-2−/− mouse embryonic fibroblast cells, and these cells were analyzed by immunostaining with an anti-cis-Golgi marker (GM130). XYLP-EGFP colocalized with the anti-cis-Golgi marker (GM130) in all cells examined (Fig. 1B), and these results indicated that XYLP localization was independent of ChGn-1 expression.
Lack of a GalNAc(4-O-sulfate) Linkage Structure in ChGn-1 Knock-out Mice
Previously, we demonstrated that the non-reducing terminal GalNAc(4-O-sulfate) linkage pentasaccharide structure of CS was associated with an increased number of CS chains when the enzyme source was any one of several complexes comprising any two of the four ChSy family members (21). In addition, we showed that the number of CS chains was regulated by the expression levels of ChGn-1 and C4ST-2 (21). We then analyzed the linkage region hexasaccharides of mature GAGs obtained from wild-type, ChGn-1−/−, and ChGn-2−/− growth plate cartilage. Samples were digested with chondroitinase ABC, and the digests were analyzed by anion exchange HPLC. A major peak was observed at the position of authentic 2AB-labeled nonsulfated hexasaccharide ΔHexUAα1–3GalNAcβ1–4GlcUAβ1–3Galβ1–3Galβ1–4Xyl-2AB (ΔHexUA represents 4-deoxy-Δ-l-threo-hex-4-enepyranosyluronic acid) in all examined samples (Fig. 2). In contrast, the 2AB-labeled 4-O-sulfated hexasaccharide ΔHexUAα1–3GalNAc(4-O-sulfate)β1–4GlcUAβ1–3Galβ1-3Galβ1–4Xyl-2AB was detected in samples from ChGn-2−/− and wild-type growth plate cartilage but not from ChGn-1−/− growth plate cartilage (Fig. 2). Moreover, we examined whether C4ST-2 could sulfate the GalNAc phosphorylated linkage residue. C4ST-2 showed no activity toward GalNAc-GlcUA-Gal-Gal-Xyl(2-O-phosphate)-TM, whereas C4ST-2 transferred sulfate to GalNAc-GlcUA-Gal-Gal-Xyl-TM (71.5 ± 5.2 pmol/mg/h). These results indicated that addition of the GalNAc residue by ChGn-1 was accompanied by rapid dephosphorylation of the Xyl residue by XYLP with 4-O-sulfate subsequently transferred to the GalNAc residue by C4ST-2 as proposed (21).
FIGURE 2.
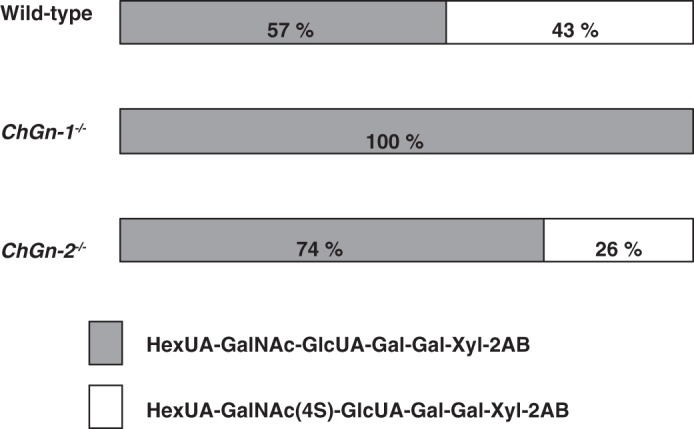
Diagrammatic presentation of the structures of the linkage region hexasaccharides prepared by chondroitinase ABC digestion of CS. The structures of CS from wild-type, ChGn-1−/−, and ChGn-2−/− are illustrated based on the findings obtained from the analyses of the GAG-protein linkage region by chondroitinase ABC digestion. The proportion of ΔHexUAα1–3GalNAcβ1–4GlcUAβ1–3Galβ1–3Galβ1–4Xyl-2AB is denoted by gray horizontal bars, and the proportion of ΔHexUAα1–3GalNAc(4S) β1–4GlcUAβ1–3Galβ1–3Galβ1–4Xyl-2AB is denoted by open boxes. 4S represents 4-O-sulfate. Values were obtained from the average of three separate experiments.
Possible Involvement of the Phosphorylated Pentasaccharide GalNAc-GlcUA-Gal-Gal-Xyl(2-O-phosphate) Structure in Chondroitin Polymerization
Previously, we reported that chondroitin polymerization did not occur on the non-reducing terminal GalNAc linkage pentasaccharide structure GalNAc-GlcUA-Gal-Gal-Xyl (21). We measured polymerization activity using GalNAc-GlcUA-Gal-Gal-Xyl(2-O-[32P]phosphate)-TM with a 2-O-phosphorylated linkage pentasaccharide as an acceptor substrate with co-expression of two of the following proteins as the enzyme source: ChSy-1, ChSy-2, ChSy-3, and ChPF. The reaction products obtained were subjected to reductive β-elimination using NaBH4/NaOH, and the released radiolabeled saccharides were analyzed by gel filtration chromatography using a Superdex 200 column. Chondroitin polymerization was induced on GalNAc-GlcUA-Gal-Gal-Xyl(2-O-[32P]phosphate)-TM when any combination of the proteins was used as the enzyme source albeit with low efficacy (Fig. 3). Notably, the chondroitin chains polymerized on GalNAc-GlcUA-Gal-Gal-Xyl(2-O-[32P]phosphate)-TM by all combinations were much shorter than those formed on GlcUA-Gal-Gal-Xyl-TM (3–5) or GalNAc(4-O-sulfate)-GlcUA-Gal-Gal-Xyl-TM (21). These findings were consistent with previous studies that reported phosphorylation was barely detected on C2 of the Xyl residue structure in mature PGs (26, 27). These results indicate that chondroitin polymerization could occur on the phosphorylated pentasaccharide, although dephosphorylation of the Xyl residue followed by 4-O-sulfation of the non-reducing terminal GalNAc residue remarkably facilitates chondroitin polymerization as described (21).
FIGURE 3.
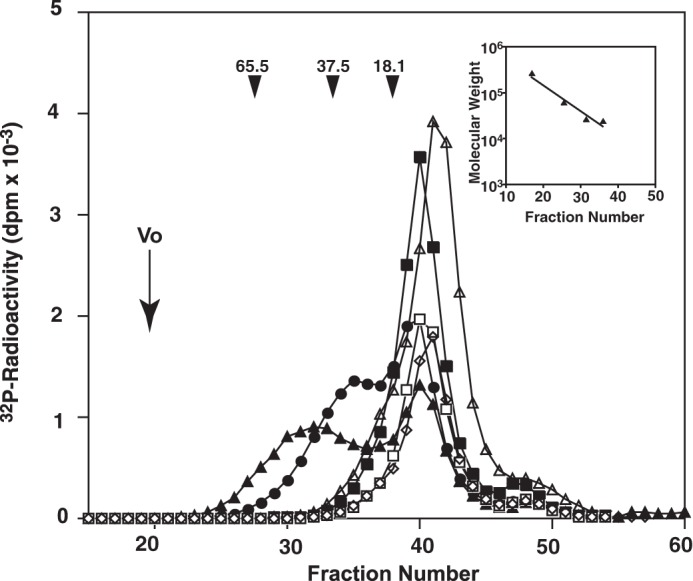
Comparison of CS chain lengths polymerized using GalNAcβ1–4GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-[32P]phosphate)-TM as an acceptor substrate. The 32P-labeled phosphorylated pentasaccharide linkage structure GalNAcβ1–4GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-[32P]phosphate)-TM was tested as an acceptor in polymerization reactions. The structure was co-expressed with the enzyme sources ChSy-1/ChPF (closed triangles), ChSy-1/ChSy-2 (open triangles), ChSy-1/ChSy-3 (closed circles), ChSy-2/ChPF (closed squares), ChSy-2/ChSy-3 (open squares), and ChSy-3/ChPF (open diamonds). 32P-labeled polymerization reaction products were first isolated by gel filtration, subjected to reductive β-elimination using NaBH4/NaOH, and then rechromatographed using a Superdex 200 column with 0.25 m NH4HCO3 and 7% 1-propanol as the eluent. Inset, the calibration curve denoting the linear relation between the log Mr and elution volume generated using the data obtained with commercial polysaccharides of known sizes (dextran average Mr, 200,000, 65,500, 37,500, and 18,100; all from Sigma). The numbered arrowheads 65.5, 37.5, and 18.1 indicate the eluted positions of the 65.5-, 37.5-, and 18.1-kDa saccharides, respectively. The total volume was a fraction of ∼60 (not shown).
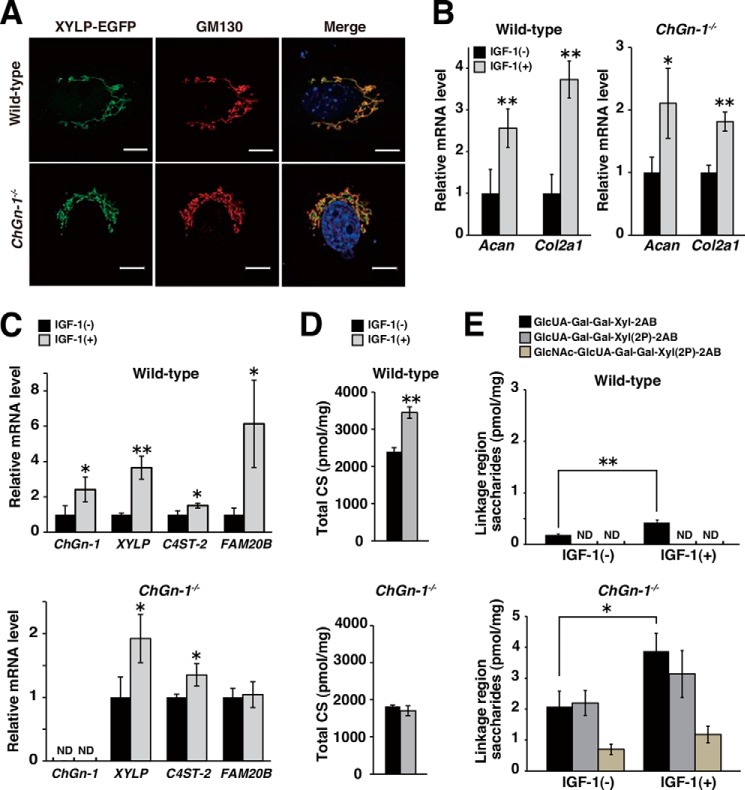
Functional Relevance of ChGn-1-mediated CS Biosynthetic Machinery in the PG Production in Chondrocytes
Among CSPGs, an aggrecan core protein is a major component of the cartilage extracellular matrix and has more than 100 putative Ser-Gly CS attachment sites, although it is reported that approximately half are not occupied by CS chains (28). The distinct nature of the aggrecan core protein might indicate that the number of CS chains attached to it is tightly associated with the ChGn-1 functions. To further evaluate the biological importance of our present findings, we investigated whether the ChGn-1-mediated CS biosynthetic machinery, most likely including XYLP and C4ST-2, is actually functional in chondrocytes, which are a primary producer of aggrecan CSPG.
Chondrocytes were isolated from long bone cartilages of newborn wild-type and ChGn-1−/− mice. Consistent with the data obtained from MEFs, XYLP was also localized in the Golgi apparatus of chondrocytes in a ChGn-1-independent fashion (Fig. 4A). In both cultures, treatment with an anabolic growth factor, IGF-1, resulted in a significant increase in the expression of cartilaginous markers Col2a1 and Acan, which encode type II collagen and aggrecan core protein, respectively (Fig. 4B). Notably, expression of ChGn-1, XYLP, C4ST-2, and FAM20B was also increased by IGF-1 treatment in wild-type chondrocyte cultures, although the expression of ChGn-1 and FAM20B in ChGn-1−/− chondrocytes was undetectable and unaltered even after IGF-1 stimulation, respectively (Fig. 4C). The apparently synchronous increase in the expression of ChGn-1, XYLP, C4ST-2, and Acan suggested a causal link of the ChGn-1-mediated machinery with boosting CS biosynthesis on aggrecan core protein in response to anabolic stimuli. In support of this notion, CS production in wild-type chondrocyte cultures was significantly augmented, whereas that in ChGn-1−/− cultures remained essentially unchanged by IGF-1 treatment (Fig. 4D). Conversely, the abundance of the truncated linkage oligosaccharides isolated from ChGn-1−/− chondrocytes was much larger than that from wild-type chondrocytes irrespective of the presence or absence of IGF-1 (Fig. 4E). Especially, as detected in growth plate cartilages, two distinct, phosphorylated linkage oligosaccharides, GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate)-2AB and GlcNAcα1–4GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate)-2AB, were also exclusive products from ChGn-1−/− chondrocytes (Fig. 4E). These results strengthen the argument that the fine-tuning of CS biosynthesis by the ChGn-1-mediated machinery is centrally involved in the increased de novo synthesis of CSPGs such as aggrecan during distinct anabolic/developmental processes.
FIGURE 4.
Functional relevance of CnGn-1-mediated fine tuning of CS biosynthesis in the increased CSPG production in chondrocytes. A, XYLP-EGFP (green) was co-localized with cis-Golgi (GM130; red) in wild-type and ChGn-1−/− chondrocytes. Scale bars, 10 μm. B and C, relative expression levels of mRNAs encoding chondrocyte marker proteins (Acan and Col2a1 in B) and CS biosynthetic enzymes (ChGn-1, XYLP, C4ST-2, and FAM20B in C) in intact (−) or IGF-1-treated (+) chondrocytes isolated from wild-type and ChGn-1−/− newborn mice (n = 3 cultures, each from an independent mouse). The expression level of respective transcripts was normalized to that of Gapdh. Data are represented as mean ± S.D. *, p < 0.05; **, p < 0.01; ND, not detected. D and E, quantification of the amounts of CS chains (estimated as the amounts of total CS disaccharides in D) and the truncated linkage oligosaccharide intermediates (E) isolated from intact (−) or IGF-1-treated (+) chondrocyte cultures derived from wild-type and ChGn-1−/− newborn mice (n = 3 cultures, each from an independent mouse). Data are represented as mean ± S.D. *, p < 0.05; **, p < 0.01; ND, not detected (<0.1 pmol/mg of dry weight). All error bars represent S.D.
DISCUSSION
Sakai et al. (29) demonstrated that overexpression of ChGn-1 in chondrosarcoma cells increased the number of CS chains attached to an aggrecan core protein, whereas overexpression of ChSy-1, ChPF, and ChSy-3 did not increase CS biosynthesis. Their observations, like ours (15, 21), indicated that ChGn-1 regulates the number of CS chains attached to the aggrecan core protein in cartilage. Here, we demonstrated that a truncated linkage tetrasaccharide, GlcUAβ1–3Galβ1–3Galβ1–4Xyl, was detected in wild-type, ChGn-1−/−, and ChGn-2−/− growth plate cartilage (Table 1). Previously, we reported that an immature, truncated GAG structure (GlcAβ1–3Galβ1–3Galβ1–4Xyl) was attached to recombinant human TM, an integral membrane glycoprotein expressed on the surface of endothelial cells (18). In the present study, we showed that PGs in growth plate cartilage and in chondrocytes, most likely aggrecan, also bear the truncated linkage tetrasaccharide. Taken together, transfer of a β-GalNAc residue to the linkage tetrasaccharide by ChGn-1 appears to play a critical role in regulating the number of CS chains.
In ChGn-1−/− growth plate cartilage and chondrocytes, the amount of truncated linkage tetrasaccharide (GlcUAβ1–3Galβ1–3Galβ1–4Xyl-2AB) was increased (Table 1). Under these conditions, considering that XYLP also interacts with GlcAT-I (3), it is likely that most of the XYLP was captured by GlcAT-I due to the lack of ChGn-1, and therefore the augmented XYLP·GlcAT-I complex may facilitate dephosphorylation of the Xyl residues of linkage structures, and the truncated linkage tetrasaccharide may accumulate in ChGn-1−/− growth plate cartilage and chondrocytes. In addition, the phosphorylated tetrasaccharide linkage structure (GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate)) and the GlcNAcα-capped phosphorylated pentasaccharide linkage structure (GlcNAcα1–4GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate)) were detected in ChGn-1−/− growth plate cartilage and chondrocytes but not in wild-type counterparts (Table 1). We recently demonstrated that GlcNAcα1–4GlcUAβ1–3Galβ1-3Galβ1–4Xyl(2-O-phosphate) is formed by EXTL2 and is considered to be a biosynthetic intermediate of an immature GAG chain (25). In fact, when the formation of the phosphorylated linkage region is excessively accelerated by FAM20B or dephosphorylation by XYLP was attenuated, the phosphorylated linkage tetrasaccharide was formed. Therefore, EXTL2 likely transferred a GlcNAc to the phosphorylated linkage tetrasaccharide (3). These results indicated that ChGn-1 might preferentially transfer GalNAc to the phosphorylated linkage tetrasaccharide in the protein linkage region of CS. Indeed, ChGn-1 transfers a GalNAc residue to the phosphorylated tetrasaccharide more efficiently than to the non-phosphorylated tetrasaccharide (Table 2). In addition, ChGn-1 and XYLP interact with each other, and GalNAc transfer by ChGn-1 was accompanied by rapid dephosphorylation by XYLP (Table 3). Therefore, we conclude that GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate) is the preferred substrate for ChGn-1 and that the number of CS chains can be cooperatively regulated by XYLP and ChGn-1.
Interestingly, IGF-1 treatment increased FAM20B expression in wild-type but not in ChGn-1−/− chondrocyte cultures (Fig. 4C). Although the molecular basis for their different responses is currently unknown, such accelerated expression of FAM20B leads to excessive production of the phosphorylated linkage tetrasaccharide that is favorable for subsequent ChGn-1-mediated CS biosynthesis in wild-type chondrocyte cultures. In contrast, despite basal level expression of FAM20B even under the stimulatory condition by IGF-1 (Fig. 4C), a marked accumulation of the phosphorylated forms of the truncated linkage oligosaccharides was detected in ChGn-1−/− chondrocytes. Given that the phosphorylated forms of linkage tetrasaccharide in ChGn-1−/− chondrocytes are generated at a constant rate during CS biosynthesis, the exclusive accumulation of the phosphorylated linkage oligosaccharides could be mainly attributed to a functional uncoupling between ChGn-1 and XYLP.
We recently demonstrated that the non-reducing terminal GalNAc(4-O-sulfate) linkage structure of CS was associated with an increased number of CS chains when the enzyme source was one of several complexes comprising any two of the four ChSy family proteins (21). In addition, C4ST-2 efficiently and selectively transferred sulfate from 3′-phosphoadenosine 5′-phosphosulfate to position 4 of non-reducing terminal GalNAc linkage residues, and the number of CS chains was regulated by the expression levels of C4ST-2 and of ChGn-1 (21). Therefore, C4ST-2 is thought to play a key role in regulating levels of CS synthesized via ChGn-1. Consistent with these findings, the 4-sulfated hexasaccharide ΔHexUA-GalNAc(4-O-sulfate)-GlcUA-Gal-Gal-Xyl-2AB was not detected in ChGn-1−/− articular cartilage (Fig. 2). Furthermore, C4ST-2 showed no activity toward GalNAc-GlcUA-Gal-Gal-Xyl(2-O-phosphate)-TM, whereas C4ST-2 transferred sulfate to GalNAc-GlcUA-Gal-Gal-Xyl-TM. These results suggest that addition of the GalNAc residue by ChGn-1 was accompanied by rapid dephosphorylation of the Xyl residue by XYLP, and 4-O-sulfate was subsequently transferred to the GalNAc residue by C4ST-2. Therefore, the number of CS chains on specific core proteins is tightly regulated during cartilage development most likely by temporal and spatial regulation of ChGn-1, C4ST-2, and XYLP expression, and progression of cartilage diseases may result from defects in these regulatory systems.
Previously, we demonstrated that ChGn-2 plays a critical role in CS chain elongation (30). However, the involvement of ChGn-2 in chain initiation and regulation of the number of CS chains is not clear. In this study, the amount of the unsaturated linkage tetrasaccharide ΔHexUA-Gal-Gal-Xyl-2AB isolated from ChGn-2−/− growth plate cartilage was slightly lower than that isolated from wild-type growth plate cartilage (Table 1). However, as in the case of wild-type growth plate cartilage, the phosphorylated tetrasaccharide linkage structure (GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate)) and the GlcNAcα-capped phosphorylated pentasaccharide linkage structure (GlcNAcα1–4GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate)) were not detected in ChGn-2−/− growth plate cartilage (Table 1). In addition, ChGn-2 and XYLP interaction was not detected (Fig. 1). These results suggest that ChGn-2 might not be primarily involved in controlling the number of CS chains as proposed previously (30).
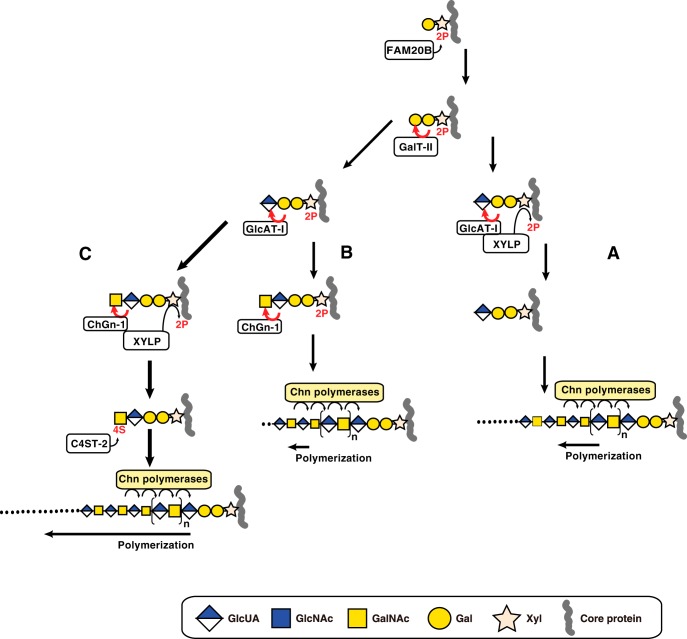
Here, we propose that CS chains can be formed via three different pathways (Fig. 5). Biosynthesis of CS is initiated by the addition of Xyl to specific serine residues in a core protein. This event is followed by the transfer of Gal residues and transient phosphorylation of Xyl residues by FAM20B (2). Next, GlcAT-I transfers GlcUA from UDP-GlcUA to the phosphorylated trisaccharide structure Galβ1–3Galβ1–4Xyl(2-O-phosphate). This final step completes the formation of the linkage region. XYLP can dephosphorylate Xyl(2-O-phosphate) structures during this last step, and, as shown in Fig. 5A, interactions between GlcAT-I and XYLP facilitate these two simultaneous steps (3). Thereafter, CS polymerization onto the linkage region tetrasaccharide can be catalyzed by chondroitin (Chn) polymerases. If ChGn-1 expression is excessively augmented, resulting in substantial amounts of XYLP being captured by ChGn-1, and free GlcAT-I is increased, biosynthetic intermediates (GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate)) may accumulate (Fig. 5B). Under these conditions, transfer of a GalNAc residue by ChGn-1 occurs on the phosphorylated linkage tetrasaccharide, and CS polymerization by Chn polymerases can occur on the phosphorylated pentasaccharides, although the chain length of Chn formed on the phosphorylated pentasaccharides is short (Fig. 3). Alternatively, the addition of the GalNAc residue (Fig. 5C) by ChGn-1 is accompanied by rapid dephosphorylation by XYLP, and subsequent 4-O-sulfation of the GalNAc residue by C4ST-2 occurs. Then CS polymerization onto the GalNAc(4-O-sulfate) linkage pentasaccharide structure is efficiently catalyzed by Chn polymerases. Therefore, the different numbers, lengths, and structures of CS chains could be synthesized through each pathway with the CS synthesized through the pathway mediated by ChGn-1, XYLP, and C4ST-2 having a specific structure that is indispensable for cartilage development.
FIGURE 5.
Three different biosynthetic pathways for CS polymerization. Synthesis of the linkage region is initiated by the addition of a Xyl residue to a specific serine residue on the core protein followed by the sequential transfer of two Gal residues and is completed by transfer of a GlcUA residue. During synthesis of the linkage region, the Xyl residue is transiently phosphorylated by FAM20B, which enhances galactosyltransferase-II (GalT-II) and GlcAT-I activities. A, after synthesis of the phosphorylated linkage region trisaccharide, GlcAT-I transfers GlcUA to the phosphorylated trisaccharide Galβ1–3Galβ1–4Xyl(2-O-phosphate). Concomitantly, Xyl dephosphorylation is induced by XYLP. Chn polymerases then induce polymerization of the linkage region tetrasaccharide. B, following complete synthesis of the phosphorylated linkage region tetrasaccharide, ChGn-1 catalyzes the transfer of a single GalNAc residue to the phosphorylated tetrasaccharide linkage region. Then Chn polymerases may utilize the phosphorylated linkage region pentasaccharide albeit with low polymerization efficacy. C, following synthesis of the phosphorylated GalNAc linkage region, dephosphorylation is induced by XYLP. C4ST-2 subsequently mediates 4-O-sulfation of the non-reducing terminal GalNAc residue. Finally, the non-reducing terminal 4-O-sulfate of the GalNAc linkage structure facilitates elongation of the CS chains via Chn polymerases. 2P and 4S represent 2-O-phosphate and 4-O-sulfate, respectively. The arrow length of polymerization represents the efficiency of polymerization.
More recently, we showed that ChGn-1−/− mice recover more completely from spinal cord injury than wild-type and chondroitinase ABC-treated mice (17). Our results indicate that the deficiency in ChGn-1 mediates excellent recovery from spinal cord injury by optimizing the counteracting effectors of axon regeneration. In addition, we recently reported two missense mutations in the human ChGn-1 gene, both of which were associated with a profound decrease in enzyme activity in two patients with neuropathy (16). As we demonstrated in this study, ChGn-1 cooperates with XYLP and C4ST-2 to regulate the biosynthetic fine-tuning of CS chains that play important roles in various biological/pathological events (31). In view of the modes of action for the ChGn-1-mediated biosynthetic machinery, its targets may include not only developing cartilage and glial scar formation after central nervous system injury (32, 33) but also inflammation processes in which excessive CSPG production often occurs (34–36). Further studies with a focus on the regulatory mechanisms involving activation of the individual enzymes are required for the clarification of generality and utility of the ChGn-1-mediated biosynthetic pathway(s). In conclusion, this study proposes a novel strategy for the treatment of degenerative cartilage disorders and recovery from spinal cord injury and minor trauma.
Acknowledgment
We thank Junko Tanaka-Umeki for synthesis of the tetrasaccharide peptide.
This work was supported in part by Grants-in-aid for Scientific Research (B) 25293014 (to H. K.), for Scientific Research (C) 24590132 (to T. M.), and for Scientific Research on Innovative Areas 23110003 (to H. K.) and by the Supported Program for the Strategic Research Foundation at Private Universities, 2012–2016 (to H. K.) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
- CS
- chondroitin sulfate
- GAG
- glycosaminoglycan
- ChSy
- chondroitin synthase
- ChGn
- chondroitin N-acetylgalactosaminyltransferase
- ChPF
- chondroitin polymerizing factor
- TM
- thrombomodulin
- GlcUA
- d-glucuronic acid
- PG
- proteoglycan
- IGF
- insulin-like growth factor
- XYLP
- 2-phosphoxylose phosphatase
- GlcAT
- glucuronyltransferase
- GalNAcT
- N-acetylgalactosaminyltransferase
- C4ST
- chondroitin 4-O-sulfotransferase
- 2AB
- 2-aminobenzamide
- ΔHexUA
- 4-deoxy-Δ-l-threo-hex-4-enepyranosyluronic acid
- Ni-NTA
- nickel-nitrilotriacetic acid
- MEF
- mouse embryonic fibroblast
- EGFP
- enhanced GFP
- Acan
- aggrecan core protein
- Col2a1
- type II collagen
- ProA
- protein A
- Chn
- chondroitin.
REFERENCES
- 1. Mikami T., Kitagawa H. (2013) Biosynthesis and function of chondroitin sulfate. Biochim. Biophys. Acta 1830, 4719–4733 [DOI] [PubMed] [Google Scholar]
- 2. Koike T., Izumikawa T., Tamura J., Kitagawa H. (2009) FAM20B is a kinase that phosphorylates xylose in the glycosaminoglycan-protein linkage region. Biochem. J. 421, 157–162 [DOI] [PubMed] [Google Scholar]
- 3. Koike T., Izumikawa T., Sato B., Kitagawa H. (2014) Identification of phosphatase that dephosphorylates xylose in the glycosaminoglycan-protein linkage region of proteoglycans. J. Biol. Chem. 289, 6695–6708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Uyama T., Kitagawa H., Tamura J., Sugahara K. (2002) Molecular cloning and expression of human chondroitin N-acetylgalactosaminyltransferase: the key enzyme for chain initiation and elongation of chondroitin/dermatan sulfate on the protein linkage region tetrasaccharide shared by heparin/heparan sulfate. J. Biol. Chem. 277, 8841–8846 [DOI] [PubMed] [Google Scholar]
- 5. Uyama T., Kitagawa H., Tanaka J., Tamura J., Ogawa T., Sugahara K. (2003) Molecular cloning and expression of a second chondroitin N-acetylgalactosaminyltransferase involved in the initiation and elongation of chondroitin/dermatan sulfate. J. Biol. Chem. 278, 3072–3078 [DOI] [PubMed] [Google Scholar]
- 6. Kitagawa H., Uyama T., Sugahara K. (2001) Molecular cloning and expression of a human chondroitin synthase. J. Biol. Chem. 276, 38721–38726 [DOI] [PubMed] [Google Scholar]
- 7. Izumikawa T., Uyama T., Okuura Y., Sugahara K., Kitagawa H. (2007) Involvement of chondroitin sulfate synthase-3 (chondroitin synthase-2) in chondroitin polymerization through its interaction with chondroitin synthase-1 or chondroitin polymerizing factor. Biochem. J. 403, 545–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Izumikawa T., Koike T., Shiozawa S., Sugahara K., Tamura J., Kitagawa H. (2008) Identification of chondroitin sulfate glucuronyltransferase as chondroitin synthase-3 involved in chondroitin polymerization: chondroitin polymerization is achieved by multiple enzyme complexes consisting of chondroitin synthase family members. J. Biol. Chem. 283, 11396–11406 [DOI] [PubMed] [Google Scholar]
- 9. Kitagawa H., Izumikawa T., Uyama T., Sugahara K. (2003) Molecular cloning of a chondroitin polymerizing factor that cooperates with chondroitin synthase for chondroitin polymerization. J. Biol. Chem. 278, 23666–23671 [DOI] [PubMed] [Google Scholar]
- 10. Mikami T., Mizumoto S., Kago N., Kitagawa H., Sugahara K. (2003) Specificities of three distinct human chondroitin/dermatan N-acetylgalactosamine 4-O-sulfotransferases demonstrated using partially desulfated dermatan sulfate as an acceptor: implication of differential roles in dermatan sulfate biosynthesis. J. Biol. Chem. 278, 36115–36127 [DOI] [PubMed] [Google Scholar]
- 11. Hiraoka N., Nakagawa H., Ong E., Akama T. O., Fukuda M. N., Fukuda M. (2000) Molecular cloning and expression of two distinct human chondroitin 4-O-sulfotransferases that belong to the HNK-1 sulfotransferase gene family. J. Biol. Chem. 275, 20188–20196 [DOI] [PubMed] [Google Scholar]
- 12. Kang H.-G., Evers M. R., Xia G., Baenziger J. U., Schachner M. (2002) Molecular cloning and characterization of chondroitin-4-O-sulfotransferase-3. A novel member of the HNK-1 family of sulfotransferases. J. Biol. Chem. 277, 34766–34772 [DOI] [PubMed] [Google Scholar]
- 13. Evers M. R., Xia G., Kang H.-G., Schachner M., Baenziger J. U. (2001) Molecular cloning and characterization of a dermatan-specific N-acetylgalactosamine 4-O-sulfotransferase. J. Biol. Chem. 276, 36344–36353 [DOI] [PubMed] [Google Scholar]
- 14. Yamauchi S., Mita S., Matsubara T., Fukuta M., Habuchi H., Kimata K., Habuchi O. (2000) Molecular cloning and expression of chondroitin 4-sulfotransferase. J. Biol. Chem. 275, 8975–8981 [DOI] [PubMed] [Google Scholar]
- 15. Watanabe Y., Takeuchi K., Higa Onaga S., Sato M., Tsujita M., Abe M., Natsume R., Li M., Furuichi T., Saeki M., Izumikawa T., Hasegawa A., Yokoyama M., Ikegawa S., Sakimura K., Amizuka N., Kitagawa H., Igarashi M. (2010) Chondroitin sulfate N-acetylgalactosaminyltransferase-1 is required for normal cartilage development. Biochem. J. 432, 47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Saigoh K., Izumikawa T., Koike T., Shimizu J., Kitagawa H., Kusunoki S. (2011) Chondroitin β-1,4-N-acetylgalactosaminyltransferase-1 missense mutations are associated with neuropathies. J. Hum. Genet. 56, 143–146 [DOI] [PubMed] [Google Scholar]
- 17. Takeuchi K., Yoshioka N., Higa Onaga S., Watanabe Y., Miyata S., Wada Y., Kudo C., Okada M., Ohko K., Oda K., Sato T., Yokoyama M., Matsushita N., Nakamura M., Okano H., Sakimura K., Kawano H., Kitagawa H., Igarashi M. (2013) Chondroitin sulphate N-acetylgalactosaminyl-transferase-1 inhibits recovery from neural injury. Nat. Commun. 4, 2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nadanaka S., Kitagawa H., Sugahara K. (1998) Demonstration of the immature glycosaminoglycan tetrasaccharide sequence GlcAβ1-3Galβ1–3Galβ1–4Xyl on recombinant soluble human α-thrombomodulin. A possible mechanism generating “part-time” proteoglycans. J. Biol. Chem. 273, 33728–33734 [DOI] [PubMed] [Google Scholar]
- 19. Sakaguchi H., Watanabe M., Ueoka C., Sugiyama E., Taketomi T., Yamada S., Sugahara K. (2001) Isolation of reducing oligosaccharide chains from the chondroitin/dermatan sulfate-protein linkage region and preparation of analytical probes by fluorescent labeling with 2-aminobenzamide. J. Biochem. 129, 107–118 [DOI] [PubMed] [Google Scholar]
- 20. Sugahara K., Okumura Y., Yamashina I. (1989) The Engelbreth-Holm-Swarm mouse tumor produces undersulfated heparan sulfate and oversulfated galactosaminoglycans. Biochem. Biophys. Res. Commun. 162, 189–197 [DOI] [PubMed] [Google Scholar]
- 21. Izumikawa T., Koike T., Kitagawa H. (2012) Chondroitin 4-O-sulfotransferase-2 regulates the number of chondroitin sulfate chains initiated by chondroitin N-acetylgalactosaminyltransferase-1. Biochem. J. 441, 697–705 [DOI] [PubMed] [Google Scholar]
- 22. Tamura J., Yamaguchi A., Tanaka J. (2002) Synthesis of betaglycan-type tetraosyl hexapeptide: a possible precursor regulating enzymatic elongation toward heparin. Bioorg. Med. Chem. Lett. 12, 1901–1903 [DOI] [PubMed] [Google Scholar]
- 23. Gosset M., Berenbaum F., Thirion S., Jacques C. (2008) Primary culture and phenotyping of murine chondrocytes. Nat. Protoc. 3, 1253–1260 [DOI] [PubMed] [Google Scholar]
- 24. Sato T., Kudo T., Ikehara Y., Ogawa H., Hirano T., Kiyohara K., Hagiwara K., Togayachi A., Ema M., Takahashi S., Kimata K., Watanabe H., Narimatsu H. (2011) Chondroitin sulfate N-acetylgalactosaminyltransferase 1 is necessary for normal endochondral ossification and aggrecan metabolism. J. Biol. Chem. 286, 5803–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nadanaka S., Zhou S., Kagiyama S., Shoji N., Sugahara K., Sugihara K., Asano M., Kitagawa H. (2013) EXTL2, a member of the EXT family of tumor suppressors, controls glycosaminoglycan biosynthesis in a xylose kinase-dependent manner. J. Biol. Chem. 288, 9321–9333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sugahara K., Mizuno N., Okumura Y., Kawasaki T. (1992) The phosphorylated and/or sulfated structure of the carbohydrate-protein-linkage region isolated from chondroitin sulfate in the hybrid proteoglycans of Engelbreth-Holm-Swarm mouse tumor. Eur. J. Biochem. 204, 401–406 [DOI] [PubMed] [Google Scholar]
- 27. Shibata S., Midura R. J., Hascall V. C. (1992) Structural analysis of the linkage region oligosaccharides and unsaturated disaccharides from chondroitin sulfate using CarboPac PA1. J. Biol. Chem. 267, 6548–6555 [PubMed] [Google Scholar]
- 28. Watanabe H., Gao L., Sugiyama S., Doege K., Kimata K., Yamada Y. (1995) Mouse aggrecan, a large cartilage proteoglycan: protein sequence, gene structure and promoter sequence. Biochem. J. 308, 433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sakai K., Kimata K., Sato T., Gotoh M., Narimatsu H., Shinomiya K., Watanabe H. (2007) Chondroitin sulfate N-acetylgalactosaminyltransferase-1 plays a critical role in chondroitin sulfate synthesis in cartilage. J. Biol. Chem. 282, 4152–4161 [DOI] [PubMed] [Google Scholar]
- 30. Izumikawa T., Okuura Y., Koike T., Sakoda N., Kitagawa H. (2011) Chondroitin 4-O-sulfotransferase-1 regulates the chain length of chondroitin sulfate in co-operation with chondroitin N-acetylgalactosaminyltransferase-2. Biochem. J. 434, 321–331 [DOI] [PubMed] [Google Scholar]
- 31. Kitagawa H. (2014) Using sugar remodeling to study chondroitin sulfate function. Biol. Pharm. Bull. 37, 1705–1712 [DOI] [PubMed] [Google Scholar]
- 32. Rhodes K. E., Fawcett J. W. (2004) Chondroitin sulphate proteoglycans: preventing plasticity or protecting the CNS? J. Anat. 204, 33–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miyata S., Kitagawa H. (2015) Mechanisms for modulation of neural plasticity and axon regeneration by chondroitin sulphate. J. Biochem. 157, 13–22 [DOI] [PubMed] [Google Scholar]
- 34. Frey H., Schroeder N., Manon-Jensen T., Iozzo R. V., Schaefer L. (2013) Biological interplay between proteoglycans and their innate immune receptors in inflammation. FEBS J. 280, 2165–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zeng-Brouwers J., Beckmann J., Nastase M. V., Iozzo R. V., Schaefer L. (2014) De novo expression of circulating biglycan evokes an innate inflammatory tissue response via MyD88/TRIF pathways. Matrix Biol. 35, 132–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nadanaka S., Kitagawa H. (2014) EXTL2 controls liver regeneration and aortic calcification through xylose kinase-dependent regulation of glycosaminoglycan biosynthesis. Matrix Biol. 35, 18–24 [DOI] [PubMed] [Google Scholar]