Background: IgG4 antibodies are able to undergo a process termed Fab arm exchange (FAE).
Results: A novel method for quantifying FAE in physiologically relevant matrices was developed.
Conclusion: A hinge mutation in IgG4 antibodies inhibits FAE to undetectable levels in the immunoassays described herein.
Significance: These methods are valuable for investigating and assessing the factors affecting and controlling IgG4 FAE.
Keywords: antibody, antibody engineering, immunology, interleukin, tumor necrosis factor (TNF), IgG4, Immunoassay, S228P mutation, affinity chromatography, fab arm exchange
Abstract
Human immunoglobulin G isotype 4 (IgG4) antibodies (Abs) are potential candidates for immunotherapy when reduced effector functions are desirable. IgG4 Abs are dynamic molecules able to undergo a process known as Fab arm exchange (FAE). This results in functionally monovalent, bispecific antibodies (bsAbs) with unknown specificity and hence, potentially, reduced therapeutic efficacy. IgG4 FAE is suggested to be an important biological mechanism that provides the basis for the anti-inflammatory activity attributed to IgG4 Abs. To date, the mechanism of FAE is not entirely understood and studies measuring FAE in ex vivo matrices have been hampered by the presence and abundance of endogenous IgG4 wild-type (WT) Abs. Using representative humanized WT IgG4 monoclonal Abs, namely, anti-IL-6 and anti-TNF, and a core-hinge stabilized serine 228 to proline (S228P) anti-IL-6 IgG4 mutant, it is demonstrated for the first time how anti-IgG4 affinity chromatography can be used to prepare physiologically relevant matrices for assessing and quantifying FAE. A novel method for quantifying FAE using a single MSD immunoassay is also reported and confirms previous findings that, dependent on the redox conditions, the S228P mutation can prevent IgG4 FAE to undetectable levels both in vitro and in vivo. Together, the findings and novel methodologies will allow researchers to monitor and quantify FAE of their own IgG4 molecules in physiologically relevant matrices.
Introduction
Immunoglobulin G (IgG)2 is the most abundant antibody (Ab) in human sera and is subdivided into four subclasses: IgG1, IgG2, IgG3, and IgG4. IgG1 and IgG4 both have two interhinge disulfide bonds (DSBs), IgG2 has four interhinge DSBs, and IgG3 has 11 interhinge DSBs. IgG1 and IgG3 are generally described as active isotypes because they elicit antibody-dependent cell-mediated cytotoxicity and complement-dependent cell-mediated cytotoxicity (1, 2). Conversely, IgG2 and IgG4 are described as inactive isotypes as they bind poorly to effector molecules, resulting in relatively low effector function induction (1–4). Among the IgG isotypes, IgG4 molecules have a unique feature in that they are able to undergo a dynamic swapping event known as “half-molecule exchange” or “Fab arm exchange” (5–11). This physiological process involves the exchange and recombination of a heavy-light chain pair (half-molecule) of one IgG4 Ab with that of another IgG4 Ab.
This inherent ability of bivalent, monospecific IgG4 Abs to undergo FAE results in the formation of functionally monovalent, bsAbs i.e. Abs unable to cross-link identical antigens (12). Exchange occurring between Abs with different and unknown V-regions results in bsAbs with unknown, and perhaps undesirable specificity. Consequently, biotherapeutic bivalent, monospecific IgG4 WT Abs undergoing FAE with endogenous IgG4 WT Abs could result in the formation of chimeric, functionally monovalent, bsAbs in which binding to the target antigen could, in time, change from an avidity to an affinity interaction. This could affect the pharmacokinetics and efficacy of the biotherapeutic and change homologous cross-linking of the originally targeted antigen to non-cross-linking behavior. Subsequently, FAE could introduce undesired pharmacodynamic unpredictability for human immunotherapy. Therefore, the propensity of WT IgG4 Abs to participate in FAE has raised question marks over choosing the, otherwise, inactive IgG4 isotype as the backbone for biotherapeutics (13, 14).
While the mechanism of FAE is not yet fully understood or characterized, the molecular determinants driving FAE in humans were identified by site-directed mutagenesis as residues Ser-228 (15, 16), located in the IgG4 core hinge, and Arg-409 in the IgG4 CH3 domain (8). To investigate and measure FAE, researchers have to date used artificial buffered matrices to mimic the physiological environment and have steered clear of using more relevant physiological matrices such as blood and plasma due to the presence of interfering endogenous IgG4 WT Abs (11, 15–17). The work detailed in this paper demonstrates how functional bridging assays and various biochemical techniques were successfully established and employed, including, for the first time, a novel method of physiological matrix preparation, to monitor and quantify FAE. Evidence to support the notion that a single amino acid mutation (S228P) in the IgG4 core-hinge region is sufficient to prevent the in vivo FAE of our anti-IL-6 IgG4 Ab, is also provided.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Humanized anti-IL-6 WT IgG4 Ab, its S228P point mutated variant (anti-IL-6 S228P IgG4 Ab), humanized anti-TNF WT IgG4 and IgG1 Abs were expressed and purified as described previously (18). Recombinant IL-6, TNF (Peprotech), and anti-human κ-light chain specific Ab (Jackson) were either biotinylated or Sulfo-tag labeled with Sulfo-NHS-LC-LC-Biotin (Thermo Scientific) or ruthenium-NHS-ester (Meso Scale Discovery, MSD), respectively, according to the manufacturers' protocols.
Human Samples-in Vivo Fab Arm Exchange
Anonymized plasma samples from healthy volunteers who received a single 3 mg/kg dose of anti-IL-6 S228P IgG4 by intravenous (IV) injection were obtained from a Phase 1, randomized, double-blind, placebo-controlled study.
Fab Arm Exchange
Anti-IL-6 WT IgG4 or anti-IL-6 S228P IgG4 Abs and their potential exchange partners, namely, either anti-TNF WT IgG4 or IgG1 Abs, were mixed as follows: for in vitro studies at 1:1 molar ratios at a total concentration of 100 μg/ml in PBS, pH 7.4. For ex vivo studies at 1:9 molar ratios at a total concentration of 600 μg/ml in either IgG4-depleted plasma or IgG4-depleted blood (see “Results”).
To allow DSB reduction, samples were supplemented with reduced glutathione (GSH) (Sigma) to a final concentration of 0.5 mm. At the start of the experiment (t = 0 h) an aliquot of the mixture was quenched (to inactivate potentially reactive thiol groups) with N-ethylmaleimide (NEM) (Sigma) to a final concentration of 10 mm and incubated alongside the rest of the reaction for 16 h at 37 °C (t = 16 h). After overnight incubation, the t = 16 h sample was quenched as above.
Non-reducing SDS-PAGE, Coomassie Staining, and Western Blotting
Ab and chromatography samples were boiled for 3 min in 1× SDS-PAGE sample buffer supplemented with NEM (to a final concentration of 10 mm) and then analyzed using 4–20% gradient Tris-glycine gels (Invitrogen). Following SDS-PAGE, gels were either stained with Coomassie or transferred to nitrocellulose membrane for Western blotting (WB). Membranes were blocked in 5% milk (w/v) in PBS/0.1% Tween-20 (PBST, blocking buffer) for 1 h at room temperature with agitation before incubation with appropriate HRP-conjugated primary Abs (AbD Serotec 1:2,000 in blocking buffer). The membranes were then washed with PBST, incubated with chemiluminescent substrate and imaged using the ImageQuant 4000 LAS analyzer (GE Healthcare). Total protein concentration was quantified by measuring absorbance at 280 nm.
IgG4 Depletion and Physiological Matrix Preparation
Whole blood from healthy human donors was collected in heparin-containing vials (BD Bioscience) and centrifuged at 1,500 × g for 10 min at room temperature to separate plasma from cells. 1.5 ml of plasma was sequentially incubated with 2 × 250 μl of Capture Select IgG4 beads (BAC Netherlands) for 1 h with agitation at room temperature. Following incubation, the unbound flow-through material (IgG4-depleted plasma) was collected and stored at 4 °C. The beads were washed three times with 1 ml of physiological salt solution (PSS, in mm: 145 NaCl, 5.6 KCl, 5.6 glucose, 1 MgCl2, 1 CaCl2, 15 HEPES; pH 7.4) before elution with 100 μl of 1× SDS-sample buffer (Invitrogen). Chromatography fractions were then analyzed by non-reducing SDS-PAGE (see below). During the incubation of plasma with affinity columns, the pelleted cells from the initial whole blood centrifugation were washed three times by resuspension in PSS and centrifugation as above. The washed blood cells were then resuspended in IgG4-depleted plasma (i.e. the flow-through from the Capture Select IgG4 columns) to yield IgG4-depleted whole blood.
Detection and Quantification of FAE in Vitro
In vitro FAE of anti-IL-6 (WT or S228P) Abs with anti-TNF WT Abs was quantified, and the presence of newly formed anti-IL-6/anti-TNF bsAbs demonstrated, using two MSD bridging methods: Method 1: indirect quantification of FAE as determined by the loss in ability of monospecific, bivalent anti-IL-6 Abs to cross-link two differently tagged variants of the same antigen. Quenched reaction samples diluted 1:4,000 in PBS/1% BSA (PB) were incubated with 2 μg/ml of a 1:1 mixture of biotinylated IL-6 and Sulfo-tag labeled IL-6 for 1 h at room temperature with agitation. After incubation, samples were transferred to, and incubated with agitation in, PB pre-blocked streptavidin-coated MSD plates (MSD) for 1 h at room temperature before being processed as per Method 2 below. Method 2: directly monitoring FAE as determined by the gain in ability of the newly formed anti-IL-6/anti-TNF bsAbs to cross-link two different antigens. 1:4,000 PB diluted reaction samples were incubated with 2 μg/ml of a 1:1 mixture of biotinylated-IL-6 and Sulfo-tag labeled-TNF in PB for 1 h at room temperature with agitation. After incubation, samples were transferred to, and incubated with agitation in, PB pre-blocked streptavidin coated MSD plates for 1 h at room temperature. After incubation with tagged antigens, wells were washed three times with PBST before signals were revealed and measured using the manufacturer's read buffer and SectorImager 6000 analyzer (MSD). Background values obtained from control parallel reactions in which non-biotinylated-IL-6 was substituted for the biotinylated variant, were subtracted from all signals. Duplicate values from at least three independent experiments were used for all calculations. Averaged data with S.E. values are plotted.
Anti-IL-6/TNF bsAb Generation
CH3 mutants of humanized anti-IL-6 and anti-TNF WT IgG1 Abs were generated using QuikChange site-directed mutagenesis kit (Agilent Technologies). F405L was introduced into the anti-IL-6 Ab and K409R was introduced into the anti-TNF Ab (19). Equal quantities of the Abs were mixed to a final concentration of 0.9 mg/ml and incubated in the presence of 50 mm 2-mercaptoethylamine HCl. Samples were quenched using a 10-fold molar excess of NEM at either, the beginning of the experiment or after 2 h of incubation at 37 °C. Samples were buffer exchanged against PBS, pH 7.4, using Zeba Spin Desalting Columns (Thermo Scientific) and subsequently analyzed using Hydrophobic interaction chromatography (HIC). Approximately, 5 μg of material was loaded onto a ProPAC HIC-10 column (Thermo Scientific), pre-equilibrated in 0.8 m ammonium sulfate, 50 mm sodium phosphate, pH 7.4, attached to a 1260 Infinity HPLC system (Agilent Technologies). Proteins were eluted using a linear gradient of 50 mm phosphate buffer, pH 7.4, and monitored using a fluorescence detector. The percentage of bsAb after FAE was determined by integration of the HIC peaks at their respective retention times.
Assay Characterization and Generation of Anti-IL-6/TNF bsAb Standard Curves
Known amounts of the anti-IL-6/TNF bsAb were serially diluted in relevant matrices (buffer, plasma, or whole blood) containing increasing amounts of parental anti-IL-6 and anti-TNF WT Abs. Serially diluted standards were assayed as in Method 1 and used to characterize the assay as per industry guidelines (20, 21). Data were plotted using GraphPad Prism software and a 4-parameter logistic fit with 1/y2 weighting.
Detection of Anti-IL-6 S228P IgG4 Fab Arm Exchange in Vivo
Plasma samples from healthy volunteers dosed with anti-IL-6 S228P IgG4 were taken at regular time intervals and assayed by two discrete MSD assays namely, total and intact. The total assay detects all monovalent Ab half-molecules (i.e. both non-exchanged and exchanged species) while the intact assay only detects monospecific, bivalent Ab molecules (i.e. only non-exchanged species). In brief, for the total assay, plasma samples were serially diluted in PB and incubated with agitation for 1 h at room temperature with 1 μg/ml of biotinylated IL-6. After incubation, samples were transferred to, and incubated with agitation in PB pre-blocked streptavidin-coated MSD plates for 1 h at room temperature. Wells were then washed three times with PBST before incubation with 1 μg/ml of Sulfo-tag-labeled goat anti-human κ-light chain Ab (to label captured anti-IL-6 S228P IgG4 κ-light chain half-molecules) in PB for another 1 h at room temperature with agitation and processed as per the intact assay below. For the intact assay, after dilution with PB, plasma samples were incubated with agitation for 1 h at room temperature with 2 μg/ml of a 1:1 mixture of biotinylated IL-6 and Sulfo-tag-labeled IL-6. After incubation, wells were washed with PBST and signals revealed and measured as previously detailed. Known amounts of serially diluted anti-IL-6 S228P IgG4 were used in parallel assays to obtain calibration curves from which the plasma anti-IL-6 S228P IgG4 concentrations were calculated. Duplicate values from two independent experiments were used for all calculations. The amount of anti-IL-6 S228P IgG4 FAE was assessed by comparing the two generated profiles. In vivo plasma anti-IL-6 S228P IgG4 half-lives were calculated using GraphPad Prism software.
RESULTS
Analysis of Abs by Non-reducing SDS-PAGE
To investigate FAE four different Abs were used throughout this work, these were: anti-IL-6 WT IgG4, anti-IL-6 S228P IgG4, anti-TNF WT IgG4, and IgG1. Their analysis by denaturing, non-reducing SDS-PAGE, and Coomassie staining is shown in Fig. 1. As can be seen from the gel, the WT IgG4 Abs are a heterogenous preparation containing two predominant species: the full-size, bivalent, monospecific Ab (Fig. 1, open arrowhead) and a lower molecular weight species corresponding to the monovalent, monospecific, heavy-light chain half-molecules (Fig. 1, closed arrowhead). In agreement with Angal et al. (6), this IgG4 WT specific heterogeneity is abolished by the introduction of a single S241P point mutation (numbered according to the Kabat system (22)) or, alternatively, as referred to hereafter, S228P, numbered according to the EU numbering system (23)). In Fig. 1, note the absence of heavy-light chain bands in lane 2 (S228P IgG4) and lane 4 (WT IgG1).
FIGURE 1.
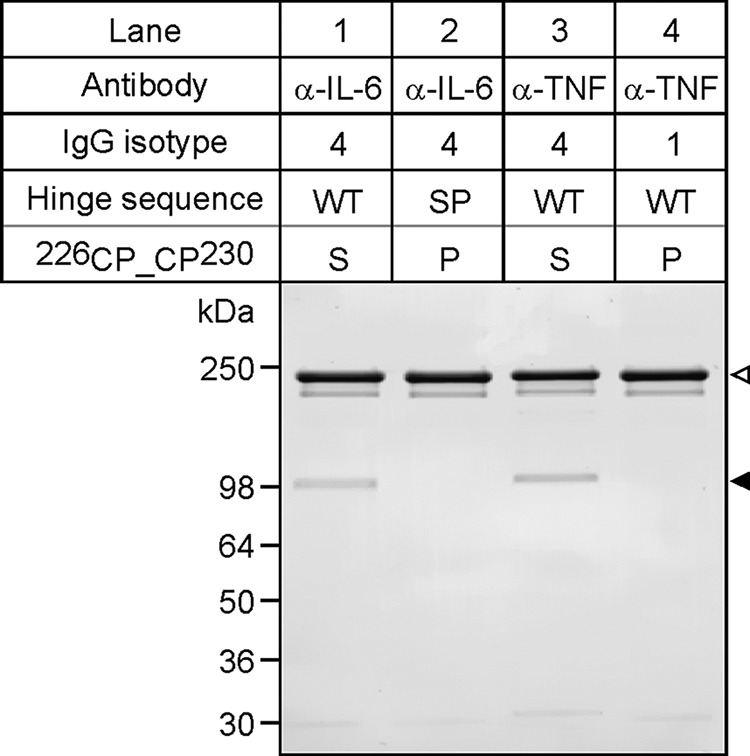
Antibody analysis by SDS-PAGE and Coomassie staining. Equivalent amounts of each Ab were analyzed by denaturing, non-reducing SDS-PAGE and Coomassie staining. The specificity, isotype, hinge sequence (WT or SP mutation) and amino acid identity at position 228 (S, Serine; P, Proline) of each Ab is specified above. Open arrowhead, indicates the position of intact, full-length Abs; closed arrowhead, indicates the position of half-molecules. Note the absence of detectable half-molecules in both the anti-IL-6 S228P and the anti-TNF WT IgG4 Ab preparations (lanes 2 and 4), respectively. The positions and molecular masses of protein markers are shown on the left.
Investigating Fab Arm Exchange in Vitro
To study the effect of the S228P mutation on anti-IL-6 IgG4 FAE in vitro, anti-IL-6 WT IgG4, or anti-IL-6 S228P IgG4 Abs were mixed with either anti-TNF WT IgG4 or IgG1 Abs in a 1:1 molar ratio and the samples were incubated at 37 °C in the presence or absence of GSH. To control for the potential detrimental effect of GSH, anti-IL-6 WT IgG4, or anti-IL-6 S228P IgG4 were incubated on their own in the presence of GSH in a parallel set of reactions. After incubation, the samples were assayed by two MSD-based bridging methods (Fig. 2A). Method 1 exploits the bivalent, monospecific nature of the parental Abs and involves incubation of the reaction mixture with differently tagged variants of the same antigen. A loss in MSD signal by this method infers the loss of parental IgG4 monospecific, bivalent Abs from the reaction mixture, possibly due to FAE with a counterpart Ab. Analysis of reaction mixtures by this method resulted in a marked loss (∼28%) in signal from the GSH supplemented anti-IL-6 WT IgG4 Ab and anti-TNF WT IgG4 Ab sample (Fig. 2B). Interestingly, no such loss was observed in the absence of GSH, the anti-IL-6 WT IgG4 alone, the WT IgG1, or any of the S228P IgG4-containing samples.
FIGURE 2.
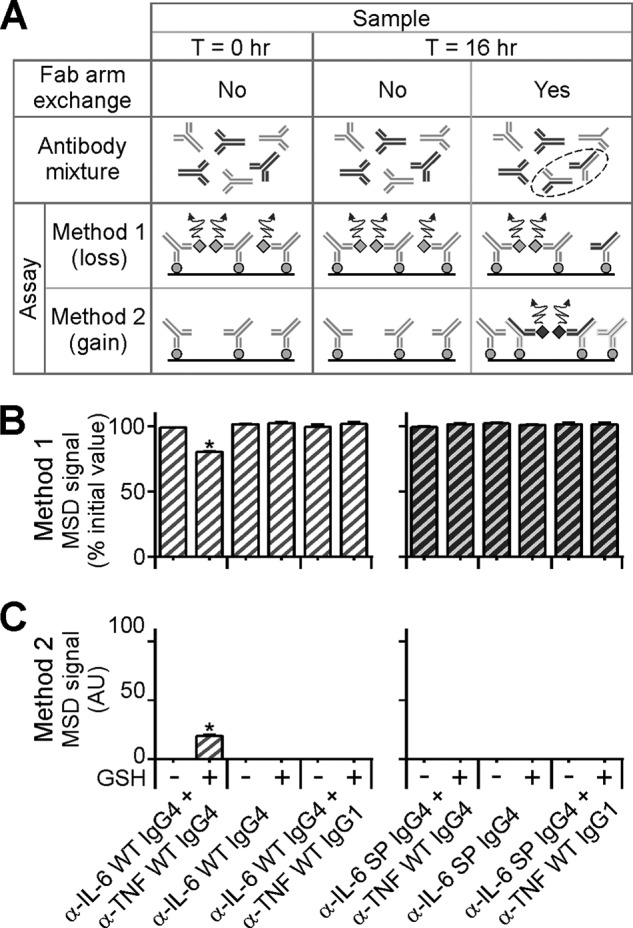
Fab arm exchange. A, schematic representation of in vitro FAE and the assays used for quantification. Method 1: samples incubated with differently tagged variants of the same antigen. A loss in signal after 16 h incubation infers FAE has taken place. Method 2: samples incubated with differently tagged variants of two different antigens. A gain in signal after 16 h incubation infers the presence of newly formed bsAbs arising from FAE. Light gray Y-shapes, anti-IL-6 (WT or S228P) Abs; dark gray Y-shapes, anti-TNF WT Abs; black broken ellipse, highlights the newly formed bispecific (anti-IL-6/anti-TNF) Abs; solid black line, streptavidin coated MSD plates; light gray circles, biotinylated IL-6; light gray diamonds, Sulfo-tag labeled IL-6; dark gray diamonds, Sulfo-tag-labeled TNF; curly arrow, denotes signal generated. B and C, quantifying and detecting FAE in vitro by two MSD-based methods. Ab samples quenched before or after overnight incubation at 37 °C (T = 0 and T = 16 h, respectively) in the presence or absence of GSH were analyzed by the two assays shown in A. Light gray hatched bars, anti-IL-6 WT IgG4 Ab containing samples; dark gray hatched bars, anti-IL-6 S228P IgG4 Ab containing samples; AU, arbitrary units. Note: Only the sample containing anti-IL-6 WT Abs and anti-TNF WT IgG4 Abs incubated in the presence of GSH shows a loss in signal when assayed by Method 1 (B) and an associated gain in signal when assayed by Method 2 (C). Data points represent mean ± S.E. values of three independent measurements. *, p < 0.05, paired t test.
Taken together these results infer that anti-IL-6 WT monospecific, bivalent, IgG4 Abs are specifically lost from the reaction mixture due to GSH-dependent FAE with WT anti-TNF IgG4 (but not WT IgG1) Abs. If true, this process ought to result in the formation of anti-IL-6/anti-TNF bsAbs.
To directly investigate the presence of anti-IL-6/anti-TNF bsAbs, equivalent aliquots of all reaction mixtures were assayed using a second MSD-based method (see Fig. 2A). Method 2 exploits the bivalent, bispecific nature of the newly formed Abs and involves incubation of the reaction mixture with a combination of antigens to first capture and then reveal the bsAbs. A gain in MSD signal by this method infers the formation of newly formed bsAbs from the FAE of disparate, monospecific parental Abs. The results of this analysis, Fig. 2C, show an associated complementary, specific and significant, gain in signal in the same sample in which a loss in signal by Method 1 was previously observed (namely, the GSH supplemented, anti-IL-6 WT IgG4 Ab and anti-TNF WT IgG4 Ab sample). No such gain in signal was detected in the absence of GSH, the anti-IL-6 WT IgG4 alone, the WT IgG1, or any of the S228P IgG4 Ab containing samples.
Taken together these results demonstrate that, under these conditions, the WT IgG4 Abs lost from solution (as demonstrated by Method 1, Fig. 2B) have participated in GSH-dependent FAE with WT IgG4 (but not WT IgG1) Abs resulting in the formation of anti-IL-6/anti-TNF bsAbs (as demonstrated by Method 2, Fig. 2C).
Assay Characterization and Generation of an Anti-IL-6/TNF bsAb Assay Calibrator
To better characterize the assay used in Method 1, an appropriate bsAb assay calibrator was required. Consequently, an anti-IL-6/TNF bsAb was generated using CH3 mutated anti-IL-6 and anti-TNF IgG1 Abs and modified methods of Labrijn et al. (19). Samples of the FAE reaction used to generate the bsAb were analyzed by HIC both, before (Fig. 3A, top panel) and after (Fig. 3A, bottom panel) incubation at 37 °C in reducing conditions. By integrating the area under the bsAb curve, it was determined that the FAE reaction resulted in an approximate 95% yield of anti-IL-6/TNF bsAb.
FIGURE 3.
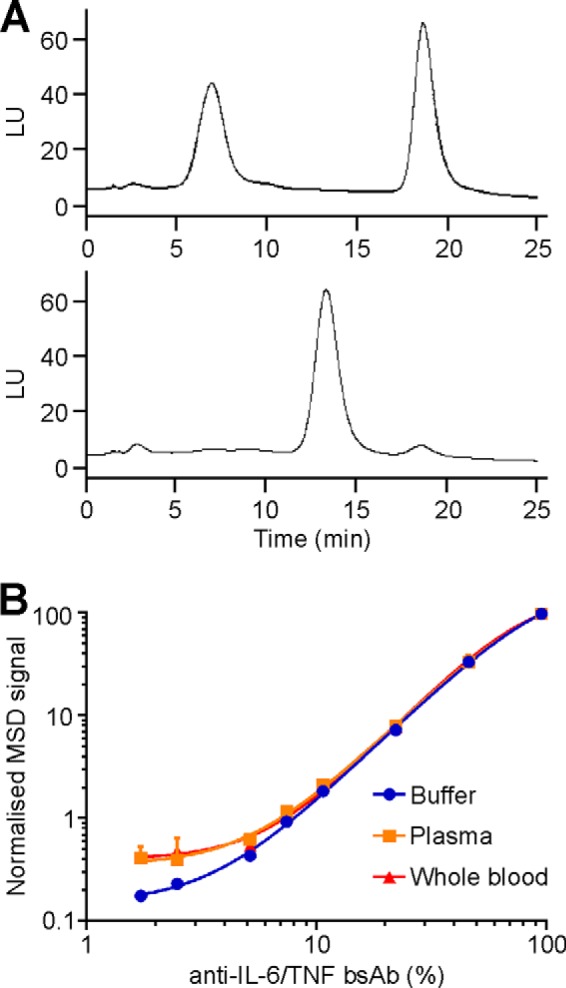
Hydrophobic interaction chromatography (HIC) analysis. A, anti-IL-6 and anti-TNF Abs before (top panel) and after (bottom panel) FAE were analyzed by HIC. Anti-IL-6, anti-TNF, and anti-IL-6/TNF bsAbs had respective retention times of 7.0, 18.7, and 13.3 min. LU: light units. B, anti-IL-6/TNF bsAb calibration curves. Anti-IL-6/TNF bsAb calibration curves were generated by serial dilution of anti-IL-6/TNF bsAbs in various matrices (detailed in figure) and subsequent analysis per Method 1 (main text).
The newly generated bsAb was used to assess the performance, precision and accuracy of the assay and to ensure the assay was fit-for-purpose. Calibrator curves of the bsAb were prepared in different matrices (Fig. 3B) and used to determine the lower limits of quantitation of the assay, which were 5 and 7.5% bsAb for, buffer and plasma/whole blood, respectively. (While beyond the scope of this report, these respective percentages corresponded to 73 and 110 ng/ml of bsAb.)
Depleting Plasma and Blood of Endogenous IgG4 Abs
While PBS provides an Ab-free and buffered environment to monitor FAE, the lack of cells and endogenous factors means this in vitro buffer is far from physiologically relevant. More appropriate physiological matrices to monitor FAE would be blood or plasma. However, the use of such matrices is hampered by the presence and abundance of endogenous IgG4 WT Abs of unknown specificity, which, due to their intrinsic ability to participate in FAE would result in dilution of the formation of the bsAbs of interest (namely, anti-IL-6/anti-TNF) and hence, an underestimation of quantified FAE. Therefore, in order to accurately monitor in vitro FAE of the Abs of interest under physiologically relevant conditions, a novel, IgG4-free, buffered matrix was established by depleting blood of endogenous IgG4 Abs using anti-IgG4 affinity chromatography, Fig. 4A. After centrifugation, the top plasma layer was harvested from the blood and, to ensure exhaustive IgG4 depletion, subjected to two sequential rounds of anti-IgG4 affinity chromatography (with a fresh aliquot of resin each time). To control for nonspecific absorption of the IgG4 Abs to the affinity matrix a control Sepharose column was used in parallel. Chromatography samples were analyzed by non-reducing SDS-PAGE and WB using HRP-conjugated anti-IgG4 Abs (Fig. 4B, left upper panel). To verify and confirm that only IgG4 Abs were being isolated during chromatography samples were additionally analyzed by SDS-PAGE and WB using anti-IgG 1, 2, and 3 HRP-conjugated Abs (Fig. 4B, left lower panels). The fractions containing IgG4 Abs were pooled, and the purity (> 99%) assessed by SDS-PAGE and Coomassie staining (Fig. 4B, right). By measuring absorbance at 280 nm the total protein content of the pooled anti-IgG4 column eluates was approximately, 600 μg/ml. The unbound, flow-through material from the anti-IgG4 column yielded IgG-4-depleted plasma and after addition of washed blood cells yielded IgG4 depleted whole blood (Fig. 4A). These two physiologically relevant, buffered, IgG4-free matrices were subsequently used to monitor FAE.
FIGURE 4.
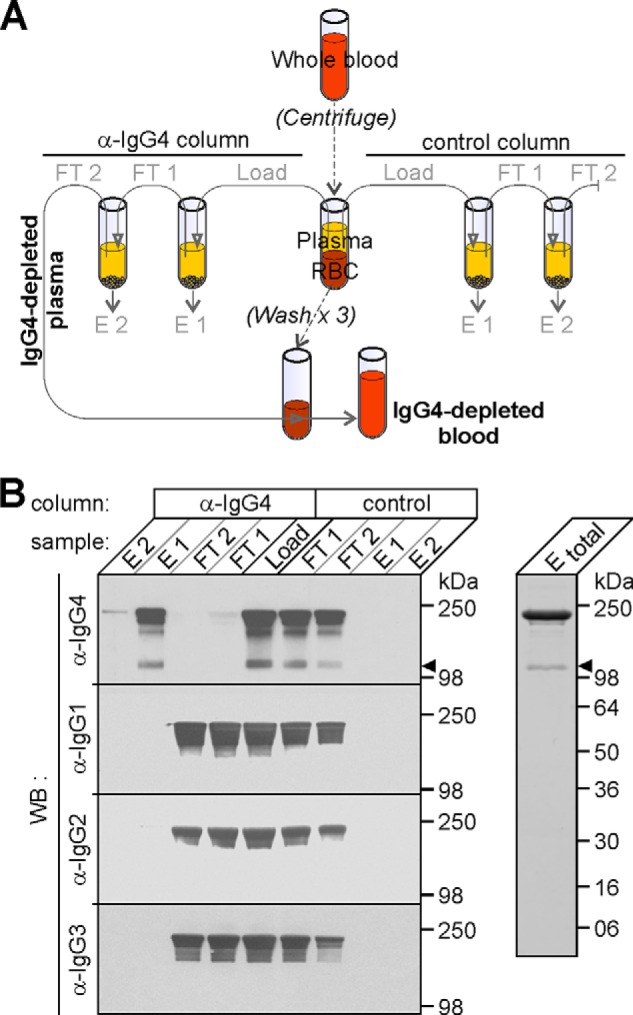
Preparation and analysis of IgG4-depleted physiological matrices. A, schematic of the anti-IgG4 (and control) affinity chromatography employed to deplete endogenous IgG4 Abs. After centrifuging blood, equivalent amounts of plasma (load) were sequentially incubated with either, two anti-Ig4 columns (left hand side) or, in parallel, with two control columns (right hand side). The unbound, flow-through (FT) material from each chromatography was collected (FT1 and 2, respectively) before columns were washed and eluted with SDS-PAGE sample buffer (E1 and 2, respectively). FT2 from the anti-IgG4 affinity chromatography was either used directly (IgG4-depleted plasma) or, was added to washed red blood cells (RBC) before use to yield, IgG4-depleted blood. B, Analysis of chromatography samples by SDS-PAGE, WB, and Coomassie staining. Chromatography samples (from A) were analyzed by SDS-PAGE and WB with HRP-conjugated anti-IgG 4, 1, 2, and 3 Fc specific Abs (specified on the left hand side of the main image). Closed arrowhead, denotes the position of half-molecules (only immunoreactive with anti-IgG4 Abs). Fractions E1 and 2 from the anti-IgG4 affinity chromatography were combined (Etotal) and analyzed by SDS-PAGE and Coomassie staining (right hand image). The positions and molecular masses of protein markers are shown on the right.
In Vitro Anti-IL-6 S228P IgG4 Fab-arm Exchange in IgG4-depleted Physiological Matrices
To ensure that physiologically relevant Ab concentrations were experimentally employed for studying FAE, the 600 μg/ml of endogenous IgG4 Abs removed from whole plasma by anti-IgG4 affinity chromatography (see above) were replaced with an equivalent amount of pre-mixed anti-IL-6 (either WT or S228P) IgG4 Abs and anti-TNF WT IgG4 Abs. To ensure clinically relevant amounts of Abs were used, 60 μg/ml of anti-IL-6 IgG4 Abs (∼equivalent to the Cmax of a 3 mg/kg IV dose)3 were mixed with 540 μg/ml (i.e. a 9-fold molar excess) of anti-TNF WT IgG4 Abs. The Abs were incubated in either of the IgG4-depleted matrices or, as a control, in buffer, in the absence or presence of GSH at 37 °C for 16 h before analysis of FAE by the previously described MSD-based methods.
Analysis of reaction mixtures by Method 1 demonstrated a marked loss in MSD signal (∼66% in IgG4-depleted blood, ∼63% in IgG4-depleted plasma and ∼69% in buffer) of the WT (but not S228P) anti-IL-6 IgG4 Abs with anti-TNF WT IgG4 Abs during GSH-dependent incubation at 37 °C (Fig. 5A).
FIGURE 5.
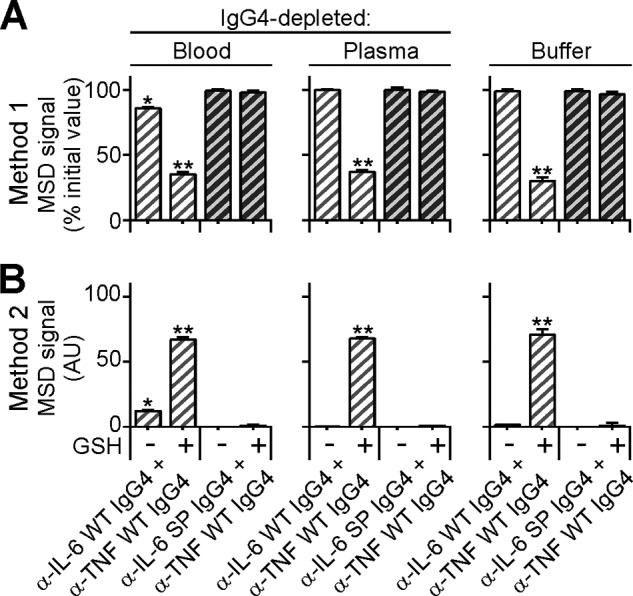
Analysis of FAE in ex vivo IgG-4-depleted physiological matrices. A and B, anti-IL-6 WT IgG4 (light gray hatched bars) or S228P Abs (dark gray hatched bars) containing samples were incubated with anti-TNF WT IgG4 Abs at a 1:9 molar ratio in either, IgG4-depleted blood (left hand side), IgG4-depleted plasma (middle) or PBS buffer (right hand side), in the presence or absence of GSH. After overnight incubation at 37 °C samples were quenched and analyzed for FAE by the two aforementioned assays. A, Method 1; samples incubated with differently tagged variants of the same antigen. B, Method 2; samples incubated with differently tagged variants of two different antigens. AU, arbitrary units. Note: Under physiologically relevant in vitro reducing conditions i.e. in the absence of supplemented GSH, FAE only occurs in the presence of blood cells but not plasma or buffer alone. Data points represent mean ± S.E. values of three independent experiments. *, p < 0.05; **, p < 0.01, paired t test.
Analysis of these same samples by Method 2 showed a specific and significant gain in MSD signal in the same reactions, which previously demonstrated a loss in signal by Method 1 (Fig. 5B). Taken together, these results suggest that the anti-IL-6 WT but not S228P IgG4 Abs undergo FAE with anti-TNF WT IgG4 Abs in the presence of GSH under these ex vivo conditions.
Interestingly, even in the absence of supplemented GSH, the WT Ab variant (but not the S228P mutant) also participates, to a significant and detectable level, in FAE in IgG4-depleted whole blood (but not in IgG4-depleted plasma) (Fig. 5). Since the only difference between these two matrices is the presence of blood cells, these data suggest that it is these, or a component thereof, which are catalyzing the FAE reaction.
In Vivo Anti-IL-6 S228P IgG4 Fab Arm Exchange
To date, the propensity of anti-IL-6 S228P IgG4 to participate in in vitro FAE by mimicking physiologically and clinically relevant in vivo conditions has been investigated. However, to directly investigate in vivo anti-IL-6 S228P IgG4 FAE, plasma samples taken from anonymized human volunteers dosed with anti-IL-6 S228P Ab were assayed using modified protocols of Stubenrauch and Shapiro (11, 17) (as detailed under “Experimental Procedures”). The first MSD assay (total) was designed to measure both exchanged and non-exchanged monovalent anti-IL-6 S228P IgG4 half-molecules able to bind IL-6 antigen while the second assay (intact) was designed to measure only non-exchanged, monospecific, bivalent anti-IL-6 S228P IgG4 molecules. Differences in the plasma concentration profiles from these two methods would be indicative of the extent of in vivo anti-IL-6 S228P IgG4 FAE, as detailed in Ref. 17. However, as can be seen from Fig. 6, analysis of anti-IL-6 S228P IgG4 Ab by the two methods produced overlapping, long-term (>100 days) plasma concentration profiles for both subjects tested inferring that the anti-IL-6 S228P IgG4 Ab does not participate, to any significant level, in in vivo FAE.
FIGURE 6.
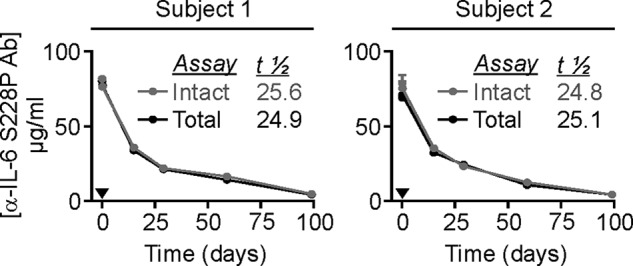
Total and intact anti-IL-6 S228P IgG4 Ab plasma concentration-time profiles of two dosed subjects. Healthy volunteers were given a 3 mg/kg IV dose of anti-IL-6 S228P IgG4 Ab and plasma samples taken at intervals were assayed by two discrete methods. Total assay; detects all monospecific, monovalent anti-IL-6 S228P IgG4 Ab half-molecules (i.e. both non-exchanged and exchanged species). Intact assay; detects only full-size, monospecific, bivalent anti-IL-6 S228P IgG4 Ab molecules (i.e. only non-exchanged species). Black arrowhead, denotes anti-IL-6 S228P IgG4 Ab administration. Black profile, total assay; gray profile, intact assay; half-lives (days) (t½).
DISCUSSION
IgG4 Abs have been shown to undergo a dynamic process of half-molecule swapping known as FAE (15, 16). The data presented herein is in good agreement with previous reports (11, 15, 24), demonstrating that the introduction of the S228P core-hinge mutation in IgG4 Abs is sufficient to limit the tendency of these Abs to form intra-DSBs (6, 11, 25, 26) as assessed by a reduction in the apparent formation of half-molecules (Fig. 1 and Refs. 6, 7, 25). While half-molecules have been shown to exist by various direct and indirect methods (6, 27–29), their precise involvement in FAE remains unclear and a focus of on-going investigation. In this report, two differently configured MSD assay formats were used to characterize and quantify IgG4 FAE in physiologically relevant in vitro matrices. The complementary nature of these assays, assessing both the loss of parental monospecific Abs from the starting material and the associated gain of newly formed bsAbs in the final sample, validates the approach and methodologies employed and provides confidence in the results obtained.
The approach of measuring FAE as a percentage loss of the starting parental, bivalent, monospecific material (Method 1), in a physiologically relevant matrix devoid of any interfering wild-type IgG4 Abs, is a novel method of in vitro FAE quantification. At equilibrium, the theoretical amount of FAE occurring between an equimolar mixture of two WT IgG4 Abs is 50% (19), however, experimentally this was calculated to be ∼28% (Fig. 2). Possible reasons for this apparent discrepancy could be that FAE was measured only under sub-optimal conditions i.e. after a single incubation time of 16 h and in the presence of a single GSH concentration of 0.5 mm. Since the specific pairing of Abs may also affect FAE4 it is also possible that the particular Abs used in this study may require extended incubation times or a different (more optimal) reducing environment to approach the theoretical limit of FAE. (Alternatively, it could be that a proportion of the heterogenous recombinant WT IgG4 Abs used in these reactions is never actually able to participate in FAE due to differences in glycosylation and/or expression). In future, samples could be incubated for different amounts of time, in the presence of different concentrations of GSH (or other reducing agents), and a comprehensive time-course of FAE investigated.
The results indicate that FAE occurred under reducing conditions in both buffer and plasma for IgG4 WT anti-IL-6 Abs but not the IgG4 S228P core-hinge mutated anti-IL-6 Abs or the IgG1 WT anti-TNF Abs (Figs. 2 and 5). These results support the notions that even at the elevated supplemented GSH concentration of 0.5 mm (∼100-fold higher than endogenous GSH concentrations of 2–6 μm (11, 30, 31)) the S228P mutation is sufficient to inhibit FAE to undetectable levels (15, 16) and that IgG1 does not participate in FAE to any significant extent (32). Interestingly, the observed ability of IgG4 WT Abs to undergo FAE in the absence of GSH in blood but not plasma or buffer alone (Fig. 5) is in good agreement with a previous study (16) and is attributed to the difference between IgG4-depleted whole blood and IgG4-depleted plasma i.e. that the redox environment is provided by blood cells.
The novel method of IgG4 Ab depletion, by anti-IgG4 affinity chromatography (Fig. 4), provides a buffered, physiologically relevant matrix to specifically monitor FAE of IgG4 Abs of interest in the absence of contaminating endogenous IgG4 WT molecules. The 600 μg/ml of endogenous IgG4 Abs depleted from plasma by anti-IgG4 affinity chromatography is in good agreement with previous reports which state that IgG4 plasma concentrations can range from 0.1 to 1.4 mg/ml in healthy individuals (33, 34). There was no significant difference in the extent of in vitro FAE in, IgG4-depleted plasma from a healthy volunteer or buffer alone (Fig. 5). These results suggest that, under the specific incubation time and non-physiological GSH concentration employed in these studies, plasma components only have a minor effect on the time course and extent of FAE. However, it is envisaged that these techniques and methodology could, in future, be used as part of a more comprehensive study to investigate the effect of physiological matrix components, from both healthy and diseased individuals, on the various factors and parameters (e.g. the identity of the V-region, Ab ratio, Ab concentration, temperature, time-course of exchange, redox environment, point mutations) implicated in controlling FAE. While previous studies have already investigated the effect of such factors on FAE (8, 28, 35), these investigations have never been carried out in healthy and diseased physiologically relevant matrices in the absence of interfering WT IgG4 Abs.
Taken together, and in good agreement with previously published reports (16, 17), these data suggest that the core-hinge mutation alone is sufficient to prevent FAE, to below the limits of detection of this assay under all conditions investigated i.e. in vitro (Fig. 2), ex vivo (Fig. 5), and in vivo (Fig. 6). The results and novel methodology presented herein pave the way, and provide valuable research tools, for future studies investigating the factors involved in controlling the propensity of IgG4 Abs (mutants or otherwise) to undergo FAE under physiologically relevant in vitro conditions. These studies will allow researchers to gain a better understanding of, and ultimately elucidate, the mechanism of physiological FAE.
J. Jose, personal communication.
H. Kirby, personal communication.
- IgG
- immunoglobulin G
- FAE
- Fab arm exchange
- bsAb
- bispecific antibody
- Ab
- antibody
- NEM
- N-ethylmaleimide
- DSB
- disulfide bond
- MSD
- Meso Scale Discovery.
REFERENCES
- 1. Salfeld J. G. (2007) Isotype selection in antibody engineering. Nat. Biotechnol. 25, 1369–1372 [DOI] [PubMed] [Google Scholar]
- 2. Natsume A., In M., Takamura H., Nakagawa T., Shimizu Y., Kitajima K., Wakitani M., Ohta S., Satoh M., Shitara K., Niwa R. (2008) Engineered antibodies of IgG1/IgG3 mixed isotype with enhanced cytotoxic activities. Cancer Res. 68, 3863–3872 [DOI] [PubMed] [Google Scholar]
- 3. Brüggemann M., Williams G. T., Bindon C. I., Clark M. R., Walker M. R., Jefferis R., Waldmann H., Neuberger M. S. (1987) Comparison of the effector functions of human immunoglobulins using a matched set of chimeric antibodies. J. Exp. Med. 166, 1351–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Riechmann L., Clark M., Waldmann H., Winter G. (1988) Reshaping human antibodies for therapy. Nature 332, 323–327 [DOI] [PubMed] [Google Scholar]
- 5. Aalberse R. C., Stapel S. O., Schuurman J., Rispens T. (2009) Immunoglobulin G4: an odd antibody. Clin. Exp. Allergy 39, 469–477 [DOI] [PubMed] [Google Scholar]
- 6. Angal S., King D. J., Bodmer M. W., Turner A., Lawson A. D., Roberts G., Pedley B., Adair J. R. (1993) A single amino acid substitution abolishes the heterogeneity of chimeric mouse/human (IgG4) antibody. Mol. Immunol. 30, 105–108 [DOI] [PubMed] [Google Scholar]
- 7. Deng L., Wylie D., Tsao Y. S., Larkin B., Voloch M., Ling W. L. (2004) Detection and quantification of the human IgG4 half-molecule, HL, from unpurified cell-culture supernatants. Biotechnol. Appl. Biochem. 40, 261–269 [DOI] [PubMed] [Google Scholar]
- 8. Labrijn A. F., Rispens T., Meesters J., Rose R. J., den Bleker T. H., Loverix S., van den Bremer E. T., Neijssen J., Vink T., Lasters I., Aalberse R. C., Heck A. J., van de Winkel J. G., Schuurman J., Parren P. W. (2011) Species-specific determinants in the IgG CH3 domain enable Fab-arm exchange by affecting the noncovalent CH3-CH3 interaction strength. J. Immunol. 187, 3238–3246 [DOI] [PubMed] [Google Scholar]
- 9. Schuurman J., Perdok G. J., Gorter A. D., Aalberse R. C. (2001) The inter-heavy chain disulfide bonds of IgG4 are in equilibrium with intra-chain disulfide bonds. Mol. Immunol. 38, 1–8 [DOI] [PubMed] [Google Scholar]
- 10. Schuurman J., Van Ree R., Perdok G. J., Van Doorn H. R., Tan K. Y., Aalberse R. C. (1999) Normal human immunoglobulin G4 is bispecific: it has two different antigen-combining sites. Immunology 97, 693–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stubenrauch K., Wessels U., Regula J. T., Kettenberger H., Schleypen J., Kohnert U. (2010) Impact of molecular processing in the hinge region of therapeutic IgG4 antibodies on disposition profiles in cynomolgus monkeys. Drug. Metab. Dispos. 38, 84–91 [DOI] [PubMed] [Google Scholar]
- 12. Margni R. A., Binaghi R. A. (1988) Nonprecipitating asymmetric antibodies. Annu. Rev. Immunol. 6, 535–554 [DOI] [PubMed] [Google Scholar]
- 13. Burton D. R., Wilson I. A. (2007) Immunology. Square-dancing antibodies. Science 317, 1507–1508 [DOI] [PubMed] [Google Scholar]
- 14. Labrijn A. F., Aalberse R. C., Schuurman J. (2008) When binding is enough: nonactivating antibody formats. Curr. Opin. Immunol. 20, 479–485 [DOI] [PubMed] [Google Scholar]
- 15. Labrijn A. F., Buijsse A. O., van den Bremer E. T., Verwilligen A. Y., Bleeker W. K., Thorpe S. J., Killestein J., Polman C. H., Aalberse R. C., Schuurman J., van de Winkel J. G., Parren P. W. (2009) Therapeutic IgG4 antibodies engage in Fab-arm exchange with endogenous human IgG4 in vivo. Nat. Biotechnol. 27, 767–771 [DOI] [PubMed] [Google Scholar]
- 16. van der Neut Kolfschoten M., Schuurman J., Losen M., Bleeker W. K., Martínez-Martínez P., Vermeulen E., den Bleker T. H., Wiegman L., Vink T., Aarden L. A., De Baets M. H., van de Winkel J. G., Aalberse R. C., Parren P. W. (2007) Anti-inflammatory activity of human IgG4 antibodies by dynamic Fab arm exchange. Science 317, 1554–1557 [DOI] [PubMed] [Google Scholar]
- 17. Shapiro R. I., Plavina T., Schlain B. R., Pepinsky R. B., Garber E. A., Jarpe M., Hochman P. S., Wehner N. G., Bard F., Motter R., Yednock T. A., Taylor F. R. (2011) Development and validation of immunoassays to quantify the half-antibody exchange of an IgG4 antibody, natalizumab (Tysabri(R)) with endogenous IgG4. J. Pharm. Biomed. Anal. 55, 168–175 [DOI] [PubMed] [Google Scholar]
- 18. Peters S. J., Smales C. M., Henry A. J., Stephens P. E., West S., Humphreys D. P. (2012) Engineering an improved IgG4 molecule with reduced disulfide bond heterogeneity and increased Fab domain thermal stability. J. Biol. Chem. 287, 24525–24533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Labrijn A. F., Meesters J. I., de Goeij B. E., van den Bremer E. T., Neijssen J., van Kampen M. D., Strumane K., Verploegen S., Kundu A., Gramer M. J., van Berkel P. H., van de Winkel J. G., Schuurman J., Parren P. W. (2013) Efficient generation of stable bispecific IgG1 by controlled Fab-arm exchange. Proc. Natl. Acad. Sci. U.S.A. 110, 5145–5150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. DeSilva B., Smith W., Weiner R., Kelley M., Smolec J., Lee B., Khan M., Tacey R., Hill H., Celniker A. (2003) Recommendations for the bioanalytical method validation of ligand-binding assays to support pharmacokinetic assessments of macromolecules. Pharm. Res. 20, 1885–1900 [DOI] [PubMed] [Google Scholar]
- 21. Viswanathan C. T., Bansal S., Booth B., DeStefano A. J., Rose M. J., Sailstad J., Shah V. P., Skelly J. P., Swann P. G., Weiner R. (2007) Quantitative bioanalytical methods validation and implementation: best practices for chromatographic and ligand binding assays. Pharm. Res. 24, 1962–1973 [DOI] [PubMed] [Google Scholar]
- 22. Kabat E. A., Wu T. T., Reid-Miller M., Perry H., Gottesman K. (1987) Sequences of Proteins of Immunological Interest, 4th Ed., United States Government Printing Office, Number 165–492 [Google Scholar]
- 23. Kabat E. A., Wu T. T., Gottesman K. S., Foeller C. (1991) Sequences of Proteins of Immunological Interest, 5th. Ed., United States Public Health Service, National Institutes of Health, Number 91–3242 [Google Scholar]
- 24. Rispens T., Vennegoor A., Wolbink G. J., Polman C. H., Killestein J. (2012) Natalizumab remains detectable in patients with multiple sclerosis long after treatment is stopped. Mult. Scler. 18, 899–901 [DOI] [PubMed] [Google Scholar]
- 25. Bloom J. W., Madanat M. S., Marriott D., Wong T., Chan S. Y. (1997) Intrachain disulfide bond in the core hinge region of human IgG4. Protein Sci. 6, 407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lewis K. B., Meengs B., Bondensgaard K., Chin L., Hughes S. D., Kjaer B., Lund S., Wang L. (2009) Comparison of the ability of wild type and stabilized human IgG(4) to undergo Fab arm exchange with endogenous IgG(4)in vitro and in vivo. Mol. Immunol. 46, 3488–3494 [DOI] [PubMed] [Google Scholar]
- 27. Rispens T., Davies A. M., Ooijevaar-de Heer P., Absalah S., Bende O., Sutton B. J., Vidarsson G., Aalberse R. C. (2014) Dynamics of inter-heavy chain interactions in human immunoglobulin G (IgG) subclasses studied by kinetic Fab arm exchange. J. Biol. Chem. 289, 6098–6109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rose R. J., Labrijn A. F., van den Bremer E. T., Loverix S., Lasters I., van Berkel P. H., van de Winkel J. G., Schuurman J., Parren P. W., Heck A. J. (2011) Quantitative analysis of the interaction strength and dynamics of human IgG4 half molecules by native mass spectrometry. Structure 19, 1274–1282 [DOI] [PubMed] [Google Scholar]
- 29. Wilkinson I. C., Fowler S. B., Machiesky L., Miller K., Hayes D. B., Adib M., Her C., Borrok M. J., Tsui P., Burrell M., Corkill D. J., Witt S., Lowe D. C., Webster C. I. (2013) Monovalent IgG4 molecules: immunoglobulin Fc mutations that result in a monomeric structure. MAbs 5, 406–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jones D. P., Liang Y. (2009) Measuring the poise of thiol/disulfide couples in vivo. Free Radic Biol. Med. 47, 1329–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Unt E., Zilmer K., Mägi A., Kullisaar T., Kairane C., Zilmer M. (2008) Homocysteine status in former top-level male athletes: possible effect of physical activity and physical fitness. Scand. J. Med. Sci. Sports 18, 360–366 [DOI] [PubMed] [Google Scholar]
- 32. Rispens T., den Bleker T. H., Aalberse R. C. (2010) Hybrid IgG4/IgG4 Fc antibodies form upon 'Fab-arm' exchange as demonstrated by SDS-PAGE or size-exclusion chromatography. Mol. Immunol. 47, 1592–1594 [DOI] [PubMed] [Google Scholar]
- 33. Divatia M., Kim S. A., Ro J. Y. (2012) IgG4-related sclerosing disease, an emerging entity: a review of a multi-system disease. Yonsei. Med. J. 53, 15–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miles J., Riches P. (1994) The determination of IgG subclass concentrations in serum by enzyme-linked immunosorbent assay: establishment of age-related reference ranges for cord blood samples, children aged 5–13 years and adults. Ann. Clin. Biochem. 31, 245–248 [DOI] [PubMed] [Google Scholar]
- 35. Rispens T., Ooijevaar-de Heer P., Bende O., Aalberse R. C. (2011) Mechanism of immunoglobulin G4 Fab-arm exchange. J. Am. Chem. Soc. 133, 10302–10311 [DOI] [PubMed] [Google Scholar]


