Abstract
Ribosomal S6 kinases (S6Ks) have been depicted as critical effectors downstream of growth factor pathways, which play an important role in the regulation of protein synthesis by phosphorylating the ribosomal protein, S6. The goal of this study was to determine whether S6Ks regulate heart size, are critical for the induction of cardiac hypertrophy in response to a pathological or physiological stimulus, and whether S6Ks are critical downstream effectors of the insulin-like growth factor 1 (IGF1)-phosphoinositide 3-kinase (PI3K) pathway. For this purpose, we generated and characterized cardiac-specific S6K1 and S6K2 transgenic mice and subjected S6K1−/−, S6K2−/−, and S6K1−/− S6K2−/− mice to a pathological stress (aortic banding) or a physiological stress (exercise training). To determine the genetic relationship between S6Ks and the IGF1-PI3K pathway, S6K transgenic and knockout mice were crossed with cardiac-specific transgenic mice overexpressing the IGF1 receptor (IGF1R) or PI3K mutants. Here we show that overexpression of S6K1 induced a modest degree of hypertrophy, whereas overexpression of S6K2 resulted in no obvious cardiac phenotype. Unexpectedly, deletion of S6K1 and S6K2 had no impact on the development of pathological, physiological, or IGF1R-PI3K-induced cardiac hypertrophy. These studies suggest that S6Ks alone are not essential for the development of cardiac hypertrophy.
Hypertrophy of cardiac myocytes plays a key role in determining the size of the heart in adult vertebrates (37), and cardiac hypertrophy is an important risk factor for cardiac morbidity and mortality (20). A key feature of cardiac hypertrophy is increased protein synthesis. Protein synthesis is regulated by molecules that interact with the translational machinery of the ribosome. An important molecule is the ribosomal S6 protein, a component of 40S ribosomal proteins, positioned at the interface between 40S and 60S ribosomal proteins and localized to regions involved in mRNA and tRNA recognition (3, 11). The growth factor-stimulated phosphorylation of S6 is believed to be mediated largely by ribosomal S6 kinases (S6Ks) (4, 11). S6Ks are ubiquitously expressed serine/threonine kinases. There are two highly homologous S6Ks in mammals: S6K1 (p70/p85) and S6K2 (p54/p56) (9, 18). S6K1 and S6K2 are reported to be regulated by a number of pathways, including phoshoinositide-3 kinase (PI3K), protein kinase C, extracellular signal-regulated kinase, and calcium pathways (22, 38). The mammalian target of rapamycin (mTOR) is the upstream kinase of S6Ks.
S6Ks have been implicated as important regulators of body and organ size. Deletion of the dS6K gene in the insect Drosophila melanogaster resulted in a high incidence of embryonic lethality, and surviving adults displayed a severe reduction in body size (27). Deletion of S6K1 in mice was not lethal, but mice were approximately 20% smaller at birth and this was maintained throughout adulthood (39). Furthermore, all organs examined were proportionately smaller. The authors suggested that the phenotype was more dramatic in Drosophila than in mice because Drosophila only expresses one form of S6K. By contrast, mice also express S6K2, and this could possibly compensate, in part, for the loss of S6K1. More recently, the characterization of S6K1−/− S6K2−/− mice was reported (30). Absence of both S6K1 and S6K2 impaired animal viability, and mice were similar in size to that described for S6K1−/−.
In vitro and in vivo models of cardiac hypertrophy have suggested that S6Ks play a key role in the stimulation of protein synthesis in the heart. In isolated cardiac myocyte models of hypertrophy (induced by angiotensin II, phenylephrine, or insulin) rapamycin, which inactivates S6Ks via mTOR, inhibited protein synthesis (2, 35, 48). In mice, our investigators previously showed that aortic banding, exercise training, or transgenic expression of insulin-like growth factor 1 receptor (IGF1R) or constitutively active phosphoinositide 3-kinase (caPI3K) induced cardiac hypertrophy (25, 40, 42). In each model, S6K1 activity and/or S6 phosphorylation was elevated in the heart. By contrast, mice expressing a dominant-negative PI3K (dnPI3K) mutant in the heart had significantly smaller hearts, and S6K1 activity and S6 phosphorylation were depressed (40). Furthermore, rapamycin can attenuate and regress pressure overload-induced cardiac hypertrophy (24, 42). Together, these studies suggest that hypertrophic stimuli regulate heart size, at least in part, by the activation of S6Ks.
Despite reasonable evidence to suggest that S6Ks play a key role in determining heart size, it was not clear whether S6Ks alone are critical regulators for the induction of cardiac hypertrophy. The aim of the present study was to determine whether S6Ks regulate heart size in vivo and whether S6Ks are critical effectors for the development of physiological or pathological cardiac hypertrophy. For this purpose we (i) generated and characterized cardiac-specific S6K1 and S6K2 transgenic mice, (ii) subjected S6K1−/−, S6K2−/−, and S6K1−/− S6K2−/− mice to a pathological stress (aortic banding) or a physiological stress (exercise training), and (iii) genetically crossed transgenic and knockout mice with IGF1R and PI3K transgenic mice.
MATERIALS AND METHODS
Generation of S6K1 and S6K2 transgenic mice.
The HA-S6K1 eukaryotic expression plasmids encoding wild-type (WT) and kinase dead (KD) alleles of rat S6K1 (70-kDa isoform) and a rapamycin-resistant (RR) mutant (E389D3E) have previously been described (10). In brief, for the RRS6K1 mutant, four phosphorylation sites within the autoinhibitory domain of S6K1 as well as T389 have been replaced with acidic residues. Together, the mutant has higher basal activity and is largely insensitive to rapamycin. A similar mutant provided the greatest degree of rescue in Drosophila with reduced dTOR activity (50).
The HA-S6K2 eukaryotic expression plasmids encoding WT and KD alleles of human S6K2 (54-kDa isoform) have also been described previously (22, 23). The cDNA inserts for WTS6K1, KDS6K1, RRS6K1, WTS6K2, and KDS6K2 were cloned into a SalI-digested α-myosin heavy chain promoter construct (clone 26; a gift from J. Robbins) (13), and cardiac-specific transgenic mice were generated as previously described (40). Animal care and experimentation were approved by the Institutional Animal Care and Use Committee of the Beth Israel Deaconess Medical Center.
Generation of S6K1−/−, S6K2−/−, and S6K1−/− S6K2−/− mice.
S6K1−/−, S6K2−/−, and S6K1−/− S6K2−/− mice were previously generated and characterized (30, 39). During postnatal life, S6K1−/− and S6K1−/− S6K2−/− mice are approximately 20% smaller by weight than wild-type mice (S6K+/+), whereas S6K2−/− mice are not significantly different from S6K+/+ mice (30, 39). Organ weights are proportionately smaller in S6K1−/− and S6K1−/− S6K2−/− mice.
Injection of IGF1.
Intravenous injection of IGF1 to mice results in the activation of S6K1 and S6 phosphorylation in the heart. To examine whether loss of S6K1, S6K2, or both altered the activation of S6K1 and S6 phosphorylation, mice were injected with IGF1. S6K+/+, S6K1−/−, S6K2−/−, and S6K1−/− S6K2−/− mice were anesthetized with intraperitoneal injections of 2,2,2-tribromoethanol (Avertin; Aldrich). IGF1 (0.5 mg/kg of body weight; rhIGF1; gift from Genentech Inc.) or the same volume of phosphate-buffered saline (PBS) was intravenously injected via a jugular vein for 15 min as described elsewhere (41). On completion, hearts were harvested.
Ascending aortic constriction.
Ascending aortic constriction was performed in male mice at 11 to 13 weeks of age, as described previously (42, 46).
Swimming training.
Groups of 14 to 16 mice at 8 to 12 weeks of age were subjected to chronic swimming training for 4 weeks as described elsewhere (26).
Administration of rapamycin.
Rapamycin is a macrocylic triene antibiotic which has effects on growth by forming a gain-of-function inhibitory complex with FKBP12 (FK506-binding protein) (4, 5, 47). This complex binds to mTOR and inhibits its function. Rapamycin is a potent inhibitor of S6K1, preventing activation of S6K1 by all known agonists at subnanomolar concentrations (4).
To confirm that any changes in heart size induced by the S6K transgenes were specifically due to changes in S6K activity, rapamycin (4 mg/kg/day; gift from Wyeth-Ayerst) was administered to S6K transgenics as previously described (41). The solvent for rapamycin was 0.1% sodium carboxymethylcellulose-0.125% polysorbate 80 in water. Rapamycin or solvent was intraperitoneally administered to nontransgenic (NTg) or S6K transgenic mice from 3 to 4 weeks of age. Mice were sacrificed at 4 weeks, heart weight was measured, and S6K1 activity and S6 phosphorylation were measured in heart lysates.
Echocardiography.
Echocardiography was performed as described elsewhere (26). For aortic banding studies, to evaluate the degree of stenosis, the pressure gradient across the constriction was assessed using Doppler echocardiography. A nonimaging Doppler pencil transducer (continuous wave) was placed at the apex and orientated towards the proximal ascending aorta. The peak velocity (in meters per second) was measured, and the maximum instantaneous gradient (millimeters of Hg) was calculated using the following Bernoulli equation: pressure gradient = 4 × (velocity)2.
Histological analysis.
Heart sections were cut and stained with Masson trichrome as described elsewhere (40).
Biochemical analysis.
Preparation of heart lysates and immunoblotting were performed as previously described (40). S6K1 activity and phosphorylation of S6 (235/236) were examined as described elsewhere (26, 40, 41). S6K2 transgene expression was confirmed with an anti-N-terminal S6K2 antibody (30). S6K2 activity was examined in the same way as S6K1 except that immunoprecipitation was performed with an antihemagglutinin (anti-HA) antibody (F-7; Santa-Cruz Biotechnology, Inc.).
Northern blot analysis.
Northern blot analysis was performed as previously described (40). Total RNA (10.0 μg) was electrophoresed in 1.3% denaturing formaldehyde agarose gels and blotted onto Hybond N membranes (Amersham). Membranes were probed with atrial natriuretic peptide (ANP), brain natriuretic peptide (BNP), α-skeletal actin, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) radiolabeled probes. The probes for ANP (40), BNP (42), α-skeletal actin (26), and GAPDH (17) have previously been described.
Cross-breeding of S6K1 transgenics and S6K knockout mice with IGF1R and PI3K transgenic mice.
Our investigators have previously generated and characterized cardiac-specific transgenic mice expressing IGF1R (25), caPI3K, or dnPI3K (40). To genetically determine the relationship of S6Ks with the IGF1R-PI3K pathway, we performed two independent analyses: (i) WTS6K1 and RRS6K1 mice were crossed with dnPI3K mice, and (ii) S6K1−/− and S6K1−/− S6K2−/− knockout mice were crossed with IGF1R and caPI3K transgenic mice.
Statistical analysis.
Results are expressed as means ± the standard errors of the means (SEM). When comparing groups, statistical significance was determined using one-way analysis of variance. If the analysis of variance showed significance (P < 0.05), it was followed by the Fisher's projected least significant difference post hoc test. The significance level was P < 0.05.
RESULTS
S6K1 and S6K2 transgenics: transgene expression and characterization.
From approximately 20 potential founders for each construct, transgenic founders were identified by Southern blot analysis of tail DNA using the human growth hormone poly(A) as a probe (the growth hormone polyadenylation site is within the α-myosin heavy chain promoter construct). Six lines of transgenic mice per gene construct were initially produced. For confirmation of transgene product in each of the transgenic lines, cardiac tissue lysates from transgenic mice were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and probed with an anti-HA antibody and an anti-S6K1 or anti-S6K2 antibody (Fig. 1). Transgenic lines with significant transgene expression were subsequently analyzed. The heart weight/body weight (HW/BW) ratios of WTS6K1 and RRS6K1 transgenic mice were approximately 10 and 15% greater, respectively, compared to NTg mice at 3 months of age (Table 1). There was no evidence of cardiomyopathic changes, such as necrosis, fibrosis, or myocyte disarray, in hearts from mice at 3 months of age (data not shown), cardiac function as assessed by echocardiography was normal (Table 2), and life span appeared normal.
FIG. 1.
Transgene expression in S6K1 and S6K2 transgenic mice. (A) Cardiac tissue lysates from NTg and S6K1 transgenic mice (WT, WTS6K1; RR, RRS6K1; KD, KDS6K1) were probed with an anti-HA epitope tag antibody (top panel), an anti-S6K1 antibody (middle panel), and an anti-GAPDH antibody (bottom panel). Two bands were present when probing with S6K1 because a start codon is present before the HA tag, in addition to one at the start of the S6K1 gene. (B) Cardiac tissue lysates from NTg and S6K2 transgenic mice (WT, WTS6K2; KD, KDS6K2) were probed with an anti-HA epitope tag antibody (top panel), an anti-S6K2 antibody (middle panel), and an anti-GAPDH antibody (bottom panel).
TABLE 1.
Body weight, organ weights, and tibial length of female S6K transgenic mice at 3 months of agea
| Mice | No. of animals | Body wt (g) | Tibial length (mm) | Organ wt (mg)b
|
HW/BW ratiob (mg/g) | ||
|---|---|---|---|---|---|---|---|
| Heart | Lung | Liver | |||||
| NTg | 14 | 23.5 ± 0.5 | 16.4 ± 0.1 | 92.1 ± 0.8 | 125.8 ± 2.8 | 933.7 ± 25.3 | 3.94 ± 0.05 |
| WTS6K1 | 9 | 24.0 ± 0.5 | 16.7 ± 0.1 | 104.9 ± 4.6* | 136.2 ± 3.9 | 989.1 ± 21.0 | 4.38 ± 0.14* |
| RRS6K1 | 13 | 22.9 ± 0.5 | 16.5 ± 0.1 | 104.2 ± 2.6* | 128.4 ± 2.3 | 940.2 ± 40.7 | 4.54 ± 0.07* |
| KDS6K1 | 9 | 23.0 ± 0.5 | 16.3 ± 0.1 | 92.0 ± 1.8 | 123.2 ± 1.9 | 1,042.5 ± 27.6 | 3.99 ± 0.06 |
| WTS6K2 | 7 | 24.8 ± 0.4 | 16.5 ± 0.1 | 94.3 ± 1.5 | 128.0 ± 4.9 | 1,046.5 ± 39.4 | 3.81 ± 0.06 |
| KDS6K2 | 5 | 25.3 ± 0.6 | 16.1 ± 0.2 | 99.4 ± 1.8 | 127.4 ± 3.0 | 1,047.4 ± 44.9 | 3.93 ± 0.04 |
Results are presented as means ± SEM.
*, P < 0.05 versus NTg.
TABLE 2.
Echocardiographic data from female S6K transgenic mice at 3 months of agea
| Mice | No. of animals | Heart rate (bpm) | IVS (mm) | LVPW (mm) | LV diam (mm)
|
Fractional shortening | |
|---|---|---|---|---|---|---|---|
| Diastolic | Systolic | ||||||
| NTg | 28 | 441 ± 12 | 0.84 ± 0.02 | 0.78 ± 0.02 | 3.18 ± 0.06 | 1.62 ± 0.05 | 50 ± 1 |
| WTS6K1 | 5 | 413 ± 33 | 0.78 ± 0.06 | 0.77 ± 0.07 | 3.59 ± 0.12 | 1.98 ± 0.15 | 45 ± 4 |
| RRS6K1 | 13 | 397 ± 14 | 0.90 ± 0.04 | 0.86 ± 0.04 | 3.25 ± 0.10 | 1.57 ± 0.08 | 52 ± 1 |
| KDS6K1 | 14 | 457 ± 19 | 0.88 ± 0.03 | 0.87 ± 0.02 | 3.08 ± 0.09 | 1.52 ± 0.10 | 51 ± 2 |
| WTS6K2 | 13 | 454 ± 18 | 0.90 ± 0.03 | 0.83 ± 0.05 | 3.31 ± 0.10 | 1.72 ± 0.11 | 48 ± 2 |
| KDS6K2 | 6 | 465 ± 32 | 0.90 ± 0.04 | 0.90 ± 0.06 | 3.18 ± 0.14 | 1.76 ± 0.13 | 45 ± 3 |
Results are presented as means ± SEM. bpm, beats per minute; IVS, interventricular septum thickness; LVPW, left ventricular posterior wall thickness; LV, left ventricular.
The HW/BW ratio of WTS6K2 transgenics was not different from that of NTg or KDS6K2 mice (Table 1), and cardiac function was normal (Table 2).
S6K activity and S6 phosphorylation in S6K1 and S6K2 transgenic mice.
To confirm the activity of the transgene products in S6K1 transgenic mice, heart tissue lysates were immunoprecipitated with a C-terminal anti-S6K1 antibody and subjected to an in vitro kinase assay using glutathione S-transferase-S6 as a substrate. S6K1 activity in heart lysates from WTS6K1 and RRS6K1 transgenics was significantly elevated compared to activity in NTg and KDS6K1 animals (Fig. 2A) (WTS6K1, 6.6-fold ± 1.2-fold, n = 7 from four separate experiments; RRS6K1, 11.4-fold ± 2.6-fold, n = 7 from three separate experiments). At the time of these experiments, S6K1 was thought to be primarily responsible for the phosphorylation of S6. Unexpectedly, phosphorylation of S6 was not significantly elevated in hearts from WTS6K1 and RRS6K1 mice (Fig. 2B).
FIG. 2.
S6K1 activity and S6 phosphorylation in S6K1 transgenic mice. (A) Representative blot showing S6K1 activity in heart lysates from NTg, WTS6K1 (WT), KDS6K1 (KD), and RRS6K1 (RR) mice. IP, immunoprecipitation; −, negative control (i.e., without antibody). (B) Phosphorylation of ribosomal S6 protein in NTg (N), WTS6K1 (W), RRS6K1 (R), and KDS6K1 (K) mice (upper panel). The membrane was stripped and reprobed with GAPDH (lower panel) for quantitation (right panel). Mean values for NTg were normalized to 1.
S6K2 activity was measured in heart lysates from S6K2 transgenic mice using an anti-HA antibody. This method was not ideal, because it did not allow us to detect basal S6K2 activity in NTg; however, at the time of the study the S6K2 antibody available was unable to detect S6K2 activity in heart lysates. S6K2 activity was approximately twofold greater in WTS6K2 transgenics than in KDS6K2 transgenics (Fig. 3A). S6 phosphorylation was not different between groups (Fig. 3B). Despite no differences in the mean values for S6 phosphorylation in S6K1 and S6K2 transgenics, there was marked variation between samples (including NTg). We considered that the feeding status of the mice just prior to sacrifice may affect S6 phosphorylation. However, we detected no significant difference in S6 phosphorylation in the hearts of mice fasted for 12 h compared to those with free access to food (data not shown).
FIG. 3.
S6K2 activity and S6 phosphorylation in S6K2 transgenic mice. (A) Blot showing S6K2 activity in heart lysates from NTg, WTS6K2 (WT), and KDS6K2 (KD) mice. IP, immunoprecipitation; −, negative control (i.e., without antibody). (B) Phosphorylation of ribosomal S6 protein in NTg (N), WTS6K2 (W), and KDS6K1 (K) mice. The membrane was stripped and reprobed with GAPDH (lower panel) for quantitation (right panel). Mean values for NTg were normalized to 1.
Effect of rapamycin on heart size of WTS6K1 and RRS6K1 mice.
To confirm that the increase in heart size observed in WTS6K1 and RRS6K1 transgenics was due to an increase in S6K1 activity, we treated NTg, WTS6K1, and RRS6K1 mice with rapamycin at 3 weeks of age for 1 week. Rapamycin is reported to have a high degree of specificity for the protein kinase mTOR (upstream of S6Ks). Rapamycin did not significantly inhibit a group of 24 protein kinases (including PKA, ERK2, protein kinase Cα, AMPK, JNK1α1, and GSK3β) at 1 μM, a concentration 10- to 20-fold higher than that required to inhibit mTOR in cell-based assays (7).
The HW/BW ratios for WTS6K1 and RRS6K1 transgenics were significantly elevated compared to that in NTg littermates at 4 weeks of age (Fig. 4A). The HW/BW ratio for rapamycin-treated WTS6K1 mice was significantly smaller than that of vehicle-treated WTS6K1 mice and was not different from that of NTg mice (Fig. 4A). Rapamycin reduced S6K1 activity and S6 phosphorylation in the hearts of WTS6K1 mice to a similar degree to rapamycin-treated NTg (Fig. 4B). These results suggest that the cardiac hypertrophy observed in WTS6K1 transgenics was due to overexpression of WTS6K1. In RRSK1 transgenics, rapamycin caused some reduction in the HW/BW ratio and S6K1 activity (Fig. 4). However, the HW/BW ratio and S6K1 activity of RRS6K1 transgenics were still significantly greater than those of NTg mice. Unexpectedly, despite the modest effect of rapamycin on S6K1 activity in hearts of RRS6K1 transgenics, S6 phosphorylation was completely inhibited (Fig. 4B). Because rapamycin also inhibits S6K2 activity (18), these data suggest that S6K2 may have a primary role in mediating S6 phosphorylation in vivo rather than S6K1.
FIG. 4.
Effect of rapamycin treatment in S6K1 transgenics. (A) HW/BW ratios of NTg and S6K1 transgenic mice treated with vehicle (NTg, n = 10; WTS6K1, n = 6; RRS6K1, n = 8) or rapamycin (NTg, n = 11; WTS6K1, n = 5; RRS6K1, n = 4) for 1 week. (B) S6K1 activity (upper panel), phosphorylation of S6 (middle panel), and GAPDH (lower panel) in NTg and S6K1 transgenic mice treated with vehicle or rapamycin. *, P < 0.05 compared with vehicle-treated NTg. #, P < 0.05 compared with vehicle treatment of the same genotype. ^, P < 0.05 compared with rapamycin-treated NTg.
Phosphorylation of S6.
It was initially thought that S6K1 was the sole in vivo kinase responsible for regulating S6 phosphorylation. However, very recent data obtained from S6K2−/− mice suggest that S6K2 is primarily responsible for the phosphorylation of S6 in vivo (30). In the present study, phosphorylation of S6 was not significantly different in heart lysates from WTS6K1, RRS6K1, and NTg mice. To investigate this further, global S6K knockout mice as well as wild-type animals (S6K+/+) were given injections of IGF1. In cardiac lysates of S6K+/+ mice, a bolus injection of IGF1 increased S6K1 activity and S6 phosphorylation and resulted in a shift in the phosphorylation state of S6K1 protein (Fig. 5). There was also an increase in S6 phosphorylation with injection of PBS but not S6K1 activity. As expected, S6K1 activity did not increase in heart lysates from S6K1 knockout mice with injection of IGF1. However, S6 phosphorylation increased to a similar extent to that observed in S6K+/+ mice. By contrast, S6 phosphorylation was almost absent in hearts of S6K2−/− mice and completely absent in hearts of S6K1−/− S6K2−/− mice at baseline or in response to IGF1 (Fig. 5). This suggests, at least in the mammalian heart, that S6K2 rather than S6K1 is the in vivo kinase responsible for regulating S6 phosphorylation. Though, it remains unclear why S6 phosphorylation was not elevated in hearts of S6K2 transgenics (Fig. 3B).
FIG. 5.
Intravenous bolus injection of IGF1 in S6K+/+, S6K1−/−, S6K2−/−, and S6K1−/− S6K2−/− mice. Hearts were collected 15 min after the injection. S6K1 activity (upper panel), immunoprecipitated S6K1 antibody in heart lysates; S6K1 protein (middle panel), immunoprecipitated S6K1 antibody; S6 phosphorylation (lower panel), direct Western blotting. N, no injection; PBS, PBS injection.
Fetal gene expression.
Cardiac hypertrophy is often associated with reexpression of the fetal gene program (16). This terminology has been used because several genes not normally expressed in the adult ventricle, but seen in the embryonic and neonatal heart, are reexpressed in the stressed heart. In the present study, the gene expression of ANP, BNP, and α-skeletal actin was elevated in hearts from RRS6K1 transgenic mice (Fig. 6A). Consistent with these findings, the expression levels of ANP and α-skeletal actin were depressed in S6K1−/− mice, and there was a trend of lower BNP (Fig. 6B). ANP but not BNP or α-skeletal actin was elevated in WTS6K1 transgenics. The reason for this finding is not clear.
FIG. 6.
Fetal gene expression in S6K1 transgenics and S6K1−/− mice. (A) Northern blot showing total RNA from ventricles of NTg, WTS6K1, and RRS6K1 mice (left panel). Expression of GAPDH was determined to verify equal loading of RNA. (Right panel) Quantitative analysis of Northern blots. Mean values for NTg mice were normalized to 1; n = 3 in each group. *, P < 0.05 compared to NTg. (B, left) Northern blot showing total RNA from ventricles of S6K+/+ and S6K1−/− mice. (Right) Quantitative analysis of Northern blots. Mean values for S6K+/+ were normalized to 1; n = 3 in each group. *, P < 0.05 compared to S6K+/+; α-sk act, α-skeletal actin.
Response of RRS6K1 transgenic mice to a pathological stimulus (pressure overload) or a physiological stimulus (chronic exercise training).
Under nonstressed conditions, WTS6K1 and RRS6K1 transgenic mice displayed a relatively mild hypertrophic phenotype. In this set of experiments we examined whether the hypertrophic responses of RRS6K1 transgenic mice to aortic banding or swimming training were similar to those of NTg littermates. The HW/BW ratios for NTg and RRS6K1 transgenics increased significantly in response to aortic banding or swimming training compared to control (non-swim or sham) (Fig. 7). Interestingly, the percent increase in HW/BW in response to exercise was similar between the two groups (NTg, ≈45%; RRS6K1, ≈41%; Fig. 7A), but RRS6K1 transgenics displayed a decreased response to aortic banding (NTg, ≈50% increase; RRS6K1, ≈20% increase; Fig. 7B). The degree of aortic stenosis, measured by the aortic pressure gradients (AoPG) across the bands of NTg and RRS6K1 transgenic mice, was similar (31.6 ± 2.9 and 26.6 ± 1.6 mm Hg, respectively). The reason for the blunted hypertrophic response in RRS6K1 transgenics subjected to aortic banding is not clear. We recently reported a similar phenomenon in IGF1R transgenics (25). This may be related, in part, to some signaling molecules playing distinct roles for the induction of pathological and physiological cardiac hypertrophy (26).
FIG. 7.
Response of NTg and RRS6K1 transgenics to a physiological or pathological stimulus. (A) Response to 4 weeks of swimming training (Sw; NTg, n = 6; RRS6K1, n = 8) compared to the non-swim group (Non-sw; NTg, n = 10; RRS6K1, n = 7). (B) Response to aortic banding for 1 week (NTg, n = 5; RRS6K1, n = 5) compared to sham (NTg, n = 6; RRS6K1, n = 5). *, P < 0.05 compared to non-swim or sham of the same genotype; #, P < 0.05 compared to NTg sham or non-swim; ^, P < 0.05 compared to NTg swim.
Effect of S6K1 and S6K2 deletion on cardiac hypertrophy induced by a pathological stimulus (pressure overload) or a physiological stimulus (exercise induced).
Aortic banding and chronic swimming training induce a significant degree of cardiac hypertrophy in mice (25, 26, 42). Unexpectedly, deletion of S6K1 and/or S6K2 had no significant effect on the increase in HW/BW ratio induced by chronic swimming training (Fig. 8A; Table 3) or aortic constriction (Fig. 8B; Table 4). For exercise training, blinded observers were unable to distinguish S6K knockout mice from S6K+/+ mice while they underwent swimming training. Because S6K knockout mice also lack S6Ks in skeletal muscle and S6K1 has been implicated for a role in protein synthesis of skeletal muscle with exercise (14), we considered that S6K knockout mice may not be able to exercise to the same degree as S6K+/+ animals. However, even if this were the case, one would expect S6K knockout mice to display a reduced hypertrophic response to exercise training. For aortic banding experiments, the degree of aortic stenosis, measured by the AoPG across the bands of S6K+/+, S6K1−/−, S6K2−/−, and S6K1−/− S6K2−/− groups was similar (Table 4).
FIG. 8.
Response of S6K+/+, S6K1−/−, S6K2−/−, and S6K1−/− S6K2−/− mice to pathological and physiological stumuli. (A) Response to chronic exercise training (swimming for 4 weeks). *, P < 0.05 compared with non-swim. (B) Response to pressure overload (ascending aortic banding for 1 week). *, P < 0.05 compared with sham-operated mice.
TABLE 3.
Response of S6K knockout mice to chronic swimming traininga
| Mice | n | BW (g) | TL (mm) | HW (mg) | Normalized HW | HW/BW (mg/g) | HW/TL (mg/mm) | Normalized HW/BW |
|---|---|---|---|---|---|---|---|---|
| Wild type S6K+/+ | ||||||||
| Non-swim | 7 | 23.5 ± 1.2 | 16.5 ± 0.2 | 113.7 ± 8.0 | 1.00 ± 0.07 | 4.81 ± 0.13 | 6.86 ± 0.44 | 1.00 ± 0.03 |
| Swim | 5 | 23.4 ± 0.8 | 16.6 ± 0.1 | 144.7 ± 7.5^ | 1.27 ± 0.07^ | 6.18 ± 0.24^ | 8.72 ± 0.48^ | 1.29 ± 0.05^ |
| S6K1−/− | ||||||||
| Non-swim | 6 | 20.7 ± 0.7*⧣ | 15.6 ± 0.2*⧣ | 95.9 ± 4.0*⧣ | 1.00 ± 0.04 | 4.64 ± 0.15 | 6.13 ± 0.25 | 0.97 ± 0.03 |
| Swim | 8 | 20.2 ± 0.7*⧣ | 15.8 ± 0.1*⧣ | 115.9 ± 4.1^ | 1.21 ± 0.04^ | 5.75 ± 0.17^ | 7.34 ± 0.25^+ | 1.20 ± 0.04^ |
| S6K2−/− | ||||||||
| Non-swim | 5 | 28.0 ± 1.2* | 16.5 ± 0.1 | 118.3 ± 2.0 | 1.00 ± 0.02 | 4.25 ± 0.15 | 7.17 ± 0.10 | 0.88 ± 0.03 |
| Swim | 6 | 24.7 ± 0.7 | 16.4 ± 0.1 | 136.7 ± 4.1^ | 1.16 ± 0.03^ | 5.56 ± 0.21^+ | 8.32 ± 0.25^ | 1.16 ± 0.04^+ |
| S6K1−/− S6K2−/− | ||||||||
| Non-swim | 3 | 20.8 ± 1.5⧣ | 15.2 ± 0.3*⧣ | 89.2 ± 2.8*⧣ | 1.00 ± 0.03 | 4.32 ± 0.28 | 5.87 ± 0.24 | 0.90 ± 0.06 |
| Swim | 2 | 19.7 ± 0.4*⧣ | 15.1 ± 0.2*⧣ | 118.6 ± 6.9^ | 1.33 ± 0.08^ | 6.03 ± 0.22^ | 7.85 ± 0.38^ | 1.25 ± 0.05^ |
Results are presented as means ± standard errors of the mean. TL, tibia length. Normalized HW, heart weights of swimming mice were normalized to those of nonswimming mice of the same genotype. Normalized HW/BW, HW/BW ratios of all groups were normalized to that of nonswimming wild-type S6K+/+ mice. *, P < 0.05 compared to S6K+/+ non-swim; ⧣, P < 0.05 compared to S6K2−/− non-swim; ^, P < 0.05 compared to non-swim mice in same group; +, P < 0.05 compared to S6K+/+ swim group.
TABLE 4.
Response of male S6K knockout mice to ascending aortic bandinga
| Mice | n | BW (g) | TL (mm) | HW (mg) | Normalized HW | HW/BW (mg/g) | HW/TL (mg/mm) | Normalized HW/BW | AoPG (mm Hg) |
|---|---|---|---|---|---|---|---|---|---|
| Wild type S6K+/+ | |||||||||
| Sham | 4 | 29.1 ± 2.2 | 17.4 ± 0.4 | 138.7 ± 6.9 | 1.00 ± 0.05 | 4.81 ± 0.17 | 7.98 ± 0.26 | 1.00 ± 0.04 | 3 ± 0 |
| Band | 10 | 28.4 ± 0.6 | 17.4 ± 0.1 | 195.7 ± 9.8^ | 1.41 ± 0.07^ | 6.90 ± 0.32^ | 11.27 ± 0.55^ | 1.43 ± 0.07^ | 32 ± 3^ |
| S6K1−/− | |||||||||
| Sham | 4 | 23.3 ± 1.3*⧣ | 16.2 ± 0.5*⧣ | 115.2 ± 6.3 | 1.00 ± 0.06 | 4.96 ± 0.20 | 7.12 ± 0.27 | 1.03 ± 0.04 | 3 ± 0 |
| Band | 12 | 23.2 ± 0.6*⧣ | 16.1 ± 0.2*⧣ | 158.9 ± 10.0^ | 1.38 ± 0.09^ | 6.83 ± 0.40^ | 9.86 ± 0.57^+ | 1.42 ± 0.08^ | 37 ± 7^ |
| S6K2−/− | |||||||||
| Sham | 3 | 28.9 ± 0.7 | 17.7 ± 0.1 | 140.1 ± 3.7 | 1.00 ± 0.03 | 4.84 ± 0.12 | 7.92 ± 0.23 | 1.01 ± 0.03 | 3 ± 0 |
| Band | 12 | 30.4 ± 1.2 | 17.5 ± 0.1 | 209.4 ± 10.0^ | 1.50 ± 0.07^ | 6.94 ± 0.31^ | 11.96 ± 0.54^ | 1.44 ± 0.07^ | 24 ± 2^ |
| S6K1−/− S6K2−/− | |||||||||
| Sham | 10 | 21.4 ± 0.9*⧣ | 15.5 ± 0.1*⧣ | 101.5 ± 1.4 | 1.00 ± 0.01 | 4.81 ± 0.21 | 6.55 ± 0.10 | 1.00 ± 0.04 | 3 ± 0 |
| Band | 14 | 21.8 ± 0.5*⧣ | 15.9 ± 0.2*⧣ | 140.8 ± 5.9^ | 1.39 ± 0.06^ | 6.50 ± 0.33^ | 8.87 ± 0.35^+ | 1.35 ± 0.07^ | 29 ± 3^ |
Results are presented as means ± standard errors of the mean. TL, tibia length. Normalized HW, heart weights of banded mice were normalized to those of sham mice of the same genotype. Normalized HW/BW, HW/BW ratios of all groups were normalized to that of sham wild-type S6K+/+ mice. *, P < 0.05 compared to S6K+/+ sham; ⧣, P < 0.05 compared to S6K2−/− sham; ^, P < 0.05 compared to sham mice in the same group; +, P < 0.05 compared to S6K+/+ band and S6K2−/− band groups.
Cross-breeding of S6K1 transgenics and S6K knockout mice with IGF1R and PI3K transgenic mice.
S6K1 is postulated to be an important downstream effector of the IGF1R-PI3K pathway. Our investigators previously reported that transgenic expression of IGF1R or a constitutively activated PI3K mutant resulted in cardiac hypertrophy which was associated with increased S6K1 activity and S6 phosphorylation (25, 40). By contrast, transgenic expression of a dnPI3K mutant resulted in a reduction in heart size, and this was associated with depressed S6K1 activity (40).
(i) Cross-breeding WTS6K1 and RRS6K1 with dnPI3K transgenic mice.
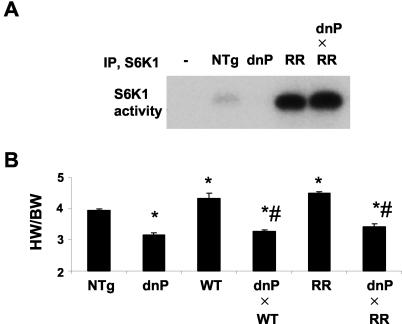
Because S6K1 is considered to be downstream of PI3K, we hypothesized that the S6K1 transgenes would rescue the fall in S6K1 activity in hearts of dnPI3K mice and would be sufficient to rescue the small heart phenotype of dnPI3K transgenics. We previously showed that expression of a constitutively active Akt mutant (Akt is a downstream effector of PI3K) was able to rescue the small heart phenotype of dnPI3K transgenics (41). In the present study, S6K1 activity was lower in dnPI3K transgenics than in NTg (Fig. 9A), as previously shown (40). S6K1 activity was elevated in double transgenic mice expressing both the dnPI3K mutant and RRS6K1 transgene and not different from that observed in RRS6K1 transgenics (Fig. 9A). A similar finding was apparent in double transgenic mice expressing the dnPI3K mutant and WTS6K1 transgene (data not shown). Rescuing S6K1 activity had no impact on heart size. The HW/BW ratio of double transgenics was not different from that of dnPI3K transgenics alone (Fig. 9B).
FIG. 9.
Genetic interaction of PI3K and S6K1 in the regulation of heart size. (A) S6K1 activity in NTg, dnPI3K transgenic (dnP), RRS6K1 (RR), and double transgenic (dnPI3K × RRS6K1 [dnP×RR]) mice. IP, immunoprecipitation; −, negative control (i.e., without antibody). (B) HW/BW ratios of dnPI3K, WTS6K1 (WT), RRS6K1, and double transgenic mice. *, P < 0.05 compared with NTg; #, P < 0.05 compared with WTS6K1 and RRS6K1. n was ≥5 in each group.
(ii) Cross-breeding S6K1−/− and S6K1−/− S6K2−/− mice with IGF1R and caPI3K transgenics.
To further examine whether S6Ks are critical for IGF1R- and PI3K-induced cardiac hypertrophy, S6K1−/− and S6K1−/− S6K2−/− mice were crossed with caPI3K and IGF1R transgenics. Deletion of S6K1 alone or together with deletion of S6K2 had no significant effect on hypertrophy induced by IGF1R or caPI3K (Table 5).
TABLE 5.
HW/BW data for caPI3K and IGF1R transgenic mice with and without deletion of S6Ksa
| Genotype | n | BW (g) | HW (mg) | Tibial length (mm) | HW/BWb |
|---|---|---|---|---|---|
| Deletion of S6K1 | |||||
| S6K1+/− | 15 | 27.8 ± 1.1 | 123.9 ± 5.2 | 16.9 ± 0.1 | 4.46 ± 0.09 |
| S6K1−/− | 10 | 24.2 ± 0.6 | 106.5 ± 3.9 | 15.8 ± 0.1 | 4.38 ± 0.08 |
| caPI3K S6K1+/− | 15 | 27.8 ± 1.2 | 148.5 ± 7.0 | 16.7 ± 0.1 | 5.34 ± 0.13* |
| caPI3K S6K1−/− | 8 | 23.7 ± 0.9 | 135.2 ± 4.5 | 16.1 ± 0.1 | 5.75 ± 0.24* |
| IGF1R S6K1+/− | 10 | 25.5 ± 1.0 | 156.0 ± 7.2 | 16.5 ± 0.2 | 6.15 ± 0.27* |
| IGF1R S6K1−/− | 9 | 22.1 ± 1.4 | 130.5 ± 10.6 | 15.8 ± 0.2 | 5.86 ± 0.18* |
| Deletion of S6K1 and S6K2 | |||||
| S6K1+/− S6K2+/− | 12 | 30.0 ± 1.6 | 130.4 ± 6.5 | 17.1 ± 0.1 | 4.35 ± 0.11 |
| S6K1−/− S6K2−/− | 9 | 25.2 ± 1.7 | 112.6 ± 9.5 | 15.9 ± 0.2 | 4.45 ± 0.14 |
| caPI3K S6K1+/− S6K2+/− | 12 | 31.2 ± 2.0 | 159.9 ± 9.5 | 17.1 ± 0.2 | 5.19 ± 0.22⧣ |
| caPI3K S6K1−/− S6K2−/− | 10 | 25.9 ± 1.3 | 124.7 ± 6.2 | 15.6 ± 0.2 | 4.82 ± 0.09⧣ |
| IGF1R S6K1+/− S6K2+/− | 5 | 30.4 ± 2.4 | 166.8 ± 7.7 | 17.3 ± 0.2 | 5.56 ± 0.23⧣ |
| IGF1R S6K1−/− S6K2−/− | 5 | 24.0 ± 1.3 | 143.6 ± 13.4 | 15.7 ± 0.2 | 6.02 ± 0.61⧣ |
Results are presented as means ± standard errors of the mean.
*, P < 0.05 compared with S6K1+/− mice; ⧣, P < 0.05 compared with S6K1+/− S6K2+/− mice.
DISCUSSION
In the present study we examined the role of S6Ks in regulating heart size (i) during normal developmental growth, (ii) in response to a pathological stimulus, (iii) in response to a physiological stimulus, and (iv) in response to IGF1R-PI3K-induced growth. Studies in Drosophila, mice, and cell culture have suggested that S6K1 is a critical effector of cell growth (10, 27, 39). Further, S6K1 appeared critical for regulating protein synthesis in cardiac myocytes (2, 35, 42, 48). However, most of these studies utilized rapamycin, an agent that inhibits S6K1 activity via mTOR, rather than directly inhibiting S6K1 itself. Rapamycin also indirectly inhibits activation of S6K2, 4E-BP1, and possibly other targets (31).
Transgenic expression of WTS6K1 induced a modest increase in heart size, which could be attenuated by treatment with rapamycin. By contrast, hypertrophy induced by overexpression of a rapamycin-resistant mutant (RRS6K1) was not significantly affected by rapamycin treatment, suggesting that the increase in heart size in S6K1 transgenics was due to overexpression of S6K1. Transgenic expression of S6K2 had no significant effect on heart size. This is consistent with previous reports that demonstrated that deletion of S6K1 reduced body size, whereas deletion of S6K2 had no effect (30, 39). When we began these studies, reports suggested that S6K1 regulated growth via the phosphorylation of S6. S6 phosphorylation was inhibited in embryonic stem cells with S6K1 deleted (45). Thus, it was somewhat surprising that S6 phosphorylation was not elevated in hearts of S6K1 transgenics. However, very recent work characterizing S6K2−/− and S6K1−/− S6K2−/− mice suggests that S6K2 rather than S6K1 is largely responsible for the phosphorylation of S6 (30). This is consistent with our findings in the heart. S6 phosphorylation was not elevated in hearts of S6K1 transgenics, and S6 phosphorylation increased normally in S6K1−/− mice in response to IGF1. By contrast S6 phosphorylation under basal conditions, as well as in response to IGF1 stimulation, was markedly depressed in S6K2−/− mice. Although, based on these data, we expected S6 phosphorylation to be elevated in hearts of S6K2 transgenics. The reason why S6 phosphorylation was not elevated in S6K2 transgenics is not clear. Though, it could be due to the relatively modest increase in S6K2 activity (approximately twofold) in our transgenic mice. Alternatively, overexpression of S6K2 alone may not be sufficient to increase S6 phosphorylation. As eluded to by Pende et al. (30), body size is reduced in S6K1−/− mice with little effect on S6 phosphorylation, whereas body size of S6K2−/− mice is normal but S6 phosphorylation is reduced. The authors suggested that S6K1 and S6K2 may exert specific functions through distinct substrates.
Cardiac hypertrophy can be induced by pathological stimuli (e.g., pressure or volume overload) or physiological stimuli (e.g., developmental growth, exercise training). Both forms of hypertrophy have distinct characteristics and appear to be mediated by distinct signaling pathways (1, 6, 8, 12, 15, 16, 19, 26, 28, 29, 36, 40, 44, 49). A number of pieces of evidence led us to hypothesize that deletion of S6Ks would modulate, at least in part, the hypertrophic response to aortic banding, exercise training, or IGF1R- or caPI3K-induced hypertrophy. Our investigators previously reported that (i) transgenic expression of IGF1R or a caPI3K mutant resulted in cardiac hypertrophy which was associated with increased S6K1 activity and S6 phosphorylation (25, 40), (ii) PI3K was critical for physiological cardiac hypertrophy induced by chronic exercise training (26), and (iii) rapamycin significantly inhibited S6K1 activity and attenuated and regressed cardiac hypertrophy induced by aortic banding (24, 42). However, in contradiction to this evidence, deletion of S6Ks had no significant effect on hypertrophy induced by aortic banding, swimming training, or the IGF1R-PI3K pathway. Some studies in Drosophila have suggested that PI3K and S6K are on distinct and parallel signaling pathways (32, 33). DS6K activation was independent of PI3K activity in cells from Drosophila. Furthermore, enlarged eyes and ommatidia size induced in transgenic flies expressing PI3K were not affected in flies lacking dS6K (33). Our data, demonstrating that S6K1 transgene expression was unable to rescue the small heart phenotype of dnPI3K transgenics, suggest that S6K1 is not responsible for the heart size defect in dnPI3K.
It is noteworthy that functional redundancy of related genes or compensatory upregulation of other genes may mask the effect of a knockout, even if the gene has an important function in the normal animal. Pende et al. reported that deletion of S6K1 and S6K2 in mice uncovered a functional redundant mitogen-activated protein kinase (MAPK)-dependent pathway in mouse embryo fibroblasts and hepatocytes which was able to phosphorylate S6 (30). However, in the present study, an intravenous injection of IGF1 (known to activate the PI3K and MAPK pathway in cardiac myocytes [21, 34]) did not phosphorylate S6 in hearts from S6K1−/− S6K2−/− mice. Thus, at least in the heart, it appears that a MAPK pathway phosphorylating S6 is not present.
While we cannot rule out the possibility that a compensatory pathway in S6K knockout mice may mask the role of S6Ks under normal conditions, data obtained from S6K1 transgenic mice also suggest that S6Ks alone are not the critical regulators of heart size. In dnPI3K transgenic mice with reduced S6K1 activity, rescuing S6K1 activity with the S6K1 transgenes was unable to rescue the small heart phenotype observed in dnPI3K mice. The heart size of double transgenic mice expressing the dnPI3K and S6K1 transgenes was not different from the heart size of mice expressing the dnPI3K mutant alone. Consistent with these data, Stolovich et al. (43) reported that transduction of growth signals into translational activation of terminal oligopyrimidine tract mRNAs was fully reliant on the PI3K pathway but independent of S6K1 and S6 phosphorylation. Finally, we recently reported that an mTOR-independent pathway is able to regulate heart size under certain conditions induced by pressure overload (24).
In summary, we have shown that overexpression of S6K1 modestly increased heart size; that deletion of S6Ks had no significant effect on exercise-, pressure overload-, IGF1R-, or caPI3K-induced cardiac hypertrophy; and that rescuing S6K1 activity in dnPI3K transgenic mice was unable to rescue the small heart phenotype. Together, these studies suggest that S6Ks alone are not critical for the induction of cardiac hypertrophy.
Acknowledgments
We thank M. Rivera for assistance swimming the mice and S. Ngoy for performing some of the mouse surgeries. We thank the Novartis Foundation and George Thomas laboratory for the use of S6K knockout mice.
This work was supported by National Institutes of Health grant RO1 HL65742 to S.I.
REFERENCES
- 1.Akhter, S. A., L. M. Luttrell, H. A. Rockman, G. Iaccarino, R. J. Lefkowitz, and W. J. Koch. 1998. Targeting the receptor-Gq interface to inhibit in vivo pressure overload myocardial hypertrophy. Science 280:574-577. [DOI] [PubMed] [Google Scholar]
- 2.Boluyt, M. O., J. S. Zheng, A. Younes, X. Long, L. O'Neill, H. Silverman, E. G. Lakatta, and M. T. Crow. 1997. Rapamycin inhibits alpha 1-adrenergic receptor-stimulated cardiac myocyte hypertrophy but not activation of hypertrophy-associated genes. Evidence for involvement of p70 S6 kinase. Circ. Res. 81:176-186. [DOI] [PubMed] [Google Scholar]
- 3.Brown, E. J., and S. L. Schreiber. 1996. A signaling pathway to translational control. Cell 86:517-520. [DOI] [PubMed] [Google Scholar]
- 4.Chou, M. M., and J. Blenis. 1995. The 70 kDa S6 kinase: regulation of a kinase with multiple roles in mitogenic signalling. Curr. Opin. Cell Biol. 7:806-814. [DOI] [PubMed] [Google Scholar]
- 5.Chung, J., C. J. Kuo, G. R. Crabtree, and J. Blenis. 1992. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell 69:1227-1236. [DOI] [PubMed] [Google Scholar]
- 6.D'Angelo, D. D., Y. Sakata, J. N. Lorenz, G. P. Boivin, R. A. Walsh, S. B. Liggett, and G. W. Dorn II. 1997. Transgenic Gαq overexpression induces cardiac contractile failure in mice. Proc. Natl. Acad. Sci. USA 94:8121-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies, S. P., H. Reddy, M. Caivano, and P. Cohen. 2000. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 351:95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duerr, R. L., S. Huang, H. R. Miraliakbar, R. Clark, K. R. Chien, and J. Ross, Jr. 1995. Insulin-like growth factor-1 enhances ventricular hypertrophy and function during the onset of experimental cardiac failure. J. Clin. Investig. 95:619-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dufner, A., and G. Thomas. 1999. Ribosomal S6 kinase signaling and the control of translation. Exp. Cell Res. 253:100-109. [DOI] [PubMed] [Google Scholar]
- 10.Fingar, D. C., S. Salama, C. Tsou, E. Harlow, and J. Blenis. 2002. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 16:1472-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fumagalli, S., and G. Thomas. 2000. S6 phosphorylation and signal transduction, p. 695-717. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 12.Geenen, D. L., A. Malhotra, and P. M. Buttrick. 1996. Angiotensin receptor 1 blockade does not prevent physiological cardiac hypertrophy in the adult rat. J. Appl. Physiol. 81:816-821. [DOI] [PubMed] [Google Scholar]
- 13.Gulick, J., A. Subramaniam, J. Neumann, and J. Robbins. 1991. Isolation and characterization of the mouse cardiac myosin heavy chain genes. J. Biol. Chem. 266:9180-9185. [PubMed] [Google Scholar]
- 14.Hernandez, J. M., M. J. Fedele, and P. A. Farrell. 2000. Time course evaluation of protein synthesis and glucose uptake after acute resistance exercise in rats. J. Appl. Physiol. 88:1142-1149. [DOI] [PubMed] [Google Scholar]
- 15.Iemitsu, M., T. Miyauchi, S. Maeda, S. Sakai, T. Kobayashi, N. Fujii, H. Miyazaki, M. Matsuda, and I. Yamaguchi. 2001. Physiological and pathological cardiac hypertrophy induce different molecular phenotypes in the rat. Am. J. Physiol. Regul. Integr. Comp. Physiol. 281:R2029-R2036. [DOI] [PubMed] [Google Scholar]
- 16.Izumo, S., B. Nadal-Ginard, and V. Mahdavi. 1988. Protooncogene induction and reprogramming of cardiac gene expression produced by pressure overload. Proc. Natl. Acad. Sci. USA 85:339-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasahara, H., S. Bartunkova, M. Schinke, M. Tanaka, and S. Izumo. 1998. Cardiac and extracardiac expression of Csx/Nkx2.5 homeodomain protein. Circ. Res. 82:936-946. [DOI] [PubMed] [Google Scholar]
- 18.Lee-Fruman, K. K., C. J. Kuo, J. Lippincott, N. Terada, and J. Blenis. 1999. Characterization of S6K2, a novel kinase homologous to S6K1. Oncogene 18:5108-5114. [DOI] [PubMed] [Google Scholar]
- 19.Lembo, G., H. A. Rockman, J. J. Hunter, H. Steinmetz, W. J. Koch, L. Ma, M. P. Prinz, J. Ross, Jr., K. R. Chien, and L. Powell-Braxton. 1996. Elevated blood pressure and enhanced myocardial contractility in mice with severe IGF-1 deficiency. J. Clin. Investig. 98:2648-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levy, D., R. J. Garrison, D. D. Savage, W. B. Kannel, and W. P. Castelli. 1990. Prognostic implications of echocardiographically determined left ventricular mass in the Framingham Heart Study. N. Engl. J. Med. 322:1561-1566. [DOI] [PubMed] [Google Scholar]
- 21.Liu, T., H. Lai, W. Wu, S. Chinn, and P. H. Wang. 2001. Developing a strategy to define the effects of insulin-like growth factor-1 on gene expression profile in cardiomyocytes. Circ. Res. 88:1231-1238. [DOI] [PubMed] [Google Scholar]
- 22.Martin, K. A., S. S. Schalm, C. Richardson, A. Romanelli, K. L. Keon, and J. Blenis. 2001. Regulation of ribosomal S6 kinase 2 by effectors of the phosphoinositide 3-kinase pathway. J. Biol. Chem. 276:7884-7891. [DOI] [PubMed] [Google Scholar]
- 23.Martin, K. A., S. S. Schalm, A. Romanelli, K. L. Keon, and J. Blenis. 2001. Ribosomal S6 kinase 2 inhibition by a potent C-terminal repressor domain is relieved by mitogen-activated protein-extracellular signal-regulated kinase kinase-regulated phosphorylation. J. Biol. Chem. 276:7892-7898. [DOI] [PubMed] [Google Scholar]
- 24.McMullen, J. R., M. C. Sherwood, O. Tarnavski, L. Zhang, A. L. Dorfman, T. Shioi, and S. Izumo. 2004. Inhibition of mTOR signaling with rapamycin regresses established cardiac hypertrophy induced by pressure overload. Circulation 3050-3055. 109: [DOI] [PubMed]
- 25.McMullen, J. R., T. Shioi, W. Y. Huang, L. Zhang, O. Tarnavski, E. Bisping, M. Schinke, S. Kong, M. C. Sherwood, J. Brown, L. Riggi, P. M. Kang, and S. Izumo. 2004. The insulin-like growth factor 1 receptor induces physiological heart growth via the phosphoinositide 3-kinase(p110α) pathway. J. Biol. Chem. 279:4782-4793. [DOI] [PubMed] [Google Scholar]
- 26.McMullen, J. R., T. Shioi, L. Zhang, O. Tarnavski, M. C. Sherwood, P. M. Kang, and S. Izumo. 2003. Phosphoinositide 3-kinase(p110α) plays a critical role for the induction of physiological, but not pathological, cardiac hypertrophy. Proc. Natl. Acad. Sci. USA 100:12355-12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montagne, J., M. J. Stewart, H. Stocker, E. Hafen, S. C. Kozma, and G. Thomas. 1999. Drosophila S6 kinase: a regulator of cell size. Science 285:2126-2129. [DOI] [PubMed] [Google Scholar]
- 28.Neri Serneri, G. G., M. Boddi, P. A. Modesti, I. Cecioni, M. Coppo, L. Padeletti, A. Michelucci, A. Colella, and G. Galanti. 2001. Increased cardiac sympathetic activity and insulin-like growth factor-I formation are associated with physiological hypertrophy in athletes. Circ. Res. 89:977-982. [DOI] [PubMed] [Google Scholar]
- 29.Neri Serneri, G. G., M. Boddi, L. Poggesi, I. Simonetti, M. Coppo, M. L. Papa, G. F. Lisi, M. Maccherini, R. Becherini, A. Boncompagni, T. Toscano, and P. A. Modesti. 2001. Activation of cardiac renin-angiotensin system in unstable angina. J. Am. Coll. Cardiol. 38:49-55. [DOI] [PubMed] [Google Scholar]
- 30.Pende, M., S. H. Um, V. Mieulet, M. Sticker, V. L. Goss, J. Mestan, M. Mueller, S. Fumagalli, S. C. Kozma, and G. Thomas. 2004. S6K1−/−/S6K2−/− mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 24:3112-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Proud, C. G. 2004. mTOR-mediated regulation of translation factors by amino acids. Biochem. Biophys. Res. Commun. 313:429-436. [DOI] [PubMed] [Google Scholar]
- 32.Radimerski, T., J. Montagne, M. Hemmings-Mieszczak, and G. Thomas. 2002. Lethality of Drosophila lacking TSC tumor suppressor function rescued by reducing dS6K signaling. Genes Dev. 16:2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radimerski, T., J. Montagne, F. Rintelen, H. Stocker, J. van der Kaay, C. P. Downes, E. Hafen, and G. Thomas. 2002. dS6K-regulated cell growth is dPKB/dPI(3)K-independent, but requires dPDK1. Nat. Cell Biol. 4:251-255. [DOI] [PubMed] [Google Scholar]
- 34.Ren, J., W. K. Samson, and J. R. Sowers. 1999. Insulin-like growth factor I as a cardiac hormone: physiological and pathophysiological implications in heart disease. J Mol. Cell. Cardiol. 31:2049-2061. [DOI] [PubMed] [Google Scholar]
- 35.Sadoshima, J., and S. Izumo. 1995. Rapamycin selectively inhibits angiotensin II-induced increase in protein synthesis in cardiac myocytes in vitro. Potential role of 70-kD S6 kinase in angiotensin II-induced cardiac hypertrophy. Circ. Res. 77:1040-1052. [DOI] [PubMed] [Google Scholar]
- 36.Scheuer, J., and P. Buttrick. 1987. The cardiac hypertrophic responses to pathologic and physiologic loads. Circulation 75:I63-I68. [PubMed] [Google Scholar]
- 37.Schluter, K. D., and H. M. Piper. 1999. Regulation of growth in the adult cardiomyocytes. FASEB J. 13(Suppl.):S17-S22. [DOI] [PubMed] [Google Scholar]
- 38.Shah, O. J., J. C. Anthony, S. R. Kimball, and L. S. Jefferson. 2000. 4E-BP1 and S6K1: translational integration sites for nutritional and hormonal information in muscle. Am. J. Physiol. Endocrinol. Metab. 279:E715-E729. [DOI] [PubMed] [Google Scholar]
- 39.Shima, H., M. Pende, Y. Chen, S. Fumagalli, G. Thomas, and S. C. Kozma. 1998. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 17:6649-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shioi, T., P. M. Kang, P. S. Douglas, J. Hampe, C. M. Yballe, J. Lawitts, L. C. Cantley, and S. Izumo. 2000. The conserved phosphoinositide 3-kinase pathway determines heart size in mice. EMBO J. 19:2537-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shioi, T., J. R. McMullen, P. M. Kang, P. S. Douglas, T. Obata, T. F. Franke, L. C. Cantley, and S. Izumo. 2002. Akt/protein kinase B promotes organ growth in transgenic mice. Mol. Cell. Biol. 22:2799-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shioi, T., J. R. McMullen, O. Tarnavski, K. Converso, M. C. Sherwood, W. J. Manning, and S. Izumo. 2003. Rapamycin attenuates load-induced cardiac hypertrophy in mice. Circulation 107:1664-1670. [DOI] [PubMed] [Google Scholar]
- 43.Stolovich, M., H. Tang, E. Hornstein, G. Levy, R. Cohen, S. S. Bae, M. J. Birnbaum, and O. Meyuhas. 2002. Transduction of growth or mitogenic signals into translational activation of TOP mRNAs is fully reliant on the phosphatidylinositol 3-kinase-mediated pathway but requires neither S6K1 nor rpS6 phosphorylation. Mol. Cell. Biol. 22:8101-8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka, N., T. Ryoke, M. Hongo, L. Mao, H. A. Rockman, R. G. Clark, and J. Ross, Jr. 1998. Effects of growth hormone and IGF-I on cardiac hypertrophy and gene expression in mice. Am. J. Physiol. 275:H393-H399. [DOI] [PubMed] [Google Scholar]
- 45.Tang, H., E. Hornstein, M. Stolovich, G. Levy, M. Livingstone, D. Templeton, J. Avruch, and O. Meyuhas. 2001. Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol. Cell. Biol. 21:8671-8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tarnavski, O., J. R. McMullen, M. Schinke, Q. Nie, S. Kong, and S. Izumo. 2004. Mouse cardiac surgery: comprehensive techniques for the generation of mouse models of human diseases and their application for genomic studies. Physiol. Genomics 16:349-360. [DOI] [PubMed] [Google Scholar]
- 47.Thomas, G., and M. N. Hall. 1997. TOR signalling and control of cell growth. Curr. Opin. Cell Biol. 9:782-787. [DOI] [PubMed] [Google Scholar]
- 48.Wang, L., X. Wang, and C. G. Proud. 2000. Activation of mRNA translation in rat cardiac myocytes by insulin involves multiple rapamycin-sensitive steps. Am. J. Physiol. Heart Circ. Physiol. 278:H1056-H1068. [DOI] [PubMed] [Google Scholar]
- 49.Wettschureck, N., H. Rutten, A. Zywietz, D. Gehring, T. M. Wilkie, J. Chen, K. R. Chien, and S. Offermanns. 2001. Absence of pressure overload induced myocardial hypertrophy after conditional inactivation of Gαq/Gα11 in cardiomyocytes. Nat. Med. 7:1236-1240. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, H., J. P. Stallock, J. C. Ng, C. Reinhard, and T. P. Neufeld. 2000. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 14:2712-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]