Background: The ribose transporter is a bacterial ABC importer with a non-canonical organization.
Results: The binding protein complexes with the membrane domain in the absence of substrate, and the ATPase dissociates from the membrane domain during transport.
Conclusion: A distinct model for transport is proposed from in vitro reassembly conditions and EPR data.
Significance: The ribose transporter typifies the diversity of ABC transporter mechanisms.
Keywords: ABC Transporter, ATPase, Bacterial Metabolism, Electron Paramagnetic Resonance (EPR), Membrane Transport, Membrane Transporter Reconstitution, Ribose Transport
Abstract
Bacterial ATP-binding cassette (ABC) importers are primary active transporters that are critical for nutrient uptake. Based on structural and functional studies, ABC importers can be divided into two distinct classes, type I and type II. Type I importers follow a strict alternating access mechanism that is driven by the presence of the substrate. Type II importers accept substrates in a nucleotide-free state, with hydrolysis driving an inward facing conformation. The ribose transporter in Escherichia coli is a tripartite complex consisting of a cytoplasmic ATP-binding cassette protein, RbsA, with fused nucleotide binding domains; a transmembrane domain homodimer, RbsC2; and a periplasmic substrate binding protein, RbsB. To investigate the transport mechanism of the complex RbsABC2, we probed intersubunit interactions by varying the presence of the substrate ribose and the hydrolysis cofactors, ATP/ADP and Mg2+. We were able to purify a full complex, RbsABC2, in the presence of stable, transition state mimics (ATP, Mg2+, and VO4); a RbsAC complex in the presence of ADP and Mg2+; and a heretofore unobserved RbsBC complex in the absence of cofactors. The presence of excess ribose also destabilized complex formation between RbsB and RbsC. These observations suggest that RbsABC2 shares functional traits with both type I and type II importers, as well as possessing unique features, and employs a distinct mechanism relative to other ABC transporters.
Introduction
ABC5 transporters comprise a large superfamily of transmembrane proteins that consists of both exporters and importers (1–9). ABC exporters are found in all domains of life and transport a variety of substrates, including lipids, peptides, toxins, antibiotics, and chemotherapeutic drugs (2). ABC importers are almost exclusively prokaryotic and are typically employed for the uptake of metabolites and cofactors (1, 4). In Gram-negative bacteria, such as Escherichia coli, the ribose transporter (RbsABC2, herein referred to as RbsABC for simplicity) is critical for the high affinity uptake of ribose and is part of an operon of genes whose products are involved in the delivery and processing of this molecular precursor for nucleic acid synthesis (10).
Despite the diverse nature of organisms and substrate chemistry, both ABC importers and exporters are thought to share a common transport mechanism involving alternating access of the transmembrane domains (TMDs) (2–4). This model for transport is based on detailed studies of type I importers (3–5), primarily the maltose transporter (11–17), where ATP binding and hydrolysis in the cytoplasmic nucleotide-binding domains (NBDs, or ABC domains) is driven by the presence of substrates. Periplasmic substrate-binding proteins (SBPs) deliver substrates to the inner membrane TMD-NBD complex, where conformational changes in the TMDs from inward to outward facing states facilitate payload acceptance. ATP hydrolysis in the NBDs leads to a reversal of these changes and subsequent substrate delivery from one leaflet of the lipid bilayer to the other (2–9).
Recent studies of type II importers, such as the vitamin B12 and molybdate II transporters, have revealed a peristaltic transport mechanism, where substrate is accepted from SBPs in a nucleotide-free state, and transport is facilitated by more modest conformational changes in the TMDs (18–20, 21). ATP hydrolysis results in rearrangement of transmembrane helices to an inward facing conformation, forcing the substrate into the cytoplasm (22). The association between the SBP and its cognate TMD-NBD complex partners is longer lived and sensitive to the availability of substrate, in stark contrast to observations made on type I systems (22).
Previous studies of the ribose transporter suggest that it shares features with both type I and type II import systems. For instance, SBPs fall into distinct structural and functional groups; those of type I importers undergo more significant conformational changes upon substrate binding than the type II variety (23). The ribose transporter SBP, RbsB, belongs to the former class based on apo- and ribose-bound crystal structures (24, 25). Structural studies of type I importers show that TMDs are normally composed of 6–8 helices/monomer, whereas type II class members consist of 10 helices/monomer (12, 18); based on in vivo labeling experiments, the ribose transporter TMD, RbsC, belongs to the latter class (26, 27). Finally, NBDs of type I importers typically have low basal rates of activity and hydrolyze ATP with high efficiency only in the presence of a substrate-loaded SBP. Many, such as the maltose transporter, also contain C-terminal regulatory domains that prevent ATP hydrolysis when bound to substrate (28). Type II importer NBDs typically have higher basal rates of ATP hydrolysis independent of substrate availability and lack C-terminal regulatory domains (18–20). The ribose transporter ABC domain, RbsA, is a single polypeptide consisting of two fused NBDs, an organization that is not typical of ABC domains in either importer class, and, like type II NBDs, lacks any sequences indicating the presence of a regulatory domain (29). Nevertheless, results from this study, based primarily upon stimulation of hydrolysis activity, suggest that RbsA is functionally more similar to the type I class.
To elucidate the RbsABC transport mechanism and to determine to which class of importer the ribose transporter belongs, we employed cofactor variation in vitro to probe conditions promoting intersubunit interactions between RbsA, RbsC, and RbsB. We purified the three components in the presence of the substrate, ribose, and the hydrolysis cofactors, ATP/ADP and Mg2+, to determine conditions promoting complex formation and activity. The results of the investigation suggest that RbsABC shares functional features with both type I and type II transporters and may belong to a distinct class of importers. These results support recent studies demonstrating that transport mechanisms in this superfamily, although consistent with the alternating access model, have significant variations and are increasingly diverse.
MATERIALS AND METHODS
Cloning of His-tagged Ribose Transporter Genes
The previously constructed (29, 30) plasmid pTATC (pT7-rbsA, pT7-rbsC, ampr) was modified to place N-terminal hexahistidine tags on either rbsA (pTHATC) or rbsC (pTATHC) to facilitate expression and affinity purification of ribose transporter complexes. pTATC was treated with NcoI in the middle of the rbsC gene, trimmed with nuclease BAL-31 to remove the remainder of rbsC, and treated with DNA polymerase I to repair the staggered ends, resulting in pT7-rbsA. The product was cut with BamHI, and the fragment was verified by agarose gel electrophoresis. pT7–6His-rbsC was cut from the previously constructed pHC (30) using PvuII and BamHI. The resultant 1.5-kb fragment containing pT7–6His-rbsC was ligated with pT7-rbsA to produce the final plasmid pTATHC, as confirmed by sequencing.
To create a plasmid with pT7–6His-rbsA and pT7-rbsC, the previously constructed plasmid pHA (29) was digested with PvuII and BamHI, which resulted in a 1.9-kb fragment that was verified by agarose gel electrophoresis and subsequently purified. The previously constructed plasmid, pC, which contains rbsC and a T7 promoter (30), was cut with HindIII, and the overhangs were filled by the Klenow reaction. The product was cut using BamHI, resulting in a gel-purified 3.3-kb fragment. The 1.9-kb and 3.3-kb fragments were ligated to produce the plasmid pTHATC (pT7–6His-rbsA, pT7-rbsC, ampr), as confirmed by sequencing.
To create a plasmid for expression of a His-tagged RbsB, rbsB was introduced into the vector pET22b(+), which contains a C-terminal hexahistidine tag to avoid interference with the signal sequence. rbsB was isolated from the previously constructed plasmid p4B1 (31, 32) using PCR with primers designed to introduce NdeI and XhoI restriction sites (GGAATTCCATATGAACATGAAAAAACTGGC and CCGCTCGAGCTGCTTAACAACCAGTTTCAG). The resulting PCR product and the vector pET22b(+) were each subsequently cut by NdeI and XhoI and produced a 0.88-kb fragment and a 5.3-kb fragment, respectively. The fragments were ligated, and the resulting plasmid was analyzed by sequencing to confirm production of pTBH (pT7-rbsB-6His, ampr).
All genes were cloned with individual T7 sequences and in the 5′–3′ orientation. All enzymes were obtained from New England Biolabs.
Expression of Membrane Protein-containing Plasmids
Plasmids containing the membrane protein rbsC with or without the cytoplasmic domain rbsA (either pTATHC, pTHATC, pTHC, or pTATC) were transformed into BL21-AI competent cells (Invitrogen) by heat shock, and proteins were expressed using the “restrained expression” method (33). Following inoculation of 1 liter of Terrific Broth medium (supplemented with 0.4% glycerol and ampicillin) with overnight starter cultures, cells were grown to A600 = 0.6 at 37 °C, subsequently induced by the addition of 0.01% (w/v) arabinose, and allowed to express overnight (16–20 h) at 16 °C. Cells were harvested by centrifugation at 6000 × g for 20 min, and the resulting pellets were collected and stored at −80 °C. Expression by this method resulted in 15–20 mg/liter of purified complexes.
Expression and Purification of RbsB and RbsA Alone
Plasmids containing rbsB (p4B1) and rbsA (pHA) were transformed into BL21(DE3) competent cells (Invitrogen) by heat shock and expressed and purified as described previously (29, 32). Plasmids containing His-tagged RbsB (pTBH) were transformed and expressed as above but purified using affinity chromatography. Following cell lysis and centrifugation at 10,000 × g for 20 min, the resulting supernatant was loaded onto 2 ml of nickel-nitrilotriacetic acid resin pre-equilibrated with binding buffer (20 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 5 mm imidazole). Following batch binding, the suspension was poured into a 10-ml Poly-Prep column (Bio-Rad). The column was washed with 10 column volumes of wash buffer (binding buffer with 60 mm imidazole), and protein was eluted using 10 column volumes of elution buffer (wash buffer with 200 mm imidazole). Samples were analyzed by SDS-PAGE.
Purification of Ribose Transporter Complexes
Cells from 1-liter cultures were resuspended in 10 ml of ice-cold lysis buffer (20 mm Tris-HCl, pH 7.5, 1 mm EDTA, 1 mm EGTA)/g of dry cell pellet. The protease inhibitors PMSF and phenanthroline were added to 1 mm each final concentration, and the suspension was lysed by three passages through a French pressure cell (Aminco). The lysate was centrifuged at 30,000 × g for 30 min. Supernatant was collected and membranes were isolated by ultracentrifugation at 150,000 × g for 1 h. The resulting supernatant was discarded, and membrane pellets were resuspended on ice by the addition of 4 ml of chilled Tris-glycerol (TG) buffer (20 mm Tris-HCl, pH 8.0, and 20% glycerol) and 6 mm each of ATP/ADP, MgCl2, and sodium orthovanadate (prepared as described (15)). 10 mg of purified RbsB were added, and the final volume was raised to 10 ml by the addition of TG buffer. The solution was gently stirred at room temperature for 1 h, after which 2 ml of 10% n-dodecyl-β-d-maltoside (DDM; Anatrace) and 8 ml of binding buffer (TG buffer + 1.5 m NaCl and 5 mm imidazole) were added. Membranes were solubilized for 1 h and centrifuged at 150,000 × g for 20 min, and the supernatant was incubated for 1 h with nickel-nitrilotriacetic acid resin equilibrated with binding buffer. Following batch binding, the suspension was poured into a 10-ml Poly-Prep column and washed with 10 column volumes of wash buffer (TG buffer with 150 mm NaCl, 60 mm imidazole, 0.02% DDM, and 1 mm each of ATP, MgCl2, and vanadate), and proteins were eluted with 10 column volumes of elution buffer (wash buffer with 200 mm imidazole). Samples were analyzed by SDS-PAGE using a 15% polyacrylamide gel.
Purification of His-tagged RbsC
The plasmid pTHC was transformed into BL21-AI cells and expressed as described above for transport complexes. Purification of RbsC was done as described above for complexes, except in the absence of cofactors and associated proteins. The detergent n-decyl-β-d-maltoside (Anatrace) was used at a final concentration of 0.1% during purification instead of DDM when preparing RbsC for reconstitution into liposomes, a consideration based upon the low critical micelle concentration of DDM.
Reconstitution of Ribose Transport Complexes into Liposomes
100 mg of E. coli polar lipids (Avanti) were dried under nitrogen and stored under vacuum overnight. Lipids were resuspended in 20 mm HEPES, pH 7.2, to a final concentration of 20 mg/ml. The resulting suspension was sonicated three times on ice for 15 s with 45-s intervals. The clarified suspension was extruded through a 1000-nm polycarbonate filter and then 10 times through a 400-nm polycarbonate membrane. Aliquots were flash-frozen and stored at −80 °C until use. For reconstitution, purified ribose transporter subunits (RbsC, RbsBC, or RbsAC) were mixed with thawed liposome aliquots in a 4:1 ratio for a final protein/lipid concentration of 1:100 (w/w), allowed to incubate on ice for 2 h, buffer-exchanged by centrifugal filtration three times into 20 mm HEPES, pH 7.2, to remove residual detergent, and then concentrated to a final 100 μg/ml of RbsC.
CD Spectroscopic Analysis
Circular dichroism measurements were made in quartz 0.01-cm path length cells using a JASCO J600 CD spectropolarimeter equipped with a temperature control system. Experimental parameters were as follows: time constant, 1–2 s; scan speed, 20–50 nm/min; step interval, 0.1 nm; bandwidth, 2–5 nm; standard sensitivity, 100 millidegrees; 4–6 accumulations; and 4 °C temperatures. RbsC was exchanged into the following buffers and concentrated to 0.2–1 mg/ml: for detergent-solubilized RbsC, 10 mm HEPES, pH 7.2, 20% glycerol, and 1.5 m ammonium sulfate; for liposome-reconstituted RbsC, 10 mm HEPES, pH 7.2, 0.05 mm EDTA, and ∼18 mg/ml E. coli polar lipids. Secondary structure content was determined by analysis of CD spectra in the 190–240-nm range as described (34–41) using PROTCD (38–41).
Loading of Liposomes with RbsB and Ribose
RbsC-containing proteoliposomes were combined with RbsB or RbsB + ribose in 1:1 molar ratios, and the resulting suspension was frozen by liquid nitrogen for 5 min and then subsequently allowed to thaw at room temperature for 15 min. The freeze-thaw cycle was repeated two additional times and resulted in RbsB- or RbsB + ribose-loaded liposomes.
Activity Assay of Proteoliposome-reconstituted Ribose Transporter
ATPase activity was measured for five different protein-substrate combinations: individually purified RbsABC 1) with and 2) without ribose; 3) co-purified RbsAC plus individually purified RbsB plus ribose; 4) co-purified RbsBC plus individually purified RbsA plus ribose; 5) purified RbsA with free liposomes; and 6) co-purified RbsAC alone. Hydrolysis activity was measured at room temperature in solutions containing 20 mm HEPES, pH 7.2, 50 μm ATP, 3 mm MgCl2, and, where appropriate, 10 μg/ml RbsC (final concentration in liposomes) and 8.2 μg/ml RbsA (except when already present). Vanadate, when present during purification, was removed prior to experiments by buffer exchange and dialysis. All reactions were started by the addition of MgCl2. Phosphate release was assayed using Malachite Green dye, as described previously (42–44). Aliquots were taken at 0, 5, 20, 40, and 60 min. Colorimetric measurements were made following the addition of fresh working dye solution (44). Color intensity was measured at 630 nm using a PerkinElmer Life Sciences Lambda 40 UV-visible spectrophotometer. Measurements were background-corrected and analyzed by linear regression in Microsoft Excel, with all curves force-fitted through the origin.
ATP Hydrolysis Activity Inhibition by Vanadate
ATP hydrolysis activity measurements in RbsAC proteoliposomes loaded with RbsB were made as described above except in the presence of 0–1.2 mm sodium orthovanadate. Measurements were made as above from aliquots taken at 0, 20, 40, 60, 80, and 100 min using the Malachite Green assay. Data were processed and analyzed as above.
Purification and Labeling of RbsBN41C
The rbsB mutation N41C was introduced to the p4B1 plasmid (31, 32) using the Stratagene QuikChange mutagenesis kit and was confirmed by sequencing, creating a single cysteine mutant for analysis by electron paramagnetic resonance (EPR) spectroscopy. The RbsBN41C mutant was expressed and purified using a modification of the protocol for wild-type RbsB described previously (32). Cells were lysed by osmotic shock, after which 10 mm dithiothreitol (DTT) was added. RbsB was precipitated by the addition of ammonium sulfate and centrifuged at 10,000 × g for 20 min. The resulting pellet was washed 10 times with 20 mm Bistris propane, pH 7.0, and resolubilized in the same buffer, after which a 10:1 molar excess of S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate (MTSL) was added to RbsB and shaken overnight at 4 °C. Unbound MTSL was removed by equilibrium dialysis against 20 mm Tris-HCl, pH 8.0.
Activities of RbsBN41C Mutant Complexes
Ribose transporter complexes were purified, as above, instead substituting RbsBN41C for wild-type RbsB. ATPase activities for detergent-solubilized wild-type and mutant complexes were measured as described above.
EPR Spectroscopy of Mutant Complexes
Purified detergent-solubilized ribose transporter complexes were exchanged into buffer containing 20 mm Tris, pH 8.0, 150 mm NaCl, 0.02% DDM, and 1 mm each of cofactors (where appropriate) before being concentrated to 40 μm. 50-μl samples were transferred to a glass capillary, which was inverted into a quartz sample tube to be placed into the instrument sample cavity (Bruker ER 4119HS). Continuous wave EPR measurements were made at room temperature using a Bruker EMX Plus instrument, and spectra were collected in the X-band. The scan width of all spectra was 250 G, and files were cropped to values stated in the figure legends. Spectral files were initially processed using the Bruker WinEPR software suite, and data were exported to MS Excel for further analysis.
Western Blot Analysis of RbsBC Complex
Primary antibodies (Bio-Rad) were diluted as follows: RbsA (1:10,000), RbsB (1:100,000), and RbsC (1:500). Following transfer onto nitrocellulose, primary antibodies were incubated for 1 h at 4 °C and then washed. Immunodetection employed alkaline phosphatase-conjugated goat anti-rabbit IgG (1:10,000; Bio-Rad), which were incubated for 1 h at 4 °C and washed. The gels were then visualized as described previously (45).
RESULTS
To study the mechanism of ribose transport by RbsABC, we utilized cofactor variation during purification to control assembly of various ribose transporter complexes and measured their respective in vitro ATPase activities in both detergent and lipid environments. Our observations suggest shared features of the ribose ABC transporter with both type I and type II importers as well as novel features that are thus far unique to RbsABC.
Isolation of Individual Ribose Transporter Subunits RbsA, RbsB, and His-RbsC
We first isolated the three components of the ribose transporter individually to facilitate in vitro assembly experiments (Fig. 1A, lanes 1–3). RbsA and RbsB are soluble proteins and were expressed and purified as described previously (10, 32). Hexahistidine-tagged (His) RbsC was expressed in E. coli and purified via affinity chromatography in buffer containing DDM, as described earlier. The low critical micelle concentration of DDM made n-decyl-β-d-maltoside the more practical detergent choice when reconstituting into liposomes.
FIGURE 1.
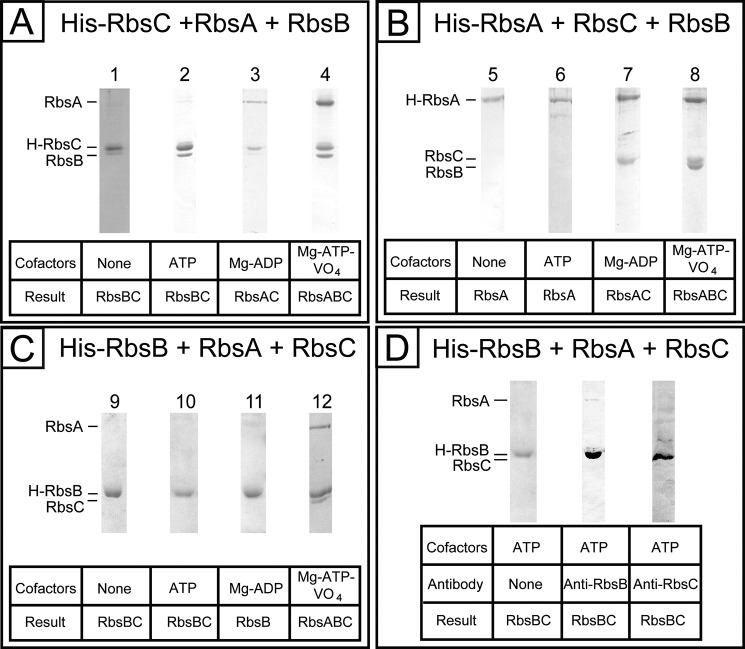
A, SDS-polyacrylamide gel of the purified ribose transporter complexes (cofactors or substrates indicated in parentheses): RbsA, 55 kDa (lane 1); RbsB, 30 kDa (lane 2); His-RbsC, 33 kDa (lane 3); RbsAC (lane 4; Mg-ATP-VO4); RbsABC (lane 5; Mg-ATP-VO4); RbsBC (lane 6; none); molecular mass marker (lane 7; all values in kDa). B, SDS-polyacrylamide gel of ribose transporter complex purified in the presence of 1 mm each of Mg2+, ATP, VO4, and ribose. C, SDS-polyacrylamide gel of ribose transporter complex purified in the presence of 1 mm ribose alone.
Isolation of the Full Complex
Previous studies of the maltose transporter have shown that vanadate trapping can be utilized to isolate full transporter complexes (15). Vanadate readily substitutes for the γ-PO4 following ATP hydrolysis and subsequent Pi release and traps the complex in a postcleavage state. The RbsABC complex was formed using this technique following extraction of His-RbsC from E. coli membranes, the addition of purified wild-type RbsA, RbsB, and requisite cofactors, and subsequent affinity purification, as described above. The results were analyzed by SDS-PAGE (Fig. 1A, lane 5). The non-hydrolyzable ATP analogues AMP-PNP and ATPγS, in combination with Mg2+, could also be readily employed to stabilize RbsABC (data not shown).
Isolation of the NBD-TMD Complex
Type I ABC importers have typical apo-states with associated NBDs and TMDs, whereas the apo-states of type II importer complexes consist of all three proteins (46–51). The isolation of RbsAC proved difficult in the absence of nucleotides and RbsB, despite the use of published protocols from similar systems. The RbsAC complex was first identified using the protocol established for RbsABC, using the cofactor and substrate combination of Mg2+, ATP, and VO4 (Fig. 1A, lane 4), and without the addition of RbsB. Because this combination leads to ATP hydrolysis, the possibility was raised that Mg2+-ADP is actually stabilizing the RbsAC complex, an observation that is consistent with the results of purification in the presence of this cofactor combination (Fig. 2A, lane 3).
FIGURE 2.
Influence of the position of the histidine tag on ribose transporter complex formation, assessed by SDS-PAGE. Each tag variant was tested in four cofactor/substrate conditions for the formation of RbsBC, RbsAC, and RbsABC complexes. Shown is complex formation using N-terminally His-tagged RbsC (A), N-terminally His-tagged RbsA (B), and C-terminally His-tagged RbsB (C). D, Western blot analysis of complex formation using His-RbsB in the cofactor condition containing ATP alone.
Isolation of the Apo-complex
Because the expected RbsAC resting state of the complex could not be isolated without hydrolysis cofactors, we tested the outcome of combining all three proteins with no cofactors present or with ATP alone. Unlike what is observed in type I and type II importers, we observed a new complex, RbsBC, consisting of the SBP and TMD alone (Fig. 1A, lane 6).
Isolation of Complexes in the Presence of Ribose
To test the effect of the substrate, ribose, on complex formation, we included 1 mm ribose in the purification conditions in addition to other cofactors. When ribose is present during conditions that otherwise produce the vanadate-trapped RbsABC complex, the RbsAC complex is observed instead (Fig. 1B). When ribose alone is present without nucleotide and Mg2+, RbsC alone is observed (Fig. 1C). This suggests that ribose loading of RbsB destabilizes its interaction with RbsC.
Assembly of Complexes Using Alternate Affinity Tag Sites
To test the possibility that assembly is affected by the location of the hexahistidine tag on RbsC, we used His tags attached to the N terminus of RbsA and the C terminus of RbsB, respectively, and a subset of the experiments was repeated. Based on the results of the His-RbsC experiments, we tested four conditions: Mg2+-ATP-VO4, Mg2+-ADP, ATP alone, and no cofactors. For experiments using His-tagged RbsA, we expressed RbsA and RbsC using the plasmid pTHATC and combined them with separately purified wild-type RbsB. The results are consistent with those observed for His-RbsC (Fig. 2B).
For experiments using His-tagged RbsB, RbsA and RbsC were expressed using the plasmid pTATC and combined with purified His-RbsB. The results were consistent with those observed for His-RbsC (Fig. 2C), albeit the analysis was complicated by the similar molecular weights of His-RbsB and RbsC. This ambiguity was addressed by employment of RbsB and RbsC antibodies in a Western blot analysis, which demonstrated the presence of both proteins following affinity purification in the cofactor-free condition (Fig. 2D), as seen before.
Reconstituted Ribose Transporter Complexes Are Active in Liposomes
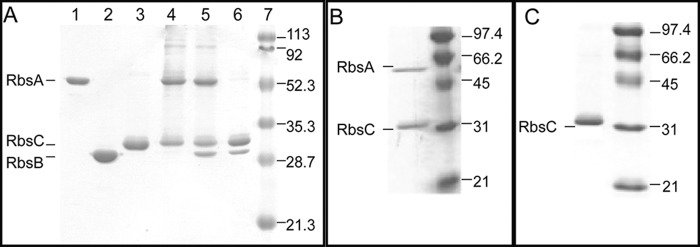
Proteoliposomes were formed as described earlier, and proper folding of RbsC in both detergent and liposomes was tested by CD spectroscopic analysis (Fig. 3). For His-RbsC in DDM, the calculated α-helical content was determined to be 57–59%, whereas that of His-RbsC in proteoliposomes was determined to be 54–61%. Both figures are in agreement with similar measurements made for both the vitamin B12 transporter (α-helical content of 62% with 20 transmembrane helices in the homodimer, BtuC) and the maltose transporter (61% with 14 transmembrane helices in the heterodimer, MalFG) (17, 18).
FIGURE 3.
Circular dichroism spectra of proteoliposome-reconstituted His-RbsC and detergent-solubilized His-RbsC. Dashed line, DDM-solubilized RbsC; solid line, RbsC reconstituted into liposomes made from polar E. coli lipids. Secondary structure content was estimated from data in the 190–240-nm spectral range using PROT-CD, as described earlier. The high concentration of lipids in the measurements for proteoliposomes introduced pronounced spectral noise in the far-UV region (190–200 nm), due to differential light scattering and differential absorption flattening effects. Both spectra yielded similar α-helical content: His-RbsC in DDM, 57–59%; His-RbsC in liposomes, 54–61%.
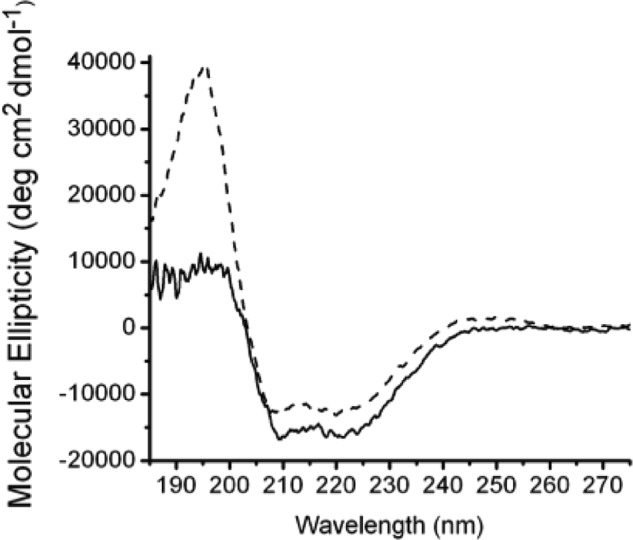
His-RbsC-containing liposomes were loaded with RbsB and mixed with RbsA and ATP, and hydrolysis was subsequently initiated by the addition of Mg2+. Maximal activity was observed with 3 mm Mg2+; further increase in magnesium concentration resulted in precipitation of phospholipids and subsequent reduction of ATPase activity. All three ribose transporter proteins plus ribose are necessary for the full substrate-stimulated ATPase activity of the membrane-reconstituted complex (Fig. 4A). The observed ATPase activity of the complex assembled and reconstituted in liposomes using individually purified components in the presence of Mg2+, ATP, and ribose (Fig. 4A, green line) was 0.27 μmol of Pi/mg of RbsA/min, a 500-fold increase in activity when compared with RbsA alone in the presence of empty liposomes (Fig. 4A, black line). The ability of the full RbsABC complex to hydrolyze ATP was increased 10-fold in the presence of ribose (Fig. 4A, green line) over measurements made in its absence (Fig. 4A, orange line), demonstrating a substrate-stimulated activity. The full complex (Fig. 4A, green line) also exhibited a 33-fold increased activity compared with uncoupled activity in the absence of both RbsB and ribose (Fig. 4A, purple line, 8.2 nmol of Pi/mg of RbsA/min). As is typical for ABC proteins, vanadate was shown to inhibit ATP hydrolysis activity of RbsABC in liposomes in a concentration-dependent manner (Fig. 4B).
FIGURE 4.
A, ATP hydrolysis activity of the reconstituted complexes. Black (○), RbsA with liposomes; purple (●), RbsAC with ribose-free proteoliposomes; orange (×), RbsA, RbsC with ribose-free proteoliposomes and RbsB; green (♦), RbsA, RbsC with ribose-loaded proteoliposomes, RbsB; red (▴), RbsAC with ribose-loaded proteoliposomes, RbsB; blue (■), RbsA, RbsBC with ribose-loaded proteoliposomes. All experiments were performed in 20 mm HEPES, pH 7.2, 50 μm ATP, and 3 mm MgCl2 in triplicate. Lines were produced by linear regression analysis (using Microsoft Excel), forcing the fit to go through the origin. B, RbsABC ATPase activity is inhibited by vanadate. All experiments were performed in triplicate in 20 mm HEPES, pH 7.2, 50 μm ATP, 3 mm MgCl2, and varying concentrations of vanadate. Measurements were made with RbsB-loaded RbsAC-proteoliposomes in the presence of 0 mm vanadate (■, black line); 0.1 mm vanadate (♦, purple line), 0.2 mm vanadate (▴, blue line); 0.4 mm vanadate (●, green line); 0.8 mm vanadate (×, orange line); 1.2 mm vanadate ( , red line).
, red line).
In addition to assembling complexes using individually purified components, we measured ATPase activities of the membrane-reconstituted partial complexes RbsAC and RbsBC in the presence of their respective cognate partners as well as ATP, Mg2+, and ribose. These experiments tested whether the partial complexes could be productive and represented meaningful steps during the ribose transport cycle. The observed ATPase activity for the reconstituted complex originating from RbsBC was 0.30 μmol of Pi/mg of RbsA/min (Fig. 4A, blue line), whereas activity for the complex originating from RbsAC was 0.26 μmol of Pi/mg of RbsA/min (Fig. 4A, red line). These measurements were within the margin of error of the activity measurements for the complex reconstituted from individually purified components, indicating that both RbsBC and RbsAC form productive complexes with their respective cognate partners.
Verification of RbsB-RbsC Interaction through EPR Spectroscopy
Complexes similar to RbsBC have not previously been reported in published studies of other bacterial ABC importers. As an additional verification of productive interactions between RbsB and RbsC in this state, we employed EPR spectroscopy to probe intersubunit contacts. The mutation N41C at the proposed binding interface of RbsB with RbsC was made for spin labeling and subsequent EPR measurements. Although no crystal structure exists for the RbsB-RbsC complex, the RbsB interaction domain has been mapped by mutational studies that abrogated ribose transport and chemotaxis (31). Structures of similar bacterial ABC importers support the involvement of this region of SBPs as the primary site of contact with their corresponding transmembrane domains (12, 20). Based on the mutational analysis and comparison with similar structures, an MTSL spin label attached to C41 would be predicted to experience significant limitations in mobility when RbsB is in complex with RbsC and be relatively unhindered when RbsB is in the unbound state. MTSL-spin-labeled RbsBN41C was prepared as described under “Materials and Methods.” Hydrolysis activity of ribose transporter complexes in surfo with RbsBN41C was comparable (7.5 nmol of Pi/mg of RbsA/min) to activity measurements made in the presence of wild-type RbsB (data not shown). EPR spectra of RbsBN41CC complexes showed significant peak broadening and splitting (Fig. 5, middle) when compared with spectra of RbsBN41C alone (Fig. 5, top). Nearly identical spectral changes are observed in the RbsABN41CC complex (Fig. 5, bottom). The spectral changes are due to immobilization of the spin label when RbsB is in complex and can be interpreted as a signature for a direct and stable interaction between RbsB and RbsC in both the RbsBC and RbsABC complexes. The data suggest that the interactions between RbsB and RbsC are canonical in what is apparently a resting state RbsBC complex.
FIGURE 5.
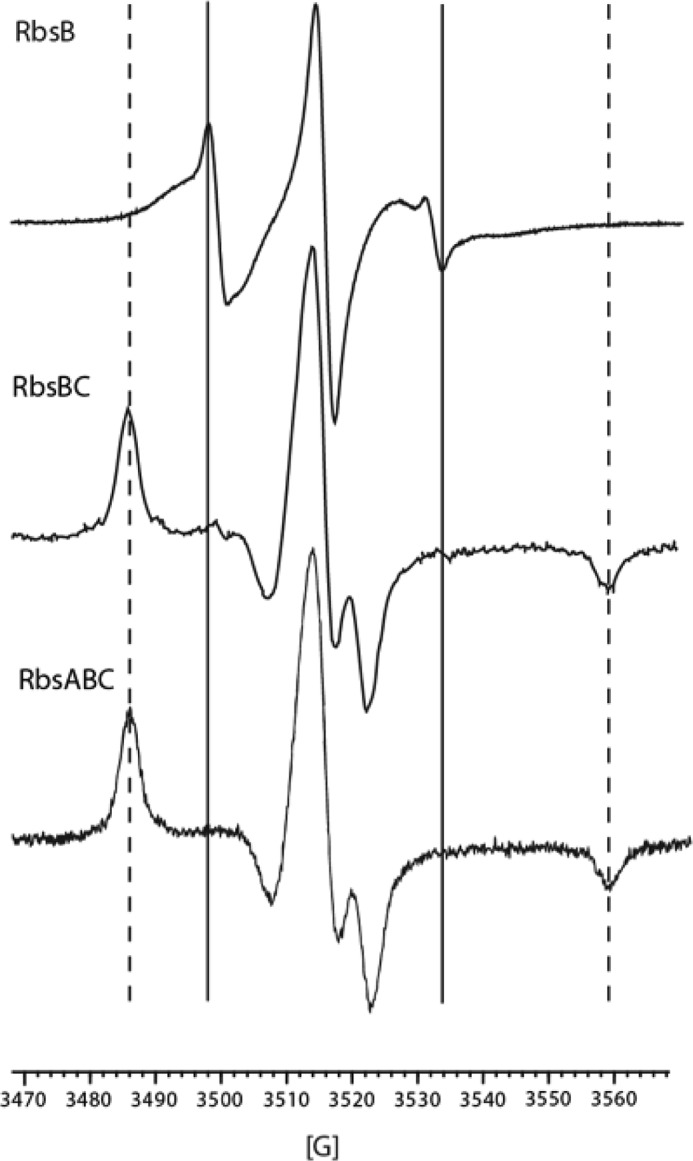
EPR spectra of RbsBN41C (top), RbsBN41CC (middle), and RbsABN41CC (bottom). All samples were taken in 20 mm Tris, pH 8, 20% glycerol, 150 mm NaCl, and, in the case of RbsABC, 1 mm ATP, MgCl2, and VO4. Solid vertical lines, low field and high field maxima and minima indicating moderate immobilization in RbsB alone. Dashed vertical lines, further splitting of maxima and minima when RbsB is in complex with RbsC.
DISCUSSION
A pathway for ribose transport by protein products of the rbs operon has previously been proposed based on observations from earlier experiments and from studies of other ABC importers (24, 25, 29). First, ribose diffuses into the periplasm of Gram-negative bacteria via outer membrane porins. Free ribose is scavenged by the SBP, RbsB, which binds its substrate with 0.3 μm affinity (32). RbsB then delivers ribose to the inner membrane complex formed by RbsAC, which subsequently flips the substrate across the membrane into the cytoplasm in an ATP-dependent manner (29, 30). Ribose is sequestered in the cell by phosphorylation by the ribokinase, RbsK (52).
To elucidate the molecular details of the E. coli ribose transporter mechanism as well as to improve understanding of ABC transporter function, we isolated rbs system complexes where the soluble components RbsA and RbsB were bound in vitro to the hexahistidine-tagged membrane protein, RbsC, and purified via affinity chromatography in the presence of substrate and cofactors. Using this method, we isolated three complexes: RbsABC, RbsAC, and the previously unobserved complex, RbsBC.
The Formation of the RbsABC Complex
We found that the full transport complex would form in the presence of the three cofactor combinations: Mg2+-AMP-PNP, Mg2+-ATPγS, or Mg2+-ATP-VO4. Based on these combinations, we determined that minimally, the following cofactors are required for stabilization of the full complex: the nucleotide, the γ-phosphate, and Mg2+. The absence of one or more of these leads to the alternate complexes discussed below. We infer from these observations that the ribose transporter forms a full and stable complex just before and after ATP hydrolysis.
The Formation of the RbsAC Complex
The TMD-NBD complex is the canonical resting state in vivo for bacterial ABC importers. This conclusion is challenged for this system based on the unexpected requirement of Mg2+-ADP for the isolation of RbsAC in vitro, even in the presence of all three proteins, suggesting that RbsAC exists only following ATP hydrolysis. This also suggests that RbsAC may not be inherently stable in vivo, although the possibility remains that some RbsA-RbsC interaction is present in the cofactor-free state but is beyond the detection limits of our experiments.
The Formation of the RbsBC Complex
The isolation of the SBP-TMD complex, heretofore unobserved in studies of other ABC importers, was unexpected and suggests that the NBD can dissociate from the TMD during the transport cycle. The absence of cofactors leads to formation of the RbsBC complex in vitro and may represent the resting state of the complex. The possibility that complex formation was impeded by the location of the histidine tag was tested and was confirmed to not play a role in this process. Verification of this observation was sought by site-directed spin-labeling EPR spectroscopy and employed placement of the cysteine-reactive MTSL spin label on the binding face of RbsB. In the presence of RbsC and no cofactors, the label showed decreased mobility compared with RbsB alone. Further, the spectrum of RbsB bound to RbsAC in the vanadate-trapped conformation was identical to that observed for RbsBC. The simplest explanation for both measurements is a stable association between the binding face of RbsB and RbsC. This observation does not require equivalent conformations of RbsBC and RbsABC but simply means that the local environment experienced by the spin label is similar or identical in both complexes. Taken together with the hydrolysis activity data, these experiments suggest that the RbsB-RbsC interaction in this complex represents a canonical binding configuration.
The Influence of Ribose on Complex Formation
When ribose was present during purification, RbsB did not associate with RbsC during conditions that otherwise promoted RbsBC or RbsABC complex formation (i.e. no cofactors or Mg2+-ATP-VO4). This observation is consistent with binding studies of the vitamin B12 transporter, which show an overall reduction in affinity of the SBP, BtuF, for its cognate TMD-NBD complex, BtuCD, in the presence of the transported substrate (51, 53). Thus, ribose is correlated with the increased ATPase activity of the complex yet also prevents co-elution of RbsB with RbsC. This suggests that ribose is released to RbsC without a requirement for ATP hydrolysis.
Ribose Transporter Complexes Are Active in Liposomes
The full RbsABC complex and partial RbsAC and RbsBC complexes were reconstituted in liposomes, and hydrolysis activity was measured. When the complexes were provided Mg2+-ATP and ribose (and, where applicable, RbsA or RbsB), a 500-fold increase in hydrolysis activity was observed when compared with activity for RbsA alone in the presence of empty liposomes. Functional analysis of the reconstituted system also confirms that ribose-bound RbsB is necessary for stimulation of hydrolysis activity, showing a 10-fold increase when substrate and the SBP are present. This result is similar to observations made from type I transporters like the maltose transporter system, where maltose-bound SBP stimulates hydrolysis activity of the reconstituted transporter 600-fold (16), but is divergent from the results of type II transporters like the vitamin B12 system, where high basal rates of hydrolysis activity are observed in a substrate- and SBP-independent manner (51). When considered in isolation, RbsA itself is far less active (0.5 nmol of Pi/mg/min) than NBD proteins from other bacterial ABC importers (60–90 nmol of Pi/mg/min). This could perhaps be attributed to the unusual architecture of RbsA relative to other bacterial ABC proteins: a single polypeptide consisting of fused NBDs, with mutations, revealed by sequence alignment, in several conserved active site amino acids critical for ATP binding and hydrolysis in one of the two domains (29).
Cofactor-dependent Purification of Complexes Suggests a Distinct Transport Mechanism
Cofactor variation in purification-based studies has led to the isolation of RbsABC, RbsAC, and RbsBC complexes. Compositions of the isolated complexes are dependent upon the presence of Mg2+, the identity of the bound nucleotide, and the addition of ribose (Fig. 6A). Based on the evidence presented in this paper and previous studies of bacterial ABC importers, we propose a scenario wherein RbsBC assembles in the absence of cofactors and subsequently interacts with Mg2+-ATP-bound RbsA to form the intact complex (Fig. 6B). Assembly of this complex leads to ATP hydrolysis and ribose uptake. Following hydrolysis, the interaction between RbsB and RbsC is destabilized, and RbsB dissociates from RbsAC. This final observation is supported by the cofactor-dependent isolation of RbsAC even in the presence of all three proteins, demonstrating that RbsB is released during the transport cycle. The physiological purpose of RbsB release may be to allow it to fulfill its dual roles as both a periplasmic ribose scavenger and a signaling molecule that interacts with the chemotaxis receptor, Trg (24, 25, 32). The physiological consequences of the observed RbsA-RbsC dissociation are less clear and merit further investigation into the mechanistic underpinnings of this process. Given that intracellular Mg2+ and ATP concentrations are typically 1–10 mm (54), the possibility exists that such dissociation is not experienced in vivo.
FIGURE 6.
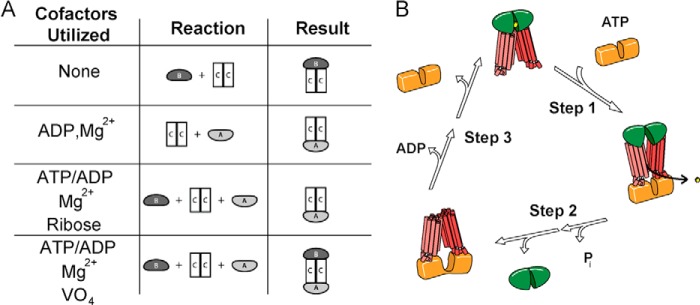
A, summary of in vitro complex formation experiments, including cofactors and/or substrates utilized, purified proteins used, and resulting complexes. All cofactors/substrates were used at 1 mm final concentrations. B, proposed transport assembly mechanism of RbsABC complex. Step 1, ATP-Mg2+ bound RbsA (orange) binds to RbsBC (green and red, respectively; bound ribose in yellow), forming RbsABC; step 2, RbsABC hydrolyzes ATP and releases ribose, changing conformation, precipitating dissociation of RbsB and subsequently the release of Pi; step 3, ADP dissociates from RbsAC, causing RbsA to dissociate, allowing loaded RbsB to again bind RbsC.
The Ribose Transporter Shares Traits with Both Type I and Type II Importers
Type I, or “small,” transporters are represented in the literature by the maltose, molybdate I (ModABC), and methionine systems (11–14, 55, 56). Type II, or “large,” transporters are represented in the literature by the vitamin B12, heme, and molybdate II (MolABC, formerly HI1470/1471) systems (18–20, 21, 22, 57). In vivo labeling experiments have suggested that the topology of RbsC is composed of 10 transmembrane helices/monomer (27), typical of type II transporters. The association of RbsB with RbsC in the absence of cofactors and the contrasting dissociation in the presence of ribose are both type II-like behaviors. The sequence of RbsA suggests that it contains no regulatory domains that have been identified in type I members MalK (maltose), ModC (type I molybdate), and MetN (methionine), much like type II transporter NBDs (11–14, 18–22, 51, 53, 55–58).
However, some ribose transporter behaviors are consistent with type I systems. The promotion of ribose transporter complex formation in the presence of ATP has also been observed in the maltose transporter (28), in contrast to ATP-dependent complex destabilization in type II transporters (53). Additionally, RbsB is a member of the SBP family typically associated with type I ABC importers (23, 59–63), which employ more significant conformational changes to facilitate substrate capture than those of the type II variety. The shared similarities of the ribose transporter with both importer types suggest that traits are not uniquely clustered, and more consideration may be merited regarding the subdivision of ABC importers. Indeed, studies have revealed that type II importers may possess divergent transport mechanisms correlated with the size of transported substrate (59–64). For instance, it was recently demonstrated that the vitamin B12 transporter has an increased affinity for the substrate-loaded SBP upon ATP binding to the transporter (65). This contrasts with previous studies that showed that the SBP associated stably only with the nucleotide-free state of the transporter and suggests a role for ATP hydrolysis beyond serving simply as a trigger for binding protein release. A similar promotion of transport is not readily apparent for the ribose transporter based on the proposed order of assembly. In addition, the impact of mutations in conserved elements of RbsA has yet to be determined for transport and may involve functional and structural asymmetry. The consequences of similar mutations have been observed in several ABC exporters, which result in asymmetric ATP binding, hydrolysis, and conformational changes (66–73). Details of this mechanism may be elucidated through further studies of the ribose transporter and other similar importers (74–76). Indeed, the structural basis of binding protein interactions has recently been demonstrated for the ECF (energy-coupling factor) transporters, a distant branch of ABC importers with multiple binding protein partners per transporter (77). These studies may provide the framework for understanding the specificity of protein interactions in type I and type II ABC importers and provide additional insight into the diverse means by which ATP is utilized for transport (78). In conclusion, we propose that, as additional ABC importers are characterized, a spectrum of traits rather than discrete combinations will be the hallmark of this protein superfamily.
Acknowledgments
We recognize Professor Amy L. Davidson posthumously for contributions to the field and thank her for inspirational and illuminating discussions. We also thank members of her group and Dr. R. Michael Everly for assistance in the application of EPR techniques.
This work was supported, in whole or in part, by National Institutes of Health Cancer Center Core Grant CA23168, Biophysics Training Grant T32-GM008296, and Public Health Service Grants GM54993 and GM64676.
- ABC
- ATP binding cassette
- AMP-PNP
- adenosine 5′-(β,γ-imido)triphosphate
- ATPγS
- adenosine 5′-O-(3-thio)triphosphate)
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- DDM
- n-dodecyl-β-d-maltoside
- MTSL
- S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate
- NBD
- nucleotide binding domain
- SBP
- substrate-binding protein
- TMD
- transmembrane domain.
REFERENCES
- 1. Davidson A. L., Dassa E., Orelle C., Chen J. (2008) Structure, function, and evolution of bacterial ATP-binding cassette system. Microbiol. Mol. Biol. Rev. 72, 317–364, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rees D.C., Johnson E., Lewinson O. (2009) ABC transporters: the power to change. Nat. Rev. Mol. Cell Biol. 10, 218–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Oldham M. L., Davidson A. L., Chen J. (2008) Structural insights into ABC transporter mechanism. Curr. Opin. Struct. Biol. 18, 726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen J. (2013) Molecular mechanism of the Escherichia coli maltose transporter. Curr. Opin. Struct. Biol. 23, 492–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Davidson A. L., Chen J. (2004) ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73, 241–268 [DOI] [PubMed] [Google Scholar]
- 6. Locher K. (2004) Structure and mechanism of ABC transporters. Curr. Opin. Struct. Biol. 14, 426–431 [DOI] [PubMed] [Google Scholar]
- 7. Borst P., Elferink R. O. (2002) Mammalian ABC transporters in health and disease. Annu. Rev. Biochem. 71, 573–592 [DOI] [PubMed] [Google Scholar]
- 8. Holland I. B., Cole S. P. C., Kuchler K., Higgins C. F. (2003) ABC Proteins from Bacteria to Man, pp. 147–203, Academic Press, London [Google Scholar]
- 9. Newstead S., Fowler P. W., Bilton P., Carpenter E. P., Sadler P. J., Campopiano D. J., Sansom M. S., Iwata S., (2009) Insights into how nucleotide-binding domains power ABC transport. Structure 17, 1213–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barroga C. F., Zhang H., Wajih N., Bouyer J. H., Hermodson M. A. (1996) The proteins encoded by the rbs operon of Escherichia coli: I. overpurification, purification, characterization, and functional analysis of RbsA. Protein Sci. 5, 1093–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Khare D., Oldham M. L., Orelle C., Davidson A. L., Chen J. (2009) Alternating access in maltose transporter mediated by rigid-body rotations. Mol. Cell 33, 528–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oldham M. L., Khare D., Quiocho F. A., Davidson A. L., Chen J. (2007) Crystal structure of a catalytic intermediate of the maltose transporter. Nature 450, 515–521 [DOI] [PubMed] [Google Scholar]
- 13. Oldham M. L., Chen J. (2011) Crystal structure of the maltose transporter in a pretranslocation intermediate state. Science 332, 1202–1205 [DOI] [PubMed] [Google Scholar]
- 14. Chen S., Oldham M. L., Davidson A. L., Chen J. (2013) Carbon catabolite repression of the maltose transporter revealed by x-ray crystallography. Nature 499, 364–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Austermuhle M. I., Hall J. A., Klug C. S., Davidson A. L. (2004) Maltose-binding protein is open in the catalytic transition state for ATP hydrolysis during maltose transport. J. Biol. Chem. 279, 28243–28250 [DOI] [PubMed] [Google Scholar]
- 16. Sharma S., Davis J. A., Ayvaz T., Traxler B., Davidson A. L. (2005) Functional reassembly of the Escherichia coli maltose transporter following purification of a MalF-MalG subassembly. J. Bacteriol. 187, 2908–2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Davidson A. L., Shuman H. A., Nikaido H. (1992) Mechanism of maltose transport in Escherichia coli: transmembrane signaling by periplasmic binding-proteins. Proc. Natl. Acad. Sci. U.S.A. 89, 2360–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Locher K. P., Lee A. T., Rees D. C. (2002) The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science 296, 1091–1098 [DOI] [PubMed] [Google Scholar]
- 19. Hvorup R. N., Goetz B. A., Niederer M., Hollenstein K., Perozo E., Locher K.P. (2007) Asymmetry in the structure of the ABC transporter-binding protein complex BtuCD-BtuF. Science 317, 1387–1390 [DOI] [PubMed] [Google Scholar]
- 20. Korkhov V. M., Mireku S. A., Locher K. P. (2012) Structure of the AMP-PNP bound vitamin B12 transporter BtuCD-F, Nature 490, 367–372 [DOI] [PubMed] [Google Scholar]
- 21. Pinkett H. W., Lee A. T., Lum P., Locher K. P., Rees D. C. (2007) An inward facing conformation of a putative metal-chelate-type ABC transporter. Science 315, 373–377 [DOI] [PubMed] [Google Scholar]
- 22. Rice A. J., Alvarez F. J. D., Schultz K. M., Klug C. S., Davidson A. L. D., Pinkett H. W. (2013) EPR spectroscopy of MolB2C2-A reveals mechanism of transport for a bacterial type II molybdate importer. J. Biol. Chem. 288, 21228–21235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Berntsson R. P., Smits S. H., Schmitt L., Slotboom D. J., Poolman B. (2010) A structural classification of substrate-binding proteins. FEBS Lett. 584, 2606–2617 [DOI] [PubMed] [Google Scholar]
- 24. Mowbray S. L., Cole L. B. (1992) 1.7 Å x-ray structure of the periplasmic ribose receptor from Escherichia coli. J. Mol. Biol. 10.1016/0022-2836(92)91033-L [DOI] [PubMed] [Google Scholar]
- 25. Björkman A. J., Mowbray S. L. (1998) Multiple open forms of ribose-binding protein trace the path of its conformational change. J. Mol. Biol. 279, 651–664 [DOI] [PubMed] [Google Scholar]
- 26. Park Y., Park C. (1999) Topology of RbsC, a membrane component of the ribose transporter, belonging to the AraH superfamily. J. Bacteriol. 181, 1039–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stewart J. B., Hermodson M. A. (2003) Topology of RbsC, the membrane component of the Escherichia coli ribose transporter. J. Bacteriol. 185, 5234–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen J., Sharma S., Quiocho F. A., Davidson A. L. (2001) Trapping the transition state of an ATP-binding cassette transporter: evidence for a concerted mechanism of maltose transport. Proc. Natl. Acad. Sci. U.S.A. 98, 1525–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Buckel S. D., Bell A. W., Rao J. K., Hermodson M. A. (1986) An analysis of the structure of the product of the rbsA gene of Escherichia coli K12. J. Biol. Chem. 261, 7659–7662 [PubMed] [Google Scholar]
- 30. Zaitseva J., Zhang H., Binnie R. A., Hermodson M. (1996) The proteins encoded by the rbs operon of Escherichia coli II: use of chimeric protein constructs to isolate and characterize RbsC. Protein Sci. 5, 1100–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Binnie R. A., Zhang H., Mowbray S., Hermodson M. A. (1992) Functional mapping of the surface of Escherichia coli ribose-binding protein: mutations that affect chemotaxis and transport. Protein Sci. 1, 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Willis R. C., Furlong C. E. (1974) Purification and properties of a ribose-binding protein from Escherichia coli. J. Biol. Chem. 249, 6926–6929 [PubMed] [Google Scholar]
- 33. Narayanan A., Ridilla M., Yernool D. (2011) Restrained expression, a method to overproduce toxic membrane proteins by exploiting operator-repressor interactions. Protein Sci. 20, 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Findley J. B. C., Evans W. H. (1987) Biological Membranes: A Practical Approach, pp. 219–277, IRL Press, Washington, D. C. [Google Scholar]
- 35. Teeters C. L., Eccles J., Wallace B. A. (1987) A theoretical analysis of the effects of sonication on differential absorption flattening in suspensions of membrane sheets. Biophys. J. 51, 527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wallace B. A., Mao D. (1984) Circular dichroism analyses of membrane proteins: an examination of differential light-scattering and absorption flattening effects in large membrane vesicles and membrane sheets. Anal. Biochem. 142, 317–328 [DOI] [PubMed] [Google Scholar]
- 37. Wallace B. A., Teeters C. L. (1987) Differential absorption flattening optical effects are significant in the circular dichroism spectra of large membrane fragments. Biochemistry 26, 65–70 [DOI] [PubMed] [Google Scholar]
- 38. Chang C. T., Wu C. S., Yang J. T. (1978) Circular dichroism analysis of protein conformation: inclusion of the β-turns. Anal. Biochem. 91, 13–31 [DOI] [PubMed] [Google Scholar]
- 39. Manavalan P., Johnson W. C. (1987) Variable selection method improves the prediction of protein secondary structure from circular dichroism spectra. Anal. Biochem. 67, 76–85 [DOI] [PubMed] [Google Scholar]
- 40. Provencher S. W., Glöckner J. (1981) Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20, 33–37 [DOI] [PubMed] [Google Scholar]
- 41. Venyaminov S. Y., Yang J. T. (1996) In Circular Dichroism and the Conformational Analysis of Biomolecules, pp. 69–107, Plenum Press, New York [Google Scholar]
- 42. Baykov A. A., Evtushenko O. A., Avaeva S. M. (1988) A Malachite Green procedure for ortho-phosphate determination and its use in alkaline phosphatase-based enzyme-immunoassay. Anal. Biochem. 171, 266–270 [DOI] [PubMed] [Google Scholar]
- 43. Chifflet S., Torriglia A., Chiesa R., Tolosa S. (1988) A method for the determination of inorganic phosphate in the presence of labile organic phosphate and high concentrations of protein: application to lens ATPases. Anal. Biochem. 168, 1–4 [DOI] [PubMed] [Google Scholar]
- 44. Geladopoulos T. P., Sotiroudis T. G., Evangelopoulos A. E. (1991) A Malachite Green colorimetric assay for protein phosphatase activity. Anal. Biochem. 192, 112–116 [DOI] [PubMed] [Google Scholar]
- 45. Towbin H., Staehelin T., Gordon J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U.S.A. 76, 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu P. Q., Ames G. F. L. (1998) In vitro disassembly and reassembly of an ABC transporter, the histidine permease. Proc. Natl. Acad. Sci. U.S.A. 95, 3495–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ames G. F. L., Nikaido K., Wang I. X., Liu P. Q., Liu C. E., Hu C. (2001) Purification and characterization of the membrane-bound complex of an ABC transporter, the histidine permease. J. Bioenerg. Biomembr. 33, 79–92 [DOI] [PubMed] [Google Scholar]
- 48. Greller G., Riek R., Boos W. (2001) Purification and characterization of the heterologously expressed trehalose/maltose ABC transporter complex of the hyperthermophilic archaeon Thermococcus litoralis. Eur. J. Biochem. 268, 4011–4018 [DOI] [PubMed] [Google Scholar]
- 49. Scheffel F., Fleischer R., Schneider E. (2004) Functional reconstitution of a maltose ATP-binding cassette transporter from the thermoacidophilic gram-positive bacterium Alicyclobacillus acidocaldarius. Biochim. Biophys. Acta 1656, 57–65 [DOI] [PubMed] [Google Scholar]
- 50. Landmesser H., Stein A., Blüschke B., Brinkmann M., Hunke S., Schneider E. (2002) Large-scale purification, dissociation and functional reassembly of the maltose ATP-binding cassette transporter (MalFGK(2)) of Salmonella typhimurium. Biochim. Biophys. Acta 1565, 64–72 [DOI] [PubMed] [Google Scholar]
- 51. Borths E. L., Poolman B., Hvorup R. N., Locher K. P., Rees D. C. (2005) In vitro functional characterization of BtuCD-F, the Escherichia coli ABC transporter for vitamin B12 uptake. Biochemistry 44, 16301–16309 [DOI] [PubMed] [Google Scholar]
- 52. Hope J. N., Bell A. W., Hermodson M. A., Groarke J. M. (1986) Ribokinase from Escherichia coli K12: nucleotide sequence and overexpression of the rbsK gene and purification of ribokinase. J. Biol. Chem. 261, 7663–7668 [PubMed] [Google Scholar]
- 53. Lewinson O., Lee A. T., Locher K. P., Rees D. C. (2010) A distinct mechanism for the ABC transporter BtuCD-BtuF revealed by the dynamics of complex formation. Nat. Struct. Mol. Biol. 17, 332–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Beis I., Newsholme E. A. (1975) The contents of adenine nucleotides, phosphagens and some glycolytic intermediates in resting muscles from vertebrates and invertebrates. Biochem. J. 152, 23–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gerber S., Comellas-Bigler M., Goetz B.A., Locher K.P. (2008) Structural basis of trans-inhibition in a molybdate/tungstate ABC transporter. Science 321, 246–250 [DOI] [PubMed] [Google Scholar]
- 56. Kadaba N. S., Kaiser J. T., Johnson E., Lee A., Rees D. C. (2008) The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation. Science 321, 250–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Woo J. S., Zeltina A., Goetz B. A., Locher K. P. (2012) X-ray structure of the Yersinia pestis heme transporter HmuUV. Nat. Struct. Mol. Biol. 19, 1310–1315 [DOI] [PubMed] [Google Scholar]
- 58. Dawson R. J., Locher K. P. (2006) Structure of a bacterial multidrug ABC transporter. Nature 443, 180–185 [DOI] [PubMed] [Google Scholar]
- 59. Karpowich N. K., Huang H. H., Smith P. C., Hunt J. F. (2003) Crystal structures of the BtuF periplasmic-binding protein for vitamin B12 suggest a functionally important reduction in protein mobility upon ligand binding. J. Biol. Chem. 278, 8429–84234 [DOI] [PubMed] [Google Scholar]
- 60. Tirado-Lee L., Lee A., Rees D. C., Pinkett H. W. (2011) Classification of Haemophilus influenzae ABC transporter HI1470/71 through its cognate molybdate periplasmic binding protein, MolA. Structure 19, 1701–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vigonsky E., Ovcharenko E., Lewinson O. (2013) Two molybdate/tungstate ABC transporters that interact very differently with their substrate binding proteins. Proc. Natl. Acad. Sci. U.S.A. 110, 5440–5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Quiocho F. A., Ledvina P. S. (1996) Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol. Microbiol. 20, 17–25 [DOI] [PubMed] [Google Scholar]
- 63. Goetz B. A., Perozo E., Locher K. P. (2009) Distinct gate conformations of the ABC transporter BtuCD revealed by electron spin resonance spectroscopy and chemical cross-linking. FEBS Lett. 583, 266–270 [DOI] [PubMed] [Google Scholar]
- 64. Joseph B., Jeschke G., Goetz B. A., Locher K. P., Bordignon E., (2011) Transmembrane gate movements in the type II ATP-binding cassette (ABC) importer BtuCD-F during nucleotide cycle. J. Biol. Chem. 286, 41008–41017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Korkhov V. M., Mireku S. A., Veprintsev D. B., Locher K. P. (2014) Structure of AMP-PNP bound BtuCD and mechanism of ATP-powered vitamin B12 transport by BtuCD-F. Nat. Struct. Mol. Biol. 21, 1097–1099 [DOI] [PubMed] [Google Scholar]
- 66. Dawson R. J., Hollenstein K., Locher K. P. (2007) Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett. 581, 935–938 [DOI] [PubMed] [Google Scholar]
- 67. Hohl M., Hürlimann L. M., Böhm S., Schöppe J., Grütter M. G., Bordignon E., Seeger M. A. (2014) Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter. Proc. Natl. Acad. Sci. U.S.A. 111, 11025–11030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hohl M., Briand C., Grütter M. G., Seeger M. A. (2012) Crystal structure of a heterodimeric ABC exporter in its inward-facing conformation. Nat. Struct. Mol. Biol. 19, 395–402 [DOI] [PubMed] [Google Scholar]
- 69. Jin M. S., Oldham M. L., Zhang Q., Chen J. (2012) Crystal structure of the multidrug transporter P-glycoprotein Caenorhabditis elegans. Nature 490, 566–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ward A., Reyes C. L., Yu J., Roth C. B., Chang G. (2007) Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc. Natl. Acad. Sci. U.S.A. 104, 19005–19010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhou Z., Wang X., Liu H. Y., Zou X., Li M., Hwang T. C. (2006) The two ATP binding sites of cystic fibrosis transmembrane conductance regulator (CFTR) play distinct roles in gating kinetics and energetics. J. Gen. Physiol. 128, 413–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mishra S., Verhalen B., Stein R. A., Wen P.-C., Tajkhorshid E., Mchaourab H. S. (2014) Conformational dynamics of the nucleotide binding domains and the power stroke of a heterodimeric ABC transporter. eLife 3, e02740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wen P. C., Verhalen B., Wilkens S., Mchaourab H. S., Tajkhorshid E., (2013) On the origin of large flexibility of P-glycoprotein in the inward-facing state. J. Biol. Chem. 288, 19211–19220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Procko E., Ferrin-O'Connell I., Ng S. L., Gaudet R. (2006) Distinct structural and functional properties of the ATPase sites in an asymmetric ABC transporter. Mol. Cell 24, 51–62 [DOI] [PubMed] [Google Scholar]
- 75. Ernst R., Kueppers P., Klein C. M., Schwarzmueller T., Kuchler K., Schmitt L. (2008) A mutation of the H-loop selectively affects rhodamine transport by the yeast multidrug ABC transporter Pdr5. Proc. Natl. Acad. Sci. U.S.A. 105, 5069–5074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dietrich D., Schmuths H., De Marcos Lousa C., Baldwin J. M., Baldwin S. A., Baker A., Theodoulou F. L., Holdsworth M. J. (2009) Mutations in the Arabidopsis peroxisomal ABC transporter COMATOSE allow differentiation between multiple functions in planta: insights from allelic series. Mol. Biol. Cell 20, 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Slotboom D. J. (2014) Structural and mechanistic insights into prokaryotic energy-coupling factor transporters. Nat. Rev. Microbiol. 12, 79–87 [DOI] [PubMed] [Google Scholar]
- 78. Rice A. J., Park A., Pinkett H. W. (2014) Diversity in ABC transporters: type I, II, and III importers. Crit. Rev. Biochem. Mol. Biol. 49, 426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]