Background: DNA primase synthesizes RNA primers and is indispensable for genome replication.
Results: We present a crystal structure of the intact human primase at 2.65 Å resolution.
Conclusion: The long linker between two domains of the large subunit is important for RNA priming.
Significance: The obtained data provide notable insight into the mechanism of primase function.
Keywords: Crystal Structure, DNA Primase, DNA Replication, Iron-Sulfur Protein, Protein Complex, Zinc
Abstract
DNA replication in bacteria and eukaryotes requires the activity of DNA primase, a DNA-dependent RNA polymerase that lays short RNA primers for DNA polymerases. Eukaryotic and archaeal primases are heterodimers consisting of small catalytic and large accessory subunits, both of which are necessary for RNA primer synthesis. Understanding of RNA synthesis priming in eukaryotes is currently limited due to the lack of crystal structures of the full-length primase and its complexes with substrates in initiation and elongation states. Here we report the crystal structure of the full-length human primase, revealing the precise overall organization of the enzyme, the relative positions of its functional domains, and the mode of its interaction with modeled DNA and RNA. The structure indicates that the dramatic conformational changes in primase are necessary to accomplish the initiation and then elongation of RNA synthesis. The presence of a long linker between the N- and C-terminal domains of p58 provides the structural basis for the bulk of enzyme's conformational flexibility. Deletion of most of this linker affected the initiation and elongation steps of the primer synthesis.
Introduction
Initiation of genome replication in eukaryotes relies on the RNA primase and DNA polymerase activities of a four-subunit primase-polymerase α (Prim2-Pol α) complex. Prim-Pol α synthesizes short RNA/DNA primers on leading and lagging strands, which are further extended by replicative DNA polymerases ϵ and δ, respectively (1). In humans, Prim consists of the small catalytic subunit (p49; also known as p48, PRIM1, Pri1, and PriS) and the large regulatory subunit (p58; also known as PRIM2, Pri2, and PriL), whereas Pol α is composed of the catalytic subunit (p180) and the accessory subunit (p70). The evolutionarily conserved C-terminal domain of p180 (p180C) plays an important structural role by tethering both p70 and primase to the Pol α catalytic subunit (2–5). The tight physical association of primase with DNA polymerase, found only in eukaryotes, ensures effective intramolecular substrate transfer between the two active sites. When primase synthesizes the unit-length RNA primers (7–10 nucleotides long), the primed template is translocated to the Pol α active site and is extended by 20–30 deoxynucleotides (5–8).
There is no structural information for full-length primase as well as primase captured at either the initiation or elongation steps of primer synthesis; therefore, the mechanism of primase action remains unclear. It has been proposed that primases initiate primer synthesis after the formation of the quaternary complex with the template DNA and two 5′-nucleoside triphosphate (NTP) molecules (7, 9). Therefore, the primase must have two NTP binding sites, referred to as the initiation site and the elongation (active) site. Eukaryotic primases have a minimal recognition site on DNA template and only require pyrimidine to template the 5′-terminal nucleotide of the primer, which means that the primase initiation site is specific for ATP or GTP (8, 10). This is different from prokaryotic primases, which usually recognize the trinucleotide sequence on the template DNA, where the middle nucleotide templates the 5′-terminal nucleotide of the primer (7).
The large subunit, which exists only in archaeal and eukaryotic primases, plays an important role in primase activity; it stabilizes the small catalytic subunit and, probably, interacts with the template DNA and the first initiating NTP (11–15). It has also been proposed that the large subunit helps to “count” the product size and participates in translocation of the primer-template to the Pol α active site (10, 12). The N-terminal part of the large subunit (p58N in human primase) provides a platform for interactions with p49 and Pol α (5, 16). Most structural elements contributing to NTP and DNA binding by the large subunit are located in the C-terminal half of the protein (p58C in human primase) (11, 17–20). The crystal structures of the C-terminal domain of the large subunit of the yeast and human primases revealed an iron-sulfur cluster (4Fe-4S) coordinated by four cysteines (17, 18, 21). The results of structural studies confirmed previously accumulated genetic and biochemical data on the role of these conserved cysteine residues in the structural stability and activity of primase (14, 19, 20, 22). It was found that the C terminus of the large subunit has structural similarity with the region of DNA photolyase/cryptochrome 3 from Arabidopsis thaliana, which encompasses its active site and includes the FAD- and DNA-binding sites (17). The first information about an overall three-dimensional architecture of eukaryotic primases was derived from the crystal structures of distantly related archaeal primases (23), including the structures of the Pyrococcus furiosus (Pfu) and Pyrococcus horikoshi (Pho) primase catalytic subunits, and the structure of the Sulfolobus solfataricus (Sso) primase catalytic subunit in complex with the N-terminal domain of its large subunit (24–26). These data and mapping of the active site of mouse p49 by site-directed mutagenesis allowed determination of the positions of three invariant catalytic aspartates (27). Recently, the crystal structures of the catalytic subunit of yeast primase and the human p49-p58(1–253) complex have been determined (16, 28). These results confirmed the early idea proposing similar organization of the active site in eukaryotic and archaeal primases and the role of three catalytic aspartates in the coordination of two divalent ions and NTP (23, 24). This implies that the primase active site is adopted for the common mechanism of nucleic acids synthesis (29). Unfortunately, the current structural information obtained for truncated primases is not sufficient to understand the mechanism of RNA primer synthesis.
Here we report the crystal structure of the wild-type human primase heterodimer containing the full-length p49 and p58 subunits at 2.65 Å resolution. The structure revealed the cashew-like shape of the complex, where the two functional domains are separated by a long flexible linker, indicating that dramatic rearrangements in the primase molecule are necessary for RNA synthesis. In a set of structure-based functional assays, we clarified the mechanism of the primer length counting and confirmed that proper length of the linker connecting the N- and C-terminal domains of p58 is required for RNA primer synthesis.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
Cloning, expression, and purification to homogeneity of the human primase heterodimer (p49-p58; p49 contains amino acids 1–420, and p58 contains amino acids 1–509), its linker mutants (Prim-Δ5, Prim-Δ15, and Prim-Ins5), as well as p49-p58(1–265), p58(266–456), and human Pol α without the catalytic core (p70-p180C; p180C contains amino acids 1265–1462) have been described elsewhere (5, 21, 30, 31). For better purity of p58(266–456), we added the chromatography step on the heparin HiTrap HP column (GE Healthcare). DNA sequences encoding for different primase mutants in the linker region between p58N and p58C were obtained by a site-directed plasmid mutagenesis protocol (32).
Primase Assay
Primase de novo activity was analyzed using DNA template-1 (5′-A15TA7) and template-2 (5′-AAA(GA)6TA7). For the primer extension assay, we used 5′-TYE665 fluorophore-modified ribooligonucleotide (5′-CTTGAAAACATAGCGA) and a 46-nucleotide DNA template-3 (5′-GATTCACACGACTAGCACACTAAGCACTCGCTATGTTTTCAAGTTT; the region complementary to the RNA primer is underlined). All oligonucleotides were from IDT Inc. The de novo activity of primase was tested in a 20-μl reaction containing 30 mm Hepes-KOH (pH 7.9), 50 mm KCl, 1 mm DTT, 2 mm MnCl2, 50 μm UTP, 50 μm CTP, 0.15 μm [γ-33P]ATP (5000 Ci/mmol; PerkinElmer Life Sciences), 2 μm template, 0.3 μm primase (wild type or mutant), and 0.4 μm p70-p180C. Reactions were assembled on ice and incubated at 35 °C for 1–20 min in a thermal cycler (Thermolyne Amplitron I). To allow complex formation, the primase sample and p70-p180C were preincubated on ice for 20 min in reaction buffer excluding MnCl2 and NTPs. Reactions were stopped by mixing with an equal volume of formamide loading buffer (90% (v/v) formamide, 5 mm EDTA, 0.02% bromphenol blue, and 0.02% xylene cyanol), heated at 65 °C for 10 min, and resolved by 20% urea-PAGE (UreaGel system (19:1 acrylamide/bisacrylamide), National Diagnostics) for 5 h at 2000 V. The gel was dried at 65 °C for 45 min using a Bio-Rad 583 gel dryer. The reaction products were visualized by phosphorimaging (Typhoon 9410, GE Healthcare). Primase activity in extending the RNA primer was tested in a 20-μl reaction containing 30 mm Hepes-KOH (pH 7.9), 50 mm KCl, 1 mm DTT, 7 mm magnesium acetate, 100 μm NTPs, 0.5 μm 5′-TYE665 fluorophore-labeled RNA primer annealed to DNA template, 0.2 μm primase (wild-type or mutant), and 0.26 μm p70-p180C. 10 μm RNA primer was annealed with 15 μm DNA template by a decrease in temperature from 70 to 25 °C at a 0.5 °C/min gradient in a thermal cycler (50 mm NaCl). Reactions were assembled, quenched, and separated as described above except for the formamide loading buffer composition (95% formamide, 0.025% Orange G, 5 mm EDTA, and 0.025% SDS). For visualization of the products, the Typhoon 9410 imager was used (emission of fluorescence at 670 nm). All activity gels were repeated at least two times.
Crystal Structure Determination and Refinement
Preparation of protein samples, crystallization, x-ray diffraction data collection, and preliminary analysis of crystals are described elsewhere (30). For the structure determination of the human p49-p58 complex by the SAD method, zinc anomalous data were collected. Self-rotation searches revealed non-crystallographic 2-fold symmetry, indicating the presence of two primase molecules in an asymmetric unit. That is why we expected to locate four metal sites, two for zinc in the p49 subunits and two for 4Fe-4S clusters in the p58C domains. However, the Patterson map inspection revealed only three metal sites, indicating that either one of the p49 subunits or one of the p58C domains is disordered. Initial phasing, density modification, and model building with three metal sites using Phenix (33) revealed some helices and strands. However, the density was not of sufficient quality. Then the structure of the 4Fe-4S cluster domain of human primase (PDB code 3Q36) was used to place it manually into the modified SAD density by keeping one of the metal positions at the center of the 4Fe-4S cluster and using the positions of the helices as a guide. Only one 4Fe-4S cluster domain could be placed, indicating that the position of the second one is not fixed. The SAD phases were recalculated with two zinc and four iron positions and combined with the phases from the 4Fe-4S cluster domain, resulting in better electron density maps, which were further improved by density-averaging for p49-p58N. About 50% of the amino acids were correctly placed by autobuilding with Phenix (33), and the remaining portion was placed manually in several cycles of model building and refinement. Turbo-Frodo software was used for manual model building and analysis. CNS version 1.1 (34, 35) was used for all crystallographic computing except for the phasing. The final model was refined at 2.65 Å resolution to an Rcryst of 22.8% and an Rfree of 27.2%. The structure has a good geometry with 83.7% of the non-glycine residues located in the most favored regions and 16.0% located in the additionally allowed regions of the Ramachandran plot. The electron density was not observed for residues 282–290, 361–377, and 416–420 in one of the p49 molecules; for residues 283–288, 361–377, and 413–420 in another p49 molecule; for residues 1–27, 84–94, and 252–509 in one of the p58 molecules; and for residues 1–21, 85–90, and 457–509 in another p58 molecule. All figures displaying the crystal structures were prepared with the PyMOL Molecular Graphics System, version 1.3, Schrödinger, LLC (New York).
Modeling
To build the model for the Prim-DNA/RNA-CTP complex, the NTP and RNA-primed DNA were superimposed on the p49-p58 molecule in two steps. At first, the UTP was superimposed on p49-p58 by aligning the catalytic subunit with that from p49-p58(1–253) (PDB code 4BPW; r.m.s. deviation 0.47 Å for the 336 α-carbon atoms). Then the DNA-RNA duplex and dCTP from the Pol α-DNA/RNA-dCTP complex (PDB code 4QCL) were modeled on p49-p58 by aligning the triphosphate and sugar moieties of UTP and dCTP by using the pair-fitting mode of PyMOL. 2′,3′-Dideoxyribose at the 3′-terminus of RNA was converted to ribose, and dCTP was converted to CTP by adding the corresponding hydroxyls. The position of the DNA-RNA duplex and the side chains of Tyr-54, Arg-56, Tyr-57, Asn-308, and Lys-311 were manually adjusted to avoid steric clashes.
RESULTS
Overall Structure
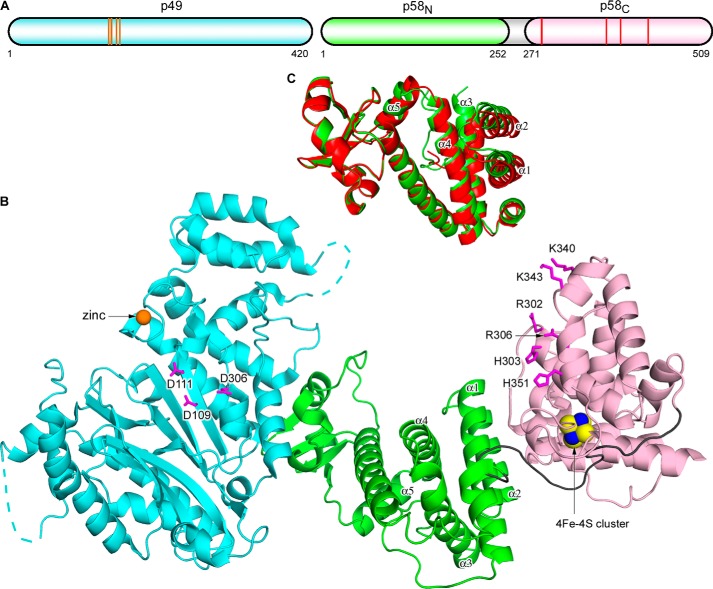
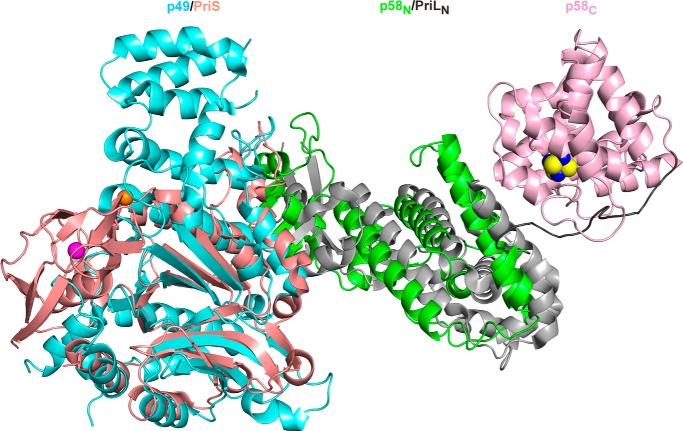
The crystal structure of the human primase p49-p58 complex was determined by the SAD method using zinc and iron anomalous diffraction data and refined at 2.65 Å resolution (Table 1). The final model has an Rcryst of 22.8% and an Rfree of 27.2%. Two independent primase molecules were found in an asymmetric unit. The molecule has an extended cashew-like shape with overall dimensions of 126 × 80 × 60 Å3 (Figs. 1 and 2). p58N is located in the middle and flanked by p49 and p58C, with the architecture of p49-p58N resembling that of Sso primase (Fig. 3A) (26). The p49 subunits from two independent molecules were similar to each other, with an r.m.s. deviation of 0.35 Å for the 389 superimposed α-carbon atoms. Each p49 subunit contained all amino acid residues, except for two loops and the C-terminal tail that are located on the edges of the molecule. Greater discrepancy was found between the p58 subunits. The structure of one p58 subunit is more complete and contains both p58N and p58C domains, except for the residues from the N- and C-terminal tails and one loop. The structure of another p58 subunit has been determined only for the p58N domain lacking the N-terminal tail and one loop. The two p58N domains are slightly different from each other, with an r.m.s. deviation of 1.0 Å for the 213 superimposed α-carbon atoms. Superimposition of the p49 subunits from two independent p49-p58 complexes revealed a significant shift of the helices α1–α5 in the helical subdomain of p58N (Fig. 1C), indicating that p58N can adopt different conformations. Such flexibility was also observed in the structures of human and Sso primases with a deleted C-terminal domain of the large subunit and can play a role in Prim-Pol α function (16, 26). The proposed NTP/DNA-binding interface of p58C (17, 18) is facing the p49 active site (Fig. 1B), which may facilitate their cooperation during RNA synthesis in the cis-position (see “Discussion”).
TABLE 1.
Data collection and refinement statistics
| Native | Anomalous | |
|---|---|---|
| Data collection | ||
| Space group | P1 | P1 |
| Cell dimensions | ||
| a, b, c (Å) | 86.20, 88.90, 94.68 | 86.12, 89.10, 94.92 |
| α, β, γ (degrees) | 93.82, 96.57, 111.72 | 93.46, 96.53, 111.63 |
| Resolution (Å) | 50-2.64 (2.69-2.64)a | 50-3.0 (3.05-3.0) |
| Unique reflections | 68,707 | 47,262 |
| Rmerge (%)b | 4.0 (58.4) | 4.8 (49.4) |
| I/σ (I) | 29.8 (1.6) | 28.3 (2.1) |
| Completeness (%) | 91.5 (93.2) | 90.9 (85.3) |
| Redundancy | 3.8 (3.4) | 3.8 (3.6) |
| Temperature (K) | 100 | 100 |
| Refinement | ||
| Resolution (Å) | 48.48-2.65 | |
| No. of reflections | 66,240 | |
| Rwork/Rfree | 22.9/27.1 | |
| No. of atoms/B-factors (Å2) | ||
| Protein | 11,709/53.5 | |
| Zinc | 2/30.6 | |
| 4Fe-4S | 8/27.5 | |
| Solvent | 88/38.9 | |
| r.m.s. deviation | ||
| Bond length (Å) | 0.009 | |
| Bond angles (degrees) | 1.47 | |
| Ramachandran plot | ||
| Favored (%) | 83.7 | |
| Allowed (%) | 16.0 | |
| Disallowed (%) | 0.2 | |
a Values in parentheses correspond to the highest resolution shell.
b Rmerge = ΣhklΣi|Iihkl − 〈Ihkl〉|/ΣhklΣIihkl, where Ihkl is the observed intensity, and 〈Ihkl〉 is the mean value of Ihkl.
FIGURE 1.
Overall structure of the human primase. A, schematic representation of the domain organization. The orange lines and red lines in the schematics present relative positions of the residues coordinating zinc (Cys-121, Cys-122, Cys-128, and Cys-131) and the 4Fe-4S cluster (Cys-287, Cys-367, Cys-384, and Cys-424), respectively. B, schematic representation of p49-p58. The p49, p58N, p58C, and the linker between p58N and p58C are colored cyan, green, light pink, and gray, respectively. Zinc is shown as an orange sphere, and the 4Fe-4S cluster is shown as a space-filled representation, with iron and sulfur atoms colored blue and yellow, respectively. The disordered regions in p49 are shown by dashed lines. Side chains of catalytic aspartates on p49 and residues forming the proposed NTP/DNA binding site on p58C are shown as sticks and colored magenta. C, comparison of the two primase molecules in an asymmetric unit by superposition of p49. The p58N domain from one molecule (with p58C) is colored green, and that from a second molecule (without p58C) is colored red. p49 and p58C are omitted for clarity. The α-helices responsible for conformational flexibility of p58N are labeled.
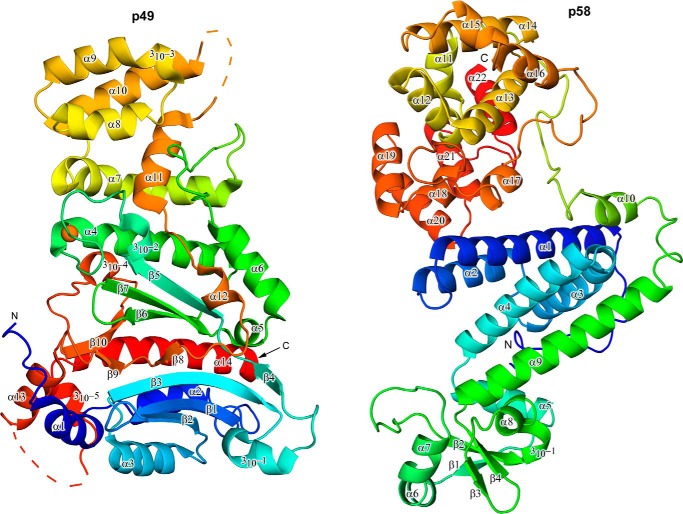
FIGURE 2.
Schematic representation of the p49 and p58 subunits with numbering of secondary structure elements. Both subunits are colored by a gradient from blue (N terminus) to red (C terminus). Zinc is shown as an orange sphere; the iron-sulfur cluster is omitted for clarity. Disordered regions in p49 are shown by dashed lines.
FIGURE 3.
Alignment of the human and Sso primase molecules. The catalytic (PriS) and large (PriL) subunits of Sso primase are colored salmon and gray, respectively. Zinc in PriS is shown as a magenta sphere.
Catalytic Subunit
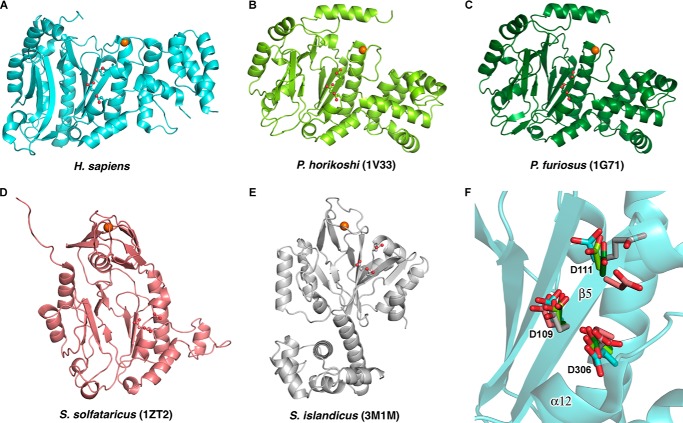
The p49 subunit of human primase has a mixed α/β Prim fold previously identified in archaeal primases (24, 25, 36). Two centrally positioned antiparallel three-strand β-sheets that are connected in the middle by a coil, two small β-strands (β4 and β8), and a small β-sheet containing β9 and β10 form a shallow cleft harboring the active site (Fig. 2). Despite low sequence identity and the larger size of p49 in comparison with the small catalytic subunits of archaeal DNA primases (Pfu, Pho, and Sso), they share some topological similarities in their central portions (Fig. 4, A–E). A Dali search (37) pulled these archaeal primases with a Z-score of greater than 18. Their putative active sites, including three invariant aspartate residues (Asp-109, Asp-111, and Asp-306 in p49), are well superimposed (Fig. 4F). A significant difference is observed in the position of the second catalytic aspartate in Sso primase. This aspartate is located on the coil rather than on the conservative β-strand, which is shortened in Sso primase. This difference may be due to different organization of the zinc-binding motif in Sso primase in comparison with Pfu, Pho, and human primases. Unlike the well superimposed central β-sheets, significant differences exist in the topologies of the helical regions surrounding the central portions (Fig. 4).
FIGURE 4.
Structural comparison of catalytic subunits from the human and archaeal primases. A–E, schematic representation of catalytic subunits from human and archaeal primases. The catalytic aspartates in active sites are highlighted as balls and sticks. The oxygen is colored red, and zinc is shown as an orange sphere. F, superimposition of catalytic aspartates from the structures of archaeal and human primases. Aspartates are depicted by a stick representation, and carbons are colored cyan (human), light green (Pho; PDB code 1V33), dark green (Pfu; PDB code 1G71), salmon (Sso; PDB code 1ZT2), and gray (Sis; PDB code 3M1M). The p49 active site is represented as a schematic where the secondary structure elements are shown with 50% transparency. The positions of catalytic aspartates in p49 are labeled.
For example, human p49 contains a bulky all-helical extension composed of helices α7–α11 (Figs. 1 and 2). This helical subdomain is inserted before the catalytic Asp-306 and stabilizes the helix α4, the coil region of zinc-binding motif, and an extended loop between helices α6 and α7, which participates in interaction with p58. Pfu and Pho primases also contain a relatively compact all-helical extension, which is positioned differently and forms the C terminus of the catalytic subunit, as in bacterial primases. In contrast to single-subunit bacterial primases, the helical subdomain in archaeal and eukaryotic primases is not flexible and plays no role in primer synthesis. Moreover, such an extension is absent in Sso and Sulfolobus islandicus (Sis) primases. Another prominent feature of human p49 is the packing of C-terminal helices 310-4, 310-5, α13, and α14, where the longest helix α14 buttresses the Prim fold opposite the active site cleft (Fig. 2A); deletion of the corresponding helix is lethal in yeast (19, 28). Corresponding C-terminal sequences in Sso primase are folded as an α/β domain adjacent to a zinc finger (Figs. 3 and 4). Interestingly, the C-terminal sequences of Pho and Pfu primases are also folded as helices, but their C-terminal helix that corresponds to helix α14 of p49 is turned away from the center of the molecule and extended closer to a zinc-binding site (Fig. 4, B and C).
Almost all primases have a zinc-binding motif with significant variability in structural organization and its position with respect to the active site (10). In prokaryotes, the zinc-binding motif is part of a separate domain and is located distant from the active site, whereas it is integrated into the Prim fold and situated near the second catalytic aspartate in archaeal and eukaryotic primases (16, 24–26, 28, 38, 39). The functional role of the zinc-binding motif in primer synthesis is not clear. However, it is proposed that its orientation determines the mode of interaction with template DNA, such as its position in regard to the active site and the size of the recognition motif (26, 40, 41). In the catalytic subunit of human primase, zinc is coordinated by four conservative cysteines (positions 121, 122, 128, and 131), which are located within the loop region between helices 310-2 and α4 (Fig. 2). The position and geometry of zinc coordination in p49 near the N terminus of helix α4 is more consistent with zinc binding in Pho and Pfu primases, where it is closer to the active site compared with Sis and especially Sso primases that adopted different geometries for zinc coordination (Fig. 4).
Large Subunit
The large p58 subunit of human primase adopts a multidomain architecture (Figs. 1 and 2). The N-terminal domain (p58N, residues 1–252) is linked to the C-terminal domain (p58C, residues 271–509) via an 18-residue linker (residues 253–270). Similar to the structure of the Sso primase large subunit, p58N is composed of two subdomains: a small subdomain that interacts with p49 and a large subdomain that is connected to p58C. The small subdomain of p58N is composed of a central four-strand antiparallel β-sheet that is surrounded by small helices α6–α8 and 310-1. The large subdomain of p58N contains seven α-helices (α1–α5, α9, and α10) and an extended N-terminal coil that is wedged into the predominantly hydrophobic cleft formed by helices α3–α5 and α9. The linker connecting the p58N and p58C domains is relatively mobile with a broadened electron density in some sections; nevertheless, an entire path of the linker was traceable. Starting from α10, it first extends toward the p58N-p58C interface and then wraps nearly half of p58C without making strong interactions. The all-helical p58C domain with an iron-sulfur cluster coordinated by Cys-287, Cys-367, Cys-384, and Cys-424 has a conformation similar to the previously reported crystal structure of p58(266–456) (21) with an r.m.s. deviation of 0.4 Å for the 213 superimposed α-carbon atoms. The p58N and p58C domains have a small interaction interface formed mostly by hydrophilic contacts between the helices α1 and α2 of p58N and helices α17 and α20 of p58C, respectively (Fig. 5).
FIGURE 5.
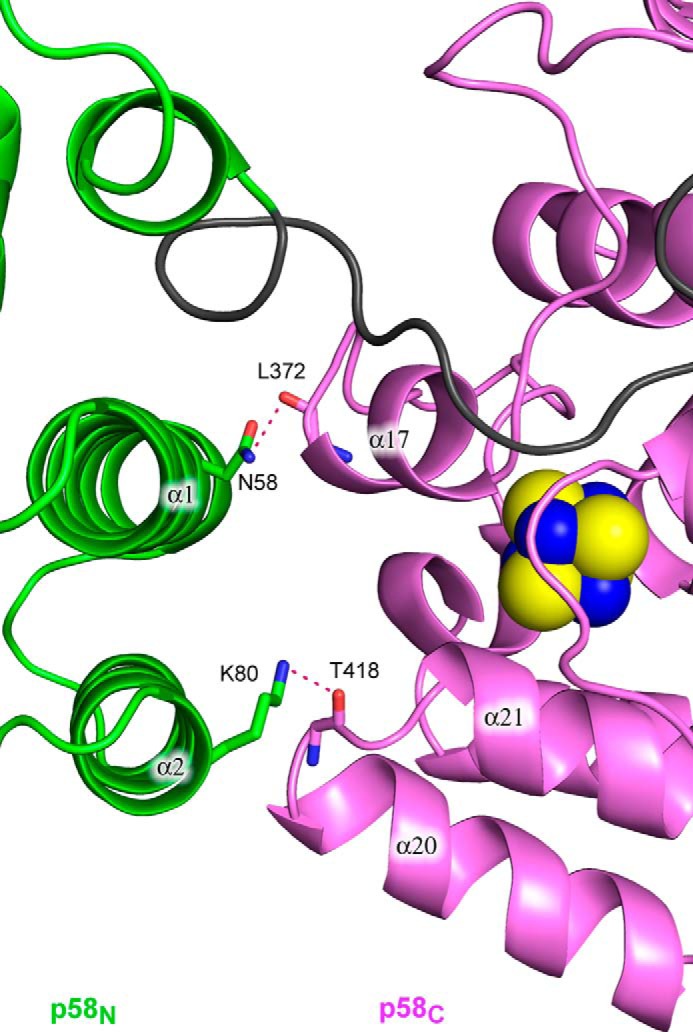
Close-up view of the p58N-p58C interaction interface. The protein is represented as a schematic and colored according to Fig. 1B. Side or main chains making the hydrogen bonds between p58N and p58C are shown as sticks. Hydrogen bonds are drawn with red dashed lines.
Interaction between Primase Subunits
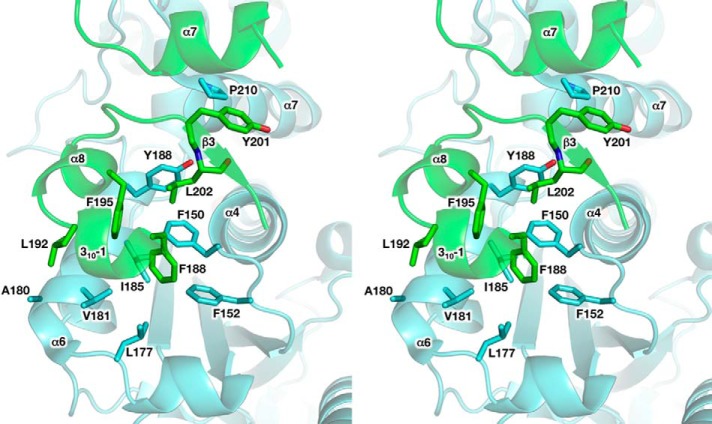
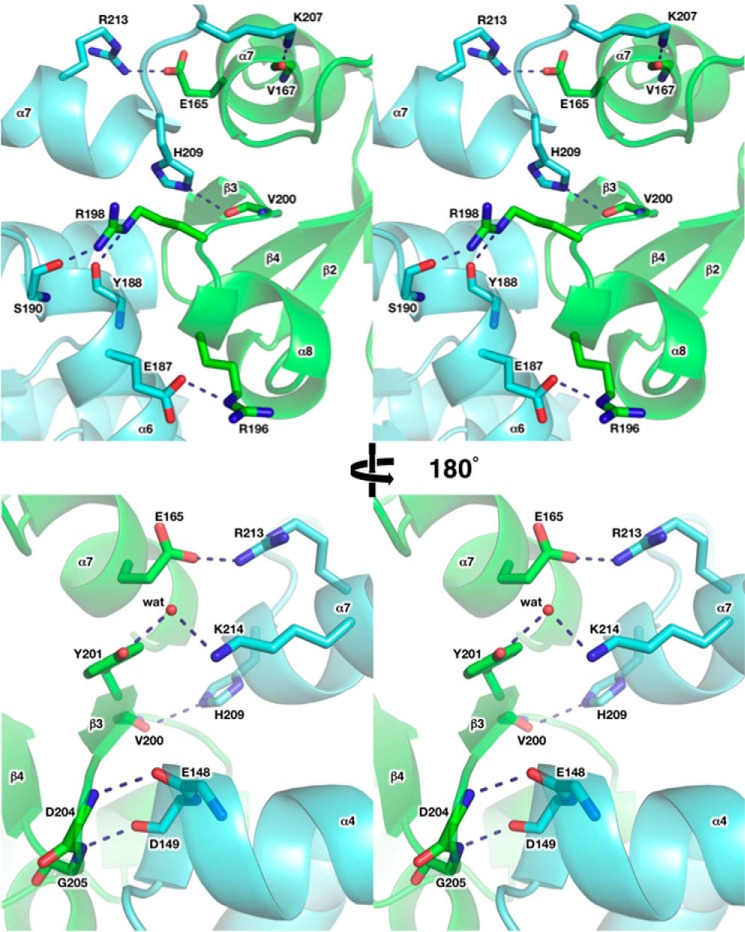
The p49-p58 interaction interface buries ∼1600 Å2 of surface area. The p49 residues Phe-150 from the C-terminal end of helix α4; Phe-152 from the loop between α4 and β6; Leu-177 from the loop between α5 and α6; Ala-180, Val-181, Ile-185, and Tyr-188 from the helix α6; and Pro-210 from the N-terminal end of helix α7 form a predominantly hydrophobic surface for the docking of p58N small subdomain hydrophobic residues Phe-188 from helix 310-1, Leu-192 and Phe-195 from helix α8, and Tyr-201 and Leu-202 from strand β3 (Fig. 6). Furthermore, the interface of human p49-p58 contains eight hydrogen bonds, including one water-mediated hydrogen bond (Fig. 7). Among these bonds, two main-chain to main-chain hydrogen bonds are observed, with donors provided by Asp-204 and Gly-205 of p58 (the loop between strands β3 and β4) and acceptors provided by Glu-148 and Asp-149 of p49 (the C-terminal end of helix α4). Interestingly, despite significant topological differences between human and Sso primases, similar hydrogen bonds are present at the Sso PriS-PriL interface (26) (Fig. 8). The positions of Glu-148 and Asp-149 in human p49 correspond to acidic residues (Glu/Asp) also in Pho and Pfu primases (24, 25). Therefore, these two main-chain to main-chain hydrogen bonds could be a signature mark of primase heterodimer formation in a broader range of species.
FIGURE 6.
Close-up stereo view of the hydrophobic interface between p49 and p58. Side chains of the residues involved in hydrophobic interactions between the two subunits are shown as sticks. Each subunit is colored according to Fig. 1B. The secondary structure elements are shown with 50% transparency.
FIGURE 7.
Close-up stereo view of the hydrophilic contacts at p49-p58 interface. Side or main chains making the hydrogen bonds between two subunits are shown as sticks. Water molecules involved in intersubunit interactions are depicted by red spheres. Each subunit is colored according to Fig. 1B. The secondary structure elements are shown with 40% transparency.
FIGURE 8.
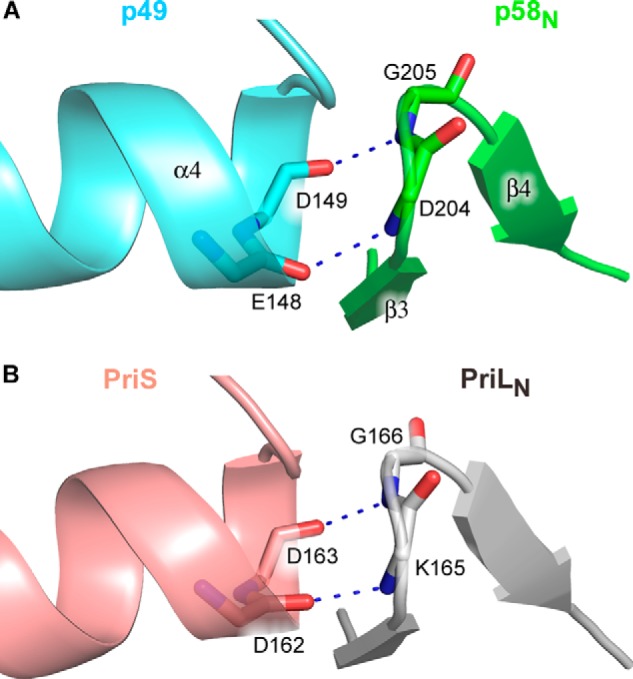
Conserved main-chain to main-chain contacts between catalytic and large subunits of the human (A) and Sso (B) primases. The color scheme is the same as in Fig. 3. Main chains of the residues involved in intersubunit interactions are shown as sticks. The hydrogen bonds are depicted by blue dashed lines.
The Role of the Linker between p58N and p58C in Primase Activity
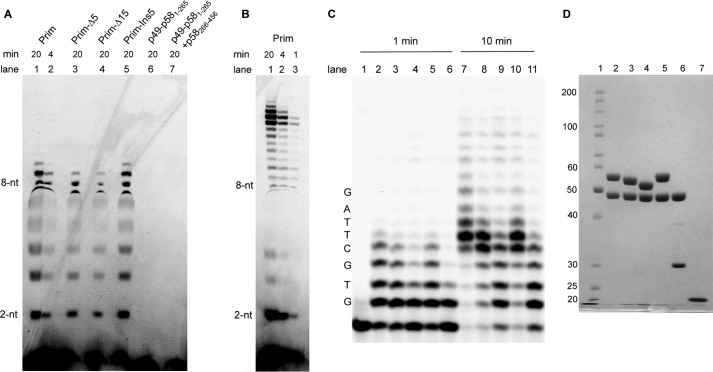
Three primase mutants were generated to investigate the role of the 18-residue-long linker (residues 253–270) between the two p58 domains in primase activity. Two of these mutants (Prim-Δ5 and Prim-Δ15) have a deletion of 5 or 15 amino acids in p58, resulting in excision of the region 256–260 or 256–270, respectively. The third mutant (Prim-Ins5) has an insertion of 5 amino acids (GSASG) after Ser-263. The activity of the full-length human primase and its linker mutants was initially tested on the template-1 (5′-A15TA7). This template was designed to provide a single start position that simplifies the analysis of product distribution and processivity. Eukaryotic primases require template thymine or cytidine in order to start the RNA primer synthesis; thus, the synthesized primers have an adenine or guanine at the 5′-end. We designed the template-1 to possess only one pyrimidine (thymine) to allow for a single unique primer start position in comparison with commonly used poly(dT) templates. It was shown recently that p70-p180C stabilizes activity of the human primase and increases its processivity (5, 16). That is why for the activity assays we used primase constructs in complex with p70-p180C. The catalytic core of Pol α was excluded from the Prim-Pol α complex to prevent substrate translocation from primase to Pol α, which can obscure the “counting” by primase. Additionally, we measured the activity at different time points that allowed us to obtain a rough estimate of to what extent the primase function was affected in the primase variants.
In a de novo RNA synthesis assay, the activity of Prim-Δ15 was decreased about 5-fold (Fig. 9A, compare lane 4 with lanes 1 and 2), whereas the enzyme with a longer linker had the same activity as wild type (lane 5). Comparison of the RNA synthesizing activity for Prim, Prim-Δ5, and Prim-Δ15 (Fig. 9A, lanes 1, 3, and 4) demonstrates its correlation with linker length. Deletion of p58C results in complete loss of the ability of primase to initiate primer synthesis (Fig. 9A, lane 6), which confirms that p58C plays a critical role in dinucleotide formation (14, 20, 22). The addition of purified p58(266–456) at a 2-fold molar excess does not recover primase activity (Fig. 9A, lane 7), indicating that p58C must be connected to the rest of the primase molecule for proper orientation relative to the p49 active site. p58(266–456) was crystallized previously (21) and contains all functional elements of p58C. The extreme C-terminal region of p58 (amino acids 458–509) is not visible in both primase molecules in asymmetric unit and is predicted to be conformationally disordered. The corresponding region is absent or significantly shortened in DNA primases from other species (17). The residual Prim-Δ15 activity means that p58C is still able to reach p49. The adjoining coil region with a small helix α10 that together span residues 241–252 is not well stabilized and can unfold, extending the linker to compensate for its shortening. This may explain the observation of residual Prim-Δ15 activity.
FIGURE 9.
Activity analysis of the human primase and its deletion mutants. A, de novo activity on template-1 (5′-A15TA7). p58(266–456) was added at a 2-fold molar excess over p49-p58(1–265). B, de novo activity of the wild-type primase on template-2 (5′-AAA(GA)6TA7). C, extension assay using fluorophore-labeled RNA primer annealed to template-3. Lane 1, control incubation (no enzyme); lanes 2 and 7, wild-type primase; lanes 3 and 8, Prim-Δ5; lanes 4 and 9, Prim-Δ15; lanes 5 and 10, Prim-Ins5; lanes 6 and 11, p49-p58(1–265). Reactions were incubated for 1 min (lanes 2–6) and 10 min (lanes 1 and 7–11) at 35 °C. The activity of primase samples was analyzed in the presence of p70-p180C. Only the 5′-end of RNA products was labeled because of using [γ-33P]ATP (A and B). D, analysis of the purity of the human primase samples. Lane 1, EZ-Run Rec protein ladder (Fisher); lane 2, Prim; lane 3, Prim-Δ5; lane 4, Prim-Δ15; lane 5, Prim-Ins5; lane 6, p49-p58(1–265); lane 7, p58(266–456). Samples were run on 10% SDS-PAGE, and proteins were detected by Coomassie Blue staining.
Next, we examined the ability of primase variants to extend preexisting RNA primers. The shortening or extension of the linker by 5 residues minimally affects the ability of primase to extend the 16-nucleotide primer (Fig. 9C), whereas the deletion of the p58C domain or 15 linker residues reduced the activity severalfold (comparison of lanes 9, 11, and 2 in Fig. 9C). Our results confirm the previous data demonstrating the important role of p58C in primer extension (14) and indicate that this domain interacts with a substrate during all steps of primer synthesis. The de novo assay allows for analysis of two steps of RNA synthesis: the dinucleotide formation and its following extension. Comparison of lanes 2 and 4 in Fig. 9A indicates that deletion of most of the linker did not affect the ratio between dinucleotide and its extension products. This conclusion is supported by a similar effect of linker deletion on primase activity in de novo and primer extension assays.
Primase Processivity Depends on DNA Template Sequence
The oligo(dT)/oligo(dA) templates have been routinely used for primase and polymerase assays, but they bend and form unusual DNA structures that may affect the interaction with enzyme (42, 43). We have recently shown that such templates result in a premature stop of human Pol α after synthesis of a 10–15-nucleotide primer, which was mistaken for an intrinsic counting ability of Pol α (5, 44). Because oligo(dA) tracks could also affect the primase processivity, we modified the template to make it more heterogeneous by replacing each second adenine with guanine (we did not add extra pyrimidines to the template to avoid initiation of synthesis from multiple sites). Primase had much higher processivity on template-2 (Fig. 9B) compared with template-1 (Fig. 9A), and the counting effect almost completely disappeared. The presence of relatively intense bands represented by 2–4-nucleotide primers indicates the high probability of primase dissociation from short primers, which is probably due to the abortive synthesis characteristic of RNA polymerases (45). We conclude that the ability of primase to count the oligo(dT) or oligo(dA) primer length depends primarily on template sequence and to a lesser extent on other factors, including structural features of the primase molecule.
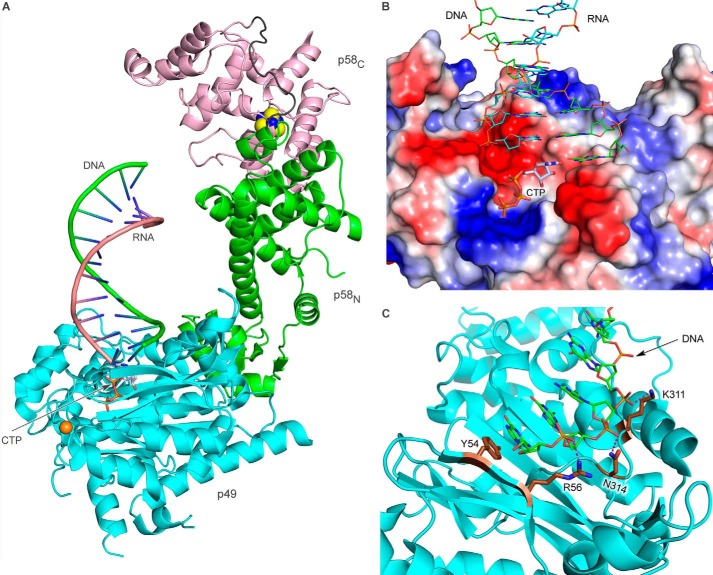
Modeling of Human Primase in Complex with NTP and DNA/RNA Substrates
The DNA-RNA duplex fits well into the active site of p49 (Fig. 10B; the modeling details are provided under “Experimental Procedures”). In the model structure, the 5′-end of the RNA primer is pointed toward p58C (Fig. 10A). The phosphate backbone of the DNA template follows the path on a positively charged surface near the active site (Fig. 10B) and could make only a few contacts with the catalytic subunit, including Arg-56, Lys-311, and Asn-314 (Fig. 10C). The 5′-overhang of DNA template whose position is not included in the model could potentially make additional contacts with p49, but this is not critical for the activity because primase reaches the 5′-end of the template during synthesis (Fig. 9B). The interaction between p49 and the DNA template features the potential stacking between Tyr-54 and the template guanine, which is complementary to the incoming nucleotide-triphosphate (Fig. 10C). This interaction could be important for catalytically productive positioning of the DNA template on p49. Consistent with our model, it has been shown recently that Arg-56 and Tyr-54 are necessary for primase activity (16).
FIGURE 10.
Modeling of Prim-DNA/RNA-CTP ternary complex. A, overall view of the ternary complex. The color scheme is the same as in Fig. 1. B, close-up view of the p49-DNA/RNA-CTP complex. The p49 surface is represented by the vacuum electrostatic potential. The carbons of CTP, DNA, and RNA are colored gray, green, and cyan, respectively. C, close-up view of the p49-DNA interaction interface. Carbons of Tyr-54, Arg-56, Lys-311, and Asn-314 are colored brown. The potential hydrogen bonds between p49 and DNA are shown by pink dashed lines.
DISCUSSION
The primase p58C domain plays a critical role in dinucleotide synthesis that requires binding of three substrates in the active site: the DNA template and two NTP molecules complementary to the two sequential nucleotides in the template. In the crystal structure of the apo-form of human primase reported here (without nucleic acid substrate), the DNA/NTP binding interface of p58C is located far away (∼60 Å, or about 20 base pairs of the DNA-RNA duplex) from the active site of p49 (Fig. 1B). How can p58C assist p49 during initiation despite the significant distance between them? If p58C is working in a cis-position, there should be dramatic conformational change in the primase molecule during the priming event. The long linker between p58N and p58C provides an opportunity for such movement. Another possibility is that p58C can work in trans (i.e. assists p49 from another molecule) and that two primase molecules are associated at least during dinucleotide synthesis. There is a precedent of that mechanism; the helical domain of prokaryotic primase Repβ′ is flexibly tethered to the catalytic domain of the same polypeptide and plays the critical role in synthesis initiation, like p58C (46). Consistent with this model, it was recently shown that the Ctf4 trimer that couples Prim-Pol α to the eukaryotic replisome can bind two Pol α molecules (47).
Several facts indicate that p58C is working in a cis-position. First, in contrast to Repβ′, the activity of human primase without the p58 C-terminal domain was not restored to any extent by the addition of that domain as a separate protein to the reaction. (Fig. 9A). Second, the relative orientation of the catalytic center on p49 and the NTP/DNA binding site on p58C as well as the position of the modeled primer-template (Fig. 10A) are favorable for intramolecular transactions (Fig. 1B). Third, the deletion of 15 amino acids in the linker connecting p58N and p58C severely affected primase activity, and we see its correlation with the linker length in the range of 3–18 residues (Fig. 9, A and C). The extension of the 18-mer linker by 5 residues had no effect on either step of RNA primer synthesis, and this result better fits the cis-model. It is most likely that primase interaction with template DNA and the first two NTPs triggers p58C relocation closer to the p49 active site. Weak contacts between p58N and p58C (Fig. 5) should not impede p58C mobility.
The length of the RNA patch in Okazaki fragments synthesized in vivo usually does not exceed 10 nucleotides (7, 48). This led to the idea of intrinsic counting by primase, which was supported by experiments on homopolymeric oligo(dT)/oligo(dA) substrates (9, 49, 50). However, such templates possess unique properties and are not relevant to most DNA templates (5). The analysis of primase activity on heterogeneous template (Fig. 9B) revealed that primase “counting” cannot explain a strict determination of RNA primer length seen in Okazaki fragments. We propose that only the whole Prim-Pol α complex is responsible for “counting,” possessing an intrinsic property to transfer the substrate from primase to the polymerase active site when RNA primer length reaches the size of 8–10 base pairs. Intriguingly, the unit length of the RNA primer corresponds to the size of the DNA-RNA duplex covered by the Pol α active site (44, 51). This may indicate that Pol α is able to capture the DNA-RNA duplex from the highly exposed primase active site. As was shown for bovine and human Prim-Pol α complexes, even in the absence of dNTPs, Pol α strongly inhibits the synthesis of RNA primers longer than 10 nucleotides (5, 6). Contrary to this, Pol α without a catalytic domain stimulated synthesis of the long RNA tracks (5).
As was suggested earlier, p58C may be involved in translocation of the RNA-primed template to the Pol α active site (12). It is possible that primer-template often dissociates from p49 during synthesis while keeping a contact with p58C. This idea seems reasonable in light of current structural data indicating weak interaction between the primer-template and p49. The C-terminal domain of p58 binds single- and double-stranded DNA with micromolar affinity (18). Photocross-linking of single-stranded DNA with human primase revealed that only p58 reacted with DNA upon photolysis (12). The structural data for primase (or Prim-Pol α) in initiation and elongation complexes with substrates are required to obtain further information about the mechanism of primase function.
This work was supported, in whole or in part, by National Institutes of Health (NIH), NIGMS, Grant GM101167 (to T. H. T.) and NIH, NCI, Grant CA129925 (to Y. I. P.). The Eppley Institute X-ray Crystallography Core Facility is supported by NIH Cancer Center Support Grant P30CA036727. This work is also based upon research conducted at the Advanced Photon Source on the NECAT beamlines, which are supported by NIH, NIGMS, Grant P41 GM103403. Use of the Advanced Photon Source, an Office of Science User Facility operated for the United States Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the United States DOE under Contract DE-AC02-06CH11357.
The atomic coordinates and structure factors (code 4RR2) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Prim
- DNA primase
- Pol α
- DNA polymerase α
- p58N and p58C
- N- and C-terminal domains of p58 subunit
- p180C
- C-terminal domain of Pol α catalytic subunit
- SAD
- single-wavelength anomalous diffraction
- r.m.s.
- root mean square.
REFERENCES
- 1. Pellegrini L. (2012) The Pol α-primase complex. Subcell. Biochem. 62, 157–169 [DOI] [PubMed] [Google Scholar]
- 2. Klinge S., Núñez-Ramirez R., Llorca O., Pellegrini L. (2009) 3D architecture of DNA Pol α reveals the functional core of multi-subunit replicative polymerases. EMBO J. 28, 1978–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kilkenny M. L., De Piccoli G., Perera R. L., Labib K., Pellegrini L. (2012) A conserved motif in the C-terminal tail of DNA polymerase α tethers primase to the eukaryotic replisome. J. Biol. Chem. 287, 23740–23747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mizuno T., Yamagishi K., Miyazawa H., Hanaoka F. (1999) Molecular architecture of the mouse DNA polymerase α-primase complex. Mol. Cell. Biol. 19, 7886–7896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang Y., Baranovskiy A. G., Tahirov T. H., Pavlov Y. I. (2014) The C-terminal domain of the DNA polymerase catalytic subunit regulates the primase and polymerase activities of the human DNA polymerase α-primase complex. J. Biol. Chem. 289, 22021–22034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sheaff R. J., Kuchta R. D., Ilsley D. (1994) Calf thymus DNA polymerase α-primase: “communication” and primer-template movement between the two active sites. Biochemistry 33, 2247–2254 [DOI] [PubMed] [Google Scholar]
- 7. Frick D. N., Richardson C. C. (2001) DNA primases. Annu. Rev. Biochem. 70, 39–80 [DOI] [PubMed] [Google Scholar]
- 8. Copeland W. C., Wang T. S. (1993) Enzymatic characterization of the individual mammalian primase subunits reveals a biphasic mechanism for initiation of DNA replication. J. Biol. Chem. 268, 26179–26189 [PubMed] [Google Scholar]
- 9. Sheaff R. J., Kuchta R. D. (1993) Mechanism of calf thymus DNA primase: slow initiation, rapid polymerization, and intelligent termination. Biochemistry 32, 3027–3037 [DOI] [PubMed] [Google Scholar]
- 10. Kuchta R. D., Stengel G. (2010) Mechanism and evolution of DNA primases. Biochim. Biophys. Acta 1804, 1180–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zerbe L. K., Kuchta R. D. (2002) The p58 subunit of human DNA primase is important for primer initiation, elongation, and counting. Biochemistry 41, 4891–4900 [DOI] [PubMed] [Google Scholar]
- 12. Arezi B., Kirk B. W., Copeland W. C., Kuchta R. D. (1999) Interactions of DNA with human DNA primase monitored with photoactivatable cross-linking agents: implications for the role of the p58 subunit. Biochemistry 38, 12899–12907 [DOI] [PubMed] [Google Scholar]
- 13. Zakharova O. D., Podust V. N., Mustaev A. A., Anarbaev R. O., Lavrik O. I. (1995) Highly selective affinity labeling of DNA polymerase α-primase from human placenta by reactive analogs of ATP. Biochimie 77, 699–702 [DOI] [PubMed] [Google Scholar]
- 14. Copeland W. C. (1997) Expression, purification, and characterization of the two human primase subunits and truncated complexes from Escherichia coli. Protein Expr. Purif. 9, 1–9 [DOI] [PubMed] [Google Scholar]
- 15. Foiani M., Lindner A. J., Hartmann G. R., Lucchini G., Plevani P. (1989) Affinity labeling of the active center and ribonucleoside triphosphate binding site of yeast DNA primase. J. Biol. Chem. 264, 2189–2194 [PubMed] [Google Scholar]
- 16. Kilkenny M. L., Longo M. A., Perera R. L., Pellegrini L. (2013) Structures of human primase reveal design of nucleotide elongation site and mode of Pol α tethering. Proc. Natl. Acad. Sci. U.S.A. 110, 15961–15966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sauguet L., Klinge S., Perera R. L., Maman J. D., Pellegrini L. (2010) Shared active site architecture between the large subunit of eukaryotic primase and DNA photolyase. PLoS One 5, e10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vaithiyalingam S., Warren E. M., Eichman B. F., Chazin W. J. (2010) Insights into eukaryotic DNA priming from the structure and functional interactions of the 4Fe-4S cluster domain of human DNA primase. Proc. Natl. Acad. Sci. U.S.A. 107, 13684–13689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Francesconi S., Longhese M. P., Piseri A., Santocanale C., Lucchini G., Plevani P. (1991) Mutations in conserved yeast DNA primase domains impair DNA replication in vivo. Proc. Natl. Acad. Sci. U.S.A. 88, 3877–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Klinge S., Hirst J., Maman J. D., Krude T., Pellegrini L. (2007) An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat. Struct. Mol. Biol. 14, 875–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Agarkar V. B., Babayeva N. D., Pavlov Y. I., Tahirov T. H. (2011) Crystal structure of the C-terminal domain of human DNA primase large subunit: implications for the mechanism of the primase-polymerase α switch. Cell Cycle 10, 926–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weiner B. E., Huang H., Dattilo B. M., Nilges M. J., Fanning E., Chazin W. J. (2007) An iron-sulfur cluster in the C-terminal domain of the p58 subunit of human DNA primase. J. Biol. Chem. 282, 33444–33451 [DOI] [PubMed] [Google Scholar]
- 23. Leipe D. D., Aravind L., Koonin E. V. (1999) Did DNA replication evolve twice independently? Nucleic Acids Res. 27, 3389–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Augustin M. A., Huber R., Kaiser J. T. (2001) Crystal structure of a DNA-dependent RNA polymerase (DNA primase). Nat. Struct. Biol. 8, 57–61 [DOI] [PubMed] [Google Scholar]
- 25. Ito N., Nureki O., Shirouzu M., Yokoyama S., Hanaoka F. (2003) Crystal structure of the Pyrococcus horikoshii DNA primase-UTP complex: implications for the mechanism of primer synthesis. Genes Cells 8, 913–923 [DOI] [PubMed] [Google Scholar]
- 26. Lao-Sirieix S. H., Nookala R. K., Roversi P., Bell S. D., Pellegrini L. (2005) Structure of the heterodimeric core primase. Nat. Struct. Mol. Biol. 12, 1137–1144 [DOI] [PubMed] [Google Scholar]
- 27. Copeland W. C., Tan X. (1995) Active site mapping of the catalytic mouse primase subunit by alanine scanning mutagenesis. J. Biol. Chem. 270, 3905–3913 [DOI] [PubMed] [Google Scholar]
- 28. Vaithiyalingam S., Arnett D. R., Aggarwal A., Eichman B. F., Fanning E., Chazin W. J. (2014) Insights into eukaryotic primer synthesis from structures of the p48 subunit of human DNA primase. J. Mol. Biol. 426, 558–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang W., Lee J. Y., Nowotny M. (2006) Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22, 5–13 [DOI] [PubMed] [Google Scholar]
- 30. Baranovskiy A. G., Gu J., Babayeva N. D., Agarkar V. B., Suwa Y., Tahirov T. H. (2014) Crystallization and preliminary x-ray diffraction analysis of human DNA primase. Acta Crystallogr. F Struct. Biol. Commun. 70, 206–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baranovskiy A. G., Lada A. G., Siebler H. M., Zhang Y., Pavlov Y. I., Tahirov T. H. (2012) DNA polymerase δ and ζ switch by sharing accessory subunits of DNA polymerase δ. J. Biol. Chem. 287, 17281–17287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu H., Naismith J. H. (2008) An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 34. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 35. Brunger A. T. (2007) Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 36. Lipps G., Weinzierl A. O., von Scheven G., Buchen C., Cramer P. (2004) Structure of a bifunctional DNA primase-polymerase. Nat. Struct. Mol. Biol. 11, 157–162 [DOI] [PubMed] [Google Scholar]
- 37. Holm L., Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kato M., Ito T., Wagner G., Richardson C. C., Ellenberger T. (2003) Modular architecture of the bacteriophage T7 primase couples RNA primer synthesis to DNA synthesis. Mol. Cell 11, 1349–1360 [DOI] [PubMed] [Google Scholar]
- 39. Corn J. E., Pease P. J., Hura G. L., Berger J. M. (2005) Crosstalk between primase subunits can act to regulate primer synthesis in trans. Mol. Cell 20, 391–401 [DOI] [PubMed] [Google Scholar]
- 40. Pan H., Wigley D. B. (2000) Structure of the zinc-binding domain of Bacillus stearothermophilus DNA primase. Structure 8, 231–239 [DOI] [PubMed] [Google Scholar]
- 41. Kato M., Ito T., Wagner G., Ellenberger T. (2004) A molecular handoff between bacteriophage T7 DNA primase and T7 DNA polymerase initiates DNA synthesis. J. Biol. Chem. 279, 30554–30562 [DOI] [PubMed] [Google Scholar]
- 42. Koo H. S., Wu H. M., Crothers D. M. (1986) DNA bending at adenine-thymine tracts. Nature 320, 501–506 [DOI] [PubMed] [Google Scholar]
- 43. Mirkin S. M., Frank-Kamenetskii M. D. (1994) H-DNA and related structures. Annu. Rev. Biophys. Biomol. Struct. 23, 541–576 [DOI] [PubMed] [Google Scholar]
- 44. Perera R. L., Torella R., Klinge S., Kilkenny M. L., Maman J. D., Pellegrini L. (2013) Mechanism for priming DNA synthesis by yeast DNA polymerase α. Elife 2, e00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Goldman S. R., Ebright R. H., Nickels B. E. (2009) Direct detection of abortive RNA transcripts in vivo. Science 324, 927–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Geibel S., Banchenko S., Engel M., Lanka E., Saenger W. (2009) Structure and function of primase RepB′ encoded by broad-host-range plasmid RSF1010 that replicates exclusively in leading-strand mode. Proc. Natl. Acad. Sci. U.S.A. 106, 7810–7815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Simon A. C., Zhou J. C., Perera R. L., van Deursen F., Evrin C., Ivanova M. E., Kilkenny M. L., Renault L., Kjaer S., Matak-Vinković D., Labib K., Costa A., Pellegrini L. (2014) A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome. Nature 510, 293–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ogawa T., Okazaki T. (1980) Discontinuous DNA replication. Annu. Rev. Biochem. 49, 421–457 [DOI] [PubMed] [Google Scholar]
- 49. Podust V. N., Vladimirova O. V., Manakova E. N., Lavrik O. I. (1992) Eukaryotic DNA primase appears to act as oligomer in DNA-polymerase-α-primase complex. Eur. J. Biochem. 206, 7–13 [DOI] [PubMed] [Google Scholar]
- 50. Singh H., Brooke R. G., Pausch M. H., Williams G. T., Trainor C., Dumas L. B. (1986) Yeast DNA primase and DNA polymerase activities. An analysis of RNA priming and its coupling to DNA synthesis. J. Biol. Chem. 261, 8564–8569 [PubMed] [Google Scholar]
- 51. Thompson H. C., Sheaff R. J., Kuchta R. D. (1995) Interactions of calf thymus DNA polymerase α with primer/templates. Nucleic Acids Res. 23, 4109–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]