Background: In vitro-produced retinal pigmented epithelial (RPE) cells represent a novel source for retinal degenerative disease healing.
Results: mRNA transfection outperformed plasmid transfection in cellular uptake. Modified-mRNA displayed negligible immune activation and functional protein expression.
Conclusion: Modified mRNA transfection can be used efficiently for the engineering of RPE cells.
Significance: The modified mRNA transfection technique offers new venues for the treatment of RPE-related diseases.
Keywords: Differentiation; Embryonic Stem Cell; Induced Pluripotent Stem Cell (iPS Cell, iPSC); mRNA; Transfection; Modified mRNA; Retinal Pigmented Epithelial Cell
Abstract
Gene- and cell-based therapies are promising strategies for the treatment of degenerative retinal diseases such as age-related macular degeneration, Stargardt disease, and retinitis pigmentosa. Cellular engineering before transplantation may allow the delivery of cellular factors that can promote functional improvements, such as increased engraftment or survival of transplanted cells. A current challenge in traditional DNA-based vector transfection is to find a delivery system that is both safe and efficient, but using mRNA as an alternative to DNA can circumvent these major roadblocks. In this study, we show that both unmodified and modified mRNA can be delivered to retinal pigmented epithelial (RPE) cells with a high efficiency compared with conventional plasmid delivery systems. On the other hand, administration of unmodified mRNA induced a strong innate immune response that was almost absent when using modified mRNA. Importantly, transfection of mRNA encoding a key regulator of RPE gene expression, microphthalmia-associated transcription factor (MITF), confirmed the functionality of the delivered mRNA. Immunostaining showed that transfection with either type of mRNA led to the expression of roughly equal levels of MITF, primarily localized in the nucleus. Despite these findings, quantitative RT-PCR analyses showed that the activation of the expression of MITF target genes was higher following transfection with modified mRNA compared with unmodified mRNA. Our findings, therefore, show that modified mRNA transfection can be applied to human embryonic stem cell-derived RPE cells and that the method is safe, efficient, and functional.
Introduction
The retinal pigmented epithelium (RPE)4 is a layer of pigmented cells located between the choroid and the photoreceptors, where it serves as a part of the barrier between the bloodstream and the retina. This monolayer of cells operates in multiple ways, all of which are crucial for visual function, including light absorption, recycling of retinoids, epithelial transport, secretion of proteins, spatial ion buffering, phagocytosis of the outer segments of the photoreceptors, and immune regulation (1).
Dysfunction, degeneration, and loss of RPE cells are major characteristics of many retinal diseases, such as Stargardt disease, Best disease, subtypes of retinitis pigmentosa, proliferative vitreoretinopathy, and age-related macular degeneration, which all lead to gradual loss of visual acuity and, eventually, in many cases, to blindness (2). A variety of therapeutic approaches to delay or repair retinal degeneration is under development, including cell-based and gene replacement therapies (2, 3). Clinical trials involving the transplantation of intact sheets or single-cell suspensions of primary RPE cells have been carried out with mixed results (4).
Human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs), as well as some somatic cell types such as mesenchymal stem cells, are all attractive cell sources for transplantation. Several reports have demonstrated that both hESCs and hiPSCs can differentiate in vitro into a functional monolayer of pigmented RPE-like cells (5–8) and that human embryonic stem cell-derived RPE can restore vision in the retinal dystrophy rat model (9). In addition, by using a mixture of transcription factors, fibroblasts can be directed to trans-differentiate toward RPE-like cells (10). Recently, the first description of transplanted human ES cell-derived RPE cells into human patients was reported (11), and, in Japan, a pilot clinical study on transplantation of autologous hiPSC-RPE cells has been initiated. Despite the great potential of these cells for future treatment of retinal degeneration, there are still some challenges regarding the degree of cell survival, immune rejection, and efficiency of engraftment. In addition, functional and molecular studies have shown that human ES cell- and hiPSC-derived RPE cells possess specific properties that are absent from currently available cell lines, such as ARPE-19, which make them useful for in vitro disease modeling or drug screening (6, 12, 13). Regardless of the application of hESC RPE or hiPSC RPE, a safe, flexible, and efficient gene delivery system is still needed. However, optimal gene delivery systems for RPE cells are limited.
The use of synthetic mRNA as a gene delivery technique holds several benefits over classical DNA-based methods. Nevertheless, because of the relatively low half-life and the strong immunogenicity of conventional mRNA, the clinical application of this technique has been delayed. However, recent groundbreaking advances have established that replacing uridine and cytidine with pseudouridine and 5-methylcytidine, respectively, allows synthetic mRNA to bypass the cellular innate immune response (14), which, in turn, opens the door to DNA-free cellular engineering strategies that would avoid any risks of genomic recombination or insertional mutagenesis. Because the transfected mRNA only has to reach the cytoplasm to achieve protein expression, the efficiency of transfection is also relatively high for cells that are considered to be difficult to transfect, such as postmitotic cells, by classical DNA-based delivery methods (because DNA must cross the nuclear envelope in addition to the plasma membrane). Modified mRNA has also been reported to have a higher translational capacity and stability than unmodified mRNA (15, 16). Since its discovery, transfection of modified mRNA has been applied successfully in different research areas, including disease treatment (17–19), vaccination (20), and regenerative medicine (21–23).
Here we demonstrate that synthetic unmodified mRNA, as well as modified mRNA, can be delivered efficiently into RPE cells independently of differentiation stage or confluence. However, administration of unmodified mRNA induces nuclear translocation of the immunogenic transcription factors IRF3 and p65/RelA and, consequently, a strong activation of their target genes, IFNβ and TNFα. In contrast, in modified mRNA-transfected cells, nuclear localization of IRF3 or p65/RelA is absent, showing minimal activation of TNFα and IFNβ. Similarly, the transcriptional activator MITF is more biologically active when expressed from modified mRNA than unmodified mRNA, as evidenced by the higher activation of its target genes. Therefore, synthetic modified mRNA offers an unprecedented opportunity for the study of RPE-related diseases and, potentially, to improve the therapeutic outcomes of degenerated RPE, including cell-based and gene-based therapies.
EXPERIMENTAL PROCEDURES
Cell Culture and Differentiation
HEK293T cells were maintained in DMEM supplemented with 10% (v/v) FBS (Invitrogen) and penicillin/streptomycin at 37 °C with 5% (v/v) CO2. The human embryonic stem cell line ES[4] (24) was cultured on Matrigel-coated (BD Biosciences) 6-well plates and maintained in mTeSR1 (Stem Cell Technologies, Grenoble, France) or in irradiated mouse embryonic fibroblast-conditioned hESC medium containing knockout DMEM (Invitrogen), 20% (v/v) knockout serum replacement (Invitrogen), non-essential amino acids (Invitrogen), 2 mm l-glutamine (Invitrogen), and 50 mm β-mercaptoethanol (Invitrogen) and supplemented with basic fibroblast growth factor (10 ng/ml). The medium was changed daily until cells were ready for passage. To initiate differentiation, after the human ES cells had grown to confluence, the human ES cell maintenance medium was replaced with human ES cell medium without basic FGF and supplemented with 10 mm nicotinamide (Sigma-Aldrich, Madrid, Spain) (herein referred to as HES-NIC medium). The medium was changed every third day. Pigmented clusters comprising RPE emerged, and these colonies were allowed to grow large enough to manually separate them from the culture. The dissected colonies were pooled and trypsinized (with 0.25% (w/v) trypsin) to obtain a single-cell suspension. Enriched putative hESC-RPE cells were then seeded on Matrigel-coated wells and cultured in RPE differentiation medium for a further 6–8 weeks. The cells could be maintained in the HES-NIC differentiation medium for up to 6 months without losing their morphology. For mRNA transfection and subculture experiments, we used EGM2 medium (Lonza, Barcelona, Spain).
mRNA Synthesis
cDNA encoding enhanced green fluorescent protein (herein referred to as GFP) was amplified by PCR and subcloned into pT7TS (Addgene, Cambridge, MA; from Paul Krieg, University of Texas at Austin, TX) containing the 5′- and 3′-untranslated region sequences of Xenopus β-globin and a dA30dC30 sequence. FLAG-MITF-M was generated by PCR and subcloned into pT7TS. Linearized GFP-pT7TS and FLAG-MITF-M-pT7TS plasmids were used as templates for the in vitro transcription reaction with the MEGAScript kit (Ambion, by Invitrogen) with T7 RNA polymerase, with a 4:1 anti-reverse cap analog:GTP ratio to give an optimal percentage of capped transcripts. For synthesis of modified mRNA, the in vitro transcription reaction substituted UTP and CTP for pseudoUTP (ψUTP) and 5-methyl-CTP. The anti-reverse cap analog) and modified NTPs were ordered from Trilink Biotechnologies. The unmodified and modified mRNAs were treated with 1 μl of DNase I (Ambion), heat-inactivated, and purified by MegaClear according to the instructions of the supplier (Ambion). Polyadenylation of the purified transcripts was performed by using recombinant yeast poly(A) polymerase (USB, Affymetrix) repurified by the MegaScript protocol. The quality and quantity of the poly(A) tailed mRNAs was subsequently analyzed by NanoDrop spectrophotometry and agarose gel electrophoresis.
mRNA and DNA Plasmid Transfection
All mRNA transfections were carried out using the Stemfect transfection reagent in accordance with the instructions of the company (Stemgent, Cambridge, MA). In summary, 4 μl of Stemfect reagent and 120 μl of Stemfect buffer were used per well of a 6-well plate. For HEK293T cells and subconfluent hESC-RPE cells, 1 μg of mRNA/well of a 6-well plate was added. For confluent and densely packed polygonal hESC-RPE cells, 2 μg of mRNA (and 8 μl of Stemfect) were used. For transfection of the pmaxGFP plasmid in HEK293 and hESC-RPE cells, FuGENE6 transfection reagent (Promega, Madrid, Spain) was used according to the instructions of the manufacturer.
Flow Cytometry Analysis
The transfection efficiencies of the pmaxGFP plasmid, unmodified and modified mRNAs into hESC-RPE and HEK293T cells were determined by means of GFP-expressing cells 24 h after transfection. Green fluorescent protein expression was measured in a fluorescence-activated cell sorter on a Beckman Coulter Gallios flow cytometer using the Kaluza flow cytometry analysis software. Propidium iodide staining was used to detect the number of dead cells.
Immunofluorescence Analysis
hESC-RPE cells were grown on plastic coverslide chambers and fixed with 4% (w/v) paraformaldehyde at room temperature for 20 min. The immunodetection was performed with TBS/0.2% (v/v) Triton X-100 for permeabilization. Primary antibodies were incubated at 4 °C overnight, and secondary antibodies were incubated at 37 °C for 2 h. The following antibodies were used: RPE65 (Novus Biologicals, Cambridge, UK, 1:200), OTX2 (catalog no. P15, Santa Cruz Biotechnology, Heidelberg, Germany, 1:50), ZO-1 (catalog no. ab2272, Millipore, Darmstadt, Germany, 1:100), BEST1 (catalog no. E6-6, Santa Cruz Biotechnology, 1:50), MITF (catalog no. C5, Santa Cruz Biotechnology, 1:50), M2-FLAG (Sigma-Aldrich, 1:200), NF-κB/P65 (Santa Cruz Biotechnology, 1:50), IRF3 (catalog no. FL-425, Santa Cruz Biotechnology, 1:50), and enhanced GFP (Invitrogen, 1:200). Images were taken on a Leica SP5 AOBS (acousto-optical beam splitter) confocal microscope.
Transmission Electron Microscopy
Samples were fixed with 2.5% (w/v) glutaraldehyde for 2 h at 4 °C, post-fixed in 1% osmium tetroxide (2 h at 4 °C), dehydrated with ethanol, and embedded in epoxy resin. Samples were examined with a JEOL 1011 transmission electron microscope (Tokyo, Japan).
Western Blot Analysis
Whole-cell extracts were prepared by using lysis buffer comprising 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1% (v/v) Triton X-100, and complete protease inhibitor mixture (Roche). Laemmli sample loading buffer was added to the lysed samples, which were incubated at 95 °C for 5–10 min and centrifuged for 10 min at 18, 000 × g. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto Immobilon polyvinilidene difluoride (Millipore). For immunochemical detection, membrane blots were incubated overnight at 4 °C with primary antibodies against OCT4 (catalog no. SC-8628, Santa Cruz Biotechnology), TTR (catalog no. A0002, Dako, Barcelona, Spain), RPE65 (Novus Biologicals), β-Actin (catalog no. A2066, Sigma-Aldrich), OTX2 (catalog no. P15, Santa Cruz Biotechnology), Bestrophin (catalog no. E6-6, Santa Cruz Biotechnology), MITF (catalog no. C5, Santa Cruz Biotechnology), and M2-FLAG (Sigma-Aldrich).
Quantitative Real-time PCR (qRT-PCR)
Cellular RNAs were isolated by using the RNeasy mini kit according to the instructions of the manufacturer (Qiagen, Hilden, Germany). The concentration of total isolated RNA was measured by NanoDrop and treated with TURBO DNase (Ambion) to remove any residual genomic DNA. cDNA synthesis from 1 μg of total RNA was carried out using the cloned AMV (avian myeloblastosis virus) first-strand cDNA synthesis kit (Invitrogen). To quantify the relative mRNA levels of typical RPE gene markers, quantitative PCR was done with Platinum SYBR Green quantitative PCR super mix (Invitrogen) and 25 ng of cDNA/well. Gene expression was normalized to that of Gapdh. Sequences of the quantitative PCR primers were as follows: Gapdh, gtcagtggtggacctgacct (forward) and aggggagattcagtgtggtg (reverse); Oct4, gttcttcattcactaaggaagg (forward) and caagagcatcattgaacttcac (reverse); Best1, tgagaccaactggattgtcg (forward) and tgaaggtggagcccataaag (reverse); Mct3, gcctgcgttgtgctaaag (forward) and cctcgcctctatttctgg (reverse); Trpm1, ccctgctgaaaggaacaaac (forward) and tctggcttcgtgctatgtc (reverse); Trpm3, atacccagcaccaaagacc (forward) and tctgaagcacggagatactg (reverse); Tyr, acttactcagcccagcatc (forward) and tggtttccaggattacgcc (reverse); Rpe65, acttctcctttcaacctcttc (forward) and ctcaaatcctttccagcagc (reverse); Cralbp, gtggaagagaagaacttgaacc (forward) and atggaagacacagagtccttg (reverse); Pedf, agatctcagctgcaagattgccca (forward) and atgaatgaactcggaggtgaggct (reverse); Ttr, gatgggatttcatgtaaccaagag (forward) and ctgcctggacttctaacatagc (reverse); Silv, gttgatggctgtggtccttg (forward) and cagtgactgctgctatgtgg (reverse); TNFα, cccaggcagtcagatcatctt (forward) and tctcagctccacgccatt (reverse); IFNβ, cattacctgaaggccaagga (forward) and cagcatctgctggttgaaga (reverse); TLR3, tcccaagccttcaacgactg (forward) and tggtgaaggagagctatccaca (reverse); Mertk, ttgcagcattcaggtcaaggaagc (forward) and ggcttgcagctgcttgatttggta (reverse); Otx2, gaccacttcgggtatggact (forward) and tggacaaggggatctgacagt (reverse); Mitf, gtgccaacttctttcatca (forward) and acctaaaccgtccattca (reverse); Pax6, tctaatcgaagggccaaatg (forward) and tgtgagggctgtgtctgttc (reverse); and Lrat, gaatgaggcatggacctgtt (forward) and tcctgcccaaaatctgtttc (reverse).
RESULTS
Generation and Characterization of hESC-RPE Cells
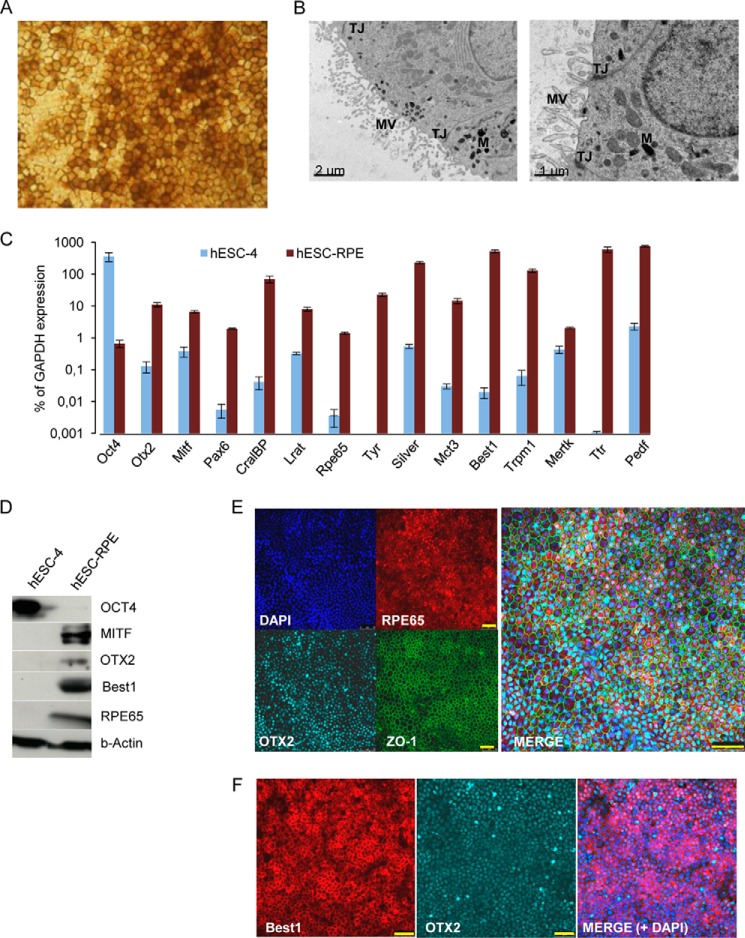
To obtain RPE cells from the human embryonic stem cell line ES4 (hESC[4]), we used a directed differentiation protocol, described by Idelson et al. (5), with minor modifications. Briefly, human ES cells were grown on Matrigel-coated 6-well plates in mTeSRTM1- or conditioned human ES cell medium (we defined conditioned medium under “Experimental Procedures”) supplemented with 10 ng/ml basic FGF. Upon reaching confluence, the human ES cell medium was replaced with differentiation medium. Four weeks later, pigmented colonies with typical RPE polygonal cell morphology were formed. After an additional 4 weeks (approximately), we manually removed the pigmented colonies from the cell cultures. The excised colonies were trypsinized, and single-cell suspensions were reseeded onto Matrigel-coated plates. Passaged hESC-RPE cells lost their epithelial morphology and pigmentation during the proliferation phase until they reached confluence, at which time the typical RPE morphology was restored, consistent with prior observations of primary RPE, iPSC-, and ESC-RPE cells (6, 7) (Fig. 1A). Transmission electron microscopy revealed the typical structural features of RPE cells, including apical microvilli, tight junctions, and melanosome granules (Fig. 1B). Moreover, as shown in Fig. 1C, the pigmented cells expressed RPE-related transcripts of genes associated with the retinal cycle (RPE-specific protein 65 (Rpe65), cellular retinaldehyde-binding protein (CralBP), and lecithin-retinol transferase (Lrat)), genes involved in melanogenesis (Silver (Silv) and Tyrosinase (Tyr)), transcripts of RPE-related transcription factors (microphthalmia-associated transcription factor (Mitf), orthodenticle homeobox 2 (Otx2), and paired box 6 (Pax6)), genes involved in phagocytosis (c-mer proto-oncogene tyrosine kinase (Mertk)) and membrane transport (Trpm1, Bestrophin 1 (Best1), and monocarboxylate transporter 3 (MCT3)), and secreted proteins (Transthyretin (TTR) and pigmented epithelium-derived factor (Pedf)). In addition, protein expression of MITF, OTX2, Best1, and RPE65 in hESC-RPE cells and OCT4 in human ES cells were confirmed by Western blot analysis (Fig. 1D), and immunofluorescence analysis of hESC-RPE cells revealed coexpression of RPE65, ZO-1, and OTX2. In addition, Best1 and OTX2 were coexpressed (Fig. 1, E and F). Down-regulation of the human ES cell marker Oct4 was confirmed by qRT-PCR (Fig. 1C), Western blotting (Fig. 1D), and immunofluorescence (data not shown).
FIGURE 1.
Characterization of hESC-derived RPE cells. Pigmented clusters of polygonal cells were selected and reseeded onto Matrigel-coated plates. After 6 weeks of proliferation into high-density cultures, the cells reacquired the morphology and pigmentation of typical RPE cells (A). B, transmission electron microscopy of hESC-derived RPE cells showing typical RPE characteristics: apical microvilli (MV), melanin granules (M), and tight junctions (TJ). Scale bar = 2 μm (left panel) and 1 μm (right panel). C, qRT-PCR analysis of human ES cells and RPE-specific gene expression. Relative expression is shown as percentage of GAPDH expression. Data are presented as mean ± S.E. of three independent experiments performed in triplicate. D, Western blot analysis on cell lysates from human ES cells and hESC-RPE cells with antibodies against OCT4, MITF, OTX2, BEST1, RPE65, and β-Actin. Immunostaining shows coexpression of RPE65, OTX2, and ZO-1 (E) and BEST1 and OTX2 (F). DAPI was used to counterstain the nuclei (blue). Scale bars = 50 μm (E and F).
Characterization of Sub- and Postconfluent hESC-RPE Cell Cultures
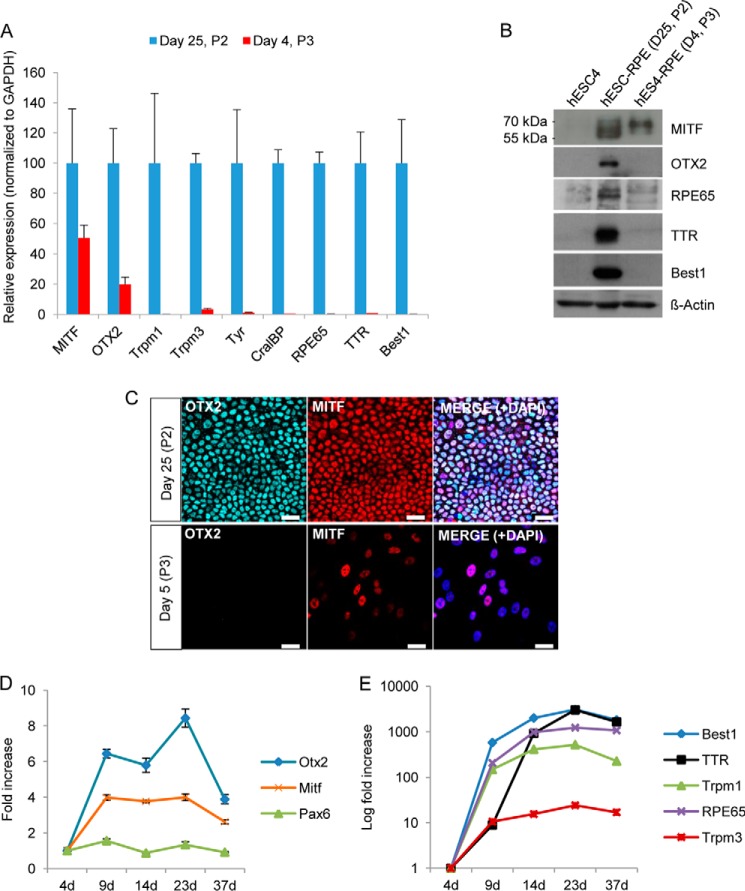
To analyze the expression of typical RPE genes in confluent polygonal pigmented cells and subconfluent recently passaged cells, we harvested hESC-derived RPE cells that had been cultured for 25 days (post-confluent culture, passage 2) and 4 days (50–70% confluence, passage 3). Differentiated cells were then analyzed for RPE-specific gene and protein expression by qRT-PCR and Western blot analysis, respectively. As shown in Fig. 2A, a drastic down-regulation of the majority of RPE-specific transcripts (Trpm1, Trpm3, Tyr, Cralbp, Rpe65, TTR, and Best1) was observed in the subconfluent culture (4 days after passage), whereas a more moderate down-regulation was detected for the two RPE-specific transcription factors Mitf and Otx2. Western blot analysis of protein expression showed a similar down-regulation of RPE-specific transcripts (Fig. 2B). Interestingly, the two cell populations displayed a different migration pattern of MITF. This might be due to different isoform expression because the MITF locus is regulated by multiple promoters (25, 26) and/or due to posttranslational modifications (i.e. phosphorylation, sumoylation, or ubiquitylation) of MITF (25, 27–29). We also performed immunostaining of postconfluent hESC-RPE cells cultured for 25 days and of subconfluent hESC-RPE cells cultured for 5 days after reseeding. Consistent with our Western blot analysis, the OTX2 protein could only be detected in polygonal hESC-RPE cells, whereas MITF could be detected in both populations, although it was differentially expressed in the 5-day cell cultures. We next performed a series of time course experiments (of the passage 3 culture) and analyzed the expression of RPE-specific genes at different time points (4, 9, 14, 23, and 37 days) during the reestablishment of the classical pigmented polygonal morphology of the cells. Consistent with the results in Fig. 2A, the transcription factors Otx2 and Mitf had a moderate increase in expression over time (Fig. 2D), whereas other RPE-specific transcripts, including Best1, TTR, Trpm1, Trpm3, and Rpe65, were highly up-regulated over time until at least day 23 (Fig. 2E). The RPE progenitor marker Pax6 had approximately similar low expression throughout the establishment of the RPE cells (Fig. 2D).
FIGURE 2.
Molecular characterization of subconfluent and postconfluent polygonal hESC-RPE cells. To compare the relative expression of RPE-related genes before (passage 2 (P2), day 25) and after (passage 3, day 4) cell passage, qRT-PCR was performed (A). The graph represents the relative gene expression of P2 cells cultured for 25 days (normalized to GAPDH). Data are presented as the mean ± S.D. of three replicates. B, Western blot analysis of the same experiment as in A shows that RPE-specific proteins are down-regulated after passage. D, day. C, immunofluorescence detection of confluent polygonal RPE cells and subconfluent cells (5 days after passage), showing expression of OTX2 and MITF. DAPI was used to counterstain the nuclei (blue). Scale bars = 25 μm. D and E, relative mRNA levels determined by qRT-PCR of RPE-specific genes at different time points after passage (P3). mRNA levels were normalized to GAPDH and plotted relative to day 4 (4d).
mRNA Transfection Is Highly Efficient in Subconfluent, Early Confluent, and Postconfluent RPE Cell Cultures
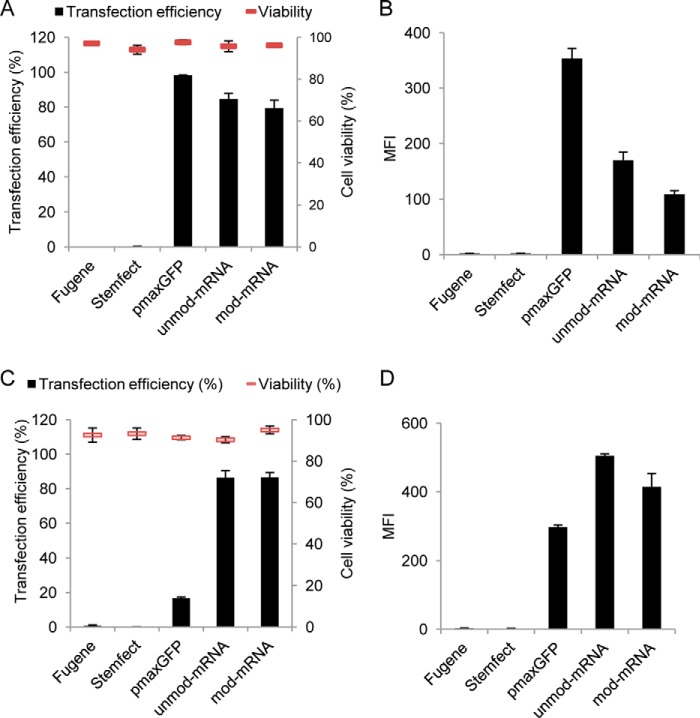
After confirming that the obtained hESC-RPE cells displayed the desired characteristics, we proceeded with the transfection experiments. To confirm that the synthetic mRNA, DNA plasmid, and transfection reagents were functional, transfections were first performed on HEK293T cells, traditionally considered easy to transfect. The cells were harvested and stained with propidium iodide (PI) 24 h after transfection, and transfection efficiency (GFP-positive cells) and cell death (PI-positive cells) were analyzed by flow cytometry. DNA transfection, making use of complexed pmaxGFP plasmid with the FuGENE6 reagent, resulted in GFP expression in 95% of the initial transfected cells, whereas both in vitro-synthesized unmodified and modified mRNA-GFP (UTP and CTP substituted with ψUTP and 5-methyl-CTP) delivered with Stemfect reagent resulted in transfection efficiencies of ∼80% (Fig. 3A). Propidium iodide staining revealed that cell viability was over 90% for all delivery approaches (Fig. 3A). The mean fluorescence intensity (mfi) was ∼2.2- and 3-fold higher in HEK293T cells transfected with pmaxGFP compared with unmodified (unmod) GFP mRNA and modified (mod) GFP mRNA, respectively (Fig. 3B).
FIGURE 3.
Efficiency of pmax-GFP plasmid and mRNA-GFP delivery to HEK293T and subconfluent hESC-RPE cells. HEK293T cells were transfected with FuGENE6-pmax-GFP, Stemfect-unmod-mRNA-GFP, or Stemfect-mod-mRNA-GFP and harvested 24 h post-transfection. Efficiency of transfection and cell viability (A) and mean fluorescence intensity (B) were quantified by flow cytometry. 4-day subconfluent hESC-RPE cell cultures were transfected in a similar way as HEK293T cells. C and D, analysis of transfection efficiency and cell viability (C) and mean fluorescence intensity (D). Error bars represent mean ± S.D., and experiments were performed at least two times in duplicate.
It has been reported previously that plasmid DNA transfection in primary cultures of RPE cells, and in RPE cell line derivatives such as ARPE19, is inefficient. To test whether this was also the case with hESC-RPE cells, we transfected subconfluent hESC-RPE cells (cultured for 4 days) with pmaxGFP. Flow cytometric analysis 24 h post-transfection revealed poor transfection efficiencies with an average of 17% GFP-positive cells. On the other hand, both unmod and mod mRNAs encoding GFP resulted in at least 80% GFP-positive cells with a cell-viability around 90% (Fig. 3C). The mean fluorescence intensity was also higher for cells transfected with either type of mRNA (400–500 mfi) than it was for those transfected with the plasmid (300 mfi) (Fig. 3D).
Next we tested the plasmid- and mRNA-based transfection approaches on early confluence hESC-RPE cell cultures (10 days in culture). The efficiency of transfection with pmaxGFP was only around 10%, whereas both unmod and mod mRNA-GFP-transfected cells showed a high efficiency (90%) 24 h after transfection (Fig. 4A). The cytotoxic effect was low (less than 10%) for both transfection approaches (Fig. 4A). The GFP expression levels from the pmaxGFP plasmid and unmod/mod mRNA-GFP were approximately equally high (∼ 400 mfi) (Fig. 4B). Representative micrographs of pmaxGFP-, unmod mRNA-GFP-, and mod mRNA-GFP-transfected cells are shown in Fig. 4C. To further evaluate the transfection capacity of the synthetic mRNA (both unmod and mod), we also performed similar experiments on postconfluence hESC-RPE cell cultures (25 days after passage, where the RPE-specific genes are highly expressed and the cells displayed the classical RPE morphology with densely packed polygonal cells, see Fig. 2). Similar trends were observed, with a decreasing efficiency of pmaxGFP plasmid and a consistently high efficiency of mRNA-transfected cells (Fig. 5A). The mean fluorescence intensity of the cultures transfected with the two types of mRNA was approximately similar (200–300 mfi), whereas the few cells that were GFP-positive after pmax plasmid transfection had an average of less than 100 mfi (Fig. 5B). Typical phase-contrast and fluorescence micrographs of postconfluent polygonal hESC-RPE cells 24 h post-transfection are shown in Fig. 5C.
FIGURE 4.
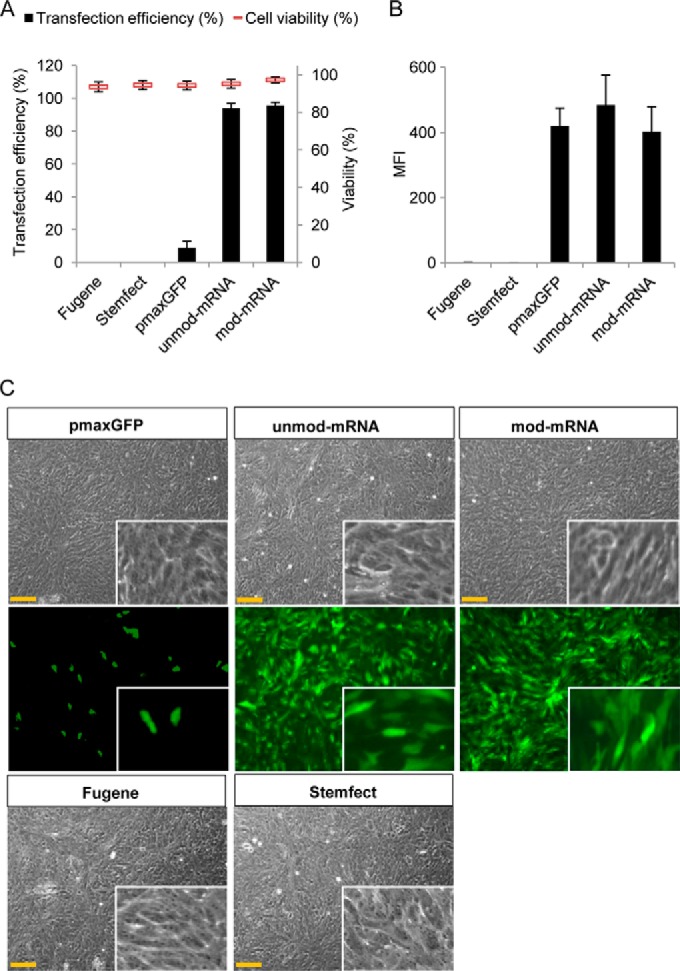
Comparison of the efficiency of plasmid-based and mRNA-based transfection on early confluence hESC-RPE cell culture. hESC-RPE cells were subcultured for 10 days to obtain the characteristics of early confluent hESC-RPE cell culture. Cells were transfected with FuGENE6-pmax-GFP, Stemfect-unmod-mRNA-GFP, or Stemfect-mod-mRNA-GFP and harvested 24 h post-transfection. Efficiency of transfection and cell viability (A) and mean fluorescence intensity (B) were monitored by flow cytometry. C, phase contrast and fluorescence microscopy analysis of early confluent hESC-RPE cells 24 h after pmaxGFP, unmod-mRNA-GFP, and mod-mRNA-GFP transfection (×20 objective). Scale bars = 75 μm. Insets show magnified images. Experiments were performed at least twice in duplicate. Data represent mean ± S.E.
FIGURE 5.
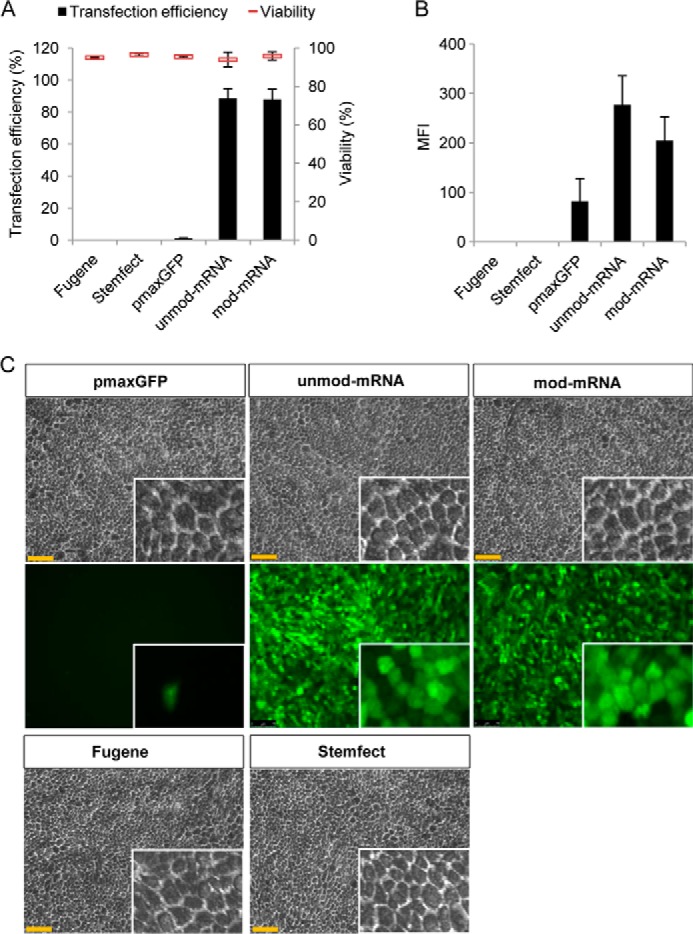
Comparison of the efficiency of plasmid-based and mRNA-based transfection on late confluence hESC-RPE cell culture. hESC-RPE cells were subcultured for 30 days to obtain the characteristics of polygonal polarized RPE cells. Cells were transfected with FuGENE6-pmax-GFP, Stemfect-unmod-mRNA-GFP, or Stemfect-mod-mRNA-GFP and harvested 24 h post-transfection. Efficiency of transfection and cell viability (A) and mean fluorescence intensity (B) were monitored by flow cytometry. C, phase-contrast and fluorescence microscopy analysis of late confluent hESC-RPE cells 24 h after pmaxGFP, unmod-mRNA-GFP, and mod-mRNA-GFP transfection, respectively (×20). Scale bars = 75 μm. Insets show magnified images. Experiments were performed at least twice in duplicate. Data represent mean ± S.E.
Functional Expression of Transfected FLAG-MITF-encoding mRNA in Confluent hESC-RPE Cells
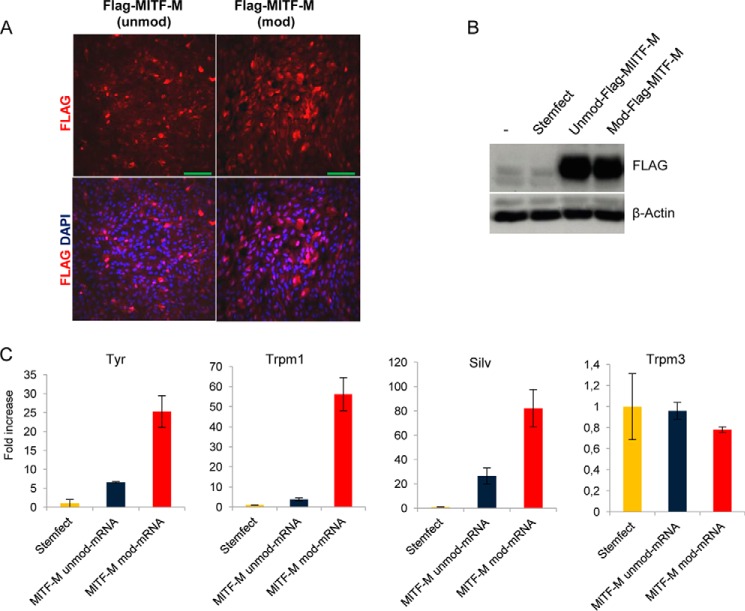
To determine whether mRNA transfection could be used to express functional RPE-related proteins, early confluent hESC-RPE cells were transfected with unmod and mod mRNA encoding the M isoform of the transcription factor MITF. To be able to distinguish between endogenously and exogenously expressed MITF, our constructs were subcloned with a FLAG tag in the N-terminal of MITF. First, we confirmed by immunostaining that transfected mRNAs (unmod and mod) encoding FLAG-MITF-M could be delivered efficiently into hESC-RPE cells and that the encoded protein was expressed and translocated to the nucleus (Fig. 6A). Western blot analysis confirmed approximately equal expression levels from unmod and mod mRNA (Fig. 6B). We next tested whether the MITF encoded by transfected unmod and mod mRNA was functional and capable of activating specific target genes. Confluent hESC-RPE cells were transfected with Stemfect-complexed FLAG-MITF-M mRNA (modified or unmodified), and cells were harvested for subsequent qRT-PCR analysis 16 h post-transfection. Importantly, FLAG-MITF-M, whether expressed from unmodified or modified mRNA, up-regulated its target genes, including SILV, TYR, and TRPM1. However, transfected FLAG-MITF-M-modified mRNA activated the transcription of MITF-regulated genes about 5-fold more than its unmodified mRNA counterpart (Fig. 6C). In addition, when analyzing other RPE genes unrelated to the MITF transcription factor, such as TRPM3, we did not find any significant differences between cells treated with only Stemfect (our negative control) and cells treated with FLAG-MITF-M mod mRNA (Fig. 6C).
FIGURE 6.
Nuclear localization and functional expression of FLAG-MITF mRNA transfected into hESC-RPE cells. A, immunofluorescence staining of confluent hESC-RPE cells transfected for 16 h with unmod and mod mRNA encoding FLAG-MITF-M. DAPI (blue) was used to stain the nucleus. Scale bars = 100 μm. B, Western blots of early confluent cells 16 h post-transfection. C, qRT-PCR analysis of a subset of RPE-specific genes, including reported MITF-target genes (TYR, SILV, and TRPM1). Data are from one representative experiment of three independent experiments. Error bars represent mean ± S.D.
Unmodified mRNA, but Not Modified mRNA, Strongly Induces the Innate Immunity of hESC-RPE Cells
It is known that exogenous single-stranded/double-stranded RNA can induce the innate immune response of many cells and that mRNA delivered to various cell types can have a cytotoxic effect. Moreover, RPE cells express TLR3 (which recognizes exogenous RNA) and are able to secrete many immune modulatory factors, such as IL-8, monocyte chemotactic protein 1 (MCP1), IFNβ, and TNFα (1, 30). On the basis of these findings and to test whether mRNA transfection has an effect upon the immune response of hESC-RPE cells, we transfected cells cultured for 10 days with either modified or unmodified mRNA encoding GFP and analyzed the expression of IFNβ, TNFα, and TLR-3. Gene expression was analyzed by qRT-PCR 16 h post-treatment. Strong up-regulation of all analyzed genes was observed when unmodified mRNA was used but not with modified mRNA or under basal conditions (Fig. 7A). Similar results were obtained after transfecting the cells with unmodified and modified mRNA encoding FLAG-MITF-M protein (data not shown). In addition, when immunostaining was performed 6 h post-transfection with GFP mRNA, our analysis revealed a strong nuclear localization of p65/RelA, a subunit of NF-κB, and interferon regulatory factor 3 (IRF3), but only when unmodified mRNA was used (Fig. 7, B and C). Both IRF3 and NF-κB are activated downstream of Toll-like receptor 3 (TLR3), whereupon they translocate to the nucleus to promote expression of IFNβ and TNFα, respectively. Such responses are consistent with our qRT-PCR data (Fig. 7A).
FIGURE 7.
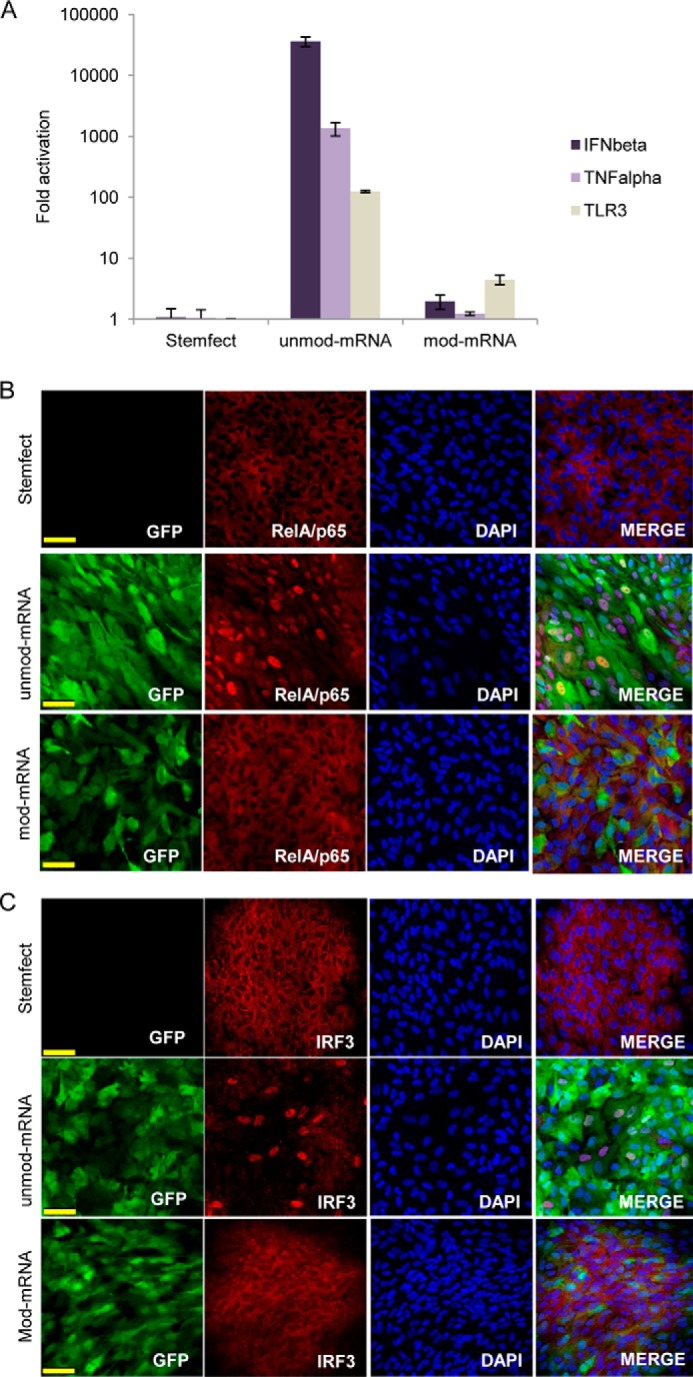
Unmod mRNA, but not mod mRNA, induces a strong innate immune response via IRF3 and p65/RelA nuclear translocation. Early confluent hESC-RPE cell cultures were transfected with synthetic mRNA (unmod or mod) encoding GFP and harvested after 16 h for qRT-PCR analysis of TNFα, IFNβ, and TLR3 (A). Early confluent hESC-RPE cells were analyzed 6 h post-transfection by immunofluorescence for expression and subcellular localization of p65/RelA (B) and IRF3 (C). Anti-GFP antibody was used to detect GFP, and DAPI was used to stain the nucleus (B and C). Scale bars = 50 μm. Data are from one representative experiment of three independent experiments. Error bars represent mean ± S.D.
DISCUSSION
Development of a safe and efficient viral-free gene delivery system is one of the major challenges for gene-based therapies of the retina. Despite recent improvements in plasmid DNA delivery, regarding efficiency and viability, the efficiency of transfection is still considered to be low or moderate (seldom over 50%) (31, 32). Moreover, the most successful in vitro plasmid DNA gene delivery studies were on RPE-derived cell lines, which are easier to transfect than primary RPE cells (33). In this report, we used hESC-derived RPE cells, because they are considered to be much more similar to primary RPE than RPE cell lines (6), to investigate the utility of using synthetic mRNA as a novel gene delivery approach for transient protein expression in primary RPE cells. hESC-RPE cells were generated using a protocol reported previously with minor modifications (5). To ensure the highest purity and maturity of our generated hESC-RPE cultures, we selected and expanded the cultures for two rounds of passage before characterizing the RPE cell cultures (34). Morphological and molecular analysis of the cells showed typical RPE features such as tight junctions, melanin granules, apical microvilli, and expression of RPE gene transcripts and proteins important for many of the typical RPE functions. In many retinal diseases, including age-related macular degeneration and proliferative vitroretinopathy, RPE cells lose their epithelial phenotype and cell-to-cell contact and become proliferative, motile fibroblast-like cells (35). A similar process also occurs when subculturing the RPE cells in vitro (6, 7, 36). However, when confluence is reestablished, the cells regain their classical pigmented polygonal RPE morphology. Consistent with similar studies on primary RPE cells, RPE cell lines, as well as iPSC- and ES cell-derived RPE cells, we found the RPE-specific genes to be drastically down-regulated in the subconfluent cell culture (37, 38). A time course study of RPE cell reestablishment showed reexpression of RPE-specific transcripts as the cells became confluent. These observations were coincident with the acquisition of the typical polygonal cell shape and the re-establishment of pigmentation. The underlying mechanisms for the loss of RPE phenotype after cell-cell dissociation are not fully understood, but studies in mouse primary RPE cells and RPE cell lines suggest that loss of contact inhibition induces activation of canonical Wnt and Smad/ZEB (Zinc finger E-box-binding homeobox) signaling and subsequent activation of an epithelial-to-mesenchymal transition and unlocking of the mitotic block (37, 39, 40). Moreover, it has been shown that ZEB can bind to the MITF promoter and, thereby, repress its transcription, which, in turn, would lead to down-regulation of MITF target genes, including many RPE-specific genes (37, 40).
Our analysis of the two transcription factors essential for RPE maintenance and function, Otx2 and Mitf, showed a moderate down-regulation of both transcripts in the subconfluent culture (4 days after passage). However, when analyzing these protein levels in 4-day subconfluent cells and 25-day cultured postconfluent RPE cells, we found that the OTX2 protein was undetectable, whereas the MITF protein could be detected readily (whereas, in 25-day postconfluent cells, both MITF and OTX2 were detectable). Because many of the analyzed RPE-related genes that were down-regulated (such as RPE65, CRALBP, and TTR) have been reported recently to be direct targets of OTX2 (41, 42), we speculate that, together with MITF, OTX2 may be responsible for the down-regulation of the RPE-specific genes after cell subculture.
Despite the great interest in hESC-RPE and hiPSC RPE cells as sources for cell therapy and in vitro disease modeling, no studies of gene delivery of these cells have, to our knowledge, been reported. We have shown that, as is the case for RPE cells, hESC-RPE cells are very difficult to transfect with plasmid DNA complexed with commercial transfection reagent (FuGENE 6). The highest efficiency of transfection with plasmid DNA was achieved on subconfluent RPE cells with an efficiency of 17%, and this decreased as the hESC-RPE cells progressed into confluence. When the cells reached a polygonal morphology, less than 1% of the cell culture was positively transfected. In contrast, synthetic unmodified and modified (UTP and CTP substituted with pseudo-UTP and 5-methyl-CTP) mRNAs were delivered efficiently into subconfluent, early confluent, and postconfluent (polygonal) RPE cells. Mean fluorescence intensities were approximately equally high in cells transfected with plasmid, unmod, and mod mRNA-GFP. This is in contrast to some reports where modified mRNA is claimed to have a higher translation efficiency than unmodified mRNA (15, 22), which should result in higher fluorescence in mod mRNA-transfected cells. The reason for this discrepancy is not clear, but because we observed a similar results in other cells (keratinocytes, HEK293T, and mesenchymal stem cells derived from adipose tissue), we believe that it is not a cell-specific event. Further optimization studies with different transfection agents, ratios of modified nucleosides, and mRNA/transfection reagent ratios are needed to understand this.
To confirm the functionality of mRNA transfection, we chose an endogenously important RPE transcription factor: MITF. This gene is expressed from several promoters in an overlapping, cell-specific manner, resulting in different isoforms of MITF. It has been shown that, during the development of the murine RPE, the N-terminal 1B1b domain of MITF is crucial (26). All MITF isoforms, except for MITF-M, contain this domain. In addition, it has been generally assumed that the M isoform is only expressed in melanocytes. However, recent reports have shown relatively high levels of the M-isoform in adult and prenatal human RPE cells as well as hESC-derived RPE cells (43, 44). Whether different isoforms have a distinct or overlapping function in the adult RPE is not yet known. However, several reports have identified the 1B1b domain to be responsible for regulating cytoplasmic shuttling of MITF (45, 46). In accordance with this, our initial experiments with the transfection of MITF-A mRNA resulted in partial cytoplasmic localization of the MITF-A protein (data not shown). On the other hand, transfection of MITF-M mRNA gave rise to a strong nuclear localization of the protein, a necessary condition for correct evaluation of the transcription-regulating activity of MITF. Because of this, we chose the M isoform for comparison of the transcriptional functionality of the transfected mRNAs. Functional validation showed that both unmodified and modified mRNAs encoding MITF-M were able to activate many of the known MITF target genes, including TYR, TRPM1, and SILV. However, the MITF-M protein expressed from modified mRNA activated these genes more strongly than that expressed from unmodified mRNA. The reason for this is not clear, but because the expression levels of the encoded MITF-M protein were approximately equal, we speculate that the immune response generated by transfection with unmodified mRNA in some way interferes with the action of MITF, either directly or indirectly. It is known that exogenous dsRNA, as well as single-stranded RNA forming double-stranded secondary structures, acts as a ligand for Toll-like receptor 3 and, as such, may induce an innate immune response (47). TLR3 activation induces the recruitment of adapter molecules and kinases that lead to the translocation of the transcription factors IRF3 and NF-κB into the nucleus, where they promote the expression of TNFα and type I interferons such as IFNβ (47, 48). Considering that TLR3 is known to be expressed in the RPE (30), our results are consistent with the current literature. The nuclear localization of RelA/p65 (an NF-κβ subunit) and IRF3, as well as the increased expression of IFNβ and TNFα in RPE cells transfected with unmodified mRNA may be attributed to TLR3 recognition of the transfected mRNA. Moreover, it has been reported that mRNA incorporating pseudouridine and/or methylated nucleosides is not efficiently recognized by TLRs (14), which explains why our transfections with modified mRNA did not greatly induce the innate immune response.
In conclusion, we have found that synthetic mRNA can be delivered effectively in hESC-RPE cells and that the expressed proteins are functional. Moreover, we have shown that synthetic modified mRNA can both bypass the innate immune response, therefore minimizing cytotoxic effects, and also generate a more biologically active protein, as shown when modified mRNA encoding MITF was transfected in hESC-RPE cells. Although the expression from the transfected mRNA is transient and, as such, might not be applicable at the present time for gene-based therapy treatments directly in patients, our study establishes a unique platform to use hESC-RPE cells and modified mRNA transfection for basic research, disease modeling, or as an approach to further develop therapeutic strategies for retinal degenerative disease treatments, such as cellular mRNA engineering, to improve successful cell transplantation.
Acknowledgments
We thank C. Tarantino, M. Díaz, L. Miquel, and the platforms at the Center of Regenerative Medicine in Barcelona for technical assistance.
Footnotes
- RPE
- retinal pigmented epithelial/epithelium
- hESC
- human embryonic stem cell
- hiPSC
- human induced pluripotent stem cell
- MITF
- microphthalmia-associated transcription factor
- TTR
- transthyretin
- qRT-PCR
- quantitative RT-PCR
- PI
- propidium iodide
- mfi
- mean fluorescence intensity
- unmod
- unmodified
- mod
- modified.
REFERENCES
- 1. Strauss O. (2005) The retinal pigment epithelium in visual function. Physiol. Rev. 85, 845–881 [DOI] [PubMed] [Google Scholar]
- 2. Sparrow J. R., Hicks D., Hamel C. P. (2010) The retinal pigment epithelium in health and disease. Curr. Mol. Med. 10, 802–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tucker B. A., Mullins R. F., Stone E. M. (2014) Stem cells for investigation and treatment of inherited retinal disease. Hum. Mol. Genet. 23, R9–R16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Falkner-Radler C. I., Krebs I., Glittenberg C., Povazay B., Drexler W., Graf A., Binder S. (2011) Human retinal pigment epithelium (RPE) transplantation: outcome after autologous RPE-choroid sheet and RPE cell-suspension in a randomised clinical study. Br. J. Ophthalmol. 95, 370–375 [DOI] [PubMed] [Google Scholar]
- 5. Idelson M., Alper R., Obolensky A., Ben-Shushan E., Hemo I., Yachimovich-Cohen N., Khaner H., Smith Y., Wiser O., Gropp M., Cohen M. A., Even-Ram S., Berman-Zaken Y., Matzrafi L., Rechavi G., Banin E., Reubinoff B. (2009) Directed differentiation of human embryonic stem cells into functional retinal pigment epithelium cells. Cell Stem Cell 5, 396–408 [DOI] [PubMed] [Google Scholar]
- 6. Klimanskaya I., Hipp J., Rezai K. A., West M., Atala A., Lanza R. (2004) Derivation and comparative assessment of retinal pigment epithelium from human embryonic stem cells using transcriptomics. Cloning Stem Cells 6, 217–245 [DOI] [PubMed] [Google Scholar]
- 7. Buchholz D. E., Hikita S. T., Rowland T. J., Friedrich A. M., Hinman C. R., Johnson L. V., Clegg D. O. (2009) Derivation of functional retinal pigmented epithelium from induced pluripotent stem cells. Stem Cells 27, 2427–2434 [DOI] [PubMed] [Google Scholar]
- 8. Carr A. J., Vugler A. A., Hikita S. T., Lawrence J. M., Gias C., Chen L. L., Buchholz D. E., Ahmado A., Semo M., Smart M. J., Hasan S., da Cruz L., Johnson L. V., Clegg D. O., Coffey P. J. (2009) Protective effects of human iPS-derived retinal pigment epithelium cell transplantation in the retinal dystrophic rat. PLoS ONE 4, e8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lund R. D., Wang S., Klimanskaya I., Holmes T., Ramos-Kelsey R., Lu B., Girman S., Bischoff N., Sauvé Y., Lanza R. (2006) Human embryonic stem cell-derived cells rescue visual function in dystrophic RCS rats. Cloning Stem Cells 8, 189–199 [DOI] [PubMed] [Google Scholar]
- 10. Zhang K., Liu G. H., Yi F., Montserrat N., Hishida T., Esteban C. R., Izpisua Belmonte J. C. (2014) Direct conversion of human fibroblasts into retinal pigment epithelium-like cells by defined factors. Protein Cell 5, 48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schwartz S. D., Hubschman J. P., Heilwell G., Franco-Cardenas V., Pan C. K., Ostrick R. M., Mickunas E., Gay R., Klimanskaya I., Lanza R. (2012) Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet 379, 713–720 [DOI] [PubMed] [Google Scholar]
- 12. Melville H., Carpiniello M., Hollis K., Staffaroni A., Golestaneh N. (2013) Stem cells: a new paradigm for disease modeling and developing therapies for age-related macular degeneration. J. Transl. Med. 11, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Strunnikova N. V., Maminishkis A., Barb J. J., Wang F., Zhi C., Sergeev Y., Chen W., Edwards A. O., Stambolian D., Abecasis G., Swaroop A., Munson P. J., Miller S. S. (2010) Transcriptome analysis and molecular signature of human retinal pigment epithelium. Hum. Mol. Genet. 19, 2468–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Karikó K., Buckstein M., Ni H., Weissman D. (2005) Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 [DOI] [PubMed] [Google Scholar]
- 15. Karikó K., Muramatsu H., Welsh F. A., Ludwig J., Kato H., Akira S., Weissman D. (2008) Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 16, 1833–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anderson B. R., Muramatsu H., Jha B. K., Silverman R. H., Weissman D., Karikó K. (2011) Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 39, 9329–9338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Y., Su H. H., Yang Y., Hu Y., Zhang L., Blancafort P., Huang L. (2013) Systemic delivery of modified mRNA encoding herpes simplex virus 1 thymidine kinase for targeted cancer gene therapy. Mol. Ther. 21, 358–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kormann M. S., Hasenpusch G., Aneja M. K., Nica G., Flemmer A. W., Herber-Jonat S., Huppmann M., Mays L. E., Illenyi M., Schams A., Griese M., Bittmann I., Handgretinger R., Hartl D., Rosenecker J., Rudolph C. (2011) Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 29, 154–157 [DOI] [PubMed] [Google Scholar]
- 19. Mays L. E., Ammon-Treiber S., Mothes B., Alkhaled M., Rottenberger J., Müller-Hermelink E. S., Grimm M., Mezger M., Beer-Hammer S., von Stebut E., Rieber N., Nürnberg B., Schwab M., Handgretinger R., Idzko M., Hartl D., Kormann M. S. (2013) Modified Foxp3 mRNA protects against asthma through an IL-10-dependent mechanism. J. Clin. Invest. 123, 1216–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Petsch B., Schnee M., Vogel A. B., Lange E., Hoffmann B., Voss D., Schlake T., Thess A., Kallen K. J., Stitz L., Kramps T. (2012) Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat. Biotechnol. 30, 1210–1216 [DOI] [PubMed] [Google Scholar]
- 21. Lui K. O., Zangi L., Silva E. A., Bu L., Sahara M., Li R. A., Mooney D. J., Chien K. R. (2013) Driving vascular endothelial cell fate of human multipotent Isl1+ heart progenitors with VEGF modified mRNA. Cell Res. 23, 1172–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Warren L., Manos P. D., Ahfeldt T., Loh Y. H., Li H., Lau F., Ebina W., Mandal P. K., Smith Z. D., Meissner A., Daley G. Q., Brack A. S., Collins J. J., Cowan C., Schlaeger T. M., Rossi D. J. (2010) Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7, 618–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zangi L., Lui K. O., von Gise A., Ma Q., Ebina W., Ptaszek L. M., Später D., Xu H., Tabebordbar M., Gorbatov R., Sena B., Nahrendorf M., Briscoe D. M., Li R. A., Wagers A. J., Rossi D. J., Pu W. T., Chien K. R. (2013) Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 31, 898–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Raya A., Rodríguez-Pizà I., Arán B., Consiglio A., Barri P. N., Veiga A., Izpisúa Belmonte J. C. (2008) Generation of cardiomyocytes from new human embryonic stem cell lines derived from poor-quality blastocysts. Cold Spring Harb. Symp. Quant. Biol. 73, 127–135 [DOI] [PubMed] [Google Scholar]
- 25. Tachibana M. (2000) MITF: a stream flowing for pigment cells. Pigment Cell Res. 13, 230–240 [DOI] [PubMed] [Google Scholar]
- 26. Bharti K., Liu W., Csermely T., Bertuzzi S., Arnheiter H. (2008) Alternative promoter use in eye development: the complex role and regulation of the transcription factor MITF. Development 135, 1169–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu W., Gong L., Haddad M. M., Bischof O., Campisi J., Yeh E. T., Medrano E. E. (2000) Regulation of microphthalmia-associated transcription factor MITF protein levels by association with the ubiquitin-conjugating enzyme hUBC9. Exp. Cell Res. 255, 135–143 [DOI] [PubMed] [Google Scholar]
- 28. Weilbaecher K. N., Motyckova G., Huber W. E., Takemoto C. M., Hemesath T. J., Xu Y., Hershey C. L., Dowland N. R., Wells A. G., Fisher D. E. (2001) Linkage of M-CSF signaling to Mitf, TFE3, and the osteoclast defect in Mitf(mi/mi) mice. Mol. Cell 8, 749–758 [DOI] [PubMed] [Google Scholar]
- 29. Miller A. J., Levy C., Davis I. J., Razin E., Fisher D. E. (2005) Sumoylation of MITF and its related family members TFE3 and TFEB. J. Biol. Chem. 280, 146–155 [DOI] [PubMed] [Google Scholar]
- 30. Kumar M. V., Nagineni C. N., Chin M. S., Hooks J. J., Detrick B. (2004) Innate immunity in the retina: Toll-like receptor (TLR) signaling in human retinal pigment epithelial cells. J. Neuroimmunol. 153, 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Puras G., Mashal M., Zárate J., Agirre M., Ojeda E., Grijalvo S., Eritja R., Diaz-Tahoces A., Martínez Navarrete G., Avilés-Trigueros M., Fernández E., Pedraz J. L. (2014) A novel cationic niosome formulation for gene delivery to the retina. J. Control. Release 174, 27–36 [DOI] [PubMed] [Google Scholar]
- 32. Sunshine J. C., Sunshine S. B., Bhutto I., Handa J. T., Green J. J. (2012) Poly(β-amino ester)-nanoparticle mediated transfection of retinal pigment epithelial cells in vitro and in vivo. PLoS ONE 7, e37543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vercauteren D., Piest M., van der Aa L. J., Al Soraj M., Jones A. T., Engbersen J. F., De Smedt S. C., Braeckmans K. (2011) Flotillin-dependent endocytosis and a phagocytosis-like mechanism for cellular internalization of disulfide-based poly(amido amine)/DNA polyplexes. Biomaterials 32, 3072–3084 [DOI] [PubMed] [Google Scholar]
- 34. Singh R., Phillips M. J., Kuai D., Meyer J., Martin J. M., Smith M. A., Perez E. T., Shen W., Wallace K. A., Capowski E. E., Wright L. S., Gamm D. M. (2013) Functional analysis of serially expanded human iPS cell-derived RPE cultures. Invest. Ophthalmol. Vis. Sci. 54, 6767–6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saika S., Yamanaka O., Flanders K. C., Okada Y., Miyamoto T., Sumioka T., Shirai K., Kitano A., Miyazaki K., Tanaka S., Ikeda K. (2008) Epithelial-mesenchymal transition as a therapeutic target for prevention of ocular tissue fibrosis. Endocr. Metab. Immune Disord. Drug Targets 8, 69–76 [DOI] [PubMed] [Google Scholar]
- 36. Aronson J. F. (1983) Human retinal pigment cell culture. In Vitro 19, 642–650 [DOI] [PubMed] [Google Scholar]
- 37. Liu Y., Ye F., Li Q., Tamiya S., Darling D. S., Kaplan H. J., Dean D. C. (2009) Zeb1 represses Mitf and regulates pigment synthesis, cell proliferation, and epithelial morphology. Invest. Ophthalmol. Vis. Sci. 50, 5080–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Adijanto J., Castorino J. J., Wang Z. X., Maminishkis A., Grunwald G. B., Philp N. J. (2012) Microphthalmia-associated transcription factor (MITF) promotes differentiation of human retinal pigment epithelium (RPE) by regulating microRNAs-204/211 expression. J. Biol. Chem. 287, 20491–20503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen H. C., Zhu Y. T., Chen S. Y., Tseng S. C. (2012) Selective activation of p120ctn-Kaiso signaling to unlock contact inhibition of ARPE-19 cells without epithelial-mesenchymal transition. PLoS ONE 7, e36864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu Y., Xin Y., Ye F., Wang W., Lu Q., Kaplan H. J., Dean D. C. (2010) Taz-tead1 links cell-cell contact to zeb1 expression, proliferation, and dedifferentiation in retinal pigment epithelial cells. Invest. Ophthalmol. Vis. Sci. 51, 3372–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Masuda T., Wahlin K., Wan J., Hu J., Maruotti J., Yang X., Iacovelli J., Wolkow N., Kist R., Dunaief J. L., Qian J., Zack D. J., Esumi N. (2014) Transcription factor SOX9 plays a key role in the regulation of visual cycle gene expression in the retinal pigment epithelium. J. Biol. Chem. 289, 12908–12921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Housset M., Samuel A., Ettaiche M., Bemelmans A., Béby F., Billon N., Lamonerie T. (2013) Loss of Otx2 in the adult retina disrupts retinal pigment epithelium function, causing photoreceptor degeneration. J. Neurosci. 33, 9890–9904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maruotti J., Thein T., Zack D. J., Esumi N. (2012) MITF-M, a “melanocyte-specific” isoform, is expressed in the adult retinal pigment epithelium. Pigment Cell Melanoma Res. 25, 641–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Capowski E. E., Simonett J. M., Clark E. M., Wright L. S., Howden S. E., Wallace K. A., Petelinsek A. M., Pinilla I., Phillips M. J., Meyer J. S., Schneider B. L., Thomson J. A., Gamm D. M. (2014) Loss of MITF expression during human embryonic stem cell differentiation disrupts retinal pigment epithelium development and optic vesicle cell proliferation. Hum. Mol. Genet. 23, 6332–6344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lu S. Y., Wan H. C., Li M., Lin Y. L. (2010) Subcellular localization of Mitf in monocytic cells. Histochem. Cell Biol. 133, 651–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Martina J. A., Puertollano R. (2013) Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J. Cell Biol. 200, 475–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. (2001) Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413, 732–738 [DOI] [PubMed] [Google Scholar]
- 48. Vercammen E., Staal J., Beyaert R. (2008) Sensing of viral infection and activation of innate immunity by toll-like receptor 3. Clin. Microbiol. Rev. 21, 13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]