Background: PTP1B is an enzyme localized to the cytoplasmic face of the ER.
Results: Calnexin binds UBC9, is SUMOylated and forms complexes with PTP1B at the ER membrane.
Conclusion: This work reveals a previously unrecognized role for calnexin in the retention of PTP1B at the ER membrane.
Significance: SUMOylation machinery and UBC9 link two ER proteins from divergent pathways.
Keywords: chaperone, endoplasmic reticulum (ER), protein folding, small ubiquitin-like modifier (SUMO), tyrosine-protein phosphatase (tyrosine phosphatase)
Abstract
Calnexin is a type I integral endoplasmic reticulum (ER) membrane protein, molecular chaperone, and a component of the translocon. We discovered a novel interaction between the calnexin cytoplasmic domain and UBC9, a SUMOylation E2 ligase, which modified the calnexin cytoplasmic domain by the addition of SUMO. We demonstrated that calnexin interaction with the SUMOylation machinery modulates an interaction with protein tyrosine phosphatase 1B (PTP1B), an ER-associated protein tyrosine phosphatase involved in the negative regulation of insulin and leptin signaling. We showed that calnexin and PTP1B form UBC9-dependent complexes, revealing a previously unrecognized contribution of calnexin to the retention of PTP1B at the ER membrane. This work shows that the SUMOylation machinery links two ER proteins from divergent pathways to potentially affect cellular protein quality control and energy metabolism.
Introduction
The endoplasmic reticulum (ER)7 is an intracellular organelle responsible for many critical cellular functions including the synthesis, folding, post-translational modification, and transport of proteins; the synthesis of lipids and steroids; assembly and trafficking of membranes; Ca2+ signaling; regulation of gene expression; energy metabolism; signaling to other cellular organelles; and stress responses (1–3). To support these versatile functions the ER takes advantage of many membrane-associated proteins including molecular chaperones, folding enzymes, oxidoreductases, and various transporters and signaling proteins (1–3). For example, calnexin is an integral ER membrane protein and molecular chaperone involved in the folding and quality control of membrane associated and secreted proteins, including many cell surface receptors regulated by tyrosine phosphorylation (4). Calnexin spans the ER membrane with a large portion of the C-terminal domain (C-tail) residing in the cytoplasm where it may play a role in substrate folding (5, 6). The C-tail undergoes distinct post-translational modifications including phosphorylation and palmitoylation (7–12). The C-tail also affects FGF-dependent STAT3 signaling (13). Palmitoylation of the C-tail of calnexin mediates its association with the ribosome-translocon complex (13) and affects calnexin association with the mitochondria associated region of the ER (10). Together, these post-translational modifications indicate that the C-tail of calnexin may play a functional role in linking ER lumenal events with cytoplasmic signaling.
Protein tyrosine phosphatase 1B (PTP1B) is a member of the protein tyrosine phosphatase family of enzymes localized to the cytoplasmic face of the ER (14, 15). PTP1B acts as a critical regulator of the insulin and leptin receptor-dependent pathways (16, 17), plays a role in cancer and modulation of the immune system, and may affect ER stress signaling pathways (17–19). PTP1B is believed to be anchored to the ER membrane via a 35 amino acid C-terminal targeting sequence (ER targeting domain) whereby the protein localizes to the cytoplasmic face of the ER membrane and the nuclear envelope (14). Despite numerous reports on the ER localization of PTP1B, to our knowledge, a contribution of ER proteins to PTP1B association with the ER membrane has received no attention.
In this study, while testing for a functional role of the calnexin C-tail, we identified several C-tail interacting proteins including UBC9, the SUMOylation E2 ligase, and showed that calnexin can be SUMOylated both in vitro and in vivo. We discovered that calnexin and PTP1B form UBC9-dependent complexes, revealing an unexpected role of calnexin in holding PTP1B to the ER membrane. This work indicated that PTP1B association with the ER membrane is not only restricted to the 35 amino acid C-terminal PTP1B “ER targeting” sequence but association also involves calnexin, an ER molecular chaperone and component of the translocon. This work shows that the SUMOylation machinery and UBC9 link two ER proteins from divergent pathways, calnexin and PTP1B, in a common pathway that may affect cellular protein quality control and energy metabolism.
EXPERIMENTAL PROCEDURES
Plasmids and Mutagenesis
The GFP-SUMO-1 and human UBC9 expression plasmids were used in this study (20). cDNA encoding canine calnexin C-tail (amino acid residues 486–573) was synthesized by PCR-driven amplification using the following primers: forward primer 5′-CAT GCC ATG GCT GGA AAG AAA CAG TCA AG-3′ and reverse primer 5′-GCT CTA GAC ACT CTC TTC GTG GCT TTC-3′. The cDNA was cloned into the pBAD His-tag vector using NcoI and XbaI restriction enzymes to generate pBAD-CNX-C vector. QuikChange Site-directed Mutagenesis (Stratagene) was used to mutate Lys506 to an Ala (consensus site VKEE) residue (forward primer; 5′-CCT CAG CCA GAT GTG GCG GAG GAG GAA GAA GAA AAG G-3′ and reverse primer 5′-CCT TTT CTT CTT CCT CCT CCG CCA CAT CTG GCT GAG G-3′). To create Lys534 (potential consensus site QKSD) or Lys554 (consensus site PKAE) to Ala residue the following DNA primers were used: for L534A forward primer 5′-CTT GAA GAG AAG CAA GCG AGT GAT GCT GAA GAA GAT GGC GG-3′ and reverse primer complement; L554A forward primer 5′-GGA CGA TAG GAA ACC TGC GGC AGA GGA GGA TG-3′ and reverse primer complement. Recombinant protein was expressed and purified as described for pBAD-CNX-C. To create pEGFP-CNX and pEGFP-PTP1B plasmids, cDNA encoding calnexin or PTP1B was cloned from Mouse Brain Quick-Clone cDNA (Clontech) using the following set of primers: calnexin forward primer 5′-TAT ACT CGA GAT GGA AGG GAA GTG GTT AC-3′ and reverse primer 5′-TAT AGA ATT CCC TCT CTT CGT GGC TTT CTG-3′; PTP1B forward primer 5′-TAT AGA ATT CGA TGG AGA TGG AGA AGG AGT T-3′ and reverse primer 5′-TAT AGT CGA CTC AGT GAA AAC ACA CCC G-3′ and subcloned in-frame with GFP into pEGFP-C1 and pEGFP-N1 vectors, respectively.
Yeast-2-hybrid and DUAL Membrane System Yeast-2-hybrid
The mouse brain Matchmaker cDNA library in pACT2 (Clontech, 638841) was transformed into the yeast strain AH109. The library was screened for interacting proteins with the C-terminal cytoplasmic region (C-tail) (Cys484-Glu571) of mouse calnexin. Comparison of amino acid sequence of mouse and human calnexin C-tail is shown in the Fig. 1F. cDNA encoding the C-tail of calnexin was obtained by PCR-driven amplification of mouse calnexin cDNA using the following primers: forward primer 5′-GGA ATT CCA TAT GTG TTC TGG AAA GAA AC-3′ and reverse primer 5′-AAG GTT CTG CAG TCA CTC TCT TCG TGG CT-3′. The PCR product was cloned in frame with the GAL4 DNA-binding domain at NdeI and PstI restriction sites to generate the clone pGBKT7-CNXC. Screening of the mouse library was carried out as recommended by the manufacturer (Clontech). Canine C-tail (Cys484-Glu573) was also used as bait to screen a human leukemia (Jurkat T cell) Matchmaker cDNA library (Clontech). Plasmid DNA from positive clones was isolated and used as a template for amplification of the insert in the library vector by the 3′ AD (activating domain) sequencing primer 5′-AGA TGG TGC ACG ATG CAC AG-3′, and the T7 yeast-2-hybrid sequencing primer, 5′-TAA TAC GAC TCA CTA TAG GG-3′.
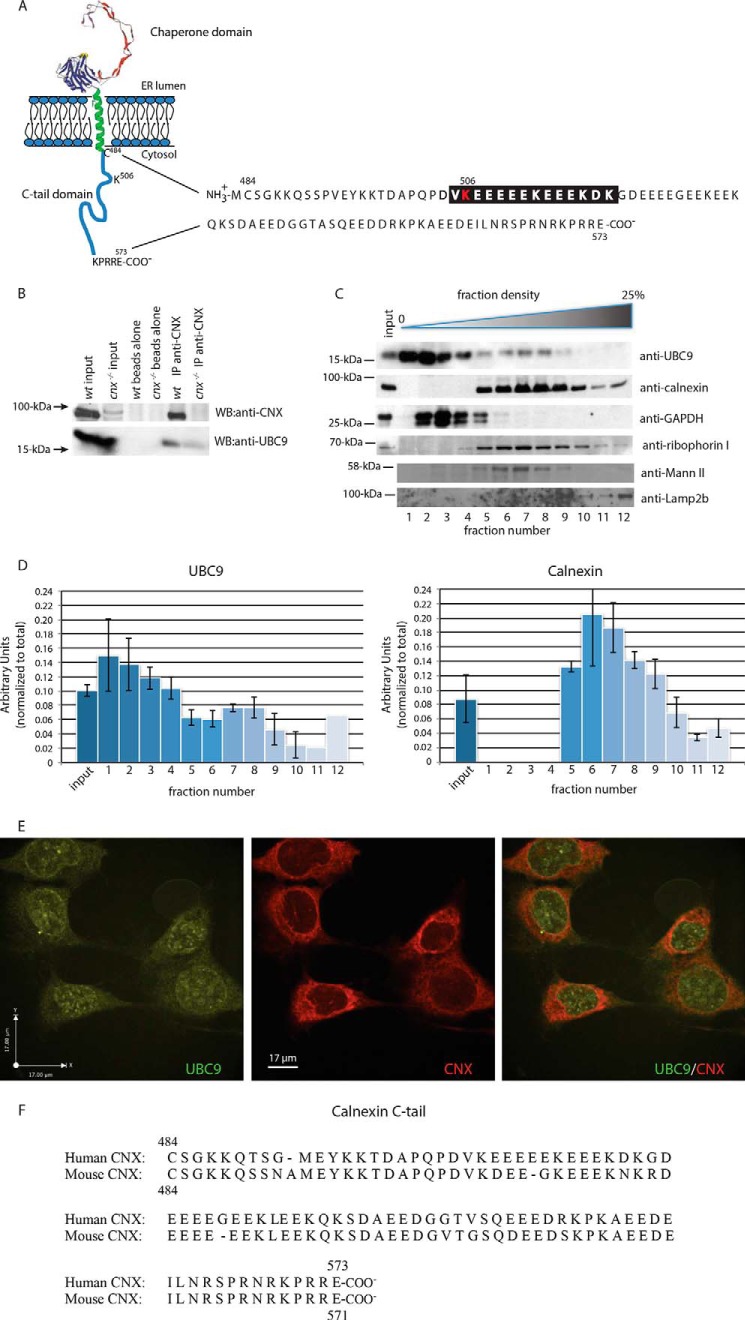
FIGURE 1.
UBC9 binds the calnexin C-tail. A, schematic representation of calnexin and the amino acid sequence of the calnexin cytoplasmic (C-tail) domain. The proposed negatively charged amino acid-dependent SUMOylation motif (NDSM) is highlighted in black with the location of Lys506 indicated in red. B, immunoprecipitation was carried out on wild-type (wt) and calnexin-deficient mouse fibroblasts with anti-calnexin antibodies followed by Western blot (WB) analysis with anti-calnexin or anti-UBC9 antibodies. The data represent three biological replicates. C, opti-prep gradient fractionation of mouse fibroblast cells. Lower density fractions (1–4) correspond to cytoplasmic components, while higher density fractions (5–12) correspond to heavier cellular compartments, including the ER. Western blot analysis was carried out with anti-UBC9, anti-calnexin, anti-GAPDH, anti-ribophorin I, anti-Mannosidase II (Mann II), and anti-Lamp2b antibodies. The data represent more than three biological replicates. D, quantitative analysis of Western blot from C. F, immunostaining of HeLa cells with anti-UBC9 antibodies (left panel) and anti-calnexin antibodies (middle panel). Right panel represents a merger of the left and the middle panels. Average threshold Pearson's co-localization coefficient of 0.558 ± 0.060, mean ± S.E. The data represent more than three biological replicates. E, amino acids sequences alignment of mouse (NP_031623) and human (NP_001737) calnexin C-tail using Clustal Omega (1.2.1).
The DUAL membrane system was used to assess yeast two-hybrid interactions (Dualsystems Biotech). The cDNA encoding full-length calnexin and PTP1B were subcloned into pBT3-SUC and pPR3 vector, respectively, and co-transformed into the yeast strain NYM51. pOst1 and pPR3 were used as a positive and negative control in the system. Yeast transformation, growth conditions, and assays for β-galactosidase activity were performed according to the manufacturer's instructions (Dualsystems Biotech).
Bimolecular Fluorescence Complementation (BiFC) Analysis
YN (amino acid 1–154 of YFP) and YC (amino acid 155–238 of YFP) fragments of YFP were amplified by PCR using the pEYFP-C1 vector as a template. The following primers were used: YN forward primer 5′-TAT AAC CGG TCA TGG TGA GCA AGG GCC AAC GTC TAT ATC ATG GCC-3′ and reverse primer 5′-TAT ATG TAC AAG GCC ATG ATA TAG ACG TTG-3′; YC forward primer 5′-TAT AAC CGG TCG ACA AGC AGA AGA ACG GC-3′ and reverse primer 5′-TAT ATG TAC AAC TTG TAC AGC TCG TCC ATG-3′. An Age I site at the 5′-end and a Bsr GI site at the 3′-end were introduced with the primers. This allowed exchange of full-length EGFP in pEGFP-PTP1B and pEGFP-CNX with the YC and YN fragments to generate YC-PTP1B and YN-CNX plasmids. cDNA encoding the PTP1B deletion mutant missing the last 35 amino acid ER targeting domain (YC-PTP1BΔ35) was synthesized by PCR-driven amplification of YC-PTP1B plasmid DNA using the following primers: forward primer 5′-ATG CGA ATT CGA TGG AGA TGG AGA AG-3′ and reverse primer 5′-ATG CGT CGA CTT CCT CCT CAG TGG AG-3′. This generated PTP1B with C-terminal 35 amino acid deletion in the YC vector. NIH3T3 cells were transfected with 1.0 μg of BiFC plasmids (NeonTM electroporation, Invitrogen) and treated with either scrambled siRNA or UBC9 siRNA (Dharmacon RNAi Technologies). BiFC (YFP, Ex514/Em527 nm) signal was detected using confocal microscopy (Leica TCS SP5). As a negative control for BiFC, YN-CNX with YC (or YC-PTP1B with YN) was co-transfected in NIH3T3 cells. DAPI (4′,6-diamidino-2-phenylindole) was used as a stain for the nucleus.
Expression and Purification of Recombinant Proteins and in Vitro SUMOylation
pBAD-CNX (encoding ER luminal domain of calnexin) and pBAD-CNX-C (encoding the C-tail of calnexin) vectors were used for expression of calnexin in TOP 10 F′ Escherichia coli. Top 10 F′ recombinant protein expression was induced with l-arabinose followed by purification of the His-tagged proteins with a Ni2+-nitriloacetic acid-agarose affinity column, concentrated by a centrifugal filter, and dissolved in a buffer containing 10 mm Tris, pH 7.0 and 1 mm EDTA. For NMR spectroscopy, expression of uniformly 15N-labeled C-tail of calnexin was performed in minimal medium supplemented with 15NH4Cl. Recombinant human UBC9 encoded by the pET28b vector was expressed in BL21 E. coli cells and purified as previously described (20). Purified C-tail protein and the C-tail SUMOylation mutant were used with an in vitro SUMOylation kit (Vaxron) as per the manufacturer's protocol.
NMR Spectroscopy
All of the NMR experiments were performed on a Varian INOVA 600 MHz NMR spectrometer using Biopack pulse sequences. The NMR samples were prepared in 90% H2O/10% D2O, 100 mm KCl, 10 mm imidazole, 0.25 mm of deuterated 2,2-dimethyl-2-silapentane-5-sulfonic acid (DSS-d6) for chemical shift reference and a pH of 6.8 according to imidazole H2 proton chemical shifts. The spectra were all processed using standard nmrPipe processing (21); prior to the Fourier transform, a sinebell function of 60 to 90° offsets was applied, and then zero filling was used to extend a maximum of twice the number of real points without using any linear prediction. The two-dimensional spectra were visualized and analyzed using NMRViewJ (One Moon Scientific Inc.).
Transfection and siRNA
Mouse fibroblasts were derived from 7-day-old wild-type and calnexin-deficient mice, respectively (22). Cells were immortalized as described previously (23) and maintained in Dulbecco's Modified Eagle's Medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C with 5% CO2. The Neon Transfection System (Invitrogen) was used for electroporation of cells as per the manufacturer's protocol with the following settings: wild-type and cnx−/− mouse fibroblasts were electroporated with a pulse voltage of 1500 V, a pulse width of 20 ms, and a pulse number of 1; HeLa cells were electroporated with a pulse voltage of 1005 V, a pulse width of 35 ms, and a pulse number of 2; and NIH 3T3 cells were electroporated with a pulse voltage of 1200 V, a pulse width of 20 ms, and a pulse number of 3.
For knockdown of UBC9 expression, NIH3T3 cells were transfected with siGENOME SMARTpool Mouse UBE2I or a negative scrambled siRNA (Thermo Scientific) using DharmaFECT 1 transfection reagent (Thermo Scientific). Co-transfection of YN-CNX with YC fusion protein (or YC-PTP1B with YN fusion protein) was used as a negative control. EGFP fluorescence and BiFC signals were detected by confocal laser scanning microscopy (Leica TCS SP5).
Isolation of Microsomal ER Membranes
Microsomes were isolated from HeLa cells grown to 80–90% confluency. A protease inhibitor mixture (0.5 mm PMSF, 0.5 mm benzamidine, 0.05 μg/ml aproprotin, 0.025 μg/ml phosphormidone, 0.05 μg/ml TLCK, 0.05 μg/ml APMSF, 0.05 μg/ml E-64, 0.025 μg/ml leupeptin, and 0.01 μg/ml pepstatin) (24) and de-SUMOylation inhibitor mixture (800 μm N-ethyl maleimide (NEM) and 200 μm iodoacetamide (IAA)) were included in all buffers. Cells were harvested, spun at 1,000 × g for 3 min at 4 °C, resuspended in 20 ml of PBS followed by transfer to LIS Buffer containing 10 mm Tris-HCl, pH 7.5, and 0.5 mm MgCl2 for 10 min. Cells were homogenized with a Dounce homogenizer in a buffer containing 0.5 m sucrose, 10 mm Tris-HCl pH 7.5, 40 μm CaCl2, 300 mm KCl, and 6.3 mm β-mercaptoethanol. The solution was spun at 8,000 × g followed by centrifugation at 120,000 × g for 1 h at 4 °C. The pellet containing the microsomal fraction was washed and resuspended in a buffer containing 5 mm Tris-HCl, pH 7.5, 20 μm CaCl2, and 150 mm KCl.
Immunoprecipitation and Western Blot Analysis
Cells at 80% confluency were lysed in a buffer containing 10 mm tris pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 1 mm EDTA pH 8.0, and 10% glycerol, with the protease/deSUMOylation inhibitor mixture or in RIPA buffer containing 50 mm Tris, pH 7.0, 150 mm NaCl, 1 mm EGTA, 1 mm EDTA, 1% Triton X-100, 0.1% SDS, and 0.5% sodium deoxycholate with the protease/deSUMOylation inhibitor mixture. After 30 min of incubation on ice, the lysed cells were spun at 15,000 × g for 15 min. at 4 °C. The supernatant was pre-cleared with protein A or a 1:1 protein A/protein G bead slurry for 1 h at 4 °C with rotation after which the primary antibodies were added to the supernatant for overnight incubation at 4 °C with rotation. Protein A or a 1:1 protein A/protein G bead slurry (10%) was added for 4–5 h at 4 °C with rotation. The beads were spun down, washed 5 times with lysis buffer, and resuspended in SDS-PAGE sample buffer containing 10% glycerol, 1% β-mercaptoethanol, 2% SDS, 65 mm Tris, pH 6.8, and 0.01% w/v bromphenol blue for analysis. Proteins were separated on 6% or 10% acrylamide SDS-PAGE gels and transferred onto nitrocellulose for Western blot analysis. Two polyclonal rabbit anti-calnexin antibodies were used: SPA-860 (Enzo Life Sciences) raised against a synthetic peptide corresponding to the C terminus of calnexin (amino acid residues 575–593) and SPA-865 (Enzo Life Sciences) raised against a synthetic peptide near the N terminus. Antibodies were used at a 1:1,000 and 1:500 dilution, respectively. Other antibodies used included anti-UBC9 (Abgent), 1:200 dilution; anti-GFP (Abcam), 1:10,000 dilution; β-tubulin, (Abcam), 1:500 dilution; anti-PTP1B (Abcam), 1:100,000 dilution; and anti-GAPDH (Abcam), 1:1,000 dilution.
Subcellular Fractionation
Fractionation was carried out using the Opti-prep (Sigma) system. Wild-type mouse fibroblasts were washed twice with PBS and resuspended in homogenization buffer (10 mm Hepes-NaOH pH 7.4, 250 mm sucrose, 1 mm EDTA, 1 mm EGTA). Cells were lysed via 25 passes through an 18 micron clearance ball bearing homogenizer (Isobiotec), then centrifuged at 800 × g for 10 min at 4 °C to remove nuclei and cellular debris. The homogenate supernatant was layered onto an eight-step gradient of 25, 22, 19, 16, 13, 10, 7, and 4% (% iodixanol) dilutions of Opti-prep in homogenization buffer. The gradient was centrifuged in a Beckman Sw50.1 rotor for 6 h at 4 °C at 150,000 × g. Twelve fractions were harvested from the top of the gradient, followed by acetone precipitation of proteins. Protein pellets were washed in 100% ethanol, then resuspended in sample buffer, separated on SDS-PAGE followed by Western blot analysis. Antibodies used included GAPDH (Pierce), 1:5,000; GFP (Abcam), 1:10,000; Lamp2b (Abcam), 1:1,000; mannosidase II (Santa Cruz Biotechnology), 1:1,000; and ribophorin I (Santa Cruz Biotechnology), 1:400.
FLT-3 Phosphorylation Assay
Expression constructs for calnexin and calnexin K506A mutant, UBC9 and pcDNA3.1 empty vector were used for calnexin rescue expression cell assay. Calnexin-deficient cells were plated at 1 × 106 cells per 6-well plate 20 h prior transfection. Cells were transfected with expression vector (3.75 μg) encoding calnexin or calnexin K506A mutant along or with vector encoding UBC9 or empty vector (3.75 μg) using Lipofectamine 2000 according to the manufacturer's recommendations. Following transfection, cells were grown in a medium containing 10% FBS. Cells were harvested in RIPA containing complete mixture of protease inhibitors. Polyclonal antibodies against phospho-FLT3 (Tyr591) and FLT3 were purchased from Cell Signaling Technology. PTP1B polyclonal antibodies were purchased from Millipore and actin polyclonal antibodies from Sigma Aldrich. Calnexin antibodies were a kind gift from Dr. John J.M. Bergeron (McGill University).
Miscellaneous
Protein concentration was estimated using a Bio-Rad DC Protein assay (25).
RESULTS
UBC9 Binds to the Calnexin Cytoplasmic Tail
We employed yeast-2-hybrid techniques to identify proteins that interact with the cytoplasmic tail (C-tail) domain (Fig. 1A) of calnexin (26). Using the 88 amino acid mus musculus calnexin C-tail (Cys484-Glu571) as bait, we screened a mouse brain cDNA library and identified nine positive clones (26), one of which encoded UBC9. A second, independent yeast-2-hybrid screen of a human T cell leukemia (Jurkat) library with canine calnexin C-tail (Cys485-Glu573) as bait identified ten putative clones, and also included UBC9.
To further examine the UBC9/calnexin C-tail interaction, we used immunoprecipitation techniques (Fig. 1B) and yeast 2-hybrid analysis (Fig. 2A). Calnexin could be efficiently immunoprecipitated with anti-calnexin antibodies from wild-type cells but not from calnexin-deficient cells (Fig. 1B). Western blot analysis with antibodies against both calnexin and UBC9 revealed that calnexin co-immunoprecipitated with UBC9 in wild-type fibroblasts (Fig. 1B), indicating that UBC9 was bound to calnexin.
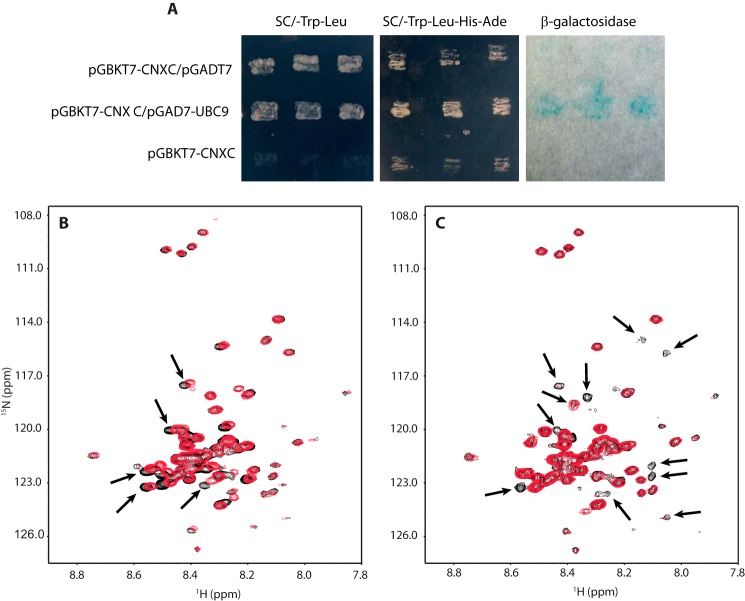
FIGURE 2.
The C-tail interacts with UBC9 in yeast 2-hybrid system, lacks structure and binds UBC9 in a Ca2+-independent manner and. A, yeast 2-hybrid analysis of interaction between calnexin and UBC9. First row: yeast AH109 strain transformed with calnexin C-tail cloned into pGBKT7 vector (pGBKT7-CNXC) and blank pGADT7. Calnexin C-tail did not interact under these conditions and consequently yeast cells did not grow on the SC/-Leu/-Trp/-His/-Ade plate and no activity of β-galactosidase was observed. Second row: yeast AH109 strain transformed with pGBKT7-CNXC and UBC9 cloned in to pGADT7 vector (pGADT7-UBC9). Yeast cells did grow in the SC/-Leu/-Trp/-His/-Ade media with activated β-galactosidase indicating the two molecules interacted. Third row: yeast AH109 strain transformed only with pGBKT7-CNXC did not on SC/-Leu/-Trp plate indicative of no interaction between calnexin C-tail alone under these experimental conditions. B, two-dimensional 1H-15N HSQC NMR spectra of the C-tail of calnexin in the apo (black) and Ca2+ (red) states. The small chemical shift dispersion indicates that the C-tail lacks nascent structure. Small chemical shift changes were observed for some amide NH cross-peaks (indicated by black arrows) after addition of 6 mm CaCl2 suggesting only a weak Ca2+ interaction. C, two-dimensional 1H-15N HSQC NMR spectra of the 15N-labeled C-tail of calnexin before (black) and after addition of unlabeled UBC9 (red). Approximately ten to twelve resonances in calnexin (indicated by black arrows) showed significant chemical shift broadening or disappeared with the addition of UBC9. This suggests that these residues were in conformational exchange, created by the binding of UBC9 to calnexin.
Next, we performed subcellular fractionation using Opti-prep gradients to assess the amount of UBC9 that co-distributed with calnexin (Fig. 1C). As expected, UBC9 was found in both the cytoplasmic fractions (using glyceraldehyde 3-phosphodehydrogenase (GAPDH) as a cytoplasmic marker) but a significant amount of the protein (>20%; Fig. 1D) was associated with ER fractions (Fig. 1C, fractions 7–9), along with calnexin and the ER marker ribophorin I (Fig. 1C), suggesting that UBC9 co-fractionated with ER fractions. We also probed for other organelle markers, including mannosidase II (Golgi) and Lamp2b (lysosomes). Finally, we used confocal microscopy techniques to investigate the co-distribution of UBC9 with calnexin. UBC9 and calnexin immunostaining were found in the nuclear envelope and at the ER (Fig. 1E). Quantitative analysis of confocal images revealed an average threshold Pearson's co-localization coefficient of 0.558 ± 0.060, indicating some but not complete co-localization of UBC9 and calnexin at the ER, consistent with UBC9 not being restricted to the ER membrane but also being localized to the nuclear membrane and throughout the cytoplasm (Fig. 1E).
The Calnexin C-tail Lacks Structure and Binds UBC9 in a Ca2+-independent Manner
To date, only the structure of the luminal domain of calnexin has been solved (27) and there is no structural information available about the calnexin C-tail. To understand the nature of the C-tail/UBC9 interaction, we employed NMR spectroscopy to examine the structural and biophysical properties of the C-tail alone and with UBC9, in the presence and absence of Ca2+ (28). The small chemical shift dispersion observed for resonances in both the amide and aliphatic regions of the one-dimensional 1H NMR spectrum, combined with the few number of NOE cross-peaks observable in a two-dimensional 1H-1H NOESY NMR spectrum, indicated that the C-tail was mostly unfolded (data not shown). The two-dimensional 1H-15N HSQC NMR spectrum of [U-15N]C-tail also showed little chemical shift dispersion for the amide NH resonances (Fig. 2B, black), confirming the absence of structure, and showed small chemical shift changes after the addition of excess Ca2+ (Fig. 2B, red), indicating weak Ca2+ binding. These results are in agreement with earlier studies on Ca2+ binding to the C-tail using Escherichia coli-expressed GST fusion calnexin fragments (29).
To further examine the nature of the C-tail/UBC9 interaction, we looked at the two-dimensional 1H-15N HSQC spectra of [U-15N]C-tail and monitored the effect of adding unlabeled UBC9. In the absence of Ca2+, the HSQC of [U-15N]C-tail (Fig. 2C, black) showed approximately ten to twelve resonances which underwent strong chemical shift broadening or disappearance after the addition of UBC9 (Fig. 2C, red). This suggested that these residues were in conformational exchange, created by the binding of UBC9 to calnexin. Several cross-peaks remained unaffected with the addition of UBC9, which was not surprising considering the size of the C-tail of calnexin. Most of the unperturbed resonances came from the resonances with the highest intensities and smaller chemical shift dispersion, suggesting that the N and C termini of calnexin most likely remained labile in the bound form. two-dimensional 1H-15N HSQC NMR spectra were acquired after adding 6 mm Ca2+ to investigate the Ca2+ requirement for UBC9 binding. Negligible changes were observed, indicating that Ca2+ saturation was not required for UBC9 interaction with the C-tail.
The C-tail of Calnexin is SUMOylated
UBC9 is a SUMO-conjugating enzyme that plays an important role in the SUMOylation of specific target proteins (30). SUMO conjugation modifies a lysine residue, typically within the core consensus motif ψKxE where ψ is a bulky hydrophobic residue (31). In addition to the core consensus motif, additional flanking amino acid sequences have been proposed to extend the SUMO consensus motif (32, 33). These include the phosphorylation-dependent SUMOylation motif (PDSM) of ψKxExxSP (32), and the negatively charged amino acid-dependent SUMOylation motif (NDSM) (33). A global search for NDSM-containing proteins in the SWISSPROT database identified that the highly acidic calnexin C-tail contains a NDSM (amino acid residues 505–518), VK506EEEEEKEEEKDK (Fig. 1A), a motif previously used successfully in determining in vivo SUMO substrates (34). To examine if the C-tail of calnexin can be a substrate for SUMOylation at this site, we used an in vitro SUMOylation system with purified canine calnexin C-tail and a K506A C-tail mutant. In the presence of SUMO-1, there was a shift in the C-tail mobility of ∼12-kDa as detected with anti-calnexin antibodies (Fig. 3A), a size shift consistent with the addition of SUMO-1. Mutation of Lys506 to an alanine abrogated in vitro SUMOylation of the C-tail (Fig. 3A), indicating that the predicted NDSM motif in the calnexin C-tail was the site of SUMO addition. Mutation of Lys534 or Lys554, two other potential SUMOylation sites in calnexin C-tail, did not eliminate in vitro SUMOylation of the C-tail (Fig. 3A). We concluded that the calnexin C-tail can be SUMOylated in vitro at Lys506 within the predicted SUMOylation NDSM consensus site.
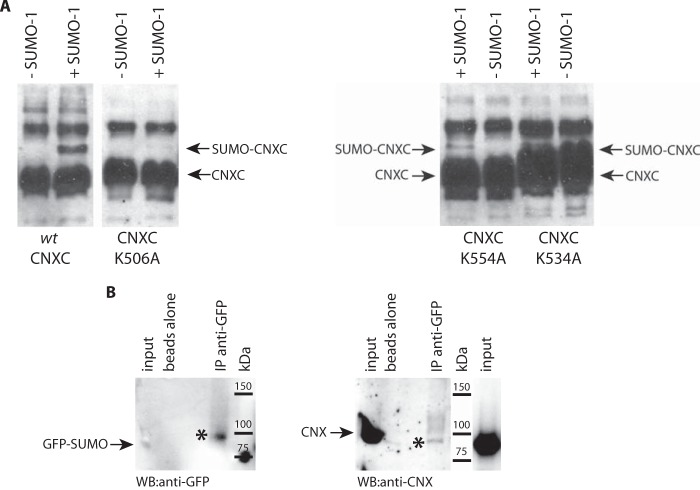
FIGURE 3.
Calnexin is SUMOylated in vitro and in vivo. A, purified recombinant calnexin C-tail (wt-CNXC) or Lys506 to Ala calnexin C-tail mutant (CNXC-K506A) (left panel) or Lys534 or Lys554 to Ala calnexin C-tail mutants (CNXC-K534A and CNXC-K554A, respectively) (right panel) were SUMOylated followed by Western blot analysis with anti-calnexin antibodies. The location of the recombinant calnexin C-tail (CNXC) and SUMO-conjugated calnexin C-tail (SUMO-CNXC) is indicated by the arrows. The location of the recombinant calnexin C-tail (CNXC) and SUMO-conjugated calnexin C-tail (SUMO-CNXC) is indicated by the arrows. The data represent more than three biological replicates. B, ER-enriched microsomes were isolated from HeLa cells expressing a GFP-SUMO-1 followed by immunoprecipitation with goat anti-GFP antibodies and Western blot analysis with anti-GFP antibodies (left panel) and anti-calnexin antibodies (right panel). The arrows indicate the location of GFP-SUMO-1 and calnexin. The asterisks indicate that GFP-SUMO-1 and calnexin co-immunoprecipitated. IP, immunoprecipitation. The data represent more than three biological replicates.
Next we examined if calnexin can be SUMOylated in vivo. The SUMO enigma is a well-documented phenomenon, where only a small portion of the total substrate is detectably SUMOylated at any given time, even when functional evidence indicates complete SUMOylation (35). To increase the probability of detecting SUMOylated calnexin (and the corresponding molecular shift) in vivo, we isolated ER microsomes from cultured cells and tested them for the presence of SUMOylated endogenous calnexin. HeLa cells were transfected with an expression vector encoding GFP-SUMO-1 followed by the isolation of ER enriched microsomes. Anti-GFP antibodies were used to pull down GFP-SUMO-1 followed by Western blot analysis with anti-GFP antibodies (to report the presence of GFP-SUMO-1) and anti-calnexin antibodies (to identify calnexin protein). Immunoprecipitation of solubilized microsomes with goat anti-GFP antibodies and immunoblotting with rabbit anti-GFP antibodies indicated that a GFP-SUMO-1 modified protein at ∼100-kDa was immunoprecipitated with anti-GFP antibodies (Fig. 3B). Western blot analysis of the same sample with anti-calnexin antibodies revealed that calnexin (detectable at ∼100-kDa) was co-immunoprecipitated with GFP-SUMO-1 from the microsomal fraction (Fig. 3B). We concluded that calnexin was SUMOylated in vivo.
PTP1B Forms Complexes with Calnexin
PTP1B is the first protein to be identified as an ER-associated SUMO modified protein (36). Since we showed here that calnexin interacted with UBC9 and was SUMOylated, we hypothesized that there may be an association between PTP1B and calnexin at the ER membrane. To test this, we carried out immunoprecipitation experiments. PTP1B was immunoprecipitated from both wild-type and calnexin-deficient fibroblasts (Fig. 4A). Next, we tested for the presence of calnexin in PTP1B immunoprecipitated complexes using anti-calnexin antibodies (Fig. 4B). Western blot analysis of immunoprecipitates from wild-type cells and cnx−/− cells (control) revealed that calnexin co-immunoprecipitated with PTP1B from wild-type cells (Fig. 4B), indicating that the two proteins formed complexes. To examine if calnexin and PTP1B interact in situ, we used two independent methods: the DUAL membrane yeast-2-hybrid method (Fig. 4C) and BiFC (Fig. 4D). For DUAL membrane yeast-2-hybrid, full-length calnexin and PTP1B were constructed into the pBT3-SUC and pPR3 vector, respectively, and co-transformed into the yeast strain NYM51. Co-transformation of calnexin and PTP1B expression vectors demonstrated comparable growth to the CNX-pBT3/Nub-Ostl positive control, as well as positive β-galactosidase activity, indicating PTP1B and calnexin formed complexes under these conditions (Fig. 4C). Next, we used BiFC to examine the in situ interactions between PTP1B and calnexin in mammalian cells (Fig. 4D). Both GFP-calnexin and GFP-PTP1B fusion proteins localized to an ER-like intracellular network (Fig. 4D). For BiFC, we constructed vectors encoding calnexin as a fusion protein with the N-terminal portion of YFP (YN-CNX) and PTP1B as a fusion protein with the C-terminal portion of YFP (YC-PTP1B). Co-transfection of the YN-CNX and YC-PTP1B constructs resulted in a robust BiFC signal (Fig. 4D), indicative of an in situ interaction between calnexin and PTP1B. Therefore, based on immunoprecipitation, yeast-2-hybrid, and BiFC techniques, we concluded that calnexin and PTP1B interacted.
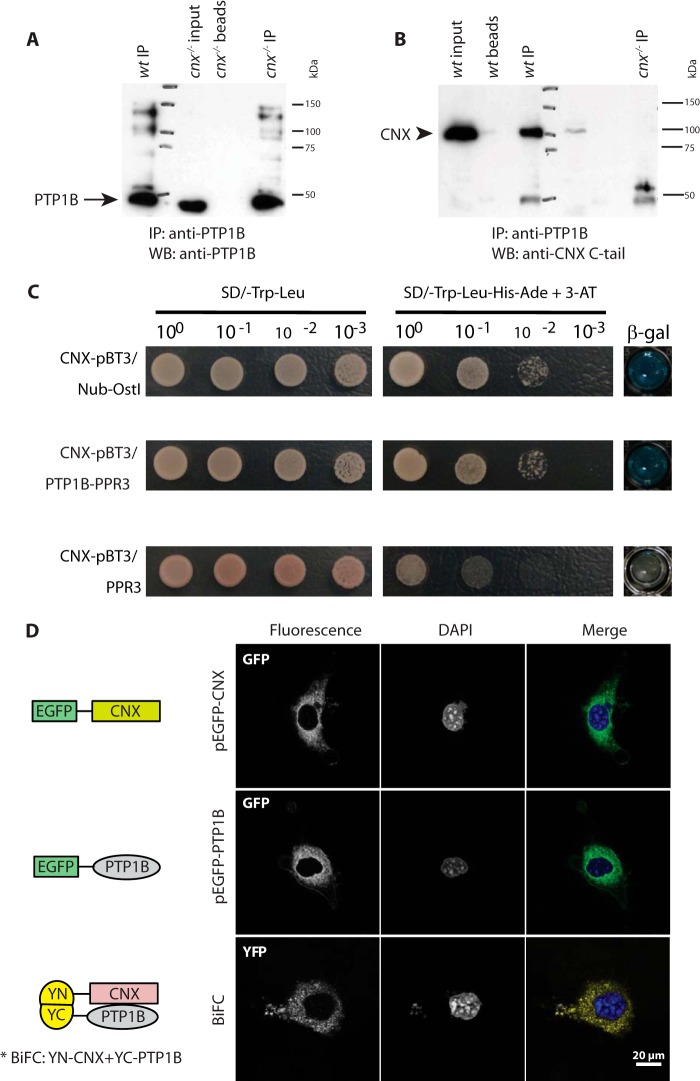
FIGURE 4.
Calnexin and PTP1B form complexes. A, immunoprecipitation was carried out on wild-type (wt) and calnexin-deficient (cnx−/−) fibroblasts with anti-PTP1B antibodies followed by Western blot analysis with anti-PTP1B antibodies. The higher molecular weight banding pattern observed is consistent with SUMO-modification of PTP1B. The arrow indicates the location of the PTP1B protein band. The data represents more than three biological replicates. IP, immunoprecipitation; WB, Western blot. B, immunoprecipitation was carried out with anti-PTP1B antibodies followed by Western blot analysis with anti-calnexin C-tail antibodies. The arrowhead indicates the location of calnexin. The data represent more than three biological replicates. C, DUAL membrane yeast-2-hybrid interaction method was used to determine if PTP1B and CNX interact in situ. Full-length calnexin and PTP1B were cloned into the pBT3-SUC and pPR3 vector, respectively, and co-transformed into the yeast strain NYM51. A positive interaction was determined by β-galactosidase activity. Co-transformation of calnexin and PTP1B demonstrated comparable growth to the CNX-pBT3/Nub-Ostl positive control as well as positive β-galactosidase activity indicating PTP1B and calnexin interact. No β-galactosidase (β-gal) activity was observed with the negative control CNX-pBT3/PPR3. The data represent more than three biological replicates. D, GFP-tagged calnexin (upper panel) and PTP1B (middle panel) are localized to the ER-like network. BiFC analysis of binding between calnexin and PTP1B in NIH3T3 cells (bottom panel). Calnexin was expressed as fusion protein with the C-terminal fragment of YFP (YN-CNX), whereas PTP1B was expressed as a fusion with the C-terminal portion of YFP (YC-PTP1B). Schematic diagrams of each of the constructs are shown in the figure. The data represent more than three biological replicates.
PTP1B Association with Calnexin Is UBC9-dependent
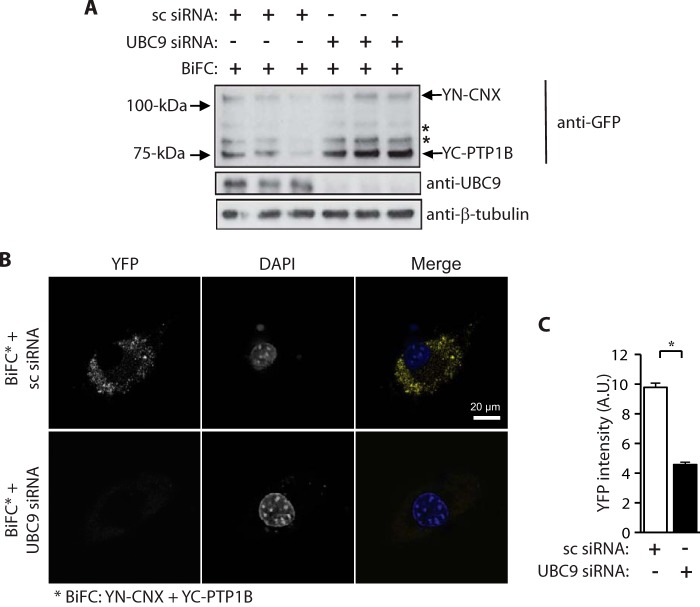
Considering that calnexin is an integral ER membrane protein, while PTP1B is a cytoplasmic protein, we envisaged that the PTP1B-calnexin interaction was most likely to occur on the cytoplasmic face of the ER and thus involve the cytoplasmic C-tail of calnexin. Since calnexin and PTP1B are SUMOylated proteins and the calnexin C-tail interacted with UBC9, we asked if the interaction between calnexin and PTP1B was dependent on UBC9 and the SUMOylation machinery. NIH3T3 cells treated with siRNA for UBC9 were transfected with YN-CNX and YC-PTP1B expression vectors followed by confocal microscope analysis. siRNA treatment reduced the abundance of UBC9 by over 90% and did not affect abundance of YN-CNX or YC-PTP1B in NIH3T3 cells (Fig. 5A). Importantly, the BiFC signal, reporting PTP1B binding to calnexin, was significantly reduced in UBC9 knockdown cells when compared with cells treated with control, scrambled siRNA (Fig. 5B). Quantitative analysis of confocal images revealed an average intensity of fluorescence with scrambled siRNA: 9.77 ± 0.3 and with UBC9 siRNA: 4.57 ± 0.18, p < 0.05 (Fig. 5C), indicating a reduction of the signal in cells with silenced UBC9. Taken together these observations indicate that silencing UBC9 expression suppressed the interaction between calnexin and PTP1B.
FIGURE 5.
Silencing of UBC9 affects interactions between calnexin and PTP1B. A, NIH3T3 cells were treated with scrambled siRNA (sc siRNA) or UBC9 specific siRNA. Cells were also transfected with YN-CNX and YC-PTP1B expression vectors for BiFC analysis. Western blot analysis was carried out with anti-GFP, anti-UBC9, and anti-β-tubulin antibodies. β-tubulin was used as a loading control. * indicates the location of nonspecific protein bands. The location of the YN-calnexin and YC-PTP1B is indicated by the arrows. The data represent more than three biological replicates. B, BiFC in NIH3T3 cells expressing YN-calnexin and YC-PTP1B fusion proteins with silenced UBC9. sc siRNA, scrambled small interference RNA; siRNA, small interference RNA; The data represent more than three biological replicates. C, quantitative analysis of BiFC (YFP) signals. The data represent more than three biological replicates. *, p < 0.05.
PTP1B Association with Calnexin Is Independent of the PTP1B ER Localization Domain
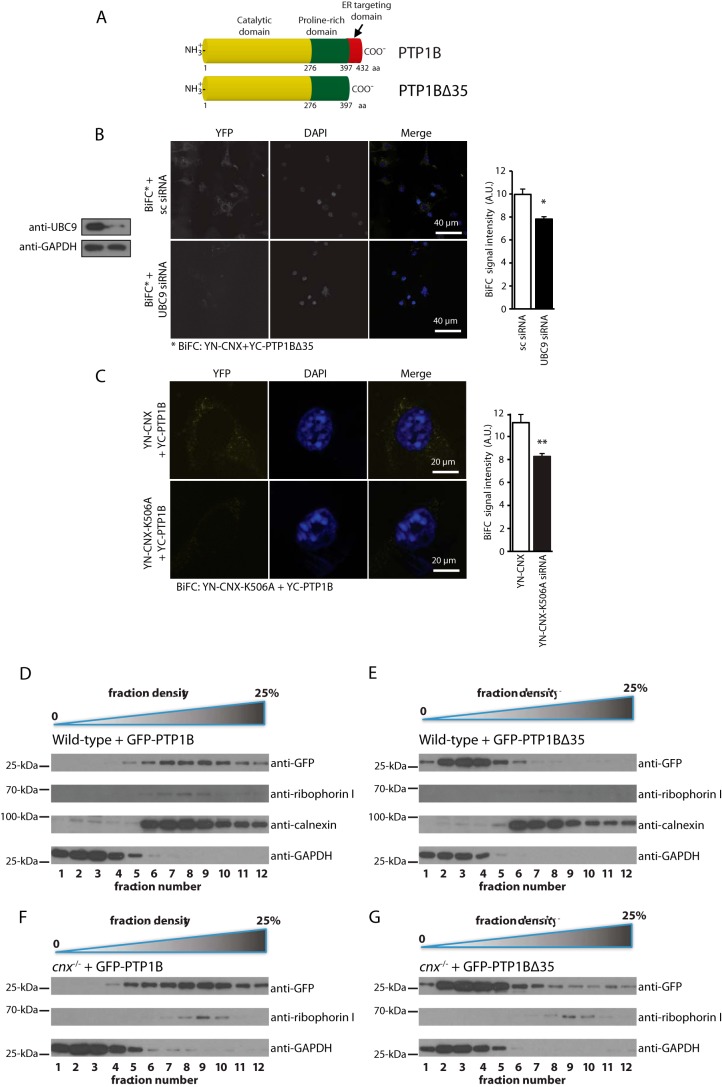
PTP1B consists of multiple functional domains including an N-terminal catalytic domain followed by a proline-rich domain and an ER localization domain (35 amino acids at the C terminus) (Fig. 6A) that is believed to anchor PTP1B to the cytoplasmic face of the ER (14, 17). To test whether PTP1B association with calnexin involved the 35 amino acid C-terminal ER targeting domain, we created an expression vector encoding full-length or truncated PTP1B missing the last 35 amino acid residues as a fusion protein with the C-terminal portion of YFP (designated YC-PTP1BΔ35). There was a strong BiFC signal in cells expressing YN-CNX and YC-PTP1BΔ35 (Fig. 6B) indicating that PTP1B and calnexin formed complexes even in the absence of the PTP1B ER targeting sequence (14). Next, we silenced UBC9 with specific siRNA followed by BiFC analysis of cells expressing YN-CNX and YC-PTP1BΔ35. Knock-down of UBC9 resulted in a significant reduction of the BiFC signal (BiFC with scrambled siRNA: 9.96 ± 0.47; BiFC with UBC9 siRNA: 7.82 ± 0.20, p < 0.05) (Fig. 6B), indicating UBC9 influenced interactions between calnexin and PTP1B.
FIGURE 6.
Interaction between calnexin and PTP1B is independent of the PTP1B 35 amino acid ER-targeting sequence. A, schematic representation of PTP1B and truncated PTP1B (PTP1BΔ35). Mouse PTP1B (NP_035331.3) consists of 432 amino acids and is composed of a catalytic phosphatase domain (yellow), a proline-rich domain (green), and a 35 amino acid C-terminal ER targeting domain (red). B, BiFC analysis was carried out in NIH3T3 cells expressing an YC-calnexin fusion protein (YN-CNX) and a PTP1B deletion mutant missing its 35 amino acid residue ER targeting domain (YC-PTP1BΔ35). DAPI was used as a nuclear stain. Cells were treated with scrambled siRNA (sc siRNA) or with UBC9 specific siRNA. The graph shows a quantitative analysis of the BiFC signal. The data represent more than three biological replicates. * p < 0.05. C, BiFC analysis in NIH3T3 cells expressing YC-calnexin fusion protein (YN-CNX) or YN-calnexin K506A mutant (YN-CNX-K506A) and YC-PTP1B. DAPI was used as a nuclear stain. The graph shows a quantitative analysis of the BiFC signal. The data represent more than three biological replicates. **, p < 0.01. D–G, Opti-prep gradient fractionation of wild-type (D, E) or cnx−/− (F, G) mouse fibroblast expressing GFP-PTP1B (D, F) or GFP-PTP1BΔ35 (E, G). Lower density fractions (1–4) correspond to cytoplasmic components, while higher density fractions (5–12) correspond to heavier cellular compartments, including the ER. Western blot analysis was carried out with anti-GFP (to visualize the location of GFP-PTP1B or GFP-PTP1BΔ35 protein), anti-calnexin, anti-GAPDH, anti-ribophorin I antibodies.
Finally, we tested if SUMOylation of the calnexin C-tail played a role in the formation of calnexin-PTP1B complexes. The K506A mutant of YN-calnexin, previously shown to be SUMOylation-deficient, was expressed in NIH3T3 cells together with YC-PTP1B followed by BiFC analysis. There was a significant reduction of the BiFC signal between YN-CNX-K506A and YC-PTP1B (BiFC with for YN-CNX: 11.11 ± 65; for YN-CNX-K506A: 8.33 ± 0.24, p < 0.01) (Fig. 6C) indicating that SUMOylated Lys506 in calnexin C-tail was important to support calnexin-PTP1B interactions.
To further investigate the subcellular localization of wild-type PTP1B and PTP1B lacking its ER targeting C terminus (PTP1BΔ35), we transfected wild-type and calnexin-deficient fibroblasts with expression vectors encoding either full-length GFP-tagged PTP1B (PTP1B-GFP) or GFP-tagged truncated PTP1B (GFP-PTP1BΔ35). 48 h post-transfection, we performed subcellular fractionations and Western blot analysis to monitor the localization of PTP1B. As expected, we observed PTP1B-GFP primarily in the same subcellular fractions as ER markers (calnexin, ribophorin I) with a small co-fractionation with a cytoplasmic marker (GAPDH) (Fig. 6D). In comparison, GFP-PTP1B-Δ35 mostly co-fractionated with GAPDH, indicative of a primarily cytoplasmic distribution (Fig. 6E). However, some GFP-PTP1BΔ35 signal was observed in the same fractions as ER markers, suggesting that PTP1B lacking its classical ER targeting sequence still has some propensity to localize to the ER membrane (Fig. 6E). Similar distribution of GFP-PTP1B was seen in fractions derived from calnexin-deficient cells (Fig. 6, F and G). In conjunction with our BiFC data, we therefore concluded that the deletion of the 35 C-terminal residues of PTP1B severely abrogated but did not completely delete the localization of PTP1B to the ER membrane. We concluded that PTP1B formed UBC9-dependent complexes with calnexin independently of the presence of a PTP1B C-terminal ER targeting domain, though this targeting domain did significantly increase the partition of PTP1B into ER-associated fractions.
Calnexin Association with PTP1B Affects Its Activity
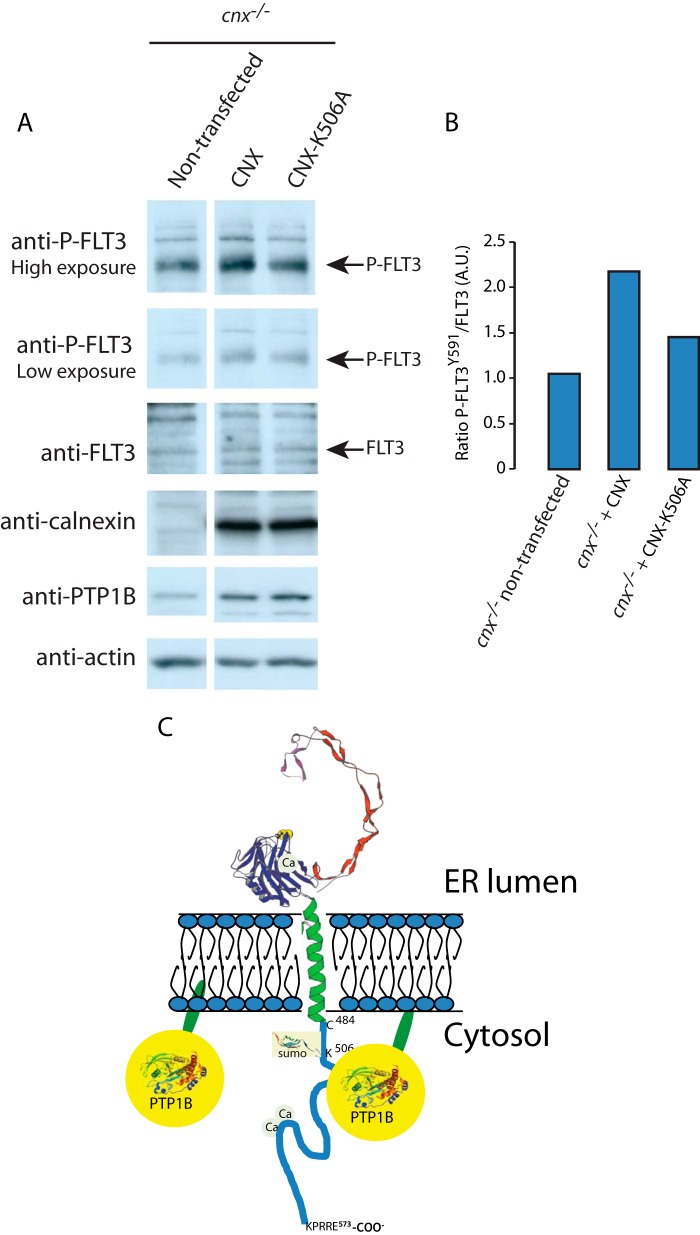
After concluding that the association of calnexin and PTP1B was not likely to serve as a mechanism for mediating the association of PTP1B with ER membranes, we next asked whether calnexin may instead alter the activity of PTP1B. We looked at the phosphorylation of FLT3, a receptor tyrosine kinase that has been shown to be dephosphoryated by PTP1B. Additionally, certain mutants of FLT3 have been shown to associate with calnexin, leading us to hypothesize the existence of a complex containing calnexin, FLT3, and PTP1B (37). We transfected calnexin-deficient cells with vectors encoding either wild-type calnexin or calnexin K506A, which cannot be SUMOylated. We found that in cnx−/− cells expression of wild-type calnexin noticeably increased the levels of phosphorylated FLT3 (P-FLT3) (Fig. 7, A and B), which suggested that PTP1B activity was reduced in the absence of calnexin. Importantly, expression of calnexin K506A mutant was not effective in a full recovery of P-FLT3 levels as compared with cells expression wild-type calnexin (Fig. 7, A and B). As calnexin K506A cannot be SUMOylated and interacts with PTP1B less efficiently, we concluded that calnexin's interaction with PTP1B may act to inhibit its phosphatase activity (at least with respect to FLT3). Further research is needed to determine if calnexin acts to inhibit global PTP1B activity or instead to regulate the substrate specificity of PTP1B.
FIGURE 7.
FLT3 phosphorylation in calnexin rescue cell lines and a proposed model for PTP1B association with ER membrane. A, FLT3 phosphorylation in calnexin rescue cell line. cnx−/− cells were transfected with CNX or calnexin K506A mutant (CNX-K506A) expression vectors. Total lysates from non-transfected and transfected cells were analyzed by immunoblotting for phosphorylated FLT3 (P-FLT3) and membrane were then stripped and re-probed for total FLT3. High and low exposure of blots probed with anti-P-FLT3 antibodies are shown. B, quantitative analysis of the Western blots shown in A. A ration between P-FLT3 to non-phosphorylated FLT3 is presented. A.U., arbitrary units. C, PTP1B can associate with ER membranes via its C-terminal 35 amino acid ER targeting domain (red), and by binding to the calnexin C-tail. The interaction between PTP1B and the calnexin C-tail is independent of the PTP1B ER targeting domain but it is sensitive to the presence of Lys506, an amino acid residue in the calnexin C-tail that can be SUMOylated. 3D structures of the calnexin ER luminal domain, PTP1B and SUMO protein are based on pdb1jhn/pdb, PTN1_Human/pdb, and 1A5R, respectively.
DISCUSSION
Calnexin is a type I integral ER membrane molecular chaperone that, in conjunction with the oxidoreductase ERp57, facilitates protein folding (38–40). Calnexin can be partitioned into distinct functional domains: the ER luminal domain, and the transmembrane and cytoplasmic domain. The ER luminal domain of the protein binds monoglucosylated glycans and consequently plays a role in the folding of virtually all glycosylated proteins including many cell surface proteins, such as receptor tyrosine kinases (4). The C-terminal domain of calnexin (C-tail) extends from the transmembrane α-helix into the cytoplasm. This unique arrangement of calnexin domains across the ER membrane (chaperone domain in the lumen of the ER and the C-tail exposed to the cytoplasm) confers the protein the ability to modulate cellular functions on both sides of the ER membrane (Fig. 7C). Indeed, the C-tail of calnexin has been proposed to play an important role in the regulation of protein-protein interactions via post-translational modification: the C-tail has been shown to be phosphorylated and palmitoylated, both of which promote calnexin interaction with the ribosome-translocon complex (6, 9, 11, 12, 42–45). Phosphorylation of the C-tail may additionally act as a molecular switch regulating calnexin-SERCA2b interactions, thereby coupling Ca2+ signaling with Ca2+-sensitive chaperone functions in the ER (7). Here, we showed a novel post-translational modification of calnexin: namely, its SUMOylation at Lys506. We also discovered that calnexin interacted with PTP1B in a UBC9 and SUMOylation-dependent manner linking ER luminal events with cytoplasmic signaling (Fig. 7C). These findings strongly support an important role for the C-tail of calnexin in maintaining ER and cellular functions.
Our discovery that the calnexin C-tail underwent SUMOylation in vitro and in vivo was consistent with previous large-scale mass spectrometry screens of SUMOylated proteins identifying calnexin as a potential SUMOylated substrate (46, 47). While UBC9 activity and SUMOylation have been extensively studied in the nuclear compartment as a constituent of transcriptional control (48, 49), examples of integral membrane proteins modified by SUMO have recently emerged, revealing a wider range of cellular SUMO substrates. SUMOylation of these proteins regulates their biological functions and/or intracellular trafficking (50, 51). With respect to ER transmembrane proteins, in addition to calnexin (this study), Scs2, an ER membrane protein that regulates phosphatidylinositol synthesis and lipid trafficking (52) and SERCA2a are SUMOylated (53). Here we identified calnexin as a new, ER associated, SUMO substrate. Importantly, SUMOylation of calnexin supports PTP1B binding to calnexin and, consequently, it contributes to the association of the phosphatase with ER membrane. This indicates a new role for the SUMOylation machinery in bringing together two functionally diverse molecules to potentially provide a link between ER chaperone function and energy metabolism.
PTP1B is a prototype non-transmembrane tyrosine phosphatase associated with the cytosolic face of the ER membrane (17). Over 20 years ago, Fragioni et al. identified a 35 amino acid C-terminal hydrophobic “ER targeting” sequence responsible for its association with the ER membrane (14). It has been widely accepted that the protein partitions into the outer leaflet of the ER phospholipid bilayer by direct insertion of the ER targeting C-terminal domain (14). Interestingly, protein-protein interactions have not been shown to play a role in PTP1B association with the ER membrane (14). With respect to the localization of PTP1B, our results were largely in agreement with the current consensus: namely, that PTP1B was associated with the cytoplasmic face of the ER membrane. Further, we confirmed that PTP1B lacking its C-terminal 35 residues had a severely reduced ability to associate with the ER membrane. However, our subcellular fractionation and BiFC experiments both indicated that even without this ER targeting sequence, a small proportion of PTP1B was still able to localize to the ER membrane. Despite forming complexes with calnexin at the ER membrane, we saw no gross differences in the subcellular distribution of PTP1B in the absence of calnexin. Our data therefore did not support a model where the binding of PTP1B to calnexin serves to anchor or recruit PTP1B to the ER membrane. Instead, we favor a model where the interaction between PTP1B and calnexin affects the pool of targets upon which PTP1B may act; calnexin-bound PTP1B preferentially targets a certain subset of proteins, while free PTP1B may have a bias toward a different subset of targets. Indeed, we found that calnexin inhibits the dephosphorylation of the receptor tyrosine kinase FLT3. This suggested that the interaction of calnexin and PTP1B inhibited its phosphatase activity, whether specific to FLT3 or as a more general mechanism to lower of PTP1B activity.
To date, PTP1B has been shown to modify 30 known substrates and has several protein binding partners (54), and undergoes several post-translational modifications including phosphorylation, oxidation, proteolytic cleavage, and SUMOylation (17). There are multiple functions ascribed to PTP1B including its role in insulin and leptin signaling (55, 56) as a part of energy metabolism. Localization of PTP1B at the ER, and its binding to calnexin, might be important for the regulation of insulin receptors during their biosynthesis (57). Calnexin is the obligate molecular chaperone for the insulin receptor, and is involved in the folding efficacy and homodimerization of the receptor (58, 59). The association of PTP1B with calnexin could provide measures during biosynthesis of the insulin receptor to prevent autoactivation. Many PTP1B substrates are membrane-associated molecules that are translated on the rough ER and are also folding substrates for calnexin (4, 54). Furthermore, PTP1B's interaction with calnexin may recruit PTP1B to the ribosome-translocon complex, ensuring proximal access to specific phosphatase targets and preventing autoactivation and aberrant cellular signaling events. PTP1B targets and dephosphorylates receptor tyrosine kinases after endocytosis, as they traffic in close proximity to the ER (15, 41, 60). Recently, we showed that the calnexin C-tail binds Src homology 3-domain growth factor receptor-bound 2-like (Endophilin) interacting protein 1 (SGIP1) and influences clathrin-dependent endocytosis due to modulation of the internalization of the receptor-ligand complexes (26). It is plausible that calnexin's interaction with PTP1B supports a similar function in clathrin-dependent endocytosis, as most of the receptor tyrosine kinases activated at the cell surface maintain an interaction with PTP1B after internalization (15).
In summary, this work demonstrated that the SUMOylation machinery and UBC9 link two ER proteins from divergent pathways, calnexin and PTP1B, in a common pathway that may affect cellular quality control and energy metabolism. SUMOylation is a novel modification of the unique calnexin C-tail that has important implications for regulation of a ubiquitous ER molecular chaperone. In addition, this study demonstrated an interaction between a major metabolic enzyme and a classical ER protein quality control chaperone. As our study linked PTP1B to calnexin as a major quality control chaperone of the secretory pathway, it is possible that the mediation of the interaction between PTP1B and calnexin by the SUMOylation machinery serves as an additional cellular measure of quality control.
Acknowledgments
We thank Robert Evans for the initial yeast-2 hybrid screen. We thank M. Dabrowska and A. Robinson for superb technical support. We thank Gianni Del Sal for the GFP-SUMO-1 expression vector and Christopher Lima for the UBC9 expression plasmid. We thank Dr. John J.M. Bergeron for a kind gift of anti-calnexin antibodies.
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) (to M. M. MOP15291; to M. L. T. MOP62887; to B. D. S. MOP37769; to N. T. MOP93659), from the Canadian Foundation for Innovation (CFI-17625) and the Natural Sciences and Engineering Research Council (NSERC) Discovery Research grants (to N. T.). A. K. was supported by a fellowship from Alberta Innovates-Health Solutions (AI-HS) and from the Multiple Sclerosis Society of Canada.
- ER
- endoplasmic reticulum
- BiFC
- bimolecular fluorescence complementation
- FTL3
- Fms-like tyrosine kinase 3
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- PTP1B
- protein tyrosine phosphatase 1
- UBC9
- ubiquitin-conjugating enzyme 9
- CNX
- calnexin
- SUMO
- small ubiquitin-like modifier.
REFERENCES
- 1. Stutzmann G. E., Mattson M. P. (2011) Endoplasmic reticulum Ca2+ handling in excitable cells in health and disease. Pharmacol. Rev. 63, 700–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Benyair R., Ron E., Lederkremer G. Z. (2011) Protein quality control, retention, and degradation at the endoplasmic reticulum. Int. Rev. Cell Mol. Biol. 292, 197–280 [DOI] [PubMed] [Google Scholar]
- 3. Fu S., Watkins S. M., Hotamisligil G. S. (2012) The role of endoplasmic reticulum in hepatic lipid homeostasis and stress signaling. Cell Metab. 15, 623–634 [DOI] [PubMed] [Google Scholar]
- 4. Hebert D. N., Molinari M. (2007) In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol. Rev. 87, 1377–1408 [DOI] [PubMed] [Google Scholar]
- 5. Ho S. C., Rajagopalan S., Chaudhuri S., Shieh C. C., Brenner M. B., Pillai S. (1999) Membrane anchoring of calnexin facilitates its interaction with its targets. Mol. Immunol. 36, 1–12 [DOI] [PubMed] [Google Scholar]
- 6. Chevet E., Smirle J., Cameron P. H., Thomas D. Y., Bergeron J. J. (2010) Calnexin phosphorylation: linking cytoplasmic signalling to endoplasmic reticulum lumenal functions. Sem. Cell Dev. Biol. 21, 486–490 [DOI] [PubMed] [Google Scholar]
- 7. Roderick H. L., Lechleiter J. D., Camacho P. (2000) Cytosolic phosphorylation of calnexin controls intracellular Ca2+ oscillations via an interaction with SERCA2b. J. Cell Biol. 149, 1235–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Myhill N., Lynes E. M., Nanji J. A., Blagoveshchenskaya A. D., Fei H., Carmine Simmen K., Cooper T. J., Thomas G., Simmen T. (2008) The subcellular distribution of calnexin is mediated by PACS-2. Mol. Biol. Cell 19, 2777–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cameron P. H., Chevet E., Pluquet O., Thomas D. Y., Bergeron J. J. (2009) Calnexin phosphorylation attenuates the release of partially misfolded α1-antitrypsin to the secretory pathway. J. Biol. Chem. 284, 34570–34579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lynes E. M., Bui M., Yap M. C., Benson M. D., Schneider B., Ellgaard L., Berthiaume L. G., Simmen T. (2012) Palmitoylated TMX and calnexin target to the mitochondria-associated membrane. EMBO J. 31, 457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lynes E. M., Raturi A., Shenkman M., Ortiz Sandoval C., Yap M. C., Wu J., Janowicz A., Myhill N., Benson M. D., Campbell R. E., Berthiaume L. G., Lederkremer G. Z., Simmen T. (2013) Palmitoylation is the Switch that Assigns Calnexin to Quality Control or ER Calcium Signaling. J. Cell Sci. 126, 3893–3903 [DOI] [PubMed] [Google Scholar]
- 12. Lakkaraju A. K., van der Goot F. G. (2013) Calnexin Controls the STAT3-Mediated Transcriptional Response to EGF. Mol. Cell 51, 386–396 [DOI] [PubMed] [Google Scholar]
- 13. Lakkaraju A. K., Abrami L., Lemmin T., Blaskovic S., Kunz B., Kihara A., Dal Peraro M., van der Goot F. G. (2012) Palmitoylated calnexin is a key component of the ribosome-translocon complex. EMBO J. 31, 1823–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frangioni J. V., Beahm P. H., Shifrin V., Jost C. A., Neel B. G. (1992) The nontransmembrane tyrosine phosphatase PTP-1B localizes to the endoplasmic reticulum via its 35 amino acid C-terminal sequence. Cell 68, 545–560 [DOI] [PubMed] [Google Scholar]
- 15. Haj F. G., Verveer P. J., Squire A., Neel B. G., Bastiaens P. I. (2002) Imaging sites of receptor dephosphorylation by PTP1B on the surface of the endoplasmic reticulum. Science 295, 1708–1711 [DOI] [PubMed] [Google Scholar]
- 16. St-Pierre J., Tremblay M. L. (2012) Modulation of leptin resistance by protein tyrosine phosphatases. Cell Metab. 15, 292–297 [DOI] [PubMed] [Google Scholar]
- 17. Feldhammer M., Uetani N., Miranda-Saavedra D., Tremblay M. L. (2013) PTP1B: A simple enzyme for a complex world. Crit. Rev. Biochem. Mol. Biol. 48, 430–445 [DOI] [PubMed] [Google Scholar]
- 18. Popov D. (2012) Endoplasmic reticulum stress and the on site function of resident PTP1B. Biochem. Biophys. Res. Commun. 422, 535–538 [DOI] [PubMed] [Google Scholar]
- 19. Yip S. C., Saha S., Chernoff J. (2010) PTP1B: a double agent in metabolism and oncogenesis. Trends Biochem. Sci. 35, 442–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yunus A. A., Lima C. D. (2005) Purification and activity assays for Ubc9, the ubiquitin-conjugating enzyme for the small ubiquitin-like modifier SUMO. Methods Enzymol. 398, 74–87 [DOI] [PubMed] [Google Scholar]
- 21. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 22. Kraus A., Groenendyk J., Bedard K., Baldwin T. A., Krause K. H., Dubois-Dauphin M., Dyck J., Rosenbaum E. E., Korngut L., Colley N. J., Gosgnach S., Zochodne D., Todd K., Agellon L. B., Michalak M. (2010) Calnexin deficiency leads to dysmyelination. J. Biol. Chem. 285, 18928–18938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakamura K., Bossy-Wetzel E., Burns K., Fadel M., Lozyk M., Goping I. S., Opas M., Bleackley R. C., Green D. R., Michalak M. (2000) Changes in endoplasmic reticulum luminal environment affect cell sensitivity to apoptosis. J. Cell Biol. 150, 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Milner R. E., Baksh S., Shemanko C., Carpenter M. R., Smillie L., Vance J. E., Opas M., Michalak M. (1991) Calreticulin, and not calsequestrin, is the major calcium binding protein of smooth muscle sarcoplasmic reticulum and liver endoplasmic reticulum. J. Biol. Chem. 266, 7155–7165 [PubMed] [Google Scholar]
- 25. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 26. Li H. D., Liu W. X., Michalak M. (2011) Enhanced clathrin-dependent endocytosis in the absence of calnexin. PLoS One 6, e21678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schrag J. D., Bergeron J. J. M., Li Y., Borisova S., Hahn M., Thomas D. Y., Cygler M. (2001) The structure of calnexin, an ER chaperone involved in quality control of protein folding. Mol. Cell 8, 633–644 [DOI] [PubMed] [Google Scholar]
- 28. Rosenbaum E. E., Hardie R. C., Colley N. J. (2006) Calnexin is essential for rhodopsin maturation, Ca2+ regulation, and photoreceptor cell survival. Neuron 49, 229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tjoelker L. W., Seyfried C. E., Eddy R. L., Jr., Byers M. G., Shows T. B., Calderon J., Schreiber R. B., Gray P. W. (1994) Human, mouse, and rat calnexin cDNA cloning: identification of potential calcium binding motifs and gene localization to human chromosome 5. Biochemistry 33, 3229–3236 [DOI] [PubMed] [Google Scholar]
- 30. Geiss-Friedlander R., Melchior F. (2007) Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 31. Melchior F. (2000) SUMO-nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16, 591–626 [DOI] [PubMed] [Google Scholar]
- 32. Hietakangas V., Anckar J., Blomster H. A., Fujimoto M., Palvimo J. J., Nakai A., Sistonen L. (2006) PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. U.S.A. 103, 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang S. H., Galanis A., Witty J., Sharrocks A. D. (2006) An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 25, 5083–5093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang S. H., Sharrocks A. D. (2006) Interplay of the SUMO and MAP kinase pathways. Ernst Schering Res Found Workshop, 193–209 [DOI] [PubMed] [Google Scholar]
- 35. Hay R. T. (2005) SUMO: a history of modification. Mol. Cell 18, 1–12 [DOI] [PubMed] [Google Scholar]
- 36. Dadke S., Cotteret S., Yip S. C., Jaffer Z. M., Haj F., Ivanov A., Rauscher F., 3rd, Shuai K., Ng T., Neel B. G., Chernoff J. (2007) Regulation of protein tyrosine phosphatase 1B by sumoylation. Nature Cell Biol. 9, 80–85 [DOI] [PubMed] [Google Scholar]
- 37. Schmidt-Arras D. E., Böhmer A., Markova B., Choudhary C., Serve H., Böhmer F. D. (2005) Tyrosine phosphorylation regulates maturation of receptor tyrosine kinases. Mol. Cell. Biol. 25, 3690–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ellgaard L., Frickel E. M. (2003) Calnexin, calreticulin, and ERp57: teammates in glycoprotein folding. Cell Biochem. Biophys. 39, 223–247 [DOI] [PubMed] [Google Scholar]
- 39. Oliver J. D., van der Wal F. J., Bulleid N. J., High S. (1997) Interaction of the thiol-dependent reductase ERp57 with nascent glycoproteins. Science 275, 86–88 [DOI] [PubMed] [Google Scholar]
- 40. Leach M. R., Cohen-Doyle M. F., Thomas D. Y., Williams D. B. (2002) Localization of the Lectin, ERp57 Binding, and Polypeptide Binding Sites of Calnexin and Calreticulin. J. Biol. Chem. 277, 29686–29697 [DOI] [PubMed] [Google Scholar]
- 41. Romsicki Y., Reece M., Gauthier J. Y., Asante-Appiah E., Kennedy B. P. (2004) Protein tyrosine phosphatase-1B dephosphorylation of the insulin receptor occurs in a perinuclear endosome compartment in human embryonic kidney 293 cells. J. Biol. Chem. 279, 12868–12875 [DOI] [PubMed] [Google Scholar]
- 42. Cala S. E., Ulbright C., Kelley J. S., Jones L. R. (1993) Purification of a 90-kDa protein (Band VII) from cardiac sarcoplasmic reticulum. Identification as calnexin and localization of casein kinase II phosphorylation sites. J. Biol. Chem. 268, 2969–2975 [PubMed] [Google Scholar]
- 43. Schué V., Green G. A., Girardot R., Monteil H. (1994) Hyperphosphorylation of calnexin, a chaperone protein, induced by Clostridium difficile cytotoxin. Biochem. Biophys. Res. Commun. 203, 22–28 [DOI] [PubMed] [Google Scholar]
- 44. Wong H. N., Ward M. A., Bell A. W., Chevet E., Bains S., Blackstock W. P., Solari R., Thomas D. Y., Bergeron J. J. (1998) Conserved in vivo phosphorylation of calnexin at casein kinase II sites as well as a protein kinase C/proline-directed kinase site. J. Biol. Chem. 273, 17227–17235 [DOI] [PubMed] [Google Scholar]
- 45. Chevet E., Wong H. N., Gerber D., Cochet C., Fazel A., Cameron P. H., Gushue J. N., Thomas D. Y., Bergeron J. J. (1999) Phosphorylation by CK2 and MAPK enhances calnexin association with ribosomes. EMBO J. 18, 3655–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schimmel J., Larsen K. M., Matic I., van Hagen M., Cox J., Mann M., Andersen J. S., Vertegaal A. C. (2008) The ubiquitin-proteasome system is a key component of the SUMO-2/3 cycle. Mol. Cell. Proteomics 7, 2107–2122 [DOI] [PubMed] [Google Scholar]
- 47. Golebiowski F., Matic I., Tatham M. H., Cole C., Yin Y., Nakamura A., Cox J., Barton G. J., Mann M., Hay R. T. (2009) System-wide changes to SUMO modifications in response to heat shock. Sci. Signal. 2, ra24. [DOI] [PubMed] [Google Scholar]
- 48. Jentsch S., Psakhye I. (2013) Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu. Rev. Genetics 47, 167–186 [DOI] [PubMed] [Google Scholar]
- 49. Flotho A., Melchior F. (2013) Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 82, 357–385 [DOI] [PubMed] [Google Scholar]
- 50. Rajan S., Plant L. D., Rabin M. L., Butler M. H., Goldstein S. A. (2005) Sumoylation silences the plasma membrane leak K+ channel K2P1. Cell 121, 37–47 [DOI] [PubMed] [Google Scholar]
- 51. Martin S., Nishimune A., Mellor J. R., Henley J. M. (2007) SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature 447, 321–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Felberbaum R., Wilson N. R., Cheng D., Peng J., Hochstrasser M. (2012) Desumoylation of the endoplasmic reticulum membrane VAP family protein Scs2 by Ulp1 and SUMO regulation of the inositol synthesis pathway. Mol. Cell. Biol. 32, 64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kho C., Lee A., Jeong D., Oh J. G., Chaanine A. H., Kizana E., Park W. J., Hajjar R. J. (2011) SUMO1-dependent modulation of SERCA2a in heart failure. Nature 477, 601–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ferrari E., Tinti M., Costa S., Corallino S., Nardozza A. P., Chatraryamontri A., Ceol A., Cesareni G., Castagnoli L. (2011) Identification of new substrates of the protein-tyrosine phosphatase PTP1B by Bayesian integration of proteome evidence. J. Biol. Chem. 286, 4173–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Elchebly M., Payette P., Michaliszyn E., Cromlish W., Collins S., Loy A. L., Normandin D., Cheng A., Himms-Hagen J., Chan C. C., Ramachandran C., Gresser M. J., Tremblay M. L., Kennedy B. P. (1999) Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 283, 1544–1548 [DOI] [PubMed] [Google Scholar]
- 56. Kushner J. A., Haj F. G., Klaman L. D., Dow M. A., Kahn B. B., Neel B. G., White M. F. (2004) Islet-sparing effects of protein tyrosine phosphatase-1b deficiency delays onset of diabetes in IRS2 knockout mice. Diabetes 53, 61–66 [DOI] [PubMed] [Google Scholar]
- 57. Boute N., Boubekeur S., Lacasa D., Issad T. (2003) Dynamics of the interaction between the insulin receptor and protein tyrosine-phosphatase 1B in living cells. EMBO Rep. 4, 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bass J., Chiu G., Argon Y., Steiner D. F. (1998) Folding of insulin receptor monomers is facilitated by the molecular chaperones calnexin and calreticulin and impaired by rapid dimerization. J. Cell Biol. 141, 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Saitoh T., Yanagita T., Shiraishi S., Yokoo H., Kobayashi H., Minami S., Onitsuka T., Wada A. (2002) Down-regulation of cell surface insulin receptor and insulin receptor substrate-1 phosphorylation by inhibitor of 90-kDa heat-shock protein family: endoplasmic reticulum retention of monomeric insulin receptor precursor with calnexin in adrenal chromaffin cells. Mol. Pharmacol. 62, 847–855 [DOI] [PubMed] [Google Scholar]
- 60. Stuible M., Abella J. V., Feldhammer M., Nossov M., Sangwan V., Blagoev B., Park M., Tremblay M. L. (2010) PTP1B targets the endosomal sorting machinery: dephosphorylation of regulatory sites on the endosomal sorting complex required for transport component STAM2. J. Biol. Chem. 285, 23899–23907 [DOI] [PMC free article] [PubMed] [Google Scholar]