Background: Little is known about metabolic pathways used by the zoonotic pathogen Streptococcus suis.
Results: Amino acid auxotrophies and biosynthetic pathways were determined from in vitro and ex vivo isotopologue studies using [13C]glucose.
Conclusion: The core metabolism of S. suis is determined by glycolysis and an essential PEP carboxylation.
Significance: The isotopologue patterns identify key metabolic features and potential targets for future drug design.
Keywords: Amino Acid, Bacterial Metabolism, Biosynthesis, Carbohydrate Metabolism, Glycolysis, Streptococcus suis, Isotopologue Profiling, Phosphoenolpyruvate Carboxylase
Abstract
Streptococcus suis is a neglected zoonotic pathogen that has to adapt to the nutritional requirements in the different host niches encountered during infection and establishment of invasive diseases. To dissect the central metabolic activity of S. suis under different conditions of nutrient availability, we performed labeling experiments starting from [13C]glucose specimens and analyzed the resulting isotopologue patterns in amino acids of S. suis grown under in vitro and ex vivo conditions. In combination with classical growth experiments, we found that S. suis is auxotrophic for Arg, Gln/Glu, His, Leu, and Trp in chemically defined medium. De novo biosynthesis was shown for Ala, Asp, Ser, and Thr at high rates and for Gly, Lys, Phe, Tyr, and Val at moderate or low rates, respectively. Glucose degradation occurred mainly by glycolysis and to a minor extent by the pentose phosphate pathway. Furthermore, the exclusive formation of oxaloacetate by phosphoenolpyruvate (PEP) carboxylation became evident from the patterns in de novo synthesized amino acids. Labeling experiments with S. suis grown ex vivo in blood or cerebrospinal fluid reflected the metabolic adaptation to these host niches with different nutrient availability; however, similar key metabolic activities were identified under these conditions. This points at the robustness of the core metabolic pathways in S. suis during the infection process. The crucial role of PEP carboxylation for growth of S. suis in the host was supported by experiments with a PEP carboxylase-deficient mutant strain in blood and cerebrospinal fluid.
Introduction
Streptococcus suis is a porcine pathogen, and S. suis infections are a major concern in the swine-producing industry because they lead to high financial losses worldwide (1). Furthermore, S. suis is considered a neglected zoonotic pathogen that can cause invasive diseases in humans (2, 3). Very severe cases of human S. suis infections causing meningitis and/or septicemia, as well as streptococcal toxic shock-like syndrome, were recorded in outbreaks occurring in China and Vietnam (4–6). Presently, S. suis is the leading causative agent of adult bacterial meningitis in Hong Kong and Vietnam (4, 7, 8). So far, 33 different serotypes have been described, based on the composition of the capsular polysaccharides. However, the S. suis strains most frequently isolated from diseased pigs and humans worldwide were classified as serotype 2 (3, 9, 10).
Disease in pigs comprises mainly arthritis, septicemia, endocarditis, and meningitis. It is thought that invasive disease is initiated after breaching the mucosal epithelial barrier of the upper respiratory tract of pigs, subsequent translocation in the bloodstream, and dissemination of the streptococci (11, 12). In cases of streptococcal meningitis, the choroid plexus epithelium, being a constituent of blood-cerebrospinal fluid (CSF)2 barrier, might be a potential entry portal for S. suis to the central nervous system (13, 14). Thus, porcine blood and CSF seem to be important host niches, to which S. suis has to adapt efficiently to survive and establish disease.
S. suis research has been primarily conducted on the identification and characterization of virulence-associated factors and their potential use as vaccine candidates (see reviews in Refs. 12 and 15). In addition to virulence factor expression, which is essential to establish an invasive infection, S. suis has to adapt its metabolism to the nutrient availability of the different host compartments. In recent years, it is becoming clearer that S. suis can sense the nutrient supply and subsequently modulate the expression of its virulence factors, e.g. by the activity of transcriptional regulators (16–19), which may also be of relevance in different host niches.
Nevertheless, very little is known about the central carbon metabolism of S. suis and the amino acid biosynthesis pathways that are linked to virulence. Whole genome sequencing data of different S. suis strains allow prediction of possible metabolic routes based on gene annotation. Regarding to these data, all S. suis strains included in the Kyoto Encyclopedia of Genes and Genomes database carry genes encoding for the Embden-Meyerhof-Parnas (EMP) pathway, the pentose phosphate pathway (PPP), and a fragmentary TCA cycle, but genes encoding for the Entner-Doudoroff pathway (ED) seem to be missing. However, this does not necessarily reflect the nature, rates, and dynamics of active metabolic reactions and metabolite fluxes.
A powerful technique to elucidate biosynthetic pathways in microorganisms grown under complex and nonstandardized conditions is 13C-isotopologue profiling (20). When 13C-labeled substrates, e.g. glucose, are used for bacterial growth, the label is distributed in downstream metabolic pathways and can be detected in de novo synthesized metabolites, such as amino acids (21). Analysis of the specific isotopologue patterns in these amino acids by GC/MS or NMR spectroscopy then provide considerable information about the carbon pathways and relative fluxes in the metabolic networks of bacteria (22). Several studies have applied this technique to pathogenic bacteria to reconstruct the central metabolic pathways, not only after cultivation in laboratory media (23–25) but also after infection of eukaryotic cells (26–28).
In the present study, we used 13C-isotopologue profiling to elucidate the de novo amino acid biosynthetic pathways and the core metabolic network of a S. suis serotype 2 strain grown in vitro. Furthermore, we expanded isotopologue profiling to S. suis grown in porcine blood and CSF to eventually identify features of its metabolic adaptation to a more complex nutrient availability encountered in the host. Our in vitro and ex vivo results indicate that S. suis has variable preferences for uptake of amino acids and de novo biosynthesis. However, under all conditions, the main pathways and relative fluxes in the core metabolism of S. suis were quite stable and were mainly dictated by the activity of the EMP pathway and an anaplerotic reaction mediated by a phosphoenolpyruvate (PEP) carboxylase.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
Bacterial strains used in this study are listed in supplemental Table S1. The highly virulent S. suis serotype 2 strain 10, the isogenic mutant 10Δppc, and the complemented strain 10cΔppc were cultured overnight at 37 °C on Columbia blood agar base (Oxoid) containing 6% (v/v) sheep blood or horse blood supplemented with spectinomycin (100 μg/ml), if necessary. For further experiments bacteria were grown in Todd Hewitt broth (THB; Becton Dickinson Diagnostics) or in chemically defined medium (CDM; see formula in supplemental Table S2). For each growth or labeling experiment, a starting culture was inoculated with a single S. suis colony in THB medium and incubated at 37 °C overnight. Bacteria were then harvested by centrifugation, washed twice in 1× PBS, and used for inoculation of respective media at an optical density A600 of ∼0.001. Amino acid auxotrophy and labeling experiments were performed in 50-ml tubes, and the optical density A600 was measured at the indicated time points or regular intervals with an Eppendorf BioPhotometer®. Growth experiments in CDM supplemented with filter sterilized casamino acids, BactoTM-Tryptone, BactoTM-Peptone (all purchased from Becton Dickinson), α-ketoglutarate, or oxaloacetate were performed in 96-well plates covered with a Breathe-Easy® sealing membrane. Growth was monitored automatically by measuring A630 in a Tecan GENios Pro microplate reader for 24 h at 37 °C. Escherichia coli strains used for subcloning were cultured on LB agar or in LB broth supplemented with spectinomycin (50 μg/ml), if required.
Growth of S. suis Serotype 2 in Porcine Blood and Cerebrospinal Fluid ex Vivo
Collection of heparinized blood from healthy pigs in our institute is registered at the Lower Saxonian State Office for Consumer Protection and Food Safety (Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit) under permit number 33.9-42502-05-11A137 and was conducted in line with the recommendations of the German Society for Laboratory Animal Science (Gesellschaft für Versuchstierkunde) and the German Veterinary Association for the Protection of Animals (Tierärztliche Vereinigung für Tierschutz e. V.). CSF samples were collected from clinically healthy pigs within the framework of the registered Lower Saxonian State Office for Consumer Protection and Food Safety permit number 33.9-42502-04-08/1612. CSF samples were sterile filtered using a 0.22-μm Rotilabo® syringe filter, and aliquots were immediately frozen at −80 °C.
For survival assays, bacteria were grown in THB medium to early exponential growth phase, then harvested by centrifugation, and washed twice in 1× PBS. Porcine blood and CSF were inoculated with S. suis WT strain 10 or strain 10Δppc at concentrations of 1 × 104 CFU/ml, and the samples were rotated overhead at 37 °C. At the indicated time points, CFU were determined by serial dilution and plating. Plating of the aliquots was performed in triplicate, and bacterial colonies were counted 24 h after incubation at 37 °C.
General Molecular Methods and Chemicals
If not stated otherwise, all chemicals were purchased from Sigma-Aldrich. The restriction enzymes, Phusion DNA polymerase, and T4 DNA ligase were purchased from New England Biolabs. The oligonucleotides and plasmids used in this study are listed in supplemental Table S1. Transformation of E. coli and S. suis, as well as isolation of chromosomal and plasmid DNA, was performed as described earlier (29). All plasmid constructs were verified by DNA sequencing. RNA isolation of S. suis, RT followed by quantitative PCR (qPCR), and evaluation of qPCR were done as described recently (17). Relative target gene transcript levels were normalized to gyrB transcript levels and expressed as 2−ΔCT values (30).
Construction of Strain 10Δppc and Its Complemented Strain 10cΔppc
Deletion of the S. suis ppc gene (SSU0479) was achieved by an adapted Cre-lox-based method as described recently (31). Briefly, a 1.2-kilobase pair fragment A resembling the ppc upstream region and a 1.2-kilobase pair fragment B identical to the region downstream of ppc were amplified by PCR from S. suis strain 10 chromosomal DNA by using the primer pairs SSU0478_Xho_f/SSU0478_Nco_r and SSU0480_Nhe_f/SSU0480_BamH_r, respectively. By this, a large part of the ppc open reading frame was not amplified. In parallel, a spectinomycin (spc) resistance cassette that is flanked by lox66 and lox71, respectively, was amplified from the plasmid pGA14 (32) using primers Spec_lox66_Sac/Spec_lox71_Nhe and cloned in pCR2.1-TOPO (Invitrogen). From the resulting vector pCR-lox-spc, the floxed spc cassette was amplified using primers Spec_lox66_NcoI and Spec_lox71_Nhe. All PCR fragments were digested with the restriction enzymes indicated and were ligated into the BamHI-XhoI-cut pCR2.1-TOPO (Invitrogen) vector. The resulting plasmid pCR-ppc::lox-spc was verified by sequencing and used for transformation of S. suis as described (32). Putative mutants (10Δppc::spc) were initially selected for spc resistance and screened using PCR. To remove the spc resistance cassette, electrocompetent 10Δppc::spc cells were transformed with pSET5s_PtufA-cre and selected for chloramphenicol resistance. After excision of the spc resistance cassette, pSET5s_PtufA-cre was removed, and possible 10Δppc deletion mutants were verified as described (31). Episomal complementation of 10Δppc was done as described earlier (16). Because the ppc gene (SSU0479) seems to form an operon with its upstream gene SSU0478, the entire operon and its native promoter were amplified from chromosomal DNA of S. suis strain 10 using primers SSU0479_comp_fw and SSU0479_comp_rev2. The EcoRI-digested PCR product was ligated into the EcoRI-cut pGA14 vector, and the resulting plasmid pGA14-ppc was introduced into strain 10Δppc.
13C Incorporation Studies
Labeling of streptococci in CDM or THB was performed by supplementation of the medium with 10 mm [U-13C6]glucose or [1,2-13C2]glucose (Sigma-Aldrich), respectively. CDM labeling experiments were performed in the absence of unlabeled glucose. For all labeling experiments, preparation of overnight cultures and inoculation of fresh media were done exactly as described above. Bacterial growth was monitored by measuring A600 in regular intervals, and bacteria were harvested until the early exponential or stationary growth phase was reached in respective media. The bacterial cells were immediately collected by centrifugation at 4,000 × g and 4 °C, rapidly washed twice with ice-cold 1× PBS, and frozen at −80 °C. Then the bacterial suspension was autoclaved at 120 °C for 20 min and lyophilized.
For ex vivo labeling experiments, porcine blood or CSF supplemented with either 5 mm [U-13C6]glucose or [1,2-13C2]glucose was inoculated with S. suis WT strain 10 and incubated as described for the survival assays. After 6 h of incubation (see Fig. 5D), bacteria were harvested. CSF samples were processed as described above, whereas blood samples were subjected to additional steps of centrifugation. These comprised two centrifugation steps at 825 × g at 4 °C for 5 min to pellet erythrocytes and leukocytes, followed by a centrifugation of the supernatant at 4,000 × g at 4 °C for 5 min to pellet the bacterial cells. The purity of the suspension was controlled by Gram staining. Then bacteria were washed twice in ice-cold 1×PBS, inactivated, and processed as described above.
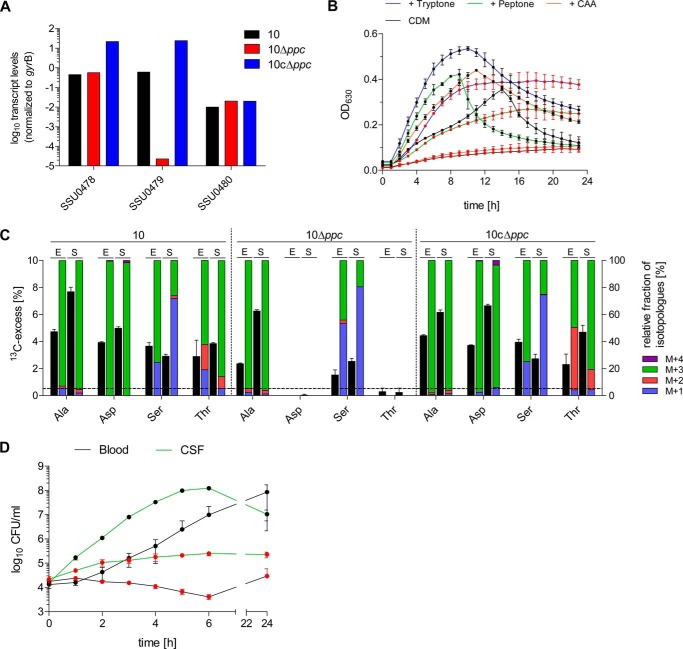
FIGURE 5.
The ppc gene of S. suis is essential for oxaloacetate production in vitro and for growth in porcine blood and CSF ex vivo. A, verification of ppc deletion by RT-qPCR. S. suis WT strain 10, the isogenic mutant 10Δppc, and the complemented strain 10cΔppc were grown in THB medium to the early exponential growth phase, RNA was extracted, and RT-qPCR was performed as described under “Experimental Procedures.” The results are shown for one representative experiment performed in duplicate and are expressed as relative transcript levels of the ppc gene (SSU0479), the upstream (SSU0478), and the downstream gene (SSU0480) normalized to gyrB. B, growth of S. suis WT strain 10 (black circles) and strain 10Δppc (red circles) in complete CDM that was optionally supplemented with 0.5% tryptone, peptone, or casamino acids (CAA). Growth was monitored by measuring the A630 in 96-well plates over 24 h. Experiments were performed in triplicate, and results are shown for two biological replicates. C, relative isotopologue composition of Ala, Asp, Ser, and Thr derived from S. suis WT strain 10, the isogenic mutant 10Δppc, and the complemented strain 10cΔppc grown to the exponential (E) and stationary (S) growth phase in THB medium supplemented with 10 mm [U-13C6]glucose. 13C excess values (mol %) are shown as black columns on the left y axis. Colored columns on the right y axis indicate the percentages of isotopologues comprising up to four 13C atoms (M+1 to M+4). Results are shown for one representative experiment, and error bars indicate S.D. of MS measurements performed in triplicate. The horizontal line indicates the detection limit set to 0.5 mol % 13C excess in these experiments. D, growth of S. suis WT strain 10 (black circles) and strain 10Δppc (red circles) in porcine blood and CSF ex vivo. Growth was monitored at the indicated time points by CFU enumeration after plating. Mean values and standard deviations are shown for three independent biological replicates.
Protein Hydrolysis and Amino Acid Derivatization
A lyophilized bacterial dry weight of ∼10 mg was resuspended in 6 m HCl and boiled at 105 °C for 24 h under an inert atmosphere. The hydrolysate was placed on a cation exchange column of Dowex 50W × 8 (H+ form, 200–400 mesh, 5 × 10 mm), which was washed with 70% methanol. The column was washed with water, and afterward developed with 4 m ammonium hydroxide. An aliquot of the eluate was dried under a nitrogen stream and then dissolved in 50 μl of water-free acetonitrile. An equal volume of N-(tert-butyldimethyl-silyl)-N-methyl-trifluoroacetamide containing 1% tert-butyl-dimethyl-silylchloride was added, and the solution was incubated at 70 °C for 30 min. The resulting N-(tert-butyldimethylsilyl) (TBDMS) derivatives of proteinogenic amino acids were then analyzed by GC/MS.
Mass Spectrometry
GC/MS analysis was performed on a GCMS-QP2010 Plus gas chromatograph/mass spectrometer (Shimadzu) equipped with a fused silica capillary column (Equity TM-5; 30 m by 0.25 mm; 0.25-μm film thickness; Supelco) working with electron impact ionization at 70 eV. One μl of a TBDMS derivative solution was injected in a 1:10 split mode at an interface temperature of 260 °C and a helium inlet pressure of 70 kPa. The column was then developed at 150 °C for 3 min with a temperature gradient of 10 °C/min to a final temperature of 260 °C that was held for 3 min. Data were collected using the LabSolutions software (Shimadzu). Selected ion monitoring data were acquired according to Eylert et al. (26) using a 0.3-s sampling rate, and the samples were analyzed in triplicate. The overall 13C excess (mol %) and the relative contribution of isotopomers were computed by an Excel-based in-house software package according to Lee et al. (33).
RESULTS
Preferred Carbon Sources and Amino Acid Auxotrophies of S. suis under Chemically Defined Conditions
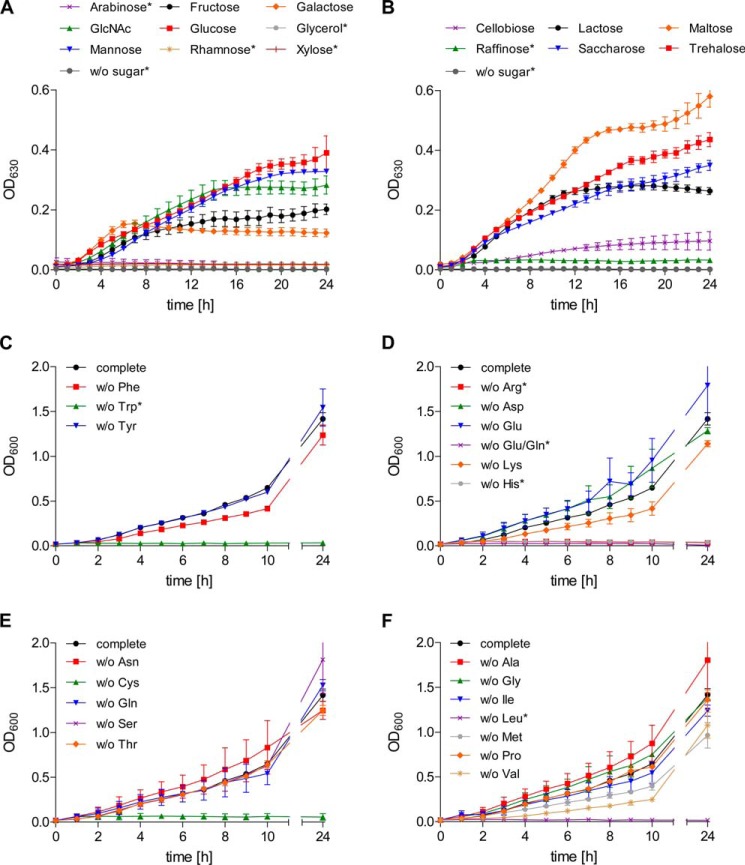
Together with clostridia and enterococci, streptococci are among the species with the highest abundance of carbohydrate uptake systems relative to their genome size (34, 35). An in silico analysis of the S. suis serotype 2 reference strain P1/7 (36) revealed the presence of 25 putative carbohydrate uptake systems including PTS or ABC transporters distributed in the S. suis genome (data not shown). Given the high number of carbohydrate transport systems, we first analyzed the ability of S. suis to use different carbohydrate sources for growth in a chemically defined medium. These experiments revealed that S. suis strain 10 utilizes a variety of monosaccharides and disaccharides for growth as depicted in Fig. 1 (A and B). Best growth results were obtained after supplementation with glucose, mannose, or the α-α′-diglucosides maltose and trehalose. Arabinose, xylose, raffinose, and rhamnose appeared to be less preferred growth substrates. All sugars except rhamnose were metabolized by S. suis strain 10 as tested in additional fermentation assays (data not shown). Because glucose seems to most efficiently support growth of S. suis, we choose this carbohydrate for studying amino acid auxotrophies. By subsequently omitting each of 20 amino acids, we found that S. suis strain 10 was not able to grow in the absence of Arg, Cys, Gln/Glu, His, Leu, or Trp, respectively (Fig. 1, C–F). Bioinformatic analysis of the S. suis P1/7 genome revealed a lack of the genes encoding for enzymes needed for the biosynthesis of Arg, His, Glu, and Trp, which may explain most of the observed auxotrophies (supplemental Table S9). Gln/Glu auxotrophy could not be restored by ammonium supplementation via (NH4)2SO4 or NH4Cl (data not shown). Because growth was not hampered when either Gln or Glu was omitted, S. suis strain 10 seems to be able to take up both amino acids for exchangeable use. Moreover, S. suis strain 10 was not able to grow when Leu was omitted, although it seems to possess all enzymes required for Leu biosynthesis. The same was found for Cys. We additionally tested whether Cys auxotrophy could be restored by thiosulfate or sulfide supplementation as described for Streptococcus mutans (37). By adding 1 mm Na2S2O3 or Na2S to the medium, we were able to restore growth, suggesting that Cys biosynthesis requires thiosulfate or sulfide as a sulfur source (data not shown). In contrast to the above mentioned amino acids, deprivation of Ala, Asp, Asn, Gly, Pro, Ser, Thr, or Tyr did not impair growth of S. suis strain 10. Growth was moderately reduced in the absence of Ile, Lys, Met, Phe, or Val. Taken together, under the conditions investigated, S. suis strain 10 seems to be auxotrophic for Arg, Gln/Glu, His, Leu, and Trp.
FIGURE 1.
Preferred carbon sources and amino acid auxotrophies of S. suis strain 10 in a CDM. A and B, S. suis strain 10 was grown in CDM medium containing 40 mm of monosaccharides (A) or di-/trisaccharides (B) indicated, and A630 nm values were recorded at 1-h intervals automatically in a thermostatic 96-well microplate reader. Results and standard deviations are shown for three biological replicates. Carbohydrate substrates that could not be used for streptococcal growth in CDM are marked by asterisks. C–F, amino acid auxotrophies of S. suis strain 10 were determined in CDM with glucose as the sole carbohydrate source and by omitting single amino acids. Growth was monitored by measuring the A600 at indicated the time points. The results are shown for two biological replicates in cultures that were individually depleted for aromatic (Phe, Tyr, or Trp) (C), charged (Arg, Asp, Glu, Glu/Gln, His, or Lys) (D), uncharged (Asn, Cys, Gln, Ser, or Thr) (E), or aliphatic and branched chain amino acids (Ala, Gly, Ile, Leu, Met, Pro, or Val) (F). Amino acid auxotrophies of S. suis strain 10 are marked by asterisks. OD600, A600; OD630, A630; w/o, without.
De Novo Amino Acid Biosynthesis of S. suis in CDM
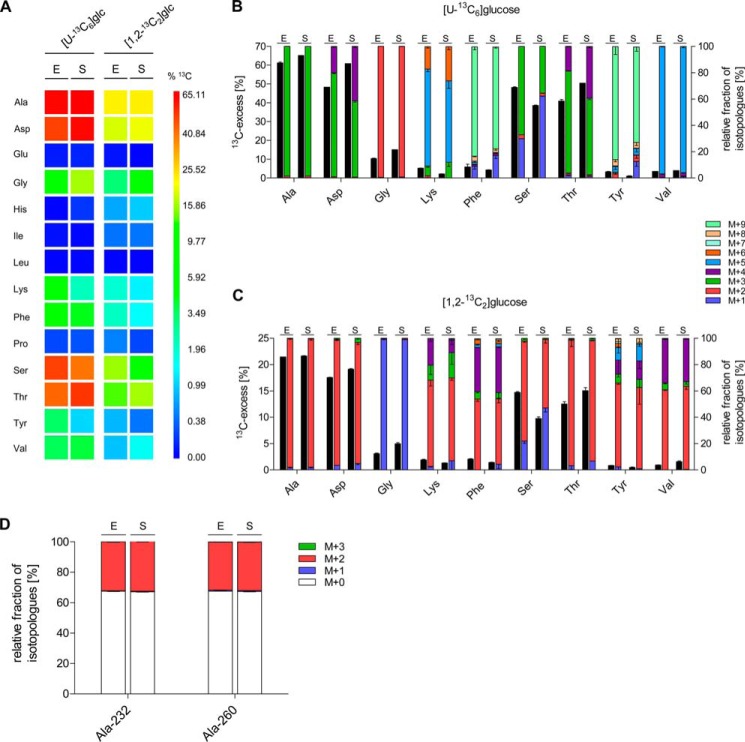
To investigate de novo synthesis of amino acids by S. suis, we performed 13C-isotopologue profiling experiments. For this, S. suis strain 10 was grown to the early exponential and early stationary growth phase in CDM containing all amino acids and either [U-13C6]glucose or [1,2-13C2]glucose as tracers. Isotopologue patterns of protein-derived amino acids were then determined by GC/MS. The acidic hydrolysis of cellular proteins did not allow the determination of Asn, Cys, Gln, Met, and Trp. Asn and Gln were converted to Asp and Glu, and therefore, values given for Asp and Glu are represented by the Asn/Asp and Gln/Glu averages, respectively (23, 26). The yields of the TBDMS-derivatives of His and Lys were low, and thus, isotopologue data of these amino acids were therefore only given when applicable. Arg did not form a suitable TBDMS derivative and was therefore omitted. Among the 14 remaining amino acids, 9 acquired significant 13C labels from both [U-13C6]glucose and [1,2-13C2]glucose, indicating their de novo biosynthesis (Fig. 2A). However, 13C incorporation (mol % 13C excess) differed among the detected amino acids, e.g. for bacteria grown to exponential growth phase in CDM supplemented with [U-13C6]glucose: Ala (61.1%) > Asp (48.2%) > Ser (48.1%) > Thr (40.9%) > Gly (10.2%) > Phe (5.8%) > Lys (5.1%) > Val (3.4%) > Tyr (3.1%). No significant 13C labeling was found in Glu, His, Ile, Leu, and Pro (Fig. 2A). For Glu, His, and Leu, this finding confirmed the respective auxotrophies observed in the growth experiments described above. In case of Ile and Pro, however, the data indicated that under these conditions S. suis seems to prefer uptake of (unlabeled) Ile and Pro from CDM or synthesis from other unlabeled precursor molecules present in CDM. At least for Pro synthesis, the latter hypothesis is likely because (unlabeled) Glu from CDM probably serves as the precursor of Pro.
FIGURE 2.
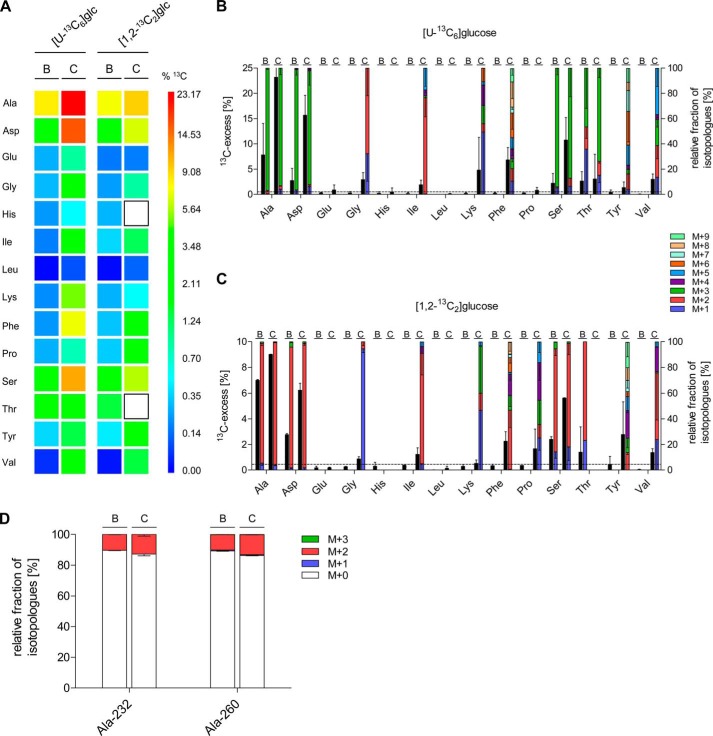
13C Profiles of de novo synthesized amino acids of S. suis strain 10 cultivated in the presence of [U-13C6]glucose or [1,2-13C2]glucose in chemically defined media. A, color map for the overall 13C excess (mol %) of labeled amino acids after growth of S. suis strain 10 in the presence [U-13C6]glucose or [1,2-13C2]glucose in chemically defined media. The results are shown for exponential (E) and stationary (S) grown bacteria. Mean values of two biological replicates for which MS measurements were performed in triplicate are given. B and C, relative isotopologue composition of amino acids derived from S. suis grown in CDM with [U-13C6]glucose (B) or [1,2-13C2]glucose (C) to the exponential (E) and stationary (S) growth phase. 13C excess values (mol %) are shown as black columns on the left y axis. Colored columns on the right y axis indicate the percentages of labeled isotopologues comprising up to nine 13C atoms (M+1 to M+9). Mean values are shown for two independent biological experiments each measured in triplicate. D, relative fractions of isotopologues in Ala-232 and Ala-260 fragments, which were relevant to discriminate glucose fluxes of the EMP pathway and the PPP are shown. S. suis strain 10 was grown to the exponential (E) and stationary (S) growth phase in CDM supplemented with 10 mm [1,2-13C2]glucose, respectively. The colored columns on the left y axis indicate the percentages of isotopologues comprising up to three 13C atoms (M+1 to M+3). Note that M+0 stands for unlabeled Ala and indicates uptake of extracellular Ala. Mean values and standard deviations are shown for two independent biological experiments each measured in triplicate.
Dissection of the Central Carbon Metabolism on the Basis of 13C-isotopologue Profiles Derived from de Novo Synthesized Asp, Ala, and Thr
A survey on the genomes of S. suis strain P1/7 and other sequenced S. suis strains revealed the presence of all genes encoding for the EMP pathway and PPP, respectively, but seems to lack the genes necessary for the ED. Furthermore, it possesses a fragmentary TCA cycle, i.e. the genes necessary for the synthesis of citrate from acetyl-CoA and oxaloacetate and further reactions to α-ketoglutarate, but no genes assigned to the steps from α-ketoglutarate to oxaloacetate. This implies an alternative way of oxaloacetate synthesis, which is essential for the formation of lysine and diaminopimelate. To elucidate this assumption, we analyzed the isotopologue pattern of Asp because it is directly synthesized from oxaloacetate via Asp transaminase (aspC, SSU0569). In fact, after feeding of [U-13C6]glucose, a high proportion of 13C3-labeled Asp was found, indicating carboxylation of a 13C3-labeled intermediate from the EMP pathway, such as PEP (Fig. 2B). Consequently, Asp synthesis must have been initiated by transferring an unlabeled CO2 to [U-13C3]PEP to synthesize [1,2,3-13C3]OAA by a PEP carboxylase (ppc, SSU0479) (Fig. 3). The minor fractions of [U-13C4]Asp most likely arose from carboxylation of [U-13C3]PEP with 13CO2, which seems to accumulate in the stationary growth phase.
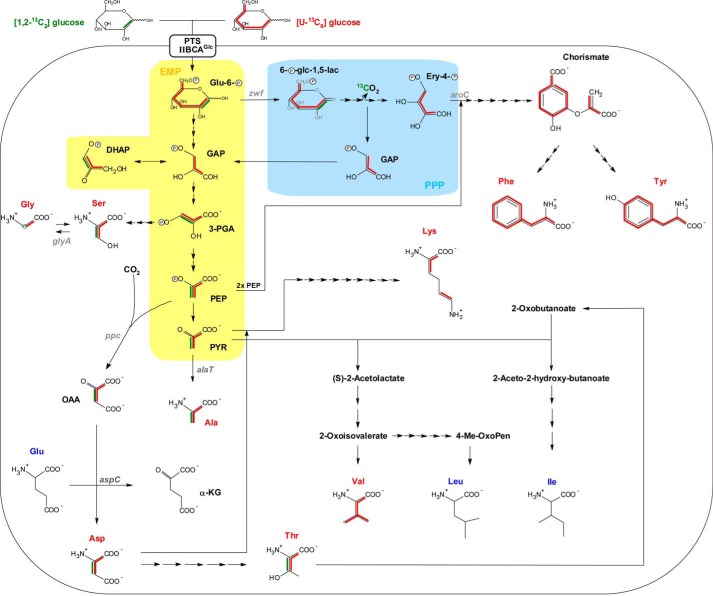
FIGURE 3.
Reconstruction of the metabolic pathways utilized for de novo syntheses of amino acids as derived from isotopologue profiling with [U-13C6]glucose and [1,2-13C2]glucose. 13C patterns of amino acid isotopologues are indicated by red and green lines, respectively, as deduced from labeling experiments performed with [U-13C6]glucose and [1,2-13C2]glucose. [1,2-13C2]Glucose-derived isotopologue patterns are shown for Ala, Asp, Thr, biosynthesis pathways, and the formation of Gly from Ser only. The three-letter abbreviation of de novo synthesized amino acids is highlighted in red, whereas that of unlabeled amino acids remains dark blue. Simplified metabolic reactions being part of the EMP pathway and PPP are highlighted by yellow and light blue shading, respectively. Note that in the case of [1,2-13C2]glucose catabolization via PPP, the formation of unlabeled GAP molecules is shown solely. Full metabolic fluxes refer to the Kyoto Encyclopedia of Genes and Genomes database, and arrows indicate the number of metabolic steps needed for biosynthesis of amino acids. Corresponding gene annotations are given in Table S9. Glu-6-P, glucose-6-phosphate; 6-P-glc-1,5-lac, 6-phospho-d-glucono-1,5-lactone; Ery-4-P, erythrose-4-phosphate; DHAP, dihydroxy-acetone-phosphate; PYR, pyruvate; α-KG, α-ketoglutarate.
All genes necessary for Thr synthesis are annotated in the S. suis genome. On this basis, Thr can be synthesized in five enzymatic reactions from Asp via homoserine. Indeed, we detected a high fraction of 13C3-labeled Thr, which confirms this metabolic route. The enrichment of [U-13C4]Thr similar to [U-13C4]Asp in the stationary growth phase further supports this. In addition to Asp and Thr, the highest molar 13C excess (>60 mol %) was detected in Ala at both growth phases of S. suis. The isotopologue distribution revealed almost solely [U-13C3]Ala, indicating its formation from universally labeled [U-13C3]pyruvate provided by glycolysis (see below). Most likely, this is linked to the gene product of alaT (SSU0319), which catalyzes the transamination of [U-13C3]pyruvate by using Glu as an amino group donor leading to [U-13C3]Ala and α-ketoglutarate, respectively. Overall, these data suggest that glucose is efficiently catabolized via the EMP pathway and indicate that PEP carboxylase is a key enzyme for OAA, Asp, and Thr biosynthesis under the tested in vitro conditions.
Glucose Is Predominantly Catabolized by the EMP
The high enrichment rates of [U-13C3]Ala caused by [U-13C6]glucose feeding reflected that glucose was mainly metabolized by EMP (or by ED, but homologues genes for the ED are missing in the genome), because single or double 13C-labeled Ala isotopologues, which would indicate a contribution of carbon cycling via the PPP, were almost undetectable. However, to unequivocally dissect whether glucose was primarily catabolized via the EMP, PPP, or the ED pathway, we performed growth experiments with [1,2-13C2]glucose. Other than using [U-13C6]glucose as a tracer molecule, which leads to the formation of identical [U-13C3]glyceraldehyde 3-phosphate (GAP) molecules (Fig. 3), the use of [1,2-13C2]glucose allows differentiation of EMP, PPP, and ED because of different labeled GAP isotopologues that are deducible from the labeling of Ala via pyruvate. In more detail, after [1,2-13C2]glucose metabolization via the PPP, GAP can occur in [1,3-13C2]GAP, [3-13C1]GAP or can remain unlabeled (Fig. 3). Hence, via the PPP, [1,2-13C2]glucose could be initially converted to [1,2-13C2]6-phospho-d-glucono-1,5-lactone by the gene product of zwf (SSU1025) and then in two steps decarboxylated to [1-13C1]d-ribulose-5-phosphate. The first 13C atom is lost in this process catalyzed by 6-phospho-gluconate dehydrogenase (gnd, SSU1541). [1-13C1]Ribulose-5-phosphate is interconvertible with [1-13C1]xylulose-5-phosphate, which could react with [1-13C1]ribose-5-phosphate to [1,3-13C2]sedoheptulose-7-phosphate and finally to unlabeled GAP in the transketolase reaction. Alternatively, the formation of [1,3-13C2]GAP and [3-13C1]GAP would result from the transaldolase and transketolase reaction of the PPP via [1,3-13C2]fructose-6-phosphate and [1-13C1]fructose-6-phosphate, respectively. In contrast, the enrichment of [1,2-13C2]GAP and, therefore, [1,2-13C2]Ala would point to an ED activity. In case of the EMP pathway, [1,2-13C2]glucose is metabolized in three steps to unlabeled GAP and [2,3-13C2]dihydroxy-acetone-phosphate. The latter two molecules are interconvertible by a triosephosphate isomerase (tpiA, SSU0483), and therefore, one half of the molecules in the GAP pool consists of [2,3-13C2]GAP, whereas the other half remains unlabeled. For Ala and Asp, both synthesized from glycolytic intermediates, this would result in the reduction of overall 13C excess by one-third when compared with [U-13C6]glucose labeling. Indeed, when comparing with the values from the experiment with [U-13C6]glucose, this factor was detected after growth of S. suis with [1,2-13C2]glucose for Ala and Asp (Fig. 2, A and C), thus indicating that glucose was mainly catabolized by the EMP pathway. In addition, a closer look to the isotopologue pattern of different Ala fragments helped to distinguish between EMP, PPP, and ED as explained above. Notably, enrichment of a completely labeled Ala-232 (m/z 232) derivative that lacks the 13C label at C-1 because of ESI fragmentation suggested that Ala was originally synthesized from [2,3-13C2]GAP precursor molecules via the EMP pathway and not via the PPP or ED pathway (Fig. 2D and supplemental Table S4), because the 13C label at C-1 of Ala would have been originated from glucose catabolization by the PPP and/or ED only. The absence of single 13C-labeled Ala-260 (m/z 260) further excludes synthesis from [3-13C1]GAP originating from labeled fructose-6-phosphate via the PPP. Taken together, these experiments revealed that glucose is predominantly metabolized by the EMP pathway instead of the PPP. Furthermore, a functional ED pathway seems not to be present in S. suis as expected from the lack of ED genes in the genome.
Reconstruction of de Novo Synthesis Pathways of Ser, Gly, Lys, Val, Phe, and Tyr by Isotopologue Profiling
Isotopologue profiling experiments with [U-13C6]glucose also showed that along with Ala, Asp, and Thr, 13C was incorporated at high rates into Ser (> 38 mol %) during both growth phases of S. suis. A relative fraction of ∼67% of [U-13C3]Ser suggests synthesis from the glycolytic precursor 3-phosphoglycerate (3-PGA). Indeed, S. suis carries all genes that are needed for a three-step Ser biosynthesis from 3-PGA. This metabolic route was further indicated by the enrichment of universally labeled [U-13C2]Gly, which is directly synthesized from [U-13C3]Ser by a Ser hydroxymethyltransferase (glyA, SSU0794). Thus, the 13C label at the C-2 of Gly must have originated from [2,3-13C2]Ser after [1,2-13C2]glucose labeling, further supporting this pathway (Fig. 2C). Ser hydroxymethyltransferase can also catalyze the reversible reaction to synthesize Ser from Gly (38, 39). Because S. suis seems not to possess a Gly synthase, which is in contrast to, for example, Listeria monocytogenes, the enrichment of [3-13C1]Ser after [U-13C6]glucose labeling indicates the reversible Ser hydroxymethyltransferase reaction of single 13C-labeled N5,N10-methylene tetrahydrofolate and unlabeled Gly (from the medium) to tetrahydrofolate and [3-13C1]Ser. 13C-Labeled N5,N10-methylene tetrahydrofolate could be generated by an initial pyruvate formate lyase (pfl, SSU0191) reaction synthesizing [U-13C1]formate and [U-13C2]acetyl-CoA from [U-13C3]Pyr. [U-13C1]formate then react with tetrahydrofolate in a two-step reaction to 13C-labeled N5,N10-methylene tetrahydrofolate. Thus, the observed labeling profile of Ser suggests its synthesis via 3-PGA and Gly as respective precursors. When reaching the stationary phase growth phase, fluxes via the Gly route seem to be increased.
When omitting Lys from CDM, S. suis was still able to grow. However, the mode of Lys biosynthesis remains unclear because S. suis apparently lacks a diaminopimelate epimerase (EC 5.1.1.7). Nevertheless, the biosynthesis rate of ∼5.2% according to our isotopologue profiling results shows that Lys can be made by the streptococci de novo, even though Lys seems to be preferentially taken up from the medium (Fig. 2B). The accumulation of 75% of 13C5-labeled Lys furthermore indicates its biosynthesis from [U-13C3]pyruvate and [1,2,3-13C3]Asp, which has also been described for L. monocytogenes (23). An enrichment of a small fraction of 13C6-labeled Lys in the stationary growth phase might reflect the biosynthesis from accumulated [U-13C4]Asp (see above). Because a BLAST search with diaminopimelate epimerase of L. monocytogenes revealed no significant hit for S. suis strain P1/7 and all other sequenced strains, the question still remains regarding how the final steps in the Lys pathway are organized.
The genome annotation also revealed the presence of all genes encoding the enzymes required for synthesizing the branched chain amino acid members Ile, Leu, and Val. Leu auxotrophy observed in the growth experiments could also be confirmed by isotopologue profiling, because we have never detected any significant 13C enrichment in Leu (supplemental Tables S3 and S4). In contrast to this, we detected a significant 13C label in Val with a biosynthesis rate of ∼3.5% with the presence of almost only [U-13C5]Val isotopologues after feeding the bacteria on [U-13C6]glucose (Fig. 2B). This isotopologue pattern indicates the biosynthesis pathway from two [U-13C3]-Pyr molecules (Fig. 3) and is consistent with the genome annotation. S. suis strain 10 was found to be not auxotrophic for Ile under the conditions tested. However, no significant 13C excess of Ile could be detected in the labeling experiments (supplemental Table S4). Generally, S. suis seems to efficiently take up all members of the branched chain amino acid family.
Within the group of aromatic amino acids, 13C incorporation rates of 5.8 and 3.3 mol % were detected in Phe and Tyr, respectively, in the exponential growth phase of S. suis (Fig. 2B). The accumulation of [U-13C9]Tyr and [U-13C9]Phe directly reflects the biosynthesis from completely labeled chorismate via the shikimate pathway (Fig. 3). Chorismate itself is synthesized in six metabolic reactions from fully labeled erythrose-4-phosphate and two molecules of PEP originating from the PPP and EMP, respectively. Furthermore, the 13C2- and 13C4-labeled isotopologues of Tyr and Phe after [1,2-13C2]glucose feeding indicate synthesis from unlabeled erythrose-4-phosphate and one or two labeled pyruvate molecules, respectively (Fig. 2C). These labeling patterns of Phe and Tyr reveal that glucose is also metabolized by the PPP and that PPP intermediates can be utilized at least for anabolic reactions like aromatic acid biosynthesis but apparently did not contribute to pyruvate formation (see above). Although Trp could not be detected, as explained above, the growth experiments and the absence of biosynthetic genes in the genome (supplemental Table S9) indicate that S. suis strain 10 cannot synthesize Trp.
Isotopologue Profiling under ex Vivo Conditions
To understand the central carbon metabolism of S. suis in ecological niches that it may encounter in natural infections, we also performed ex vivo isotopologue profiling experiments. For this, S. suis strain 10 was incubated in freshly drawn blood and CSF from clinically healthy pigs supplemented with 5 mm [U-13C6]glucose or [1,2-13C2]glucose for 6 h. Isotopologue measurements revealed that the overall 13C excess obtained after incubation in blood or CSF was lower when compared with the labeling experiments in CDM (Fig. 4A), most probably because of naturally occurring unlabeled glucose in these body fluids. Accordingly, glucose levels in porcine serum and CSF have been reported to be in the ranges of ∼4–8 and 1–4 mm, respectively (40–42). On average, 13C excess was higher in CSF than in blood, suggesting different rates of amino acid de novo biosynthesis, as well as different uptake rates of unlabeled amino acids provided by both body fluids. However, similar to the experiments in CDM medium, the highest relative labeling intensities were detected in Ala, Asp, Ser, and Thr under both conditions. [U-13C6]Glucose-derived biosynthesis rates for Ala, Asp, and Ser were in the ranges of 2.5–8 and 11–23% in porcine blood and CSF, respectively (Fig. 4B). In particular, the high proportion of [1,2,3-13C3]Asp and [2,3-13C2]Asp detected after labeling with [U-13C6]glucose and [1,2-13C2]glucose, respectively, indicated the central role of the PEP carboxylase reaction in both blood and in CSF (Fig. 4, B and C).
FIGURE 4.
13C profiles of de novo synthesized amino acids of S. suis strain 10 cultivated in porcine blood and cerebrospinal fluid ex vivo. A, color map for the overall 13C excess (mol %) of labeled amino acids after growth of S. suis strain 10 in porcine blood (B columns) and cerebrospinal fluid (C columns) in the presence of [U-13C6]glucose or [1,2-13C2]glucose. White boxes indicate amino acids that could not be detected in these experiments. Mean values of five (blood) and six (CSF) biological replicates for which MS measurements were performed in triplicate are given. B and C, relative isotopologue composition of amino acids derived from S. suis grown in porcine blood (B columns) and cerebrospinal fluid (C columns) with [U-13C6]glucose (B) or [1,2-13C2]glucose (C). 13C excess values (mol %) are shown as black columns on the left y axis. Colored columns on the right y axis indicate the percentages of labeled isotopologues comprising up to nine 13C atoms (M+1 to M+9). Mean values are shown for five (blood) and six (CSF) biological experiments each measured in triplicate. The horizontal line indicates the detection limit set to 0.5 mol % 13C excess in these experiments. D, relative fractions of isotopologues in the Ala-232 and Ala-260 fragments, which were relevant to discriminate glucose fluxes of the EMP pathway and the PPP, are shown. S. suis strain 10 was incubated for 6 h in porcine cerebrospinal fluid (C columns) or porcine blood (B columns) supplemented with 5 mm [1,2-13C2]glucose, respectively. Colored columns on the left y axis indicate the percentages of isotopologues comprising up to three 13C atoms (M+1 to M+3). Note that M+0 stands for unlabeled Ala and indicates uptake of extracellular Ala. Mean values and standard deviations are shown for two independent biological experiments, each measured in triplicate.
To elucidate the relative contribution of EMP and PPP to the central carbon metabolism of S. suis under the selected ex vivo conditions, we analyzed the Ala-232 and Ala-260 fragments (Fig. 4D). In analogy to the results from the CDM experiments, the isotopologue patterns of Ala-232 again indicated its synthesis from [2,3-13C2]GAP via the EMP pathway and not via the PPP. This is further supported by the absence of single 13C-labeled Ala-232 and Ala-260 (m/z 260) that would have been synthesized from labeled [1,3-13C2]fructose-6-phosphate and [1-13C1]fructose-6-phosphate originating from the PPP, respectively. In conclusion, glucose degradation into pyruvate is generally afforded via the EMP, but not via the PPP when growing S. suis in CDM, blood, or CSF. However, as found in vitro, a 13C carbon flux through the PPP could be detected by 13C enrichments in the aromatic acids Phe and Tyr, at least after cultivation of S. suis in CSF, revealing glucose metabolization to erythrose-4-phosphate for anabolic purposes.
The role of the EMP pathway is additionally underlined by high levels of [U-13C3]Ser (Fig. 4B). Ser biosynthesis originating from glycolytic 3-PGA seems to be even preferred in blood and CSF when compared with CDM. Further evidence is given by our results from growth experiments in CSF, which revealed high percentages of [U-13C2]Gly and single 13C-labeled Gly after [U-13C6]glucose and [1,2-13C2]glucose labeling, respectively. In addition, significant 13C enrichments were detected in Ile, Val, Tyr, Phe, and Lys after incubation of S. suis in CSF, although reconstruction of exact biosynthesis pathways for the latter four is arguable because of high standard deviations considering biological replicates. This may be explained by more complex recycling in biosynthesis pathways of labeled and unlabeled precursors that might be provided by the CSF. Nevertheless, double 13C-labeled Ile was observed in both [U-13C6]glucose and [1,2-13C2]glucose labeling experiments, suggesting its biosynthesis from labeled pyruvate and unlabeled Thr, which is in accordance with the Ile synthesis pathway predicted from the genome annotation. In good correlation with CDM labeling experiments, Glu, His, and Leu were again detected in unlabeled states under ex vivo conditions.
In conclusion, the labeling patterns provide strong evidence that S. suis in general catabolizes glucose mainly via the EMP pathway. However, they also indicate that the rates of de novo amino acid are dictated by the physiological composition of porcine blood and CSF.
Functional Characterization of a Phosphoenolpyruvate Carboxylase (ppc) Mutant
The isotopologue results suggest a dominant role of the glycolytic pathway in S. suis metabolism. In addition, the inability to synthesize Glu/Gln indicates a substantial impact of OAA/Asp on amino acid synthesis, as evidenced by a high proportion of 13C3-labeled Asp, and suggested an important role of ppc (SSU0479) for the central carbon metabolism under in vitro and ex vivo conditions. Thus, this anaplerotic reaction seems to be crucial for oxaloacetate synthesis in S. suis (Fig. 3), because the TCA cycle is incomplete.
To prove this hypothesis, we constructed a S. suis ppc deletion mutant (10Δppc) and the complemented strain 10cΔppc. Successful deletion and complementation were confirmed by RT-qPCR, respectively. Possible polar effects on the expression of upstream (SSU0478) and downstream (SSU0480) genes could be excluded (Fig. 5A).
First, we phenotypically characterized strain 10Δppc in CDM growth experiments. Notably, in contrast to the wild type, the ppc mutant showed strongly reduced growth in complete CDM containing Asp, even when casamino acids containing all amino acids except Trp were added (Fig. 5B). Also the supplementation with Asp, OAA, and α-ketoglutarate did not compensate the OAA deficiency in strain 10Δppc, most probably because of the lack of respective transporters in S. suis (data not shown). In contrast, supplementation with tryptone and peptone partially restored growth to the wild type level, indicating that Asp containing oligopeptides provided by both supplements might compensate for the Asp and OAA deficiencies. Also the reversible reaction of the aspartate transaminase (aspC, SSU0569) to synthesize OAA from Asp may not be favored under these conditions. Because the growth defect of strain 10Δppc was almost abolished in commercial THB media (data not shown), we performed isotopologue profiling experiments under these conditions in the presence of [U-13C6]glucose. Notably, 13C incorporation was not detected in Asp and Thr in strain 10Δppc, which was in striking contrast to the wild type strain and the complemented mutant strain 10cΔppc (Fig. 5C). As expected, formation of 13C-labeled Ala and Ser, synthesized from the glycolytic intermediates pyruvate and 3-PGA, respectively, was not influenced by the ppc deletion (supplemental Table S5). These experiments demonstrate that the ppc gene is essential for de novo biosynthesis of oxaloacetate and its products Asp and Thr.
The ex vivo isotopologue experiments suggested the major importance of the PEP carboxylase reaction for streptococcal proliferation in porcine blood and CSF, but the growth experiments with 10Δppc in CDM also suggested that certain supplements may compensate for ppc deficiency. To prove whether this effect also occurs under ex vivo conditions, we monitored the growth of 10Δppc in porcine blood and CSF over 24 h. In comparison with the wild type strain, the ppc mutant showed a significantly reduced growth in blood and CSF (Fig. 5D). Although strain 10Δppc survived during the tested 24 h, it seemed not to be able to use the nutrients in blood or CSF for substantial replication. This experiment clearly points toward an important central role of the PEP carboxylase for bacterial fitness in host environments.
DISCUSSION
In the present study, isotopologue profiling was applied to elucidate the metabolic pathways used by S. suis for glucose utilization and amino acid de novo biosynthesis. The data obtained substantially contribute to a better understanding of the in vitro and ex vivo metabolic network in S. suis and are summarized in the schematic model depicted in Fig. 6.
FIGURE 6.
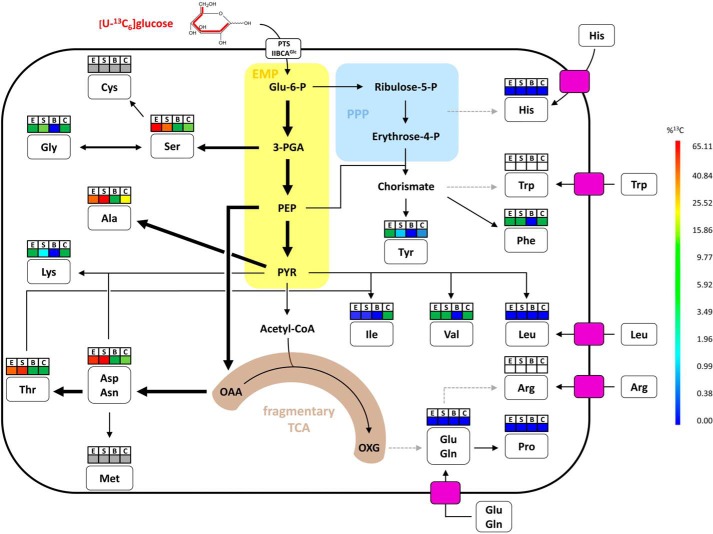
Simplified metabolic model for amino acid biosynthesis pathways of S. suis in vitro and ex vivo. This model is based on the information obtained from isotopologue profiling experiments, amino acid auxotrophy experiments, and S. suis P1/7 genome annotation. Relative metabolic fluxes are shown for isotopologue experiments conducted for S. suis grown in CDM to the exponential (E) growth phase, in CDM to the stationary (S) growth phase, in porcine blood (B), and in porcine CSF (C). Higher relative metabolic fluxes are displayed by thick arrows. Dotted gray arrows indicate that genes required for the biosynthesis of the amino acid are absent in the S. suis P1/7 genome. The colored boxes above amino acids correspond to the mean overall 13C enrichments described for Figs. 2A and 4A. Gray and white boxes stand for amino acids that could not be measured by our protocol but were found to be nonessential and essential, respectively, for growth of S. suis in CDM in vitro. Furthermore, the necessity for uptake of essential amino acids by streptococcal transporters is indicated by pink rectangles. Glu-6-P, glucose-6-phosphate; Acetyl-CoA, acetyl coenzyme A; OXG, oxoglutarate (α-ketoglutarate).
Like all other streptococci, S. suis is a facultative anaerobe bacterium that lacks a respiratory metabolism. The inability of respiration in streptococci is associated with the absence of TCA genes or, in case of S. suis and other oral streptococci, with an incomplete TCA (43). Thus, S. suis mainly metabolizes carbohydrates via homolactic or mixed acid fermentation. Correspondingly, S. suis expresses several PTS or ABC transporter systems that allow the uptake of a broad spectrum of carbohydrates (refer to Fig. 1) and ensure an efficient substrate supply for the glycolysis.
Our data show that in S. suis glucose is preferentially catabolized by the EMP pathway, although the PPP seems to provide S. suis with intermediates needed for anabolic reactions like aromatic acid biosynthesis. The importance of a glycolytic flux, at least for sugars that can be easily metabolized, such as glucose, is reflected by our in vitro and ex vivo 13C isotopologue profiling results. Although the overall mol % 13C excess was different after growth in CDM, blood, and CSF, in general the highest 13C enrichments were detected under all conditions in the EMP pathway-derived amino acids Ala, Asp, Ser, and Thr. In addition, the isotopologue profiles of Asp and Thr indicated synthesis of oxaloacetate via the carboxylation of PEP, which corresponds to studies on L. monocytogenes and Streptococcus pneumoniae (23, 25), although L. monocytogenes possesses a pyruvate carboxylase instead of a PEP carboxylase. However, in contrast to these studies, we detected high enrichments of [U-13C4]Asp and [U-13C4]Thr isotopologues after growth of S. suis in CDM, which must be a result of the incorporation of 13CO2. Putative sources of 13CO2 are probably the decarboxylation reactions of the fragmentary TCA (pdhABC, pyruvate dehydrogenase complex; icd, isocitrate dehydrogenase) or the oxidative part of the PPP (gnd, 6-phosphogluconate dehydrogenase). A minor contribution of the PPP to glucose metabolization in vitro can be deduced from universally labeled [U-13C9]Tyr and [U-13C9]Phe, which were formed from [U-13C6]glucose precursors via biosynthesis of fully labeled PEP and erythrose-4-phosphate of the nonoxidative part of the PPP. However, the isotopologue patterns of Ala-260 and Ala-232 did not suggest such a contribution of the PPP. Explanations for this contradiction could be that the amount of [1,3-13C2]Ala and [3-13C1]Ala from uniquely labeled [1,3-13C2]fructose-6-phosphate and [1-13C1]fructose-6-phosphate provided by the PPP was under the detection limit of our method or that these fructose molecules were not efficiently catabolized to pyruvate and Ala. The latter case would suggest that glucose catabolized by the PPP will predominantly fuel anabolic reactions like the aromatic acid biosynthesis instead of the catabolic EMP pathway. Such modular subnetwork structures have been postulated for central metabolic pathways like EMP or PPP in E. coli (44, 45), whereas in S. suis this awaits further experiments.
Our data revealed that oxaloacetate synthesis by carboxylation of PEP is a central reaction in glucose catabolism of S. suis. Oxaloacetate is an indispensable precursor not only for the biosynthesis of Asp and Thr, but also for Lys, β-alanine (pantothenate), nicotinamide, nicotinate, α-ketoglutarate, and purines (46, 47). In fact, the growth deficiency of strain 10Δppc in CDM may be caused by the lack of oxaloacetate production. Interestingly, growth was even abolished when the purines adenine and guanine were omitted from CDM (data not shown), suggesting that oxaloacetate is exhausted in strain 10Δppc during growth. Considering that S. suis seems not to encode additional proteins for gluconeogenesis or anaplerotic reactions like pyruvate carboxylase, decarboxylating malate dehydrogenase, or PEP carboxykinase (48), our results emphasize an essential role of ppc for the central carbon metabolism of S. suis. Indeed, this metabolic feature is conserved within all sequenced S. suis strains and was confirmed in our additional experiments with another serotype 2 strain 05ZYH33, which had been isolated from a human outbreak in China (supplemental Table S8).
Isotopologue profiling of S. suis grown in porcine blood or CSF ex vivo clearly indicated that the central metabolism of S. suis is well adapted to nutrient sources provided by the host. Overall, our isotopologue data show an efficient glucose flux through the glycolysis in vitro and ex vivo and therefore emphasize the relevance of these reactions for survival of S. suis in vivo. Porcine blood and CSF contain glucose levels of ∼4–8 and 1–4 mm, respectively (40–42). The high glucose levels in blood and CSF might explain the lower 13C overall excess observed ex vivo because of naturally occurring unlabeled glucose. However, they do not sufficiently explain the labeling differences observed between blood and CSF. This rather suggests that the supply with other nutrients, most probably carbohydrates or oligopeptides, also regulates the metabolic fate of glucose through glycolysis and PPP ex vivo. Indeed, S. suis encodes several peptidases, glycosidases, as well as carbohydrate and peptide transporters that would allow an efficient utilization of available, alternative substrates in blood and CSF. Especially porcine blood may provide S. suis with alternative (unlabeled) carbohydrate sources or oligopeptides that can be used for proliferation and therefore might explain the reduced ability to detect [U-13C6]glucose-dependent de novo amino acid biosynthesis by isotopologue profiling. This assumption may be also supported by the lower overall 13C enrichment rates observed in blood when compared with CDM, although the concentrations of certain amino acids in porcine serum (1–10 mg/dl) and CDM (10 mg/dl) are not strongly differing from each other. In accordance, our data show that S. suis performs de novo biosynthesis and uptake of amino acids at the same time. Thus, very efficiently synthesized amino acids like Asp, Ala, Ser, and Thr were never detected with 13C excess rates of ∼100% in CDM, as well as in porcine body fluids, indicating a simultaneously occurring uptake of unlabeled amino acids under all conditions. This holds true in particular for amino acids found to be labeled in even lower percentages like Lys, Phe, Tyr, or Val, indicating that uptake rather than de novo biosynthesis is favored by S. suis. The amino acid concentrations of porcine serum (1–10 mg/dl) and CSF (0.1–2 mg/dl) differ and even vary in the composition of the single amino acids (49, 50). This implies that porcine blood and CSF provide sufficient amounts of amino acids for S. suis at least for the time period investigated. Nevertheless, the higher 13C excess in amino acids found after cultivation in CSF compared with blood suggests an enhanced demand of de novo biosynthesis by S. suis. Further evidence for this metabolic fine-tuning is given by the higher de novo biosynthesis rates for Lys, Phe, and Ile after incubation of S. suis in CSF compared with CDM. Ile and Phe are amino acids that were found in rather low concentrations in CSF (49, 50). In particular, Ile biosynthesis seems to be the result of exhausting Ile amounts in CSF because de novo biosynthesis of Ile could not be detected in vitro.
Regarding the overall amino acid composition of porcine serum and CSF, Gln is among the most abundant amino acids present. S. suis is auxotrophic for Gln/Glu but to satisfy a high demand on Glu, which is also needed for transamination reactions, it is not surprising that five different Gln transporters (GlnQ1–GlnQ5) were annotated in the S. suis P1/7 genome (36). This will ensure an efficient Gln/Glu supply for the bacterial cell and may reflect an evolutionary mechanism for a specific adaptation of S. suis to its host niches. The Gln/Glu auxotrophy also raises the question on the need of the fragmentary TCA cycle for S. suis fitness. In contrast to L. monocytogenes, which utilizes its incomplete TCA for Glu synthesis via α-ketoglutarate (23), S. suis obviously lacks the genes encoding for a Glu synthetase. Even though S. suis possesses a Glu dehydrogenase (gdh, SSU0234), S. suis Gdh was reported to preferentially utilize Glu rather than α-ketoglutarate as the substrate (51). It is conceivable that this fragmentary pathway allows synthesis of additional NADH that is not only used for homolactic fermentation of pyruvate to lactate but also contributes to a balanced NADH/NAD+ ratio (52). This ratio is implicated in controlling the shift from homolactic to mixed acid fermentation in Lactococcus lactis subsp. lactis and has been discussed in the context of pneumococcal fitness (53, 54). Interestingly, in vitro we only detected unlabeled α-ketoglutarate (data not shown), indicating its production predominately by transamination reactions from unlabeled Glu. Because we were not able to measure α-ketoglutarate from ex vivo experiments, the role of the incomplete TCA for S. suis fitness is still speculative and awaits further analysis.
Overall, with respect to the central carbon catabolic pathways, EMP and PPP, as well as most of the amino acid biosynthesis pathways, our experimental data corresponded well with the genome annotation. Nevertheless, we were not able to detect [U-13C6]glucose dependent de novo biosynthesis of α-ketoglutarate and Glu under our conditions, contradicting whole genome data because the respective S. suis genes SSU1040–1042 and gdh, which suggest potential biosynthesis pathways, were present in all sequenced S. suis strains. This holds also true for Leu and Lys biosynthesis, where genome annotation does not correspond to the phenotypes we observed. These inconsistencies have to be addressed in further experiments.
Finally, it is important to note that despite the knowledge gained in the identification of virulence-associated factors of S. suis, no serotype cross-protective vaccine is available. A more in-depth understanding of bacterial metabolism might help in the design of new strategies for vaccine development to control S. suis infection and to overcome this dilemma. In more detail, our data suggest that transporter proteins for the essential amino acids (Arg, His, Leu, and Trp), the disruption of the anaplerotic reaction, or other biosynthetic pathways may be promising strategies for vaccine development or rational drug design for treatment. In line with this, a S. suis strain deficient in the biosynthesis pathway for aromatic acids has been postulated to be a potential live attenuated vaccine in swine (55). In conclusion, our study denotes the importance of the central carbon metabolism and related amino acid biosynthesis for S. suis fitness and emphasizes its metabolic adaptation to the nutrient availability in the host.
Acknowledgments
We are grateful to Hilde Smith (Lelystad, The Netherlands) and Jiaqi Tang (Research Institute for Medicine of Nanjing Command, Nanjing, China) for providing S. suis strains 10 and 05ZYH33, respectively. We thank Anna Drees, Petra Gruening, and Alexandra von Altrock for collecting porcine CSF samples. Kira van Vorst is acknowledged for laboratory support.
This work was supported by Grants GO-983-3/1 and EI-384/5-1 from the Deutsche Forschungsgemeinschaft within the Priority Programme SPP1316 and by the Niedersachsen-Research Network on Neuroinfectiology of the Ministry of Science and Culture of Lower Saxony.

This article contains supplemental Tables S1–S9.
- CSF
- cerebrospinal fluid
- ppc
- phosphoenolpyruvate carboxylase
- EMP
- Embden-Meyerhof-Parnas
- PPP
- pentose phosphate pathway
- OAA
- oxaloacetate
- PEP
- phosphoenolpyruvate
- CDM
- chemically defined medium
- ED
- Entner-Doudoroff pathway
- THB
- Todd Hewitt broth
- qPCR
- quantitative PCR
- TBDMS
- N-(tert-butyldimethylsilyl)
- GAP
- glyceraldehyde 3-phosphate
- 3-PGA
- 3-phosphoglycerate.
REFERENCES
- 1. Varela N. P., Gadbois P., Thibault C., Gottschalk M., Dick P., Wilson J. (2013) Antimicrobial resistance and prudent drug use for Streptococcus suis. Anim. Health Res. Rev. 14, 68–77 [DOI] [PubMed] [Google Scholar]
- 2. Gottschalk M., Xu J., Calzas C., Segura M. (2010) Streptococcus suis: a new emerging or an old neglected zoonotic pathogen? Future Microbiol. 5, 371–391 [DOI] [PubMed] [Google Scholar]
- 3. Huong V. T., Ha N., Huy N. T., Horby P., Nghia H. D., Thiem V. D., Zhu X., Hoa N. T., Hien T. T., Zamora J., Schultsz C., Wertheim H. F., Hirayama K. (2014) Epidemiology, clinical manifestations, and outcomes of Streptococcus suis infection in humans. Emerg. Infect. Dis. 20, 1105–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mai N. T., Hoa N. T., Nga T. V., Linh le D., Chau T. T., Sinh D. X., Phu N. H., Chuong L. V., Diep T. S., Campbell J., Nghia H. D., Minh T. N., Chau N. V., de Jong M. D., Chinh N. T., Hien T. T., Farrar J., Schultsz C. (2008) Streptococcus suis meningitis in adults in Vietnam. Clin. Infect. Dis. 46, 659–667 [DOI] [PubMed] [Google Scholar]
- 5. Tang J., Wang C., Feng Y., Yang W., Song H., Chen Z., Yu H., Pan X., Zhou X., Wang H., Wu B., Wang H., Zhao H., Lin Y., Yue J., Wu Z., He X., Gao F., Khan A. H., Wang J., Zhao G. P., Wang Y., Wang X., Chen Z., Gao G. F. (2006) Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLoS Med. 3, e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yu H., Jing H., Chen Z., Zheng H., Zhu X., Wang H., Wang S., Liu L., Zu R., Luo L., Xiang N., Liu H., Liu X., Shu Y., Lee S. S., Chuang S. K., Wang Y., Xu J., Yang W. (2006) Human Streptococcus suis outbreak, Sichuan, China. Emerg. Infect. Dis. 12, 914–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wertheim H. F., Nguyen H. N., Taylor W., Lien T. T., Ngo H. T., Nguyen T. Q., Nguyen B. N., Nguyen H. H., Nguyen H. M., Nguyen C. T., Dao T. T., Nguyen T. V., Fox A., Farrar J., Schultsz C., Nguyen H. D., Nguyen K. V., Horby P. (2009) Streptococcus suis, an important cause of adult bacterial meningitis in northern Vietnam. PLoS One 4, e5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hui A. C., Ng K. C., Tong P. Y., Mok V., Chow K. M., Wu A., Wong L. K. (2005) Bacterial meningitis in Hong Kong: 10-years' experience. Clin. Neurol. Neurosurg. 107, 366–370 [DOI] [PubMed] [Google Scholar]
- 9. Silva L. M., Baums C. G., Rehm T., Wisselink H. J., Goethe R., Valentin-Weigand P. (2006) Virulence-associated gene profiling of Streptococcus suis isolates by PCR. Vet. Microbiol. 115, 117–127 [DOI] [PubMed] [Google Scholar]
- 10. Staats J. J., Feder I., Okwumabua O., Chengappa M. M. (1997) Streptococcus suis: past and present. Vet. Res. Commun. 21, 381–407 [DOI] [PubMed] [Google Scholar]
- 11. Gottschalk M., Segura M. (2000) The pathogenesis of the meningitis caused by Streptococcus suis: the unresolved questions. Vet. Microbiol. 76, 259–272 [DOI] [PubMed] [Google Scholar]
- 12. Fittipaldi N., Segura M., Grenier D., Gottschalk M. (2012) Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol. 7, 259–279 [DOI] [PubMed] [Google Scholar]
- 13. Tenenbaum T., Adam R., Eggelnpöhler I., Matalon D., Seibt A., Novotny G. E., Galla H. J., Schroten H. (2005) Strain-dependent disruption of blood-cerebrospinal fluid barrier by Streptoccocus suis in vitro. FEMS Immunol. Med. Microbiol. 44, 25–34 [DOI] [PubMed] [Google Scholar]
- 14. Tenenbaum T., Papandreou T., Gellrich D., Friedrichs U., Seibt A., Adam R., Wewer C., Galla H. J., Schwerk C., Schroten H. (2009) Polar bacterial invasion and translocation of Streptococcus suis across the blood-cerebrospinal fluid barrier in vitro. Cell. Microbiol. 11, 323–336 [DOI] [PubMed] [Google Scholar]
- 15. Baums C. G., Valentin-Weigand P. (2009) Surface-associated and secreted factors of Streptococcus suis in epidemiology, pathogenesis and vaccine development. Anim. Health Res. Rev. 10, 65–83 [DOI] [PubMed] [Google Scholar]
- 16. Willenborg J., Fulde M., de Greeff A., Rohde M., Smith H. E., Valentin-Weigand P., Goethe R. (2011) Role of glucose and CcpA in capsule expression and virulence of Streptococcus suis. Microbiology 157, 1823–1833 [DOI] [PubMed] [Google Scholar]
- 17. Willenborg J., de Greeff A., Jarek M., Valentin-Weigand P., Goethe R. (2014) The CcpA regulon of Streptococcus suis reveals novel insights into the regulation of the streptococcal central carbon metabolism by binding of CcpA to two distinct binding motifs. Mol. Microbiol. 92, 61–83 [DOI] [PubMed] [Google Scholar]
- 18. Pan X., Ge J., Li M., Wu B., Wang C., Wang J., Feng Y., Yin Z., Zheng F., Cheng G., Sun W., Ji H., Hu D., Shi P., Feng X., Hao X., Dong R., Hu F., Tang J. (2009) The orphan response regulator CovR: a globally negative modulator of virulence in Streptococcus suis serotype 2. J. Bacteriol. 191, 2601–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li M., Wang C., Feng Y., Pan X., Cheng G., Wang J., Ge J., Zheng F., Cao M., Dong Y., Liu D., Wang J., Lin Y., Du H., Gao G. F., Wang X., Hu F., Tang J. (2008) SalK/SalR, a two-component signal transduction system, is essential for full virulence of highly invasive Streptococcus suis serotype 2. PLoS One 3, e2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eisenreich W., Dandekar T., Heesemann J., Goebel W. (2010) Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat. Rev. Microbiol. 8, 401–412 [DOI] [PubMed] [Google Scholar]
- 21. Tang J. K., You L., Blankenship R. E., Tang Y. J. (2012) Recent advances in mapping environmental microbial metabolisms through 13C isotopic fingerprints. J. R. Soc. Interface 9, 2767–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fuchs T. M., Eisenreich W., Kern T., Dandekar T. (2012) Toward a systemic understanding of Listeria monocytogenes metabolism during infection. Front. Microbiol. 3, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eisenreich W., Slaghuis J., Laupitz R., Bussemer J., Stritzker J., Schwarz C., Schwarz R., Dandekar T., Goebel W., Bacher A. (2006) 13C isotopologue perturbation studies of Listeria monocytogenes carbon metabolism and its modulation by the virulence regulator PrfA. Proc. Natl. Acad. Sci. U.S.A. 103, 2040–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eylert E., Herrmann V., Jules M., Gillmaier N., Lautner M., Buchrieser C., Eisenreich W., Heuner K. (2010) Isotopologue profiling of Legionella pneumophila: role of serine and glucose as carbon substrates. J. Biol. Chem. 285, 22232–22243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Härtel T., Eylert E., Schulz C., Petruschka L., Gierok P., Grubmüller S., Lalk M., Eisenreich W., Hammerschmidt S. (2012) Characterization of central carbon metabolism of Streptococcus pneumoniae by isotopologue profiling. J. Biol. Chem. 287, 4260–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eylert E., Schär J., Mertins S., Stoll R., Bacher A., Goebel W., Eisenreich W. (2008) Carbon metabolism of Listeria monocytogenes growing inside macrophages. Mol. Microbiol. 69, 1008–1017 [DOI] [PubMed] [Google Scholar]
- 27. Gillmaier N., Götz A., Schulz A., Eisenreich W., Goebel W. (2012) Metabolic responses of primary and transformed cells to intracellular Listeria monocytogenes. PLoS One 7, e52378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Götz A., Eylert E., Eisenreich W., Goebel W. (2010) Carbon metabolism of enterobacterial human pathogens growing in epithelial colorectal adenocarcinoma (Caco-2) cells. PLoS One 5, e10586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gruening P., Fulde M., Valentin-Weigand P., Goethe R. (2006) Structure, regulation, and putative function of the arginine deiminase system of Streptococcus suis. J. Bacteriol. 188, 361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schmittgen T. D., Livak K. J. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 31. Koczula A., Willenborg J., Bertram R., Takamatsu D., Valentin-Weigand P., Goethe R. (2014) Establishment of a Cre recombinase based mutagenesis protocol for markerless gene deletion in Streptococcus suis. J. Microbiol. Methods 107, 80–83 [DOI] [PubMed] [Google Scholar]
- 32. Smith H. E., Wisselink H. J., Vecht U., Gielkens A. L., Smits M. A. (1995) High-efficiency transformation and gene inactivation in Streptococcus suis type-2. Microbiology 141, 181–188 [DOI] [PubMed] [Google Scholar]
- 33. Lee W. N., Byerley L. O., Bergner E. A., Edmond J. (1991) Mass isotopomer analysis: theoretical and practical considerations. Biol. Mass Spectrom. 20, 451–458 [DOI] [PubMed] [Google Scholar]
- 34. Bidossi A., Mulas L., Decorosi F., Colomba L., Ricci S., Pozzi G., Deutscher J., Viti C., Oggioni M. R. (2012) A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PLoS One 7, e33320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Paulsen I. T., Nguyen L., Sliwinski M. K., Rabus R., Saier M. H., Jr. (2000) Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 301, 75–100 [DOI] [PubMed] [Google Scholar]
- 36. Holden M. T., Hauser H., Sanders M., Ngo T. H., Cherevach I., Cronin A., Goodhead I., Mungall K., Quail M. A., Price C., Rabbinowitsch E., Sharp S., Croucher N. J., Chieu T. B., Mai N. T., Diep T. S., Chinh N. T., Kehoe M., Leigh J. A., Ward P. N., Dowson C. G., Whatmore A. M., Chanter N., Iversen P., Gottschalk M., Slater J. D., Smith H. E., Spratt B. G., Xu J., Ye C., Bentley S., Barrell B. G., Schultsz C., Maskell D. J., Parkhill J. (2009) Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS One 4, e6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sperandio B., Gautier C., McGovern S., Ehrlich D. S., Renault P., Martin-Verstraete I., Guédon E. (2007) Control of methionine synthesis and uptake by MetR and homocysteine in Streptococcus mutans. J. Bacteriol. 189, 7032–7044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schirch V., Hopkins S., Villar E., Angelaccio S. (1985) Serine hydroxymethyltransferase from Escherichia coli: purification and properties. J. Bacteriol. 163, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Blakley R. L. (1955) The interconversion of serine and glycine: participation of pyridoxal phosphate. Biochem. J. 61, 315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kaiser G. M., Frühauf N. R. (2007) Method of intracranial pressure monitoring and cerebrospinal fluid sampling in swine. Lab. Anim. 41, 80–85 [DOI] [PubMed] [Google Scholar]
- 41. Montgomery G. W., Flux D. S., Greenway R. M. (1980) Tryptophan deficiency in pigs: changes in food intake and plasma levels of glucose, amino acids, insulin and growth hormone. Horm. Metab. Res. 12, 304–309 [DOI] [PubMed] [Google Scholar]
- 42. van Hulst R. A., Haitsma J. J., Klein J., Lachmann B. (2003) Oxygen tension under hyperbaric conditions in healthy pig brain. Clin. Physiol. Funct. Imaging 23, 143–148 [DOI] [PubMed] [Google Scholar]
- 43. Poolman B. (1993) Energy transduction in lactic acid bacteria. FEMS Microbiol. Rev. 12, 125–147 [DOI] [PubMed] [Google Scholar]
- 44. Noor E., Eden E., Milo R., Alon U. (2010) Central carbon metabolism as a minimal biochemical walk between precursors for biomass and energy. Mol. Cell 39, 809–820 [DOI] [PubMed] [Google Scholar]
- 45. Chubukov V., Gerosa L., Kochanowski K., Sauer U. (2014) Coordination of microbial metabolism. Nat. Rev. Microbiol. 12, 327–340 [DOI] [PubMed] [Google Scholar]
- 46. Schär J., Stoll R., Schauer K., Loeffler D. I., Eylert E., Joseph B., Eisenreich W., Fuchs T. M., Goebel W. (2010) Pyruvate carboxylase plays a crucial role in carbon metabolism of extra- and intracellularly replicating Listeria monocytogenes. J. Bacteriol. 192, 1774–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sauer U., Eikmanns B. J. (2005) The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 29, 765–794 [DOI] [PubMed] [Google Scholar]
- 48. Fuchs T. M., Eisenreich W., Heesemann J., Goebel W. (2012) Metabolic adaptation of human pathogenic and related nonpathogenic bacteria to extra- and intracellular habitats. FEMS Microbiol. Rev. 36, 435–462 [DOI] [PubMed] [Google Scholar]
- 49. Rosenling T., Slim C. L., Christin C., Coulier L., Shi S., Stoop M. P., Bosman J., Suits F., Horvatovich P. L., Stockhofe-Zurwieden N., Vreeken R., Hankemeier T., van Gool A. J., Luider T. M., Bischoff R. (2009) The effect of preanalytical factors on stability of the proteome and selected metabolites in cerebrospinal fluid (CSF). J. Proteome. Res. 8, 5511–5522 [DOI] [PubMed] [Google Scholar]
- 50. Hickman R., Alp M. H. (1986) A predictable pathophysiological model of porcine hepatic failure. Eur. Surg. Res. 18, 283–292 [DOI] [PubMed] [Google Scholar]
- 51. Okwumabua O., Persaud J. S., Reddy P. G. (2001) Cloning and characterization of the gene encoding the glutamate dehydrogenase of Streptococcus suis serotype 2. Clin. Diagn. Lab. Immunol. 8, 251–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Neijssel O. M., Snoep J. L., Teixeira de Mattos M. J. (1997) Regulation of energy source metabolism in streptococci. Soc. Appl. Bacteriol. Symp. Ser. 26, 12S–19S [PubMed] [Google Scholar]
- 53. Auzat I., Chapuy-Regaud S., Le Bras G., Dos Santos D., Ogunniyi A. D., Le Thomas I., Garel J. R., Paton J. C., Trombe M. C. (1999) The NADH oxidase of Streptococcus pneumoniae: its involvement in competence and virulence. Mol. Microbiol. 34, 1018–1028 [DOI] [PubMed] [Google Scholar]
- 54. Garrigues C., Loubiere P., Lindley N. D., Cocaign-Bousquet M. (1997) Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ ratio. J. Bacteriol. 179, 5282–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fittipaldi N., Harel J., D'Amours B., Lacouture S., Kobisch M., Gottschalk M. (2007) Potential use of an unencapsulated and aromatic amino acid-auxotrophic Streptococcus suis mutant as a live attenuated vaccine in swine. Vaccine 25, 3524–3535 [DOI] [PubMed] [Google Scholar]