Background: 15-Hydroxyprostaglandin dehydrogenase (15PGDH) catalyzes the oxidation of hydroxylated polyunsaturated fatty acids to α,β-unsaturated carbonyl-containing electrophiles.
Results: Hydroxylated docosahexaenoic acid (DHA) metabolites are substrates for 15PGDH, yielding electrophilic oxoDHA products that are anti-inflammatory.
Conclusion: Hydroxylated Ω-3 fatty acid species are conferred with cell signaling capabilities after oxidation by 15PGDH.
Significance: Formation of biologically active metabolites by 15PGDH contributes to the salutary signaling actions of Ω-3 fatty acids.
Keywords: Eicosanoid, Fatty Acid Oxidation, Lipid Oxidation, Lipid Signaling, Signaling
Abstract
15-Hydroxyprostaglandin dehydrogenase (15PGDH) is the primary enzyme catalyzing the conversion of hydroxylated arachidonic acid species to their corresponding oxidized metabolites. The oxidation of hydroxylated fatty acids, such as the conversion of prostaglandin (PG) E2 to 15-ketoPGE2, by 15PGDH is viewed to inactivate signaling responses. In contrast, the typically electrophilic products can also induce anti-inflammatory and anti-proliferative responses. This study determined that hydroxylated docosahexaenoic acid metabolites (HDoHEs) are substrates for 15PGDH. Examination of 15PGDH substrate specificity was conducted in cell culture (A549 and primary human airway epithelia and alveolar macrophages) using chemical inhibition and shRNA knockdown of 15PGDH. Substrate specificity is broad and relies on the carbon position of the acyl chain hydroxyl group. 14-HDoHE was determined to be the optimal DHA substrate for 15PGDH, resulting in the formation of its electrophilic metabolite, 14-oxoDHA. Consistent with this, 14-HDoHE was detected in bronchoalveolar lavage cells of mild to moderate asthmatics, and the exogenous addition of 14-oxoDHA to primary alveolar macrophages inhibited LPS-induced proinflammatory cytokine mRNA expression. These data reveal that 15PGDH-derived DHA metabolites are biologically active and can contribute to the salutary signaling actions of Ω-3 fatty acids.
Introduction
15-Hydroxyprostaglandin dehydrogenase (15PGDH)4 is a key enzyme in prostaglandin metabolism. It is responsible for the conversion of prostaglandin (PG) E2, a hydroxylated prostaglandin, to its corresponding oxidized counterpart, 9,15-dioxo-11α-hydroxy-prosta-5Z,13E-dien-1-oic acid (15-ketoPGE2). PGE2 is a pro-proliferative prostaglandin whose activity is increased in cancer because of an increase in the expression of cyclooxygenase 2 (COX-2), an inducible enzyme that contributes to PGE2 production, and a corresponding decrease in 15PGDH expression. 15PGDH renders PGE2 biologically inactive by oxidizing the hydroxyl group at the C15 position to a carbonyl (1–4). This mechanism of oxidation is not specific to PGE2 as 15PGDH is also responsible for the conversion of 15(S)-hydroxy-5Z,8Z,11Z,13E-eicosatetraenoic acid and 11(R)-hydroxy-5Z,8Z,12E,14Z-eicosatetraenoic acid to their corresponding keto metabolites, 15- and 11-oxoeicosatetraenoic acid (15- and 11-oxoETE), respectively (5, 6). Also, oxidation of the C15 hydroxyl group of the trihydroxy arachidonic acid metabolite lipoxin A4 (LXA4) putatively inactivates LXA4 by inhibiting G protein-coupled receptor ligand activity (7).
Numerous hydroxylated Ω-3 and Ω-6 fatty acids are produced through COX, lipoxygenase, and cytochrome P450 metabolism or by reactive oxygen species. However, the dehydrogenase(s) responsible for the oxidation of the hydroxyl group to a carbonyl has not been identified for many of these metabolites. 5-Hydroxyeicosatetraenoic acid (5-HETE) is oxidized by 5-hydroxyeicosanoid dehydrogenase (8, 9), and there is some evidence of a 13-hydroxyoctadecadienoic acid (13-HODE) dehydrogenase that converts 13-HODE and 9-HODE to their corresponding oxo-octadecadienoic acid metabolites (10). Notably, all of the 15PGDH substrates thus described are metabolites of arachidonic acid, with the exception of (5(S),12(R),18(R))-trihydroxy-6Z,8E,10E,14Z,16E-eicosapentaenoic acid (resolvin E1) (11).
From a different perspective, oxo metabolites of hydroxy-fatty acids are α,β-unsaturated ketones that can mediate significant cell signaling and functional responses. For example, 15-ketoPGE2 formation not only decreases the levels of the pro-proliferative PGE2 but also inhibits growth of hepatocellular carcinoma and cholangiocarcinoma by peroxisome proliferator activator receptor γ (PPARγ) activation (12, 13). Studies in human umbilical vein endothelial cells affirm that 11-oxoETE and 15-oxoETE mediate anti-proliferative properties (5, 6).
Electrophilic lipids such as fatty acid nitroalkene derivatives and Ω-6 and Ω-3-derived α,β-unsaturated ketones are products of metabolic and oxidative inflammatory reactions that also act as signaling mediators. Mechanisms of action include induction of the expression of anti-inflammatory genes regulated by the transcription factor nuclear factor-erythroid 2-related factor 2 (Nrf2). Electrophilic fatty acids form Michael adducts with cysteines, for example primarily the Cys273 and Cys288 in KEAP1 (Kelch-like ECH-associated protein 1), which is responsible for sequestering Nrf2 in the cytosol and regulating its proteasomal degradation (14). This adduction promotes the release of Nrf2 and its translocation to the nucleus, where it promotes antioxidant response element gene expression (14, 15). Electrophilic fatty acids also induce heat shock factor expression and suppress proinflammatory gene expression through Cys38 alkylation and inhibition of DNA binding by the p65 subunit of NF-κB (14, 16–18). By these mechanisms, electrophilic fatty acids can act as pleiotropic signaling molecules that form reversible Michael addition adducts with nucleophilic species, including low molecular weight thiols and cysteine residues of proteins (16). In this context, 17-oxoDHA and 7Z,10Z,13Z,15E,19Z-17-keto-docosapentaenoic acid (17-oxoDPA) and 15-oxoETE can induce Nrf2 expression, activate Nrf2-dependent gene expression (heme oxygenase-1 (HO-1), glutamate cysteine ligase modifier (GCLM), NAPDH oxidoreductase 1 (NQO1)), decrease the protein expression of inducible nitric-oxide synthase and COX-2 in Raw264.7 cells, and suppress NF-κB-mediated mRNA cytokine expression in THP-1 cells (19, 20).
This study evaluated the substrate specificity of 15PGDH for hydroxylated DHA (HDoHE) species and revealed the signaling actions of 15PGDH-derived electrophilic products in human primary alveolar macrophages. These results support two key precepts. First, a broad array of hydroxy-polyunsaturated fatty acids are substrates for 15PGDH. Second, the products of hydroxy-fatty acid dehydrogenation are electrophilic and can thus be conferred with pleiotropic anti-inflammatory signaling activities.
EXPERIMENTAL PROCEDURES
Materials
(±)-5-HETE, 5-oxoETE-d7, PGE2-d9, PGE2, 15-ketoPGE2, 13,14-dihydro-15-ketoPGE2, (±)-4-HDoHE, (±)-7-HDoHE, (±)-8-HDoHE (±)-10-HDoHE, (±)-11-HDoHE, (±)-13-HDoHE, (±)-14-HDoHE, (±)-16-HDoHE, (±)-17-HDoHE, (±)-20-HDoHE, docosahexaenoic acid (DHA), arachidonic acid (AA), and 15PGDH inhibitor, CAY10397, were all purchased from Cayman Chemical Co. (Ann Arbor, MI). Solvents for liquid chromatography mass spectrometry (LC-MS) including pure water and acetonitrile were purchased from Burdick and Jackson (Morristown, NJ). Glacial acetic acid, Optima grade methanol, and chloroform were purchased from Fisher. Anhydrous diethyl ether and Dess Martin periodinane were from Sigma.
Cell Culture
A549 (CCL-185) cells were purchased from American Type Culture Collection (Manassas, VA). A549 cells were maintained at 37 °C in 5% CO2 and grown in Ham's F12K media containing 10% FBS and 1% penicillin/streptomycin.
Bronchoscopy and Primary Cell Culture
Bronchoscopy of mild to moderate asthmatics with endobronchial epithelial brushing and primary airway epithelial cell (AEC) culture was performed as described previously and with University of Pittsburgh IRB approval (PRO11010186) (21, 22). Briefly, a total of six brushings were obtained, and cells were placed into 10 ml of phosphate-buffered saline, centrifuged, and resuspended in 1 ml of serum-free hormonally supplemented bronchial epithelial growth medium (Clonetics, San Diego, CA) containing 50 μg/ml gentamicin and 50 μg/ml amphotericin. The cells were plated at ∼3 × 105/plate in bronchial epithelial growth medium in 60-mm tissue culture dishes coated with rat tail type I collagen (BD Discovery Labware, Bedford, MA). Cells were cultured at 37 °C in a 5% CO2 environment. When cells were almost 100% confluent they were either plated at the air-liquid interface in 12-well transwell plates as described previously (21) or frozen for future use. When needed, frozen primary AECs were thawed, recounted, and plated at ∼5 × 105 cells per 60-mm collagen-coated dish and grown to confluency in Lifeline (Lifeline Cell Technology, Frederick, MD) serum-free media containing bronchial life factors. Upon 80% confluency, cells were trypsinized (0.25%) and plated on collagen-coated 6-well plates (Corning, Corning, NY) in serum-free Lifeline Bronchial Life Media with growth factors and 1% penicillin/streptomycin for metabolism studies (Lifeline Cell Technology, Frederick, MD). Primary alveolar macrophages were separated from bronchoalveolar lavage (BAL) fluid at the time of bronchoscopy, counted, and plated at 5 × 105 cells/well in a 24-well plate in RPMI 1640 supplemented with 10% FBS and penicillin/streptomycin. After 90 min, media were changed to remove non-adherent cells (23–25).
15PGDH shRNA Knockdown in A549 Cells
Lentiviral particles were generated by co-transfection of four plasmids (control plasmid (pLKO.1-SCRshRNA-Puro) or one of the five different 15PGDH-specific shRNA expressing plasmids, pLKO.1-shRNA-15PGDH.1-.5) together with pMD2.g (VSVG), pVSV-REV, and pMDLg/pRRE) into 293-FT cells using FuGENE 6 transfection reagent with support from the UPCI Lentiviral Facility. The 15PGDH-specific shRNAs target the following sequences in the 15PGDH gene: 1) CACAGCCATCCTTGAATCAAT, 2) GAAGGCGGCATCATTATCAAT, 3) GCAACAACTGAGAGACACTTT, 4) CTGGAGTGAATAATGAGAAA, and 5) ACTCATAACAACACAGACATA. The collection and isolation of lentiviral particles and transduction of A549 cells were performed as described previously (26). Stable cell lines were selected in puromycin (1 μg/ml) for 2 weeks. Knockdown of 15PGDH was confirmed by immunoblot.
Cell Treatment
Primary AECs were treated with 10 μm DHA for 2 h or with 2 μm HDoHEs, 2 μm 5-HETE, or 2 μm PGE2 for 4 h in cell growth media. After determining optimal inhibitor concentrations, treatments were conducted with and without 50 μm CAY10397. Media was extracted for LC-MS analysis as described below. A549 and A549 shRNA cell growth media were replaced with serum-free Hanks' balanced salt solution for cell treatments. Cells were treated for 4 h with either 10 μm AA, 10 μm DHA or 2 μm PGE2, 5-HETE, or HDoHEs for 4 h with or without 50 μm CAY10397 inhibitor. Primary alveolar macrophages were treated in RPMI with 1% FBS for 4 h and with 2 μm 14-HDoHE with and without 50 μm CAY10397 inhibitor. Media were collected for LC-MS analysis as described below. Results are an average of ≥3 individual treatments, and statistical significance was determined by either t test or one-way analysis of variance as appropriate.
BAL Cell Pellets
BAL obtained from bronchoscopies of 6 mild to moderate asthmatics was centrifuged to separate fluid from the cellular content. BAL cell pellets (1.5 × 106 cells/sample) were lysed briefly in liquid nitrogen, and 10 ng of 5-oxoETE-d7 was added as an internal standard. The cell pellet was incubated with 500 mm β-mercaptoethanol (BME) in 20 μl of 50 mm phosphate buffer, pH 7.4, for 1 h at 37 °C. The reaction was stopped with 80 μl of cold acetonitrile with 1% formic acid. The samples were centrifuged at 10,000 × g for 5 min, and the supernatant was used for LC-MS analysis of HDoHEs and electrophilic BME adducts (27).
RT-PCR
Primary alveolar macrophage media was replaced with 1% FBS in RPMI and treated for 6 h with vehicle, 100 ng/ml LPS, 10 μm 14-oxoDHA, or the combination of LPS and 14-oxoDHA. Results are representative of three individual treatments, and statistical significance was determined by one-way analysis of variance.
LC-Single Reaction Monitoring/MS
Free fatty acid metabolites in the media were extracted using diethyl ether. Briefly, media were collected, and 20 ng of 5-oxoETE-d7 and 10 ng of PGE2-d9 internal standards were added to each sample. The samples were allowed to equilibrate for 5 min before shaking for 10 min. Next, samples were centrifuged at 2800 × g for 10 min. The top layer (organic) was transferred to a clean vial and dried under a stream of nitrogen. Samples were reconstituted in 100 μl methanol before analysis. A Shimadzu HPLC (Columbia, MD) coupled to an AB Sciex (Framingham, MA) 5000 triple quadrupole mass spectrometer was used for the quantification of fatty acids. Sample (10 μl) was separated on a Phenomenex (Torrence, CA) C18(2) ODS column, 2.1 × 150 mm, 5-μm bead size, 100 Å pore size. The solvent system employed aqueous 0.1% acetic acid (A) and 0.1% acetic acid in acetonitrile (B). The 60-min gradient with a flow rate of 0.25 ml/min started at 35% B and ramped to 90% B over 46 min. This was followed by a wash using 100% B for 6 min. The gradient then returned to starting conditions at 35% B for 8 min. MS analyses by electrospray ionization were run in negative mode with the collision gas set at 4 units, curtain gas at 40 units, ion source gas 1 and 2 at 40 units, ion spray voltage −4500 V, and temperature at 550 °C. The declustering potential was set to −50, entrance potential −5, collision energy −17, and the collision exit potential −18.4. Single reaction monitoring was used for sample analysis and quantification. The following transitions were used: HDoHE and oxoDPA 343.2 → 299.2, 4-HDoHE 343.2 → 101.2, 7-HDoHE 343.2 → 141.2, 8-HDoHE 343.2 → 109.2, 10-HDoHE 343 → 153.2, 11-HDoHE 343.3 → 149.2, 13-HDoHE 343.2→ 193.2, 14-HDoHE 343.2 → 205.2, 16-HDoHE 343.2 → 233.2, 17-HDoHE 343.2→ 273.2, 20-HDoHE 343.2 → 187.2, oxoDHA 341.2 → 297.2, 14-oxoDHA 341.2 → 205.2, PGE2 351.2 → 271.2, 15-ketoPGE2 349.2 → 161.2, 13,14-dihydro-15-ketoPGE2 351.2 → 235.2, 5-oxoETE-d7 324.2 → 210.2, and PGE2-d9 360.2 → 280.2.
Structural characterization and retention time confirmation of 14-oxoDHA and 14-oxoDPA were confirmed using an Orbitrap Velos accurate mass, high resolution mass spectrometer (Thermo Scientific, Waltham, MA). The LC conditions were the same as described above for the quantification on the triple quadrupole mass spectrometer except the injection volume was 20 μl. Both scans were performed in Fourier transform mass spectrometry negative ion mode at 30,000 resolution with an acquisition time of 30 ms, an ActQ of 0.25, and collision energy of 35. The ion spray voltage was maintained at 4 kV, and the heater and capillary temperatures were set to 300 °C and 270 °C, respectively. The sheath, auxiliary, and sweep gases were adjusted to 20, 18, and 12 arbitrary units. Mass accuracy was <5 ppm for all confirmed structures.
Identification of Lipid Electrophiles
Confirmation of electrophilicity was determined using the nucleophile BME (27). Media were incubated with 50 mm BME for 1 h at 37 °C and then extracted with diethyl ether and dried under nitrogen before LC-MS analysis. Fatty acid BME adducts were analyzed on the AB Sciex 5000 triple quadrupole mass spectrometer described above. The oxoDPA species was monitored at 421.2 → 343.2 for the loss of 78 atomic mass units (BME), and the electrophilic 5-oxoETE-d7 was used as a positive control for BME adduct formation (402.2 → 324.2).
RESULTS
Formation of Electrophilic Species from DHA
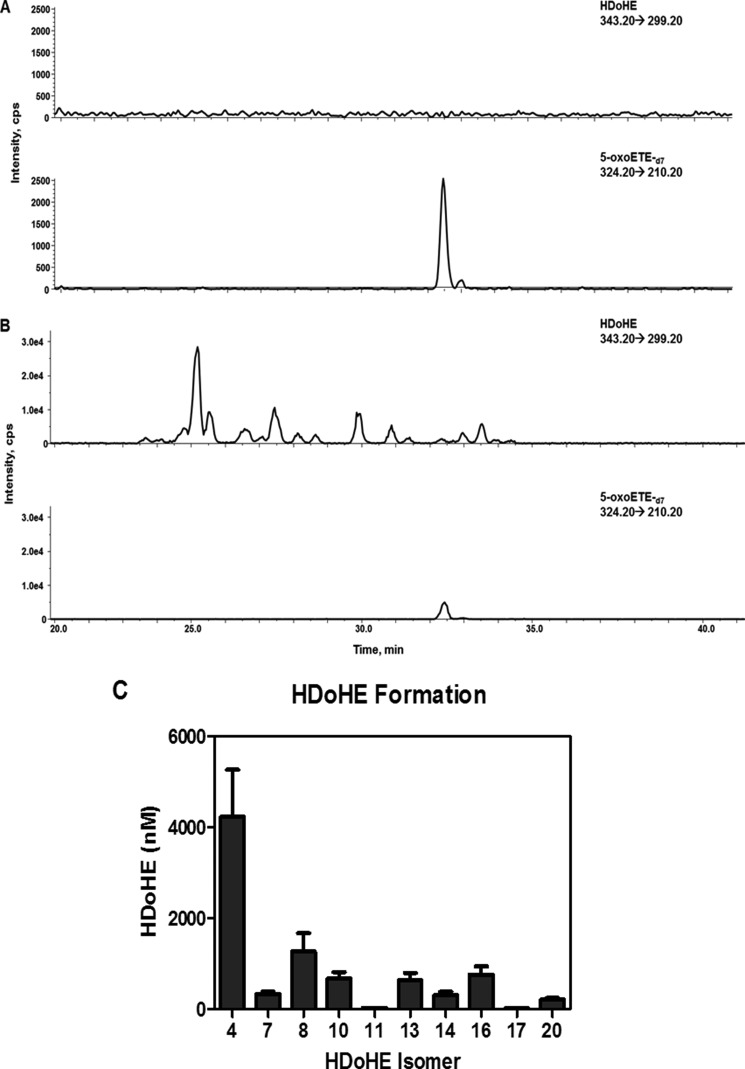
Primary AECs were treated with 10 μm DHA for 2 h and the formation of HDoHEs and the corresponding electrophilic oxoDHA species was determined. Analysis of control cell lysate and media did not result in detectable levels of endogenous DHA-derived HDoHE metabolites monitored by the nonspecific transition (loss of CO2) 343.2 → 299.2 (Fig. 1A). Additionally, the incubation of cell-free Hanks' balanced salt solution or media with DHA resulted in minimal generation of HDoHEs from DHA autoxidation (data not shown), whereas the addition of exogenous DHA to primary AEC resulted in the generation of HDoHEs (Fig. 1B) via enzymatic and non-enzymatic reactions that could be quantitated in the media using 5-oxoETE-d7 as the internal standard (Fig. 1C) (28–33).
FIGURE 1.
HDoHE formation in primary human airway epithelial cell cultures. Primary AEC were treated with vehicle control showing no endogenous HDoHE (A) 10 μm DHA for 2 h resulting in HDoHE formation monitored in the media (B). HDoHE regioisomers were quantified using 5-oxoETE-d7 as an internal standard (C).
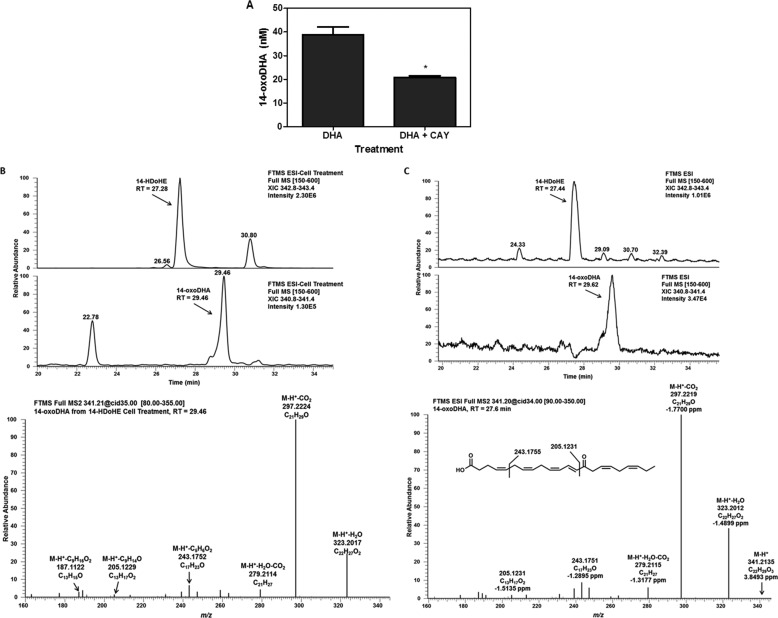
Regardless of the route of HDoHE formation, these DHA-derived species are potential substrates of 15-PGDH. Simultaneous treatment of primary AEC with DHA and the 15PGDH inhibitor CAY10397 (50 μm) resulted in a significant decrease in what was suspected to be 14-oxoDHA, the product of 15PGDH oxidation of 14-HDoHE (Fig. 2A). The identity of 14-oxoDHA was confirmed using the 14-HDoHE standard that was oxidized to its corresponding electrophile using the Dess Martin periodinane reaction as no commercial standard was available. Characterization of the oxidized product confirmed the identity of the 14-oxoDHA species using retention time and MS2 spectra from high resolution accurate mass LC-MS/MS (Fig. 2, B and C).
FIGURE 2.
Primary AEC treated for 2 h with 10 μm DHA and 50 μm CAY10397 resulted in a significant decrease in 14-oxoDHA; *, p ≤ 0. 05 (A). To confirm the identification of 14-oxoDHA, a standard was made from 14-HDoHE oxidation using the Dess Martin periodinane reagent. 14-OxoDHA formed from cell treatment with 2 μm 14-HDoHE (B) and the standard (C) were characterized by retention time and full scan MS2 on a high resolution accurate mass Orbi Velos. FTMS-ESI, Fourier transform mass spectrometry-electrospray ionization.
15PGDH Substrate Specificity
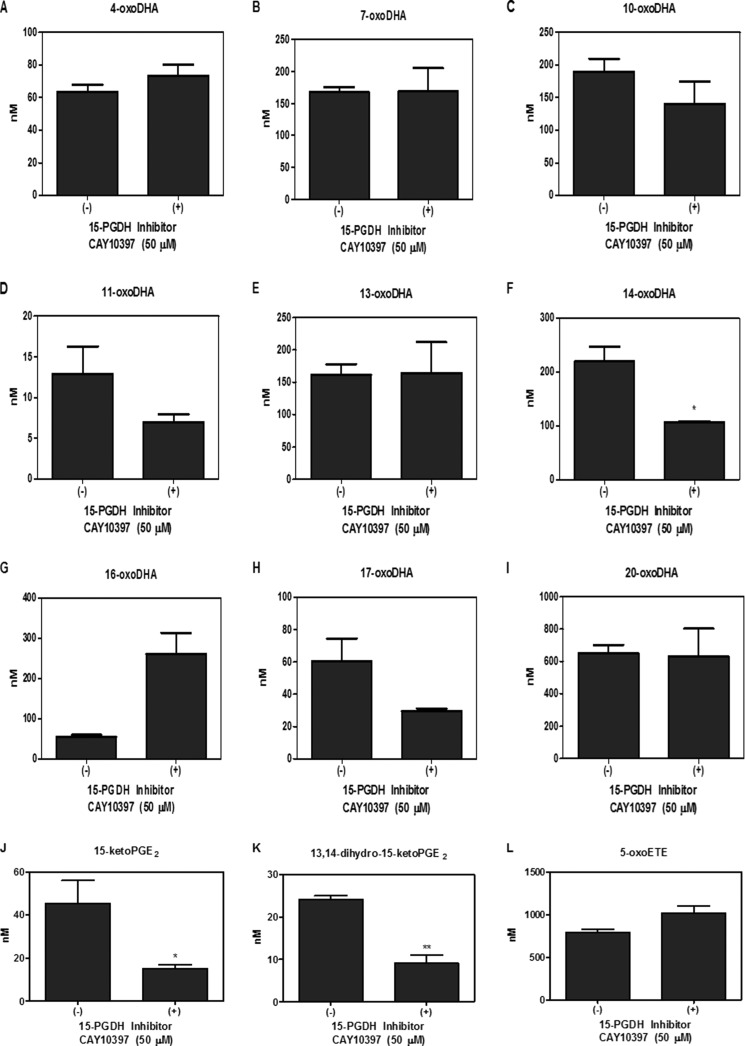
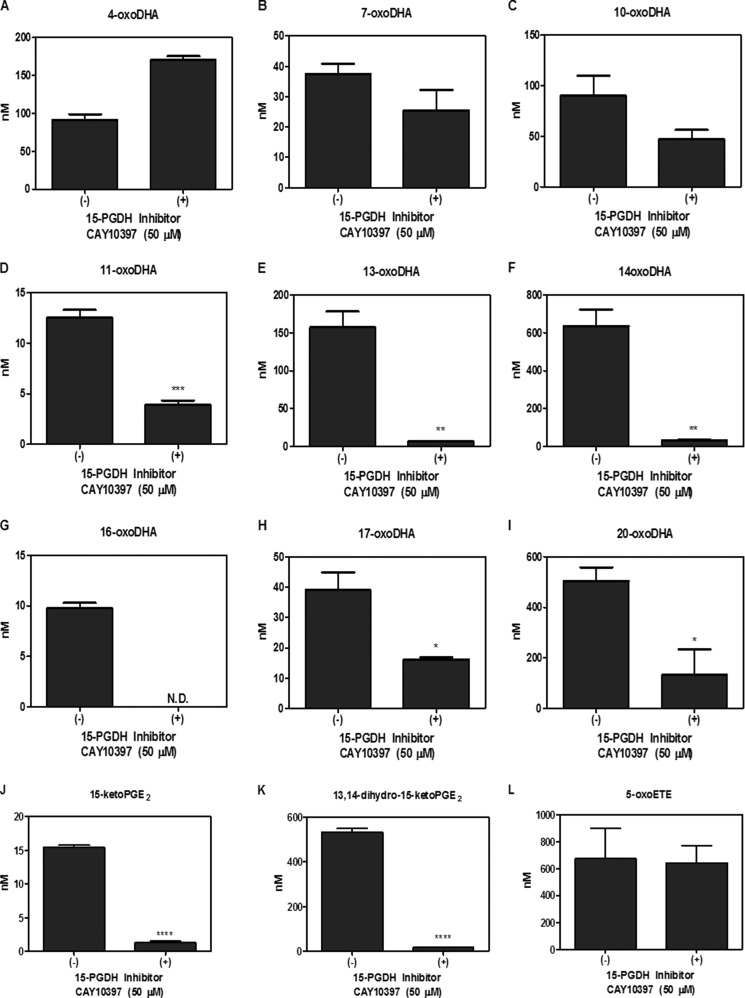
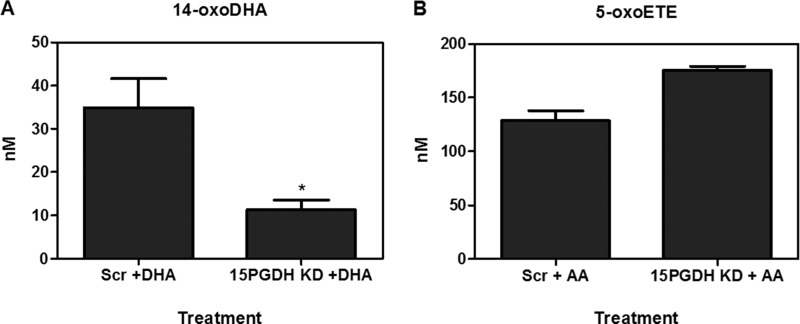
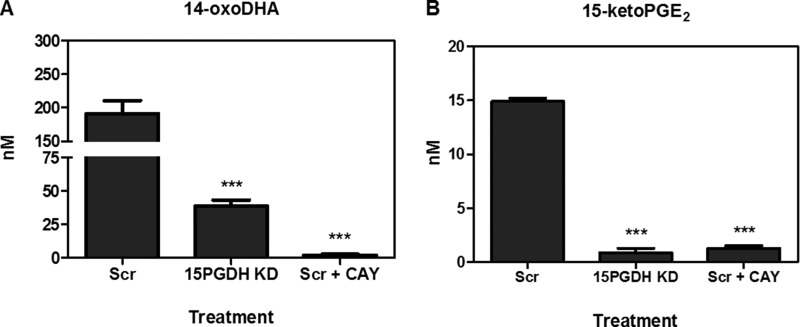
Metabolism was further investigated in primary AECs using HDoHE species (2 μm) having the hydroxyl group at C4, C7, C8, C10, C11, C13, C14, C16, C17, and C20. HDoHEs were added to cells cultured in Hanks' balanced salt solution (HBSS) with and without 50 μm CAY10397 for 4 h. Both HBSS and cell lysates were extracted; however, HDoHEs and downstream metabolites were predominantly detected in the media. The corresponding oxoDHA species could be detected for all HDoHE substrates except for 8-HDoHE (Fig. 3, A–I). HDoHE metabolism to their corresponding oxoDHA metabolite was only significantly inhibited by CAY10397 for the oxidation of 14-HDoHE to 14-oxoDHA, consistent with the inhibition of 15PGDH after exogenous DHA addition (Fig. 3F). PGE2 and 5-HETE were used as positive and negative controls, respectively. CAY10397 significantly inhibited the formation of 15-ketoPGE2 and its non-electrophilic metabolite 13,14-dihydro-15-ketoPGE2, whereas 5-oxoETE formation did not change as 5-HETE is a substrate for 5-hydroxyeicosanoid dehydrogenase, not 15PGDH (Fig. 3, J–L). The same analyses were repeated in A549 cells in which results varied significantly from the primary AEC (Fig. 4, A–L). 14-oxoDHA was abundantly formed in A549 cells, and its formation was significantly inhibited with CAY10397. However, CAY10397 also significantly inhibited oxoDHA formation for substrates with the hydroxyl group position ranging from 11-HDoHE to 20-HDoHE (Fig. 4, D–I). These data provide a clear indication that (a) dehydrogenase expression is cell line-specific, (b) CAY10397 is not specific for 15PGDH, and (c) transformed cell lines do not recapitulate what is observed in primary cells. To further confirm that 14-oxoDHA is a product of 15PGDH in a more specific manner, A549 cells were transduced with lentiviruses expressing either a scrambled or 15PGDH-specific shRNA. Loss of expression was validated by immunoblot (data not shown). Scrambled (Scr) control and 15PGDH knockdown (KD) cells were treated with 10 μm DHA for 4 h, and AA-derived 5-HETE was used as a control. The formation of 14-oxoDHA was significantly decreased in the 15PGDH KD cells compared with the Scr control, whereas there was no decrease in 5-oxoETE formation (Fig. 5). Scr A549 cells were treated with 2 μm 14-HDoHE with and without 50 μm CAY10397 and 15PGDH KD cells were treated with 2 μm 14-HDoHE for 4 h (Fig. 6A). The PGE2 metabolite 15-ketoPGE2 was used as the positive control. 15-ketoPGE2 was significantly decreased to the same extent in Scr A549 cells treated with 15PGDH inhibitor and 15PGDH KD cells (Fig. 6B). 14-oxoDHA formation was also significantly decreased with inhibitor in Scr A549 cells and in 15PGDH KD cells (Fig. 6A). In both 15PGDH KD treatments 14-oxoDHA formation was not completely inhibited (Figs. 5A and 6A). In fact, Scr A549 cells treated with CAY10397 resulted in lower levels of 14-oxoDHA compared with the 15PGDH KD cells (Fig. 6A). These differences underscore the lower specificity of the CAY10397 inhibitor and highlight that other dehydrogenases may also contribute to 14-oxoDHA formation.
FIGURE 3.
Electrophile formation in primary AEC cultures. Primary AEC were treated with 2 μm HDoHE regioisomers (A–I) for 4 h with and without 50 μm CAY10397, and oxoDHA formation was quantified. Formation of oxoDHA metabolites corresponding to each HDoHE regioisomer was detected for all except 8-HDoHE. 2 μm PGE2 (positive) and 5-HETE (negative) treatments were used as controls for 15PGDH substrate specificity, and 15-ketoPGE2 (J), 13, 14-dihydro-15-ketoPGE2 (K), and 5-oxoETE (L) formation were quantified. *, p ≤ 0.05; **, p ≤ 0.01.
FIGURE 4.
Electrophile formation in A549 cells. A549 cells were treated with 2 μm HDoHE regioisomers (A–I) with and without 50 μm CAY10397 for 4 h, and oxoDHA formation was quantified. Formation of oxoDHA metabolites corresponding to each HDoHE regioisomer was detected for all except 8-HDoHE. 2 μm PGE2 (positive) and 5-HETE (negative) treatments were used as controls for 15PGDH substrate specificity, and 15-ketoPGE2 (J), 13,14-dihydro-15-ketoPGE2 (K), and 5-oxoETE (L) formation were quantified. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001. ND, not detected.
FIGURE 5.
Knockdown of 15PGDH. A549 cells transfected with 15PGDH shRNA or scramble shRNA were treated with 10 μm DHA or 10 μm AA for 4 h. 14-oxoDHA (A) and 5-oxoETE (ctrl) (B) levels were quantified. *, p ≤ 0.05.
FIGURE 6.
15PGDH knockdown versus 15PGDH inhibition. A549 cells transfected with either 15PGDH shRNA or scramble shRNA were treated with 2 μm 14-HDoHE or PGE2 (ctrl) for 4 h. Scramble A549 cells were also treated with 50 μm CAY10397 (CAY). 14-oxoDHA (A) and 15-ketoPGE2 (B) formation was quantified. ***, p ≤ 0.001.
Electrophilic Metabolites
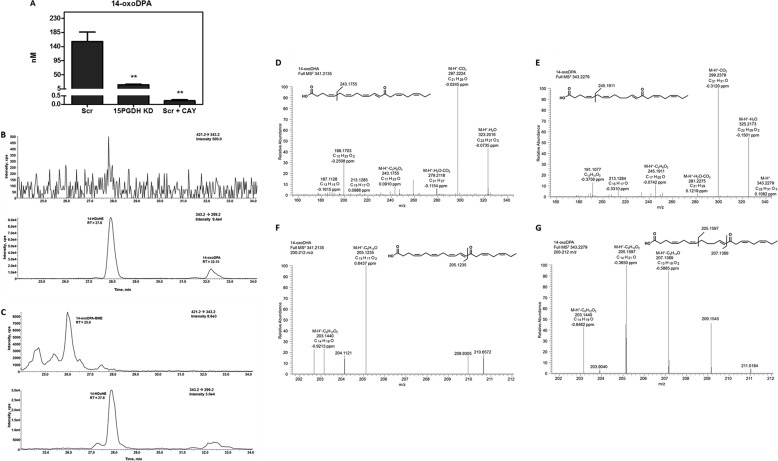
The formation of oxoDHA metabolites is also affected by 15PGDH. LC-MS analysis revealed a key metabolite of 14-oxoDHA is its reduced metabolite, 14-oxoDPA. This product was detected in the transition 343.2 → 299.2, which is used to monitor the loss of CO2 from any HDoHE species; however, reduced keto species can also be detected in this transition. The retention time of 14-oxoDPA is increased compared with 14-HDoHE due to the increase in hydrophobicity. The formation of 14-oxoDPA significantly decreased with either CAY10397 inhibition or 15PGDH KD (Fig. 7A).
FIGURE 7.
Saturation products of 14-oxoDHA. The concentration of the 14-oxoDHA reduced product, 14-oxoDPA, was decreased in Scr A549 treated with 2 μm 14-HDoHE and 50 μm CAY10397 for 4 h and in 15PGDH KD cells. *, p ≤ 0.05; **, p ≤ 0.01 (A). To test the electrophilicity of 14-oxoDPA, cell media were incubated with BME. The transition 421.2 → 343.2 shows an absence of product in the top chromatogram of B before β-mercaptoethanol addition. The bottom chromatogram shows peaks for 14-HDoHE (retention time = 27.8 min) and 14-oxoDPA (retention time = 31.1 min) in the 343.2 → 299.2 transition (B). The incubation of 14-oxoDPA + 50 mm BME form a hydrophilic Michael adduct that can be monitored at 421.2 → 343.2 with a concomitant disappearance of 14-oxoDHA from the343.2 → 299.2 transition (C). MS2 characterization of 14-oxoDHA (D and F) and 14-oxoDPA (E and G) using high resolution accurate mass spectrometry. 14-oxoDHA MS2 shows 323.2016 (M-H+-H2O), 297.2224 (M-H+-CO2), 279.2118 (M-H+-H2O-CO2), 243.1755 (M-H+-C5H6O2) (D). 14-oxoDPA MS2 shows 343.2279 (M-H+), 325.2173 (M-H+-H2O), 299.2379 (M-H+-CO2), 281.2275 (M-H+-H2O-CO2), and 245.1911 (M-H+-C5H6O2) (E). 14-oxoDHA MS2 zoomed in to see the m/z window 200–212 that shows 205.1235 (M-H+-C13H17O2) and 203.1440 (M-H+-C8H12O2) (F). 14-oxoDPA MS2 zoomed in to see the m/z window 200–212 that shows 207.1389 (M-H+-C9H14O) corresponding to the 205.1235 fragment of 14-oxoDHA (G). The 205.1597 (M-H+-C8H10O2) is a fragment from the loss of C8H10O2 from the Ω end of the aliphatic chain.
Prostaglandin reductase (PTGR1) is the enzyme responsible for the conversion of the 15-ketoPGE2 to its reduced metabolite 13,14-dihydro-15ketoPGE2 (34). This reduction occurs at the double bond adjacent to the carbonyl group rendering 13,14-dihydro-15-ketoPGE2 non-electrophilic. To test the electrophilicity of 14-oxoDPA, media were incubated with the nucleophile BME yielding a Michael adduct with the electrophilic α, β-unsaturated carbon (27). The top panel of Fig. 7B shows a transition for 421.2 → 343.2 that represents the loss of BME from an electrophilic metabolite with a parent mass of 343.2. In A549 cell-derived media without BME, there is no peak in the 421.2 → 343.2 transition. The bottom panel of Fig. 7B shows the transition (343.2 → 299.2) for the loss of CO2 from a metabolite with the parent mass of 343.2. In this panel 14-HDoHE is present at a retention time of 27.8 min, and a more hydrophobic metabolite, 14-oxoDPA, is present at a retention time of 32.1 min (Fig. 7B). However, upon incubation with BME (Fig. 7C), the peak at 32.1 min disappears concomitantly with the appearance of a more hydrophilic peak in the 421.2 → 343.2 transition for the 14-oxoDPA-BME Michael adduct, monitoring for a loss of BME (m/z 78). It is proposed that prostaglandin reductase is not the main reductase responsible for double-bond saturation because 14-oxoDPA maintains electrophilic characteristics. Characterization of the 14-oxoDPA species using high accuracy MS/MS resulted in similar fragment ions as seen for 14-oxoDHA (Fig. 7, D and F), except that they were 2 mass units higher and the parent mass was 343.2279 (<0.5 ppm). The 14-oxoDHA fragment ion with m/z 205.1235 (M-H+-C9H14O, <1 ppm), representing cleavage between the carbonyl and α,β-unsaturated carbon, was not a major fragment in the 14-oxoDPA MS2; rather, m/z 207.1389 (M-H+-C9H12O, <1ppm) was detected. This is indicative of saturation on the Ω side of the carbonyl (Fig. 7, F and G). The fragment with m/z 205.1597 (M-H+-C8H10O2) coming from the MS2 of 14-oxoDPA provides evidence that the two saturated double bonds closest to the Ω end are present. Additionally, 14-oxoDHA has a characteristic fragmentation of 243.1751 (M-H+-C5H6O2), whereas the MS2 of 14-oxoDPA contains a fragment ion with m/z 245.1911 (M-H+-C5H6O2) representing the same loss, indicating that the double bond closest to the carbonyl is not reduced. Therefore, it can be deduced from the MS2 comparisons that the saturation occurs at the γ,Δ-unsaturated double bond (Fig. 7, F and G).
Detection of HDoHEs and oxoDHA Metabolites in Bronchoalveolar Lavage Cells
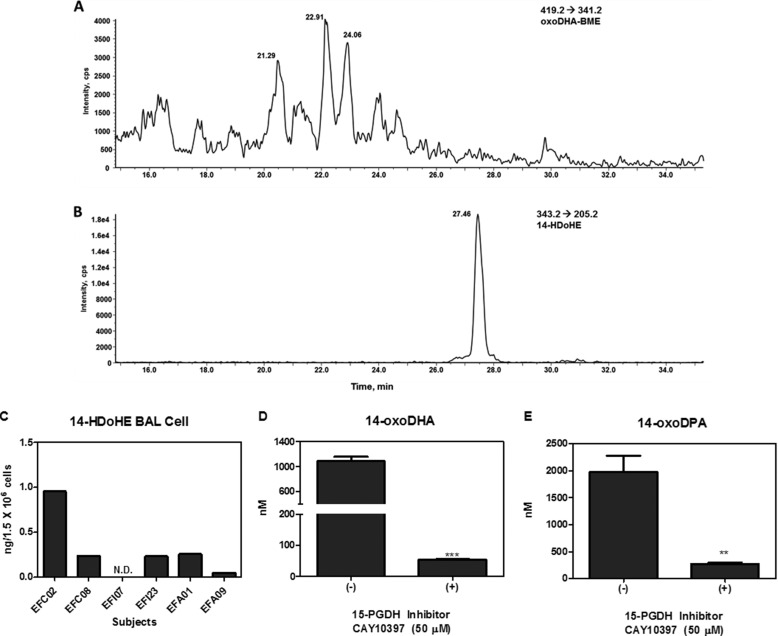
The clinical generation of electrophilic oxoDHA metabolites and HDoHE precursors occurred in cells from the BAL of six mild to moderate asthmatics. Electrophilic species were captured via reaction with added BME and then analyzed by LC-MS, where electrophilic species were monitored by the loss of BME. The chromatogram of a BAL cell pellet extracted after BME incubation reveals electrophilic species when monitored at 419.2 → 341.2 (Fig. 8A), with the 5-oxoETE-d7 internal standard serving as the BME control monitored at 402.2 → 324.2 (not shown). Electrophilic BME adducts are more hydrophilic, and their retention shifts to the left by ∼5.5 min. The non-electrophilic HDoHEs do not adduct to BME and could be detected at their specific single reaction monitoring transitions without a shift in retention time. 14-HDoHE was detected in the clinical samples using the transition 343.2 → 205.2 at 27.4 min (Fig. 8B). The levels of 14-HDoHE detected in BAL cell pellets ranged from not detectable to 0.95 ng/1.5 × 106 cells (Fig. 8C). Primary alveolar macrophages were also separated from BAL fluid at the time of bronchoscopy from mild to moderate asthmatics and plated. Macrophages were treated with 2 μm 14-HDoHE with and without CAY10397, and metabolites were monitored after 4 h. 14-oxoDHA and 14-oxoDPA were both detected, and their formation was significantly inhibited with the CAY10397 (Fig. 8, D and E).
FIGURE 8.
Detection of oxoDHA-BME adducts and 14-HDoHE in BAL cells. Bronchoalveolar lavage cell pellets were incubated with 500 mm BME and extracted for LC-MS analysis. The loss of BME (78 atomic mass units) was monitored for oxoDHA-BME adducts (419.2 → 341.2) (A). Endogenous levels of 14-HDoHE (343.3 → 205.2) were detected in BAL cell pellets (B). Endogenous 14-HDoHE levels ranged from not detectable to 0.95 ng/106 cells in the clinical samples (C). Primary alveolar macrophages taken from bronchoscopies were plated at a confluency of 5 × 105 cells/well in a 24 well plate. Alveolar macrophages were incubated for 4 h with 2 μm 14-HDoHE ± 50 μm CAY10397, and 14-oxoDHA (D) and 14-oxoDPA (E) metabolites were quantified. **, p ≤ 0.01; ***, p ≤ 0.001.
15PGDH Regulation of Inflammatory Signaling
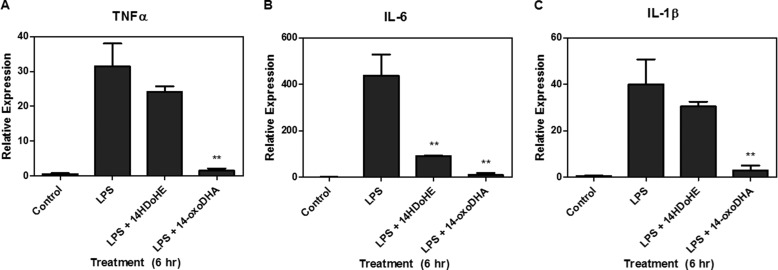
Electrophilic lipids mediate a broad array of cell signaling responses (16). Primary alveolar macrophages were treated for 6 h with vehicle control, 10 μm 14-oxoDHA, 100 ng/ml LPS, or concurrent treatment of 100 ng/ml LPS and 10 μm 14-HDoHE or 100 ng/ml LPS and 10 μm 14-oxoDHA (Fig. 9). LPS treatment significantly increased the mRNA expression of NF-κB-driven cytokines, TNFα, IL-6, and IL-1β. Concurrent LPS treatment with 14-oxoDHA resulted in significant abrogation of cytokine mRNA expression. Interestingly, IL-6 mRNA expression was also significantly blunted by 14-HDoHE (Fig. 9).
FIGURE 9.
14-oxoDHA inhibits NF-κB-derived cytokine mRNA expression. Primary alveolar macrophages were treated with 100 ng/ml LPS, 100 ng/ml LPS, and 10 μm 14-HDoHE or 100 ng/ml LPS and 10 μm 14-oxoDHA for 6 h. LPS induced cytokine mRNA expression, which was abrogated by 14-oxoDHA for TNFα (A), IL-6 (B), and IL-1β (C). ** p ≤ 0.01. 14-HDoHE treatment resulted in a significant inhibition of IL-6.
DISCUSSION
15PGDH is noted for inactivating the pro-proliferative PGE2 and the polyhydroxylated fatty acid derivatives termed lipoxin A4 and resolvin E1. These oxidations catalyzed by 15PGDH are deemed inactivation reactions because of putative attenuation of G protein-coupled receptor ligand activity and downstream signaling responses. Notably, the oxidized products of 15PGDH become electrophilic species that can also mediate anti-inflammatory and anti-proliferative actions via pleiotropic G protein-coupled receptor-independent signaling mechanisms.
The inverse relationship between COX-2 and 15PGDH that leads to an excess of pro-proliferative signaling in many cancers highlights the importance of 15PGDH in the regulation of fatty acid metabolism and signaling (3, 4, 35). However, these studies only demonstrated that 15PGDH metabolites were products of the metabolism of pro-proliferative signaling molecules and not potentially conferred with alternative signaling potential. More recent studies of hepatocellular carcinoma and cholangiocarcinoma show that 15-ketoPGE2 formation not only decreases the levels of the pro-proliferative PGE2 but also inhibits cell growth via activation of PPARγ (12, 13). The hepatocellular carcinoma model determined that cell proliferation was inhibited by an increase in p21WAF1/Cip1 (cyclin-dependent kinase inhibitor 1) expression, leading to downstream association with cyclin-dependent kinase (CDK2), CDK4, and proliferating cell nuclear antigen (12). In the cholangiocarcinoma model, inhibition of proliferation is acquired through PPARγ-, (SMAD2/3)-, and Tap63 (tumor protein)-mediated signaling actions (13). This study also linked 15-ketoPGE2 generation with the induction of SMAD 2/3 dissociation from PPARγ, thus promoting SMAD 2/3 complex formation with transforming growth factor B receptor 1 and SMAD anchor receptor for activation. In a related context, 15-oxoETE significantly up-regulated Nrf2-activated heme oxygenase-1 and NAPDH oxidoreductase 1 (NQO1) protein expression and inhibited TNFα, IL-6, and IL-1β mRNA expression in THP-1 cells (20). Moreover, 11-oxoETE and 15-oxoETE demonstrated anti-proliferative properties in human umbilical vein endothelial cells using a bromodeoxyuridine-based cell proliferation assay. The IC50 of 11-oxoETE for the inhibition of human umbilical vein endothelial cell proliferation was 2.1 μm, similar to that of the electrophilic prostaglandin 15-deoxy prostaglandin J2 (6). In aggregate, these observations indicate that 15PGDH and the oxo-fatty acid products it generates can profoundly influence cell function.
The present study reveals that hydroxylated Ω-3 fatty acids are substrates for 15PGDH and potentially other dehydrogenases. These species, like 15-ketoPGE2 and AA-derived oxoETEs, are electrophilic and possess anti-inflammatory signaling properties. Primary human AEC and A549 cell-based studies indicated that the most abundant 15-PGDH-derived oxoDHA metabolite is 14-oxoDHA, which was characterized by high resolution accurate mass LC-MS/MS (Fig. 2). When individual HDoHE metabolites were added to cell culture, the position of the hydroxyl group on the acyl chain dictated substrate specificity for 15PGDH in primary AEC (Fig. 3). Consistent with this, 15PGDH inhibition led to the most extensive decrease in 14-oxoDHA formation by A549 and primary AEC (Figs. 3D and 4D). 11, 13, 16, 17, and 20-oxoDHA formation was also significantly inhibited in A549 cells, but not in primary AEC. This could be a result of differences in the expression of cell-specific dehydrogenase activities or the specificity of the CAY inhibitor for 15PGDH. Additionally, A549 cells are metabolically more active and in a more oxidative state, which may influence the extent of formation of various electrophilic oxoDHA species. This also highlights the concern of using transformed carcinogenic cell lines for metabolism studies as the results do not always correlate with primary cell lines.
The specificity of CAY10397 for 15PGDH inhibition of 14-oxoDHA formation was confirmed in A549 cells transduced with lentiviruses expressing either 15PGDH-specific shRNA or a scrambled shRNA control. 14-OxoDHA generation was significantly decreased in 15PGDH KD cells supplemented with either DHA or 14-HDoHE (Figs. 5 and 6). Interestingly, CAY10397 treatment of Scr A549 cells resulted in a greater inhibition of 14-oxoDHA formation compared with 15PGDH KD cells. This reiterates that CAY10397 is not specific for 15PGDH and that other cellular dehydrogenases are capable of forming oxoDHA species.
Electrophilic oxoDHA species are also substrates for further metabolism, particularly the reduction of a double bond. In the case of PGE2 metabolism, 15-ketoPGE2 is reduced by prostaglandin reductase at the α,β-unsaturated bond, yielding the non-electrophilic metabolite 13,14-dihydro-15ketoPGE2. The 14-oxoDHA metabolite was also reduced to its corresponding oxoDPA species; however, this metabolite retained its electrophilic nature, as indicated by retention of BME reactivity (Fig. 7, B and C). Further characterization by MS2 and comparison of fragmentation patterns point to saturation of the γ,Δ-double bond (Fig. 7, D–G). These data support that although prostaglandin reductase is expressed in human lung epithelium (36), 14-oxoDHA is not a substrate for prostaglandin reductase reduction. The identity of an enzyme responsible for 14-oxoDHA reduction remains to be investigated.
The observation that oxoDHA species abrogate proinflammatory cytokine expression (Fig. 9) reinforces the concept that electrophilic 15PGDH metabolites can acquire additional and unique biological activities (5, 6, 13, 19). Herein and in other 15-oxoETE, 17-oxoDHA, and 15d-PGJ2 signaling studies, higher than physiological concentrations of the electrophile have been added to culture systems. In each instance, the intracellular concentrations of electrophile are in the nm range, affirming that these model systems are biologically relevant (19, 20, 37). These and other electrophilic fatty acids signal in vitro and in vivo via Michael adduction to key nucleophilic cysteines in transcriptional regulatory proteins that regulate the inflammatory response including Nrf2 and NF-κB (16, 19, 20, 38–41). We have demonstrated that 15PGDH-derived 14-oxoDHA can inhibit NF-κB-mediated cytokine expression after LPS treatment. Similar results are obtained using FAT-1 mice, where the inflammatory response to methacholine after ovalbumin challenge is suppressed. FAT-1 mice express a Caenorhabditis elegans desaturase that converts Ω-6 to Ω-3 fatty acids, providing endogenously elevated levels of DHA and eicosapentaenoic acid in lung tissue (42) and presumably elevated oxoDHA products described in the present study. Compared with wild type mice, FAT-1 mice that underwent ovalbumin challenge showed a decrease in allergic airway inflammation as measured by proinflammatory cytokine and chemokine levels and a reduced response to methacholine (42).
In summary, the Ω-3 class of fatty acids instigates anti-inflammatory signaling actions by modulating multiple target molecules and signaling pathways. Most commonly, Ω-3 fatty acids are viewed to compete with their Ω-6 fatty acids as substrates for enzymes such as COX, lipoxygenase, and cytochrome P450 to limit the formation of proinflammatory Ω-6 metabolites. The tri-hydroxylated lipid mediators termed resolvins, maresins, and protectins are also proposed to transduce signaling responses of Ω-3 fatty acids through ligand activity toward the G-protein-coupled chemerin, ALX, and orphan GPR23 receptors (11, 32, 43). We show herein that additional signaling events are operative downstream of Ω-3 and Ω-6 fatty acid hydroxylation. These actions are mediated by the further oxidation of mono-, di-, and tri-hydroxylated species to electrophilic product. This represents a mechanism whereby metabolic and inflammatory fatty acid oxygenation yields a common class of oxo metabolites that mediate adaptive anti-inflammatory effects through less restrictive and more pleiotropic mechanisms. Specifically, this involves the kinetically facile oxo-fatty acid formation of Michael adducts with nucleophilic cysteines present in established redox-sensing transcriptional regulatory proteins that regulate metabolism and inflammation. Thus, 15PGDH and related dehydrogenases not only regulate fatty acid catabolism but also yield electrophilic metabolites with metabolic, anti-proliferative, and anti-inflammatory signaling activities.
Acknowledgments
We thank Meghan Delmastro-Greenwood for critical review of the manuscript. Support for the UPCI Lentiviral Facility was provided by National Institutes of Health Cancer Center Support Grant CA047904.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-HL058115 (to B. A. F.), R01-HL64937 (to B. A. F.), P01-HL103455 (to B. A. F.), CA148629 (to R. W. S.), GM087798 (to R. W. S.), ES019498 (to R. W. S.), ES021116 (to R. W. S.), and GM099213 (to R. W. S.). B. A. F. acknowledges financial interest in Complexa, Inc., and R. W. S. is a scientific consultant for Trevigen, Inc.
- 15PGDH
- 15-hydroxyprostaglandin dehydrogenase
- PGE2
- prostaglandin E2
- 5-HETE
- (±)-5-hydroxyeicosatetraenoic acid
- 5-oxoETE
- 5-oxo-6E,8Z,11Z,14Z-eicosatetraenoic acid
- CAY10397
- 5-[[4-(ethoxycarbonyl)phenyl]azo]-2-hydroxy-benzeneacetic acid
- HDoHE
- hydroxylated docosahexaenoic acid metabolite
- 13,14-dihydro-15-ketoPGE2
- 9,15-dioxo-11α-hydroxy-prost-5Z-en-1-oic acid
- 7-HDoHE
- (±)7-hydroxy-4Z,8E,10Z,13Z,16Z,19Z-docosahexaenoic acid; (±)10-hydroxy-4Z,7Z,11E,13Z,16Z,19Z-docosahexaenoic acid (10-HDoHE); (±)13-hydroxy-4Z,7Z,10Z,14E,16Z,19Z-docosahexaenoic acid (13-HDoHE); (±)14-hydroxy-4Z,7Z,10Z,12E,16Z,19Z-docosahexaenoic acid (14-HDoHE)
- 17-HDoHE
- (±)17-hydroxy-4Z,7Z,10Z,13Z,15E,19Z-docosahexaenoic acid
- 20-HDoHE
- (±)20-hydroxy-4Z,7Z,10Z,13Z,16Z,18E-docosahexaenoic acid
- DHA
- docosahexaenoic acid
- 17-oxoDHA
- 4Z,7Z,10Z,13Z,15E,19Z-17-keto-DHA
- BME
- β-mercaptoethanol
- Nrf2
- nuclear factor (erythroid-derived 2)-like 2
- NF-κB
- nuclear factor κ light chain enhancer of activated B cells
- PPARγ
- peroxisome proliferator-activated receptor γ
- AEC
- primary airway epithelial cell
- SMAD
- mothers against decapentaplegic homolog; tumor protein
- AA
- arachidonic acid
- BAL
- bronchoalveolar lavage
- Scr
- Scrambled
- KD
- knockdown.
REFERENCES
- 1. Liu Z., Wang X., Lu Y., Han S., Zhang F., Zhai H., Lei T., Liang J., Wang J., Wu K., Fan D. (2008) Expression of 15-PGDH is down-regulated by COX-2 in gastric cancer. Carcinogenesis 29, 1219–1227 [DOI] [PubMed] [Google Scholar]
- 2. Hughes D., Otani T., Yang P., Newman R. A., Yantiss R. K., Altorki N. K., Port J. L., Yan M., Markowitz S. D., Mazumdar M., Tai H. H., Subbaramaiah K., Dannenberg A. J. (2008) NAD+-dependent 15-hydroxyprostaglandin dehydrogenase regulates levels of bioactive lipids in non-small cell lung cancer. Cancer Prev. Res. (Phila) 1, 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Myung S. J., Rerko R. M., Yan M., Platzer P., Guda K., Dotson A., Lawrence E., Dannenberg A. J., Lovgren A. K., Luo G., Pretlow T. P., Newman R. A., Willis J., Dawson D., Markowitz S. D. (2006) 15-Hydroxyprostaglandin dehydrogenase is an in vivo suppressor of colon tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 12098–12102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yan M., Rerko R. M., Platzer P., Dawson D., Willis J., Tong M., Lawrence E., Lutterbaugh J., Lu S., Willson J. K., Luo G., Hensold J., Tai H. H., Wilson K., Markowitz S. D. (2004) 15-Hydroxyprostaglandin dehydrogenase, a COX-2 oncogene antagonist, is a TGF-β-induced suppressor of human gastrointestinal cancers. Proc. Natl. Acad. Sci. U.S.A. 101, 17468–17473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wei C., Zhu P., Shah S. J., Blair I. A. (2009) 15-oxo-Eicosatetraenoic acid, a metabolite of macrophage 15-hydroxyprostaglandin dehydrogenase that inhibits endothelial cell proliferation. Mol. Pharmacol. 76, 516–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu X., Zhang S., Arora J. S., Snyder N. W., Shah S. J., Blair I. A. (2011) 11-Oxoeicosatetraenoic acid is a cyclooxygenase-2/15-hydroxyprostaglandin dehydrogenase-derived antiproliferative eicosanoid. Chem. Res. Toxicol. 24, 2227–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clish C. B., Levy B. D., Chiang N., Tai H. H., Serhan C. N. (2000) Oxidoreductases in lipoxin A4 metabolic inactivation: a novel role for 15-onoprostaglandin 13-reductase/leukotriene B4 12-hydroxydehydrogenase in inflammation. J. Biol. Chem. 275, 25372–25380 [DOI] [PubMed] [Google Scholar]
- 8. Powell W. S., Rokach J. (2005) Biochemistry, biology, and chemistry of the 5-lipoxygenase product 5-oxo-ETE. Prog. Lipid Res. 44, 154–183 [DOI] [PubMed] [Google Scholar]
- 9. Patel P., Cossette C., Anumolu J. R., Erlemann K. R., Grant G. E., Rokach J., Powell W. S. (2009) Substrate selectivity of 5-hydroxyeicosanoid dehydrogenase and its inhibition by 5-hydroxy-Δ6-long-chain fatty acids. J. Pharmacol. Exp. Ther. 329, 335–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bull A. W., Bronstein J. C., Earles S. M., Blackburn M. L. (1996) Formation of adducts between 13-oxooctadecadienoic acid (13-OXO) and protein-derived thiols, in vivo and in vitro. Life Sci. 58, 2355–2365 [DOI] [PubMed] [Google Scholar]
- 11. Arita M., Bianchini F., Aliberti J., Sher A., Chiang N., Hong S., Yang R., Petasis N. A., Serhan C. N. (2005) Stereochemical assignment, antiinflammatory properties, and receptor for the omega-3 lipid mediator resolvin E1. J. Exp. Med. 201, 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lu D., Han C., Wu T. (2014) 15-PGDH inhibits hepatocellular carcinoma growth through 15-keto-PGE/PPARγ-mediated activation of p21. Oncogene 33, 1101–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lu D., Han C., Wu T. (2013) 15-hydroxyprostaglandin dehydrogenase (15-PGDH)-derived 15-keto-PGE2 inhibits cholangiocarcinoma cell growth through interaction with PPARγ, SMAD2/3 and TAP63. J. Biol. Chem. 288, 19484–19502 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14. Kansanen E., Bonacci G., Schopfer F. J., Kuosmanen S. M., Tong K. I., Leinonen H., Woodcock S. R., Yamamoto M., Carlberg C., Ylä-Herttuala S., Freeman B. A., Levonen A. L. (2011) Electrophilic nitro-fatty acids activate NRF2 by a KEAP1 cysteine 151-independent mechanism. J. Biol. Chem. 286, 14019–14027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Villacorta L., Zhang J., Garcia-Barrio M. T., Chen X. L., Freeman B. A., Chen Y. E., Cui T. (2007) Nitro-linoleic acid inhibits vascular smooth muscle cell proliferation via the Keap1/Nrf2 signaling pathway. Am. J. Physiol. Heart Circ. Physiol. 293, H770–H776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schopfer F. J., Cipollina C., Freeman B. A. (2011) Formation and signaling actions of electrophilic lipids. Chem. Rev. 111, 5997–6021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schopfer F. J., Cole M. P., Groeger A. L., Chen C. S., Khoo N. K., Woodcock S. R., Golin-Bisello F., Motanya U. N., Li Y., Zhang J., Garcia-Barrio M. T., Rudolph T. K., Rudolph V., Bonacci G., Baker P. R., Xu H. E., Batthyany C. I., Chen Y. E., Hallis T. M., Freeman B. A. (2010) Covalent peroxisome proliferator-activated receptor γ adduction by nitro-fatty acids: selective ligand activity and anti-diabetic signaling actions. J. Biol. Chem. 285, 12321–12333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kansanen E., Jyrkkänen H. K., Volger O. L., Leinonen H., Kivelä A. M., Häkkinen S. K., Woodcock S. R., Schopfer F. J., Horrevoets A. J., Ylä-Herttuala S., Freeman B. A., Levonen A. L. (2009) Nrf2-dependent and -independent responses to nitro-fatty acids in human endothelial cells: identification of heat shock response as the major pathway activated by nitro-oleic acid. J. Biol. Chem. 284, 33233–33241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Groeger A. L., Cipollina C., Cole M. P., Woodcock S. R., Bonacci G., Rudolph T. K., Rudolph V., Freeman B. A., Schopfer F. J. (2010) Cyclooxygenase-2 generates anti-inflammatory mediators from omega-3 fatty acids. Nat. Chem. Biol. 6, 433–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Snyder N. W., Golin-Bisello F., Gao Y., Blair I. A., Freeman B. A., Wendell S. G. (2014) 15-Oxoeicosatetraenoic acid is a 15-hydroxyprostaglandin dehydrogenase-derived electrophilic mediator of inflammatory signaling pathways. Chem. Biol. Interact. 10.1016/j.cbi.2014.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chu H. W., Balzar S., Seedorf G. J., Westcott J. Y., Trudeau J. B., Silkoff P., Wenzel S. E. (2004) Transforming growth factor-β2 induces bronchial epithelial mucin expression in asthma. Am. J. Pathol. 165, 1097–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Trudeau J., Hu H., Chibana K., Chu H. W., Westcott J. Y., Wenzel S. E. (2006) Selective downregulation of prostaglandin E2-related pathways by the Th2 cytokine IL-13. J. Allergy Clin. Immunol. 117, 1446–1454 [DOI] [PubMed] [Google Scholar]
- 23. Davies J. Q., Gordon S. (2005) Isolation and culture of human macrophages. Methods Mol. Biol. 290, 105–116 [DOI] [PubMed] [Google Scholar]
- 24. Hempel S. L., Monick M. M., Hunninghake G. W. (1994) Lipopolysaccharide induces prostaglandin H synthase-2 protein and mRNA in human alveolar macrophages and blood monocytes. J. Clin. Invest. 93, 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cohen A. B., Cline M. J. (1971) The human alveolar macrophage: isolation, cultivation in vitro, and studies of morphologic and functional characteristics. J. Clin. Invest. 50, 1390–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tang J. B., Goellner E. M., Wang X. H., Trivedi R. N., St Croix C. M., Jelezcova E., Svilar D., Brown A. R., Sobol R. W. (2010) Bioenergetic metabolites regulate base excision repair-dependent cell death in response to dna damage. Mol. Cancer Res. 8, 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schopfer F. J., Batthyany C., Baker P. R., Bonacci G., Cole M. P., Rudolph V., Groeger A. L., Rudolph T. K., Nadtochiy S., Brookes P. S., Freeman B. A. (2009) Detection and quantification of protein adduction by electrophilic fatty acids: mitochondrial generation of fatty acid nitroalkene derivatives. Free Radic. Biol. Med. 46, 1250–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. VanRollins M., Baker R. C., Sprecher H. W., Murphy R. C. (1984) Oxidation of docosahexaenoic acid by rat liver microsomes. J. Biol. Chem. 259, 5776–5783 [PubMed] [Google Scholar]
- 29. VanRollins M., Murphy R. C. (1984) Autooxidation of docosahexaenoic acid: analysis of ten isomers of hydroxydocosahexaenoate. J. Lipid Res. 25, 507–517 [PubMed] [Google Scholar]
- 30. O'Flaherty J. T., Hu Y., Wooten R. E., Horita D. A., Samuel M. P., Thomas M. J., Sun H., Edwards I. J. (2012) 15-Lipoxygenase metabolites of docosahexaenoic acid inhibit prostate cancer cell proliferation and survival. PLoS ONE 7, e45480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hamberg M., Samuelsson B. (1967) On the specificity of the oxygenation of unsaturated fatty acids catalyzed by soybean lipoxidase. J. Biol. Chem. 242, 5329–5335 [PubMed] [Google Scholar]
- 32. Chiang N., Takano T., Arita M., Watanabe S., Serhan C. N. (2003) A novel rat lipoxin A4 receptor that is conserved in structure and function. Br. J. Pharmacol. 139, 89–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arnold C., Markovic M., Blossey K., Wallukat G., Fischer R., Dechend R., Konkel A., von Schacky C., Luft F. C., Muller D. N., Rothe M., Schunck W. H. (2010) Arachidonic acid-metabolizing cytochrome P450 enzymes are targets of Ω-3 fatty acids. J. Biol. Chem. 285, 32720–32733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pace-Asciak C. (1975) Activity profiles of prostaglandin 15- and 9-hydroxydehydrogenase and 13-reductase in the developing rat kidney. J. Biol. Chem. 250, 2795–2800 [PubMed] [Google Scholar]
- 35. Na H. K., Park J. M., Lee H. G., Lee H. N., Myung S. J., Surh Y. J. (2011) 15-Hydroxyprostaglandin dehydrogenase as a novel molecular target for cancer chemoprevention and therapy. Biochem. Pharmacol. 82, 1352–1360 [DOI] [PubMed] [Google Scholar]
- 36. Tai H. H., Ensor C. M., Tong M., Zhou H., Yan F. (2002) Prostaglandin catabolizing enzymes. Prostaglandins Other Lipid. Mediat. 68, 483–493 [DOI] [PubMed] [Google Scholar]
- 37. Oh J. Y., Giles N., Landar A., Darley-Usmar V. (2008) Accumulation of 15-deoxy-Δ(12,14)-prostaglandin J2 adduct formation with Keap1 over time: effects on potency for intracellular antioxidant defence induction. Biochem. J. 411, 297–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bonacci G., Baker P. R., Salvatore S. R., Shores D., Khoo N. K., Koenitzer J. R., Vitturi D. A., Woodcock S. R., Golin-Bisello F., Cole M. P., Watkins S., St Croix C., Batthyany C. I., Freeman B. A., Schopfer F. J. (2012) Conjugated linoleic acid is a preferential substrate for fatty acid nitration. J. Biol. Chem. 287, 44071–44082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang J., Villacorta L., Chang L., Fan Z., Hamblin M., Zhu T., Chen C. S., Cole M. P., Schopfer F. J., Deng C. X., Garcia-Barrio M. T., Feng Y. H., Freeman B. A., Chen Y. E. (2010) Nitro-oleic acid inhibits angiotensin II-induced hypertension. Circ. Res. 107, 540–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Villacorta L., Chang L., Salvatore S. R., Ichikawa T., Zhang J., Petrovic-Djergovic D., Jia L., Carlsen H., Schopfer F. J., Freeman B. A., Chen Y. E. (2013) Electrophilic nitro-fatty acids inhibit vascular inflammation by disrupting LPS-dependent TLR4 signalling in lipid rafts. Cardiovasc. Res. 98, 116–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Charles R. L., Rudyk O., Prysyazhna O., Kamynina A., Yang J., Morisseau C., Hammock B. D., Freeman B. A., Eaton P. (2014) Protection from hypertension in mice by the Mediterranean diet is mediated by nitro fatty acid inhibition of soluble epoxide hydrolase. Proc. Natl. Acad. Sci. U.S.A. 111, 8167–8172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bilal S., Haworth O., Wu L., Weylandt K. H., Levy B. D., Kang J. X. (2011) Fat-1 transgenic mice with elevated omega-3 fatty acids are protected from allergic airway responses. Biochim Biophys Acta 1812, 1164–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Arita M., Ohira T., Sun Y. P., Elangovan S., Chiang N., Serhan C. N. (2007) Resolvin E1 selectively interacts with leukotriene B4 receptor BLT1 and ChemR23 to regulate inflammation. J. Immunol. 178, 3912–3917 [DOI] [PubMed] [Google Scholar]