Abstract
The gonadotropin-releasing hormone receptor (GnRHR) is expressed primarily in the gonadotropes of the anterior pituitary. Pituitary homeobox 1 (Pitx-1) has been shown to activate pituitary-specific gene expression by direct DNA binding and/or protein-protein interaction with other transcription factors. We hypothesized that Pitx-1 might also dictate tissue-specific expression of the mouse GnRHR (mGnRHR) gene in a similar manner. Pitx-1 activated the mGnRHR gene promoter, and transactivation was localized to sequences between −308 and −264. Pitx-1 bound to this region only with low affinity. This region includes an activating protein 1 (AP-1) site, which was previously shown to be important for mGnRHR gene expression. Further characterization indicated that an intact AP-1 site was required for full Pitx-1 responsiveness. Furthermore, Pitx-1 and AP-1 were synergistic in the activation of the mGnRHR gene promoter. A Pitx-1 homeodomain (HD) point mutation, which eliminated DNA binding ability, caused only a partial reduction of transactivation, whereas deletion of the HD completely prevented transactivation. Pitx-1 interacted directly with c-Jun, and the HD was sufficient for this interaction. While the point mutation in the Pitx-1 HD did not affect interaction with c-Jun, deletion of the HD eliminated the interaction. Taken together, our studies indicate that Pitx-1 can direct transactivation of the mGnRHR gene, in part by DNA binding and in part by an action of Pitx-1 as a cofactor for AP-1, augmenting AP-1 activity through a novel protein-protein interaction between c-Jun and the HD of Pitx-1.
Gonadotropin-releasing hormone (GnRH), released from the hypothalamus of the brain, is a critical neurohormone in maintaining the integrity of mammalian reproductive physiology. GnRH acts on the GnRH receptor (GnRHR) in gonadotropes of the anterior pituitary gland to stimulate the expression of genes encoding the subunits of the gonadotropins, luteinizing hormone (LH) and follicle-stimulating hormone (FSH), and GnRHR itself (2, 12, 16, 26). The GnRHR is a cell membrane receptor that is expressed primarily in the gonadotropes of the anterior pituitary. GnRHR cDNA has been cloned; mouse (36, 48), rat (10, 18), sheep (14), bovine (20), and human (7, 19) GnRHRs consist of seven putative transmembrane domains and are coupled to G-proteins (18, 36, 41, 48, 50). A 1.2-kb 5′-flanking region of the mouse GnRHR (mGnRHR) gene promoter has been cloned and characterized (1). Several functional cis-elements have been defined, including a steroidogenic factor 1 (SF-1) binding site at position −181 to −173 relative to the major transcriptional start site (8), an activating protein 1 (AP-1) binding site at −276 to −269 (9, 30), a SMAD binding site also known as GnRHR activating sequence (GRAS) at −329 to −318 (9, 32), and an element referred to as sequence underlying responsiveness to GnRH-1 (SURG-1) at −292 to −285 (30). We and others have shown previously that GnRH-stimulated GnRHR gene expression is induced primarily through the activation of Jun and Fos that bind to the AP-1 site (11, 30, 31, 51).
The mechanism underlying tissue-specific expression of the GnRHR gene in gonadotropes, however, is not yet fully understood. A previous report has shown that the GRAS, the AP-1 site, and the SF-1 site all contribute to cell-specific expression of the mGnRHR gene (9). A pan-pituitary homeobox transcription factor, pituitary homeobox 1 (Pitx-1), is primarily expressed in the anterior pituitary (22). It first appears in Rathke's pouch during early stages of development, and its expression in the anterior pituitary persists thereafter (43). Many anterior pituitary-derived cell lines also express Pitx-1; among them, murine gonadotrope-derived cell lines αT3-1 and LβT2 contain high levels of Pitx-1 compared with other cell lines (46, 53). It has been demonstrated that the expression of many anterior pituitary cell type-specific genes, including α-glycoprotein subunit (3, 43, 46), proopiomelanocortin (POMC) (22, 43, 46), growth hormone (GH) and prolactin (43, 46), and thyroid-stimulating hormone β (46), is increased by Pitx-1, suggesting an important role for Pitx-1 in maintaining tissue-specific gene expression in differentiated pituitary cell subtypes. The expression levels of the gonadotrope-specific genes, LHβ, FSHβ, and GnRHR, are all increased by Pitx-1 (35, 42, 46, 53). Furthermore, a synergistic effect has been shown for Pitx-1 with SF-1 and early growth response protein 1 (Egr-1) in the activation of the LHβ gene (45-47). These reports have led us to hypothesize that Pitx-1 may also have an important role in tissue-specific expression of the mGnRHR gene.
To investigate the mechanisms of action of Pitx-1 in mediating tissue-specific expression of the mGnRHR gene, we have localized Pitx-1 responsiveness in the mGnRHR gene promoter, analyzed Pitx-1 binding activity to the promoter, and demonstrated direct protein-protein interaction between Pitx-1 and c-Jun that is important for mGnRHR gene promoter activation.
MATERIALS AND METHODS
Generation of expression plasmids and fusion proteins.
The mouse Pitx-1 cDNA was generated by reverse transcription-PCR (RT-PCR) and subcloned into pcDNA1/Amp and pcDNA3 (Invitrogen, Carlsbad, Calif.). The RT reaction was performed using total RNA from the murine αT3-1 gonadotrope-derived cell line and an oligo(dT) primer, and the RT-generated first-strand poly(A) cDNA products were subjected to PCR using Pitx-1 cDNA-specific primers. PCR primers (sense, 5′-ACGTAAAGCTTTCTCTTGTCCCCAACAGGTC-3′; antisense, 5′-ACGTAGAATTCGGTCAGCTGTTGTACTGGCA-3′) were designed based on the murine Pitx-1 cDNA sequence (GenBank accession number U71206) (22) to include the Pitx-1 open reading frame region from +401 to +1435 of the cDNA, and a HindIII or EcoRI restriction enzyme site was incorporated into the 5′ end of each primer for the purpose of subcloning. Site-directed mutagenesis was performed by PCR to introduce a point mutation in the position of amino acid 139, to change Lys (AAG) to Arg (AGG), using primers including mutations and Pfu Turbo DNA polymerase (Stratagene, La Jolla, Calif.). Deletion mutants of Pitx-1 were also generated by PCR. Bidirectional sequence analyses were performed using the dideoxynucleotide chain termination method (39) to ensure the correct Pitx-1 cDNA sequence.
Glutathione S-transferase (GST)/Pitx-1 plasmids were generated by the insertion of the Pitx-1 or mutant Pitx-1 cDNA fragment downstream of the GST coding sequence in the expression vector pGEX-4T-2 (Amersham Pharmacia Biotech, Piscataway, N.J.) in the BamHI/NotI sites. These plasmids were introduced into bacterial stocks of Escherichia coli BL21(DE3) (Stratagene) and induced with isopropyl-β-d-thiogalactoside (Sigma, St. Louis, Mo.) to express the fusion proteins. Precipitated fusion proteins were purified from inclusion bodies as described previously (23). Briefly, proteins in the inclusion bodies were denatured with 4 M guanidine in 25 mM HEPES buffer (pH 7.6) containing 100 mM KCl, 100 μM EDTA, 12.5 mM MgCl2, 1 mM dithiothreitol, 10% glycerol, and 0.1% Igepal CA-630 (Sigma) and were refolded in vitro with ice-cold 50 mM Tris buffer (pH 7.9) containing 1 M l-Arg, 1 mM EDTA, 1 mM reduced glutathione, and 800 μM oxidized glutathione. Refolded fusion proteins were purified using a glutathione-Sepharose column (Amersham Pharmacia Biotech), and the identity of purified proteins was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analyses (see below). Human c-Jun (hc-Jun; Promega, Madison, Wis.) protein was also used in select experiments.
GAL4 DNA binding domain (DBD) fusion constructs were generated for c-Jun and c-Fos basic leucine zipper (bZIP) regions (amino acids 237 to 334 and 137 to 216, respectively) (54). Each bZIP domain was fused downstream of the GAL4 DBD in the pM vector (Clontech, Palo Alto, Calif.). c-Jun and c-Fos expression plasmids in pCMV5 were kindly provided by William L. Miller (North Carolina State University, Raleigh, N.C.).
Generation of Pitx-1 antibody.
A polyclonal anti-Pitx-1 (α-Pitx-1) antibody was raised in rabbits against a peptide corresponding to amino acids 24 to 52 of murine Pitx-1 and conjugated to keyhole limpet hemocyanin as a hapten carrier (Covance, Richmond, Calif.). This region is unique to Pitx-1 based on homology analysis and possesses high hydrophilicity and antigenicity. Prepared α-Pitx-1 antiserum was further purified to obtain an α-Pitx-1 immunoglobulin G (IgG) fraction using a serum IgG purification kit (Econo-Pac; Bio-Rad, Hercules, Calif.). Commercial α-c-Jun and α-Pitx-1 antibodies (Santa Cruz Biotechnology, Santa Cruz, Calif.) were also used in select experiments.
Northern blot analysis.
αT3-1 cells were treated with 100 nM GnRH agonist (des-Gly10, [d-Ala6]-LH-releasing hormone ethylamide; Sigma) for up to 8 h, and total RNA was purified from each group of cells using the total RNA isolation (TRI) reagent (Molecular Research Center, Cincinnati, Ohio). Total RNA (10 μg) from each group was subjected to Northern gel separation, blotted onto a nylon membrane (Schleicher & Schuell, Keene, N.H.), and hybridized with a Pitx-1 cRNA probe labeled with [α-32P]UTP (NEN Life Sciences, Boston, Mass.) by using an RNA polymerase (New England BioLabs, Beverly, Mass.). The signals were quantitated using a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). Cyclophilin mRNA signal was used for normalization.
Transient-transfection analyses.
Construction of the mGnRHR reporter plasmids using a luciferase (Luc) reporter plasmid, pXP2 (29), has been reported previously (17, 30). For mammalian one-hybrid analyses, a Luc/pT109 reporter plasmid containing a thymidine kinase minimal promoter and five copies of the upstream activator sequence (5xUAS), the GAL4 DBD binding site, was used. Transient-transfection analyses were performed using the calcium-phosphate coprecipitation method (30) in CV-1 cells (derived from African green monkey kidney fibroblasts), a nongonadotropic cell line that does not express endogenous Pitx-1 (46, 53). Cells were transfected with or without cotransfection of a Pitx-1, c-Jun, and/or c-Fos expression vector (1 μg/well) for 24 h in six-well plates and were maintained for an additional 24 h after washing and changing to fresh medium. Additional transfection assays were performed in αT3-1 cells, which express endogenous Pitx-1 (37, 46, 53). The αT3-1 cells were transfected by calcium-phosphate coprecipitation for 16 h, followed by harvest. The empty reporter was used as a negative control, and the appropriate empty expression vector was used for cotransfection controls. The total amount of DNA transfected in each well was kept constant. Luc activity was normalized to the activity of a cotransfected plasmid encoding a β-galactosidase reporter gene under the control of a Rous sarcoma virus promoter. All transfection analyses were performed in at least three independent experiments, each performed in triplicate. A human embryonic kidney-derived cell line, 293T, was transfected with Pitx-1 mutant constructs. Cells were transfected by calcium-phosphate coprecipitation for 16 h, and after changing to fresh medium they were maintained for an additional 24 h, followed by harvest and extraction of nuclear proteins (see below).
EMSA.
The ability of Pitx-1 to bind to the mGnRHR promoter was characterized by electrophoretic mobility shift assays (EMSA) using GST/Pitx-1 fusion protein, nuclear extracts from 293T cells transfected with wild-type or mutant Pitx-1 constructs, or nuclear extracts from LβT2 or αT3-1 cell lines. GST protein was used as a negative control, and hc-Jun was used when appropriate. Extraction of nuclear proteins was performed as previously reported (44). The region of interest of the mGnRHR gene promoter (−300 to −259), either wild type or containing a point mutation in the AP-1 site at position −269 (C-269T), was used as a probe. Previously defined Pitx-1 binding elements from the POMC gene promoter (CE3) (22) and a murine LHβ promoter sequence (mLHβ) (45) were used as probes for positive controls. To generate probes, sense and antisense oligonucleotides corresponding to −300 to −259 of the mGnRHR promoter, C-269T, CE3, or mLHβ were annealed, gel purified, and end labeled with [γ-32P]ATP (NEN Life Sciences) by using T4 polynucleotide kinase (New England BioLabs). For EMSA studies, the amount of probe in each lane was 200 kcpm. Each protein-DNA reaction mixture was incubated on ice for 1 h. Competition studies were performed using excess unlabeled oligonucleotides. Cold competitor oligonucleotides were added 5 min prior to the addition of probe. For supershift EMSA studies, antibody or antiserum was incubated with protein on ice for 1 h, after which probe was added and the reaction mixtures were incubated on ice for an additional 1 h. Preimmune serum derived from the same rabbit was used as a negative control for Pitx-1 antiserum. All gel separations were performed using 5% polyacrylamide gels. In some cases, signal densities of Pitx-1-DNA complexes were quantitated using a PhosphorImager (Molecular Dynamics).
ChIP analysis.
Chromatin immunoprecipitation (ChIP) analysis was performed as previously reported (5, 40). Briefly, LβT2 cells were cross-linked with 1% formaldehyde at room temperature for 10 min and were harvested after cross-linking was terminated by the addition of 125 mM Gly. Cells were sonicated eight times for 20 s each with 1-min intervals on ice. After sonication, the chromatin solutions were precleared with protein A-agarose (Upstate, Charlottesville, Va.) and salmon sperm DNA and subjected to immunoprecipitation (IP) by incubating with α-c-Jun or α-Pitx-1 antibody (Santa Cruz Biotechnology), followed by incubation with protein A-agarose-salmon sperm DNA. Preimmune IgG was used as a negative control. After serial washes, precipitated chromatin was eluted and cross-links were reversed with 0.3 M NaCl. After protein removal with proteinase K (Roche Applied Science, Indianapolis, Ind.), chromatin was purified by phenol-chloroform extraction and ethanol precipitation. PCR was performed using primers (sense, 5′-GTATCTGTCTAGTCACAACAG-3′; antisense, 5′-TCCTGAAGGCCAAGTGTAACC-3′) that spanned −337 to −170 of the mGnRHR gene promoter, which includes the GRAS, SURG-1, AP-1, and SF-1 sites.
Uracil and methylation interference analyses.
Uracil and methylation interference analyses were performed using GST/Pitx-1 and hc-Jun, which was used for detection of the AP-1 site as a positive control. For uracil interference, PCR primers for −387 to −220 of the mGnRHR gene promoter were end labeled with [γ-32P]ATP using T4 polynucleotide kinase, and thymidine nucleotides were randomly replaced by uracil in a PCR. For methylation interference, a cDNA corresponding to −387 to −220 was labeled with [α-32P]dCTP (NEN Life Sciences) using Klenow enzyme (Promega). After random methylation by dimethyl sulfate (Sigma), each probe (500 kcpm/reaction mixture) was incubated with either GST/Pitx-1 (1 μg/reaction mixture) or hc-Jun (1 footprinting unit [fpu]/reaction mixture; 640 ng). EMSAs were performed, free and bound probes from each reaction mixture were extracted from the polyacrylamide gels, and samples were subjected to cleavage by using piperidine (Sigma). Uracil probes were pretreated with uracil-N-glycosylase (New England BioLabs) prior to piperidine treatment to make them susceptible to cleavage. After cleavage, each probe (4 kcpm/lane) was separated on a 6% polyacrylamide sequencing gel. A set of sequencing reactions for −387 to −220 was also run in parallel as a size marker. Thymidines were visualized in uracil interference, and guanines were visualized in methylation interference.
GST pull-down and Western blot analyses.
GST pull-down analyses were performed using intact or mutant GST/Pitx-1 proteins and hc-Jun. Intact or mutant GST/Pitx-1 proteins (1 μg/reaction mixture) bound to glutathione-Sepharose beads were incubated with hc-Jun (0.1 fpu/reaction mixture) at 4°C for 16 h. The same amount of GST was used as a negative control. After extensive washing, bead pellets from each reaction were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis separation, and proteins on the gel were transferred onto a nitrocellulose membrane (Schleicher & Schuell) for Western blot analyses. After blotting, c-Jun signal was detected with α-c-Jun antibody (1:5,000) and α-rabbit IgG-horseradish peroxidase (HRP) (1:10,000; Pierce, Rockford, Ill.). To ensure that even amounts of GST/Pitx-1 proteins were loaded, the blot was strip washed with a stripping buffer (Pierce) and subjected to GST detection using an α-GST antibody from goat (1:10,000; Amersham Pharmacia Biotech) and α-goat IgG-HRP (1:10,000; Santa Cruz Biotechnology). Western blot analysis was also performed using nuclear extracts (10 μg/lane) from 293T cells transfected with the Pitx-1 mutant constructs. Expression of Pitx-1 proteins was determined using α-Pitx-1 antibody (1:2,000) and α-rabbit IgG-HRP.
Co-IP analysis.
Coimmunoprecipitation (co-IP) analysis was performed based on a previously reported method (21). Briefly, LβT2 cells were treated with 100 nM GnRH agonist for 1 h, and nuclear proteins were extracted. Nuclear extracts (245 μg/reaction mixture) were immuno-cleared by incubating with preimmune IgG and protein G-agarose (Santa Cruz Biotechnology) and were subjected to IP by incubating with α-c-Jun or α-Pitx-1 antibody. Preimmune IgG was used as a negative control. After incubating with protein G-agarose, bound proteins were washed and subjected to Western blot analysis.
Data analysis.
All results were analyzed by one- or two-way analysis of variance (ANOVA) followed by post hoc comparisons with Fisher's protected least significant difference test. A P value less than 0.05 was considered statistically significant. Data are presented as the mean ± the standard error of the mean (SEM).
RESULTS
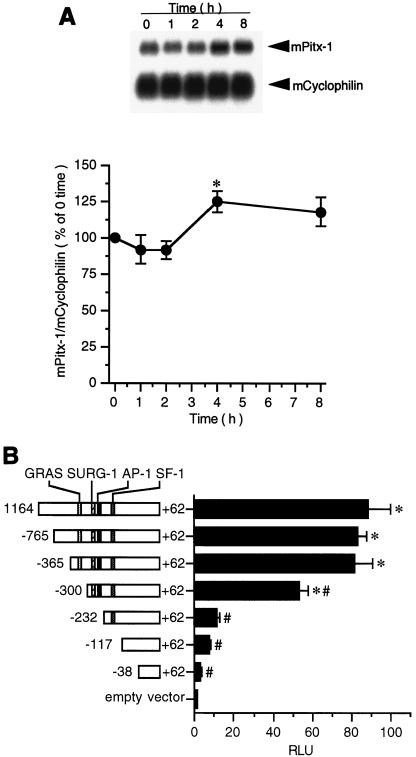
GnRH has little effect on Pitx-1 mRNA levels in αT3-1 cells.
Expression of many transcription factors, including Jun/Fos (6), activating transcription factor 2 (28), and Egr-1 (45), has been shown to be increased by GnRH in anterior pituitary or in gonadotrope-derived cell lines. To determine if Pitx-1 expression is similarly modulated by GnRH, total RNA from GnRH agonist-treated αT3-1 cells was subjected to Northern blot analysis (Fig. 1A). Pitx-1 mRNA levels were not changed during GnRH agonist treatment except at 4 h, when a small (25%) but statistically significant increase was observed. These data are consistent with a previous report (45) and suggest that Pitx-1 gene expression is not primarily under the control of GnRH, but rather that it is constitutively expressed in gonadotrope cells and may be more important for gonadotrope-specific expression of the mGnRHR gene than for its GnRH response.
FIG. 1.
mGnRHR gene promoter activity is localized primarily to −365 to −232 in αT3-1 cells. (A) Northern blot analysis was performed as described in Materials and Methods, and the signals were quantitated and plotted. Results were analyzed by one-way ANOVA. Data are presented as the mean ± SEM of four independent experiments. Time indicates GnRH agonist treatment duration. mPitx-1, mouse Pitx-1 mRNA; mCyclophilin, mouse cyclophilin mRNA; *, P < 0.05 versus time zero. (B) A series of 5′-deletion constructs of the mGnRHR gene promoter fused to a Luc reporter plasmid (pXP2) were transfected into αT3-1 cells. Results were analyzed by two-way ANOVA followed by post hoc comparisons with Fisher's protected least significant difference test. Data are presented as the mean ± SEM of at least three independent experiments. RLU, relative light units; *, P < 0.05 versus pXP2 empty vector; #, P < 0.05 versus −1164/+62/pXP2.
mGnRHR gene promoter activity is localized primarily to −365 to −232 in αT3-1 cells.
It had been shown previously that a tripartite cis element, comprising GRAS, AP-1, and SF-1 binding elements, is necessary for basal and tissue-specific expression of the mGnRHR gene (9). We similarly analyzed the activity of the mGnRHR gene promoter (Fig. 1B). 5′-deletion constructs of the mGnRHR gene promoter fused to a Luc reporter gene (pXP2) were transiently transfected into αT3-1 cells. The activity of the 1.2-kb promoter (−1164/+62) was 88-fold ± 11-fold higher than pXP2, the promoterless control. This activity was maintained despite 5′ deletion to −365. Further 5′ deletion to −300 resulted in a decrease in activity, to 61% of the activity of −1164/+62, with a further reduction in activity to 11% with further 5′ deletion to −232. These data demonstrate the importance of sequences between −365 and −232, especially −300 to −232, which contain the SURG-1 and AP-1 sites, for transactivation in the gonadotrope-derived αT3-1 cell line.
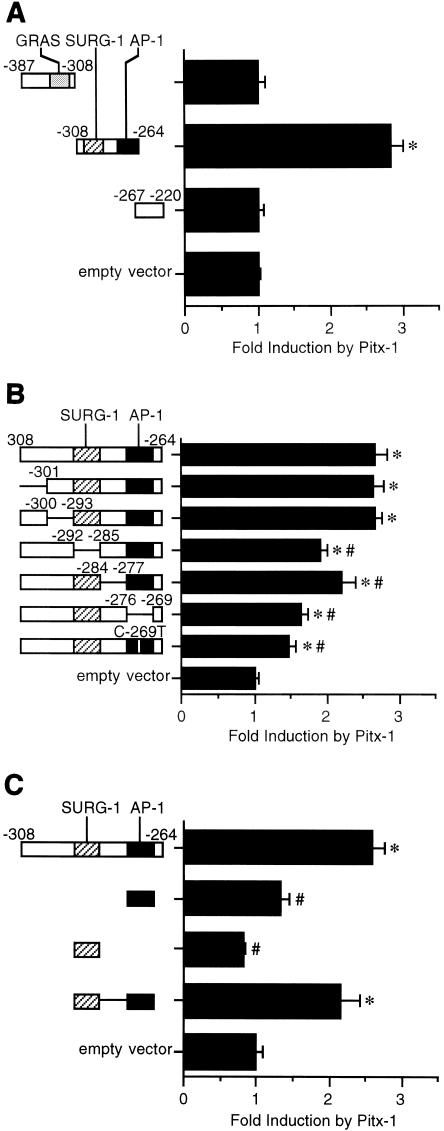
Pitx-1 responsiveness is localized to −308 to −264 of the mGnRHR gene promoter.
Based on the data in Fig. 1B and a previous report suggesting a possible Pitx-1 consensus sequence between the SURG-1 and AP-1 sites (46), we focused on the sequences between −365 and −232 for our next studies. To determine Pitx-1 responsiveness of this region, we performed transfection analyses using a series of fragments of the −387 to −220 region of mGnRHR gene promoter fused upstream of the heterologous rat GH gene minimal promoter (GH50) in pXP2 (30) in CV-1 cells, with cotransfection of either Pitx-1 or an empty control expression vector (Fig. 2A). Transactivation by Pitx-1 was observed only for the −308/−264 construct, which spans the SURG-1 and AP-1 sites, with 2.8- ± 0.1-fold induction relative to GH50/pXP2.
FIG. 2.
Pitx-1 responsiveness is localized to −308 to −264 of the mGnRHR gene promoter. Luc reporter genes fused to GH50 with a series of deletion mutations of −387/−220 (A), with a series of NotI linker-scanner mutations of −308/−264 or C-269T (B), or with AP-1 and/or SURG-1 sites (C) of the mGnRHR gene were transfected into CV-1 cells with Pitx-1 cotransfection. In panel C, the SURG-1/AP-1 construct has NotI restriction sequences between the two sites. The NotI restriction sequences are shown as lines in the constructs in panels B and C. Data are plotted as the induction by Pitx-1 over the empty expression vector control response for each reporter. Data are presented as the mean ± SEM of at least three independent experiments. *, P < 0.05 versus GH50/pXP2 empty vector; #, P < 0.05 versus −308/−264/GH50/pXP2.
To further localize the Pitx-1-responsive element, we performed transfection analyses using a series of NotI restriction site (5′-GCGGCCGC-3′) linker-scanner mutations of the −308 to −264 sequences, as well as a construct containing a point mutation in the AP-1 site (C-269T), fused upstream of the GH50-Luc reporter in CV-1 cells, again with cotransfection of Pitx-1 (Fig. 2B). The C-269T point mutation has been shown previously to abolish AP-1 binding in EMSA (30). A significant reduction in Pitx-1 responsiveness was observed for the Δ(−292/−285) (by 44%) and Δ(−276/−269) (by 61%) constructs, in which the replaced sequence eliminated the SURG-1 and the AP-1 sites, respectively. The C-269T mutant showed a reduction of Pitx-1 responsiveness (by 72%) similar to that of the Δ(−276/−269) construct. A modest but significant reduction was also observed for the Δ(−284/−277) construct (by 27%). However, none of these mutant constructs completely eliminated Pitx-1 responsiveness, suggesting contributions from each element and a requirement for intact sequences between −292 and −269.
To further investigate the importance of the AP-1 and SURG-1 elements for Pitx-1 activity, we carried out transfection analyses using the AP-1 and/or SURG-1 elements fused upstream of the GH50-Luc reporter in CV-1 cells (Fig. 2C). Pitx-1 was able to significantly induce activity only for the SURG-1/AP-1 construct, to a similar extent as the −308/−264 construct (2.2- ± 0.3- and 2.6- ± 0.2-fold relative to GH50/pXP2, respectively) and failed to induce Luc activity when only the AP-1 or the SURG-1 element was present. Taken together, these data suggest that intact AP-1 and SURG-1 sites are required for Pitx-1 to enhance transactivation of the mGnRHR gene promoter.
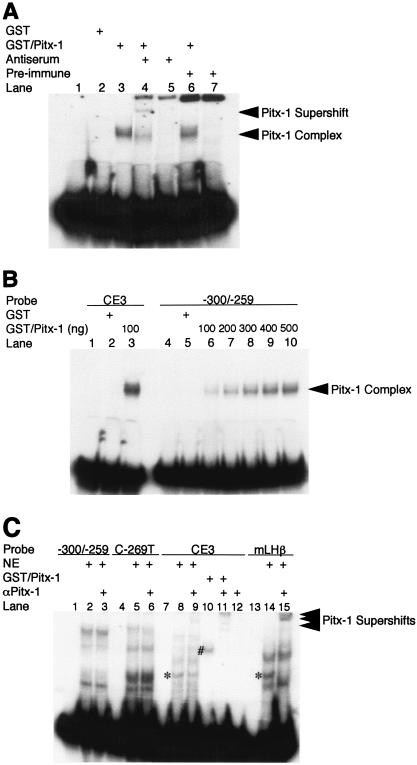
Pitx-1 binds to sequences between −300 and −259 of the mGnRHR gene promoter with low affinity.
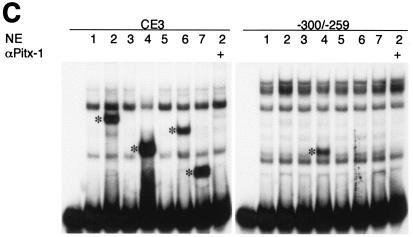
One likely mechanism for Pitx-1 transactivation of the mGnRHR gene promoter is by direct DNA binding to the promoter, near the SURG-1 or AP-1 site. Pitx-1 has been shown to transactivate the POMC, LHβ, and FSHβ genes by binding directly to promoter elements (22, 45, 53) and, as mentioned earlier, a putative Pitx-1 consensus binding sequence has been identified previously in the mGnRHR gene promoter, between the SURG-1 and AP-1 sites (46). To test whether Pitx-1 can bind to this region, EMSA analyses were performed, using either a GST/Pitx-1 fusion protein or nuclear extracts from the gonadotrope-derived LβT2 cell line (Fig. 3). GST/Pitx-1 formed a complex with an oligonucleotide probe spanning −300 to −259 of the mGnRHR gene promoter (Fig. 3A). The migration of this complex was further retarded by α-Pitx-1 antiserum (lane 4), but not by preimmune serum (lane 6). This binding activity was increased in a dose-dependent manner by increasing amounts of GST/Pitx-1 (Fig. 3B). However, the intensity of the complex was lower than that formed when CE3, a previously defined Pitx-1 binding element in the POMC gene promoter, was used as probe; even 500 ng of GST/Pitx-1 per lane (lane 10) formed a less intense complex with −300 to −259 than that of 100 ng of GST/Pitx-1 per lane with the CE3 probe (lane 3). Nonetheless, despite the known expression of Pitx-1 by LβT2 gonadotrope cells (37, 53) and the ability of Pitx-1 from LβT2 cell nuclear extract to bind to CE3 and to the mLHβ promoter (Fig. 3C, lanes 8 and 14), we were unable to detect binding of Pitx-1 to mGnRHR gene promoter sequences −300 to −259 (lanes 2, 3, 5, and 6) using nuclear extracts from LβT2 cells. Similar results were observed using αT3-1 cell nuclear extracts (data not shown). These data suggest that Pitx-1 can bind to sequences between −300 and −259 of the mGnRHR gene promoter, but with low affinity that is not sufficient to result in detectable binding at concentrations of Pitx-1 present in LβT2 or αT3-1 nuclear extracts.
FIG. 3.
Pitx-1 binds to sequences between −300 and −259 of the mGnRHR gene promoter with low affinity. EMSA analyses were performed as described in Materials and Methods. (A) Purified Pitx-1 protein fused downstream of GST (GST/Pitx-1; 100 ng/lane) was incubated with −300/−259 probe with or without α-Pitx-1 antiserum. GST (100 ng/lane) and preimmune serum were used as negative controls. (B) Increasing amounts of GST/Pitx-1 (100 to 500 ng/lane) were incubated with −300/−259 probe and compared to 100 ng of GST/Pitx-1 per lane incubated with CE3 probe. GST protein (500 ng/lane) was used as a negative control. (C) Nuclear extracts from LβT2 cells (10 μg/lane) or GST/Pitx-1 (20 ng/lane) were incubated with −300/−259, C-269T, CE3, or mLHβ probe with or without purified α-Pitx-1 antibody. NE, nuclear extract; *, Pitx-1-DNA complexes; #, GST/Pitx-1-CE3 complex.
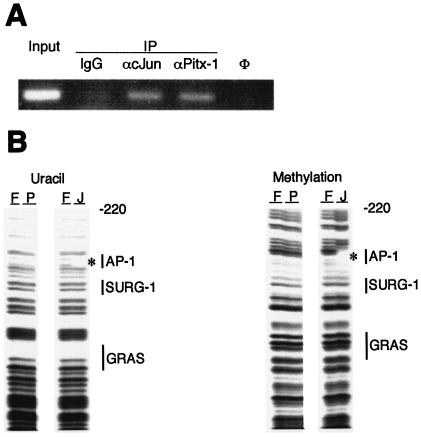
Pitx-1 binds to the mGnRHR gene promoter in vivo but does not have distinct binding sites.
To determine whether DNA binding by Pitx-1 to this region of the mGnRHR gene promoter can occur in vivo in gonadotrope cells, we performed ChIP analysis using the LβT2 cell line (Fig. 4A). After cross-linking, lysis, and sonication, chromatin was immunoprecipitated with α-Pitx-1 antibody. DNA was extracted and subjected to PCR amplification, using primers flanking the regulatory region of interest in the mGnRHR gene promoter. The resulting PCR product indicated that Pitx-1 protein can bind to the mGnRHR gene promoter in vivo in the gonadotrope-derived LβT2 cell line. α-c-Jun antibody was used as a positive control, based on the known AP-1 consensus binding site in this region of the mGnRHR gene promoter. Preimmune IgG, used as a negative control, did not give rise to detectable PCR product, confirming the specificity of the ChIP analysis results.
FIG. 4.
Pitx-1 binds to the mGnRHR gene promoter in vivo but does not have distinct binding sites. (A) ChIP analysis was performed using LβT2 gonadotrope cells. After cross-linking, IP was performed by incubating with preimmune IgG, α-c-Jun, or α-Pitx-1 antibody. Input sample was prepared without IP and was further diluted to 1:100 for PCR. IgG, preimmune IgG; φ, water control for PCR. (B) Uracil (left panel) and methylation (right panel) interference analyses were performed using GST/Pitx-1 as described in Materials and Methods. hc-Jun was used as a positive control, binding to the AP-1 site. Locations of designated cis-elements are marked with vertical bars. F, free probe; P, GST/Pitx-1; J, c-Jun; *, c-Jun footprint.
To further identify and localize possible Pitx-1 binding elements within the mGnRHR gene promoter, we performed uracil and methylation interference analyses, using the −387 to −220 region of the promoter and GST/Pitx-1 protein (Fig. 4B). hc-Jun protein was used as a positive control. While the AP-1 site was clearly evident by c-Jun binding in both analyses, no distinct binding site for Pitx-1 was identified in either uracil (left panel) or methylation (right panel) interference analysis. These results suggest that Pitx-1 does not have a specific binding site in −387 to −220 of the mGnRHR gene promoter, but rather that it may bind to a number of sequences in this region with low affinity.
Pitx-1 interacts directly with c-Jun.
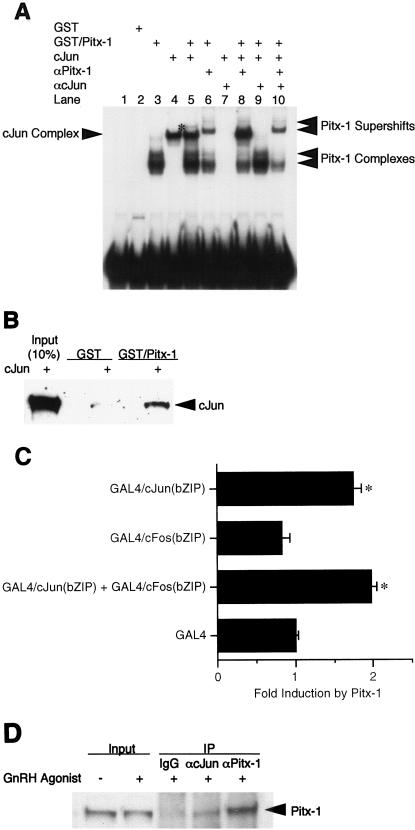
Based on our transfection studies (Fig. 2), the AP-1 site appears to be necessary for Pitx-1 transactivation of the mGnRHR gene promoter. We hypothesized that Pitx-1 may modulate AP-1 complex formation on its binding site. To test this possibility, we performed supershift EMSA with combinations of GST/Pitx-1 and/or hc-Jun, as well as their antibodies, using −300/−259 as a probe (Fig. 5A). When GST/Pitx-1 and c-Jun were coincubated with the probe, a new, faint complex appeared (lane 5). This new complex disappeared in the presence of either α-Pitx-1 antibody (lane 8), α-c-Jun antibody (lane 9), or both (lane 10). The intensities of the Pitx-1 and c-Jun complexes (lanes 3 and 4) did not change in the presence of both proteins (lane 5). These data indicate that a complex containing both Pitx-1 and AP-1 can form with −300/−259, but that Pitx-1 does not modulate the binding pattern or affinity of c-Jun to the AP-1 site.
FIG. 5.
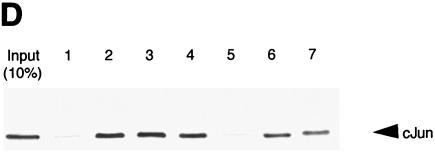
Pitx-1 interacts directly with c-Jun. (A) EMSA was performed using GST/Pitx-1 and hc-Jun. −300/−259 was used as the probe. Each combination of GST/Pitx-1 (500 ng/lane), hc-Jun (0.1 fpu/lane), α-Pitx-1 antibody, and/or α-c-Jun antibody was incubated with probe. GST (500 ng/lane) was used as a negative control. *, new protein-DNA complex in the presence of both Pitx-1 and c-Jun. (B) GST pull-down analysis using hc-Jun (0.1 fpu/lane) and GST/Pitx-1 (1 μg/lane) was performed as described in Materials and Methods. GST (1 μg/lane) was used as a negative control. For the 10% input lane, 0.01 fpu of hc-Jun was loaded. (C) Luc reporter (pT109) containing 5xUAS was cotransfected into CV-1 cells with either Pitx-1 or empty expression vector and the indicated GAL4 DBD fusion constructs. Data are plotted as induction by Pitx-1 over the empty expression vector control response for each GAL4 DBD fusion construct. Data are presented as the mean ± SEM of three independent experiments. *, P < 0.05 versus GAL4 empty vector. (D) Co-IP analysis was performed using nuclear extracts (245 μg/lane) from LβT2 gonadotrope cells treated with 100 nM GnRH agonist for 1 h. IP was performed by incubating nuclear extracts with preimmune IgG, α-c-Jun antibody, or α-Pitx-1 antibody, followed by Western blot analysis with α-Pitx-1 antibody. For the input lanes, 10 μg of each nuclear extract with or without GnRH agonist treatment was loaded. IgG, preimmune IgG.
Another possible mechanism for Pitx-1 transactivation of the mGnRHR promoter via the AP-1 site is that Pitx-1 may have direct protein-protein interaction with Jun/Fos to modulate AP-1 activity. To characterize a possible physical interaction between Pitx-1 and c-Jun proteins, we performed GST pull-down analysis using GST/Pitx-1 and hc-Jun (Fig. 5B). c-Jun was able to bind directly to GST/Pitx-1, but not to GST. These data indicate that Pitx-1 and c-Jun can interact directly with each other.
To test whether this protein-protein interaction between Pitx-1 and c-Jun occurs in vivo in cultured cells, we adapted a mammalian one-hybrid system, using two fusion constructs encoding c-Jun and c-Fos bZIP domains, each fused downstream of the GAL4 DBD. These fusion constructs were cotransfected with the Pitx-1 expression vector and a Luc reporter gene containing 5xUAS (5xUAS-Luc) in CV-1 cells (Fig. 5C). While Pitx-1 did not appear to interact with GAL4/c-Fos(bZIP), it interacted with GAL4/c-Jun(bZIP), as reflected by increased transactivation of the 5xUAS-Luc reporter by 1.7- ± 0.1-fold. No further transactivation of the 5xUAS-Luc reporter was observed in the presence of both GAL4/c-Jun(bZIP) and GAL4/c-Fos(bZIP). These results suggest that Pitx-1 may interact specifically with c-Jun but not with c-Fos. Thus, it appears that Pitx-1 binds to the AP-1 complex and may thereby modulate its activity to increase transactivation of the mGnRHR gene promoter. These results suggest that Pitx-1 may increase mGnRHR gene transactivation through direct protein-protein interaction with c-Jun.
To confirm whether this protein-protein interaction between Pitx-1 and c-Jun occurs in vivo in gonadotrope cells, we performed co-IP analysis using LβT2 nuclear extracts (Fig. 5D). IP with α-c-Jun antibody followed by Western blot analysis with α-Pitx-1 antibody clearly demonstrated the presence of Pitx-1 among the immunoprecipitated proteins. Preimmune IgG was used as a negative control, and α-Pitx-1 served as a positive control in the IP reaction. These results confirm that the interaction between Pitx-1 and c-Jun occurs in vivo in gonadotrope cells.
The Pitx-1 HD is necessary for transactivation of the mGnRHR gene promoter.
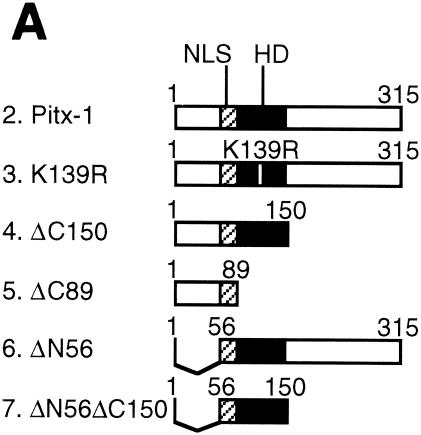
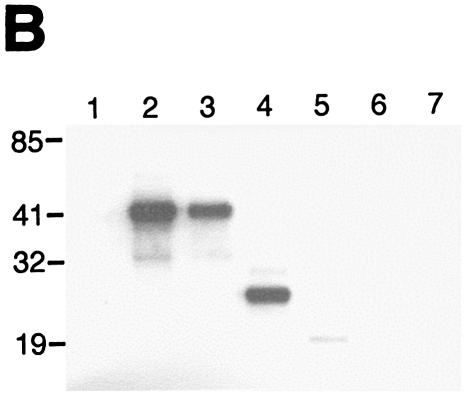
If the mechanism of Pitx-1 transactivation of the mGnRHR gene promoter is solely through interaction with c-Jun, then Pitx-1 action should be independent of its DNA binding capability. To test this hypothesis and to identify the Pitx-1 domain(s) responsible for interaction with c-Jun and for mGnRHR gene promoter transactivation, we generated a series of Pitx-1 mutant constructs (Fig. 6A). To first verify the expression of Pitx-1 mutant constructs in vivo, 293T cells were transfected with these Pitx-1 mutant constructs and Western blot analysis and EMSA were performed with the nuclear proteins (Fig. 6B and C). Intact Pitx-1 (lane 2), K139R (lane 3), ΔC150 (lane 4), and ΔC89 (lane 5) proteins were all detected by Western blot analysis (Fig. 6B). ΔN56 (lane 6) and ΔN56ΔC150 (lane 7) proteins were not able to be detected by our α-Pitx-1 antibody, since the epitope for antibody generation was located in the N terminus (see Materials and Methods) and was thus absent in these mutants. The expression level of ΔC89 protein appeared to be lower than that of the other detected proteins. It is not clear whether this was due to inefficient nuclear translocation (the nuclear localization signal of Pitx-1 is in its N terminus), low expression levels, protein instability, or reduced recognition by the α-Pitx-1 antibody. The N-terminal mutant proteins were readily detected by EMSA with the CE3 probe, as were the other Pitx-1 proteins with an intact homeodomain (HD) (Fig. 6C, left gel), thereby verifying their nuclear expression in the transfected cells. Interestingly, ΔC150 protein was able to bind to −300/−259 (right gel).
FIG. 6.
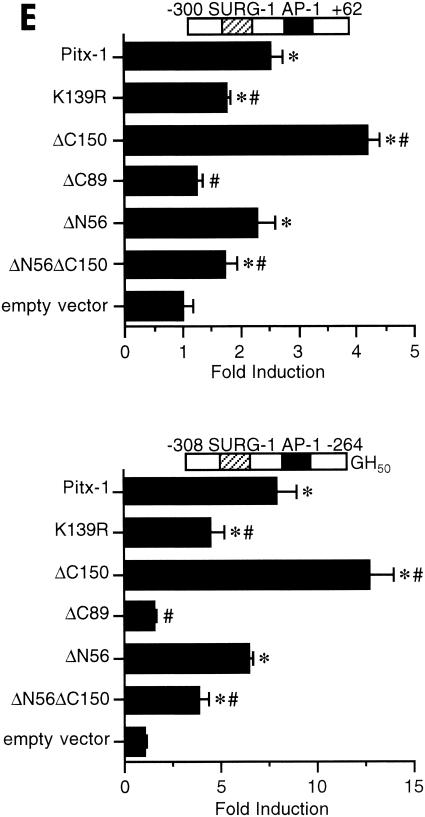
The Pitx-1 HD is necessary for transactivation of the mGnRHR gene promoter. (A) Schematic illustration of structures for intact Pitx-1 and the mutant Pitx-1 constructs generated. One set of constructs was generated in the pcDNA3 expression vector, and another set was generated in the pGEX-4T-2 expression vector for GST fusion proteins. NLS, nuclear localization signal. (B) Western blot analysis was performed using nuclear extracts (10 μg/lane) of 293T cells transfected with each Pitx-1 construct. Lanes: 1, empty pcDNA3 vector; 2 to 7, Pitx-1 constructs corresponding to the structures illustrated in panel A. Molecular masses (in kilodaltons) of protein markers are indicated on the left side of the gel. (C) EMSA analyses were performed using nuclear extracts (20 μg/lane) of 293T cells transfected with each Pitx-1 construct. CE3 or −300/−259 was used as probe. Numbers for each lane denote the Pitx-1 constructs as indicated for panels A and B. In the last lane of each gel, α-Pitx-1 antibody was added to the EMSA reaction mixture with nuclear extract containing intact Pitx-1. NE, nuclear extract; *, Pitx-1-DNA complexes. (D) GST pull-down analysis using hc-Jun (0.1 fpu/lane) and intact or mutant GST/Pitx-1 proteins (1 μg/lane) was performed as described in Materials and Methods. Lanes: 1, GST; 2 to 7, GST/Pitx-1 protein constructs corresponding to the structures illustrated in panel A. For the 10% input lane, 0.01 fpu of hc-Jun was loaded. (E) Luc reporter genes fused to −300/+62 or −308/−264/GH50 were cotransfected into CV-1 cells with either intact or mutant Pitx-1 expression vectors, or with empty vector, pcDNA3. Data are plotted as the induction by Pitx-1 over the empty expression vector control response for each reporter. Data are presented as the mean ± SEM of three independent experiments. *, P < 0.05 versus pcDNA3 empty vector; #, P < 0.05 versus intact Pitx-1.
To determine the Pitx-1 domain responsible for protein-protein interaction with c-Jun, we performed GST pull-down analysis using a series of GST/Pitx-1 mutant proteins (Fig. 6D). c-Jun was able to bind directly to all of the Pitx-1 mutant proteins except GST/ΔC89 (lane 5), in which Pitx-1 lacked its HD (Fig. 6A). The same amount of GST/ΔC89 protein (1 μg) as other GST/Pitx-1 fusion proteins was input into the pull-down reaction mixture. The GST/Pitx-1 protein with a point mutation in the HD (GST/K139R, lane 3), which eliminates binding to DNA (Fig. 6C), and the GST/ΔN56ΔC150 protein (lane 7), which contains only the HD (Fig. 6A), were still able to interact with c-Jun. These data suggest that the Pitx-1 HD is both necessary and sufficient for protein-protein interaction with c-Jun.
We then performed transfection analyses in CV-1 cells, cotransfecting the Pitx-1 mutant constructs along with Luc reporters containing either −300 to +62 of the mGnRHR gene or −308 to −264 fused upstream of the GH50 promoter (Fig. 6E). Pitx-1 significantly increased the activity of −300/+62, by 2.5- ± 0.2-fold. The Pitx-1 mutant protein harboring a point mutation in the HD (K139R), which prevented DNA binding (Fig. 6C), remained able to partially transactivate the reporter, although the level of activity was reduced by 50% compared with intact Pitx-1. This suggests that about half of the mGnRHR gene promoter transactivation by Pitx-1 is dependent on DNA binding and half is independent of DNA binding activity. Consistent with this result, the ΔC150 protein, which had increased DNA binding activity (Fig. 6C), also had increased transactivation function compared with intact Pitx-1. In contrast to the effects of the point mutation in the Pitx-1 HD, deletion of the entire HD (ΔC89) completely prevented transactivation. The N-terminal (ΔN56) or C-terminal (ΔC150) deletion of Pitx-1 retained transactivation activity. Indeed, the HD alone (ΔN56ΔC150) was sufficient to confer transactivation, although to a reduced extent compared with intact Pitx-1. The effects of the intact and mutant Pitx-1 proteins on the −308/−264-GH50 reporter were similar to those on the homologous promoter (−300/+62), although the magnitude of the Pitx-1 response was greater (7.8- ± 1.0-fold for −308/−264-GH50, versus 2.5- ± 0.2-fold for −300/+62). Again, deletion of the HD (ΔC89) completely eliminated transactivation, whereas a point mutation in the HD (K139R) only partially reduced transactivation of the reporter. Transfection of 10-fold-larger amounts (10 μg/well) of the ΔC89 construct, to overcome the low protein level in the nuclear extracts (Fig. 6B), was not able to rescue the lack of transactivation by this Pitx-1 mutant protein, despite increased levels of protein expression (data not shown). Thus, for both reporters tested, deletion of the HD (ΔC89) completely abolished the Pitx-1 response, whereas C-terminal (ΔC150) or N-terminal (ΔN56) deletions retained it. Transfection analyses using 293T cells produced similar results (data not shown). Overall, these results suggest that Pitx-1 transactivation of the mGnRHR gene promoter occurs in part through direct DNA binding and in part through protein-protein interaction with AP-1. The loss of Pitx-1 interaction with c-Jun with the deletion of the HD (ΔC89) paralleled the complete loss of transactivation of the mGnRHR gene promoter by this mutant Pitx-1 construct; a point mutation in the Pitx-1 HD that eliminated DNA binding but did not prevent interaction with c-Jun only partially reduced transactivation of the mGnRHR gene promoter. Thus, the Pitx-1 HD is necessary for both DNA binding and protein-protein interaction with c-Jun; Pitx-1-induced transactivation of the mGnRHR gene promoter originates from the Pitx-1 HD.
Pitx-1 synergizes with c-Jun and c-Fos in transactivation of the mGnRHR gene promoter.
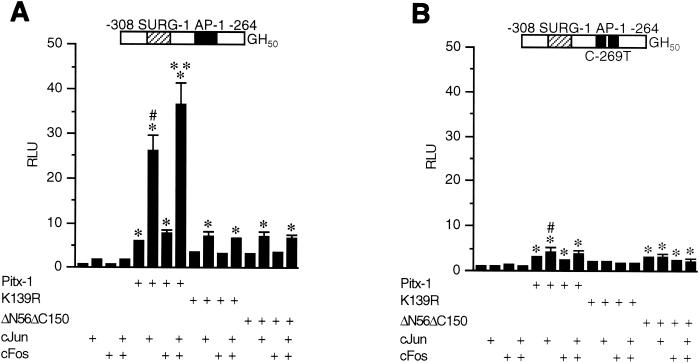
To further characterize the functional consequences of interactions between Pitx-1 and AP-1 in the context of the mGnRHR gene, CV-1 cells were cotransfected with intact or mutant Pitx-1 constructs, c-Jun and/or c-Fos expression vectors, and the −308/−264-GH50 reporter or the corresponding C-269T AP-1 mutant (Fig. 7).
FIG. 7.
Pitx-1 synergizes with c-Jun/c-Fos in transactivation of the mGnRHR gene promoter. A Luc reporter gene fused to −308/−264-GH50 (A) or −308/C-269T/−264-GH50 (B) was cotransfected into CV-1 cells with either intact or mutant Pitx-1 expression vectors, or with empty vector, pcDNA3. c-Jun and/or c-Fos expression vectors or empty vector, pCMV5, were also cotransfected. Data are plotted in comparison with the empty expression vector control for each reporter. Data are presented as the mean ± SEM of three independent experiments. *, P < 0.05 versus the corresponding group in the absence of Pitx-1 cotransfection; #, P < 0.05 versus intact Pitx-1 alone; **, P < 0.05 versus intact Pitx-1/c-Jun.
As noted earlier, intact Pitx-1 alone was able to transactivate −308/−264 (6.1- ± 0.2-fold) (Fig. 7A). c-Jun alone was also able to transactivate −308/−264, although to a lesser extent (1.7- ± 0.1-fold). Synergy was observed in the presence of both Pitx-1 and c-Jun, and further with c-Jun/c-Fos, with a resultant 36.6- ± 4.8-fold increase in Luc activity. As before, a point mutation in the HD of Pitx-1 (K139R) reduced transactivation of −308/−264 by 56% compared to that with intact Pitx-1. This point mutation in the Pitx-1 HD almost entirely eliminated synergism between Pitx-1 and AP-1 and reduced transactivation of −308/−264 in the presence of both factors by 85% compared to that with intact Pitx-1. Despite the reduction in synergism, however, this mutant was still able to further increase the transactivation in the presence of c-Jun, suggesting that Pitx-1 K139R interacted with c-Jun to enhance transactivation. Similar effects were observed with the Pitx-1 HD construct (ΔN56ΔC150). These results indicate that the DNA binding by Pitx-1, although low affinity in nature, is necessary for full synergism between Pitx-1 and AP-1. Furthermore, although the HD alone is sufficient for protein-protein interaction with c-Jun, additional domains of Pitx-1 are important for full synergistic transactivation of the mGnRHR gene promoter by Pitx-1 and AP-1.
When the AP-1 site was mutated (−308/C-269T/−264), transactivation by c-Jun and/or c-Fos was eliminated as expected (Fig. 7B). In addition, transactivation by Pitx-1 was reduced by 63%, and the synergism of intact Pitx-1 with c-Jun/c-Fos was almost entirely eliminated. The K139R mutation of Pitx-1 completely prevented transactivation of −308/C-269T/−264, in both the presence and absence of overexpressed AP-1 proteins. Taken together, these results strongly suggest that DNA binding of both Pitx-1 and c-Jun/c-Fos are necessary for full activity of the mGnRHR gene promoter, and that Pitx-1 is functionally important for AP-1 activity in the regulation of mGnRHR gene expression.
Multiple mutant oligonucleotides of −300 to −259 of the mGnRHR gene promoter reveal several Pitx-1 binding sites.
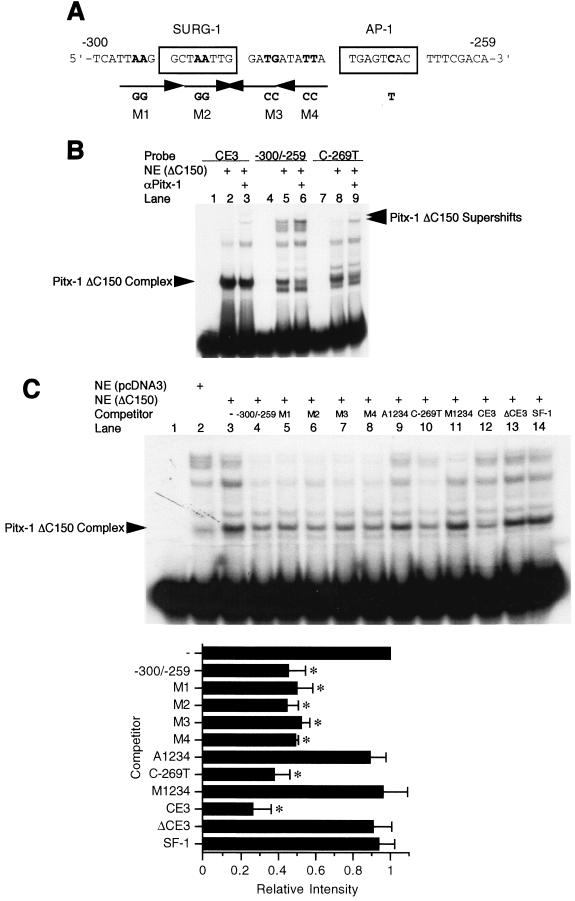
The consensus sequence for the Pitx-1 binding element has been defined as 5′-TAA(T/G)CC-3′ (46). Within our area of interest, −300 to −259 of the mGnRHR gene promoter, there are at least four sequences with partial homology to the Pitx-1 consensus sequence (Fig. 8A). Since the Pitx-1 ΔC150 mutant protein was able to bind to the −300/−259 construct with higher affinity than intact Pitx-1 (Fig. 6C), we sought to take advantage of this construct to localize possible Pitx-1 binding sites. Pitx-1 ΔC150 protein was able to bind to both CE3 (Fig. 8B, lane 2) and −300/−259 (lane 5) when they were used as probes, as well as to C-269T (lane 8), indicating that Pitx-1 binding to DNA does not require an intact AP-1 site. The migration of the complexes was further retarded by α-Pitx-1 antibody (lanes 3, 6, and 9), confirming their identity as Pitx-1-containing complexes. Furthermore, Pitx-1 ΔC150 protein did not bind to oligonucleotides encoding a consensus AP-1 or SF-1 site (data not shown), confirming that the binding to −300/−259 is specific. To determine which of the putative Pitx-1 elements (identified in Fig. 8A) is bound by the Pitx-1 ΔC150 protein, oligonucleotides spanning wild-type −300 to −259 or harboring mutations in each of the four putative Pitx-1 elements, individually (M1-M4) or in combination (M1234), and/or in the AP-1 site (A1234, C-269T) were used as unlabeled competitors in EMSA (Fig. 8C). Oligonucleotides encompassing CE3 (lane 12), ΔCE3 (harboring a mutation in the Pitx-1 element, lane 13), or the SF-1 binding site from the rat LHβ promoter (13) (lane 14) were also used as competitors, as positive and negative controls. A 1,000-fold excess of the wild-type −300/−259 oligonucleotide was able to compete effectively for Pitx-1 binding to the −300/−259 probe (lane 4), as expected. The M1 to M4 oligonucleotides were able to compete to a similar extent as the wild-type oligonucleotide (lanes 5 to 8), indicating that none of the four individual putative Pitx-1 elements was solely responsible for the binding of Pitx-1 to −300/−259. However, when combined, the resultant oligonucleotides were no longer able to effectively compete for Pitx-1 binding to −300/−259, regardless of whether the AP-1 site was intact (M1234, lane 11) or mutated (A1234, lane 9). Combinations of any three mutations out of the four mutation sites failed to prevent competition (data not shown). These results indicate that Pitx-1 binding is not restricted to one specific element in −300/−259, but rather that it may bind to all four of the putative Pitx-1 elements.
FIG. 8.
Multiple mutant oligonucleotides of −300/−259 of the mGnRHR gene promoter reveal several Pitx-1 binding sites. (A) mGnRHR gene promoter sequence between −300 and −259. The SURG-1 and AP-1 elements are boxed, and putative Pitx-1 binding sites (M1 to M4) are indicated with arrows to indicate the direction. Mutated sequences (AA, CA→GG) in each putative Pitx-1 binding site (M1 to M4) are indicated in bold. The location of a point mutation in the AP-1 site (C→T) is also indicated in bold. (B and C) Nuclear extract (10 μg/lane) from 293T cells transfected with Pitx-1 ΔC150 expression vector was incubated with either CE3, −300/−259, or C-269T with or without α-Pitx-1 antibody. The same amount of 293T cell nuclear extract transfected with empty pcDNA3 vector was used as a negative control. NE, nuclear extract. In panel C, excess (1,000-fold) unlabeled oligonucleotides comprising wild-type −300/−259, multiple mutant derivatives thereof (M1-M4, A1234, C-269T, and M1234), CE3, ΔCE3 (a mutant CE3 oligonucleotide in which Pitx-1 consensus sequences were deleted), or SF-1 (SF-1 binding element derived from the rat LHβ gene promoter) were used as competitors. The A1234 mutant oligonucleotide includes all of the M1-M4 mutations and an AP-1 site point mutation (C-269T); the M1234 mutant oligonucleotide includes all of the M1-M4 mutations but has an intact AP-1 site. Quantitation of the signals was performed using a PhosphorImager, and the signals were plotted as relative intensities compared with results with no competition (−). Data are presented as the mean ± SEM of three independent experiments. *, P < 0.05 versus no competition.
DISCUSSION
In this report, we have demonstrated that Pitx-1 transactivates the mGnRHR gene promoter, and we have localized Pitx-1 responsiveness to sequences between −308 and −264 of the promoter. We have further shown that this Pitx-1-induced transactivation derives in part from direct protein-protein interaction between Pitx-1 and c-Jun, mediated by the Pitx-1 HD, and in part from direct interaction between Pitx-1 and DNA sequences immediately upstream of the AP-1 consensus binding site. Moreover, Pitx-1 and AP-1 synergize in the activation of the mGnRHR gene promoter, further supporting the functional importance of these interactions.
More precisely, Pitx-1 responsiveness is localized to sequences between −292 and −269 of the mGnRHR gene promoter, based on functional transfection analyses (Fig. 2B). Both the AP-1 and SURG-1 sites lie within this region and appear to be necessary for full Pitx-1 responsiveness. Although the AP-1 site is necessary for Pitx-1 activity, as evidenced by the reduced transactivation of Δ(−276/−269) and C-269T mutant reporter constructs, it alone is not sufficient for full activity of Pitx-1. In this regard, it is notable that the SURG-1 element (−292/−285) and the intervening sequences between SURG-1 and AP-1 (−284/−277) are also required for maximum Pitx-1-induced transactivation of the mGnRHR gene promoter. Although a complex has been suggested to form on the SURG-1 site (30), the factor(s) that can functionally interact with SURG-1 has yet to be identified.
The relative importance of the DNA sequences immediately upstream of the AP-1 site is likely due to direct DNA binding of Pitx-1 to elements in this area. Several sites with homology to the Pitx-1 consensus element have been identified in this region. Binding affinity of intact Pitx-1 to this region is too low to be detected using nuclear extracts from the Pitx-1-expressing αT3-1 or LβT2 cell lines (Fig. 3C), or from 293T cells transiently transfected with a Pitx-1 expression vector (Fig. 6C), but it can be detected using a GST/Pitx-1 fusion protein (Fig. 3A and B and 5A). However, ChIP studies demonstrate interaction of Pitx-1 with the mGnRHR gene promoter in vivo in LβT2 cells (Fig. 4A). Furthermore, deletion of the C-terminal domain of Pitx-1 (ΔC150) appears to increase DNA binding affinity to both the mGnRHR gene promoter and to CE3 (Fig. 6C). We have taken advantage of this construct to localize Pitx-1 binding sites. Competition EMSA with Pitx-1 ΔC150 protein and oligonucleotides with mutations in the putative Pitx-1 binding sites, individually or in combination, reveals that only mutation of all four putative Pitx-1 binding sites between −300 and −269 effectively reduces competition for Pitx-1 ΔC150 protein binding (Fig. 8C), suggesting that Pitx-1 can bind to multiple sequences in this region. Although we cannot rule out the possibility that the DNA binding specificity of the Pitx-1 ΔC150 protein is distinct from that of intact Pitx-1, our data suggest that direct Pitx-1 binding to DNA sequences between −300 and −269 is necessary for maximal Pitx-1 transactivation of the mGnRHR gene promoter. The presence of multiple Pitx-1 binding sites in this region, coupled with the relatively low affinity of the interaction, likely explains the failure to localize a distinct binding element by uracil and methylation interference analyses (Fig. 4B). Nonetheless, the evidence of low-affinity binding of Pitx-1 to this region (Fig. 3B), coupled with functional transfection studies localizing Pitx-1 responsiveness to this region (Fig. 2B) and the partial loss of Pitx-1 transactivation by the introduction of a point mutation (K139R) into the Pitx-1 HD that eliminates DNA binding activity (Fig. 6C and E), together indicate that Pitx-1 transactivation of the mGnRHR gene promoter occurs in part by direct binding to DNA sequences between −300 and −269. The ChIP analysis also suggests direct DNA binding activity by Pitx-1 in vivo in gonadotrope cells (Fig. 4A). However, we cannot rule out the possibility that this observation may be the result of protein-protein interaction between Pitx-1 and c-Jun, with only c-Jun binding directly to DNA.
The AP-1 site in the mGnRHR gene promoter is necessary for maximal Pitx-1 activity; mutation of the AP-1 site reduces Pitx-1 responsiveness (Fig. 2B). However, Pitx-1 does not bind directly to the AP-1 site (Fig. 8C). Rather, Pitx-1 can interact with the AP-1 complex, in that we have demonstrated direct protein-protein interaction between Pitx-1 and c-Jun (Fig. 5B). The functional importance of this interaction is supported by evidence that it can occur in vivo in a mammalian one-hybrid system as well as in gonadotrope cells (Fig. 5C and D), that the Pitx-1 HD is necessary for this interaction (Fig. 6D), and that deletion of the Pitx-1 HD completely eliminates the ability of Pitx-1 to transactivate the mGnRHR gene promoter, whereas a point mutation in the Pitx-1 HD that eliminates DNA binding but preserves interaction with c-Jun (K139R) retains partial transactivation function (Fig. 6E). Thus, it appears that in addition to binding directly to DNA sequences immediately upstream of the AP-1 site to mediate transactivation, Pitx-1 also mediates its effects by interacting directly with the AP-1 complex, possibly thereby increasing AP-1 activity. Given the fact that the point mutation (K139R) retains the interaction with c-Jun, it is notable that the subdomain(s) within the HD responsible for this protein-protein interaction appears to be different from the subdomain important for DNA binding.
The ability of Pitx-1 to bind to c-Jun is not unprecedented. There have been other reports demonstrating that Pitx-1 can interact directly with several other transcription factors. Pitx-1 has been identified as a Pit-1-associated factor and has been shown to interact with Pit-1 through its C-terminal domain (amino acids 150 to 281) to synergize in the transactivation of the GH gene promoter (43). Similarly, Pitx-1 can interact with CLIM1a (cofactor of LIM homeodomain protein) and PLim through its C-terminal domain (amino acids 150 to 192) to synergistically activate the α-glycoprotein subunit gene promoter (3). The protein-protein interactions of Pitx-1 on these genes appear to require Pitx-1 binding to their promoter elements. Pitx-1 has also been shown to interact with SF-1 and Egr-1 through its C-terminal domain (amino acids 197 to 234) to synergize in the transactivation of the LHβ gene promoter, both by direct DNA binding and by protein-protein interaction with these factors (45, 47). For the Müllerian-inhibiting substance gene, however, Pitx-1 appears to synergize with SF-1 in the transactivation solely by direct protein-protein interaction with SF-1, without binding directly to DNA (47). Furthermore, a recent report has demonstrated that the Pitx-1 HD is sufficient for interaction with Pan1 in the synergistic transactivation of the POMC gene promoter (34). Thus, Pitx-1 is able to functionally interact with other tissue-specific transcription factors, primarily through its C-terminal domain, although also through the HD. In the case of interaction with Pan1 to increase POMC gene expression, the Pitx-1 HD appears to be sufficient for Pan1 interaction and transactivation of the POMC gene promoter, even in the absence of Pitx-1 binding to the gene promoter. In our case, Pitx-1 appears to interact functionally with the mGnRHR gene promoter, in addition to its interaction with c-Jun. The biological significance of these interactions in the regulation of mGnRHR remains to be confirmed in a physiologic model. In this regard, Pitx-1 knock-down experiments or generation of a targeted pituitary gonadotrope Pitx-1 null mouse model will be an important avenue of future investigation.
The ability of intact Pitx-1 to synergize with c-Jun/c-Fos in the transactivation of the mGnRHR gene promoter supports the importance of Pitx-1 for full induction of the mGnRHR gene promoter activity by AP-1. In this report, we have used only c-Jun and c-Fos. It is possible that other members of the Jun/Fos families may be equally involved in synergy with Pitx-1. It has been shown recently that JunD and FosB in particular are able to bind to the AP-1 site in the mGnRHR gene promoter in αT3-1 gonadotrope cells (11). Likewise, we cannot rule out possible interactions of Pitx-1 with other additional transcription factors and/or cofactors to regulate mGnRHR gene promoter transactivation.
Protein-protein interaction in a DNA-independent manner is a common phenomenon among transcription factors and cofactors in the regulation of target gene expression (27, 52), and this interaction is in some cases mediated by the factor's DBD (21, 24). Indeed, the AP-1 complex has been shown to interact with the DBD of several other factors in a DNA-independent manner. Estrogen receptor α or β binds to the AP-1 complex through its DBD as a nonclassical pathway (15, 33); Ets2 binds to the AP-1 complex through its DBD (4); and C/EBP homologous protein 10 (CHOP) binds to the AP-1 complex through its leucine zipper dimerization domain (49).
Recently, Pitx-1 has been shown to form homodimers and to bind to multiple response elements in the Chinook salmon LHβ gene promoter but not to interact with the general coactivators CREB-binding protein and P-300 (25). Similarly, Pitx-2a, another member of the Pitx subfamily and an important factor for pituitary gland development, has also been shown to form homodimers, and a point mutation of its HD (K88E or K88R, the corresponding position in the HD as K139R in our study) has been shown to act as a dominant negative form with increased dimerization activity with intact Pitx-2a (38), leading to a clinical disorder, Axenfeld-Rieger syndrome. Based on our results in this study and the results of others, we speculate that Pitx-1 may transactivate mGnRHR gene expression by forming homodimers that bind simultaneously to the multiple elements between −300 and −269 of the mGnRHR gene promoter and act as a cofactor or coactivator for AP-1 by interacting with c-Jun bound to its cognate site, thereby recruiting the basal transcriptional machinery for the initiation and maintenance of mGnRHR gene transcription.
In summary, in the present study we have elucidated a novel molecular mechanism of Pitx-1 action in the transactivation of the mGnRHR gene promoter, demonstrating the importance of the Pitx-1 HD both in protein-protein interaction with c-Jun and in DNA binding. These results broaden our understanding of the regulation of mGnRHR gene expression and also provide new insights into the broad range of mechanisms of action of homeobox factors.
Acknowledgments
We thank Guemalli R. Cardona, Anthony N. Hollenberg, Paul M. Yen, Akira Takeshita, Lan Ko, Errol R. Norwitz, and Lisa M. Halvorson for helpful discussions.
This work was supported in part by National Institutes of Health grant HD19938 (U.B.K.) and by a Lalor Foundation Research Fellowship Grant (K.-H.J.).
REFERENCES
- 1.Albarracin, C. T., U. B. Kaiser, and W. W. Chin. 1994. Isolation and characterization of the 5′-flanking region of the mouse gonadotropin-releasing hormone receptor gene. Endocrinology 135:2300-2306. [DOI] [PubMed] [Google Scholar]
- 2.Ando, H., C. L. Hew, and A. Urano. 2001. Signal transduction pathways and transcription factors involved in the gonadotropin-releasing hormone-stimulated gonadotropin subunit gene expression. Comp. Biochem. Physiol. B 129:525-532. [DOI] [PubMed] [Google Scholar]
- 3.Bach, I., C. Carrière, H. P. Ostendorff, B. Andersen, and M. G. Rosenfeld. 1997. A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes Dev. 11:1370-1380. [DOI] [PubMed] [Google Scholar]
- 4.Basuyaux, J. P., E. Ferreira, D. Stéhelin, and G. Butticè. 1997. The Ets transcription factors interact with each other and with the c-Fos/c-Jun complex via distinct protein domains in a DNA-dependent and -independent manner. J. Biol. Chem. 272:26188-26195. [DOI] [PubMed] [Google Scholar]
- 5.Boyd, K. E., and P. J. Farnham. 1999. Coexamination of site-specific transcription factor binding and promoter activity in living cells. Mol. Cell. Biol. 19:8393-8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cesnjaj, M., K. J. Catt, and S. S. Stojilkovic. 1994. Coordinate actions of calcium and protein kinase-C in the expression of primary response genes in pituitary gonadotrophs. Endocrinology 135:692-701. [DOI] [PubMed] [Google Scholar]
- 7.Chi, L., W. Zhou, A. Prikhozhan, C. Flanagan, J. S. Davidson, M. Golembo, N. Illing, R. P. Millar, and S. C. Sealfon. 1993. Cloning and characterization of the human GnRH receptor. Mol. Cell. Endocrinol. 91:R1-R6. [DOI] [PubMed] [Google Scholar]
- 8.Duval, D. L., S. E. Nelson, and C. M. Clay. 1997. A binding site for steroidogenic factor-1 is part of a complex enhancer that mediates expression of the murine gonadotropin-releasing hormone receptor gene. Biol. Reprod. 56:160-168. [DOI] [PubMed] [Google Scholar]
- 9.Duval, D. L., S. E. Nelson, and C. M. Clay. 1997. The tripartite basal enhancer of the gonadotropin-releasing hormone (GnRH) receptor gene promoter regulates cell-specific expression through a novel GnRH receptor activating sequence. Mol. Endocrinol. 11:1814-1821. [DOI] [PubMed] [Google Scholar]
- 10.Eidne, K. A., R. E. Sellar, G. Couper, L. Anderson, and P. L. Taylor. 1992. Molecular cloning and characterisation of the rat pituitary gonadotropin-releasing hormone (GnRH) receptor. Mol. Cell. Endocrinol. 90:R5-R9. [DOI] [PubMed] [Google Scholar]
- 11.Ellsworth, B. S., B. R. White, A. T. Burns, B. D. Cherrington, A. M. Otis, and C. M. Clay. 2003. c-Jun N-terminal kinase activation of activator protein-1 underlies homologous regulation of the gonadotropin-releasing hormone receptor gene in αT3-1 cells. Endocrinology 144:839-849. [DOI] [PubMed] [Google Scholar]
- 12.Gharib, S. D., M. E. Wierman, M. A. Shupnik, and W. W. Chin. 1990. Molecular biology of the pituitary gonadotropins. Endocr. Rev. 11:177-199. [DOI] [PubMed] [Google Scholar]
- 13.Halvorson, L. M., U. B. Kaiser, and W. W. Chin. 1996. Stimulation of luteinizing hormone β gene promoter activity by the orphan nuclear receptor, steroidogenic factor-1. J. Biol. Chem. 271:6645-6650. [DOI] [PubMed] [Google Scholar]
- 14.Illing, N., G. F. Jacobs, I. I. Becker, C. A. Flanagan, J. S. Davidson, A. Eales, W. Zhou, S. C. Sealfon, and R. P. Millar. 1993. Comparative sequence analysis and functional characterization of the cloned sheep gonadotropin-releasing hormone receptor reveal differences in primary structure and ligand specificity among mammalian receptors. Biochem. Biophys. Res. Commun. 196:745-751. [DOI] [PubMed] [Google Scholar]
- 15.Jakacka, M., M. Ito, J. Weiss, P.-Y. Chien, B. D. Gehm, and J. L. Jameson. 2001. Estrogen receptor binding to DNA is not required for its activity through the nonclassical AP1 pathway. J. Biol. Chem. 276:13615-13621. [DOI] [PubMed] [Google Scholar]
- 16.Kaiser, U. B., P. M. Conn, and W. W. Chin. 1997. Studies of gonadotropin-releasing hormone (GnRH) action using GnRH receptor-expressing pituitary cell lines. Endocr. Rev. 18:46-70. [DOI] [PubMed] [Google Scholar]
- 17.Kaiser, U. B., E. Sabbagh, R. A. Katzenellenbogen, P. M. Conn, and W. W. Chin. 1995. A mechanism for the differential regulation of gonadotropin subunit gene expression by gonadotropin-releasing hormone. Proc. Natl. Acad. Sci. USA 92:12280-12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaiser, U. B., D. Zhao, G. R. Cardona, and W. W. Chin. 1992. Isolation and characterization of cDNAs encoding the rat pituitary gonadotropin-releasing hormone receptor. Biochem. Biophys. Res. Commun. 189:1645-1652. [DOI] [PubMed] [Google Scholar]
- 19.Kakar, S. S., L. C. Musgrove, D. C. Devor, J. C. Sellers, and J. D. Neill. 1992. Cloning, sequencing, and expression of human gonadotropin-releasing hormone (GnRH) receptor. Biochem. Biophys. Res. Commun. 189:289-295. [DOI] [PubMed] [Google Scholar]
- 20.Kakar, S. S., C. H. Rahe, and J. D. Neill. 1993. Molecular cloning, sequencing, and characterizing the bovine receptor for gonadotropin releasing hormone (GnRH). Domest. Anim. Endocrinol. 10:335-342. [DOI] [PubMed] [Google Scholar]
- 21.Ko, L., G. R. Cardona, A. Henrion-Caude, and W. W. Chin. 2002. Identification and characterization of a tissue-specific coactivator, GT198, that interacts with the DNA-binding domains of nuclear receptors. Mol. Cell. Biol. 22:357-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamonerie, T., J. J. Tremblay, C. Lanctôt, M. Therrien, Y. Gauthier, and J. Drouin. 1996. Ptx1, a bicoid-related homeo box transcription factor involved in transcription of the pro-opiomelanocortin gene. Genes Dev. 10:1284-1295. [DOI] [PubMed] [Google Scholar]
- 23.Lilie, H., E. Schwarz, and R. Rudolph. 1998. Advances in refolding of proteins produced in E. coli. Curr. Opin. Biotechnol. 9:497-501. [DOI] [PubMed] [Google Scholar]
- 24.Mathur, M., P. W. Tucker, and H. H. Samuels. 2001. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol. Cell. Biol. 21:2298-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melamed, P., M. Koh, P. Preklathan, L. Bei, and C. Hew. 2002. Multiple mechanisms for Pitx-1 transactivation of a luteinizing hormone β subunit gene. J. Biol. Chem. 277:26200-26207. [DOI] [PubMed] [Google Scholar]
- 26.Mercer, J. E., and W. W. Chin. 1995. Regulation of pituitary gonadotrophin gene expression. Hum. Reprod. Update 1:363-384. [DOI] [PubMed] [Google Scholar]
- 27.Moilanen, A.-M., H. Poukka, U. Karvonen, M. Häkli, O. A. Jänne, and J. J. Palvimo. 1998. Identification of a novel RING finger protein as a coregulator in steroid receptor-mediated gene transcription. Mol. Cell. Biol. 18:5128-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulvaney, J. M., and M. S. Roberson. 2000. Divergent signaling pathways requiring discrete calcium signals mediate concurrent activation of two mitogen-activated protein kinases by gonadotropin-releasing hormone. J. Biol. Chem. 275:14182-14189. [DOI] [PubMed] [Google Scholar]
- 29.Nordeen, S. K. 1988. Luciferase reporter gene vectors for analysis of promoters and enhancers. BioTechniques 6:454-458. [PubMed] [Google Scholar]
- 30.Norwitz, E. R., G. R. Cardona, K.-H. Jeong, and W. W. Chin. 1999. Identification and characterization of the gonadotropin-releasing hormone response elements in the mouse gonadotropin-releasing hormone receptor gene. J. Biol. Chem. 274:867-880. [DOI] [PubMed] [Google Scholar]
- 31.Norwitz, E. R., K.-H. Jeong, and W. W. Chin. 1999. Molecular mechanisms of gonadotropin-releasing hormone receptor gene regulation. J. Soc. Gynecol. Investig. 6:169-178. [DOI] [PubMed] [Google Scholar]
- 32.Norwitz, E. R., S. Xu, J. Xu, L. B. Spiryda, J. S. Park, K.-H. Jeong, E. A. McGee, and U. B. Kaiser. 2002. Direct binding of AP-1 (Fos/Jun) proteins to a SMAD binding element facilitates both gonadotropin-releasing hormone (GnRH)- and activin-mediated transcriptional activation of the mouse GnRH receptor gene. J. Biol. Chem. 277:37469-37478. [DOI] [PubMed] [Google Scholar]
- 33.Paech, K., P. Webb, G. G. J. M. Kuiper, S. Nilsson, J.-A. Gustafsson, P. J. Kushner, and T. S. Scanlan. 1997. Differential ligand activation of estrogen receptors ERα and ERβ at AP1 sites. Science 277:1508-1510. [DOI] [PubMed] [Google Scholar]
- 34.Poulin, G., M. Lebel, M. Chamberland, F. W. Paradis, and J. Drouin. 2000. Specific protein-protein interaction between basic helix-loop-helix transcription factors and homeoproteins of the Pitx family. Mol. Cell. Biol. 20:4826-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quirk, C. C., K. L. Lozada, R. A. Keri, and J. H. Nilson. 2001. A single Pitx1 binding site is essential for activity of the LHβ promoter in transgenic mice. Mol. Endocrinol. 15:734-746. [DOI] [PubMed] [Google Scholar]
- 36.Reinhart, J., L. M. Mertz, and K. J. Catt. 1992. Molecular cloning and expression of cDNA encoding the murine gonadotropin-releasing hormone receptor. J. Biol. Chem. 267:21281-21284. [PubMed] [Google Scholar]
- 37.Rosenberg, S. B., and P. L. Mellon. 2002. An Otx-related homeodomain protein binds an LHβ promoter element important for activation during gonadotrope maturation. Mol. Endocrinol. 16:1280-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saadi, I., A. Kuburas, J. J. Engle, and A. F. Russo. 2003. Dominant negative dimerization of a mutant homeodomain protein in Axenfeld-Rieger syndrome. Mol. Cell. Biol. 23:1968-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 41.Stanislaus, D., S. Ponder, T. H. Ji, and P. M. Conn. 1998. Gonadotropin-releasing hormone receptor couples to multiple G proteins in rat gonadotrophs and in GGH3 cells: evidence from palmitoylation and overexpression of G proteins. Biol. Reprod. 59:579-586. [DOI] [PubMed] [Google Scholar]
- 42.Szeto, D. P., C. Rodriguez-Esteban, A. K. Ryan, S. M. O'Connell, F. Liu, C. Kioussi, A. S. Gleiberman, J. C. Izpisúa-Belmonte, and M. G. Rosenfeld. 1999. Role of the bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes Dev. 13:484-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szeto, D. P., A. K. Ryan, S. M. O'Connell, and M. G. Rosenfeld. 1996. P-OTX: a PIT-1-interacting homeodomain factor expressed during anterior pituitary gland development. Proc. Natl. Acad. Sci. USA 93:7706-7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Therrien, M., and J. Drouin. 1993. Cell-specific helix-loop-helix factor required for pituitary expression of the pro-opiomelanocortin gene. Mol. Cell. Biol. 13:2342-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tremblay, J. J., and J. Drouin. 1999. Egr-1 is a downstream effector of GnRH and synergizes by direct interaction with Ptx1 and SF-1 to enhance luteinizing hormone β gene transcription. Mol. Cell. Biol. 19:2567-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tremblay, J. J., C. Lanctôt, and J. Drouin. 1998. The pan-pituitary activator of transcription, Ptx1 (pituitary homeobox 1), acts in synergy with SF-1 and Pit1 and is an upstream regulator of the Lim-homeodomain gene Lim3/Lhx3. Mol. Endocrinol. 12:428-441. [DOI] [PubMed] [Google Scholar]
- 47.Tremblay, J. J., A. Marcil, Y. Gauthier, and J. Drouin. 1999. Ptx1 regulates SF-1 activity by an interaction that mimics the role of the ligand-binding domain. EMBO J. 18:3431-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsutsumi, M., W. Zhou, R. P. Millar, P. L. Mellon, J. L. Roberts, C. A. Flanagan, K. Dong, B. Gillo, and S. C. Sealfon. 1992. Cloning and functional expression of a mouse gonadotropin-releasing hormone receptor. Mol. Endocrinol. 6:1163-1169. [DOI] [PubMed] [Google Scholar]
- 49.Ubeda, M., M. Vallejo, and J. F. Habener. 1999. CHOP enhancement of gene transcription by interactions with Jun/Fos AP-1 complex proteins. Mol. Cell. Biol. 19:7589-7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ulloa-Aguirre, A., D. Stanislaus, V. Arora, J. Väänänen, S. Brothers, J. A. Janovick, and P. M. Conn. 1998. The third intracellular loop of the rat gonadotropin-releasing hormone receptor couples the receptor to Gs- and Gq/11-mediated signal transduction pathways: evidence from loop fragment transfection in GGH3 cells. Endocrinology 139:2472-2478. [DOI] [PubMed] [Google Scholar]
- 51.White, B. R., D. L. Duval, J. M. Mulvaney, M. S. Roberson, and C. M. Clay. 1999. Homologous regulation of the gonadotropin-releasing hormone receptor gene is partially mediated by protein kinase C activation of an activator protein-1 element. Mol. Endocrinol. 13:566-577. [DOI] [PubMed] [Google Scholar]
- 52.Yen, P. M., E. C. Wilcox, and W. W. Chin. 1995. Steroid hormone receptors selectively affect transcriptional activation but not basal repression by thyroid hormone receptors. Endocrinology 136:440-445. [DOI] [PubMed] [Google Scholar]
- 53.Zakaria, M. M., K.-H. Jeong, C. Lacza, and U. B. Kaiser. 2002. Pituitary homeobox 1 activates the rat FSHβ (rFSHβ) gene through both direct and indirect interactions with the rFSHβ gene promoter. Mol. Endocrinol. 16:1840-1852. [DOI] [PubMed] [Google Scholar]
- 54.Zhou, X.-F., X.-Q. Shen, and L. Shemshedini. 1999. Ligand-activated retinoic acid receptor inhibits AP-1 transactivation by disrupting c-Jun/c-Fos dimerization. Mol. Endocrinol. 13:276-285. [DOI] [PubMed] [Google Scholar]