Abstract
The mouse gene Zfp36L1 encodes zinc finger protein 36-like 1 (Zfp36L1), a member of the tristetraprolin (TTP) family of tandem CCCH finger proteins. TTP can bind to AU-rich elements within the 3′-untranslated regions of the mRNAs encoding tumor necrosis factor (TNF) and granulocyte-macrophage colony-stimulating factor (GM-CSF), leading to accelerated mRNA degradation. TTP knockout mice exhibit an inflammatory phenotype that is largely due to increased TNF secretion. Zfp36L1 has activities similar to those of TTP in cellular RNA destabilization assays and in cell-free RNA binding and deadenylation assays, suggesting that it may play roles similar to those of TTP in mammalian physiology. To address this question we disrupted Zfp36L1 in mice. All knockout embryos died in utero, most by approximately embryonic day 11 (E11). Failure of chorioallantoic fusion occurred in about two-thirds of cases. Even when fusion occurred, by E10.5 the affected placentas exhibited decreased cell division and relative atrophy of the trophoblast layers. Although knockout embryos exhibited neural tube abnormalities and increased apoptosis within the neural tube and also generalized runting, these and other findings may have been due to deficient placental function. Embryonic expression of Zfp36L1 at E8.0 was greatest in the allantois, consistent with a potential role in chorioallantoic fusion. Fibroblasts derived from knockout embryos had apparently normal levels of fully polyadenylated compared to deadenylated GM-CSF mRNA and normal rates of turnover of this mRNA species, both sensitive markers of TTP deficiency in cells. We postulate that lack of Zfp36L1 expression during mid-gestation results in the abnormal stabilization of one or more mRNAs whose encoded proteins lead directly or indirectly to abnormal placentation and fetal death.
The tristetraprolin (TTP) family of CCCH tandem zinc finger proteins consists of three known members in mammals and a fourth more distantly related protein in frogs and fish (reviewed in reference 2). The best-studied family member is TTP, which is the product of the immediate-early response gene Zfp36 in the mouse (ZFP36 in humans). TTP appears to act in normal physiology to bind to AU-rich elements (ARE) in certain mRNAs and to destabilize those transcripts. Specifically, the mRNAs encoding tumor necrosis factor (TNF) and granulocyte-macrophage colony-stimulating factor (GM-CSF) are stabilized in TTP knockout (KO) mice and in cells derived from them. This results in excessive secretion of these cytokines from the appropriate cell types, resulting in a severe systemic inflammatory phenotype with myeloid hyperplasia in the TTP KO mice (37).
The two other known mammalian TTP family members contain CCCH tandem zinc finger (TZF) domains that are very similar to that of TTP; these TZF domains represent the ARE-binding domains of the proteins. One of these proteins (Zfp36L1, for Zfp36-like 1) was actually the first member of this family in which the TZF domain was recognized (15). Its cDNA was originally cloned as an epidermal growth factor (EGF)-inducible gene in rat intestinal epithelial cells (15), where it was called cMG1; other aliases include TIS11b, ERF1, BRF1, or Berg36 (5, 26, 27, 35). The third family member is Zfp36L2 (also known as TIS11D, ERF2, or BRF2 [26, 28, 39]). The TZF domain of human TTP, including the 6-amino-acid lead-in sequence, is 71% identical to the TZF domain of human ZFP36L1 and 73% identical to that of human ZFP36L2, but outside this region the similarities among the three proteins are more limited. The human ZFP36L1 and ZFP36L2 TZF domains are more closely related to each other than either is to TTP, sharing 93% amino acid identity. All three proteins are evolutionarily conserved, with orthologues having been identified in many mammalian species as well as in Xenopus laevis and Danio rerio (1, 12, 34, 38).
In contrast to that of TTP, little is known about the physiological function of Zfp36L1 protein. Its mRNA can be induced by mitogens and growth factors in appropriate cell types with induction kinetics markedly different from those of TTP (11, 15, 39). Zfp36L1 can act like TTP in cell-free RNA binding and cellular transfection assays (23) as well as in a cell-free deadenylation assay (24). Stoecklin and colleagues have identified defects in Zfp36L1 as the causes of abnormal stabilization of TNF and interleukin-3 mRNAs in cultured cells (35). Taken together, these data suggest that Zfp36L1 could play a role in normal physiology similar to that of TTP, i.e., it may be responsible for the destabilization of one or more ARE-containing mRNAs in normal cells and tissues.
To address this hypothesis, we created mice from two independent clones of embryonic stem (ES) cells in which Zfp36L1 had been disrupted by using homologous recombination. These mice died in utero, generally before approximately embryonic day 11 (E11). Most embryos died because of failure of chorioallantoic fusion; when fusion did occur, by E10.5 there was apparent decreased cellular proliferation and atrophy in the trophoblastic placental layers. Thus, unlike Zfp36, Zfp36L1 in mice is critical for normal feto-placental development and fetal survival. By analogy with TTP, the phenotype suggests that Zfp36L1 may normally destabilize one or more mRNAs whose protein products accumulates abnormally in the KO feto-placental unit in mid-gestation, leading to abnormalities of placentation and the death of the embryo.
MATERIALS AND METHODS
Creation of targeting vector.
A genomic clone for Zfp36L1 was obtained by screening a mouse genomic 129SV phage library (Stratagene) by standard techniques with a random-primed, α-32P-labeled probe consisting of nucleotides 1 to 1190 of the rat cMG1 cDNA (15). Four positive phage clones were identified in this screen, one of which contained both 5′ and 3′ sequences from the Zfp36L1 cDNA as ascertained by Southern blotting using both 5′ and 3′ cDNA probes. Analysis by restriction enzyme mapping and Southern blotting revealed that the genomic clone contained a HindIII fragment of approximately 7.5 kb that included sequences from both the 5′ and 3′ probes described above. This fragment was excised and cloned into the HindIII site of the pBluescript cloning vector (Stratagene, La Jolla, Calif.) to create the plasmid pZfp36L1-7.5H3. Portions of this plasmid were sequenced by dRhodamine Terminator Cycle sequencing (PerkinElmer Life Sciences), and it was found to contain ∼3.2 kb of sequence of the Zfp36L1 promoter, the entire Zfp36L1 protein coding region, an intron of ∼2.2 kb, and 2.1 kb of the 3′-untranslated region.
The plasmid pPGKneobpAlox2PGKDTA (generously provided by Philip Soriano, University of Washington), containing the selection genes NEO and DT, each driven by the PGK promoter, was used as a backbone for the Zfp36L1 targeting vector. To interrupt the Zfp36L1 coding region at the unique Eco47III site in exon 2 (at nucleotides 157 to 163 of the DNA sequence with GenBank accession number M58566), first a ∼5 kb NotI-Eco47III fragment was excised from pZfp36L1-7.5H3, ligated to NotI linker oligos, digested with NotI, and cloned into the NotI site of pPGKneobpAlox2PGKDTA, upstream of and in the same orientation as the PGK-NEO cassette. The resulting plasmid, pGZfp36L1-LA, contained the long arm of Zfp36L1. Next, a 2-kb Eco47III-EcoRV fragment from pZfp36L1-7.5H3 was excised and cloned into the EcoRV site of pGZfp36L1-LA, which is downstream of the PGK-NEO gene and upstream of the PGK-DT gene. The resulting plasmid was designated Zfp36L1-TV and was used as a targeting vector.
A positive control vector for PCR screening for homologous recombination events was constructed by first excising a 449-nucleotide HindIII-EcoRI fragment (nucleotides 2210 to 2659) from the mouse EST clone corresponding to the DNA sequence under GenBank accession number AW912232. The overhangs were filled in with Klenow (Life Sciences, Inc.) according to the manufacturer's instructions, and it was then cloned into Zfp36L1-TV at the unique EcoRV site. Resulting clones were checked for correct orientation of the insert by restriction mapping, and the resulting plasmid was designated Zfp36L1-TVCON.
Screening ES cells.
ES cells were electroporated with the XhoI-linearized Zfp36L1-TV targeting vector by standard methods. To screen for clones in which homologous recombination events had occurred, DNA was isolated from ES cells growing in 96-well plates as described previously (17) and was resuspended in 50 μl of TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA). Five microliters of this mixture was used as a template for PCR. The primers used were 5′-ATACGCTTGATCCGGCTACCT-3′ (corresponding to nucleotides 1675 to 1655 of the neomycin gene in plasmid pPGKneobpAlox2PGKDTA) and 5′-CTATGGATTTGTCATCCCG-3′ (corresponding to nucleotides 2299 to 2281 of the DNA sequence with accession number M58566). For Southern blotting, DNA from positive PCR samples was digested with either HindIII and XbaI or with BamHI alone, separated by electrophoresis on 0.7% agarose gels, transferred to Nytran (Schleicher & Schuell, Keene, N.H.), and hybridized to random-primed, α-32P-labeled probes. The probe used to detect the 5′ region immediately upstream of the targeted sequence was a 300-nucleotide KpnI fragment excised from the pZfp36L1-7.5H3 plasmid, and the probe used to detect the 3′ region immediately downstream of the targeted sequence was a 449-nucleotide HindIII-EcoRI fragment (nucleotides 2210 to 2659) excised from expressed sequence tag clone with GenBank accession number AW912232. Two separate clones of ES cells were selected by antibiotic resistance, and correct targeting was confirmed by PCR and Southern blotting, as described below. Both cell lines were used to generate KO mice according to standard procedures (17).
Mouse genotyping.
Genomic DNA was isolated from mouse tail pieces by incubating them for 4 to 24 h with 0.5 mg of Proteinase K (Boehringer Mannheim GnbH, Mannheim, Germany)/ml in high-salt digestion buffer (100 mM Tris-HCl [pH 8.0], 5 mM EDTA, 200 mM NaCl, 0.2% [wt/vol] sodium dodecyl sulfate). Genomic DNA was precipitated by adding absolute ethanol followed by centrifugation, rinsed with 70% ethanol, dried, and resuspended in TE buffer. Approximately 100 ng of mouse genomic DNA was used as a template for PCR. The primers used were 5′-ATACGCTTGATCCGGCTACCT-3′ (corresponding to nucleotides 1675 to 1655 of the neomycin gene in plasmid pPGKneobpAlox2PGKDTA), 5′-TCCTTTGTGGACACCATGCC-3′ (corresponding to the single Zfp36L1 intron, 57 nucleotides upstream of exon 2 to 37 nucleotides upstream of exon 2), and 5′-CTGAGAAGCTGGTTCTGATGGAAC-3′ (corresponding to nucleotide 273 to 249 of the DNA sequence with accession number M58566). For Southern blotting, DNA was digested with restriction enzymes and were hybridized as described above.
Northern blot analysis.
Total cellular RNA was isolated from primary MEFs, prepared from E12.5 embryos as described previously (10) with the RNEasy system (Qiagen, Valencia, Calif.) according to the manufacturer's instructions. Northern blots were prepared as described elsewhere (22) and were hybridized with random-primed, α-32P-labeled probes. The probe used for detecting Zfp36L1 mRNA consisted of nucleotides 920 to 1320 of the DNA sequence with GenBank accession number X52590. Mouse Zfp36L2 mRNA was detected by using a 0.43-kb PstI fragment of the DNA sequence under GenBank accession number AA021952 (nucleotides 72 to 502). The probe for detecting mouse TTP mRNA consisted of a 2.4-kb EcoRI-SstI fragment from the mouse TTP cDNA (25). Blots were also hybridized to a random-primed, α-32P-labeled glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA probe (8) or a cyclophilin cDNA probe (7) to monitor gel loading.
Whole-mount in situ hybridization histochemistry.
To determine Zfp36L1 mRNA expression patterns in early mouse embryos, embryos were fixed in 4% paraformaldehyde overnight at 4°C and then were placed into diethyl pyrocarbonate-treated phosphate-buffered saline (PBS) until they could be dehydrated through a methanol gradient into 100% methanol. They were then treated with Proteinase K (10 μg/ml) for 10 to 12 min. Embryos were postfixed in 4% paraformaldehyde and 0.2% glutaraldehyde for 15 min at room temperature and were rinsed for 5 min in diethyl pyrocarbonate-treated PBT (PBS plus 0.1% Tween 20) at room temperature, and then they were preblocked in a solution of 1× KTBT (50 mM Tris HCl [pH 7.5], 150 mM NaCl, 10 mM KCl, 0.5% Triton X-100), 20% heat-inactivated sheep serum, and 2% blocking reagent (Roche) for at least 2 h at room temperature. An anti-digoxigenin antibody (Roche) was added to the blocking solution at a 1:2,000 dilution and was incubated overnight at 4°C. Embryos were then washed five times in 1× KTBT for 1 h each at room temperature. Color was then developed by using the BM purple alkaline phosphatase substrate (Roche). Embryos were then washed in a 1:1 solution of PBT:hybridization buffer (hybridization buffer contained 50% formamide, 1.3× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 5 mM EDTA, 50 μg of yeast RNA/ml, 0.2% Tween 20, 0.5% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], and 100 μg of heparin/ml) for 5 min at room temperature and then were washed in 100% hybridization buffer for 5 min at room temperature followed by incubation in fresh hybridization buffer in a 70°C heat block for at least 1 h. Following preincubation in hybridization buffer, embryos were incubated with digoxigenin-labeled probes (0.1 μg/ml) overnight at 70°C in hybridization buffer. Embryos were then washed with 2× SSC-0.1% CHAPS and 0.2× SSC-0.1% CHAPS (three times each for 20 min at 70°C) and finally were washed with 1× KTBT (200 mM Tris [pH 7.5], 1 M NaCl, 10 mM KCl, 1% Triton X-100).
Several probes were tested, but the probe that consistently produced the best signal with the least background was 538 bp in length. The cDNA insert for the probe was made by PCR using the Zfp36L1 cDNA as a template; the forward primer contained a BamHI restriction enzyme site and aligned with bp 1155 to 1177 of the DNA sequence under GenBank accession number M58566 (5′ cgcaGGATCCcaccccctaactttccctcaaac 3′, in which the uppercase letters represent the BamHI restriction site), and the reverse primer contained a HindIII restriction site and aligned with bp 1692 to 1673 of the sequence under accession number M58566 (5′ gatgAAGCTTacccctccccccaaaaaaac 3′, in which the uppercase letters represent the HindIII site). The final probe spanned bp 1155 to 1692 of the DNA sequence under accession number M58566. PCR was conducted with Platinum Pfx DNA polymerase (Invitrogen Life Technologies) with 10 ng of plasmid, following the manufacturer's instructions. The PCR product was cut with BamHI and HindIII, gel purified, and then ligated with BamHI- and HindIII-cut Bluescript KS+ (Stratagene).
Digoxigenin-labeled antisense probes for in situ hybridization were generated by linearizing 10 μg of plasmid with BamHI and transcribing it with T7 RNA polymerase (Roche) for 2 h at 37°C. Transcription reactions were performed with the following in combination with DIG RNA labeling mix (Roche): 2 μl of 10× transcription buffer, 2 μl of digoxigenin RNA label, 1 μl of Protector RNase inhibitor (Roche), 2 μl of RNA polymerase, 1.5 μl of template DNA, 11.5 μl of H20. Transcription reactions were stopped by adding 5 μl of H20. Three subsequent LiCl-ethanol extractions were performed to clean the probe prior to use. Control sense probes were generated by HindIII-mediated linearization and were transcribed using T3 RNA polymerase (Roche) as described above. Antisense and sense in situ hybridizations were performed in tandem to ensure validity of expression patterns.
Fibroblast growth assays.
To determine proliferation rates, 105 cells were plated into each well of 6-well plates in Dulbecco's modified Eagle's medium supplemented with a solution of 15% fetal calf serum, 2 mM glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. On days 1, 3, 5, 7, 9, and 11 the cells were trypsinized and counted with a hemocytometer. The growth medium was replaced with fresh medium on the same days. The cells were used at passage three or four; three dishes per point were used, and two cell lines per genotype were analyzed.
Fibroblast mRNA stability assays.
Fibroblast mRNA stability assays were conducted in mouse embryonic fibroblasts (MEFs) as described previously for bone marrow-derived stromal cells (9), except that the stimuli for GM-CSF induction were EGF (20 ng/ml for 30 min) followed by treatment with actinomycin D (10 g/ml) or with mouse TNF (10 ng/ml for 60 min) followed by actinomycin D (5 μg/ml). GM-CSF mRNA stability was assessed by Northern blotting as described previously (9).
Fibroblast apoptosis assays. (i) Measurement of cell membrane changes.
Changes in phosphatidylserine symmetry were determined by using Annexin conjugated to fluorescein isothiocyanate (FITC) (Trevigen, Gaithersburg, Md.) and flow cytometry as described previously (33). Briefly, 5 × 105 control or serum-starved MEFs were washed with PBS and then incubated with 3 μl of Annexin-FITC and propidium iodide (PI) for 15 min at room temperature according to the manufacturer's instructions. Annexin-FITC-PI-labeled cells were diluted in 400 μl of 1× Annexin binding buffer and were examined immediately using a Becton Dickinson (BD) FACSort flow cytometer with CellQuest software. Ten thousand cells were excited at 488 nm and were examined at 530 and 585 nm for Annexin-FITC and PI fluorescence, respectively.
(ii) Caspase activity
Caspase activity was determined by using the CaspaTag activity kits (Intergen, Purchase, N.Y.) for the fluorescent detection of caspase-3, -8, and -9 according to the manufacturer's instructions. Briefly, 5 × 105 control or serum-starved MEFs were added to the various caspase substrates and incubated for 30 min at 37°C in 7% CO2. PI was added to each sample immediately before flow analysis at a final concentration of 10 μg/ml. Ten thousand cells were analyzed with a BD FACSort flow cytometer with an excitation of 488 nm and with emission at 530 and 585 nm for caspase activity and PI, respectively.
(iii) DNA analysis
The DNA content for each sample was determined by flow cytometry as described previously (3). Briefly, 5 × 105 control or serum-starved MEFs were fixed by the slow addition of cold 70% ethanol to a volume of approximately 1.5 ml with gentle agitation. The volume was adjusted to 5 ml with cold 70% ethanol and was stored overnight at 4°C. For flow analysis, the fixed cells were pelleted and washed once in PBS and stained in 1 ml of a solution containing 20 μg of PI/ml and 1 mg of RNase/ml in 1× PBS for 20 min. Cells (7,500) were analyzed by flow cytometry using a BD FACSort by gating on a PI area-versus-width dot plot to exclude cell debris and cell aggregates. The percentage of degraded DNA was determined by the number of cells with subdiploid DNA divided by the total number of cells examined under each experimental condition.
Histology.
Embryos were fixed in Bouin's solution for various times at room temperature, washed extensively in 70% (vol/vol) ethanol, and then processed for paraffin embedding and staining with hematoxylin and eosin by standard techniques. Apoptotic cells were identified in hematoxylin- and eosin-stained slides by the presence of nuclear condensation and fragmentation accompanied by cell shrinkage, increased cytoplasmic eosinophilia, and a clear halo around apoptotic cells; apoptosis was confirmed in some cases by terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining. Paraffin sections of embryos or placentas were stained for apoptotic bodies by the TUNEL technique and for the proliferation marker Ki67 as described previously (4, 30).
Whole-mount histochemistry.
The vital lysosomal dye LysoTracker Red (Molecular Probes, Eugene, Oreg.) was used to assess apoptosis in whole mounts of embryos as described previously (13, 44); it was also used as a generalized fluorescent label in fixed embryos for anatomical assessment, in both cases using confocal laser-scanning microscopy.
RESULTS
Generation of Zfp36L1-deficient mice.
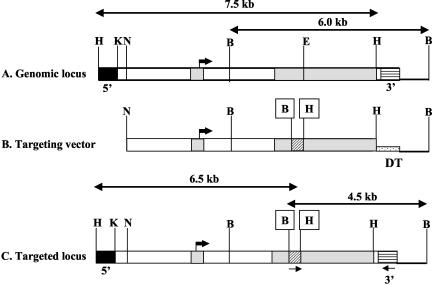
The targeting vector was designed to interrupt the coding region of exon 2 in Zfp36L1 with a PGKNEO cassette. The vector was designed to introduce HindIII and BamHI restriction sites flanking PGKNEO, as depicted schematically in Fig. 1, for screening purposes. The intron-exon structure of the Zfp36L1 genomic locus, the untargeted regions used as probes in Southern blotting, relevant restriction sites, and PCR primers are indicated in Fig. 1.
FIG. 1.
Gene-targeting strategy for Zfp36L1 inactivation. (A) Schematic representation of the normal mouse genomic locus for Zfp36L1. The two exons are represented by light gray shading, and the translational start site in exon 1 is indicated by the large black arrow. Genomic 5′ (black boxes) and 3′ (horizontal hatching) probes outside the targeted region are shown. (B) The targeting vector contained an interruption of the 5′ portion of exon 2 with a neomycin marker cassette (diagonal cross hatch), with the creation of new restriction sites for HindIII (box labeled H) and BamHI (box labeled B). A diptheria toxin resistance element (speckled hatching labeled DT) was also inserted into the vector. (C) The final targeted locus. The locations of PCR primers used to detect homologous recombination are indicated by the small arrows underneath the targeted locus. Cleavage sites for the following restriction enzymes are indicated: B, BamHI; E, Eco47III; H, HindIII; K, KpnI; N, NotI.
Of 600 G418-resistant ES cell clones screened, homologous recombination of the Zfp36L1 targeting vector was detected in 7. A Southern blot of DNA digested with XbaI and HindIII from wild-type (WT) (Fig. 2A, lane 1) and targeted (lane 2) ES cells was hybridized to the 5′ probe indicated in Fig. 1. In DNA from WT cells the probe recognized a band of 7.5 kb, whereas in DNA from targeted cells it recognized a smaller band of 6.5 kb, as predicted. A Southern blot of DNA digested with BamHI from WT (Fig. 2B, lane 3) and targeted (Fig. 2B, lane 4) cells showed hybridization of the probe to the 3′ band of 6 kb, whereas in DNA from targeted cells it recognized a smaller band of 4.5 kb, as predicted.
FIG. 2.
Southern and Northern analysis confirming Zfp36L1 targeting in ES cells. (A and B) Southern blots of mouse tail DNA from WT and Het. mice. (A) HindIII/XbaI cleavage of DNA and hybridization with the Zfp36L1 5′-flanking probe generated a diagnostic 7.5-kb fragment from WT (+/+) DNA (lane 1) and a 6.5-kb targeted Het. (+/−) DNA fragment (lane 2) (arrows). (B) BamHI cleavage of DNA and hybridization with the Zfp36L1 3′-flanking probe generated a diagnostic 6.0-kb WT DNA fragment (lane 3) and a 4.5-kb targeted (Het.) DNA fragment (lane 4) (arrows). Northern blots of total cellular RNA isolated from cultured MEFs are shown in panels C to F; each lane contained 15 μg of total cellular RNA from cells that were WT, Het., or KO for the targeted Zfp36L1 allele. The blots were probed with the indicated 32P-labeled cDNA probes for Zfp36L1 (C), GAPDH (D), TTP and cyclophilin (Cyclo) (E), and Zfp36L2 (F). Note that the apparently increased TTP and ZFP36L2 mRNA expression seen in the RNA from the KO cells in panels E and F is largely due to gel overloading in those lanes, as evidenced by the larger amounts of GAPDH and cyclophilin mRNA present in the same lanes in panels D and E.
Targeted ES cells were injected into blastocysts for germ line transmission according to standard procedures (17). Litters from matings of founder mice were genotyped by Southern blotting and PCR. Matings between heterozygote animals failed to generate the expected Mendelian ratios of homozygous KO mice. From the first five litters, 9 out of 19 pups were +/+ (or WT) and 10 out of 19 were +/− (or heterozygous [Het.]) for the targeted allele, with no living −/− (or KO) pups detected. Subsequent timed pregnancy experiments revealed that most of the Zfp36L1 KO embryos died in utero between days 8 and 13 of gestation. Of 423 pups from 84 litters allowed to go to term from mice derived from both ES cell lines, 149 (35%) were WT and 274 (65%) were Het., with no living KO mice being born.
Zfp36L1 mRNA expression in fibroblasts from control and KO mice.
To confirm the absence of Zfp36L1 mRNA expression, MEFs were derived from individual WT, Het., and KO embryos at E12.5. Total cellular RNA from the MEFs was subjected to Northern blotting. Zfp36L1 mRNA was present in cells derived from WT and Het. mice (Fig. 2C), with the expression in the Het. cells being approximately half that of the WT cells; the mRNA was not detectable in RNA from the KO cells (Fig. 2C). These data confirmed that the KO mice did not express Zfp36L1 mRNA. The same blot probed with the control GAPDH probe showed that the gels contained larger amounts of KO RNA than the other two genotypes (Fig. 2D).
To determine whether there was compensatory increased or decreased expression of the mRNAs for the other two known members of this protein family, we probed the same blot with cDNA probes for TTP and Zfp36L2 mRNA. TTP expression was barely detectable in the RNA from these unstimulated cells from the WT and Het. mice (Fig. 2E). Its expression was apparently increased in the KO sample; however, this was probably due to relative overloading of this lane of the blot (see cyclophilin [Fig. 2E] and GAPDH [Fig. 2D] mRNA expression in the same blot). PhosphorImager analysis of these data revealed that TTP expression was the same in the WT, Het., and KO lanes after correction for the amount of either GAPDH or cyclophilin mRNA in the same blot. Zfp36L2 expression was readily detectable in these mRNA samples (Fig. 2F); however, there was no apparent increase in its expression in the KO samples compared to expression of the others after normalization for the expression of either control probe.
Zfp36L1 mRNA expression in mouse embryos and tissues.
To compare expression of Zfp36L1 mRNA in normal mouse tissues to that of TTP (25), RNA from a panel of mouse tissues (Ambion, Inc., Austin, Tex.) was subjected to Northern blotting and was hybridized to probes for Zfp36L1 and TTP mRNAs (Fig. 3A). Neither TTP nor Zfp36L1 was highly expressed in brain (lane 3), liver (lane 6), or testis (lane 10). Both TTP and Zfp36L1 mRNA were readily detectable in heart (lane 4), kidney (lane 5), lung (lane 6), spleen (lane 9), and thymus (lane 11). Expression of Zfp36L1 mRNA, but not TTP mRNA, was readily detectable in ES cells (lane 1), E12.5 embryos (lane 2), and adult ovary (lane 8).
FIG. 3.
Tissue distribution and developmental expression of Zfp36L1 mRNA. (A) Total cellular RNA from adult mouse tissues as well as from whole embryos at E12.5 and from ES cells were subjected to electrophoresis and Northern blotting using probes for Zfp36L1 and TTP mRNA, as indicated by the arrows. Each lane was loaded with 10 μg of total RNA. (B) A different Northern blot that used total RNA isolated from whole embryos at the embryonic day indicated or from the specified adult mouse tissues (B, brain; L, liver; T, testis). The blot was probed with either Zfp36L1 or cyclophilin (cyclo) cDNA probes, as indicated. Each lane contained 15 μg of total cellular RNA. (C) Northern blot using total cellular RNA from embryo head and placenta at E14.5. The same blot was probed for both TTP and GAPDH mRNAs, as indicated by the arrows.
Further analysis of embryonic expression is shown in Fig. 3B. In the Northern blots shown in Fig. 3B, equal amounts of RNA were loaded in each gel lane; the same blots were also probed with a cyclophilin probe as a loading control. At E7.5, Zfp36L1 mRNA was barely detectable. However, by E9.5 there was readily detectable mRNA expression, which was at a similar level at E13.5. There was an apparent decrease in overall expression at later stages of gestation (Fig. 3B). These expression levels should be compared with those seen in several organs from adult mice probed in the same blot (Fig. 3B). These data indicate that barely detectable expression of Zfp36L1 mRNA at E7.5 turns into relatively high-level expression by E9.5. In whole mouse placenta at E14.5, Zfp36L1 mRNA was expressed at about half the level of whole embryo head from the same developmental stage (Fig. 3C).
Because of the embryonic lethality of the phenotype in mid-gestation, we also analyzed the expression of ZFP36L1 mRNA in intact embryos at E8.0 to E8.5 by in situ hybridization using a 538-bp antisense probe labeled with digoxigenin. As shown in Fig. 4, some Zfp36L1 mRNA expression at approximately E8.0 occurred diffusely throughout the embryo; this is particularly obvious in Fig. 4C, comparing a WT to a KO control embryo in the same experiment. Within the WT embryos at E8.0, higher-level expression was seen in the allantois (Fig. 4A to D) and throughout the neurectoderm, with apparent increased expression in the neural folds, particularly in the anterior neural ridge and presumptive midbrain (Fig. 4A to C). The embryos were also more strongly labeled in the paraxial mesoderm and along the anterior and posterior margins of the somites. Expression stopped at the lateral extent of the embryo where it joined the amnion. That the hybridization was specific with this probe was demonstrated by using Zfp36L1 KO embryos from the same litter as a negative control (Fig. 4C); the corresponding sense probe also produced no signal (data not shown).
FIG. 4.
Whole-mount in situ hybridization histochemistry of Zfp36L1 mRNA expression at approximately E8.0. WT embryos were processed for in situ hybridization histochemistry as described in Materials and Methods. Specific probe hybridization is indicated by the purple color. Note the higher-level expression in the neural folds, neurectoderm, and anterior neural ridge (ANR) as well as the patchy but readily detectable expression in the allantois (A to C). The specificity of staining with the Zfp36L1 probe was demonstrated by comparing staining of a WT embryo with that of its KO littermate embryo processed at the same time; no staining was seen (C). A control using a sense version of the probe was also negative (not shown). (D) Color development in the whole conceptus was stopped at an earlier time, demonstrating high-level expression in the allantois.
We also performed whole-mount in situ hybridization histochemistry on the entire feto-placental unit at E8.0 (Fig. 4D); in this experiment, staining was stopped earlier than in experiments depicted in Fig. 4A to C. By far the highest level of expression was seen in the allantois, which should fuse with the chorion by E8.5. There also appeared to be a ring of expression where the yolk sac joined the placenta, with minimal expression in the rest of the chorionic plate and the yolk sac under these conditions.
Phenotype of the Zfp36L1 KO embryos.
Most aspects of the phenotype of the KO embryos appeared to be due to failure of chorioallantoic fusion, which resulted in embryo death. In 14 out of 64 (21.9%) KO embryos examined at E9.5 there was persistent attachment of an allantoic remnant (Fig. 5A and B); this was associated with severe embryo runting and death. The presence of an allantoic remnant was not a good indicator of the frequency of failed chorioallantoic fusion, because these remnants frequently became detached from the embryos during processing. There was clear failure of chorioallantoic fusion in 8 out of 12 placentas that could be evaluated at E9.5 and E10.5.
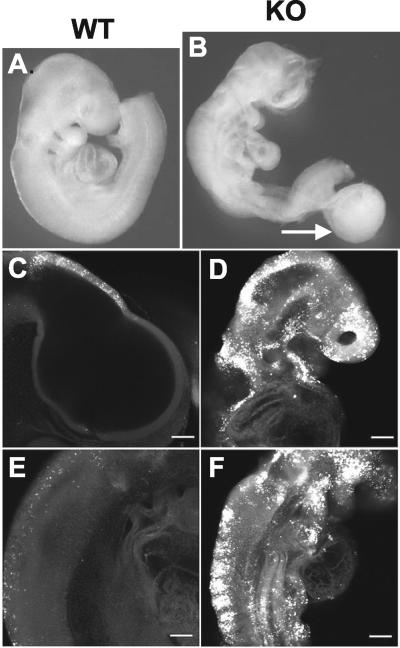
FIG. 5.
Phenotype of Zfp36L1-deficient embryos at E9.5. Right lateral views of an E9.5 WT embryo (A) and a KO littermate (B). Both embryos were photographed in the unfixed state. Note the abnormalities of the cranial neural tube in the KO embryo; the general underdevelopment of the abdominal organs and uncoiling of the heart tube; and the generally decreased size, abnormal body axis, and the presence of a large allantoic remnant or cyst (arrow).The head region (C and D) and thoracic region (E and F) of lateral confocal images of Lysotracker staining, indicating apoptosis. Note the widespread and striking staining in the neural tube of the KO embryo (D and F) compared to that of the WT embryo (C and E).
In embryos in which chorioallantoic fusion had occurred, there was often generalized runting and persistent neural tube opening in the forebrain region as well as severe diffuse apoptosis throughout the neural tube, as evidenced by staining with the TUNEL and hematoxylin and eosin techniques (data not shown) and with Lysotracker (Fig. 5C to F). The single embryo that survived until E13.5 exhibited a dramatic form of exencephaly, while other facial structures were largely intact (Fig. 6). Other common abnormalities were consistently shorter cardiac outflow tracts and reduced or absent epicardial layers. Whether these embryo defects represent primary defects or are secondary to placental failure (see below) is not known.
FIG. 6.
Phenotype of a Zfp36L1-deficient embryo at E13.5. (A and B) External appearance of the heads from WT (+/+) and KO (−/−) littermate mice at E13.5. The KO embryo was the longest surviving embryo in this series. Note the relatively normal facial structures in the KO mouse but the extreme abnormalities at the surface of the head. (C and D) Hematoxylin- and eosin-stained coronal sections through the heads of the same embryos at approximately the same level; one eye is present in each section (arrows). The extreme disorganization of the KO brain (D) should be contrasted with the brain from the WT littermate (C). Magnification, ×1.
Placental defects.
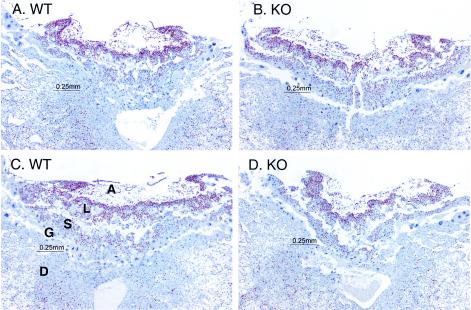
Because of the high frequency of apparently failed chorioallantoic fusion and the general failure-to-thrive phenotype of many of the KO embryos, we examined placentas in which choriallantoic fusion had occurred from the KO and WT embryos at E9.5. Figure 7 shows low-power images of two WT and two KO placentas in which fusion had occurred. The red-brown nuclear staining is from the Ki67 antigen, an indicator of cell proliferation (4). At E9.5, the KO placentas (Fig. 7B and D) closely resembled their WT counterparts (Fig. 7A and C). All of the major placental layers were present, as labeled in Fig. 7C. Higher-power views of the same and adjacent sections showed that all major cell types were present in the KO placentas. In addition, there were similar numbers and depths of the early invaginations of the allantois into the chorion and similar extents of cell proliferation in the trophoblast layers between the WT and KO placentas.
FIG. 7.
Placental histology and cell proliferation at E9.5. Shown are sections through placentas from two WT and two KO embryos at E9.5 in which chorioallantoic fusion had occurred. The layers labeled include the maternal decidua (D), giant cell layer (G), spongiotrophoblast layer (S), labryrinthine layer (L), and allantois (A). The red-brown nuclear staining is for the Ki67 cell proliferation antigen; the counterstain was hematoxylin.
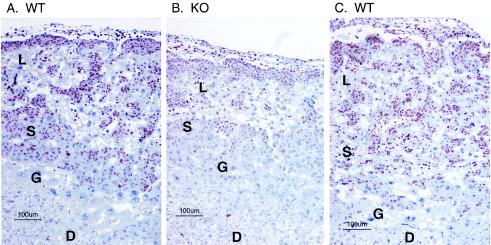
However, the appearance of the placenta was quite different by E10.5 (Fig. 8), which shows placentas from three littermates, two WT and one KO, in all of which chorioallantoic fusion had occurred. These sections were again stained with the Ki67 proliferation marker. In the KO placenta there was marked generalized atrophy of both the spongiotrophoblast and labyrinthine layers, and there was also a marked decrease in the number and intensity of cells stained by the Ki67 proliferation marker (Fig. 8B). Similar findings were noted in other KO placentas in which fusion had occurred. There was no apparent corresponding increase in apoptosis in these layers in the KO placenta, as assessed by hematoxylin and eosin staining (data not shown).
FIG. 8.
Placental histology and cell proliferation at E10.5. Shown are sections through three littermate placentas at E10.5, two WT (A and C) and one KO (B). The KO placenta was one in which chorioallantoic fusion had occurred. The sections were stained with an antibody to the cell proliferation antigen Ki67 (red-brown color) and counterstained with hematoxylin. The abbreviations are the same as those described in the legend for Fig. 7. Note the near absence of Ki67 staining in the decidual layer from all placentas; the relative atrophy of the embryonic layers in the KO placenta (B); and the markedly diminished Ki67 staining in the embryonic layers from the KO placenta (B).
These data suggest that, in the minority of placentas in which chorioallantoic fusion had occurred, there was secondary placental failure due to atrophy of the trophoblast layers, apparently secondary to decreased cell proliferation. This placental failure presumably resulted in the deaths of the embryos that had successfully undergone chorioallantoic fusion.
Growth rates, apoptosis, and mRNA stability in MEFs from WT and KO embryos.
To determine whether cells derived from the Zfp36L1 KO embryos had an intrinsic defect in either proliferative ability or susceptibility to apoptotic stimuli, growth rates of MEFs in serum-containing medium were evaluated. A total of five separate growth experiments were conducted with passage-three and -four MEFs derived from two embryos of each genotype. There were no significant differences in growth rates between the WT and KO cells at any of six time points (1, 3, 5, 7, 9, and 11 days) after plating. These data suggest that, at least for fibroblasts prepared in this way, there was no major intrinsic difference in the growth response to serum of the KO cells.
Because of the apparent increase in neural tube apoptosis in the KO embryos compared to that of the controls, we also examined the MEFs for apoptosis in response to various stimuli. In none of the cases tested was there an apparent difference in susceptibility to apoptotic stimuli among the WT, Het., and KO genotypes.
Finally, we evaluated the stability in these cells of one of the transcripts that was previously noted to be stabilized in bone marrow-derived stromal cells from TTP KO mice versus WT mice, that encoding GM-CSF (9). This transcript represents a sensitive marker of TTP expression and action in the cells for two reasons: (i) in the TTP KO stromal cells there was virtually complete stabilization of the mRNA compared to that of cells from the WT mice in which the mRNA turned over with a half-life of about 100 min, and (ii) the transcript in the KO cells was almost entirely in the larger, fully polyadenylated form, whereas that seen in the WT cells was composed of approximately equal amounts of fully polyadenylated and deadenylated transcripts, differing in apparent size by approximately 200 bp (9). Thus, the effect of TTP on this mRNA is to promote its deadenylation and ultimately its more rapid turnover in these stromal cells.
For these reasons, we analyzed the turnover rates and polyadenylation status of the GM-CSF mRNA in MEFs derived from Zfp36L1 WT and KO fibroblasts. In two pairs of matched littermate MEF cells derived from the Zfp36L1-deficient embryos, GM-CSF transcript accumulation was stimulated for 30 min with EGF, followed by treatment with actinomycin D; total cellular RNA was extracted from the cells and was used for Northern blots at intervals thereafter (Fig. 9A to D). In both experiments the rate of degradation of the GM-CSF transcripts was similar between RNA from the cells derived from the WT and that from KO mice (Fig. 9C and D). Of equal importance was the fact that there were no apparent differences in the relative levels of the fully polyadenylated and deadenylated transcripts in the WT and KO cells (Fig. 9A and B). By using a different stimulus, a similar pair of MEF cells was treated with mouse TNF for 60 min; actinomycin D was then added, and the cells were processed for RNA purification and Northern blotting at intervals thereafter. Again, there was no apparent difference between the cells of the two genotypes in terms of transcript accumulation after stimulation, transcript decay after actinomycin D treatment, or the relative proportions of the fully polyadenylated and deadenylated transcripts (Fig. 9E and F). Therefore, at least using this well-defined TTP-sensitive, ARE-containing transcript in this cell type, we were unable to document a similar effect of Zfp36-L1 deficiency to either stabilize the GM-CSF transcript or promote the accumulation of its fully polyadenylated form.
FIG. 9.
GM-CSF mRNA stability in MEFs derived from Zfp36L1 WT and KO embryos. MEFs of the indicated genotype were stimulated with EGF (20 ng/ml for 30 min) (A to D) or mouse TNF (10 ng/ml for 60 min) (E and F) followed by actinomycin D (Act. D) (10 μg/ml in A to D, 5 μg/ml in E and F). The cells were then processed at different times for Northern blotting using a GM-CSF-specific probe. The indicated times represent the total elapsed time (in minutes) after the initial stimulation. (A and B) Comparison of results from WT and KO cells after EGF stimulation in one pair of MEFs derived from littermates of the WT and KO genotypes; after PhosphorImager normalization for the levels of GAPDH mRNA, the decay curves for the GM-CSF mRNAs after addition of actinomycin D (arrows) from this and a second identical experiment are shown in panels C and D. The solid lines are from the WT cells, and the dashed lines are from the KO cells. (E and F) Northern blots from WT and KO fibroblasts after stimulation with mouse TNF for 60 min followed by actinomycin D treatment. Note the similar relative expression levels of the two bands of the GM-CSF transcript in the two genotypes as well as the similar rapid decay rates of the mRNA in both genotypes. GAPDH mRNA levels are also shown in the same blots as loading controls.
DISCUSSION
Our data from two independent lines of Zfp36L1-deficient mice on a mixed C57Bl6 and 129 background indicate that this protein plays an essential role in mouse development. Failure of expression of Zfp36L1 mRNA led to universal intrauterine death, generally between approximately E8 and E12, a time when Zfp36L1 mRNA was normally expressed in the developing mouse embryo. Failure of chorioallantoic fusion was frequent and was presumably the cause of death in approximately two-thirds of the embryos. However, even the KO embryos that had undergone chorioallantoic fusion exhibited a general failure-to-thrive phenotype, with runting, frequent neural tube defects, increased neural tube apoptosis, and abnormalities of the cardiac outflow tract and epicardium. Mutant placentas were also abnormal by E10.5, with thinning of the spongiotrophoblast and labyrinthine layers and decreased rates of cell proliferation in these layers. Although it is possible that some of the embryo defects were due to the lack of embryonic expression of Zfp36L1, it seems likely that most if not all aspects of the embryonic phenotype were due to placental insufficiency, even in those cases in which chorioallantoic fusion had occurred.
This suggestion of a primary placental phenotype is supported by preliminary microarray analyses of WT and KO embryos at E10.5 (D. J. Stumpo and P. J. Blackshear, unpublished data). Many of the transcripts most highly overexpressed in the KO embryos were stress response genes, whereas many of the genes relatively underexpressed in the KO embryos were related to hemoglobin synthesis or erythroid hematopoiesis. These findings suggest that the KO embryos were under significant stress and that their perfusion from the placenta was subnormal. Because the allantois appeared to express the highest levels of Zfp36L1 in the embryo immediately before chorioallantoic fusion, future microarray studies will focus on this structure at this point in development.
To date, the Zfp36L1 protein has mimicked the biochemical effects of TTP in all of our experimental systems, including (i) binding to ARE-containing RNA probes, as assayed by gel shift analysis (23); (ii) UV light-induced cross-linking to ARE-containing RNA probes (23); (iii) zinc finger-dependent Zfp36L1-induced decay of ARE-containing target RNAs in intact cell cotransfection experiments (23); and (iv) ARE-dependent, TTP-like stimulation of deadenylation in cell-free assays (24). Given that TTP's role in normal physiology seems to be to promote the deadenylation and turnover of the ARE-containing mRNAs coding for TNF and GM-CSF (2), it seems likely that the absence of Zfp36L1 protein expression in early development leads to the abnormal stabilization of one or more mRNA species whose protein product is directly or indirectly toxic to the developing placenta and possibly the fetus. However, our analysis of one transcript that is a useful and sensitive TTP target, the GM-CSF mRNA, did not show any evidence of increased mRNA stability or decreased deadenylation in MEFs derived from KO embryos. Thus, the putative Zfp36L1 targets stabilized in the KO embryos and placentas are unknown, again suggesting that a search should be made for transcripts abnormally stabilized by the absence of Zfp36L1 protein in the allantois immediately before chorioallantoic fusion.
Mouse gene KO models with mid-gestational lethality often exhibit failures of chorioallantoic fusion and/or of labyrinth and spongiotrophoblast development (19, 32, 40). For example, in knockouts for α4 integrin and vascular cell adhesion molecule 1 (VCAM-1) there were both placentation failures and associated epicardial defects, as in the present case. In both cases, about half of the embryos died from failure of chorioallantoic fusion, while the surviving embryos had epicardial defects (21, 43). These and other studies have suggested that the α4 integrin is expressed in the chorion, whereas VCAM-1 is expressed in the allantois; expression of both at the appropriate place and time appears to be required for fusion to occur. Interestingly, one group found that there were persistent defects in the placenta even after successful chorioallantoic fusion in the VCAM-1 KO mice (16), whereas no such defects were apparent in the α4 integrin mouse (43). A small number of male VCAM-1-deficient mice also survived to a fertile adulthood (16), which was not seen in the present study.
Although there are several other examples of KO mice with failed chorioallantoic fusion in the literature (19, 32, 40), one of the most strikingly parallel phenotypes to that of the present case is that of the fibroblast growth factor receptor 2 KO (42). About a third of these mice had failed chorioallantoic fusion; the remaining embryos lacked the labyrinthine portion of the placenta. There was markedly diminished cell proliferation within the placental trophoblast cells, while there was no apparent increase in apoptosis in these layers. These authors concluded that the primary defect in the fibroblast growth factor receptor 2 embryo is the block on proliferation of trophoblast cells that leads to failure of the labyrinthine portion of the placenta.
It seems possible that the Zfp36L1 KO affects one or more of the pathways involved in chorioallantoic fusion and/or trophoblast proliferation. However, it is not clear how a presumed mRNA-destabilizing factor, whose deficiency should result in an increase in one or more specific transcripts and their translated proteins, would lead to a phenotype similar to those of these gene disruptions. One possibility might be that the Zfp36L1 disruption results in the accumulation of a factor that is toxic to placentation; known TTP targets, such as TNF, might be candidates for such a factor (6, 14, 41). However, mice whose endogenous, unstable TNF mRNA was replaced by a stable mRNA (ΔARE) by gene knock-in techniques did not exhibit universal prenatal death; instead, these animals developed dramatic failure of weight gain and a severe systemic inflammatory syndrome, leading to death between 5 and 12 weeks of postnatal life (20).
The analogous experiment has not been done with the GM-CSF mRNA ARE to our knowledge. However, a transgenic mouse was recently described that overexpressed a stabilized GM-CSF mRNA driven by a cytomegalovirus promoter (18). These mice apparently exhibited universal prenatal lethality, accompanied by increases in granulocytes and macrophages in ectopic locations. Neither placental histology nor chorioallantoic fusion frequency was commented on in that report, but it remains possible that abnormally stabilized GM-CSF mRNA, perhaps in a specific cell type, contributed to the mid-gestational lethality exhibited by the ZFP36L1 KO embryos in the present study.
Regardless of mechanism, the present studies allow for the firm conclusion that the Zfp36L1 protein, when expressed in its normal tissues and at its normal developmental times, is critical for fetal survival. However, the limitations of the conventional KO technology used here do not identify the physiological role of this protein in cells or tissues of the adult mouse.
Two recent studies shed some light on this issue. In the first, Storch et al. (36) identified Zfp36L1 as a gene whose expression levels exhibited circadian rhythm regulation in both the heart and the liver of the normal mouse. This might indicate a potential role in the established circadian variability of cytokine levels in the blood, as shown in humans for TNF (31). In the second, Stoecklin et al. (35) identified Zfp36L1 as a gene that was mutated in cell lines in which interleukin-3 mRNA was abnormally stabilized; this mRNA contains a typical class II ARE and is known to be destabilized by Zfp36L1 and TTP in cell transfection studies (22). Elucidation of a role for Zfp36L1 and its encoded protein in tissues of the adult animal will probably await the use of conditional or cell-specific KO strategies.
Acknowledgments
We are very grateful to Carl Bortner for the flow cytometry analyses, to Julie Foley for immunohistochemistry, to Gail Pearse for help with the histology, to Manas Ray for reviewing the manuscript, and especially to Jay Cross for help with evaluating the placenta sections.
This work was supported in part by NIH grant NICHD HD39948 to E.N.M.
REFERENCES
- 1.Blackshear, P. J. 2002. Xenopus laevis genomic biomarkers for environmental toxicology studies, p. 339-353. In S. Wilson and W. A. Suk (ed.), Biomarkers of environmentally associated disease. CRC Press, Inc., Boca Raton, Fla.
- 2.Blackshear, P. J. 2002. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 30:945-952. [DOI] [PubMed] [Google Scholar]
- 3.Bortner, C. D., and J. A. Cidlowski. 1996. Absence of volume regulatory mechanisms contributes to the rapid activation of apoptosis in thymocytes. Cell Physiol. 40:C950-C961. [DOI] [PubMed] [Google Scholar]
- 4.Bulmer, J. N., L. Morrison, and P. M. Johnson. 1988. Expression of the proliferation markers Ki67 and transferrin receptor by human trophoblast populations. J. Reprod. Immunol. 14:291-302. [DOI] [PubMed] [Google Scholar]
- 5.Bustin, S. A., X. F. Nie, R. C. Barnard, V. Kumar, J. C. Pascall, K. D. Brown, L. M. Leigh, N. S. Williams, and I. A. McKay. 1994. Cloning and characterization of ERF-1, a human member of the Tis11 family of early-response genes. DNA Cell Biol. 13:449-459. [DOI] [PubMed] [Google Scholar]
- 6.Byrne, A. T., J. Southgate, D. R. Brison, and H. J. Leese. 2002. Effects of insulin-like growth factors I and II on tumour-necrosis-factor-alpha-induced apoptosis in early murine embryos. Reprod. Fertil. Dev. 14:79-83. [DOI] [PubMed] [Google Scholar]
- 7.Carballo, E., and P. J. Blackshear. 2001. Roles of tumor necrosis factor-alpha receptor subtypes in the pathogenesis of the tristetraprolin-deficiency syndrome. Blood 98:2389-2395. [DOI] [PubMed] [Google Scholar]
- 8.Carballo, E., W. S. Lai, and P. J. Blackshear. 1998. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281:1001-1005. [DOI] [PubMed] [Google Scholar]
- 9.Carballo, E., W. S. Lai, and P. J. Blackshear. 2000. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95:1891-1899. [PubMed] [Google Scholar]
- 10.Carballo, E., D. M. Pitterle, D. J. Stumpo, R. T. Sperling, and P. J. Blackshear. 1999. Phagocytic and macropinocytic activity in MARCKS-deficient macrophages and fibroblasts. Am. J. Physiol. 277:C163-C173. [DOI] [PubMed] [Google Scholar]
- 11.Corps, A. N., and K. D. Brown. 1995. Insulin and insulin-like growth factor I stimulate expression of the primary response gene cMG1/TIS11b by a wortmannin-sensitive pathway in RIE-1 cells. FEBS Lett. 368:160-164. [DOI] [PubMed] [Google Scholar]
- 12.De, J., W. S. Lai, J. M. Thorn, S. M. Goldsworthy, X. Liu, T. K. Blackwell, and P. J. Blackshear. 1999. Identification of four CCCH zinc finger proteins in Xenopus, including a novel vertebrate protein with four zinc fingers and severely restricted expression. Gene (Amsterdam) 228:133-145. [DOI] [PubMed] [Google Scholar]
- 13.Dunty, W. C., R. M. Zucker, Jr., and K. K. Sulik. 2002. Hindbrain and cranial nerve dysmorphogenesis result from acute maternal ethanol administration. Dev. Neurosci. 24:328-342. [DOI] [PubMed] [Google Scholar]
- 14.Gendron, R. L., F. P. Nestel, W. S. Lapp, and M. G. Baines. 1991. Expression of tumor necrosis factor alpha in the developing nervous system. Int. J. Neurosci. 60:129-136. [DOI] [PubMed] [Google Scholar]
- 15.Gomperts, M., J. C. Pascall, and K. D. Brown. 1990. The nucleotide sequence of a cDNA encoding an EGF-inducible gene indicates the existence of a new family of mitogen-induced genes. Oncogene 5:1081-1083. [PubMed] [Google Scholar]
- 16.Gurtner, G. C., V. Davis, H. Li, M. J. McCoy, A. Sharpe, and M. I. Cybulsky. 1995. Targeted disruption of the murine VCAM1 gene: essential role of VCAM-1 in the chorioallantoic fusion and placentation. Genes Dev. 9:1-14. [DOI] [PubMed] [Google Scholar]
- 17.Hogan, B., R. Beddington, F. Constantini, and L. Lacy. 1994. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 18.Houzet, L., D. Morello, P. Defrance, P. Mercier, G. Huez, and V. Kruys. 2001. Regulated control by granulocyte-macrophage colony-stimulating factor AU-rich element during mouse embryogenesis. Blood 98:1281-1288. [DOI] [PubMed] [Google Scholar]
- 19.Ihle, J. N. 2000. The challenges of translating knockout phenotypes into gene function. Cell 102:131-134. [DOI] [PubMed] [Google Scholar]
- 20.Kontoyiannis, D., M. Pasparakis, T. T. Pizarro, F. Cominelli, and G. Kollias. 1999. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10:387-398. [DOI] [PubMed] [Google Scholar]
- 21.Kwee, L., H. S. Baldwin, H. M. Shen, C. L. Stewart, C. Buck, C. A. Buck, and M. A. Labow. 1995. Defective development of the embryonic and extraembryonic circulatory systems in vascular cell adhesion molecule (VCAM-1) deficient mice. Development 121:489-503. [DOI] [PubMed] [Google Scholar]
- 22.Lai, W. S., E. Carballo, J. R. Strum, E. A. Kennington, R. S. Phillips, and P. J. Blackshear. 1999. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 19:4311-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai, W. S., E. Carballo, J. M. Thorn, E. A. Kennington, and P. J. Blackshear. 2000. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to AU-rich elements and destabilization of mRNA. J. Biol. Chem. 275:17827-17837. [DOI] [PubMed] [Google Scholar]
- 24.Lai, W. S., E. A. Kennington, and P. J. Blackshear. 2003. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol. Cell. Biol. 23:3798-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai, W. S., D. J. Stumpo, and P. J. Blackshear. 1990. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. J. Biol. Chem. 265:16556-16563. [PubMed] [Google Scholar]
- 26.Maclean, K. N., I. A. McKay, and S. A. Bustin. 1998. Differential effects of sodium butyrate on the transcription of the human TIS11 family of early-response genes in colorectal cancer cells. Br. J. Biomed. Sci. 55:184-191. [PubMed] [Google Scholar]
- 27.Maclean, K. N., C. G. See, I. A. McKay, and S. A. Bustin. 1995. The human immediate early gene BRF1 maps to chromosome 14q22-q24. Genomics 30:89-90. [DOI] [PubMed] [Google Scholar]
- 28.Nie, X. F., K. N. Maclean, V. Kumar, I. A. McKay, and S. A. Bustin. 1995. ERF-2, the human homologue of the murine Tis11d early response gene. Gene 152:285-286. [DOI] [PubMed] [Google Scholar]
- 29.Ning, Z. Q., J. D. Norton, J. Li, and J. J. Murphy. 1996. Distinct mechanisms for rescue from apoptosis in Ramos human B cells by signaling through CD40 and interleukin-4 receptor: role for inhibition of an early response gene, Berg36. Eur. J. Immunol. 26:2356-2363. [DOI] [PubMed] [Google Scholar]
- 30.Nyska, A., R. R. Maronpot, S. R. Eldridge, J. K. Haseman, and J. R. Hailey. 1997. Alteration in cell kinetics in control B6C3F1 mice infected with Helicobacter hepaticus. Toxocol. Pathol. 25:591-596. [DOI] [PubMed] [Google Scholar]
- 31.Petroysky, N., and L. C. Harrison. 1998. The chronobiology of human cytokine production. Int. Rev. Immunol. 16:635-649. [DOI] [PubMed] [Google Scholar]
- 32.Rossant, J., and J. C. Cross. 2001. Placental development: lessons from mouse mutants. Nat. Rev. Genet. 2:538-548. [DOI] [PubMed] [Google Scholar]
- 33.Scoltock, A. B., C. D. Bortner, G. St. J. Bird, J. W. Putney, and J. A. Cidlowski. 2000. A selective requirement for elevated calcium in DNA degradation, but not early events in anti-Fas-induced apoptosis. J. Biol. Chem. 275:30586-30596. [DOI] [PubMed] [Google Scholar]
- 34.Stevens, C. J., H. Schipper, J. Samallo, H. W. Stroband, and T. te Kronnie. 1998. Blastomeres and cells with mesendodermal fates of carp embryos express cth1, a member of the TIS11 family of primary response genes. Int. J. Dev. Biol. 42:181-188. [PubMed] [Google Scholar]
- 35.Stoecklin, G., M. Colombi, I. Raineri, S. Leuenberger, M. Mallaun, M. Schmidlin, B. Gross, M. Lu, T. Kitamura, and C. Moroni. 2002. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 21:4709-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storch, K. F., O. Lipan, I. Leykin, N. Viswanathan, F. C. Davis, W. H. Wong, and C. J. Weitz. 2002. Extensive and divergent circadian gene expression in liver and heart. Nature 417:78-83. [DOI] [PubMed] [Google Scholar]
- 37.Taylor, G. A., E. Carballo, D. M. Lee, W. S. Lai, M. J. Thompson, D. D. Patel, D. I. Schenkman, G. S. Gilkeson, H. E. Broxmeyer, B. F. Haynes, and P. J. Blackshear. 1996. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4:445-454. [DOI] [PubMed] [Google Scholar]
- 38.te Kronnie, G., H. Stroband, H. Schipper, and J. Samallo. 1999. Zebrafish CTH1, a C3H zinc finger protein, is expressed in ovarian oocytes and embryos. Dev. Genes Evol. 209:443-446. [DOI] [PubMed] [Google Scholar]
- 39.Varnum, B. C., Q. F. Ma, T. H. Chi, B. Fletcher, and H. R. Herschman. 1991. The TIS11 primary response gene is a member of a gene family that encodes proteins with a highly conserved sequence containing an unusual Cys-His repeat. Mol. Cell. Biol. 11:1754-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ward, J. M. and D. E. Devor-Henneman. 2000. Gestational mortality in genetically engineered mice: evaluating the extraembryonal and embryonic placenta and membranes, p. 103-122. In J. M. Ward, J. F. Mahler, R. R. Maronpot, J. P. Sundberg, and R. M. Frederickson (ed.), Pathology of genetically engineered mice. Iowa State University Press, Ames, Iowa.
- 41.Wride, M. A., P. H. Lapchak, and E. J. Sanders. 1994. Distribution of TNF alpha-like proteins correlates with some regions of programmed cell death in the chick embryo. Int. J. Dev. Biol. 38:673-682. [PubMed] [Google Scholar]
- 42.Xu, X., M. Weinstein, C. Li, M. Naaski, R. I. Cohen, D. M. Ornitz, P. Leder, and C. Deng. 1998. Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development 125:753-765. [DOI] [PubMed] [Google Scholar]
- 43.Yang, J. T., H. Rayburn, and R. O. Hynes. 1995. Cell adhesion events mediated by alpha 4 integrins are essential in placental and cardiac development. Development 121:549-560. [DOI] [PubMed] [Google Scholar]
- 44.Zucker, R. M., S. Hunter, and J. M. Rogers. 1998. Confocal laser scanning microscopy of apoptosis in organogenesis-stage mouse embryos. Cytometry 33:348-354. [DOI] [PubMed] [Google Scholar]