Abstract
Intestinal epithelial cell-derived interleukin (IL)-7 functions as a pleiotropic and nonredundant cytokine in the human intestinal mucosa; however, the molecular basis of its production has remained totally unknown. We here showed that human intestinal epithelial cells both constitutively and when induced by gamma interferon (IFN-γ) produced IL-7, while several other factors we tested had no effect. Transcriptional regulation via an IFN regulatory factor element (IRF-E) on the 5′ flanking region, which lacks canonical core promoter sequences, was pivotal for both modes of IL-7 expression. IRF-1 and IRF-2, the latter of which is generally known as a transcriptional repressor, were shown to interact with IRF-E and transactivate IL-7 gene expression in an IFN-γ-inducible and constitutive manner, respectively. Indeed, tetracycline-inducible expression experiments revealed that both of these IRF proteins up-regulated IL-7 protein production, and their exclusive roles were further confirmed by small interfering RNA-mediated gene silencing systems. Moreover, these IRFs displayed distinct properties concerning the profile of IL-7 transcripts upon activation and expression patterns within human colonic epithelial tissues. These results suggest that the functional interplay between IRF-1 and IRF-2 serves as an elaborate and cooperative mechanism for timely as well as continuous regulation of IL-7 production that is essential for local immune regulation within human intestinal mucosa.
Intestinal epithelial cells (IECs) function as active participants in local immune regulation via the secretion of a variety of cytokines. Among these, interleukin-7 (IL-7) is particularly important in terms of its pleiotropic functions in the intestinal immune system. Studies have demonstrated that IEC-derived IL-7 stimulates the proliferation of lamina propria lymphocytes and intraepithelial lymphocytes (IELs) (5, 30) and also enhances cytokine release from lamina propria lymphocytes in humans (20). In addition, analyses in mice have revealed the nonredundant functions of IL-7, because inactivation of IL-7 or the IL-7 receptor gene resulted in severely impaired development of γδ-IELs, Peyer's patches, and cryptopatches, all of which play critical roles in mucosal immune regulation (13, 21, 29). These findings suggest that IL-7 production from IECs might be tightly controlled for variable levels of production that properly respond to the altered status of mucosal lymphocytes and also for the constitutive levels of secretion that might support the nonredundant functions of IL-7, for example, on the development of gut-associated lymphoid tissues. Previously, our group has demonstrated that the mRNA and protein of IL-7 are expressed throughout the epithelial layer of human colonic tissues, and the epithelial goblet cells are the type of cells where the expression of IL-7 is relatively abundant (30). To date, however, the mechanisms of IL-7 production in human IECs are poorly defined.
Lack of knowledge about the mechanism of IL-7 production is not confined to IECs but is also the case with other tissue-derived cells of human origin. Previous reports demonstrated that IL-7 production from human bone marrow (BM) stromal cells, the major cell type from which IL-7 is produced in vivo, was regulated by several cytokines such as IL-1, tumor necrosis factor alpha (TNF-α) and transforming growth factor beta (TGF-β) (27, 34); however, the intracellular mechanisms of these regulations have remained unclear. In addition, little is known about the mechanisms by which IL-7 is constitutively produced, while such cells as BM stromal cells exhibited the ability to produce a substantial amount of IL-7 even in the absence of specific cytokines in vitro (27, 34). Moreover, studies on murine tissue-derived cells rather complicated the question as to the mechanisms of IL-7 production in human cells, since these studies implied a different mechanism for murine IL-7 gene expression (3), despite a high degree of conservation in the 5′ flanking region of the IL-7 genes of both species (3, 8, 23). For example, in murine keratinocytes Pam 212 cells, expression of the IL-7 gene was not influenced by IL-1, TNF-α, or TGF-β but was up-regulated by another cytokine, gamma interferon (IFN-γ) (3), indicating that murine cells respond differently than human BM stromal cells to these cytokines (27, 34). These collective findings suggest that IL-7 production might be under the control of a tissue-specific and/or a species-specific regulatory mechanism. Therefore, it seems crucial to clarify the mechanisms of IL-7 production in human IECs to gain a better understanding of the functions of this cytokine on local immune regulation.
In this study, using human colonic epithelial cell lines, we showed that IL-7 protein was produced both constitutively and in response to IFN-γ in human IECs. The transcriptional regulation via an interferon regulatory factor element (IRF-E) was important for IL-7 production in human IECs, which is consistent with the previous report on murine keratinocytes. Of note, it was found that not only IRF-1 but also IRF-2, generally known as a transcriptional repressor, up-regulated IL-7 production. Intriguingly, IRF-1 and IRF-2 exclusively exerted their functions in an IFN-γ-inducible and constitutive manner, respectively, with properties to induce different sets of IL-7 transcript upon activation. Along with the demonstration that both IRF-1 and IRF-2 were expressed in normal human colonic epithelial cells, these data suggest that the functional interplay between IRF-1 and IRF-2 might serve as an elaborate mechanism for the finely tuned regulation of IL-7 production that is indispensable for local immune regulation within the human intestinal mucosa.
MATERIALS AND METHODS
Cell culture.
Human colon carcinoma-derived DLD-1 and HT29-18N2 cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Except where indicated otherwise, cells were seeded at a density of 3 × 105 cells/ml in the medium 36 h prior to each experiment.
ELISA.
Cells at a density of 8 × 105 cells per ml of culture medium were seeded onto 24-well plates. After 36 h of culture, the medium was removed, and the cells were washed twice with phosphate-buffered saline. Following the addition of 1 ml of culture medium alone, or medium containing either human IL-1β, TNF-α, TGF-β, IFN-γ (PeproTec), or doxycycline (DOX; Clontech), cells were cultured for 24 h, and human IL-7 protein levels in the culture supernatants were measured by a human IL-7 enzyme-linked immunosorbent assay (ELISA) kit (R&D Systems).
Semiquantitative reverse transcription (RT)-PCR.
Total RNA was isolated by using Trizol reagent (Invitrogen) according to the manufacturer's instructions. Aliquots of 5 μg of total RNA were used for cDNA synthesis in 21 μl of reaction volume. One microliter of cDNA was amplified with 0.25 U of LA Taq polymerase (TaKaRa) in a 25-μl reaction. Sense (S) and antisense (AS) primers used here were as follows: S1, 5′-AGCTTGCTCCTGCTCCAGTT-3′; S2, 5′-GAGATCATCTGGGAAGTCTTTTACC-3′; S3, 5′-ACTTGTGGCTTCCGTGCACACATTA-3′; AS1, 5′-TGCATTTCTCAAATGCCCTAATCCG-3′; and AS2, 5′-ATCCGCCAGCAGTGTACTTTCAGTT-3′ for human IL-7 (see Fig. 2A). For glyceraldehyde 3-phosphate dehydrogenase (G3PDH) amplification, the primers were5′-TGAAGGTCGGAGTCAACGGATTTGGT-3′ (S) and and 5′-CATGTGGGCCATGAGGTCCACCAC-3′ (AS). Each cycle of PCR amplification consisted of denaturation at 94°C for 30 sec, annealing at 61°C for 30 sec, and extension at 72°C for 30 sec. Twenty-seven cycles were performed for IL-7, and 17 cycles were performed for G3PDH, and the amplification for each gene was in the linear curve under these conditions. PCR products were separated on 1.5% agarose gels, stained by ethidium bromide, and visualized by using a Lumi-Imager F1 system (Roche).
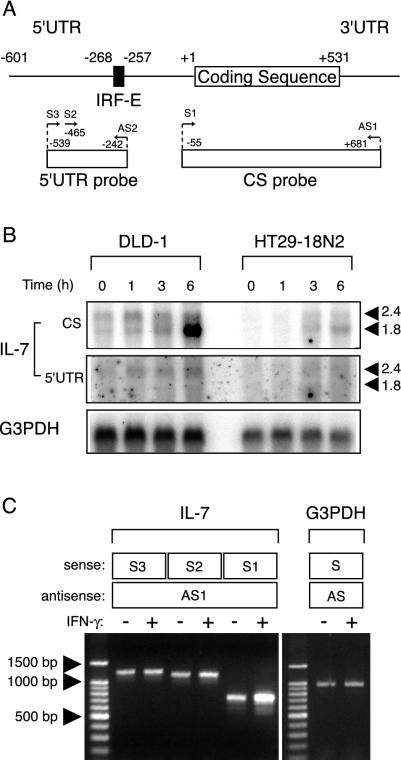
FIG. 2.
IFN-γ-dependent and -independent IL-7 production is distinctively regulated by expression of IL-7 transcripts that differ in their 5′ UTR. (A) Schematic drawing of human IL-7 mRNA, with the primers and cDNA probes used in this study. The nucleotide number was designated with respect to the translation start site (+1). An IRF-E located at the region from position −268 to −257 is also indicated. (B) DLD-1 and HT29-18N2 cells were stimulated with IFN-γ (50 ng/ml) for the indicated time periods. Fifteen micrograms of poly(A)+ mRNA was subjected to Northern blotting for IL-7 mRNA by using the 32P-labeled CS probe or the 5′ UTR probe. The bottom panel indicates the level of G3PDH mRNA as a control. (C) DLD-1 cells were treated with IFN-γ (50 ng/ml) or left untreated for 6 h, collected for total RNA isolation, and then subjected to semiquantitative RT-PCR for IL-7 mRNA. PCR amplification was performed by using either the S1, S2, or S3 primer along with AS1 primer depicted in panel A. As controls, samples from IFN-γ-treated and untreated cells were amplified with a primer set for G3PDH.
Northern blotting.
Poly(A)+ mRNA was isolated by using a FastTrack 2.0 kit (Invitrogen) according to the manufacturer's instructions. Northern blotting was performed as described previously (22) by using 15 μg of poly(A)+ mRNA. The cDNA probe corresponding to nucleotides at positions −55 to +681 (coding sequence [CS] probe) and −539 to −242 (5′ untranslated region [UTR] probe) for human IL-7 were generated by RT-PCR by using the primers S1/AS1 and S3/AS2, respectively, from an RNA sample of DLD-1 cells as described above. The probe for G3PDH was also generated by RT-PCR by using the primers described above. Hybridization was carried out at 42°C overnight for IL-7 and at 55°C for 2 h for G3PDH.
RLM-RACE.
Determination of the transcription initiation sites of the human IL-7 gene was accomplished by RNA ligase-mediated [RLM] 5′ rapid amplification of cDNA ends [RACE] by using a GeneRacer Kit (Invitrogen). In brief, poly(A)+ mRNAs extracted from IFN-γ-stimulated (6 h) DLD-1 cells were treated with calf intestinal phosphatase to eliminate 5′ phosphates from truncated mRNA without affecting 5′ capped intact mRNA. The dephosphorylated RNA was then treated with tobacco acid pyrophosphatases to remove the 5′ cap structure. The GeneRacer RNA Oligo was ligated to the 5′ end of the decapped mRNA by using T4 RNA ligase. First-strand cDNA synthesis was performed by reverse-transcribing the ligated mRNA in the presence of the GeneRacer oligo dT primer. Sequential PCRs were performed by using a primer set of the GeneRacer 5′ primer and 3′ reverse IL-7 gene-specific primer 1 (GSP-1) and then by using the nested primer set of the GeneRacer 5′ nested primer and 3′ reverse IL-7 GSP-2 to amplify only the cDNAs that have the GeneRacer RNA Oligo ligated to the 5′ end. As a control, PCR with a primer set for amplifying the 5′ part of the human β-actin gene was also performed in parallel, according to the manufacturer's recommendation. The primers used were 5′-TGCCCTAATCCGTTTTGACCATGGTG-3′ (IL-7 GSP-1) and 5′-GCAACAGAACAAGGATCAGGGGAGG-3′ (IL-7 GSP-2). PCR products of around 600 and 300 bp were gel purified and cloned into the pGEM-T vector (Promega) independently, and then 10 clones of each were sequenced. All the clones contained the IL-7 gene sequence along with the adapter sequences, indicating these clones to be derived from mRNAs retaining complete 5′ ends.
Plasmids.
The human IL-7 DNA fragment between either position −3194, −1322, −609, or −282 and −3 was amplified from human genomic DNA by PCR and ligated into the pGL3 Basic luciferase reporter plasmid (Promega) to create −3194-Luc, −1322-Luc, −609-Luc, and −282-Luc. The nucleotide position number was assigned relative to the translation start site (+1). A series of 5′ deletions of the −609-Luc, shown as −362-Luc, −251-Luc, and −215-Luc, was constructed by unidirectional digestion by using an exonuclease III. An internal deletion mutant −609-Luc-Δ-282/-251 was constructed by PCR-mediated mutagenesis. Plasmids −609-mtIRF-E-Luc and −282-mtIRF-E-Luc, both of which contain a 4-bp mutation within IRF-E, were also constructed by PCR-mediated mutagenesis. Introduced mutations and the wild-type sequences within the region of positions −280 to −253 were given with top strand sequences as follows: mutant, 5′-AAGCGCAAAGTAGAGGCTGAGGGTACAC-3′ (underlined residues indicate introduced mutations); wild type, 5′-AAGCGCAAAGTAGAAACTGAAAGTACAC-3′. Expression vectors pcDNA3-IRF-1 and pcDNA3-IRF-2 were prepared by subcloning the PCR-amplified open reading frame of human IRF-1 and IRF-2 cDNA into a pcDNA3 (Invitrogen). To construct tetracycline (TET)-inducible expression plasmids, the open reading frames of IRF-1 and IRF-2 were subcloned into a pcDNA4/TO/Myc/His (Invitrogen) in frame. All constructs were verified by DNA sequencing.
Transient transfection and reporter assays.
DLD-1 cells seeded in a 60-mm culture dish were transfected with 3 μg of reporter plasmid along with 10 ng of pRL-tk plasmid (Promega) as described previously (22). Cells were harvested 24 h after transfection, lysed by three cycles of freezing and thawing, and then luciferase activities were measured by a luminometer (Turner Designs). Luciferase activities as indicated by arbitrary unit were normalized by renilla luciferase activities in each sample.
EMSA.
The preparation of nuclear extracts and electrophoretic mobility shift assays (EMSA) were performed essentially as described previously (22), except for the use of 0.5 μg of poly(dI-dC) · poly(dI-dC) per binding reaction. A DNA probe and its mutated version were prepared by annealing oligonucleotides as follows: top strand, 5′-AAGCGCAAAGTAGAAACTGAAAGT-3′, and bottom strand, 5′-GTGTACTTTCAGTTTCTACTTTG-3′, for the wild-type probe; and top strand, 5′-AAGCGCAAAGTAGAGGCTGAGGGT-3′, bottom strand, 5′-GTGTACCCTCAGCCTCTACTTTG-3′, for the mutant probe. For competition experiments, a 20-fold excess of unlabeled double-stranded probe or its mutated version was added prior to the labeled probe. In supershift experiments, antibodies (Santa Cruz Biotechnology) against either IRF-1 (catalogue no. sc-497), IRF-2 (sc-498), IRF-3 (sc-9082), IRF-4 (sc-6059), IRF-7 (sc-9083), IRF-8 (sc-6058), or IRF-9 (sc-496) were used.
Immunoblotting.
Immunoblotting was performed as described elsewhere (22). Twenty-five micrograms of nuclear extracts was analyzed by using anti-IRF-1 (catalogue no. sc-497), anti-IRF-2 (sc-498), and anti-upstream factor (USF)-2 (sc-861) antibodies (all from Santa Cruz Biotechnology) at a 1:500 dilution as a primary antibody. Proteins were visualized with an enhanced chemiluminescence detection system (Amersham Bioscience).
Establishing tetracycline-regulated IRF-1- and IRF-2-expressing DLD-1 cell lines.
Sublines of DLD-1 cells, in which the expression of either IRF-1 or IRF-2 is inducible under the control of the addition of TET, were established by using a T-REx System (Invitrogen). In brief, a DLD-1-derived subclone that constitutively expresses the TET repressor (TR) was created by transfecting parental DLD-1 cells with a plasmid pcDNA6/TR (Invitrogen). Several clones were selected in the culture medium containing blasticidin (7.5 μg/ml; Invitrogen). An appropriate clone was isolated, designated as DLD-1/TR cells, and then transfected with either expression plasmid pcDNA4/TO/IRF-1-Myc/His or pcDNA4/TO/IRF-2-Myc/His. Cells stably expressing each of these genes were selected in the presence of 750 μg of Zeocin (Invitrogen) per ml to establish the sublines designated as DLD-1/TR/IRF-1-tag or DLD-1/TR/IRF-2-tag cells. In all experiments, we used DOX as an alternative inducer of gene expression because it has a longer half-life than TET.
siRNA experiments.
All small interfering RNA (siRNA) duplex oligonucleotides were synthesized and subsequently annealed for use. DLD-1 cells were seeded at a density of 3 × 105 cells per ml onto a 24-well plate or a 100-mm culture dish. After 36 h, cells were transfected with 100 nM siRNA oligonucleotides as described previously (37), and the siRNA-containing medium was removed after 12 h of transfection. Cells were cultured for an additional 12 h under the usual conditions, and then the medium was exchanged with either the medium alone or medium containing IFN-γ. For immunoblotting analysis, cells were collected from the 100-mm dishes after 12 h of medium exchange, and the nuclear extracts were isolated. For the ELISA, the culture supernatants were collected from the 24-well plates after 24 h of medium exchange. The sequences of siRNAs used here were as follows (S strand only): IRF-1, CCAAGAACCAGAGAAAAGATT; IRF-2, CUCUUUAGAAACUGGGCAATT; and negative control (G85R mutant superoxide dismutase), UGUUGGAGACUUCGGCAAUTT. Italicized letters indicate deoxynucleotides.
ChIP assays.
A chromatin immunoprecipitation (ChIP) assay was performed essentially as described previously (24) with some modifications. DLD-1 cells seeded onto a 150-mm dish were stimulated with IFN-γ or left untreated for 6 h, cross-linked with 1% formaldehyde for 5 min at room temperature, and then quenched by adding glycine. Cells were washed with phosphate-buffered saline, resuspended in 1 ml of lysis buffer (10 mM Tris-HCl [pH 8.0], 0.25% Triton X-100, 10 mM EDTA, and 0.5 mM EGTA) and left on ice for 10 min. After centrifugation, the nuclei were washed with 1 ml of wash buffer (10 mM Tris-HCl [pH 8.0], 200 mM NaCl, 10 mM EDTA, and 0.5 mM EGTA, 10 mM sodium butyrate, 20 mM β-glycerophosphate, 100 μM sodium orthovanadate, 1 μM microcystin, and the protease inhibitor cocktail) and resuspended in 400 μl of sonication buffer (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, and 0.5 mM EGTA). The sonication was performed in two steps by using a VP-152 system (TAITEC); the first step was carried out for 5 min, followed by the addition of 50 μl of 10% sodium dodecyl sulfate (SDS) and incubation for 1 h to solubilize the chromatin, and then the second sonication was performed for 4 min. This yielded genomic fragments with an average size of 500 bp. Aliquots (100-μl) of sheared chromatin were diluted into 1 ml of radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl [pH 8.0], 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, 1 mM EDTA, 0.5 mM EGTA, and 140 mM NaCl) and precleared with 50 μl of protein G-Sepharose (50% slurry in RIPA buffer) for 1 h at 4°C. Immunoprecipitation was performed overnight at 4°C with 10 μg of an anti-IRF-1 (catalogue no. sc-497), anti-IRF-2 (sc-498), normal mouse immunoglobulin G (IgG; sc-2025) (all from Santa Cruz Biotechnology), or an antihistone H3 antibody (Abcam, Inc.). A 20-μl aliquot of 50% protein G-Sepharose slurry (same as above but containing 2 mg of herring sperm DNA per ml and 2 mg of bovine serum albumin per ml) was added to each and incubated for 1 h at 4°C. Precipitates were washed sequentially in RIPA buffer three times, in 0.5 M NaCl RIPA buffer (same as RIPA buffer but with 500 mM NaCl) three times, in LiCl wash buffer (10 mM Tris-HCl [pH 8.0], 0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 1 mM EGTA, 10 mM sodium butyrate, 100 μM sodium orthovanadate, and the protease inhibitor cocktail) twice, and in TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) twice, for 3 min for each wash. Samples were extracted twice with 50 μl of elution buffer (1% SDS, 0.1 M NaHCO3, 10 mM dithiothreitol) and digested with 2 μg of proteinase K at 37°C for 4 h. Then 4 μl of 5 M NaCl was added, and the samples were incubated at 65°C overnight to reverse cross-linking. DNA fragments were recovered by phenol-chloroform extraction and ethanol precipitation.
The genomic DNA fragments in the immunoprecipitated samples were analyzed by PCR by using a primer set for amplifying the −539 to −159 region of the human IL-7 gene (5′-ACTTGTGGCTTCCGTGCACACATTA-3′ and 5′-GACTGCAGTTTCATCCATCCCAAG-3′) to detect the IRF-E-containing fragment, and another set for the +976 to +1337 region (5′-GCTCTTTCTTTTGATGGCTACTCCG-3′ and 5′-TAGCCCATGATTCATATAACTGTGC-3′; numbers indicate the positions on the genomic DNA relative to the translation start site) (see Fig. 8A) as controls. Initially, quantitative PCR on a LightCycler system (Roche) was performed to quantify the immunoprecipitated DNA. A 5-μl aliquot from a total of 100 μl of DNA solution was amplified and the threshold cycle was obtained from each amplification curve. In practice, DNA fragments were nonspecifically and reproducibly recovered after the ChIP assay in the absence of specific antibody but were usually amplified 5 to 6 cycles later than specifically recovered fragments. By using software provided by the manufacturer, the amount of DNA fragment in each sample was calculated relative to the standard curve obtained by the three different dilutions of input DNAs (10, 1, and 0.1%). Three independent chromatin preparations were made, and the average value obtained for each sample was indicated as a percentage of total input DNA. The same amounts of DNA samples or the diluted inputs were also analyzed by conventional PCR in parallel with the following parameters: denaturation at 94°C for 15 s, annealing at 61°C for 30 s, and extension at 72°C for 30 s for 37 cycles. The products were resolved by agarose gel electrophoresis, stained with ethidium bromide, and visualized by using a Lumi-Imager F1 system (Roche).
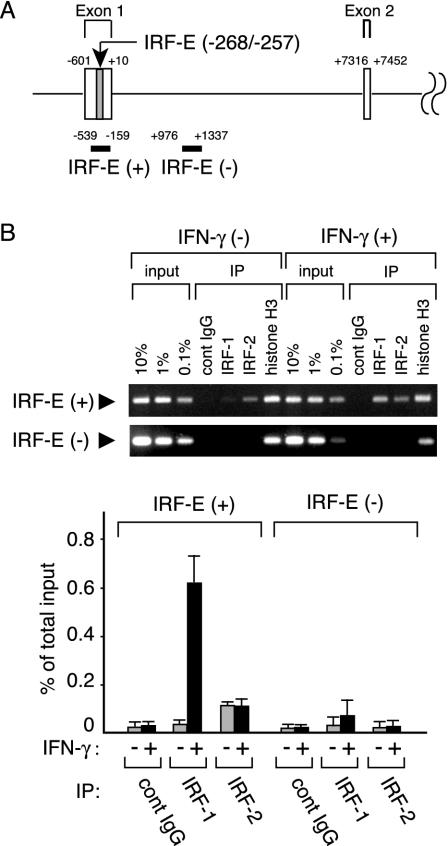
FIG. 8.
IRF-1 and IRF-2 bind to IRF-E without mutual exclusion in vivo. (A) Schematic representation of the 5′ part of the human IL-7 gene. Exon 1, 2, and the intervening intron are shown with the nucleotide number relative to the translation initiation site (+1). IRF-E located in exon 1 (shaded) and the DNA regions analyzed by ChIP assays [IRF-E(+) and IRF-E(-)] are also indicated. (B) DLD-1 cells were treated with IFN-γ or left untreated for 6 h and processed for ChIP assays by using anti-IRF-1, anti-IRF-2, and anti-histone H3 antibodies (positive control) or control Ig (negative control). Precipitated DNA was subjected to both conventional PCR (top) and quantitative PCR (bottom) to amplify either the IL-7 gene fragment (−539 to −159) containing the IRF-E on its 5′ UTR [IRF-E(+)] or the intronic fragment (+976 to +1337) [IRF-E(-)]. The amount of immunoprecipitated IL-7 gene fragment relative to that present in total input chromatin (% of total input) was calculated as described in Materials and Methods. Data are shown as the means ± standard deviations of three independent chromatin immunoprecipitations (bottom). IP, immunoprecipitation; cont, control.
Immunohistochemistry.
Normal colonic mucosae were obtained from three patients with colorectal cancer who underwent colectomy. Written informed consent was obtained from all patients, and these experiments were approved by the Tokyo Medical and Dental University Hospital Committee on Human Subjects. Samples fixed by 4% paraformaldehyde were cut into 8 μm-thick sections, treated with 0.5% hydrogen peroxide in methanol solution, blocked for 45 min, and then incubated with either an anti-IRF-1 (catalogue no. sc-497; Santa Cruz Biotechnology), an anti-IRF-2 (sc-13042), or purified rabbit IgG (10 mg/ml; negative control) overnight at 4°C. The sections were incubated with biotinylated goat antirabbit IgG for 60 min and reacted with streptavidin-enzyme conjugates (Vector Laboratories Inc), and then the peroxidase activities were developed by diaminobenzidin. After the samples were counterstained with hematoxylin, the localization of IRF-1 or IRF-2 was examined by light microscopy.
RESULTS
Human IECs constitutively produce IL-7, and IL-1, TNF-α, and TGF-β have no influence on the levels of IL-7 production, but IFN-γ does.
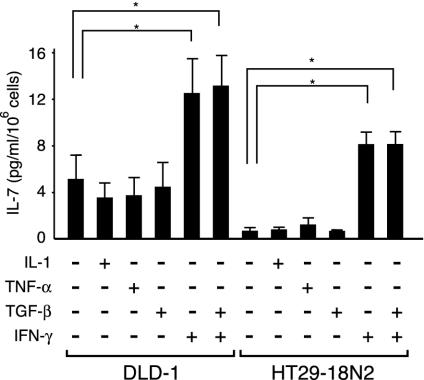
To investigate the mechanisms of IL-7 production in human IECs, human colonic epithelial cell lines, DLD-1 and HT29-18N2 cells, were analyzed. Previous reports showed that both IL-1 and TNF-α had enhancing effects on IL-7 production in human BM stromal cells or osteoblasts (34), while TGF-β had suppressive effects on IL-7 production in BM stromal cells (27). In contrast, shown with murine keratinocytes, none of these cytokines had any effect, while IFN-γ solely exhibited enhancing effects on IL-7 mRNA expression among various factors (3). Therefore, to test whether IL-7 production in human IECs is also diversely regulated by these cytokines, DLD-1 and HT29-18N2 cells were incubated for 24 h with either IL-1, TNF-α, TGF-β, IFN-γ, or the medium alone, and IL-7 production was measured by ELISA. As shown in Fig. 1, both of these cells constitutively produced substantial amounts of IL-7 protein, with a higher concentration per cell number in DLD-1 than in HT29-18N2 cells. In addition, treatment with IFN-γ significantly enhanced IL-7 production in both cell types, while stimulation with IL-1, TNF-α, or TGF-β had no effect (Fig. 1). We also tested the possibility that TGF-β might act as an inhibitory factor not for the constitutive but for the inducible production of IL-7, but treatment with TGF-β did not affect IFN-γ-inducible IL-7 production (Fig. 1). These data indicated that human IECs produce IL-7 both constitutively and in response to IFN-γ, while several other cytokines have no regulatory effect on this process.
FIG. 1.
Human IECs constitutively produce IL-7, and IL-1, TNF-α, and TGF-β do not influence the levels of IL-7 production, whereas IFN-γ does. DLD-1 and HT29-18N2 cells were cultured in medium alone or in medium containing 50 ng of either IL-1, TNF-α, TGF-β, IFN-γ, or IFN-γ plus TGF-β per ml for 24 h. The supernatants were collected and assayed for IL-7 production by ELISA. Results are the means ± standard deviations of three independent experiments. *, P < 0.05 by a paired Student t test.
Transcription start sites of the human IL-7 gene are clustered within two distinct regions, and IFN-γ preferentially induces short species of mRNA via a selective usage of downstream initiation sites.
Human tissues have been shown to express two major IL-7 mRNAs of ∼1.8 and ∼2.4 kb, and this has been inferred as a result of alternative polyadenylation (8). To examine whether constitutive and IFN-γ-inducible IL-7 protein production is regulated at the mRNA level, we next assessed the expression of IL-7 transcripts by Northern blot analysis by using a cDNA probe covering the IL-7 protein coding sequences (Fig. 2A, CS probe). In DLD-1 cells, two major mRNA species were clearly observed in the absence of IFN-γ (Fig. 2B, left). Since each of these bands migrated somewhat heterogeneously, it was difficult to determine the precise size of these transcripts. However, the analysis of mRNAs extracted from SK-Hep1 cells, human hepatocellular carcinoma cells originally used for the cloning of the human IL-7 gene (8), showed equally migrating bands (data not shown). Thus, we tentatively equated these transcripts with those described previously (8). When DLD-1 cells were treated with IFN-γ, ∼1.8-kb mRNA was significantly induced within 6 h, whereas the increase in ∼2.4-kb mRNA was modest (Fig. 2B, left). Although the basal level of IL-7 mRNA was lower in HT29-18N2 cells than could be visualized, these cells displayed a similar pattern of IL-7 mRNA expression: IFN-γ significantly induced expression of ∼1.8-kb and, to a lesser extent, ∼2.4-kb of mRNAs (Fig. 2B, right). These data indicated that the levels of IL-7 protein production correlate well with those of mRNA expression and that IFN-γ treatment predominantly induces the short mRNA species of the IL-7 gene. Interestingly, in murine keratinocytes, IFN-γ treatment was demonstrated to induce the expression of relatively short species of IL-7 mRNA through the use of alternative transcription start sites (3). Given an analogy in IFN-γ-dependent induction of selective IL-7 transcripts between human IECs and murine keratinocytes, it seemed possible that the mechanisms of IFN-γ-dependent IL-7 gene expression are, at least in part, conserved between these two cell types.
To date, a detailed analysis of the transcription start sites for the human IL-7 gene has not been reported. We thus attempted to precisely map the 5′ end of IL-7 mRNA by using a RLM-RACE method that ensures the amplification of only full-length transcripts via the elimination of truncated mRNAs (see Materials and Methods). When poly(A)+ RNA extracted from IFN-γ-treated (6 h) DLD-1 cells was analyzed, fragments around 600 and 300 bp were obtained by PCR by using the 5′ nested primer and the 3′ reverse IL-7 GSP-2 (corresponding to nucleotides 36 to 60 of the IL-7 gene) (Fig. 3A). No product was obtained when RNA was not treated with tobacco acid pyrophosphatases, indicating that these products were derived from full-length mRNA (data not shown). Both of these products appeared to migrate somewhat diffusely when subjected to gel electrophoresis (Fig. 3A). This result was not attributable to experimental artifacts, because PCR amplification with another set of primers, designed for detecting the 5′ part of the human β-actin gene, yielded products of the expected size (872 bp) that migrated as a single band from the same sample (Fig. 3A). The ∼600- and ∼300-bp fragments were independently isolated and cloned, and then 10 clones of each were sequenced. All 20 clones contained the IL-7 gene sequence along with the adapter sequences, showing these clones to be derived from mRNAs retaining complete 5′ ends. Alternative splicing appeared to be infrequent in this region (upstream of the sequences corresponding to IL-7 GSP-2), because no nucleotide deletion was observed in any of the sequenced clones. As depicted in Fig. 3B, the 5′ ends of longer fragments were located within the −601 to −515 region upstream of the translation start site (+1), while the 5′ ends of shorter fragments were mapped within the −197 to −131 region. These results demonstrated that the human IL-7 gene is transcribed from multiple transcription start sites that are clustered within two distinct regions approximately 300 to 500 bp apart from each other.
FIG. 3.
Transcription initiation sites for the human IL-7 gene were clustered within two separate regions upstream from the translation start site. (A) RLM-RACE analysis was performed by using poly(A)+ RNAs from IFN-γ-treated (6 h) DLD-1 cells as described in Materials and Methods. PCR products amplified by a primer set for the IL-7 or β-actin gene, respectively, were run on 1.5% agarose gel, stained with ethidium bromide, and visualized. (B) Two major fragments of ∼600 and ∼300 bp shown in panel A were independently cloned, and then 10 clones of each were sequenced. The 5′ end of each clone is shown by a filled triangle (clones derived from ∼600-bp fragments) or an open triangle (clones from ∼300-bp fragments) on the first 650 bp of sequence upstream of the translation start site. The authentic translation start site is indicated in bold. Numbering in base pairs is indicated to the left, with negative numbers representing nucleotides upstream of the ATG. Consensus sequences for the IRF-E are boxed and labeled. Potential translation initiation codons (ATG) are underlined. Consensus sequences for MED-1 are also underlined.
We then tested whether ∼1.8- and ∼2.4-kb IL-7 mRNAs might indeed differ in their 5′ UTR stretches by Northern blot analysis using a 5′ UTR probe corresponding to nucleotides at positions −539 to −242 (Fig. 2A). Interestingly, this probe exhibited subtle but substantial hybridization with ∼2.4-kb but not with ∼1.8-kb mRNA, showing the different lengths of 5′ UTR between these mRNA species (Fig. 2B). To further confirm this, RT-PCR with any of the S primers (S1, S2, or S3) along with the AS1 primer (Fig. 2A) was carried out. When RNAs from untreated and IFN-γ-treated DLD-1 cells were examined, amplification with the primer sets of S3/AS1 and S2/AS1 showed no difference in the amount of products before and after IFN-γ treatment (Fig. 2C). In contrast, amplification with the primer set of S1/AS1 displayed a significant increase in the amounts of PCR products in response to IFN-γ (Fig. 2C). Therefore, it was demonstrated that stimulation with IFN-γ preferentially induces relatively short-form IL-7 mRNA expression via the selective usage of transcription initiation sites within the region −197 to −131. Of note, the maximum difference in 5′ UTR lengths among clones obtained in RLM-RACE was less than 500 bp and did not match that between the ∼2.4- and ∼1.8-kb transcripts seen in Northern blot analysis. We assumed that this discrepancy might have in some part resulted from the difficulties in determining the precise size of transcripts by Northern blot analysis; however, this issue was not further examined in the present study.
IFN-γ-dependent and -independent IL-7 gene expression is mediated through IRF-E within its 5′ UTR.
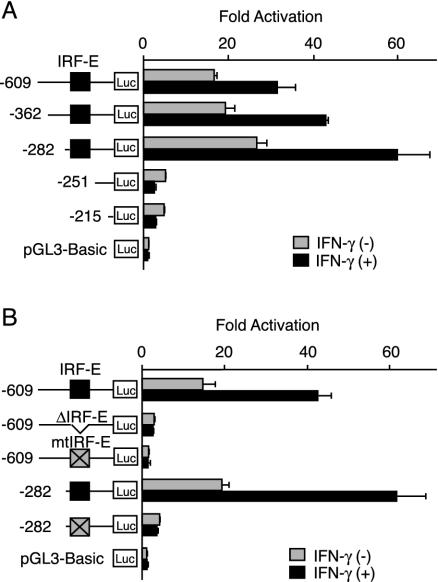
To characterize cis-acting regulatory elements in the 5′ flanking or intragenic regions of the human IL-7 gene, we constructed a reporter plasmid in which the luciferase gene expression was under the control of the upstream region of the human IL-7 gene. When a reporter plasmid with the region −3194 to −3 (−3194-Luc) was transiently transfected in DLD-1 cells, a significant increase of luciferase activity was observed in both untreated and IFN-γ-treated cells, when compared to that seen with the control reporter plasmid (data not shown). A series of 5′ deletions from the −3194-Luc to position −609 (−609-Luc) showed no apparent difference in reporter activities (data not shown), and, thus, we constructed another series of 5′ deletion clones to be analyzed. As shown in Fig. 4A, the reporter activity of the −609-Luc was approximately 15-fold higher than that of the control in the absence of IFN-γ. In addition, the reporter activity exhibited approximately a twofold induction in response to IFN-γ. Deletions from the 5′ end to −282 showed a slight increase in both basal and IFN-γ-inducible reporter activities. In contrast, further deletion up to position −251 resulted in a dramatic decrease of reporter activities not only of uninduced levels but also with regard to IFN-γ-dependent induction (Fig. 4A). These results suggested that the critical enhancer element for both IFN-γ-dependent and -independent up-regulation of IL-7 gene expression might be located within the region from position −282 to −251. Substantial activities in the constitutive levels of reporter gene expression were observed further in the −215 to −3 region, while the IFN-γ-dependent induction of the reporter activity almost diminished by deleting up to position −251 (Fig. 4A). Since the region from position −282 to −251 contained an IRF-E at position −268 to −257 (Fig. 3B), we postulated that IL-7 gene activation might be mediated through this site. As expected, when an internal deletion mutant of the −282 to −251 sequence (−609-ΔIRF-E-Luc) was assayed, a drastic decrease in reporter activities in untreated as well as IFN-γ-treated cells was observed (Fig. 4B). In addition, introduction of a 4-bp mutation into the IRF-E sequences of −609-Luc and −282-Luc similarly culminated in a marked decrease in reporter activities (Fig. 4B). These findings suggested that the region from position −282 to −251 and, in particular, the IRF-E sequences within this region play a critical role in determining both IFN-γ-dependent and -independent enhancer activities in human IECs.
FIG. 4.
IFN-γ-dependent and -independent IL-7 gene activation is mediated through IRF-E within its 5′ UTR. (A) DLD-1 cells were transiently transfected with a −609-Luc or either of its 5′ deletion mutant plasmids, cultured in the presence or the absence of IFN-γ for 12 h, and then assayed for reporter activities. (B) DLD-1 cells were transiently transfected with a −609-Luc or either of its mutated versions of plasmids, cultured as described for panel A, and then reporter activities were assayed. Luciferase activities were normalized and indicated as increases in activation compared with activity levels of cells transfected with pGL3-Basic plasmid and left untreated. Results are the means ± standard deviations of three independent experiments. Xs in boxes indicate mutated IRF-E sequences.
IRF-1 is an inducible, while IRF-2 is a constitutive, binding protein to the IRF-E.
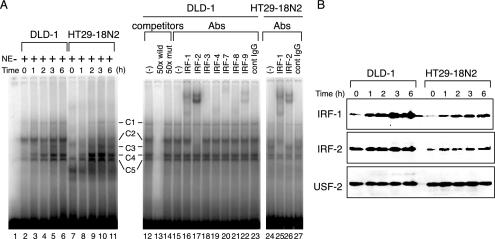
To identify nuclear factors that interact with the regulatory element of the IL-7 gene, we performed an EMSA by using a DNA probe corresponding to the sequences of the region −280 to −253. When nuclear extracts of DLD-1 and HT29-18N2 cells before and after IFN-γ addition were examined, several DNA-protein complexes were observed (Fig. 5A, C1 through C5). Among these, two (C2 and C5) displayed constitutive complex formation, and others (C1, C3, and C4) were induced by IFN-γ in both of these cell types (Fig. 5A, lanes 1 to 11). The formation of these complexes was sequence specific, since the nonlabeled DNA probe competed out the binding of nuclear proteins with the labeled probe (Fig. 5A, lane 13). In addition, a nonlabeled mutant probe did not affect complex formation, showing these complexes to be composed of proteins that specifically recognized IRF-E sequences (Fig. 5A, lane 14). Consistently, an anti-IRF-1 antibody shifted complex C3 (Fig. 5A, lanes 16 and 25), one of the inducible complexes in both cell types. In addition, complex C2, continuously observed with higher intensity in DLD-1 cells than in HT29-18N2 cells, was completely shifted with an anti-IRF-2 antibody (Fig. 5A, lanes 17 and 26). These observations were further supported by immunoblotting, since the nuclear expression of these IRF proteins correlated well with the results of the EMSA analysis (Fig. 5B). It should be noted that in the supershift experiments, nuclear complexes containing other IRF family proteins such as IRF-4, IRF-8, and IRF-9 were also present to some extent (Fig. 5A, lanes 19, 21, and 22). However, the degree of their occupancy on IRF-E sequences remained unclear, since antibodies against these molecules could neither shift nor disrupt the protein-DNA complexes. Together, these observations indicated that IRF-1 and IRF-2 bind to the IRF-E in an IFN-γ-inducible and constitutive manner, respectively, and then transcriptionally regulate IL-7 gene expression. Again, this notion showed an analogy with the previously described mechanisms of IL-7 gene transcription in murine keratinocytes in terms of the IFN-γ-inducible DNA binding of IRF-1 to IRF-E (2). Our data, however, raised the further possibility that IRF-2, generally known as a transcriptional repressor, also acts as a positive regulator of IL-7 gene expression, since the constitutive DNA binding and relative abundance of the IRF-2-containing nuclear complex in DLD-1 compared to HT29-18N2 cells, closely paralleled the levels of IL-7 mRNA expression.
FIG. 5.
IRF-1 is an inducible, while IRF-2 is a constitutive, binding protein to the IRF-E. (A) DLD-1 and HT29-18N2 cells were stimulated with IFN-γ (50 ng/ml) for the indicated time periods. Nuclear extracts (NE) were prepared, and 10 μg of each was subjected to EMSA by using 32P-labeled oligonucleotide probes corresponding to the sequence −280 to −253 of the IL-7 gene (left). Competition assays were performed by adding a 50-fold molar excess of unlabeled specific oligonucleotide (wild) or the mutant (mut) probe to the reaction mixture containing the extracts from cells treated with IFN-γ for 6 h (right). Supershift assays were performed by preincubating the reaction mixture of 6 h-treated nuclear extracts with either 2 μg of antibodies (Abs) against the indicated proteins or 2 μg of mouse IgG. (B) Twenty-five micrograms of nuclear extracts as described in panel A was separated on an SDS-10% polyacrylamide gel and immunoblotted with an anti-IRF-1 antibody (IRF-1), and the blot was sequentially reprobed with an anti-IRF-2 (IRF-2), and an anti-USF-2 antibody (USF-2, loading control).
IRF-1 and IRF-2 distinctively up-regulate IL-7 protein production via IRF-E-mediated transcription.
Given these observations, we next analyzed the functional effects of IRF-1 and IRF-2 on IL-7 gene expression. To this end, a −609-Luc plasmid was cotransfected with an expression plasmid for either IRF-1 or IRF-2 into DLD-1 cells. Intriguingly, introduction of not only IRF-1 but also IRF-2 significantly enhanced the reporter activities (Fig. 6A). The effects of these IRF proteins were mediated via IRF-E, since neither IRF-1 nor IRF-2 affected gene expression from a mutant reporter plasmid (Fig. 6A). These results clearly showed that both IRF-1 and IRF-2 positively regulate expression of the IL-7 gene through IRF-E on its 5′ UTR.
FIG. 6.
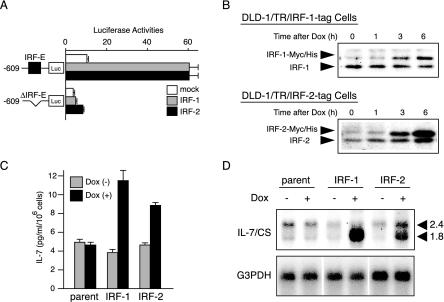
IRF-1 and IRF-2 distinctively up-regulate IL-7 protein production via IRF-E-mediated transcription. (A) Either a −609-Luc plasmid or −609-mtIRF-E-Luc was transiently transfected into DLD-1 cells with 0.1 μg of the expression vector encoding either IRF-1 or IRF-2 (pcDNA3-IRF-1 or-IRF-2) or with an empty vector (mock). Cells were then cultured for 12 h and assayed for reporter activities. Luciferase activities were normalized and indicated as the means ± standard deviations of three independent experiments. (B) DLD-1-derived cells in which either IRF-1-tag or IRF-2-tag protein is inducible upon DOX were established. DLD-1/TR/IRF-1-tag and DLD-1/TR/IRF-2-tag cells were treated with DOX (100 ng/ml) for the indicated time periods. Nuclear extracts were prepared, and 25 μg of each was subjected to immunoblot analysis by using either anti-IRF-1 antibody for DLD-1/TR/IRF-1-tag cells or anti-IRF-2 antibody for DLD-1/TR/IRF-2-tag cells, respectively. (C) Parental DLD-1 (parent), DLD-1/TR/IRF-1-tag (IRF-1) and DLD-1/TR/IRF-2-tag (IRF-2) cells were treated with 100 ng of DOX (+) per ml or left untreated (−) for 24 h, and the supernatant was assayed for IL-7 production by ELISA. Results are the means ± standard deviations of three independent experiments. (D) Parental DLD-1 (parent), DLD-1/TR/IRF-1-tag (IRF-1) and DLD-1/TR/IRF-2-tag (IRF-2) cells were treated with DOX (100 ng/ml) and then collected at the time point of the addition (−) of IFN-γ or 6 h (+) after stimulation. Poly(A)+ mRNA was extracted, and Northern blotting of IL-7 mRNA was performed as described in the legend of Fig. 2B by using a 32P-labeled CS probe.
To examine whether such transcriptional regulation leads to IL-7 protein production, we next assessed DLD-1-derived cells in which a gene encoding either IRF-1 or IRF-2 was stably transfected. Since several studies showed that IRF-1 suppresses and that IRF-2 promotes cellular proliferation, respectively (10), we employed the TET-on inducible system to achieve conditional expression of these IRF proteins, thereby excluding the possibilities that such growth-regulating functions might affect the direct effects of IRF-mediated transcription. In our system, tagged IRF proteins were expressed upon the addition of DOX, which relieved the repressive effects of TR proteins. When each clone of DLD-1/TR/IRF-1-tag or DLD-1/TR/IRF-2-tag cells was examined, each IRF protein was efficiently induced upon DOX treatment (Fig. 6B). In addition, when analyzed by transient transfection of −609-Luc, both clones displayed marked enhancement of reporter activities in response to DOX, suggesting that these tagged IRF proteins are transcriptionally competent (data not shown). When the culture supernatants of these cells before and after the DOX addition were assayed for IL-7 by ELISA, a marked induction of IL-7 proteins was observed in both DLD-1/TR/IRF-1-tag and DLD-1/TR/IRF-2-tag cells but not in parental DLD-1 cells (Fig. 6C), indicating that activation of IRF-1 or IRF-2 could induce IL-7 protein production. We further took advantages of this system to examine how inducible expression of each IRF protein influences the profile of IL-7 mRNAs. Interestingly, when Northern blotting with the IL-7 CS probe was performed, DOX-dependent IRF-1 expression exclusively induced generation of ∼1.8-kb IL-7 mRNA (Fig. 6D). In contrast, the expression of IRF-2 significantly enhanced the expression of both ∼2.4- and ∼1.8-kb IL-7 transcripts (Fig. 6D). These results not only demonstrated the up-regulatory functions of IRF-1 and IRF-2 on IL-7 gene expression but also further reinforced our hypothesis that IRF-1 is an inducible, while IRF-2 is a constitutive, regulator of IL-7 gene expression; that is, these data coincided with the observation that IFN-γ-dependent IRF-1 expression was followed by the predominant expression of ∼1.8-kb IL-7 mRNA, whereas constitutive IRF-2 expression was accompanied with the expression of both sizes of transcripts (Fig. 2B).
Inducible and constitutive IL-7 production is suppressed by siRNAs for IRF-1 and IRF-2, respectively.
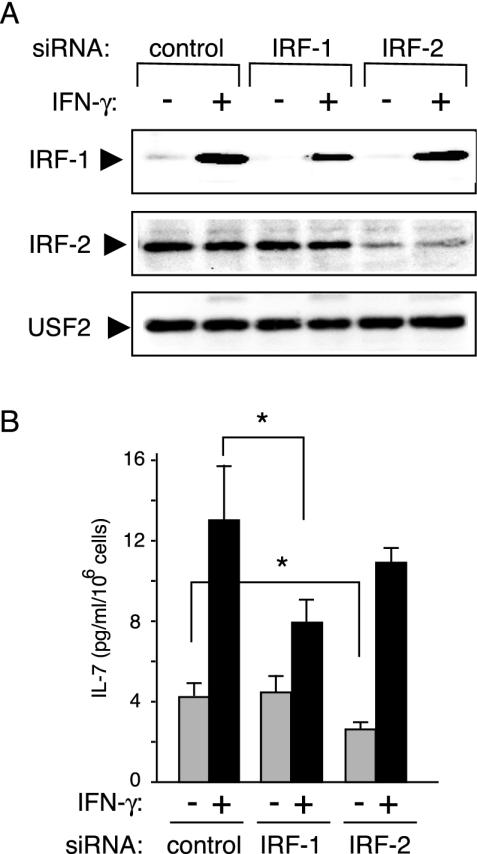
To clarify the distinct roles of IRF-1 and IRF-2 in IL-7 production directly, we designed siRNAs for either of these IRFs and then examined how these siRNAs affect IL-7 production. When DLD-1 cells were transiently transfected with either of the siRNAs, IFN-γ-dependent expression levels of IRF-1 and constitutive levels of IRF-2 were suppressed by 50 and 80% at the protein levels, respectively (Fig. 7A). In parallel, in cells treated with siRNA for IRF-1, IFN-γ-dependent IL-7 production was significantly inhibited by 50%, whereas constitutive IL-7 secretion was barely affected (Fig. 7B). In striking contrast, basal levels of IL-7 protein production in cells depleted of IRF-2 by specific siRNA were reduced by 40%, while these cells displayed an efficient induction of IL-7 secretion in response to IFN-γ (Fig. 7B). These data clearly demonstrated that IRF-1 functions as an inducible factor for IL-7 production, primarily in response to cellular stimuli such as IFN-γ, while IRF-2 predominantly serves as a factor that maintains the constitutive levels of IL-7 production.
FIG. 7.
Inducible and constitutive IL-7 production is suppressed by siRNAs for IRF-1 and IRF-2, respectively. DLD-1 cells were transfected with either siRNA duplex oligonucleotides targeting IRF-1, IRF-2, or control siRNA. After transfection, cells were cultured under the usual conditions for an additional 12 h, washed twice, and then cultured in the presence (+) or absence (−) of IFN-γ (50 ng/ml). The cells collected before (−) IFN-γ treatment and after 6 h (+) of IFN-γ treatment were subsequently subjected to immunoblotting (A) as described in the legend of Fig. 4B. In parallel, cells were identically treated as described for panel A, and the supernatants were collected after 24 h of culture in the presence (+) or the absence (−) of IFN-γ (B). Measurement of IL-7 was performed by ELISA, and results are indicated as the means ± standard deviations of three independent experiments. *, P < 0.05 by a paired Student t test.
IRF-1 and IRF-2 bind to the IRF-E without competition in vivo.
Our data showing the distinct functions of IRF-1 and IRF-2 on gene expression and the production of IL-7 raised the question of how the binding of each IRF to the IRF-E is regulated in vivo. To examine this, ChIP assays were performed with DLD-1 cell chromatin extracts. We designed two sets of PCR primers to amplify DNA fragments corresponding to the regions −539 to −159 and +976 to +1337 (positions on the genomic sequences relative to the translation start site), respectively (Fig. 8A). The former set of primers was used to amplify a fragment containing the IRF-E [IRF-E(+)] and the latter to amplify a distal genomic fragment within the intronic sequences more than 1.2 kb downstream of the IRF-E [IRF-E(-)]. As shown in Fig. 8B, anti-IRF-1 antibody precipitated only 0.03% of the IRF-E(+) DNA in total input chromatin in the absence of IFN-γ and, upon stimulation with IFN-γ, the levels of IRF-1 occupancy in this region significantly increased (0.6%) (Fig. 8B). By contrast, the binding of IRF-2 to the IRF-E remained unchanged before (0.11%) and after (0.10%) stimulation with IFN-γ (Fig. 8B). The specificity of immunoprecipitation with antibodies against each IRF protein was verified by the finding that the amounts of IRF-E(+) fragments in the immunoprecipitates by control antibodies were at significantly low levels (0.02% in both IFN-γ-untreated and -treated chromatins) (Fig. 8B). In addition, specific binding of IRF-1 and IRF-2 to the IRF-E was confirmed because immunoprecipitation of IRF-1 or IRF-2 recovered only a small amount of the genomic region located far downstream of the IRF-E [IRF-E(-)], while that of histone H3 recovered approximately 10% of the total input IRF-E(-) fragments (Fig. 8B). Since there may exist variations in cross-linking and immunoprecipitation efficiency between these IRF proteins, it seems difficult to directly compare the absolute levels of promoter occupancy of these IRFs. However, from these observations, it was formally suggested that IRF-Es on the IL-7 genes are constitutively but not fully occupied by IRF-2, regardless of the extracellular stimuli, and are further bound by IRF-1 upon stimulation with IFN-γ.
IRF-1 and IRF-2 proteins are expressed in human colonic epithelial cells with distinct patterns of distribution.
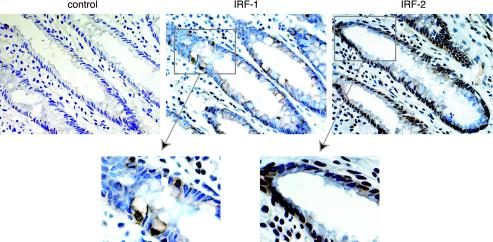
Finally, to clarify the issue as to whether IRF-1 and IRF-2 proteins are physiologically expressed in human IECs in vivo, sections of adult human colonic tissues were immunostained with a specific antibody against IRF-1 or IRF-2. As shown in Fig. 9, both IRF proteins were expressed in colonic epithelial cells, as well as in nonepithelial cells in the lamina propria (Fig. 9). Furthermore, immunoreactivities against these factors preferentially exhibited nuclear patterns, indicating that these IRF proteins function as transcriptional regulators in human IECs in vivo. Interestingly, staining with anti-IRF-2 antibody distributed throughout the epithelial layer (Fig. 9). In contrast, IRF-1 was expressed with a patchy distribution, irrespective of the cellular configuration within the crypt (Fig. 9). Remarkably, most of the IRF-1-positive cells were shown to be epithelial goblet cells, as judged by their expanded shape at the apical portion (Fig. 9). We confirmed this finding by double staining with anti-IRF-1 antibody and acidic mucus staining with alcian blue on the same section (data not shown). These results indicated that both IRF-1 and IRF-2 proteins are expressed in colonic epithelial cells with quite distinct patterns of distribution. Moreover, together with our previous demonstration that IL-7 is substantially expressed throughout the epithelial layer, with the most abundant expression in the goblet cells (30), it was suggested that these distinct patterns of distribution might be associated with the diffuse but nonuniform expression of IL-7 in human IECs in vivo.
FIG. 9.
IRF-1 and IRF-2 proteins are expressed in human colonic epithelial cells with distinct patterns of distribution. Sections of human colonic mucosal tissues were subjected to immunohistochemical analysis. Tissue sections (8 μm) were stained with either anti-IRF-1 (IRF-1), anti-IRF-2 (IRF-2) antibody, or purified rabbit IgG (control) (original magnification, ×400).
DISCUSSION
Recent evidence has implicated the profound effects of IL-7 on developing and mature lymphocytes not only in systemic (6) but also in local immune regulations in humans. However, the mechanisms of IL-7 production in human tissue-derived cells have remained unclear. In this study, using human IEC lines, we investigated the molecular mechanisms of IL-7 production and showed that IRF-1 and IRF-2 serve as critical factors for gene expression and the production of IL-7. Furthermore, IRF-1 and IRF-2 were demonstrated to play different roles in this process, suggesting that the IL-7 production might be regulated via finely coordinated mechanisms mediated by these IRF proteins.
Concerning the potentials of various cellular stimuli to influence IL-7 production, IL-1, TNF-α, and TGF-β had no effect and only IFN-γ was capable of regulating IL-7 production from IECs. These findings contrasted to results obtained with other tissue-derived human cells, since previous studies revealed that IL-1 and TNF-α enhance (34), while TGF-β suppresses IL-7 production in BM stromal cells (27). It seems unlikely that human IECs failed to respond to IL-1, TNF-α, or TGF-β merely due to low expression levels of the specific receptors for each factor, because most of these cytokines were proved to induce multiple biological responses within the human IEC lines or their sublines examined in this study (4, 33, 35). In addition, because the influences of these cytokines on IL-7 expression in human IECs were quite similar to those observed in murine keratinocytes (3), we favor the idea that IL-7 production in human IECs may be regulated by a tissue-specific mechanism which differs at least from that in human BM stromal cells but resembles that in murine keratinocytes. Although the mechanisms accounting for the diversity of IL-7 production in different cell types have remained unknown, it would be of importance to clarify this issue to understand a variety of biological functions exerted by IL-7 in systemic immune regulation in humans.
In this study, we demonstrated that the transcription of the human IL-7 gene begins from multiple sites distributed within two separate regions at positions −601 to −515 and −197 to −131 bp upstream of the translation start site. Utilization of many transcription start sites is frequently observed in the regulation of genes whose promoters lack common core promoter sequences (26). Consistent with this, analysis of the human IL-7 gene revealed that none of the consensus sequences for the canonical TATA box, the initiator element YYAN(T/A)YY, or the downstream core promoter element occur within or in the vicinity of the region −601 to +1 (Fig. 3B). Instead, as initially documented in an earlier report (18), the 5′ flanking region of the human IL-7 gene displays an unusually high number of CpG dinucleotides within a ∼700-bp region. This is also in accordance with the fact that a number of promoters within CpG islands lack all these classes of core elements (26). In addition, recent studies identified a new class of promoter motif on several genes that utilize multiple start sites in their TATA-less promoters. This motif, called MED-1 (multiple start site element downstream), was defined as the sequence GCTCC(C/G) and was shown to lie 20 to 45 bp downstream of the multiple transcription initiation window of various TATA-less promoters (11). Interestingly, an identical sequence to MED-1 occurs on the human IL-7 gene at position −498 (Fig. 3B), 17 to 103 bp downstream of one of two separate windows of the IL-7 gene transcription start sites. The functional relevance of these promoter structures to the start site selection of the IL-7 gene has remained undetermined; however, our work provides evidence that the expression of human IL-7 is regulated through a unique and unusual promoter architecture.
In addition to the promoter structure, unusual features of regulatory mechanisms were also found in IL-7 gene transcription. We showed that, by use of TET-inducible expression systems for each IRF protein, IRF-1 selectively induces transcription of the IL-7 gene from the relatively downstream region, while IRF-2 up-regulates transcription from two regions both upstream and downstream of IRF-E. These results suggest that utilization of the aforementioned two separate promoters is regulated by distinct as well as overlapping properties of IRF-1 and IRF-2 via binding to the same IRF-E. Recently, growing evidence has revealed the existence of alternative promoters on various human genes, suggesting the regulatory roles of alternative promoter usages in tissue-specific or developmentally controlled gene expression (15). Among these, however, the mechanism of IL-7 gene regulation seems to be unique, because there has been no report of such a gene whose alternative promoter usage is regulated by the differential binding of transcription factors to a single cis-element so far. At this time, it remains unclear why IRF-2 simultaneously promotes transcription from two regions but IRF-1 does not. One possible explanation for the IRF-2-mediated dual promoter usage is that the binding of IRF-2 to the IRF-E might alter the chromatin architecture to a more relaxed configuration than that of IRF-1. For example, it was shown that the proto-oncogene c-myc is transcribed from two distinct promoters that are located 160 bp apart, and an element called ME1a1, located between these two promoters, is required for the simultaneous opening of the chromatin configuration for both promoters (1). Therefore, it could be speculated that the IRF-E, like ME1a1 on the c-myc gene, might allow the transcription from two promoters only when it is bound by IRF-2 in conjunction with the specifically assembled transcription machinery.
Despite the alternative promoter usage, no variation in the resulting IL-7 proteins has been reported. Interestingly, as is the case with the murine IL-7 transcripts (8), human transcripts contain multiple potential initiation codons (nine in total throughout the −601 to −1 region) upstream from the authentic initiation codon (Fig. 3B). As these potential upstream sites generally decrease the translational efficiency (14) and as the removal of the 5′ noncoding sequence improves the translational efficiency of murine IL-7 mRNA (23), it is postulated that the human IL-7 transcripts with a shorter 5′ UTR might also be translationally more active than a transcript with a longer one.
In the present study, we demonstrated the physiological roles of IRF-1 and IRF-2 in IL-7 production by human IECs. IRF-1, originally identified as a transcriptional regulator for the human IFN-β gene (19), is induced upon various stimuli and activates target gene expression (9, 19). Likewise, we here showed that nuclear expression of IRF-1 and its binding to the IRF-E were also induced upon IFN-γ treatment in human IECs. These data recapitulated the mechanisms existing in murine keratinocytes, since it was shown that IFN-γ-dependent IL-7 gene expression was preceded by increased binding of IRF-1 to the IRF-E in these cells, and the inhibition of IRF-1 mRNA expression by UV light suppressed this IFN-γ-inducible IL-7 expression (2). In addition, our observations of DOX-inducible expression and siRNA-mediated suppression of IRF-1 were of importance, since these confirmed that such IRF-1-mediated transcription is indeed important for the inducible production of IL-7 protein. IRF-1 protein is substantially expressed with a patchy distribution in normal human intestinal epithelia, and the predominant expression of IRF-1 in goblet cells was consistent with the fact that relatively abundant expression of IL-7 is observed in these cells (30). Considering that IFN-γ is constitutively expressed in IELs (16), IRF-1 expression in vivo might be the result of the IFN-γ action that is locally produced from a certain type of cells such as IELs. Alternatively, expression of IRF-1 might be additionally regulated by stimuli other than IFN-γ, since IRF-1 is up-regulated by a variety of cellular stimuli such as various cytokines (7) and viral infection (19). At present, it is unclear which type of stimuli is responsible for IRF-1 expression in vivo and why goblet cells are prone to express IRF-1 proteins. However, our study provides not only the molecular basis accounting for the cell type-dependent variable expression of IL-7 in human IECs but also a clue for the better understanding of a well-coordinated network system within the intestinal mucosa: IL-7 production from IECs is regulated by sensing and responding to the immunological status such as the microenvironmental cytokine milieu, primarily utilizing IRF-1 as a transcriptional activator.
Although IRF-2 was originally described as a transrepressor with its potential for competing with IRF-1 (9), studies have shown that IRF-2 also functions as a transcriptional activator for several genes (12, 17, 28, 36). We here demonstrated that IRF-2 also acts as a transactivator for the IL-7 gene, but its up-regulatory functions in the production of IL-7 is quite different from those of IRF-1. Silencing IRF-2 expression by its specific siRNA resulted in suppression not of the IFN-γ-inducible but of the basal levels of IL-7 production, and DOX-regulated expression of IRF-2 enhanced IL-7 protein production via expression of both ∼2.4- and ∼1.8-kb IL-7 mRNAs that are constitutively expressed. Concerning the fact that IRF-2 was ubiquitously expressed throughout the epithelial layer of human colonic tissues, these findings strongly suggest that IRF-2, in contrast to IRF-1, serves as a critical regulator for IL-7 production from wide-ranging areas of human IECs in vivo. Furthermore, it was previously shown that IRF-1 is a short-lived protein with a half-life of about 30 min, while IRF-2 protein has a relatively longer half-life of more than 8 h (32). Given this, we may postulate that IRF-1 might act as a transient regulator of IL-7 production in response to cellular stimuli and, in contrast, that IRF-2 might serve as a critical factor to ensure the basal and steady-state levels of IL-7 production, not only at the cellular level but also in the tissue configuration within human intestinal mucosa.
We have previously reported that intestinal inflammation occurred in IL-7 transgenic mice (31). These mice spontaneously developed acute colitis at 1 to 3 weeks of age, which was followed by a chronic phase of colitis that histopathologically mimicked the human inflammatory bowel diseases. In these diseased mice, expression of IL-7 was increased in the acute phase while it was decreased in the chronic phase of colitis at the sites of inflammation (31). These results suggested that aberrant production of IL-7 might directly lead to the dysregulation of the local immune network in the intestinal mucosa. Based on these observations, the present study also raises the issue of a potential role of IRF proteins in human diseases such as inflammatory bowel disease. We indicated that physiological expression of IRF-1 in vivo was dominated in epithelial goblet cells, depletion of which is one of the most prominent features of ulcerative colitis in humans. Meanwhile, it is well known that the inflamed mucosa in Crohn's disease exhibit increased levels of proinflammatory cytokines including IFN-γ (25). Therefore, it is possible for us to speculate that functional alteration or a decrease in the number of goblet cells in ulcerative colitis or the altered local cytokine milieu in Crohn's disease might lead to the escape from appropriate function or expression of these IRF proteins in IECs, linking improper production of IL-7 to the dysregulation of mucosal lymphocytes within the sites of inflammation.
In summary, we here show that IRF-1 and IRF-2 serve as activators for IL-7 gene expression and protein production, while their respective roles are quite different. These results not only provide a molecular basis for understanding the profound functions of IEC-derived IL-7 in human intestinal mucosa but also suggest that the functional interplay between IRF-1 and IRF-2 is an exquisite mechanism that regulates the timely as well as continuous production of IL-7. We believe that the present work raises several interesting issues for further studies on the biological and pathological significance of these IRF proteins, especially in human IECs, in terms of their relationship with the pleiotropic functions of IL-7 in intestinal immune regulation.
Acknowledgments
This study was supported in part by grants-in-aid for Scientific Research, Scientific Research on Priority Areas, Exploratory Research, and Creative Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science and Technology; the Japanese Ministry of Health, Labor and Welfare; the Japan Medical Association; the Foundation for Advancement of International Science; Terumo Life Science Foundation; Ohyama Health Foundation; Yakult Bio-Science Foundation; and the Research Fund of Mitsukoshi Health and Welfare Foundation.
REFERENCES
- 1.Albert, T., J. Wells, J. O. Funk, A. Pullner, E. E. Raschke, G. Stelzer, M. Meisterernst, P. J. Farnham, and D. Eick. 2001. The chromatin structure of the dual c-myc promoter P1/P2 is regulated by separate elements. J. Biol. Chem. 276:20482-20490. [DOI] [PubMed] [Google Scholar]
- 2.Aragane, Y., A. Schwarz, T. A. Luger, K. Ariizumi, A. Takashima, and T. Schwarz. 1997. Ultraviolet light suppresses IFN-gamma-induced IL-7 gene expression in murine keratinocytes by interfering with IFN regulatory factors. J. Immunol. 158:5393-5399. [PubMed] [Google Scholar]
- 3.Ariizumi, K., Y. Meng, P. R. Bergstresser, and A. Takashima. 1995. IFN-gamma-dependent IL-7 gene regulation in keratinocytes. J. Immunol. 154:6031-6039. [PubMed] [Google Scholar]
- 4.Bartke, T., D. Siegmund, N. Peters, M. Reichwein, F. Henkler, P. Scheurich, and H. Wajant. 2001. p53 upregulates cFLIP, inhibits transcription of NF-kappaB-regulated genes and induces caspase-8-independent cell death in DLD-1 cells. Oncogene 20:571-580. [DOI] [PubMed] [Google Scholar]
- 5.Bilenker, M., A. I. Roberts, R. E. Brolin, and E. C. Ebert. 1995. Interleukin-7 activates intestinal lymphocytes. Dig. Dis. Sci. 40:1744-1749. [DOI] [PubMed] [Google Scholar]
- 6.Fry, T. J., and C. L. Mackall. 2002. Interleukin-7: from bench to clinic. Blood 99:3892-3904. [DOI] [PubMed] [Google Scholar]
- 7.Fujita, T., L. F. Reis, N. Watanabe, Y. Kimura, T. Taniguchi, and J. Vilcek. 1989. Induction of the transcription factor IRF-1 and interferon-beta mRNAs by cytokines and activators of second-messenger pathways. Proc. Natl. Acad. Sci. USA 86:9936-9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodwin, R. G., S. Lupton, A. Schmierer, K. J. Hjerrild, R. Jerzy, W. Clevenger, S. Gillis, D. Cosman, and A. E. Namen. 1989. Human interleukin 7: molecular cloning and growth factor activity on human and murine B-lineage cells. Proc. Natl. Acad. Sci. USA 86:302-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harada, H., T. Fujita, M. Miyamoto, Y. Kimura, M. Maruyama, A. Furia, T. Miyata, and T. Taniguchi. 1989. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell 58:729-739. [DOI] [PubMed] [Google Scholar]
- 10.Harada, H., M. Kitagawa, N. Tanaka, H. Yamamoto, K. Harada, M. Ishihara, and T. Taniguchi. 1993. Anti-oncogenic and oncogenic potentials of interferon regulatory factors-1 and -2. Science 259:971-974. [DOI] [PubMed] [Google Scholar]
- 11.Ince, T. A., and K. W. Scotto. 1995. A conserved downstream element defines a new class of RNA polymerase II promoters. J. Biol. Chem. 270:30249-30252. [DOI] [PubMed] [Google Scholar]
- 12.Jesse, T. L., R. LaChance, M. F. Iademarco, and D. C. Dean. 1998. Interferon regulatory factor-2 is a transcriptional activator in muscle where it regulates expression of vascular cell adhesion molecule-1. J. Cell Biol. 140:1265-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanamori, Y., K. Ishimaru, M. Nanno, K. Maki, K. Ikuta, H. Nariuchi, and H. Ishikawa. 1996. Identification of novel lymphoid tissues in murine intestinal mucosa where clusters of c-kit+ IL-7R+ Thy1+ lympho-hemopoietic progenitors develop. J. Exp. Med. 184:1449-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozak, M. 1984. Selection of initiation sites by eucaryotic ribosomes: effect of inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Res. 12:3873-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landry, J. R., D. L. Mager, and B. T. Wilhelm. 2003. Complex controls: the role of alternative promoters in mammalian genomes. Trends Genet. 19:640-648. [DOI] [PubMed] [Google Scholar]
- 16.Lundqvist, C., S. Melgar, M. M. Yeung, S. Hammarstrom, and M. L. Hammarstrom. 1996. Intraepithelial lymphocytes in human gut have lytic potential and a cytokine profile that suggest T helper 1 and cytotoxic functions. J. Immunol. 157:1926-1934. [PubMed] [Google Scholar]
- 17.Luo, W., and D. G. Skalnik. 1996. Interferon regulatory factor-2 directs transcription from the gp91phox promoter. J. Biol. Chem. 271:23445-23451. [DOI] [PubMed] [Google Scholar]
- 18.Lupton, S. D., S. Gimpel, R. Jerzy, L. L. Brunton, K. A. Hjerrild, D. Cosman, and R. G. Goodwin. 1990. Characterization of the human and murine IL-7 genes. J. Immunol. 144:3592-3601. [PubMed] [Google Scholar]
- 19.Miyamoto, M., T. Fujita, Y. Kimura, M. Maruyama, H. Harada, Y. Sudo, T. Miyata, and T. Taniguchi. 1988. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-beta gene regulatory elements. Cell 54:903-913. [DOI] [PubMed] [Google Scholar]
- 20.Monteleone, G., T. Parrello, F. Luzza, and F. Pallone. 1998. Response of human intestinal lamina propria T lymphocytes to interleukin 12: additive effects of interleukin 15 and 7. Gut 43:620-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore, T. A., U. von Freeden-Jeffry, R. Murray, and A. Zlotnik. 1996. Inhibition of gamma delta T cell development and early thymocyte maturation in IL-7 −/− mice. J. Immunol. 157:2366-2373. [PubMed] [Google Scholar]
- 22.Nakamura, T., R. Ouchida, T. Kodama, T. Kawashima, Y. Makino, N. Yoshikawa, S. Watanabe, C. Morimoto, T. Kitamura, and H. Tanaka. 2002. Cytokine receptor common beta subunit-mediated STAT5 activation confers NF-kappa B activation in murine proB cell line Ba/F3 cells. J. Biol. Chem. 277:6254-6265. [DOI] [PubMed] [Google Scholar]
- 23.Namen, A. E., S. Lupton, K. Hjerrild, J. Wignall, D. Y. Mochizuki, A. Schmierer, B. Mosley, C. J. March, D. Urdal, and S. Gillis. 1988. Stimulation of B-cell progenitors by cloned murine interleukin-7. Nature 333:571-573. [DOI] [PubMed] [Google Scholar]
- 24.Orlando, V. 2000. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 25:99-104. [DOI] [PubMed] [Google Scholar]
- 25.Podolsky, D. K. 2002. Inflammatory bowel disease. N. Engl. J. Med. 347:417-429. [DOI] [PubMed] [Google Scholar]
- 26.Smale, S. T., and J. T. Kadonaga. 2003. The RNA polymerase II core promoter. Annu. Rev. Biochem. 72:449-479. [DOI] [PubMed] [Google Scholar]
- 27.Tang, J., B. L. Nuccie, I. Ritterman, J. L. Liesveld, C. N. Abboud, and D. H. Ryan. 1997. TGF-beta down-regulates stromal IL-7 secretion and inhibits proliferation of human B cell precursors. J. Immunol. 159:117-125. [PubMed] [Google Scholar]
- 28.Vaughan, P. S., F. Aziz, A. J. van Wijnen, S. Wu, H. Harada, T. Taniguchi, K. J. Soprano, J. L. Stein, and G. S. Stein. 1995. Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature 377:362-365. [DOI] [PubMed] [Google Scholar]
- 29.von Freeden-Jeffry, U., P. Vieira, L. A. Lucian, T. McNeil, S. E. Burdach, and R. Murray. 1995. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J. Exp. Med. 181:1519-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe, M., Y. Ueno, T. Yajima, Y. Iwao, M. Tsuchiya, H. Ishikawa, S. Aiso, T. Hibi, and H. Ishii. 1995. Interleukin 7 is produced by human intestinal epithelial cells and regulates the proliferation of intestinal mucosal lymphocytes. J. Clin. Invest. 95:2945-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe, M., Y. Ueno, T. Yajima, S. Okamoto, T. Hayashi, M. Yamazaki, Y. Iwao, H. Ishii, S. Habu, M. Uehira, H. Nishimoto, H. Ishikawa, J. Hata, and T. Hibi. 1998. Interleukin 7 transgenic mice develop chronic colitis with decreased interleukin 7 protein accumulation in the colonic mucosa. J. Exp. Med. 187:389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe, N., J. Sakakibara, A. G. Hovanessian, T. Taniguchi, and T. Fujita. 1991. Activation of IFN-beta element by IRF-1 requires a posttranslational event in addition to IRF-1 synthesis. Nucleic Acids Res. 19:4421-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weaver, S. A., M. P. Russo, K. L. Wright, G. Kolios, C. Jobin, D. A. Robertson, and S. G. Ward. 2001. Regulatory role of phosphatidylinositol 3-kinase on TNF-alpha-induced cyclooxygenase 2 expression in colonic epithelial cells. Gastroenterology 120:1117-1127. [DOI] [PubMed] [Google Scholar]
- 34.Weitzmann, M. N., S. Cenci, L. Rifas, C. Brown, and R. Pacifici. 2000. Interleukin-7 stimulates osteoclast formation by up-regulating the T-cell production of soluble osteoclastogenic cytokines. Blood 96:1873-1878. [PubMed] [Google Scholar]
- 35.Wilson, L., C. Szabo, and A. L. Salzman. 1999. Protein kinase C-dependent activation of NF-kappaB in enterocytes is independent of IkappaB degradation. Gastroenterology 117:106-114. [DOI] [PubMed] [Google Scholar]
- 36.Xi, H., B. Goodwin, A. T. Shepherd, and G. Blanck. 2001. Impaired class II transactivator expression in mice lacking interferon regulatory factor-2. Oncogene 20:4219-4227. [DOI] [PubMed] [Google Scholar]
- 37.Yokota, T., N. Sakamoto, N. Enomoto, Y. Tanabe, M. Miyagishi, S. Maekawa, L. Yi, M. Kurosaki, K. Taira, M. Watanabe, and H. Mizusawa. 2003. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Rep. 4:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]