Abstract
To understand the mechanisms involved in the transition from protists to multicellular animals (metazoans), studying unicellular relatives of metazoans is as important as studying metazoans themselves. However, investigations remain poor on the closest unicellular (or colonial) relatives of Metazoa, i.e., choanoflagellates, filastereans and ichthyosporeans. Molecular-level analyses on these protists have been severely limited by the lack of transgenesis tools. Their genomes, however, contain several key genes encoding proteins important for metazoan development and multicellularity, including those involved in cell–cell communication, cell proliferation, cell differentiation, and tissue growth control. Tools to analyze their functions in a molecular level are awaited. Here we report techniques of cell transformation and gene silencing developed for the first time in a close relative of metazoans, the ichthyosporean Creolimax fragrantissima. We propose C. fragrantissima as a model organism to investigate the origin of metazoan multicellularity. By transgenesis, we demonstrate that its colony develops from a fully-grown multinucleate syncytium, in which nuclear divisions are strictly synchronized. It has been hypothesized that metazoan multicellular development initially occurred in the course of evolution through successive rounds of cell division, which were not necessarily be synchronized, or alternatively through cell aggregation. Our findings point to another possible mechanism for the evolution of animal multicellularity, namely, cellularization of a syncytium in which nuclear divisions are synchronized. We believe that further studies on the development of ichthyosporeans by the use of our methodologies will provide novel insights into the origin of metazoan multicellularity.
Keywords: Evolution of multicellularity, Creolimax fragrantissima, Syncytium, Nuclear division, Transgenesis, Gene silencing
Introduction
The acquisition of multicellularity was a crucial step in the evolution of animals (or metazoans), allowing the development of highly specialized cells that interact with each other (Bonner, 1998; Grosberg and Strathmann, 2007). This led to a variety of complex and mobile forms that we see now in extant animals. It has been hypothesized that the metazoan multicellular development initially occurred in the course of evolution through serial incomplete cell divisions, or alternatively through cell aggregation (Fairclough et al., 2010; Grosberg and Strathmann, 2007; Willmer, 1990). The striking morphological resemblance between the colonies of choanoflagellates, the closest relatives of metazoans, and the choanocyte chamber of sponges, the most basal metazoan lineage, has for example lent a support to such hypothesis (Fairclough et al., 2010; Willmer, 1990). Another hypothesis claims that, as seen in some species of slime molds and fungi, the ancestral metazoan cell formed syncytium, where nuclear division occurred without cell division. It would then cellularize by generating a membrane around each nucleus, forming a colony-like structure (Bonner, 1998; Grosberg and Strathmann, 2007; Willmer, 1990). In the Metazoa, this strategy is typically seen in insect embryogenesis, where a syncytium (or coenocyte) cellularizes prior to the blastoderm formation. Multi-nucleate cells formed through cell fusion are more often seen and play crucial roles during metazoan development, for instance, in various muscle tissues (Chen et al., 2007), nematode hypodermis (Sommer, 2000), and the large multinucleate tissues of glass sponge embryos (Leys et al., 2006).
At a molecular level, the metazoan multicellularity and their developmental processes are enabled by several elaborate mechanisms such as cell–cell communication, cell proliferation, cell differentiation, and tissue growth control. These mechanisms have been mainly studied by the use of metazoan models. However, the investigations only on metazoans do not answer the question on how the metazoan multicellularity initially evolved. To elucidate this key evolutionary transition, studies on the extant closest relatives of metazoans, which can be unicellular or colonial, is essential. Such group of protists includes choanoflagellates, filastereans, and ichthyosporeans (or mesomycetozoans) (Carr et al., 2008; Lang et al., 2002; Ruiz-Trillo et al., 2008; Shalchian-Tabrizi et al., 2008; Steenkamp and Baldauf, 2004; Torruella et al., 2011). Understanding the developmental mechanism of the colony formation on these closest relatives of metazoans should provide some insights into the origin of metazoan multicellularity (Dayel et al., 2011). For instance, studies on the colony-forming choanoflagellate Salpingoeca rosetta have shown that its colony formation is carried out only through cell divisions that are not synchronized, triggered by a sphingolipid-like molecule secreted by the prey bacteria (Alegado et al., 2012; Fairclough et al., 2010).
Choanoflagellates are excellent models for the study of multicellularity evolution in the context of their closer relationship to metazoans than any other pre-metazoan lineage and their characteristic morphology reminiscent of sponge choanocytes (King, 2004). However, the traditional hypothesis that an ancestral choanoflagellates-like protist gave rise to the choanocyte-bearing metazoan ancestor has been challenged by the presence of choanocyte-like cells in non-sponge metazoans (Cantell et al., 1982; Lyons, 1973; Nerrevang and Wingstrand, 1970). This hypothesis is also contradicted by some molecular and histological evidence indicating that sponge choanocytes are rather specialized cells that arise from non-collared cells during embryogenesis (Maldonado, 2005). It is thus possible that the evolution of metazoan multicellularity may not be best shaped by the colonial choanoflagellates.
More importantly, the lack of methodologies for functional assays, e.g. cell transformation and gene silencing, limits further investigation on choanoflagellates at a molecular level. Recent analyses on the genomes of the unicellular or colonial relatives of metazoans have shown that they contain several genes encoding key proteins to the metazoan multicellularity and development, such as the cell adhesion proteins cadherins and integrins, the cell–cell communication and differentiation-controlling proteins tyrosine kinases, a variety of transcription factors, and the organ growth-controlling pathway (King et al., 2008; Manning et al., 2008; Nichols et al., 2012; Sebé-Pedrós et al., 2010, 2011, 2012; Suga et al., 2008, 2012). However, the functions of these proteins in the non-metazoan lineages remain unclear. It is therefore critical to establish, among the closest metazoan relatives, new model organisms where functional molecular assays are possible. We here propose the ichthyosporean C. fragrantissima as a model organism to understand the origin of Metazoa, demonstrating the possibility of functional assays on it by transgenesis and gene silencing.
Materials and methods
Cultures of ichthyosporeans
Live cultures of C. fragrantissima (Marshall et al., 2008) and S. arctica (Jøstensen et al., 2002) were kindly provided by Wyth Marshall (University of British Columbia, Vancouver, Canada) and by Bjarne LandFald (University of Tromsø, Tromsø, Norway), respectively. They were maintained at 12 1C in the MAP medium (18.6 g/L Difco marine broth 2166 (BD), 20 g/L Bacto peptone (BD) and 10 g/L NaCl).
DNA constructs
C. fragrantissima and S. arctica histone H2B sequences were obtained from their draft genome sequences. C. fragrantissima genomic fragment containing β-tubulin gene (Cfrβtub) was cloned according to GenomeWalker (Clontech) protocol. 972 bp of the upstream sequence and 493 bp of the downstream sequence of the coding region were combined with Venus (RIKEN Brain Science Institute) yellow fluorescent protein by PCR, and cloned into pBluescript (Agilent Technologies). To examine the movement of nuclei in live cells, we made another construct by combining the Cfrβtub promoter and terminator with the coding sequence of S. arctica histone H2B (SarH2B) fused to mCherry (Clontech) red fluorescent protein. A similar construct was made by the use of histone H2B2 of C. fragrantissima (CfrH2B2), which is one of the two duplicated paralogs found in the draft genome sequence. These fusion proteins were designated SarH2B-mCherry and CfrH2B2-mCherry, respectively. The sequences newly identified in this study are deposited in DNA Data Bank of Japan (DDBJ) with the accession numbers AB636312, AB636313, AB666311, and AB666312.
Electroporation and live cell imaging
Using Neon (Life Technologies), we applied two pulses of 1000 V (33 V/mm) for 30 ms in 10 μL tip with R buffer (Life Technologies). Two DNA constructs expressing Venus and histone-mCherry fusion protein (80 ng/μl each) were co-electroporated, and then cells were directly transferred into the MAP medium and incubated overnight at 12 °C. All examined Venus-positive cells expressed also mCherry, thus we used the Venus signal as a transgenesis marker. The transformants were transferred to a glass bottom dish (MatTek) and time-lapse imaging was performed in the MAP medium with AF7000 fluorescence microscope (Leica) and Temperable Insert P Lab-Tek (PeCon), which keeps the live specimen at the appropriate temperature. The images were processed with ImageJ program (http://imagej.nih.gov/ij/).
Gene silencing
siRNAs (Sigma) were designed from the coding sequence of mCherry and that of S. arctica H2B (designated siRNA-mCherry and siRNA–SarH2B, respectively). Their concentrations were adjusted at the maximum available (200 μM) and 1 μL was mixed with 10 μL cell suspension immediately before electroporation.
A Morpholino antisense oligo (Gene Tools) was designed on the basis of the SarH2B coding sequence including the start codon and two bases prior to it. We determined the position of the Morpholino target sequence such that it includes as small 5′ UTR sequence as possible. By this strategy, we eliminated the possible influence to the translation of endogenous β-tubulin genes and to that of the transgenesis marker Venus, in both of which the same promoter sequence (C. fragrantissima β-tubulin promoter) as that of the Morpholino target construct is used. However, the native SarH2B sequence was not optimum for designing Morpholino at this location because of its biased nucleotide composition. We thus modified the SarH2B sequence at two nucleotide positions (described in the Results section) and designed Morpholino against this modified target sequence. These modifications do not affect the mCherry signal at nuclei (see the Results section). The modified sequence is called SarH2Bm (S. arctica H2B modified). Accordingly, the Morpholino oligo is called MO-SarH2Bm.
The sequences of siRNAs and Morpholino are in Table 1. Note that neither the siRNA nor the Morpholino were designed from the sequences of C. fragrantissima genes in order to avoid knocking down their native expression from the nuclear genome. After electroporation, we measured the ratio of fluorescence intensity of mCherry to that of Venus for all transformants found under the microscope.
Table 1.
siRNA and Morpholino sequences.
| Molecule | Target | Strand | Sequence |
|---|---|---|---|
| siRNA | mCherry | Sense | 5′-CAUGGCCAUCAUCAAGGAG[dT][dT]-3′ |
| Antisense | 5′-CUCCUUGAUGAUGGCCAUG[dT][dT]-3′ | ||
| siRNA | S. arctica | Sense | 5′-CUCGUUCGUUAUUGACGUG[dT][dT]-3′ |
| H2B | Antisense | 5′-CACGUCAAUAACGAACGAG[dT][dT]-3′ | |
| Morpholino | S. arctica | Antisense | 5′-CTTGATGGTACTTTAGGATGCATGA-3′ |
| H2Bm |
Immunostaining
Cells were fixed by 4% paraformaldehyde freshly prepared in artificial sea water for 10 min. After washing with PBS, they were permeabilized by rapid freeze–thaw procedure performed twice with liquid nitrogen. This permeabilization step is unnecessary for staining only with DAPI. Cells were incubated with primary antibodies (rabbit anti-actin; Sigma A-2066, and mouse anti-α-tubulin; Sigma T-9026) for overnight at 4 °C. and then, after washing, with secondary antibodies (anti-mouse Alexa Fluor 488 and anti-rabbit Alexa Fluor 568; Invitrogen) for 1 h at room temperature. After the secondary antibodies were washed off, DAPI (Roche) was applied and washed two more times. Cells were then mounted with Fluorescence Mounting Medium (Dako) and observed under TCS SP5 confocal microscope (Leica). The images were processed with the ImageJ program.
Results
Development of C. fragrantissima
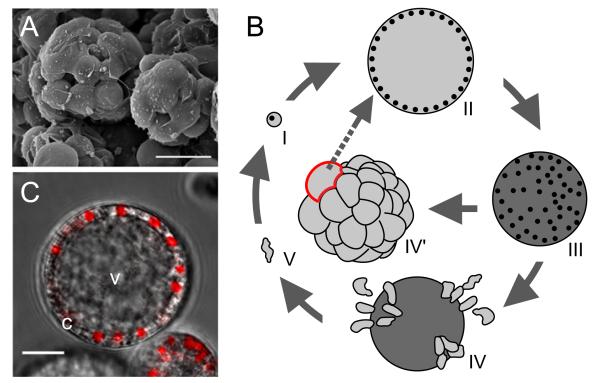
The ichthyosporean C. fragrantissima (Fig. 1A) spends a unique lifecycle starting from a minute uninucleate cell of 6–8 μm in diameter (Marshall et al., 2008) (Fig. 1B). The cell matures after growing to 30–70 μm in diameter, producing numerous uninucleate motile amoebae in the mother cell. The amoebae then migrate out through tears of the cell wall. The maturation step is a dynamic process taking around 2–3 h, including rapid rearrangement of the cytoplasm and amoeba formation (Video 1). The amoeba settles and encysts after various periods of traveling (Video 2). When the culture matures, the amoebae usually stay and encyst within the mother cell and large colonies reminiscent of metazoan blastula embryos become prominent (Fig. 1A). A C. fragrantissima growth-stage cell is a syncytium, containing multiple nuclei in a single cytoplasm (Marshall et al., 2008). The nuclei are always arranged near the cell surface in the growth stage, and usually one large vacuole occupies the central cavity (Fig. 1C).
Fig. 1.
Creolimax fragrantissima-an emerging metazoan-relative model. (A) Scanning electron microscopy of a mature C. fragrantissima culture (Photo courtesy of Arnau Sebé-Pedrós). (B) Lifecycle of C. fragrantissima, comprising growth (I–II), maturation (II–III), dissemination (IV), colonial (IV’) and amoeboid (V) stages. Nuclei (black dots) are depicted for I–III. (C) Nuclear staining with DAPI (red) overlaid on bright field image of a growth-stage cell of C. fragrantissima. c—cytoplasm. v—vacuole.
Transformation of C. fragrantissima by electroporation
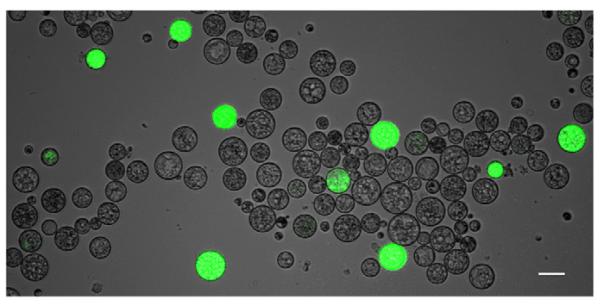
We succeeded in transforming C. fragrantissima by electroporating DNA constructs (Fig. 2). A capillary tip with a wire-type electrode, which applies a uniform electric field to the sample (Kim et al., 2008), was used for the electroporation. The conventional cuvette-based electroporation chamber did not yield any positive result even when a similar electric field condition was applied. The transformation efficiency was around 7% at maximum. No stable transformant was obtained. However, the short generation time of C. fragrantissima (~2 days) allowed the expressed protein to be maintained until after the next generation starts (Video 3).
Fig. 2.
C. fragrantissima transgenesis. Cells were observed approximately at 16 h after electroporation with the marker construct Cfrβtub:Venus, which expresses Venus yellow fluorescent protein in the cytoplasm. The bright field image and the fluorescence microscopic image (in green channel) are merged. Scale bar: 30 μm.
Synchronized nuclear division in a growing syncytium prior to the colony formation in ichthyosporeans
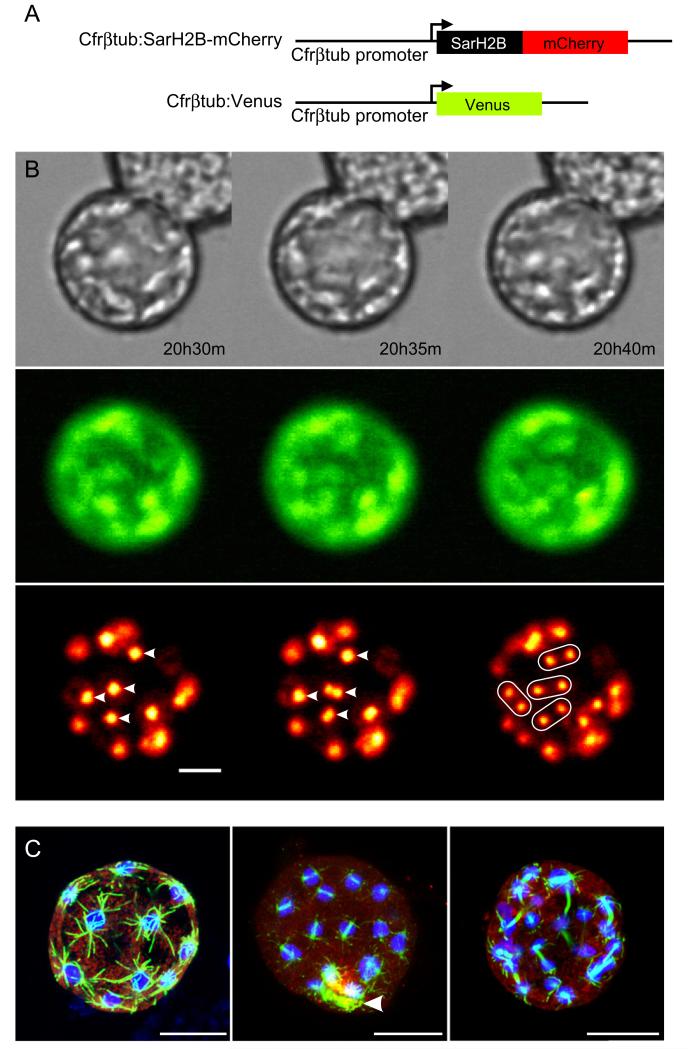
Using the developed transformation technique, we traced the movement of nuclei in a C. fragrantissima cell in the growth stage. We transformed the cells with a DNA construct (Fig. 3A), in which C. fragrantissima β-tubulin (Cfrβtub) promoter was combined with the histone H2B (SarH2B) coding sequence of Sphaeroforma arctica, another ichthyosporean (Jøstensen et al., 2002). SarH2B was tagged with mCherry red fluorescent protein (the expressed fusion protein is thus called SarH2B-mCherry). Another construct that expresses Venus yellow fluorescent protein by the same promoter was co-electroporated as a transformation marker. The obtained transformants visualized nuclear movement and division in live cells. Interestingly, the time-lapse movie reveals that the nuclei divide strictly synchronously (Video 4 and Fig. 3B).
Fig. 3.
C. fragrantissima transformation reveals the synchronized nuclear division. (A) Schemes of constructs. (B) Stills of time-lapse movie showing synchronized nuclear divisions in a live growth-stage transformant. Bright field images (top), fluorescence in cytoplasm from the Venus protein (middle), and that in nuclei from mCherry protein (bottom), which are both expressed by the C. fragrantissima β-tubulin (Cfrβtub) promoter, are shown. Four examples of synchronized nuclear divisions are highlighted by arrowheads and ovals. Time from the start of the movie is shown. Full movie available in Video 4. (C) Synchronized nuclear division of S. arctica suggested by antibody staining. Nuclei (blue: stained with DAPI), spindle bodies (green: α-tubulin antibody) and cytoplasm (red: actin antibody) of three S. arctica growth stage cells were stained. Z-axis projection was applied to each hemisphere. An artifact signal present at a breakage on the cell wall (arrowhead), which was introduced by the permeabilization step of the protocol. Scale bar: 10 μm.
To confirm this in other ichthyosporean species, we examined the subcellular structures of S. arctica in its growth stage by antibody staining. The lifecycle of S. arctica is very similar to that of C. fragrantissima, but lacks the amoeboid stage (Jøstensen et al., 2002). The structures of spindle bodies, which represent the stages of nuclear division, indicate that the nuclei also divide synchronously in this ichthyosporean species (Fig. 3C)
Gene silencing by RNA interference and Morpholino
Gene silencing (or knockdown) by the mechanism of RNA interference (RNAi) or by blocking translation with antisense oligonucleotides such as Morpholino is a powerful tool for functional assays. We thus tested gene silencing by both RNAi and Morpholino in C. fragrantissima.
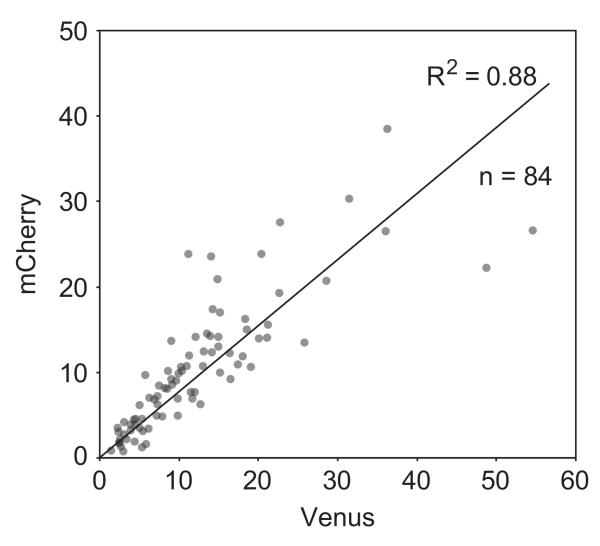
Evaluating the effect of silencing in C. fragrantissima is not as straightforward as in metazoans, due to the low transformation efficiency and the difficulty in quantifying the marker expression in the unicellular context. From the previous experiment (Fig. 3B), however, we realized that the signal intensity of mCherry in the nuclei appears to be well correlated with that of Venus, the transformation marker. Indeed, a graph plotting the intensities of these two fluorescent proteins in individual transformant cells shows a good correlation (R2=0.88) between these two values even though the fluorescence intensity itself greatly differs depending on the cells (Fig. 4). This means that the ratio of the mCherry expression to the Venus expression is fairly constant regardless of the absolute expression level, likely due to the constant ratio of the amounts of constructs brought into the cells. We thus used this ratio to evaluate the effect of silencing.
Fig. 4.
Correlation between Venus and mCherry signals in transformants. Intensities of fluorescence signals of Venus and mCherry are plotted. A point represents one transformant. The coefficient of determination R2 and the sample number n are shown. Axis values are arbitrary.
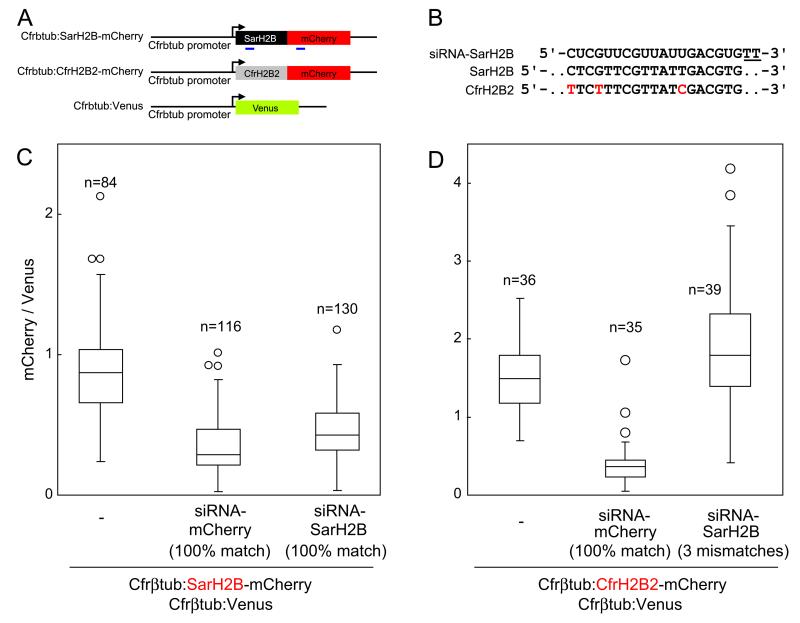
We first designed two synthetic small interfering RNAs (siRNAs) from the coding sequence of mCherry and from that of SarH2B. Each of these siRNAs was then co-electroporated together with the two constructs Cfrβtub:Venus and Cfrβtub:SarH2B-mCherry. In either experiment, the siRNA down-regulated the expression of SarH2B-mCherry (Fig. 5A–C). This silencing effect disappeared when the target sequence contained three mismatches (one terminal mismatch and two non-terminal mismatches), demonstrating the sequence specificity of the silencing effect (Fig. 5B and D).
Fig. 5.
RNA interference on C. fragrantissima and the influence of sequence mismatches. (A) Scheme of constructs. The sequences where the siRNAs were designed are indicated by blue lines. The effect of mismatches between the sequence of siRNA and that of the target construct onto silencing was tested by the second construct Cfrβtub:CfrH2B2-mCherry. SarH2B, S. arctica histone H2B; CfrH2B2, C. fragrantissima histone H2B2. (B) Sequence alignment of siRNA–SarH2B sense strand, SarH2B, and CfrH2B2. Nucleotide positions 183–201 of the histone H2B coding sequences are shown. Two dT overhang of the siRNA are underlined. The mismatched sites between the siRNA and target sequences are in red. (C) RNA interference by siRNAs against mCherry (middle) and against SarH2B (right). Used siRNAs and the co-electroporated constructs are indicated at the graph bottom. Ratio of mCherry to Venus signal intensity (values arbitrary) was measured for each transformant. (n, sample number) In box plots, the outliers beyond 1.5 Interquartile range (IQR) but still within 3 IQR are shown by circles. (D) siRNA is nonfunctional with three mismatches (right) while the effect of siRNA-mCherry is unchanged (middle).
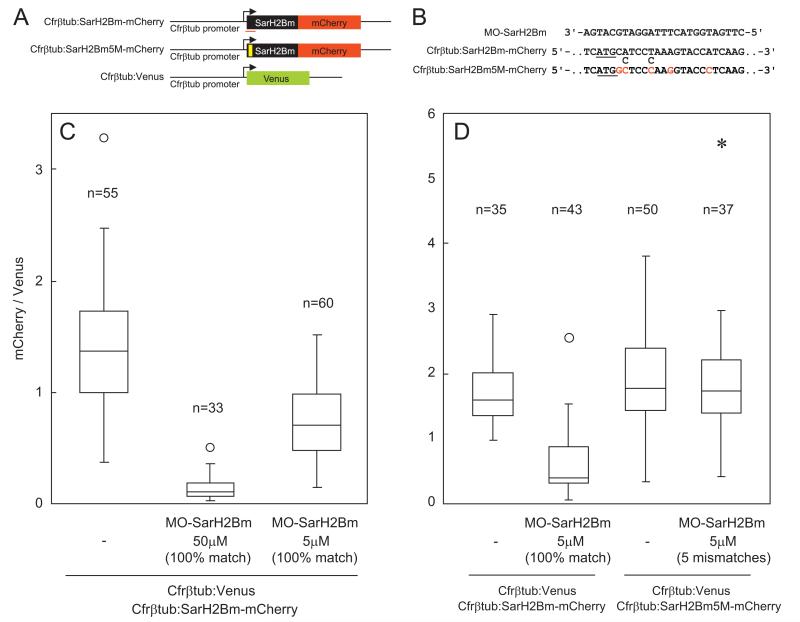
Next, we tested the translational block by a Morpholino antisense oligo designed from the SarH2B sequence. We slightly modified this target sequence (designated SarH2Bm) in order to optimize the Morpholino sequence design at the location where the target contains as small 5′UTR as possible (see Materials and Methods section). Similar to siRNA, this Morpholino oligo (MO-SarH2Bm) knocked down the expression of the construct encoding SarH2Bm-mCherry fusion protein (Fig. 6A–C). A dose-dependency was observed in the effect (Fig. 6C). The effect totally disappeared when five mismatches were introduced in the target sequence (Fig. 6D).
Fig. 6.
Silencing by Morpholino and the influence of sequence mismatches. (A) Schemes of constructs. The sequences where the Morpholino was designed are indicated by red lines. To test the effect of the sequence mismatches, we made another construct (Cfrβtub:SarH2Bm5M-mCherry), in which five mismatches (position indicated by a yellow box) were incorporated. (B) Sequences of the Morpholino (MO-SarH2Bm) and the two target constructs. To optimize the design of the Morpholino sequence at the position where the included 5′ UTR sequence is minimized, we modified the native SarH2B sequence at two sites (designated SarH2Bm). The original nucleotides (two cytosines) of SarH2B are shown below the sequence of Cfrβtub:SarH2Bm-mCherry. The five mismatches introduced in the construct Cfrβtub:SarH2Bm5M-mCherry are highlighted by red letters. Start codons are underlined. (C) Silencing by Morpholino in two different concentrations. The final Morpholino concentration in the cell suspension and the coelectroporated constructs are indicated at the graph bottom. Ratio of mCherry to Venus signal intensity was measured for each transformant cell (n, sample number). (D) Silencing by Morpholino with or without sequence mismatches. Note that MO-SarH2Bm at this concentration did not suppress the translation when five mismatches were introduced in the target (right). The outliers beyond 3 IQR are shown by asterisks and those beyond 1.5 IQR but still within 3 IQR are shown by circles.
Discussion
Implications for the origin of metazoan multicellularity
Regarding the origin of metazoan multicellularity, three possible mechanisms have been traditionally discussed: first, successive rounds of incomplete cell division; second, spontaneous aggregation of individual cells; and third, cellularization of a syncytium (Bonner, 1998; Grosberg and Strathmann, 2007; Willmer, 1990). At a cellular level, the second hypothesis appears not to have been well credited (Willmer, 1990) because metazoan embryos do not usually allow genetic heterogeneity. The first hypothesis is supported, for instance, by the recent observation of the colonial choanoflagellate S. rosetta (Fairclough et al., 2010). Interestingly, the cell division rendering its colony formation loses the synchrony after several divisions (Fairclough et al., 2010). On the other hand, our findings in ichthyosporeans suggest the third mechanism. Unlike the choanoflagellate cell division, the nuclear divisions in ichthyosporean cells are strictly synchronized during their whole growth stage.
It has been considered that the mechanism of choanoflagellate colony formation implies a possible strategy for developing metazoan multicellularity. Their clearly closer relationship to metazoans than any other pre-metazoan lineage advantageously lends support to this hypothesis. However, only a minor fraction of choanoflagellate species form colonies, whose morphology is diverse (Carr et al., 2008; Leadbeater and Thomsen, 1989; Thomsen and Buck, 1991). Moreover, recent phylogenetic studies suggest the paraphyly of the colony-forming species (Nitsche et al., 2011; Paps et al., 2013). It is therefore possible that choanoflagellates gained their colony-forming property several times within the choanoflagellate lineage, independently of metazoan evolution. Unlike choanoflagellates, many ichthyosporeans (especially the order Ichthyophonida) spend a similar life-cycle with a characteristic colonial stage (Jøstensen et al., 2002; Marshall and Berbee, 2010; Marshall et al., 2008; Mendoza et al., 2002; Whisler, 1960). It is noteworthy that the colony formation through syncytium is also seen in some slime molds and fungal species (Bonner, 1998; Grosberg and Strathmann, 2007), which are the closest relatives of metazoans besides choanoflagellates, filastereans, and ichthyosporeans. Thus, the colony development through syncytium cellularization clearly represents an important mechanism for developing multi-cell-like structures among the metazoan relatives. We therefore consider the cellularization of a syncytium with synchronized nuclear divisions to be a feasible strategy for the origin and evolution of metazoan multicellularity, in addition to the successive rounds of incomplete cell division.
It is possible, given the diversity of syncytium formation during metazoan embryogenesis (Chen et al., 2007), that the ichthyosporean colony formation does not have a direct evolutionary relationship to any of the metazoan syncytial embryogenesis or development that we see now in extant animals. In the Metazoa, syncytium formation during development may have evolved several times, independently of the metazoan ancestor, similarly to the collar cells, which also seem to have been invented many times during metazoan evolution (Cantell et al., 1982; Lyons, 1973; Nerrevang and Wingstrand, 1970). If the morphological analogies do not contribute to elucidating the origin of metazoan multicellularity, it is important to investigate the close metazoan relatives in a molecular level.
Future perspectives
Recent genomic studies on the closest living relatives of metazoans have revealed the presence of several key genes encoding proteins important for metazoan development and multicellularity (King et al., 2008; Manning et al., 2008; Nichols et al., 2012; Sebé-Pedrós et al., 2010, 2011, 2012; Suga et al., 2008,2012). These proteins seem unnecessary for their principally unicellular lifestyles. Consequently, gene co-option has been suggested to be an important mechanism for the evolution of multicellularity (Abedin and King, 2010; Sebé-Pedrós et al., 2011; Suga et al., 2012). However, how they have been co-opted at the transition from unicellular to multicellular organisms remains unclear because their original functions in the ancestor protists are unknown. To infer the ancestral functions of these genes, one has to know their functions in the extant closest metazoan relatives. In this regard, our success in transgenesis and gene silencing for the first time among the closest relatives of metazoans represents an important progress toward understanding the origin of animals. Moreover, although the genome sequencing of C. fragrantissima is still ongoing, genes that are involved in some important regulatory mechanisms for metazoan multicellularity have already been identified in ichthyosporeans, for instance, the integrin-mediated adhesion and signaling pathway (Sebe-Pedros and Ruiz-Trillo, 2010). We therefore propose C. fragrantissima as an ideal model organism to investigate the origin of metazoan multicellularity.
Supplementary Material
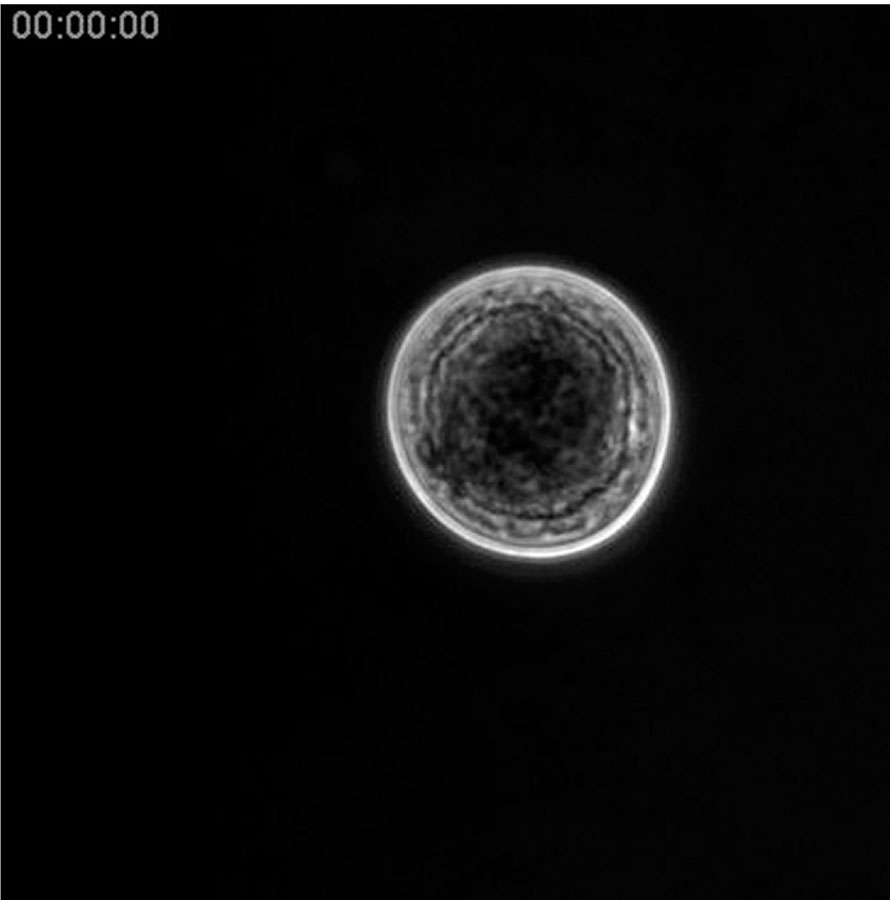
Video 1. Time-lapse movie (600x) of the growth–maturation–dissemination stages of C. fragrantissima cell
Movie starts at the end of the growth stage. A large vacuole at the cell center is being filled by the moving cytoplasm during the maturation stage. Amoebae are subsequently formed in the mother cell and migrate through tears of the cell wall. Elapsed time is shown. Movie size,131 μm × 131 μm.
A video clip is available online. Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.ydbio.2013.01.009.
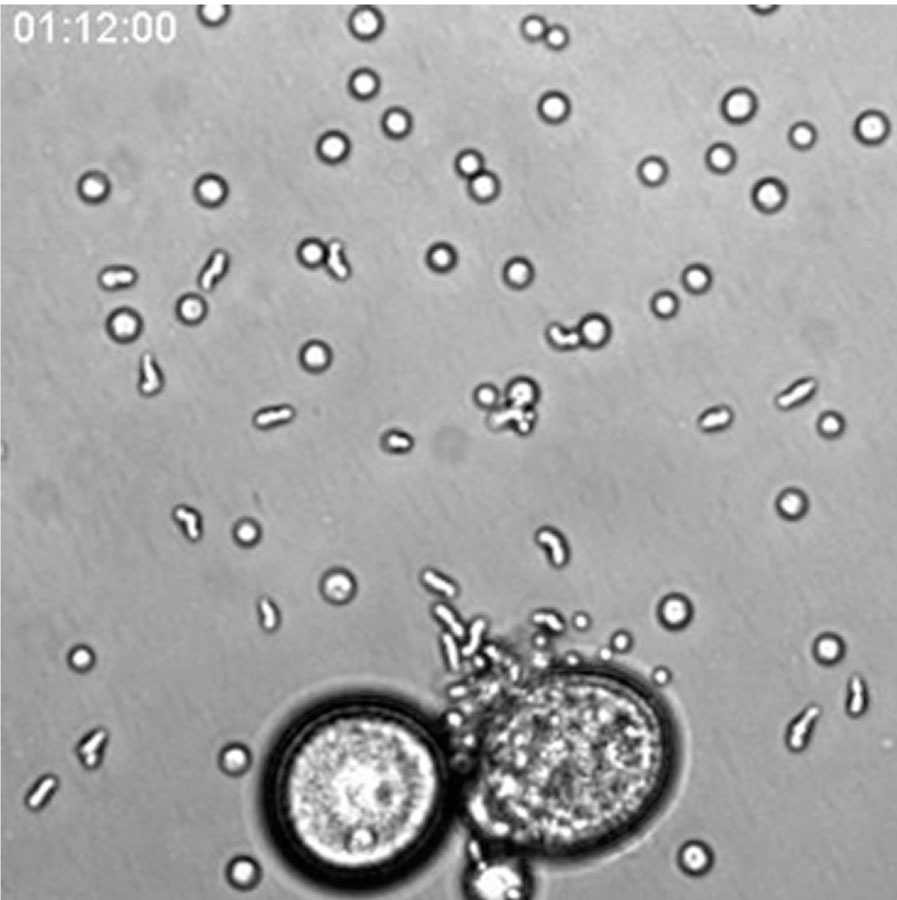
Video 2. Time-lapse movie of migrating amoebae
Movie is focused on amoebae, which migrate out of the mother cell and settle down after creeping on the dish bottom. Elapsed time is shown. Movie size, 239 μm × 239 μm.
A video clip is available online. Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.ydbio.2013.01.009.
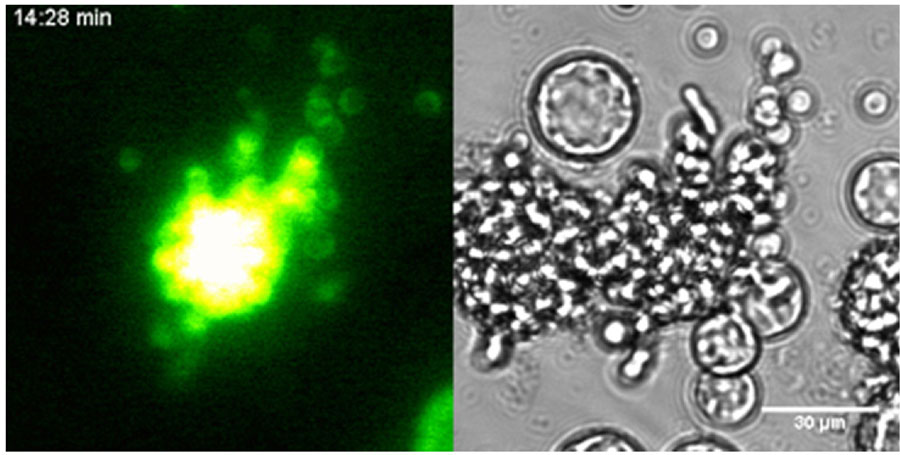
Video 3. Time-lapse movie of a transformed C. fragrantissima cell and its offspring amoebae
C fragrantissima. β-tubulin promoter is driving the expression of Venus fluorescent protein (left), which are carried over to the next generation. The bright field image is on the right. Elapsed time is shown.
A video clip is available online. Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.ydbio.2013.01.009.

Video 4. Time-lapse movie of a transformed C. fragrantissima cell showing the movement of its nuclei
C. fragrantissima β-tubulin promoter is driving the expression of SarH2B-mCherry fusion protein (left) and Venus fluorescent protein (middle). On the right is bright field image. The cell in the first frame contains four nulei (only two in focus). Stronger Venus fluorescence is colocalized with nuclei likely because the cytoplasm thickens at where the nuclei are (see the right panel). A non-transformed cell is attached to the transformant (top-right). Such cell aggregation is common in this species, and is thought to arise from adjacent endospores or amoebae sticking together, but not from budding (Marshall et al., 2008). Elapsed time is shown.
A video clip is available online. Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.ydbio.2013.01.009.
Acknowledgments
This work was supported by the European Research Council (ERC-2007-StG-206883; Iñaki Ruiz-Trillo), Ministerio Economía y Competitividad (BFU2011-23434; Iñaki Ruiz-Trillo), and the Marie Curie Intra-European Fellowship (FP7-PEOPLE-IEF-2008–236635; Hiroshi Suga).
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2013.01.009.
References
- Abedin M, King N. Diverse evolutionary paths to cell adhesion. Trends Cell Biol. 2010;20:734–742. doi: 10.1016/j.tcb.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alegado RA, Brown LW, Cao S, Dermenjian RK, Zuzow R, Fairclough S, Clardy J, King N. Bacterial regulation of colony development in the closest living relatives of animals. eLife. 2012;1:e00013. doi: 10.7554/eLife.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner J. The origins of multicellularity. Integrative Biol. 1998;1:27–36. [Google Scholar]
- Cantell CE, Franzén Å, Sensenbaugh T. Ultrastructure of multiciliated collar cells in the pilidium larva of Lineus bilineatus (Nemertini) Zoomorphol. 1982;101:1–15. [Google Scholar]
- Carr M, Leadbeater BS, Hassan R, Nelson M, Baldauf SL. Molecular phylogeny of choanoflagellates, the sister group to Metazoa. Proc. Natl. Acad. Sci. USA. 2008;105:16641–16646. doi: 10.1073/pnas.0801667105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EH, Grote E, Mohler W, Vignery A. Cell–cell fusion. FEBS Lett. 2007;581:2181–2193. doi: 10.1016/j.febslet.2007.03.033. [DOI] [PubMed] [Google Scholar]
- Dayel MJ, Alegado RA, Fairclough SR, Levin TC, Nichols SA, McDonald K, King N. Cell differentiation and morphogenesis in the colony-forming choanoflagellate Salpingoeca rosetta. Dev. Biol. 2011;357:73–82. doi: 10.1016/j.ydbio.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairclough SR, Dayel MJ, King N. Multicellular development in a choanoflagellate. Curr. Biol. 2010;20:R875–R876. doi: 10.1016/j.cub.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosberg EK, Strathmann RR. The evolution of multicellularity: a minor major transition? Annu. Rev. Ecol. Evol. Syst. 2007;38:621–654. [Google Scholar]
- Jøstensen J-P, Sperstad S, Johansen S, Landfald B. Molecular-phylogenetic, structural and biochemical features of a cold-adapted, marine ichthyosporean near animal–fungal divergence, described from in vitro cultures. Eur. J. Protistol. 2002;38:93–104. [Google Scholar]
- Kim JA, Cho K, Shin MS, Lee WG, Jung N, Chung C, Chang JK. A novel electroporation method using a capillary and wire-type electrode. Biosensors Bioelectronics. 2008;23:1353–1360. doi: 10.1016/j.bios.2007.12.009. [DOI] [PubMed] [Google Scholar]
- King N. The unicellular ancestry of animal development. Dev. Cell. 2004;7:313–325. doi: 10.1016/j.devcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, Marr M, Pincus D, Putnam N, Rokas A, Wright KJ, Zuzow R, Dirks W, Good M, Goodstein D, Lemons D, Li W, Lyons JB, Morris A, Nichols S, Richter DJ, Salamov A, Sequencing J, Bork P, Lim WA, Manning G, Miller WT, McGinnis W, Shapiro H, Tjian R, Grigoriev IV, Rokhsar D. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang BF, O’Kelly C, Nerad T, Gray MW, Burger G. The closest unicellular relatives of animals. Curr. Biol. 2002;12:1773–1778. doi: 10.1016/s0960-9822(02)01187-9. [DOI] [PubMed] [Google Scholar]
- Leadbeater BS, Thomsen HA. Choanoflagellata. In: Lee JJ, Leedale GF, Bradbury P, editors. An Illustrated Guide to the Protozoa, 2nd Edition Society of Protozoologists. Lawrence; 1989. pp. 14–38. [Google Scholar]
- Leys SP, Cheung E, Boury-Esnault N. Embryogenesis in the glass sponge Oopsacas minuta: formation of syncytia by fusion of blastomeres. Integrative Comp. Biol. 2006;46:104–117. doi: 10.1093/icb/icj016. [DOI] [PubMed] [Google Scholar]
- Lyons KM. Collar cells in planula and adult tentacle ectoderm of the solitary coral Balanophyllia regia (anthozoa eupsammiidae) Cell Tissue Res. 1973;145:54–74. doi: 10.1007/BF00307189. [DOI] [PubMed] [Google Scholar]
- Maldonado M. Choanoflagellates, choanocytes, and animal multicellularity. Invertebrate Biol. 2005;123:1–22. [Google Scholar]
- Manning G, Young SL, Miller WT, Zhai Y. The protist, Monosiga brevicollis, has a tyrosine kinase signaling network more elaborate and diverse than found in any known metazoan. Proc. Natl. Acad. Sci. USA. 2008;105:9674–9679. doi: 10.1073/pnas.0801314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WL, Berbee ML. Facing unknowns: living cultures (Pirum gemmata gen. nov., sp. nov., and Abeoforma whisleri, gen. nov., sp. nov.) from invertebrate digestive tracts represent an undescribed clade within the unicellular opisthokont lineage Ichthyosporea (Mesomycetozoea) Protist. 2010;162:33–57. doi: 10.1016/j.protis.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Marshall WL, Celio G, McLaughlin DJ, Berbee ML. Multiple isolations of a culturable, motile ichthyosporean (Mesomycetozoa, Opisthokonta), Creolimax fragrantissima n. gen., n. sp., from marine invertebrate digestive tracts. Protist. 2008;159:415–433. doi: 10.1016/j.protis.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Mendoza L, Taylor JW, Ajello L. The class Mesomycetozoea: a heterogeneous group of microorganisms at the animal-fungal boundary. Annu. Rev. Microbiol. 2002;56:315–344. doi: 10.1146/annurev.micro.56.012302.160950. [DOI] [PubMed] [Google Scholar]
- Nerrevang A, Wingstrand KG. On the occurrence and structure of choanocyte-like cells in some echinoderms. Acta Zool. 1970;51:249–270. [Google Scholar]
- Nichols SA, Roberts BW, Richter DJ, Fairclough SR, King N. Origin of metazoan cadherin diversity and the antiquity of the classical cadherin/beta-catenin complex. Proc. Natl. Acad. Sci. USA. 2012;109:13046–13051. doi: 10.1073/pnas.1120685109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsche F, Carr M, Arndt H, Leadbeater BS. Higher level taxonomy and molecular phylogenetics of the Choanoflagellatea. J. Eukaryotic Microbiol. 2011;58:452–462. doi: 10.1111/j.1550-7408.2011.00572.x. [DOI] [PubMed] [Google Scholar]
- Paps J, Medina-Chacón LA, Marshall WL, Suga H, Ruiz-Trillo I. Molecular phylogeny of unikonts: new insights into the position of apusomonads and ancyromonads and the internal relationships of opisthokonts. Protist. 2013;164:2–12. doi: 10.1016/j.protis.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Roger AJ, Burger G, Gray MW, Lang BF. A phylogenomic investigation into the origin of Metazoa. Mol. Biol. Evol. 2008;25:664–672. doi: 10.1093/molbev/msn006. [DOI] [PubMed] [Google Scholar]
- Sebé-Pedrós A, de Mendoza A, Lang BF, Degnan BM, Ruiz-Trillo I. Unexpected repertoire of metazoan transcription factors in the unicellular holozoan Capsaspora owczarzaki. Mol. Biol. Evol. 2011;28:1241–1254. doi: 10.1093/molbev/msq309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Roger AJ, Lang FB, King N, Ruiz-Trillo I. Ancient origin of the integrin-mediated adhesion and signaling machinery. Proc. Natl. Acad. Sci. USA. 2010;107:10142–10147. doi: 10.1073/pnas.1002257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Zheng Y, Ruiz-Trillo I, Pan D. Premetazoan origin of the Hippo signaling pathway. Cell Rep. 2012;1:13–20. doi: 10.1016/j.celrep.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebe-Pedros A, Ruiz-Trillo I. Integrin-mediated adhesion complex: cooption of signaling systems at the dawn of Metazoa. Communicative Integrative Biol. 2010;3:475–477. doi: 10.4161/cib.3.5.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalchian-Tabrizi K, Minge MA, Espelund M, Orr R, Ruden T, Jakobsen KS, Cavalier-Smith T. Multigene phylogeny of Choanozoa and the origin of animals. PloS One. 2008;3:e2098. doi: 10.1371/journal.pone.0002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer RJ. Evolution of nematode development. Curr. Opin. Genet. Dev. 2000;10:443–448. doi: 10.1016/s0959-437x(00)00110-6. [DOI] [PubMed] [Google Scholar]
- Steenkamp ET, Baldauf SL. Origin and evolution of animals, fungi and their unicellular allies (Opisthokonta) In: Hirt RP, Horner DS, editors. Organelles, Genomes and Eukaryote Phylogeny: An Evolutionary Synthesis in the Age of Genomics. CRC Press; London: 2004. pp. 109–129. [Google Scholar]
- Suga H, Dacre M, de Mendoza A, Shalchian-Tabrizi K, Manning G, Ruiz-Trillo I. Genomic survey of pre-metazoans shows deep conservation of cytoplasmic tyrosine kinases and multiple radiations of receptor tyrosine kinases. Sci. Signaling. 2012;5:ra35. doi: 10.1126/scisignal.2002733. [DOI] [PubMed] [Google Scholar]
- Suga H, Sasaki G, Kuma K, Nishiyori H, Hirose N, Su ZH, Iwabe N, Miyata T. Ancient divergence of animal protein tyrosine kinase genes demonstrated by a gene family tree including choanoflagellate genes. FEBS Lett. 2008;582:815–818. doi: 10.1016/j.febslet.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Thomsen HA, Buck KR. Choanoflagellate diversity with particular emphasis on the Acanthoecidae. In: Patterson DJ, Larsen J, editors. The Biology of Free-living Heterotrophic Flagellates. Clarendon Press; Oxford: 1991. pp. 285–294. [Google Scholar]
- Torruella G, Derelle R, Paps J, Lang BF, Roger AJ, Shalchian-Tabrizi K, Ruiz-Trillo I. Phylogenetic relationships within the Opisthokonta based on phylogenomic analyses of conserved single copy protein domains. Mol. Biol. Evol. 2011;29:531–544. doi: 10.1093/molbev/msr185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisler HC. Culture and nutrition of Amoebidium parasiticum. Am. J. Bot. 1960;49:193–199. [Google Scholar]
- Willmer P. The origin of the Metazoa, Invertebrate Relationships: Pattern in Animal Evolution. Cambridge University Press; Cambridge: 1990. pp. 163–198. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials

Video 1. Time-lapse movie (600x) of the growth–maturation–dissemination stages of C. fragrantissima cell
Movie starts at the end of the growth stage. A large vacuole at the cell center is being filled by the moving cytoplasm during the maturation stage. Amoebae are subsequently formed in the mother cell and migrate through tears of the cell wall. Elapsed time is shown. Movie size,131 μm × 131 μm.
A video clip is available online. Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.ydbio.2013.01.009.

Video 2. Time-lapse movie of migrating amoebae
Movie is focused on amoebae, which migrate out of the mother cell and settle down after creeping on the dish bottom. Elapsed time is shown. Movie size, 239 μm × 239 μm.
A video clip is available online. Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.ydbio.2013.01.009.

Video 3. Time-lapse movie of a transformed C. fragrantissima cell and its offspring amoebae
C fragrantissima. β-tubulin promoter is driving the expression of Venus fluorescent protein (left), which are carried over to the next generation. The bright field image is on the right. Elapsed time is shown.
A video clip is available online. Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.ydbio.2013.01.009.

Video 4. Time-lapse movie of a transformed C. fragrantissima cell showing the movement of its nuclei
C. fragrantissima β-tubulin promoter is driving the expression of SarH2B-mCherry fusion protein (left) and Venus fluorescent protein (middle). On the right is bright field image. The cell in the first frame contains four nulei (only two in focus). Stronger Venus fluorescence is colocalized with nuclei likely because the cytoplasm thickens at where the nuclei are (see the right panel). A non-transformed cell is attached to the transformant (top-right). Such cell aggregation is common in this species, and is thought to arise from adjacent endospores or amoebae sticking together, but not from budding (Marshall et al., 2008). Elapsed time is shown.
A video clip is available online. Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.ydbio.2013.01.009.