Abstract
How animals (metazoans) originated from their single-celled ancestors remains a major question in biology. As transcriptional regulation is crucial to animal development, deciphering the early evolution of associated transcription factors (TFs) is critical to understanding metazoan origins. In this study we uncovered the repertoire of seventeen metazoan TFs in the amoeboid holozoan Capsaspora owczarzaki, a representative of a unicellular lineage that is closely related to choanoflagellates and metazoans. Phylogenetic and comparative genomic analyses with the broadest possible taxonomic sampling allowed us to formulate new hypotheses regarding the origin and evolution of developmental metazoan TFs. We show that the complexity of the TF repertoire in Capsaspora owczarzaki is strikingly high, pushing back further the origin of some TFs formerly thought to be metazoan-specific, such as T-box or Runx. Nonetheless, TF families whose beginnings antedate the origin of the animal kingdom, such as homedomain or bHLH, underwent significant expansion and diversification along metazoan and eumetazoan stems.
Keywords: multicellularity, T-box, homeodomain, brachyury, transcription factors, Capsaspora owczarzaki
Introduction
What genomic changes took place at the dawn of the Metazoa remains a major biological question. Transcriptional regulation appears to be one of the most crucial aspects of animal development. Thus, understanding the early evolution of the transcriptional regulatory machinery is critical for drawing a complete picture of metazoan origins. Transcription factors (TF) act as regulators of cell fate, cell cycle, patterning, proliferation, development and differentiation in metazoans (Larroux et al. 2008). Previous studies have shown that most transcription factors that play important roles in bilaterian development originated before the divergence of extant animal phyla (Larroux et al. 2006, 2008; King et al. 2008; Degnan et al. 2009; Srivastava et al. 2010). However, the complexity of most TF families appears to have increased during early eumetazoan evolution, with cnidarians having a TF gene repertoire typically being two to three times larger than that of sponges and placozoans (Putnam et al. 2007; Degnan et al. 2009; Srivastava et al. 2010). Based on comparative analyses, it has been hypothesized that the metazoan TF ‘toolkit’ included members of the bHLH, Mef2, Fox, Sox, T-box, Ets, nuclear receptor, Rel/NF-kappaB, bZIP, and Smad families and a range of homebox-containing classes, including ANTP, Prd-like, Pax, POU, LIM-HD, Six, and TALE (for a review see Degnan et al. 2009).
Comparative analyses including the holozoan choanoflagellate Monosiga brevicollis, the putative sister-group to metazoans, is greatly improving our understanding of metazoan TF evolution. The genome of M. brevicollis contains the standard set of TFs observed across eukaryotes but lacks most of the well-known metazoan TFs, except p53, Myc and a putative Sox (King et al. 2008; Degnan et al. 2009). Under this scenario, metazoan-specific TFs appear to include ANTP, Prd-like, POU, LIM-HD, and Six homeobox genes, group I Fox, most bHLH groups (except B), some bZIP families, Ets, Runx, Mef2, and nuclear receptor families (Degnan et al. 2009).
To gain further insight into the evolution of TFs leading to the metazoan lineage, we characterized and analyzed all of the TFs that supposedly constitute the metazoan TF ‘toolkit’ in another close unicellular relative of animals, the amoeboid holozoan Capsaspora owczarzaki, putatively the sister-group to metazoans and choanoflagellates (Ruiz-Trillo et al. 2004, 2008; Shalchian-Tabrizi et al. 2008; Brown, Spiegel and Silberman 2009; see Figure 1). The complete genome sequence of Capsaspora owczarzaki (hereafter “Capsaspora”) has recently been obtained under the ‘UNICORN project’ at the Broad Institute (Ruiz-Trillo et al. 2007). In addition to the TFs outlined above, our survey of the Capsaspora genome in this study also included other TFs known to be important to animal development, including Churchill, p53, Stat, and LSF/GRH (Figure 1). Comparative genomic analyses were performed on holozoan genomes, and in some cases other recently sequenced opisthokont and apusozoan genomes, namely Allomyces macrogynus, Spizellomyces punctatus and Thecamonas trahens (see Materials & Methods, Figure 1). These results show that the complexity of TFs in Capsaspora is very high, indicating that some TFs thought to be metazoan-specific evolved prior to the metazoan and choanoflagellate divergence, and were subsequently lost in the choanoflagellate lineage.
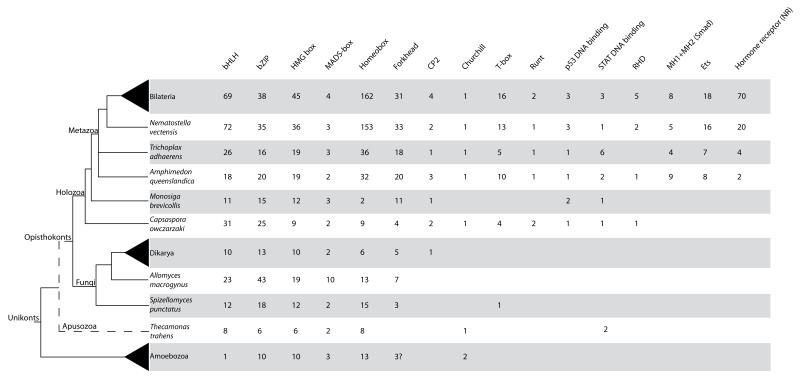
Figure 1.
Table of domain presence and number of genes across unikonts. Columns represent all the PFAM domains analyzed in this study. The number of genes in each TF family was inferred from each organisms’s proteome by PfamScan using the PfamScan default parameters. For M. brevicollis and C. owczarzaki the analyses were performed by HMMER 3.0 searches. For Smad proteins, containing one MH1 and one MH2 domain, the number shown is the minimal number of either MH1 or MH2.
For Bilateria, Dikarya and Amoebozoa the average number is shown. Bilateria includes Homo sapiens, Ciona intestinalis, Drosophila melanogaster, Anopheles gambiae, Caenorhabditis elegans, Helobdella robusta, and Lottia gigantea. Dikarya includes Saccharomyces cerevisiae, Schizosaccharomyces pombe, Cryptococcus neoformans, Yarrowia lypolitica, Ustilago maydis, Aspergillus niger, Neurospora crassa, and Phanerochaete chrysosporium. Amoebozoa includes Dictyostellium discoideum, Dictyostellium purpureum, Entamoeba histolytica, Entamoeba dispar, and Acanthamoeba castellanii. The phylogenetic relationships are based on several recent phylogenomic studies (Burki et al. 2008; Ruiz-Trillo et al. 2008; Brown, Spiegel and Silberman 2009; Liu et al. 2009; Minge et al. 2009).
Materials and methods
Taxonomic sampling
We surveyed, and characterized, a list of metazoan TFs in Capsaspora owczarzaki. In some cases we extended our searches to the widest possible set of eukaryotic taxa. This was case for those TF families with specific and unique domains: T-box (T-box DNA binding domain), Runx (Runt DNA binding domain), NF-kappaB (RHD domain), Mef2 (MADS box+Mef2 domain), p53 (p53 DNA binding domain), Stat (Stat DNA binding domain), Churchill (Churchill domain), Smad (MH1+MH2 domains), Ets (Ets domain), and nuclear receptor. Our extended searches included published and publicly available eukaryotic genomes, and other UNICORN taxa, such as the basal fungi Allomyces macrogynus and Spizellomyces punctatus and the apusozoan Thecamonas trahens (see http://www.broadinstitute.org/annotation/genome/multicellularity_project/MultiHome.html). For the remaining TF families (i.e. bZIP (bZIP domain), Fox (forkhead domain), Sox (HMG box), homeobox (homeodomain), bHLH (bHLH domain), and LSF/GRH (CP2 domain)) we classified those Capsaspora genes with homology to metazoan genes. To this end we used published fungal, metazoan and choanoflagellate homologs. We also characterized bZIPs and Mef2 in M. brevicollis.
Gene searches
A primary search was performed using the basic local alignment sequence tool (BLAST: blastp and tblastn) using Homo sapiens proteins as queries against Protein and Genome databases with the default BLAST parameters and an e-value threshold of 10-05 at the National Center for Biotechnology Information (NCBI) and against completed or on-going genome project databases at the Joint Genome Institute (JGI), the Broad Institute, as well as the A. queenslandica genome database (www.metazome.net/amphimedon). In the case of T. trahens, A. macrogynus and A. castellanii, we assembled the trace data using the WGS assembler (“http://sourceforge.net/apps/mediawiki/wgs-assembler/index.php?title=Main_Page”http://sourceforge.net/apps/mediawiki/wgs-assembler/index.php?title=Main_Page). We then annotated the genes of interest using both Genescan (Burge and Karlin 1997) and Augustus (Stanke and Morgenstern 2005) and performed local BLAST searches. When the BLAST searches of the genome data described above returned significant ‘hits’, the sequences obtained were then reciprocally searched against the NCBI protein database by BLAST in order to confirm the validity of the sequences retrieved with the initial search. Hmmer searches using HMMER3.0b2 (Eddy 1998) were also performed, with standard PFAM profiles in the case of widespread domains or with home-made profiles in the case of specific domains.
Protein domain arrangements
For all proteins, the presence of specific protein domains was further checked by searching the Pfam (“http://pfam.sanger.ac.uk/search”http://pfam.sanger.ac.uk/search) and SMART (“http://smart.embl-heidelberg.de/”http://smart.embl-heidelberg.de/) databases.
Polymerase Chain Reaction (PCR) confirmation of Capsaspora owczarzaki T-box, Runx and NF-kappaB genes
We confirmed the presence of the three Capsaspora TFs that were formerly considered to be metazoan-specific TFs now identified in Capsaspora (Runx, T-box and NF-kappaB), using gene-specific oligonucleotide primers. mRNA was extracted using a Dynabeads mRNA purification kit (Invitrogen, Carlsbad, CA) and subsequent reverse transcriptase PCR (RT PCR) was performed using a Superscript III First Strand Synthesis kit (Invitrogen, Carlsbad, CA). The full sequence of the 5′ and 3′ ends of the cited Capsaspora TF cDNAs were obtained by RACE, using a nested PCR and with specific oligonucleotide primers designed from the original genome data. Both coding and non-coding strands were sequenced using an ABI PRISM BigDye Termination Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). New sequences were deposited in GenBank under the following accession numbers: GU985459 (Capsaspora Bra-like), GU985460 (Capsaspora Double-tbox), GU985461 (Capsaspora Tbox3), GU985462 (Capsaspora Runx1), GU985463 (Capsaspora Runx2) and GU985464 (Capsaspora NF-kappaB).
Phylogenetic analyses
Alignments were constructed for the following gene families and classes: T-box, homeobox, Fox, Sox, bHLH, bZIP, LASS, STAT, Mef2, p53, NF-kappaB, Churchill, HMG-box, GRH/LSF and Runx. Alignments were obtained using the MAFFT v.6 online server (Katoh et al. 2005a, 2005b) and then manually inspected and edited in Geneious. Only those species and those positions that were unambiguously aligned were included in the final analyses. Maximum likelihood (ML) phylogenetic trees were estimated by RaxML (Stamatakis 2006) using the PROTGAMMAWAGI model, which uses the Whelan and Goldman amino acid exchangeabilities and accounts for among-site rate variation with a four category discrete gamma approximation and a proportion of invariable sites (WAG+ Γ+I). Statistical support for bipartitions was estimated by performing 1000-bootstrap replicates using RaxML with the same model. Bayesian analyses were performed with MrBayes 3.1 (Ronquist and Huelsenbeck 2003), using the WAG+Γ+I model of evolution, with four chains, a subsampling frequency of 100 and two parallel runs. Runs were stopped when the average standard deviation of split frequencies of the two parallel runs was < 0.01, usually at around 1,000,000 generations. The two LnL graphs were checked and an appropriate burn-in length established; stationarity of the chain typically occurred after ~15% of the generations. Bayesian posterior probabilities (BPP) were used to assessing the confidence values of each bipartition.
Homeodomain gene assignment
An alignment with members of the ANTP, Paired-like, POU and LIM homeodomain classes was constructed using published data from Amphimedon, Drosophila and Nematostella and other already classified sequences (Larroux et al. 2008). A RaxML best tree resulting from this phylogeny was produced to obtain the fixed topology, which recovered monophyly for all four classes. From this tree we manually created constrained topologies that represented all the possible positions of Capsaspora non-TALE homeodomains. Site-wise log-likelihoods were calculated for all the generated topologies with RaxML. Best-scoring ML trees were chosen using the likelihood-based approximately unbiased (AU) test as implemented in CONSEL (Shimodaira and Hasegawa 2001). The positions of Capsaspora homeodomain genes that could not be statistically excluded (p>=0.05) were taken into account. Whenever the significant positions fell in the branches that connect the different classes of homeodomains (POU, LIM, …), the homeodomain was not classified. When Capsaspora hits fell inside just one cluster (e.g., Capsaspora6 inside Paired-like), they were classified accordingly.
Quantitative TF analyses in unikont taxa
To quantify the number of genes in each TF family we used PfamScan using the PfamScan default parameters. The predicted proteomes used for PfamScan analysis were Amphimedon queenslandica (JGI), Trichoplax adhaerens (JGI), Nematostella vectensis (JGI), Homo sapiens (NCBI), Ciona intestinalis (JGI), Drosophila melanogaster (NCBI), Anopheles gambiae (NCBI), Caenorhabditis elegans (NCBI), Helobdella robusta (JGI), Lottia gigantea (JGI), Saccharomyces cerevisiae (NCBI), Schizosaccharomyces pombe (NCBI), Cryptococcus neoformans (JGI), Yarrowia lypolitica (NCBI), Ustilago maydis (JGI), Aspergillus niger (JGI), Neurospora crassa (NCBI), Phanerochaete chrysosporium (JGI), Allomyces macrogynus (Broad Institute), Spizellomyces punctatus (Broad Institute), Dictyostellium discoideum (NCBI), Dictyostellium purpureum (JGI), Entamoeba histolytica (NCBI), Entamoeba dispar (NCBI) and Acanthamoeba castellanii (home-made prediction). For M. brevicollis and Capsaspora the analyses were performed by HMMER 3.0 searches. For Smad proteins, containing one MH1 and one MH2 domain, the number was inferred by taking the minimal number of either MH1 or MH2 present in the proteomes.
Results and discussion
Rel/NF-kappaB
The Rel homology domain (RHD) is a conserved DNA binding and dimerization domain that is present in the N-terminal region of two protein families: nuclear factor activated T-cells (NFAT) and Rel/nuclear factor-kappaB (NF-kappaB). NFATand Rel/NF-kappaB are involved in immune system processes in metazoans (Macian 2005). Rel/NF-kappaB also plays different roles in development and cell differentiation, receiving inputs from several signaling pathways (Hayden and Ghosh 2004). Until now, the RHD domain has not been identified outside metazoans and was thus considered a metazoan innovation (Gauthier and Degnan 2008).
However, we identified a single RHD domain in Capsaspora, but failed to recover RHD from any other sequenced non-metazoan taxa (Figure 1). Our phylogenetic analysis of the RHD domain shows the Capsaspora homolog branching off as sister-group of all metazoan Rel/NF-kappaB (Figure S1). Furthermore, the Capsaspora RHD-domain-containing protein shares several key features with metazoan Rel and NF-kappaB homologs, such as 1) a highly conserved and specific recognition loop located within the RHD domain, which is involved in dimerization; 2) an IPTG or RHD2 domain, which confers binding specificity; 3) a basic nuclear-localization sequence (NLS); 4) a glycine-serine rich region; and 5) several ankyrin repeats, which are exclusive to metazoan NF-kappaB proteins (Figure S2).
Thus, our data show that the RHD domain is not exclusive to metazoans as previously thought, but rather it originated prior to the divergence of Capsaspora from choanoflagellates and metazoans. This implies that the RHD domain was subsequently lost in the choanoflagellate lineage.
Runx
The Runt DNA binding domain defines a family of metazoan TFs (Runx) with essential roles in animal development (Coffman 2003; Robertson et al. 2009). They can act as transcriptional activators or repressors, in the latter case usually via co-repressors of the Groucho/TLE family (Wheeler et al. 2000). Runx genes encode the Runt DNA binding domain and heterodimerization domain and a C-terminal WRPY motif that interacts with the Groucho/TLE co-repressor (Coffman 2003), except in the demosponge Amphimedon queenslandica and some bilaterian paralogs (specifically one of the two leech and planarian paralogs) which all lack the C-terminal WRPY motif (Robertson et al. 2009). A sigle Runx gene is present in A. queenslandica, N. vectensis, and T. adhaerens, although most bilaterians have several copies as a result of independent duplications (Rennert et al. 2003). Runx was previously considered to be metazoan-specific (Robertson et al. 2009).
We failed to recover Runx genes from any other sequenced non-metazoan genome except Capsaspora, which has two genes (Figure 1). Both Capsaspora Runxs possess key DNA-binding amino acids in the Runt motif (Wheeler et al. 2000; Sullivan et al. 2008), although only one of the paralogs (Co_Runx1) has the two Cys residues involved in redox regulation (Akamatsu et al. 1997) (Figure S3). Interestingly, as in A. queenslandica, and one of the two leech and planarian paralogs both Capsaspora Runx lack the specific C-terminal WRPY Groucho-interacting motif. In contrast to A. queenslandica, however, Capsaspora does not encode Groucho in its genome. Neither does Capsaspora encode CBFβ, the heterodimeric binding partner of the Runt domain that enhances its DNA affinity (Sullivan et al. 2008). This suggests that the Runt domain acts independently from CBFβ in Capsaspora. Our results show that Runx originated prior to the divergence of Capsaspora from choanoflagellates and metazoans, being secondarily lost in the choanoflagellate lineage. We hypothesize that Runx originally functioned independently of Groucho and CBFβ proteins, and that the WRPY Groucho-interacting motif appeared in the eumetazoan lineage, as previously suggested (Robertson et al. 2009).
T-box
T-box TFs are characterized by an evolutionary conserved DNA binding motif of 180-200 amino acids, the T-box domain (Smith 1999). They are key regulators of metazoan development (Muller and Herrmann 1997). The most well-known type of T-box is Brachyury, which has a key role in mesoderm specification (Marcellini et al. 2003), although its ancestral function may have been blastopore determination and gastrulation (Scholz and Technau 2003). T-box genes were previously generally considered to be metazoan-specific (King et al. 2008; Larroux et al. 2008; Rokas 2008).
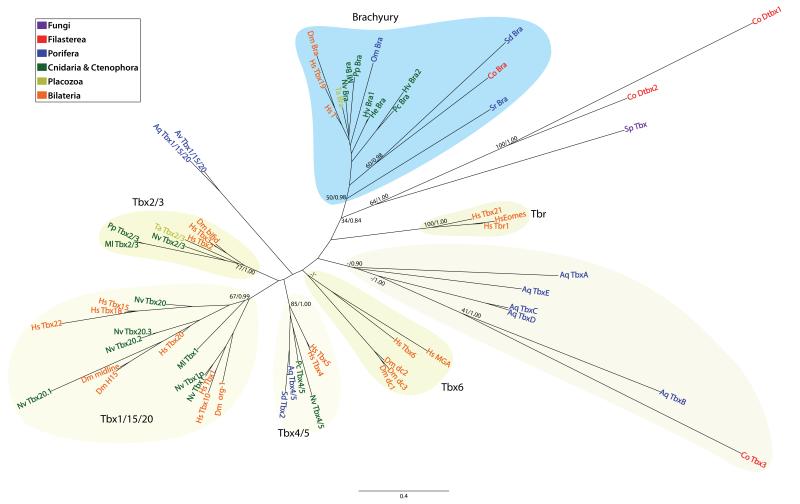
Here we report the discovery of T-box genes in two non-metazoan species. Three T-box genes are present in Capsaspora (one containing two consecutive T-box domains), and one gene exists in the basal chytrid fungus Spizellomyces punctatus (Figure 1). Our searches, however, failed to recover T-box homologs from any other fungi (including the chytrids Allomyces macrogynus and Batrachochytrium dendrobatidis), or other eukaryote (including the choanoflagellate M. brevicollis). Remarkably, all the T-box homologs from both Capsaspora and S. punctatus contain most of the key DNA-binding and dimerization amino acids of the metazoan T-box (Muller and Herrmann 1997; Bielen et al. 2007) (Figure S4). The phylogenetic analysis of T-box domains (Figure 2) places one Capsaspora homolog (Co-Bra) inside the Brachyury family (BV=50%). The two T-box domains in the Capsaspora “double-tbox” (Co-Dtbx1 and Co-Dtbx2) and the S. punctatus T-box clearly cluster together adjacent to the Brachyury family. The third Capsaspora homolog (Co-Tbx3) clusters within a group of unclassified T-box genes from the sponge A. queenslandica that may represent an independent and novel class of T-box genes. Our general topology supports the hypothesis that Brachyury is probably the ancestral class within the T-box family (Adell et al. 2003; Adell and Muller 2005; Larroux et al. 2008). Moreover, our findings imply that T-box genes appeared not in metazoans, but in the common ancestor of opisthokonts and were subsequently lost in most fungi and in choanoflagellates.
Figure 2.
Maximum likelihood tree of T-box domains showing the different T-box families. The tree is rooted using the midpoint-rooted tree option. Statistical support was obtained by RAxML with 1000-bootstrap replicates (bootstrap value, BV) and Bayesian Posterior Probabilities (BPP). Both values are shown on key branches. Colors show different taxonomic assignments. Aq (A. queenslandica), Av (Axinella verrucosa), Co (Capsaspora owzarzaki), Dm (Drosophila melanogaster), He (Hydractinia echinata), Hs (Homo sapiens), Hv (Hydra vulgaris), Ml (Mnemiopsis leydi), Nv (Nematostella vectensis), Om (Oopsacas minuta), Pc (Podocoryne carnea), Pp (Pleurobrachia pileus), Sd (Suberites domuncula), Sp (Spizellomyces punctatus), Sr (Sycon raphanus), Ta (Trichoplax adhaerens). Co-Dtbx1 and Co-Dtbx2 are the two T-box domains of the same T-box Capsaspora gene (see main text for further details).
Churchill
Churchill is a zinc-finger TF that is involved in cell movement and cell fate determination (Londin, Mentzer and Sirotkin 2007). In Xenopus and chick, Churchill appears to regulate the T-box gene brachyury (Sheng et al. 2003). We have found orthologs of Churchill in Capsaspora, in T. trahens, and, interestingly, also in the amoebozoan Acanthamoeba castellanii (Figures 1, and S5). This finding indicates a deeper origin of this gene than previously thought, probably in the common ancestor of unikonts. This suggests that Churchill was secondarily lost in apusozoans, fungi and choanoflagellates, as well as in other amoebozoans. What role the Churchill orthologs play in Capsaspora, T. trahens, or A. castellanii, and whether, in Capsaspora, it is related at all its T-box genes is unknown.
P53
The P53 tumor suppressor protein is a multifaceted TF that is involved in different cellular responses to DNA damage, such as DNA repair, cell cycle arrest, senescence and apoptosis (Coutts and La Thangue 2005; Espinosa 2008). The p53 family includes p53, p63 and p73, the last two being more closely related to each other than to p53. The three p53 members have some differences in function and in the protein domain architecture. p63 and p73 share an additional C-terminal sterile alpha motif (SAM) domain, while all three share a transcriptional activation domain, a DNA-binding domain, and C-terminal tetramerization domain (Nedelcu and Tan 2007). Choanoflagellates have both a p53 and a p63/73 ortholog (Nedelcu and Tan 2007).
Here we characterize a unique member of the p53 gene family in Capsaspora (Figure 1 and S6), the gene encodes a SAM domain. The phylogenetic analysis places Capsaspora-p53/63/73 close to the choanoflagellate group (Figure S7). The tree topology implies that the last common ancestor of holozoans had a single p53/63/73 gene, which followed independent divergences in vertebrates and choanoflagellates. In the absence of DNA damage, p53 appears to be downregulated by ubiquitination, which in vertebrates is carried out by the vertebrate-exclusive Mdm2 protein. However, other mechanisms of regulation have been proposed, such as ubiquitin ligases or CBP/p300 (Shi et al. 2009). Interestingly, we identified CBP/p300 both in Capsaspora and M. brevicollis (see below), although whether CBP/p300 down-regulates p53 in these holozoans remains unknown.
Stat
Signal transducer and activator of transcription (STAT) proteins are TFs that, in response to a wide variety of extracellular signaling proteins, regulate the action of several genes that are involved in cell growth and homeostasis (Bromberg 2002; Levy and Darnell 2002). Structurally, STAT proteins have a N-terminal interacting domain, a STAT alpha domain with a coiled-coil structure involved in protein-protein interactions (e.g., it recruits HATs, specially CBP/p300), a STAT DNA-binding domain, a SH2 domain and a C-terminal transactivation domain (Levy and Darnell 2002) (Figure S8). The activation of STAT is mediated by the phosphorylation of a key tyrosine residue, located after the SH2 domain (Levy and Darnell 2002). Our searches identified well-conserved STAT proteins in Capsaspora, M. brevicollis and the apusozoan T. trahens (Figure 1). The STAT proteins from the latter two taxa appear, however, to be slightly truncated at the 5′ end (see Figure S8). STAT proteins had previously been identified in amoebozoans (Kawata et al. 1997; Lee et al. 2008; Araki et al. 2010), but the protein domain analysis clearly showed that amoebozoan STAT are quite different from metazoan STAT proteins (Figure S8). In contrast, the homologs from M. brevicollis, Capsaspora, and T. trahens are very similar to metazoan STATs. Moreover, a phylogenetic analysis of STATs using amoebozoan CudA proteins as outgroup (Yamada et al. 2008), showed amoebozoan-specific STATs as a sister-group to the holozoan+apusozoan clade (BV=92%) (Figure S9). As STAT proteins are present in extant apusozoans, these are likely to have been lost early in the fungal lineage.
Metazoan STAT proteins form part of the JAK signaling pathway, which is absent in non-metazoan lineages (King et al. 2008). However, STAT proteins can interact with other receptor and non-receptor tyrosine kinases (Kawata et al. 1997; Levy and Darnell 2002). Indeed, the distribution of STATs coincides with the distribution of tyrosine kinases among eukaryotes, being present in amoebozoans (Kawata et al. 1997; Goldberg et al. 2006), apusozoans and Capsaspora (Ruiz-Trillo, unpublished data), choanoflagellates (King et al. 2008; Manning et al. 2008; Suga et al. 2008) and metazoans (Mayer 2008), all of which also have tyrosine kinases.
bZIP
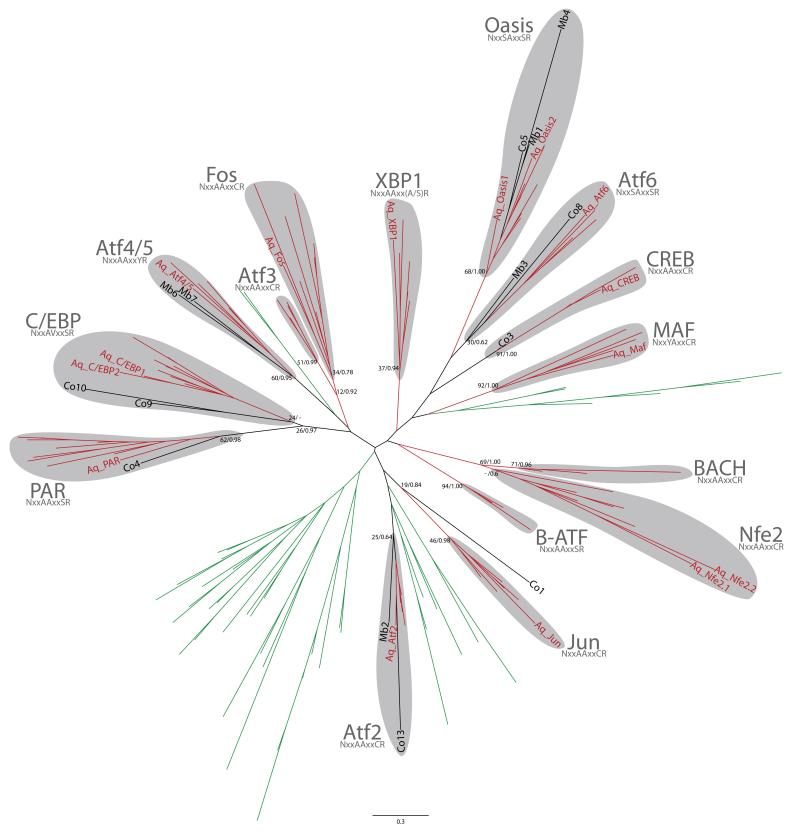
Basic-region leucine zipper (bZIP) TFs are named after the highly conserved structure containing a basic region (BR) and a leucine zipper (LZ) (Hurst 1994). bZIP proteins are ubiquitous among eukaryotes, and are involved in several processes such as environmental sensing and development (Deppmann, Alvania and Taparowsky 2006). We have identified 25 and 15 bZIP proteins in Capsaspora and M. brevicollis respectively. A. queenslandica has 20, and the average biltaterian 38 (Figure 1). Interestingly, the chytrid fungus A. macrogynus has 43 bZIP genes, while most Dikarya have approximately 13 (Figure 1). We could only classified unambiguously seven and six of the bZIP proteins present in Capsaspora and M. brevicollis respectively. A phylogenetic analysis including only the classified proteins showed that Capsaspora bZIPs correspond to PAR, C/EBP, Atf2, Oasis, Atf6 and CREB families, while the Monosiga homologs correspond to Atf4/5, Atf2, Oasis and Atf6 families (Figure 3, see Figure S10 for a tree with all Capsaspora genes). Based on these analyses, we hypothesize that most, if not all, current metazoan bZIP families were present in the holozoan ancestor, with some of them subsequently being lost in choanoflagellates and Capsaspora. Some families, such as CREB, most likely underwent a protein domain re-arrangement within metazoans, similarly to that described in other gene families (King et al. 2008; de Mendoza, Suga and Ruiz-Trillo 2010). Interestingly, all bZIP proteins that we identified in the unicellular relatives of metazoans belong to families that act strictly (Atf6, PAR, CREB, Oasis) or facultatively (Atf4/5, Atf2, C/EBP) as homodimers. This suggests that bZIP proteins in unicellular organisms may work mostly as homodimers, as already seen in yeast bZIP interactions (Deppmann, Alvania and Taparowsky 2006). Our data suggest that although bZIP originated before the dawn of the Metazoa, their connectivity and combinatorial interactions may have increased in animals. For example, the Capsaspora homolog of CREB does not have the kinase-inducible activation domain (KID) that allows its interaction with p300/CBP (Giebler, Lemasson and Nyborg 2000), even thought p300/CBP is present in the Capsaspora genome.
Figure 3.
Maximum likelihood tree of bZIP genes including the unambiguosly assigned Capsaspora bZIP homologs. The tree is rooted using the midpoint-rooted tree option. Statistical support was obtained by RAxML with 1000-bootstrap replicates (BV) and Bayesian Posterior Probabilities (BPP). Both values are shown on key branches. Aq (A. queenslandica), Co (Capsaspora), Mb (M. brevicollis). Metazoan branches depicted in red and fungal branches in green. For each family, the signature sequence for DNA recognition is indicated and only proteins with this conserved motif are included in the family (Fujii et al. 2000). A tree including all Capsaspora bZIP genes is shown in Figure S10.
bHLH
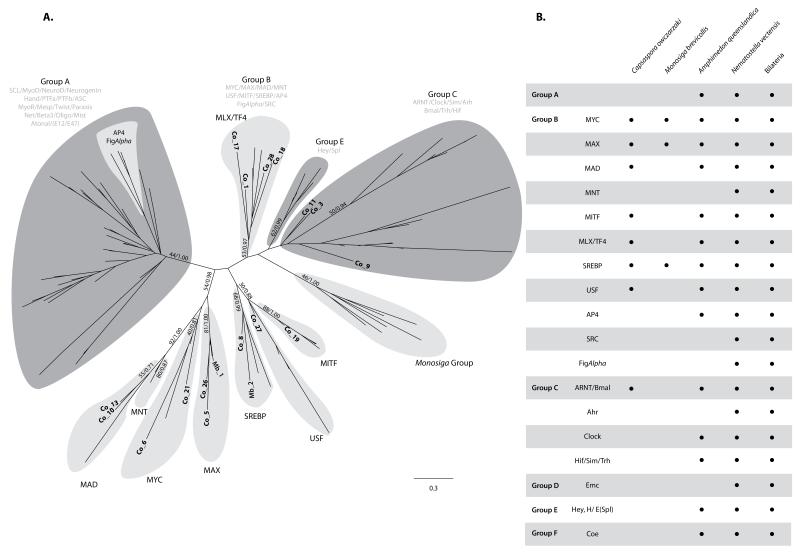
Basic helix-loop-helix (bHLH) is a domain that is present in a large superfamily of TFs that are widespread among eukaryotes. In metazoans they regulate critical developmental processes, such as neurogenesis, sex determination, myogenesis, and hematopoiesis (Jones 2004). This family of TFs has a DNA-binding basic region followed by two alpha-helices separated by a variable loop region. Many bHLH proteins also include other domains that are involved in protein-protein interactions (Simionato et al. 2007). bHLH proteins can act as homodimers or heterodimers to regulate gene expression. Metazoan bHLH have been grouped into six different higher order clades (A to F) (Simionato et al. 2007; Degnan et al. 2009) (see Figure 4 for a general overview). Group A, which includes genes such as MyoD and neurogenin, has only a bHLH domain and is exclusive to metazoans. Group B, which includes Myc or SREBP, has a leucine-zipper 3′ to the bHLH domain and is found throughout the eukaryotes. Group C, which includes Clock and ARNT, has two PAS domains (PAS and PAS3) 3′ to the bHLH domain, and is also thought to be exclusive to metazoans. Group D, which is metazoan-specific, lacks the DNA-binding basic region and, hence, their members are unable to bind to DNA, acting as antagonists to Group A members. Most group E proteins include a metazoan-specific orange domain and a WRPW peptide in their carboxyl terminal part. Finally, Group F lacks the DNA-binding basic region but possesses a COE domain, which is involved both in dimerization and binding (Simionato et al. 2007).
Figure 4.
A) Maximum likelihood tree of the bHLH domain including the unambiguosly assigned Capsaspora bHLH homologs. The tree is rooted using the midpoint-rooted tree option. Statistical support was obtained by RAxML with 100-bootstrap replicates (BV) and Bayesian Posterior Probabilities (BPP). Both values are shown on key branches. Groups and families are defined by the classificaiton of Simionato et al. (Simionato et al. 2007). The taxa used were H. sapiens, N. vectensis, A. queenslandica, M. brevicollis (Mb) and Capsaspora (Co). For the sake of clarity only the last two are specifically shown. B) Table of the presence/absence of bHLH groups and families in some key taxa. A. queenslandica, N. vectensis and Bilateria data obtained from Simionato et al (Simionato et al. 2007). A tree including all Capsaspora bHLH genes is shown in Figure S11.
We identified 31 bHLH proteins in Capsaspora, including orthologs of Myc, Mad, Max, SREBP, Mlx/TF4, MITF and USF group B families, nonspecific homologs (ARNT-like) of group C, and some unclassifiable proteins (see Figures 1, 4, S11, S12, and S13). This is more than double the bHLH genes found in Monosiga (11) and most fungi (around 10 in Dikarya). Compared to Metazoa, Capsaspora has a wider bHLH repertoire than the sponge A. queenslandica (18), but half what is present in cnidarians (72 in N. vectensis) or bilaterians (average of 69) (Figure 1). The choanoflagellate M. brevicollis contains Max, SREBP, and Myc and a lineage-specific group of bHLH genes. Thus, homologs of group C of bHLH were already present in the common ancestor of Capsaspora, choanoflagellates, and metazoans. Our data reveals that a basic Myc, MAX, Mxd/Mnt network of bHLH TFs was already present in the common ancestor of metazoans and Capsaspora, and became more complex in multicellular life-styles, incorporating, for example, Mnt and Mga. Interestingly, Capsaspora bHLH proteins are all homologs of those implicated in cell cycle and metabolism and none of those are involved in differentiation. From our survey we can also corroborate the two expansions periods (of bHLH groups and classes) in bHLH evolution previously inferred by Simionato et al. (2007) and later revised by Degnan et al. (2009), one before the divergence between Capsaspora and choanoflagellates+metazoans, and another early in eumetazoan evolution (Figure 1). In contrast to bZIP TFs, there are some putative heterodimeric TF interactions among Capsaspora bHLH. For example, Myc, Mad, MAX, and Mlx can act as heterodimers in metazoans.
Mef2
Myocite enhancer factors 2 (Mef2) are the metazoan representatives of Type II MADS-box genes. They are characterized by the presence of a Mef2 domain following the N-terminal MADS domain (Alvarez-Buylla et al. 2000). Mef2 genes play important roles in metazoan development, especially in the mesoderm (Potthoff and Olson 2007). Some authors considered Mef2 to be metazoan-specific (Larroux et al. 2006; Degnan et al. 2009), although other authors had proposed Saccharomyces Smp1 and Rlm1 genes to be fungal homologs of metazoan Mef2 (Dodou and Treisman 1997). We identified canonical metazoan-type Mef2 in Capsaspora and in the cythrid fungi S. punctatus and A. macrogynus (Figure 1). The protein structure of Capsaspora and S. punctatus Mef2 closely resembles the canonical metazoan Mef2 (Figure S14). We also identified a putative Mef2 homolog in M. brevicollis and in the amoeobozoans Dictyostelium discoideum, Dictyostelium purpureum, Entamoeba histolytica and A. castellanii as well as in the oomycetes Phytophthora sojae, P. ramorum, P. infestans and P. caspis, although their sequences are divergent and have little similarity to the canonical metazoan Mef2 (Figure S14). The fact that Phytophthora species encodes a Mef2 homolog may be explained by a lateral gene transfer (LGT) event, since they are the only analyzed eukaryotes outside opisthokonts and amoebozoans to have a mef2 gene. In fact, it has already been shown that some Phytophthora genes have a close relationship with amoebozoans genes (Tyler et al. 2006; Torruella et al. 2009). A phylogenetic analysis using fungal sequences as outgroup yields a clade that comprises all the taxa with a canonical metazoan-type Mef2 domain, that is all metazoans plus Capsaspora, A. macrogynus and S. punctatus (Figure S15). Our data show that the canonical metazoan Mef2 domain has a deeper origin than previously thought, with a conserved Mef2 domain present at least in the common ancestor of opisthokonts.
Fox
Fox genes TFs are characterized by the presence of a DNA binding domain known as Forkhead box. Fox genes play important roles as regulators of both development and metabolism, and they seem to be specific to opisthokonts (Tuteja and Kaestner 2007a, 2007b; Shimeld, Degnan and Luke 2009). We identified four Fox genes in Capsaspora, none of them being part of the metazoan-specific class I, but rather present in the supposedly opisthokont specific class II that also includes fungi (Larroux et al. 2008) (Figure 1 and S16). Interestingly, we identified three putative Fox genes in the amoebozoan A. castellanii (Figure 1), although their sequences are divergent compared to opisthokont ones. Thus, our results show that Fox genes are not specific to opisthokonts and were already present before the divergence of amoebozoans and opisthokonts.
HMG box genes
HMG-box containing genes are TFs that are involved in genome stability, chromatin structure and gene regulation (Stros, Launholt and Grasser 2007). Metazoan specific families are Sox and Tcf/Lef (Larroux et al. 2008). We characterized nine HMG-box-containing proteins in Capsaspora (Figures 1 and S17), a similar number as those found in Monosiga (12) and Amoebozoa (average of 10), and significantly less than those found in Bilateria (average of 45). Two of Capsaspora HMG box genes have strong similarities to MATalpha box, typical sex-determinant genes that are present in Ascomycota (Fraser and Heitman 2003; Fraser et al. 2004). Capsaspora also encodes a HMG-B, a SSRP-1, and a SWI/SNF homolog, plus some HMG-box containing genes that can not confidently be assigned to any HMG-box class.
Homeobox genes
Homeobox genes encode an acid helix-turn-helix DNA-binding motif known as the homeodomain. Homeobox genes are known to have key roles in animal, plant, fungal and amoebozoan development, such as regional patterning, regulation of cell proliferation, differentiation, adhesion, and migration (Gehring, Affolter and Burglin 1994; Derelle et al. 2007). There are two large superfamilies, the canonical (non-TALE) class with a 60-amino-acids homeodomain and the Three-Amino acid-Loop Extension (TALE) superclass characterized by an insertion of three amino acids between helix 1 and 2 of the homeodomain (Mukherjee and Burglin 2007). Both TALE and non-TALE superclasses were already present in the ancestor of eukaryotes (Derelle et al. 2007). The two homeobox genes of the choanoflagellate M. brevicollis have already been characterized, both of them belonging to the TALE superclass, although they can not confidently be assigned to any major metazoan homeobox family (King et al. 2008; Larroux et al. 2008). We identified nine homeodomain-containing genes in Capsaspora: three TALE and six non-TALE (Figures 1, and S18-S22). A phylogenetic analysis of these genes including members of all major families of homeodomains from metazoans, amoebozoans and fungi failed to confidently assign Capsaspora homeobox genes to any of the major metazoan classes, except for one clear ortholog to the longevity assurance homolog (LAG-1) class. To further improve the resolution and classify the remaining Capsaspora homeobox genes, we performed phylogenetic analyses specific for TALE or non-TALE genes. This allowed us to assign one Capsaspora TALE homeobox gene to the PBC family, althought it lacks the PBC N-terminal domain and support is not very high. The remaining two Capsaspora TALE genes have an unclear phylogenetic relationship to other TALEs although they appear to be closely related to the two M. brevicollis homeobox genes (Figure S19). Interestingly, the sponge A. queenslandica appears not to have a homolog of PBC (Figure S19) (Larroux et al. 2008). Capsaspora non-TALE genes appeared in unclear phylogenetic positions even with a restricted non-TALE-only dataset, although there is a potential homolog of LIM and two potential homologs of POU (Figure S20). Thus, in order to classify them we constructed different phylogenetic trees in which Capsaspora genes were forced to be members of a specific family and then we compared the likelihood values among all possible trees (for further details see Material & methods). Four Capsaspora non-TALE homologs appear to be at the root of the tree. Another one (Capsaspora-6) falls within the paired-like (Prd-like) clade with significant statistical support, this gene product possesses five of the six diagnostic amino acids of Prd-like genes (Galliot, de Vargas and Miller 1999) (Figure S21). However, it does not have the typical Q or K amino acid at position 50, and its intron it is not located in the typical position (between codons 46 and 47), as consistently observed in metazoans. A specific phylogeny of ANTP, prd-like, LIM, and POU also supports this assignment but is not statistically significant (Figure S21). The last Capsaspora non-TALE homeobox gene has a C-terminal TRAM LAG1 CLN8 (TLC) domain and a transmembrane domain, the characteristic domain architecture of the lass (Longevity assurance homologs of yeast (Lag-1)) genes, which are considered to be homologs to fungal Lag genes. Interestingly, phylogenetic analysis of the TLC domain showed that LAG genes with homedomain are exclusive to metazoans and Capsaspora, while genes with the TLC domains and TRAM1 domain are found in amoebozoans, fungi and metazoans (Figure S22). Lass genes, however, are implicated in ceramide synthesis, the function of their homeodomain being unclear and their specific transcription factor activity unknown (Teufel et al. 2009). Our data show that the repertoire of homeobox genes in metazoan unicellular relatives is larger than previously thought (see Figure 1), however some specific homeobox gene classes, such as ANTP appear to be exclusive to the Metazoa. Genome data from additional unicellular relatives of metazoans will be needed to corroborate this.
CBP/p300
The CREB-binding protein(CBP)/p300 is a ubiquitous metazoan transcriptional coactivator that interacts with several TFs, acting as an acetyltransferase (Coutts and La Thangue 2005) and being involved in cell growth and development (Goodman and Smolik 2000). Specifically, CBP/p300 interacts with such TFs such as NF-kappaB (Perkins et al. 1997), Stat (Levy and Darnell 2002; Wojciak et al. 2009), Runx (Jin et al. 2004; Makita et al. 2008), p53 (Grossman 2001), CREB (Manna, Dyson and Stocco 2009) and C/EBP (Manna, Dyson and Stocco 2009). For example, CBP/p300 acetylates Runx genes (Jin et al. 2004) and ubiquitinates p53 (Shi et al. 2009). We have identified CBP/p300 homologs in both Capsaspora and M. brevicollis. This implies that CBP/p300 originated prior to the divergence of Capsaspora from choanoflagellates and metazoans. It is worth mentioning that this multifunctional cofactor seems to have evolved concomitant to the emergence of several holozoan TFs, such as Runx and NF-kappaB. This suggests that a relatively high level of regulatory complexity was already emerging on early in the holozoan lineage, well before the divergence of metazoan and choanoflagellate lineages.
LSF/Grainyhead
The LSF/Grainyhead family of TFs is characterized by the CP2 domain and its members play important roles in bilaterians, being involved in vertebrate organogenesis, cell cycle progression and cell survival and differentiation (Bray and Kafatos 1991; Uv et al. 1997; Veljkovic and Hansen 2004; Traylor-Knowles et al. 2010). LSF/Grainyhead can be divided into two groups, the LSF/CP2 and the GRH subfamilies (Shirra and Hansen 1998; Traylor-Knowles et al. 2010). Members of the LSF/CP2 subfamily act as tetramers and possess an extra sterile alpha motif (SAM) domain C-terminal to the specific CP2 DNA-binding domain. Members of the Grainyhead (GRH) subfamily do not have the SAM domain and act as dimers.
The CP2 domain is present throughout the opisthokonts, including choanoflagellates (Traylor-Knowles et al. 2010), and seems to be a synapomorphy of this group of eukaryotes. Interestingly, it has been hypothesized that the GRH subfamily originated by duplication of an ancestral LSF/GRH-like gene at the origin of the Metazoa and was co-opted to epidermal determination in metazoans (Traylor-Knowles et al. 2010). We identified two LSF/Grainyhead genes in Capsaspora, a LSF-like and a GRH-like (Figures 1 and S23), although the Capaspora LSF-like gene lacks the characteristic C-terminal SAM domain found in metazoan LSF proteins. This domain may have been gained by domain shuffling before the split between metazoans and choanoflagellates, although the loss of this domain in Capsaspora cannot be ruled out. Our findings imply that the duplication of the LSF/GRH gene occurred before Capsaspora diversified from choanoflagellates and metazoans, and that GRH was lost in choanoflagellates. Thus, the presence of a GRH gene antedates the origin of the metazoan epithelium.
Nuclear receptors (NR), Smad and Ets
Our data show that these three TFs families remain, at this time, metazoan-specific since we did not identify any homologs in non-metazoan taxa. The complete genome sequences of additional non-metazoan taxa are needed to corroborate this hypothesis.
Origin and early evolution of metazoan TFs
The repertoire of TFs in the holozoan Capsaspora owczarzaki reported here and its comparison with metazoan, fungal and choanoflagellate TFs provides important insights into the origin and evolution of TFs that are essential for metazoan multicellularity. This allows us to propose a new hypothesis regarding the origin of key metazoan TFs (see Figure 5). Some metazoan TF domains have deep origins being widespread in eukaryotes, such as HMGbox, homeodomain (both TALE and non-TALE), bHLH, bZIP, or Mef2-like (see also Degnan et al. 2009). However, major diversifications of genes encoding some of these domains took place along metazoan and fungal stems (see Figure 1), generating lineage-specific classes and subfamilies. In regards to metazoan-specific TF gene families, there appears to have been two major expansions (Figure 5): one prior to the divergence of Capsaspora, choanoflagellates and the Metazoa (for example, in bZIP and bHLH); and another within the metazoan lineage (such as Sox, homedomains, and further diversification of bZIP and bHLH). Several other TF domains, such as Churchill, STAT, and, most likely, Fox, were already present in the common ancestor of unikonts (i.e. amoebozoans, apusozoans and opisthokonts). This finding changes previous views in which Churchill was considered exclusive to metazoans and Fox exclusive to opisthokonts (although the assignment of A. castellanii hits to Fox remain contentious). Although STAT domains were present in the common ancestor of unikonts, our data show that canonical metazoan-type STAT seem to be exclusive to apusozoans (T. trahens) and opisthokonts. A major challenge to previous proposals that T-box genes are metazoan innovations is the discovery of T-box genes in S. punctatus and Capsaspora. This means that T-box genes appeared before the divergence of fungi and holozoans. What role are these T-box genes playing in these non-metazoan lineages remains to be studied.
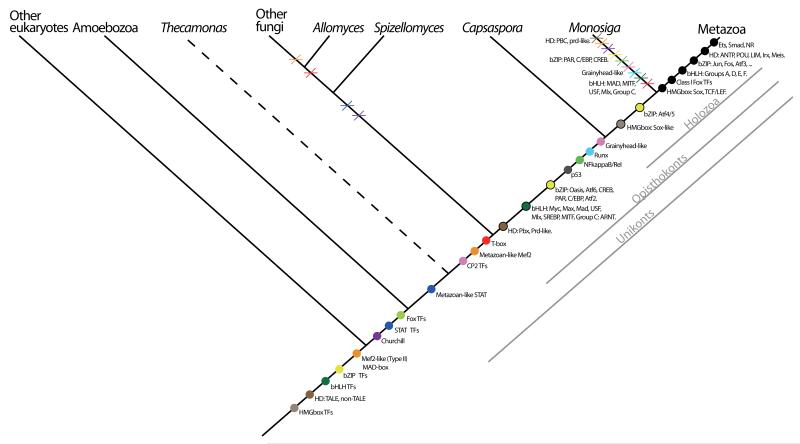
Figure 5.
Cladogram representing TF evolution among the analyzed taxa. Colors are unique for each domain class. A colored dot means the hypothetical origin of the domain. A black circled dot indicates where a specific protein family appears in our taxon sampling. A cross means the loss of the domain or specific protein family in a lineage. Metazoan apomorphies are shown as black dots. The phylogenetic relationships are based on several recent studies (Burki et al. 2008; Ruiz-Trillo et al. 2008; Brown, Spiegel and Silberman 2009; Liu et al. 2009; Minge et al. 2009).
Interestingly, some TFs appear to have evolved prior to the divergence of Capsaspora from choanoflagellates and metazoans, such as p53, Runx and NF-kappaB; the latter two previously being considered metazoan-specific. This pattern of gene families that are relevant to metazoan multicellularity evolving prior to the emergence of the metazoan stem lineage is not new, and has been observed in other cases such as tyrosine kinases, cadherins, MAGUKs, and integrins (Abedin and King 2008; King et al. 2008; Manning et al. 2008; Suga et al. 2008; Degnan et al. 2009; de Mendoza, Suga and Ruiz-Trillo 2010; Sebe-Pedros et al. 2010). Finally, there are some TF domains that, under the current taxon sampling, appear to be metazoan innovations. These are ETS, Smad, and nuclear receptors. Moreover, some specific homeobox genes (ANTP, LIM, POU, Irx, Meis, Tgif, Six), bZIP classes (for example, Jun, Fos), bHLH classes (A, D-F groups) and HMGbox classes (Sox, TCL/lef) appear also to be metazoan-specific (Figure 5), although we can not rule out the possibility that some of these may have a more ancient origin and secondarily lost in non-metazoan lineages. This new evolutionary scenario implies that significant lineage-specific TFs losses occurred within the choanoflagellate lineage. For example, Runx, T-box, RHD domain, GRH-like and Churchill appear to have been lost in M. brevicollis. Whether this is specific to one choanoflagellate lineage (that of M. brevicollis) or to choanoflagellates in general remains unknown. Only genomic data from additional choanoflagellate taxa will resolve this issue. A similar pattern of lineage-specific loss in choanoflagellates has recently been shown for the integrin-mediated adhesion machinery (Sebe-Pedros et al. 2010).
A quantitative analysis (Figure 1) of TFs evolution suggests that several expansions occurred in Eumetazoa, such as bHLH and homeobox gene families, and to a lesser degree HMGbox and bZIP families. Specific domain expansions have already been reported in the Viridiplantae for bHLH and homedomain proteins (Mukherjee, Brocchieri and Burglin 2009; Pires and Dolan 2010). There are several theories about the correlation of these expansions with the transition to multicellularity (Derelle et al. 2007; Pires and Dolan 2010). On the other hand, some TF domains, such as CP2, Runt, MADSbox, Churchill, p53 and STAT, have similar number of members in unicellular and multicellular holozoans. Capsaspora TF complexity is quite high, with a wider range of bHLH and bZIP domain-containing proteins than in some early-branching metazoans such A. queenslandica or T. adhaerens. Since Capsaspora has a complex (and not fully understood) life cycle, in which there is a symbiotic stage within the mollusc Biomphalaria glabrata, one may wonder whether the complexity of TFs identified in Capsaspora is due to lateral gene transfer from the host, or even from the trematode flatworm S. mansoni, a metazoan parasite of B. glabrata. Based on our phylogenetic analyses, we do not favor this hypothesis, None of the phylogenetic trees shown (all including bilaterians; some even B. glabrata homologs), show the Capsaspora homolog grouping closer to bilaterians than to other metazoans. Instead, we hypothesize that the common ancestor of Capsaspora, choanoflagellates, and metazoans had a richer TF repertoire than previously believed, and that some TFs were subsequently lost in the choanoflagellate lineage (or at least in M. brevicollis).
In summary, our results show that the evolution of metazoan TFs includes the acquisition of new genes (some of them via domain shuffling), gene co-option, and the diversification of ancestral domains increasing the combinatorial complexity. How these metazoan developmental TFs are functioning in unicellular organisms and how they were exapted into new functions in multicellular animals remains to be answered.
Supplementary Material
Acknowledgments
The genome sequences of C. owczarzaki, A. macrogynus, S. punctatus, and T. trahens are being determined by the Broad Institute of MIT/Harvard University under the auspices of the National Human Genome Research Institute (NHGRI) and within the UNICORN initiative. We thank JGI, BI, and BCM for making data publicly available. We thank Kim C. Worley and the team of the A. castellanii genome project for accession to the genome data. We thank Peter W. H. Holland for help in the assignment of homeodomain-containing proteins and Romain Derelle, Jordi Paps and Hiroshi Suga for helpful insights. This work was supported by an ICREA contract, an European Research Council Starting Grant (ERC-2007-StG-206883), and a grant (BFU2008-02839/BMC) from Ministerio de Ciencia e Innovación (MICINN) to IR-T. AS’s salary was supported by a pre-graduate FPU grant and AdM’s salary from a FPI grant both from MICINN. BFL’s and BMD’s contributions were supported by the Canadian Research Chair Program and the Australian Research Council, respectively.
Footnotes
See supplementary material file 1 for figures S1-S7; file 2 for figures S8 and S17, file 3 for figures S18-S23.
Supplementary material file 4 includes the annotation of the Capsaspora sequences included in this study.
Literature cited
- Abedin M, King N. The premetazoan ancestry of cadherins. Science. 2008;319:946–948. doi: 10.1126/science.1151084. [DOI] [PubMed] [Google Scholar]
- Adell T, Grebenjuk VA, Wiens M, Muller WE. Isolation and characterization of two T-box genes from sponges, the phylogenetically oldest metazoan taxon. Dev Genes Evol. 2003;213:421–434. doi: 10.1007/s00427-003-0345-5. [DOI] [PubMed] [Google Scholar]
- Adell T, Muller WE. Expression pattern of the Brachyury and Tbx2 homologues from the sponge Suberites domuncula. Biol Cell. 2005;97:641–650. doi: 10.1042/BC20040135. [DOI] [PubMed] [Google Scholar]
- Akamatsu Y, Ohno T, Hirota K, Kagoshima H, Yodoi J, Shigesada K. Redox regulation of the DNA binding activity in transcription factor PEBP2. The roles of two conserved cysteine residues. J Biol Chem. 1997;272:14497–14500. doi: 10.1074/jbc.272.23.14497. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Pelaz S, Liljegren SJ, Gold SE, Burgeff C, Ditta GS, Ribas de Pouplana L, Martinez-Castilla L, Yanofsky MF. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc Natl Acad Sci U S A. 2000;97:5328–5333. doi: 10.1073/pnas.97.10.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki T, van Egmond WN, van Haastert PJ, Williams JG. Dual regulation of a Dictyostelium STAT by cGMP and Ca2+ signalling. J Cell Sci. 2010;123:837–841. doi: 10.1242/jcs.064436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielen H, Oberleitner S, Marcellini S, Gee L, Lemaire P, Bode HR, Rupp R, Technau U. Divergent functions of two ancient Hydra Brachyury paralogues suggest specific roles for their C-terminal domains in tissue fate induction. Development. 2007;134:4187–4197. doi: 10.1242/dev.010173. [DOI] [PubMed] [Google Scholar]
- Bray SJ, Kafatos FC. Developmental function of Elf-1: an essential transcription factor during embryogenesis in Drosophila. Genes Dev. 1991;5:1672–1683. doi: 10.1101/gad.5.9.1672. [DOI] [PubMed] [Google Scholar]
- Bromberg J. Stat proteins and oncogenesis. J Clin Invest. 2002;109:1139–1142. doi: 10.1172/JCI15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MW, Spiegel FW, Silberman JD. Phylogeny of the “forgotten” cellular slime mold, Fonticula alba, reveals a key evolutionary branch within Opisthokonta. Mol Biol Evol. 2009;26:2699–2709. doi: 10.1093/molbev/msp185. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Burki F, Shalchian-Tabrizi K, Pawlowski J. Phylogenomics reveals a new ‘megagroup’ including most photosynthetic eukaryotes. Biol Lett. 2008;4:366–369. doi: 10.1098/rsbl.2008.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman JA. Runx transcription factors and the developmental balance between cell proliferation and differentiation. Cell Biol Int. 2003;27:315–324. doi: 10.1016/s1065-6995(03)00018-0. [DOI] [PubMed] [Google Scholar]
- Coutts AS, La Thangue NB. The p53 response: emerging levels of co-factor complexity. Biochem Biophys Res Commun. 2005;331:778–785. doi: 10.1016/j.bbrc.2005.03.150. [DOI] [PubMed] [Google Scholar]
- de Mendoza A, Suga H, Ruiz-Trillo I. Evolution of the MAGUK protein gene family in premetazoan lineages. BMC Evol Biol. 2010;10:93. doi: 10.1186/1471-2148-10-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan BM, Vervoort M, Larroux C, Richards GS. Early evolution of metazoan transcription factors. Curr Opin Genet Dev. 2009;19:591–599. doi: 10.1016/j.gde.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Deppmann CD, Alvania RS, Taparowsky EJ. Cross-species annotation of basic leucine zipper factor interactions: Insight into the evolution of closed interaction networks. Mol Biol Evol. 2006;23:1480–1492. doi: 10.1093/molbev/msl022. [DOI] [PubMed] [Google Scholar]
- Derelle R, Lopez P, Guyader HL, Manuel M. Homeodomain proteins belong to the ancestral molecular toolkit of eukaryotes. Evol Dev. 2007;9:212–219. doi: 10.1111/j.1525-142X.2007.00153.x. [DOI] [PubMed] [Google Scholar]
- Dodou E, Treisman R. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol Cell Biol. 1997;17:1848–1859. doi: 10.1128/mcb.17.4.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Espinosa JM. Mechanisms of regulatory diversity within the p53 transcriptional network. Oncogene. 2008;27:4013–4023. doi: 10.1038/onc.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, Diezmann S, Subaran RL, Allen A, Lengeler KB, Dietrich FS, Heitman J. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2004;2:e384. doi: 10.1371/journal.pbio.0020384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, Heitman J. Fungal mating-type loci. Curr Biol. 2003;13:R792–5. doi: 10.1016/j.cub.2003.09.046. [DOI] [PubMed] [Google Scholar]
- Fujii Y, Shimizu T, Toda T, Yanagida M, Hakoshima T. Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nat Struct Biol. 2000;7:889–893. doi: 10.1038/82822. [DOI] [PubMed] [Google Scholar]
- Galliot B, de Vargas C, Miller D. Evolution of homeobox genes: Q50 Paired-like genes founded the Paired class. Dev Genes Evol. 1999;209:186–197. doi: 10.1007/s004270050243. [DOI] [PubMed] [Google Scholar]
- Gauthier M, Degnan BM. The transcription factor NF-kappaB in the demosponge Amphimedon queenslandica: insights on the evolutionary origin of the Rel homology domain. Dev Genes Evol. 2008;218:23–32. doi: 10.1007/s00427-007-0197-5. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Affolter M, Burglin T. Homeodomain proteins. Annu Rev Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- Giebler HA, Lemasson I, Nyborg JK. p53 recruitment of CREB binding protein mediated through phosphorylated CREB: a novel pathway of tumor suppressor regulation. Mol Cell Biol. 2000;20:4849–4858. doi: 10.1128/mcb.20.13.4849-4858.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg JM, Manning G, Liu A, Fey P, Pilcher KE, Xu Y, Smith JL. The dictyostelium kinome--analysis of the protein kinases from a simple model organism. PLoS Genet. 2006;2:e38. doi: 10.1371/journal.pgen.0020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- Grossman SR. p300/CBP/p53 interaction and regulation of the p53 response. Eur J Biochem. 2001;268:2773–2778. doi: 10.1046/j.1432-1327.2001.02226.x. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- Hurst HC. Transcription factors. 1: bZIP proteins. Protein Profile. 1994;1:123–168. [PubMed] [Google Scholar]
- Jin YH, Jeon EJ, Li QL, Lee YH, Choi JK, Kim WJ, Lee KY, Bae SC. Transforming growth factor-beta stimulates p300-dependent RUNX3 acetylation, which inhibits ubiquitination-mediated degradation. J Biol Chem. 2004;279:29409–29417. doi: 10.1074/jbc.M313120200. [DOI] [PubMed] [Google Scholar]
- Jones S. An overview of the basic helix-loop-helix proteins. Genome Biol. 2004;5:226. doi: 10.1186/gb-2004-5-6-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Miyata T, Toh H. Improvement in the accuracy of multiple sequence alignment program MAFFT. Genome Inform. 2005a;16:22–33. [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005b;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata T, Shevchenko A, Fukuzawa M, Jermyn KA, Totty NF, Zhukovskaya NV, Sterling AE, Mann M, Williams JG. SH2 signaling in a lower eukaryote: a STAT protein that regulates stalk cell differentiation in dictyostelium. Cell. 1997;89:909–916. doi: 10.1016/s0092-8674(00)80276-7. [DOI] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larroux C, Luke GN, Koopman P, Rokhsar DS, Shimeld SM, Degnan BM. Genesis and expansion of metazoan transcription factor gene classes. Mol Biol Evol. 2008;25:980–996. doi: 10.1093/molbev/msn047. [DOI] [PubMed] [Google Scholar]
- Larroux C, Fahey B, Liubicich D, Hinman V,F, Gauthier M, Gongora M, Green K, WoÃàrheide G, Leys S,P, Degnan B,M. Developmental expression of transcription factor genes in a demosponge: Insights into the origin of metazoan multicellularity. Evolution and Development. 2006;8:150–173. doi: 10.1111/j.1525-142X.2006.00086.x. [DOI] [PubMed] [Google Scholar]
- Lee NS, Rodriguez M, Kim B, Kim L. Dictyostelium kinase DPYK3 negatively regulates STATc signaling in cell fate decision. Dev Growth Differ. 2008;50:607–613. doi: 10.1111/j.1440-169X.2008.01058.x. [DOI] [PubMed] [Google Scholar]
- Levy DE, Darnell JEJ. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- Liu Y, Steenkamp ET, Brinkmann H, Forget L, Philippe H, Lang BF. Phylogenomic analyses predict sistergroup relationship of nucleariids and fungi and paraphyly of zygomycetes with significant support. BMC Evol Biol. 2009;9:272. doi: 10.1186/1471-2148-9-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londin ER, Mentzer L, Sirotkin HI. Churchill regulates cell movement and mesoderm specification by repressing Nodal signaling. BMC Dev Biol. 2007;7:120. doi: 10.1186/1471-213X-7-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- Makita N, Suzuki M, Asami S, Takahata R, Kohzaki D, Kobayashi S, Hakamazuka T, Hozumi N. Two of four alternatively spliced isoforms of RUNX2 control osteocalcin gene expression in human osteoblast cells. Gene. 2008;413:8–17. doi: 10.1016/j.gene.2007.12.025. [DOI] [PubMed] [Google Scholar]
- Manna PR, Dyson MT, Stocco DM. Role of basic leucine zipper proteins in transcriptional regulation of the steroidogenic acute regulatory protein gene. Mol Cell Endocrinol. 2009;302:1–11. doi: 10.1016/j.mce.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Young SL, Miller WT, Zhai Y. The protist, Monosiga brevicollis, has a tyrosine kinase signaling network more elaborate and diverse than found in any known metazoan. Proc Natl Acad Sci U S A. 2008;105:9674–9679. doi: 10.1073/pnas.0801314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcellini S, Technau U, Smith JC, Lemaire P. Evolution of Brachyury proteins: identification of a novel regulatory domain conserved within Bilateria. Dev Biol. 2003;260:352–361. doi: 10.1016/s0012-1606(03)00244-6. [DOI] [PubMed] [Google Scholar]
- Mayer BJ. Clues to the evolution of complex signaling machinery. PNAS. 2008;105:9453–9454. doi: 10.1073/pnas.0804669105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minge MA, Silberman JD, Orr RJ, Cavalier-Smith T, Shalchian-Tabrizi K, Burki F, Skjaeveland A, Jakobsen KS. Evolutionary position of breviate amoebae and the primary eukaryote divergence. Proc Biol Sci. 2009;276:597–604. doi: 10.1098/rspb.2008.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K, Brocchieri L, Burglin TR. A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol Biol Evol. 2009;26:2775–2794. doi: 10.1093/molbev/msp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K, Burglin TR. Comprehensive analysis of animal TALE homeobox genes: new conserved motifs and cases of accelerated evolution. J Mol Evol. 2007;65:137–153. doi: 10.1007/s00239-006-0023-0. [DOI] [PubMed] [Google Scholar]
- Muller CW, Herrmann BG. Crystallographic structure of the T domain-DNA complex of the Brachyury transcription factor. Nature. 1997;389:884–888. doi: 10.1038/39929. [DOI] [PubMed] [Google Scholar]
- Nedelcu AM, Tan C. Early diversification and complex evolutionary history of the p53 tumor suppressor gene family. Dev Genes Evol. 2007;217:801–806. doi: 10.1007/s00427-007-0185-9. [DOI] [PubMed] [Google Scholar]
- Perkins ND, Felzien LK, Betts JC, Leung K, Beach DH, Nabel GJ. Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- Pires N, Dolan L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol Biol Evol. 2010;27:862–874. doi: 10.1093/molbev/msp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Rennert J, Coffman JA, Mushegian AR, Robertson AJ. The evolution of Runx genes I. A comparative study of sequences from phylogenetically diverse model organisms. BMC Evol Biol. 2003;3:4. doi: 10.1186/1471-2148-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson AJ, Larroux C, Degnan BM, Coffman JA. The evolution of Runx genes II. The C-terminal Groucho recruitment motif is present in both eumetazoans and homoscleromorphs but absent in a haplosclerid demosponge. BMC Res Notes. 2009;2:59. doi: 10.1186/1756-0500-2-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A. The origins of multicellularity and the early history of the genetic toolkit for animal development. Annu Rev Genet. 2008;42:235–251. doi: 10.1146/annurev.genet.42.110807.091513. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. (Oxford, England) [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Burger G, Holland PW, King N, Lang BF, Roger AJ, Gray MW. The origins of multicellularity: a multi-taxon genome initiative. Trends Genet. 2007;23:113–118. doi: 10.1016/j.tig.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Inagaki Y, Davis LA, Sperstad S, Landfald B, Roger AJ. Capsaspora owczarzaki is an independent opisthokont lineage. Curr Biol. 2004;14(22):R946–947. doi: 10.1016/j.cub.2004.10.037. [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Roger AJ, Burger G, Gray MW, Lang BF. A phylogenomic investigation into the origin of metazoa. Mol Biol Evol. 2008;25:664–672. doi: 10.1093/molbev/msn006. [DOI] [PubMed] [Google Scholar]
- Scholz CB, Technau U. The ancestral role of Brachyury: expression of NemBra1 in the basal cnidarian Nematostella vectensis (Anthozoa) Dev Genes Evol. 2003;212:563–570. doi: 10.1007/s00427-002-0272-x. [DOI] [PubMed] [Google Scholar]
- Sebe-Pedros A, Roger AJ, Lang FB, King N, Ruiz-Trillo I. Ancient origin of the integrin-mediated adhesion and signaling machinery. Proc Natl Acad Sci U S A. 2010;107:10142–10147. doi: 10.1073/pnas.1002257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalchian-Tabrizi K, Minge MA, Espelund M, Orr R, Ruden T, Jakobsen KS, Cavalier-Smith T. Multigene phylogeny of choanozoa and the origin of animals. PLoS ONE. 2008;3:e2098. doi: 10.1371/journal.pone.0002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng G, dos Reis M, Stern CD. Churchill, a zinc finger transcriptional activator, regulates the transition between gastrulation and neurulation. Cell. 2003;115:603–613. doi: 10.1016/s0092-8674(03)00927-9. [DOI] [PubMed] [Google Scholar]
- Shi D, Pop MS, Kulikov R, Love IM, Kung AL, Grossman SR. CBP and p300 are cytoplasmic E4 polyubiquitin ligases for p53. Proc Natl Acad Sci U S A. 2009;106:16275–16280. doi: 10.1073/pnas.0904305106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimeld SM, Degnan B, Luke GN. Evolutionary genomics of the Fox genes: Origin of gene families and the ancestry of gene clusters. Genomics. 2009 doi: 10.1016/j.ygeno.2009.08.002. doi.org/10.1016/j.ygeno.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Shirra MK, Hansen U. LSF and NTF-1 share a conserved DNA recognition motif yet require different oligomerization states to form a stable protein-DNA complex. J Biol Chem. 1998;273:19260–19268. doi: 10.1074/jbc.273.30.19260. [DOI] [PubMed] [Google Scholar]
- Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, Coornaert D, Degnan BM, Vervoort M. Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol Biol. 2007;7:33. doi: 10.1186/1471-2148-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. T-box genes: what they do and how they do it. Trends Genet. 1999;15:154–158. doi: 10.1016/s0168-9525(99)01693-5. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier MEA, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stanke M, Morgenstern B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005;33:W465–7. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stros M, Launholt D, Grasser KD. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell Mol Life Sci. 2007;64:2590–2606. doi: 10.1007/s00018-007-7162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H, Sasaki G, Kuma K, Nishiyori H, Hirose N, Su ZH, Iwabe N, Miyata T. Ancient divergence of animal protein tyrosine kinase genes demonstrated by a gene family tree including choanoflagellate genes. FEBS Lett. 2008;582:815–818. doi: 10.1016/j.febslet.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Sullivan JC, Sher D, Eisenstein M, Shigesada K, Reitzel AM, Marlow H, Levanon D, Groner Y, Finnerty JR, Gat U. The evolutionary origin of the Runx/CBFbeta transcription factors--studies of the most basal metazoans. BMC Evol Biol. 2008;8:228. doi: 10.1186/1471-2148-8-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teufel A, Maass T, Galle PR, Malik N. The longevity assurance homologue of yeast lag1 (Lass) gene family (review) Int J Mol Med. 2009;23:135–140. [PubMed] [Google Scholar]
- Torruella G, Suga H, Riutort M, Pereto J, Ruiz-Trillo I. The Evolutionary History of Lysine Biosynthesis Pathways Within Eukaryotes. J Mol Evol. 2009;69:240–248. doi: 10.1007/s00239-009-9266-x. [DOI] [PubMed] [Google Scholar]
- Traylor-Knowles N, Hansen U, Dubuc TQ, Martindale MQ, Kaufman L, Finnerty JR. The evolutionary diversification of LSF and Grainyhead transcription factors preceded the radiation of basal animal lineages. BMC Evol Biol. 2010;10:101. doi: 10.1186/1471-2148-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja G, Kaestner KH. Forkhead transcription factors II. Cell. 2007a;131:192. doi: 10.1016/j.cell.2007.09.016. [DOI] [PubMed] [Google Scholar]
- Tuteja G, Kaestner KH. SnapShot: forkhead transcription factors I. Cell. 2007b;130:1160. doi: 10.1016/j.cell.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Tyler BM, Tripathy S, Zhang X, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- Uv AE, Harrison EJ, Bray SJ. Tissue-specific splicing and functions of the Drosophila transcription factor Grainyhead. Mol Cell Biol. 1997;17:6727–6735. doi: 10.1128/mcb.17.11.6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veljkovic J, Hansen U. Lineage-specific and ubiquitous biological roles of the mammalian transcription factor LSF. Gene. 2004;343:23–40. doi: 10.1016/j.gene.2004.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler JC, Shigesada K, Gergen JP, Ito Y. Mechanisms of transcriptional regulation by Runt domain proteins. Semin Cell Dev Biol. 2000;11:369–375. doi: 10.1006/scdb.2000.0184. [DOI] [PubMed] [Google Scholar]
- Wojciak JM, Martinez-Yamout MA, Dyson HJ, Wright PE. Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains. EMBO J. 2009;28:948–958. doi: 10.1038/emboj.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Wang HY, Fukuzawa M, Barton GJ, Williams JG. A new family of transcription factors. Development. 2008;135:3093–3101. doi: 10.1242/dev.026377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.