Abstract
The antiapoptotic function of the interleukin-7 (IL-7) receptor is related to regulation of three members of the Bcl2 family: synthesis of Bcl2, phosphorylation of Bad, and cytosolic retention of Bax. Here we show that, in an IL-7-dependent murine T-cell line, different regions of the IL-7 receptor initiate the signal transduction pathways that regulate these proteins. Both Box1 and Y449 are required to signal Bcl2 synthesis and Bax cytosolic retention. This suggests a sequential model in which Jak1, which binds to Box1, is first activated and then phosphorylates Y449, leading to Bcl2 and Bax regulation, accounting for approximately 90% of the survival function. Phosphorylation of Bad required Box1 but not Y449, suggesting that Jak1 also initiates an additional signaling cascade that accounts for approximately 10% of the survival function. Stat5 was activated from the Y449 site but only partially accounted for the survival signal. Proliferation required both Y449 and Box1. Thymocyte development in vivo showed that deletion of Y449 eliminated 90% of αβ T-cell development and completely eliminated γδ T-cell development, whereas deleting Box 1 completely eliminated both αβ and γδ T-cell development. Thus the IL-7 receptor controls at least two distinct pathways, in addition to Stat5, that are required for cell survival.
Interleukin-7 (IL-7) plays an essential role in the development and maintenance of T lymphocytes. This requirement was first demonstrated by treating mice with anti-IL-7 antibodies, which severely interrupted development of T cells (4, 11). Similar T-cell deficiencies were noted in mice with knockouts of IL-7 or of either of its two receptor chains, IL-7Rα and -γc, or of Jak3, the kinase that is associated with IL-7Rγc (5, 9, 18, 23, 26-28, 37, 41). In humans, genetic defects in either IL-7Rα (30, 31), γc (21), or Jak3 (17, 32) produced a block in T-cell development and are a major cause of the severe combined immunodeficiency disease (SCID) phenotype. More recently, it was appreciated that the T cell also requires IL-7 after leaving the thymus for homeostatic survival and proliferation (33).
The IL-7Rα chain is shared with the receptor for TSLP, whereas the γc chain is shared by the cytokine receptors for IL-2, IL-4, IL-9, IL-15, and IL-21. The IL-7Rα chain belongs to the type I cytokine receptor family and is a membrane glycoprotein with a single 25-amino-acid (aa) transmembrane domain. The intracellular domain of 195 aa (39) does not have the features of a signaling kinase but includes potential docking sites for signaling molecules. One region is rich in acidic residues (A region), one region is rich in serine residues (S region), and a tyrosine (T) region contains three tyrosine residues, Tyr401, Tyr449, and Tyr456, which are conserved in evolution between the murine and human IL-7Rα chains. In addition, proximal to the membrane, an 8-aa motif termed Box1 (20) is homologous within the type I cytokine receptor family and can bind the signaling kinase Jak1.
In previous studies, the activity of IL-7 on T cells was partly attributed to a survival or trophic effect mediated by the balance of pro- versus antiapoptotic members of the Bcl2 family. IL-7 induced synthesis of antiapoptotic Bcl2 (15), whereas it blocked the proapoptotic proteins Bad and Bax by posttranslational mechanisms. IL-7 signals constrained Bax to the cytosol, stopping it from entering mitochondria and initiating apoptosis (13). IL-7 signals induced phosphorylation of Bad (14; W. Q. Li et al., submitted for publication), inducing its binding to 14-3-3, which prevents it from translocating to mitochondrion and inactivating Bcl2. The importance of the balance of Bcl2/Bax in the IL-7 response has been demonstrated in vivo; these results show that the deficiency in T-cell development in IL-7R−/− mice can be rescued by overexpressing Bcl2 (1, 19) or deleting Bax (14).
It has been reported that IL-7 stimulates several signaling molecules, including the Stats (38), phosphatidylinositol 3-kinase (PI3K) (6, 7), src family kinases (25, 34), and Pyk2 (3). However, based on the phenotype of mice in which each of these candidate signaling molecules has been knocked out, no single one of these components can thus far be shown to be essential to the IL-7 signal; in contrast, IL-7Rα, γc, and Jak3 must be critically required to initiate the signaling since mice with each individually knocked out show the thymic deficiency of IL-7 knockouts (reviewed by Jiang et al. [12a]). Assuming that the next stage in signaling begins with proteins docking to the intracellular domain of the IL-7Rα chain, as an initial step in identifying the signaling pathways that regulate the Bcl2 family, we focused on this intracellular domain. Deletion of various regions of the intracellular domain of the IL-7Rα chain was performed, and the effects on the induction of Bcl2, Bax cytosolic retention, and Bad phosphorylation were examined. The results suggest that at least two distinct pathways, in addition to the Stat5 pathway, are involved in the IL-7 survival effects.
MATERIALS AND METHODS
Cell lines.
The IL-7-dependent cell line D1 (15) was maintained in RPMI 1640 (Mediatech Inc.) supplemented with 10% fetal bovine serum (FBS; HyClone), 50 μM β-mercaptoethanol (Invitrogen), 100 U of penicillin per ml, 100 μg of streptomycin (Mediatech Inc.) per ml, and 50 ng of mouse IL-7 (PeproTech) per ml. The Phoenix-Eco and Phoenix amphotrophic retrovirus packaging cell lines (22) were maintained in Dulbecco’s modification of Eagle medium (Mediatech Inc.) supplemented with 10% FBS. C1498, a murine acute myeloid leukemia cell line, and BW5147, a murine thymic lymphoma cell line, were maintained in RPMI 1640 supplemented with 10% FBS.
Retroviral chimeric receptor plasmid construction and D1 cell infection.
The full-length murine IL-7R (mIL-7R) was cloned from the D1 cell line by reverse transcription-PCR. The extracellular domain of human IL-4R (hIL-4R) was amplified from pDL-hIL-4R, generously provided by Immunex Corp. The extracellular domain of hIL-4R and the transmembrane and intracellular domains of mIL-7R were fused and cloned into the pMIG vector (a gift from J. Keller, National Cancer Institute), designated pMIG-hIL-4R/mIL-7R(WT). Different mutations and deletions of the intracellular domain were prepared by PCR strategies. All constructs were verified by DNA sequencing.
Constructs were transfected into the Phoenix-Eco packaging cell line. The retrovirus-containing supernatant was collected and loaded onto a RetroNectin (TaKaRa)-coated plate, and then D1 cells were added and infected overnight. One week later, green fluorescent protein (GFP)-positive cells were sorted, stable cell lines expressing different chimeric receptors were established, and cells were maintained in mIL-7.
D1 cell infection with Stat5A amphitrophic retroviruses.
A retrovirus vector (LZRSpBMN-IRES-EGFP) containing an active form of Stat5 (24) was prepared for transfection into cells with FuGene-6 transfection reagent (Roche Diagnostics Corporation) according to the manufacturer's instructions. The LZRS vector replicates episomally via the EBNA-1 protein and also contains a puromycin resistance gene (16). Phoenix amphitrophic packaging cells were plated at 1.5 to 2 million cells per 60-mm-diameter plate 18 to 24 h prior to transfection. After transfection, the Phoenix cells were selected with 2 μg of puromycin for day 7, at which time the population was highly enriched (95%) for enhanced GFP (EGFP)-expressing cells. The retrovirus supernatant was collected and used to infect D1 cells. Two to 3 days postinfection, D1 cells were sorted by flow cytometry and analyzed by Western blotting to verify the presence of the transduced recombinant genes.
Cytometric analysis of surface expression of chimeric receptors.
To detect expression of chimeric hIL-4R-mIL-7R constructs, D1 cells were stained with a monoclonal antibody (R&D Systems) specific for hIL-4R, followed by incubation with a phycoerythrin (PE)-labeled second antibody (Sigma). Stained cells were analyzed on a FACscan flow cytometer.
MTT assay.
D1 cells were plated in 96-well plate at a density of 5 × 104 cells/well and incubated for various times. Eight microliters of MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide; 5 mg/ml; Sigma) was added, and cells were incubated for 4 to 6 h. Then 100 μl of solubilization solution (Promega) was added, and cells were incubated overnight at 37°C. Plates were read by spectrophotometer at wavelengths 570 and 620 nm.
DNA content analysis.
DNA content analysis was carried out according to the Telford procedure. Briefly, cells were resuspended in a detergent buffer. Then, an equal volume of detergent buffer containing 50 μg of propidium iodide (PI; Sigma)/ml and 50 μg of RNase (Puregene)/ml was added. Cells were mixed by inversion, incubated at room temperature in the dark for 1 h, and analyzed by using a FACscan flow cytometer.
RPAs.
Total RNA was prepared with the TRIzol reagent (Invitrogen). The multiprobe RNase protection assay (RPA) (mAPO-2 template set) was performed by following the manufacturer's (Pharmingen) directions with modifications as previously described (14, 15).
Mitochondrial protein preparation.
D1 or mutant cells were first homogenized in isotonic buffer (200 mM mannitol, 70 mM sucrose, 1 mM EGTA, 10 mM HEPES, pH 6.9). Unbroken cells, nuclei, and heavy membranes were spun down at about 1,000 × g and discarded. The mitochondrial fraction was produced by pelleting at 12,000 × g for 20 min and washing the pellet once in isotonic buffer, followed by resuspension in radioimmunoprecipitation assay lysis buffer (1× phosphate-buffered saline [PBS], 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, protease inhibitor mixture [Calbiochem]).
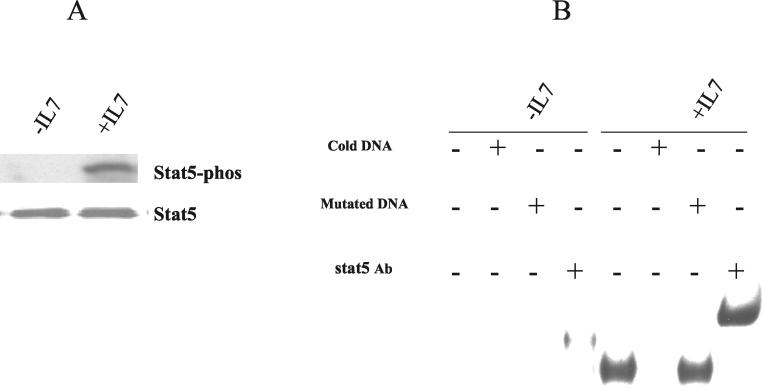
EMSA.
Nuclear extracts were prepared as described previously (2). Electrophoretic mobility shift assays (EMSA) were performed according to the Promega protocol. Briefly, a Stat5 consensus oligonucleotide (Santa Cruz Biotechnology) was end labeled with T4 polynucleotide kinase and purified with ProbeQuant G-50 microcolumns (Amersham Biosciences). The 32P-labeled probe was incubated with 10 μg of nuclear extracts on ice for 15 min. The reaction mixtures were resolved in a 5% native polyacrylamide gel electrophoresis (PAGE) gel, and specific protein complexes were detected by autoradiography. For competition experiments, a 100-fold molar excess of cold Stat5 or mutated Stat5 oligonucleotides (Santa Cruz Biotechnology) was included in the binding reaction mixture. For supershifting experiments, 1 μl of Stat5 antibody (Santa Cruz Biotechnology) was included.
Western blotting.
For Triton X-100 lysates, cells were lysed for 15 min on ice in a 1% Triton buffer containing 62.5 mM Tris-HCl, 10% glycerol, 50 mM dithiothreitol supplement with protease inhibitor mixtures (Calbiochem). Extracts were cleared by centrifugation at 14,000 × g for 10 min at 4°C.
Triton X-100 lysates or nuclear extract proteins were resolved on a 12% PAGE gel (Invitrogen) by blotting with antibodies against phospho-Jak1 (Biosource), Jak1 (BD Biosciences), phospho-Bad (Ser112) (monoclonal; Cell Signaling), Bad (Calbiochem), phospho-Stat1 (Cell Signaling), phospho-Stat3 (Cell Signaling), phospho-Stat5 (Cell Signaling), Stat5 (Santa Cruz Biotechnology), β-actin (Sigma), and Flag (Stratagene).
For detection of mitochondrial protein, a rabbit polyclonal antibody specific for Bax (N20; Santa Cruz Biotechnology) and a hamster monoclonal antiserum against Bcl2 (PharMingen) were used, followed by the appropriate secondary antibodies conjugated to horseradish peroxidase (Santa Cruz Biotechnology), and then protein was revealed by a BM chemiluminescence blotting system (Roche).
Reconstitution of IL-7R−/− bone marrow stem cells using retroviruses.
Retroviral plasmid construction was performed as follows. The coding sequence for the full-length mIL-7R was cloned into the pMIG vector, designated pMIG-mIL-7R(mWT). Different mutations were introduced into the mIL-7R intracellular domain by PCR strategies. All mutated genes were cloned into the pMIG vector and were verified by DNA sequencing. Different retroviral plasmids were transiently transfected into the Phoenix ecotropic packaging line, and the supernatants were collected.
As a control to show that all constructs were expressed on the cell surface, C1498 or BW5147 cells were infected with retrovirus supernatants for 24 h and stained with a tricolor-conjugated anti-mIL-7R antibody (eBioscience). Expression of mIL-7R and GFP was analyzed by a FACScan flow cytometer.
IL-7R−/− mice (B6.129S7-IL-7rtm1lm) and Rag1−/− mice were purchased from the Jackson Laboratory (Bar Harbor, Maine) and maintained by homozygous breeding at the National Cancer Insitute-Frederick Cancer Research and Development Center animal facility.
Cycling bone marrow stem cells were enriched by injecting IL-7R−/− mice with 5-fluorouracil (150 mg/kg of body weight; Schein Pharmaceuticals) in Dulbecco's PBS 4 days prior to harvest of bone marrow from tibia and femur. Bone marrow was cultured (106 cells/ml) in X-vivo 10 medium (BioWhittaker) supplemented with 5% fetal calf serum, murine stem cell factor (SCF; 100 ng/ml), mIL-6 (50 ng/ml), and Flt-3 ligand (100 ng/ml). After 48 h, bone marrow cells were infected overnight with different retroviral supernatants from the packaging line and then the infection was repeated after 72 h. On the fourth day, 2 × 106 infected bone marrow cells were injected intravenously into Rag1−/− recipients previously given 3 Gy of whole-body irradiation.
Four weeks after bone marrow reconstitution, mice were sacrificed and thymus and spleen cell suspensions were stained with PE-conjugated anti-CD4, CyChrome-conjugated anti-CD8, PE-conjugated anti-T-cell receptor γδ (TCR-γδ) and CyChrome-conjugated anti-TCR-β antibodies (PharMingen). Stained cells were analyzed on a FACScan flow cytometer.
RESULTS
Expression of chimeric receptors on D1 cells.
The D1 thymocyte line requires IL-7 for survival and proliferation and therefore contains the signaling molecules downstream of mIL-7R. To identify functional regions of the intracellular domain of IL-7Rα, we could not simply introduce a mutated IL-7Rα chain into the cell because its effects would be masked by the endogenous IL-7Rα chain. We therefore used a strategy of coupling the intracellular domain of IL-7Rα to the extracellular domain of hIL-4Rα. The parent murine D1 cell does not respond to hIL-4, which is incapable of binding the mIL-4R, whereas transfecting this chimeric hIL-4R-mIL-7R construct confers a vigorous response to hIL-4. This enabled us to observe the effect of mutations in the intracellular domain of the IL-7R. The chimeric receptor constructs were stably introduced into D1 cells in a retroviral vector with a packaging cell line.
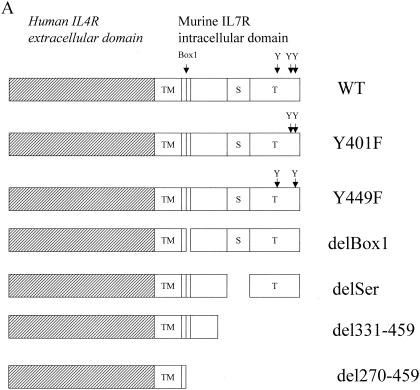
Figure 1A is a summary of the chimeric receptor constructs, beginning with the wild type (WT). The mutations Y401F or Y449F represent a change of Tyr to Phe at these sites. Deletion constructs are shown for Box1 (aa 272 to 279) and the serine-rich region (delSer; aa 359 to 394). Truncations of the C terminus were performed by deleting regions distal to aa 331 or 270. The pMIG retroviral vector (Fig. 1B) expresses a bicistronic mRNA corresponding to the genes for the chimeric receptor and GFP. Retrovirus containing the chimeric receptors were produced in packaging cell lines and used to infect D1 cells, establishing stable cell lines.
FIG. 1.
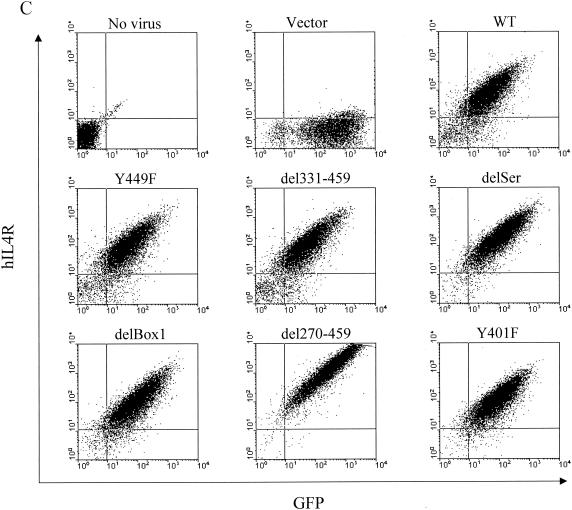
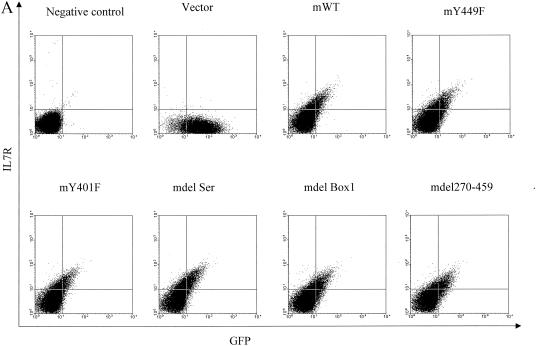
Construction of chimeric IL-7 receptors and expression on D1 cells. (A) Structures of the chimeric mutant receptors used in this study. Constructs contain the hIL-4R extracellular domain, mIL-7R transmembrane domain (TM), and mIL-7R intracellular region. S, serine-rich domain; T, tyrosine-containing domain. (B) Structure of the pMIG vector used to generate retroviruses. LTR, long terminal repeat; IRES, internal ribosomal entry site; MCS, multiple cloning site. (C) Flow-cytometric analysis of D1 cells infected with the parental retrovirus (Vector) or retroviruses. Cells were sorted for GFP and then stained for surface expression of hIL-4R.
First, the surface expression of the chimeric receptor was evaluated to determine whether or not the protein was processed and displayed. Figure 1C shows two-color analysis for GFP on the x axis versus hIL-4Rα on the y axis. WT showed substantial GFP and surface hIL-4Rα expression. The expression levels of the different chimeric receptors were quite similar in all the mutant constructs except for that of del270-459, which was more highly expressed for unknown reasons, perhaps because shorter constructs are more abundantly expressed or because it affects internalization. Although del270-459 was more highly expressed, it did not signal, as will be shown; therefore, its higher expression did not affect interpretation of the results.
MTT analysis of cell survival and proliferation.
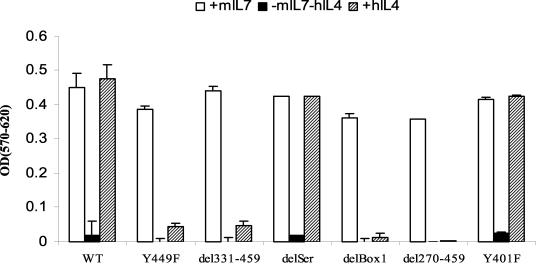
MTT analysis (Fig. 2) was performed to measure the effect of mutation and deletion of the IL-7Rα intracellular domain on viability and cell growth in response to hIL-4. Cells transfected with WT showed a response to hIL-4 that was equivalent to that to mIL-7, indicating that the chimeric construct functioned well in delivering a survival signal to the IL-7-dependent cell line. Upon stimulation with hIL-4, the delSer and Y401F mutants showed no defect compared to stimulation with mIL-7, indicating that these sites are dispensable. The Y449F, del331-459, delBox1, and del270-459 mutants failed to thrive. These results identify two small regions in the intracellular domain of IL-7R that are critical for proliferation and/or survival, Box 1 and Y449. We then analyzed the effect of deleting these two critical regions in more detail.
FIG. 2.
MTT analysis of the response of cells to various mutations and deletions of the intracellular domain of IL-7R. Transfected D1 cells were maintained in mIL-7, washed, and cultured for 48 h with 50 ng of mIL-7/ml or 50 ng of hIL-4/ml. Results are from one representative experiment of three performed. Chimeric constructs imparted a response to hIL-4. Deletion of Box1 or mutation of Y449 in the intracellular domain of IL-7R severely impaired the response. OD(570-620), absorbance at 570 nm minus background absorbance at 620 nm.
DNA content analysis.
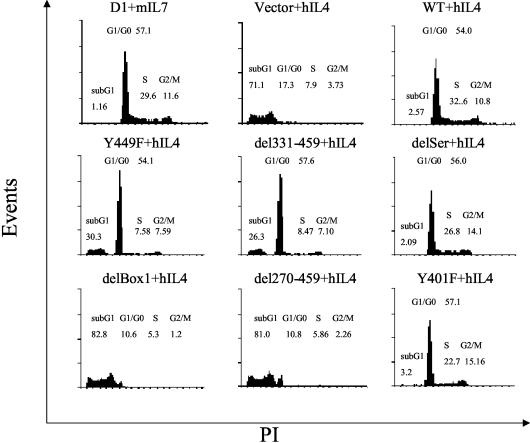
To distinguish the signal for cell proliferation from the signal for protection from apoptosis, we assayed DNA content (Fig. 3). The vector control showed that hIL-4 failed to support survival of most cells which underwent apoptotic DNA fragmentation. WT transfectants showed that the chimeric receptor imparted a response to hIL-4 that was equivalent to the response to mIL-7, both in terms of inducing progression through S phase and G2/M, as well as in protection from apoptosis. delSer and Y401F mutants showed normal cell cycle and survival, consistent with the MTT analysis. del270-459 and delBox1 mutants failed to survive and proliferate, much like the empty vector. Y449F and del331-459 mutants showed G1 arrest, and about 40% of the cells underwent apoptosis at the time point shown, which was 3 days following exchange of mIL-7 for hIL-4; by 6 days all cells in these two groups were dead (not shown). These data show that both Box1 and Y449 are important for signaling survival and proliferation but that deletion of Box1 gave more-rapid death than mutation of Y449, and this difference will be discussed further.
FIG. 3.
Cell cycle profile of the response of cells to various mutations and deletions of the intracellular domain of IL-7R. Transfected D1 cells were maintained in mIL-7, washed, and cultured for 72 h in the presence of 50 ng of hIL-4/ml and then stained with PI, followed by flow-cytometric analysis. Results are from one representative experiment of two performed. Percentages of G1/G0, S, G2/M, and sub-G1 cells are shown in the figure. Deletion of Box1 resulted in rapid apoptosis, whereas Y449 mutation induced G1 cell cycle arrest followed by apoptosis.
Jak1 activation.
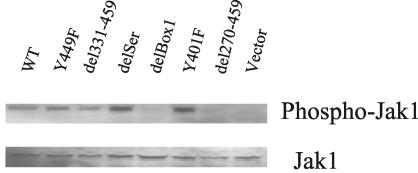
The intracellular domain of the γc chain of IL-7R binds Jak3, whereas the intracellular domain of IL-7Rα binds Jak1. Although it is clear that IL-7Rα signaling activates Jak1, there was controversy concerning the Jak1 binding site on the intracellular domain of the IL-7Rα chain (20, 29). To determine whether Box1 or the serine-rich region was required to activate Jak1, we evaluated Jak1 phosphorylation following hIL-4 stimulation (Fig. 4). Phosphorylation of Jak1 was observed in WT and the Y449F, del331-449, delSer, and Y401F mutants, whereas Jak1 activation clearly failed in the delBox1 mutant. This indicates that Jak1 phosphorylation required Box1, but not Y449 or the serine-rich region, consistent with Box1 serving as the anchor site for Jak1.
FIG. 4.
Jak1 activation versus mutations and deletions of the intracellular domain of IL-7R. Transfected D1 cells were maintained in mIL-7, washed, and stimulated with 50 ng of hIL-4/ml for 15 min. Total Triton X-100 cell lysates were resolved by sodium dodecyl sulfate-PAGE, and Western blotting was performed with an antibody against phospho-Jak1 or Jak1. The activation of Jak1 required Box1 but not Y449.
bcl2 mRNA induction.
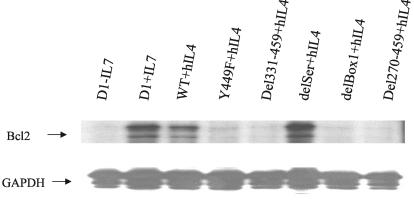
Bcl2 is an important mediator of survival induced by IL-7R; therefore we examined which mutant constructs could induce bcl2 transcription. RPAs were performed to quantify bcl2 mRNA induction (Fig. 5). WT showed strong bcl2 induction, and delSer did not reduce this induction. Deletion of Box1 or mutation of Y449 sharply curtailed the ability of the receptor to induce bcl2 synthesis. Combining this finding with the preceding analyses of survival and replication, the relationship of Box1 to Y449 could be that Box1 first binds Jak1 which then phosphorylates Y449, creating a docking site for signaling molecules that induce the bcl2 gene as well as inducing cell division. We then examined other responses to IL-7 to determine if all IL-7R signals required both Box1 and Y449.
FIG. 5.
bcl2 mRNA induction versus mutations and deletions in the intracellular domain of IL-7R. Transfected or nontransfected D1 cells were maintained in mIL-7, washed, and stimulated with either mIL-7 or hIL-4 for 4 h. Total RNA was extracted, and bcl2 transcripts were analyzed by RPA. Induction of bcl2 required both Box1 and Y449. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Regulation of Bax distribution.
Another important survival function of IL-7R is to inactivate the death protein Bax by retaining it in cytosol and blocking its mitochondrial translocation. To determine whether Box1 and Y449 were required for Bax cytosolic retention, transfected cells were cultured in hIL-4 for 8 h and mitochondrial protein was extracted. The mitochondrial Bax protein increased after IL-7 withdrawal (Fig. 6), whereas WT transfectants stimulated with hIL-4 showed Bax cytosolic retention comparable to that for IL-7-stimulated D1 cells. In Box1, del270-459, and Y449F mutants, mitochondrial Bax protein levels increased compared to those in WT. Bcl2 was used as a control for mitochondrial proteins because the level of Bcl2 protein remains rather stable for 8 h following IL-7 withdrawal (13). These data suggest that Box1 and Y449 were both required for Bax cytosolic retention as well as for bcl2 induction and cell division.
FIG. 6.
Bax cytosolic retention versus mutations and deletions in the intracellular domain of IL-7R. Transfected or nontransfected D1 cells were maintained in mIL-7, washed, and stimulated with either mIL-7 or hIL-4 for 8 h. Cells were disrupted, and mitochondrial protein fractions were prepared and immunoblotted with an anti-Bax or anti-Bcl2 antibody. Cytosolic retention of Bax required both Box1 and Y449.
Regulation of Bad phosphorylation.
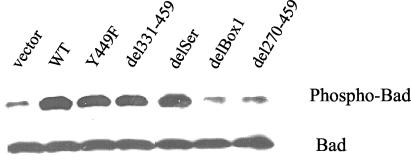
Having shown that Box1 and Y449 were both involved in regulating two survival factors, Bcl2 and Bax, we then examined a third survival factor that is regulated by IL-7, Bad. During apoptosis, Bad translocates from cytosol to mitochondria, where it blocks the survival function of Bcl2. In the healthy cell, IL-7R induces Bad phosphorylation, which results in its cytosolic sequestration. Surprisingly, Bad phosphorylation required Box1 but not Y449. Thus, stimulation with hIL-4 for 4 h of WT and Y449F, del331-459, and delSer mutants showed strong Bad phosphorylation in each case, whereas delBox1 and del270-459 mutants showed no Bad phosphorylation (Fig. 7). This suggested that Jak1, which binds to Box1, initiates two survival pathways, one via phosphorylation of Y449, regulating Bcl2 and Bax, and a second Y449-independent pathway leading to Bad phosphorylation. The concept that Box 1 controls two survival pathways and that Y449 controls one pathway could explain the preceding result (Fig. 3) that Box1 deletion caused death more rapidly than Y449 mutation. Thus Bad could accelerate cell death by neutralizing residual Bcl2 after it stops being synthesized following IL-7 withdrawal.
FIG. 7.
Bad phosphorylation versus mutations and deletions of the intracellular domain of IL-7R. Transfected D1 cells were maintained in mIL-7, washed, and stimulated with hIL-4 for 4 h. Total Triton X-100 cell lysates were analyzed by Western blotting with an antibody against phospho-Bad or Bad. Phosphorylation of Bad required Box1 but not Y449.
Stat activation in different mutants.
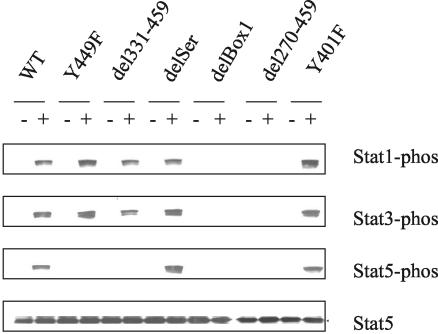
We then examined whether the two critical IL-7R regions, Box1 and Y449, were involved in activating Stats. As shown in Fig. 8, Stat1, -3, and -5 were all phosphorylated following hIL-4 stimulation through the chimeric WT receptor in D1 cells. Mutation of Y449 eliminated phosphorylation of Stat5 but not Stat1 and -3, whereas deletion of Box1 eliminated phosphorylation of all three Stats. Stat5 was then evaluated further, and we determined that, in addition to being phosphorylated following receptor activation (Fig. 9A), it also translocated to the nucleus and bound DNA (Fig. 9B). Controls verified that the DNA binding was specific, and assays with anti-Stat5 showed that the protein was bone fide Stat5.
FIG. 8.
Stat activation versus mutations and deletions of the intracellular domain of IL-7R. Transfected D1 cells were deprived of IL-7 overnight and then stimulated with hIL-4 (+) or without hIL-4 (−) for 10 min. Total Triton X-100 cell lysates were analyzed by Western blotting with antibodies against phospho-Stat1, phospho-Stat3, phospho-Stat5, or Stat5. Phosphorylation of Stat5 required both Box 1 and Y449. Phosphorylation of Stat1 and -3 required Box1 but not Y449.
FIG. 9.
Activation of Stat5 in D1 cells following IL-7 stimulation. (A) D1 cells were deprived of IL-7 overnight and then stimulated with mIL-7 for 10 min. (B) D1 cells were deprived of IL-7 overnight and then stimulated with mIL-7 (50 ng/ml) for 2 h. Nuclear extracts were analyzed by EMSA using a labeled Stat5 consensus oligonucleotide. Specificity was evaluated by competition with cold DNA or with an antibody (Ab) against Stat5. Results show that IL-7 induced Stat5 nuclear translocation and DNA binding.
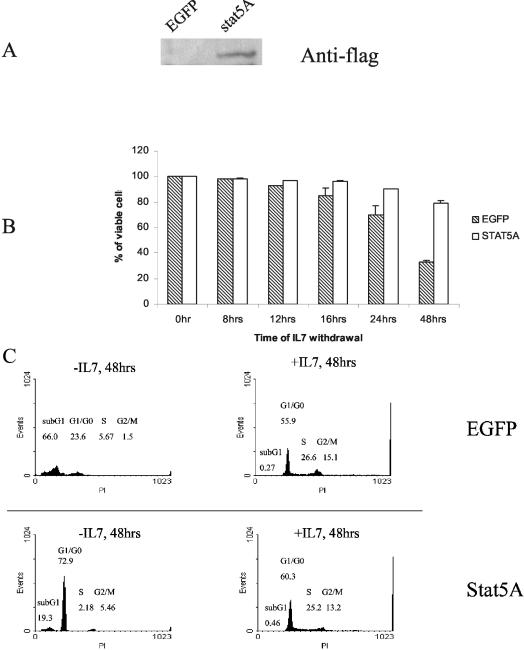
Survival effect of active Stat5A in D1 cells.
Since Y449 was required for induction of bcl2, survival, and the cell cycle and since Y449 also activated Stat5, we examined whether Stat5 mediated all the effects of Y449 by introducing a constitutively activated Stat5A into D1 cells. First we confirmed expression of the Flag-tagged active Stat5A construct (Fig. 10A). Active Stat5A slowed the cell death process following IL-7 withdrawal by about a day but did not keep D1 cells alive indefinitely; in contrast, overexpression of Bcl2 maintains survival (without proliferation) of D1 cells indefinitely in the absence of IL-7 (W. Q. Li and S. K. Durum, unpublished data). Thus Stat5, although activated by IL-7R, does not account for all of the survival signaling, and there must be additional pathways. Since Y449 also regulated cell proliferation, in addition to survival, we examined the effect of the active Stat5A on the cell cycle. As shown in Fig. 10C, G1 arrest occurred in Stat5A-transfected cells deprived of IL-7, indicating that Stat5 also cannot account for the proliferative effects of IL-7 emanating from Y449.
FIG. 10.
Survival effect of expression of active Stat5A on D1 cells following IL-7 withdrawal. (A) Nuclear proteins were prepared from D1 cells infected with parental retrovirus (EGFP) or with retrovirus harboring active Stat5A (stat5A) and subjected to Western blotting with a Flag antibody. (B) EGFP- or Stat5A-expressing cells were washed twice with PBS and cultured without mIL-7 for the indicated times. Percentages of viable cells were determined by PI staining. (C) EGFP (top)- or Stat5A (bottom)-expressing infected D1 cells were cultured with or without mIL-7 (50 ng/ml) for 48 h and stained with PI following flow-cytometric analysis. Percentages of G1/G0, S, G2/M.and sub-G1 cells are shown. Results show that activated Stat5 delayed death of D1 cells deprived of IL-7 but did not induce cell division.
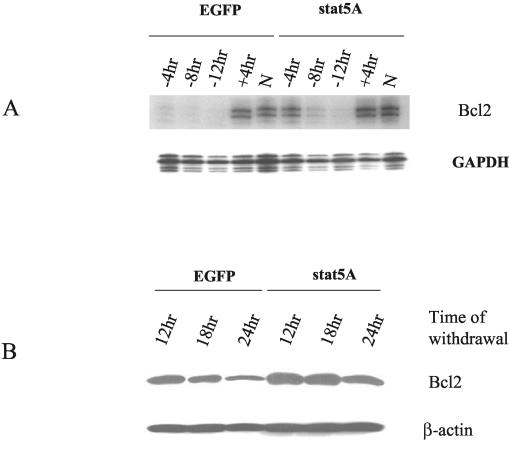
Up-regulation of Bcl2 by active Stat5A.
Because active Stat5A prolonged D1 cell survival following IL-7 withdrawal, we examined whether active Stat5A increased the levels of bcl2 transcripts and protein. Figure 11A shows that, 4 h after IL-7 withdrawal, Bcl2 synthesis had already ceased in EGFP-expressing cells (although the Bcl2 protein is stable over 8 h). In Stat5A-expressing cells, bcl2 transcripts remained at 4 h but eventually declined. This suggests that active Stat5A could sustain bcl2 4 to 8 h longer than it was sustained in controls but could not replace IL-7, suggesting that it may amplify the activity of another IL-7-induced transcription factor on the bcl2 promoter. In EGFP-expressing cells, Bcl2 protein levels showed a pattern that paralleled that of mRNA although the decrease of protein was delayed several hours compared to the decrease of mRNA. In Stat5A cells, at the 24-h time point, the Bcl2 protein level only decreased slightly. Thus, the prolongation of survival by active Stat5 may due to the higher Bcl2 levels.
FIG. 11.
Active Stat5A versus bcl2 expression following IL-7 withdrawal. D1 cells infected with control EGFP or Stat5A retrovirus supernatant were deprived of IL-7 for the indicated times. (A) Total RNA was prepared and subjected to RPA for expression of bcl2 or GAPDH as a loading control. As positive controls, cells were stimulated with mIL-7 for 4 h after overnight starvation (+4hr) or were continuously cultured in the presence of mIL-7 (N). (B) Total Triton X-100 extracts were prepared. Western blotting was performed with either a Bcl2 or β-actin antibody. Results show that activated Stat5 was insufficient to maintain high expression of Bcl2 in the absence of IL-7. However, activated Stat5 increased the expression of bcl2 in the presence of IL-7 and sustained bcl2 expression for 4 h following IL-7 withdrawal. Bcl2 protein levels have a long half-life in D1 cells, and activated Stat5 extended the levels of Bcl2 following IL-7 withdrawal.
Reconstitution of IL-7Rα−/− T-cell development by IL-7Rα constructs.
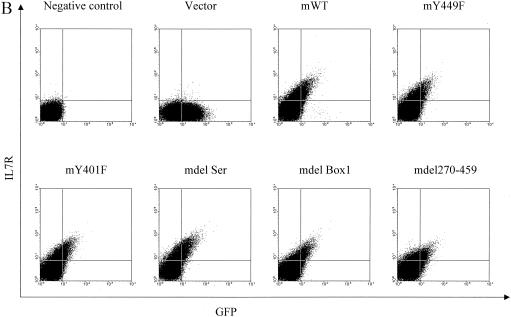
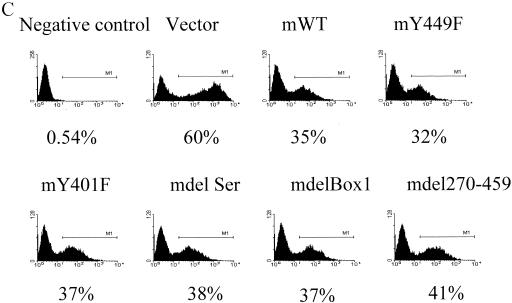
Having identified Box1 and Y449 as critical in the IL-7-responsive cell line D1, we evaluated the effect of these regions in vivo on T-cell development. First, we showed that, in the hematopoietic cell lines C1498 and BW5147, different constructs expressed murine (rather than chimeric) IL-7R on the cell surface, correlating with GFP expression (Fig. 12A and B). IL-7Rα−/− hematopoietic stem cells were then infected with retroviruses expressing mIL-7R constructs together with GFP. Figure 12C shows that transfection efficiencies for different receptor constructs are similar, except that for the vector control, which was typically somewhat higher.
FIG. 12.
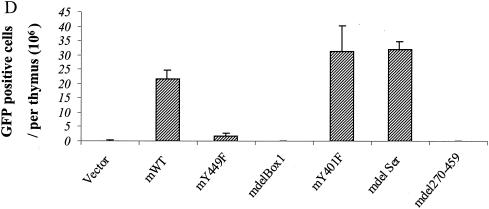
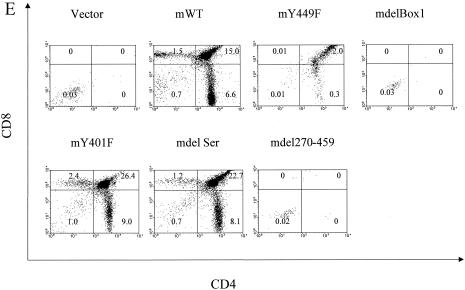
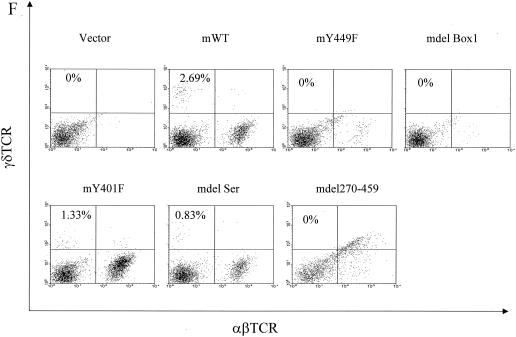
Reconstitution of IL-7R−/− T-cell development in vivo. mIL-7R (rather than chimeric) constructs (and GFP) were tested for expression by introducing them into hematopoietic cell lines C1498 and BW5147 by retrovirus infection. Twenty-four hours later, surface expression of mIL-7R (together with GFP expression) was analyzed by fluorescence-activated cell sorting. (A) GFP versus surface expression of mIL-7R in infected C1498 cells. (B) GFP versus surface expression of mIL-7R in infected BW5147 cells. Receptor constructs (and GFP) were introduced by retroviral transfer into IL-7R−/− hematopoietic stem cells, which were injected into Rag1−/− recipients. One month later, thymocytes and spleen cells were analyzed by flow microfluorimetry, gating on GFP+ cells. (C) Transfection efficiency in bone marrow stem cells. Forty-eight hours after retroviral infection, hematopoietic stem cells were analyzed for expression of GFP. Percentages of GFP-positive cells are indicated and are similar for different receptor constructs. (D) Numbers of GFP-positive cells in different groups were obtained based on percentages of GFP-positive cells and total cells in each thymus. Data are from three experiments using six individual mice. (E) Thymocytes were stained for CD4 and CD8. The numbers shown indicate the number (106) of cells of each subset per thymus. Deleting Box1 completely eliminated thymocyte development, whereas Y449F eliminated approximately 90%. There was no effect of deleting the serine region or Y401F. (F) Spleen cells were stained for αβ and γδ T cells. Percentages of γδ T cell are shown. mY449F completely eliminated γδ T-cell development, as did deleting Box1 or aa 270 to 459. No effect was seen for mY401F or deletion of the serine region.
Infected stem cells were used to reconstitute Rag1−/− mice, and the development of T cells in thymus and spleen was evaluated 1 month later. As shown in Fig. 12D, 20 to 30 million GFP-positive cells were recovered from the thymus in murine WT, Y401F, and delSer groups, while about 2 million were recovered in the Y449F group and virtually no GFP-positive cells were recovered in other groups. The numbers of each subset in the thymus are shown in Fig. 12E. Development of the αβ lineage showed a response proportional to that of the D1 cell, which could be interpreted as indicating that 90% was controlled by a Box1-Y449 pathway and that 10% was controlled by an additional Box1 pathway. A total of six experiments of this type have been performed with results similar to those shown. The development of the γδ T-cell lineage appeared to be completely inhibited by Y449F in both spleen (Fig. 12F) and thymus (not shown). This could indicate that the proposed Box1-Y449 pathway is the exclusive pathway for γδ cells, whereas αβ cells also have a secondary pathway that leads from Box1. Alternatively, because the number of γδ cells is normally much lower than the number of αβ cells, the same secondary pathway may exist but the resulting γδ cells were below the level of detection.
DISCUSSION
The IL-7R delivers critical signals for lymphocyte development and homeostasis. The current model of this antiapoptotic effect relates to the balance of Bad and Bax versus Bcl2 in mitochondria. In the presence of IL-7, Bcl2 is synthesized, Bax is constrained to cytosol, and Bad is inactivated. Following IL-7 withdrawal, Bcl2 synthesis ceases, Bax translocates to mitochondria, and Bad is dephosphorylated and binds to Bcl2, inactivating it. During the first several hours of IL-7 withdrawal, residual Bcl2 can still function and protect cells from Bax-induced damage. However, when the level of active Bad exceeds the level of Bcl2 and Bax accumulates in mitochondria with increasing time of IL-7 withdrawal, cells begin to die. The importance of the balance of Bcl2/Bax in the IL-7 response has been demonstrated in vivo by experiments showing that the deficiency in T-cell development in IL-7 R−/− mice can be rescued by overexpressing Bcl2 (1, 19) or deleting Bax (14). Introducing activated Bad kills D1 cells, and this was countered by overexpressing Bcl2 (Li et al., submitted).
To begin to identify the signaling pathways that connect the IL-7R with these apoptosis regulators, we sought to determine the critical regions of the intracellular domain of IL-7Rα using the D1 thymocyte cell line, which depends on IL-7 for survival and proliferation. Two small regions of the intracellular domain of IL-7Rα were shown to be critical, Box1 and Y449. The same two regions were also shown to be critical in vivo in thymocyte development, as examined by reconstitution of IL-7Rα−/− stem cells by retrovirus expressing various receptor constructs.
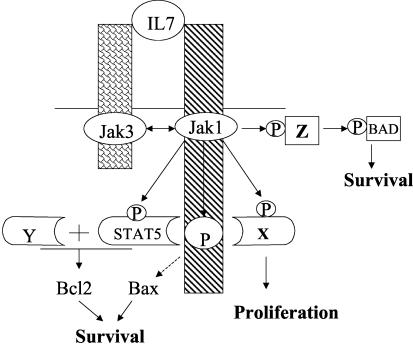
Based on our findings and that of others, we propose the model illustrated in Fig. 13. IL-7 ligation brings together the intracellular domains of IL-7Rα and -γc, bearing their associated kinases, Jak1 (associated with Box1) and Jak3, respectively. Jak1 and -3 mutually phosphorylate one another, increasing their kinase activities. Jak1 (or Jak3) then phosphorylates Y449, creating a docking site for a signaling cascade leading to bcl2 induction, Bax cytosolic retention, and cell cycling. Part of the signal from Y449 to bcl2 may be Stat5, but another signal from Y449 is also required to fully activate the bcl2 promoter and to induce cell division. Jak1 (or Jak3) also triggers a Y449-independent pathway leading to Bad phosphorylation, which also promotes cell survival. The Y449 pathway is necessary for most IL-7-induced effects, although our experiments have not proven that it is sufficient.
FIG. 13.
Current model for IL-7R regulation of survival and proliferation. IL-7 binding to the receptor brings together the α and γc chains and their associated kinases, Jak1 (bound to Box1) and Jak3, respectively. The two kinases phosphorylate one another and increase their enzymatic activity. These kinases then phosphorylate Y449, which serves as a docking site for Stat5 and additional unknown proteins that are required for synthesis of Bcl2, cytosolic retention of Bax, and cell proliferation. A second pathway leads from Jak1 (independent of Y449) and induces phosphorylation of Bad, Stat1, and Stat3.
One candidate for a Y449-induced pathway would be PI3K-AKT which is a well-known mediator of receptor signals for cell survival. In the IL-7 system, the PI3K pathway was previously observed to operate in B cells responding to IL-7R signaling and to depend on Y449 (40). However PI3K does not appear to be critical for IL-7-induced survival of D1 cells since inhibitors of PI3K did not kill these cells or reduce bcl2 expression, nor did active AKT protect from IL-7 withdrawal (W. Q. Li et al., unpublished data). We have verified that AKT is activated by IL-7 in the D1 T-cell line, but, surprisingly, there was no difference in activation of AKT by chimeric receptors among the deletion mutants (data not shown). Since deletion of the entire intracellular domain of IL-7Rα still retained AKT activation following ligand stimulation, perhaps in these cells the signal to activate PI3K-AKT comes from the γc chain or possibly from some other receptor that is collaterally activated by the external or transmembrane regions of the IL-7Rα chain. Taking this evidence together, the postulated survival pathway that emanates from Y449 does not appear to be the PI3K-AKT pathway.
Another candidate pathway emanating from Y449 is the Stat5 pathway, which has previously been implicated in IL-7R signaling (10). However, knockout of the two isoforms of Stat5 did not produce a deficiency in thymic development (36), indicating that Stat5 is dispensable for IL-7 signaling in vivo. Our findings confirm that Stat5 is activated by the IL-7R in D1 cells but that an activated Stat5 did not confer long-term survival in the absence of IL-7; moreover, a dominant negative Stat5, at least in experiments so far, has not shown an inhibitory effect on survival in the presence of IL-7. Activated Stat5 prolonged the life of D1 cells by a few days in the absence of IL-7 and appeared to sustain expression of bcl2 for about that length of time. The bcl2 promoter does not show an apparent Stat5 consensus binding site, so the effect of Stat5 on the bcl2 gene may be indirect; perhaps IL-7R first induces production of a transcription factor that in turn amplifies the action of other transcription factors that are essential to expression of the bcl2 gene. It is also possible that the stability of bcl2 transcripts is enhanced by a Stat5 signal.
Other Stats, in addition to Stat5, were activated by ligand stimulation of the IL-7R. However, both Stat1 and Stat3 were activated in the absence of Y449, indicating that this is not the docking site for their activation and showing that the significance of Y449 cannot be explained based on docking any of these Stats. Note that the activation of Stat1 and -3 occurs in the absence of any IL-7Rα tyrosines. Unlike Stat5, which apparently docks to phosphorylated Y449 and becomes phosphorylated, Stat1 and -3 are phosphorylated without docking to the IL-7Rα via its own phosphotyrosine. At this time, we do not know how (or if) these Stats associate with the receptor complex. Their phosphorylation requires Box1, which could indicate direct phosphorylation by a Jak or by a kinase downstream of the Jaks. Because they are rapidly phosphorylated (10 min) there would not seem to be enough time to induce synthesis of other cytokines that would act on other receptors that could in turn activate Stat1 and -3.
To identify the critical signaling cascade from the IL-7 receptor, it will be helpful to identify Y449 as a critical residue regulating survival via Bcl2 synthesis and Bax cytosolic retention. Assuming that this residue is phosphorylated following IL-7 binding, future studies will be aimed at identifying the protein, distinct from Stat5, that binds this phosphotyrosine motif.
Several kinases have been proposed to phosphorylate Bad at Ser 112, including AKT, protein kinase A, and Rsk in some cell types (8, 12, 35). However, we found that a PI3K inhibitor could not block Bad phosphorylation at Ser112 in D1 cells (Li et al., submitted). Studies are ongoing to identify the kinase related to the Box1 region which phosphorylates Bad at Ser112.
The development of γδ T cells depends on the IL-7 pathway. Here we show that this development depends on the Box1-Y449 axis. A possible distinction from the αβ T-cell pathway was observed in that, whereas the αβ T-cell pathway could still develop to 10% of normal in the absence of Y449, the development or survival or both of γδ T cells were completely blocked. This could indicate that IL-7 signals more than survival in the γδ pathway and also gives a unique signal for γδ T cells coming from Y449. This would be consistent with the ability of a bcl2 transgene to reconstitute αβ T-cell development, but not γδ T-cell development.
Acknowledgments
We thank K. Noer, R. Matthai, and L. Finch for flow cytometry; R. Wyles for technical work; M. Sanford for performing RNase protection analyses; S. Spence for help with retroviral vectors; and J. Oppenheim for comments on the manuscript.
REFERENCES
- 1.Akashi, K., M. Kondo, U. von Freeden-Jeffry, R. Murray, and I. L. Weissman. 1997. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor-deficient mice. Cell 89:1033-1041. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benbernou, N., K. Muegge, and S. K. Durum. 2000. Interleukin (IL)-7 induces rapid activation of Pyk2, which is bound to Janus kinase 1 and IL-7Rα. J. Biol. Chem. 275:7060-7065. [DOI] [PubMed] [Google Scholar]
- 4.Bhatia, S. K., L. T. Tygrett, K. H. Grabstein, and T. J. Waldschmidt. 1995. The effect of in vivo IL-7 deprivation on T cell maturation. J. Exp. Med. 181:1399-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao, X., E. W. Shores, L. J. Hu, M. R. Anver, B. L. Kelsall, S. M. Russell, J. Drago, M. Noguchi, A. Grinberg, E. T. Bloom, et al. 1995. Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity 2:223-238. [DOI] [PubMed] [Google Scholar]
- 6.Corcoran, A. E., F. M. Smart, R. J. Cowling, T. Crompton, M. J. Owen, and A. R. Venkitaraman. 1996. The interleukin-7 receptor alpha chain transmits distinct signals for proliferation and differentiation during B lymphopoiesis. EMBO J. 15:1924-1932. [PMC free article] [PubMed] [Google Scholar]
- 7.Dadi, H., S. Ke, and C. M. Roifman. 1994. Activation of phosphatidylinositol-3 kinase by ligation of the interleukin-7 receptor is dependent on protein tyrosine kinase activity. Blood 84:1579-1586. [PubMed] [Google Scholar]
- 8.Datta, S. R., H. Dudek, X. Tao, S. Masters, H. Fu, Y. Gotoh, and M. E. Greenberg. 1997. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 91:231-241. [DOI] [PubMed] [Google Scholar]
- 9.DiSanto, J. P., W. Muller, D. Guy-Grand, A. Fischer, and K. Rajewsky. 1995. Lymphoid development in mice with a targeted deletion of the interleukin 2 receptor gamma chain. Proc. Natl. Acad. Sci. USA 92:377-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foxwell, B. M., C. Beadling, D. Guschin, I. Kerr, and D. Cantrell. 1995. Interleukin-7 can induce the activation of Jak 1, Jak 3 and STAT 5 proteins in murine T cells. Eur. J. Immunol. 25:3041-3046. [DOI] [PubMed] [Google Scholar]
- 11.Grabstein, K. H., T. J. Waldschmidt, F. D. Finkelman, B. W. Hess, A. R. Alpert, N. E. Boiani, A. E. Namen, and P. J. Morrissey. 1993. Inhibition of murine B and T lymphopoiesis in vivo by an anti-interleukin 7 monoclonal antibody. J. Exp. Med. 178:257-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harada, H., B. Becknell, M. Wilm, M. Mann, L. J. Huang, S. S. Taylor, J. D. Scott, and S. J. Korsmeyer. 1999. Phosphorylation and inactivation of BAD by mitochondria-anchored protein kinase A. Mol. Cell 3:413-422. [DOI] [PubMed] [Google Scholar]
- 12a.Jiang, Q., W. Q. Li, F. B. Aiello, A. R. Khaled, and S. K. Durum. Cell biology of IL-7, a key lymphotrophin. Semin. Immunol., in press. [DOI] [PubMed]
- 13.Khaled, A. R., K. Kim, R. Hofmeister, K. Muegge, and S. K. Durum. 1999. Withdrawal of IL-7 induces Bax translocation from cytosol to mitochondria through a rise in intracellular pH. Proc. Natl. Acad. Sci. USA 96:14476-14481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khaled, A. R., W. Q. Li, J. Huang, T. J. Fry, A. S. Khaled, C. L. Mackall, K. Muegge, H. A. Young, and S. K. Durum. 2002. Bax deficiency partially corrects interleukin-7 receptor alpha deficiency. Immunity 17:561-573. [DOI] [PubMed] [Google Scholar]
- 15.Kim, K., A. R. Khaled, D. Reynolds, H. A. Young, C. K. Lee, and S. K. Durum. 2003. Characterization of an interleukin-7-dependent thymic cell line derived from a p53−/− mouse. J. Immunol. Methods 274:177-184. [DOI] [PubMed] [Google Scholar]
- 16.Kinsella, T. M., and G. P. Nolan. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 7:1405-1413. [DOI] [PubMed] [Google Scholar]
- 17.Macchi, P., A. Villa, S. Giliani, M. G. Sacco, A. Frattini, F. Porta, A. G. Ugazio, J. A. Johnston, F. Candotti, and J. J. O'Shea. 1995. Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID). Nature 377:65-68. [DOI] [PubMed] [Google Scholar]
- 18.Maki, K., S. Sunaga, Y. Komagata, Y. Kodaira, A. Mabuchi, H. Karasuyama, K. Yokomuro, J. I. Miyazaki, and K. Ikuta. 1996. Interleukin 7 receptor-deficient mice lack γδ T cells. Proc. Natl. Acad. Sci. USA 93:7172-7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maraskovsky, E., L. A. O'Reilly, M. Teepe, L. M. Corcoran, J. J. Peschon, and A. Strasser. 1997. Bcl-2 can rescue T lymphocyte development in interleukin-7 receptor-deficient mice but not in mutant rag-1−/− mice. Cell 89:1011-1019. [DOI] [PubMed] [Google Scholar]
- 20.Murakami, M., M. Narazaki, M. Hibi, H. Yawata, K. Yasukawa, M. Hamaguchi, T. Taga, and T. Kishimoto. 1991. Critical cytoplasmic region of the interleukin 6 signal transducer gp130 is conserved in the cytokine receptor family. Proc. Natl. Acad. Sci. USA 88:11349-11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noguchi, M., Y. Nakamura, S. M. Russell, S. F. Ziegler, M. Tsang, X. Cao, and W. J. Leonard. 1993. Interleukin-2 receptor gamma chain: a functional component of the interleukin-7 receptor. Science 262:1877-1880. [DOI] [PubMed] [Google Scholar]
- 22.Nolan, G. P., and A. R. Shatzman. 1998. Expression vectors and delivery systems. Curr. Opin. Biotechnol. 9:447-450. [DOI] [PubMed] [Google Scholar]
- 23.Nosaka, T., J. M. van Deursen, R. A. Tripp, W. E. Thierfelder, B. A. Witthuhn, A. P. McMickle, P. C. Doherty, G. C. Grosveld, and J. N. Ihle. 1995. Defective lymphoid development in mice lacking Jak3. Science 270:800-802. [DOI] [PubMed] [Google Scholar]
- 24.Onishi, M., T. Nosaka, K. Misawa, A. L. Mui, D. Gorman, M. McMahon, A. Miyajima, and T. Kitamura. 1998. Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol. Cell. Biol. 18:3871-3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Page, T. H., F. V. Lali, and B. M. Foxwell. 1995. Interleukin-7 activates p56lck and p59fyn, two tyrosine kinases associated with the p90 interleukin-7 receptor in primary human T cells. Eur. J. Immunol. 25:2956-2960. [DOI] [PubMed] [Google Scholar]
- 26.Park, S. Y., K. Saijo, T. Takahashi, M. Osawa, H. Arase, N. Hirayama, K. Miyake, H. Nakauchi, T. Shirasawa, and T. Saito. 1995. Developmental defects of lymphoid cells in Jak3 kinase-deficient mice. Immunity 3:771-782. [DOI] [PubMed] [Google Scholar]
- 27.Peschon, J. J., B. C. Gliniak, P. J. Morrissey, and E. Maraskovsky. 1998. Lymphoid development and function in IL-7R-deficient mice, p. 37-52. In S. K. Durum and K. Muegge (ed.), Cytokine knockouts. Humana Press Inc., Totowa, N.J.
- 28.Peschon, J. J., P. J. Morrissey, K. H. Grabstein, F. J. Ramsdell, E. Maraskovsky, B. C. Gliniak, L. S. Park, S. F. Ziegler, D. E. Williams, C. B. Ware, et al. 1994. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J. Exp. Med. 180:1955-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Porter, B. O., P. Scibelli, and T. R. Malek. 2001. Control of T cell development in vivo by subdomains within the IL-7 receptor alpha-chain cytoplasmic tail. J. Immunol. 166:262-269. [DOI] [PubMed] [Google Scholar]
- 30.Puel, A., S. F. Ziegler, R. H. Buckley, and W. J. Leonard. 1998. Defective IL-7R expression in T−B+NK+ severe combined immunodeficiency. Nat. Genet. 20:394-397. [DOI] [PubMed] [Google Scholar]
- 31.Roifman, C. M., J. Zhang, D. Chitayat, and N. Sharfe. 2000. A partial deficiency of interleukin-7R alpha is sufficient to abrogate T-cell development and cause severe combined immunodeficiency. Blood 96:2803-2807. [PubMed] [Google Scholar]
- 32.Russell, S. M., N. Tayebi, H. Nakajima, M. C. Riedy, J. L. Roberts, M. J. Aman, T. S. Migone, M. Noguchi, M. L. Markert, and R. H. Buckley. 1995. Mutation of Jak3 in a patient with SCID: essential role of Jak3 in lymphoid development. Science 270:797-800. [DOI] [PubMed] [Google Scholar]
- 33.Schluns, K. S., W. C. Kieper, S. C. Jameson, and L. Lefrancois. 2000. Interleukin-7 mediates the homeostasis of naive and memory CD8 T cells in vivo. Nat. Immunol. 1:426-432. [DOI] [PubMed] [Google Scholar]
- 34.Seckinger, P., and M. Fougereau. 1994. Activation of src family kinases in human pre-B cells by IL-7. J. Immunol. 153:97-109. [PubMed] [Google Scholar]
- 35.Tan, Y., H. Ruan, M. R. Demeter, and M. J. Comb. 1999. p90(RSK) blocks bad-mediated cell death via a protein kinase C-dependent pathway. J. Biol. Chem. 274:34859-34867. [DOI] [PubMed] [Google Scholar]
- 36.Teglund, S., C. McKay, E. Schuetz, J. M. van Deursen, D. Stravopodis, D. Wang, M. Brown, S. Bodner, G. Grosveld, and J. N. Ihle. 1998. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell 93:841-850. [DOI] [PubMed] [Google Scholar]
- 37.Thomis, D. C., C. B. Gurniak, E. Tivol, A. H. Sharpe, and L. J. Berg. 1995. Defects in B lymphocyte maturation and T lymphocyte activation in mice lacking Jak3. Science 270:794-797. [DOI] [PubMed] [Google Scholar]
- 38.van der Plas, D. C., F. Smiers, K. Pouwels, L. H. Hoefsloot, B. Lowenberg, and I. P. Touw. 1996. Interleukin-7 signaling in human B cell precursor acute lymphoblastic leukemia cells and murine BAF3 cells involves activation of STAT1 and STAT5 mediated via the interleukin-7 receptor alpha chain. Leukemia 10:1317-1325. [PubMed] [Google Scholar]
- 39.Venkitaraman, A. R., and R. J. Cowling. 1992. Interleukin 7 receptor functions by recruiting the tyrosine kinase p59fyn through a segment of its cytoplasmic tail. Proc. Natl. Acad. Sci. USA 89:12083-12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venkitaraman, A. R., and R. J. Cowling. 1994. Interleukin-7 induces the association of phosphatidylinositol 3-kinase with the alpha chain of the interleukin-7 receptor. Eur. J. Immunol. 24:2168-2174. [DOI] [PubMed] [Google Scholar]
- 41.von-Freeden-Jeffry, U., P. Vieira, L. A. Lucian, T. McNeil, S. E. Burdach, and R. Murray. 1995. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J. Exp. Med. 181:1519-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]