Abstract
We previously demonstrated the critical role of RNA polymerase I (Pol I)-associated factor PAF53 in mammalian rRNA transcription. Here, we report the isolation and characterization of another Pol I-associated factor, PAF49. Mouse PAF49 shows striking homology to the human nucleolar protein ASE-1, so that they are considered orthologues. PAF49 and PAF53 were copurified with a subpopulation of Pol I during purification from cell extracts. Physical association of PAF49 with Pol I was confirmed by a coimmunoprecipitation assay. PAF49 was shown to interact with PAF53 through its N-terminal segment. This region of PAF49 also served as the target for TAFI48, the 48-kDa subunit of selectivity factor SL1. Concomitant with this interaction, the other components of SL1 also coimmunoprecipitated with PAF49. Specific transcription from the mouse rRNA promoter in vitro was severely impaired by anti-PAF49 antibody, which was overcome by addition of recombinant PAF49 protein. Moreover, overexpression of a deletion mutant of PAF49 significantly reduced pre-rRNA synthesis in vivo. Immunolocalization analysis revealed that PAF49 accumulated in the nucleolus of growing cells but dispersed to nucleoplasm in growth-arrested cells. These results strongly suggest that PAF49/ASE-1 plays an important role in rRNA transcription.
Initiation of transcription is a complex biological process that critically determines gene expression. In order to understand this process, it is important to know the core component molecules participating in it. Enormous efforts over decades have disclosed a set of proteins essential for initiation by each class of eukaryotic RNA polymerase. For RNA polymerase I (Pol I), which is dedicated to the transcription of the large rRNA precursor, two transcription factors have been defined in mammals. One is the selectivity factor SL1, which plays a critical role in recognition of the core promoter element (56). SL1 consists of the TATA-binding protein (TBP) and three TBP-associated factors (TAFIs), TAFI110/95, TAFI63/68, and TAFI48, for the human and murine rRNA transcription systems (6, 17, 56). The other is the upstream binding factor (UBF), which interacts with the upstream control element (UCE) to facilitate the assembly of the transcription initiation complex including SL1 and Pol I (29, 57). Other transcription factors, such as factor C* (4), p70 (49, 50), TFIC (22), TIF-IA (37), and TIF-IC (38), were also identified by biochemical analyses. However, the molecular nature of these factors is still to be determined.
Recent identification and subsequent functional characterization of Saccharomyces cerevisiae Rrn3 and its mammalian homologue hRRN3 have greatly promoted our understanding of the growth-dependent regulation of rRNA synthesis (28, 51). Rrn3 is essential for promoter-directed rRNA transcription in S. cerevisiae (51). Only a small population of Pol I was found to be tightly associated with Rrn3; however, it was in the form that was competent for transcription (26). Importantly, the association of Rrn3 with Pol I is cell growth dependent, that is, the Rrn3-Pol I complex was found in extracts from exponentially growing S. cerevisiae but not in stationary-phase cells (26). This association was mediated by the interaction between Rrn3 and the A43 subunit of Pol I (33). Rrn3 was also shown to bind to Rrn6, one of the subunits of the core factor essential for core element recognition of yeast ribosomal DNA (32, 33).
Interestingly, the mammalian homologue of Rrn3 was reported to interact directly with the TAFI110/95 and TAFI63/68 subunits of SL1, although no apparent sequence homology was evident between human TAFs and yeast Rrn6 (27, 32, 54). These results suggest that Rrn3has functionally evolved to recruit the polymerase to the transcription initiation complex by bipartite interactions with Pol I and the promoter recognition factors. On the other hand, it has also been reported that Rrn3 may not function in Pol I recruitment in S. cerevisiae but is rather involved in a later step of initiation (2). In addition, Rrn3 was phosphorylated in both S. cerevisiae and mammalian cells (5, 8). Phosphorylation of Rrn3 was required for the association with Pol I core enzyme in mammalian cells (5), while in S. cerevisiae, both phosphorylated and unphosphorylated forms of Rrn3 were able to form a transcriptionally competent Pol I-Rrn3 complex (8). The role of phosphorylation on Rrn3 thus remains somewhat obscure, but posttranslational modification(s) of Rrn3 seems to be a major cue for the growth-dependent transcription of rRNA (3, 57).
Affinity purification and conventional column chromatography revealed that, at least in samples from a rat, mouse, frog, and broccoli, a portion of cellular Pol I activity existed as a holoenzyme (14, 34, 41). Size fractionation experiments indicated that the Pol I holoenzyme was >2 MDa and that the holoenzyme was self-competent for promoter-specific transcription as it contained preassembled SL1 (1, 13, 41). Among >50 polypeptides included in the holoenzyme, casein kinase II and histone acetyltransferase activities, topoisomerase I, and Ku70/80 have been identified (1, 13, 14, 35), although their functional significance in rRNA transcription has yet to be established.
To understand the roles of these potential regulatory factors, it is important to define the genuine components of RNA polymerase I. In this regard, we have demonstrated that the Pol I core enzyme consists of at least 11 polypeptides, as judged from silver-stained sodium dodecyl sulfate (SDS)-polyacrylamide gels of the enzyme fractions purified to near homogeneity (42). Actually, molecular cloning of the cDNA encoding the 40-kDa polypeptide (referred to as RPA40) revealed that it was the mammalian orthologue of AC40, a shared subunit of yeast Pol I and Pol III (23, 42). Two-hybrid screening with RPA40 as the bait identified RPA16, which was also a mammalian orthologue of another S. cerevisiae shared subunit, AC19 (7), and was shown to be present in the purified enzyme (53). These results strongly suggest that the established purification procedure for Pol I yields genuine enzyme. In the course of the purification, however, we found that Pol I activity was also recovered in biochemically different fractions and that some particular polypeptides were missing from the polymerase in these fractions.
We therefore isolated a cDNA encoding one of these polypeptides and characterized it as Pol I-associated factor PAF53 (11). PAF53 was shown to interact with UBF. Anti-PAF53 antibody inhibited promoter-directed rRNA transcription but had no effect on nonspecific random RNA synthesis. Immunolocalization studies indicated that PAF53 was present in the nucleoli of exponentially growing cells but dispersed in serum-starved cells (11). Moreover, the cellular content of PAF53 decreased after serum starvation and increased in response to insulin or serum refeeding (12). These results indicate that PAF53 plays a critical role in the initiation of rRNA transcription by mediating protein-protein interaction between Pol I and UBF and suggest that it also participates in the growth-dependent regulation of rRNA transcription.
Here we report the isolation and characterization of another Pol I-associated factor, PAF49. As shown for PAF53, PAF49 also exists in subpopulation of Pol I and accumulates in the nucleolus of exponentially growing cells. PAF49 interacts with the SL1 complex through direct binding to the TAFI48 subunit. Antibodies against PAF49 inhibited promoter-dependent rRNA transcription in vitro, and overexpression of a deletion mutant of PAF49 reduced pre-rRNA synthesis in vivo. These results indicate that PAF49 is involved in the regulation of rRNA transcription.
MATERIALS AND METHODS
Isolation and characterization of PAF49 cDNA.
Purification of RNA polymerase I and microsequencing of PAF49 were done as described previously (11, 42). Degenerate sense and antisense primers were deduced from peptide sequences 1 and 2 described in the text and used for PCR amplification with a random-primed mouse F9 cDNA library (kindly provided by A. Fukushima, Kagawa Nutrition University). The PCR-based screening was carried out either with the sense primer from sequence 1 and the antisense primer from peptide 2 or the antisense primer from sequence 1 and the sense primer from peptide 2. The successful combination of primers was the sense primer of sequence 1, 5′-GA(C/T) ACI CA(A/G) GA(A/G) GCI GTI AA(C/T) (A/C)G-3′, and an antisense primer of sequence 2, 5′-GG (C/T)TC IGT (A/G)TG (C/T)TC IAC (C/T)TG-3′, where I represents an inosine nucleotide. PCR products were sequenced, and the DNA fragment encoding a proper internal sequence of PAF49 was used to screen full-length cDNA clones with the MH134 λZAPII cDNA library by conventional plaque hybridization techniques. cDNAs from the positive clones were excised as plasmids from the phage vector to verify the sequences as described previously (36). The complete sequence of the PAF49 cDNA was aligned with the sequences of human ASE-1 (48) and the lamprey ASE-1/CAST (45) homologues with Clustal W multiple sequence alignment software, and a phylogenetic tree was drawn by the neighbor-joining method on the GenomeNet homepage of the Bioinformatic Center, Institute for Chemical Research, Kyoto University (http://clustalw.genome.jp/).
Construction of plasmids.
To create conventional restriction enzyme cutting sites at the outside of the initiation codon of PAF49 cDNA, the junction between the 5′ end of PAF49 cDNA and the vector was excised with KpnI and then replaced with a compatible linker (5′-GTAAAGCTTGGATCCATATGGCGGGTAC-3′/5′-CCGCCATATGGATCCAAGCTTTACGTAC-3′). The resultant plasmid contained a KpnI site recreated in the PAF49 coding region but not in the vector sequence and unique HindIII and BamHI sites followed by an NdeI site at the initiation codon. A portion of the 3′ untranslated region was removed from the AccI site in PAF49 cDNA and the EcoRI site of the vector, followed by ClaI linker ligation after a 3′-fill-in reaction (PAF49BSKII). Then an epitope tag sequence, Flag or hemagglutinin (HA), was attached in-frame via the NdeI site to create plasmids pBSKIIFlagPAF49 and pBSKIIHAPAF49, respectively. The tagged PAF49 sequence was transferred to a bacterial or mammalian expression vector such as pET, pGEX, or pcDNA3 via compatible restriction enzyme cutting sites. Murine TAFI48 cDNA was kindly provided by I. Grummt (German Cancer Research Center, Heidelberg, Germany) and subcloned for expression in mammalian cells (pcDNA3/FlagTAFI48).
Antibody preparation.
A synthetic peptide corresponding to amino acids 184 to 196 or a glutathione S-transferase (GST) fusion protein containing amino acids 21 to 271 of PAF49 (PAF49ΔC3) was used to immunize rabbits, and PAF49-specific antibodies were purified from the rabbit antiserum by affinity resin coupled with the synthetic peptide or hexahistidine-tagged PAF49ΔC3, according to standard procedures (15). The specificity of the affinity-purified antibodies was verified by Western blotting (see supplemental data at http://www.med.nagasaki-u.ac.jp/mmi/bogyo/supdata.html). The preparation of affinity-purified antibodies against PAF53, RPA40, RPA16, and RPB14 was done as described previously (11, 31, 42, 53). Anti-RPA194 antiserum was a gift from L. I. Rothblum (Geisinger Clinic, Danville, Pa). Anti-UBF mouse monoclonal antibody F-9, anti-TAFI95 antibody M-18, and anti-TAFI68 antibody N-15 (goat polyclonal antibodies) were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). Monoclonal antibody against the C-terminal domain of the largest subunit of RNA polymerase II was purchased from Promega Corp. (Madison, Wis.). Anti-Flag M2 monoclonal antibody was purchased from Sigma Chemical Co. (St. Louis, Mo.).
Preparation of Pol IA and Pol IB.
Separation of Pol IA and Pol IB was done essentially as described previously (11) with slight modifications. Whole-cell extracts were prepared and fractionated by phosphocellulose as reported previously (49). Fraction C, which was step eluted from a phosphocellulose column at 0.3 to 0.6 M KCl, was dialyzed against buffer DM [20 mM HEPES-KOH (pH 7.9), 0.1 mM EDTA, 20% glycerol, 0.2 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, and 5 mM MgCl2] containing 100 mM KCl (referred to as DM100) and then loaded onto a DEAE-Sepharose FF column (25 by 50 mm) at the flow rate of 0.5 ml/min. After extensive washing with buffer DM100, bound proteins were eluted with a linear gradient of 0.1 M to 0.8 M KCl in buffer DM.
The protein fractions were checked for Pol I activity by a random transcription assay, and the peak fractions were collected and dialyzed against buffer DM100. The protein pool was loaded onto a CM-Sepharose FF column (15 by 70 mm) at 0.4 ml/min. The flowthrough fraction served as the source of Pol IA. Proteins bound to the CM-Sepharose column were eluted with a linear gradient of 0.1 M to 0.6 M KCl in buffer DM. The peak Pol I fractions were collected, diluted with buffer DM to reduce the KCl concentration to 250 mM, and then loaded onto a Mono-Q HR 5/5 column (Amersham Biosciences Corp., Piscataway, N.J.) equilibrated with buffer DM250. Bound proteins were eluted with a linear gradient of 0.25 M to 0.5 M KCl in buffer DM. Active Pol I fractions were collected and concentrated by Ultrafree-4 (Millipore Corp., Bedford, Mass.), layered on 5 ml of a 25 to 50% glycerol gradient containing 20 mM HEPES-KOH (pH 7.9), 0.1 mM EDTA, 100 mM KCl, 0.2 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, and 5 mM MgCl2, and centrifuged at 60,000 rpm for 16 h in a bucket of a Beckman SW60Ti rotor at 4°C. After sedimentation, proteins were collected in 100- to 200-μl fractions and then assayed for Pol I activity.
Random transcription assay.
The Pol I activity in the fractions was assayed by the electrophoresis-based quantitation method described by Jackson et al. (19) with several modifications. Aliquots (1 μl) of protein fractions were incubated in a 10-μl in vitro transcription assay mixture containing 12 mM HEPES-KOH (pH 7.9), 12% glycerol, 80 mM KCl, 6 mM MgCl2, 0.6 mM dithiothreitol, 0.3 mM EDTA, 100 μg of α-amanitin per ml, 500 μM ATP, 100 μM UTP, 2 μCi of [α-32P]UTP (400 mCi/mmol; Amersham Biosciences Corp.), and 0.1 mg of poly(dA-dT)-poly(dA-dT) per ml as the template for 30 min at 30°C. The reactions were terminated by adding 10 μl of stop solution containing 2% SDS, 10 mM EDTA, 80% formamide, and 0.05% bromophenol blue, and then 5 μl of each terminated reaction mixture was electrophoresed on a 5% polyacrylamide-7 M urea gel (15 by 15 cm) for 20 min at 500 V. The gels were dried, and the radioactivity migrating at areas greater than 100 nucleotides was quantitated with a BAS2000 image analyzer (Fujifilm, Tokyo, Japan).
Immunoprecipitation assay.
For immunoprecipitation, 200 μg of nuclear extracts was incubated with 2 μg of the affinity-purified antibodies in immunoprecipitation buffer [20 mM HEPES-KOH (pH 7.9), 12.5 mM MgCl2, 0.2 mM EDTA, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 10% glycerol, and 0.1% NP-40] containing 100 mM KCl for 1 h on ice, and then immune complexes were precipitated with 10 μl of protein A-Sepharose 4FF (Amersham Biosciences Corp.). The beads were washed extensively with immunoprecipitation buffer containing 400 mM KCl, and the proteins retained were recovered by boiling the beads in SDS sample buffer. The eluted proteins were loaded onto an SDS-polyacrylamide gel and analyzed by Western blotting on a polyvinylidene difluoride membrane with enhanced chemiluminescent detection (Amersham Biosciences Corp.).
Far-Western blotting.
Purification of Pol I and GST-PAF53 and preparation of 35S-labeled proteins were done as described previously (11). The 35S-labeled proteins were further purified by Sepharose G-25 (Amersham Biosciences Corp.) for removal of free [35S]methionine and buffer exchange. About 500 ng of pure Pol I or 400 ng of GST-PAF53 was subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose filters (BA-S 85; Schleicher & Schuell, Dassel, Germany). The protein blots were then denatured in 6 M guanidine-HCl in renaturation buffer [20 mM HEPES-KOH (pH 7.9), 10% glycerol, 60 mM KCl, 6 mM MgCl2, 0.6 mM EDTA, and 2 mM dithiothreitol] for 5 min at 4°C and renatured in successive twofold dilution of guanidine-HCl with renaturation buffer for 5 min at 4°C five times. The blots were then blocked with 0.25% skim milk in renaturation buffer for 10 min at 4°C. Then each blot was soaked in 10 ml of blocking buffer supplemented with 35S-labeled protein probe (in 500 μl of the main fraction of Sepharose G-25) for 16 h at 4°C with gentle shaking. The blots were washed twice in 20 mM HEPES-KOH (pH 7.9)-100 mM KCl-6 mM MgCl2-0.2 mM EDTA-2 mM dithiothreitol for 10 min at room temperature and then analyzed with a BAS2000 image analyzer (Fujifilm, Tokyo, Japan).
GST pulldown assay.
The DNA fragment containing the PAF49 coding region was excised from plasmid PAF49BSKII with BamHI and SmaI and then subcloned into the BamHI and SmaI sites of plasmid pGEX-3X (Amersham Biosciences Corp.). For production of N-terminal and C-terminal truncation mutants, a 0.7-kb NcoI-SmaI fragment or 0.75-kb NdeI-Eco81I fragment of PAF49 cDNA was used for subcloning in the pGEX vector, respectively. The construction of plasmids producing the GST-PAF53 and GST-RPA40 proteins was described previously (11). The resulting plasmids as well as the parental plasmid pGEX-3X were introduced into Escherichia coli strain TG1, and cell lysates were prepared according to the manufacturer's instructions. The lysates were rocked for 1 h at 4°C with glutathione-Sepharose 4B (Amersham Biosciences Corp.). The beads were washed three times with a buffer (0.5XDBT) containing 10 mM HEPES-KOH (pH 7.9), 50 mM KCl, 0.1 mM EDTA, 0.5 mM dithiothreitol, 10% glycerol, and 0.2% Triton X-100 and then suspended in 0.5XDBT buffer.
PAF53, UBF, TAFI95, TAFI68, and TAFI48 were labeled with [35S]methionine with the TNT T7 coupled reticulocyte lysate system (Promega Corp.), purified by Sepharose G-75 (Amersham Biosciences Corp.), and precleared by glutathione-Sepharose 4B. Typical binding reactions were performed with 60 μl of the precleared labeled protein and 20 μl of the beads liganded by GST fusion protein in a 100-μl reaction with 0.5XDBT by rocking at 4°C for 3 h. The beads were then washed three times with buffer 0.5XDBT containing 300 mM KCl, and the bound proteins were recovered by boiling the beads in SDS sample buffer for denaturing polyacrylamide gel electrophoresis. The gels were dried and analyzed by autoradiography.
In vitro and in vivo transcription assays.
The in vitro transcription assay was carried out at 30°C for 60 min in a 25-μl reaction mixture containing 12 mM HEPES-KOH (pH 7.9), 12% glycerol, 80 mM KCl, 4 mM MgCl2, 0.6 mM dithiothreitol, 0.3 mM EDTA, and 100 μg of α-amanitin per ml. Nucleotide concentrations were 600 μM for ATP, GTP, and CTP and 30 μM for UTP containing 5 μCi of [α-32P]UTP (400 mCi/mmol; Amersham Biosciences Corp.). The reaction mixture included 6 μl of nuclear extract and 0.5 μg of template DNA (pMrBKSP2 digested with EcoRV) (49) or 1.65 to 3.3 μg of purified immunoglobulin G (IgG) and 4.2 μg of hexahistidine-tagged PAF49ΔC3, where indicated. Reactions were terminated by adding 200 μl of stop mixture (0.1% SDS, 10 mM EDTA, and 100 μg of yeast tRNA per ml). The transcripts were extracted once with phenol and once with phenol-chloroform, precipitated with ethanol, and analyzed by electrophoresis on a 5% polyacrylamide-7 M urea gel.
For in vivo analysis, NIH 3T3 cells were transfected with expression plasmids for Flag-tagged PAF49 or deletion mutants with the TransFast reagent (Promega Corp.) according to the manufacturer's instruction. After incubation for 24 h, transfected cells were recovered and divided into two parts; one was used for the extraction of total RNA by means of Isogen (Nippon Gene), and the other was used for nuclear extract preparation by the method of Shreiber et al. (39). Total RNAs were analyzed by a primer extension system (Promega Corp.) with a 5′-labeled pre-rRNA primer (5′-TGGACAGCAAAACAGCCTTAAATCG-3′, complementary to +80 to +56 of murine 45S pre-rRNA) and β-actin primer (5′-GGACCGGCAACGAAGGAGCTGC-3′, complementary to +60 to +39 of murine β-actin mRNA) as instructed by the manufacturer.
Immunofluorescence microscopy.
NIH 3T3 cells were seeded on sterilized coverslips in a six-well plate (5 × 104/well) and grown for 24 h in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated calf serum, penicillin G (10 U/ml), and streptomycin (50 μg/ml) in a humidified incubation chamber containing 5% CO2 in air at 37°C. Then the cells were washed and incubated in Dulbecco's modified Eagle's medium supplemented with 0.1% calf serum and antibiotics for 48 h. During this incubation, NIH 3T3 cells were seeded in another six-well plate and grown in 10% calf serum-containing medium. Both the growing and serum-restricted cells on the coverslips were fixed in 1% paraformaldehyde in Dulbecco's modified Eagle's medium at 37°C for 15 min and permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS) at room temperature for 10 min. After blocking with 1.5% bovine serum albumin and 1.5% skim milk in PBS for 1 h, the cells were incubated overnight with antigen-purified anti-PAF49 rabbit polyclonal IgG (7 μg/ml) and anti-UBF mouse monoclonal IgG1 F-9 (2 μg/ml) in PBS supplemented with 2% bovine serum albumin at 4°C in a humidified chamber. The cells were washed three times with PBS and then incubated with Alexa Fluor 488-chicken anti-rabbit IgG (1:500; Molecular Probes) and Alexa Fluor 594-chicken anti-mouse IgG (1:500; Molecular Probes) at room temperature in a humidified chamber for 1 h. After washing three times with PBS, the coverslips were mounted in PBS supplemented with 50% glycerol and sealed with clear nail polish. Cells were viewed with a Carl Zeiss LSM510META confocal laser scanning microscope (Nagasaki University Center for Frontier Life Sciences).
Nucleotide sequence accession number.
The nucleotide sequence of PAF49 has been deposited in DDBJ/EMBL/GenBank under accession number AB091121.
RESULTS
Molecular cloning of PAF49.
Pol I was purified from the whole-cell extract of murine MH134 ascites cells, and the partial amino acid sequence of the 49-kDa polypeptide, which we refer to as PAF49, was determined as described previously (11, 42). Three peptide sequences were obtained from Achromobacter proteinase 1 (AP-1) digestion: sequence 1, GTDASSDTQEAVNRH; sequence 2, GTETSQVEHTEP; and sequence 3, YFMQEEMEA. Sequences 1 and 2 were used to design degenerate primers for PCR amplification with a cDNA stock prepared from mouse embryonal carcinoma F9 cells as the template. A 224-bp PCR product was selectively amplified, the predicted amino acid sequence of which initiated from sequence 1, ended at sequence 2, and contained sequence 3 (Fig. 1).
FIG. 1.
Amino acid sequence alignment of PAF49 with the human and lamprey orthologues. (A) Residues identical between murine PAF49 (mPAF49) and its potential human counterpart ASE-1 (hASE-1) (48) and lamprey CAST (45) are indicated by dark shading. Similar residues are also highlighted by gray shading, according to the grouping (A, I, L, V), (F, W, Y), (E, D), (G, P), (K, R), (N, Q), (S, T), (C), (H), and (M). Sequences missing in PAF49 are represented by dashes. The sequences obtained by peptide microsequencing of PAF49 are indicated above the sequence of PAF49 as sequence 1, 2, or 3. Asterisks under the sequence of CAST indicate clusters of basic aminoacids. The position of the amino acid insertion (GE) found in human CAST is indicated by an open triangle above the sequence of PAF49 (see text for details). (B) Neighbor-joining tree of PAF49/ASE-1 sequences. The tree is based on the amino acid alignment in panel A and is rooted at the midpoint.
To obtain full-length cDNA, the MH134 cDNA library was screened with the PCR product as the probe. Five independent clones were isolated, each of which contained a 3′ poly(A) tail but varied in the length of the 5′ end. The longest cDNA was 1,320 nucleotides long and contained an open reading frame for a 411-amino-acid protein with a predicted molecular weight of 44,000. Database search revealed that PAF49 has striking homology with human ASE-1 (Fig. 1). ASE-1 was identified as a human autoimmune antigen that locates to fibrillar centers in interphase and to nucleolar organizer regions during cell division (48). It was also reported that ASE-1 colocalizes with UBF throughout the cell cycle and that the two proteins bind each other. However, the precise functions of ASE-1 have not yet been clarified.
We searched the databases comprehensively and found a sequence deposited as a lamprey homologue of the human protein (45). However, we could not obtain functionally annotated related sequences from any other species in either expressed sequence tag or genome databases. Sequence alignment of PAF49 with the possible human and lamprey counterparts revealed that the N-terminal half of the protein was well conserved among the species (Fig. 1). The C-terminal part of each protein also had highly conserved sequences, but the human and lamprey homologues had various amino acid insertions and a C-terminal extension. Notably, there were several stretches of repeated lysine and arginine residues in this region. Each of these basic repeats was followed by a shorter stretch of acidic amino acids. The arrangement of these alternating basic and acidic regions was also conserved among the species.
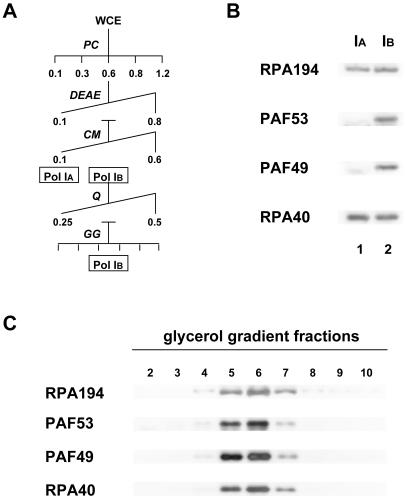
PAF49 is present in a subpopulation of Pol I.
We demonstrated previously that mouse Pol I can be separated into two populations during purification by column chromatography (24). The one referred to as Pol IB stays bound on a CM-Sepharose column, but the other (Pol IA) flows through the column under 0.1 M KCl (Fig. 2A) (11). To determine the relationship of PAF49 to these forms of Pol I, CM-Sepharose fractions of Pol IA and Pol IB were analyzed by Western blotting (Fig. 2B). The flowthrough and 0.3 M KCl fractions contained comparable amounts of the Pol I core subunits RPA194 and RPA40, whereas PAF53 was found only in the 0.3 M KCl fraction, consistent with the previous report (11). Reprobing the blot with anti-PAF49 antibody indicated that PAF49 was also present exclusively in the 0.3 M KCl fraction. To rule out the possibility that PAF49 and PAF53 were fortuitously eluted with Pol I core enzyme in the 0.3 M KCl fraction, it was further purified by a linear salt gradient on a Mono-Q column, followed by glycerol gradient fractionation (Fig. 2A). In this highly purified Pol I preparation, both PAF49 and PAF53 still copurified with the Pol I subunits (Fig. 2C). On the other hand, Pol IA fractions purified further by Mono-Q and glycerol gradient fractionation contained neither PAF49 nor PAF53 (data not shown). These results clearly indicate that PAF49 and PAF53 are definite components of Pol IB.
FIG. 2.
PAF49 is contained in Pol IB. (A) Schematic diagram of the purification procedure for Pol IA and Pol IB from whole-cell extracts (WCE). Abbreviations: PC, phosphocellulose; DEAE, DEAE-Sepharose; CM, CM-Sepharose; Q, Mono-Q; GG, glycerol gradient fractionation. The values represent the molar concentration of KCl used to develop the column. (B) The flowthrough fraction (Pol IA; lane 1) and 0.3 M KCl fraction (Pol IB; lane 2) from CM-Sepharose were analyzed by Western blotting with anti-RPA194, anti-PAF53, anti-PAF49, and anti-RPA40 antibodies. (C) The glycerol gradient fractions (2 to 10, recovered from the bottom to the top of the sediments, respectively) were analyzed by Western blotting with anti-RPA194, anti-PAF53, anti-PAF49, and anti-RPA40 antibodies.
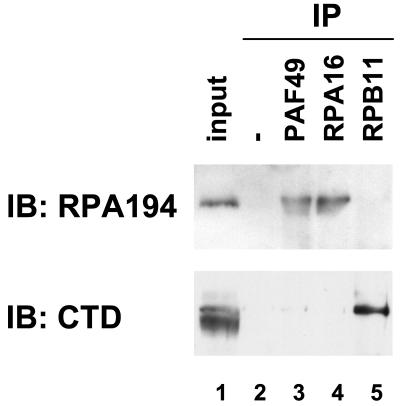
PAF49 is physically associated with Pol I.
To examine whether PAF49 physically associates with Pol I core enzyme, a coimmunoprecipitation assay was performed. Nuclear extracts were incubated with a variety of affinity-purified antibodies as well as control IgG, and then the immune complexes were precipitated with protein A-Sepharose beads. As reported previously (53), the largest Pol I subunit, RPA194, was coimmunoprecipitated by an antibody against the RPA16 subunit (Fig. 3, lane 4, upper panel). RPA194 was also detected in the immune complex precipitated by anti-PAF49 antibody (lane 3, upper panel). Neither the control IgG nor an antibody against the Pol II-specific subunit RPB11 (31) precipitated RPA194 (lanes 2 and 5, upper panel). On the other hand, the anti-RPB11 antibody effectively precipitated the largest subunit of RNA polymerase II, supporting the specificity of the antibodies used. These results indicate that PAF49 physically associates with the Pol I core enzyme.
FIG. 3.
Physical association of PAF49 with Pol I core enzyme. Mouse nuclear extracts (indicated in lane 1 as input) were incubated with a control IgG (lane 2), affinity-purified anti-PAF49 antibody (lane 3), anti-RPA16 antibody (lane 4), or anti-RPB11 antibody (lane 5), and then precipitated with protein A-Sepharose beads. The presence of the Pol I-specific largest subunit (upper panel) or the Pol II-specific largest subunit (lower panel) in each immune complex was determined by Western blotting with anti-RPA194 antiserum and anti-CTD monoclonal antibody, respectively. IB, immunoblot.
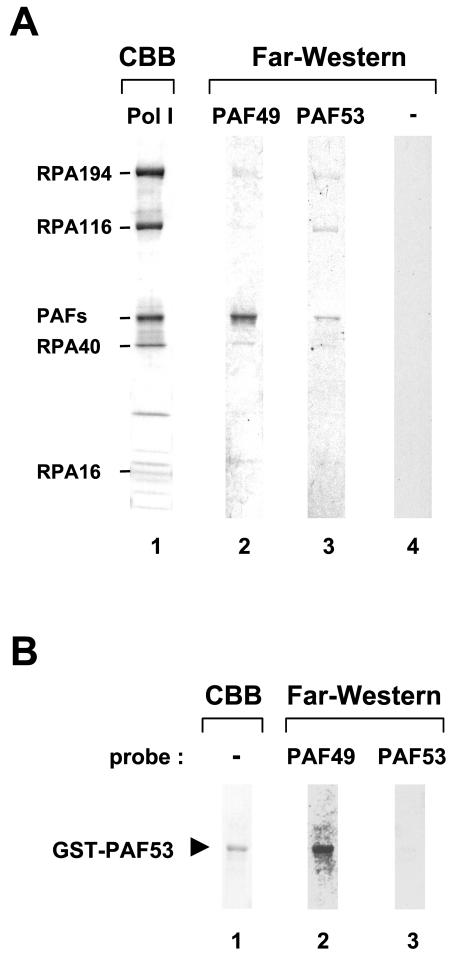
Examination of direct protein-protein contacts in the Pol I complex by PAF49.
To determine whether PAF49 directly binds to Pol I subunits, we used the far-Western blotting technique. Purified Pol I was electrophoresed, blotted onto a nitrocellulose membrane, and then incubated with a protein probe labeled with [35S]methionine by in vitro coupled transcription-translation. The Pol I sample was also visualized by Coomassie brilliant blue staining to assign individual far-Western signals (Fig. 4A, lane 1). The largest and the second largest subunits (RPA194 and RPA116) were identified by size, and some of the others (RPA40 and RPA16) were identified by immunoblotting beforehand. When PAF49 was used as the probe, a strong signal was observed at the position corresponding to the third largest polypeptide band of Pol I (lane 2). Although it seemed to be an apparently single band, this region has been shown to contain PAF49 as well as PAF53 and its close relative, PAF51 (11). The PAF53 probe also reacted with a polypeptide(s) in this region (lane 3), raising the question of whether PAF49 and PAF53 interact with each other (see below). To a lesser extent, PAF53 bound to RPA194 and RPA116. RPA40 and RPA194 also showed very weak far-Western signals with the PAF49 probe. The functional significance of these interactions needs to be examined further (see Discussion).
FIG. 4.
Far-Western analysis of PAF49 and PAF53. (A) Purified Pol IB was blotted onto a nitrocellulose membrane and then incubated with 35S-labeled PAF49 (lane 2) or PAF53 (lane 3) prepared by in vitro coupled transcription-translation reaction. A control labeling reaction with an empty plasmid template was used as control probe (lane 4). Same Pol I sample was also loaded onto the SDS-polyacrylamide gel, and individual subunits of Pol I were stained by Coomassie brilliant blue (lane 1). The positions of formerly identified subunits and PAFs are indicated at the left. (B) The far-Western blotting experiment was performed with purified GST-PAF53 and 35S-labeled PAF49 (lane 2) or PAF53 (lane 3). The purity of GST-PAF53 was indicated by Coomassie staining (lane 1).
PAF49 interacts with PAF53.
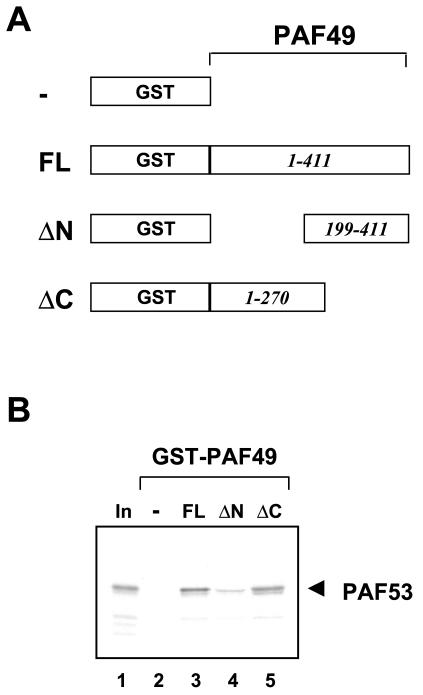
The far-Western experiment described above indicated that both PAF49 and PAF53 interacted with a protein(s) in the region of the third largest polypeptide. However, it could not distinguish heteromeric interaction between PAF49 and PAF53 from self-interaction by the individual factors. To address this issue, recombinant fusion protein GST-PAF53 was purified from bacterial cells, and its ability to interact with PAF53 or PAF49 was tested by far-Western blotting. As shown in Fig. 4B, blotted GST-PAF53 was specifically bound by 35S-labeled PAF49 (lane 2), whereas no obvious signal was detected with 35S-labeled PAF53 (lane 3). These results strongly suggest that the PAF-PAF interactions detected in the primary far-Western blot with purified Pol I were due to the heteromeric interaction between PAF49 and PAF53. To confirm this, we performed a GST pulldown assay with PAF49 deletion mutants (Fig. 5A). Consistent with the results of far-Western blotting, PAF53 bound to full-length PAF49 fused to GST (Fig. 5B, lane 3) but not to GST alone (lane 2). Deletion of the N-terminal 198 residues from PAF49 abolished PAF53 binding (lane 4), whereas the C-terminal 141 residues were dispensable for the interaction (lane 5). In contrast, PAF49 failed to interact with any form of the GST-PAF49 fusion proteins (data not shown). These results demonstrate that PAF49 binds to PAF53 through its N-terminal region.
FIG. 5.
PAF49 binds to PAF53 through its N-terminal region. (A) Schematic presentation of GST-PAF49 fusion proteins: —, GST protein alone; FL, full-length PAF49 fused to GST; ΔN, GST fusion with PAF49 lacking the N-terminal 198 residues; ΔC, GST fusion with PAF49 lacking the C-terminal 141 residues. (B) GST pulldown assay with GST-PAF49 mutants and PAF53. The recombinant fusion proteins GST-PAF49FL (lane 3), ΔN (lane 4), and ΔC (lane 5) as well as GST alone (lane 2) were immobilized on glutathione beads and then incubated with 35S-labeled PAF53. The bound protein was detected by autoradiography after SDS-PAGE. Ten percent of the input PAF53 was electrophoresed in lane 1.
PAF49 interacts with TAFI48.
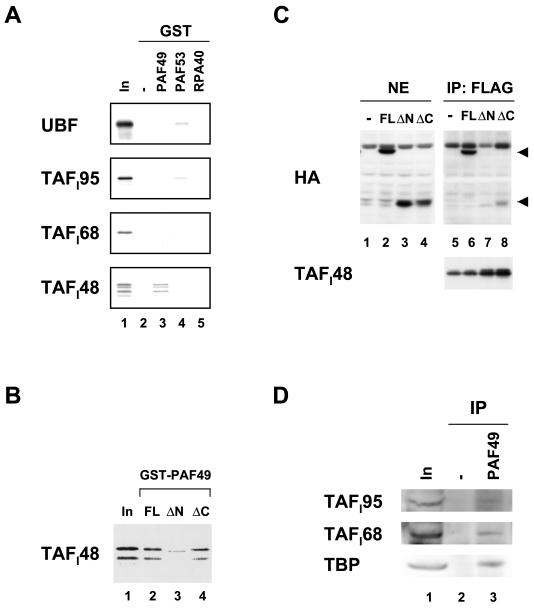
Next we examined whether PAF49 and PAF53 interact with other components of the rRNA transcription machinery. GST fusion proteins with PAF49 and PAF53 were incubated with 35S-labeled UBF1 and the TBP-associated factors of the SL1 complex (TAFI95, TAFI68, and TAFI48). Consistent with the previous report (11), UBF1 interacted with PAF53 (Fig. 6A, uppermost panel, lane 4). However, UBF1 could not bind to PAF49 (lane 3). Among the proteins tested, TAFI48 exhibited the best interaction with PAF49 (lowermost panel, lane 3). A weak signal of TAFI95 was detected with PAF53 (second panel, lane 4), suggesting a possible interaction between PAF53 and TAFI95. In contrast, RPA40 did not bind to any of the proteins tested (all panels, lane 5).
FIG. 6.
PAF49 interacts with TAFI48 in vitro and in vivo. (A) GST-PAF49 (lane 3), GST-PAF53 (lane 4), and GST-RPA40 (lane 5) as well as GST alone (lane 2) were immobilized on glutathione beads and then incubated with 35S-labeled UBF1 and the SL1 subunits TAFI95, TAFI68, and TAFI48. After extensive washing of the beads, bound proteins were eluted by boiling in SDS sample buffer and applied to an SDS-polyacrylamide gel. Ten percent of the input labeled protein was also included in lane 1. (B) GST-PAF49FL (lane 2), ΔN (lane 3), and ΔC (lane 4) were immobilized on glutathione beads and then incubated with 35S-labeled TAFI48. The bound protein was detected by autoradiography. Ten percent of the input labeled TAFI48 was also included (lane 1). (C) NIH 3T3 cells were cotransfected with plasmids expressing Flag-tagged TAFI48 and various forms of HA-tagged PAF49 as well as the empty vector. The expression of HA-tagged full-length PAF49 (FL, lane 2), the N-terminal truncation mutant (ΔN, lane 3), and the C-terminal truncation mutant (ΔC, lane 4) was confirmed in nuclear extracts by Western blotting. The extracts were then immunoprecipitated with anti-Flag M2 affinity beads (lanes 5 to 8). The positions of the full-length and C-terminally truncated forms of PAF49 in the immune complexes are indicated by arrowheads at the right. The amounts of Flag-tagged TAFI48 in the immune complexes are shown in the lower panel (lanes 5 to 8). (D) Nuclear extracts were incubated with (lane 3) or without (lane 2) anti-PAF49 antibodies, followed by precipitation with protein A beads. Each immune complex was analyzed by Western blotting with anti-TAFI95 antibody (upper panel), anti-TAFI68 antibody (middle panel), or anti-TBP antibody (lower panel). About 6% of the input nuclear extracts were also examined on the left (lane 1).
To localize the region of PAF49 required for TAFI48 binding, labeled TAFI48 was incubated with the N-terminal or C-terminal truncation mutant of PAF49 (Fig. 6B). Deletion of the N-terminal half severely impaired the binding of PAF49 to TAFI48 (lane 3), whereas the C-terminal deletion mutant of PAF49 showed affinity comparable to that of TAFI48 (lane 4). These results suggest that the N-terminal region of PAF49 is important for the interaction with TAFI48. To confirm these observations, a coimmunoprecipitation assay was performed. HA-tagged full-length and truncation mutants of PAF49 were coexpressed with Flag-tagged TAFI48 in culture cells, and the nuclear extracts were precipitated with anti-Flag monoclonal antibody. Full-length PAF49 was coprecipitated as well with TAFI48 (Fig. 6C, lane 6). In contrast, the N-terminal deletion mutant was barely detectable in the immune complex (lane 7), even though the protein was expressed equally well with full-length PAF49 (compare lane 3 with lane 2). Deletion of the C-terminal half of PAF49 moderately influenced the ability to bind TAFI48 (lane 8). These results indicate that the N-terminal region of PAF49 is important for the interaction with TAFI48 and that the C-terminal region may also contribute, to a lesser extent, to the interaction in vivo.
To confirm the physiological relevance of this interaction, we tested whether the SL1 complex could be coprecipitated with PAF49. Nuclear extracts were incubated with anti-PAF49 antibodies, and then immune complexes were precipitated with protein A beads. As shown in Fig. 6D, all other SL1 subunits (TAFI95, TAFI68, and TBP) were also coprecipitated with PAF49 (lane 3). These results strongly suggest that TAFI48 interacts with PAF49 not only as an isolated polypeptide but also as an integrated component of SL1. These results support the “holoenzyme” hypothesis in that the basic components required for rRNA transcription are preassembled in vivo (14, 34, 41).
Involvement of PAF49 in rRNA transcription.
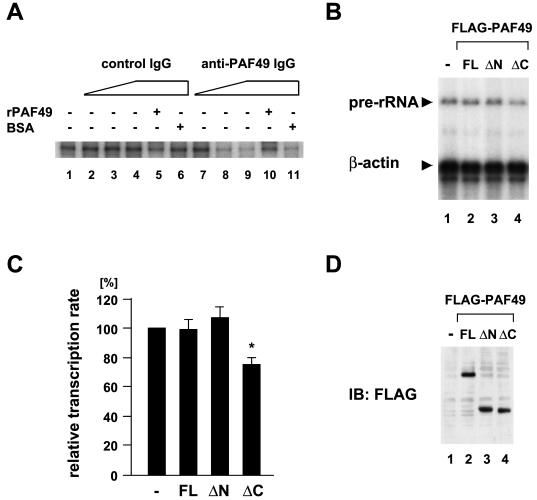
We then asked whether PAF49 functioned in rRNA transcription. PAF49-specific IgG (anti-PAF49 IgG) was purified from anti-PAF49 antiserum by protein A-Sepharose and PAF49 affinity column chromatography. rRNA promoter-specific transcription in vitro was performed in the presence of anti-PAF49 IgG or control IgG. Anti-PAF49 IgG inhibited rRNA transcription in a dose-dependent fashion (Fig. 7A, lanes 7 to 9), whereas the control IgG did not affect transcription (lanes 2 to 4). The transcriptional inhibition by anti-PAF49 IgG was relieved by the addition of recombinant PAF49 (lane 10) but not by an equal amount of bovine serum albumin (lane 11). Therefore, anti-PAF49 IgG specifically interfered with rRNA transcription in vitro.
FIG. 7.
Inhibition of rRNA transcription in vitro by anti-PAF49 antibody and in vivo by overexpression of the PAF49 mutant. (A) Nuclear extracts (30 μg) were incubated with affinity-purified anti-PAF49 IgG (lane 7, 0.8 μg; lane 8, 1.6 μg; lanes 9 to 11, 3.2 μg) or control IgG (lane 2, 0.8 μg; lane 3, 1.6 μg; lanes 4 to 6, 3.2 μg) in the absence (lanes 1 to 4 and 7 to 9) or the presence of 4.2 μg of recombinant PAF49 (lanes 5 and 10) or bovine serum albumin (lanes 6 and 11). Then, the template DNA containing the rRNA promoter and nucleotides was added for in vitro transcription. The promoter-specific transcripts were analyzed on a denaturing polyacrylamide gel and visualized by autoradiography. (B) NIH 3T3 cells were transfected with expression plasmids for Flag-tagged PAF49 or its deletion mutants. Total RNA were recovered after incubation for 24 h and analyzed by primer extension assay with a 5′-labeled pre-rRNA primer and β-actin primer. (C) Summary of the primer extension assay results. The relative transcription rate of pre-rRNA was obtained by normalizing the amounts of pre-rRNA to those of β-actin mRNA and compared with the control value. Values are represented as averages of three independent experiments with the standard deviation. *, P < 0.01, Student's t test (ΔC versus full-length PAF49). (D) The amounts of ectopically expressed PAF49 and its deletion mutants were determined by Western blotting with anti-Flag M2 monoclonal antibody.
The role of PAF49 on rRNA transcription was also examined in vivo. NIH 3T3 cells were transfected with a plasmid expressing full-length PAF49 or its N- or C-terminal truncation mutant, and the amounts of unprocessed pre-rRNA transcripts were quantitated by primer extension assay to determine the rate of rRNA transcription initiation in the transfected cells. Pre-rRNA synthesis was barely affected in the cells expressing either full-length PAF49 or the N-terminal truncation mutant (Fig. 7B, lanes 2 and 3). On the other hand, we reproducibly detected a 20 to 30% reduction in cellular pre-rRNA in cells expressing the C-terminal truncation mutant (Fig. 7B, lane 4; see also Fig. 7C for quantitative representation, P < 0.01). The amounts of the ectopically expressed proteins were comparable among the transfected cells (Fig. 7D). Together, these results strongly suggest that PAF49 functions in rRNA transcription.
Growth-dependent nucleolar accumulation of PAF49.
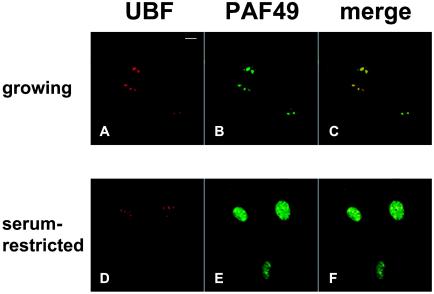
rRNA synthesis and subsequent processing occur in the nucleolus (reviewed in reference 44). It is also known that the rRNA transcription highly correlates with cell growth and metabolism (reference 43 and references therein). We thus compared the subcellular distribution of PAF49 in growing cells and growth-inhibited cells. In exponentially growing NIH 3T3 cells, a few large discrete nucleoli were observed in cell nuclei, where the ribosomal transcriptional activation factor UBF accumulated (Fig. 8A). PAF49 was colocalized with UBF in this culture (Fig. 8B and C). When cells were cultured in serum-restricted medium, some PAF49 dispersed into the nucleoplasm, though some remained in the nucleoli (Fig. 8E). The nucleoli became smaller and increased in number, and UBF remained in these tiny nucleoli (Fig. 8D). A small population of PAF49 also localized in nucleoli as well as in the nucleoplasm (Fig. 8F). Similar distribution of PAF49 was also observed in the nuclei of density-arrested cells (data not shown). These results clearly demonstrate that PAF49 accumulates in the nucleolus in accordance with cell proliferation.
FIG. 8.
PAF49 accumulates in the nucleolus of growing cells. NIH 3T3 cells were grown in culture medium containing 10% (A to C) or 0.1% (D to F) calf serum. The cells were then fixed and incubated with anti-UBF mouse monoclonal antibody and anti-PAF49 rabbit polyclonal antibodies, followed by fluorescent secondary antibodies. Images were obtained separately for UBF (red; A and D) and for PAF49 (green; B and E) or merged (C and F); yellow spots indicate colocalization of UBF and PAF49.
DISCUSSION
We reported the RNA polymerase I-associated factor PAF53 previously (11). Here we describe the characterization of another Pol I-associated factor, PAF49. PAF49 shows strong homology to the human protein ASE-1. Initially, ASE-1 was predicted as an open reading frame present in the antisense orientation to the gene coding for the DNA repair enzyme ERCC-1 and was thus given the name ASE-1, standing for antisense to ERCC-1 (46). Subsequently, another group identified the gene product as a nucleolar protein by screening a HeLa cDNA library with a human autoimmune serum (48). They showed that ASE-1 occurred at the fibrillar center in interphase and then in the nucleolar organizer regions during cell division. The N-terminal 197 amino acids of PAF49 show 66% identity with that of ASE-1. Although the C-terminal half of PAF49 also shows substantial homology (48% identity and 63% similarity), there are several extra sequences within the C-terminal half and at the C terminus of ASE-1.
Among the conserved sequences in the C-terminal region, a motif rich in lysine and arginine residues was repeatedly observed. All or some of the basic motifs may serve as a nuclear and/or nucleolar localization signal, although they do not resemble the representative nuclear or nucleolar localization signals. Three of the five basic repeats are also conserved in the lamprey homologue. Phylogenetic analysis revealed that the mouse and human sequences are closer homologues than the lamprey sequence. Interestingly, when we used the lamprey sequence in the search of the database by the PSI-BLAST algorithm, S. cerevisiae gene RPA34 was detected as a related sequence, with 9.8% identity. RPA34 encodes A34.5, a 34.5-kDa phosphoprotein that copurifies with S. cerevisiae Pol I (9). Although RPA34 showed more superficial similarity to PAF49/ASE-1, it is tempting to speculate on a potential link between these proteins (see below).
A human protein that is quite similar to ASE-1 has been reported to function in the signal transduction pathway for T-cell activation. This protein, named CAST for a CD3ɛ-associated signal transducer (52), was identified by West-Western screening of a λgt11 expression library with a 32P-labeled protein probe containing the cytoplasmic domain of CD3ɛ. The primary sequence of CAST differs from that of ASE-1 only by insertion of two amino acids (glycine-glutamic acid) at position 8 of ASE-1 (Fig. 1). When a T-cell hybridoma overexpressing HA-tagged CAST was stimulated by cross-linking with anti-CD3ɛ monoclonal antibody, a unique tyrosine residue at position 82 became phosphorylated. The mutant form of CAST, having a substitution of this tyrosine residue to phenylalanine, or truncations from the N and C termini leaving the minimal CD3ɛ-binding region intact, dominantly inhibited the NFAT-dependent induction of interleukin-2 upon T-cell receptor stimulation (52). Our preliminary experiments indicated that T-cell receptor stimulation did not phosphorylate the conserved tyrosine residue of PAF49 (data not shown). Moreover, PAF49 was localized to the nucleolus in growing cells, while CAST has been reported to be located in both the cytoplasm and nucleus (52). PAF49 showed sharply different characteristics from CAST. Although it is not known at present whether ASE-1 and CAST functionally represent two sides of a single gene product, we at least favor the idea that ASE-1 is the likely human orthologue of PAF49.
We have shown here that PAF49 has multiple protein-protein interactions in the Pol I transcription machinery. Far-Western blotting experiments showed PAF49 binding to a set of the Pol I subunits with different affinities. Among them, we further examined the interaction of PAF49 with RPA40 by a yeast two hybrid-system. However, no activation of the reporter genes was observed (data not shown). Thus, more detailed analysis should be required to fully describe the PAF49-Pol I subunit interactions. Isolation of cDNAs encoding these candidate Pol I subunits is in progress. The most prominent far-Western signal was observed from the interaction with the third largest polypeptide, which was then shown to be the specific interaction between PAF49 and PAF53. Given the stoichiometric composition of the purified Pol I used here, it is plausible that the association of PAF49 with Pol I may depend primarily on PAF53. Actually, the cellular content of PAF49 was far less than that of PAF53, raising the possibility that the association of PAF49 with Pol I confers an extra potential on the Pol I enzyme (see below).
PAF49 was also shown to interact with murine TAFI48 (Fig. 6). It was demonstrated both in vitro and in vivo that the N-terminal domain of PAF49 was required for this interaction. This N-terminal region also participated in the binding to PAF53 (Fig. 5). This region is highly conserved between mouse PAF49 and the human counterpart ASE-1, whereas the C-terminal halves of the molecules show less conservation. In contrast, both PAF53 and TAFI48 have overall similarity between mouse and human in the entire primary sequences (17) (data not shown). Thus, the N-terminal region of PAF49 might be conserved to fold into a proper structure for protein-protein interactions with PAF53 and TAFI48, while the C-terminal half might have evolved so that it may participate in a species-specific function (16, 50). Actually, the anti-PAF49 polyclonal antibody, which was raised against the N-terminal region of PAF49, abolished promoter-specific transcription in vitro (Fig. 7A), indicating that the protein-protein interactions between the N-terminal region of PAF49 and PAF53/TAFI48 are fundamentally important in rRNA transcription. More detailed structure-function relationships must be examined to understand the roles of the C-terminal region of PAF49 and the meaning of its divergence among different species.
We have reported previously that there are free and transcriptionally engaged Pol I in mammalian cells (25). In accordance with this observation, CM-Sepharose column chromatography resolved Pol I into two forms during purification from cell extracts, one (Pol IB) containing PAF53 and the other (Pol IA) not (11). Both forms of Pol I were equally active in the random transcription assay, whereas only Pol IB directed promoter-specific transcription in vitro. Indeed, anti-PAF53 antibody inhibited promoter-specific transcription but did not have any inhibitory effects on random transcription in in vitro assays. In addition, PAF53 accumulated in the nucleolus of growing cells but not in serum-starved cells (11). These findings led us to the idea that the association of PAF53 is required for the commitment of Pol I to the initiation of rRNA transcription. Seither et al. (40), however, claimed that PAF53 is not an associated regulatory factor but a bona fide subunit of Pol I. Although it may depend on the definition of the core Pol I, we demonstrated here again that PAF53-containing Pol IB could be separated from PAF53-lacking Pol IA and further showed that another molecule, PAF49, was also present exclusively in Pol IB (Fig. 2).
Because the experimental approaches of Seither et al. were significantly different from ours, it is difficult to interpret the discrepancy at present. However, growth-dependent nucleolar accumulation and redistribution to the nucleoplasm (Fig. 8) strongly support the idea that PAF49 actually changes its state to engage in rRNA transcription in vivo. As mentioned above, it is interesting that short stretches of amino acids in the PAF49 sequence show similarities to the A34.5 subunit of S. cerevisiae Pol I. Cells devoid of A34.5 are viable but generate a structurally modified Pol I which lacks the A49 subunit in addition to A34.5 upon in vitro purification (9). S. cerevisiae A49 and the related Schizosaccharomyces pombe RPA51 have slight similarity to mouse PAF53 (11, 21, 30). The yeast HMG-box protein Hmo1 genetically interacts with A49 (10).
It has been speculated that the Pol I* form, which lacks A49 and A34.5 (18), is physiologically relevant and coexists in vivo with the complete enzyme (9). Thus, the situation of A49 and A34.5 in S. cerevisiae is reminiscent of that of PAF53 and PAF49 in the mouse. To understand the physiological meanings of dissociation and association of PAF53 and PAF49/ASE-1 with mammalian Pol I, reconstitution of Pol IB by recombinant PAFs and Pol IA is certainly required. We have been attempting this but have not succeeded because of the instability of full-length PAF49 recombinant protein in both bacterial and insect cells. Moreover, we found that PAF49 was likely to have a sort of posttranslational modifications which were important for the association of PAF49 with the Pol I core enzyme (K. Yamamoto and T. Matsuyama, unpublished data).
Recently, Zomerdijk and colleagues reported alternative forms of Pol I from human cells: the transcriptionally inactive bulk complex (Pol Iα) and the subpopulation (Pol Iβ) that is competent for specific transcription (27). The latter form of Pol I has been shown to contain the human homologue of Rrn3 (hRRN3), an essential transcription factor of Pol I-dependent transcription in S. cerevisiae (20, 27, 28, 51). hRRN3 mediates the recruitment of Pol I to the preinitiation complex by interacting with hTAFI110 and hTAFI63. Importantly, it was noted that PAF53 was found in both the Pol Iα and Pol Iβ preparations (27). We examined the integration of RRN3 in our Pol I preparations with anti-hRRN3 polyclonal antibodies, but unfortunately the antibodies did not cross-react with murine RRN3 even in nuclear extracts (data not shown). Thus, it is not clear at present whether PAF49 is contained in either Pol Iα or Pol Iβ. However, our preliminary experiments suggested that not all the PAF53-containing Pol I population possessed PAF49. Taking into account the fact that PAF49 binds to SL1 (Fig. 6) and chromatin-associated transcription factors (K. Yamamoto, T. Matsuyama, and M. Muramatsu, unpublished observations), PAF49 may confer additional transcription potency on Pol I in concert with RRN3.
Acknowledgments
We thank Larry Rothblum and Ingrid Grummt for the generous gift of anti-RPA194 antiserum and mTAFI48 cDNA, respectively. We are grateful to Namiko Hihara and Fumiyo Tsujita for technical assistance and express deep gratitude to Koji Hisatake for critical reading of the manuscript and valuable comments.
This work was supported by grants-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan.
REFERENCES
- 1.Albert, A.-C., M. Denton, M. Kermekchiev, and C. S. Pikaard. 1999. Histone acetyltransferase and protein kinase activities copurify with a putative Xenopus RNA polymerase I holoenzyme self-sufficient for promoter-dependent transcription. Mol. Cell. Biol. 19:796-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aprikian, P., B. Moorefield, and R. H. Reeder. 2001. New model for the yeast RNA polymerase I transcription cycle. Mol. Cell. Biol. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodem, J., G. Dobreva, U. Hoffmann-Rohrer, S. Iben, H. Zentgraf, H. Delius, M. Vingron, and I. Grummt. 2002. TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis, is the mammalian homolog of yeast Rrn3p. EMBO Rep. 1:171-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brun, R. P., K. Ryan, and B. Sollner-Webb. 1994. Factor C*, the specific initiation component of the mouse RNA polymerase I holoenzyme, is inactivated early in the transcription process. Mol. Cell. Biol. 14:5010-5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavanaugh, A. H., I. Hirschler-Laszkiewicz, Q. Hu, M. Dundr, T. Smink, T. Misteli, and L. I. Rothblum. 2002. Rrn3 phosphorylation is a regulatory checkpoint for ribosome biogenesis. J. Biol. Chem. 277:27423-27432. [DOI] [PubMed] [Google Scholar]
- 6.Comai, L., J. C. B. Zomerdijk, H. Beckmann, S. Zhou, A. Admon, and R. Tjian. 1994. Reconstitution of transcription factor SL1: exclusive binding of TBP by SL1 or TFIID subunits. Science 266:1966-2018. [DOI] [PubMed] [Google Scholar]
- 7.Dequard-Chblat, M., M. Riva, C. Carles, and A. Sentenac. 1991. RPC19, the gene for a subunit common to yeast RNA polymerase A (I) and C (III). J. Biol. Chem. 15:15300-15307. [PubMed] [Google Scholar]
- 8.Fath, S., P. Milkereit, G. Peyroche, M. Riva, C. Carles, and H. Tschochner. 2001. Differential roles of phosphorylation in the formation of transcriptional active RNA polymerase I. Proc. Natl. Acad. Sci. USA 98:14334-14339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gadal, O., S. Mariotte-Labarre, S. Chedin, E. Quemeneur, C. Carles, A. Sentenac, and P. Thuriaux. 1997. A34.5, a nonessential component of yeast RNA polymerase I, cooperates with subunit A14 and DNA topoisomerase I to produce a functional rRNA synthesis machine. Mol. Cell. Biol. 17:1787-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gadal, O., S. Labarre, C. Boschiero, and P. Thuriaux. 2002. Hmo1, an HMG-box protein, belongs to the yeast ribosomal DNA transcription system. EMBO J. 21:5498-5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanada, K., C. Z. Song, K. Yamamoto, K. Yano, Y. Maeda, K. Yamaguchi, and M. Muramatsu. 1996. RNA polymerase I associated factor 53 binds to the nucleolar transcription factor UBF and functions in specific rDNA transcription. EMBO J. 15:2217-2226. [PMC free article] [PubMed] [Google Scholar]
- 12.Hannan, K. M., L. I. Rothblum, and L. S. Jefferson. 1998. Regulation of ribosomal DNA transcription by insulin. Am. J. Physiol. 275:C130-C138. [DOI] [PubMed] [Google Scholar]
- 13.Hannan, R. D., A. Cavanaugh, W. M. Hempel, T. Moss, and L. Rothblum. 1999. Identification of a mammalian RNA polymerase I holoenzyme containing components of the DNA repair/replication system. Nucleic Acids Res. 27:3720-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hannan, R. D., W. M. Hempel, A. Cavanaugh, T. Arino, S. I. Dimitrov, T. Moss, and L. Rothblum. 1998. Affinity purification of mammalian RNA polymerase I: identification of an associated kinase. J. Biol. Chem. 273:1257-1267. [DOI] [PubMed] [Google Scholar]
- 15.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Heix, J., and I. Grummt. 1995. Species-specificity of transcription by RNA polymerase I. Curr. Opin. Genet. Dev. 5:652-656. [DOI] [PubMed] [Google Scholar]
- 17.Heix, J., J. C. B. M. Zomerdijk, A. Ravanpay, R. Tjian, and I. Grummt. 1997. Cloning of murine RNA polymerase I-specific TAF factors: conserved interactions between the subunits of the species-specific transcription initiation factor TIF-IB/SL1. Proc. Natl. Acad. Sci. USA 94:1733-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huet, J., J. M. Buhler, A. Sentenac, and P. Fromageot. 1975. Dissociation of two polypeptide chains from yeast RNA polymerase A. Proc. Natl. Acad. Sci. USA 72:3034-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson, A. J., M. Ittmann, and B. F. Pugh. 1995. The BN51 protein is a polymerase (Pol)-specific subunit of RNA Pol III which reveals a link between Pol III transcription and pre-rRNA processing. Mol. Cell. Biol. 15:94-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keener, J., C. A. Josaitis, J. A. Dodd, and M. Nomura. 1998. Reconstitution of yeast RNA polymerase I transcription in vitro from purified components. TATA-binding protein is not required for basal transcription. J. Biol. Chem. 273:33795-33802. [DOI] [PubMed] [Google Scholar]
- 21.Liljelund, P., S. Mariotte, J.-M. Buhler, and A. Sentenac. 1992. Characterization and mutagenesis of the gene encoding A49 subunit of RNA polymerase A in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 89:9302-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahajan, P. B., and E. A. Thompson. 1991. Glucocorticoid regulation of rRNA synthesis. Mol. Cell. Biochem. 104:195-200. [DOI] [PubMed] [Google Scholar]
- 23.Mann, C., J.-M. Buhler, I. Treich, and A. Sentenac. 1987. RPC40, a unique gene for a subunit shared between yeast RNA polymerase A and C. Cell 48:627-637. [DOI] [PubMed] [Google Scholar]
- 24.Matsui, T., T. Onishi, and M. Muramatsu. 1976. Nucleolar DNA-dependent RNA polymerase from rat liver. 1. Purification and subunit structure. Eur. J. Biochem. 71:351-360. [DOI] [PubMed] [Google Scholar]
- 25.Matsui, T., T. Onishi, and M. Muramatsu. 1976. Nucleolar DNA-dependent RNA polymerase from rat liver. 2. Two forms and their physiological significance. Eur. J. Biochem. 71:361-368. [DOI] [PubMed] [Google Scholar]
- 26.Milkereit, P., and H. Tschochner. 1998. A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J. 17:3692-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, G., K. I. Panov, J. K. Friedrich, L. Trinkle-Mulcahy, A. I. Lamond, and J. C. B. M. Zomerdijk. 2001. hRRN3 is essential in the SL1-mediated recruitment of RNA polymerase I to rRNA gene promoter. EMBO J. 20:1373-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moorefield, B., E. A. Greene, and R. H. Reeder. 2000. RNA polymerase I transcription factor Rrn3 is functionally conserved between yeast and human. Proc. Natl. Acad. Sci. USA 97:4724-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moss, T., V. Y. Stefanovsky, and G. Pelletier. 1998. The structural and architectural role of upstream binding factor, UBF, p. 75-94. In M. R. Paule (ed.), Transcription of ribosomal RNA genes by eukaryotic RNA polymerase I. Springer-Verlag, Berlin, Germany.
- 30.Nakagawa, K., K. Hisatake, Y. Imazawa, A. Ishiguro, M. Matsumoto, L. Pape, A. Ishihama, and Y. Nogi. 2003. The fission yeast RPA51 is a functional homolog of the budding yeast A49 subunit of RNA polymerase I and required for maximizing transcription of ribosomal DNA. Genes Genet. Syst. 78:199-209. [DOI] [PubMed] [Google Scholar]
- 31.Nishi, Y., K. Yamamoto, Y. Yao, M. Yamamoto, Y. Nogi, H. Matsuo, and M. Muramatsu. 1997. Isolation and characterization of cDNA encoding mouse RNA polymerase II subunit RPB14. Gene 187:165-170. [DOI] [PubMed] [Google Scholar]
- 32.Nomura, M. 1998. Transcription factors used by Saccharomyces cerevisiae RNA polymerase I and the mechanism of initiation, p. 155-172. In M. R. Paule (ed.), Transcription of ribosomal RNA genes by eukaryotic RNA polymerase I. Springer-Verlag, Berlin, Germany.
- 33.Peyroche, G., P. Milkereit, N. Bischler, H. Tschochner, P. Schultz, A. Sentenac, C. Carles, and M. Riva. 2000. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J. 19:5473-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saez-Vasquez, J., and C. S. Pikaard. 1997. Extensive purification of a putative RNA polymerase I holoenzyme from plants that accurately initiates rRNA gene transcription in vitro. Proc. Natl. Acad. Sci. USA 94:11869-11874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saez-Vasquez, J., M. Meissner, and C. S. Pikaard. 2001. RNA polymerase I holoenzyme-promoter complexes include an associated CK2-like protein kinase. Plant Mol. Biol. 47:449-459. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Schnapp, A., G. Schnapp, B. Emy, and I. Grummt. 1993. Function of the growth-regulated transcription initiation factor TIF-IA in initiation complex formation at the murine ribosomal gene promoter. Mol. Cell. Biol. 13:6723-6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnapp, G., F. Santoni, C. Carles, M. Riva, and I. Grummt. 1994. TIF-IC, a factor involved in both transcription initiation and elongation of RNA polymerase I. EMBO J. 13:190-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schreiber, E., P. Matthias, M. M. Müller, and W. Shaffner. 1989. Rapid detection of octamer binding proteins with ‘mini-extracts', prepared from a small number of cells. Nucleic Acids Res. 17:6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seither, P., O. Zatsepina, M. Hoffmann, and I. Grummt. 1997. Constitutive and strong association of PAF53 with RNA polymerase I. Chromosoma 106:216-225. [DOI] [PubMed] [Google Scholar]
- 41.Seither, P., S. Iben, and I. Grummt. 1998. Mammalian RNA polymerase I exists as a holoenzyme with associated basal transcription factors. J. Mol. Biol. 275:43-53. [DOI] [PubMed] [Google Scholar]
- 42.Song, C. Z., K. Hanada, K. Yano, Y. Maeda, K. Yamamoto, and M. Muramatsu. 1994. High conservation of subunit composition of RNA polymerase I(A) between yeast and mouse and the molecular cloning of mouse RNA polymerase I 40-kDa subunit RPA40. J. Biol. Chem. 269:26976-26981. [PubMed] [Google Scholar]
- 43.Stefanovsky, V. Y., G. Pelletier, R. Hannan, T. Gagnon-Kugler, L. I. Rothblum, and T. Moss. 2001. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell 8:1063-1073. [DOI] [PubMed] [Google Scholar]
- 44.Tschochner, H., and E. Hurt. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13:255-263. [DOI] [PubMed] [Google Scholar]
- 45.Uinuk-ool, T., W. E. Mayer, A. Sato, R. Dongak, M. D. Cooper, and J. Klein. 2002. Lamprey lymphocyte-like cells express homologs of genes involved in immunologically relevant activities of mammalian lymphocytes. Proc. Natl. Acad. Sci. USA 99:14356-14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Duin, M., J. van den Tol, J. H. J. Hoeijmakers, D. Bootsma, I. P. Rupp, P. Reynolds, L. Prakash, and S. Prakash. 1989. Conserved pattern of antisense overlapping transcription in the homologous human ERCC-1 and yeast RAD10 DNA repair gene regions. Mol. Cell. Biol. 9:1794-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437-440. [DOI] [PubMed] [Google Scholar]
- 48.Whitehead, C. M., R. J. Winkfein, M. J. Fritzler, and J. B. Rattner. 1997. ASE-1: a novel protein of the fibrillar centres of the nucleolus and nucleolus organizer region of mitotic chromosomes. Chromosoma 106:493-502. [DOI] [PubMed] [Google Scholar]
- 49.Yamamoto, K., A. Koga, M. Yamamoto, Y. Nishi, T. Tamura, Y. Nogi, and M. Muramatsu. 2000. Identification of a novel 70 kDa protein that binds to the core promoter element and is essential for ribosomal DNA transcription. Nucleic Acids Res. 28:1199-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamamoto, K., M. Yamamoto, Y. Nogi, and M. Muramatsu. 2001. Species-specific interaction of transcription factor p70 with the rDNA core promoter. Biochem. Biophys. Res. Commun. 281:1001-1005. [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto, R. T., Y. Nogi, J. A. Dodd, and M. Nomura. 1996. RRN3 gene of Saccharomyces cerevisiae encodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO J. 15:3964-3973. [PMC free article] [PubMed] [Google Scholar]
- 52.Yamazaki, T., Y. Hamano, H. Tashiro, K. Itoh, H. Nakano, S. Miyatake, and T. Saito. 1999. CAST, a novel CD3ɛ-binding protein transducing activation signal for interleukin-2 production in T cells. J. Biol. Chem. 274:18173-18180. [DOI] [PubMed] [Google Scholar]
- 53.Yao, Y., K. Yamamoto, Y. Nishi, Y. Nogi, and M. Muramatsu. 1996. Mouse RNA polymerase I 16-kDa subunit able to associate with 40-kDa subunit is a homolog of yeast AC19 subunit of RNA polymerases I and III. J. Biol. Chem. 271:32881-32885. [DOI] [PubMed] [Google Scholar]
- 54.Yuan, X., J. Zhao, H. Zentgraf, U. Hoffmann-Rohrer, and I. Grummt. 2002. Multiple interactions between RNA polymerase I, TIF-IA and TAFI subunits regulate preinitiation complex assembly at the ribosomal gene promoter. EMBO Rep. 3:1082-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao, J., X. Yuan, M. Frödin, and I. Grummt. 2003. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol. Cell 11:405-413. [DOI] [PubMed] [Google Scholar]
- 56.Zomerdijk, J. C. B. M., and R. Tjian. 1998. Structure and assembly of human selectivity factor SL1, p. 67-73. In M. R. Paule (ed.), Transcription of ribosomal RNA genes by eukaryotic RNA polymerase I. Springer-Verlag, Berlin, Germany.
- 57.Zomerdijk, J. C. B. M., and R. Tjian. 1998. Initiation of transcription on human rRNA gene, p. 121-134. In M. R. Paule (ed.), Transcription of ribosomal RNA genes by eukaryotic RNA polymerase I. Springer-Verlag, Berlin, Germany.