ABSTRACT
Net1 isoform A (Net1A) is a RhoA GEF that is required for cell motility and invasion in multiple cancers. Nuclear localization of Net1A negatively regulates its activity, and we have recently shown that Rac1 stimulates Net1A relocalization to the plasma membrane to promote RhoA activation and cytoskeletal reorganization. However, mechanisms controlling the subcellular localization of Net1A are not well understood. Here, we show that Net1A contains two nuclear localization signal (NLS) sequences within its N-terminus and that residues surrounding the second NLS sequence are acetylated. Treatment of cells with deacetylase inhibitors or expression of active Rac1 promotes Net1A acetylation. Deacetylase inhibition is sufficient for Net1A relocalization outside the nucleus, and replacement of the N-terminal acetylation sites with arginine residues prevents cytoplasmic accumulation of Net1A caused by deacetylase inhibition or EGF stimulation. By contrast, replacement of these sites with glutamine residues is sufficient for Net1A relocalization, RhoA activation and downstream signaling. Moreover, the N-terminal acetylation sites are required for rescue of F-actin accumulation and focal adhesion maturation in Net1 knockout MEFs. These data indicate that Net1A acetylation regulates its subcellular localization to impact on RhoA activity and actin cytoskeletal organization.
KEY WORDS: Net1, RhoA, Acetylation, Localization, EGF, F-actin
INTRODUCTION
Rho GTPases are small GTP-binding proteins that cycle between their active, GTP-bound, and inactive, GDP-bound states. There are over 20 Rho proteins in humans, with Rac1, Cdc42 and RhoA being the most highly studied. Once activated Rho proteins interact with downstream effectors to control cell signaling pathways that impact upon a wide variety of phenotypes including actin cytoskeletal organization and cell motility (Burridge and Wennerberg, 2004; Bustelo et al., 2007; Jaffe and Hall, 2005).
Rho GTPase activation is controlled by a family of proteins known as Rho guanine-nucleotide-exchange factors (Rho GEFs), which stimulate GDP release to allow Rho proteins to bind GTP (Rossman et al., 2005). The neuroepithelial transforming gene 1 (Net1) is a RhoA- and RhoB-directed GEF that was first discovered as a transforming gene in NIH 3T3 cells (Chan et al., 1996). Two isoforms of Net1 exist in most cells, Net1 and Net1A, which are identical except for their N-terminal regulatory domains. Previous studies have shown that Net1 expression is required for cell motility and extracellular matrix (ECM) invasion (Hayashi et al., 2013; Leyden et al., 2006; Murray et al., 2008), and this phenotype is mainly due to the actions of the Net1A isoform (Carr et al., 2013; Dutertre et al., 2010; Papadimitriou et al., 2012). Net1A accomplishes this by controlling RhoA-dependent phosphorylation of the myosin light chain regulatory subunit and focal adhesion kinase activation (Carr et al., 2013).
Net1 proteins are unusual among Rho GEFs in that they localize to the nucleus in resting cells (Carr et al., 2013a; Qin et al., 2005; Schmidt and Hall, 2002). However, it is clear that Net1 isoforms must be exported to the plasma membrane to stimulate RhoA activity and initiate actin cytoskeletal reorganization (Qin et al., 2005; Schmidt and Hall, 2002). Thus, identification of mechanisms controlling the subcellular localization of Net1 proteins is crucial to understanding how they control cell motility and ECM invasion. Recently we have shown that activation of Rac1 downstream of integrin ligation causes a robust, temporally regulated relocalization of the Net1A isoform to the plasma membrane. Moreover, Net1A relocalization is required for focal adhesion maturation and cell spreading in breast cancer cells (Carr et al., 2013a). However, the mechanism by which Rac1 stimulates Net1A relocalization is not known.
The nuclear localization of many proteins depends on the presence of nuclear localization signal (NLS) sequences, which consist of linear or bipartite groups of positively charged lysine and arginine residues (Schmidt et al., 2007). Because the arrangement of charged residues within NLS sequences is so important for binding to nuclear importins, NLS sequences are often regulated by post-translation modifications that alter this charge. For example, phosphorylation of residues in or near NLS sequences commonly alters their function (Moll et al., 1991; Rihs et al., 1991). In addition, acetylation of lysine residues within an NLS can inhibit their function, as addition of neutral acetyl groups to the ε-amino group of lysine residues negates their positive charge (Close et al., 2010; Yang, 2004). A prominent example is the tyrosine kinase c-Abl, which is blocked from entering the nucleus by acetylation of residues in its NLS sequence (Dietschy et al., 2009). Previously it has been shown that the Net1 isoform contains two NLS sequences in its unique N-terminal regulatory domain (Schmidt and Hall, 2002). However, NLS sequences within Net1A have not been identified and it has been unclear why Net1A localizes to the nucleus. Mechanisms controlling its cytosolic localization are similarly not well understood.
In the present work we show that Net1A contains two NLS sequences in its N-terminus, and that it is subject to Rac1-stimulated acetylation. Net1A acetylation occurs on multiple sites within its N-terminus and DH domain. Importantly, we demonstrate that acetylation of the N-terminal sites is required for ligand-stimulated accumulation of Net1A outside the nucleus, and for rescue of F-actin polymerization and focal adhesion maturation in Net1-knockout mouse embryonic fibroblasts (MEFs). These data represent the first instance of regulation of a Rho GEF by acetylation and shed light on molecular mechanisms controlling the subcellular localization of Net1A.
RESULTS
Identification of NLS sequences within Net1 and Net1A
Net1 isoforms localize within the nucleus in quiescent cells, thereby preventing them from activating RhoA (Carr et al., 2013a; Qin et al., 2005; Schmidt and Hall, 2002). Previously it has been shown that the longer Net1 isoform contains two NLS sequences within its unique N-terminal regulatory domain. However, additional NLS sequences might exist in Net1, as deletion of these NLS sequences did not wholly preclude Net1 localization to the nucleus (Schmidt and Hall, 2002). NLS sequences within Net1A have not been identified. As nuclear localization of Net1 proteins is a key determinant of their function in cells, we determined whether Net1 isoforms contain unrecognized NLS sequences.
To identify additional NLS motifs in Net1 isoforms we analyzed the primary amino acid sequences of mouse Net1 and Net1A using the Eukaryotic Linear Motif (ELM) resource (http://elm.eu.org/). This analysis identified one of the NLS sequences previously recognized in Net1 between amino acids 66–72 (Fig. 1A, NLS1), as well as two new monopartite NLS sequences located at amino acids 89–94 and 147–153 (Fig. 1A, NLS3 and NLS4, respectively). These additional NLS sequences were contained within a portion of the Net1A N-terminal regulatory domain that is common to both isoforms (Fig. 1A, denoted as NLS1 and NLS2, respectively). When coupled with previously published work, this analysis suggests that Net1 contains four NLS sequences within its N-terminal regulatory domain, whereas Net1A only contains two NLS sequences (Fig. 1A). Importantly, all four putative NLS sequences are conserved in human Net1 isoforms (data not shown).
Fig. 1.
Identification of NLS sequences in Net1 isoforms. (A) Schematic of Net1 isoform domains and NLS sequences. Numbers refer to amino acids in mouse Net1 proteins. DH, Dbl homology domain; PH, pleckstrin homology domain; PDZ, PDZ-domain-binding site. (B) Subcellular fractionation of HeLa cells. Expression of endogenous Net1 and RhoA in the nuclear and cytosolic fractions was detected by western blotting. Lamin A/C and SOD1 were used as controls for purity of the nuclear and cytosolic fractions, respectively. Shown is a representative experiment from three independent experiments. (C) Subcellular localization of Net1 NLS mutants. HeLa cells were transfected with plasmids for HA-tagged wild-type and mutant Net1 cDNAs, fixed and stained for HA (green) and DNA (blue). Shown are representative images. (D) Quantification of subcellular localization of Net1 NLS mutants. Ratios are cytosolic staining divided by nuclear staining. Shown is the mean±s.e.m. of three independent experiments. At least 20 cells per experiment were analyzed. ***P<0.001. (E) Subcellular localization of Net1A NLS mutants in HeLa cells. Net1A proteins are in green, DNA is in blue. Shown are representative images. Scale bars: 20 µm. (F) Quantification of the subcellular localization of Net1A NLS mutants. Shown is the mean±s.e.m. of three independent experiments. ***P<0.001. cyto, cytoplasmic; nuc, nuclear.
NLS sequences are defined by the presence of adjacent, positively charged lysine and arginine residues, which are required for binding to nuclear importins (Lange et al., 2007). To determine the relative contributions of the putative NLS sequences to the nuclear localization of Net1 isoforms, all of the lysine and arginine residues within each NLS sequence were mutated to alanine residues (Fig. 1A). Plasmids encoding these mutant Net1 proteins were then transfected into HeLa cells and the subcellular localization of the expressed proteins was assessed by immunofluorescence microscopy. Localization of Net1 isoforms was quantified as the ratio of cytoplasmic staining divided by nuclear staining. Because overexpression of Net1A has been reported to cause aberrant cytoplasmic localization (Qin et al., 2005), transfected cells in each experiment were analyzed to establish the range of expression where localization of wild-type Net1 isoforms was independent of expression level (supplementary material Fig. S1). Only cells with transgene expression within this range were quantified.
As expected, overexpressed wild-type Net1 and Net1A were predominantly nuclear, similar to the localization of endogenous Net1 isoforms in HeLa cells (Fig. 1B,C,E). Importantly, mutation of the previously identified NLS1 and NLS2 sequences within Net1 (denoted NLS1&2m) only partially impaired its nuclear localization. Similarly, mutation of the newly identified NLS3 and NLS4 (denoted NLS3&4m) sequences caused only a low level of Net1 relocalization. However, mutation of all four NLS sequences together (denoted NLS1,2,3&4m) caused a dramatic relocalization of Net1 outside the nucleus (Fig. 1C,D). These data indicate that all four of the NLS sequences in Net1 play important roles in directing it to the nucleus. These findings fit with published data indicating that Net1ΔN, which contains the previously unrecognized NLS4, still exhibits partial nuclear localization (Schmidt and Hall, 2002).
Using a similar approach we assessed the importance of the two NLS sequences identified in Net1A. This analysis showed that both NLS1 (residues 35–40) and NLS2 (residues 93–98) contributed to the nuclear localization of Net1A, with NLS1 being more important than NLS2 (Fig. 1E,F). Notably, mutation of both NLS sequences together caused a significant relocalization of Net1A outside the nucleus that was as strong as that caused by co-expression of constitutively active V12Rac1, which potently stimulates Net1A localization to the cytosol and plasma membrane (Fig. 1E,F) (Carr et al., 2013a). Taken together, these data indicate that Net1 isoforms contain additional, unrecognized NLS sequences that contribute significantly to their nuclear localization.
Identification of acetylated residues within Net1A
It has been shown previously that breast cancer cells mainly require Net1A, but not Net1, for adhesion, motility and ECM invasion (Carr et al., 2013a; Carr et al., 2013b; Dutertre et al., 2010). Moreover, Rac1 preferentially stimulates relocalization of Net1A, but not Net1, to the plasma membrane (Carr et al., 2013a). To understand how Rac1 promotes Net1A relocalization, we searched for changes in post-translational modifications of Net1A caused by co-expression of V12Rac1 using liquid chromatography tandem mass spectrometry (LC-MS/MS; Taplin Mass Spectrometry Core Facility). This analysis did not show any differences in Net1A phosphorylation, however, it did identify a potential acetylation site at K83. To determine whether additional acetylation sites exist within Net1A, we treated Net1A-transfected HeLa cells overnight with the pan-histone deacetylase (HDAC) inhibitor trichostatin A (TSA). Net1A was then immunoprecipitated and analyzed by LC-MS/MS for acetylated residues. This analysis identified five acetylation sites within Net1A (K83, K95, K226, K247 and K263; Fig. 2A). To determine whether endogenous Net1A was acetylated, we treated MDA-MB-231 cells, which express high levels of Net1A, with TSA for 2 h. Endogenous Net1A was then immunoprecipitated and tested for acetylation by western blotting with an antibody specific for acetylated lysine. We found that endogenous Net1A was acetylated in these cells (Fig. 2B). To determine whether we had identified all of the acetylation sites within Net1A stimulated by deacetylase inhibition, we mutated all five lysine residues to arginine. We also mutated the residue adjacent to K226 (K227) to arginine to preclude promiscuous acetylation of the adjacent residue. HeLa cells were then transfected with wild-type Net1A or the Net1A 6K/R mutant and treated overnight with TSA and nicotinamide. Nicotinamide inhibits the Sirtuin family of deacetylases. After immunoprecipitation the Net1A proteins were tested for acetylation by Western blotting. This analysis showed that mutation of all six lysine to arginine residues effectively eliminated Net1A acetylation after deacetylase inhibitor treatment (Fig. 2C). To test whether active Rac1 stimulated Net1A acetylation, HeLa cells were transfected with wild-type Net1A, with or without constitutively active V12Rac1, immunoprecipitated and tested for acetylation. We observed that Net1A exhibited a basal level of acetylation that was significantly increased by co-expression of V12Rac1 (Fig. 2D). Taken together, these data indicate that Net1A is acetylated on multiple residues, and that inhibition of cellular deacetylase activity or expression of constitutively active Rac1 stimulates this modification.
Fig. 2.

Identification of Net1A acetylation sites. (A) Schematic of Net1A domain structure and acetylation sites. Numbers refer to amino acids in mouse Net1A. (B) Acetylation of endogenous Net1A in MDA-MB-231 cells. Cell lysates were subject to immunoprecipitation (IP) with control or anti-Net1 antibodies. Net1A was tested for acetylation by western blotting. Shown is a representative experiment from two independent experiments. (C) Arginine replacement of the identified acetylated lysine residues prevents Net1A acetylation. HeLa cells were transfected with wild-type Net1A and or Net1A with K83R, K95R, K226R, K227R, K247R and K263R mutations (Net1A 6K/R). Cells were treated overnight with TSA and nicotinamide. Net1A proteins were immunoprecipitated and tested for acetylation by western blotting. Shown is a representative experiment from three independent experiments. (D) Constitutively active Rac1 stimulates Net1A acetylation. HeLa cells were transfected with wild-type Net1A, with or without V12Rac1. Net1A proteins were then immunoprecipitated and tested for acetylation by western blotting. Shown is a representative experiment from three independent experiments.
Net1A acetylation stimulates its relocalization outside the nucleus
Acetylation of sites in and around NLS sequences has been shown to inhibit the nuclear localization of a small number of proteins, including c-Abl, RECQL4, E1A, Skp2 and IFI16 (di Bari et al., 2006; Dietschy et al., 2009; Inuzuka et al., 2012; Li et al., 2012; Madison et al., 2002). Given that K83 and K95 are adjacent to or contained within the NLS2 of Net1A, respectively, we examined whether treatment of cells with TSA stimulated Net1A relocalization. We observed that TSA treatment for 2 h stimulated relocalization of Net1A outside the nucleus (Fig. 3A). The degree of relocalization was not increased by longer treatments (data not shown), and was equivalent to that observed when NLS2 alone was mutated (Fig. 1E,F). Importantly, this effect required the N-terminal acetylation sites within Net1A, as mutation of K83 and K95 to arginine residues largely prevented Net1A relocalization by TSA (Fig. 3A,B).
Fig. 3.
TSA-stimulated Net1A relocalization requires the N-terminal acetylation sites. (A) TSA-stimulated Net1A relocalization requires K83 and K95. HeLa cells were transfected with wild-type (wt) HA–Net1A or HA–Net1A with K83R and K95R mutations (K88/95R), exposed to TSA for 2 h, fixed and stained for HA–Net1A, DNA and F-actin. Shown are representative images of HA–Net1A-expressing cells. (B) Quantification of wild-type and K83/95R Net1A localization. Show is the mean±s.e.m. of four independent experiments. *P<0.05; **P<0.01; ***P<0.001. (C) The DH domain acetylation sites are not required for TSA-stimulated Net1A relocalization. Shown are representative images of cells expressing the indicated HA–Net1A. Scale bars: 20 µm. (D) Quantification of wild-type and mutant Net1A localization. Shown is the mean±s.e.m. of three independent experiments. *P<0.05; **P<0.01. Cyto, cytoplasmic; Nuc, nuclear.
To determine whether acetylation of Net1A was sufficient for relocalization, the Net1A acetylation sites in the N-terminus or the DH domain were mutated into the acetylation-mimic glutamine residues. The subcellular localization of these mutant Net1A proteins was then tested. In these experiments we found that mutation of K83 and K95 to glutamine residues (Net1A 2K/Q) caused a modest but significant relocalization of Net1A outside the nucleus (Fig. 3C,D). This was in contrast to glutamine replacement of the DH domain acetylation sites (Net1A 4K/Q), which had no effect on Net1A localization. In addition, mutation of all six lysine residues into glutamine residues did not result in greater Net1A relocalization than glutamine replacement of the N-terminal sites alone (Net1A 6K/Q) (Fig. 3C,D). These data suggest that acetylation of the N-terminal sites, but not the DH domain sites, is sufficient to relocalize Net1A outside the nucleus.
The N-terminal acetylation sites within Net1A are required for EGF-stimulated relocalization
As Rac1 stimulates Net1A relocalization outside the nucleus, we reasoned that growth factors that stimulate Rac1 activity, such as EGF, might also cause Net1A relocalization. To test this, MCF7 cells were transfected with control or Rac1 small interfering RNAs (siRNAs), and then re-transfected with HA–Net1A. After starvation overnight, the cells were stimulated with EGF for 15 min and Net1A localization was assessed. We observed that EGF stimulated Net1A relocalization outside the nucleus, and that this was completely dependent upon Rac1 expression (Fig. 4A,B).
Fig. 4.
The N-terminal acetylation sites within Net1A are required for EGF-stimulated relocalization. (A) EGF stimulates Net1A relocalization in a Rac1-dependent manner. MCF7 cells were transfected with control or Rac1 siRNAs. After 1 day, the cells were re-transfected with HA–Net1A, starved overnight and stimulated with EGF. Cells were then fixed and stained for HA, DNA and F-actin. Shown are representative images of HA–Net1A-expressing cells. (B) Quantification of Net1A localization. Shown is the mean±s.e.m. of three independent experiments. **P<0.01. Inset shows a western blot for cells transfected with control and Rac1 siRNA from a representative experiment. (C) EGF-stimulated Net1A relocalization requires K83 and K95. MCF7 cells were transfected with HA–Net1A (wt) or HA–Net1A with K83R and K95R mutations (K88/95R), starved overnight and stimulated with EGF. Shown are representative images of HA–Net1A-expressing cells. (D) Quantification of wild-type and K83/95R Net1A localization. Shown is the mean±s.e.m. of five independent experiments. *P<0.05. (E) Mutation of the DH domain acetylation sites does not affect EGF-stimulated relocalization of Net1A. MCF7 cells were transfected with the HA-epitope tagged wild type Net1A or Net1A with K226R, K227R, K247R and K263R mutations (Net1A 4K/R), starved overnight and stimulated with EGF. Shown are representative images of HA–Net1A-expressing cells. Scale bars: 20 µm. (F) Quantification of Net1A localization. Shown is the mean±s.e.m. of four independent experiments. ***P<0.001. Cyto, cytoplasmic; Nuc, nuclear.
We then examined whether the N-terminal acetylation sites of Net1A were required for the EGF-stimulated relocalization. MCF7 cells were transfected with wild-type Net1A or Net1A K83/95R, starved and then stimulated for different periods of time with EGF. We observed that EGF stimulated a time-dependent relocalization of wild-type Net1A outside the nucleus that peaked at 15 min and was completed by 30 min. However, replacement of the N-terminal acetylation sites with arginine residues in Net1A abrogated EGF-stimulated relocalization (Fig. 4C,D). By contrast, arginine replacement of the DH domain acetylation sites did not affect EGF-stimulated Net1A relocalization (Fig. 4E,F). Taken together, these data indicate that Net1A acetylation on the N-terminal sites, but not the DH domain sites, is required for EGF-stimulated relocalization outside the nucleus.
Acetylation of the N-terminal sites is necessary for Net1A-dependent RhoA activation, actin cytoskeletal reorganization and focal adhesion tyrosine phosphorylation
We and others have found that Net1A must relocalize outside the nucleus to stimulate RhoA activation and F-actin accumulation (Carr et al., 2013a; Qin et al., 2005; Schmidt and Hall, 2002). Given that replacement of the N-terminal acetylation sites within Net1A with glutamine residues increased its extranuclear localization, we tested whether this also resulted in an increase in RhoA activity. MCF7 cells were transfected with empty vector, wild-type Net1A or Net1A K83/95Q. The cells were then starved overnight, lysed, and RhoA activity was measured using a G-LISA assay. In these experiments overexpression of wild-type Net1A did not cause a significant increase in basal RhoA activity. This is consistent with the predominantly nuclear localization of wild-type Net1A in unstimulated MCF7 cells. By contrast, expression of Net1A K83/95Q caused a significant increase in RhoA activation (Fig. 5A,B). To confirm that Net1A K83/95Q was more active towards RhoA than wild-type Net1A, we measured signaling outputs downstream of RhoA. An important effector molecule of RhoA is Rho kinase (ROCK1 and ROCK2), which phosphorylates and inactivates the myosin phosphatase regulatory subunit MYPT1. This, in turn, leads to an accumulation of phosphorylated myosin regulatory light chain (MLC2, also known as MYL9) (Kimura et al., 1996). HeLa cells were transfected with empty vector, wild-type Net1A or Net1A K83/95Q, starved overnight and tested for phosphorylation of MYPT1 and MLC2 by western blotting. These assays demonstrated that Net1A K83/95Q expression stimulates MYPT1 and MLC2 phosphorylation more strongly that wild-type Net1A (Fig. 5C).
Fig. 5.
The N-terminal acetylation sites are required for Net1A-stimulated RhoA activation. (A) Expression of Net1A with K83Q and K95Q mutations (K83/95Q) enhances cellular RhoA activity. MCF7 cells were transfected with empty vector, HA–Net1A or HA–Net1A K83/95Q. The cells were then starved overnight and RhoA activity was measured using a G-LISA assay. Shown is the mean±s.e.m. of three independent experiments. **P<0.01. (B) Representative western blots of Net1A-transfected cells used for RhoA G-LISA assays. (C) Net1A K83/95Q expression stimulates MYPT1 and MLC2 phosphorylation. HeLa cells were transfected with the vectors shown, starved overnight in 0% FBS and tested for phosphorylation of MYPT1 and MLC2 by western blotting. Shown is a representative experiment from three independent experiments. wt, wild type. (D) The intrinsic activity of Net1A K83/95Q is similar to wild-type Net1A. MCF7 cells were transfected with wild-type Net1A or Net1A K83/95Q. Binding of Net1A proteins to GST–A17RhoA was detected by western blotting. Shown is a representative experiment. (E) Quantification of wild-type and K83/95Q Net1A activity, as assessed by binding to GST–A17RhoA. Net1A bound to GST–A17RhoA was normalized to expression in the lysate and to the amount of GST–A17RhoA in each pulldown. Shown is the mean±s.e.m. of three independent experiments. *P<0.05. a.u., arbitrary units.
To determine whether the intrinsic GEF activity of Net1A was altered by mutation of the N-terminal acetylation sites, we performed GST–A17RhoA pulldown assays. A17RhoA is a nucleotide-deficient form of RhoA that binds tightly to activated RhoA GEFs (Carr et al., 2013a; Garcia-Mata et al., 2006). We observed that wild-type Net1A and Net1A K83/95Q bound similarly to A17RhoA, with Net1A K83/95Q displaying slightly less binding activity (Fig. 5D,E). However, this was not a large difference and would not be expected to significantly affect its ability to stimulate RhoA activity in the cell. Thus, these data support the idea that relocalization of Net1A outside the nucleus is a key determinant of its activity towards RhoA, and indicate that the increased extranuclear localization of Net1A K83/95Q accounts for the observed elevation in RhoA activation and downstream signaling.
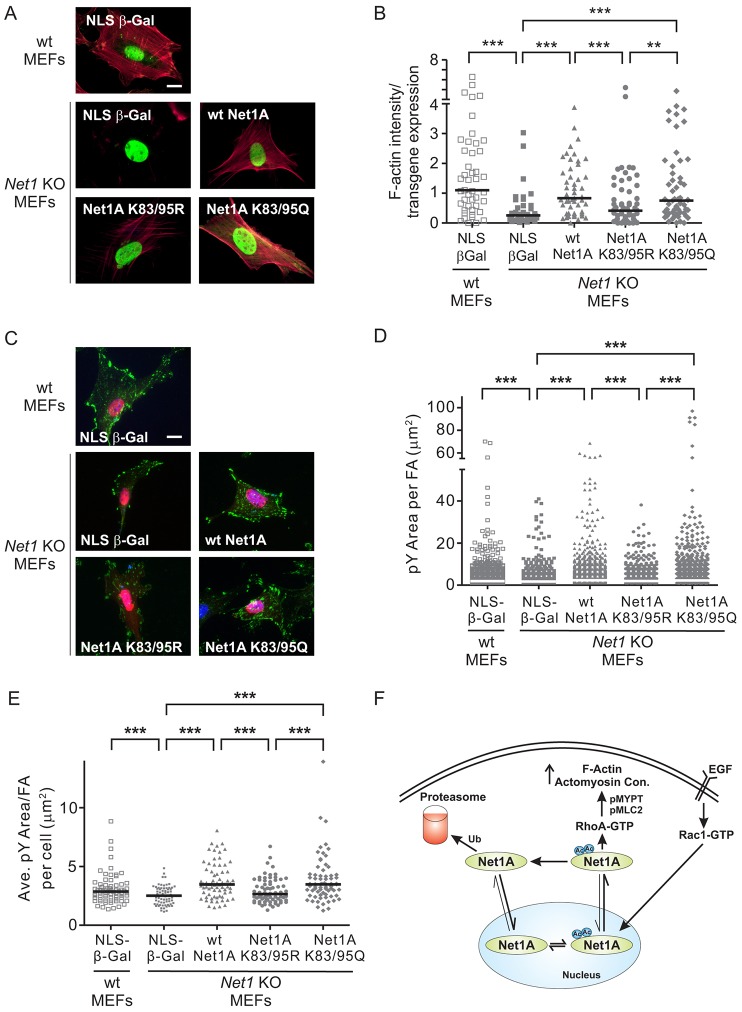
Because RhoA activation stimulates the accumulation of F-actin in cells, we examined whether the N-terminal acetylation sites within Net1A were required for regulation of cellular F-actin content. For these experiments we made use of immortalized MEFs derived from Net1-knockout mice, which do not express either Net1 isoform (Zuo et al., 2014). Lack of Net1 isoform expression in Net1-knockout MEFs was confirmed by real-time quantitative PCR (data not shown). To assess effects on F-actin content, Net1-knockout MEFs were transfected with wild-type Net1A, Net1A K83/95R or Net1A K83/95Q. As a negative control, both wild-type and Net1-knockout MEFs were transfected with Myc-epitope tagged β-galactosidase containing the NLS sequence from SV40 Large T antigen (NLS-β-Gal). The cells were then starved overnight, fixed and stained for NLS-β-Gal or Net1A expression, as well as F-actin. The amount of F-actin staining was quantified and normalized to the amount of expressed protein. In these experiments, we observed that wild-type MEFs contained significantly more F-actin than Net1-knockout MEFs, and that expression of NLS-β-Gal did not rescue F-actin content (Fig. 6A,B). This is consistent with the established role of Net1 isoforms in controlling RhoA activation in other cell types (Carr et al., 2013a; Carr et al., 2013b; García-Mata et al., 2007; Lee et al., 2010; Murray et al., 2008; Papadimitriou et al., 2012). By contrast, expression of wild-type Net1A restored F-actin content close to that observed in wild-type MEFs. This is consistent with a predominant role for Net1A over the Net1 isoform in controlling cellular RhoA activity (Carr et al., 2013a; Carr et al., 2013b; Papadimitriou et al., 2012). Importantly, expression of Net1A K83/95R did not efficiently rescue F-actin accumulation, whereas Net1A K83/95Q restored F-actin staining (Fig. 6A,B). These data indicate that the N-terminal acetylation sites within Net1A are required for it to rescue F-actin polymerization in Net1-knockout MEFs.
Fig. 6.
The N-terminal acetylation sites are required for Net1A-stimulated F-actin accumulation and focal adhesion maturation. (A) Expression of Net1A K83/95R cannot restore F-actin content in Net1-knockout (KO) MEFs. Wild-type (wt) or Net1-knockout MEFs were transfected with plasmids encoding the proteins shown, starved overnight, and fixed and stained for the expressed protein (green) and F-actin (red). Shown are representative images. (B) Quantification of F-actin staining in transfected MEFs. F-actin intensity in transfected cells was quantified using Image J and normalized to the expression of the transfected cDNA. Shown are data from five independent experiments. Bars are median values. **P<0.01; ***P<0.001 (Mann–Whitney test). (C) Expression of Net1A with K83R and K95R mutations (K83/95Q) cannot rescue focal adhesion maturation in Net1-knockout MEFs. Wild-type or Net1-knockout MEFs were transfected with plasmids encoding the proteins shown, starved overnight, and fixed and stained for the expressed protein (red), phospho-tyrosine (pY, green) and DNA (blue). Shown are representative images. Scale bars: 20 µm. (D) Quantification of pY area per focal adhesion (FA). Shown is the combined data from four independent experiments. White bars show the median. ***P<0.001. (E) Quantification of average pY area per focal adhesion in each cell. A total of 70 cells from four independent experiments were analyzed. ***P<0.001. (F) Model for regulation of Net1A localization and signaling by acetylation. Blue circles, acetyl groups.
To further support a role for N-terminal acetylation of Net1A in its cellular function, we tested the requirement for these sites in focal adhesion development. The maturation of nascent focal contacts to focal adhesions is a RhoA-driven process that is typified by an enlargement in adhesion size and is accompanied by an accumulation of tyrosine phosphorylated proteins within the adhesion (Burridge and Wennerberg, 2004; Chrzanowska-Wodnicka and Burridge, 1996; Huveneers and Danen, 2009; Parsons et al., 2010). Thus, we tested the ability of transfected Net1A to increase focal adhesion size and phosphorylated tyrosine (phospho-tyrosine, pY) content in Net1-knockout MEFs. As expected, Net1-knockout MEFs displayed fewer pY-containing focal adhesions that were smaller in size than wild type MEFs, and this was not altered by expression of NLS-β-GAL (Fig. 6C–E). However, expression of wild-type Net1A effectively increased pY area within individual focal adhesions (Fig. 6C,D), as well as the average pY area per focal adhesion within a given cell (Fig. 6C,E). Notably, expression of Net1A K83/95R did not rescue pY content within focal adhesions, whereas expression of Net1A K83/95Q potently restored pY staining (Fig. 6C–E). As RhoA activation promotes both F-actin accumulation and focal adhesion maturation, these data further support the premise that acetylation of the N-terminal sites within Net1A controls its relocalization outside the nucleus and, therefore, its cellular activity.
DISCUSSION
Relocalization of Net1A from the nucleus to the plasma membrane is a crucial determinant of its ability to activate RhoA and stimulate actin cytoskeletal reorganization. However, mechanisms controlling the subcellular localization of Net1A are not well understood. In the present work, we have shown that Net1A contains two NLS sequences in its N-terminal regulatory domain, and that acetylation near the second of these sequences negatively regulates Net1A nuclear localization. Furthermore, we demonstrated that EGF-stimulated export of Net1A from the nucleus requires the presence of the N-terminal acetylation sites, and that these sites are necessary for Net1A-dependent regulation of RhoA activity, F-actin accumulation and focal adhesion maturation. These data show for the first time regulation of Rho GEF function by acetylation, and offer insight into the mechanism by which Rac1 controls Net1A relocalization outside the nucleus.
As a single NLS sequence is normally sufficient to promote nuclear import of a protein, it is curious that multiple NLS sequences are present in both Net1 isoforms. Presumably this provides the opportunity for concurrent interaction with multiple nuclear importins to ensure rapid nuclear import, as well as allowing for modulation of the rate of nuclear import by acetylation of an NLS sequence. This suggests that it is extremely important for a cell to control the length of time that Net1 isoforms are available in the cytosol to stimulate RhoA activation. Indeed, multiple mechanisms have been shown to limit the cytoplasmic accumulation and activity of Net1 proteins. For example, in addition to nuclear sequestration, cells can downregulate Net1 enzymatic activity by Pak1-dependent phosphorylation (Alberts et al., 2005). Net1A is also subject to ubiquitylation and proteasome-mediated degradation (Carr et al., 2009; Papadimitriou et al., 2012), and inhibition of proteasome function extends the duration of Net1A localization outside the nucleus following integrin ligation (Carr et al., 2013a). In this regard, it is not clear whether proteasomal degradation and acetylation-dependent regulation of Net1A nuclear re-import are competing mechanisms to control Net1A activity, or if they are functionally related. For example, the ubiquitylation sites within Net1A have not been identified, and it is possible that Net1A acetylation prevents its ubiquitylation by modifying the residues that are normally targeted for ubiquitylation, or by blocking interaction with ubiquitin E3 ligases that target Net1A. Acetylation has been shown to prevent ubiquitylation of other proteins such as Myc and β-catenin, thereby inhibiting their degradation (Ge et al., 2009; Patel et al., 2004). Thus, Net1A acetylation might serve the dual purpose of impeding nuclear re-import as well as preventing Net1A degradation.
A important finding of this work is the identification of EGF as a potent regulator of Net1A cytosolic accumulation. Previously we have shown that stimulation of integrin activity by re-plating cells on collagen causes a Rac1-dependent relocalization of Net1A to the cytoplasm and plasma membrane (Carr et al., 2013a). As EGF stimulation of Net1A relocalization is also Rac1-dependent, this suggests that multiple extracellular ligands that stimulate Rac1 activity might also cause cytoplasmic accumulation of Net1A. In support of this, we have observed that lysophosphatidic acid (LPA) also stimulates Net1A cytoplasmic accumulation, and that this similarly requires Rac1 expression (not shown). Rac1 is typically thought of as inhibiting RhoA activity through stimulation of p190-RhoGAP-A and p190-RhoGAP-B activity (Leeuwen et al., 1997; Nimnual et al., 2003; Sander et al., 1999). Thus, it is surprising that Rac1 would promote relocalization of a RhoA GEF such as Net1A. However, it is now apparent that Rho GTPase activation is highly localized within a cell, and the timing of activation of different Rho proteins is precisely regulated. For example, both RhoA and Rac1 are activated at the leading edge in migrating cells (Itoh et al., 2002; Kraynov et al., 2000; Pertz et al., 2006), and both GTPases contribute to leading edge dynamics in a temporally and spatially regulated manner (Machacek et al., 2009).
It is probable that N-terminal acetylation of Net1A is only part of the mechanism controlling its extranuclear localization. Others have shown that N-terminally truncated Net1 (Net1ΔN) accumulates in the nucleus when cells are treated with leptomycin B, which is an inhibitor of the nuclear exportin protein CRM1, and that the PH domain of Net1ΔN is required for its nuclear export. However, this domain does not contain a canonical nuclear export signal (NES) sequence, suggesting that Net1 interacts with another NES-containing protein to allow for nuclear export (Schmidt and Hall, 2002). These data imply that Net1 proteins can cycle between the nucleus and plasma membrane, and that in an unstimulated cell they accumulate in the nucleus owing to the predominance of NLS function. Our data indicate that stimulation of cells with ligands such as EGF causes N-terminal acetylation of Net1A, thereby slowing its nuclear re-import and causing the accumulation of Net1A outside the nucleus.
Taken together, these data support a model (Fig. 6F) in which an extracellular ligand, such as EGF, stimulates Rac1 activity. This results in an increase in Net1A acetylation, which slows nuclear import of Net1A and tips the balance towards its cytoplasmic accumulation. Once in the cytosol Net1A can access RhoA at the plasma membrane to stimulate its activity. This results in an increase in MYPT1 and MLC2 phosphorylation, thereby promoting actomyosin contraction and F-actin accumulation. Deacetylation of Net1A by cytosolic deacetylases would allow for cessation of Net1A signaling, either by promoting increased nuclear re-import or by allowing for Net1A degradation by the proteasome. Thus, important issues for future study will be to identify mechanisms that permit Net1A nuclear export, and to better understand the relationship between acetylation and proteasomal degradation of Net1A. An improved understanding of mechanisms controlling Net1A localization and activity has important implications for processes that require cell motility, such as cancer cell metastasis. Net1 isoforms are overexpressed in many cancers, including breast cancer, and this might be a mechanism by which cancer cells maintain elevated RhoA activity to promote invasion into the ECM. In the future it will be important to determine whether mechanisms promoting Net1A acetylation also stimulate cancer cell metastasis.
MATERIALS AND METHODS
Cell culture, reagents and plasmids
HeLa and MCF7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with high glucose and glutamine (Hyclone), supplemented with 10% fetal bovine serum (FBS; Invitrogen) and 100 U/ml penicillin and 100 µg/ml streptomycin (HyClone). MDA-MB-231 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with Ham's F12 (1∶1) (Hyclone), supplemented with 10% fetal bovine serum (FBS; Invitrogen) and 100 U/ml penicillin and 100 µg/ml streptomycin (HyClone). Mouse embryonic fibroblasts (MEFs) were grown in DMEM plus 10% FBS and 100 U/ml penicillin, 100 µg/ml streptomycin. Plasmids were transfected with Lipofectamine Plus reagent (Life Technologies) according to the manufacturer's instructions. siRNA transfections were performed with INTERFERin (PolyPlus) according to the manufacturer's instructions. For dual siRNA and plasmid transfections, plasmids were transfected 24 h after the siRNA. Cells were grown for an additional 48 h before being analyzed. Recombinant human epidermal growth factor (EGF) (R&D Systems) was used at 100 ng/ml. Hemagglutinin (HA)-tagged mouse Net1A and Myc-tagged, constitutively active V12Rac1 were as previously described (Carr et al., 2013a; Qin et al., 2005). Net1 and Net1A NLS mutants, and Net1A acetylation site mutants were created by site-directed mutagenesis using Pfu polymerase (Agilent). The entire cDNA inserts were sequenced to confirm correct amplification. Net1 NLS1 mutants were as follows: NLS1m, A12PAAASAA19; NLS2m, A66AAAAEK72; NLS3m, N89AAVAP94; NLS4m, P147TAAASS153. Net1A NLS mutants were as follows: NLS1m, N35AAVAP40; NLS2m, P93TAAASS99.
Isolation and immortalization of MEFs
The generation and characterization of Net1-knockout mice is described elsewhere (Zuo et al., 2014). All animal studies were approved by the Institutional Animal Care and Use Committee (protocol AWC 13-073) and were conducted in accordance with the guidelines of the US Public Health Service Policy for Humane Care and Use of Laboratory Animals. MEFs from Net1+/+ and Net1−/− C57BL/6 mice were isolated from embryos at 12.5–13.5 dpc. Embryo limbs, internal organs and brains were removed and the remaining carcasses were rinsed once with sterile PBS and then three times with DMEM without serum. Minced carcasses were incubated in 10 ml of trypsin (0.05%) with EDTA at 37°C for 30 min with shaking. The supernatant was collected and carcasses were incubated with 10 ml trypsin with EDTA two more times. Supernatants were pooled and cells released into the supernatant were collected by centrifugation at 500 g for 5 min at room temperature. Cells were resuspended in DMEM plus 10% FBS and penicillin-streptomycin (100 U/ml; 100 µg/ml), and plated on a 150-mm tissue culture dish. The culture medium was changed the next day to remove dead cells, at which time only the MEFs survived. Immortal wild-type and Net1-knockout MEFs were isolated by serial passage. Expression of Net1 isoforms in wild-type and Net1 knockout MEFs was tested by quantitative real-time PCR using primers that recognize both Net1 isoforms, as described previously (Zuo et al., 2014).
Cell fractionation
Nuclear and cytoplasmic fractions were extracted from asynchronous HeLa cells using a NE-PER kit (Thermo-Scientific), according to the manufacturer's instructions. Endogenous Net1 and RhoA were detected by western blotting using mouse anti-Net1 (Santa Cruz Biotechnology) and mouse anti-RhoA antibodies (Santa Cruz Biotechnology). Rabbit anti-Lamin A/C (Santa Cruz Biotechnology) and rabbit anti-SOD1 antibodies (Santa Cruz Biotechnology) were used as controls to assess the purity of the nuclear and cytoplasmic fractions, respectively.
Immunofluorescence microscopy
Cells were plated on acid-washed glass coverslips. Cells were fixed in 4.0% paraformaldehyde (Thermo Fisher Scientific) in PBS for 5 min at 37°C, and then permeabilized with 0.2% Triton X-100 in PBS for 5 min at room temperature. After washing in PBS plus 0.1% Tween 20 (PBST), cells were incubated with mouse anti-HA antibody (2 µg/ml) (Santa Cruz Biotechnology) in PBST plus 1% BSA for 1 h at 37°C. Mouse anti-phospho-tyrosine (clone 4G10, Millipore) was used for visualization of phospho-tyrosine-containing focal adhesions. Cells were then incubated for 1 h at 37°C with anti-mouse-IgG antibody conjugated to Alexa Fluor 488 (Life Technologies) and 4′,6-diamidino-2-phenylindole (DAPI) (1 mg/ml) (Sigma-Aldrich) diluted 1∶1000 in PBST plus 1% BSA, with or without Alexa-Fluor-647–phalloidin (Life Technologies). After washing with PBST, the cells were mounted on glass slides with Fluormount reagent (EMD Millipore Chemicals). Cells were visualized with a Zeiss Axiophot epifluorescence microscope, and images were acquired using Axiovision software. The intensity of HA–Net1A in the cytosolic and nuclear compartments was measured using Image J software, and calculated as described previously (Carr et al., 2013a). For each experiment the range of Net1 isoform expression that did not cause its relocalization in the absence of stimulus was determined and data analysis was restricted to those cells expressing Net1 isoforms within that range. Linear regression analysis of data sets was performed with GraphPad Prism 5 software. F-actin intensity, Myc–NLS-βGal and HA–Net1 expression in transfected MEFs was measured using Image J software. The size and intensity of phospho-tyrosine-containing focal adhesions was assessed using the particle analysis function of Image J, with a cutoff of 1 µm2 to eliminate non-specific background staining. Unless otherwise stated, statistical significance was determined by an unpaired Student's t-test.
Net1A acetylation
For identification of acetylation sites within Net1A, HeLa cells were transfected with HA–Net1A, with or without V12Rac1. After overnight incubation with 400 ng/ml trichostatin A (TSA) (Enzo Life Sciences), cells were lysed in RIPA buffer (1.0% Triton X-100, 0.5% sodium deoxycholate, 1% SDS, 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 80 mM β-glycerophosphate, 50 mM NaF, 1 mM sodium orthovanadate, 1 mM EDTA, 10 µg/ml leupeptin, 10 µg/ml pepstatin A, 2 µg/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride) and HA–Net1A was immunoprecipitated, resolved by SDS-PAGE and visualized by Coomassie Blue staining. The Net1A band was excised and analyzed by LC-MS/MS by the Taplin Mass Spectrometry Facility at the Harvard Medical School, on a fee-for-use basis.
For confirmation of Net1A acetylation sites by western blotting, HeLa cells transfected with Net1A plasmids were treated overnight with 400 ng/ml TSA and 20 mM nicotinamide (Spectrum Chemicals and Laboratory Products). Cells were lysed in RIPA buffer plus 400 ng/ml TSA, 20 mM nicotinamide and 5 mM N-ethylmaleimide (NEM) (Sigma-Aldrich), incubated on ice for 10 min, and insoluble proteins were pelleted by centrifugation (16,100 g, 10 min., 4°C). Equal amounts of soluble lysate were incubated for 1 h at 4°C with mouse anti-HA antibody followed by protein-A–Sepharose (Rockland Immunochemicals) for 1 h at 4°C. Precipitates were washed with buffer containing 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5% Triton X-100, TSA, nicotinamide and NEM, resolved by SDS-PAGE, transferred onto a PVDF membrane (GE Healthcare), and analyzed by western blot using rabbit anti-acetylated lysine (Cell Signaling Technology) and mouse anti-HA antibodies. For detection of Net1A acetylation regulated by V12Rac1, cells were not treated prior to lysis with deacetylase inhibitors.
RhoA activation assay
For measurement of RhoA activity, MCF7 cells were transfected with empty vector, HA-Net1A or HA-Net1A K83/95Q, and starved overnight in DMEM plus 1% FBS and penicillin/streptomycin. Cells were lysed and RhoA activity was measured using a G-LISA assay kit (Cytoskeleton Inc.) according to the manufacturer's instructions. Statistical significance was determined by an unpaired Student's t-test.
For measurement of MYPT1 and MLC2 phosphorylation, HeLa cells were transfected with empty vector, HA–Net1A or HA–Net1A K83/95Q. The cells were then starved overnight in DMEM plus 0% FBS and penicillin-streptomycin, and then lysed in 2% SDS buffer (2% SDS, 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 80 mM β-glycerophosphate, 50 mM NaF, 1 mM sodium orthovanadate, 1 mM EDTA, 10 µg/ml leupeptin, 10 µg/ml pepstatin A, 2 µg/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride). Lysates were sonicated for 20 s and resolved by SDS-PAGE. After transfer onto PVDF membrane, blots were probed for pMYPT1 (Cell Signaling Technology), MYPT1 (Santa Cruz Biotechnology), pMLC2 (Cell Signaling Technology), MLC2 (GeneTex Inc.) and the HA epitope.
GST–A17RhoA pulldown assay
Assays were performed essentially as described previously (Carr et al., 2013a). Briefly, transfected HeLa cells were washed with PBS and lysed in buffer A (20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1% Triton X-100, 1 mM DTT, 1 mM PMSF, and 10 µg/ml each of aprotinin, leupeptin, and pepstatin A), sonicated for 30 s, and clarified by centrifugation (16,100 g, 10 min., 4°C). Lysate concentrations were determined by a BCA assay (Thermo Fisher Scientific) and equal amounts of lysate were mixed for 1 h at 4°C with 20 µg of GST or GST–A17RhoA beads. GST and GST–A17RhoA were expressed in E. coli and purified as described previously (Carr et al., 2013a). Beads were pelleted by centrifugation and washed three times in buffer A, resuspended in 25 µl Laemmli sample buffer, boiled for 5 min, separated by SDS-PAGE and transferred onto PVDF membrane for western blot analysis. Net1A activity was quantified as the intensity of Net1A in the pulldown divided by that in the lysate, and normalized to the amount of GST in the pulldown. Statistical significance was determined by an unpaired Student's t-test.
Supplementary Material
Acknowledgments
We thank members of the Frost laboratory for support and advice, as well as the Cheng, Cunha, Dessauer, Denicourt, and Du laboratories for helpful discussions. We would also like to thank Rebecca Berdeaux, Richard Clark, and Catherine Denicourt for critical reading of the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
All authors contributed to the writing of the manuscript. E.H.S. was responsible for the majority of experimental work and assembly of figures; W.O. performed subcellular fractionation, examined focal adhesion maturation in Net1 knockout MEFs and aided in data analysis; A.U. examined subcellular localization of Net1 DH domain mutants and aided in data analysis; H.S.C. identified Net1A acetylation sites and tested for acetylation of endogenous Net1A in MDA-MB-231 cells; Y.Z. produced the immortal Net1 knockout MEFs; J.A.F. contributed to experimental design and analysis, and writing of the manuscript.
Funding
This work was supported by grants from the National Cancer Institute [grant numbers CA116356 and CA172129 to J.A.F.]; Cancer Prevention and Research Institute of Texas (CPRIT) [grant numbers RP100502 to J.A.F.]; and a Department of Defense (DOD) Breast Cancer Research Program (BRCP) fellowship [number BC112395 to W.O.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.158121/-/DC1
References
- Alberts A. S., Qin H., Carr H. S., Frost J. A. (2005). PAK1 negatively regulates the activity of the Rho exchange factor NET1. J. Biol. Chem. 280, 12152–12161 10.1074/jbc.M405073200 [DOI] [PubMed] [Google Scholar]
- Burridge K., Wennerberg K. (2004). Rho and Rac take center stage. Cell 116, 167–179 10.1016/S0092-8674(04)00003-0 [DOI] [PubMed] [Google Scholar]
- Bustelo X. R., Sauzeau V., Berenjeno I. M. (2007). GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. BioEssays 29, 356–370 10.1002/bies.20558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr H. S., Cai C., Keinänen K., Frost J. A. (2009). Interaction of the RhoA exchange factor Net1 with discs large homolog 1 protects it from proteasome-mediated degradation and potentiates Net1 activity. J. Biol. Chem. 284, 24269–24280 10.1074/jbc.M109.029439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr H. S., Morris C. A., Menon S., Song E. H., Frost J. A. (2013a). Rac1 controls the subcellular localization of the RhoGEF Net1A to regulate focal adhesion formation and cell spreading. Mol. Cell. Biol. 33, 622–634 10.1128/MCB.00980-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr H. S., Zuo Y., Oh W., Frost J. A. (2013b). Regulation of FAK activation, breast cancer cell motility and amoeboid invasion by the RhoA GEF Net1. Mol. Cell. Biol. 33, 2773–2786 10.1128/MCB.00175-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A. M., Takai S., Yamada K., Miki T. (1996). Isolation of a novel oncogene, NET1, from neuroepithelioma cells by expression cDNA cloning. Oncogene 12, 1259–1266. [PubMed] [Google Scholar]
- Chrzanowska-Wodnicka M., Burridge K. (1996). Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J. Cell Biol. 133, 1403–1415 10.1083/jcb.133.6.1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close P., Creppe C., Gillard M., Ladang A., Chapelle J. P., Nguyen L., Chariot A. (2010). The emerging role of lysine acetylation of non-nuclear proteins. Cell. Mol. Life Sci. 67, 1255–1264 10.1007/s00018-009-0252-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Bari M. G., Ciuffini L., Mingardi M., Testi R., Soddu S., Barilà D. (2006). c-Abl acetylation by histone acetyltransferases regulates its nuclear-cytoplasmic localization. EMBO Rep. 7, 727–733 10.1038/sj.embor.7400700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietschy T., Shevelev I., Pena-Diaz J., Hühn D., Kuenzle S., Mak R., Miah M. F., Hess D., Fey M., Hottiger M. O. et al. (2009). p300-mediated acetylation of the Rothmund-Thomson-syndrome gene product RECQL4 regulates its subcellular localization. J. Cell Sci. 122, 1258–1267 10.1242/jcs.037747 [DOI] [PubMed] [Google Scholar]
- Dutertre M., Gratadou L., Dardenne E., Germann S., Samaan S., Lidereau R., Driouch K., de la Grange P., Auboeuf D. (2010). Estrogen regulation and physiopathologic significance of alternative promoters in breast cancer. Cancer Res. 70, 3760–3770 10.1158/0008-5472.CAN-09-3988 [DOI] [PubMed] [Google Scholar]
- Garcia-Mata R., Wennerberg K., Arthur W. T., Noren N. K., Ellerbroek S. M., Burridge K. (2006). Analysis of activated GAPs and GEFs in cell lysates. Methods Enzymol. 406, 425–437. [DOI] [PubMed] [Google Scholar]
- García-Mata R., Dubash A. D., Sharek L., Carr H. S., Frost J. A., Burridge K. (2007). The nuclear RhoA exchange factor Net1 interacts with proteins of the Dlg family, affects their localization, and influences their tumor suppressor activity. Mol. Cell. Biol. 27, 8683–8697 10.1128/MCB.00157-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Jin Q., Zhang F., Yan T., Zhai Q. (2009). PCAF acetylates beta-catenin and improves its stability. Mol. Biol. Cell 20, 419–427 10.1091/mbc.E08-08-0792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi A., Hiatari R., Tsuji T., Ohashi K., Mizuno K. (2013). p63RhoGEF-mediated formation of a single polarized lamellipodium is required for chemotactic migration in breast carcinoma cells. FEBS Lett. 587, 698–705 10.1016/j.febslet.2013.01.043 [DOI] [PubMed] [Google Scholar]
- Huveneers S., Danen E. H. (2009). Adhesion signaling - crosstalk between integrins, Src and Rho. J. Cell Sci. 122, 1059–1069 10.1242/jcs.039446 [DOI] [PubMed] [Google Scholar]
- Inuzuka H., Gao D., Finley L. W., Yang W., Wan L., Fukushima H., Chin Y. R., Zhai B., Shaik S., Lau A. W. et al. (2012). Acetylation-dependent regulation of Skp2 function. Cell 150, 179–193 10.1016/j.cell.2012.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh R. E., Kurokawa K., Ohba Y., Yoshizaki H., Mochizuki N., Matsuda M. (2002). Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol. Cell. Biol. 22, 6582–6591 10.1128/MCB.22.18.6582-6591.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A. B., Hall A. (2005). Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21, 247–269 10.1146/annurev.cellbio.21.020604.150721 [DOI] [PubMed] [Google Scholar]
- Kimura K., Ito M., Amano M., Chihara K., Fukata Y., Nakafuku M., Yamamori B., Feng J., Nakano T., Okawa K. et al. (1996). Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science 273, 245–248 10.1126/science.273.5272.245 [DOI] [PubMed] [Google Scholar]
- Kraynov V. S., Chamberlain C., Bokoch G. M., Schwartz M. A., Slabaugh S., Hahn K. M. (2000). Localized Rac activation dynamics visualized in living cells. Science 290, 333–337 10.1126/science.290.5490.333 [DOI] [PubMed] [Google Scholar]
- Lange A., Mills R. E., Lange C. J., Stewart M., Devine S. E., Corbett A. H. (2007). Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 282, 5101–5105 10.1074/jbc.R600026200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Moon H. J., Lee J. M., Joo C. K. (2010). Smad3 regulates Rho signaling via NET1 in the transforming growth factor-beta-induced epithelial-mesenchymal transition of human retinal pigment epithelial cells. J. Biol. Chem. 285, 26618–26627 10.1074/jbc.M109.073155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeuwen F. N., Kain H. E., Kammen R. A., Michiels F., Kranenburg O. W., Collard J. G. (1997). The guanine nucleotide exchange factor Tiam1 affects neuronal morphology; opposing roles for the small GTPases Rac and Rho. J. Cell Biol. 139, 797–807 10.1083/jcb.139.3.797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyden J., Murray D., Moss A., Arumuguma M., Doyle E., McEntee G., O'Keane C., Doran P., MacMathuna P. (2006). Net1 and Myeov: computationally identified mediators of gastric cancer. Br. J. Cancer 94, 1204–1212 10.1038/sj.bjc.6603054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Diner B. A., Chen J., Cristea I. M. (2012). Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc. Natl. Acad. Sci. USA 109, 10558–10563 10.1073/pnas.1203447109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machacek M., Hodgson L., Welch C., Elliott H., Pertz O., Nalbant P., Abell A., Johnson G. L., Hahn K. M., Danuser G. (2009). Coordination of Rho GTPase activities during cell protrusion. Nature 461, 99–103 10.1038/nature08242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison D. L., Yaciuk P., Kwok R. P., Lundblad J. R. (2002). Acetylation of the adenovirus-transforming protein E1A determines nuclear localization by disrupting association with importin-alpha. J. Biol. Chem. 277, 38755–38763 10.1074/jbc.M207512200 [DOI] [PubMed] [Google Scholar]
- Moll T., Tebb G., Surana U., Robitsch H., Nasmyth K. (1991). The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell 66, 743–758. [DOI] [PubMed] [Google Scholar]
- Murray D., Horgan G., Macmathuna P., Doran P. (2008). NET1-mediated RhoA activation facilitates lysophosphatidic acid-induced cell migration and invasion in gastric cancer. Br. J. Cancer 99, 1322–1329 10.1038/sj.bjc.6604688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimnual A. S., Taylor L. J., Bar-Sagi D. (2003). Redox-dependent downregulation of Rho by Rac. Nat. Cell Biol. 5, 236–241 10.1038/ncb938 [DOI] [PubMed] [Google Scholar]
- Papadimitriou E., Vasilaki E., Vorvis C., Iliopoulos D., Moustakas A., Kardassis D., Stournaras C. (2012). Differential regulation of the two RhoA-specific GEF isoforms Net1/Net1A by TGF-β and miR-24: role in epithelial-to-mesenchymal transition. Oncogene 31, 2862–2875 10.1038/onc.2011.457 [DOI] [PubMed] [Google Scholar]
- Parsons J. T., Horwitz A. R., Schwartz M. A. (2010). Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11, 633–643 10.1038/nrm2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J. H., Du Y., Ard P. G., Phillips C., Carella B., Chen C. J., Rakowski C., Chatterjee C., Lieberman P. M., Lane W. S. et al. (2004). The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol. Cell. Biol. 24, 10826–10834 10.1128/MCB.24.24.10826-10834.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertz O., Hodgson L., Klemke R. L., Hahn K. M. (2006). Spatiotemporal dynamics of RhoA activity in migrating cells. Nature 440, 1069–1072 10.1038/nature04665 [DOI] [PubMed] [Google Scholar]
- Qin H., Carr H. S., Wu X., Muallem D., Tran N. H., Frost J. A. (2005). Characterization of the biochemical and transforming properties of the neuroepithelial transforming protein 1. J. Biol. Chem. 280, 7603–7613 10.1074/jbc.M412141200 [DOI] [PubMed] [Google Scholar]
- Rihs H. P., Jans D. A., Fan H., Peters R. (1991). The rate of nuclear cytoplasmic protein transport is determined by the casein kinase II site flanking the nuclear localization sequence of the SV40 T-antigen. EMBO J. 10, 633–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman K. L., Der C. J., Sondek J. (2005). GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 6, 167–180 10.1038/nrm1587 [DOI] [PubMed] [Google Scholar]
- Sander E. E., ten Klooster J. P., van Delft S., van der Kammen R. A., Collard J. G. (1999). Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J. Cell Biol. 147, 1009–1022 10.1083/jcb.147.5.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Hall A. (2002). The Rho exchange factor Net1 is regulated by nuclear sequestration. J. Biol. Chem. 277, 14581–14588 10.1074/jbc.M111108200 [DOI] [PubMed] [Google Scholar]
- Schmidt A., Durgan J., Magalhaes A., Hall A. (2007). Rho GTPases regulate PRK2/PKN2 to control entry into mitosis and exit from cytokinesis. EMBO J. 26, 1624–1636 10.1038/sj.emboj.7601637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. J. (2004). The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 32, 959–976 10.1093/nar/gkh252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y., Berdeaux R., Frost J. A. (2014). The RhoGEF Net1 is required for normal mammary gland development. Mol. Endocrinol. 28, 1948–1960 10.1210/me.2014-1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.