Abstract
Extracellular signal-regulated kinase 3 (ERK3) is an unstable mitogen-activated protein kinase homologue that is constitutively degraded by the ubiquitin-proteasome pathway in proliferating cells. Here we show that a lysineless mutant of ERK3 is still ubiquitinated in vivo and requires a functional ubiquitin conjugation pathway for its degradation. Addition of N-terminal sequence tags of increasing size stabilizes ERK3 by preventing its ubiquitination. Importantly, we identified a fusion peptide between the N-terminal methionine of ERK3 and the C-terminal glycine of ubiquitin in vivo by tandem mass spectrometry analysis. These findings demonstrate that ERK3 is conjugated to ubiquitin via its free NH2 terminus. We found that large N-terminal tags also stabilize the expression of the cell cycle inhibitor p21 but not that of substrates ubiquitinated on internal lysine residues. Consistent with this observation, lysineless p21 is ubiquitinated and degraded in a ubiquitin-dependent manner in intact cells. Our results suggests that N-terminal ubiquitination is a more prevalent modification than originally recognized.
The ubiquitin/proteasome proteolytic pathway is an evolutionarily conserved regulatory system that controls a host of cellular processes, including transcription, cell cycle progression, differentiation and development, tumor suppression, and immune responses (17). Malfunctioning of the system, as a result of either loss-of-function mutations or abnormal activity, has been implicated in the pathogenesis of cancer and of many other human diseases (17, 28). Proteins targeted for degradation by the 26S proteasome are generally tagged with multiple copies of ubiquitin, which serve as a recognition signal for the 19S regulatory particle. The formation of ubiquitin conjugates is a highly regulated process that requires the sequential action of three enzymes (20). The last step, which is catalyzed by a large family of E3 ubiquitin ligases, confers specificity to the reaction.
For most proteins, the first ubiquitin molecule is attached via an isopeptide bond formed between its C-terminal glycine residue and the ɛ-NH2 group of an internal lysine of the substrate. The polyubiquitin chain is then synthesized by the successive conjugation of ubiquitin molecules to an internal lysine of the previously conjugated ubiquitin. There is no consensus about the positioning of internal lysines that are conjugated to ubiquitin, although a number of studies have highlighted the importance of specific lysine residues. For example, study of Sic1 ubiquitination has revealed that some lysines are more efficiently ubiquitinated than others and, most importantly, that only 6 N-terminal lysines out of 20 support efficient degradation by the 26S proteasome (31). Structural analysis of β-catenin ubiquitination by SCFβ-TrCP has shown that the position of the lysine upstream of the β-transducin repeat-containing protein (β-TrCP) binding site greatly influences the rate of ubiquitin ligation (45). The sites of lysine ubiquitination have been mapped for some well-characterized protein substrates, such as p53 (34), SOCS3 (36), and IκBα (1, 37). Replacement of these specific lysines stabilizes the mutant protein, suggesting that they are at least necessary for degradation. However, for the majority of proteasome substrates, the precise ubiquitination site(s) has not been characterized.
Alternative modes of substrate recognition and targeting have also been described. In the case of the transcriptional activator MyoD, the free α-NH2 terminus of the protein serves as the conjugation site for ubiquitin (7). For a small number of proteins, such as ornithine decarboxylase (13), the cell cycle inhibitor p21 (40), and α-synuclein (41), it has been suggested that degradation by the proteasome occurs in an ubiquitin-independent manner. However, although purified recombinant p21 and α-synuclein can be efficiently degraded by the proteasome in vitro (25), it remains unclear if degradation of these substrates in vivo is independent of a functional ubiquitin system. The observation that p21-ubiquitin conjugates are formed in vivo is also intriguing (8, 26, 35, 40).
Extracellular signal-regulated kinase 3 (ERK3) is a distantly related member of the mitogen-activated protein (MAP) kinase superfamily (30, 43). Although the exact physiological functions of ERK3 remain to be established, accumulating evidence points to a role for the kinase in the control of cell differentiation. ERK3 transcripts are upregulated during differentiation of P19 embryonal carcinoma cells into neuronal or muscle cells (6). ERK3 protein also markedly accumulates during differentiation of C2C12 myoblasts into muscle cells, with kinetics parallel to that of p21 (14). Notably, overexpression of ERK3 in fibroblasts causes cell cycle arrest. Unlike conventional MAP kinases, ERK3 is a highly unstable protein that is constitutively degraded by the ubiquitin-proteasome pathway in proliferating cells (14). We have identified two degrons in the N-terminal lobe of the kinase domain that are both necessary and sufficient to promote ERK3 degradation.
To further understand the mechanism of ERK3 degradation, we have mapped the ubiquitination site(s) of the protein. We report here that ERK3 is ubiquitinated and degraded by the proteasome in a lysine-independent fashion. We show that addition of N-terminal sequence tags of increasing sizes efficiently inhibits N-terminal but not lysine-dependent ubiquitination of protein substrates. In the course of these studies, we also found that proteasome degradation of p21 is independent of the presence of lysine residues but is dependent on a functional ubiquitination pathway. Our results suggest that N-terminal ubiquitination may be a more widespread phenomenon than originally anticipated.
MATERIALS AND METHODS
Reagents and antibodies.
The source of reagents has been described previously (14). Tumor necrosis factor alpha (TNF-α) was obtained from R&D Systems. Polyclonal anti-hemagglutinin (HA) antibody was from Santa-Cruz Biotechnology. The polyclonal antibody to the C terminus of ERK3 (E3-CTE4) was described previously (14).
Plasmid constructs and mutagenesis.
Mutations were introduced into human ERK3 (27) and human p21 (16) cDNAs by using the Altered Sites In Vitro Mutagenesis system (Promega). pcDNA3-HA-ERK3, HA-ERK3Δ, and HA-ERK1 have already been described (14). To construct the C-terminal His6-tagged expression vectors, two oligonucleotides coding for His6 tag sequence were inserted into the EcoRI/XbaI sites of pcDNA3-HA. The coding sequences of ERK3Δ, ERK3Δ-0K, p21, and p21-0K were subcloned in frame into the EcoRI site. To construct the N-terminal His6-tagged ERK3, two oligonucleotides encoding the sequence MRGSHHHHHHGIS were introduced downstream of a Kozak sequence. Myc6-ERK3 was constructed by first subcloning the Myc6 tag sequence derived from pCS3MT (a gift from A. J. Waskiewicz) into the BamHI/EcoRI sites of pcDNA3 to yield pcDNA3-Myc6. The ERK3 coding sequence was then subcloned into pcDNA3-Myc6. Enhanced green fluorescent protein (EGFP)-ERK3 was constructed by first inserting the EGFP sequence (Clontech) into the KpnI/EcoRI sites of pcDNA3. The ERK3 coding sequence was then subcloned into the EcoRI site of the resulting vector. To construct Mycn-ERK3 expression vectors, a pair of oligonucleotides coding for a unique Myc epitope was sequentially cloned into the EcoRI/XbaI sites of pcDNA3-Koz, which was designed to contain a Met initiator codon into a consensus Kozak sequence. ERK3-EGFP was constructed by first subcloning the EGFP coding sequence into pcDNA3-Koz. The full-length ERK3 was then ligated in frame upstream of EGFP. Glutathione S-transferase (GST)-tagged ERK3-EGFP and EGFP-ERK3 were constructed by subcloning the GST sequence, derived from pGEX-KG (18), into the EcoRI/XbaI sites of the respective pcDNA3 vectors. To generate the M5HA expression vector, the sixth Myc epitope was first replaced by an HA tag by inserting a set of oligonucleotides into the NcoI/EcoRI sites of pCS3MT. The resulting M5HA tag was then subcloned into the BamHI/EcoRI sites of pcDNA3 to yield pcDNA3-M5HA. The coding sequences of hamster ERK1, human ERK3, human SOCS3 (gift from A. Yoshimura), human p53 (gift from G. Ferbeyre), human IκBα (gift from J. Hiscott), and human p21 were subcloned into the EcoRI/XbaI sites of pcDNA3-M5HA. pMT123 vector encoding HA-tagged ubiquitin was kindly provided by Dirk Bohmann (42). All mutations were confirmed by DNA sequencing. Sequence of the primers used for PCR and details about the cloning strategies are available upon request.
Cell culture and transfections.
Human embryonic kidney 293 (HEK 293), A31, and ts20 cells were cultured as described previously (14). HEK 293 cells were transfected by the calcium phosphate coprecipitation method. A31 and ts20 cells were transfected with Lipofectamine reagent (Invitrogen).
Protein biochemistry methods and immunofluorescence.
Cell lysis, protein precipitation, and immunoblot analysis were performed as previously described (14, 39). Protein concentrations were measured by using the bicinchoninic acid assay (Pierce), and equal amounts of lysate proteins were subjected to sodium dodecyl sulfate (SDS)-gel electrophoresis. In vivo phosphorylation analysis and immunofluorescence studies were based on techniques previously described (33). In vitro transcription and translation was carried out using the TNT Coupled Reticulocyte Lysate system (Promega) according to the manufacturer's instructions. The reaction products were detected by fluorography with Amplify (Amersham).
In vivo ubiquitination assays.
HEK 293 cells were cotransfected with the indicated expression vectors (4 μg) together with HA-tagged ubiquitin (2 μg). After 36 h, the cells were treated with 25 μM MG-132 for 12 h. For purification of C-terminal His6-tagged proteins, the cells were lysed in buffer lacking EDTA and were supplemented with 20 mM imidazole and 10 mM N-ethyl maleimide (NEM). Cell lysates (750 μg of protein) were incubated with 20 μl of Ni-nitrilotriacetic acid agarose beads (QIAGEN) for 2 h at 4°C with constant agitation. The beads were washed three times with lysis buffer without NEM. Proteins were separated by SDS-gel electrophoresis and were detected by immunoblotting with anti-HA monoclonal antibody 12CA5. C-terminal GST-tagged proteins were purified by using glutathione-conjugated agarose beads (Pharmacia). The cells were lysed in Triton X-100 lysis buffer supplemented with 10 mM NEM, and the clarified lysates were further incubated for 10 min with 2.5 mM dithiothreitol to quench free NEM. Cell lysates (750 μg of protein) were incubated for 2 h at 4°C with 20 μl of glutathione-agarose resin. The bound proteins were washed four times in lysis buffer and were resolved by SDS-gel electrophoresis.
MS analysis.
Twenty 10-cm-diameter culture dishes of HEK 293 cells were transfected with expression vectors encoding ERK3Δ or p21 tagged at the N terminus with HA and tagged at the C terminus with GST. After 36 h, the cells were treated with 25 μM MG-132 for 12 h. The cells were lysed in NEM-containing buffer, and the transfected proteins were precipitated with glutathione-agarose beads. The purified proteins (4 μg) immobilized on beads were resuspended in 50 mM ammonium bicarbonate (pH 8.1). Modified trypsin (Promega) was then added at an enzyme:substrate ratio of 1:25, and the samples were incubated overnight at 37°C. Supernatants were collected and the beads were washed three times with 100 μl of 50 mM ammonium bicarbonate prior to evaporation in a Speedvac. The protein digests were reconstituted in 40 μl of aqueous formic acid (0.2%) containing 5% acetonitrile, and 10 μl was loaded on the chromatographic system. All mass spectrometry (MS) analyses were conducted on a Waters CapLC coupled to a Q-TOF Ultima via a nanoLC interface (Waters). The on-line two-dimensional liquid chromatography-mass spectrometry (LC-MS) chromatographic system comprised a homemade strong cation exchange (SCX) column, 5 mm by 360 μm (PolyLC), a 300-μm by 5-mm Waters Symmetry C18 precolumn, and a 150-μm by 10-cm homemade Jupiter C18 (Phenomenex) analytical column. After sample injection, peptides were eluted sequentially from the SCX column with 20-μl salt plugs of ammonium acetate of increasing concentrations, ranging from 0, 60, 70, 80, 100, 150, 200, 400, to 1,000 mM at pH 3.0. Peptides eluted from the SCX column were retained on the C18 precolumn and subsequently were separated on the analytical column with a linear gradient (10 to 60%) of 0.2% formic acid in acetonitrile over 63 min. The mass spectrometer was operated in a data-dependent acquisition mode whereby the instrument cycled through a survey scan (1s) and a product ion spectrum (2s). Collisional activation of selected precursors was obtained by using nitrogen as a target gas, with collision energies ranging from 30 to 80 eV (laboratory frame of reference) scaled according to m/z and the charge of the precursor ion. Fragment ions formed in the replicative form-only quadrupole were recorded by a time-of-flight mass analyzer. Mascot (Matrix Science) database searches of the acquired MS-MS spectra were performed against the National Center for Biotechnology Information nonredundant protein database.
RESULTS
In vivo degradation of ERK3 by the proteasome is independent of lysines.
To further characterize the mechanism of ERK3 ubiquitination and to possibly obtain stable mutants, we decided to map the ubiquitination site(s) of the kinase. It was previously shown that ERK3 ubiquitination is confined within the first 365 amino acids of the protein (14). We therefore used this truncated ERK3 mutant (ERK3Δ) as a backbone to replace the 16 lysines residues with arginine (Fig. 1A). The effect of the mutations on the stability of ERK3 was estimated from the steady-state levels of the ectopically expressed protein. An initial series of nine double, triple, or quadruple lysine mutants of ERK3Δ was transiently transfected in HEK 293 cells and was analyzed by immunoblotting. All these constructs were expressed at levels similar to that of the wild-type protein, suggesting that none of the individual lysines is absolutely essential for ERK3 ubiquitination in vivo (data not shown). To test the possible redundancy of these lysines, we then analyzed the different mutants in combination. The progressive elimination of up to 14 of the 16 putative lysine ubiquitination sites had no significant effect on ERK3 steady-state levels (data not shown). We finally constructed a lysineless (0K) mutant of ERK3Δ in an attempt to completely abolish ubiquitination. Surprisingly, we observed that ERK3Δ-0K was expressed at a level comparable to that of wild-type ERK3, which is much lower than that of the stable ERK1 protein (Fig. 1B). All three constructs were translated with similar efficiency in vitro (Fig. 1B, lower panel). To determine whether the weak expression of ERK3Δ and ERK3Δ-0K was due to rapid turnover of the proteins, we measured their half-lives by cycloheximide chase experiments (Fig. 1C). We found that both ERK3Δ and ERK3Δ-0K are rapidly degraded, with half-lives of approximately 45 min in exponentially proliferating cells, a value similar to the half-life of the full-length ERK3 protein (14).
FIG. 1.
A lysineless mutant of ERK3 is unstable. (A) Schematic representation of ERK3Δ constructs showing the position of lysines mutated in this study. (B) HEK 293 cells were transfected with the indicated constructs. After 48 h, cell lysates were analyzed by immunoblotting with anti-HA antibody. Two different exposures of the gel are shown (upper two panels). The same constructs were translated in vitro in the presence of 35S-labeled amino acids and were analyzed by fluorography (lower panel). (C) HEK 293 cells transfected with the different ERK3 constructs were treated with cycloheximide (100 μg/ml) for the indicated times. Ectopically expressed ERK3 was detected by HA immunoblotting. (D) HEK 293 cells were transfected with Myc6-tagged ERK3Δ, ERK3Δ kinase dead (KD), ERK3Δ Ser189Ala (S189A), ERK3Δ-0K, or ERK3Δ1-296. The cells were metabolically labeled with 32P, and the transfected ERK3 proteins were immunoprecipitated with anti-Myc antibody. Phosphorylation was revealed by autoradiography (upper panel). Aliquots of cell lysates were analyzed by immunoblotting to monitor expression of the ectopic proteins (lower panel). exp., exposure; IB, immunoblot; IP, immunoprecipitation.
To exclude the possibility that the rapid degradation of ERK3Δ-0K is due to misfolding of the mutated protein, we compared the in vivo phosphorylation status of wild-type ERK3Δ with that of various mutants. ERK3 has been previously shown to be phosphorylated on the activation loop Ser-189 in intact cells (9, 14). Metabolic labeling experiments confirmed that the truncated ERK3Δ is also phosphorylated in vivo (Fig. 1D). Replacement of Ser-189 by alanine almost completely suppressed 32P incorporation, indicating that Ser-189 is the major phosphorylation site of ERK3. The catalytically inactive mutant ERK3Δ-KD was phosphorylated to the same extent as the wild-type protein, consistent with the idea that phosphorylation of Ser-189 is executed by a cellular ERK3 kinase (10). MAP kinase kinases are remarkably specific enzymes that recognize their targets only in the native conformation (38). In agreement with this, deletion of the extreme C-terminal end of the ERK3 kinase domain (ERK3Δ1-296) is sufficient to abolish its phosphorylation by ERK3 kinase (Fig. 1D). Importantly, we observed that ERK3Δ-0K is phosphorylated in vivo, albeit to a lesser extent than the wild-type protein (Fig. 1D). We conclude from these results that the ERK3Δ-0K mutant is correctly folded.
As previously reported for the native ERK3 protein (14), treatment with the structurally unrelated proteasome inhibitors MG-132 and lactacystin β-lactone induced the accumulation of both ERK3Δ and the lysineless mutant (Fig. 2A). Cycloheximide chase experiments further confirmed that incubation with MG-132 strongly stabilizes ERK3Δ and ERK3Δ-0K proteins (Fig. 2B). These results indicate that ERK3 is actively degraded by the proteasome in vivo in the complete absence of lysine residues.
FIG. 2.
ERK3 is degraded by the proteasome in a lysine-independent manner. (A) HEK 293 cells were transfected with HA-tagged ERK3Δ or ERK3Δ-0K constructs. After 36 h, the cells were treated for 12 h with the protease inhibitors MG-132 (25 μM), E-64 (25 μM), lactacystin β-lactone (20 μM), or vehicle (0.1% dimethyl sulfoxide [DMSO]). Ectopically expressed ERK3 was detected by HA immunoblotting. (B) HEK 293 cells were transfected as described for panel A. After 44 h, the cells were pretreated with dimethyl sulfoxide or MG-132 for 30 min. Cycloheximide was then added for a period of 4 h. Cell lysates were analyzed by HA immunoblotting. IB, immunoblot.
Lysineless ERK3 is ubiquitinated and requires a functional ubiquitin pathway for its efficient degradation in vivo.
Because previous reports have suggested that some proteins may be targeted to the proteasome without ubiquitination (44), we sought to evaluate the role of the ubiquitin pathway in ERK3Δ-0K turnover. To this end we measured the steady-state levels of ERK3Δ and ERK3Δ-0K in the ts20 cell line that harbors a temperature-sensitive ubiquitin-activating enzyme, E1 (11). As shown in Fig. 3A, both ERK3Δ and ERK3Δ-0K accumulated when ts20 cells were shifted to the restrictive temperature (39°C). No change in expression was observed in parental A31 cells. Thus, degradation of a lysineless mutant of ERK3 is still dependent on a functional ubiquitin conjugation pathway.
FIG. 3.
Lysineless ERK3 is ubiquitinated in vivo and requires a functional ubiquitin conjugation pathway for its efficient degradation. (A) Parental BALB/c 3T3 (A31) and E1-thermosensitive mutant (ts20) cells were transfected with the indicated ERK3 constructs or empty vector (V). After 24 h, the cells were maintained at the permissive temperature (34°C) or shifted to the restrictive temperature (39°C) for the indicated times. Ectopically expressed ERK3 was detected by HA immunoblotting. (B) HEK 293 cells were cotransfected with expression vectors encoding ERK1, ERK3Δ, or ERK3Δ-0K together with HA-tagged ubiquitin. The protein kinase constructs contain an N-terminal HA tag and a C-terminal His6 affinity tag. After 36 h, the cells were treated with MG-132 for 12 h. His6-tagged proteins were purified from cell lysates with nickel-agarose beads and were analyzed by immunoblotting with anti-HA antibody (upper panel). Asterisks mark nonspecific bands. Total cell lysates were analyzed for global ubiquitination activity by HA immunoblotting (lower panel). IB, immunoblot.
The above findings prompted us to ask whether ERK3Δ-0K is ubiquitinated in vivo. To address this question, C-terminal His6-tagged constructs of ERK3 were cotransfected with HA-tagged ubiquitin and the kinase was purified from cell lysates with Ni-agarose beads. Ubiquitination was monitored by immunoblotting with anti-HA antibody. A ladder of high-molecular-size bands corresponding to polyubiquitinated species of ERK3 was clearly detected for both ERK3Δ and the lysineless ERK3Δ-0K mutant (Fig. 3B). ERK1 was used as a negative control and shows no specific polyubiquitin adducts under these conditions.
Addition of large N-terminal sequence tags stabilizes ERK3 through inhibition of N-terminal ubiquitination.
With the exception of E1, E2, and HECT-type E3 enzymes that transiently become conjugated to monoubiquitin via an internal cysteine residue, ubiquitination of substrates absolutely requires an acceptor amino group (17, 20, 32). Moreover, the thiol ester bond formed between ubiquitin and cysteine is labile and highly sensitive to the reducing conditions of Laemmli's sample buffer (15, 21). Thus, the only possible conjugation site of ERK3Δ-0K to ubiquitin is the free N-terminal NH2 of the kinase. In the case of MyoD, it was clearly shown that chemical blocking of the α-NH2 group completely stabilizes the protein in an in vitro degradation assay (7). Notably, these authors reported that addition of a Myc6 epitope tag at the N terminus of MyoD also results in the stabilization of the protein by a poorly understood mechanism. To further test this idea, we generated a series of constructs of the full-length ERK3 (containing all 45 lysine residues) containing different tags at their N termini. As was previously observed (14), addition of small (≤1.5 kDa) epitope tags does not influence ERK3 expression. The His6 (10 residues)- and HA (13 residues)-tagged ERK3 constructs were expressed at levels similar to that of the untagged ectopic protein (Fig. 4A). Note that in these experimental conditions, these short epitope-tagged proteins are expressed at levels comparable to that of the endogenous ERK3 protein. Interestingly, addition of a Myc6 epitope or EGFP (yielding fusion proteins of 120 and 130 kDa, respectively) dramatically enhanced ERK3 expression (Fig. 4A, upper panel) without influencing the rate of in vitro translation (lower panel). This result suggested that the observed differences in steady-state levels likely reflect changes in the half-lives of the protein constructs. Indeed, cycloheximide chase experiments confirmed that EGFP-ERK3 is at least 10 times more stable than His6-ERK3 (Fig. 4B). Treatment with MG-132 induced a marked accumulation of His6-ERK3, whereas it had little effect on the expression of EGFP-ERK3 (Fig. 4C). Because Myc6 and EGFP do not show significant sequence homology, the similar effect of the two tags suggests that the size of the sequence tag influences ERK3 protein stability. To directly test this idea, we constructed ERK3 versions containing 1, 3, 5, or 7 Myc epitope tags tandemly arrayed at the N terminus. Results of this experiment confirmed that the size of the tag has a strong influence on ERK3 accumulation; addition of three repeats of the Myc epitope was sufficient to increase the expression of the kinase (Fig. 4D). Cycloheximide chase experiments clearly showed that these differences in steady-state expression result from changes in protein turnover (Fig. 4E). We conclude that addition of large N-terminal tags (larger than 5 kDa) is sufficient to stabilize ERK3. The presence of a large N-terminal tag also stabilized the lysineless mutant of ERK3 (Fig. 4F), further substantiating the idea that N-terminal ubiquitination of ERK3Δ-0K is necessary for its degradation by the proteasome. The stabilization of ERK3Δ-0K by the Myc6 tag provides additional evidence that this mutant is properly folded in vivo.
FIG. 4.
Addition of large N-terminal tags stabilizes ERK3 expression. (A) HEK 293 cells were transfected with the indicated constructs. After 48 h, endogenous and ectopic ERK3 were detected by immunoblotting with antibody E3-CTD4, which specifically recognizes the ERK3 C terminus (upper panel). The asterisk marks a nonspecific band. The same constructs were translated in vitro in the presence of 35S-labeled amino acids and were analyzed by fluorography (lower panel). (B) HEK 293 cells were transfected with ERK3 constructs or empty vector (V). After 48 h, the cells were treated with cycloheximide for the indicated times. His6-ERK3 was purified from cell lysates with nickel-agarose beads and detected by immunoblotting with antibody E3-CTD4 (upper panel). EGFP-ERK3 was analyzed by immunoblotting using anti-GFP antibody (lower panel). (C) HEK 293 cells were transfected as described for panel B and were treated with MG-132 (25 μM) for 12 h. (D) N-terminal tags larger than ∼5 kDa enhance ERK3 expression. HEK 293 cells were transfected with ERK3 constructs tagged at their N termini with an increasing number of copies of the Myc epitope. After 48 h, endogenous and transfected ERK3 was detected by immunoblotting with E3-CTD4 antibody (upper panel). The same constructs were in vitro translated and detected by fluorography (lower panel). (E) The half-lives of ectopically expressed Myc1-ERK3 (upper panel) and Myc3-ERK3 (lower panel) were evaluated by cycloheximide chase. (F) The half-lives of Myc6-ERK3Δ (upper panel) and Myc6-ERK3Δ-0K (lower panel) were evaluated as described for panel E. IB, immunoblot; IP, immunoprecipitation.
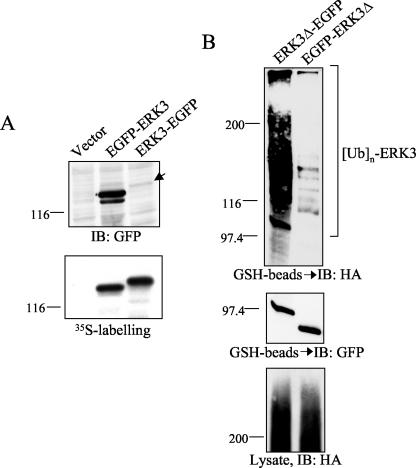
To better understand the mechanism by which fusion of large sequence tags stabilizes ERK3 protein, we tested whether the position of the tag is important. We found that addition of EGFP to the C terminus of ERK3 is completely inefficient in stabilizing the protein, again highlighting the importance of the N-terminal domain in the regulation of ERK3 degradation (Fig. 5A). We also compared the ubiquitination status of the two fusion proteins made between ERK3 and EGFP. Addition of EGFP to the N terminus of ERK3 drastically inhibited the ubiquitination of the kinase (Fig. 5B), suggesting that the stabilizing effect of large N-terminal tags results in part from their ability to interfere with N-terminal ubiquitination.
FIG. 5.
The presence of a large N-terminal tag inhibits ERK3 ubiquitination. (A) HEK 293 cells were transfected with ERK3 constructs fused to the EGFP sequence at the N-terminal or C-terminal extremity. The transfected proteins were detected by immunoblotting with anti-GFP antibody (upper panel). The same constructs were in vitro translated and detected by fluorography (lower panel). (B) HEK 293 cells were cotransfected with the indicated ERK3-EGFP fusion constructs together with HA-ubiquitin. The ERK3 constructs contain the GST sequence at the C terminus to allow for specific and quantitative recovery of the transfected proteins. After 36 h, the cells were treated with MG-132 for 12 h. Transfected proteins were purified from cell lysates with glutathione-agarose beads, and comparable amounts of ERK3-EGFP fusion proteins were analyzed. Ubiquitin conjugates were detected by anti-HA immunoblotting (upper panel). The membrane was reprobed with anti-EGFP antibody to confirm that an equivalent amount of ERK3 fusion proteins was loaded on the gel (middle panel). Total cell lysates were analyzed for global ubiquitination activity by HA immunoblotting (lower panel). IB, immunoblot.
The stabilizing effect of large N-terminal tags is specific to N-terminal ubiquitinated proteins.
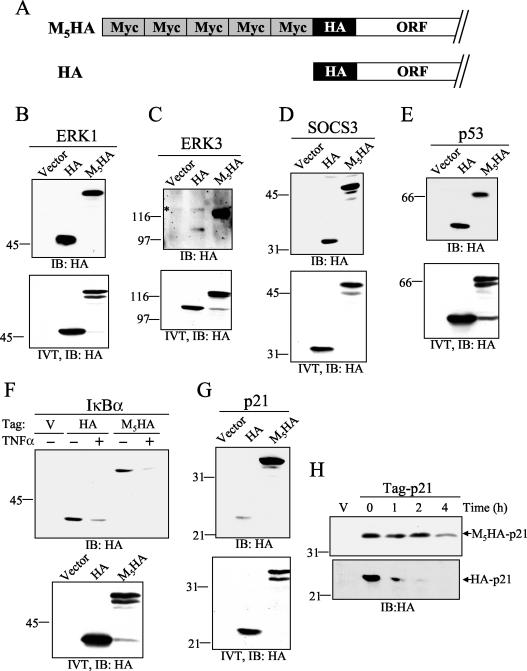
We next wanted to determine if the addition of large N-terminal tags specifically stabilizes N-terminal ubiquitinated proteins or also affects proteins ubiquitinated on internal lysine residues. To this end, we constructed a dual set of tagged vectors: one vector expresses the target protein with a single HA epitope tag at the N terminus, while the second vector contains a larger tag of Myc5 epitopes placed upstream of the HA epitope sequence (Fig. 6A). Transfection of cells with each of the two vectors, followed by simple HA immunoblot analysis, will reflect the steady-state levels of the tagged proteins and should reveal whether addition of a larger N-terminal tag has a stabilizing effect. We first validated our assay by using the stable ERK1 protein and ERK3. ERK1, which is not ubiquitinated under these conditions, was expressed to the same extent with both vectors (Fig. 6B). In contrast, expression of ERK3 was greatly augmented by addition of the larger N-terminal tag (Fig. 6C).
FIG. 6.
Addition of large N-terminal tags stabilizes expression of ERK3 and p21 but not that of proteins ubiquitinated on internal lysine residues. (A) Schematic representation of the constructs used in these experiments. Note that proteins expressed from these two vectors are tagged with a single HA epitope. (B to E) HEK 293 cells were transfected with empty vector or with either HA- or Myc5-HA (M5HA)-tagged expression vectors encoding ERK1 (B), ERK3 (C), SOCS3 (D), and p53 (E). After 48 h, the HA-tagged proteins were detected by immunoblotting with anti-HA antibody (upper panel). An asterisk denotes a nonspecific band. The same constructs were translated in vitro (IVT) and were analyzed by anti-HA immunoblotting (lower panel). (F) HEK 293 cells were transfected with the same vectors encoding IκBα. After 24 h, the cells were serum starved for 24 h and then left untreated or treated with TNF-α (50 ng/ml) for 30 min in the presence of cycloheximide. HA-tagged proteins were detected as described for panel B. (G) HEK 293 cells were transfected with HA- or M5HA-tagged p21. After 48 h, the expression of ectopic p21 was monitored by HA immunoblotting. (H) HEK 293 cells transfected with p21 expression vectors were treated with cycloheximide for the indicated times. HA-tagged proteins were detected as described for panel B. ORF, open reading frame; IB, immunoblot.
We next tested our approach on additional substrates known to be ubiquitinated on internal lysines: p53, SOCS3, and IκBα. p53 is ubiquitinated by MDM2 on C-terminal lysines, notably lysines 370, 372, 373, 381, 382, and 386 (34). SOCS3 and IκBα are ubiquitinated on lysine 6 and on lysines 21 and 22, respectively (1, 36, 37). Mutation of these specific lysines is sufficient to stabilize the corresponding protein. Both SOCS3 and p53 are constitutively ubiquitinated in exponentially proliferating cells (26, 36). Addition of a large N-terminal tag did not result in the accumulation of either protein under these conditions (Fig. 6D and E). In the case of IκBα, degradation of the protein is induced by proinflammatory stimuli like TNF-α (2). We found that TNF-α induces the degradation of both HA- and Myc5-HA-tagged IκBα to a similar extent (Fig. 6F). In all of these examples, the proteins were in vitro translated with similar efficiency regardless of the N-terminal tag (Fig. 6B to F, lower panels). Importantly, the subcellular localization of the proteins tested was not affected by the nature of the tag (data not shown). These results suggest that stability of proteins ubiquitinated on lysine residues is not affected by the presence of large N-terminal tags.
We made the hypothesis that this assay could be used as a mean to identify proteins ubiquitinated on the N terminus. As mentioned above, the cyclin-dependent kinase inhibitor p21, like MyoD and ERK3, is degraded by the proteasome in a lysine-independent manner (40). The precise ubiquitination site of the protein has not been characterized, despite the fact that p21 is known to be polyubiquitinated in vivo. We recently observed that differentiation of C2C12 myoblasts into muscle cells is accompanied by a time-dependent accumulation of ERK3, which results in large part from the stabilization of the protein (14). Interestingly, the upregulation of ERK3 shows a striking parallel to that of p21, a known marker of muscle differentiation (19, 29). These observations prompted us to test the effect of adding a large N-terminal tag on the expression of p21. Immunoblot analysis revealed that Myc5-HA-tagged p21 is expressed at levels at least 10 times higher than that of HA-p21 (Fig. 6G). Cycloheximide chase experiments confirmed that the accumulation of p21 upon addition of the large N-terminal tag results from stabilization of the protein (Fig. 6H). These results suggest that p21 is ubiquitinated on the NH2 terminus in vivo and that this site is important for its efficient degradation.
N-terminal ubiquitination of p21 is necessary for its degradation in vivo.
To investigate the role of the ubiquitin pathway in the proteasome-mediated degradation of p21 in vivo, we monitored by immunoblot analysis the steady-state levels of endogenous p21 in ts20 and parental A31 cells. We found that p21 accumulates to very high levels when ts20 cells are grown at the restrictive temperature (Fig. 7A). Because p21 is a direct transcriptional target of p53, the upregulation of p21 may be a consequence of the accumulation of p53 in ts20 cells as previously documented (11). To directly test the importance of ubiquitination in regulating the turnover of p21, we measured the half-life of the protein by cycloheximide chase experiments in ts20 cells grown at the permissive or restrictive temperature. Inactivation of the E1 enzyme clearly stabilized the endogenous p21 protein (Fig. 7B). The half-life of p21 is ∼15 to 30 min in exponentially proliferating ts20 cells cultured at 34°C and increases to more than 4 h when the temperature is shifted to 39°C. The ectopically expressed p21 protein also accumulated at the restrictive temperature in ts20 cells (Fig. 7C). These results indicate that degradation of p21 is dependent on a functional ubiquitin conjugation pathway.
FIG. 7.
Degradation of lysineless p21 is dependent on a functional ubiquitin conjugation pathway. (A) Parental A31 and E1-mutant ts20 cells were cultured at 34 or 39°C for the indicated times. Expression of endogenous p21 was monitored by immunoblotting with anti-p21 antibody. (B) Half-life of endogenous p21 was measured by cycloheximide chase experiments in ts20 cells grown for 12 h at the permissive or restrictive temperature. (C) A31 or ts20 cells were transfected with the indicated p21 constructs or empty vector (V). The p21 constructs contain an N-terminal HA tag and a C-terminal His6 tag. After 24 h, the cells were maintained at 34 or 39°C for the indicated times. Ectopically expressed p21 proteins were purified from cell lysates with nickel-agarose beads and were analyzed by immunoblotting with anti-HA antibody. (D) HEK 293 cells were cotransfected with the indicated p21 expression vectors together with HA-tagged ubiquitin. After 36 h, the cells were treated with MG-132 for 12 h. His6-tagged proteins were purified from cell lysates with nickel-agarose beads. Half of the purified material was separated on a 7.5% gel (upper panel), while the other half was loaded on a 12% gel (middle panel). Ubiquitin conjugates were detected by HA immunoblotting. The arrow indicates the position of the nonubiquitinated p21 protein. An asterisk marks a nonspecific band. Total cell lysates were analyzed for global ubiquitination activity by HA immunoblotting (lower panel). IB, immunoblot.
Results presented in Fig. 6 suggest that p21 might be ubiquitinated on its N terminus in intact cells. If this is indeed the case, a lysineless version of p21 should accumulate to the same extent as the wild-type protein in ts20 cells grown at 39°C. We therefore constructed a lysineless mutant of p21 by replacing all six lysines with arginine residues. In A31 and HEK 293 cells, p21-0K was expressed at steady-state levels comparable to that of the wild-type protein (Fig. 7C and D). Moreover, the p21-0K mutant similarly accumulated to high levels in ts20 cells grown at the restrictive temperature (Fig. 7C), thereby indicating that degradation of lysineless p21 is also dependent on ubiquitination. We next sought to determine if p21 is ubiquitinated in a lysine-independent fashion in vivo. Cotransfection experiments with HA-ubiquitin confirmed previous observations (26, 40) that p21 is polyubiquitinated in intact cells (Fig. 7D). Importantly, we also detected the presence of high-molecular-mass immunoreactive species (∼66 to 200 kDa) of p21-0K, indicative of polyubiquitin conjugates. Together, our results are consistent with the idea that N-terminal ubiquitination of p21 targets the protein for proteasomal degradation.
Identification of N-terminal ubiquitin conjugates of ERK3 and p21 by MS.
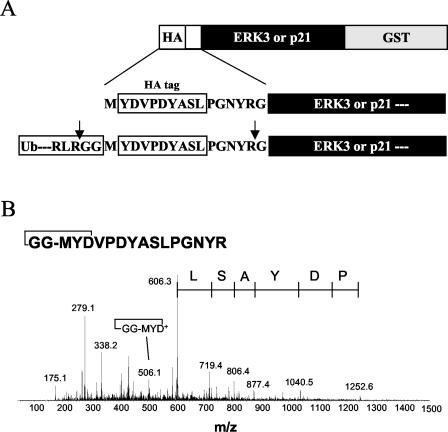
To unequivocally demonstrate that ERK3 and p21 are ubiquitinated at the N terminus, we analyzed whole tryptic digests of purified HA-ERK3Δ-GST and HA-p21-GST fusion proteins (Fig. 8A) by tandem MS. A two-dimensional separation approach using on-line SCX and reverse-phase chromatography was devised in order to simplify the peptide distribution obtained from the nanoLC-MS analyses. Approximately 3,500 MS-MS spectra were acquired for each set of the nine SCX salt fractions. These MS-MS analyses enabled the identification of ERK3Δ and p21 fusion proteins with sequence coverage of 37 and 42%, respectively. Both proteins comprised an N-terminal HA peptide sequence that could not be identified directly from the database search. Rather, manual identification of the peptide bearing the desired HA sequence was facilitated by searching the list of MS-MS spectra for characteristic y-type fragment ion series. More specifically, the presence of two prolines in the expected HA-containing tryptic peptide MYDVPDYASLPGNYR gave rise to abundant y5 and y9 fragment ions at m/z 606.3 and 1,252.6, respectively. The search of all MS-MS spectra containing these characteristic ions revealed several peptide candidates, all comprising the N-terminal methionine residue. Importantly, the tryptic HA peptide was detected under three distinct forms: unmodified free amine (1,759.6 Da), Nα-acetylated (1,801.6 Da), and conjugated to a short Gly-Gly ubiquitin tag (1,855.6 Da) at the N terminus. The latter peptide corresponded to a dehydrated form whereby the amine of the terminal glycine is cyclized to the carboxylic group of the neighboring aspartate residue. This condensation is possibly a reaction by-product of the tryptic digestion, during which the free amine of the diglycine ubiquitin tag is formed. This assignment was supported by the MS-MS spectrum of the doubly charged precursor ion at m/z 928.8, where a distinct b5 ion at m/z 506.1 was assigned to the cyclized GG-MYD fragment ion (Fig. 8B). It is noteworthy that this ion was unique to the cyclized diglycine HA peptide and was not observed for either the Nα-acetylated or the free amine form of the same peptide (data not shown). Rationalization of the MS-MS spectrum of the cyclized diglycine HA peptide is shown in Fig. 8B along with the y-type fragment ion series. Essentially the same spectra were obtained for ERK3 and p21. These data provide the first direct evidence for N-terminal ubiquitination of a protein substrate.
FIG. 8.
Identification of N-terminally ubiquitinated ERK3 and p21 by MS-MS. (A) Schematic representation of the constructs used in this study. The ERK3Δ and p21 fusion proteins are tagged with an N-terminal HA peptide, which upon tryptic digestion releases the sequence MYDVPDYASLPGNYR (boldface characters identify the HA peptide) together with an extra diglycine derived from the C terminus of ubiquitin. (B) HEK 293 cells were transfected with the indicated constructs and treated for 12 h with MG-132. The transfected proteins were purified from cell lysates with glutathione-agarose beads and were analyzed by LC-MS-MS. The fragmentation pattern of the ubiquitinated N-terminal HA peptide is shown. The tandem mass spectrum of the doubly protonated precursor ion at m/z 928.8 shows a series of y-type fragment ions arising from peptide cleavage with charge retention at the C terminus, thus confirming the expected HA peptide sequence containing the methionine residue. The cyclization of the amine of the terminal glycine to the neighboring aspartate residue is supported by a characteristic b5 fragment ion at m/z 506.1, corresponding to the cleavage of the amide bond with charge retention at the N terminus.
DISCUSSION
We demonstrate that the atypical MAP kinase homologue ERK3 and the cyclin-dependent kinase inhibitor p21 are ubiquitinated on their NH2 termini. With the transcriptional activator MyoD (7), ERK3 and p21 are the second and third examples of mammalian proteins ubiquitinated in that fashion. This conclusion is based on the following evidence. First, lysineless versions of both proteins are degraded by the proteasome with kinetics similar to that of the wild-type protein in vivo. In contrast, lysineless mutants of cyclin E (12) or Sic1 (31) are very stable in vivo, indicating that these proteins require an internal lysine(s) for efficient ubiquitination and degradation. Second, in vivo degradation of ERK3 and p21 lysineless mutants is absolutely dependent on a functional ubiquitin conjugation pathway. Third, we clearly detected polyubiquitin conjugates of lysineless ERK3 and p21 proteins. Fourth, we showed that addition of N-terminal tags of increasing size stabilizes ERK3 and p21 by specifically interfering with their ubiquitination. Finally, and most importantly, we demonstrated the existence of a fusion peptide between the N-terminal methionine of ERK3 and p21 constructs and the C-terminal glycine of ubiquitin in intact cells.
Recent studies have shown that positioning of the ubiquitin attachment site is critical for efficient polyubiquitination and proteasome recognition. Petroski and Deshaies have shown that the context of the ubiquitin acceptor site in Sic1 affects the targeting property of the polyubiquitin chain (31). Ubiquitination of C-terminal Sic1 lysines occurs slowly, while producing substrates poorly recognized by the 26S proteasome. In contrast, the six N-terminal lysine residues are more efficiently ubiquitinated in vitro and assemble ubiquitin chains that are more competent for degradation by the proteasome. Interestingly, these authors also showed that a single polyubiquitin chain can support efficient proteasomal degradation in vitro. The results presented here suggest that the N-terminal amine of ERK3 and p21 is both necessary and sufficient to sustain efficient degradation by the proteasome in intact cells. Accordingly, lysineless mutants of ERK3 and p21 have half-lives similar to those of their wild-type counterparts. Our findings further substantiate the idea that a single ubiquitin chain is sufficient to efficiently target a protein substrate to the proteasome in vivo.
A recent structural study has also highlighted the importance of ubiquitin acceptor site positioning. The X-ray structure of the ternary complex Skp1/β-trcp1/β-catenin has revealed that the amino group, provided by a lysine in this case, must be properly placed to be efficiently ubiquitinated by SCFβ-trcp1 (45). For example, displacing the lysine acceptor by 19 residues can decrease the apparent in vitro ubiquitination rate by a factor of 3. Here we show that addition of sequence tags at the N terminus greatly affects the ubiquitination status and stability of ERK3 and p21 in vivo. Most importantly, we show that the size of the tag and not its primary sequence is important for this effect. Small tags comprising less than 13 residues have a minimal effect, whereas tags larger than 5 kDa very efficiently inhibit protein ubiquitination and degradation. The observation that three Myc epitopes are sufficient to stabilize ERK3 is in good agreement with the results of Sheaff et al. (40), who showed that addition of Myc3 tag stabilizes expression of p21 by about threefold. Thus, our results support the notion that the E3 ligase active site has a limited range of action and that displacing the NH2 acceptor site can greatly influence the rate of catalysis. Alternatively, the presence of large N-terminal tags may sterically interfere with the binding of the E3 to the substrate. More detailed in vitro studies with purified E3 ligases will allow for discrimination between these possibilities.
It has been previously reported that the cell cycle inhibitor p21 is degraded by the proteasome in a ubiquitin-independent manner (3, 23, 40). This conclusion was based mainly on two sets of arguments. First is the apparent lack of involvement of the ubiquitin conjugation pathway in regulating p21 turnover based on the fact that a lysineless mutant of p21 was degraded at the same rate as wild-type p21 in intact cells. In addition, Sheaff et al. (40) reported that overexpression of a ubiquitin mutant (UbR7) that stabilizes cyclin E expression had no apparent effect on the steady-state levels of p21. However, the half-life of p21 was not measured in UbR7-transfected cells and it is not known if this mutant ubiquitin is used by all classes of E3 ligases with similar efficiency in vivo. Here we used a robust genetic approach with an E1 temperature-sensitive cell line to evaluate the importance of the ubiquitin conjugation pathway in the in vivo degradation of p21. We unambiguously demonstrate that both endogenous and ectopically expressed wild-type or lysineless p21 is degraded in a ubiquitin-dependent manner. The second piece of evidence for ubiquitin-independent degradation of p21 was the inability to detect ubiquitin conjugates of a lysineless p21 mutant in vivo. In contrast to these studies, we clearly detected polyubiquitin conjugates of this mutant. Why was polyubiquitination of p21-0K not detected earlier? Our results reveal the presence of two apparent populations of ubiquitinated species of ERK3 and p21 with high- and low-molecular-size ubiquitin adducts (Fig. 3 and 7). However, we noticed the absence of low-molecular-size ubiquitin conjugates for both the ERK3-0K and p21-0K mutants, whereas higher-molecular-size adducts are detected. Thus, the lack of detection of low-molecular-size ubiquitination species could lead to the wrong conclusion that a lysineless mutant is not ubiquitinated in vivo (for example, see Fig. 7D, middle panel).
The physiological significance of N-terminal ubiquitination remains to be established. In the case of ERK3, accumulating evidence suggests that N-terminal ubiquitination plays a critical role in regulating the effect of the kinase on cell proliferation. In a recent study, it was shown that expression of stable ERK3 chimeras (in which ERK3 degrons were replaced by analogous ERK1 sequences) strongly inhibits S-phase entry in fibroblasts (14). In contrast, expression of unstable wild-type ERK3 protein had no significant effect on cell cycle progression. Notably, it was observed that ectopic expression of Myc6-ERK3 also potently inhibits entry of cells into S phase (24). These observations, together with the findings reported here, suggest that N-terminal ubiquitination is essential to repress the negative regulatory effect of ERK3 on cell cycle progression. However, our results do not exclude the possibility that ubiquitination of internal lysine residues may contribute in some way to the regulation of ERK3 or p21. Indeed, the absence of low-molecular-size ubiquitin adducts in lysineless mutants suggests that these ubiquitin chains are normally conjugated to internal lysine residues. Based on their apparent size, these conjugates may contain one to five ubiquitin molecules, although it is not known if these adducts occur on one or multiple lysine(s). Based on the fact that lysineless mutants of ERK3 and p21 have half-lives very similar to that of the wild-type protein, we conclude that these low-molecular-size ubiquitin conjugates play a minor role, if any, in proteasomal targeting. Modification by mono- or short ubiquitin species may serve functions other than proteolysis, such as regulation of subcellular localization (22).
During the course of this work, Bloom and coworkers (4) have independently shown that p21 is ubiquitinated on its NH2 terminus in vivo. In agreement with the findings reported here, these authors have shown that p21 degradation is dependent on a functional ubiquitin system and that addition of a Myc6 tag stabilizes the protein. However, contrary to what we observed for ERK3, they suggested that the stabilization effect of the Myc6 tag does not result from inhibition of p21 ubiquitination. This discrepancy may be intrinsic to the studied proteins or may result from differences in experimental procedures. The identification of additional proteins ubiquitinated on the NH2 terminus will help clarify this issue.
One of the key issues that needs to be addressed is the identity of the ubiquitin ligase(s) responsible for the polyubiquitination of ERK3 and p21. In the case of p21, different E3 ligases have been implicated in the regulation of its degradation. The SCFSkp2 ligase was found to catalyze the ubiquitination of p21 in vitro and to participate in the degradation of p21 specifically during S phase of the cell cycle (5). SCFSkp2 was also implicated in the rapid degradation of p21 in response to low doses of UV light (3). Consistent with these findings, treatment of cells with antisense oligonucleotides to Cul1, Skp1, or Skp2 leads to the accumulation of p21 (46), and physical complexes of Skp2 and p21 can be found in intact cells (3). However, p21 half-life is unchanged in asynchronous Skp2−/− mouse embryonic fibroblasts (3), and the inhibitor is also unstable in postmitotic cells in the absence of Skp2 expression. These observations argue that additional ubiquitin ligases contribute to the proteasomal degradation of p21 that may be specific to the cell cycle phase or cellular context. The ring finger MDM2 is another E3 ligase that binds to p21 and has been shown to promote its degradation by the proteasome (23).
Acknowledgments
We thank D. Bohmann, G. Ferbeyre, J. Hiscott, H. Ozer, B. Vogelstein, A. J. Waskiewicz, and A. Yoshimura for reagents.
This work was supported by grants from the Canadian Institutes for Health Research. P.C. is the recipient of a studentship from the National Cancer Institute of Canada. G.R. is the recipient of the Anna D. Barker Fellowship of the American Association for Cancer Research. S.M. holds a Canada Research Chair in Cellular Signaling.
REFERENCES
- 1.Baldi, L., K. Brown, G. Franzoso, and U. Siebenlist. 1996. Critical role for lysines 21 and 22 in signal-induced, ubiquitin-mediated proteolysis of I kappa B-alpha. J. Biol. Chem. 271:376-379. [DOI] [PubMed] [Google Scholar]
- 2.Beg, A. A., T. S. Finco, P. V. Nantermet, and A. S. Baldwin, Jr. 1993. Tumor necrosis factor and interleukin-1 lead to phosphorylation and loss of IκBα: a mechanism for NF-κB activation. Mol. Cell. Biol. 13:3301-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bendjennat, M., J. Boulaire, T. Jascur, H. Brickner, V. Barbier, A. Sarasin, A. Fotedar, and R. Fotedar. 2003. UV irradiation triggers ubiquitin-dependent degradation of p21(WAF1) to promote DNA repair. Cell 114:599-610. [DOI] [PubMed] [Google Scholar]
- 4.Bloom, J., V. Amador, F. Bartolini, G. DeMartino, and M. Pagano. 2003. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell 115:71-82. [DOI] [PubMed] [Google Scholar]
- 5.Bornstein, G., J. Bloom, D. Sitry-Shevah, K. Nakayama, M. Pagano, and A. Hershko. 2003. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J. Biol. Chem. 278:25752-25757. [DOI] [PubMed] [Google Scholar]
- 6.Boulton, T. G., S. H. Nye, D. J. Robbins, N. Y. Ip, E. Radziejewska, S. D. Morgenbesser, R. A. DePinho, N. Panayotatos, M. H. Cobb, and G. D. Yancopoulos. 1991. ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell 65:663-675. [DOI] [PubMed] [Google Scholar]
- 7.Breitschopf, K., E. Bengal, T. Ziv, A. Admon, and A. Ciechanover. 1998. A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 17:5964-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cayrol, C., and B. Ducommun. 1998. Interaction with cyclin-dependent kinases and PCNA modulates proteasome-dependent degradation of p21. Oncogene 17:2437-2444. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, M., T. G. Boulton, and M. H. Cobb. 1996. ERK3 is a constitutively nuclear protein kinase. J. Biol. Chem. 271:8951-8958. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, M., E. Zhen, M. J. Robinson, D. Ebert, E. Goldsmith, and M. H. Cobb. 1996. Characterization of a protein kinase that phosphorylates serine 189 of the mitogen-activated protein kinase homolog ERK3. J. Biol. Chem. 271:12057-12062. [DOI] [PubMed] [Google Scholar]
- 11.Chowdary, D. R., J. J. Dermody, K. K. Jha, and H. L. Ozer. 1994. Accumulation of p53 in a mutant cell line defective in the ubiquitin pathway. Mol. Cell. Biol. 14:1997-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clurman, B. E., R. J. Sheaff, K. Thress, M. Groudine, and J. M. Roberts. 1996. Turnover of cyclin E by the ubiquitin-proteasome pathway is regulated by cdk2 binding and cyclin phosphorylation. Genes Dev. 10:1979-1990. [DOI] [PubMed] [Google Scholar]
- 13.Coffino, P. 2001. Regulation of cellular polyamines by antizyme. Nat. Rev. Mol. Cell Biol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 14.Coulombe, P., G. Rodier, S. Pelletier, J. Pellerin, and S. Meloche. 2003. Rapid turnover of extracellular signal-regulated kinase 3 by the ubiquitin-proteasome pathway defines a novel paradigm of mitogen-activated protein kinase regulation during cellular differentiation. Mol. Cell. Biol. 23:4542-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deffenbaugh, A. E., K. M. Scaglione, L. Zhang, J. M. Moore, T. Buranda, L. A. Sklar, and D. Skowyra. 2003. Release of ubiquitin-charged Cdc34-S-Ub from the RING domain is essential for ubiquitination of the SCF(Cdc4)-bound substrate Sic1. Cell 114:611-622. [DOI] [PubMed] [Google Scholar]
- 16.el Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 17.Glickman, M. H., and A. Ciechanover. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82:373-428. [DOI] [PubMed] [Google Scholar]
- 18.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262-267. [DOI] [PubMed] [Google Scholar]
- 19.Halevy, O., B. G. Novitch, D. B. Spicer, S. X. Skapek, J. Rhee, G. J. Hannon, D. Beach, and A. B. Lassar. 1995. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science 267:1018-1021. [DOI] [PubMed] [Google Scholar]
- 20.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 21.Hershko, A., H. Heller, S. Elias, and A. Ciechanover. 1983. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 258:8206-8214. [PubMed] [Google Scholar]
- 22.Hicke, L. 2001. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2:195-201. [DOI] [PubMed] [Google Scholar]
- 23.Jin, Y., H. Lee, S. X. Zeng, M. S. Dai, and H. Lu. 2003. MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. EMBO J. 22:6365-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Julien, C., P. Coulombe, and S. Meloche. 2003. Nuclear export of ERK3 by a CRM1-dependent mechanism regulates its inhibitory action on cell cycle progression. J. Biol. Chem. 278:42615-42624. [DOI] [PubMed] [Google Scholar]
- 25.Liu, C. W., M. J. Corboy, G. N. DeMartino, and P. J. Thomas. 2003. Endoproteolytic activity of the proteasome. Science 299:408-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maki, C. G., and P. M. Howley. 1997. Ubiquitination of p53 and p21 is differentially affected by ionizing and UV radiation. Mol. Cell. Biol. 17:355-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meloche, S., B. G. Beatty, and J. Pellerin. 1996. Primary structure, expression and chromosomal locus of a human homolog of rat ERK3. Oncogene 13:1575-1579. [PubMed] [Google Scholar]
- 28.Pagano, M., and R. Benmaamar. 2003. When protein destruction runs amok, malignancy is on the loose. Cancer Cell. 4:251-256. [DOI] [PubMed] [Google Scholar]
- 29.Parker, S. B., G. Eichele, P. Zhang, A. Rawls, A. T. Sands, A. Bradley, E. N. Olson, J. W. Harper, and S. J. Elledge. 1995. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science 267:1024-1027. [DOI] [PubMed] [Google Scholar]
- 30.Pearson, G., F. Robinson, G. T. Beers, B. E. Xu, M. Karandikar, K. Berman, and M. H. Cobb. 2001. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 22:153-183. [DOI] [PubMed] [Google Scholar]
- 31.Petroski, M. D., and R. J. Deshaies. 2003. Context of multiubiquitin chain attachment influences the rate of Sic1 degradation. Mol. Cell. 11:1435-1444. [DOI] [PubMed] [Google Scholar]
- 32.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 33.Rodier, G., A. Montagnoli, L. Di Marcotullio, P. Coulombe, G. F. Draetta, M. Pagano, and S. Meloche. 2001. p27 cytoplasmic localization is regulated by phosphorylation on Ser10 and is not a prerequisite for its proteolysis. EMBO J. 20:6672-6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez, M. S., J. M. Desterro, S. Lain, D. P. Lane, and R. T. Hay. 2000. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol. Cell. Biol. 20:8458-8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rousseau, D., D. Cannella, J. Boulaire, P. Fitzgerald, A. Fotedar, and R. Fotedar. 1999. Growth inhibition by CDK-cyclin and PCNA binding domains of p21 occurs by distinct mechanisms and is regulated by ubiquitin-proteasome pathway. Oncogene 18:4313-4325. [DOI] [PubMed] [Google Scholar]
- 36.Sasaki, A., K. Inagaki-Ohara, T. Yoshida, A. Yamanaka, M. Sasaki, H. Yasukawa, A. E. Koromilas, and A. Yoshimura. 2003. The N-terminal truncated isoform of SOCS3 translated from an alternative initiation AUG codon under stress conditions is stable due to the lack of a major ubiquitination site, Lys-6. J. Biol. Chem. 278:2432-2436. [DOI] [PubMed] [Google Scholar]
- 37.Scherer, D. C., J. A. Brockman, Z. Chen, T. Maniatis, and D. W. Ballard. 1995. Signal-induced degradation of I kappa B alpha requires site-specific ubiquitination. Proc. Natl. Acad. Sci. USA 92:11259-11263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seger, R., N. G. Ahn, J. Posada, E. S. Munar, A. M. Jensen, J. A. Cooper, M. H. Cobb, and E. G. Krebs. 1992. Purification and characterization of mitogen-activated protein kinase activator(s) from epidermal growth factor-stimulated A431 cells. J. Biol. Chem. 267:14373-14381. [PubMed] [Google Scholar]
- 39.Servant, M. J., P. Coulombe, B. Turgeon, and S. Meloche. 2000. Differential regulation of p27(Kip1) expression by mitogenic and hypertrophic factors: involvement of transcriptional and posttranscriptional mechanisms. J. Cell Biol. 148:543-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheaff, R. J., J. D. Singer, J. Swanger, M. Smitherman, J. M. Roberts, and B. E. Clurman. 2000. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol. Cell. 5:403-410. [DOI] [PubMed] [Google Scholar]
- 41.Tofaris, G. K., R. Layfield, and M. G. Spillantini. 2001. Alpha-synuclein metabolism and aggregation is linked to ubiquitin-independent degradation by the proteasome. FEBS Lett. 509:22-26. [DOI] [PubMed] [Google Scholar]
- 42.Treier, M., L. M. Staszewski, and D. Bohmann. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787-798. [DOI] [PubMed] [Google Scholar]
- 43.Turgeon, B., B. F. Lang, and S. Meloche. 2002. The protein kinase ERK3 is encoded by a single functional gene: genomic analysis of the ERK3 gene family. Genomics 80:673-680. [DOI] [PubMed] [Google Scholar]
- 44.Verma, R., and R. J. Deshaies. 2000. A proteasome howdunit: the case of the missing signal. Cell 101:341-344. [DOI] [PubMed] [Google Scholar]
- 45.Wu, G., G. Xu, B. A. Schulman, P. D. Jeffrey, J. W. Harper, and N. P. Pavletich. 2003. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol. Cell. 11:1445-1456. [DOI] [PubMed] [Google Scholar]
- 46.Yu, Z. K., J. L. Gervais, and H. Zhang. 1998. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21(CIP1/WAF1) and cyclin D proteins. Proc. Natl. Acad. Sci. USA 95:11324-11329. [DOI] [PMC free article] [PubMed] [Google Scholar]