Abstract
A major component of the vessel wall of large arteries and veins is the extracellular matrix (ECM), which consists of collagens, elastin, and proteoglycans. Collagen type I is one of the most abundant of the ECM proteins. We have previously shown that the pro-collagen type I alpha 2 gene contains an enhancer which confers tissue-specific expression in the majority of collagen-producing cells, including blood vessels. In this paper, we delineate a specific vascular smooth muscle cell (vSMC) element: a 100-bp sequence around −16.6 kb upstream of the transcription start site that regulates collagen expression exclusively in vSMCs. Furthermore, we show that the expression is activated through the binding of the homeodomain protein Nkx2.5, which is further potentiated in the presence of GATA6. In contrast, this element was repressed by the binding of the zinc-finger protein δEF1/ZEB1. We propose a model of regulation where the activating transcription factor Nkx2.5 and the repressor δEF1/ZEB1 compete for an overlapping DNA binding site. This element is important in understanding the molecular mechanisms of vessel remodeling and is a potential target for intervention in vascular diseases where there is excessive deposition of collagen in the vessel wall.
Blood vessels deliver oxygen, nutrients, and immune cells to all the tissues of the body. The walls of large vessels are composed of three layers (tunicae), which contain different cell types embedded in an extracellular matrix (ECM). The tunica intima is located nearest the lumen and is lined by endothelial cells. The tunica media is the elastic and contractile layer composed of highly ordered vascular smooth muscle cells (vSMCs). The tunica adventitia is the outer layer and is primarily composed of connective tissue and resident fibroblasts. The ECM in blood vessels consists of collagens, elastin, fibrillins, proteoglycans, and others. Collagen type I is the most abundant of the collagen fibrils and is found in all three tunicae and especially around the smooth muscle cells of the media, where they provide the necessary mechanical strength and contractility. Collagen type I is also found in the outer layer (adventitia), where it forms large bundles of fibrils which increase progressively in size from its innermost component, closest to the media, to its outermost aspect (24). Collagen type I is a fibrillar protein consisting of two alpha 1 chains (pro-col1a1) and one alpha 2 chain (pro-col1a2) coiled together into a triple helix (20). Collagen type I fibers provide the structural support for all tissues and organs of the body and play a critical role in morphogenesis and growth, as well as homeostasis and repair. Little is known about the regulation of collagen type I expression in blood vessels and especially the transcriptional control of this process either during development or in adult tissue. Although the pro-collagen type I chains are transcribed from two different chromosomes, their expression is coordinately regulated at the transcriptional level (46). In the pro-col1a1 gene, a close homology exists between human and mouse promoters. Brenner and colleagues demonstrated by progressively deleting the mouse pro-col1a1 gene from its 5′ end that sequences downstream of −181 bp are needed for high-level transient transfection (4). However, in transgenic mice, the mouse minimal proximal promoter (−220 to +110 bp) is silent (30), showing that it is not sufficient to drive expression in vivo. By increasing the length of this promoter, evidence from different groups using human, rat, and mouse promoters showed that the first 3.6 kb drive expression in skin, tendon, bone, and teeth (2, 18, 23, 35). Indeed, the work on the mouse 3.6 kb has further characterized these elements. An element directing expression in the skin was between −220 and −900 (30). The bone element was delineated to 117 bp between −1656 and −1540 (31), and an element with tendon specificity was found to be a combination of sequences between −3.2 and −3.6 kb that have been termed TSE1 and TSE2 (43). From these experiments, a modular arrangement of separate cis-acting elements that activate the pro-col1a1 gene in different type I collagen-producing cells was hypothesized. In contrast, examination of the pro-col1a2 gene proximal promoter found no such modular arrangement of regulatory sequences in the proximal region, which only directed weak reporter gene expression in transgenic mice (21). However, expression was significantly increased with the identification of an enhancer region located around −17 kb away from the transcription site. Detailed analysis using transgenesis revealed that this enhancer sequence conferred a high level of expression in almost all collagen type I-producing cells, including those of the blood vessels (3).
We hypothesized, based on the pro-col1a1 data, that there must be a separate cis-regulatory element(s) within the pro-col1a2 enhancer sequence which controls the expression of collagen type I in mesenchymal cells, including vSMCs, during embryonic development. Such an element(s) may be important not only in the development of blood vessels but also in adult vessels during remodeling, repair, and vascular pathology, such as atherosclerosis and pulmonary hypertension, where excessive levels of matrix proteins are deposited within the vessel (reviewed in references 17 and 29). With this in mind, our aim was to delineate the element that confers expression in vSMCs and elucidate the mechanism of its regulation by identifying the transcription factors that bind the DNA within this element.
In this study, we present data that identify, for the first time, a vSMC-specific collagen type I regulatory element located within the pro-col1a2 enhancer region. We have determined the minimal sequence within this enhancer able to drive the expression of a reporter gene exclusively in vSMCs. Furthermore, we show that the homeodomain transcription factor Nkx2.5 activates this sequence and that the zinc-finger transcriptional repressor δEF1 (δEF1/ZEB1) inhibits transcriptional activation, by a mechanism involving competition for an overlapping DNA binding site, which characterizes the vSMC-specific sequence.
MATERIALS AND METHODS
DNA constructions.
The clone λ3-3 derived from a λ FIX II library contained 5′-flanking sequences of the pro-col1a2 collagen gene from kb −23.3 to −11.0 relative to the transcriptional start site. pLacRM350 (construct I) and pLacES350 vectors contained the mouse pro-col1a2 gene minimal promoter (bp +54 to −350) 5′ of the LacZ gene (10). Constructs II (pGB −17.0/−13.5), III (pGB −17.0/−16.4), and VI (pGB −16.4/−13.5) were previously described (3). All of the smaller constructs were generated by using proofreading Pfu DNA polymerase (Stratagene, La Jolla, Calif.), using construct II as template and with various primers (designed to include sites for endonucleases XhoI and NotI). The PCR fragments were originally cloned into TOPO-Blunt vector (Invitrogen), sequenced, and then excised and inserted into the pLacES expression vector. pMP −16.8/−16.5 (construct V) contains the 364-bp fragment from kb −16.82 to −16.476. pMP −16.7/−16.5 (construct VI) contains the 202-bp fragment from kb −16.698 to −16.476. pMP −16.65/−16.5 (construct VII) contains the 149-bp sequence between kb −16.644 and −16.476. pMP −16.6/−16.5 (construct VIII) contains the 140-bp sequence between kb −16.616 and −16.476. pMP −16.7/−16.55 (construct IX) contains the fragment which extends from kb −16.698 to −16.576 (120 bp). pMP −16.65/−16.55 (construct X) contains sequences between kb −16.644 and −16.576 (70 bp).
Mutant constructs were synthesized using a PCR approach. Primers (5′-CTCGAGCAGGTACTCCATGTG-3′, 5′-CTCGAGCAGAATCACCATGTG-3′, and 5′-GCGGCCGCATAATAACCACTTTGTAGT-3′) were designed for the pdEF1WT, pdEF1mut 1, and pdEF1mut 2 constructs, respectively. These were amplified as the wild-type sequences described above and cloned in the pLacES350 vector. Complementary oligonucleotides with appropriate overhangs containing point mutations were synthesized and annealed before being cloned into the pLacES350 vector. For the pMP −16.65/−16.55 Nkx mutant construct, the following sequence was synthesized: 5′-TCGAGAGTCTTGTGGATTTCTCCACAACCTAATGGGTTAGGATGCTGACCCGAGGGTGTGCCATTGGTGGGA-3′ (with mutations underlined). Expression vectors for rat Nkx2.5, mouse Nkx2.5, pcmvDef1, and pGATA4/GATA6 were kind gifts from S. Evans, R. Harvey, H. Kondoh, and R. Schwartz, respectively.
Cell lines.
All cells were cultured in Dulbecco's modified Eagle's medium (Life Technologies) supplemented with 10% heat-inactivated fetal calf serum and 2 mM l-glutamine. A7r5, smooth muscle cells derived from the thoracic aorta of an embryonic BDIX rat, were obtained from the European Collection of Animal Cell Culture. Human vSMCs (HvSMCs) were obtained from human circumflex coronary artery by explant outgrowth as described in reference 13 (kind gift of the Department of Cardiothoracic Surgery, National Heart and Lung Institute, Royal Brompton and Harefield Hospitals) or from the human aorta (TCS Cellworks). ROS17/2, a rat osteosarcoma cell line, was used. The EL4 cells are a mouse lymphoblast cell line grown in suspension (European Collection of Animal Cell Culture). L929 is a mouse fibroblastic cell line derived from subcutaneous connective tissue of a C3H/An mouse (European Collection of Animal Cell Culture).
Transient transfections.
Transient-transfection assays were performed using the FuGENE 6 transfection reagent according to the manufacturer's instructions. Briefly, various cell lines (2 × 105 cells per transfection mixture) were cultured in vitro and transfected with 2 μg of total plasmid DNA and 6 μl of FuGENE 6 reagent. pRSVLuc was cotransfected (0.2 μg) to check the efficiency of transfection. The reagent was left on the cells and harvested 72 h posttransfection using lysis buffer. All transfections were performed in triplicate within each experiment, and each experiment was repeated on more than five separate occasions.
The activities of reporter genes, beta-galactosidase and luciferase, used in the transient-transfection assays were measured using the Dual-Light chemiluminescent reporter gene assay system (TROPIX) according to the manufacturer's instructions.
EMSAs.
Nuclear extracts were prepared from 25 large flasks of A7r5 cells according to a method described elsewhere (1). The nuclear extracts were used in electrophoretic mobility shift assays (EMSAs) according to the method described in reference 16. As Nkx2.5 and δEF1 are zinc-finger proteins, zinc chloride (2 mM final concentration) was added to the binding reaction mixture. Super-shift assays were carried out using specific antibodies raised against Nkx2.5, δEF1 (ZEB), and GATA6 (all from Santa Cruz). Double-stranded oligonucleotides (70 bp) were synthesized and used for EMSAs. The wild-type sequence was mutated at key residues in order to corrupt specific sites (described below) as follows: 5′-GTTCCTCCATGTGTTTAGTTAAGGACCTCATGAGTCTTGTGGATTTCTCCACAACCTAAGTGGTTATT-3′, the wild-type sequence; 5′-GTTCCTCCATGTGTTTAGTTAAGGACCTCATGAGTCTTGTGGATTTCTCCACAACCTAATGGGTTATTATG-3′, the mutant Nkx2.5 site d; and 5′-GTTCCTCCATGTGTTTAGTTAAGGACCTCATGAGTCTTGTGGATTTCTCCACACACTAAGTGGTTATT-3′, the mutant δEF1 overlapping site.
RT-PCR.
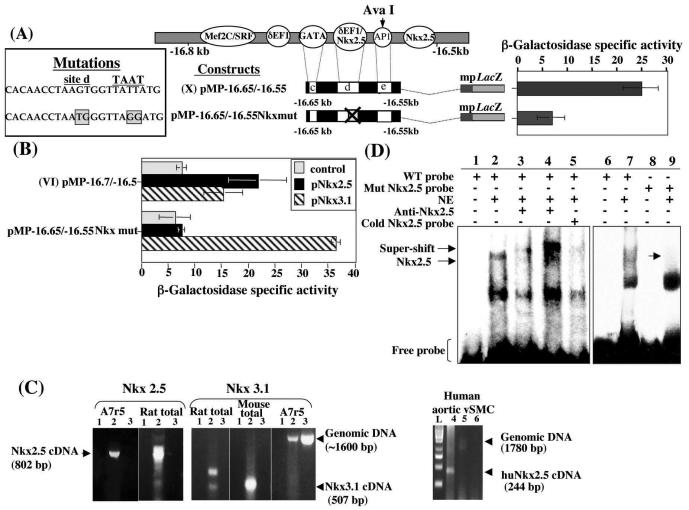
RNA was extracted from cells (107) grown in 10-cm petri dishes according to the methods described in reference 8. Reverse transcription-PCRs (RT-PCRs) were performed using the QIAGEN OneStep RT-PCR kit according to the manufacturer's instructions. Primers were designed for mouse Nkx3.1 (accession number U88542), giving rise to a 507-bp RT-PCR product (forward primer, base 3 to 22, 5′-GCGGAGGAGGGATGCTTA-3′; reverse primer, base 490 to 509, 5′-CGGTGAGTTTGAGGTTCTTG-3′) or an approximately 1,600-bp genomic DNA product. The primers designed for the Nkx2.5 RT-PCR (forward primer, from base 5677 to 5696, 5′-TCCCAGCCGCCCCCACATTT-3′; reverse primer, from base 7837 to 7856, 5′-CGAGGGACAGGGCATAGTGG-3′) were designed using the mouse sequence (accession number AF083133) but were identical to the rat sequence. The resulting RT-PCR product was 802 bp in length, while a genomic DNA would lead to a 2,180-bp product.
The primers for the RT-PCR carried out in human aortic vSMCs were obtained from the human Nkx2.5 sequence (accession number NT023133; forward primer, from base 805 to 824, 5′-CAAGTGTGCGTCTGCCTTTC-3′; reverse primer, from base 2584 to 2602, 5′-GGTACCGCTGCTGCTTGAA-3′) and gave rise to an RT-PCR product of 243 bp and a genomic DNA product of 1,798 bp.
Briefly, RT was performed at 50°C for 30 min in the presence or absence of RNA template (2 μg). A further control was performed by the addition of RNA template after the RT step, during the PCR activation step (15 min at 95°C). A total of 35 cycles of PCR were performed, with denaturation for 30 s at 94°C, annealing for 1 min at 58 to 60°C depending on the primers, and extension for 1 min at 72°C. The final extension was carried out for 10 min at 72°C. For the δEF1, RT-PCR was carried out on A7r5 mRNA as described above using two primers designed from the mouse sequence (accession number NM_011546; forward primer, from base 2443 to 2462, 5′-CGCCAACAAGCAGACTATTC-3′; reverse primer, from base 2729 to 2748, 5′-TGAGGCCTCTTACCTGTGTG-3′). These primers and PCR conditions were previously described (47).
RESULTS
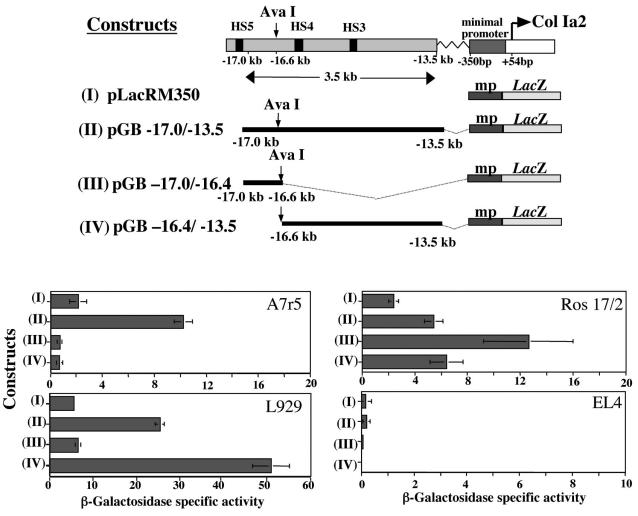
In order to identify the sequences which contained a vSMC-specific regulatory element, plasmid constructs were generated harboring different sequences within the 3.5-kb enhancer region between −13.3 and −17.0 kb upstream of the transcription start site, which has been shown in transgenic mice to induce reporter gene expression in almost all collagen-producing cells, including vSMCs in vessels (3). These enhancer sequences were placed 5′ of the proximal pro-col1a2 promoter (350 bp), driving the expression of the β-galactosidase reporter gene, and were transiently transfected in different cell lines. Figure 1 shows a diagrammatic representation of the full enhancer (construct II) and the pro-col1a2 proximal promoter (350 bp) alone (construct I). A convenient restriction site at −16.6 kb (AvaI) divided the 3.65-kb enhancer into two fragments of 0.4 kb (construct III) and 3.1 kb (construct IV).
FIG. 1.
Diagrammatic representation of the far-upstream enhancer sequence required for expression in most of the collagen-expressing cell lineages. Constructs I to IV were generated using fragments of the enhancer-promoter region fused to the LacZ reporter gene. Transient-transfection assays showed differential expression of the constructs used in various cell lineages. In the vSMC line A7r5, construct II was most active, whereas in L929 fibroblasts construct IV was more active. In Ros17/2 osteoblasts, construct III showed the highest activity. Control, non-collagen-expressing EL4 lymphocytes did not show any beta-galactosidase reporter gene activity. HS, hypersensitive site.
Among many collagen-expressing cell lines used were the vSMC line A7r5, the embryonic mouse fibroblast cell line L929, and the osteoblast cell line Ros17/2. The non-collagen-expressing lymphocyte cell line EL4 was used as a control cell line. The transgene was expressed in all cell lines (A7r5, L929, and Ros17) except the non-collagen-producing cell line, EL4. β-Galactosidase activity was enhanced by the addition of the 3.5-kb far-upstream enhancer. The 0.4-kb fragment (construct III) showed elevated β-galactosidase activity in the osteoblast cell line (Ros17/2), suggesting that it may contain important sequences for collagen expression in this cell type. In contrast, this construct produced very low β-galactosidase activity in the fibroblast L929 cell line, whereas the 3.1-kb fragment, construct IV, conferred the highest levels of β-galactosidase activity, suggesting that the 0.4-kb 5′ region of the enhancer is not involved in the regulation of pro-col1a2 in this cell type.
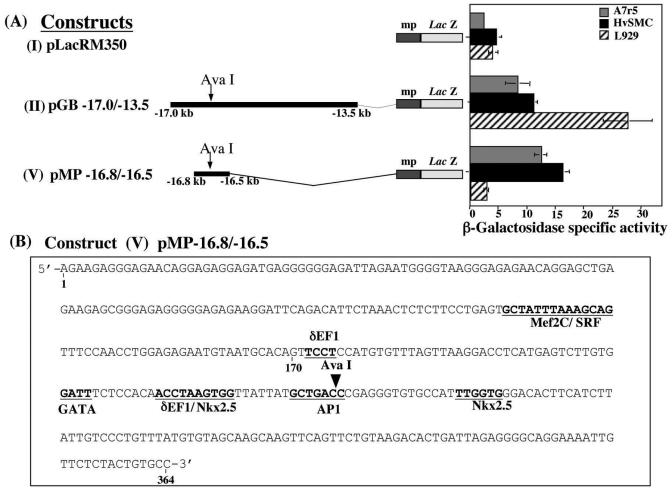
Of greater interest to this study are the data obtained from the vSMC line A7r5, showing that neither the 0.4-kb fragment (construct III) nor the 3.1-kb fragment (construct IV) generated any β-galactosidase activity compared with the full enhancer sequence. On the basis of these results, we hypothesized that in vSMCs, the enhancer-specific cis-acting sequence is disrupted by the AvaI restriction enzyme digest. To test this, we generated construct V, a 364-bp construct which includes the AvaI restriction enzyme site (Fig. 2A). This was fused to the minimal promoter and the LacZ reporter gene as with the rest of the constructs. Transient transfection in A7r5 cells, HvSMCs, and L929 cells produce specific increases of beta-galactosidase activity in A7r5 and HvSMCs (Fig. 2A), but not in the L929 fibroblasts or any of the other cell lines (data not shown), suggesting a specificity of this regulatory sequence in vSMCs.
FIG. 2.
(A) Transient transfections in A7r5, HvSMC, and L929 fibroblast cells showed the similarities in reporter gene activities in primary HvSMCs and in A7r5 cells. The 364-bp sequence located 16.8 kb upstream of the transcriptional start site of pro-col1a2 overlapping the AvaI restriction enzyme site was exclusively active in vSMCs. (B) Putative transcription factor binding sites within the 364-bp enhancer sequence are shown.
Delineation of the vSMC element.
The 364-bp DNA sequence surrounding the AvaI site (construct V) was analyzed using a transcription factor site program (Transfac/Matinspector) (27), which revealed several putative binding sites (Fig. 2B). Among these were muscle-specific transcription factor Mef2C, a serum response element (SRE), a delta crystalin elongation factor 1 (δEF1/ZEB1), GATA, and the homeobox transcription factor Nkx2.5, as well as activation protein 1 (AP1).
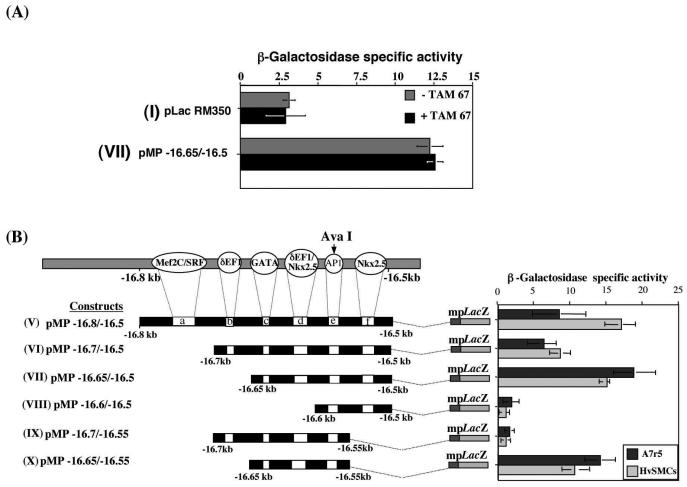
Since the AP1 site overlapped the Ava1 site, we tested whether the disruption of the AP1 binding site had an effect on the vSMC element. Transient cotransfections using a trans-activation domain mutant of AP1 (c-Jun; TAM67) (15) with construct VII resulted in no change in their overall activities in vitro (Fig. 3A). A functional role of AP1 in vitro would have predicted a decrease in overall activity.
FIG. 3.
(A) Histogram showing that the AP1 dominant-negative expression vector (TAM67) cotransfected in A7r5 cells with the active construct VII had no effect. (B) Series of deletion constructs (V to X) where one or a combination of putative transcription factor binding sites was removed. Data (mean ± the standard error of the mean) from several transient-transfection experiments in A7r5 cells and HvSMCs are shown (n = 10).
In order to define the minimal functional sequence which drives the vSMC element, smaller deletion constructs were made and tested in transient-transfection assays to ascertain the importance of each of the putative binding sites in vSMCs (Fig. 3B). In this experiment, the transfections were repeated on HvSMCs. Removal of the Mef2C/SRE site, construct VI, produced a small decrease in beta-galactosidase activity in A7r5 cells and HvSMCs. This decrease in activity was not statistically significant (P > 0.05) in A7r5 cells but was significant in HvSMCs (P < 0.01), suggesting that the Mef2C/SRE site may be species dependent but not essential for the overall regulation of this element. However, when both the Mef2C/SRE (site a) and the δEF1/ZEB1 (site b) sites were removed in construct VII, beta-galactosidase activity was significantly increased (P < 0.001) in both A7r5 cells and HvSMCs. Taken together, these results imply that while the Mef2C/SRE site may not be functionally essential in the vSMC element, the δEF1/ZEB1 site exerts a repressive effect on reporter gene activity, since its removal leads to significantly increasing activity levels compared with those of construct VI (P < 0.001). Removal of site d, which contains the putative overlapping δEF1/ZEB1 and Nkx2.5 sites in construct VIII, caused the complete loss of reporter gene activity in both A7r5 cells and HvSMCs, indicating the potential importance of this site within the vSMC element in pro-col1a2 expression.
Moreover, the data obtained from transfections with construct VIII suggested that the second Nkx2.5 (site f) cannot drive activation in isolation but did seem to cooperate with the main activation site (site d). Construct X was still active in vitro in vSMCs despite the absence of site f, though it was significantly less than construct VII (P < 0.01). These results were also supported by the data obtained with constructs VI and IX. Construct VI, although active, is partially repressed by δEF1/ZEB1 binding. However construct IX, which only differs from construct VI by the absence of site f, is significantly less active (P < 0.001). All the constructs described above are inactive in the fibroblastic cell line L929 (data not shown).
δEF1/ZEB1 represses the vSMC element.
The transient-transfection data presented above implied that the binding of transcription factor to site b has an overall repressive effect on reporter gene expression in vSMCs. Sites b and d have been identified as putative binding sites for zinc-finger DNA binding proteins of the δEF1/ZEB1 family (25, 26, 28, 34). These contain two zinc-finger clusters, each binding independently to a C(A/T)CCT(G) sequence in a bipartite manner, with the separating distance and orientation between the two sequences being variable (28). The objective in the following experiment was to determine whether sites b and d were functional binding sites for δEF1/ZEB1 and whether they were part of the transcriptional regulatory mechanism of the vSMC element.
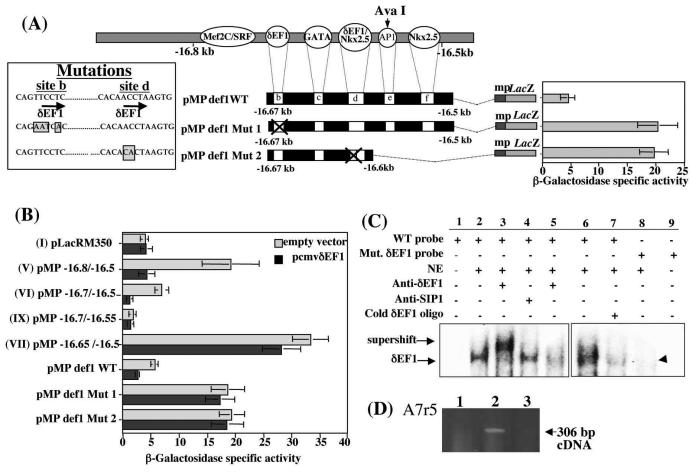
Point mutations were introduced in either site b or site d using a PCR-based method (Fig. 4A) in order to abolish possible binding of δEF1/ZEB1. The mutations in site d, which includes an overlapping putative Nkx2.5 binding site, were designed to alter only the δEF1/ZEB1 site, leaving the putative Nkx2.5 site unchanged. According to the reported mechanism of binding of δEF1/ZEB1, the destruction of either sequence on which the zinc-finger clusters can bind should prevent association of δEF1/ZEB1 with the DNA (28). The data obtained using mutant constructs in A7r5cells, pMPdef1 Mut 1 and pMPdef1 Mut 2, clearly showed that the repression of reporter gene activity observed with constructs VI and pMP def1WT was alleviated when either site b or site d was mutated (Fig. 4A).
FIG. 4.
Mutagenesis of putative δEF1/ZEB1 sites leads to the abolition of repression and confirms the requirement of both δEF1/ZEB1 sites for repression. Point mutations were introduced in the putative δEF1/ZEB1 sites (b and d) to generate constructs (pMPdef1 Mut1 and pMPdef1 Mut2) and used in transient transfections. (B) Effect of δEF1/ZEB1 mutant constructs in the presence of an overexpression vector (pcmv δEF1). (C) A 70-bp double-stranded oligonucleotide starting on base 170 of the sequence presented in Fig. 2B and containing the two putative δEF1/ZEB1 consensus sites b and d was used in an EMSA (lanes 1 to 5) in the presence or absence of A7r5 nuclear extracts. The bound probe-DNA complex was incubated with an anti-δEF1/ZEB1-specific antibody (lane 3) and with anti-SIP1-specific antibody (lane 4). Preincubation of the A7r5 nuclear extract with the δEF1/ZEB1 antibody for 15 min prior to the addition of the labeled probe is shown in lane 5. The wild-type (WT) probe control was analyzed in the absence (lane 1) and presence (lanes 2 and 6) of nuclear extracts. Competition was performed using a 10 M excess of unlabeled double-stranded consensus oligonucleotide for δEF1/ZEB1 (lane 7). The same 70 bp of the core sequence with a mutation in the putative δEF1/ZEB1 consensus site d abolished binding (lanes 8 and 9). (D) RT-PCR showed the presence of δEF1/ZEB1 in A7r5 cells. Controls included no reverse transcriptase (lane 1) and water instead of A7r5 mRNA (lane 3).
Cotransfections of the wild-type and mutant constructs with a pCMVdEF1 overexpression vector were also carried out in A7r5 cells. In the presence of pCMVdEF1, the normally active construct V was repressed, as was the control construct pMPdef1WT (Fig. 4B). Constructs VI and IX, which contain both putative δEF1/ZEB1 sites (sites b and d), are already repressed, and the additional δEF1/ZEB1 expression had little effect, although their activity was lower. Additional δEF1/ZEB1 expression did not alter the activity levels of construct VII (which contains only site d) or the levels of the two mutant constructs pMPdef1 Mut 1 and pMPdef1 Mut 2 (Fig. 4B). These data supported our hypothesis that δEF1/ZEB1 repressed the active pro-col1a2 vSMC element.
To confirm that δEF1/ZEB1 actually binds to the DNA sequence, we carried out DNA EMSAs. Figure 4C shows that the incubation of the A7r5 nuclear extracts with wild-type probe followed by incubation with a δEF1-specific antibody led to a supershift of the δEF1/ZEB1-DNA complex (lane 3). In addition, when Smad-interactive protein 1 (SIP1) antibody was incubated with the binding mixture, no effect was observed on the δEF1/ZEB1-DNA complex (lane 4). SIP1 belongs to the same family of zinc finger transcription factors and is known to preferentially bind the same DNA sequence (CACCT).
Depletion of δEF1/ZEB1 by preincubation of nuclear extracts with CACCT fragments prior to the addition of the labeled probe led to a decrease in the intensity of the δEF1/ZEB1 complex (lane 5). In addition, cold competitor oligonucleotides of the putative δEF1/ZEB1 (site b) successfully competed for the formation of the labeled probe-δEF1/ZEB1 complex (lane 7). In control experiments when a probe (in which the putative δEF1/ZEB1 site d had been mutated) was used, the DNA-δEF1 complex was completely abolished (lane 8). Similar results were obtained with the probe in which the δEF1/ZEB1 site b was mutated (data not shown). The presence of δEF1/ZEB1 in vSMCs and specifically in the A7r5 line has never been formally demonstrated. Therefore, RT-PCR was carried out on mRNA obtained from A7r5 cells and using primers already published (47), and it showed that δEF1/ZEB1 is expressed in this cell line (Fig. 4D).
Activation of the vSMC element.
Having shown that the vSMC element was repressed by δEF1/ZEB1 binding to the DNA at sites b and d, we focused on the transcription factors that are involved in the activation of this element. Transient-transfection data presented above in Fig. 3 showed that both constructs VII and X have the highest levels of reporter gene activity in vSMCs. However, construct X does not include site f (a putative Nkx2.5 binding site), but it does contain site d, a putative site for the Nkx2.5 homeodomain transcription factor which has been shown to be involved in the regulation of cardiovascular genes (41). However, other homeodomain NK family members are capable of binding the same sequence, including Nkx3.1 (5), which is also expressed in vessels (42) and has been shown to be involved in the transcriptional regulation of vSMC genes (5).
To verify the involvement of Nkx2.5, a mutant construct was generated, pMP−16.65/−16.55 Nkx mut 1, which has four point mutations of essential bases which impair Nkx2.5 binding (two in the Nkx2.5 consensus site and two in the TAAT box). The TAAT box is a complementary site which, together with the Nkx2.5 consensus site, forms the Nkx2.5 response element (7). The mutant construct has intact CACTTAG sequences and can, therefore, bind Nkx3.1 but not Nkx2.5. Transient transfection using this construct (Fig. 5A) clearly showed that when the consensus binding Nkx2.5 sites were mutated, the levels of reporter gene activity decreased significantly compared with that of the wild type (P < 0.01). Furthermore, when the Nkx2.5 site mutant construct was cotransfected with an Nkx2.5 overexpression vector, it remained inactive (Fig. 5B). In contrast, when an Nkx3.1 overexpression vector was added, the expression of the reporter gene returned to its highest level. These data suggested that, although Nkx3.1 can activate the vSMC element in vitro, it may not be binding in the A7r5 cell line. Indeed, RT-PCR of A7r5 mRNA using specific RT-PCR primers for Nkx2.5 and Nkx3.1 showed that Nkx2.5 but not Nkx3.1 mRNA was present in the A7r5 cell line (Fig. 5C). Moreover, primary human aortic SMCs also expressed Nkx2.5 (Fig. 5C).
FIG. 5.
Nkx2.5 activates the vSMC element. (A) Point mutations in the putative Nkx2.5 (site d) and TAAT box effectively abolished Nkx2.5 binding and transient activity compared with that in construct X. (B) Construct VI in the presence of pNkx2.5 and pNkx3.1 overexpression vectors (0.5 μg of each) can activate the vSMC element despite the presence of δEF1/ZEB1. Mutated Nkx2.5 blocks the activity of Nkx2.5 but not of Nkx3.1. (C) RT-PCR on mRNA isolated from A7R5 cells showed an 802-bp PCR cDNA product for NKx2.5 in A7r5 mRNA (lane 2), the controls lacking reverse transcriptase (lane 3), and water substituted for cDNA (lane 1). The rat total RNA confirmed the 802-bp product. No Nkx3.1 (507-bp) product was observed in A7r5, although the 1,600-bp genomic PCR product was seen in lane 2. RT-PCR was also carried out on mRNA from primary human aortic vSMCs. The 244-bp cDNA product (lane 4) showed Nkx2.5 expression in these cells, while only a faint product (1,780 bp) was observed in the genomic control (lane 5). Lane 6 shows the control, in which water was substituted for cDNA. L, the DNA ladder (GeneRuler 100-bp ladder) for size reference. (D) A 70-bp double-stranded oligonucleotide starting from base 170 of the sequence presented in Fig. 2B containing the putative Nkx2.5 (site d) (lanes 1 to 5) was used as a radiolabeled probe in an EMSA with nuclear extracts from A7r5 cells. Two probe-DNA complexes were observed (lane 2). Preincubation of nuclear extract with an Nkx2.5-specific antibody prior to the addition of the radiolabeled probe diminished binding of the probe-DNA complexes (lane 3). A supershift of the upper band was observed when Nkx2.5-specific antibody was incubated with the probe-DNA complex mixture after it had been formed (lane 4). Competition using a 5 M excess of unlabeled double-stranded consensus oligonucleotide for Nkx2.5 is shown in lane 5. The same 70-bp core sequence with a TAAGTG motif mutated in site d to TAATGG (lanes 8 and 9) can be compared to the wild-type (WT) probe (lanes 6 and 7).
In order to confirm that site d is a functional Nkx2.5 binding site, EMSAs were carried out using 32P-labeled double-stranded oligonucleotides (Fig. 5D). The wild-type probe was incubated in the presence of A7r5 nuclear extracts (lanes 2 and 7). Nkx2.5 present in the nuclear extract was then depleted by preincubation with anti-Nkx2.5-specific antibody prior to the addition of the labeled probe (lane 3). It was clear that only one of the DNA-protein complexes had diminished in intensity. This same complex was supershifted by the addition of the Nkx2.5-specific antibody to the DNA-protein mixture (lane 4). This supershift was not seen with Nkx3.1-specific antibody (data not shown). The addition of a cold competitor oligonucleotide for the Nkx2.5 site resulted in the abolition of the putative Nkx2.5-probe complex (lane 5). In order to confirm the presence of the Nkx2.5-probe complex, another probe was made which had two point mutations within the putative Nkx2.5 site, resulting in the corruption of this site. EMSAs carried out using this probe resulted in the absence of only the putative Nkx2.5-probe complex (lane 9).
Synergy between Nkx2.5 and GATA6.
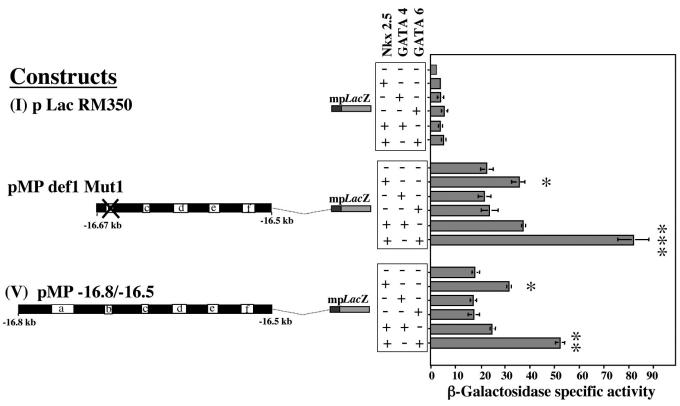
The regulatory sequence reported in this study contains a putative GATA binding site, although it is a noncanonical site (GATT). This putative GATA site is present in the 5′ end of construct VI, but it is notably absent from constructs VII and X. Using overexpression vectors for GATA4 and GATA6, we carried out cotransfection experiments with construct I, pMPdef1 Mut 1, and construct V. The data obtained in Fig. 6 revealed that only GATA6 cooperated with Nkx2.5 to enhance reporter gene activity.
FIG. 6.
Synergy between GATA6 and Nkx2.5 in the activation of the vSMC element. Transient transfections were carried out in A7r5 cells using constructs I, V, and pMPDef1 Mut1. Cotransfections with expression vectors for Nkx2.5, GATA4, and GATA6 (all added at 1 μg) showed significant increases in reporter gene activity when the Nkx2.5 and GATA6 transcription factors were added together. *, P < 0.01; **, P < 0.005; ***, P < 0.001.
DISCUSSION
This paper presents evidence for a specific vSMC element in the mouse pro-col1a2 enhancer located approximately −16.6 kb upstream of the transcription start site. It also supports the idea that transcriptional regulation by this element is achieved by a finely tuned repression-activation mechanism in which the repressing transcription factor δEF1/ZEB1 and the activating homeodomain factor Nkx2.5 compete for an overlapping site.
The first piece of evidence for a specific vSMC regulatory element was provided by transient-transfection data in various cell lines. The interruption of a sequence at an AvaI (−16.6 kb) site resulted in the loss of reporter gene expression only in vSMCs. Further investigation confirmed that this sequence was exclusively activated in A7R5 cells and human primary vSMCs. Deletion experiments carried out on both the 5′ and 3′ ends of the sequence identified a core functional sequence located 12 bp upstream of the AvaI site and spanning a further 50 bp upstream. In silico analysis of this sequence provided putative transcription factor binding sites, which were investigated further individually and in combination for their potential role in the regulation of this element.
One of these putative transcription factors is δEF1/ZEB1, a DNA binding protein containing two zinc-finger clusters separated by a homeodomain. Each of the clusters binds independently to a 5′-CACCT or 5′-CCTCCT sequence, both of which are necessary for binding, and if either is mutated binding is abolished (28, 34, 36).
In this paper we have identified two functional δEF1 sites, 43 bp apart, within the core sequence. Together, these sites repress reporter gene activity in transient transfections of vSMCs, and if either site is mutated, repression of reporter gene activity is lost. Moreover, we have provided evidence for the presence of endogenous δEF1 expression in nuclear extracts of vSMCs and showed that δEF1 binds to the naked DNA containing the core sequence. This protein complex bound to the DNA sequence was further characterized by antibody raised against δEF1. The protein-DNA complex did not contain other members of the δEF1/ZEB-1 transcription factor family, such as SIP1 (ZEB-2), which is also a repressor and binds to the same DNA motif (28). Thus, δEF1/ZEB-1 is the primary candidate for the repression of the pro-col1a2 element in vSMCs. It is of interest in this respect that the pro-col1a1 gene was shown to be negatively regulated by δEF1/ZEB-1 in osteoblasts (44). Expression of other collagen genes, namely, col2a1 (19) and col11a2, in cartilage also appears to be repressed by δEF1/ZEB-1 (40).
The 3′ δEF1 site within this element overlaps another putative transcription factor binding site belonging to the NK-2 homeodomain family (14), which includes several members which are highly conserved across vertebrate species, notably Nkx2.5 (32) and Nkx3.1, which have been reported to be expressed in vessels (42). Our data provide evidence that the homeodomain transcription factor Nkx2.5 binds directly to the vSMC-specific col1a2 enhancer sequence, resulting in the activation of the reporter gene. Nkx2.5 is a homeobox gene that has been identified as the vertebrate homologue of tinman, and it is required for drosophila and mammalian cardiac development (reviewed by Harvey [14] and Schwartz and Olson [32]). It is also involved in the transcriptional regulation of vascular smooth muscle genes (11, 48). However, Nkx2.5 expression has not been reported in vSMCs. Nkx2.5 null mice die between embryonic days 9.5 and 11.5 embryonic development from arrest of cardiac development after cardiac looping and poor development of blood vessels (41). In this study, Nkx2.5 but not Nkx3.1 expression was found in the A7r5 cell line by using RT-PCR. Interestingly, a combination of transfection and mutation analysis showed that Nkx3.1 is able to activate the element when cotransfected in the cells but that it is not the endogenous activator, since EMSA data showed a supershift when using anti-Nkx 2.5 antibodies but not with anti-Nkx3.1 antibodies. We conclude that Nkx2.5 is the most likely candidate responsible for the activation of the vSMC element in the pro-col1a2 promoter.
In order to test whether all the factors that are necessary for the functional activity of the core element have been accounted for, transfection experiments in fibroblasts were carried out. The Nkx2.5 expression vector was cotransfected with the active core element (in which the δEF1/ZEB1 binding motif was mutated) in L929 fibroblasts. They produced no reporter gene activity, indicating that other factors are necessary which may be absent in these cells. Since GATA6 transcripts have been detected in human and rat vSMCs (38) and GATA4 has been reported in pulmonary artery smooth muscle cells (39), we investigated their contributions to the functional activity of the core vSMC element. When the homeodomain transcription factors GATA4 and GATA6 were cotransfected with Nkx2.5 in fibroblasts, a significant increase of reporter gene activity was observed (data not shown). When repeated in the A7r5 cell line, this experiment resulted in a considerable increase in reporter gene activity in the presence of GATA6 but not GATA4. Cooperation between Nkx2.5 and GATA family members has been reported widely (9, 11, 22). GATA6 is a transcriptional regulatory protein with a distinct developmental and tissue-specific profile. It is thought to regulate cell-specific expression patterns and may mediate growth factor stimulation in vSMCs (38). We hypothesize that GATA6 binds to the core element on a noncanonical GATT motif 11 bp upstream of the functional Nkx2.5 binding site. Experiments are under way to evaluate the interaction(s) of GATA6, Nkx2.5, and other transcriptions factors such as LIM proteins (6) in the regulation of collagen type I.
We also examined the potential role of AP1, which overlaps the AvaI restriction site, and we found that at least in transient transfections, where chromatin structure is not involved, the ubiquitous AP1 transcription factor is not essential for the functional activity of this sequence in vSMCs. However, this does not rule out its potential for a role in vivo. Careful analysis of the sequence also revealed that a series of Nkx2.5 sites downstream of the AvaI site, although not essential for activation in vitro, may cooperate in vivo to increase the potential of the main activation site (Nkx2.5, site d).
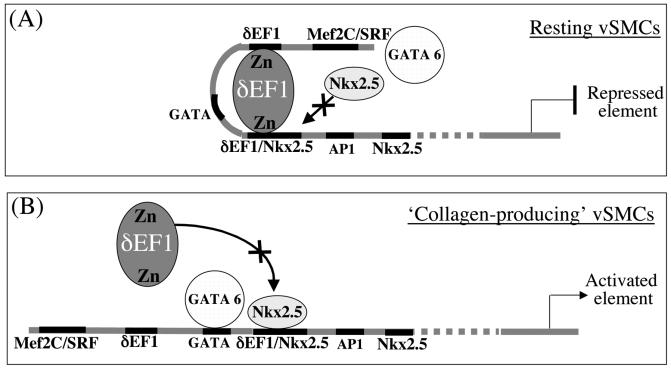
Based on the data presented in this study, we propose a simple mechanism of action in vSMCs (Fig. 7) whereby, during the resting state, collagen type I is repressed by δEF1/ZEB1. Binding by δEF1/ZEB1 on the putative overlapping site shared with Nkx2.5 would impair the access of the homeodomain Nkx2.5 and hinder the binding of the synergistic activator GATA6. Indeed, δEF1/ZEB1 has been previously reported to cause repression by competing for an activating site as well as possessing an inhibitory domain (33). When vSMCs are stimulated to produce collagen type I during vascular remodeling or injury, chromatin modification coupled with altered concentrations of relevant transcription factors leads to increased occupation by Nkx2.5 of the overlapping site to the relative exclusion of δEF1/ZEB1. Once Nkx2.5 is bound, it may recruit GATA6 to enhance its activity, as has been shown in the developing heart (9).
FIG. 7.
Proposed model for the regulation of the pro-col1a2 vSMC element. (A) In the resting state, when the vSMC is not producing collagen, the δEF1/ZEB1 homeodomain repressor is bound on two adjacent sites on a vSMC element located at approximately −16.6 kb upstream of the transcriptional start site. The 3′ δEF1/ZEB1 DNA binding site overlaps a Nkx2.5 homeodomain activator binding site. When the δEF1/ZEB1 repressor is occupying both of its sites, the Nkx2.5 activator is unable to access its binding motif. GATA6 is also unable to access its binding site when δEF1/ZEB1 is bound to the DNA. These conditions result in a repressed element. (B) When the vSMC is activated and collagen type I is synthesized, we propose that δEF1/ZEB1 is displaced from its binding site by Nkx2.5, thus opening the DNA and allowing GATA6 to bind and synergize with Nkx2, leading to transcriptional activation of pro-col1a2.
The biological significance of such an on-off switch regulatory mechanism for the production of collagen type I in vSMCs is very attractive and with much therapeutic potential, especially since SMCs play an important role in vascular pathological processes, such as essential hypertension and atherosclerosis, where major changes take place in arterial wall structure. Human atherosclerotic lesions start off as preexisting intimal thickening, which is defined by the accumulation of SMCs in the intima in the absence of lipid or macrophage foam cells (45). ECM proteins are typical components of the arterial intima, and it is estimated that up to 60% of the volume of the intima consists of ECM proteins such as collagens and proteoglycans (37). The importance of matrix proteins for intimal maturation was recently confirmed by gene expression profiling of the maturing neo-intima and the underlying media. Thirteen genes were upregulated in the neo-intima. Seven of these 13 genes encoded matrix proteins (six collagens and one versican), and two genes encoded known inducers of matrix synthesis (Cbfa1/Runx2 and connective tissue growth factor) (12).
Since there is a high degree of homology between the mouse and human pro-COL1A2 far-upstream enhancer sequences, this model may be applicable to human vascular disease. Indeed, two footprints between kb −20.1 and −19.9 contain similar transcription factor binding motifs (1).
In summary, this is the first study to provide evidence for a lineage-specific element within the far-upstream enhancer of pro-col1a2, namely, a vascular smooth muscle transcriptional element. The core element consists of a 70-bp DNA sequence on which the repressor δEF1/ZEB1 competes with the activator Nkx2.5 for an overlapping site, providing a finely balanced mechanism for the regulation of collagen type I in vSMCs.
Acknowledgments
This work was supported by the Arthritis Research Campaign UK, the Medical Research Council UK, and the Raynaud's and Scleroderma Association.
We thank S. Evans, R. Harvey, J. Rossert, H. Kondoh, and R. Schwartz for expression vectors used in this project. We also thank V. Lefebvere for guidance with the EMSA.
REFERENCES
- 1.Antoniv, T., S. De Val, D. Wells, C. P. Denton, C. Rabe, B. de Crombrugghe, F. Ramirez, and G. Bou-Gharios. 2001. Characterization of an evolutionarily conserved far-upstream enhancer in the human alpha 2(I) collagen (COL1A2) gene. J. Biol. Chem. 276:21754-21764. [DOI] [PubMed] [Google Scholar]
- 2.Bogdanovic, Z., A. Bedalov, P. H. Krebsbach, D. Pavlin, C. O. Woody, S. H. Clark, H. F. Thomas, D. W. Rowe, B. E. Kream, and A. C. Lichtler. 1994. Upstream regulatory elements necessary for expression of the rat COL1A1 promoter in transgenic mice. J. Bone Miner. Res. 9:285-292. [DOI] [PubMed] [Google Scholar]
- 3.Bou-Gharios, G., L. A. Garrett, J. Rossert, K. Niederreither, H. Eberspaecher, C. Smith, C. Black, and B. Crombrugghe. 1996. A potent far-upstream enhancer in the mouse pro alpha 2(I) collagen gene regulates expression of reporter genes in transgenic mice. J. Cell Biol. 134:1333-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenner, D. A., R. A. Rippe, and L. Veloz. 1989. Analysis of the collagen alpha 1(I) promoter. Nucleic Acids Res. 17:6055-6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carson, J. A., R. A. Fillmore, R. J. Schwartz, and W. E. Zimmer. 2000. The smooth muscle gamma-actin gene promoter is a molecular target for the mouse bagpipe homologue, mNkx3-1, and serum response factor. J. Biol. Chem. 275:39061-39072. [DOI] [PubMed] [Google Scholar]
- 6.Chang, D. F., N. S. Belaguli, D. Iyer, W. B. Roberts, S. P. Wu, X. R. Dong, J. G. Marx, M. S. Moore, M. C. Beckerle, M. W. Majesky, and R. J. Schwartz. 2003. Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev. Cell 4:107-118. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. Y., and R. J. Schwartz. 1995. Identification of novel DNA binding targets and regulatory domains of a murine tinman homeodomain factor, nkx-2.5. J. Biol. Chem. 270:15628-15633. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 9.Davis, D. L., A. Wessels, and J. B. Burch. 2000. An Nkx-dependent enhancer regulates cGATA-6 gene expression during early stages of heart development. Dev. Biol. 217:310-322. [DOI] [PubMed] [Google Scholar]
- 10.De Val, S., M. Ponticos, T. Antoniv, D. Wells, D. Abraham, T. Partridge, and G. Bou-Gharios. 2002. Identification of the key regions within the mouse pro a2(I) collagen gene far-upstream enhancer. J. Biol. Chem. 277:9286-9292. [DOI] [PubMed] [Google Scholar]
- 11.Durocher, D., F. Charron, R. Warren, R. J. Schwartz, and M. Nemer. 1997. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 16:5687-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geary, R. L., J. M. Wong, A. Rossini, S. M. Schwartz, and L. D. Adams. 2002. Expression profiling identifies 147 genes contributing to a unique primate neointimal smooth muscle cell phenotype. Arterioscler. Thromb. Vasc. Biol. 22:2010-2016. [DOI] [PubMed] [Google Scholar]
- 13.Hafizi, S. A. S. P., A. T. Goodwin, A. H. Chester, and M. H. Yacoub. 1999. Endothelin-1 stimulates proliferation of human coronary smooth muscle cells via the ETA receptor and is co-mitogenic with growth factors. Atherosclerosis 146:351-359. [DOI] [PubMed] [Google Scholar]
- 14.Harvey, R. P. 1996. NK-2 homeobox genes and heart development. Dev. Biol. 178:203-216. [DOI] [PubMed] [Google Scholar]
- 15.Kim, S., P. H. Brown, and M. J. Birrer. 1996. The inhibitory activity of a transdominant c-jun mutant fused to the ligand binding domain of the estrogen receptor. Oncogene 12:1043-1053. [PubMed] [Google Scholar]
- 16.Lefebvre, V., W. Huang, V. R. Harley, P. N. Goodfellow, and B. de Crombrugghe. 1997. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol. Cell. Biol. 17:2336-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Libby, P., P. M. Ridker, and A. Maseri. 2002. Inflammation and atherosclerosis. Circulation 105:1135-1143. [DOI] [PubMed] [Google Scholar]
- 18.Liska, D. J., M. J. Reed, E. H. Sage, and P. Bornstein. 1994. Cell-specific expression of alpha 1(I) collagen-hGH minigenes in transgenic mice. J. Cell Biol. 125:695-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray, D., P. Precht, R. Balakir, and W. E. Horton, Jr. 2000. The transcription factor δEF1 is inversely expressed with type II collagen mRNA and can repress Col2a1 promoter activity in transfected chondrocytes. J. Biol. Chem. 275:3610-3618. [DOI] [PubMed] [Google Scholar]
- 20.Myllyharju, J., and K. I. Kivirikko. 2001. Collagens and collagen-related diseases. Ann. Med. 33:7-21. [DOI] [PubMed] [Google Scholar]
- 21.Niederreither, K., R. D'Souza, M. Metsaranta, H. Eberspaecher, P. D. Toman, E. Vuorio, and B. de Crombrugghe. 1995. Coordinate patterns of expression of type I and III collagens during mouse development. Matrix Biol. 14:705-713. [DOI] [PubMed] [Google Scholar]
- 22.Nishida, W., M. Nakamura, S. Mori, M. Takahashi, Y. Ohkawa, S. Tadokoro, K. Yoshida, K. Hiwada, K. Hayashi, and K. Sobue. 2002. A triad of serum response factor and the GATA and NK families governs the transcription of smooth and cardiac muscle genes. J. Biol. Chem. 277:7308-7317. [DOI] [PubMed] [Google Scholar]
- 23.Pavlin, D., A. C. Lichtler, A. Bedalov, B. E. Kream, J. R. Harrison, H. F. Thomas, G. A. Gronowicz, S. H. Clark, C. O. Woody, and D. W. Rowe. 1992. Differential utilization of regulatory domains within the alpha 1(I) collagen promoter in osseous and fibroblastic cells. J. Cell Biol. 116:227-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plenz, G. A., M. C. Deng, H. Robenek, and W. Volker. 2003. Vascular collagens: spotlight on the role of type VIII collagen in atherogenesis. Atherosclerosis 166:1-11. [DOI] [PubMed] [Google Scholar]
- 25.Postigo, A. A. 2003. Opposing functions of ZEB proteins in the regulation of the TGFβ/BMP signaling pathway. EMBO J. 22:2443-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Postigo, A. A., and D. C. Dean. 1999. ZEB represses transcription through interaction with the corepressor CtBP. Proc. Natl. Acad. Sci. USA 96:6683-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quandt, K., K. Frech, H. Karas, E. Wingender, and T. Werner. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23:4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Remacle, J. E., H. Kraft, W. Lerchner, G. Wuytens, C. Collart, K. Verschueren, J. C. Smith, and D. Huylebroeck. 1999. New mode of DNA binding of multi-zinc finger transcription factors: δEF1 family members bind with two hands to two target sites. EMBO J. 18:5073-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ross, R. 1999. Atherosclerosis—an inflammatory disease. N. Engl. J. Med. 340:115-126. [DOI] [PubMed] [Google Scholar]
- 30.Rossert, J., H. Eberspaecher, and B. de Crombrugghe. 1995. Separate cis-acting DNA elements of the mouse pro-alpha 1(I) collagen promoter direct expression of reporter genes to different type I collagen-producing cells in transgenic mice. J. Cell Biol. 129:1421-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossert, J. A., S. S. Chen, H. Eberspaecher, C. N. Smith, and B. de Crombrugghe. 1996. Identification of a minimal sequence of the mouse pro-alpha 1(I) collagen promoter that confers high-level osteoblast expression in transgenic mice and that binds a protein selectively present in osteoblasts. Proc. Natl. Acad. Sci. USA 93:1027-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz, R. J., and E. N. Olson. 1999. Building the heart piece by piece: modularity of cis-elements regulating Nkx2-5 transcription. Development 126:4187-4192. [DOI] [PubMed] [Google Scholar]
- 33.Sekido, R., K. Murai, Y. Kamachi, and H. Kondoh. 1997. Two mechanisms in the action of repressor δEF1: binding site competition with an activator and active repression. Genes Cells 2:771-783. [DOI] [PubMed] [Google Scholar]
- 34.Sekido, R., T. Takagi, M. Okanami, H. Moribe, M. Yamamura, Y. Higashi, and H. Kondoh. 1996. Organization of the gene encoding transcriptional repressor δEF1 and cross-species conservation of its domains. Gene 173:227-232. [DOI] [PubMed] [Google Scholar]
- 35.Slack, J. L., D. J. Liska, and P. Bornstein. 1991. An upstream regulatory region mediates high-level, tissue-specific expression of the human alpha 1(I) collagen gene in transgenic mice. Mol. Cell. Biol. 11:2066-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sooy, K., and M. B. Demay. 2002. Transcriptional repression of the rat osteocalcin gene by δEF1. Endocrinology 143:3370-3375. [DOI] [PubMed] [Google Scholar]
- 37.Stary, H. C., D. H. Blankenhorn, A. B. Chandler, S. Glagov, W. Insull, Jr., M. Richardson, M. E. Rosenfeld, S. A. Schaffer, C. J. Schwartz, W. D. Wagner, et al. 1992. A definition of the intima of human arteries and of its atherosclerosis-prone regions. A report from the Committee on Vascular Lesions of the Council on Arteriosclerosis, American Heart Association. Arterioscler. Thromb. 12:120-134. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki, E., T. Evans, J. Lowry, L. Truong, D. W. Bell, J. R. Testa, and K. Walsh. 1996. The human GATA-6 gene: structure, chromosomal location, and regulation of expression by tissue-specific and mitogen-responsive signals. Genomics 38:283-290. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki, Y. J., R. M. Day, C. C. Tan, T. H. Sandven, Q. Liang, J. D. Molkentin, and B. L. Fanburg. 2003. Activation of GATA-4 by serotonin in pulmonary artery smooth muscle cells. J. Biol. Chem. 278:17525-17531. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka, K., N. Tsumaki, C. A. Kozak, Y. Matsumoto, F. Nakatani, Y. Iwamoto, and Y. Yamada. 2002. A Kruppel-associated box-zinc finger protein, NT2, represses cell-type-specific promoter activity of the α2(XI) collagen gene. Mol. Cell. Biol. 22:4256-4267. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Tanaka, M., Z. Chen, S. Bartunkova, N. Yamasaki, and S. Izumo. 1999. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development 126:1269-1280. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka, M., G. E. Lyons, and S. Izumo. 1999. Expression of the Nkx3.1 homeobox gene during pre- and postnatal development. Mech. Dev. 85:179-182. [DOI] [PubMed] [Google Scholar]
- 43.Terraz, C., G. Brideau, P. Ronco, and J. Rossert. 2002. A combination of cis-acting elements is required to activate the pro-alpha 1(I) collagen promoter in tendon fibroblasts of transgenic mice. J. Biol. Chem. 277:19019-19026. [DOI] [PubMed] [Google Scholar]
- 44.Terraz, C., D. Toman, M. Delauche, P. Ronco, and J. Rossert. 2001. δEf1 binds to a far upstream sequence of the mouse pro-alpha 1(I) collagen gene and represses its expression in osteoblasts. J. Biol. Chem. 276:37011-37019. [DOI] [PubMed] [Google Scholar]
- 45.Virmani, R., F. D. Kolodgie, A. P. Burke, A. Farb, and S. M. Schwartz. 2000. Lessons from sudden coronary death: a comprehensive morphological classification scheme for atherosclerotic lesions. Arterioscler. Thromb. Vasc. Biol. 20:1262-1275. [DOI] [PubMed] [Google Scholar]
- 46.Vuust, J., M. E. Sobel, and G. R. Martin. 1985. Regulation of type I collagen synthesis. Total pro alpha 1(I) and pro alpha 2(I) mRNAs are maintained in a 2:1 ratio under varying rates of collagen synthesis. Eur. J. Biochem. 151:449-453. [DOI] [PubMed] [Google Scholar]
- 47.Yang, G. H., and J. J. Pestka. 2002. Vomitoxin (deoxynivalenol)-mediated inhibition of nuclear protein binding to NRE-A, an IL-2 promoter negative regulatory element, in EL-4 cells. Toxicology 172:169-179. [DOI] [PubMed] [Google Scholar]
- 48.Zilberman, A., V. Dave, J. Miano, E. N. Olson, and M. Periasamy. 1998. Evolutionarily conserved promoter region containing CArG*-like elements is crucial for smooth muscle myosin heavy chain gene expression. Circ. Res. 82:566-575. [DOI] [PubMed] [Google Scholar]