Abstract
Cell culture models are used widely to study the effects of dengue virus (DENV) on host cell function. Current methods of identification of cells infected with an unmodified DENV requires fixation and permeablization of cells to allow DENV-specific antibody staining. This method does not permit imaging of viable cells over time. In this report, a plasmid-based reporter was developed to allow non-destructive identification of DENV-infected cells. The plasmid-based reporter was demonstrated to be broadly applicable to the four DENV serotypes, including low-passaged strains, and was specifically cleaved by the viral protease with minimal interference on viral production. This study reveals the potential for this novel reporter system to advance the studies of virus-host interactions during DENV infection.
Keywords: Dengue virus, viral reporter, protease, fluorescence microscopy
1. Introduction
Dengue virus (DENV) is a mosquito-borne human pathogen of global medical importance (Bhatt et al., 2013; Simmons et al., 2012). DENV causes an acute febrile illness that, in some patients, is associated with a life-threatening plasma leakage syndrome, dengue hemorrhagic fever (DHF) (Anonymous, 2009). While there is ongoing debate regarding the contribution of different mechanisms in dengue illness, there is substantial evidence supporting both viral and host factors in disease pathogenesis. Most models propose a cascade involving systemic infection of cells of the myeloid cell lineage and cytokine production through both direct (infected cells) and indirect (bystander and immune cells) pathways (Halstead et al., 2010; Rothman, 2010; Whitehorn and Farrar, 2010). There is ongoing scientific need and interest in examining the direct effects of DENV infection on cellular metabolic processes and gene expression, which will give insight into the initial events that may impact disease severity.
Given the limitations of clinical studies and existing animal models, cell culture models remain an important approach to studying DENV infection and host responses (Hibberd et al., 2006; Ramirez-Ortiz et al., 2006; Becerra et al., 2009; Heaton and Randall, 2010; Chase et al., 2011; Pena and Harris, 2012). These studies have involved both continuous cell lines and primary cells from human and arthropod hosts. The analysis of host cell responses to infection has been accomplished through imaging, immunoassays, and RT-PCR. These methods are subject to at least two technical limitations- difficulty in discriminating infected and uninfected cells in the same culture, and collection of static data from individual time points. Detection of individual DENV-infected cells generally relies on immunostaining. Because most DENV proteins are not expressed on the cell surface, sensitive detection of infected cells requires fixation and permeabilization of the cells, an approach that is incompatible with ongoing observation of living cells and potentially introduces artifactual changes in cellular ultrastructure. One approach to investigate early events during DENV infection in living cells used virus labeled with a lipophilic fluorescent probe that fluoresces upon viral fusion with membranes of acidified vesicles. This approach is limited to analysis of events prior to viral fusion (van der Schaar et al., 2007; van der Schaar et al., 2008). Several groups have reported recently the construction of DENV replicons or infectious viruses that encode reporter molecules such as luciferase or GFP (Mattia et al., 2011; Zou et al., 2011; Leardkamolkarn and Sirigulpanit, 2012; Leardkamolkarn et al., 2012; Schoggins et al., 2012; Yang et al., 2013;). These constructs can effectively label cells infected with the engineered virus or replicon. However, recombinant modified DENV genomes are invariably attenuated in comparison to wild-type DENV, which is likely to complicate the interpretation of virus-host interactions. Furthermore, these strategies will require further extensive manipulation to permit the study of other DENV variants, limiting the generalizability of the findings.
To facilitate studies of live cells infected with DENV, a novel plasmid-based reporter, p4B5-EGFP, was created that utilize the viral protease cleavage site resulting in nuclear localization of GFP in DENV-infected cells. The 4B5-EGFP reporter was shown to be effective in detecting infection of cells by all four DENV serotypes, including low-passaged viral strains. The results demonstrate the capability of the 4B5-EGFP reporter to identify DENV-infected cells without destruction of the cell.
2. Materials and Methods
2.1. Cell lines and virus
Vero cells were obtained from the ATCC and maintained in Dulbecco’s Modification of Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 U/ml streptomycin, with cell media also containing nonessential amino acid (1×, HyClone). Cells were incubated in a humidified chamber at 37 °C and 5% CO2.
Prototype DENV strains DENV-1 Hawaii, DENV-2 16681, DENV-2 New Guinea C (NGC), DENV-3 CH53489, and DENV-4 814669 were originally obtained from the American Type Culture Collection and the Walter Reed Army Institute of Research, and were passaged multiple times in C6/36 cells (ATCC, USA). Low passaged DENV-2 strains C0102/96 and C0112/96 were isolated from plasma collected in 1996 as part of a prospective study (Vaughn et al., 2000); these viruses were initially isolated by intrathoracic injection of Toxorhynchites splendens mosquitoes and then passaged up to 4 times in C6/36 cells. Virus titers were determined by immunostained plaque assay on Vero cells based on the method of Liu et al with minor modifications (Liu et al., 2012). Briefly, Vero cells (1×105 cells in 50 µl/well) were added to replicate wells of 96-well white-bottom plates with 50 µl of serial 0.5 log dilutions of virus. Plates were incubated for 2 h and then 100 µl of overlay containing 1% carboxymethylcellulose was added. Plates were stained after 3 d incubation using anti-DENV antibody MAB8705 (EMD Millipore, Billerica, MA, 1:1000), horseradish peroxidase-conjugated anti-mouse Ig (Southern Biotech, 1:2000), and TMB substrate (Mabtech, Cincinnati, OH). Stained regions were read using an ELISpot plate reader to give focus-forming units per ml (ffu/ml). The ffu/ml was log transformed and graphed using Graph Pad Prism 6.0 software.
2.2. Construction of the DENV reporter plasmid
The DENV reporter plasmid, p4B5-EGFP, was constructed to encode the full-length DENV-2 NS4B protein (without sequences encoding the 2k peptide) and the first 10 amino acids of the DENV-2 NS5 protein fused to the SV40 nuclear localization signal sequence (NLS, PKKKRKVG (Cressman et al., 2001)) and the enhanced GFP (EGFP) protein in the pcDNA3.1 vector (Life Technologies, Grand Island, NY). The primers used for PCR synthesis are shown in Table 1. The DENV sequences were originally amplified from a DENV-2 NGC infectious clone, which was kindly provided by Dr. Barry Falgout (Polo et al., 1997). A plasmid generated in our lab containing DENV-2 sequences from nucleotides 6757 to 7599, which includes NS4B and the first 30 nucleotides of NS5, was used to insert the SV40 NLS and GFP sequences downstream of the NS4B-5 cleavage site. Briefly, to generate a fragment containing the SV40 NLS upstream of GFP, a forward primer ‘NLSGFP-EcoRI’ that incorporated a 5’ EcoRI restriction site and the SV40 NLS sequence and the reverse primer ‘GFP XhoI’ that contained a 3’XhoI restriction site were used to amplify from the pTRE-eGFP plasmid (Clontech) by PCR. The PCR fragment was digested with EcoRI and XhoI, gel purified, and ligated into the vector downstream of nucleotide 7599. To generate the p4B5-EGFP, the ‘NS4B HindIII’ forward primer and the ‘GFP XhoI’ reverse primer was used to amplify the reporter sequence by PCR. The product of the PCR reaction and pcDNA 3.1 (Life Technologies, Grand Island, NY) were then digested with HindIII and XhoI, gel purified and ligated together. The identities of the clones were confirmed by DNA sequencing.
TABLE 1.
Oligonucleotide primers used for PCR amplification.
| Oligonucleotide | Sequencea |
|---|---|
| NS4B HindIII F | 5'-CATTGGCAAAGCTTGCCACCATGGCGAACGAGATGGGTTTCCTAGAAAAAACGAAG-3' |
| NS5(10aa) EcoRI R | 5'-CATTTCTCGAATTCTCCAAGCGTCTCTCCTATGTTGCCAGTTCCCCTTC-3' |
| SV40NLS-eGFP EcoRI F | 5'-CGCGGAATTCGCCACCATGCCGAAGAAAAAGCGGAAGGTTGGCGTGAGCAAGGGCGAGGAGCTGTTCACCGGGGTGGTGCCCATCCTGGT-3' |
| eGFP XhoI R | 5'-CGCGCTGCCTCGAGTTACTTGTACAGCTCGTCCATGCCGAGAGTGATC-3' |
| NS2B3 HindIII F | 5'-CAAGAAAAGGAAGCTTGCCACCATGAGCTGGCCATTAAATGAGGCTATCATG-3' |
| NS2B3 XbaI R | 5'-GGTCAGAGATCTAGACTTTCTTCCGGCTGCAAATTC-3' |
underlined text = the Kozak sequence, bold text = restriction endonuclease, italics = SV40 NLS
The plasmid pNS2B3 expressing the DENV-2 NS2B3 protease was constructed using DENV-2 NGC RNA as a template. Sense and antisense primers (Table 1) were designed to generate a cDNA fragment encompassing nucleotides 4132 to 6375 of DENV-2 NGC using SuperScript® One-Step RT-PCR for long templates (Life Technologies, Grand Island, NY). The PCR fragment and the pcDNA3.1 V5-His vector (Life Technologies, Grand Island, NY) were digested with HindIII and XbaI, gel purified and ligated together. The identities of the clones were confirmed by DNA sequencing.
2.3. Transfection and DENV infection
Vero cells were transfected using GeneJuice® Transfection Reagent (EMD Millipore, Billerica, MA) following the manufacturer’s instructions. Briefly, cells were seeded in an 8-chambered Nunc Lab-Tek slide (Thermo Fisher Scientific, Rockford, IL) with a glass coverslip bottom at 2×104 cells per well 24 hrs prior to transfection. For transfection, 1.2 µl of GeneJuice® Transfection Reagent was diluted in 15µl serum-free media and incubated at room temperature for 5 minutes, and then 0.55µg of plasmid were added to the diluted GeneJuice® Transfection Reagent and incubated for 15 minutes at room temperature. The complex was then added to the cells. Vero cells were infected with DENV at a multiplicity of infection of 1 as previously described (Medin and Rothman, 2006).
For cotransfection with p4B5-EGFP and pNS2B3, Vero cells were transfected with 22.5µg of each plasmid.
2.4. Western Blot
Whole cell extracts were prepared using lysis buffer (10% glycerol, 20 mM Tris (pH 7), 150 mM NaCl, 0.5 mM EDTA, 1% Nonidet P-40) freshly supplemented with a protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO) and 25 U of the Pierce Universal Nuclease (Thermo Fisher Scientific, Rockford, IL). Cells were placed on ice for 15 min and centrifuged for 15 min at 14,000 rpm. Protein concentrations were determined using Bradford reagent in the Envision plate reader (Perkin Elmer, Waltham, Massachusetts) using bovine serum albumin as a standard. 15μg of protein were separated using a 12% SDS-PAGE precast gel (Biorad, Hercules, CA) and then transferred onto nitrocellulose membranes using the Trans-Blot® Turbo™ Transfer System (Biorad, Hercules, CA). Blots were blocked with 5% skim milk and subjected to immunoblot analysis using anti-GFP (Thermo Fisher Scientific, Rockford, IL, cat. 22169) diluted 1:3000 and anti-β-actin (Abcam, Cambridge, MA, cat. 8226) diluted 1:5000 antibodies as indicated in the figure legends.
2.5. Staining and immunofluorescence microscopy
In experiments analyzing nuclear localization of GFP, nuclear DNA was stained immediately prior to imaging with NucBlue™ Live Cell Stain or NucBlue™ Fixed Cell Stain following the manufacturer’s protocol (Life Technologies, Grand Island, NY). For detection of ER membranes, cells were washed with 1× DPBS and stained with 500nM ER-Tracker Blue-White DPX solution (Life Techonologies, Grand Island, NY) for 30 minutes following the manufacturer’s protocol. Staining solution was replaced with cell culture media and immediately imaged by fluorescence microscopy using a DAPI filter.
Fluorescence images were obtained using an EVOS® fl digital inverted fluorescence microscope (Advanced Microscopy Group, USA). To obtain a time series of images of nuclear localization of GFP after DENV infection, Vero cells transfected with p4B5-EGFP were infected with DENV-2 16681. The wells were washed after two hours and fresh media was added to each well. The plate was placed into Cytation3 Cell Imager (Biotek, Winooski, VT) that contains a temperature and CO2 controlled chamber. The images were acquired every 45 minutes starting at 4 h after adding virus to the cell culture using a 20× objective.
Individual cells were analyzed for total cellular and nuclear GFP fluorescence using imageJ’s Analyze and Measure function. Nuclear and cytoplasmic GFP fluorescence was defined by setting parameters based on NucBlue staining of the nucleus. Background fluorescence was measured from three regions outside of the cell. The product of the average background and area of the cell was subtracted from the integrated density (sum of the intensity of the pixels) for both the total fluorescence and nuclear fluorescence giving a corrected fluorescence as described previously (Gavet and Pines, 2010). The ratios of nuclear to total fluorescence were calculated.
Total fluorescence data were not distributed normally necessitating use of nonparametric methods. Wilcoxon-Mann-Whitney rank sum test was used to quantify differences between groups. Data presented includes analyses of individual cells with n = 24–36 cells. All statistical analyses were performed utilizing Graph Pad Prism 6.0 software.
To confirm DENV infection, cells were stained for DENV antigen at 24 and 48 hrs post-infection. Cells were washed once with PBS, fixed with 4% paraformaldehyde for 30 min at room temperature, and permeabilized with Cytoperm (BD Biosciences, Franklin Lakes, NJ) for 15 min at 4°C. DENV antigen was detected by indirect immunofluorescence staining using DENV complex-specific monoclonal antibody (EMD Millipore, Billerica, MA, cat. MAB8705) diluted 1:300 as the primary antibody and allophycocyanin (APC)-conjugated goat anti-mouse immunoglobulin G antibody (BD Biosciences, Franklin Lakes, NJ, cat. 558026) diluted 1:100 as the secondary antibody.
To confirm NS2B3 expression, cells were stained for NS3 at 48hrs post transfection using an anti-NS3 primary antibody (Genetex, Irvine, CA) and APC-conjugated goat anti-mouse IgG as the secondary antibody.
3. Results
3.1. Construction and characterization of DENV reporter plasmid
The DENV reporter, p4B5-EGFP, was designed to take advantage of two events that occur during DENV infection- localization of the DENV NS4B protein to the endoplasmic reticulum (ER) and proteolytic processing of the cleavage site between NS4B and NS5 by the NS2B3 protease (Miller et al., 2006; Bera et al., 2007) (Figure 1a). The processing of the cleavage site by NS2B3 is expected to release the GFP component (with the small fragment of NS5) to relocalize from the cytoplasm to the nucleus.
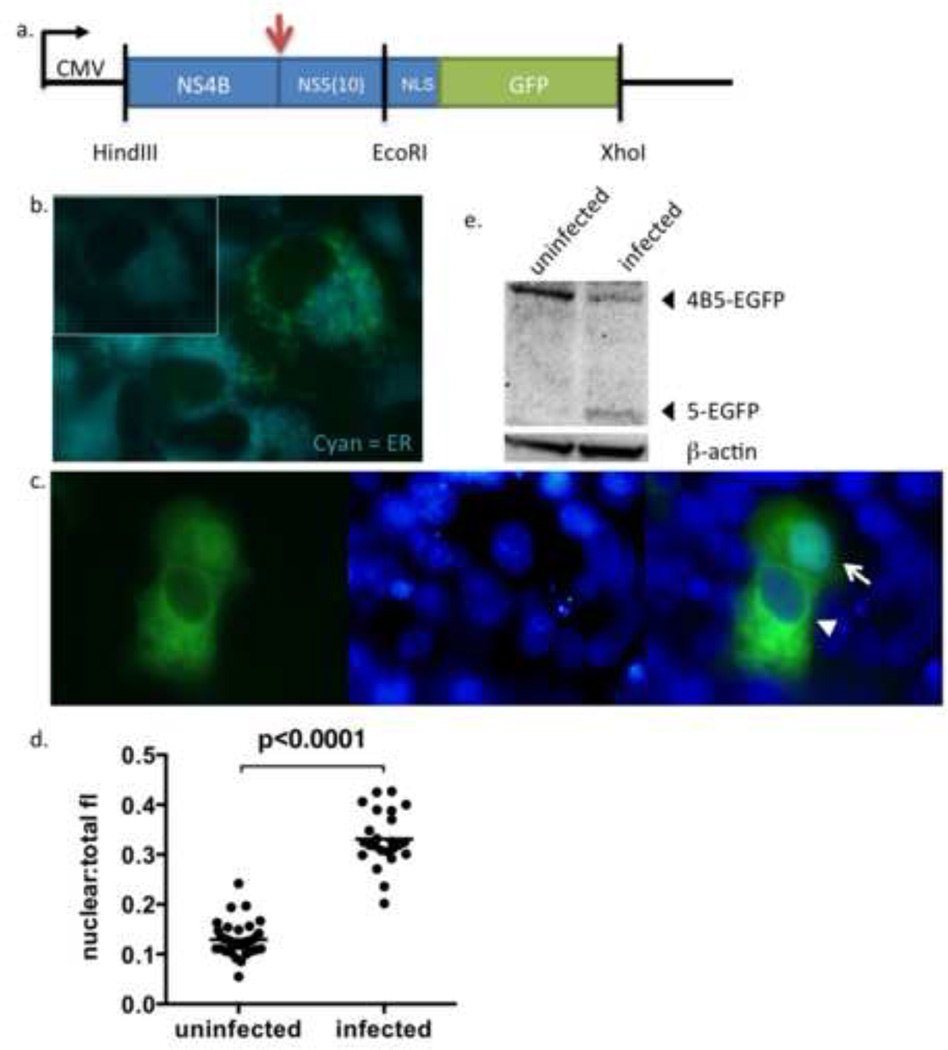
Figure 1. Construction and characterization of p4B5-EGFP.
(a) Schematic of the p4B5-EGFP construct containing NS4B and the first 30 nucleotides of NS5 of the DENV-2 genome tagged with the SV40 NLS and eGFP. The arrow indicates the cleavage site utilized by the DENV NS2B3 protease. The vertical lines represent restriction endonuclease sites.
(b) Unfixed Vero cells transfected with p4B5-EGFP were stained with ER-Tracker Blue-White DPX dye (Cyan) 24hrs post-transfection and immediately imaged to detect colocalization of NS4B5-EGFP (reporter, green) with ER membranes (blue, magnification, ×100). Inset reflects ER staining of cells alone. (c) Unfixed Vero cells transfected with p4B5-EGFP were infected with DENV-2 16681. Nuclei were counterstained with NucBlue for live cells (Invitrogen) (blue) to detect nuclear localization of GFP (green, magnification, ×100). Cytoplasmic expression of GFP is indicated by the arrowhead and nuclear expression of GFP is indicated by an arrow (magnification, ×100). (d) Analysis of nuclear to total fluorescence intensity ratios of GFP (nuclear:total fl) in uninfected and DENV infected cells at 24 h was performed using ImageJ software. Each symbol represents analysis of a single cell. Statistical analysis was performed using nonparametric Wilcoxon-Mann-Whitney rank sum test. Straight line represents the median. (e) Cellular lysates were prepared 24 h post infection and subjected to Western blotting for GFP or β-actin. The 5-EGFP fragment represents the cleavage product resulting from cleavage between NS4B and NS5-EGFP during viral infection.
The expression and cellular distribution of the reporter construct in living cells was first evaluated by transfecting cells with the reporter plasmid and staining with an ER membrane stain. In the absence of infection with DENV (Figure 1b), transfected cells showed cytoplasmic green fluorescence that colocalized with the ER membrane stain, indicating ER retention of the reporter.
To determine whether infection with DENV would induce proteolytic cleavage between NS4B and NS5(10) and mobilize GFP to the nucleus, cells were transfected with p4B5-EGFP and infected with DENV-2 16681. Figure 1c shows a DENV-infected Vero cell culture expressing 4B5-EGFP and stained with the nuclear dye NucBlue for live-cell labeling (Invitrogen). The two cells show different phenotypes of GFP localization at 24hrs after infection, with one cell showing cytoplasmic GFP (arrowhead) and the other cell showing nuclear GFP (arrow). Analysis of nuclear to total cellular fluorescence for individual cells at 24 h showed at least a 2.4 fold increase in infected cells when compared to uninfected cells and were significantly different (p=0.0001) (Figure 1d). These results confirmed that GFP localized to the nucleus in infected cells. Since these images were taken on a standard fluorescence microscope, the small number of cells in the uninfected 24h condition showing an increase in fluorescence intensity was due to the fluorescence bleed from cytoplasmic region into the nuclear region.
To verify cleavage of the 4B5-EGFP, Western blot analysis was performed on cell lysates using an antibody against GFP. As seen in Figure 1e, uninfected cells transfected with p4B5-EGFP showed a single band localizing to approximately 60kD, which is the expected size of the expressed fusion protein. In cells infected with DENV-2 at an m.o.i. of 1 for 24 hours, the 60kD band was reduced in intensity and a lower 30kD band appeared. The 30kD band corresponds to the expected size of the NS5 fragment fused to eGFP. These results confirm that the 4B5-EGFP is cleaved during DENV infection.
3.2. Kinetics of nuclear localization of GFP after DENV infection
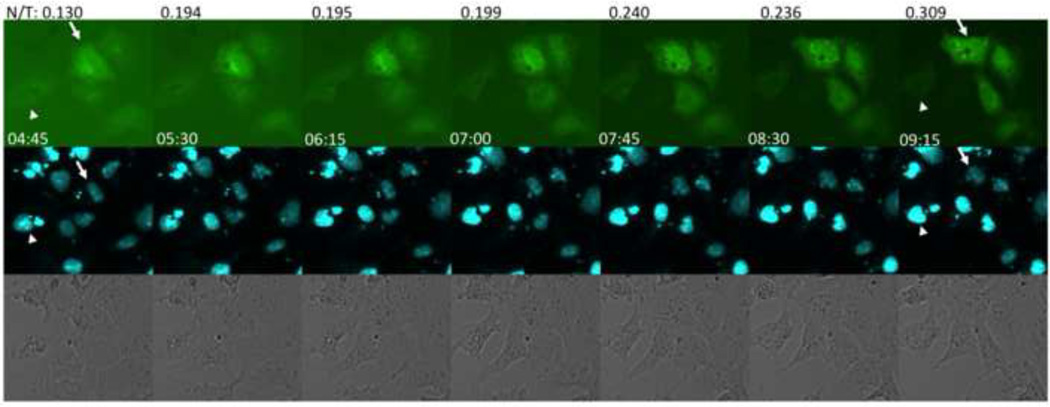
To determine the earliest time point that the 4B5-EGFP reporter could detect DENV infection, transfected cells infected with DENV were analyzed by fluorescence microscopy. Relocation of green fluorescence into the nucleus was seen as early as 8 h post-infection (data not shown). Time-lapse images of Vero cells in a DENV-infected culture were taken to further determine the time required to visually identify GFP movement from the cytoplasm to the nucleus (Figure 2). Movement of cytoplasmic GFP to the nucleus was visible in four of the five cells in Figure 2 by 7 h after addition of virus. One cell remained uninfected during this analysis (Figure 2, arrowhead). Calculation of the ratio of nuclear to total fluorescence at each time point for a representative cell (Figure 2, arrow) showed successive increase in nuclear fluorescence over time.
Figure 2.
Kinetics of nuclear localization of GFP after DENV infection. Vero cells transfected with p4B5-EGFP were infected with DENV-2 16681 and sequential images were acquired to assess the time for GFP to localize into the nucleus. Each set of images shows the expression of GFP in p4B5-EGFP transfected cells (top row), nuclear stain using NucBlue (middle row) and transmitted images (bottom row) at each time point. Time after addition of virus to the culture is located at the lower left of each image (magnification, ×20). Nuclear/total fluorescence (N/T) was calculated at each time point for a representative cell (arrow). The arrowhead shows a cell that remains uninfected over time. Data are representative of at least five experiments.
3.3. Nuclear GFP correlates with DENV antigen staining
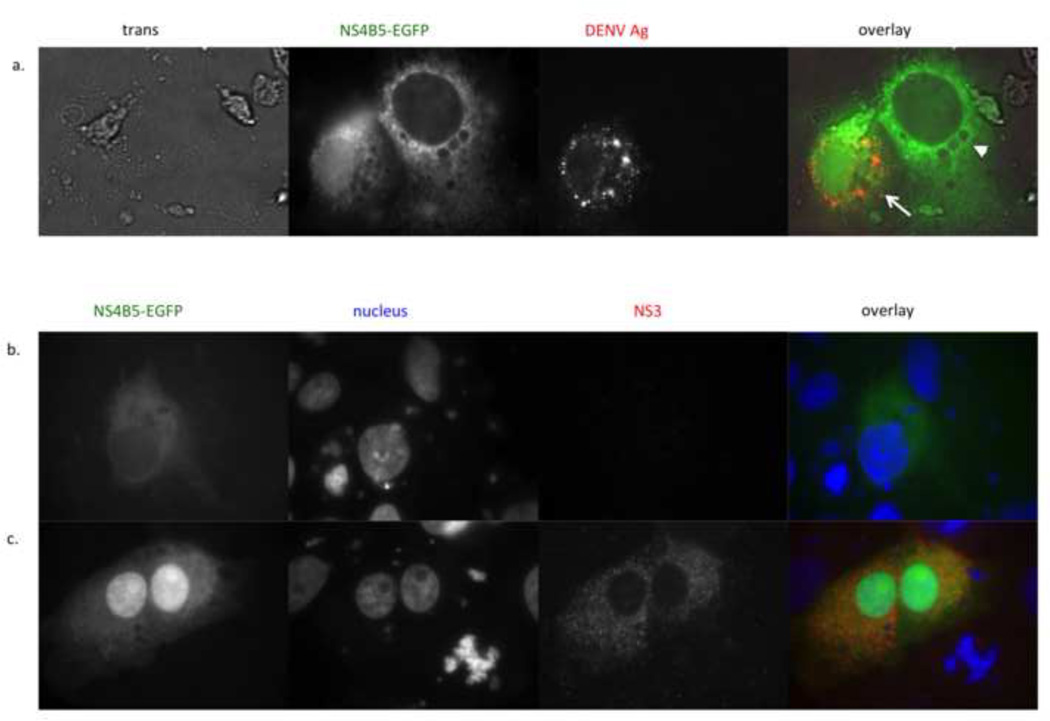
To verify that the cells displaying nuclear localization of GFP were infected with DENV, cells were fixed, permeabilized and stained using a DENV-specific monoclonal antibody. Figure 3a shows a pair of cells in a DENV-infected cell culture. The arrow shows a cell with nuclear GFP, indicating viral infection, whereas the arrowhead shows a cell with only cytoplasmic GFP, indicating no viral infection. As seen in the third and fourth panels of Figure 3a, the bottom left cell (with nuclear GFP) stained with the anti-DENV antibody whereas the upper cell (with only cytoplasmic GFP) did not.
Figure 3.
Nuclear localization of GFP correlates with DENV antigen and co-expression of the NS2B3 protease. (a) Vero cells were transfected with p4B5-EGFP (green) and infected with DENV-2 16681 at an m.o.i of 1. 24 hours post-infection, cells were fixed, permeabilized and stained with antibody against DENV complex. The NS4B5-EGFP panel shows cytoplasmic expression of GFP (right, arrow neighboring cytoplasmic and nuclear expression of GFP (left, arrowhead) (magnification, ×100). DENV Ag panel shows DENV antigen staining in cells infected with DENV. Overlay of GFP and antigen staining (red) shows that nuclear localization of GFP correlates with DENV antigen staining (magnification, ×100). (b and c) Vero cells transfected with the p4B5-EGFP alone (b) or cotransfected with pNS2B3 (c) were analyzed for nuclear localization of GFP at 48hrs post-transfection (magnification, ×100). NS4B5-EGFP panel shows location of GFP within the cells (green). The nucleus panel shows the nucleus stained with NucBlue (blue, Life Technologies). The NS2B3 panel shows indirect antibody staining for NS3 (red). The overlay shows that nuclear GFP expression correlates with NS2B3 expression. Data are representative of at least six (a) and two (b) experiments.
3.4. Expression of the DENV NS2B-3 protease is sufficient for cleavage of 4B5-EGFP
Previous reports have shown that both NS2B and NS3 are required for efficient proteolytic cleavage of the DENV polyprotein (Falgout et al., 1991; Brinkworth et al., 1999). A construct that expresses DENV-2 NS2B3 was created to determine if the NS2B3 protease alone could induce nuclear localization of GFP. Cells were transfected with the p4B5-EGFP alone or cotransfected with the NS2B3 plasmid. As seen in Figure 3b, cells transfected with p4B5-EGFP showed cytoplasmic GFP expression only. In contrast, cells expressing NS2B3 and 4B5-EGFP showed nuclear localization of GFP (Figure 3c). In these experiments, nuclear localization was detected 48 and 72 hs post-transfection. This delay in protease activity may reflect a lower level of protease expression from the plasmid compared to DENV-infected cells. Alternatively, NS2B3 may require additional viral factors to cleave efficiently the NS4B-5 junction.
3.5. DENV serotype and strain specificity of the 4B5-EGFP reporter
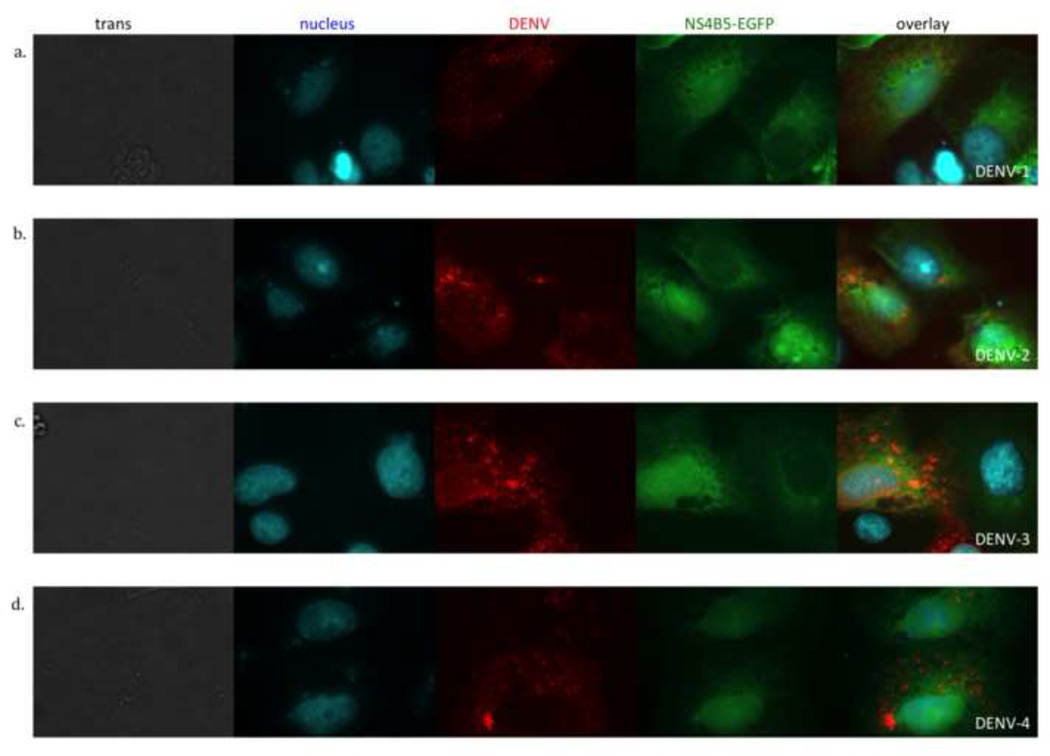
The viral gene segment in p4B5-EGFP was derived from DENV-2 NGC, a laboratory-adapted virus originally isolated in 1944 and was tested using DENV-2 16681, another laboratory-adapted virus that was isolated in 1964 (Vaughn et al., 1996). Each DENV serotype can be subdivided into specific genotypes, which can have up to 6% genetic divergence (Rico-Hesse, 1990; Twiddy et al., 2002). To assess whether the construct would be processed by other DENV-2 strains, low passaged recent clinical isolates were used to infect cells. Transfected cells were infected with two viral isolates, C0102/96 and C0112/96, which were originally isolated from the plasma of patients with dengue in 1996. Nuclear localization of GFP and DENV antigen staining was analyzed at 24 hours post-infection. As seen with DENV-2 16681, DENV antigen staining correlated with nuclear GFP for both C0102/96 (data not shown) and C0112/96 (Figure 4b).
Figure 4.
All four DENV serotypes induce cleavage of p4B5-EGFP to localize GFP to the nucleus. Vero cells transfected with p4B5-EGFP (green) were infected with each of the four DENV serotypes at an MOI of 1. Cells were fixed, permeabilized and stained for DENV antigen (red) and nuclear DNA (cyan). Cells were analyzed at 24 hours post-infection by fluorescence microscopy. Each row is a representative image of DENV infected cells in bright field (trans) and fluorescence images showing nuclear stain (cyan), DENV antigen stain (red), 4B5-EGFP expression (green). The overlay is a composite of the nucleus, DENV and NS4B-EGFP images. (a) DENV-1 Hawaii, (b) DENV-2 C0112/96, (c) DENV-3 CH53489 and (d) DENV-4 814669. Data are representative of at least four experiments each.
The four serotypes of DENV have up to 70% sequence homology across their genomes (Blok, 1985). Previous reports have shown that the DENV protease is less selective than the proteases of other flaviviruses and can tolerate amino acid changes within the cleavage site (Shiryaev et al., 2007). Additionally, the NS2B3 protease cleaves at least five sites with little sequence homology within the same DENV polyprotein (Bera et al., 2007). Therefore, to analyze whether this reporter plasmid could be used to detect infection with other DENV serotypes, cells transfected with the reporter plasmid were infected with each of the four DENV serotypes. For all four serotypes, nuclear localization of GFP was detected at 24 hours post- infection (Figure 4). The specificity of nuclear GFP for DENV-infected cells was verified by co-staining for DENV antigen (red).
3.6. Effects of 4B5-EGFP on DENV replication
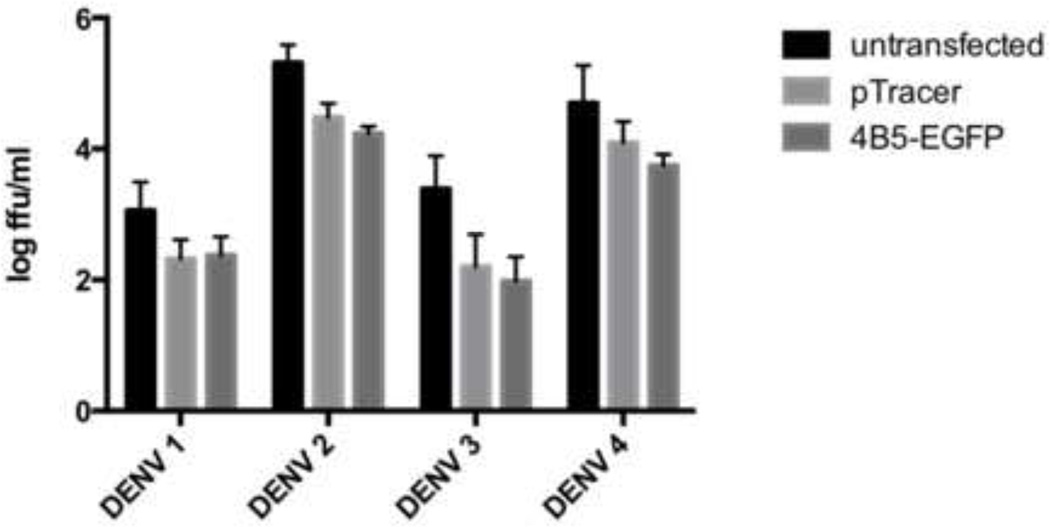
Full length NS4B contains a cleavable 2K signal peptide at the N terminus, and has been shown to be required for IFN antagonism (Munoz-Jordan et al., 2005). The NS4B in the pNS4B5-EGFP was designed to be expressed without the 2K region and therefore, should have no effect on inhibiting the antiviral response. To determine whether transfection of the p4B5-EGFP had an effect on viral production compared untransfected cells, viral titers in supernatants of DENV-infected Vero cells transfected with p4B5-EGFP or a control plasmid, pTracer, expressing GFP alone were assessed plaque assay. Viral titers in virally infected cells without transfection were compared cells that were transfected. Since Vero cells do not have an intact innate immune system (Mosca and Pitha, 1986), parallel experiments with CV-1 cells, which do elicit an innate immune response to dsRNA was also performed (Thacore et al., 1990). Cells transfected with pNS4B5-EGFP and pTracer showed a 3 to 7-fold reduction in viral when compared to infected cells that were untransfected depending on the serotype used for infection (Figure 5). There were minimal differences in viral titers between transfected with either the control plasmid or cells transfected with p4B5-EGFP. These results indicate that transfection alone reduced viral titers. Additionally, cells transfected with p4B5-EGFP and pTracer were comparable at both 24 and 48h post-infection in Vero and CV-1 cells indicating that the reporter did not affect viral replication (data not shown).
Figure 5. Expression of 4B5-EGFP reporter has minimal effects on DENV replication.
Vero cells were transfected with pTracer or p4B5-EGFP or untransfected and infected at an m.o.i. of 1 with DENV 24h later. After 24 h supernatants were collected for viral titration. Each time point represents the geometric mean and 95% confidence interval (CI) of six experiments.
4. Discussion
In the present study, a DENV-specific plasmid-based reporter system, 4B5-EGFP, was characterized for detection of infected cells in live cell culture for all four 14 DENV serotypes. Although systems for analyzing DENV replication in culture have expanded in recent years, detection methods applicable to imaging and detection of cells infected with native viral isolates in unfixed cells have not been available. The 4B5-EGFP reporter does not require genetic modification of the viral genome.
The 4B5-EGFP reporter also showed detection of cellular infection with each of the DENV serotypes. Previous reports have analyzed sequence variation within the NS4B-NS5 junction between various flaviviruses (Lin et al., 1993; Bera et al., 2007; Shiryaev et al., 2007). The amino acids in the P1, P2 and P1’ positions are similar for all four DENV serotypes as well as Yellow Fever virus, however, amino acids occupying P2’ and P3’ are different (Shiryaev et al., 2007). Those experiments were done with synthetically produced peptides; the role of surrounding amino acids and membrane tethering of the proteins may be an additional factor in the selectivity of trans cleavage by flaviviral proteases (Clum et al., 1997). Further analysis using other flaviviruses would be useful to establish the specificity of this reporter system.
The advantage of using the 4B5-EGFP reporter the detection of GFP can be carried out with fluorescence microscopy. Additionally, the ability to monitor infection by fluorescence relocalization in living cells also suggests the possibility of continuous visualization of DENV infection. The 4B5-EGFP reporter was proteolytically processed as early as 8 hrs post-infection, which correlates with synthesis of new viral protein (Shrivastava et al., 2011). The ability to detect viral infection during initial translation of the viral genome will extend analysis of cellular events to early time points.
Numerous studies have demonstrated that in vitro infection with DENV induces significant changes in cellular gene expression and metabolic processes (Hibberd et al., 2006; Ramirez-Ortiz et al., 2006; Becerra et al., 2009; Heaton and Randall, 2010; Chase et al., 2011; Pena and Harris, 2012;). The experiments conducted in those studies processed unsorted cultures of cells exposed to DENV. Therefore, the effects measured in those studies reflect both direct effects of viral infection on the infected cell as well as secondary effects on both infected and uninfected cells in the same culture (Palmer et al., 2005; Nightingale et al., 2008). The 4B5-EGFP reporter in this study has the potential to identify effects on individually infected and uninfected cells within the same culture.
Live-cell analysis of virus-infected cells by fluorescence microscopy represents a promising approach to investigate virus-cell interactions (van der Schaar et al., 2007; van der Schaar et al., 2008; Jones et al., 2010; Lee-Huang et al., 2011; Paloheimo et al., 2011; Sivaraman et al., 2011; Coller et al., 2012; Rand et al., 2012). This approach has been used for other viruses to study virion entry and assembly and the dynamics of movement of viral proteins and cell structures within the infected cell. For several viruses, live cell imaging has revealed new mechanisms critical to viral replication or cell-to-cell spread and has been used for antiviral drug screening.
A theoretical concern with this approach was that the DENV protein segment expressed by the plasmid might itself affect cellular function. For example, DENV NS4B has been reported to inhibit type I IFN signaling (Munoz-Jordan et al., 2003). Munoz-Jordan et al reported, however, that deletion of the N-terminal 2K segment eliminated the inhibition of IFN without affecting ER localization (Munoz-Jordan et al., 2005). The 4B5-EGFP reporter plasmid did not contain the 2K segment, and, consistent with the published results, was not different from a control plasmid in its effects on DENV replication. Viral replication was lower in transfected cells than in untransfected cells, likely due at least in part to induction of type I IFN (Park et al., 2003). A cell line stably transfected with the 4B5-EGFP reporter would be expected to overcome this limitation.
The reagents developed in this project have some limitations for other types of analyses. To this effect the 4B5-EGFP transfected cells fluoresce green regardless of infection status, therefore the cells are not amenable to cell sorting for downstream analyses. Since eGFP is a commonly used fluorescence molecule, exchanging eGFP for a less commonly used color such as BFP or YFP will permit multicolor fluorescence target analyses. Additionally, there are potential applications of the 4B5-EGFP reporter in drug screening and the development of novel animal or cell culture models of DENV infection.
The 4B5-EGFP reporter described is a promising strategy for identifying live DENV-infected cells by fluorescence microscopy. Potential applications of the 4B5-EGFP reporter include identifying infectious virus directly from patient samples. To this effect, nuclear GFP was seen in transfected cell cultures infected with low-passaged isolates.
Highlights.
A dengue viral cleavage site between NS4B and NS5 was cloned upstream of an SV40 nuclear localization sequences and GFP to produce a DENV reporter, pNS4B5-GFP.
Dengue serotypes 1 – 4 cleaved the GFP from NS4B and GFP was localized to the nucleus of virally infected cells.
Nuclear GFP correlated with dengue antigen staining
Time-lapse imaging showed nuclear localization occurred as early as 8 h post infection.
Acknowledgements
We thank Drs. Kate Fitzgerald and Shanaka Rodrigo for helpful discussions, Diane Lang, Susannah Colt and Jurand Janus for technical assistance, Dr. Barry Falgout for providing the DENV-2 infectious clone and Dr. Jennifer Friedman for statistical analysis. This work was supported by NIH grants P01 AI034533, P20 GM103430-12 and U01 AI070484.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anonymous. Dengue: guidelines for diagnosis, treatment, prevention and control - new edition. Geneva: World Health Organization; 2009. [PubMed] [Google Scholar]
- Becerra A, Warke RV, Martin K, Xhaja K, de Bosch N, Rothman AL, Bosch I. Gene expression profiling of dengue infected human primary cells identifies secreted mediators in vivo. Journal of medical virology. 2009;81:1403–1411. doi: 10.1002/jmv.21538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera AK, Kuhn RJ, Smith JL. Functional characterization of cis and trans activity of the Flavivirus NS2B-NS3 protease. The Journal of biological chemistry. 2007;282:12883–12892. doi: 10.1074/jbc.M611318200. [DOI] [PubMed] [Google Scholar]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blok J. Genetic relationships of the dengue virus serotypes. The Journal of general virology. 1985;66(Pt 6):1323–1325. doi: 10.1099/0022-1317-66-6-1323. [DOI] [PubMed] [Google Scholar]
- Brinkworth RI, Fairlie DP, Leung D, Young PR. Homology model of the dengue 2 virus NS3 protease: putative interactions with both substrate and NS2B cofactor. The Journal of general virology. 1999;80(Pt 5):1167–1177. doi: 10.1099/0022-1317-80-5-1167. [DOI] [PubMed] [Google Scholar]
- Chase AJ, Medina FA, Munoz-Jordan JL. Impairment of CD4+ T cell polarization by dengue virus-infected dendritic cells. J Infect Dis. 2011;203:1763–1774. doi: 10.1093/infdis/jir197. [DOI] [PubMed] [Google Scholar]
- Clum S, Ebner KE, Padmanabhan R. Cotranslational membrane insertion of the serine proteinase precursor NS2B-NS3(Pro) of dengue virus type 2 is required for efficient in vitro processing and is mediated through the hydrophobic regions of NS2B. The Journal of biological chemistry. 1997;272:30715–30723. doi: 10.1074/jbc.272.49.30715. [DOI] [PubMed] [Google Scholar]
- Coller KE, Heaton NS, Berger KL, Cooper JD, Saunders JL, Randall G. Molecular determinants and dynamics of hepatitis C virus secretion. PLoS Pathog. 2012;8:e1002466. doi: 10.1371/journal.ppat.1002466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cressman DE, O'Connor WJ, Greer SF, Zhu XS, Ting JP. Mechanisms of nuclear import and export that control the subcellular localization of class II transactivator. Journal of immunology. 2001;167:3626–3634. doi: 10.4049/jimmunol.167.7.3626. [DOI] [PubMed] [Google Scholar]
- Falgout B, Pethel M, Zhang YM, Lai CJ. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. Journal of virology. 1991;65:2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavet O, Pines J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Developmental cell. 2010;18:533–543. doi: 10.1016/j.devcel.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead SB, Mahalingam S, Marovich MA, Ubol S, Mosser DM. Intrinsic antibody-dependent enhancement of microbial infection in macrophages: disease regulation by immune complexes. Lancet Infect Dis. 2010;10:712–722. doi: 10.1016/S1473-3099(10)70166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8:422–432. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd ML, Ling L, Tolfvenstam T, Mitchell W, Wong C, Kuznetsov VA, George J, Ong SH, Ruan Y, Wei CL, Gu F, Fink J, Yip A, Liu W, Schreiber M, Vasudevan SG. A genomics approach to understanding host response during dengue infection. Novartis Found Symp. 2006;277:206–214. doi: 10.1002/0470058005.ch15. discussion 214–207, 251–203. [DOI] [PubMed] [Google Scholar]
- Jones CT, Catanese MT, Law LM, Khetani SR, Syder AJ, Ploss A, Oh TS, Schoggins JW, MacDonald MR, Bhatia SN, Rice CM. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol. 2010;28:167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leardkamolkarn V, Sirigulpanit W. Establishment of a stable cell line coexpressing dengue virus-2 and green fluorescent protein for screening of antiviral compounds. Journal of biomolecular screening. 2012;17:283–292. doi: 10.1177/1087057111426903. [DOI] [PubMed] [Google Scholar]
- Leardkamolkarn V, Sirigulpanit W, Chotiwan N, Kumkate S, Huang CY. Development of Dengue type-2 virus replicons expressing GFP reporter gene in study of viral RNA replication. Virus research. 2012;163:552–562. doi: 10.1016/j.virusres.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Huang S, Lin Huang P, Lee Huang P. Live-cell real-time imaging reveals role of mitochondria in cell-to-cell transmission of HIV-1. Biochem Biophys Res Commun. 2011;415:384–389. doi: 10.1016/j.bbrc.2011.10.078. [DOI] [PubMed] [Google Scholar]
- Lin C, Chambers TJ, Rice CM. Mutagenesis of conserved residues at the yellow fever virus 3/4A and 4B/5 dibasic cleavage sites: effects on cleavage efficiency and polyprotein processing. Virology. 1993;192:596–604. doi: 10.1006/viro.1993.1076. [DOI] [PubMed] [Google Scholar]
- Liu L, Wen K, Li J, Hu D, Huang Y, Qiu L, Cai J, Che X. Comparison of plaque- and enzyme-linked immunospot-based assays to measure the neutralizing activities of monoclonal antibodies specific to domain III of dengue virus envelope protein. Clin Vaccine Immunol. 2012;19:73–78. doi: 10.1128/CVI.05388-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattia K, Puffer BA, Williams KL, Gonzalez R, Murray M, Sluzas E, Pagano D, Ajith S, Bower M, Berdougo E, Harris E, Doranz BJ. Dengue reporter virus particles for measuring neutralizing antibodies against each of the four dengue serotypes. PloS one. 2011;6:e27252. doi: 10.1371/journal.pone.0027252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medin CL, Rothman AL. Cell type-specific mechanisms of interleukin-8 induction by dengue virus and differential response to drug treatment. The Journal of infectious diseases. 2006;193:1070–1077. doi: 10.1086/502630. [DOI] [PubMed] [Google Scholar]
- Miller S, Sparacio S, Bartenschlager R. Subcellular localization and membrane topology of the Dengue virus type 2 Non-structural protein 4B. The Journal of biological chemistry. 2006;281:8854–8863. doi: 10.1074/jbc.M512697200. [DOI] [PubMed] [Google Scholar]
- Mosca JD, Pitha PM. Transcriptional and post transcriptional regulation of exogenous human beta interferon gene in simian cells defective in interferon synthesis. Molecular and cellular biology. 1986;6:2279–2283. doi: 10.1128/mcb.6.6.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Jordan JL, Laurent-Rolle M, Ashour J, Martinez-Sobrido L, Ashok M, Lipkin WI, Garcia-Sastre A. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. Journal of virology. 2005;79:8004–8013. doi: 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Jordan JL, Sanchez-Burgos GG, Laurent-Rolle M, Garcia-Sastre A. Inhibition of interferon signaling by dengue virus. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:14333–14338. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale ZD, Patkar C, Rothman AL. Viral replication and paracrine effects result in distinct, functional responses of dendritic cells following infection with dengue 2 virus. Journal of leukocyte biology. 2008;84:1028–1038. doi: 10.1189/jlb.0208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DR, Sun P, Celluzzi C, Bisbing J, Pang S, Sun W, Marovich MA, Burgess T. Differential effects of dengue virus on infected and bystander dendritic cells. J Virol. 2005;79:2432–2439. doi: 10.1128/JVI.79.4.2432-2439.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paloheimo O, Ihalainen TO, Tauriainen S, Valilehto O, Kirjavainen S, Niskanen EA, Laakkonen JP, Hyoty H, Vihinen-Ranta M. Coxsackievirus B3-induced cellular protrusions: structural characteristics and functional competence. J Virol. 2011;85:6714–6724. doi: 10.1128/JVI.00247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MS, Shaw ML, Munoz-Jordan J, Cros JF, Nakaya T, Bouvier N, Palese P, Garcia-Sastre A, Basler CF. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. Journal of virology. 2003;77:1501–1511. doi: 10.1128/JVI.77.2.1501-1511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena J, Harris E. Early dengue virus protein synthesis induces extensive rearrangement of the endoplasmic reticulum independent of the UPR and SREBP-2 pathway. PloS one. 2012;7:e38202. doi: 10.1371/journal.pone.0038202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S, Ketner G, Levis R, Falgout B. Infectious RNA transcripts from full-length dengue virus type 2 cDNA clones made in yeast. Journal of virology. 1997;71:5366–5374. doi: 10.1128/jvi.71.7.5366-5374.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Ortiz ZG, Warke RV, Pacheco L, Xhaja K, Sarkar D, Fisher PB, Shaw SK, Martin KJ, Bosch I. Discovering innate immunity genes using differential display: a story of RNA helicases. Journal of cellular physiology. 2006;209:636–644. doi: 10.1002/jcp.20797. [DOI] [PubMed] [Google Scholar]
- Rand U, Rinas M, Schwerk J, Nohren G, Linnes M, Kroger A, Flossdorf M, Kaly-Kullai K, Hauser H, Hofer T, Koster M. Multi-layered stochasticity and paracrine signal propagation shape the type-I interferon response. Mol Syst Biol. 2012;8:584. doi: 10.1038/msb.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico-Hesse R. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology. 1990;174:479–493. doi: 10.1016/0042-6822(90)90102-w. [DOI] [PubMed] [Google Scholar]
- Rothman AL. Cellular immunology of sequential dengue virus infection and its role in disease pathogenesis. Current topics in microbiology and immunology. 2010;338:83–98. doi: 10.1007/978-3-642-02215-9_7. [DOI] [PubMed] [Google Scholar]
- Schoggins JW, Dorner M, Feulner M, Imanaka N, Murphy MY, Ploss A, Rice CM. Dengue reporter viruses reveal viral dynamics in interferon receptor-deficient mice and sensitivity to interferon effectors in vitro. Proc Natl Acad Sci U S A. 2012;109:14610–14615. doi: 10.1073/pnas.1212379109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiryaev SA, Kozlov IA, Ratnikov BI, Smith JW, Lebl M, Strongin AY. Cleavage preference distinguishes the two-component NS2B-NS3 serine proteinases of Dengue and West Nile viruses. The Biochemical journal. 2007;401:743–752. doi: 10.1042/BJ20061136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava N, Sripada S, Kaur J, Shah PS, Cecilia D. Insights into the internalization and retrograde trafficking of Dengue 2 virus in BHK-21 cells. PloS one. 2011;6:e25229. doi: 10.1371/journal.pone.0025229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons CP, Farrar JJ, Nguyen vV, Wills B. Dengue. The New England journal of medicine. 2012;366:1423–1432. doi: 10.1056/NEJMra1110265. [DOI] [PubMed] [Google Scholar]
- Sivaraman D, Biswas P, Cella LN, Yates MV, Chen W. Detecting RNA viruses in living mammalian cells by fluorescence microscopy. Trends Biotechnol. 2011;29:307–313. doi: 10.1016/j.tibtech.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Thacore HR, Lin HY, Davis PJ, Schoenl M. Effect of protein kinase C inhibitors on interferon-beta production by viral and non-viral inducers. The Journal of general virology. 1990;71(Pt 12):2833–2839. doi: 10.1099/0022-1317-71-12-2833. [DOI] [PubMed] [Google Scholar]
- Twiddy SS, Farrar JJ, Vinh Chau N, Wills B, Gould EA, Gritsun T, Lloyd G, Holmes EC. Phylogenetic relationships and differential selection pressures among genotypes of dengue-2 virus. Virology. 2002;298:63–72. doi: 10.1006/viro.2002.1447. [DOI] [PubMed] [Google Scholar]
- van der Schaar HM, Rust MJ, Chen C, van der Ende-Metselaar H, Wilschut J, Zhuang X, Smit JM. Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog. 2008;4:e1000244. doi: 10.1371/journal.ppat.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Schaar HM, Rust MJ, Waarts BL, van der Ende-Metselaar H, Kuhn R, Wilschut J, Zhuang X, Smit JM. Characterization of the early events in dengue virus cell entry by biochemical assays and single-virus tracking. Journal of virology. 2007;81:12019–12028. doi: 10.1128/JVI.00300-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn DW, Green S, Kalayanarooj S, Innis BL, Nimmannitya S, Suntayakorn S, Endy TP, Raengsakulrach B, Rothman AL, Ennis FA, Nisalak A. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. The Journal of infectious diseases. 2000;181:2–9. doi: 10.1086/315215. [DOI] [PubMed] [Google Scholar]
- Vaughn DW, Hoke CH, Jr, Yoksan S, LaChance R, Innis BL, Rice RM, Bhamarapravati N. Testing of a dengue 2 live-attenuated vaccine (strain 16681 PDK 53) in ten American volunteers. Vaccine. 1996;14:329–336. doi: 10.1016/0264-410x(95)00167-y. [DOI] [PubMed] [Google Scholar]
- Whitehorn J, Farrar J. Dengue. Br Med Bull. 2010;95:161–173. doi: 10.1093/bmb/ldq019. [DOI] [PubMed] [Google Scholar]
- Yang CC, Tsai MH, Hu HS, Pu SY, Wu RH, Wu SH, Lin HM, Song JS, Chao YS, Yueh A. Characterization of an efficient dengue virus replicon for development of assays of discovery of small molecules against dengue virus. Antiviral research. 2013 doi: 10.1016/j.antiviral.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Zou G, Xu HY, Qing M, Wang QY, Shi PY. Development and characterization of a stable luciferase dengue virus for high-throughput screening. Antiviral Res. 2011;91:11–19. doi: 10.1016/j.antiviral.2011.05.001. [DOI] [PubMed] [Google Scholar]