Abstract
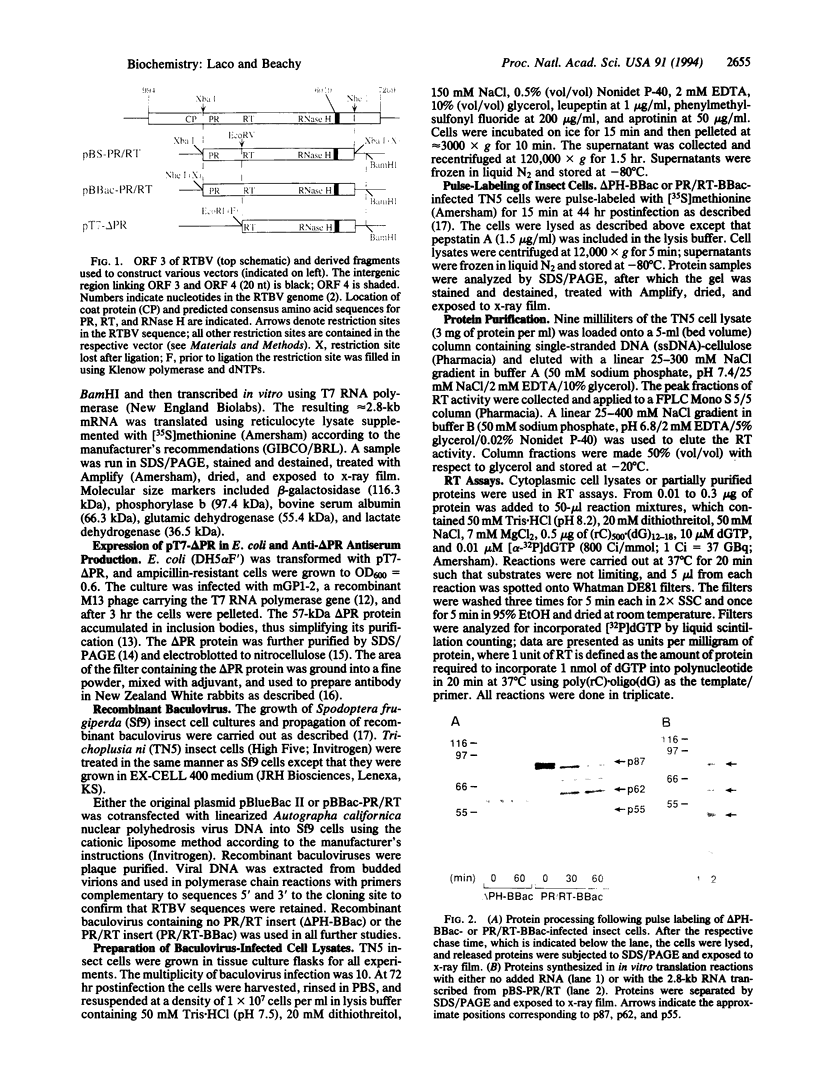
Rice tungro bacilliform virus (RTBV) is a newly described badnavirus and proposed member of the plant pararetrovirus group. RTBV open reading frame 3 is predicted to encode a capsid protein, protease (PR), and reverse transcriptase (RT) and has the capacity to encode other proteins of as yet unknown function. To study the possible enzymatic activities encoded by open reading frame 3, a DNA fragment containing the putative PR and RT domains was used to construct the recombinant baculovirus PR/RT-BBac. Trichoplusia ni insect cells infected with PR/RT-BBac were used in pulse-labeling experiments and demonstrated synthesis of an 87-kDa polyprotein that corresponds in molecular mass to that predicted from the PR/RT DNA coding sequence. The 87-kDa polyprotein was processed with concomitant accumulation of 62-kDa (p62) and 55-kDa (p55) proteins. Amino-terminal sequencing of p62 and p55 determined that they mapped to the PR/RT domain and shared common amino termini. p62 and p55 were purified and exhibited both RT and DNA polymerase activities using synthetic primer/template substrates. Only p55 had detectable ribonuclease H activity, an activity intrinsic to all reverse transcriptases studied to date. Characterization of the RTBV RT provides a biochemical basis for classifying RTBV as a pararetrovirus and will lead to further studies of these proteins and their role in virus replication.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bhattacharyya-Pakrasi M., Peng J., Elmer J. S., Laco G., Shen P., Kaniewska M. B., Kononowicz H., Wen F., Hodges T. K., Beachy R. N. Specificity of a promoter from the rice tungro bacilliform virus for expression in phloem tissues. Plant J. 1993 Jul;4(1):71–79. doi: 10.1046/j.1365-313x.1993.04010071.x. [DOI] [PubMed] [Google Scholar]
- Chang L. J., Pryciak P., Ganem D., Varmus H. E. Biosynthesis of the reverse transcriptase of hepatitis B viruses involves de novo translational initiation not ribosomal frameshifting. Nature. 1989 Jan 26;337(6205):364–368. doi: 10.1038/337364a0. [DOI] [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Johnson M. S., McClure M. A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989 Mar;64(1):1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- Gordon K., Pfeiffer P., Fütterer J., Hohn T. In vitro expression of cauliflower mosaic virus genes. EMBO J. 1988 Feb;7(2):309–317. doi: 10.1002/j.1460-2075.1988.tb02814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell S. H., Hull R. Replication of cauliflower mosaic virus and transcription of its genome in turnip leaf protoplasts. Virology. 1978 May 15;86(2):468–481. doi: 10.1016/0042-6822(78)90086-7. [DOI] [PubMed] [Google Scholar]
- Huberman J. A. New views of the biochemistry of eucaryotic DNA replication revealed by aphidicolin, an unusual inhibitor of DNA polymerase alpha. Cell. 1981 Mar;23(3):647–648. doi: 10.1016/0092-8674(81)90426-8. [DOI] [PubMed] [Google Scholar]
- Jacobo-Molina A., Arnold E. HIV reverse transcriptase structure-function relationships. Biochemistry. 1991 Jul 2;30(26):6351–6356. doi: 10.1021/bi00240a001. [DOI] [PubMed] [Google Scholar]
- Jones M. C., Gough K., Dasgupta I., Rao B. L., Cliffe J., Qu R., Shen P., Kaniewska M., Blakebrough M., Davies J. W. Rice tungro disease is caused by an RNA and a DNA virus. J Gen Virol. 1991 Apr;72(Pt 4):757–761. doi: 10.1099/0022-1317-72-4-757. [DOI] [PubMed] [Google Scholar]
- Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992 Jun 26;256(5065):1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Le Grice S. F., Grüninger-Leitch F. Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur J Biochem. 1990 Jan 26;187(2):307–314. doi: 10.1111/j.1432-1033.1990.tb15306.x. [DOI] [PubMed] [Google Scholar]
- Le Grice S. F., Naas T., Wohlgensinger B., Schatz O. Subunit-selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 1991 Dec;10(12):3905–3911. doi: 10.1002/j.1460-2075.1991.tb04960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu R. D., Bhattacharyya M., Laco G. S., De Kochko A., Rao B. L., Kaniewska M. B., Elmer J. S., Rochester D. E., Smith C. E., Beachy R. N. Characterization of the genome of rice tungro bacilliform virus: comparison with Commelina yellow mottle virus and caulimoviruses. Virology. 1991 Nov;185(1):354–364. doi: 10.1016/0042-6822(91)90783-8. [DOI] [PubMed] [Google Scholar]
- Rucheton M., Lelay M. N., Jeanteur P. Evidence from direct visualization after denaturing gel electrophoresis that RNase H is associated with MSV-MuLV reverse transcripase. Virology. 1979 Aug;97(1):221–223. doi: 10.1016/0042-6822(79)90392-1. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanese N., Goff S. P. Domain structure of the Moloney murine leukemia virus reverse transcriptase: mutational analysis and separate expression of the DNA polymerase and RNase H activities. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1777–1781. doi: 10.1073/pnas.85.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M., Hull R., Bryant J. A., Maule A. J. Isolation of a fraction from cauliflower mosaic virus-infected protoplasts which is active in the synthesis of (+) and (-) strand viral DNA and reverse transcription of primed RNA templates. Nucleic Acids Res. 1985 Jun 25;13(12):4557–4576. doi: 10.1093/nar/13.12.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torruella M., Gordon K., Hohn T. Cauliflower mosaic virus produces an aspartic proteinase to cleave its polyproteins. EMBO J. 1989 Oct;8(10):2819–2825. doi: 10.1002/j.1460-2075.1989.tb08428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]