Abstract
The cellular response to oxygen deprivation is governed largely by a family of transcription factors known as Hypoxia Inducible Factors (HIFs). This review focuses on the molecular mechanisms by which HIFs regulate the transcriptional apparatus to enable the cellular and organismal response to hypoxia. We discuss here how the various HIF polypeptides, their post-translational modifications, binding partners and transcriptional cofactors affect RNA polymerase II activity to drive context-dependent transcriptional programs during hypoxia.
Keywords: HIF1A, HIF2A, HIF3A, ARNT, hydroxylation, CBP, p300, CDK8, Mediator, PKM2, SWI/SNF, histones, RNA polymerase II, chromatin, hypoxia response element
The cellular adaptation to hypoxia sustains the growth of rapidly proliferating tissues such as developing embryos and solid tumors. Many of the cellular responses to a hypoxic environment are orchestrated by a group of DNA binding proteins known as Hypoxia Inducible Factors (HIFs). These transcription factors induce the expression of genes that mediate a metabolic rewiring of the hypoxic cell, induce formation of new blood vessels around the hypoxic tissue, and can promote cell survival in this scenario. Consequently, HIFs are essential for embryonic development, but also exploited by cancer cells during the progression of many solid tumors (Iyer et al., 1998a, Ryan et al., 1998, Tian et al., 1998, Compernolle et al., 2002, Scortegagna et al., 2003, Tang et al., 2004, Koh et al., 2008b, Wong et al., 2011, Huang et al., 2013). More recently, activation of HIFs in fat adipose tissue was implicated in the development of dietary obesity, a key risk factor for type II diabetes (Krishnan et al., 2012). HIFs have also been linked to other pathologies, such as atherosclerosis, pulmonary hypertension and abdominal aortic aneurysm formation (Erdozain et al., 2011, Semenza, 2012a). Thus, a detailed understanding of the molecular mechanism of action of HIFs will advance multiple areas of biomedical research. This review focuses on the available knowledge about the molecular mechanisms by which HIFs regulate gene expression at the transcriptional level, with the goal of identifying key venues of future research.
1. The HIF family of transcription factors
HIFs are basic helix-loop-helix DNA binding proteins of the PER-ARNT-SIM family (bHLH-PAS). These transcription factors function as heterodimers composed of an oxygen-regulated alpha subunit and a stably expressed beta subunit (Semenza and Wang, 1992, Wang et al., 1995, Tian et al., 1997, Gu et al., 1998). In mammals, the alpha subunits are encoded by three genes: HIF1A, EPAS1 (also known as and referred to hereafter as HIF2A), and HIF3A (Ema et al., 1997, Gu et al., 1998, Iyer et al., 1998b). Under conditions of normal oxygen tension, the alpha subunits are hydroxylated at key proline and asparagine residues, which inhibits their transactivation function and targets them for proteasomal degradation (Huang et al., 1998, O'Rourke et al., 1999, Ivan et al., 2001, Jaakkola et al., 2001, Mahon et al., 2001, Lando et al., 2002a, Lando et al., 2002b). The HIF1 beta subunits (HIF1B), also known as aryl hydrocarbon receptor nuclear translocators (ARNT), are encoded by two genes ARNT1 and ARNT2. HIF1B is stably expressed and is also an obligate partner for the aryl hydrocarbon receptor (AHR) (Hoffman et al., 1991, Wang et al., 1995, McIntosh et al., 2010).
Upon hypoxia, the HIF alpha subunits are stabilized and accumulate in the nucleus, where they dimerize with HIF1B, allowing them to bind to DNA and stimulate the transcription of their target genes. This allows the coordinate activation of genes essential in the adaptive response to hypoxia including pathways that decrease the cellular demand for O2 and increase O2 delivery and tissue re-oxygenation (Majmundar et al., 2010, Greer et al., 2012). For example, the first HIF target gene identified was erythropoietin (EPO), a glycoprotein hormone that stimulates the generation of new red blood cells, thereby increasing O2 delivery. Investigation of the induction of EPO in response to hypoxia led to the identification of HIF1 as a factor binding to an enhancer in the 3’ region of this gene (Semenza and Wang, 1992). In a waterfall of subsequent publications, Semenza and colleagues further characterized HIF1A as a bHLH-PAS transcription factor, determined its dimerization partner, HIF1B, and demonstrated that the DNA binding activity of HIF1A under hypoxia was a general cellular response to decreased oxygen tension (Wang and Semenza, 1993b, Wang and Semenza, 1993a, Wang et al., 1995, Wang and Semenza, 1995). The discovery of a second oxygen-sensitive nuclear factor by multiple independent groups followed soon thereafter. HIF2A shares high sequence homology with HIF1A and functions in a similar manner. HIF2A also dimerizes with HIF1B upon hypoxic induction and stimulates the expression of a distinct set of target genes (Ema et al., 1997, Flamme et al., 1997, Tian et al., 1997). Although identified via homology searches for additional bHLH-PAS and HIF-related proteins, the third known alpha subunit, HIF3A displays less similarity as well as distinct functional characteristics compared to HIF1A and HIF2A (Gu et al., 1998, Makino et al., 2001). Interestingly, there are at least six HIF3A splicing variants and these display variable oxygen-sensitivity, ability to dimerize with HIF1B, and differing transcription regulatory functions (Makino et al., 2002, Maynard et al., 2003, Pasanen et al., 2010, Heikkilä et al., 2011). The splice variant HIF3A4 is highly similar in sequence and function to the murine Inhibitory PAS domain protein 1 (IPAS). Notably, this variant is neither oxygen-sensitive nor possesses transactivation capacity but rather acts as a dominant negative regulator of hypoxia-induced HIF gene expression by sequestering HIF1A to prevent HIF1B dimerization and DNA binding (Makino et al., 2002, Maynard et al., 2005).
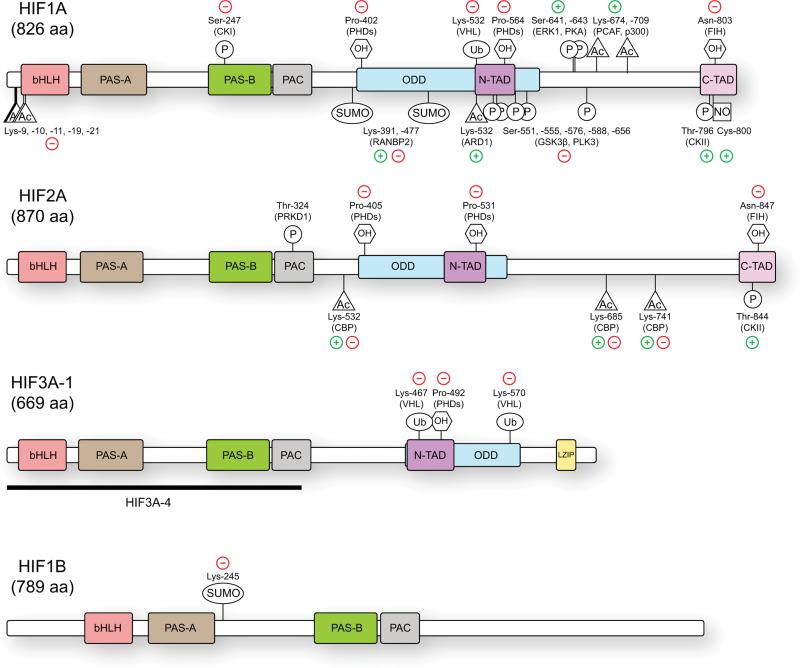
Domain organization of the HIF polypeptides
Overall, the HIF family possesses a conserved protein domain structure (Figure 1). The three alpha isoforms as well as HIF1B carry an amino-terminal bHLH that is necessary for DNA binding as well as PAS-A and PAS-B domains that are required for heterodimerization. Both the bHLH and PAS domains exhibit strong sequence and functional conservation among the HIFs. In fact, while the PAS domains between HIF1A and HIF2A exhibit approximately 70% identity, their bHLH domains share 85% identity with the basic region consisting of almost identical sequences. Consequently, HIF1A and HIF2A are able to bind indistinguishable DNA sequences (Tian et al., 1997). Comparatively, the bHLH and PAS domains of HIF3A (common to most isoforms) share only 74% and 52-58% identity with HIF1A and HIF2A, respectively, which reveals the more divergent nature of this paralog (Hara et al., 2001).
Figure 1.
HIF protein domains and post-translational modifications. The HIF proteins are comprised of several conserved domains that are involved in DNA binding (basic Helix-Loop-Helix, bHLH), protein-protein interactions and dimerization (PER-ARNT-SIM, PAS-A, PAS-B, and PAS-associated C-terminal domain), oxygen-dependent degradation (ODD) and transcriptional activation (N-TAD, C-TAD). Numerous HIF3A isoforms exist, with several longer forms possessing transactivation and leucine zipper (LZIP) domains (HIF3A-1) while others lack any known transactivation domains and act as negative regulators (HIF3A-4). Multiple post-translational modifications are known to modulate HIF protein stability and transcriptional activity. Selected modifications are shown here along with the enzyme responsible and the overall positive (+) or negative (−) effects on HIF transcriptional function.
HIF1A and HIF2A also carry N- and C-terminal transactivation domains (N-TAD and C-TAD) that are required for activation of HIF target genes. Oxygen-dependent degradation domains (ODD) within the alpha subunits confer oxygen-regulated turnover and overlap the N-TADs (Jiang et al., 1996, Jiang et al., 1997, Huang et al., 1998, O'Rourke et al., 1999, Lendahl et al., 2009). The ODD is a highly conserved domain that controls the activity and stability of the alpha subunits, as it contains the key asparagine (N) and proline (P) residues targeted for hydroxylation in normoxic conditions. As explained below, these hydroxylation events effectively shut down the HIF transcriptional program.
Many of the various HIF3A splicing isoforms carry an N-TAD but lack a C-TAD with several harboring a C-terminal leucine zipper motif of unknown function (Maynard et al., 2003, Pasanen et al., 2010). Although the HIF3A splicing variants have been shown to physically associate with HIF1A, HIF2A, and HIF1B (Heikkilä et al., 2011) and many variants were found to reduce activity of HRE reporters, HIF3A1 has been shown to weakly transactivate target genes (Yamashita et al., 2008, Huang et al., 2013). Interestingly, the negative regulation was only apparent under HIF1B-limiting conditions suggesting that their negative function requires competition with HIF1A or HIF2A. The best-characterized isoform, HIF3A4 lacks the known transactivation domains and acts as a negative regulator of HIF1A (Makino et al., 2001, Maynard et al., 2005). HIF3A4 is thought to be expressed in specific tissues, such as corneal lens epithelium, that require stringent inhibition of neovascularization upon hypoxia (Makino et al., 2001).
The presence of two distinct TADs confers the alpha subunits with both common and unique regulatory properties. Although the C-TAD exhibits the highest degree of sequence divergence, it acts to regulate the transactivation of target genes common to both HIF1A and HIF2A through coactivator recruitment (Ema et al., 1999, Dayan et al., 2006, Hu et al., 2007). On the other hand, the N-TADs of HIF1A and HIF2A confer target gene selectivity between these two family members and this could be explained by the action of distinct transcriptional cofactors recognizing the N-TAD of HIF1A versus HIF2A (see the following section HIF Transcriptional Cofactors for further discussion)(Aprelikova et al., 2006, Hu et al., 2007).
Post-translational modifications of HIFs
Under normoxic conditions, HIF alpha subunits are continually transcribed and translated. Sufficient oxygen tension however leads to their proteasomal degradation. In normoxia, a family of prolyl-4-hydroxylases (PHD1-4), most prominently PHD2 (also known as EGLN1), hydroxylate the oxygen-sensitive alpha subunits (Berra et al., 2003, Appelhoff, 2004, Landazuri et al., 2006, Koivunen et al., 2007). The PHDs require iron, α-ketoglutarate, ascorbate and molecular oxygen in order to modify conserved proline residues (Pro-402 and Pro-564 in HIF1A; Pro-405 and Pro-531 in HIF2A; Pro-492 in HIF3A; Figure 1) (Ivan et al., 2001, Jaakkola et al., 2001). These prolines all exist within LXXLAP motifs found in the ODDs of HIF1A, HIF2A, and HIF3A (Masson et al., 2001, Maynard et al., 2003). Notably, some HIF3A isoforms contain only a single prolyl hydroxylation site, while others are missing this region entirely and are therefore not subject to regulation in response to oxygen. Proline hydroxylation allows binding of the von Hippel Lindau (VHL) E3 ubiquitin ligase complex, which poly-ubiquitinates the alpha subunits triggering their degradation by the proteasome (Huang et al., 1998, Ohh et al., 2000, Tanimoto et al., 2000, Foxler et al., 2012). It is worth mentioning that PHD2 and PHD3 (EGLN3) are HIF target genes, which sets up possibly negative feedback loops within the network.
HIF1A and HIF2A are subject to an additional regulatory hydroxylation. An enzyme known as Factor Inhibiting HIF (FIH) hydroxylates an asparagine (Asn-803 for HIF1A, Asn-847 for HIF2A) within the C-TADs of HIF1A and HIF2A, thereby disrupting the transactivation potential of these factors (Figure 1) (Mahon et al., 2001, Lando et al., 2002a, Lando et al., 2002b). The mechanism by which this hydroxylation inhibits HIF transactivation appears to be by interfering with the binding of cofactors to the C-TAD. For example, it has been shown that the interaction between the C-TAD and CBP/p300 involves conformational changes that bury this asparagine deep within the binding interface explaining how hydroxylation is able to block the CBP/p300 interaction (Dames et al., 2002, Freedman et al., 2003). Importantly, FIH has a lower Km for oxygen than the PHDs, meaning that it is able to maintain activity at lower oxygen tensions and suggesting that even upon stabilization, HIFs are subject to oxygen-dependent regulation of their activities. Notably, FIH appears to more efficiently hydroxylate HIF1A than HIF2A (Koivunen et al., 2004, Bracken et al., 2006).
Multiple oxygen-independent mechanisms can also lead to HIF stabilization. Importantly, growth factor signaling results in stabilized HIF due to the activation of mammalian target of rapamycin (mTOR) and a subsequent increase in the rate of HIF translation (Laughner et al., 2001). As a result, the abundance of HIF alpha proteins rises disproportionately to PHDs, thus leading to stabilized HIF proteins. Additionally, various metabolites such as fumarate and succinate inhibit PHDs often by deregulating their enzymatic activity or outcompeting them for necessary cofactors, including iron and ascorbate (Salnikow et al., 2004). Nutrient conditions have also been found to regulate HIF stability independent of oxygen tension. For example, accumulation of lactate and pyruvate has been found to result in oxygen-independent HIF stabilization (Lu et al., 2002). Additionally, it has been shown that low glucose conditions led to increased expression of HIFA target genes in HIF1A +/+ but not HIF1A −/− embryonic stem (ES) cells (Carmeliet et al., 1998). Notably, the effect of nutrient conditions on HIFA stability may be dependent on genetic background. In non-tumor cell lines, low glucose conditions were found to increase HIF1A stabilization (Dehne, 2010), whereas in cancer cells high glucose levels resulted in stabilized HIF1A (Lu et al., 2005).
Several additional proteins associated with E3 ubiquitin ligase activity, including receptor of activated protein kinase C (RACK1), carboxyl terminus of Hsp70-interacting protein (CHIP) and hypoxia-associated factor (HAF), also regulate the stability of alpha subunits via ubiquitination and proteasomal degradation; however, they do so independently of oxygen tension (Liu et al., 2007, Koh et al., 2008a, Luo et al., 2010). For example, Bento and colleagues discovered that in the presence of high glucose concentrations, the accumulation of methylglyoxal results in the O2/PHD/VHL-independent degradation of HIF1A by Hsp70-associated recruitment of CHIP (Bento et al., 2010). In proliferating cells, HAF ubiquitinates HIF1A, but not HIF2A, targeting it for destruction regardless of oxygen tension (Koh et al., 2008a). It remains to be defined if these proteins simply regulate HIF stability or directly modulate transactivation function.
Phosphorylation of HIF1A and HIF2A subunits has been demonstrated to enhance transactivation of target genes by either disrupting interaction with VHL and thereby stabilizing HIF, or by increasing the affinity of HIF for transcriptional coactivators (Richard et al., 1999). Hypoxia-induced phosphorylation by casein kinase II (CKII) has been demonstrated in the C-TAD at conserved threonine residues (Thr796 for HIF1A, Thr844 for HIF2A) and mutation of these residues decreased reporter activity, possibly by increasing HIF1A affinity for FIH (Conrad et al., 1999, Gradin et al., 2002, Lancaster et al., 2004, Cho et al., 2007). HIF1A phosphorylation also seems to be regulated in a cell type- and stimulus-specific fashion. In endothelial cells, distinct kinases phosphorylate HIF1A dependent on the duration of hypoxia. In response to acute hypoxia (1% O2), ERK1 directly phosphorylates the C-terminal domain of HIF1A (Ser-641 and Ser-643). These modifications mask a nuclear export signal leading to increased nuclear accumulation and transcriptional activity (Mylonis et al., 2006). Interestingly, in response to intermittent hypoxia (1 hr 1% O2 followed by 30 min re-oxygenation), PKA (protein kinase A) phosphorylates HIF1A even though both ERK isoforms (p42/p44) are activated and capable of phosphorylation in this setting (Minet et al., 2000, Toffoli et al., 2007).
Other phosphorylation events however lead to decreased HIF stability or activity. For example, casein kinase 1 (CKI), a constitutively active monomeric enzyme that regulates proteins involved in cellular proliferation, differentiation and circadian rhythm, phosphorylates HIF1A within the PAS-B domain (Ser-247) inhibiting its ability to associate with ARNT, thus decreasing HIF1A induced gene expression (Knippschild et al., 2005, Kalousi et al., 2010). Negative regulation of HIF1A also occurs by glycogen synthase kinase 3β (GSK3β) downstream of AKT/PI3K signaling under longer periods of hypoxia (Mottet et al., 2003). GSK3β phosphorylates HIF1A at several serine residues within the ODD (Figure 1; Ser551, 555, and 589) leading to decreased HIF1A stability and degradation by the proteasome in a VHL-independent manner (Flügel et al., 2007). Similarly, polo-like kinase 3 (PLK3), an important regulator of cell cycle progression, destabilizes HIF1A by phosphorylation at Ser576 and 659 (Xu et al., 2010).
S-Nitrosylation is the redox-related addition of nitric oxide (NO) moiety to sulfhydryl groups and is implicated in many pathologic and physiologic responses (Foster et al., 2003). S-nitrosylation of HIF1A at Cys-800 appears to increase the transcriptional activity of the protein (Yasinska and Sumbayev, 2003), however the addition of a polar NO group to this buried residue may disrupt the interaction between HIF1A and the coactivators CBP/p300 (Cho et al., 2007). It is possible that this modification increases the transcriptional output independently of CBP/p300 such as by interaction with other coactivators. Similarly, the effect of HIF1A sumoylation is a matter of debate (Núñez-O'Mara and Berra, 2013). Analyses by different groups report sumoylation at the same residues, Lys-391 and Lys-477, by RanBP2; however, they disagree on whether these modifications enhance or disrupt the transactivation potential of HIF1A (Bae et al., 2004, Berta et al., 2007).
Acetylation status appears to regulate HIF1A and HIF2A both positively and negatively. Several investigations of HIFA protein acetylation demonstrate that acetylation can have different downstream effects depending on the location of the modified residue. Geng et al. (2011) have linked acetylation of amino-terminal HIF1A lysines (K9, K10, K11, K19 and K21) to decreased protein stability and impaired activation of a subset of HIF1A target genes. Correspondingly, de-acetylation of these residues by HDAC4 appears to increase HIF1A protein levels and increase activation of target genes (Geng et al, 2011).
Conversely, acetylation of lysines located in the carboxy-terminal region of HIF1A (K674 and K709) boosts HIF1A protein levels and enhances target gene activation by distinct mechanisms (Lim et al, 2010, Geng et al, 2012). The p300/CBP-associated factor (PCAF) can acetylate K674 of HIF1A leading to increased HIF1A protein levels and binding of the transcriptional co-activator p300, and to enhanced transactivation of certain HIF1A target genes (Lim et al, 2010). Acetylation of K709 was shown to require the acetyl-transferase activity of p300 (Geng et al, 2012). As this residue is also subject to poly-ubiquitination, the positive effect of acetylation could be explained by blocking degradation of HIF1A.
Lysine acetylation of HIF1A at K532 has been demonstrated (Xenaki et al., 2008, Jeong et al., 2002) but remains controversial. Jeong et al. (2002) found that the protein acetyl-transferase ARD1 directly interacted with HIF1A ODD to modify K532 in an oxygen-dependent manner. In an in vitro pull-down assay, acetylation of the HIF1A ODD produced an enhanced association with VHL, and resulted in decreased HIF1A protein stability. The authors posit acetylation at K532 as necessary for maximal degradation of HIF1A under normoxia (Jeong et al, 2002). However several other groups have failed to detect acetylation of K532, even in the same cell lines (Arnesen et al., 2005, Murray-Rust et al., 2006, Zhong et al., 2010).
HIF2A can be acetylated at K385, K685, and K741 but there is contradictory evidence concerning the role of these modifications. It has been reported that in Hep3B cells, CBP modifies these residues under hypoxic conditions resulting in increased EPO gene expression (Chen et al., 2012). In contrast, deacetylation of these residues by the redox-sensitive Sirtuin1 (SIRT1) was also found to augment the transcriptional activity of HIF2A target genes (Dioum et al., 2009). Furthermore, deacetylation by SIRT1 at K674 was shown to have a repressive effect on HIF1A transcriptional regulation (Lim et al., 2010). Clearly, much work remains in order to clearly define the role that many of these acetylation events play in the regulation of both HIF protein stability and transcriptional activity.
2. The HIF transcriptional program
The hypoxia response element
Upon translocation to the nucleus, HIF-A/B heterodimers bind the core consensus sequence 5’-(A/G)CGTG-3’ within the hypoxia-response element (HRE). Although the HRE consensus sequence is highly abundant across the genome, less than 1% of potential sites are bound by HIFs in response to hypoxia, as demonstrated by a number of recent genome-wide analyses of HIF chromatin occupancy (Schödel et al. , 2011, Mole et al. , 2009, Xia and Kung, 2009, Schödel et al., 2013). These analyses revealed that bound HREs showed a general preference for A over G at position one and T at the position immediately upstream of the core HRE (Wenger et al., 2005, Mole et al., 2009, Xia and Kung, 2009, Schödel et al., 2011). However, no other features of the sequences surrounding the core HRE were identified at the HIF-bound sites, suggesting that there are determinants other than DNA sequence contributing to HIF binding.
In addition to the core HRE, the promoters of several well-established and newly identified HIF1A target genes possess an inverted, imperfect repeat of the HRE (5’-CAGGT-3’) known as the HIF ancillary sequence (HAS). Using site-directed mutagenesis and luciferase-based reporter assays, Kimura and colleagues demonstrated that alteration or deletion of this sequence severely compromised VEGF and EPO reporter induction in response to hypoxia or nitric oxide (Kimura et al., 1999, Kimura et al., 2001). Interestingly, a spacing requirement of 8-9 nucleotides downstream of the HRE is necessary for the HAS to manifest a functional effect. Other than this, the function of the HAS remains unknown (Zagórska, 2004, Wenger et al., 2005).
Functional HREs tend to localize at the proximal promoters of target genes, though they may also act over long-range distances. Importantly, HIFs appear to preferentially bind HREs within regions of permissive chromatin that display DNAse I hypersensitivity, RNAPII enrichment, histone modifications and basal transcriptional activity under normoxic conditions (Mole et al., 2009, Xia and Kung, 2009, Schödel et al., 2011, Galbraith et al., 2013). This may enable the HIF proteins to mount a rapid transcriptional response upon their stabilization by hypoxia and may also explain cell type-specific activation of HIF target genes. This phenomenon appears to be widespread, and has been demonstrated for other stimulus-responsive transcription programs (Gilchrist et al., 2008, Espinosa, 2010, Rahl et al., 2010). Additional regulatory mechanisms must be at work however as only a small percentage of consensus sequences at so-called “permissive loci” are bound by HIF during hypoxia (Xia and Kung, 2009, Schödel et al., 2013).
Functional interplay between HIFs and other transcription factors
Sequence analysis of regions surrounding functional HREs revealed motifs characteristic of other transcription factors, including FOS, CREB, CEBPB, NFY, MIF and E2F among others, suggesting that a functional interplay among DNA-binding proteins may qualify the HIF transcriptional response (Mole et al., 2009, Villar et al., 2012). Indeed, functional analysis testing this notion in a cervical carcinoma cell line (HeLa) revealed that cooperativity of transcription factors with HIF1A fine-tunes the expression of HIF target genes. Utilizing luciferase reporter assays, the authors compared the induction of well-characterized HIF1A target genes in response to hypoxia when either the HRE or neighboring transcription factor binding sites were mutated. While altering the HRE completely abrogated the transcriptional response, modification of cooperative transcription factor binding sites tested significantly reduced the transcriptional output (Villar et al., 2012). This indicates that HIF1A may stimulate transcription either by means of cooperative DNA binding or cooperative recruitment of coactivators. Along these lines, a recent investigation by Pawlus and colleagues demonstrated that STAT3 functions cooperatively with HIF1A to recruit RNAPII to HIF1A target genes, including VEGF, CA9, and PGK1 (Pawlus et al., 2013). Several reports indicate that HIF2A also functions through partnership with other DNA binding proteins, such as USF, SP1, and ELK acting at the same enhancer (Hu et al., 2007, Salnikow et al., 2008, Wang et al., 2010b, Koizume et al., 2012, Pawlus et al., 2012). Several studies have revealed cell type-specific HIF-dependent gene expression profiles (Chi et al., 2006, Warnecke et al., 2008, Xia and Kung, 2009, Ortiz-Barahona et al., 2010), thus highlighting the importance of contributions by other transcription factors acting with HIFs. Consequently, their availability and/or activation may in part explain the variable nature of the HIF transcriptional program (Pawlus and Hu, 2013).
Interestingly, HIF1A has been shown to contribute to transcriptional control independently of its DNA binding activity, working instead in partnership with other DNA binding proteins to affect other cellular pathways. Both HIF and Notch signaling are known to inhibit differentiation of certain cell types (Fortini, 2009, Guruharsha et al., 2012) and several lines of evidence suggest that they are able to work together. The Notch intracellular domain (ICD) is stabilized during hypoxia and HIF1A appears to act as a coactivator at the Notch target genes HEY and HES in mouse cells (Gustafsson et al., 2005). The Notch ICD interacts with the N-terminal region of HIF1A, but the C-TAD is required for coactivation. HIF1A recruitment to the HES promoter was observed only upon both Notch activation and hypoxia. Furthermore, HIF1A DNA binding is probably not required as a synthetic Notch-responsive promoter without HREs was strongly activated in the presence of HIF1A.
Several additional observations demonstrate further crosstalk of hypoxia and Notch signaling. The APH-1A component of γ-secretase, the protease that releases the Notch ICD, is itself a HIF1A target gene (Wang et al., 2006). Furthermore, FIH interacts with and hydroxylates the Notch ICD (Gustafsson et al., 2005, Zheng et al., 2008), which inhibits Notch signaling resulting in accelerated cell differentiation. Finally, FIH appears to have a higher affinity for Notch ICD than for HIF1A, suggesting that the ability of Notch signaling to enhance HIF1A binding and transactivation during differentiation and in tumors may be partly due to sequestration of FIH (Bhattacharya et al., 1999, Sahlgren et al., 2008, Zheng et al., 2008).
Dang et al. have recently reported that HIF1A can also act as a coactivator for RORγt, an important regulator of pro-inflammatory TH17 cells, and is important for increased expression of key genes under hypoxic conditions (Dang et al., 2011). A HIF1A mutant unable to bind DNA retained the ability to induce an IL-17 promoter reporter. HIF1A and RORγt were shown to physically interact with each other and HIF1A was recruited along with RORγt to the IL-17 and CNS2 promoters, suggesting that HIF1A is recruited by RORγt rather than by direct DNA binding. The authors also observed concurrent recruitment of p300 and increased histone H3 and H4 acetylation. A HIF1A C-TAD deletion was unable to coactivate the IL-17 promoter reporter, and although this was taken to indicate that HIF1A functions by recruiting p300, it is important to point out that this region also exhibits a strong interaction with CDK8-Mediator (Galbraith et al., 2013).
The HIF target genes
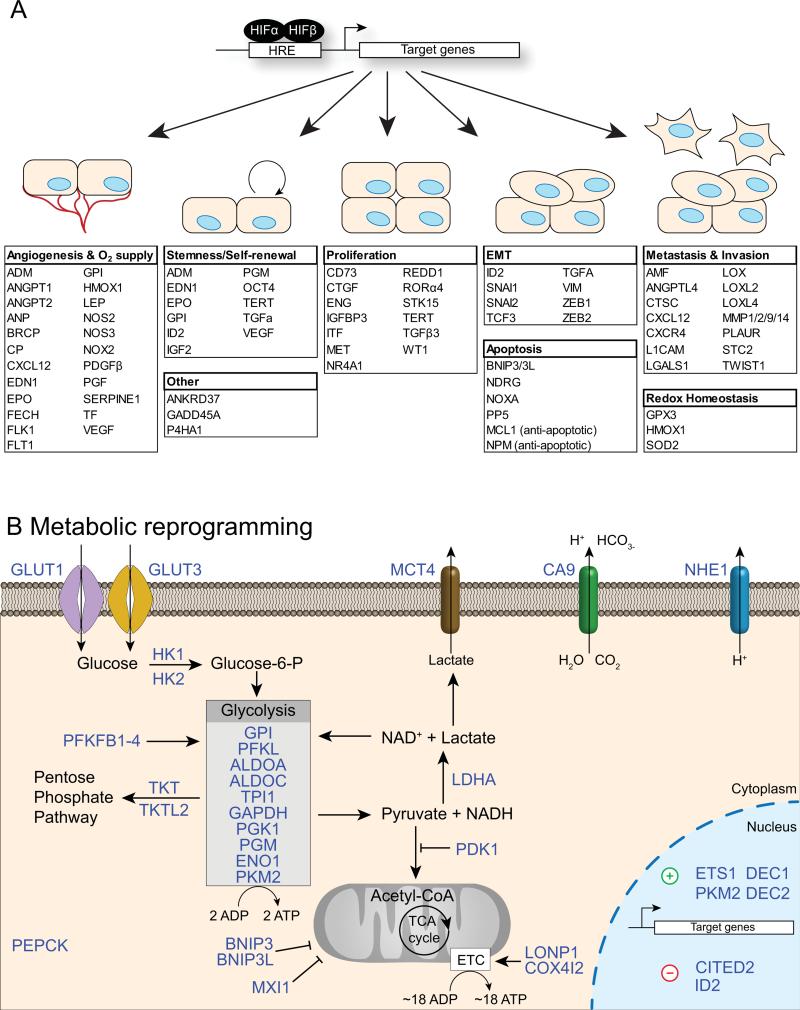
The HIFs regulate target genes in diverse biological pathways. Many of the most prominent and well-characterized targets are involved in the regulation of oxygen supply and utilization via angiogenesis and metabolic reprogramming. In order to coordinate the most efficient use of oxygen by the cell, HIFs activate genes central to a metabolic rewiring that shifts energy dependence away from high oxygen demand and towards glycolysis. The genes encoding essentially all glycolytic enzymes are directly upregulated by HIFs (Figure 2) (Benita et al., 2009, Semenza, 2012b). HIFs also regulate targets that increase the distribution of the available oxygen supply such as EPO, VEGF and its receptors FLT1 and FLK1, as well as END1 and ANGPT1 (Takeda et al., 2004).
Figure 2.
HIFs regulate target genes in diverse biological pathways. (A) There are over 70 known direct HIF target genes that function in numerous aspects of both normal physiology and disease processes, including angiogenesis and oxygen supply, stemness/self-renewal, proliferation, epithelial to mesenchymal transition (EMT), metastasis and invasion, redox homeostasis and apoptosis. (B) One of the largest functional categories of HIF target genes include those responsible for the metabolic rewiring that shifts cellular energy metabolism from oxidative phosphorylation and towards glycolysis. The list of HIF target genes included here is intended to be illustrative rather than complete.
In addition to pathways important for maintaining oxygen homeostasis however are HIF targets involved in autophagy, apoptosis, redox homeostasis, inflammation and immunity, stemness and self-renewal, and metastasis and invasion (Figure 2; (Wenger et al., 2005, Chi et al., 2006, Mole et al., 2009, Semenza, 2012b). HIFs regulate several genes that confer stemness properties including the well-known OCT4 (POU5F1), as well as many genes that are involved in epithelial to mesenchymal transition (EMT) during development and utilized by cancer cells in metastasis and invasion such as LOX, MMP1 and TWIST (Petrella et al., 2005, Covello et al., 2006, Erler et al., 2006, Yang et al., 2008, Semenza, 2012,). For a more in-depth perspective, we refer readers to several other reviews of the HIF transcriptional program (Wenger et al., 2005, Keith and Simon, 2007, Tsai and Wu, 2012).
HIF1A versus HIF2A target gene selectivity
Though HIF1A and HIF2A are able to bind the same consensus sequence, their target genes can differ in a context-dependent manner. The selectivity of HIF1A versus HIF2A target genes may be driven by several mechanisms and, furthermore, may change with chronic exposure to hypoxia and/or the abundance of stabilized HIF-alpha proteins. Although several studies have attempted to define the isoform-specific transcriptional programs, few common themes have emerged from these investigations, thus highlighting the complex nature of this cellular response. Variables such as cell type, severity, duration and variety of stimulation, the presence of functional VHL, and even culture conditions reportedly influence the transcriptional output mediated by HIF1A versus HIF2A. Furthermore, many of these studies have only examined either HIF1A or HIF2A, and untangling HIF-dependent from HIF-independent hypoxia-induced responses has proved challenging.
In addition to HIF2A having more restricted expression across tissues, there is evidence supporting a temporal organization of the HIF1A- and HIF2A-dependent responses. Under conditions of acute hypoxia (i.e. < 24 hours stimulation), transactivation of target genes occurs primarily by virtue of HIF1A, whereas following longer periods HIF2A begins to exert more of an influence (Koh et al., 2011, Koh and Powis, 2012). Per its role in mediating acute responses, HIF1A regulates the activation of glycolytic genes, thereby allowing cells to survive under conditions of decreased oxygen by switching the metabolic scheme from one of oxidative phosphorylation to anaerobic glycolysis. Several studies demonstrate that HIF2A plays no role in glycolytic gene regulation, but rather induces the expression of genes involved in pluripotent stem cell maintenance and angiogenesis (Hu et al., 2003, Takeda et al., 2004, Koh et al., 2011). Though solid tumors exist in a state of chronic hypoxia, very few studies follow the HIF-transcriptional response beyond 24 hours hypoxia.
Interestingly, the bHLH DNA-binding domains do not determine target gene selectivity between HIF1A and HIF2A. In fact, HIF1A and HIF2A recognize the same core HRE consensus sequence. Instead, the N-TADs appear to contribute to target gene selectivity (Hu et al., 2007, Lau et al., 2007). In an attempt to elucidate the mechanism of target gene selectivity between HIF1A and HIF2A, independent groups performed domain exchange experiments demonstrating that the C-terminal portions of these proteins, rather than the DNA-binding domains, can confer target gene selectivity. Utilizing fusion proteins composed of reciprocal sections of HIF1A and HIF2A, Lau and colleagues demonstrated that for induction of the HIF1A-specific target gene carbonic anhydrase IX (CA9), regions between amino acids 96-390 and 411-574 are necessary, whereas for induction of the HIF2A predominant target locus PHD3 (EGLN3), a single region between amino acids 543-870 is necessary and sufficient for the appropriate response (Lau et al., 2007).
Following in close succession, Hu and colleagues corroborated the efforts by Lau et al by showing that at distinct HIF1A-specific, HIF2A-specific, or HIF1A/HIF2A common targets the DNA binding domain is irrelevant for target gene selectivity (2007). Furthermore, using a series of domain-deletion and domain-exchange mutants, they elegantly honed target gene selectivity predominantly to the N-TAD (Hu et al., 2007).
Many of the studies attempting to elucidate HIF-isoform specific transcriptional activities point to the functional importance of specific co-activators, particularly other transcription factors that may bind cis-regulatory regions in the vicinity of active HREs as well as protein-protein interactions that mediate specific activity of a given isoform.
Cell type-specific regulation by HIFs
In addition to target gene selectivity exhibited by HIF1A and HIF2A there is evidence also for cell type-specific regulation. For example, recent examination of the hypoxic response in neuroblastoma cell lines identified RGS4 as a direct HIF1A/HIF2A common transcriptional target that is unique to this tissue type (Olechnowicz et al., 2012).
Though at least 70 direct HIF targets have been identified, a meta-analysis examining 19 publically available HIF-regulated gene expression data sets found that only 17 genes were commonly upregulated across all of the studies (Wenger et al., 2005, Ortiz-Barahona et al., 2010). This specificity may be conferred by differences in HIF protein expression and sensitivity to HIF induction, pre-conditioning to hypoxia, and chromatin status and the availability of cooperative transcription factors.
A comparative study analyzing the HIF transcriptional response in several cell types found the degree of induction of HIF targets was higher in epithelial cells than mesenchymal cells and additionally the number of genes induced in these cell types was significantly higher (Chi et al., 2006). Interestingly, the same study found a set of genes induced specifically in a single cell type, renal proximal tube epithelial cells (RPTEC). Many of the unique targets, involved in immune regulation, solute transport, and genomic integrity, are associated with recovery to ischemic injury due to loss of perfusion as well as tumor formation and progression. The authors speculate that the differences seen in these cells may be due to enhanced hypoxic response, as HIF1A mRNA was induced up to five times more in RPTECs than the other cell types examined (Chi et al., 2006).
Increased HIF protein abundance however is likely not the primary cause for cell type-specific differences. Increasingly, cell type-specific differences and HIF1A versus HIF2A target gene selectivity can be attributed to chromatin status, RNAPII activity, and the availability of partner transcription factors or coactivators (Xia and Kung, 2009, Villar et al., 2012, Galbraith et al., 2013, Pawlus et al., 2013).
3. HIF transcriptional cofactors
Upon binding to their cognate response elements on the DNA, transcription factors can modulate the transcriptional output of their target genes by a myriad of mechanisms including: 1) Effects on the recruitment of the general transcription factors (GTFs) that mediate pre-initiation complex (PIC) assembly and RNAPII recruitment, 2) Chromatin-based mechanisms such as nucleosome remodeling, post-translational modification of histones and three-dimensional rearrangements of the chromatin fiber, and 3) Modulation of RNAPII activity at post-recruitment steps, such as promoter escape and elongation. This regulatory diversity is driven by the availability and recruitment of transcriptional coactivators and corepressors acting in a combinatorial fashion at various stages of the transcription cycle. It is unclear at this point to what extent HIFs employ these various mechanisms. Importantly, a better understanding of HIF transcriptional cofactors may reveal novel strategies to manipulate HIF function for therapeutic purposes. We review here the salient findings about HIF transcriptional cofactors.
CBP and p300
Until recently, the HIF transactivation mechanism has been almost entirely understood as ultimately leading to recruitment of the coactivators CBP and p300 to the HREs of HIF target genes. These coactivators are mainly assumed to be required due to their ability to modify the local chromatin environment via their lysine acetyl-transferase activity and also their ability to interact with the core transcription machinery (Bedford et al., 2010). The interaction between HIF1A and p300 was initially identified during a search for p300 binding proteins as part of an effort to better understand the mechanism of p300 action (Arany et al., 1996). Many subsequent HIF studies confirmed the interaction between HIF1A/2A and CBP/p300, and established that the interaction occurs through the HIF C-TADs and is regulated by hydroxylation (Huang et al., 1996, Ema et al., 1997, Ebert and Bunn, 1998, Carrero et al., 2000, Lando et al., 2002b, Ruas et al., 2002, Ziel et al., 2004). Many of these studies utilized transient transfection of reporter constructs to examine the ability of CBP and p300 to increase transcriptional activity of HIFs towards a limited number of target genes (chiefly EPO, VEGF and LDHA). Although enhanced CBP/p300 binding was often observed to be concurrent with activation of HIF target genes, neither the CBP/p300 binding events nor the reporter constructs are in an endogenous chromatin context. It is also worth pointing out that in most cases these types of analyses do not provide clear evidence that CBP/p300 are sufficient by themselves to drive transcription of HIF target genes, as many other transcriptional cofactors are present in these assays.
CBP and p300 are among the most interconnected proteins in the cell nucleus, with at least 400 interaction partners, and due to their requirement for cell viability it remains difficult to determine in vivo which of their established functions is required at specific genes (Bedford et al., 2010). The C-TADs of HIF1A/2A bind to the CH1 domains of CBP and p300 (Bhattacharya et al., 1999, Ruas et al., 2002). In order to test the requirement for CBP and p300 for HIF-activated transcription in vivo, Kasper et al. (2005) generated MEFs in which the CH1 domains of both CBP and p300 alleles were deleted. The CH1 deletions almost completely abrogated the transcriptional activity of a C-TAD-GAL4 chimera in a reporter assay. However, microarray analyses revealed that the CH1 domains of CBP and p300 are essential for only a few HIF target genes, partially required for many and completely dispensable for others. Importantly, although the N-TAD of HIF1A has been reported to interact with the CH3 domains of CBP and p300 (Ruas et al., 2010), deletion of the CH1 domain abolishes recruitment of CBP and p300 to HREs (Kasper et al., 2005), indicating that the CH3-mediated interaction is not sufficient for coactivator recruitment.
Several other coactivators with lysine acetyl-transferase activity have also been implicated as coactivators of HIF target genes. Three members of the p160 family of coactivators, NCOA1 (SRC-1/KAT13A), NCOA2 (SRC-2/KAT13C/TIF2/GRIP1) and NCOA3 (SRC-3/KAT13C/ACTR/TRAM-1) have been shown to interact with and enhance HIF1A transcriptional activity at specific target genes or reporters (Carrero et al., 2000, Ruas et al., 2005, Wang et al., 2010a). The ability of NCOA1 and NCOA2 to act as coactivators of HIF1A was shown be hypoxia-dependent and synergistic with CBP/p300, the latter likely due to their ability to bind to both HIF1A TADs and to CBP/p300 (Carrero et al., 2000). Later work suggested that the interaction of NCOA1 with HIF1A is dependent on CBP (Ruas et al., 2005, Wang et al., 2010a). However, it remains unclear if the lysine acetyl-transferase activity of the p160 coactivators is required in this context.
PKM2
A recent study has demonstrated a surprising role for a metabolic enzyme as a HIF coactivator. Pyruvate kinase (PK) catalyzes the final step in glycolysis and has been previously implicated in the Warburg effect (Christofk et al., 2008). Luo et al. (2011) found that the M2 isoform (PKM2) is itself a HIF target gene and that it is able to interact with both HIF1A and HIF2A, via the M2-specific exon 10 region. In cotransfection assays with HIF1A and HIF2A, PKM2 was able to increase activity of an ENO1 HRE reporter independently of its catalytic activity. Interestingly, PKM2 possesses a LXXLAP motif that is hydroxylated by PHD3 (EGLN3). Mutation of these residues or PHD3 knockdown reduced interaction with and coactivation of HIF. Knockdown of PKM2 reduced HIF1A binding and p300 recruitment to the HREs of select target genes. Interestingly, PKM2 and PHD3 were also recruited to the LDHA and PDK1 loci during hypoxia. An interaction between PKM2 and p300 was detected, and the knockdown of PKM2 reduced occupancy of p300 and histone H3 acetylation at the LDHA HRE in both normoxia and hypoxia. Taken together these data suggest that PHD3 hydroxylation of PKM2 stimulates its interaction with HIF transcriptional complexes containing p300 where it can act as a coactivator.
The fact that PKM2 acts as a coactivator for HIF target genes whose products contribute to the shift away from oxidative towards glycolytic metabolism (SLC2A1/GLUT1, LDHA, PDK1) may partly explain its role in the Warburg effect. Of note, PKM2 enzymatic activity was not required for its coactivator function, and others have shown that phosphorylation of Y105, which inhibits its enzymatic activity, is important for Warburg type effects (Hitosugi et al., 2009). This phosphorylation event could potentially act as a switch between the metabolic and coactivator roles of PKM2. It will be important to determine the exact mechanism by which PKM2 coactivates HIF and to what extent PKM2 affects HIF action at a global scale. Interestingly, PKM2 has also been implicated as a coactivator in β-catenin-dependent transcriptional program (Yang et al., 2011).
CDK8-Mediator
We recently reported that the Mediator-associated kinase CDK8, but not its paralog CDK19, is an important coactivator of HIF1A target genes (Galbraith et al., 2013). We found that CDK8 is a major regulator of the transcriptional program induced by hypoxia and is enriched at the regulatory regions of ~65% of hypoxia-inducible genes in hypoxia. A detailed analysis of two CDK8-dependent HIF1A target genes (ANKRD37 and STC2) confirmed that CDK8 was indeed recruited to their promoter-proximal regions in response to hypoxia, while HIF1A binding was unaffected by knockdown of CDK8. Strikingly, the hypoxia-induced increases in histones H3 and H4 acetylation at these regions were also unaffected by depletion of CDK8, indicating that histone acetylation alone is not sufficient for induction of these genes by HIF1A. These results revealed a CDK8-dependent coactivation event acting in addition to recruitment of histone acetyl-transferases. Mechanistically, CDK8 is required for steps subsequent to PIC formation, RNAPII binding and transcription initiation. Instead, CDK8 is essential for the hypoxia-induced recruitment of the Super Elongation Complex (SEC). Knockdown of CDK8 led to reduced recruitment of the SEC components CDK9 and AFF4, along with a reduction in RNAPII transit through the gene body of both ANKRD37 and STC2. This indicates that CDK8 is essential for the transition from paused RNAPII into productive elongation.
Analysis of HIF1A−/− cells indicated that HIF1A itself is largely dispensable for RNAPII recruitment, but essential for binding of CDK8 and the SEC, as well as histone acetylation. This suggested that HIF1A target genes as a group might be regulated at the pause-release stage, rather than by chromatin remodeling or transcription initiation. Importantly, genome-wide analysis of both total and transcriptionally-active RNAPII revealed that hypoxia-inducible genes are predominantly bound by active but paused RNAPII prior to their induction by hypoxia. This is consistent with the previous report by Xia et al., (2009) demonstrating that HIF1A preferentially binds to permissive loci and activates genes with basal transcriptional activity. Furthermore, we found that a region of HIF1A comprised of both TADs exhibits strong interactions with CDK8-Mediator subunits in addition to p300. Several of these interactions were preserved using the C-TAD alone but were undetectable with the N-TAD. Together, our findings indicate that HIF1A employs CDK8-Mediator to stimulate transcriptional elongation at its target genes and demonstrate a physical and mechanistic link between these two oncogenes.
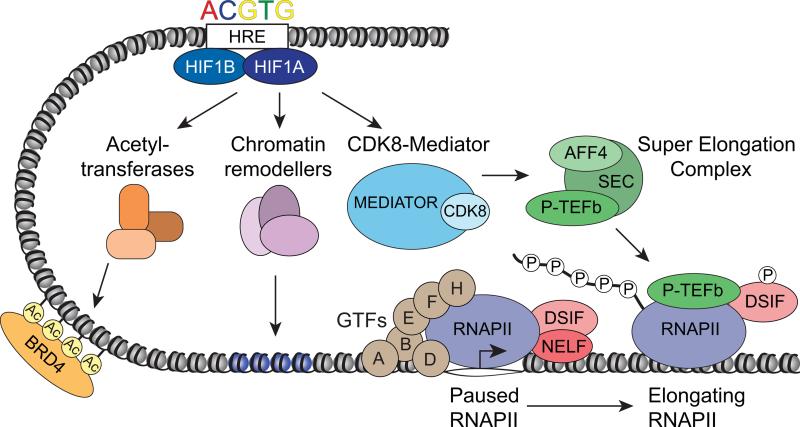
Based on these results, we propose a revised model for HIF1A-dependent transcription activation (Figure 3). In normoxia, HIF target loci carry transcriptionally engaged but paused RNAPII within an open chromatin environment. Upon hypoxia, HIF1A is stabilized and binds to chromatin along with HIF1B. This leads to at least two coactivation events: 1) Recruitment of histone acetyl-transferases (CBP/p300 or others) followed by histone acetylation and binding of the Bromodomain protein BRD4; and 2) Recruitment of CDK8-Mediator along with the SEC, which leads to release of paused RNAPII and productive elongation.
Figure 3.
A model of HIF1A-dependent transactivation. During normoxia, almost all HIF1A target genes display an open chromatin environment and harbor transcriptionally active but paused RNA polymerase II (RNAPII). Upon stabilization by hypoxia, HIF1A dimerizes with HIF1B and binds to the permissive HREs of these target genes. HIF1A then recruits various coactivators including acetyl-transferases and chromatin remodelers as well as CDK8-Mediator along with the Super Elongation Complex (SEC) containing AFF4 and P-TEFb. These events result in increased histone acetylation and BRD4 binding and the P-TEFb-triggered release of paused RNAPII into productive elongation. Importantly, without CDK8, HIF1A is still able to bind to the HRE and induce histone acetylation but fails to recruit Mediator and the SEC or to stimulate RNAPII elongation.
A number of important questions regarding the exact mechanism of HIF-dependent transactivation remain unanswered and we hope that this will stimulate further investigations. What is the nature of the interaction between HIF1A and CDK8-Mediator? Is the kinase activity of CDK8 required for its role as a coactivator of HIF1A? How is the recruitment of CDK8-Mediator affected by post-translational modification of HIF1A? To what extent is the action of p300/CBP or other acetyl-transferases necessary at genes that require CDK8-Mediator? Is CDK8-Mediator also required for HIF2A transactivation?
Pontin
The ATPase/helicase chromatin-remodeling factor Pontin (RUVBL1) was recently found to potentiate activation of a subset of HIF1A target genes (Lee et al., 2011). The lysine methyl-transferases G9a and GLP were found to methylate Pontin during hypoxia at six lysine residues. Pontin was also found to interact with HIF1A during hypoxia. Knockdown of Pontin led to reduced activity of a HRE reporter while expression of wild type but not a lysine-to-alanine mutant (KA) increased activity, suggesting that these lysine residues are indeed important for HIF1A transactivation potential. Microarray analysis in MCF7 cells in normoxia/hypoxia revealed that Pontin knockdown affected ~24% of differentially expressed genes and that out of 36 hypoxia-induced Pontin-dependent genes, 6 were known HIF targets with others containing putative HREs. Although the effect of Pontin on chromatin remodeling was not examined, the ATPase activity of Pontin was found not to be required for coactivation in this context. Pontin was found to be recruited to the HIF1A target gene ETS1 after hypoxia, and Pontin knockdown affected p300 but not HIF1A recruitment. Wild type but not KA Pontin was able to rescue p300 recruitment which lead the authors to conclude that Pontin methylation is crucial for p300 association at HIF1A target loci. Work by the same group has demonstrated that the related remodeling factor, Reptin (RUVBL2), is also methylated under hypoxic conditions. Interestingly, in this case it appears to be mainly acting as a repressor of hypoxia-inducible genes (Lee et al., 2010).
SWI/SNF
The SWI/SNF nucleosome remodeling complex has also been implicated in coactivation of HIF1A target genes. The alternative ATPase subunits within SWI/SNF, BRM and BRG1, were shown to increase expression of a HRE reporter in an ATPase-dependent manner. It was shown that both ATPases associate with the EPO 3’ enhancer and the VEGF promoter in vivo in a complex with HIF1A (Wang et al., 2004a). Knockdown of BRM and BRG1 reduced expression of endogenous EPO mRNA but not VEGF mRNA, indicating gene-specific requirements for this cofactor. A later report demonstrated that HIF1A itself is a direct target of SWI/SNF, suggesting that any role of SWI/SNF in the wider HIF transcriptional program may be mostly explained by regulation of HIF1A levels (Kenneth et al., 2009).
Interestingly, a genome-wide RNAi screen in Drosophila, using a mouse LDHA enhancer HRE reporter, identified several subunits of chromatin remodeling complexes as regulators of HIF-dependent transcription in hypoxia (Dekanty et al., 2010). Among the hits of this screen were the Drosophila homologs of both Pontin and Reptin, as well as five subunits of the SWI/SNF complex. Of note, Pontin and Reptin are shared subunits of multiple chromatin remodeling/modifying complexes including SRCAP, which is involved in exchange of the histone variant H2A.Z, the INO80 complex, and the acetyl-transferase TIP60 complex (Huen et al., 2010). It would be interesting to define which if any of these complexes participate in HIF-dependent transcriptional control.
ARNT as a transcriptional coactivator
Although insufficient to drive transcription in the absence of its HIFA dimerization partners, several recent studies suggest that HIF1B/ARNT is not simply a partner for heterodimerization and DNA binding but can directly contribute to transactivation by HIF. The HIF1B/ARNT PAS-B domain (which is also required for heterodimerization) has been shown to recruit several coiled-coil coactivator proteins. TRIP230 is a HIF1B interactor that can coactivate both AHR- and HIF-dependent reporters, and is recruited to the VEGF and EPO loci upon hypoxia along with HIF1A and SRC1 (Beischlag et al., 2004). CoCoA interacts with multiple bHLH domain proteins and is able to enhance the transcriptional activity of HIF1B (Kim and Stallcup, 2004). TACC3 (AINT) was originally identified as a HIF1B interactor able to stimulate an EPO HRE reporter, and later found to share a HIF1B interaction interface with TRIP230, CoCoA and HIFA (Sadek et al., 2000, Partch and Gardner, 2011). Interestingly, point mutations in the PAS-B domain of HIF1B altered the specificity of coactivator recruitment and target gene induction, suggesting a gene-specific requirement for these coactivators (Partch and Gardner, 2011).
Finally, interaction between AHR and the Mediator subunits MED1 (TRAP220) and MED23 (Med130) has been demonstrated along with recruitment of the Mediator subunits MED1 and CDK8 to the AHR/HIF1B-regulated mouse CYP1A1 enhancer but not its promoter (Wang et al., 2004b). However, an interaction between Mediator subunits and HIF1B alone was not detected, suggesting that its AHR and HIFA binding partners are responsible for this aspect of transactivation.
Negative regulation of HIF transactivation by CITED2, SIRT6 and Reptin
In addition to negative regulation by FIH, a number of other factors have been reported to repress HIF transactivation potential. We make a distinction between factors that regulate HIFs at the protein stability level and limit our discussion here to those that play a direct role in the transactivation function of HIF.
CITED2 is a member of the CBP/p300-interacting transactivator with Glu/Asp (ED)-rich (CITED) family. These proteins function as coactivators for a number of transcription factors such as AP-2 and PPARα/γ by virtue of their ability to interact with CBP/p300 (Braganca et al., 2003, Tien et al., 2004). In contrast, CITED2 acts to inhibit HIF1A transactivation by competing with HIF1A for interaction with CBP/p300 (Bhattacharya et al., 1999, Freedman et al., 2003, Yoon et al., 2011). CITED2 is able to interact with the CH1 and CH3 domains of CBP/p300 to block interaction with both the N-TAD and C-TAD of HIF and is able to displace the HIF C-TAD from p300 in vitro (Freedman et al., 2003, Yoon et al., 2011). CITED2 is itself a HIF target gene and may therefore function in part to provide negative feedback to suppress HIF-mediated transcription upon return to normal oxygen. In addition, CITED4 has also been identified as a negative regulator of HIF1A (Fox et al., 2004).
A number of Sirtuins have been found to have roles in regulating the HIF proteins but, to date, only SIRT6 has a demonstrated direct role as a corepressor of HIF1A. SIRT6 is able to interact with HIF1A and associates with the promoters of several glycolytic HIF target genes under normal glucose and oxygen conditions, where it appears to function as a repressor via its histone H3K9 deacetylase activity (Zhong et al., 2010). Although the SIRT6 / HIF1A interaction was not confirmed to be direct, knockdown of HIF1A reduced recruitment of SIRT6 to these promoters. Interestingly SIRT6 also appears to regulate HIF1A at both the protein synthesis and stability levels, with SIRT6 knock out leading to detectable levels of HIF1A in normoxic conditions. Together these data suggest that SIRT6 is able to prevent an inappropriate shift towards glycolytic metabolism by suppressing HIF1A-mediated transcription under normal oxygen and nutrient conditions.
Final remarks
Given the undisputed biological importance of HIFs, a deeper understanding of the molecular mechanisms by which they regulate the transcriptional apparatus will advance multiple fields of research. In particular, a better understanding of the impact of HIF post-translational modifications, the functional differences arising from specialization of HIF alpha subunits, the combinatorial use of DNA binding partners, and the contribution of diverse cofactors to the global hypoxia-induced transcriptional program will undoubtedly advance the field.
With regards to post-translational modifications, it would be of great utility to define the global extent to which specific phosphorylation and acetylation events affect the HIF transcriptional program. Ideally, these investigations will involve the creation of knock-in mouse models to create HIF alleles lacking specific modification sites. It is imperative that results in this area that were obtained from in vitro cell cultures be tested at the organismal level, during both normal homeostasis and cancer development. These studies could be complemented with knock-in human cell lines to test the impact of these modifications in tumor biology. With the advent of more effective genome editing techniques utilizing zinc-finger directed nucleases and TALENs, a significant number of alleles could be generated for these studies.
A deeper understanding of the molecular mechanisms differentiating HIF1A versus HIF2A may reveal strategies to specifically manipulate the activity of each factor. Since their specialization seems to be partly determined by their N-TADs, it will be critical to define what cofactors interact with these domains, what target genes are affected by said cofactors and whether these differential cofactors could be subjected to pharmacological manipulation.
Finally, the precise characterization of other transcription factors and coactivators working in cooperation with HIFs may reveal strategies to tame this highly pleiotropic transcriptional program for therapeutic purposes. Given that a normal HIF response has beneficial effects (e.g. enhanced EPO production to maintain a healthy red blood cell count), but at the same time can be exploited during cancer development (e.g. enhanced glycolytic activity), it could be very useful to identify DNA binding partners or coactivators that exert pathway-specific effects. This knowledge could then be employed to skew the HIF transcriptional program toward a specific cellular response during therapeutic intervention.
Footnotes
DECLARATION OF INTEREST
The authors report no declarations of interest.
REFERENCES
- Appelhoff RJ. Differential Function of the Prolyl Hydroxylases PHD1, PHD2, and PHD3 in the Regulation of Hypoxia-inducible Factor. J Biol Chem. 2004;279:38458–38465. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- Aprelikova O, Wood M, Tackett S, et al. Role of ETS transcription factors in the hypoxia-inducible factor-2 target gene selection. Cancer Res. 2006;66:5641–5647. doi: 10.1158/0008-5472.CAN-05-3345. [DOI] [PubMed] [Google Scholar]
- Arany Z, Huang LE, Eckner R, et al. An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci. 1996;93:12969–12973. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnesen T, Kong X, Evjenth R, et al. Interaction between HIF-1 alpha (ODD) and hARD1 does not induce acetylation and destabilization of HIF-1 alpha. FEBS Lett. 2005;579:6428–32. doi: 10.1016/j.febslet.2005.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S-H, Jeong J-W, Park JA, et al. Sumoylation increases HIF-1α stability and its transcriptional activity. Biochem Biophys Res Commun. 2004;324:394–400. doi: 10.1016/j.bbrc.2004.09.068. [DOI] [PubMed] [Google Scholar]
- Bedford DC, Kasper LH, Fukuyama T, et al. Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics : official journal of the DNA Methylation Society. 2010;5:9–15. doi: 10.4161/epi.5.1.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beischlag TV, Taylor RT, Rose DW, et al. Recruitment of thyroid hormone receptor/retinoblastoma-interacting protein 230 by the aryl hydrocarbon receptor nuclear translocator is required for the transcriptional response to both dioxin and hypoxia. J Biol Chem. 2004;279:54620–54628. doi: 10.1074/jbc.M410456200. [DOI] [PubMed] [Google Scholar]
- Benita Y, Kikuchi H, Smith AD, et al. An integrative genomics approach identifies Hypoxia Inducible Factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res. 2009;37:4587–4602. doi: 10.1093/nar/gkp425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento CF, Fernandes R, Ramalho J, et al. The chaperone-dependent ubiquitin ligase CHIP targets HIF-1α for degradation in the presence of methylglyoxal. PLoS ONE. 2010;5:e15062. doi: 10.1371/journal.pone.0015062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berra E, Benizri E, Ginouvès A, et al. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1 in normoxia. EMBO J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berta MA, Mazure N, Hattab M, et al. SUMOylation of hypoxia-inducible factor-1α reduces its transcriptional activity. Biochem Biophys Res Commun. 2007;360:646–652. doi: 10.1016/j.bbrc.2007.06.103. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, Michels CL, Leung MK, et al. Functional role of p35srj, a novel p300/CBP binding protein, during transactivation by HIF-1. Genes Dev. 1999;13:64–75. doi: 10.1101/gad.13.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken CP, Fedele AO, Linke S, et al. Cell-specific regulation of hypoxia-inducible factor (HIF)-1α stabilization and transactivation in a graded oxygen environment. J Biol Chem. 2006;281:22575–22585. doi: 10.1074/jbc.M600288200. [DOI] [PubMed] [Google Scholar]
- Bragança J, Eloranta JJ, Bamforth SD, et al. Physical and functional interactions among AP-2 transcription factors, p300/CREB-binding protein, and CITED2. J Biol Chem. 2003;278:16021–9. doi: 10.1074/jbc.M208144200. [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Dor Y, Herbert JM, et al. Role of HIF-1α in hypoxia-mediated apoptosis, cell proliferation and tumor angiogenesis. Nature. 1998;394:485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- Carrero P, Okamoto K, Coumailleau P, et al. Redox-regulated recruitment of the transcriptional coactivators CREB-binding protein and SRC-1 to hypoxia-inducible factor 1alpha. Mol Cell Biol. 2000;20:402–415. doi: 10.1128/mcb.20.1.402-415.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Xu M, Hogg RT, et al. The Acetylase/Deacetylase Couple CREB-binding Protein/Sirtuin 1 Controls Hypoxia-inducible Factor 2 Signaling. J Biol Chem. 2012;287:30800–30811. doi: 10.1074/jbc.M111.244780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi JT, Wang Z, Nuyten D, et al. Gene expression programs in response to hypoxia: Cell type specificity and prognostic significance in human cancers. PLoS Med. 2006;3:395–409. doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Ahn D-R, Park H, et al. Modulation of p300 binding by posttranslational modifications of the C-terminal activation domain of hypoxia-inducible factor-1α. FEBS Lett. 2007;581:1542–1548. doi: 10.1016/j.febslet.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Compernolle V, Brusselmans K, Acker T, et al. Loss of HIF-2α and inhibition of VEGF impair fetal lung maturation, whereas treatment with VEGF prevents fatal respiratory distress in premature mice. Nat Med. 2002;8:703–710. doi: 10.1038/nm721. [DOI] [PubMed] [Google Scholar]
- Covello KL, Kehler J, Yu H, et al. HIF-2α regulates OCT-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 2006;20:557–570. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad PW, Freeman LT, Beitner-Johnson D, et al. EPAS1 trans-Activation during Hypoxia Requires p42/p44 MAPK. J Biol Chem. 1999;274:33709–33713. doi: 10.1074/jbc.274.47.33709. [DOI] [PubMed] [Google Scholar]
- Dames SA, Martinez-Yamout M, De Guzman RN, et al. Structural basis for Hif-1a/CBP recognition in the cellular hypoxic response. Proc Natl Acad Sci. 2002;99:5271–5276. doi: 10.1073/pnas.082121399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang EV, Barbi J, Yang H-Y, et al. Control of TH17/Treg Balance by Hypoxia-Inducible Factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayan F, Roux D, Brahimi-Horn MC, et al. The oxygen sensor factor-inhibiting hypoxia-inducible factor-1 controls expression of distinct genes through the bifunctional transcriptional character of hypoxia-inducible factor-1α. Cancer Res. 2006;66:3688–3698. doi: 10.1158/0008-5472.CAN-05-4564. [DOI] [PubMed] [Google Scholar]
- Dehne N, Hintereder G, Brüne B. High glucose concentrations attenuate hypoxia-inducible factor-1α expression and signaling in non-tumor cells. Exp Cell Res. 2010;316:1179–1189. doi: 10.1016/j.yexcr.2010.02.019. [DOI] [PubMed] [Google Scholar]
- Dekanty A, Romero NM, Bertolin AP, et al. Drosophila Genome-Wide RNAi Screen Identifies Multiple Regulators of HIF-Dependent Transcription in Hypoxia. PLOS Genet. 2010;6:e1000994. doi: 10.1371/journal.pgen.1000994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dioum EM, Chen R, Alexander MS, et al. Regulation of Hypoxia-Inducible Factor 2 α Signaling by the Stress-Responsive Deacetylase Sirtuin 1. Science. 2009;324:1289–1293. doi: 10.1126/science.1169956. [DOI] [PubMed] [Google Scholar]
- Ebert BL, Bunn HF. Regulation of transcription by hypoxia requires a multiprotein complex that includes hypoxia-inducible factor 1, an adjacent transcription factor, and p300/CREB binding protein. Mol Cell Biol. 1998;18:4089–4096. doi: 10.1128/mcb.18.7.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema M, Hirota K, Mimura J, et al. Molecular mechanisms of transcription activation by HLF and HIF1α in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 1999;18:1905–1914. doi: 10.1093/emboj/18.7.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema M, Taya S, Yokotani N, et al. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1alpha regulates the VEGF expression and is potentially involved in lung and vascular development. Proc Natl Acad Sci USA. 1997;94:4273–4278. doi: 10.1073/pnas.94.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdozain OJ, Pegrum S, Winrow VR, et al. Hypoxia in Abdominal Aortic Aneurysm Supports a Role for HIF-1α and Ets-1 as Drivers of Matrix Metalloproteinase Upregulation in Human Aortic Smooth Muscle Cells. J Vasc Res. 2011;48:163–170. doi: 10.1159/000318806. [DOI] [PubMed] [Google Scholar]
- Erler JT, Bennewith KL, Nicolau M, et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nat Lett. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- Espinosa JM. The meaning of pausing. Mol Cell. 2010;40:507–508. doi: 10.1016/j.molcel.2010.11.025. [DOI] [PubMed] [Google Scholar]
- Flamme I, Fröhlich T, Von Reutern M, et al. HRF, a putative basic helix-loop-helix-PAS-domain transcription factor is closely related to hypoxia-inducible factor-1α and developmentally expressed in blood vessels. Mech Dev. 1997;63:51–60. doi: 10.1016/s0925-4773(97)00674-6. [DOI] [PubMed] [Google Scholar]
- Flügel D, Görlach A, Michiels C, et al. Glycogen Synthase Kinase 3 Phosphorylates Hypoxia-Inducible Factor 1 and Mediates Its Destabilization in a VHL-Independent Manner. Mol Cell Biol. 2007;27:3253–3265. doi: 10.1128/MCB.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Dev Cell. 2009;16:633–47. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Foster MW, Mcmahon TJ, Stamler JS. S-nitrosylation in health and disease. Trends Mol Med. 2003;9:160–168. doi: 10.1016/s1471-4914(03)00028-5. [DOI] [PubMed] [Google Scholar]
- Fox SB, Bragança J, Turley H, et al. CITED4 inhibits hypoxia-activated transcription in cancer cells, and its cytoplasmic location in breast cancer is associated with elevated expression of tumor cell hypoxia-inducible factor 1α. Cancer Res. 2004;64:6075–6081. doi: 10.1158/0008-5472.CAN-04-0708. [DOI] [PubMed] [Google Scholar]
- Foxler DE, Bridge KS, James V, et al. The LIMD1 protein bridges an association between the prolyl hydroxylases and VHL to repress HIF-1 activity. Nat Cell Biol. 2012;14:201–208. doi: 10.1038/ncb2424. [DOI] [PubMed] [Google Scholar]
- Freedman SJ, Sun Z-YJ, Kung AL, et al. Structural basis for negative regulation of hypoxia-inducible factor-1alpha by CITED2. Nat Struct Biol. 2003;10:504–512. doi: 10.1038/nsb936. [DOI] [PubMed] [Google Scholar]
- Galbraith MD, Allen MA, Bensard CL, et al. HIF1A Employs CDK8-Mediator to Stimulate RNAPII Elongation in Response to Hypoxia. Cell. 2013;153:1327–1339. doi: 10.1016/j.cell.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H, Harvey CT, Pittsenbarger J, et al. HDAC4 Protein Regulates HIF1α Protein Lysine Acetylation and Cancer Cell Response to Hypoxia. J Biol Chem. 2011;286:38095–38102. doi: 10.1074/jbc.M111.257055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H, Liu Q, Xue C, et al. HIF1α protein stability is increased by acetylation at lysine 709. J Biol Chem. 2012;287:35496–35505. doi: 10.1074/jbc.M112.400697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee C, et al. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradin K, Takasaki C, Fujii-Kuriyama Y, et al. The transcriptional activation function of the HIF-like factor requires phosphorylation at a conserved threonine. J Biol Chem. 2002;277:23508–23514. doi: 10.1074/jbc.M201307200. [DOI] [PubMed] [Google Scholar]
- Greer SN, Metcalf JL, Wang Y, et al. The updated biology of hypoxia-inducible factor. EMBO J. 2012;31:2448–2460. doi: 10.1038/emboj.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YZ, Moran SM, Hogenesch JB, et al. Molecular characterization and chromosomal localization of a third alpha-class hypoxia inducible factor subunit, HIF3α. Gene Expr. 1998;7:205–213. [PMC free article] [PubMed] [Google Scholar]
- Guruharsha KG, Kankel MW, Artavanis-Tsakonas S. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet. 2012;13:654–666. doi: 10.1038/nrg3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MV, Zheng X, Pereira T, et al. Hypoxia Requires Notch Signaling to Maintain the Undifferentiated Cell State. Dev. Cell. 2005;9:617–628. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Hara S, Hamada J, Kobayashi C, et al. Expression and Characterization of Hypoxia-Inducible Factor (HIF)-3α in Human Kidney: Suppression of HIF-Mediated Gene Expression by HIF-3α. Biochem Biophys Res Commun. 2001;287:808–813. doi: 10.1006/bbrc.2001.5659. [DOI] [PubMed] [Google Scholar]
- Heikkilä M, Pasanen A, Kivirikko KI, et al. Roles of the human hypoxia-inducible factor (HIF)-3α variants in the hypoxia response. Cell Mol Life Sci. 2011;68:3885–3901. doi: 10.1007/s00018-011-0679-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T, Kang S, Vander Heiden MG, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009:2. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman EC, Reyes H, Chu FF, et al. Cloning of a factor required for activity of the Ah (dioxin) receptor. Science. 1991;252:954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- Hu CH, Wang L-Y, Chodosh LA, et al. Differential Roles of Hypoxia-Inducible Factor 1 (HIF-1 ) and HIF-2 in Hypoxic Gene Regulation. Mol Cell Biol. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CJ, Sataur A, Wang L, et al. The N-Terminal Transactivation Domain Confers Target Gene Specificity of Hypoxia-inducible Factors HIF-1 and HIF-2. Mol Biol Cell. 2007;18:4528–4542. doi: 10.1091/mbc.E06-05-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LE, Arany Z, Livingston DM, et al. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J Biol Chem. 1996;271:32253–9. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- Huang LE, Gu J, Schau M, et al. Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Kapere Ochieng J, Kempen MB-V, et al. Hypoxia inducible factor 3α plays a critical role in alveolarization and distal epithelial cell differentiation during mouse lung development. PLoS ONE. 2013;8:e57695. doi: 10.1371/journal.pone.0057695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen J, Kakihara Y, Ugwu F, et al. Rvb1-Rvb2: essential ATP-dependent helicases for critical complexes. Biochem Cell Biol. 2010;88:29–40. doi: 10.1139/o09-122. [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, et al. HIFα Targeted for VHL-Mediated Destruction by Proline Hydroxylation: Implications for O2 Sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Iyer NV, Kotch LE, Agani F, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev. 1998a;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer NV, Leung SW, Semenza GL. The Human Hypoxia-Inducible Factor 1α Gene: HIF1AStructure and Evolutionary Conservation. Genomics. 1998b;52:159–165. doi: 10.1006/geno.1998.5416. [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, et al. Targeting of HIF-α to the von Hippel-Lindau Ubiquitylation Complex by O2-Regulated Prolyl Hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Jeong J-W, Bae M-K, Ahn M-Y, et al. Regulation and Destabilization of HIF-1α by ARD1-Mediated Acetylation. Cell. 2002;111:709–720. doi: 10.1016/s0092-8674(02)01085-1. [DOI] [PubMed] [Google Scholar]
- Jiang BH, Zheng JZ, Leung SW, et al. Transactivation and inhibitory domains of hypoxia-inducible factor 1α: Modulation of transcriptional activity by oxygen tension. J Biol Chem. 1997;272:19253–19260. doi: 10.1074/jbc.272.31.19253. [DOI] [PubMed] [Google Scholar]
- Jiang BH, Rue E, Wang GL, et al. Dimerization, DNA Binding, and Transactivation Properties of Hypoxia-inducible Factor 1. J Biol Chem. 1996;271:17771–17778. doi: 10.1074/jbc.271.30.17771. [DOI] [PubMed] [Google Scholar]
- Kaelin WG, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Kalousi A, Mylonis I, Politou AS, et al. Casein kinase 1 regulates human hypoxia-inducible factor HIF-1. J Cell Sci. 2010;123:2976–2986. doi: 10.1242/jcs.068122. [DOI] [PubMed] [Google Scholar]
- Kasper LH, Boussouar F, Boyd K, et al. Two transactivation mechanisms cooperate for the bulk of HIF-1-responsive gene expression. EMBO J. 2005;24:3846–3858. doi: 10.1038/sj.emboj.7600846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenneth NS, Mudie S, Van Uden P, et al. SWI/SNF regulates the cellular response to hypoxia. J Biol Chem. 2009;284:4123–4131. doi: 10.1074/jbc.M808491200. [DOI] [PubMed] [Google Scholar]
- Kim JH, Stallcup MR. Role of the coiled-coil coactivator (CoCoA) in aryl hydrocarbon receptor-mediated transcription. J Biol Chem. 2004;279:49842–49848. doi: 10.1074/jbc.M408535200. [DOI] [PubMed] [Google Scholar]
- Kimura H, Weisz A, Kurashima Y, et al. Hypoxia response element of the human vascular endothelial growth factor gene mediates transcriptional regulation by nitric oxide: control of hypoxia-inducible factor-1 activity by nitric oxide. blood. 1999;95:189–197. [PubMed] [Google Scholar]
- Kimura H, Weisz A, Ogura T, et al. Identification of Hypoxia-inducible Factor 1 Ancillary Sequence and Its Function in Vascular Endothelial Growth Factor Gene Induction by Hypoxia and Nitric Oxide*. J Biol Chem. 2001;276:2292–2298. doi: 10.1074/jbc.M008398200. [DOI] [PubMed] [Google Scholar]
- Knippschild U, Gocht A, Wolff S, et al. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cellular Signalling. 2005;17:675–689. doi: 10.1016/j.cellsig.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Koh MY, Darnay BG, Powis G. Hypoxia-associated factor, a novel E3-ubiquitin ligase, binds and ubiquitinates hypoxia-inducible factor 1α, leading to its oxygen-independent degradation. Mol Cell Biol. 2008a;28:7081–7095. doi: 10.1128/MCB.00773-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh MY, Spivak-Kroizman TR, Powis G. HIF-1 regulation: not so easy come, easy go. Trends Biochem Sci. 2008b;33:526–534. doi: 10.1016/j.tibs.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Koh MY, Lemos R, Liu X, et al. The Hypoxia-Associated Factor Switches Cells from HIF-1 - to HIF-2 -Dependent Signaling Promoting Stem Cell Characteristics, Aggressive Tumor Growth and Invasion. Cancer Res. 2011;71:4015–4027. doi: 10.1158/0008-5472.CAN-10-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh MY, Powis G. Passing the baton: the HIF switch. Trends Biochem Sci. 2012;37:364–372. doi: 10.1016/j.tibs.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen P, M H, Günzler V, et al. Catalytic properties of the asparaginyl hydroxylase (FIH) in the oxygen sensing pathway are distinct from those of its prolyl 4-hydroxylases. J Biol Chem. 2004;279:9899–9904. doi: 10.1074/jbc.M312254200. [DOI] [PubMed] [Google Scholar]
- Koivunen P, Tiainen P, Hyvarinen J, et al. An endoplasmic reticulum transmembrane prolyl 4-hydroxylase is induced by hypoxia and acts on hypoxia-inducible factor alpha. J Biol Chem. 2007;282:30544–30552. doi: 10.1074/jbc.M704988200. [DOI] [PubMed] [Google Scholar]
- Koizume S, Ito S, Miyagi E, et al. HIF2α-Sp1 interaction mediates a deacetylation-dependent FVII-gene activation under hypoxic conditions in ovarian cancer cells. Nucleic Acids Res. 2012;40:5389–5401. doi: 10.1093/nar/gks201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan J, Danzer C, Simka T, et al. Dietary obesity-associated Hif1α activation in adipocytes restricts fatty acid oxidation and energy expenditure via suppression of the Sirt2-NAD+ system. Genes Dev. 2012;26:259–270. doi: 10.1101/gad.180406.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster DE, McNeill LA, McDonough MA, et al. Disruption of dimerization and substrate phosphorylation inhibit factor inhibiting hypoxia-inducible factor (FIH) activity. Biochem J. 2004;383:429–437. doi: 10.1042/BJ20040735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landázuri MO, Vara-Vega A, Vitón M, et al. Analysis of HIF-prolyl hydroxylases binding to substrates. Biochem Biophys Res Comm. 2006;351:313–320. doi: 10.1016/j.bbrc.2006.09.170. [DOI] [PubMed] [Google Scholar]
- Lando D, Peet DJ, Gorman JJ, et al. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002a;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando D, Peet DJ, Whelan DA, et al. Asparagine Hydroxylation of the HIF Transactivation Domain: A Hypoxic Switch. Science Signaling. 2002b;295:858. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- Lau KW, Tian YM, Raval RR, et al. Target gene selectivity of hypoxia-inducible factor-alpha in renal cancer cells is conveyed by post-DNA-binding mechanisms. Br. J. Cancer. 2007;96:1284–1292. doi: 10.1038/sj.bjc.6603675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughner E, Taghavi P, Chiles K, et al. HER2 (neu) Signaling Increases the Rate of Hypoxia-Inducible Factor 1α (HIF-1α) Synthesis: Novel Mechanism for HIF-1-Mediated Vascular Endothelial Growth Factor Expression. Mol Cell Biol. 2001;21:3995–4004. doi: 10.1128/MCB.21.12.3995-4004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Kim Y, Bhin J, et al. Hypoxia-induced methylation of a pontin chromatin remodeling factor. Proc Natl Acad Sci. 2011;108:13510–13515. doi: 10.1073/pnas.1106106108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Kim Y, Kim IS, et al. Negative regulation of hypoxic responses via induced Reptin methylation. Mol Cell. 2010;39:71–85. doi: 10.1016/j.molcel.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendahl U, Lee KL, Yang H, et al. Generating specificity and diversity in the transcriptional response to hypoxia. Nat Rev Genet. 2009;10:821–832. doi: 10.1038/nrg2665. [DOI] [PubMed] [Google Scholar]
- Lim J-H, Lee Y-M, Chun Y-S, et al. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1α. Mol Cell. 2010;38:864–878. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- Liu YV, Baek JH, Zhang H, et al. RACK1 competes with HSP90 for binding to HIF-1α and is required for O2-independent and HSP90 inhibitor-induced degradation of HIF-1α. Mol Cell. 2007;25:207–217. doi: 10.1016/j.molcel.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Forbes RA, Verma A. Hypoxia-inducible Factor 1 Activation by Aerobic Glycolysis Implicates the Warburg Effect in Carcinogenesis. J Biol Chem. 2002;277:23111–23115. doi: 10.1074/jbc.M202487200. [DOI] [PubMed] [Google Scholar]
- Lu H, Dalgard CL, Mohyeldin A, et al. Reversible Inactivation of HIF-1 Prolyl Hydroxylases Allows Cell Metabolism to Control Basal HIF-1. J Biol Chem. 2005;280:41928–41939. doi: 10.1074/jbc.M508718200. [DOI] [PubMed] [Google Scholar]
- Luo W, Hu H, Chang R, et al. Pyruvate Kinase M2 Is a PHD3-Stimulated Coactivator for Hypoxia-Inducible Factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Zhong J, Chang R, et al. Hsp70 and CHIP Selectively Mediate Ubiquitination and Degradation of Hypoxia-inducible Factor (HIF)-1 but Not HIF-2. J Biol Chem. 2010;285:3651–3663. doi: 10.1074/jbc.M109.068577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majmundar AJ, Wong WJ, Simon MC. Hypoxia-Inducible Factors and the Response to Hypoxic Stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino Y, Cao R, Svensson K, et al. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature. 2001;414:550–554. doi: 10.1038/35107085. [DOI] [PubMed] [Google Scholar]
- Makino Y, Kanopka A, Wilson WJ, et al. Inhibitory PAS domain protein (IPAS) is a hypoxia-inducible splicing variant of the hypoxia-inducible factor-3α locus. J Biol Chem. 2002;277:32405–32408. doi: 10.1074/jbc.C200328200. [DOI] [PubMed] [Google Scholar]
- Masson N, Willam C, Maxwell PH, et al. Independent function of two destruction domains in hypoxia-inducible factor-α chains activated by prolyl hydroxylation. EMBO J. 2001;20:5197–5206. doi: 10.1093/emboj/20.18.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard MA, Evans AJ, Hosomi T, et al. Human HIF-3α4 is a dominant-negative regulator of HIF-1 and is down-regulated in renal cell carcinoma. FASEB J. 2005;19:1396–1406. doi: 10.1096/fj.05-3788com. [DOI] [PubMed] [Google Scholar]
- Maynard MA, Qi H, Chung J, et al. Multiple Splice Variants of the Human HIF-3α Locus Are Targets of the von Hippel-Lindau E3 Ubiquitin Ligase Complex. J Biol Chem. 2003;278:11032–11040. doi: 10.1074/jbc.M208681200. [DOI] [PubMed] [Google Scholar]
- McIntosh BE, Hogenesch JB, Bradfield CA. Mammalian Per-Arnt-Sim Proteins in Environmental Adaptation. Annu Rev Physiol. 2010;72:625–645. doi: 10.1146/annurev-physiol-021909-135922. [DOI] [PubMed] [Google Scholar]
- Minet E, Arnould T, Michel G, et al. ERK activation upon hypoxia: involvement in HIF-1 activation. FEBS Lett. 2000;468:53–58. doi: 10.1016/s0014-5793(00)01181-9. [DOI] [PubMed] [Google Scholar]
- Mole DR, Blancher C, Copley RR, et al. Genome-wide association of hypoxia-inducible factor (HIF)-1α and HIF-2α DNA binding with expression profiling of hypoxia-inducible transcripts. J Biol Chem. 2009;284:16767–16775. doi: 10.1074/jbc.M901790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottet D, Dumont V, Deccache Y, et al. Regulation of hypoxia-inducible factor-1α protein level during hypoxic conditions by the phosphatidylinositol 3-kinase/Akt/glycogen synthase kinase 3β pathway in HepG2 cells. J Biol Chem. 2003;278:31277–31285. doi: 10.1074/jbc.M300763200. [DOI] [PubMed] [Google Scholar]
- Murray-Rust TA, Oldham NJ, Hewitson KS, et al. Purified recombinant hARD1 does not catalyse acetylation of Lys532 of HIF-1α fragments in vitro. FEBS Lett. 2006;580:1911–1918. doi: 10.1016/j.febslet.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Mylonis I, Chachami G, Samiotaki M, et al. Identification of MAPK phosphorylation sites and their role in the localization and activity of hypoxia-inducible factor-1α. J Biol Chem. 2006;281:33095–33106. doi: 10.1074/jbc.M605058200. [DOI] [PubMed] [Google Scholar]
- Núñez-O’Mara A, Berra E. Deciphering the emerging role of SUMO conjugation in the hypoxia-signaling cascade. Biol Chem. 2013;394:459–469. doi: 10.1515/hsz-2012-0319. [DOI] [PubMed] [Google Scholar]
- O'Rourke JF, Tian Y-M, Ratcliffe PJ, et al. Oxygen-regulated and Transactivating Domains in Endothelial PAS Protein 1: Comparison with Hypoxia-inducible Factor-1α. J Biol Chem. 1999;274:2060–2071. doi: 10.1074/jbc.274.4.2060. [DOI] [PubMed] [Google Scholar]
- Ohh M, Kaelin WG, Park CW, et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the β-domain of the von Hippel-Lindau protein. Nat. Cell Biol. 2000;2:423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- Olechnowicz SWZ, Fedele AO, Peet DJ. Hypoxic Induction of the Regulator of G-Protein Signalling 4 Gene Is Mediated by the Hypoxia-Inducible Factor Pathway. PLoS ONE. 2012;7:e44564. doi: 10.1371/journal.pone.0044564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Barahona A, Villar D, Pescador N, et al. Genome-wide identification of hypoxia-inducible factor binding sites and target genes by a probabilistic model integrating transcription-profiling data and in silico binding site prediction. Nucleic Acids Res. 2010;38:2332–2345. doi: 10.1093/nar/gkp1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partch CL, Gardner KH. Coactivators necessary for transcriptional output of the hypoxia inducible factor, HIF, are directly recruited by ARNT PAS-B. Proc Natl Acad Sci. 2011;108:7739–7744. doi: 10.1073/pnas.1101357108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasanen A, Heikkilä M, Rautavuoma K, et al. Hypoxia-inducible factor (HIF)-3α is subject to extensive alternative splicing in human tissues and cancer cells and is regulated by HIF-1 but not HIF-2. Int J Biochem Cell Biol. 2010;42:1189–1200. doi: 10.1016/j.biocel.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Pawlus MR, Hu CJ. Enhanceosomes as integrators of hypoxia inducible factor (HIF) and other transcription factors in the hypoxic transcriptional response. Cellular Signalling. 2013;25:1895–1903. doi: 10.1016/j.cellsig.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlus MR, Wang L, Hu CJ. STAT3 and HIF1α cooperatively activate HIF1 target genes in MDA-MB-231 and RCC4 cells. Oncogene. 2013 doi: 10.1038/onc.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlus MR, Wang L, Ware K, et al. Upstream stimulatory factor 2 and hypoxia-inducible factor 2α (HIF2α) cooperatively activate HIF2 target genes during hypoxia. Mol Cell Biol. 2012;32:4595–4610. doi: 10.1128/MCB.00724-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella BL, Lohi J, Brinckerhoff CE. Identification of membrane type-1 matrix metalloproteinase as a target of hypoxia-inducible factor-2α in von Hippel-Lindau renal cell carcinoma. Oncogene. 2005;24:1043–1052. doi: 10.1038/sj.onc.1208305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, et al. c-Myc Regulates Transcriptional Pause Release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard DE, Berra E, Gothié E, et al. p42/p44 Mitogen-activated Protein Kinases Phosphorylate Hypoxia-inducible Factor 1α (HIF-1α) and Enhance the Transcriptional Activity of HIF-1. J Biol Chem. 1999;274:32631–32637. doi: 10.1074/jbc.274.46.32631. [DOI] [PubMed] [Google Scholar]
- Ruas JL, Berchner-Pfannschmidt U, Malik S, et al. Complex regulation of the transactivation function of hypoxia-inducible factor-1α by direct interaction with two distinct domains of the CREB-binding protein/p300. J Biol Chem. 2010;285:2601–2609. doi: 10.1074/jbc.M109.021824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruas JL, Poellinger L, Pereira T. Functional analysis of hypoxia-inducible factor- 1α-mediated transactivation: Identification of amino acid residues critical for transcriptional activation and/or interaction with CREB-binding protein. J Biol Chem. 2002;277:38723–38730. doi: 10.1074/jbc.M205051200. [DOI] [PubMed] [Google Scholar]
- Ruas JL, Poellinger L, Pereira T. Role of CBP in regulating HIF-1-mediated activation of transcription. J Cell Sci. 2005;118:301–311. doi: 10.1242/jcs.01617. [DOI] [PubMed] [Google Scholar]
- Ryan HE, Lo J, Johnson RS. HIF-1α is required for solid tumor formation and embryonic vascularization. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadek CM, Jalaguier S, Feeney EP, et al. Isolation and characterization of AINT: a novel ARNT interacting protein expressed during murine embryonic development. Mech Dev. 2000;97:13–26. doi: 10.1016/s0925-4773(00)00415-9. [DOI] [PubMed] [Google Scholar]
- Sahlgren C, Gustafsson MV, Jin S, et al. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc Natl Acad Sci. 2008;105:6392–6397. doi: 10.1073/pnas.0802047105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikow K, Aprelikova O, Ivanov S, et al. Regulation of hypoxia-inducible genes by ETS1 transcription factor. Carcinogenesis. 2008;29:1493–1499. doi: 10.1093/carcin/bgn088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikow K, Donald SP, Bruick RK, et al. Depletion of Intracellular Ascorbate by the Carcinogenic Metals Nickel and Cobalt Results in the Induction of Hypoxic Stress. J Biol Chem. 2004;279:40337–40344. doi: 10.1074/jbc.M403057200. [DOI] [PubMed] [Google Scholar]
- Schödel J, Mole DR, Ratcliffe PJ. Pan-genomic binding of hypoxia-inducible transcription factors. Biol Chem. 2013;394:507–517. doi: 10.1515/hsz-2012-0351. [DOI] [PubMed] [Google Scholar]
- Schödel J, Oikonomopoulos S, Ragoussis J, et al. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. blood. 2011;117:e207–e217. doi: 10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scortegagna M, Morris MA, Oktay Y, et al. The HIF family member EPAS1/HIF-2α is required for normal hematopoiesis in mice. blood. 2003;102:1634–1640. doi: 10.1182/blood-2003-02-0448. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Hypoxia-Inducible Factors in Physiology and Medicine. Cell. 2012a;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci. 2012b;33:207–214. doi: 10.1016/j.tips.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N, Maemura K, Imai Y, et al. Endothelial PAS domain protein 1 gene promotes angiogenesis through the transactivation of both vascular endothelial growth factor and its receptor, Flt-1. Circ. Res. 2004;95:146–153. doi: 10.1161/01.RES.0000134920.10128.b4. [DOI] [PubMed] [Google Scholar]
- Tang N, Wang L, Esko J, et al. Loss of HIF-1α in endothelial cells disrupts a hypoxia-driven VEGF autocrine loop necessary for tumorigenesis. Cancer Cell. 2004;6:485–495. doi: 10.1016/j.ccr.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Tanimoto K, Makino Y, Pereira T, et al. Mechanism of regulation of the hypoxia-inducible factor-1α by the von Hippel-Lindau tumor suppressor protein. EMBO J. 2000;19:4298–4309. doi: 10.1093/emboj/19.16.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Hammer RE, Matsumoto AM, et al. The hypoxia-responsive transcription factor EPAS1 is essential for catecholamine homeostasis and protection against heart failure during embryonic development. Genes Dev. 1998;12:3320–3324. doi: 10.1101/gad.12.21.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Mcknight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- Tien ES, Davis JW, Vanden Heuvel JP. Identification of the CREB-binding protein/p300-interacting protein CITED2 as a peroxisome proliferator-activated receptor alpha coregulator. J Biol Chem. 2004;279:24053–63. doi: 10.1074/jbc.M401489200. [DOI] [PubMed] [Google Scholar]
- Toffoli S, Feron O, Raes M, et al. Intermittent hypoxia changes HIF-1α phosphorylation pattern in endothelial cells: Unravelling of a new PKA-dependent regulation of HIF-1α. BBA-Mol Cell Res. 2007;1773:1558–1571. doi: 10.1016/j.bbamcr.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Tsai Y-P, Wu K-J. Hypoxia-regulated target genes implicated in tumor metastasis. J Biomed Sci. 2012;19:102. doi: 10.1186/1423-0127-19-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D, Ortiz-Barahona A, Gómez-Maldonado L, et al. Cooperativity of stress-responsive transcription factors in core hypoxia-inducible factor binding regions. PloS ONE. 2012;7:e45708. doi: 10.1371/journal.pone.0045708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhang R, Beischlag TV, et al. Roles of Brahma and Brahma/SWI2-related gene 1 in hypoxic induction of the erythropoietin gene. J Biol Chem. 2004a;279:46733–46741. doi: 10.1074/jbc.M409002200. [DOI] [PubMed] [Google Scholar]
- Wang S, Ge K, Roeder RG, et al. Role of Mediator in Transcriptional Activation by the Aryl Hydrocarbon Receptor. J Biol Chem. 2004b;279:13593–13600. doi: 10.1074/jbc.M312274200. [DOI] [PubMed] [Google Scholar]
- Wang F, Zhang R, Wu X, et al. Roles of coactivators in hypoxic induction of the erythropoietin gene. PloS ONE. 2010a;5:e10002. doi: 10.1371/journal.pone.0010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, H JB, Rue EA, et al. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Semenza GL. Characterization of hypoxia-inducible factor 1 and regulation of DNA binding activity by hypoxia. J Biol Chem. 1993a;268:21513–21518. [PubMed] [Google Scholar]
- Wang GL, Semenza GL. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc Natl Acad Sci. 1993b;90:4304–4308. doi: 10.1073/pnas.90.9.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Semenza GL. Purification and characterization of hypoxia-inducible factor 1. J Biol Chem. 1995;270:1230–1237. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- Wang R, Zhang YW, Zhang X, et al. Transcriptional regulation of APH-1A and increased gamma-secretase cleavage of APP and Notch by HIF-1 and hypoxia. FASEB J. 2006;20:1275–1277. doi: 10.1096/fj.06-5839fje. [DOI] [PubMed] [Google Scholar]
- Wang V, Davis DA, Veeranna RP, et al. Characterization of the activation of protein tyrosine phosphatase, receptor-type, Z polypeptide 1 (PTPRZ1) by hypoxia inducible factor-2 alpha. PLoS ONE. 2010b doi: 10.1371/journal.pone.0009641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke C, Weidemann A, Volke M, et al. The specific contribution of hypoxia-inducible factor-2α to hypoxic gene expression in vitro is limited and modulated by cell type-specific and exogenous factors. Exp Cell Res. 2008;314:2016–2027. doi: 10.1016/j.yexcr.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Wenger RH, Stiehl DP, Camenisch G. Integration of oxygen signaling at the consensus HRE. Science's STKE : signal transduction knowledge environment. 2005:re12. doi: 10.1126/stke.3062005re12. 2005. [DOI] [PubMed] [Google Scholar]
- Wong CC-L, Gilkes DM, Zhang H, et al. Hypoxia-inducible factor 1 is a master regulator of breast cancer metastatic niche formation. Proc Natl Acad Sci. 2011;108:16369–16374. doi: 10.1073/pnas.1113483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xenaki G, Ontikatze T, Rajendran R, et al. PCAF is an HIF-1α cofactor that regulates p53 transcriptional activity in hypoxia. Oncogene. 2008;27:5785–5796. doi: 10.1038/onc.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Kung AL. Preferential binding of HIF-1 to transcriptionally active loci determines cell-type specific response to hypoxia. Genome biology. 2009;10:R113. doi: 10.1186/gb-2009-10-10-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Yao Y, Lu L, et al. Plk3 Functions as an Essential Component of the Hypoxia Regulatory Pathway by Direct Phosphorylation of HIF-1. J Biol Chem. 2010;285:38944–38950. doi: 10.1074/jbc.M110.160325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Ohneda O, Nagano M, et al. Abnormal heart development and lung remodeling in mice lacking the hypoxia-inducible factor-related basic helix-loop-helix PAS protein NEPAS. Mol Cell Biol. 2008;28:1285–1297. doi: 10.1128/MCB.01332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MH, Wu MZ, Chiou SH, et al. Direct regulation of TWIST by HIF-1α promotes metastasis. Nat Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- Yang W, Xia Y, Ji H, et al. Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasinska IM, Sumbayev VV. S-nitrosation of Cys-800 of HIF-1α protein activates its interaction with p300 and stimulates its transcriptional activity. FEBS Lett. 2003;549:105–109. doi: 10.1016/s0014-5793(03)00807-x. [DOI] [PubMed] [Google Scholar]
- Yoon H, Lim J-H, Cho C-H, et al. CITED2 controls the hypoxic signaling by snatching p300 from the two distinct activation domains of HIF-1α. Biochim Biophys Acta. 2011;1813:2008–2016. doi: 10.1016/j.bbamcr.2011.08.018. [DOI] [PubMed] [Google Scholar]
- Zagórska A, Dulak J. HIF-1: the knowns and unknowns of hypoxia sensing. Acta Biochimica Polonica. 2004;51:563–585. [PubMed] [Google Scholar]
- Zheng X, Linke S, Dias JM, et al. Interaction with factor inhibiting HIF-1 defines an additional mode of cross-coupling between the Notch and hypoxia signaling pathways. Proc Natl Acad Sci. 2008;105:3368–3373. doi: 10.1073/pnas.0711591105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, D'Urso A, Toiber D, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1α. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziel KA, Campbell CC, Wilson GL, et al. Ref-1/Ape is critical for formation of the hypoxia-inducible transcriptional complex on the hypoxic response element of the rat pulmonary artery endothelial cell VEGF gene. FASEB J. 2004;18:986–988. doi: 10.1096/fj.03-1160fje. [DOI] [PubMed] [Google Scholar]