Abstract
Plant-specific NAC transcription factors (TFs) constitute a large family and play important roles in regulating plant developmental processes and responses to environmental stresses, but only some of them have been investigated for effects on disease reaction in cereal crops. Virus-induced gene silencing (VIGS) is an effective strategy for rapid functional analysis of genes in plant tissues. In this study, TaNAC1, encoding a new member of the NAC1 subgroup, was cloned from bread wheat and characterized. It is a TF localized in the cell nucleus, and contains an activation domain in its C-terminal. TaNAC1 was strongly expressed in wheat roots and was involved in responses to infection by the obligate pathogen Puccinia striiformis f. sp. tritici and defense-related hormone treatments such as salicylic acid (SA), methyl jasmonate, and ethylene. Knockdown of TaNAC1 with barley stripe mosaic virus-induced gene silencing (BSMV-VIGS) enhanced stripe rust resistance. TaNAC1-overexpression in Arabidopsis thaliana plants gave enhanced susceptibility, attenuated systemic-acquired resistance to Pseudomonas syringae DC3000, and promoted lateral root development. Jasmonic acid-signaling pathway genes PDF1.2 and ORA59 were constitutively expressed in transgenic plants. TaNAC1 overexpression suppressed the expression levels of resistance-related genes PR1 and PR2 involved in SA signaling and AtWRKY70, which functions as a connection node between the JA- and SA-signaling pathways. Collectively, TaNAC1 is a novel NAC member of the NAC1 subgroup, negatively regulates plant disease resistance, and may modulate plant JA- and SA-signaling defense cascades.
Keywords: wheat, stripe rust, disease resistance, BSMV-VIGS, jasmonic acid
INTRODUCTION
Crop plants provide most of the world’s food intake. A plethora of pathogens including viruses, bacteria, fungi, oomycetes, and nematodes cause severe yield losses in crop production. Fortunately, plants have evolved sophisticated defense mechanisms to cope with biotic stresses (Jones and Dangl, 2006; Howe and Jander, 2008). Multiple defense mechanisms, including basal resistance, PAMP-triggered immunity (PTI) and effector-triggered immunity (ETI), are activated at different stages of pathogen infection. Plant defense systems translate pathogen-induced early signaling events into activation of effective defense responses, most of which depend on the action of plant phytohormones such as salicylic acid (SA), jasmonates (JAs), ethylene (ET), abscisic acid (ABA), auxins, cytokinins (CKs), and gibberellins (GAs; Pieterse et al., 2012). The actions of JAs and SA as signals in the regulation of plant immune responses are well established. The JA-signaling pathway is primarily effective in mediating resistance against necrotrophic pathogens and herbivores, while the SA-signaling pathway primarily functions in mediating resistance to biotrophic pathogens. The JA and SA defense pathways generally antagonize each other; thus, elevated resistance against necrotrophs is often correlated with increased susceptibility to biotrophs (Glazebrook, 2005; Derksen et al., 2013; Vos et al., 2013).
In addition to hormones, plant defense responses require diverse types of transcription factors (TFs) involved in downstream signaling pathways. TF families such as bZIP, bHLH, ERF/AP2, MYB, MYC, NAC, and WRKY play important roles in disease resistance (Singh et al., 2002). Since the first NAC (NAM, ATAF1/2, and CUC1)-type TF NAM (no apical meristem) was identified in Petunia (Souer et al., 1996), many other NAC-type genes have been found in different plant species and constitute a large plant-specific TF family. Based on several whole plant genome sequences, genome-wide identifications of NAC-type transcription factors (NAC-TFs) have been carried out. These studies predicted 117 NAC-TFs in Arabidopsis thaliana (Ooka et al., 2003), 163 in Populus trichocarpa (Hu et al., 2010), 151 in Glycine max (Le et al., 2011), 152 in Solanum tuberosum (Singh et al., 2013), 147 in Vitis vinifera (Wang et al., 2013), 110 in Malus domestica (Su et al., 2013), 74 in Oryza sativa (Nuruzzaman et al., 2010), 180 in Setaria italica (Puranik et al., 2013), and 145 in Gossypium raimondii Ulbr (Shang et al., 2013). Typically, most NAC proteins share a highly conserved N-terminal NAC domain, which usually consists of five subdomains (A–E) and a well-diversified C-terminal transcription regulatory (TR) region.
NAC-type transcription factors are involved in diverse plant biological regulation processes, including developmental programs, plant senescence control, secondary cell wall formation, and various biotic and abiotic stress responses (Olsen et al., 2005; Nakashima et al., 2012; Puranik et al., 2012). In addition to being involved in plant responses to biotic stresses some NAC proteins have dual roles as positive or negative regulators of response to different pathogens (Seo et al., 2010). For example, ATAF1 and its barley homolog HvNAC6 enhanced resistance to the biotrophic fungus Blumeria graminis f. sp. graminis (Bgh; Chen et al., 2013), but attenuated resistance to pathogens such as Pseudomonas syringae, Botrytis cinerea and Alternaria brassicicola (Wang et al., 2009; Wu et al., 2009). Most NACs that function in biotic stress regulation belong to the SNAC (stress-responsive NAC) subgroup. Some members of the NAC-TF family are multifunctional mediators of both development and stress signaling, and 20–25% of NAC genes function in at least one or more stress response(s) (Fang et al., 2008). For example, members of the NAC1 subgroup have roles in both development and stress response. It was clearly shown that NAC1 (ANAC021/ANAC022) directed lateral root formation in Arabidopsis through the auxin signaling pathway whereas TaNAC7 was induced by dehydration, salinity and cold (Xie et al., 2000, 2002; Tang et al., 2012). However, knowledge of the functions of most members in the NAC1 subgroup in disease response is limited.
Bread wheat (Triticum aestivum, genomes AABBDD, 2n = 6X = 42) is a globally important crop. As with other plants, stress factors such as drought, high salinity and disease limit productivity and reduce yield. Several characterized NAC-TFs in the wheat genome function in regulating nutrient transportation, development, disease resistance, and stress tolerance (Xie et al., 1999; Uauy et al., 2006; Xia et al., 2010a,b; Xue et al., 2011; Mao et al., 2012). Recently, the genomic sequences of diploid wild einkorn wheat (Triticum urartu, AA, 2n = 2X = 14) and diploid goat grass (Aegilops tauschii, DD, 2n = 2X = 14) were sequenced, and 76 and 117 NAC-TFs were predicted in the T. urartu and A. tauschii genomes, respectively. As bread wheat evolved from natural hybridization between cultivated tetraploid emmer wheat and diploid goat grass, the number of NAC-TFs in bread wheat should be much higher than in the parental diploid species and could show greater genetic diversity. However, the exact roles of most wheat NAC-TFs in response to biotic stresses remain unknown.
Previously we isolated an EST sequence encoding a putative NAC-TF, which was differentially expressed during stripe rust infection. The full length of this EST sequence was isolated in the present study. It belongs to the NAC1 subgroup and was named TaNAC1. Its expression patterns in response to exogenous hormone applications were analyzed. The biological functions of TaNAC1 in wheat and transgenic Arabidopsis plants were also investigated. TaNAC1 was involved in both plant disease resistance and development regulation.
MATERIALS AND METHODS
PLANT GROWTH, BIOTIC STRESS, AND CHEMICAL TREATMENTS
Wheat near-isogenic line (NIL) Yr10∗6/Taichung 29 containing the Yr10 gene is resistant to most races of P. striiformis f. sp. tritici in China, whereas its backcross parent Taichung 29 is highly susceptible. A full length cDNA library of near-isogenic line (NIL) Yr10∗6/Taichung 29 infected by P. striiformis race CYR17 was constructed. P. striiformis races CYR17 and G22, respectively avirulent and virulent to the resistance gene Yr10 were used in comparative analyses. Wheat seedlings were grown in 8 cm pots and cultivated under 14/10 h day/night photoperiods at 20°C. Seven-day-old seedlings were inoculated with urediniospores of P. striiformis as previously described (Xia et al., 2010a), and leaf samples were collected at 0, 8, 12, 24, and 48 h post-inoculation (hpi). To profile the expression patterns of TaNAC1 responding to exogenous plant hormones, 2-week-old seedlings of Yr10∗6/Taichung 29 were treated with 1 mM SA, 0.1 mM MeJA (both SA and MeJA were dissolved in 0.1% ethanol) and ET released from 0.2 mM ethephon, respectively (Zhang et al., 2004), and leaf samples were collected at 0, 1, 3, 6, 12, 24 h post-treatment (hpt). Intact root, stem, leaf, and spike tissues at flowering were sampled separately for tissue-specific expression analyses of TaNAC1. After collection all samples were immediately frozen in liquid nitrogen and then stored at -80°C for RNA isolation.
Arabidopsis thaliana accession Col-0 was used for genetic transformation. Arabidopsis seeds were grown in 8 cm pots containing a mixture of soil and vermiculite (2:1) in a 16 h light/8 h darkness photoperiod (150 μmol m-2s-1) and 70% relative humidity at 22°C, four plants per pot.
Arabidopsis TRANSFORMATION AND LATERAL ROOT ANALYSIS
To obtain 35S::TaNAC1 transgenic plants, TaNAC1 cDNA was cloned into the binary vector pCAMBIA-1301 under the control of the cauliflower mosaic virus (CaMV) 35S promoter and transformed into Arabidopsis Col-0 by the floral dip method (Bechtold et al., 2003) using Agrobacterium tumefaciens strain GV3101. The gene-specific primer pairs included forward primer: 5′-GGTACCCGATCCGACCGAGAAGATG-3′ (Kpn I site in italics) and reverse primer: 5′-TCTAGAGCCATTCCACTAATCTACTG-3′ (Xba I site in italics). Positive transgenic lines were firstly screened on half-strength MS plates (with 3% sucrose and 1.2% agar) containing 30 mg L-1 hygromycin and then identified by reverse-transcription PCR. Homozygous T3 lines were used for further analyses. Col-0 and TaNAC1 overexpressing lines were planted on half-strength MS plates under a 16 h light/8 h darkness photoperiod at 22°C in a growth chamber for 10 days. Root lengths were then measured and the numbers of lateral roots per plant were recorded. SPSS 16.0 (SPSS Inc., USA1) software was used to determine significant differences by one-way ANOVA, taking P < 0.05 level as significant according to Duncan’s multiple range test.
GENE EXPRESSION ANALYSES
Total RNA from each wheat and Arabidopsis sample was extracted using TRIZOL reagent according to the manufacturer’s protocol (Invitrogen, USA). Five micrograms (μg) of total RNA per sample was used to synthesize first-strand cDNA with the TransScript II One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (TransGen Biotech, Beijing). Quantitative RT-PCR was performed using GoTaq® qPCR Master Mix (Promega, Wisconsin) and signals were detected with an ABI7500 Real-Time PCR System (Applied BioSystems, New York). Dissociation curves were generated for each reaction to ensure specific amplification. Every reaction was performed three times. Threshold values (CT) generated from the ABI PRISM 7500 software tool (Applied Biosystems) were employed to quantify relative gene expression using the comparative threshold (2-ΔΔCT) method (Schmittgen and Livak, 2008). ADP-RF (ADP-ribosylation factor, Ta2291; Paolacci et al., 2009) and PP2A (protein phosphatase 2A subunit A3; Czechowski et al., 2005) transcripts were used as controls in quantitative reverse transcription–PCR (qRT–PCR) analyses of expression patterns of TaNAC1 and its regulated genes in transgenic Arabidopsis plants. Three independent biological replicates were performed for each experiment. SPSS 16.0 (SPSS Inc., USA2) software was used to determine significant differences by one-way ANOVA, taking P < 0.05 level as significant according to Duncan’s multiple range test, either between expression levels in the same treatment at different times or between experimental and mock treatments. The sequences of qRT-PCR primers are listed in Table 1.
Table 1.
Primers used in qRT-PCR.
| Primer name | Forward sequence | Reverse sequence |
|---|---|---|
| TaNAC1 | TCGTCTTCTACCAGGGGAGG | AGAACACCCTGCATAGCACC |
| TaADP-RF | GCTCTCCAACAACATTGCCAAC | GCTTCTGCCTGTCACATACGC |
| PR1 | ATGAATTTTACTGGCTTCTCG | TTAGTATGGCTTCTCGTTCACAT |
| PR2 | CAGATTCCGGTACATCAACG | AGTGGTGGTGTCAGTGGCTA |
| PDF1.2 | AAGTGGGACATGGTCAGGGGTT | ACTTGTGTGCTGGGAAGACATAGTT |
| ICS1 | GGCAGGGAGACTTACG | AGGTCCCGCATACATT |
| LEC | GTTTCGTCTCTGGGTCATGGA | GCAGCAACTTGTTATTCCTTGGA |
| VSP2 | TCAGTGACCGTTGGAAGTTGTG | GTTCGAACCATTAGGCTTCAATATG |
| ERF1 | CGAGAAGCTCGGGTGGTAGT | GCCGTGCATCCTTTTCC |
| ORA59 | CATACAGAGGAGTGAGGAAACG | AATTGAGTACTGCGAGGCTG |
| MYC2 | TCATACGACGGTTGCCAGAA | AGCAACGTTTACAAGCTTTGATTG |
| ANAC019 | GCATCTCGTCGCTCAG | CTCGACTTCCTCCTCCG |
| ANAC055 | GCGCTGCCTCATAGTC | CGAGGAATCCCCTCAGT |
| ANAC072 | TGGGTGTTGTGTCGAAT | ATCGTAACCACCGTAACT |
| AtWRKY70 | CGTCATCATGGTTCGTCCA | CCACCTCCAAACACCATGAGAT |
| PP2A | TAACGTGGCCAAAATGATGC | GTTCTCCACAACCGCTTGGT |
SEQUENCE ANALYSES AND PHYLOGENETIC TREE CONSTRUCTION
Gene sequences were analyzed using DNAMAN software (Lynnon Biosoft, USA), BLAST and ORF Finder on the NCBI website. Conserved domains and motifs were predicted using the PROSITE Scan tool3. The coding sequence of TaNAC1 was deduced from its isolated cDNA clone, and the closest homologs in other plants were identified using TBLASTX4. Multiple alignments of protein sequences were performed with the program Clustal X version 2.0 program (Larkin et al., 2007). The phylogenetic comparison of isolated full-length TaNAC1 and characterized NAC proteins based on the literature (Ooka et al., 2003; Jamar et al., 2010; Christiansen et al., 2011; D’haeseleer et al., 2011; Tang et al., 2012; Feng et al., 2014) and GenBank5 were constructed with the neighbor-joining algorithm using MEGA4 program (Tamura et al., 2007). Bootstrap values were calculated from 1000 bootstrap replicates.
SUBCELLULAR LOCALIZATION
An expression vector containing a complete encoding region of TaNAC1 cDNA was constructed to confirm nuclear localization of TaNAC1. Kpn I and Xba I linkers were added to the encoding region and the stop codon was deleted by PCR amplification. The forward primer 5′-GGTACCCGATCCGACCGAGAAGATG-3′ (Kpn I site in italics) and reverse primer 5′-TCTAGATGACAAGCCGTTCTCCTG-3′ (Xba I site in italics) were used. High-fidelity DNA polymerase HIFI Taq (TransGen Biotech, Beijing) was used, and the PCR product was cloned into Kpn I and Xba I sites of the binary vector pCaMV35S::GFP to produce fusion vector pCaMV35S::TaNAC1-GFP. The expression plasmid and vector control were transformed into onion epidermal cells by particle bombardment using a Biolistic PDS-1000/He gene gun system (BIO-RAD, Hercules, CA, USA). After 24 h of incubation at 25°C, the fluorescence of DAPI and GFP images of the transformed onion epidermal cells were observed under a confocal microscope (Zeiss LSM 510 Meta Confocal Microscope).
TRANSCRIPTIONAL ACTIVATION ANALYSES IN YEAST
To investigate the transcriptional regulation domains, the complete ORF (S1, 1/290, primer TF1 5′-TCGAATTCATGAGCTCTA- TTGGCATGAT-3′ and primer TR1 5′-TACTGCAGTCAAAGGG- GTGTCCACATCT-3′, EcoR I and Pst I sites in italics), one C-terminal truncated (S2: 1/180, primer TF1 and primer TR2 5′-TACTGCAGTCTGCTCTTGTAGAACACCCT-3′, Pst I site in italics) and two N-terminal truncated (S3: 107/290, primer TF3 5′-TCGAATTCGGCGTTGGTGGGGATGCGCA-3′ and TR1; S4: 178/290, primer TF4 5′-TCGAATTCCAGAACAAGCAGCCCAA- GGC1-3′ and TR1, EcoR I sites in italics) fragments of TaNAC1 were fused in-frame with encoding region for the GAL4 DNA-binding domain in expression vector pGBKT7. pGBKT7 was used as a negative control. These different constructs were transformed into yeast strain AH109. The transformants were streaked on SD/Trp- and SD/Trp-/His-/Ade- medium plates. After incubation at 28°C for 3 days, the growth status of the transformants was evaluated. The α-galactosidase filter assay was carried out according to the manufacturer’s instructions.
FUNCTIONAL ANALYSES OF TaNAC1 IN RESPONSE TO P. striiformis INFECTION USING VIGS
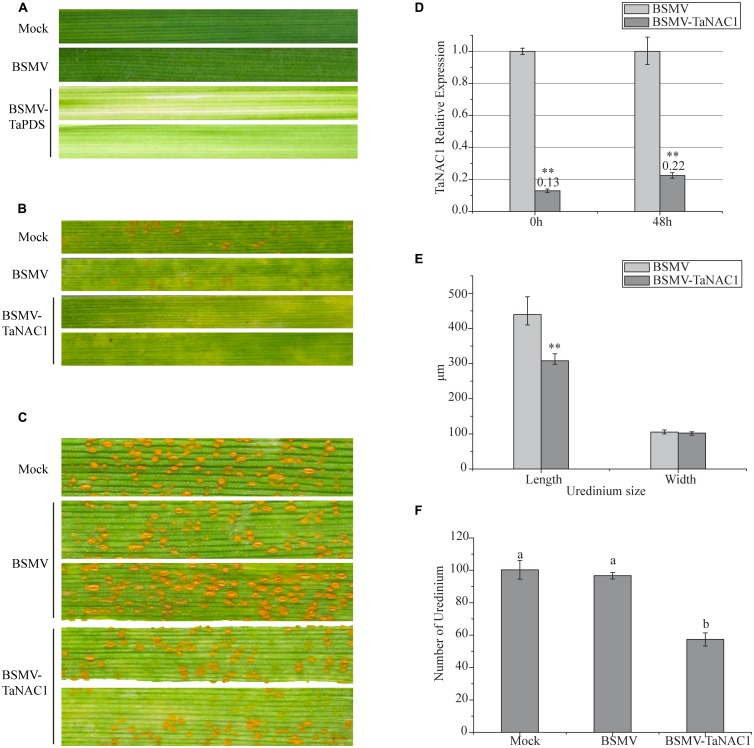
BSMV-induced gene silencing assays were conducted as described (Yuan et al., 2011). Specific sequences of target genes TaPDS (with primers pairs, vTaPDSf1, 5′-AAGGAAGTTTAACTGCATAAACG- CTTAAAAG-3′ and vTaPDSr1, 5′-AACCACCACCACCGTTCT- CCAGTTATTTGAG-3′, LIC adapters in italics) and TaNAC1 (with primers pairs, vTaNACf1, 5′-AAGGAAGTTTAATAGCGATCCG- ACCGAGAAGA-3′ and vTaNACr1, 5′-AACCACCACCACCGTG- CTTGTGGAGGAGGTAGTCG-3′, LIC adapters in italics) were inserted into pCa-cbLIC. Briefly, 2-week-old wheat seedlings of cultivar Funo were cultured in a growth chamber under a 16/8 h day/night photoperiod at 16 ± 2°C, and watered as needed. The entire second leaf surfaces of two-leaf stage wheat seedlings were inoculated with Nicotiana benthamiana leaf sap containing BSMV particles by gently sliding pinched fingers from the leaf base to the tip three times. Control plants were mock-inoculated in the same manner with sterile water. Three independent sets of plants were prepared for each treatment (Mock, BSMV, and BSMV-TaNAC1). Inoculated seedlings were placed in a growth chamber in darkness and relative humidity of 60% for 24 h at 25 ± 2°C, and then kept under a 16/8 h day/night photoperiod under the same temperature and humidity conditions. Virus phenotypes were observed and photographed after 9 days. At 14 days post virus inoculation, fresh urediniospores of race G22 were collected and inoculated onto the surfaces of third leaves with a paintbrush. Three independent biological replications were performed for each treatment. Stripe rust phenotypes were recorded and photographed 10–15 dpi. The lengths and widths of 60 random uredinia were measured with ocular micrometers at 15 dpi. SPSS 16.0 (SPSS Inc., USA6) software was used to determine the significance of differences in length and width between the BSMV and BSMV-TaNAC1 samples, taking P < 0.05 level as significant according to Student’s t-tests.
IN PLANTA DISEASE RESPONSE ASSAYS
Homozygous T3 generation seeds of transgenic Arabidopsis lines were used for pathogen infection assays. Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) and Pst DC3000 (avrRpt2) were cultivated on King’s B medium plates with corresponding antibiotics at 30°C for 2 days. Pathogen infection and the determination of in planta pathogen population growth were conducted as described (Cao et al., 1997). For the systemic acquired resistance assay, three lower leaves were inoculated with the avirulent Pst strain DC3000 (avrRpt2; OD600nm = 0.1), and plants infiltrated with 10 mM MgCl2 served as mock controls. Infected leaves were removed at 3 days post primary infection and two upper leaves were pressure-infiltrated with the virulent Pst strain DC3000 (OD600nm = 0.0001). Colony-forming units were scored 3 days later. Plants inoculated only with Pst DC3000 were used as controls. Data were subjected to ANOVA using SPSS 16.0 (SPSS Inc., USA7), and the significance of differences between different bacterial populations were assessed by the Student t-tests.
RESULTS
SEQUENCE ANALYSIS OF PUTATIVE TaNAC1
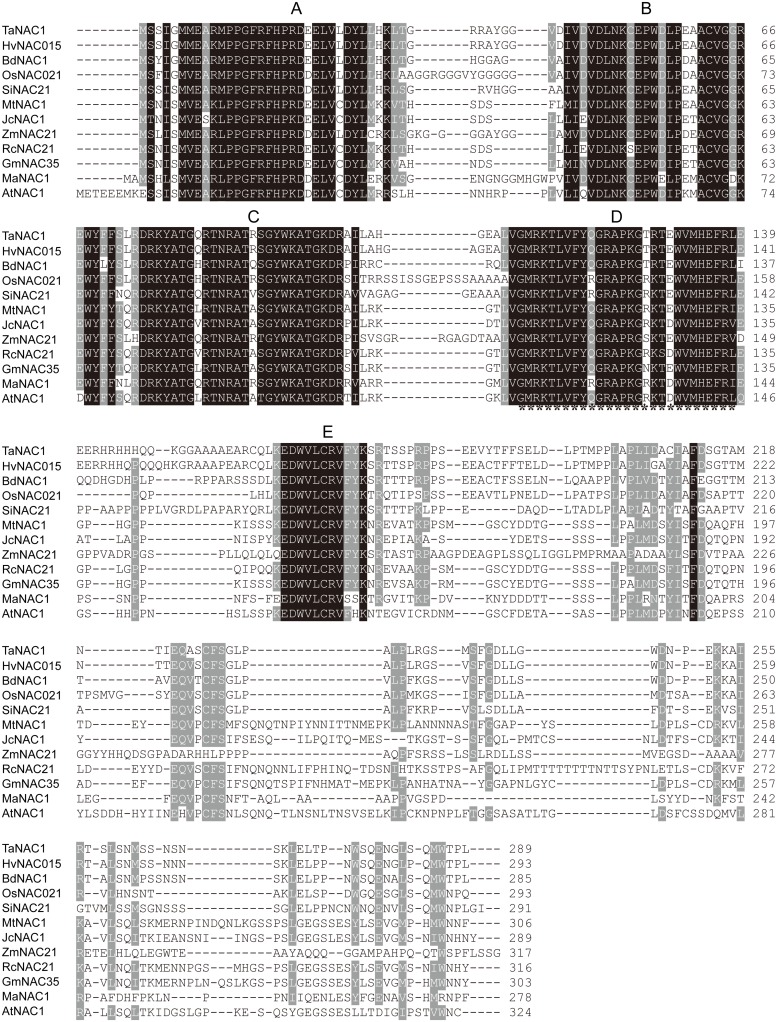
Based on the EST sequence of a differentially expressed NAC1 gene during stripe rust infection, the corresponding full length gene was cloned from a cDNA library of wheat near-isogenic line (NIL) Yr10∗6/Taichung 29 by PCR with FastPfu DNA Polymerase. The 1,227 bp cDNA clone contained an 870 bp open reading frame, encoding a putative NAC TF of 289 amino acid residues. Its calculated molecular weight was 32.75 kDa and pI 8.14. Sequence alignment with NAC factors from different plant species showed that this NAC factor contains a conserved NAC DNA-binding domain comprising five subdomains (A–E; Figure 1). According to NCBI blastp results the gene was most similar to HvNAC015 with an identity of 92.7%, followed by BdNAC21-22-2 (76.8%), OsNAC21 (73.9%), and AtNAC1. It was named TaNAC1. A NAC transcription repression domain (NARD) containing a conserved hydrophobic ‘LVFY’ motif (Hao et al., 2010) was present in TaNAC1 (Figure 1). Accordingly, it may function as a transcription repressor.
FIGURE 1.
TaNAC1 sequence analysis. Amino acid alignment of TaNAC1 with other NAC family members from selected plant species. The numbers on the left indicate amino acid positions. Identical amino acid residues are shaded black representing 100% similarity and dark gray for 75% similarity of conserved amino acids. A putative transcription repression region containing a ‘LVFY’ motif is underlined with asterisks.
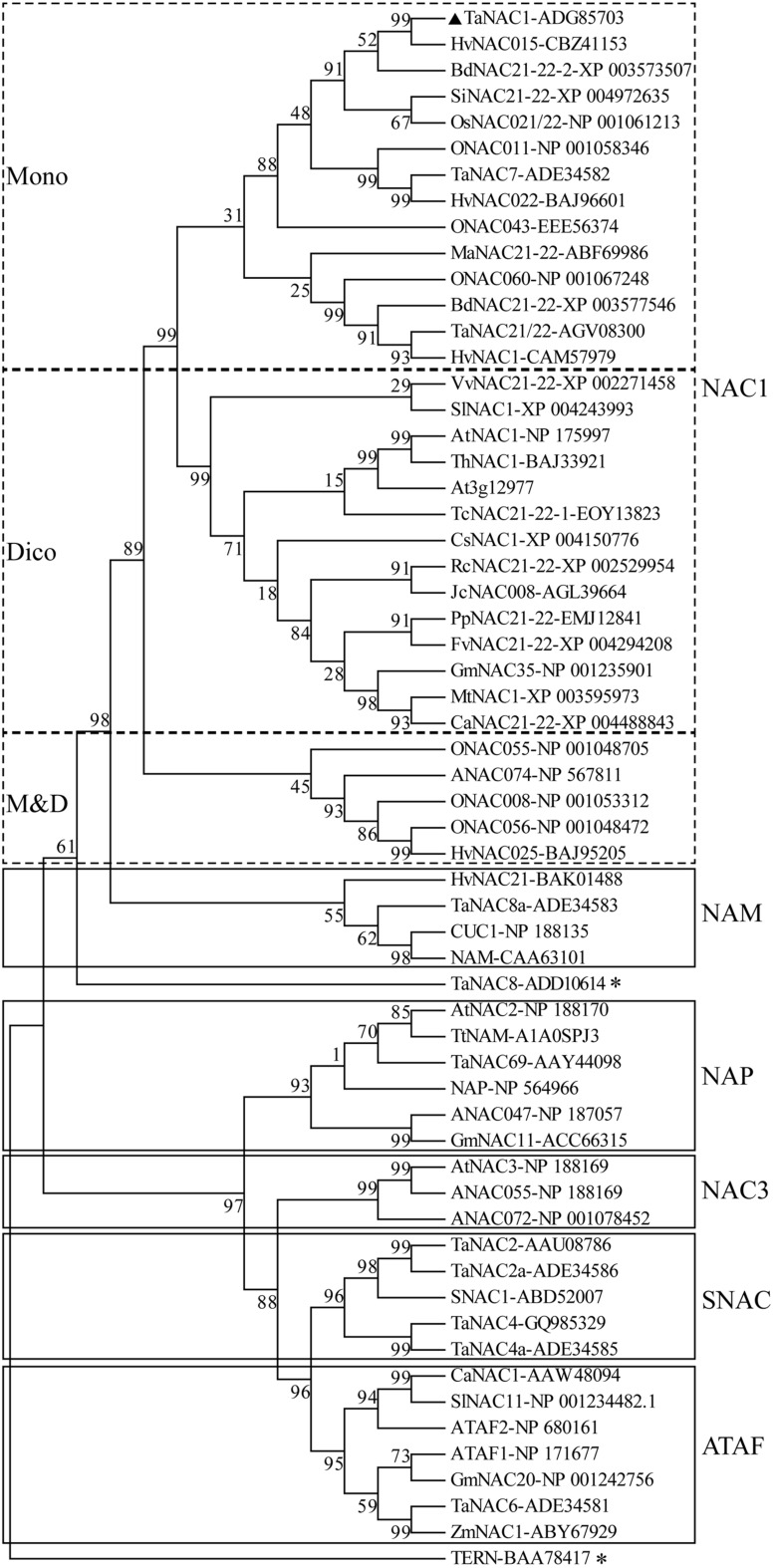
A phylogenetic tree was constructed with the putative amino acid sequences of TaNAC1 and NAC family members from other plant species. Phylogenetic analysis revealed that TaNAC1 clustered with the NAC1 subgroup (ANAC021/22) containing 33 members, and those from monocotyledons or dicotyledons clearly clustered into different branches (Figure 2).
FIGURE 2.
Phylogenetic tree of NAC transcription factors (NAC TFs). Phylogenetic tree of all reported NAC TFs from wheat, selected NAC TFs from barley (HvNACs), Brachypodium distachyon (BdNACs), Oryza sativa (ONACs), Arabidopsis thaliana (ANACs), and a few selected NAC factors from other species. TaNAC8 and TERN marked with an asterisk do not belong to any subgroup in this phylogenetic tree. Accession numbers follow the gene names.
SUBCELLULAR LOCALIZATION OF TaNAC1
Expression vector pCaMV35S::TaNAC1–GFP was constructed by fusing full-length TaNAC1 cDNA with the green fluorescent protein (GFP) encoding region, and introduced into onion epidermal cells by particle bombardment in an attempt to determine the subcellular localization of TaNAC1. In the onion cells transformed with GFP vector control, green fluorescence was observed throughout the entire cells (Figures 3A–D). In contrast, fluorescence was predominantly detected in the nuclei of cells transformed with the plasmid encoding pCaMV35S::TaNAC1–GFP fusion protein (Figure 3E–H), indicating that TaNAC1 is a nuclear-localized protein with roles confines to the cell nucleus.
FIGURE 3.
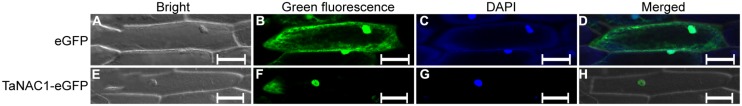
Subcellular localization of TaNAC1 protein. eGFP vector control and fusion gene TaNAC1–eGFP were transiently expressed in onion epidermal cells. The control eGFP protein (A–D) and the TaNAC1::eGFP fusion protein (E–H) were observed under a confocal microscope. (A,E) Bright field images; (B,F) Green fluorescent images (C,G). Onion cells were stained with the DNA-binding dye DAPI; (D,H) Merged images of bright, green fluorescence, and DAPI signal. eGFP, enhanced the green fluorescent protein; DAPI, 4′, 6-diamidino-2-phenylindole. Bars, 100 μm.
TaNAC1 HAS A TRANSCRIPTION ACTIVATION DOMAIN IN ITS C-TERMINAL
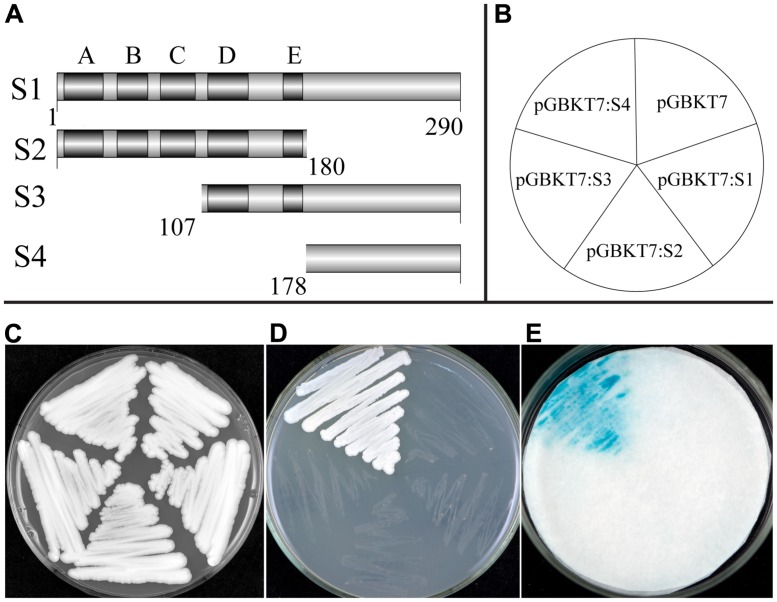
A transcriptional activation assay was investigated using the yeast one-hybrid system. Transformed yeast cells containing the fusion plasmid of full-length cDNA (S1) or the vector control pGBKT7 grew well on selective SD medium without tryptophane (Trp-). However, growth was largely inhibited when streaked on SD medium lacking tryptophane, histidine, and adenine (Trp-/His-/Ade-), indicating that TaNAC1 could not function as a transcriptional activator. We made another three fusion constructs containing truncated fragments, S2, the N-terminal containing the NAC DNA-binding domain (amino acid no. 1/180), S3 and S4, the C-terminals (amino acid no. 107/290, 178/290) containing the putative activation domain, with the pGBKT7 vector (Figure 4A), and tested transcriptional activation on Trp-/His-/Ade- medium plates and an α-galactosidase activity assay (Figure 4B–E). Only the yeast cells containing fusion construct of S4 could grew fast on selective medium (Trp-/His-/Ade-) showing a positive blue color indicative of strong α-galactosidase activity, and suggesting that this region has a strong transcriptional activation domain. On the contrary, constructs S1 (full length), S2 (N-terminal containing subdomains A–E), and S3 (C-terminal containing subdomains D and E) gave no transactivation activity. We thus confirm that the C-terminal region (amino acid No. 178/290) contained a transcriptional activation domain in TaNAC1.
FIGURE 4.
Transcription activation activity of TF TaNAC1. Full coding region (S1), N-terminal part (S2), or C-terminal parts (S3 and S4) of TaNAC1 were fused with the GAL4 DNA-binding domain in vector pGBKT7 (A). Plasmids containing the fusion genes and the empty vector control pGBKT7 were introduced into yeast cells of strain AH109. Yeast transformants of different vectors were plated as shown in the diagram (B) and incubated for 2 days at 28°C on an SD/Trp- plate (C) and SD/Trp-/His-/Ade- plate (D). α-galactosidase activities were examined by X-α-Gal staining and kept at 28°C for 8 h (E).
ORGAN-SPECIFIC EXPRESSION ANALYSIS OF TaNAC1
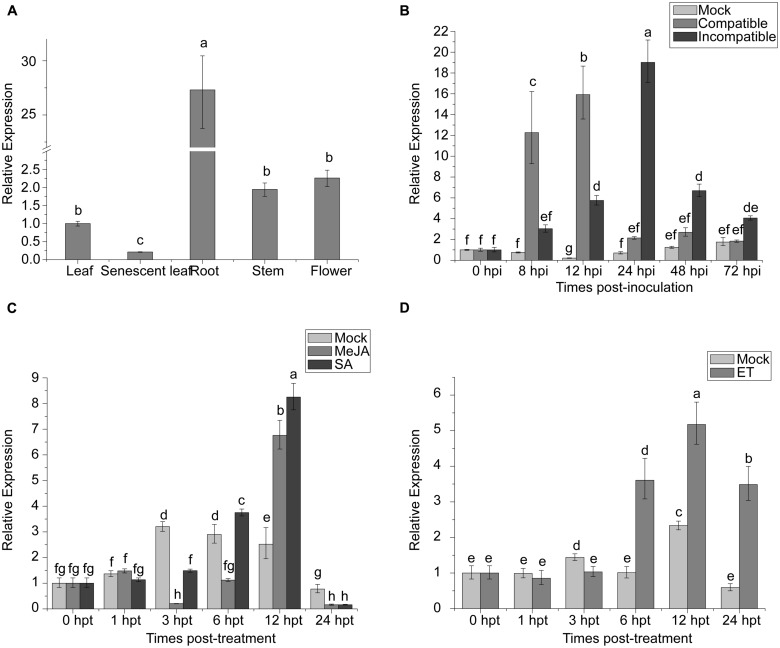
NAC proteins can be involved in regulation of plant development. The expression of TaNAC1 in different wheat tissues was analyzed by qRT-PCR. TaNAC1 was expressed in all wheat tissues tested, but most strongly in roots, at 27-fold that in young leaves, and least in senescent leaves (Figure 5A).
FIGURE 5.
Expression profiling of TaNAC1 in wheat. (A) Relative expression of TaNAC1 in young or senescent wheat leaves, stems, roots, and flowers; (B) Expression patterns of TaNAC1 in wheat seedlings after inoculation with Puccinia striiformis f. sp. tritici races CYR17 and G22, which were avirulent and virulent to Yr10∗6/Taichung29, respectively. The relative expression of TaNAC1 was calculated from its transcript level in infected seedling leaves compared to that in the mock control across sampling time-points; (C,D) Expression patterns of TaNAC1 in wheat seedling leaves treated with signaling chemicals MeJA, SA, and a mock control of 0.1% (v/v) ethanol, or with ET and its mock control. Results were from three technical replicates. Values are means ± SD (n = 3). Similar results were seen in all three biological replicates. Different letter types indicate significant differences by one-way ANOVA, taking P < 0.05 level according to Duncan’s multiple range tests of comparisons between different treatment times. MeJA, methyl jasmonate; SA, salicylic acid; ET, ethylene.
TaNAC1 RESPONDS TO BIOTIC STRESS AND HORMONE TREATMENTS
To elucidate whether TaNAC1 is involved in interaction between wheat and P. striiformis, Yr10∗6/Taichung 29 seedlings were inoculated with races CYR17 and G22. The peak expression level of TaNAC1 appeared earlier and increased faster in the compatible interaction (Figure 5B). During the early stages of infection (8–12 hpi) TaNAC1 expression level in the compatible interaction was significantly higher than in the incompatible interaction, but the inverse relationship occurred in the later stages (24–72 hpi). Expression patterns of TaNAC1 with exogenous applications of SA, MeJA, ET, and mock treatments were analyzed. For SA treatment compared with the mock control, transcripts of TaNAC1 initially decreased at 3 hpt, and then increased at 6 hpt to reach a peak level at 12 hpt, before declining sharply at 24 hpt (Figure 5C). After MeJA treatment, the expression of TaNAC1 was decreased at 3 and 6 hpt, then rapidly increased and peaked at 12 hpt (Figure 5C). In response to the ET application, expression was slightly induced at 6 hpt, peaked at 12 hpt, and then decreased to a lower level at 24 hpt, although still considerably higher than the mock control at this time point (Figure 5D).
TaNAC1-KNOCKDOWN PLANTS SHOW IMPROVED STRIPE RUST RESISTANCE
In order to determine the role of TaNAC1 in interaction between the wheat host and the stripe rust pathogen, we constructed a BSMV-TaNAC1 (barley stripe mosaic virus with a specific fragment of TaNAC1) recombinant vector to silence TaNAC1 expression. The BSMV-inoculated plants displayed mild chlorotic mosaic symptoms at 14 days post-inoculation (dpi), but no obvious defects were observed during further leaf growth. As shown in Figure 6A, about 14 days after virus inoculation, typical photobleaching occurred on the leaves of wheat pre-inoculated with BSMV-TaPDS (barley stripe mosaic virus with a specific fragment of the wheat phytoene desaturase gene PDS) indicating that a genetic interference system applicable to this study was feasible (Figures 6A,D). In addition, the virulent P. striiformis race G22 successfully infected wheat seedlings previously inoculated with BSMV. Fifteen days after pre-inoculation with the virus, third leaves of these seedlings were inoculated with fresh urediniospores of race G22. At 11 dpi with P. striiformis pustules erupted on the leaves of the Mock and viral control plants, but no uredinia appeared on the TaNAC1-knockdown seedling leaves (Figure 6B). At 15 dpi uredinia were visible (Figure 6C). However, there were less uredinial development on TaNAC1-knockdown seedling leaves than on the Mock and viral controls (Figure 6F). The lengths of P. striiformis uredinia on TaNAC1-knockdown seedling leaves were significantly shorter than those on the Mock and viral controls at 15 dpi, but there was no difference in uredinial width ((Figures 6C,E).
FIGURE 6.
Functional characterization of TaNAC1 in response to stripe rust infection using the BSMV-VIGS system. (A) Phenotypic changes of third seedling leaves of wheat cultivar Funo pre-inoculated with water (Mock), empty BSMV vectors and positive control combination vectors (BSMV-TaPDS) at 14 days post-virus treatment. (B, C) Phenotypes of third leaves inoculated with P. striiformis race G22 were observed at 11 and 15 dpi; the second leaves of these seedlings were pre-inoculated with water (Mock), empty BSMV vectors and BSMV-TaNAC1. (D) Silence efficiency of TaNAC1 in TaNAC1-knockdown leaves determined by qRT-PCR; third leaves were sampled at 0 and 48 hpi. (E) Statistics of 60 uredinia (lengths and widths) on third leaves inoculated at 15 dpi. (F) Statistics of uredinia on the 2 cm leaf segments of the third leaves inoculated at 15 dpi. Representative experiment (n = 3), each including 60 uredinia and six segments from empty BSMV vectors and BSMV-TaNAC1. Error bars indicate SD, asterisks indicate significant differences between BSMV and BSMV-TaNAC1 samples determined by Student’s t-tests (P = 0.01) and one-way ANOVA, taking P < 0.05 level according to Duncan’s multiple range tests.
TaNAC1 REGULATES ROOT DEVELOPMENT IN TRANSGENIC Arabidopsis
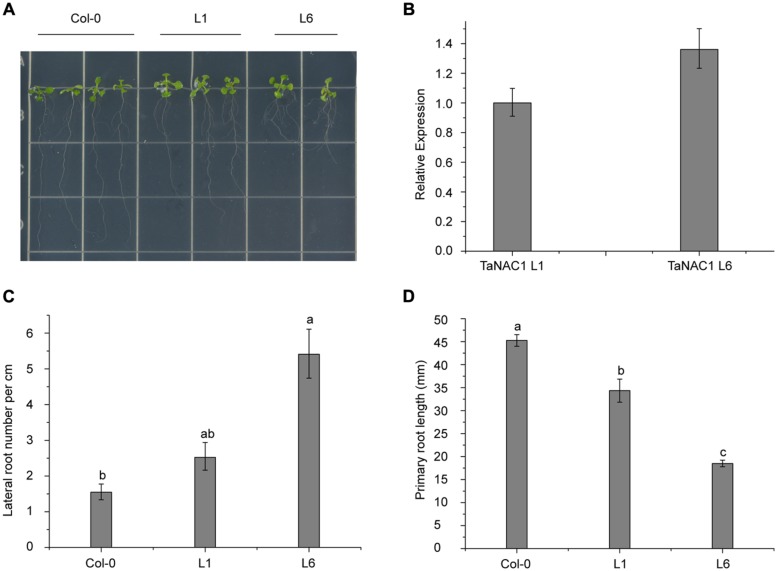
To analyze its biological functions, TaNAC1 under the control of the CaMV 35S promoter was transformed into Arabidopsis Col-0 wild-type plants using Agrobacterium tumefaciens strain GV3101. Six positive lines were obtained and two homozygous T3 generation transgenic lines (L1 and L6) with different TaNAC1 expression levels were selected for further biological function analyses. Ten-day-old Col-0 and transgenic seedlings grown vertically on half-strength MS (0.5 × MS) plates were used for observing lateral and primary root development. L6 plants had nearly twice as many lateral roots as L1; and both transgenic lines developed more lateral roots than the wild-type plants (Figures 7A,C). However, the primary root lengths of L1 (3.43 cm) and L6 (1.85 cm) plants were 1.10 and 2.65 cm shorter, respectively, than those of wild-type plants (4.52 cm; Figure 7D). TaNAC1 expression level in L6 plants was almost 1.35-fold higher than in L1 plants (Figure 7B). Therefore, when TaNAC1 was overexpressed in Arabidopsis, lateral root development in transgenic lines was promoted, but primary root elongation was suppressed. The phenotypes of transgenic Arabidopsis root development were closely related to TaNAC1 expression level.
FIGURE 7.
TaNAC1 regulation of root development on transgenic Arabidopsis plants. (A) TaNAC1 overexpression promoted 10-day-old transgenic Arabidopsis plants of L1 and L6 to produce more lateral roots than the wild type; (B) Relative TaNAC1 expression levels of L1 and L6 analyzed by qRT-PCR; (C) Comparison of lateral root density (number of emerged lateral roots per cm of main root) in Col-0 and transgenic lines of L1 and L6; (D) Primary root lengths of 10-day-old Col-0 and TaNAC1-overexpressing seedlings of L1 and L6. Data show average and SD of 25–30 seedlings and are representative of at least three independent experiments. Different letter types indicate significant differences by one-way ANOVA, taking P < 0.05 level according to Duncan’s multiple range tests of comparisons between Col-0 and transgenic lines L1 and L6.
TaNAC1 OVEREXPRESSION ENHANCES SUSCEPTIBILITY OF Arabidopsis TO VIRULENT STRAIN Pst DC3000
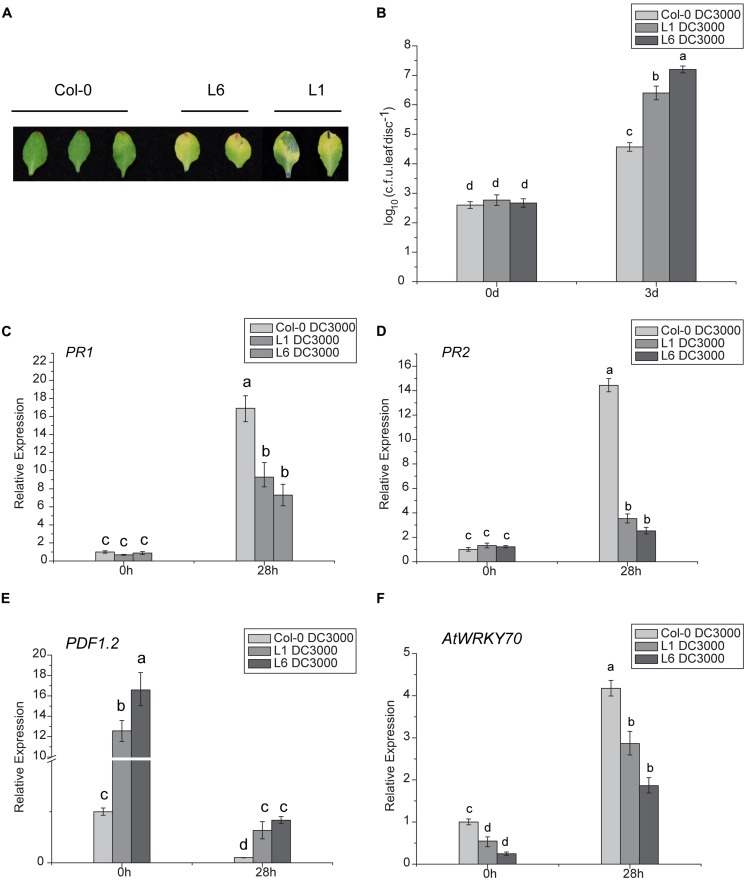
The DC3000 strain of Pseudomonas syringae pv. tomato (Pst DC3000) was used as a model virulent pathogen for assessing disease susceptibility in transgenic Arabidopsis lines. Significant symptom differences developed after infiltrating a low-dose of inoculum (OD600nm = 0.002) of Pst DC3000 into leaves of TaNAC1-overexpressing transgenic plants L1 and L6, and Col-0. At 48 h post bacterial infiltration larger light yellow lesions were present on infected leaves of L1 and L6 plants compared to slight chlorosis on Col-0, and the disease-associated chlorotic lesions on the leaves of the two transgenics were significantly larger than the control at 3 days post infiltration (Figure 8A). Furthermore, transgenic plants supported higher bacterial growth rates than the wild-type plants (Figure 8B).
FIGURE 8.
Analyses of susceptibility of transgenic Arabidopsis plants to Pst DC3000 and expression patterns of plant defense-related marker genes. (A) Lesion development on infected leaves of L1, L6, and Col-0 plants 3 days after infiltration with Pst DC3000; (B) Bacterial populations in infected leaves of wild-type and transgenic lines L1 and L6. Leaves of 4-week-old plants were infiltrated with bacterial suspensions. Nine leaf disks (8 mM diameter) from three different plants were collected 3 days after inoculation. Defense-related marker genes PR1 (C), PR2 (D), PDF1.2 (E), and AtWRKY70 (F) expressions in TaNAC1-overexpressing lines L1 and L6 and Col-0 plants at 28 hpi with Pst strain DC3000 and in the corresponding controls were analyzed by qRT-PCR. Error bars indicate SD of three technical replicates. Similar results were seen in three biological replicates. Different letter types indicate significant differences by one-way ANOVA, taking P < 0.05 level according to Duncan’s multiple range tests.
TaNAC1 AFFECTS THE EXPRESSION OF DEFENSE-RELATED MARKER GENES
To determine whether enhanced pathogen susceptibility of TaNAC1 transgenic plants was linked to changes in defense-related gene expression and to gain further insight into possible regulation pathways, the expression patterns of several well-known defense-related marker genes in TaNAC1-overexpressing and Col-0 plants infiltrated with Pst DC3000 were compared. There was no significant difference in basal expression levels of SA-responsive genes PR1 and PR2 between L1, L6 and Col-0 plants. However, both PR1 and PR2 in TaNAC1 transgenic plants and Col-0 greatly increased after infiltration with Pst DC3000, and Col-0 plants had a higher expression level than L1 and L6 plants (Figures 8C,D). ET- and JA-regulated defense-related marker gene PDF1.2 significantly increased in transgenic plants and the basal transcript levels in L1 and L6 plants were nearly 13-fold and 16-fold higher than that in Col-0 plants (Figure 8E). PDF1.2 expression was suppressed at 28 hpi, but was still significantly higher in L1 and L6 plants than in the wild-type control.
The AtWRKY70 factor functions as a node between the JA- and SA-signaling pathways; expression of AtWRKY70 is repressed by JA, but activated by SA, and AtWRKY70 acts as a negative regulator of expression of JA-responsive downstream genes and as a positive regulator of SA-induced genes such as PR1 (Li et al., 2006). AtWRKY70 expression levels in the present study were lower in TaNAC1 transgenic lines than Col-0 plants before and after Pst DC3000 infection, although its transcription could be induced by Pst DC3000 infection. Its expression was apparently suppressed in L1 and L6 plants compared to Col-0 plants (Figure 8F). As a consequence, overexpression of TaNAC1 resulted in significantly higher levels of expression of the ET- and JA-regulated marker gene PDF1.2.
TaNAC1 INVOLVES ET- AND JA-SIGNALING PATHWAYS
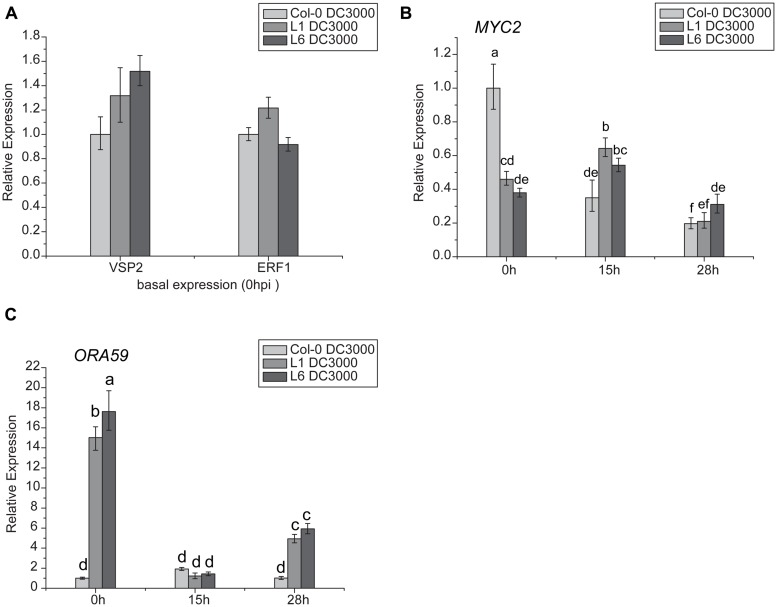
Jasmonates signaling occurs via two different branches, and is regulated by MYC2 or ERF1/ORA59. MYC2 is also involved in the ABA- and JA-signaling pathways. In this study, VSP2 was used as a marker gene for the MYC2 pathway, and PDF1.2 for the ERF1/ORA59 pathway. Results indicated that there was no significant difference in VSP2 and ERF1 transcription levels between L1, L6, and Col-0 plants (Figure 9A), but basal transcript levels of ORA59 were significantly higher in L1 and L6 plants than Col-0 at 0 h (Figure 9C). Moreover, MYC2 expression was highly suppressed in TaNAC1 transgenic plants compared with Col-0 plants before infection (Figure 9B).
FIGURE 9.
Jasmonates (JA)-signaling pathways genes showed different expression patterns between TaNAC1 transgenic and Col-0 plants. (A) VSP2 and ERF1 expressions in uninoculated Col-0 and TaNAC1-overexpressing lines L1 and L6; (B) MYC2 and (C) ORA59 expressions in Col-0 and TaNAC1-overexpressing L1 and L6 plants inoculated with Pst strain DC3000 (OD600nm = 0.001) at 0, 15, and 28 hpi. Error bars indicate SD of three technical replicates. Similar results were seen in three biological replicates. Different letter types indicate significant differences by one-way ANOVA, taking P < 0.05 level according to Duncan’s multiple range tests.
Because ORA59 and MYC2 expression levels were significantly changed in transgenic plants, the expression patterns of these two genes were further analyzed following infection with Pst DC3000. The expression level of ORA59 in Col-0 plants, remained similar after infiltration whereas in L1 and L6 plants it was significantly reduced at 15 hpi and then partially recovered, but was still much higher than in Col-0 at 28 hpi (Figure 9C). The MYC2 transcript level in Col-0 decreased significantly during incubation after infiltration, but in L1 and L6 plants it was slightly increased at 15 hpi and then decreased at 28 hpi (Figure 9B), suggesting that JA- and ET-signaling pathways were affected by TaNAC1 overexpression.
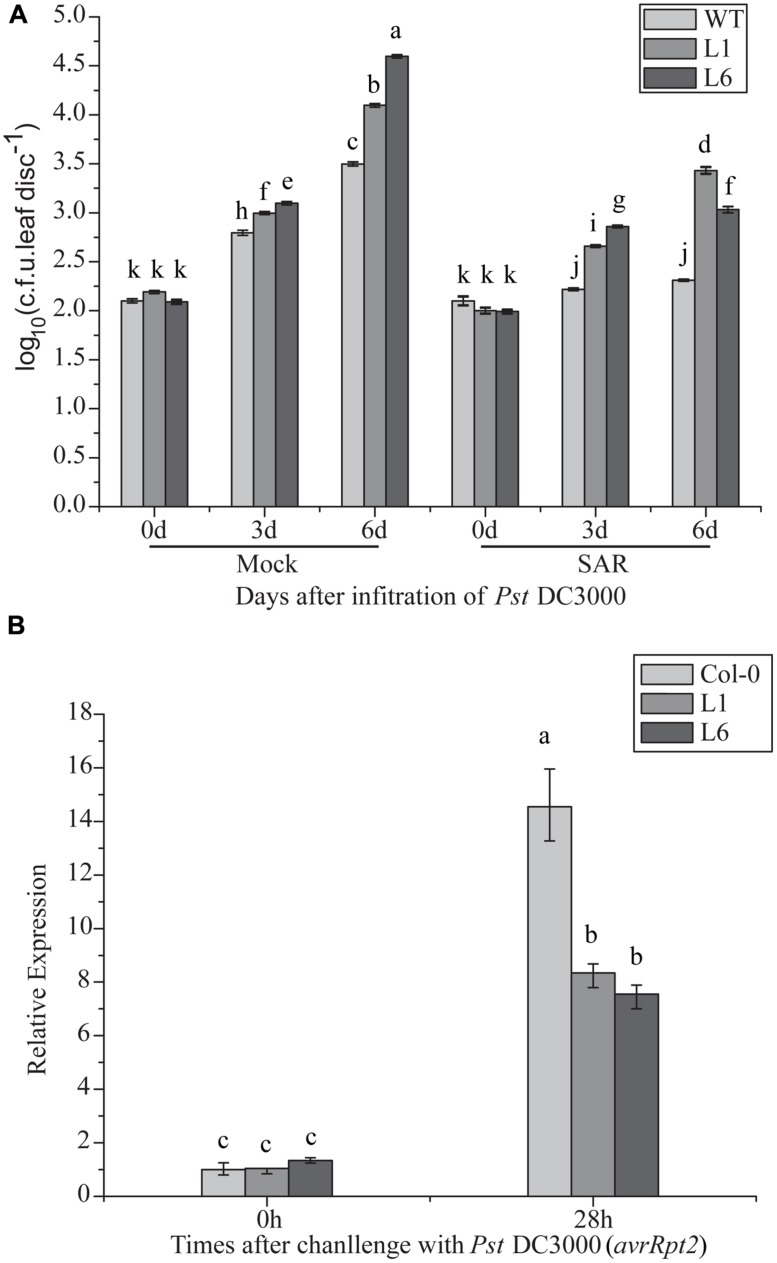
TaNAC1 OVEREXPRESSION ATTENUATED SYSTEMIC-ACQUIRED RESISTANCE
Compromised disease resistance in TaNAC1-overexpressing plants may affect SAR establishment. To test this hypothesis, changes in the bacterial population with or without SAR treatment were analyzed. Transgenic TaNAC1 plants supported larger DC 3000 bacterial populations than the control (Figure 10A). PR1 expression level was significantly increased in the transgenic lines after SAR treatment, indicating that TaNAC1 overexpression did not block SAR establishment, but the TaNAC1 transgenic plants had a lower PR1 transcription level than Col-0 when pre-inoculated with an avirulent strain of Pst DC3000 (avrRpt2; Figure 10B). Therefore, SAR treatment induced disease resistance in both TaNAC1-overexpressing lines and Col-0 plants compared with the corresponding mock controls. TaNAC1 overexpression in Arabidopsis did not block SAR but compromised its induction of resistance.
FIGURE 10.
Induction of systemic acquired resistance (SAR) in TaNAC1 overexpressing plants. (A) Three lower leaves on each plant were inoculated with Pst strain DC3000 (avrRpt2; OD600nm = 0.1; SAR) and plants infiltrated with 10 mM MgCl2 served as mock controls. Three days later, two upper uninfected leaves were pressure-infiltrated with virulent Pst strain DC3000 (OD600nm = 0.0001). Plants inoculated only with Pst DC3000 served as controls for SAR analysis. The in planta bacterial titers were determined immediately and 3, 6 days after challenge inoculation. Data represent the mean of 4–10 independent samples with SD. Different letter types indicate significant differences by one-way ANOVA, taking P < 0.05 level according to Duncan’s multiple range test of comparisons between populations from different plants or different times. (B) PR1 expression in uninoculated Col-0 and TaNAC1-overexpressing lines L1 and L6. Three lower leaves on each plant were inoculated with Pst DC3000 (avrRpt2; OD600nm = 0.1; SAR). 28 h later, total RNA was extracted from upper non-infected leaves and analyzed for gene expression using qRT-PCR. Error bars indicate SD of three technical replicates. Similar results were observed in three biological replicates. Different letter types indicate significant differences by one-way ANOVA, taking P < 0.05 level according to Duncan’s multiple range tests.
DISCUSSION
NAC-type transcription factors form one of the largest plant-specific TF families, regulating plant development and responses to environmental stimuli (Nakashima et al., 2012; Puranik et al., 2012). Several NAC-TFs have been characterized in wheat (Xie et al., 1999; Uauy et al., 2006; Xue et al., 2006, 2011; Xia et al., 2010a,b; Mao et al., 2012, 2014; Tang et al., 2012; Avni et al., 2014; Feng et al., 2014). For example, TaNAC21/22, a member of the NAC1 subgroup, takes part in regulation of resistance to stripe rust (Feng et al., 2014). The first characterized member of NAC1 subgroup, NAC1, was identified in Arabidopsis and primarily regulates plant root development (Xie et al., 2000).
THE C-TERMINAL OF TaNAC1 HAS TRANSCRIPTIONAL ACTIVATION ABILITY
In this study, a novel member to the NAC-TF family was isolated from bread wheat. Basedonn a neighbor-joining cluster tree of known NAC proteins from Arabidopsis, wheat, rice, barley and other species, this NAC factor was classified into the NAC1 subgroup together with NAC1 and At3g12977.1 from Arabidopsis and TaNAC21/22 and TaNAC7 from wheat.
We performed transactivation analyses of TaNAC1 in yeast, and the results showed that the full length and N-terminal region of TaNAC1 lacked transcriptional activation ability. But the C-terminal possessed strong transactivation capacity. The N terminus of TaNAC1 was predicted to contain a NARD domain (Figure 1), which was recently reported to be a conserved and active repression domain in NAC proteins (Hao et al., 2010). GmNAC20 and TaNAC7, which both belong to the NAC1 subgroup (Figure 2), did not have transcriptional activation ability (Tang et al., 2012). So, the NARD domain may remarkably attenuate the transcriptional activation of TaNAC1 factor.
TaNAC1 IS INVOLVED IN RESPONSE TO FUNGAL AND BACTERIAL PATHOGENS
Phytohormones such as SA, MeJA, and ET are believed to play key roles in signaling pathways involved in plant responses biotic and abiotic stresses. Each of these molecules, especially SA, transiently induced TaNAC1 expression (Figure 5). Until now, more than 10 wheat NAC-TFs have been reported, but only TaNAC4 and TaNAC8, belonging to the SNAC and OsNAC8 subgroups, respectively, were reported to be induced by MeJA, ET, and pathogen infection. However, SA did not affect the expression of those two genes (Xia et al., 2010a,b).
NAC-type transcription factors are proved to be involved in responses to pathogen attack. For example, ATAF1-overexpressing plants increased susceptibility to Pst DC3000 and the necrotrophic pathogen Botrytis cinerea. HvNAC6 (Chen et al., 2013), ANAC019, ANAC055, and ANAC072 (Zheng et al., 2012), ATAF2 (Delessert et al., 2005), OsNAC4 (Kaneda et al., 2009), and membrane-tethered NAC gene NTL6 (Seo et al., 2010) were also involved in plant–pathogen interactions. Generally, three phases of plant defense are defined, the first pre-invasive defense barrier of the plant (Phase I), the second barrier of early post-invasive defense (Phase II), and transcriptomic and metabolomic re-programming, resulting from a hypersensitive response (Phase III). Phase III is associated with production of intra- and intercellular signals, including defense hormones (Ton et al., 2009). In the present research, TaNAC1 expression was closely related to the wheat defense response to P. striiformis, and was significantly upregulated at an early stage of infection (Phases I and II) in the compatible interaction. The expression levels of TaNAC1 were partially synchronic with pathogenesis phases described previously (Figure 5B). Furthermore, when TaNAC1 was knocked down, uredinia produced by a virulent P. striiformis race appeared later and were smaller on knockdown plants. Therefore, suppression of TaNAC1 transcription apparently delayed the infection process and partially increased resistance to the virulent P. striiformis race.
TaNAC1 overexpression in Arabidopsis enhanced susceptibility of transgenic plants to the virulent Pst strain DC3000, promoting population growth of the bacterial pathogen and accelerating the pathogenesis process. But the SAR of TaNAC1 transgenic plants remained almost intact. These results strongly confirmed that TaNAC1 functioned as a negative regulator in the basal disease resistance pathway to Pst strain DC3000. In Arabidopsis, several genes were characterized as negative disease resistance regulators to pathogenic bacteria, including NAC TFs (Kim et al., 2012), ERF1 (Berrocal-Lobo et al., 2002), WRKY factors (Journot-Catalino et al., 2006; Kim et al., 2006, 2008; Zheng et al., 2007), kinase and kinase phosphatase MKP (Anderson et al., 2011), CBRLK1 (Kim et al., 2009), and MPK4 (Colcombet et al., 2013).
TaNAC1 MAY SUPPRESS RESISTANCE-RELATED GENE EXPRESSION IN SA SIGNALING
Cross-talk between JA, ET, and SA signaling is thought to operate as a mechanism to fine-tune induced defenses activated in response to multiple biotic attackers. SA appears primarily for signaling in plant resistance to biotrophic and hemibiotrophic pathogens such as P. syringae. JA plays a role in plant defense against insects and necrotrophic pathogens and may suppress some defense responses through antagonism with SA. Arabidopsis plants defective in SA biosynthesis or its signaling were more susceptible to hemibiotrophic bacterial pathogens than wild-type plants, whereas plants with enhanced JA biosynthesis or signaling also support bacterial pathogen population growth. SA levels are usually tightly regulated in dicotyledonous plants. When infiltrated with the virulent Pst strain DC3000, SA-regulated PR gene expression level decreased in TaNAC1-overexpressing plants. Furthermore, infection in the lower leaves of TaNAC1 transgenic plants by an avirulent P. syringae strain induced PR1 gene expression in upper uninoculated leaves and increased Pst DC3000 population growth after inoculation of transgenic plants relative to Col-0 plants. These results strongly proved that TaNAC1 is a negative regulator of P. syringae-induced PR gene expression in Arabidopsis.
ORA59 acts as an integrator of JA- and ET-signaling pathways and is the key regulator of JA- and ET-responsive gene PDF1.2 (Pre et al., 2008). A recent study showed that the SA pathway inhibits the SCF(COI1)-JAZ complex of JA downstream signaling via a negative effect on the transcriptional activator ORA59 but without influence on ERF1 expression (Van der Does et al., 2013). When higher levels of ORA59 transcripts accumulated in plants infiltrated with Pst DC3000, additional free SA was needed to reduce ORA59, thus priming the SA-signaling resistance pathway. In the present study, ET- and JA-regulated defense-related marker genes PDF1.2 and ORA59 were expressed at significantly higher levels in transgenic plants than in wild-type, and there was no significant difference in ERF1 transcription levels. In addition, AtWRKY70 functions as a connection node between JA- and SA-signaling pathways, and is activated by SA but suppressed by JA. This gene was greatly suppressed in the TaNAC1 overexpression lines, and hence may lead to lower expression levels of SA-regulated genes in TaNAC1-overexpressing plants.
In Arabidopsis, two major branches of the JA signaling pathway are recognized. One is the MYC branch with marker gene VSP2, and another is the ERF branch requires both JA and ET signaling with the marker gene PDF1.2. In general, the ERF branch is associated with enhanced resistance to necrotrophic pathogens (Berrocal-Lobo et al., 2002; Lorenzo et al., 2003), whereas the MYC branch is associated with the wound response and defense against insect herbivores (Lorenzo et al., 2004; Kazan and Manners, 2008). In wheat under treatment with MeJA, TaNAC1 also showed a rapid increase and peaked at 12 hpt. However, the roles of JA in biotrophic and necrotrophic resistances are unclear in many monocot pathosystems. There is no dichotomy between the effectiveness of the JA pathway and the lifestyle of the invading pathogen in rice (De Vleesschauwer et al., 2014), and some reports implicate roles of JA in resistance against (hemi)biotrophic pathogens Xanthomonas oryzae and Magnaporthe oryzae (Mei et al., 2006; Yamada et al., 2012; Taniguchi et al., 2014), necrotrophic rice pathogens (Taheri and Tarighi, 2010; Peng et al., 2012) and insect herbivores (Zhou et al., 2009). Thus TaNAC1 may play a positive role in resistance to the necrotrophic pathogens both in Arabidopsis and wheat.
Therefore enhanced susceptibility of TaNAC1-overexpressing plants to Pst DC3000 may result from suppression of resistance-related gene expression in SA signaling by regulation of the JA-signaling pathway.
TaNAC1 IS AN IMPORTANT REGULATOR OF ROOT DEVELOPMENT
Several members of the NAC1 subgroup are involved in regulation of plant root development. Arabidopsis NAC1 functioned in auxin-mediated signaling and increased the number of lateral roots in transgenic overexpressing plants (Xie et al., 2000; Hao et al., 2011). GmNAC11 and GmNAC20 were reported to be the members of the NAC1 subgroup and increased lateral root number (Hao et al., 2011). In this study, however, GmNAC11 and GmNAC20 clustered with the NAC3 and ATAF1 subgroups, respectively. For MtNAC1, another factor of the NAC1 subgroup, no changes in lateral root development or nodulation occurred on transgenic plants (D’haeseleer et al., 2011). As a novel member of NAC1 subgroup, TaNAC1 has high similarity to its homolog NAC1 of Arabidopsis, and its organ-specific expression pattern highly resembles that of NAC1 (Xie et al., 2000) in being more abundantly expressed in roots than in other organs. Moreover, TaNAC1-overexpressing Arabidopsis plants developed more lateral roots but primary root elongation was considerably suppressed.
Lateral root development in higher plants is influenced by a wide range of environmental cues, but auxin plays a dominant role (Fukaki et al., 2007; Petricka et al., 2012). JA is also involved in this process by activating auxin biosynthesis and has effects on auxin transport into the root basal meristem (Sun et al., 2009). Application of exogenous JA inhibited plant root growth (Staswick et al., 1992). Arabidopsis mutant JA-induced defective lateral root1 (jdl1/asa1-1) repressed lateral root formation, while anthranilate synthase α1 (ASA1) was needed for JA-induced auxin biosynthesis. Combined with positive regulation roles in the JA- and ET-signaling pathways TaNAC1 functions in regulating root development may depend on the JA-signaling pathway.
In brief, TaNAC1 encodes a new NAC-type TF. Based on the results of the present study, we concluded that TaNAC1 might negatively regulate wheat defense against P. striiformis and Arabidopsis response to Pseudomonas syringae, as well as being involved in regulation of plant root development.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Robert A. McIntosh from the University of Sydney for critically reading and commenting on the manuscript, Dr. DaWei Li from China Agricultural University for providing BSMV vectors. We are grateful to financial support of National Key Basic Research Program of China (No. 2013CB127700), National Natural Science Foundation of China (No. 31261140370 and No. 31171838) and China Postdoctoral Science Foundation (No. 2012M520475).
Footnotes
REFERENCES
- Anderson J. C., Bartels S., Gonzalez Besteiro M. A., Shahollari B., Ulm R., Peck S. C. (2011). Arabidopsis MAP Kinase Phosphatase 1 (AtMKP1) negatively regulates MPK6-mediated PAMP responses and resistance against bacteria. Plant J. 67 258–268 10.1111/j.1365-313X.2011.04588.x [DOI] [PubMed] [Google Scholar]
- Avni R., Zhao R., Pearce S., Jun Y., Uauy C., Tabbita F., et al. (2014). Functional characterization of GPC-1 genes in hexaploid wheat. Planta 239 313–324 10.1007/s00425-013-1977-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N., Jolivet S., Voisin R., Pelletier G. (2003). The endosperm and the embryo of Arabidopsis thaliana are independently transformed through infiltration by Agrobacterium tumefaciens. Transgenic Res. 12 509–517 10.1023/A:1024272023966 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M., Molina A., Solano R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29 23–32 10.1046/j.1365-313x.2002.01191.x [DOI] [PubMed] [Google Scholar]
- Cao H., Glazebrook J., Clarke J. D., Volko S., Dong X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88 57–63 10.1016/S0092-8674(00)81858-9 [DOI] [PubMed] [Google Scholar]
- Chen Y. J., Perera V., Christiansen M. W., Holme I. B., Gregersen P. L., Grant M. R., et al. (2013). The barley HvNAC6 transcription factor affects ABA accumulation and promotes basal resistance against powdery mildew. Plant Mol. Biol. 83 577–590 10.1007/s11103-013-0109-1 [DOI] [PubMed] [Google Scholar]
- Christiansen M. W., Holm P. B., Gregersen P. L. (2011). Characterization of barley (Hordeum vulgare L.) NAC transcription factors suggests conserved functions compared to both monocots and dicots. BMC Res. Notes 4:302 10.1186/1756-0500-4-302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J., Berriri S., Hirt H. (2013). Constitutively active MPK4 helps to clarify its role in plant immunity. Plant Signal. Behav. 8:e22991 10.4161/psb.22991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M. K., Scheible W. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139 5–17 10.1104/pp.105.063743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delessert C., Kazan K., Wilson I. W., Van Der Straeten D., Manners J., Dennis E. S., et al. (2005). The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J. 43 745–757 10.1111/j.1365-313X.2005.02488.x [DOI] [PubMed] [Google Scholar]
- Derksen H., Rampitsch C., Daayf F. (2013). Signaling cross-talk in plant disease resistance. Plant Sci. 207 79–87 10.1016/j.plantsci.2013.03.004 [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer D., Xu J., Hofte M. (2014). Making sense of hormone-mediated defense networking: from rice to Arabidopsis. Front. Plant Sci. 5:611 10.3389/fpls.2014.00611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’haeseleer K., Den Herder G., Laffont C., Plet J., Mortier V., Lelandais-Briere C., et al. (2011). Transcriptional and post-transcriptional regulation of a NAC1 transcription factor in Medicago truncatula roots. New Phytol. 191 647–661 10.1111/j.1469-8137.2011.03719.x [DOI] [PubMed] [Google Scholar]
- Fang Y., You J., Xie K., Xie W., Xiong L. (2008). Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genomics 280 547–563 10.1007/s00438-008-0386-6 [DOI] [PubMed] [Google Scholar]
- Feng H., Duan X., Zhang Q., Li X., Wang B., Huang L., et al. (2014). The target gene of tae-miR164, a novel NAC transcription factor from the NAM subfamily, negatively regulates resistance of wheat to stripe rust. Mol. Plant Pathol. 15 284–296 10.1111/mpp.12089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H., Okushima Y., Tasaka M. (2007). Auxin-mediated lateral root formation in higher plants. Int. Rev. Cytol. 256 111–137 10.1016/S0074-7696(07)56004-3 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43 205–227 10.1146/annurev.phyto.43.040204.135923 [DOI] [PubMed] [Google Scholar]
- Hao Y. J., Song Q. X., Chen H. W., Zou H. F., Wei W., Kang X. S., et al. (2010). Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta 232 1033–1043 10.1007/s00425-010-1238-2 [DOI] [PubMed] [Google Scholar]
- Hao Y. J., Wei W., Song Q. X., Chen H. W., Zhang Y. Q., Wang F., et al. (2011). Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 68 302–313 10.1111/j.1365-313X.2011.04687.x [DOI] [PubMed] [Google Scholar]
- Howe G. A., Jander G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59 41–66 10.1146/annurev.arplant.59.032607.092825 [DOI] [PubMed] [Google Scholar]
- Hu R., Qi G., Kong Y., Kong D., Gao Q., Zhou G. (2010). Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 10:145 10.1186/1471-2229-10-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamar C., Loffet F., Frettinger P., Ramsay L., Fauconnier M. L., Du Jardin P. (2010). NAM-1 gene polymorphism and grain protein content in Hordeum. J. Plant Physiol. 167 497–501 10.1016/j.jplph.2009.10.014 [DOI] [PubMed] [Google Scholar]
- Jones J. D., Dangl J. L. (2006). The plant immune system. Nature 444 323–329 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Journot-Catalino N., Somssich I. E., Roby D., Kroj T. (2006). The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18 3289–3302 10.1105/tpc.106.044149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda T., Taga Y., Takai R., Iwano M., Matsui H., Takayama S., et al. (2009). The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death. EMBO J. 28 926–936 10.1038/emboj.2009.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J. M. (2008). Jasmonate signaling: toward an integrated view. Plant Physiol. 146 1459–1468 10.1104/pp.107.115717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. S., Jung M. S., Lee S. M., Kim K. E., Byun H., Choi M. S., et al. (2009). An S-locus receptor-like kinase plays a role as a negative regulator in plant defense responses. Biochem. Biophys. Res. Commun. 381 424–428 10.1016/j.bbrc.2009.02.050 [DOI] [PubMed] [Google Scholar]
- Kim H. S., Park H. C., Kim K. E., Jung M. S., Han H. J., Kim S. H., et al. (2012). A NAC transcription factor and SNI1 cooperatively suppress basal pathogen resistance in Arabidopsis thaliana. Nucleic Acids Res. 40 9182–9192 10.1093/nar/gks683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. C., Fan B., Chen Z. (2006). Pathogen-induced Arabidopsis WRKY7 is a transcriptional repressor and enhances plant susceptibility to Pseudomonas syringae. Plant Physiol. 142 1180–1192 10.1104/pp.106.082487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. C., Lai Z., Fan B., Chen Z. (2008). Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20 2357–2371 10.1105/tpc.107.055566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., Mcgettigan P. A., Mcwilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23 2947–2948 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Le D. T., Nishiyama R., Watanabe Y., Mochida K., Yamaguchi-Shinozaki K., Shinozaki K., et al. (2011). Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 18 263–276 10.1093/dnares/dsr015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Brader G., Kariola T., Palva E. T. (2006). WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 46 477–491 10.1111/j.1365-313X.2006.02712.x [DOI] [PubMed] [Google Scholar]
- Lorenzo O., Chico J. M., Sanchez-Serrano J. J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16 1938–1950 10.1105/tpc.022319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Piqueras R., Sanchez-Serrano J. J., Solano R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15 165–178 10.1105/tpc.007468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Chen S., Li A., Zhai C., Jing R. (2014). Novel NAC transcription factor TaNAC67 confers enhanced multi-abiotic stress tolerances in Arabidopsis. PLoS ONE 9:e84359 10.1371/journal.pone.0084359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Zhang H., Qian X., Li A., Zhao G., Jing R. (2012). TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 63 2933–2946 10.1093/jxb/err462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei C., Qi M., Sheng G., Yang Y. (2006). Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol. Plant Microbe Interact. 19 1127–1137 10.1094/MPMI-19-1127 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Takasaki H., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819 97–103 10.1016/j.bbagrm.2011.10.005 [DOI] [PubMed] [Google Scholar]
- Nuruzzaman M., Manimekalai R., Sharoni A. M., Satoh K., Kondoh H., Ooka H., et al. (2010). Genome-wide analysis of NAC transcription factor family in rice. Gene 465 30–44 10.1016/j.gene.2010.06.008 [DOI] [PubMed] [Google Scholar]
- Olsen A. N., Ernst H. A., Leggio L. L., Skriver K. (2005). NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10 79–87 10.1016/j.tplants.2004.12.010 [DOI] [PubMed] [Google Scholar]
- Ooka H., Satoh K., Doi K., Nagata T., Otomo Y., Murakami K., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10 239–247 10.1093/dnares/10.6.239 [DOI] [PubMed] [Google Scholar]
- Paolacci A. R., Tanzarella O. A., Porceddu E., Ciaffi M. (2009). Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol. Biol. 10:11 10.1186/1471-2199-10-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., Hu Y., Tang X., Zhou P., Deng X., Wang H., et al. (2012). Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta 236 1485–1498 10.1007/s00425-012-1698-7 [DOI] [PubMed] [Google Scholar]
- Petricka J. J., Winter C. M., Benfey P. N. (2012). Control of Arabidopsis root development. Annu. Rev. Plant Biol. 63 563–590 10.1146/annurev-arplant-042811-105501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C. M., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S. C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28 489–521 10.1146/annurev-cellbio-092910-154055 [DOI] [PubMed] [Google Scholar]
- Pre M., Atallah M., Champion A., De Vos M., Pieterse C. M., Memelink J. (2008). The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147 1347–1357 10.1104/pp.108.117523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik S., Sahu P. P., Mandal S. N., Venkata Suresh B., Parida S. K., Prasad M. (2013). Comprehensive genome-wide survey, genomic constitution and expression profiling of the NAC transcription factor family in foxtail millet (Setaria italica L.). PLoS ONE 8:e64594 10.1371/journal.pone.0064594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik S., Sahu P. P., Srivastava P. S., Prasad M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17 369–381 10.1016/j.tplants.2012.02.004 [DOI] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3 1101–1108 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Seo P. J., Kim M. J., Park J. Y., Kim S. Y., Jeon J., Lee Y. H., et al. (2010). Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 61 661–671 10.1111/j.1365-313X.2009.04091.x [DOI] [PubMed] [Google Scholar]
- Shang H., Li W., Zou C., Yuan Y. (2013). Analyses of the NAC transcription factor gene family in Gossypium raimondii Ulbr.: chromosomal location, structure, phylogeny, and expression patterns. J. Integr. Plant Biol. 55 663–676 10.1111/jipb.12085 [DOI] [PubMed] [Google Scholar]
- Singh K., Foley R. C., Onate-Sanchez L. (2002). Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 5 430–436 10.1016/S1369-5266(02)00289-3 [DOI] [PubMed] [Google Scholar]
- Singh A. K., Sharma V., Pal A. K., Acharya V., Ahuja P. S. (2013). Genome-wide organization and expression profiling of the NAC transcription factor family in potato (Solanum tuberosum L.). DNA Res. 20 403–423 10.1093/dnares/dst019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souer E., Van Houwelingen A., Kloos D., Mol J., Koes R. (1996). The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85 159–170 10.1016/S0092-8674(00)81093-4 [DOI] [PubMed] [Google Scholar]
- Staswick P. E., Su W., Howell S. H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. U.S.A. 89 6837–6840 10.1073/pnas.89.15.6837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H., Zhang S., Yuan X., Chen C., Wang X. F., Hao Y. J. (2013). Genome-wide analysis and identification of stress-responsive genes of the NAM-ATAF1,2-CUC2 transcription factor family in apple. Plant Physiol. Biochem. 71 11–21 10.1016/j.plaphy.2013.06.022 [DOI] [PubMed] [Google Scholar]
- Sun J., Xu Y., Ye S., Jiang H., Chen Q., Liu F., et al. (2009). Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 21 1495–1511 10.1105/tpc.108.064303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri P., Tarighi S. (2010). Riboflavin induces resistance in rice against Rhizoctonia solani via jasmonate-mediated priming of phenylpropanoid pathway. J. Plant Physiol. 167 201–208 10.1016/j.jplph.2009.08.003 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- Tang Y., Liu M., Gao S., Zhang Z., Zhao X., Zhao C., et al. (2012). Molecular characterization of novel TaNAC genes in wheat and overexpression of TaNAC2a confers drought tolerance in tobacco. Physiol. Plant. 144 210–224 10.1111/j.1399-3054.2011.01539.x [DOI] [PubMed] [Google Scholar]
- Taniguchi S., Hosokawa-Shinonaga Y., Tamaoki D., Yamada S., Akimitsu K., Gomi K. (2014). Jasmonate induction of the monoterpene linalool confers resistance to rice bacterial blight and its biosynthesis is regulated by JAZ protein in rice. Plant Cell Environ. 37 451–461 10.1111/pce.12169 [DOI] [PubMed] [Google Scholar]
- Ton J., Flors V., Mauch-Mani B. (2009). The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14 310–317 10.1016/j.tplants.2009.03.006 [DOI] [PubMed] [Google Scholar]
- Uauy C., Distelfeld A., Fahima T., Blechl A., Dubcovsky J. (2006). A NAC Gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314 1298–1301 10.1126/science.1133649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Does D., Leon-Reyes A., Koornneef A., Van Verk M. C., Rodenburg N., Pauwels L., et al. (2013). Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25 744–761 10.1105/tpc.112.108548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos I. A., Pieterse C. M. J., Van Wees S. C. M. (2013). Costs and benefits of hormone-regulated plant defences. Plant Pathol. 62 43–55 10.1111/ppa.12105 [DOI] [Google Scholar]
- Wang N., Zheng Y., Xin H., Fang L., Li S. (2013). Comprehensive analysis of NAC domain transcription factor gene family in Vitis vinifera. Plant Cell Rep. 32 61–75 10.1007/s00299-012-1340-y [DOI] [PubMed] [Google Scholar]
- Wang X., Basnayake B. M., Zhang H., Li G., Li W., Virk N., et al. (2009). The Arabidopsis ATAF1, a NAC transcription factor, is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens. Mol. Plant Microbe Interact. 22 1227–1238 10.1094/MPMI-22-10-1227 [DOI] [PubMed] [Google Scholar]
- Wu Y., Deng Z., Lai J., Zhang Y., Yang C., Yin B., et al. (2009). Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 19 1279–1290 10.1038/cr.2009.108 [DOI] [PubMed] [Google Scholar]
- Xia N., Zhang G., Liu X. Y., Deng L., Cai G. L., Zhang Y., et al. (2010a). Characterization of a novel wheat NAC transcription factor gene involved in defense response against stripe rust pathogen infection and abiotic stresses. Mol. Biol. Rep. 37 3703–3712 10.1007/s11033-010-0023-4 [DOI] [PubMed] [Google Scholar]
- Xia N., Zhang G., Sun Y. F., Zhu L., Xu L. S., Chen X. M., et al. (2010b). TaNAC8, a novel NAC transcription factor gene in wheat, responds to stripe rust pathogen infection and abiotic stresses. Physiol. Mol. Plant Pathol. 74 394–402 10.1016/j.pmpp.2010.06.005 [DOI] [Google Scholar]
- Xie Q., Frugis G., Colgan D., Chua N. H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14 3024–3036 10.1101/gad.852200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Guo H. S., Dallman G., Fang S., Weissman A. M., Chua N. H. (2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419 167–170 10.1038/nature00998 [DOI] [PubMed] [Google Scholar]
- Xie Q., Sanz-Burgos A. P., Guo H., Garcia J. A., Gutierrez C. (1999). GRAB proteins, novel members of the NAC domain family, isolated by their interaction with a geminivirus protein. Plant Mol. Biol. 39 647–656 10.1023/A:1006138221874 [DOI] [PubMed] [Google Scholar]
- Xue G., Bower N. I., Mcintyre C. L., Riding G. A., Kazan K., Shorter R. (2006). TaNAC69 from the NAC superfamily of transcription factors is up-regulated by abiotic stresses in wheat and recognises two consensus DNA-binding sequences. Funct. Plant Biol. 33 43–57 10.1071/FP05161 [DOI] [PubMed] [Google Scholar]
- Xue G. P., Way H. M., Richardson T., Drenth J., Joyce P. A., Mcintyre C. L. (2011). Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol. Plant 4 697–712 10.1093/mp/ssr013 [DOI] [PubMed] [Google Scholar]
- Yamada S., Kano A., Tamaoki D., Miyamoto A., Shishido H., Miyoshi S., et al. (2012). Involvement of OsJAZ8 in jasmonate-induced resistance to bacterial blight in rice. Plant Cell Physiol. 53 2060–2072 10.1093/pcp/pcs145 [DOI] [PubMed] [Google Scholar]
- Yuan C., Li C., Yan L., Jackson A. O., Liu Z., Han C., et al. (2011). A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS ONE 6:e26468 10.1371/journal.pone.0026468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhang D., Chen J., Yang Y., Huang Z., Huang D., et al. (2004). Tomato stress-responsive factor TSRF1 interacts with ethylene responsive element GCC box and regulates pathogen resistance to Ralstonia solanacearum. Plant Mol. Biol. 55 825–834 10.1007/s11103-004-2140-8 [DOI] [PubMed] [Google Scholar]
- Zheng X. Y., Spivey N. W., Zeng W., Liu P. P., Fu Z. Q., Klessig D. F., et al. (2012). Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11 587–596 10.1016/j.chom.2012.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Mosher S. L., Fan B., Klessig D. F., Chen Z. (2007). Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol. 7:2 10.1186/1471-2229-7-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G., Qi J., Ren N., Cheng J., Erb M., Mao B., et al. (2009). Silencing OsHI-LOX makes rice more susceptible to chewing herbivores, but enhances resistance to a phloem feeder. Plant J. 60 638–648 10.1111/j.1365-313X.2009.03988.x [DOI] [PubMed] [Google Scholar]