Abstract
Background
Most animal species exhibit sexually dimorphic behaviors, many of which are linked to reproduction. A number of these behaviors, including blood feeding in female mosquitoes, contribute to the global spread of vector-borne illnesses. However, knowledge concerning the genetic basis of sexually dimorphic traits is limited in any organism, including mosquitoes, especially with respect to differences in the developing nervous system.
Methods
Custom microarrays were used to examine global differences in female vs. male gene expression in the developing pupal head of the dengue vector mosquito, Aedes aegypti. The spatial expression patterns of a subset of differentially expressed transcripts were examined in the developing female vs. male pupal brain through in situ hybridization experiments. Small interfering RNA (siRNA)-mediated knockdown studies were used to assess the putative role of Doublesex, a terminal component of the sex determination pathway, in the regulation of sex-specific gene expression observed in the developing pupal brain.
Results
Transcripts (2,527), many of which were linked to proteolysis, the proteasome, metabolism, catabolic, and biosynthetic processes, ion transport, cell growth, and proliferation, were found to be differentially expressed in A. aegypti female vs. male pupal heads. Analysis of the spatial expression patterns for a subset of dimorphically expressed genes in the pupal brain validated the data set and also facilitated the identification of brain regions with dimorphic gene expression. In many cases, dimorphic gene expression localized to the optic lobe. Sex-specific differences in gene expression were also detected in the antennal lobe and mushroom body. siRNA-mediated gene targeting experiments demonstrated that Doublesex, a transcription factor with consensus binding sites located adjacent to many dimorphically expressed transcripts that function in neural development, is required for regulation of sex-specific gene expression in the developing A. aegypti brain.
Conclusions
These studies revealed sex-specific gene expression profiles in the developing A. aegypti pupal head and identified Doublesex as a key regulator of sexually dimorphic gene expression during mosquito neural development.
Keywords: Aedes aegypti, Mosquito, Vector, Pupae, Brain, Nervous system, Dimorphism, Doublesex, Development, Optic lobe
1 Background
Vector mosquitoes inflict more human suffering than any other organism—and kill more than one million people each year. Aedes aegypti is the primary vector for dengue, the most widespread and significant arboviral disease in the world. Dengue is presently a threat to >2.5 billion people in the tropics with a yearly incidence of approximately 50 million cases resulting in approximately 22,000 deaths annually worldwide. The A. aegypti genome project [1] greatly facilitated efforts to study the biology of this mosquito, but the genetic regulation of mosquito developmental biology is still poorly understood. Our laboratory has begun to address this need by pursuing functional developmental genetic studies in A. aegypti[2]-[8]. Here, we examine the genetic basis of sexual dimorphism during A. aegypti development.
Most animal species exhibit sexually dimorphic behaviors, many of which are linked to sexual reproduction [9]. Mosquitoes, which display innate sexually dimorphic behaviors that contribute to the spread of human disease, are excellent subjects for studies that explore the biological basis of sexual dimorphism. For example, only female adult mosquitoes, which require a blood meal for reproduction, bite humans and transmit disease. If identified, the genes which regulate the development of adult female blood feeding behavior could represent novel targets for vector control. To date, the analysis of mosquito sexual dimorphism has primarily focused on understanding differences between adult female and male mosquitoes, particularly with respect to sex-specific behaviors related to disease transmission (for example [10]). However, sexually dimorphic phenotypes, including behaviors, are the products of differential gene expression that initiates during development and therefore must also be studied during development. We presently lack knowledge concerning which genes regulate the development of sexually dimorphic traits in mosquitoes.
Our extremely limited knowledge of the developmental genetic basis for sexual dimorphism in insects is largely restricted to Drosophila melanogaster, a genetically tractable—albeit highly derived—dipteran insect that exhibits innate sexually dimorphic behaviors. The genetic tractability of the Drosophila system has facilitated analysis of the developmental genetic basis for sexual dimorphism in this species. The Drosophila doublesex (dsx) gene encodes a terminal transcription factor in the sex determination pathway (reviewed by [11],[12]). The pre-mRNAs of dsx are sex-specifically spliced [13],[14], giving rise to male (DsxM) and female (DsxF) proteins. Male and female Dsx splice variants have a common DNA-binding domain, but distinct C termini that differentially direct sex-specific gene expression (reviewed by [11],[12]). Although research has begun to elucidate the genetic basis of sexually dimorphic development in Drosophila, more work is necessary not only in Drosophila, but also in non-model insects, including disease vector mosquitoes. Moreover, our detection of numerous differences between mosquito and Drosophila development underscores the importance of analyzing dsx gene function directly in mosquito species of interest [3]-[5],[15].
Male and female splice variants of the dsx gene have been detected in A. aegypti[16]. Although these sex-specific dsx splice forms are believed to differentially regulate target gene transcription and sex-specific development in males and females, this hypothesis has yet to be functionally tested in mosquitoes. Unfortunately, exploration of the roles of dsx and the developmental genetic basis for sexual dimorphism in mosquitoes has been hampered by a lack of methodology to pursue functional genetic studies during mosquito development. Here, we examine differences in gene expression in the developing A. aegypti pupal brain. We then use small interfering RNA (siRNA)-mediated gene targeting to demonstrate that Dsx regulates dimorphic gene expression in the developing A. aegypti central nervous system.
2 Methods
2.1 Animal rearing
The A. aegypti Liverpool-IB12 (LVP-IB12) strain, from which the current genome sequence [1] was generated, was used in these studies. These mosquitoes were reared as described [17], except that an artificial membrane blood feeding system was employed. Mosquitoes were maintained in an insectary at 26°C, approximately 80% humidity, under a 12-h light and 12-h dark cycle with 1-h crepuscular periods at the beginning and end of each light cycle. Mosquito larvae were fed a suspension of dried beef liver powder, while adults were provided cotton soaked with 10% sugar solution.
2.2 Microarray experiments
Tissues for the microarray studies were prepared as follows: age-matched (±15 min) male and female pupae were raised to 24 h after puparium formation (APF) and sexed on the basis of differing tail morphology as previously described [18]. Although our own primary interest is the pupal brain, dimorphic gene expression was assessed in male vs. female head tissue since removal of the head (as opposed to the more time-consuming dissection of the brain) can be performed quickly and promotes minimization of temporal changes in gene expression between males and females or among replicate experiments. Twenty male and female heads were microdissected for each of four replicates during daylight hours at roughly the same time of day (approximately 11 a.m.) so that circadian-related changes in gene expression would not be a confounding factor. Following dissection, head tissue was immediately stored in RNAlater (Ambion, Austin, TX, USA).
For each of four replicates, RNA extraction was performed with the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. Total RNA was quantified spectrophotometrically, and RNA quality was assessed using an Agilent 2100 Bioanalyzer (Santa Clara, CA, USA). Both the quantity and quality of the RNA preps were deemed to be suitable. RNA samples were then submitted to the Notre Dame Genomics and Bioinformatics Core Facility (Notre Dame, IN, USA), which performed hybridization experiments using the A. aegypti 12-plex microarray design: 090305_Aedes_aegypti_TEfam_expr.ndf (Gene Expression Omnibus platform GPL18530) developed by Susanta K. Behura and David W. Severson in conjunction with Roche NimbleGen. On this microarray, 16,581 genes/ORFs are represented. Each gene/ORF is represented by 1 to 3 unique 60-mer probe sequences that are synthesized in triplicate on the microarray (effectively 3 to 9 probes per gene). RNA labeling and hybridization experiments were performed according to the manufacturer's instructions. Four unique replicates (male 1, 2, 3, 4 and female 1, 2, 3, 4) and two repeat replicates (male 5, which is a repeat hybridization of male 1; male 6, a repeat hybridization of male 4; female 5, a repeat hybridization of female 1; female 6, a repeat hybridization of female 2) were assessed in the hybridization experiment. Microarrays were scanned using a NimbleGen MS 200 Microarray Scanner (Roche NimbleGen, Madison, WI, USA). Data were extracted and analyzed using NimbleScan Version 2.5 software (Roche NimbleGen). Microarray data pre-processing and normalization were performed using the Bioconductor packages in R Version 3.0.1. Raw gene expression data were log2-transformed and normalized using the quantile normalization method [19]. The Student's t test was used for analysis of statistical significance. Raw p values were adjusted using the Bonferroni procedure for control of false discovery rate. Microarray data were evaluated with DAVID [20] and GenMAPP2 [21] to identify GO terms and KEGG pathways (Koyota Encyclopedia of Genes and Genomes). Data were deposited at Gene Expression Omnibus (accession number GSE56521).
2.3 Whole mount in situ hybridization
In situ hybridization was performed as described [22] on male vs. female pupal brain tissue prepared as discussed in [6],[8],[23]. Riboprobes were synthesized per the Patel [24] protocol to the following genes: arrow (VB:AAEL009806), caspase 7 (VB:AAEL012143), cyclin-dependent kinase 4/6 (cdk4/6; VB:AAEL001407), cubitus interruptus (ci; VB:AAEL012039), dsx (VB:AAEL009114), geko (VB:AAEL006310), odorant binding protein 10 (obp10; VB:AAEL007603), odorant binding protein 13 (obp13; VB:AAEL002591), p53 (VB:AAEL007594), rab6 (VB:AAEL006091), synaptojanin (synj; VB:AAEL011417), and takeout (VB:AAEL011966). These gene numbers correspond to Vectorbase (VB) [25], which was also the source for all orthology assignments in this study. Based on information provided in Salvemini et al. [16], probes corresponding to the following dsx exons were synthesized: (i) exon 2, which is common to both the male and female splice forms, (ii) exon 5a, which is female-specific, and (iii) exons 4 combined with 6, which are spliced together in males, but separated by exons 5a/b in females.
At least 25 tissue specimens from each sex were used in each in situ hybridization reaction, and experiments were replicated minimally three times. In addition to confirming dimorphic gene expression at 24 h APF, which corresponds to the time in which the pupal brains were gathered for microarray analyses, dimorphic gene expression was also verified at both 22 and 26 h APF. This allowed for confirmation that the dimorphic expression observed was not simply a result of slight age differences between male and female pupae. As discussed above, such differences were experimentally minimized through use of narrow 30-min animal collection windows. Sense riboprobes served as controls in all hybridization experiments, which routinely yielded unlabeled brains. Following processing, colorimetrically stained tissues were mounted and analyzed using a Zeiss Axioimager equipped with a Spot Flex camera (Oberkochen, Germany). Images were processed with Adobe Photoshop software. Double labeling experiments were performed with gene-specific probes in conjunction with anti-HRP (Jackson Immunoresearch, West Grove, PA, USA) detected with goat anti-rabbit FITC (Jackson Immunoresearch) as described [22]. Imaging of these specimens was performed with a Zeiss 710 confocal microscope using Zen software, and scanned images were analyzed using FIJI and Adobe Photoshop software.
For in situ hybridization experiments on sectioned brain tissues, paraffin embedding and sectioning was performed generally as described in [26]. Blocks were mounted and sectioned at 12 μm thickness on a Leica RM2155 Rotary Microtome (Leica Microsystems GmbH, Nussloch, Germany). Following deparaffinization, which was performed as described in the Abcam IHC-Paraffin Protocol (IHC-P), slides were placed in LockMailer Microscope Slide Jars (Simport Scientific, Beloeil, QC, USA), rehydrated, and then processed as described above.
2.4 Immunohistochemical staining
Immunohistochemical staining was performed as described earlier [6],[23],[27]. Rat anti-DN-cadherin developed by T. Uemura was obtained from the Developmental Studies Hybridoma Bank, which was created by the NICHD-NIH and is maintained at the University of Iowa, and was used at a concentration of 1:50 to visualize neuropile domains in the A. aegypti brain. Alexa Fluor 568 goat anti-rat IgG secondary antibody (Life Technologies) was used at a concentration of 1:200. Nuclear counterstaining was performed with TOTO-3 iodide (Molecular Probes, Grand Island, NY, USA). Double labeling immunohistochemical experiments combined with in situ hybridization were performed with gene-specific probes in conjunction with anti-HRP (Jackson Immunoresearch) detected with goat anti-rabbit FITC (Jackson Immunoresearch) as described in Haugen et al. [22]. Imaging of fluorescently stained specimens was performed with a Zeiss 710 confocal microscope using Zen software, and scanned images were analyzed using FIJI and Adobe Photoshop software.
2.5 Analysis of Dsx consensus binding sites
A. aegypti scaffold locations that differ by no more than one mismatch to the Dsx binding site consensus GCAACAATGTTGC [28] were identified using a custom Perl script, which is available at https://bitbucket.org/NDBL/bindingsearch/. First, the consensus and all 13-mers that differ at a single base (3 alternative bases × 13 total positions = 39 alternative 13-mers) were placed into a hash data structure. Next, each 13-mer in the A. aegypti genome was checked against the hash to quickly determine valid locations. Genes in which the consensus sites resided, as well as open reading frames located within 10,000 kB upstream or downstream of the consensus sites, were identified using the gene browser tool in Vectorbase [25]. GO terms associated with these genes were identified with DAVID [20].
2.6 RNAi knockdown experiments
Knockdown of dsx was performed with two different siRNA duplexes, dsx KD A and dsx KD B, both of which were designed to target the male and female dsx splice forms. The two siRNAs correspond to different target sequences in exon 2, which is common to both the male and female dsx splice variants. The sequences of the siRNA duplexes are as follows: dsx KD A: 5′AGAGAUGAUCCAUAAUUCUCAGCAG3′/5′UUUCUCUACUAGGUAUUAAGAGUCGUC3′ and dsx KD B: 5′CAGGAACAGACGACGAACUUGUCAA3′/5′GAGUCCUUGUCUGCUGCUUGAACAGUU3′. These siRNAs were designed using Integrated DNA Technology (IDT) software and chemically synthesized by IDT. The siRNAs were confirmed through Blast searches to have no significant homology to A. aegypti genes other than dsx. All phenotypes were confirmed following knockdown with both dsx KD A and dsx KD B, suggesting that none of the phenotypes reported here were the result of off-site targeting by either siRNA. A scrambled version of dsx KD B, an siRNA duplex lacking significant sequence homology to any genes in the A. aegypti genome, was used in control experiments: 5′ACAGUACCAGUGGACACAUACGG 3′/5′AUAGCCACAGACAUGAGCGGCAU3′. None of the phenotypes reported were observed in control-injected animals, which resembled wild-type animals.
siRNAs were delivered using a modification of our embryo microinjection protocol [29] and the Blitzer et al. [30] larval injection protocol. In summary, age-synchronized freshly emerged pupae were anesthetized in ice water for 5 min prior to injection and placed on dry filter paper. Animals were injected with control or KD siRNAs vertical to the body axis in the thoracic region. Following injection, pupae were placed in water and maintained through 24 h APF. Fifty individuals were injected per treatment for each replicate, and experiments to confirm each phenotype reported here were repeated in triplicate. Knockdown was verified through in situ hybridization according to the protocol described above and through use of riboprobes corresponding to the common, male-specific, and female-specific splice forms (see above).
3 Results
3.1 Detection of global gene expression differences in the pupal head
A cDNA microarray experiment was conducted to profile and compare pupal head transcript expression in female vs. male A. aegypti pupae. Head tissues from 24 h pupae were selected for these studies, as our recent work has identified this time point, which follows pupal histolysis, to be a critical period for nervous system development [6],[8],[23]. Microarray analysis of 16,581 genes/ORFs revealed differential gene expression among a number of transcripts in the male vs. female pupal head transcriptome. The dimorphism microarray experiment uncovered 2,527 significant differentially expressed transcripts (DETs) (Additional file 1). A volcano plot comparing significance vs. fold change for these data is displayed in Figure 1. One hundred eighty-nine DETs were found to be significantly upregulated in males (p < 0.05) with a log2 fold change greater than 2.0, while 201 DETs were significantly upregulated in females (p < 0.05) with a log2 fold change greater than 2.0.
Figure 1.
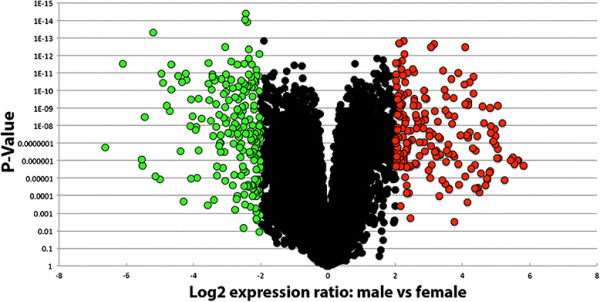
Sex-biased gene expression in the pupal head. A volcano plot comparing significance vs. fold change for male vs. female 24 h pupal head transcriptome data is shown. The red dots denote 189 genes that were found to be significantly upregulated in males (p < 0.05) with a log2 fold change greater than 2. Green dots represent the 201 genes that were found to be significantly upregulated in females (p < 0.05) with a log2 fold change greater than 2. Black dots represent genes with a log2 fold change less than 2.
Microarray data were evaluated with DAVID [20] and GenMAPP [21] to identify gene ontology terms and KEGG pathways significantly associated with sex-specific DETs. Significantly upregulated DETs in the female pupal head are primarily linked to the proteolytic, metabolic, catabolic, protein dephosphorylation, homeostatic, cell cycle, and microtubule-based movement GO terms (Figure 2A, Table 1). A large number of the DETs significantly upregulated in females are related to proteolysis (Figure 2A), with many of the transcripts encoding serine proteases (Table 2). KEGG pathway analysis of DETs significantly upregulated in female pupal heads identified the proteasome, glycosphingolipid biosynthesis, glycosaminoglycan, and other glycan degradation, as well as glycine/serine/threonine metabolism as significant pathways (Figure 3).
Figure 2.
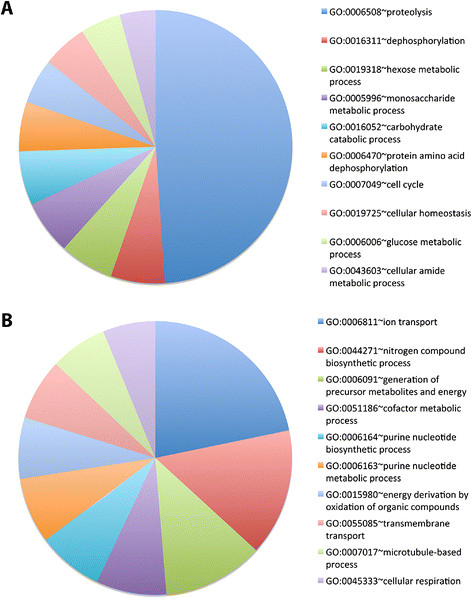
Over-represented GO categories among sex-biased genes expressed in the pupal head. The top ten significant GO terms (p < 0.05) associated with genes that were significantly (p < 0.05) upregulated in female (A) or male (B) 24 h pupal heads are indicated.
Table 1.
Significantly over-represented GO categories associated with DETs upregulated in female pupal heads
| Term | Count | p value |
|---|---|---|
| GO:0006508—proteolysis | 92 | 3.17E − 06 |
| GO:0016311—dephosphorylation | 12 | 0.002031467 |
| GO:0019318—hexose metabolic process | 12 | 0.009445235 |
| GO:0005996—monosaccharide metabolic process | 12 | 0.013314328 |
| GO:0016052—carbohydrate catabolic process | 12 | 0.020208459 |
| GO:0006470—protein amino acid dephosphorylation | 11 | 0.001514357 |
| GO:0007049—cell cycle | 10 | 0.002262897 |
| GO:0019725—cellular homeostasis | 10 | 0.033082 |
| GO:0006006—glucose metabolic process | 9 | 0.025805435 |
| GO:0043603—cellular amide metabolic process | 8 | 0.00250685 |
| GO:0006733—oxidoreduction coenzyme metabolic process | 8 | 0.009081419 |
| GO:0045454—cell redox homeostasis | 8 | 0.039387289 |
| GO:0006098—pentose-phosphate shunt | 7 | 6.34E − 04 |
| GO:0006739—NADP metabolic process | 7 | 6.34E − 04 |
| GO:0019748—secondary metabolic process | 7 | 0.002617106 |
| GO:0006769—nicotinamide metabolic process | 7 | 0.002617106 |
| GO:0009820—alkaloid metabolic process | 7 | 0.002617106 |
| GO:0046496—nicotinamide nucleotide metabolic process | 7 | 0.002617106 |
| GO:0019362—pyridine nucleotide metabolic process | 7 | 0.007431131 |
| GO:0006007—glucose catabolic process | 7 | 0.047566184 |
| GO:0019320—hexose catabolic process | 7 | 0.047566184 |
| GO:0006913—nucleocytoplasmic transport | 6 | 0.0364855 |
| GO:0051169—nuclear transport | 6 | 0.0364855 |
| GO:0046700—heterocycle catabolic process | 5 | 0.026037743 |
| GO:000716—enzyme linked receptor protein signaling pathway | 5 | 0.026037743 |
| GO:0009070—serine family amino acid biosynthetic process | 4 | 0.008178934 |
| GO:0006564—L-serine biosynthetic process | 3 | 0.017716355 |
| GO:0042176—regulation of protein catabolic process | 3 | 0.017716355 |
| GO:0009894—regulation of catabolic process | 3 | 0.017716355 |
| GO:0019439—aromatic compound catabolic process | 3 | 0.017716355 |
Over-represented GO categories among sex-biased genes expressed in the female 24 h pupal head are shown. The number of DETs associated with each GO term (count) and corresponding Fisher's exact test p values are indicated.
Table 2.
Genes upregulated in the female pupal head are associated with proteolysis
| Gene | Description | Expression |
|---|---|---|
| AAEL000028 | CLIPB34 Clip-domain serine protease family B | 2.5 |
| AAEL018109 | Hypothetical protein | 6.0 |
| AAEL000037 | CLIPB35 Clip-domain serine protease family B | 4.0 |
| AAEL000038 | CLIPB6 Clip-domain serine protease family B | 4.3 |
| AAEL000074 | CLIPB1 Clip-domain serine protease family B | 4.0 |
| AAEL000224 | Serine protease | 2.2 |
| AAEL001077 | CLIPB45 Clip-domain serine protease family B | 8.8 |
| AAEL001084 | CLIPB21 Clip-domain serine protease family B | 9.4 |
| AAEL001098 | Clip-domain serine protease | 3.7 |
| AAEL002124 | CLIPD6 Clip-domain serine protease family D | 11.2 |
| AAEL002126 | CLIPA15 Clip-domain serine protease family A | 5.3 |
| AAEL002585 | Serine protease | 2.1 |
| AAEL002590 | Serine protease | 2.7 |
| AAEL002593 | Serine protease | 3.0 |
| AAEL002595 | Serine protease | 5.0 |
| AAEL002600 | Serine protease | 2.0 |
| AAEL002629 | Serine protease | 5.1 |
| AAEL002767 | Conserved hypothetical protein | 5.1 |
| AAEL003243 | CLIPB13A Clip-domain serine protease family B | 3.1 |
| AAEL003251 | Serine protease snake | 2.7 |
| AAEL003276 | Hypothetical protein | 3.3 |
| AAEL003280 | CLIPB26 Clip-domain serine protease family B | 3.8 |
| AAEL003610 | CLIPB9 Clip-domain serine protease family B | 2.3 |
| AAEL004518 | CLIPC5A Clip-domain serine protease family C | 6.0 |
| AAEL004524 | CLIPC5B Clip-domain serine protease family C | 8.2 |
| AAEL004540 | CLIPC6 Clip-domain serine protease family C | 3.1 |
| AAEL004979 | CLIPD2 Clip-domain serine protease family D | 2.1 |
| AAEL005064 | CLIPB5 Clip-domain serine protease family B | 4.4 |
| AAEL006136 | Serine protease | 3.0 |
| AAEL006151 | Serine protease | 3.7 |
| AAEL006434 | Serine protease | 4.5 |
| AAEL006708 | Hedgehog | 2.3 |
| AAEL007107 | Serine protease | 2.3 |
| AAEL007511 | Serine protease | 8.5 |
| AAEL007597 | CLIPC3 Clip-domain serine protease family C | 2.8 |
| AAEL007602 | Trypsin | 3.5 |
| AAEL007969 | Serine protease | 4.5 |
| AAEL010267 | Serine protease | 3.4 |
| AAEL010270 | Hypothetical protein | 9.1 |
| AAEL010780 | Carboxypeptidase | 3.6 |
| AAEL010866 | Serine protease | 4.0 |
| AAEL010867 | Serine protease | 10.8 |
| AAEL011324 | Hypothetical protein | 3.0 |
| AAEL011991 | CLIPC1 Clip-domain serine protease family C | 2.0 |
| AAEL012143 | CaspaseS7 | 3.3 |
| AAEL012500 | Ubiquitin-protein ligase | 2.2 |
| AAEL012713 | CLIPC16 Clip-domain serine protease family C | 2.9 |
| AAEL012775 | Serine protease | 3.3 |
| AAEL012777 | Serine protease snake | 3.5 |
| AAEL012780 | Trypsin | 7.4 |
| AAEL012785 | CLIPB23 Clip-domain serine protease family B | 4.1 |
| AAEL012797 | Serine protease | 3.4 |
| AAEL013245 | CLIPB28 Clip-domain serine protease family B | 10.1 |
| AAEL013299 | Serine protease | 2.6 |
| AAEL014344 | Adam (a disintegrin and metalloprotease) | 5.3 |
| AAEL014567 | Oviductin | 10.1 |
| AAEL015439 | CLIPD7 Clip-domain serine protease family D | 2.7 |
| AAEL015527 | Hypothetical protein | 2.4 |
| AAEL015533 | Conserved hypothetical protein | 4.1 |
KEGG pathway analyses indicated that proteolysis is the most represented pathway corresponding to DETs upregulated in the female pupal head. The gene numbers, descriptions, and log2 fold change with respect to males is indicated for each gene corresponding to proteolysis.
Figure 3.
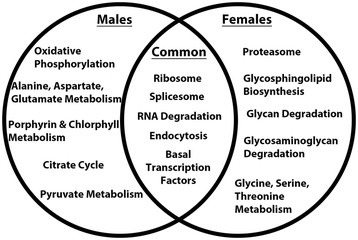
Differential distribution of KEGG pathway genes in male vs. female pupal heads. Pathways in which the number of significant DETs is significantly (p < 0.05) higher in male or female 24 h pupal heads are indicated. A number of pathways were commonly activated in females and males without statistically significant bias (common pathways). An itemized list of genes corresponding to each pathway is provided in Additional file 3.
Significant DETs upregulated in male pupal heads are significantly linked to ion transport, biosynthetic, and metabolic GO terms (Figure 2B, Table 3). A majority of the DETs associated with ion transport (Figure 2B), the GO term linked to the largest number of male-specific upregulated transcripts, encode ion channels (Table 4). Pathway analysis of DETs significantly upregulated in male pupal heads identified oxidative phosphorylation, alanine/aspartate/glutamate metabolism, porphyrin and chlorophyll metabolism, the citric acid cycle, and pyruvate metabolism as significant pathways (Figure 3). Non-differentially expressed head transcripts common to both the male and female brain grouped under a number of significant pathways that were generally associated with the regulation of gene and protein expression (Figure 3).
Table 3.
Significantly over-represented GO categories associated with DETs upregulated in male pupal heads
| Term | Count | p value |
|---|---|---|
| GO:0006811—ion transport | 42 | 0.010896197 |
| GO:0044271—nitrogen compound biosynthetic process | 29 | 0.007748843 |
| GO:0006091—generation of precursor metabolites and energy | 23 | 2.70E − 04 |
| GO:0051186—cofactor metabolic process | 16 | 0.033341803 |
| GO:0006164—purine nucleotide biosynthetic process | 15 | 0.021253849 |
| GO:0006163—purine nucleotide metabolic process | 15 | 0.023426935 |
| GO:0015980—energy derivation by oxidation of organic compounds | 14 | 1.86E − 04 |
| GO:0055085—transmembrane transport | 14 | 0.011896142 |
| GO:0007017—microtubule-based process | 13 | 0.01594331 |
| GO:0045333—cellular respiration | 12 | 5.71E − 04 |
| GO:0018130—heterocycle biosynthetic process | 12 | 0.001152569 |
| GO:0006119—oxidative phosphorylation | 12 | 0.004475745 |
| GO:0046394—carboxylic acid biosynthetic process | 11 | 0.040301387 |
| GO:0016053—organic acid biosynthetic process | 11 | 0.040301387 |
| GO:0007005—mitochondrion organization | 9 | 8.07E − 07 |
| GO:0022900—electron transport chain | 9 | 0.003534463 |
| GO:0016044—membrane organization | 8 | 0.001738542 |
| GO:0042773—ATP synthesis coupled electron transport | 7 | 0.001700687 |
| GO:0022904—respiratory electron transport chain | 7 | 0.004925969 |
| GO:0046942—carboxylic acid transport | 7 | 0.017801566 |
| GO:0015849—organic acid transport | 7 | 0.017801566 |
| GO:0009187—cyclic nucleotide metabolic process | 7 | 0.026556712 |
| GO:0009190—cyclic nucleotide biosynthetic process | 7 | 0.026556712 |
| GO:0070585—protein localization in mitochondrion | 6 | 1.36E − 04 |
| GO:0006626—protein targeting to mitochondrion | 6 | 1.36E − 04 |
| GO:0006839—mitochondrial transport | 6 | 6.95E − 04 |
| GO:0046068—cGMP metabolic process | 6 | 0.005142832 |
| GO:0006182—cGMP biosynthetic process | 6 | 0.005142832 |
| GO:0009064—glutamine family amino acid metabolic process | 6 | 0.028832748 |
| GO:0007006—mitochondrial membrane organization | 5 | 3.74E − 04 |
| GO:0007007—inner mitochondrial membrane organization | 5 | 3.74E − 04 |
| GO:0045039—protein import into mitochondrial inner membrane | 5 | 3.74E-04 |
| GO:0065002—intracellular protein transmembrane transport | 5 | 0.002241034 |
| GO:0042775—mitochondrial ATP synthesis coupled electron transport | 5 | 0.010659342 |
| GO:0006612—protein targeting to membrane | 5 | 0.037429854 |
| GO:0009084—glutamine family amino acid biosynthetic process | 5 | 0.037429854 |
| GO:0006561—proline biosynthetic process | 4 | 0.03331591 |
| GO:0033014—tetrapyrrole biosynthetic process | 4 | 0.046519627 |
| GO:0006560—proline metabolic process | 4 | 0.046519627 |
| GO:0006493—protein amino acid O-linked glycosylation | 3 | 0.025391936 |
| GO:0006122—mitochondrial electron transport | 3 | 0.047602115 |
Over-represented GO categories among sex-biased genes expressed in the male 24 h pupal head are shown. The number of DETs associated with each GO term (count) and corresponding Fisher's exact test p values are indicated.
Table 4.
Genes upregulated in male pupal heads are associated with ion transport
| Gene | Description | Expression |
|---|---|---|
| AAEL001123 | Hypothetical protein | 10.6 |
| AAEL001198 | Sodium/solute symporter | 2.3 |
| AAEL001646 | Kir3 inward-rectifying potassium channel | 2.1 |
| AAEL002299 | High affinity copper transporter | 2.1 |
| AAEL004664 | Hypothetical protein | 2.0 |
| AAEL004919 | Hypothetical protein | 3.5 |
| AAEL005014 | Transient receptor potential channel | 2.0 |
| AAEL007770 | Voltage and ligand gated potassium channel | 2.3 |
| AAEL008338 | Ion channel NompC | 2.5 |
| AAEL009813 | Glutamate receptor 7 | 2.0 |
| AAEL009856 | Sodium/dicarboxylate cotransporter | 3.0 |
| AAEL011109 | Hypothetical protein | 3.3 |
| AAEL011679 | Ion channel NompC | 2.7 |
| AAEL015091 | Hypothetical protein | 3.3 |
KEGG pathway analyses indicated that ion transport is the most represented pathway corresponding to DETs upregulated in male pupal heads. The gene numbers, descriptions, and log2 fold change with respect to females is indicated for each ion transport-related gene.
3.2 Examination of the expression of DETs in the pupal brain
Whole mount in situ hybridization was performed to assess the expression of a subset of DETs in male vs. female pupal brains. A variety of genes were selected for these studies. Choices included genes linked to significant GO terms and pathways noted above (see further explanation in the Discussion) and genes of interest to our laboratory. Several genes were selected on their basis of their proximity to Dsx binding sites (see below). First, we performed whole mount in situ hybridization experiments in an effort to validate the data set. Differential gene expression was confirmed in the pupal brain for takeout, ci, cdk4/6, obp10, arrow, caspase 7 (Figure 4A,B,C,D,E,F; all upregulated in females), synj, geko, p53, and obp13 (Figure 4G,H,I,J; all upregulated in males). For all of these genes, the female or male sex-specific upregulation detected through in situ hybridization in the 24 h pupal brain (Figure 4) was in agreement with the sex-biased expression results detected in the whole head microarray experiments (Additional file 1). To be certain that the dimorphic expression observed did not result from slight age differences between female and male pupal samples, sexually dimorphic patterns of gene expression were also confirmed at both 22 and 26 h of development for all of these genes; representative data for obp13 (Figure 5A), caspase 7 (Figure 5B), obp10 (Figure 5C), and ci (Figure 5D) are shown.
Figure 4.
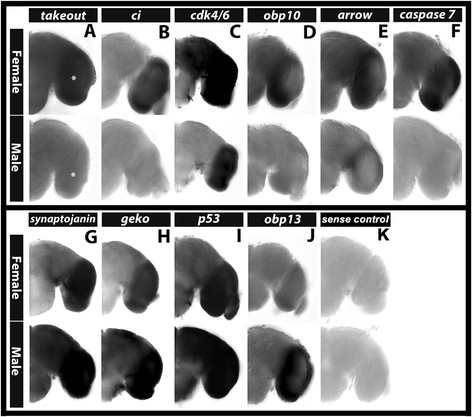
Sex-specific expression of DETs in theA. aegyptipupal brain. Differential expression of the indicated genes was confirmed in female vs. male 24 h pupal brains through whole mount in situ hybridization experiments. In agreement with the whole head microarray experimental data, these experiments demonstrated that takeout, ci, cdk4/6, obp10, arrow, and caspase7 (top row, A-F respectively) are upregulated in the female 24 h pupal brain, while synj, geko, p53, and obp13 are upregulated in the male 24 h pupal brain (G-J, respectively). A brain processed with a sense control probe lacks staining (K). For a number of the genes assayed (takeout, ci, obp10, arrow, caspase 7, and obp13), dimorphic gene expression localized to the optic lobe, which is marked by an asterisk in (A) and located at a similar position in other panels. Brain hemisegments are oriented dorsal upward in each panel.
Figure 5.
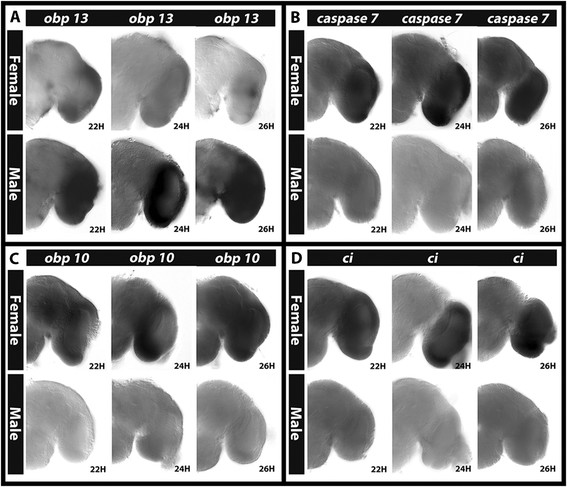
Sex-specific gene expression patterns are maintained from 22 to 26 h of pupal brain development. Differential male vs. female obp13(A), caspase7(B), obp10(C), and ci(D) expression patterns in the brain identified at 24 h APF were also confirmed at 22 and 26 h APF. Brain hemisegments are oriented dorsal upward in each panel.
For many of the genes analyzed, dimorphic expression localized to the optic lobe of the brain. For example, takeout, ci, obp10, arrow, and caspase 7 (Figure 4A,B,D,E,F, respectively) are upregulated in the female optic lobes with respect to males in which little if any transcripts were detected through in situ hybridization. Expression of obp13 is upregulated in the male optic lobe with respect to females (Figure 4J). To be certain that this intense optic lobe staining was not an artifact of the tissue preparation or whole mount in situ hybridization process, we confirmed that negative sense control probes gave little background staining in the optic lobes or in other regions of the brain (Figure 4K). We also confirmed optic lobe expression for a number of DETs by performing in situ hybridization experiments on sectioned brain tissues (Figure 6).
Figure 6.
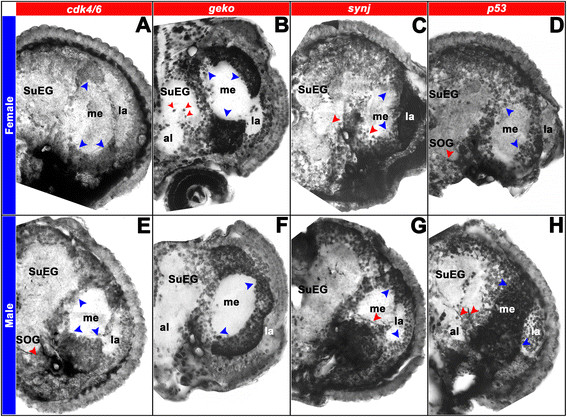
Sex-specific expression patterns of DETs in sectionedA. aegyptipupal brains. Differential expression of cdk4/6(A, E), geko(B, F), synj(C, G), and p53(D, H) was detected in 12 μ sections through 24 hr female (A-D) and male (E-H) pupal brains. Hemisegments oriented dorsal upward are shown. Hybridizations with a sense control probe detected no signal in comparable brain sections (not shown). cdk4/6 is commonly expressed in the optic lobe (blue arrowheads in A, E), but males have an additional cdk4/6 expression domain in the ventral suboesophageal ganglion (red arrowhead in E). geko, which is commonly expressed in the female and male optic lobe (blue arrowheads in B, F), is expressed in additional large cell bodies near the female midbrain and antennal lobe (red arrowheads in B). synj expression is detected in the optic lobe (blue arrowheads in C, G) and in a subset of midbrain neurons (red arrowheads in C, G). The red arrowhead in G marks sex-specific synj optic lobe expression in males, and midbrain synj levels are generally higher in males (compare expression adjacent to red arrowheads in C, G). p53 is expressed in the optic lobe and suboesophageal ganglion of females (blue and red arrowheads, respectively in D). p53 is also expressed in the male optic lobe (blue arrowheads in H), but not in the male subesophageal ganglion (H). Male-specific p53 expression is detected in neurons adjacent to the antennal lobe (red arrowheads in H). These data are consistent with the results presented in Figure 7. al antennal lobe, la lamina, me medulla, SuEG, supraesophageal ganglion.
Experiments on sectioned brain tissue (Figure 6), as well as co-labeling experiments with anti-HRP, which marks various neuropil regions in the developing mosquito brain [23] (Figure 7C,D,E,F,G,H,I,J), helped us to pinpoint the location of dimorphic gene expression for a number of DETs in which dimorphic expression was noted outside of the optic lobe region. These studies revealed upregulation of cdk4/6 expression in the ventral region of the male subesophageal ganglion (Figure 6E, compare to Figure 6A; Figure 7D, compare to Figure 7C), as well as upregulation of geko transcripts near the midbrain and antennal lobes of females (Figure 6B, compare to Figure 6F; Figure 7E, compare to Figure 7F). Higher levels of synj expression were detected in the midbrain of males (Figure 6G, compare to Figure 6C; Figure 7H, compare to Figure 7G), and sex-specific patterns of synj expression were detected in the optic lobe (Figures 6C,G and 7G,H). Finally, p53 is expressed in the female, but not the male subesophageal ganglion (Figure 6D, compare to Figure 6H; Figure 7I, compare to Figure 7J), while male-specific p53 expression was detected in neurons adjacent to the antennal lobe region (Figures 6H and 7J).
Figure 7.
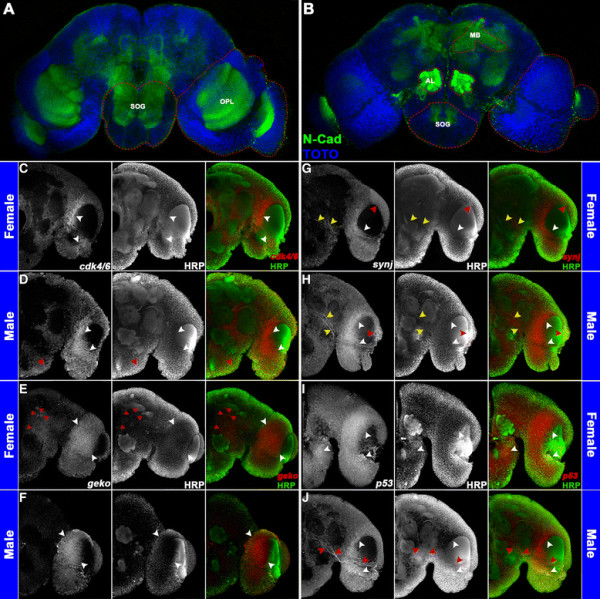
Sex-specific expression patterns of DETs in theA. aegyptipupal brain. The optic lobe (OPL), antennal lobe (AL), suboesophageal ganglion (SOG), and mushroom body (MB) regions are highlighted by red dots in a whole brain labeled with anti-N-Cadherin (green in A, B) and TOTO nuclear stain (blue in A, B). These regions were assessed through confocal imaging following whole mount in situ hybridization and anti-HRP staining. Five merged Z-stacks (totaling 5 μ) of brain hemisegments oriented dorsal upward (C-J) labeled with anti-HRP staining (center panels in C-J; green in overlays at right) and riboprobes corresponding to the indicated transcripts (left panels in C-J; red in overlays at right) are shown. Differential expression of cdk4/6(C, D), geko(E, F), synj(G, H), and p53(I, J) was detected in 24 h female (C, E, G, I) and male (D, F, H, J) pupal brains. cdk4/6 is commonly expressed in the optic lobe (white arrowheads in C, D), but males have an additional cdk4/6 expression domain in the ventral suboesophageal ganglion (red arrowhead in D). geko, which is commonly expressed in the female and male optic lobe (white arrowheads in E, F), is expressed in additional large cell bodies near the female midbrain and antennal lobe (red arrowheads in E). synj expression is detected in the optic lobe (white/red arrowheads in G, H) and in a subset of midbrain neurons (yellow arrowheads in G, H). Red arrowheads mark sex-specific synj optic lobe expression (G, H), and midbrain synj levels are generally higher in males (compare expression adjacent to yellow arrowheads in G, H). p53 is expressed in the optic lobe and suboesophageal ganglion of females (white arrowheads in I). p53 is also expressed in the male optic lobe (white arrowheads in J), but not in the male subesophageal ganglion. Male-specific p53 expression is detected in neurons adjacent to the antennal lobe (red arrowheads in J). These data are consistent with the results presented in Figure 6.
3.3 Putative Dsx consensus binding sites associated with DETs
The consensus binding site sequence for Dsx, GCAACAATGTTGC, was deduced by Luo et al. [28], who reported that the sequence of this binding site is well-conserved in many dipteran species and is detected 26-fold over expected in A. aegypti. Both male and female Dsx splice variants share this DNA-binding domain but have different C termini that differentially direct sex-specific gene expression [11],[12],[16]. Search of the A. aegypti genome for the Dsx consensus sequence uncovered 732 genes located in proximity to putative Dsx binding sites (Additional file 2). A. aegypti genes with intragenic, upstream, or downstream Dsx consensus sites grouped under a number of significant GO-terms, which are itemized in Table 5. Many of these are linked to neurological processes or neural development. These include a variety of GO terms related to the sensory system and sensory development, particularly the compound eye and compound eye development. Additional neural GO terms associated with these genes include neuron development, neuron differentiation, neuron fate commitment, and neurological system processes. Moreover, 48 of the 732 genes associated with Dsx binding sites correspond to genes identified in the dimorphism microarray experiments (Additional file 2). Together, these data support the hypothesis that Dsx is a regulator of sexually dimorphic gene expression in the A. aegypti nervous system. This hypothesis was examined in the developing A. aegypti pupal brain.
Table 5.
Over-represented GO categories among genes associated with Dsx consensus binding sites
| Term | Count | p value |
|---|---|---|
| GO:0007166—cell surface receptor linked signal transduction | 14 | 0.026157815 |
| GO:0030182—neuron differentiation | 12 | 0.00682396 |
| GO:0007423—sensory organ development | 12 | 0.006946573 |
| GO:0003002—regionalization | 11 | 0.034377574 |
| GO:0007389—pattern specification process | 11 | 0.04735023 |
| GO:0001745—compound eye morphogenesis | 10 | 0.002178058 |
| GO:0048592—eye morphogenesis | 10 | 0.003563147 |
| GO:0048749—compound eye development | 10 | 0.008521908 |
| GO:0001654—eye development | 10 | 0.013504773 |
| GO:0048666—neuron development | 10 | 0.01756628 |
| GO:0007350—blastoderm segmentation | 8 | 0.01232515 |
| GO:0009880—embryonic pattern specification | 8 | 0.016503315 |
| GO:0035282—segmentation | 8 | 0.029278118 |
| GO:0001751—compound eye photoreceptor cell differentiation | 7 | 0.001660725 |
| GO:0001754—eye photoreceptor cell differentiation | 7 | 0.00207928 |
| GO:0046530—photoreceptor cell differentiation | 7 | 0.003406961 |
| GO:0048232—male gamete generation | 7 | 0.008583448 |
| GO:0007283—spermatogenesis | 7 | 0.008583448 |
| GO:0045165—cell fate commitment | 7 | 0.049850932 |
| GO:0022604—regulation of cell morphogenesis | 6 | 0.011632368 |
| GO:0042051—compound eye photoreceptor development | 5 | 0.003084585 |
| GO:0042462—eye photoreceptor cell development | 5 | 0.003302533 |
| GO:0042461—photoreceptor cell development | 5 | 0.005753226 |
| GO:0009968—negative regulation of signal transduction | 5 | 0.029343023 |
| GO:0010648—negative regulation of cell communication | 5 | 0.03026886 |
| GO:0040007—growth | 5 | 0.037232894 |
| GO:0048193—Golgi vesicle transport | 4 | 0.012925753 |
| GO:0016049—cell growth | 4 | 0.017389354 |
| GO:0046552—photoreceptor cell fate commitment | 4 | 0.020430767 |
| GO:0048663—neuron fate commitment | 4 | 0.02374654 |
| GO:0007507—heart development | 4 | 0.044413566 |
| GO:0007479—leg disc proximal/distal pattern formation | 3 | 0.01405488 |
| GO:0035223—leg disc pattern formation | 3 | 0.01405488 |
| GO:0035051—cardiac cell differentiation | 3 | 0.021604074 |
| GO:0035287—head segmentation | 3 | 0.023702413 |
| GO:0007449—proximal/distal pattern formation, imaginal disc | 3 | 0.023702413 |
| GO:0009954—proximal/distal pattern formation | 3 | 0.028137457 |
| GO:0035215—genital disc development | 3 | 0.040521397 |
| GO:0006869—lipid transport | 3 | 0.040521397 |
| GO:0045466—R7 cell differentiation | 3 | 0.045952969 |
| GO:0007419—ventral cord development | 3 | 0.048763797 |
| GO:0048052—R1/R6 cell differentiation | 2 | 0.034007252 |
| GO:0007462—R1/R6 cell fate commitment | 2 | 0.034007252 |
| GO:0031887—lipid particle transport along microtubule | 2 | 0.045086826 |
A. aegypti genes associated with Dsx consensus sites (defined as genes with Dsx consensus sites located within or 10 kB upstream/dowstream of the genes) grouped under a number of significant GO terms (p < 0.05). The number of genes associated with each GO term (count) and corresponding Fisher's exact test p values are indicated.
3.4 Dsx is a regulator of sex-specific gene expression in the developing A. aegypti pupal brain
Sex-specific dsx expression was examined in the A. aegypti pupal brain. For these experiments, probes corresponding to the male and female splice forms, as well as exon 2 which is common to both splice forms [16] (see Methods for details) were synthesized and utilized in whole mount in situ hybridization experiments. Expression of the male splice form was detected in the male brain, and expression of the female-specific splice form was detected in the female brain, while the dsx common probe marked expression of dsx transcripts in both female and male pupal brains (Figure 8A). In females and males, the highest levels of dsxF and dsxM transcripts, respectively, are detected in the optic lobes (Figure 8A). To more precisely map the locations of sex-specific dsx expression outside of the optic lobe region, we again performed in situ hybridization experiments on sectioned head tissue (Figure 9) and also performed co-labeling experiments with anti-HRP antibody (Figure 10). These experiments revealed dimorphic expression patterns of dsx in the A. aegypti antennal lobe and mushroom body. Expression of dsx is detected in the 24 h female antennal lobe (Figures 9C and 10C). In males, although dsx expression is detected in the antennal lobe (Figures 9F and 10B), it is restricted to the ventral-most cells. Furthermore, although dsx expression is detected in the female mushroom body (Figures 9A and 10E), dsx expression is not detected in the male mushroom body (Figures 9D and 10F). These expression studies suggest that dsx may regulate sex-specific gene expression in the A. aegypti pupal brain. We conducted dsx knockdown experiments to test this hypothesis.
Figure 8.

Expression and knockdown ofdsxin theA. aegyptipupal brain. (A) Detection of dsx expression in developing wild-type female (top) and male (bottom) pupal brain hemisegments with riboprobes corresponding to exon 2 (dsx common), DsxM (dsx male), and DsxF (dsx female). At right in (A), a brain processed with a sense control probe lacks staining. Knockdown of both splice variants was detected through use of the dsx common (B), dsxF (C, top), and dsxM (C, bottom) probes in female (top in B, C) and male (bottom in B, C) pupal brain hemisegments following injection of dsx KD A or KD B siRNAs. Control siRNA-injected animals (control in B, C) resemble wild-type animals. Brain hemisegments are oriented dorsal upward in each panel.
Figure 9.
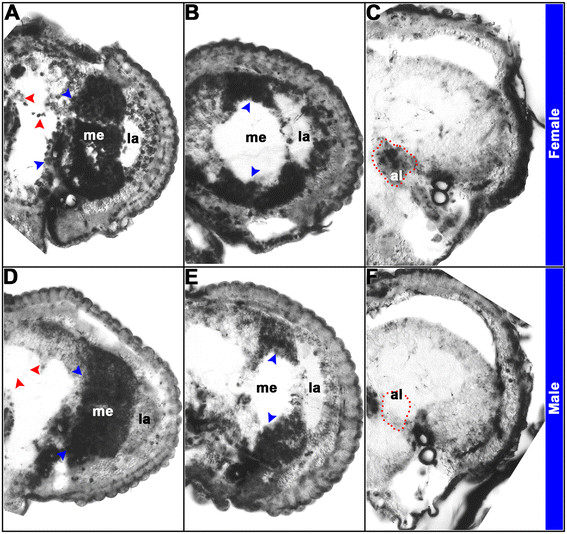
Sex-specific expression patterns ofdsxin sectionedA. aegyptipupal brains. The detailed expression pattern of dsx was analyzed through in situ hybridization experiments (using the dsx common probe) that were performed following preparation of paraffin sections of female (A-C) and male (D-F) heads. Twelve micron sections through different portions of the brain reveal the following structures, which are shown in brain hemisegments oriented dorsal upward in (A-F): antennal lobe (al), lamina (la), and medulla (me). Expression of dsx is detected in the visual system of both females and males (blue arrowheads in A, B, D, E). However, dsx is dimorphically expressed in the antennal lobe (highlighted by red dots in C, F) and the mushroom bodies (red arrowheads in A, D) of females and males. Hybridizations with a sense control probe detected no signal in comparable brain sections (not shown). These data are consistent with the results presented in Figure 10.
Figure 10.

Sex-specific expression patterns ofdsxin theA. aegyptipupal brain. The precise locations of dsx expression in the 24 h female (A, C, E) and male (B, D, F) pupal brain were also mapped through co-labeling experiments with dsx common probe (left in panels A-F; red in overlays at right) and anti-HRP staining (center panels of A-F; green in overlays at right), which marks various neuropil regions in the developing A. aegypti brain. Confocal imaging was performed following staining of whole mount brains. Five merged Z-stacks of confocal sections (totaling 5 μ) through the optic lobe (A, B), antennal lobe (C,D), and mushroom body (E, F) regions of brain hemisegments oriented dorsal upward are shown. dsx expression is detected in the 24 h female pupal optic lobe (A), throughout the antennal lobe (C), and in the mushroom body (E). dsx expression is detected in the 24 h male pupal optic lobe (B) and a group of ventral-most cells in the antennal lobe (D), but not in the mushroom body (F). These data are consistent with the results presented in Figure 9.
siRNA-mediated gene targeting was used to knockdown dsx in A. aegypti pupae (Figure 7B,C). Two dsx knockdown siRNAs, dsx KD A and dsx KD B, which correspond to two different target sequences in exon 2, an exon common to both the female and male splice forms, were used in these experiments. dsx KD A and dsx KD B effectively target pupal brain expression of both the male and female dsx splice forms when microinjected into the pupal thorax (Figure 8B,C). In these experiments, control injections were performed with a scrambled version of dsx KD B, an siRNA which lacks significant sequence homology to any genes in the A. aegypti genome (Figure 8B,C).
The impact of dsx knockdown on the developmental expression of putative Dsx target genes was then assessed. p53, a DET in the microarray study that is upregulated in male heads (Additional file 1), is associated with a Dsx binding site (Additional file 2) and is differentially expressed in wild-type and control-fed female and male pupal brain hemisegments (Figures 4I and 11A). Whole mount in situ hybridization experiments revealed that levels of p53 are reduced to nearly undetectable levels in female and male pupae injected with dsx KD A or dsx KD B siRNAs (Figure 11A). Comparable results were obtained for synj (Figures 4G and 11B), geko (Figures 4H and 11C), and rab6 (Figure 11E), all of which are genes associated with Dsx consensus binding sites (Additional file 2) that correspond to DETs noted in microarray studies (Additional file 1). Expression of cdk4/6 (Figures 4C and 11D), which contains a Dsx binding site, was reduced but not completely eliminated in dsx knockdown females and males, in which cdk4 transcripts could still be detected in the optic lobes, albeit at reduced levels. Together, these results indicate that Dsx is required for sex-specific gene expression in the developing A. aegypti pupal central nervous system.
Figure 11.
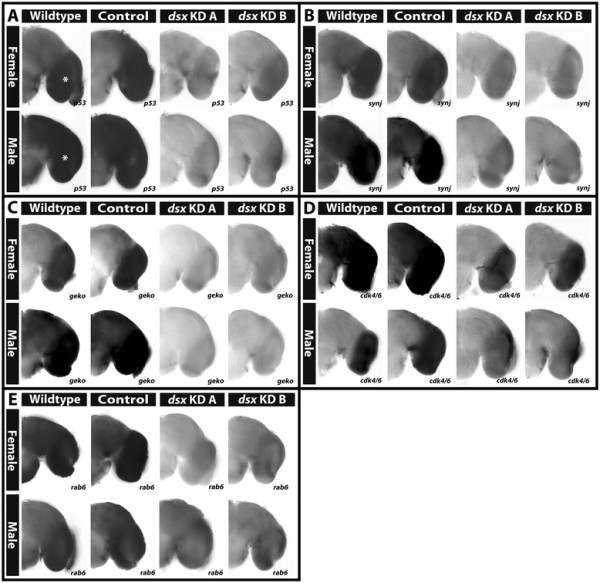
Dsx is a regulator of sex-specific gene expression in theA. aegyptipupal brain. Sex specific patterns of p53(A), synj(B), geko(C), cdk4/6(D), and rab6(E) mRNA transcripts were detected in female (upper portion of each panel) and male (lower portion of each panel) wild-type and control-fed 24 h pupal brain hemisegments, which are oriented dorsal upward in each panel. The position of the optic lobe is marked by an asterisk in (A) and located at a similar position in the other panels. The sex-specific expression patterns of each gene were disrupted following injection of either dsx KD A or KD B siRNAs.
4 Discussion
4.1 Sex-specific gene expression in the developing A. aegypti pupal head
This investigation revealed global patterns of sex-specific gene expression in the A. aegypti pupal head transcriptome, which was assessed through custom A. aegypti microarray experiments (Additional file 1). These studies provided insight regarding the genes, pathways, and processes impacted by sex-specification in the 24 h pupal head transcriptome. The data collected may be of interest to those studying a number of tissues of vector importance, including the fat bodies, antennae, maxillary palp, and proboscis. However, the primary focus of the present study is the brain, and so the results of the microarray experiments were validated through in situ hybridization experiments performed on female and male pupal brain tissue (Figures 4, 5, 6, 7, 8, 9, 10). The 24 h pupal brain was selected, as our recent work has identified this time point to be a critical period for nervous system development. At this stage, which follows periods of extensive proliferative activity and pupal histolysis, the brain has an anatomical organization structure that is similar to that of the adult A. aegypti brain. The neuropils that are characteristic of the adult brain, including the antennal lobe, central complex, and three optic lobe neuropils, have begun to form. At 24 h APF, extensive neural process outgrowth, targeting of higher order brain neurons, synapse formation, and arborization occur, ultimately generating the increased neuropil density observed in the adult [6],[8],[23].
These studies detected sexually dimorphic expression of obp 10 and takeout through both head transcriptome microarray experiments and through analysis of transcript expression in the developing brain (Additional file 1, Figure 4A,D). Although expression of these genes had not previously been spatially assessed in the developing brain, dimorphic expression of both genes had been noted in A. aegypti by Bohbot and Vogt [31], who hypothesized that the genes might regulate differential olfactory and feeding behaviors in mosquitoes. It was subsequently demonstrated through knockdown experiments in Anopheles gambiae that Takeout regulates blood-feeding propensity [32]. Our own ability to detect dimorphic expression of these genes further validates the effectiveness of the methodological approach employed, while also raising the question of whether their dimorphic expression in the brain is functionally significant. This is a topic to be explored in future studies, which would be greatly enhanced through technical advancements that permit the manipulation of gene expression in a brain-specific manner in mosquitoes.
Components of critical developmental signaling cascades, including cubitus interruptus (ci), a member of the Hedgehog signaling pathway (reviewed by [33]), as well as arrow, a component of the Wnt/Wingless (Wg) signaling pathway [34], are upregulated in the developing female pupal brain (Figure 4B,E, Additional file 1). It will be interesting to functionally examine the roles of these genes during brain development, particularly given that Drosophila Ci has both transcriptional activation and transcriptional repressor forms (reviewed by [33]) that might differentially shape male and female development. Furthermore, although the function of Wg signaling in the regulation of sex-specific development has not yet been assessed in the insect nervous system, Wg signaling has been shown to regulate sexually dimorphic abdomen development in Drosophila. In flies, dimorphic Wg regulation in conjunction with monomorphic segment-specific cell death generates sex-specific abdomen shape [35].
More broadly, the results of this investigation suggest that underlying differences in proteolytic, metabolic, catabolic, and biosynthetic processes, as well as cell growth/proliferation and ion transport underlie differences in A. aegypti males vs. females (Tables 1, 2, 3, 4; Figures 2 and 3). Here, we discuss the potential relevance of these findings, focusing this discussion on development of the nervous system.
4.2 Proteases
Proteases, especially serine proteases, are significantly upregulated in female pupal heads (Figure 3, Table 2). These include 23 Clip-domain serine proteases that are significantly overexpressed in females. Clip-domain serine proteases have been well studied with regard to their multiple roles in insect immunity, including antimicrobial peptide synthesis, hemolymph coagulation, and melanization of pathogen surfaces [36]. In Anopheles gambiae, Clip-domain proteases are known for their function in the regulation of malaria parasite melanization [37]. In reference to insect development, two Clip-domain proteases function as components of the Toll pathway in the regulation of dorso-ventral axis specification (reviewed in [38]). Although Clip proteases have not been well studied in the context of the developing nervous system, the results of this investigation suggest that they are dimorphically expressed in female and male pupal heads. Other types of serine proteases have important CNS functions, including the regulation of cell migration, neurite outgrowth, and pathfinding, glial and neuronal cell survival, and synaptic remodeling [39]-[42]. Thus, it is possible that these Clip proteases might function as modifiers of the extracellular matrix in the context of neural migration.
4.3 Cell cycle/cell death
Cell cycle components (Figure 2A) were noted as a significant GO term among DETs upregulated in females (Table 1). For example, cdk4/6, a known regulator of cellular growth in D. melanogaster[43],[44], is upregulated in female pupal heads (Figures 4C and 11D). In flies, overexpression of Cyclin D-Cdk4/6 promotes cellular growth, and loss of cdk4 function results in flies with a smaller body size [43],[44]. It is possible that increased expression of cdk4 in A. aegypti females upregulates the growth of A. aegypti female pupae/heads with respect to males, which have a significantly smaller body and head (but not brain) size [18]. In relation to the developing nervous system, it has been shown that cdk4 expression is positively regulated in response to axon guidance gene signaling [45]. Thus, upregulation of cdk4 in the developing A. aegypti female brain might contribute to differential neural outgrowth. Furthermore, the microtubule-based movement GO term was significantly associated with DETs upregulated in females (Figure 2A; Table 1), suggesting that spindle fiber movement may be regulated in a sex-specific manner.
p53, a regulator of G1-S transition (reviewed by [46]), is also differentially expressed in pupal heads (Additional file 1), in which increased p53 expression can be detected in the male 24 h pupal brain (Figures 4I and 11A). In flies, p53 regulates apoptosis during development (reviewed by [46]). The regulation of cell death is a critical aspect of nervous system development which has also been linked to the development of sexual dimorphism in fruit flies (reviewed by [9]). For example, Dsx regulates death of the female TN1 neurons during Drosophila metamorphosis. In males, these neurons survive and are believed to function in the neural circuitry that is involved in production of the male courtship song (reviewed by [9]). These findings, in conjunction with the results of our investigation, suggest that regulation of p53 expression by Dsx may generate differences in programmed cell death that might shape the development of sex-specific traits in mosquitoes. In further support of this notion, caspase 7, an ortholog of Drosophila death caspase 1 and Ice, was also found to be expressed in a sex-specific manner (Figure 4F; Additional file 1). In addition to regulation of apoptosis, p53 has also been shown to regulate neurite outgrowth and axonal regeneration [47], suggesting that it may impact these processes in the mosquito brain. Finally, p53 has been linked to the regulation of life span in Drosophila[48]. Overexpression of p53 in the female nervous system results in increased life span in females. Thus, it is possible that female-specific upregulation of p53 expression in the A. aegypti female brain may contribute to the relatively longer lifespan of A. aegypti females with respect to males.
4.4 Ion transport
A wide variety of ion transporters are upregulated in male pupal heads (Figure 2B, Table 4), which suggests that sexually dimorphic differences in the developing mosquito brain may result from differential ion channel expression. As discussed by Yamamoto and Lopez-Bendito [49], the interaction of neural activity and genetic programs specifies neural circuit composition and organization during development. Changes in electrical activity regulate neural growth, axonogenesis, and the branching of axons. The sex-specific differences in ion transport-related genes noted here are therefore of potential interest with respect to the development of sexually dimorphic neural circuitries in mosquitoes. Furthermore, the activation and inactivation properties of ion channels in axons determine the short-term dynamics of axonal propagation and synaptic transmission [50].
Our microarray experiments detected upregulated male-specific expression of glutamate receptor 7 (Table 4), an ortholog of D. melanogaster ionotropic receptor (IR) 25a. In relation to this, glutamate metabolism was also noted as a significant pathway among DETs upregulated in males (Figure 3; Additional file 3). In Drosophila, IRs, which have evolved from ionotropic glutamate receptors (iGluRs), are expressed in a combinatorial fashion in sensory neurons and respond to many distinct odors [51]. Sensory neurons expressing IR84a in fruit flies innervate a glomerulus that expresses male-specific isoforms of the sex determination gene fruitless (fru), one of three sexually dimorphic glomeruli in the antennal lobe. Mutation of IR84a reduces male courtship (reviewed by [52],[53]). These observations suggest that differential expression of glutamate receptors and differential glutamate metabolism may impact sex-specific behaviors in mosquitoes.
4.5 Proteasome
DETs upregulated in females were significantly associated with the proteasome (Figure 3, Additional file 3), which is well known for its role in the intracellular degradation of ubiquitinated proteins. Protein synthesis and degradation are particularly important to neuronal development and function because of the large distances between synapses and soma. The ubiquitin proteasome system influences synaptic protein levels through the regulation of protein function and localization, as well as endocytosis (reviewed by [54]). It should be noted that synj (Figures 4G, 6C,G, 7G,H, 11B, Additional file 1), which also regulates endocytosis at the Drosophila synapse [55], is dimorphically expressed in the A. aegypti pupal brain. Elements of the ubiquitin proteasome system have also been found to regulate synaptic strength, homeostatic plasticity, axon growth and guidance, and dendrite morphogenesis (reviewed by [54]). Thus, differential expression of ubiquitin proteasome pathway components in female and male mosquitoes may have significant impacts on neural development and function that contribute to the development of sexually dimorphic behaviors.
4.6 Biosynthetic and catabolic processes
Significantly upregulated DETs in the female pupal head were also associated with biosynthetic and catabolic processes (Figure 3, Additional file 3). For example, glycosphingolipid biosynthesis is significantly over-represented among DETs upregulated in females. The functions of glycosphingolipids have been well studied in the context of development and neurobiology (reviewed by [56],[57]). For example, gangliosides, sialic acid-containing glycosphingolipids, are abundant in the nervous system, where they function in cell-cell recognition, adhesion, and signal transduction [57]. Work in Drosophila has shown that sphingolipid regulators affect cell survival, growth, and specification, as well as the control of lipid storage and responses to nutrient availability [56]. Thus, the differential regulation of sphingolipids in the developing male and female brain could have significant impact on the development of sex-specific behaviors in A. aegypti.
DETs significantly upregulated in female pupal heads are also associated with the glycosaminoglycan (GAG) degradation pathway (Figure 3, Additional file 3). In A. aegypti, cell surface GAGs have primarily been studied in relation to their potential roles as dengue virus receptors (reviewed by [58]). It will therefore be interesting to determine if there is differential GAG expression in other tissues, such as the midgut, a subject for future investigations. In reference to the brain, extracellular proteoglycans, which have one or more covalently attached GAGs, are known to impact many aspects of neural development and CNS maintenance. For example, during development, proteoglycans regulate cell adhesion, neurite formation, axon growth and guidance, and synapse formation. Proteoglycans have also been implicated in axon regeneration and sprouting following injury [59],[60]. Thus, differential GAG degradation could impact multiple aspects of neural development and function in A. aegypti females vs. males.
4.7 Dsx is a regulator of dimorphic gene expression in the A. aegypti pupal nervous system
Our search of the A. aegypti genome for the Luo et al. [28] Dsx consensus sequence uncovered 732 putative Dsx binding sites (Additional file 2), 48 of which correspond to genes identified in our microarray experiments (Additional file 3). Our search algorithm allowed for one mismatch in the consensus binding site (see Methods). This was useful to us since it permitted the most rigorous selection of putative targets prior to pursuing the dsx knockdown studies, which are labor-intensive. However, it is quite possible/probable that more mismatches are permissible and that additional binding sites are present in the genome. To more fully pursue the identification of Dsx binding sites throughout the genome, future studies might include a ‘wet lab’ approach, for example chromatin immunoprecipitation with anti-Dsx antibodies, or perhaps through use of the DamID approach utilized by Luo et al. (2011) to assess D. melanogaster Dsx binding sites.
Few of the targets identified in our study correspond to known Dsx transcriptional targets in D. melanogaster[28], suggesting that the targets of Dsx have evolved rapidly within the Diptera and highlighting the need to pursue analysis of dsx function in mosquitoes. To this end, we used siRNA-mediated gene targeting to examine the function of dsx in the developing pupal brain (Figure 11). These experiments indicated that Dsx is a key regulator of sex-specific gene expression during A. aegypti neural development. The detection of Dsx consensus binding sites associated with the p53, synj, geko, cdk4/6, and rab6 genes (Additional file 2), all of which have sexually dimorphic expression in the pupal brain (Additional file 1, Figures 4, 6, 7, 11) that is disrupted by dsx knockdown (Figure 11A-E), suggests that Dsx directly regulates expression of these genes. However, Dsx of course may function to regulate expression of these genes and its other targets through direct and/or indirect mechanisms, a question for future studies. Moreover, studies of the regulation of yolk protein genes, which are direct targets of Dsx in D. melanogaster, suggested that DsxF acts as a transcriptional activator, while DsxM functions as a repressor [61],[62]. Interestingly, none of our dsx knockdown experiments in A. aegypti resulted in upregulation of putative target gene expression in males or females (Figure 11), but it will be interesting to explore this subject further in future investigations.
The roles of dsx in development of the nervous system have been explored in D. melanogaster. In flies, Dsx regulates differences in programmed cell death, which is known to contribute to sexually dimorphic nervous system development [9]. It is therefore possible that control of p53 expression by Dsx in the A. aegypti CNS (Figure 11A) may be required for regulation of sex-specific cell death patterns in the mosquito brain, an interesting subject for future investigations. Dsx has also been associated with dimorphic cell proliferation in the Drosophila CNS, which is interesting in light of the observation that Dsx regulates expression of the cell cycle regulator cdk4/6 in the developing A. aegypti brain (Figure 11D). Moreover, Rideout et al. [63] detected sex-specific differences in the numbers, axonal projections, and synaptic density of Dsx-expressing neurons in Drosophila, and Dsx may regulate these aspects of neural development in mosquitoes. Finally, a recent study demonstrated that in female fruit flies, the neural circuitry associated with female post-mating behavior is specified by Dsx function. During copulation, this female circuitry senses male sex peptides and relays the signal to higher order circuits in the brain that generate post-mating physiological and behavioral output responses in females [64]. It will be interesting to determine if Dsx plays similar roles in mosquitoes.
We note that in comparison to D. melanogaster, in which sex-specific expression of dsx is detected in only several subsets of neurons [63],[65], dsx is expressed much more broadly within the A. aegypti female and male pupal brain (Figures 8A, 9, 10). For example, although dsx expression is not detected in the optic lobe of the D. melanogaster pupal brain, A. aegypti dsx is expressed in the male and female pupal optic lobes (Figures 8A, 9A,D, 10A,B). Moreover, we detect sex-specific differences in dsx expression in the antennal lobe (Figures 9C,F, 10C,D) and mushroom bodies (Figure 9A,D, 10E,F) of A. aegypti, differences which have not been reported in D. melanogaster pupae. These findings suggest that Dsx may play a prominent role in the regulation of sex-specific neural development in A. aegypti, in which its dimorphic expression pattern suggests that it may contribute to sex-specific differences in the visual and olfactory systems, the processing of sensory information, as well as learning and memory.
Previous studies have demonstrated that Dsx and Fru act in the same neurons to generate neuronal wiring and behaviors [63],[66],[67]. Fru targets, which were recently identified in D. melanogaster[68],[69], include many genes that regulate neural processes, including neurotransmission, ion-channel signaling, and neuron development. Furthermore, Neville et al. [68], who detected over-representation of known Dsx targets in their FruM target data set, speculate that Drosophila Dsx and Fru act together, either in a physical complex or through coregulation of target genes, to specify sex-specific neural development. Sex-specific Fru splice forms have been detected in A. aegypti[70], but the expression patterns of these transcripts have not yet been reported in the developing brain, and fru function has not been investigated in mosquitoes. In future studies, it will be interesting to functionally assess the roles of fru isoforms during A. aegypti female and male neural development.
4.8 Sexual dimorphism in the visual system
Differential expression of a number of the transcripts detected in the sex-specific pupal head transcriptome localized to the optic lobe of the brain (Figures 4 and 11), where high levels of the female and male dsx splice forms were also detected (Figures 8A, 9A,D, 10A,B). Moreover, many of the over-represented GO categories among genes with Dsx consensus binding sites are related to the compound eye or compound eye development (Table 5). Combined, these findings suggest that male and female compound eyes and higher order visual processing may differ in A. aegypti. Such differences could potentially contribute to female-specific behaviors, such as detection of blood meal hosts or oviposition sites. It is also possible that dependence on vision for mating success differs in males and females. This has been observed in D. melanogaster, in which males have a strong dependence on visual function for finding and tracking females, while female receptivity is based on responses to non-visual cues, such as male courtship song quality [71]. Exploration of these topics will prove interesting in future investigations.
Sexually dimorphic visual development has been particularly well-studied in stalk-eyed flies, which as a result of their elongated and sexually dimorphic eye-stalks are a model system for investigating sexual selection [72]. Wilkinson et al. [72] compared sex-specific global patterns of gene expression in Teleopsis dalmanni, in which they identified 415 female-biased and 482 male-biased transcripts associated with dimorphic eyestalk development. Many of the genes upregulated in developing female eyestalks are associated with cell differentiation and patterning, which was also the case for genes flanking Dsx consensus binding sites in A. aegypti (Table 5). DETs upregulated in males are disproportionately associated with Teleopsis dalmanni growth, which is to be expected given the male-specific eye stalk elongation observed in this species. Genes flanking Dsx consensus sites in A. aegypti (Additional file 2) are also significantly associated with growth (Table 5), and as discussed above, DETs upregulated in females are associated with the cell cycle (Figure 2). It is likely that these genes are associated with overgrowth of female tissues in A. aegypti given that female pupae of this species are larger than males. In support of this notion, cdk4, a positive regulator of cellular growth in D. melanogaster[43],[44], is upregulated in the A. aegypti female pupal brain.
The results of this investigation indicate that rab6, which is upregulated in females, is a target of Dsx (Figure 11E). In Drosophila, Rab6 functions as a GTP-binding protein regulator of vesicular trafficking that has been implicated in the transport of rhodopsin [73]. It will be interesting to determine if Rab6 expression is dimorphic in the eye and in adult A. aegypti, in which it could potentially regulate Rhodopsin transport. The A. aegypti genome project uncovered 10 Rhodopsins [1]. Based on studies in D. melanogaster, one would expect that changes in Rab6 levels would differentially impact transport of the 10 Rhodopsin proteins [73]. If this is the case in A. aegypti, differential Rab6-mediated transport of the 10 Rhodopsin proteins might be of significant consequence to male and female vision. This question, as well as functional analysis of many other DETs in the developing visual system, will be explored in future investigations.
5 Conclusions
This investigation revealed sex-specific gene expression profiles in the developing A. aegypti pupal head (Figure 1, Additional file 1) and identified Dsx as a key regulator of sexually dimorphic gene expression during mosquito CNS development (Figure 11). Analysis of DETs suggests that dimorphic expression of genes linked to proteolysis, the proteasome, metabolism, catabolic and biosynthetic processes, cell growth and proliferation, as well as ion transport underlie differences in developing A. aegypti males vs. females (Tables 1, 2, 3, 4, Figures 2 and 3, Additional files 1 and 3). In situ hybridization experiments for a subset of DETs in the pupal brain validated the data set and revealed that the differential expression of a number of the transcripts localized to the optic lobe of the brain (Figures 4 and 11). These results, combined with analysis of dsx expression in the developing pupal brain (Figures 8, 9, 10) and the detection of numerous compound eye and eye development GO categories among genes associated with Dsx consensus binding sites (Table 5; Additional file 2), indicate that male and female compound eyes and higher order visual processing may differ in A. aegypti. Sex-specific differences in gene expression also localized to the antennal lobe and mushroom body of the 24 h pupal brain (Figures 6 and 7), in which expression of dsx was also found to be sexually dimorphic (Figures 9 and 10). These findings suggest that Dsx may contribute to underlying differences in the visual and olfactory systems, the processing of sensory information, as well as learning and memory in A. aegypti females and males.
As discussed by Wilkinson et al. [72], although sex-biased gene expression has been assessed in several insects, few studies have compared global patterns of gene expression in developing somatic tissue with the goal of understanding a trait that is sexually dimorphic in adults. Their discussion highlights the need for studies like their sex-specific analysis of stalk-eyed fly eye development and our analysis of the A. aegypti female vs. male pupal head transcriptome. In addition to performing more studies of this type, it will be critical to functionally assess the roles of the dimorphically expressed genes identified in order to understand if and how these genes contribute to the development of sexually dimorphic traits. This is of course a large task that can be particularly daunting with respect to non-model organisms. However, siRNA-mediated knockdown approaches such as the one employed in this investigation are making this challenging task much more feasible.
Abbreviations
APF: after puparium formation
ci: cubitus interruptus
cdk4/6: cyclin-dependent kinase 4/6
DETs: differentially expressed transcripts
Dsx: Doublesex
DsxF: female-specific Dsx
Fru: Fruitless
GO: gene ontology
GAG: glycosaminoglycan
DsxM: male-specific Doublesex
IDT: Integrated DNA Technology
IGluR: ionotropic glutamate receptor
IR: ionotropic receptor
KD: knockdown
KEGG: Koyota Encyclopedia of Genes and Genomes
LVP-IB12: Liverpool-IB12
obp10: odorant binding protein 10
Synj: synaptojanin
Wg: Wingless
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MT, KM, PL, SE, DWS, and MDS conceived and designed the experiments. MT, LS, KM, PL, and SE performed the experiments. MT, PL, KM, DWS, and MDS analyzed the data. SE and DWS contributed reagents and analysis tools. MT, KM, SE, DWS, and MDS prepared the manuscript. All authors read and approved the final manuscript.
Additional files
Supplementary Material
Full list of significant DETs in the male vs. female 24 h pupal head. Four unique replicates (male 1, 2, 3, 4 and female 1, 2, 3, 4) as well as two repeat replicates (male 5, which is a repeat hybridization of male 1; male 6, a repeat hybridization of male 4; female 5, a repeat hybridization of female 1; female 6, a repeat hybridization of female 2) were assessed. The raw intensity values for each male or female replicate or repeat replicate are indicated. The mean male intensity, mean female intensity, log2 (male/female) fold change, and fold change (male/female) values are indicated for each probe. Raw p values adjusted using the Bonferroni procedure for control of false discovery rate are indicated (Bonferroni p value) for significant (p < 0.05) DETs. File format: Excel spreadsheet (XLSX).
List of Dsx consensus binding sites identified in theA. aegyptigenome. The locations of Dsx consensus binding sites, as well as the genes in which they are contained or flank are indicated. Genes corresponding to significant (p < 0.05) DETs in the 24 h female vs. male pupal head microarray data set are noted. File format: Excel spreadsheet (XLSX).
Genes corresponding to KEGG pathways that are significantly over-represented among DETs significantly upregulated in females or males. Gene numbers and descriptions for genes corresponding to each significant (p < 0.05) KEGG pathway are indicated. File format: Excel spreadsheet (XLSX).
Contributor Information
Michael Tomchaney, Email: mtomchan@alumni.nd.edu.
Keshava Mysore, Email: kmysore@iu.edu.
Longhua Sun, Email: Longhua.Sun.15@nd.edu.
Ping Li, Email: pl2@iupui.edu.
Scott J Emrich, Email: semrich@nd.edu.
David W Severson, Email: David.W.Severson.1@nd.edu.
Molly Duman-Scheel, Email: mscheel@nd.edu.
Acknowledgements
We thank the Notre Dame Genomics and Bioinformatics Core Facility for their assistance. Many thanks to Mayra Sandoval-Cooper for her help with sectioning and the Keck Center for Transgene Research for use of equipment. Confocal imaging was performed at the Indiana University School of Medicine, South Bend, IN, Imaging and Flow Cytometry Core Facility. Thanks to the members of the lab and the Eck Institute for Global Health, particularly Charles Tessier, Susanta Behura, Zain Syed, and Karen Cowden-Dahl for their advice. The dimorphism microarray study was funded by a University of Notre Dame Eck Institute for Global Health Pilot Study grant to M.D.S. Development and optimization of whole mount in situ hybridization and siRNA-mediated gene targeting in A. aegypti pupal brains was supported by NIH/NIAID Award R01-AI081795 to M.D.S.
References
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, Ren Q, Zdobnov EM, Lobo NF, Campbell KS, Brown SE, Bonaldo MF, Zhu J, Sinkins SP, Hogenkamp DG, Amedeo P, Arensburger P, Atkinson PW, Bidwell S, Biedler J, Birney E, Bruggner RV, Costas J, Coy MR, Crabtree J, Crawford M. et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316(5832):1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons A, Haugen M, Flannery E, Tomchaney M, Kast K, Jacowski C, Le C, Mori A, Simanton Holland W, Sarro J, Severson DW, Duman-Scheel M: Aedes aegypti:an emerging model for vector mosquito development.Cold Spring Harb Protoc 2010, 2010(10). pdb emo141. [DOI] [PMC free article] [PubMed]
- Clemons A, Haugen M, Le C, Mori A, Tomchaney M, Severson DW, Duman-Scheel M. siRNA-mediated gene targeting in Aedes aegypti embryos reveals that frazzled regulates vector mosquito CNS development. PLoS One. 2011;6(1):e16730. doi: 10.1371/journal.pone.0016730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen M, Flannery E, Tomchaney M, Mori A, Behura SK, Severson DW, Duman-Scheel M. Semaphorin-1a is required for Aedes aegypti embryonic nerve cord development. PLoS One. 2011;6(6):e21694. doi: 10.1371/journal.pone.0021694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C, Andrews E, Le C, Sun L, Annan Z, Clemons A, Severson DW, Duman-Scheel M. Functional genetic characterization of salivary gland development in Aedes aegypti. Evodevo. 2013;4(1):9. doi: 10.1186/2041-9139-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysore K, Flannery EM, Tomchaney M, Severson DW, Duman-Scheel M. Disruption of Aedes aegypti olfactory system development through chitosan/siRNA nanoparticle targeting of semaphorin-1a. PLoS Negl Trop Dis. 2013;7(5):e2215. doi: 10.1371/journal.pntd.0002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarro J, Andrews E, Sun L, Behura SK, Tan JC, Zeng E, Severson DW, Duman-Scheel M. Requirement for commissureless2 function during dipteran insect nerve cord development. Dev Dyn. 2013;242(12):1466–1477. doi: 10.1002/dvdy.24059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysore K, Andrews E, Li P, Duman-Scheel M. Chitosan/siRNA nanoparticle targeting demonstrates a requirement for single-minded during larval and pupal olfactory system development of the vector mosquito Aedes aegypti. BMC Dev Biol. 2014;14(1):9. doi: 10.1186/1471-213X-14-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K. Role of cell death in the formation of sexual dimorphism in the Drosophila central nervous system. Dev Growth Differ. 2011;53(2):236–244. doi: 10.1111/j.1440-169X.2010.01223.x. [DOI] [PubMed] [Google Scholar]
- Pitts RJ, Rinker DC, Jones PL, Rokas A, Zwiebel LJ. Transcriptome profiling of chemosensory appendages in the malaria vector Anopheles gambiae reveals tissue- and sex-specific signatures of odor coding. BMC Genomics. 2011;12:271. doi: 10.1186/1471-2164-12-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen AE, Keisman EL, Ahmad SM, Baker BS. Sex comes in from the cold: the integration of sex and pattern. Trends Genet. 2002;18(10):510–516. doi: 10.1016/S0168-9525(02)02769-5. [DOI] [PubMed] [Google Scholar]
- Camara N, Whitworth C, Van Doren M. The creation of sexual dimorphism in the Drosophila soma. Curr Top Dev Biol. 2008;83:65–107. doi: 10.1016/S0070-2153(08)00403-1. [DOI] [PubMed] [Google Scholar]
- Ryner LC, Goodwin SF, Castrillon DH, Anand A, Villella A, Baker BS, Hall JC, Taylor BJ, Wasserman SA. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell. 1996;87(6):1079–1089. doi: 10.1016/S0092-8674(00)81802-4. [DOI] [PubMed] [Google Scholar]
- Burtis KC, Baker BS. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989;56(6):997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- Behura SK, Haugen M, Flannery E, Sarro J, Tessier CR, Severson DW, Duman-Scheel M. Comparative genomic analysis of Drosophila melanogaster and vector mosquito developmental genes. PLoS One. 2011;6(7):e21504. doi: 10.1371/journal.pone.0021504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvemini M, Mauro U, Lombardo F, Milano A, Zazzaro V, Arca B, Polito LC, Saccone G. Genomic organization and splicing evolution of the doublesex gene, a Drosophila regulator of sexual differentiation, in the dengue and yellow fever mosquito Aedes aegypti. BMC Evol Biol. 2011;11:41. doi: 10.1186/1471-2148-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons A, Mori A, Haugen M, Severson DW, Duman-Scheel M: Culturing and egg collection ofAedes aegypti.Cold Spring Harb Protoc 2010, 2010(10). pdb prot5507. [DOI] [PMC free article] [PubMed]
- Christophers SR. Aedes aegypti, the Yellow Fever Mosquito: Its Life History, Bionomics, and Structure. Cambridge University Press, Cambridge, UK; 1960. [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Huang Da W, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, Lempicki RA: Extracting biological meaning from large gene lists with DAVID.Curr Protoc Bioinformatics 2009, 13(Unit 13 11). [DOI] [PubMed]
- Salomonis N, Hanspers K, Zambon AC, Vranizan K, Lawlor SC, Dahlquist KD, Doniger SW, Stuart J, Conklin BR, Pico AR. GenMAPP 2: new features and resources for pathway analysis. BMC Bioinformatics. 2007;8:217. doi: 10.1186/1471-2105-8-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen M, Tomchaney M, Kast K, Flannery E, Clemons A, Jacowski C, Simanton Holland W, Le C, Severson D, Duman-Scheel M. Whole-mount in situ hybridization for analysis of gene expression during Aedes aegypti development. Cold Spring Harb Protoc. 2010;2010(10):ᅟ. doi: 10.1101/pdb.prot5509. pdb prot5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysore K, Flister S, Muller P, Rodrigues V, Reichert H. Brain development in the yellow fever mosquito Aedes aegypti: a comparative immunocytochemical analysis using cross-reacting antibodies from Drosophila melanogaster. Dev Genes Evol. 2011;221(5–6):281–296. doi: 10.1007/s00427-011-0376-2. [DOI] [PubMed] [Google Scholar]
- Patel N. In situ hybridization to whole mount Drosophila embryos. Wiley-Liss, New York; 1996. [Google Scholar]
- Megy K, Emrich SJ, Lawson D, Campbell D, Dialynas E, Hughes DS, Koscielny G, Louis C, Maccallum RM, Redmond SN, Sheehan A, Topalis P, Wilson D. VectorBase: improvements to a bioinformatics resource for invertebrate vector genomics. Nucleic Acids Res. 2012;40(Database issue):D729–D734. doi: 10.1093/nar/gkr1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobysheva D, Ameel K, Welch B, Ellison E, Chaichana K, Hoang B, Sharma S, Neckameyer W, Srinakevitch I, Murphy KJ, Schmid A. An optimized method for histological detection of dopaminergic neurons in Drosophila melanogaster. J Histochem Cytochem. 2008;56(12):1049–1063. doi: 10.1369/jhc.2008.951137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons A, Flannery E, Kast K, Severson D, Duman-Scheel M: Immunohistochemical analysis of protein expression duringAedes aegyptidevelopment.Cold Spring Harb Protoc 2010, 2010(10). pdb prot5510. [DOI] [PMC free article] [PubMed]
- Luo SD, Shi GW, Baker BS. Direct targets of the D. melanogaster DSXF protein and the evolution of sexual development. Development. 2011;138(13):2761–2771. doi: 10.1242/dev.065227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons A, Haugen M, Severson D, Duman-Scheel M: Functional analysis of genes inAedes aegyptiembryos.Cold Spring Harb Protoc 2010, 2010(10). pdb prot5511. [DOI] [PMC free article] [PubMed]
- Blitzer EJ, Vyazunova I, Lan Q. Functional analysis of AeSCP-2 using gene expression knockdown in the yellow fever mosquito, Aedes aegypti. Insect Mol Biol. 2005;14(3):301–307. doi: 10.1111/j.1365-2583.2005.00560.x. [DOI] [PubMed] [Google Scholar]
- Bohbot J, Vogt RG. Antennal expressed genes of the yellow fever mosquito (Aedes aegypti L.); characterization of odorant-binding protein 10 and takeout. Insect Biochem Mol Biol. 2005;35(9):961–979. doi: 10.1016/j.ibmb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Das S, Dimopoulos G. Molecular analysis of photic inhibition of blood-feeding in Anopheles gambiae. BMC Physiol. 2008;8:23. doi: 10.1186/1472-6793-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aza-Blanc P, Kornberg TB. Ci: a complex transducer of the hedgehog signal. Trends Genet. 1999;15(11):458–462. doi: 10.1016/S0168-9525(99)01869-7. [DOI] [PubMed] [Google Scholar]
- Tolwinski NS, Wehrli M, Rives A, Erdeniz N, DiNardo S, Wieschaus E. Wg/Wnt signal can be transmitted through arrow/LRP5,6 and Axin independently of Zw3/Gsk3beta activity. Dev Cell. 2003;4(3):407–418. doi: 10.1016/S1534-5807(03)00063-7. [DOI] [PubMed] [Google Scholar]
- Wang W, Kidd BJ, Carroll SB, Yoder JH. Sexually dimorphic regulation of the Wingless morphogen controls sex-specific segment number in Drosophila. Proc Natl Acad Sci U S A. 2011;108(27):11139–11144. doi: 10.1073/pnas.1108431108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman MJ, Paskewitz SM. Serine proteases as mediators of mosquito immune responses. Insect Biochem Mol Biol. 2001;31(3):257–262. doi: 10.1016/S0965-1748(00)00145-4. [DOI] [PubMed] [Google Scholar]
- Barillas-Mury C. CLIP proteases and Plasmodium melanization in Anopheles gambiae. Trends Parasitol. 2007;23(7):297–299. doi: 10.1016/j.pt.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Jang IH, Nam HJ, Lee WJ. CLIP-domain serine proteases in Drosophila innate immunity. BMB Rep. 2008;41(2):102–107. doi: 10.5483/BMBRep.2008.41.2.102. [DOI] [PubMed] [Google Scholar]
- Monard D. Cell-derived proteases and protease inhibitors as regulators of neurite outgrowth. Trends Neurosci. 1988;11(12):541–544. doi: 10.1016/0166-2236(88)90182-8. [DOI] [PubMed] [Google Scholar]
- Seeds NW, Haffke S, Christensen K, Schoonmaker J. Cerebellar granule cell migration involves proteolysis. Adv Exp Med Biol. 1990;265:169–178. doi: 10.1007/978-1-4757-5876-4_16. [DOI] [PubMed] [Google Scholar]
- Liu Y, Fields RD, Fitzgerald S, Festoff BW, Nelson PG. Proteolytic activity, synapse elimination, and the Hebb synapse. J Neurobiol. 1994;25(3):325–335. doi: 10.1002/neu.480250312. [DOI] [PubMed] [Google Scholar]
- Houenou LJ, Turner PL, Li L, Oppenheim RW, Festoff BW. A serine protease inhibitor, protease nexin I, rescues motoneurons from naturally occurring and axotomy-induced cell death. Proc Natl Acad Sci U S A. 1995;92(3):895–899. doi: 10.1073/pnas.92.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datar SA, Jacobs HW, de la Cruz AF, Lehner CF, Edgar BA. The Drosophila cyclin D-Cdk4 complex promotes cellular growth. EMBO J. 2000;19(17):4543–4554. doi: 10.1093/emboj/19.17.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer CA, Jacobs HW, Datar SA, Du W, Edgar BA, Lehner CF. Drosophila Cdk4 is required for normal growth and is dispensable for cell cycle progression. EMBO J. 2000;19(17):4533–4542. doi: 10.1093/emboj/19.17.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannery E, VanZomeren-Dohm A, Beach P, Holland WS, Duman-Scheel M. Induction of cellular growth by the axon guidance regulators netrin A and semaphorin-1a. Dev Neurobiol. 2010;70(7):473–484. doi: 10.1002/dneu.20788. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JE, Korenjak M, Brehm A. Tumour suppressors–a fly's perspective. Eur J Cancer. 2003;39(10):1355–1362. doi: 10.1016/S0959-8049(03)00263-6. [DOI] [PubMed] [Google Scholar]
- Di Giovanni S, Knights CD, Rao M, Yakovlev A, Beers J, Catania J, Avantaggiati ML, Faden AI. The tumor suppressor protein p53 is required for neurite outgrowth and axon regeneration. EMBO J. 2006;25(17):4084–4096. doi: 10.1038/sj.emboj.7601292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Tower J. Drosophila foxo acts in males to cause sexual-dimorphism in tissue-specific p53 life span effects. Exp Gerontol. 2010;45(2):97–105. doi: 10.1016/j.exger.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N, Lopez-Bendito G. Shaping brain connections through spontaneous neural activity. Eur J Neurosci. 2012;35(10):1595–1604. doi: 10.1111/j.1460-9568.2012.08101.x. [DOI] [PubMed] [Google Scholar]
- Debanne D, Daoudal G, Sourdet V, Russier M. Brain plasticity and ion channels. J Physiol Paris. 2003;97(4–6):403–414. doi: 10.1016/j.jphysparis.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136(1):149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson BJ. Wired for sex: the neurobiology of Drosophila mating decisions. Science. 2008;322(5903):904–909. doi: 10.1126/science.1159276. [DOI] [PubMed] [Google Scholar]
- Rytz R, Croset V, Benton R. Ionotropic receptors (IRs): chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem Mol Biol. 2013;43(9):888–897. doi: 10.1016/j.ibmb.2013.02.007. [DOI] [PubMed] [Google Scholar]
- Hamilton AM, Zito K. Breaking it down: the ubiquitin proteasome system in neuronal morphogenesis. Neural Plast. 2013;2013:196848. doi: 10.1155/2013/196848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstreken P, Koh TW, Schulze KL, Zhai RG, Hiesinger PR, Zhou Y, Mehta SQ, Cao Y, Roos J, Bellen HJ. Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron. 2003;40(4):733–748. doi: 10.1016/S0896-6273(03)00644-5. [DOI] [PubMed] [Google Scholar]
- Kraut R. Roles of sphingolipids in Drosophila development and disease. J Neurochem. 2011;116(5):764–778. doi: 10.1111/j.1471-4159.2010.07022.x. [DOI] [PubMed] [Google Scholar]
- Yu RK, Tsai YT, Ariga T. Functional roles of gangliosides in neurodevelopment: an overview of recent advances. Neurochem Res. 2012;37(6):1230–1244. doi: 10.1007/s11064-012-0744-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidari KI, Abe T, Suzuki T. Carbohydrate-related inhibitors of dengue virus entry. Viruses. 2013;5(2):605–618. doi: 10.3390/v5020605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Freeman C, Jacobson GA, Small DH. Proteoglycans in the central nervous system: role in development, neural repair, and Alzheimer's disease. IUBMB Life. 2013;65(2):108–120. doi: 10.1002/iub.1118. [DOI] [PubMed] [Google Scholar]
- Song YS, Kim E. Presynaptic proteoglycans: sweet organizers of synapse development. Neuron. 2013;79(4):609–611. doi: 10.1016/j.neuron.2013.07.048. [DOI] [PubMed] [Google Scholar]
- Coschigano KT, Wensink PC. Sex-specific transcriptional regulation by the male and female doublesex proteins of Drosophila. Genes Dev. 1993;7(1):42–54. doi: 10.1101/gad.7.1.42. [DOI] [PubMed] [Google Scholar]
- An W, Wensink PC. Integrating sex- and tissue-specific regulation within a single Drosophila enhancer. Genes Dev. 1995;9(2):256–266. doi: 10.1101/gad.9.2.256. [DOI] [PubMed] [Google Scholar]
- Rideout EJ, Dornan AJ, Neville MC, Eadie S, Goodwin SF. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat Neurosci. 2010;13(4):458–466. doi: 10.1038/nn.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaval C, Pavlou HJ, Dornan AJ, Chan YB, Kravitz EA, Goodwin SF. Neural circuitry underlying Drosophila female postmating behavioral responses. Curr Biol. 2012;22(13):1155–1165. doi: 10.1016/j.cub.2012.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Hall JC, Park JH. Doublesex gene expression in the central nervous system of Drosophila melanogaster. J Neurogenet. 2002;16(4):229–248. doi: 10.1080/01677060216292. [DOI] [PubMed] [Google Scholar]
- Rideout EJ, Billeter JC, Goodwin SF. The sex-determination genes fruitless and doublesex specify a neural substrate required for courtship song. Curr Biol. 2007;17(17):1473–1478. doi: 10.1016/j.cub.2007.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Hachiya T, Koganezawa M, Tazawa T, Yamamoto D. Fruitless and doublesex coordinate to generate male-specific neurons that can initiate courtship. Neuron. 2008;59(5):759–769. doi: 10.1016/j.neuron.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Neville MC, Nojima T, Ashley E, Parker DJ, Walker J, Southall T, Van de Sande B, Marques AC, Fischer B, Brand AH, Russell S, Ritchie MG, Aerts S, Goodwin SF. Male-specific fruitless isoforms target neurodevelopmental genes to specify a sexually dimorphic nervous system. Curr Biol. 2014;24(3):229–241. doi: 10.1016/j.cub.2013.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernes SC. Genome wide identification of Fruitless targets suggests a role in upregulating genes important for neural circuit formation. Sci Rep. 2014;4:4412. doi: 10.1038/srep04412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvemini M, D'Amato R, Petrella V, Aceto S, Nimmo D, Neira M, Alphey L, Polito LC, Saccone G. The orthologue of the fruitfly sex behaviour gene fruitless in the mosquito Aedes aegypti: evolution of genomic organisation and alternative splicing. PLoS One. 2013;8(2):e48554. doi: 10.1371/journal.pone.0048554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn WG, Greenspan RJ. Learning and courtship in Drosophila: two stories with mutants. Annu Rev Neurosci. 1984;7:67–93. doi: 10.1146/annurev.ne.07.030184.000435. [DOI] [PubMed] [Google Scholar]
- Wilkinson GS, Johns PM, Metheny JD, Baker RH. Sex-biased gene expression during head development in a sexually dimorphic stalk-eyed fly. PLoS One. 2013;8(3):e59826. doi: 10.1371/journal.pone.0059826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty KM, Kurada P, O'Tousa JE. Rab6 regulation of rhodopsin transport in Drosophila. J Biol Chem. 1998;273(32):20425–20430. doi: 10.1074/jbc.273.32.20425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full list of significant DETs in the male vs. female 24 h pupal head. Four unique replicates (male 1, 2, 3, 4 and female 1, 2, 3, 4) as well as two repeat replicates (male 5, which is a repeat hybridization of male 1; male 6, a repeat hybridization of male 4; female 5, a repeat hybridization of female 1; female 6, a repeat hybridization of female 2) were assessed. The raw intensity values for each male or female replicate or repeat replicate are indicated. The mean male intensity, mean female intensity, log2 (male/female) fold change, and fold change (male/female) values are indicated for each probe. Raw p values adjusted using the Bonferroni procedure for control of false discovery rate are indicated (Bonferroni p value) for significant (p < 0.05) DETs. File format: Excel spreadsheet (XLSX).
List of Dsx consensus binding sites identified in theA. aegyptigenome. The locations of Dsx consensus binding sites, as well as the genes in which they are contained or flank are indicated. Genes corresponding to significant (p < 0.05) DETs in the 24 h female vs. male pupal head microarray data set are noted. File format: Excel spreadsheet (XLSX).
Genes corresponding to KEGG pathways that are significantly over-represented among DETs significantly upregulated in females or males. Gene numbers and descriptions for genes corresponding to each significant (p < 0.05) KEGG pathway are indicated. File format: Excel spreadsheet (XLSX).


