Abstract
The timing of cold acclimation and deacclimation, dormancy, and budbreak play an integral role in the life cycle of woody plants. The molecular events that regulate these parameters have been the subject of much study, however, in most studies these events have been investigated independently of each other. Ectopic expression of a peach CBF (PpCBF1) in apple increases the level of both non-acclimated and acclimated freezing tolerance relative to the non-transformed control, and also inhibits growth, induces early bud set and leaf senescence, and delays bud break in the spring. The current study examined differences in the seasonal expression of genes (CBF, DAM, RGL, and EBB) that have been reported to be associated with freezing tolerance, dormancy, growth, and bud break, respectively, in the PpCBF1 T166 transgenic apple line and the non-transformed M.26 control. Results indicated that expression of several of these key genes, including MdDAM, MdRGL, and MdEBB was altered in transgenic T166 trees relative to non-transformed M.26 trees. In particular, several putative MdDAM genes, associated with the dormancy-cycle in other species of woody plants in the Rosaceae, exhibited different patterns of expression in the T166 vs. M.26 trees. Additionally, for the first time a putative APETALA2/Ethylene-responsive transcription factor, originally described in poplar and shown to regulate the timing of bud break, was shown to be associated with the timing of bud break in apple. Since the overexpression of PpCBF1 in apple results in a dramatic alteration in cold acclimation, dormancy, and growth, this transgenic line (T166) may represent a useful model for studying the integration of these seasonal life-cycle parameters.
Keywords: freezing tolerance, fruit trees, DAM genes, CBF genes, bud break, DELLA genes, EBB genes, Malus × domestica
Introduction
The timing of cold acclimation and deacclimation, the onset and release from dormancy, as well as the timing of bud break play an integral role in the life cycle of woody plants and their adaptation to the external environment (Rios et al., 2014). These parameters are especially important for perennial fruit crops where the timing and regulation of these events play a critical role in the selection of cultivars that are appropriate for use in a specific growing region and where the inability of a cultivar to adequately respond to regional environmental conditions can result in the complete loss of a harvestable fruit crop or an entire planting. The molecular events that regulate these parameters have been the subject of much study and numerous reviews on these topics have been published (Rohde et al., 2007; Ruttink et al., 2007; Hänninen and Tanino, 2011; Cooke et al., 2012; Rios et al., 2014). In most studies, however, these events have been investigated independently of each other, with the focus being on either dormancy, cold acclimation, or growth regulation, and the systems used have been advantageous for providing information specific to one of these events. While this approach has provided a wealth of essential information, it is also important to note that these events do not occur in isolation but rather as an integrated part of the entire life cycle of the plant species being investigated. For instance, it has been well established that woody plants cannot cold acclimate to their full potential unless they are dormant and that it is difficult for plants to reacclimate to any substantial degree once the plant is released from endodormancy. The loss of freezing tolerance is also intimately associated with the onset of growth (bud break), a parameter that lies outside the realm of endodormancy and is regulated by the accumulation of heat units, a requirement which is determined by an interaction between the genetics of a species and the environment. Epigenetics, in addition to genetic makeup, has also been recognized to play a significant role in the regulation of these phenological events (Leida et al., 2012; Rios et al., 2014), further adding to the complexity.
Several specific transcription factors and regulatory genes have been demonstrated to play an important role in one or more of the processes mentioned above. CBF genes have been shown to play an integral role in the induction of freezing tolerance of both herbaceous and woody plants (Gilmour et al., 1998, 2000; Thomashow et al., 2001; Welling and Palva, 2006; Wisniewski et al., 2014). The regulation of freezing tolerance by CBF in woody plants, however, appears to be more complex than in herbaceous plants and the role of specific CBF genes can also vary (Benedict et al., 2006; Xiao et al., 2006; Welling and Palva, 2008; Wisniewski et al., 2014). Additionally, CBF expression can alter parameters other than freezing tolerance, such as growth and the timing and development of flowering (Achard et al., 2008). Collectively, these reports add to the complexity of the regulation of freezing tolerance by CBF genes and also suggest that integration, cross-talk, and some degree of overlap may exist in the regulation of key developmental aspects of plants.
A variety of genetic components that contribute to the intricate regulation of dormancy have been reported (Rohde and Bhalerao, 2007; Ruttink et al., 2007; Cooke et al., 2012). Among these regulatory components, DAM (Dormancy Associated MADS-box) genes have been reported to play an intimate role in controlling dormancy in fruit trees within the Rosaceae, and other plant species (Bielenberg et al., 2008; Li et al., 2009; Horvath et al., 2010; Falavigna et al., 2014). In particular, DAM5 and DAM6 have been highlighted as being associated with the onset and release of dormancy in peach (Prunus persica L. Batsch) (Yamane et al., 2011). A natural mutation of one of the DAM genes in peach has resulted in a non-dormant evergreen peach (Rodriguez et al., 1994; Bielenberg et al., 2008). Several DAM genes have also been associated with the dormancy in apricot, Prunus mume (Sasaki et al., 2011) Epigenetic regulation of DAM genes has been reported (Leida et al., 2012). The role of DAM genes in the regulation of dormancy, however, does not appear to be universal (Rios et al., 2014). For example, DAM genes do not map to the QTL associated with dormancy in Rohde et al. (2011). It is also important to note that genes directly regulating endormancy may not be the same genetic components regulating time to bud break, once plants have acquired sufficient chill units and become ecodormant. In this regard, Yordanov et al. (2014) have recently reported that the expression of an early bud-break1 (EBB1) gene in poplar plays a major role in regulating the timing of bud break.
RGL genes, which code for DELLA proteins, act to restrain growth, whereas GA promotes growth by overcoming DELLA-mediated growth restraint (Achard and Genschik, 2009; Claeys et al., 2014). CBF genes have been reported to influence the expression of RGL genes and this interrelationship has been used to explain the impact of CBF genes on growth (Achard et al., 2008). Thus, the interaction of CBF with RGL genes may play a role in the interaction between growth and deacclimation. While plausible, this still needs to be demonstrated and how one process (cold acclimation vs. growth) becomes dominant still remains to be explored.
Wisniewski et al. (2011) reported that the ectopic expression of a peach CBF (PpCBF1) in apple not only increased the level of both non-acclimated and acclimated freezing tolerance in the transgenic apple (T166), relative to the non-transformed control (M.26), but also inhibited growth, and surprisingly rendered the T166 plants sensitive to short day (SD) photoperiod, and induced early leaf senescence and bud set, again relative to the non-transformed M.26 plants. The observed sensitivity to SD was novel and unexpected since apples are typically not sensitive SD in terms of inducing growth cessation (Heide and Prestrud, 2005). Three years of field studies with the T166 plants further confirmed that, relative to the control, the transgenic apple line had increased level of freezing tolerance, reduced growth (current year and main stem diameter growth), set bud earlier, experienced earlier leaf senescence, and later bud break in the spring (Artlip et al., 2014). Thus, this transgenic line may serve as a model for studying the integration of the regulation of freezing tolerance, dormancy, bud break, and growth in woody plants. The current study examined differences in the seasonal expression of genes (CBF, DAM, RGL, and EBB) that have been reported to be associated with freezing tolerance, dormancy, bud break, and growth in the T166 line of transgenic apple and its non-transformed M.26 control. The purpose of the present study was to characterize the expression of all five different apple CBF genes in response to a cold acclimating conditions and to determine if the overexpression of CBF also modified the expression of genes that have been reported in the literature to modify dormancy, bud break, and growth.
Materials and methods
Plant material
“Malling 26” (M.26) is a standard dwarfing apple rootstock. T166 (PpCBF1) transgenic line was initially described by Wisniewski et al. (2011). Briefly, M.26 leaves underwent Agrobacterium-mediated transformation with a vector consisting of a pBINPLUSARS (Belknap et al., 2008) backbone and the peach (Prunus persica) PpCBF1 gene driven by a dual 35 s enhancer segment derived from pRTL2 (Restrepo et al., 1990). Plants were maintained in tissue culture, roots initiated, plantlets grown successively in growth chambers and greenhouse, before being planted in October, 2010 at the Appalachian Fruit Research Station, USDA-ARS, Kearneysville, WV per Artlip et al. (2014). A commercial scion cultivar, “CrimsonCrisp,” was used in a limited fashion to compare expression of RGL genes in a scion variety vs. the non-transformed (M.26) and transformed (T166) rootstock cultivar. The trees were also located on the grounds of the research station and were planted 07 May, 2007 on Bud-9 rootstock and subjected to conventional management practices.
Growth and phenology of field-grown M.26 and T166 trees
Growth measurements were taken monthly during the growing season (March to November) during 2013. Caliper (stem diameter) data were taken at a point 30 cm above the ground on the main stem, and current season's shoot lengths were taken from the terminal bud scar of the previous year's growth to the tip of the main stem; cumulative data are presented. Dates of bud break for each tree were recorded during spring 2013. Percent bud break was determined as follows: three shoots on each of three trees of M.26 and T166 were tagged and bud break from 20 individual lateral buds from the terminal bud were tracked. The range of dates of leaf loss were recorded in autumn, 2012. Two-sample independent t-tests were used to determine significance between the calculated means. For the current year shoot data, n = 10 for the transgenic T166 line and n = 7 for the non-transformed M.26 trees. For the stem caliper data, n = 13 for the transgenic T166 line and n = 7 for the non-transformed M.26 trees. Differences in sample number between the lines were due to different numbers of trees in the original planting and the loss of some trees, mainly M.26, over the last 4 years.
Tissue collection
Small branches were removed from the trees for bark or axillary bud collection. Bark tissue (phloem, cambium and epidermis) was destructively sampled from M.26 and T166 trees on a monthly basis in 2013. Axillary buds were collected bi-weekly from M.26 and T166 trees from January through April, 2013. The tissues were flash-frozen in liquid N2, lyophilized, and stored at −20°C until use.
M.26 cold acclimation experiment
One-year-old M.26 trees were propagated in tissue culture, rooted, and grown in a glass house as per Wisniewski et al. (2011). Ten tress were transferred to a PGV36 growth chamber (Conviron, Winnipeg, MN, Canada) at 4°C, 200 μmole photons M−2 s−1, 8 h light/16 h dark for cold treatment and acclimation. Leaves were removed from three trees each at 0, 15, 30 min, 1, 2, 4, 8, 12, 24, 48, 96 h, 1 and 3 weeks. The leaves were immediately flash frozen in liquid N2 and stored at −80°C until use.
RT-qPCR
Total RNA was isolated from leaf and bark tissues using Concert Plant RNA Reagent (Invitrogen, Carlsbad, CA, USA), treated with DNase (Turbo DNA-free Kit; Ambion, Austin, TX, USA) and then were diluted based on preliminary testing for optimal response. Reverse transcriptase, quantitative polymerase chain reaction (RT-qPCR) analysis was performed using appropriate quantities of total RNA (per preliminary testing) as a template with the Power SYBR Green RNA-to-Ct 1-Step Kit (Applied Biosystems, Foster City, CA, USA) and 2.0 pmol of each primer per reaction; no-RT control reactions were included to test for residual DNA contamination. A ViiA 7 Real-Time PCR System (Applied Biosystems) was set to cycle as follows: cDNA synthesis at 48.0°C for 30 min; 95.0°C denaturation for 10 min; 40 cycles of 95.0°C for 15 s followed by 52.0–57.0°C (depending on primers used; Table S1) for 1 min; followed by ABI-specified hold and melt curve stages. Primers were verified for specificity by using genomic DNA template and assessing the resulting amplicon by agarose gel electrophoresis and qPCR with genomic DNA on the ViiA 7 Real-Time PCR System; all primers had a single band and single peak. Primer efficiency was also verified for all primer sets by qPCR analysis of a standard curve, constructed by serially diluting RNAs from the sample set starting at some concentration above what was used in unknown samples and ending at a concentration well below it. Three technical replicates were used for each biological replicate (tree, N = 3). The standard curve method was used to calculate transcript abundance relative to EF1-α as a reference gene (user bulletin no. 2; Applied Biosystems http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_040980.pdf). Other endogenous reference genes were also examined, but EF1-α was determined to be the best overall reference gene using NormFinder (Anderson et al., 2004). Normalized data were then re-normalized to the respective values at time 0, and the means taken from the biological replicates. Standard errors (SEs) were derived by dividing the standard deviations by the square root of n, where n = 9 (3 biological replicates × 3 technical replicates).
Bioinformatic analyses
In order to identify putative gene families, apple CBF (Wisniewski et al., 2014), peach (Li et al., 2009), and pear (Saito et al., 2013) DAM, and poplar (Yordanov et al., 2014) EBB genes were used as queries in BLASTn (Thompson et al., 1994) analyses of the Malus × domestica genome v 1.0 at the Genome Database for Rosaceae (GDR; http://www.rosaceae.org). Sequences for apple DELLA(RGL) were based on the report by Foster et al. (2007). In order to identify cis-regulatory elements within putative promoter regions, in silico analysis of the 5′-UTRs (up to 1000 bp upstream of the putative translational start site) was conducted using PAN (http://plantpan.mbc.nctu.edu.tw/gene_group/index.php) (Chang et al., 2008), PLACE (http://www.dna.affrc.go.jp/PLACE/) (Higo et al., 1999), and PLANTCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002).
Results
Phenology and growth
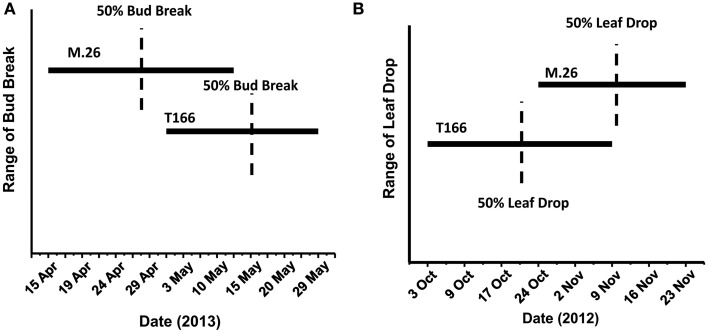
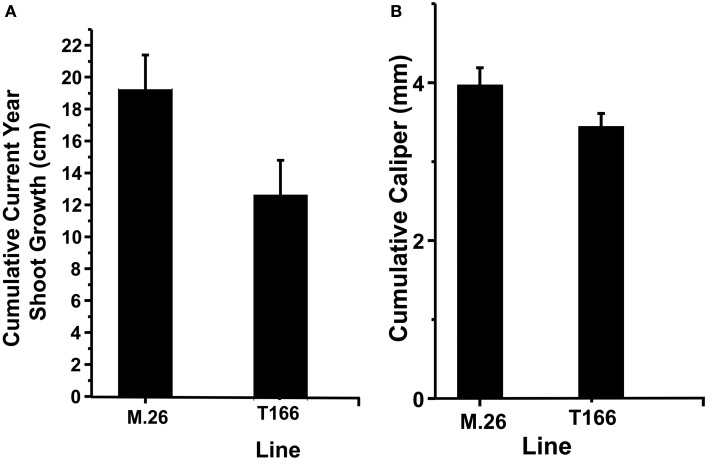
As documented in a recent study (Artlip et al., 2014), field-planted, transgenic apple trees (T166) overexpressing a peach (Prunus persica L. Batsch.) CBF gene continued to exhibit delayed bud break and early senescence relative to the non-transformed, parent clone M.26 (Figure 1). The difference in the time of bud break and the onset of leaf senescence was very prominent between the two lines, being offset by approximately 2 weeks (Figure 2). Both current-year shoot growth (extension growth) and stem diameter (caliper growth) were reduced in T166 trees (Figure 3), as previously documented (Wisniewski et al., 2011; Artlip et al., 2014). Additionally, T166 trees typically had fewer lateral branches. The impact of the differences in growth between M.26 and T166 trees accumulated over several years resulting in T166 trees that were much smaller than the non-transformed M.26 trees (Figure 4). Average height for the T166 and M. 26 trees was 130 and 190 cm, respectively. This observation is significant since M.26 is known to be a dwarfing rootstock.
Figure 1.
M26 trees break bud earlier and enter dormancy later than T166 trees. (A) Bud break. Photographs were taken in spring, 2012. (B–D) Leaf senescence and dormancy. Photographs were taken in September, October, and November and illustrate that Line T166 trees enter dormancy sooner as evidenced by leaf senescence and leaf drop occurring sooner than non-transformed M.26 trees.
Figure 2.
Phenological disparities between non-transformed M.26 and Line T166. (A) Bud break. Bud break was quantitatively assessed as the percent of 20 lateral buds from the terminal bud on three shoots and qualitatively assessed as the dates on which the terminal buds on these shoots broke dormancy. (B) Leaf drop. A quantitative assessment of percent leaf loss was made by estimating leaf loss over the entire tree compared to early September, when no leaf loss was evident, qualitatively assessed by date range when leaf loss was observed.
Figure 3.
T166 trees display reduced growth compared to M.26. (A) Cumulative current year shoot growth. Growth was measured on the central axis from the previous season bud scar to the current terminal bud. The cumulative current year shoot growth was significantly different between the genotypes at the 0.05 level as assessed by a two-sample independent t-test. (B) Cumulative caliper (stem diameter) growth as measured 30 cm from the ground. The cumulative caliper growth was significantly different between the genotypes at the 0.05 level as assessed by a two-sample independent t-test.
Figure 4.
Representative photographs of growth disparities between non-transformed M.26 and Line T166. Non-transformed M.26 trees were 190 ± 10.2 cm overall height (n = 7 trees) while Line T166 trees were 130 ± 3.2 cm overall height (n = 12 trees).
Expression of apple CBF genes in response to cold acclimation
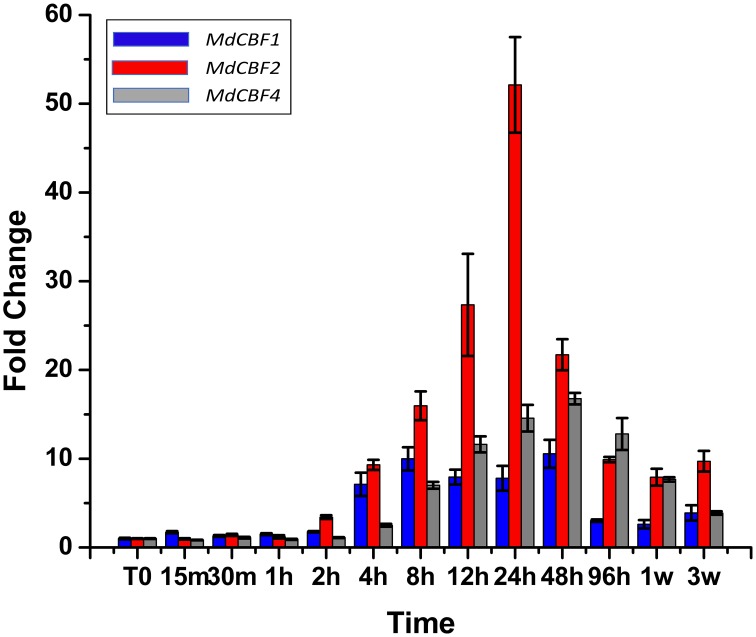
To date, the presence of five CBF genes (MdCBFs1-5) have been documented in the apple genome (Wisniewski et al., 2014; Figure S1). Previous research found that transcript abundance of two apple CBF genes (MdCBF1 and MdCBF2) normally induced in response to low temperature are unaffected by ectopic expression of PpCBF1 (Wisniewski et al., 2011). In the present analysis, an attempt was made to characterize the response of all five apple CBF genes to low temperatures over a short (96 h) and an extended (3 week) period of time (Figure 5) in non-transformed trees. Despite the use of numerous different primers (Table S1) and protocol adjustments, expression of MdCBF3 and MdCBF5 in response to low temperature (4°C, short day photoperiod) could not be documented. Only MdCBF1, MdCBF2, and MdCBF4 appeared to be responsive to low temperature with MdCBF2 exhibiting by far the strongest response as measured by fold change (Figure 5). A measurable induction was observed for MdCBF1 and MdCBF2 within 2 h after exposure of the M.26 trees to low temperature and peaked at 24 h. The pattern of induction of MdCBF4 was slightly offset from the expression of the other two CBF genes. MdCBF4 was induced after 4 h and appeared to peak at 48 h. After 24 h (MdCBF1 and MdCBF2) or 48 h (MdCBF4) expression levels of all three induced MdCBF genes began to decrease, however, a measurable level of induction, relative to time 0, was still observable after 3 weeks.
Figure 5.
MdCBF transcript accumulation kinetics during cold-shock and cold-acclimation vary between genes. Non-transformed M.26 trees were shifted to 4°C, 8 h light/16 h dark conditions, and leaves harvested at short time intervals (hours to days) or longer time intervals (weeks). MdCBF1, 2, and 4 are shown; MdCBF3 and 5 could not be detected by the primer sets used.
Dormancy and bud break candidate gene expression
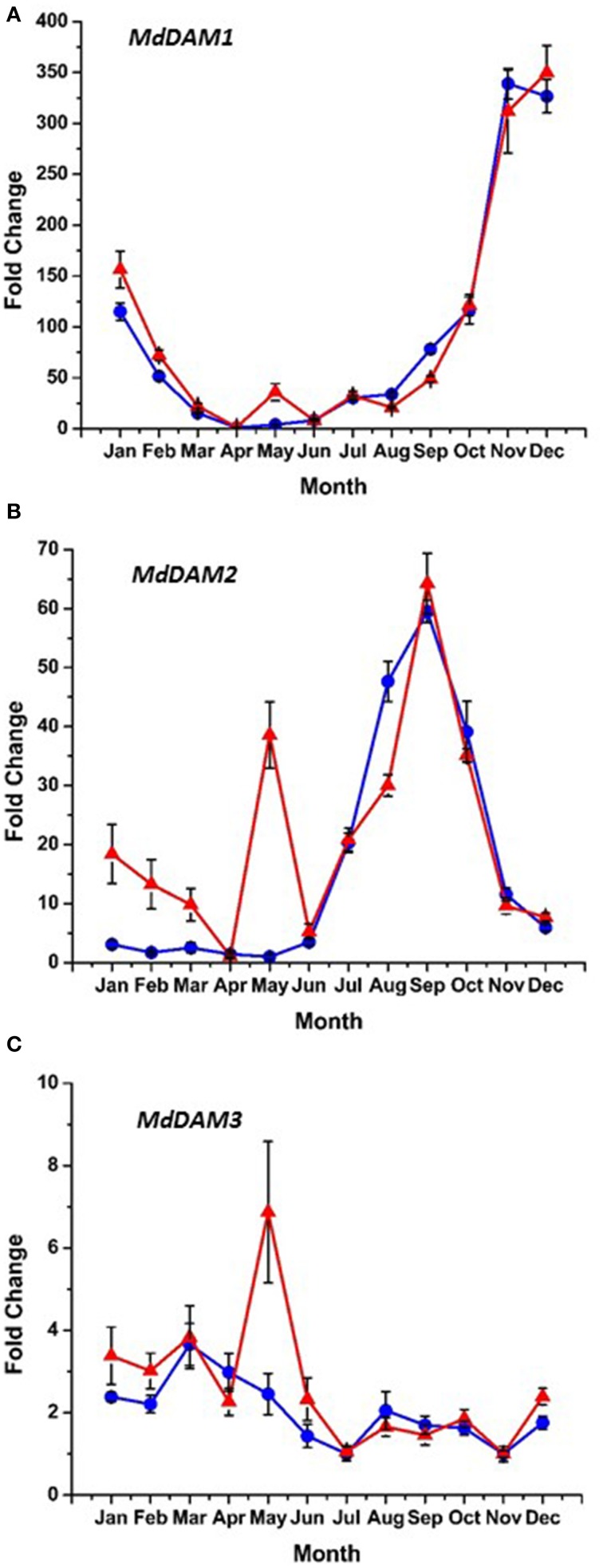
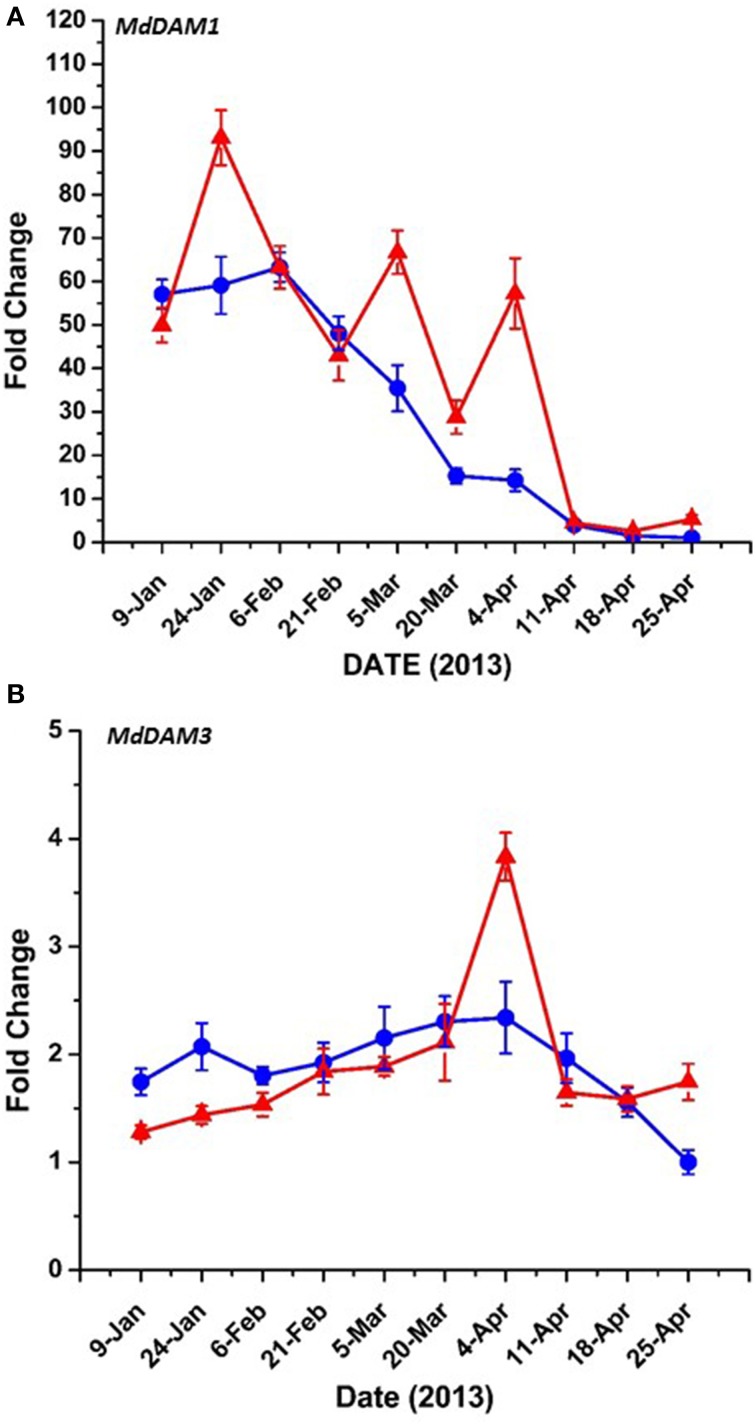
Bioinformatic analysis identified four putative DAM genes in the apple genome as listed in Genomic Database for the Rosaceae (GDR) (http://www.rosaceae.org) (Figure S2). Three (MDP0000527190, MDP0000322567, MDP0000259294,) bear high similarity to reported DAM genes in pear (Saito et al., 2013), and an additional predicted gene (MDP0000209705) was also considered due to its similarity to the other putative DAM genes. We have annotated three of these genes thusly: MDP0000322567 = MdDAM1 (KP164996), MDP0000259294 = MdDAM2 (KP164997), and MDP0000209705 = MdDAM3 (KP164998). Despite the use of several primers (Table S1) and protocol adjustments no measurable expression was observed for MDP0000527190 in either bark or bud tissues. As such, it was not assigned an MdDAM designation. Seasonal expression of the other three putative DAM genes in bark tissues collected from trees of M.26 and T166 are presented in Figure 6. Similar patterns of expression were observed for MdDAM1 in both genotypes (Figure 6A). Levels of expression rose in the fall, reached a maximum in November/December and then declined reaching a minimum in April. In contrast, levels of expression for MdDAM2 began to increase in mid-summer, reached a maximum in September/October and declined and remained low throughout the winter and spring months (Figure 6B). Notably, MdDAM2 expression was higher in January–March in the T166 trees, and a brief rapid increase in the expression level of MdDAM2 was observed during April/May (Figure 6B). The pattern of MdDAM3 was similar in both genotypes except for a single spike in expression in the T166 trees during April/May (Figure 6C). This was similar to the spike in expression observed for MdDAM2 (Figure 6B). In bud tissues, expression of MdDAM1 and MdDAM3 were the only DAM genes for which products could be obtained by RT-qPCR (Figure 7). In contrast to bark tissues, where expression levels of MdDAM1 were similar, the level of expression differed significantly in bud tissues (Figure 7A) collected from the transgenic (T166) and non-transformed (M.26) trees. Overall the expression level of MdDAM1 was higher during the winter months and then declined during the spring in both genotypes. In buds of T166 plants, however, several spikes in expression were observed in early spring. A similar trend was observed for the level of expression of MdDAM3 in bud tissues of both genotypes, however, in the case of buds from T166 trees only a single spike in expression was observed (Figure 7B).
Figure 6.
Seasonal MdDAM transcript accumulation kinetics from bark tissue vary between genes. (A) MdDAM1. (B) MdDAM2. (C) MdDAM3. Blue circles: non-transformed M.26; Red triangles: Line T166.
Figure 7.
MdDAM transcript accumulation kinetics from bud tissue during winter and early spring vary between genes. (A) MdDAM1. (B) MdDAM3. MdDAM2 was not detected. Blue circles: non-transformed M.26; Red triangles: Line T166.
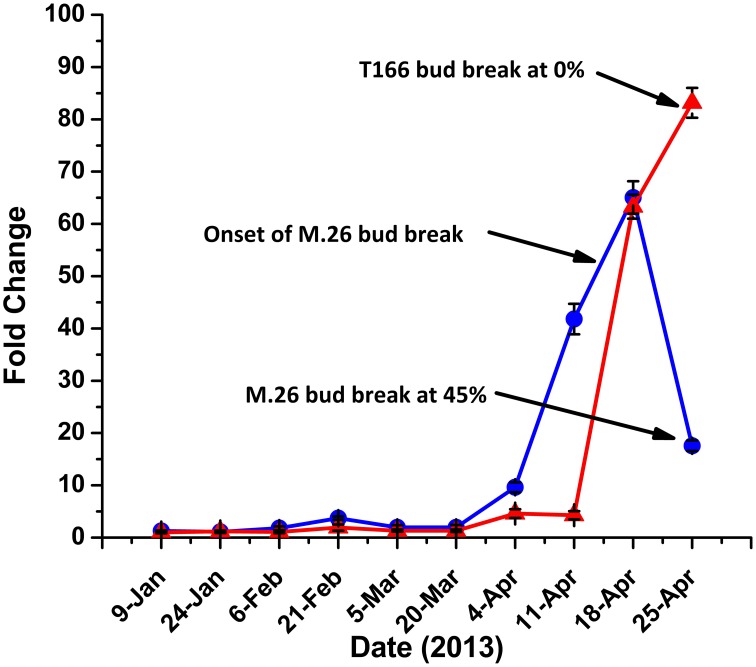
Yordanov et al. (2014) recently demonstrated the functional role of EEB1, a putative APETALA2/Ethylene responsive transcription factor, in determining the time of bud break in poplar (Populus tremuloides). BLAST analysis of the apple genome revealed two homologs, MDP0000827400 and MDP0000123172, of the poplar EBB1 gene (Figure S3). Expression of either gene was not observed in bark tissues (data not shown), however, expression of MDP0000827400 was observed in bud tissues, exhibiting a pattern of expression that could be associated with the timing of bud break observed in the two genotypes (Figure 8). As such, we define MDP0000827400 = MdEBB1 (KP164995). Induction of MdEBB1 began earlier in the non-transformed M.26 trees, as did the occurrence of bud break. In contrast, expression in buds of T166 trees was induced about 2 weeks later and rose to higher relative levels. This delay was in agreement with the delay in bud break observed in the T166 trees. The onset of bud break in M.26 trees occurred just prior to peak expression of MdEBB1, however, a similar connection could not be determined in T166 trees due to a limited collection of buds.
Figure 8.
MdEBB1 transcript accumulation kinetics in winter through spring correlate with delayed bud break in T166. Blue circles: non-transformed M.26; Red triangles: Line T166.
DELLA gene expression and growth
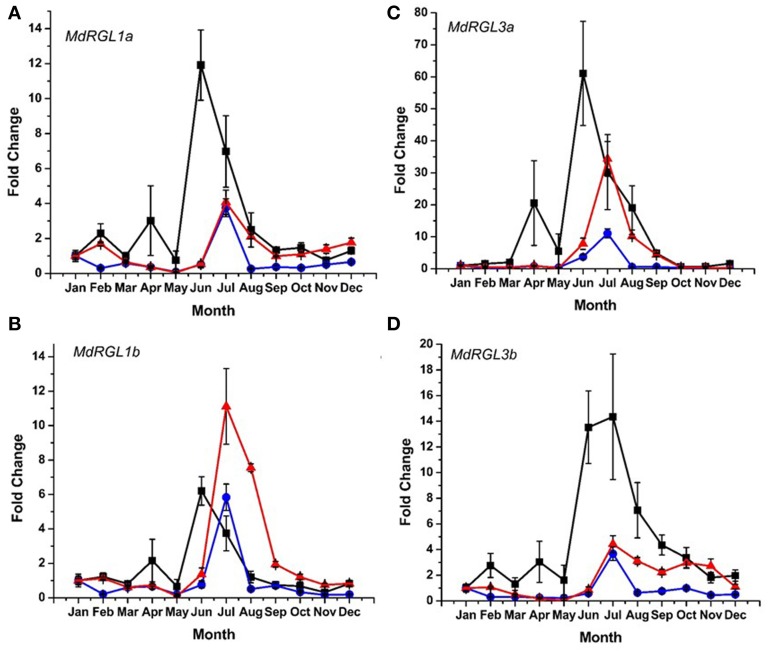
DELLA protein abundance is inversely related to the amount of bioactive forms of gibberellic acid (GA) (Achard and Genschik, 2009). Since distinct differences in seasonal patterns and overall levels of growth were observed in T166 vs. M.26 trees, an analysis of RGL gene expression was conducted in the two genotypes. The pattern of RGL gene expression was also examined in a scion genotype (“CrimsonCrisp”) for comparative purposes. Six DELLA genes have been identified in the apple genome (Foster et al., 2007); Figure S4. Using various primer sets (Table S1), the seasonal pattern of expression of four of the apple DELLA genes (MdRGL1a, 1b, 3a, and 3b) could be characterized. Expression of MdRGL2a and 2b could not be discerned. In all three genotypes, the highest level of expression of the four DELLA genes was observed during the summer months of July–August (Figure 9). Except for MdRGL1b the highest level of expression was observed in the scion genotype, “CrimsonCrisp.” The exception was MdRGL1b where T166 trees exhibited the highest fold change. In comparing just the non-transformed (M.26) genotype with the transgenic (T166) line, the transcript abundance of the four RGL genes was higher in the T166 samples than in the M.26 samples. An extended period of elevation for MdRGL3b, lasting through the fall months (September–November) was observed in T166 trees.
Figure 9.
Seasonal MdRGL (MdDELLA) transcript accumulation kinetics from bark tissue of three apple genotypes vary between genes and genotypes. (A) MdRGL1a. (B) MdRGL1b. (C) MdRGL3a. (D) MdRGL3b. Blue circles: non-transformed M.26; Red triangles: Line T166; Black squares: “CrimsonCrisp.”
Promoter analyses
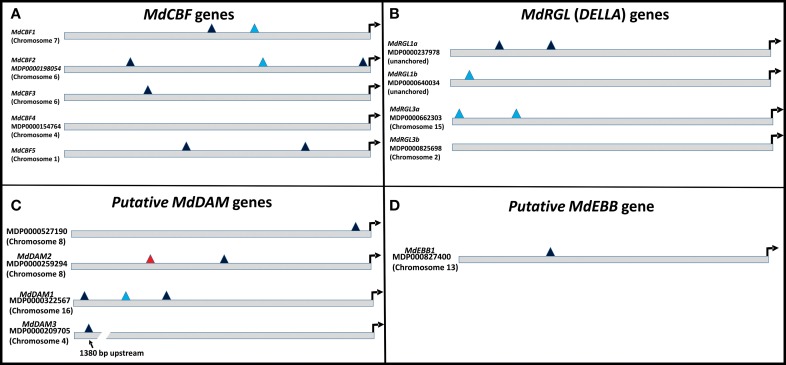
Several phenotypic characteristics were altered in T166 trees, and these changes were, to some degree, reflected in differences in expression of candidate genes associated with cold hardiness, dormancy, bud break, and growth. Therefore, 1000 bp upstream to the start codon of the examined genes were subjected to a bioinformatics analysis using various web-based regulatory element search tools. The presence or absence of the C-repeat (LTRE, Low Temperature Response Element) in these genes was of particular interest, since it could potentially help to explain how overexpression of CBF could lead to changes in these other processes. Results of for the presence of C-repeat elements are presented in Figure 10 and a more detailed and complete analysis is presented in Table S2. The canonical dicot C-repeat element is present in four of the five MdCBF genes, suggesting some degree of self- or cross-regulation. Each of the three putative MdDAM genes had at least one C-repeat element, with MdDAM1 having three, one of which represented a C-repeat typical of monocots. Additionally, the C-repeat present in MdDAM3 was approximately 1500 bp upstream from the coding region. A C-repeat element was also observed in the promoter region of MdEBB1, which was more highly expressed in T166 buds, and MdRGL1a and b and MdRGL3a.
Figure 10.
Schematic of C-repeats present in the 1000 bp upstream of the translational start sites of the genes examined in this report. C-repeats are binding sites for CBF transcription factors. (A) MdCBF genes. (B) MdRGL genes. (C) MdDAM genes. (D) MdEBB1 gene. Dark blue triangles: canonical dicot C-repeats; light blue triangles: monocot C-repeats: red triangles: monocot DRE binding site (nearly identical to C-repeat). Arrow denotes translational start site.
Additional regulatory elements were also observed (Table S2). These included dehydration related elements (Abscissic Acid Response Elements, ABREs, and sites associated with certain MYB transcription factors), light-related elements (PIF, Evening Element, GATA and circadian rhythm) and flowering-related elements (agamous and certain RAV1 transcription factors). MdCBF genes were also examined for the presence of ICEr1 (MYC core), ICEr2, and CAMTA binding sites (Table S2).
Discussion
Cold acclimation, dormancy, and the timing of bud break are parameters that play a critical role in the life cycle of temperate woody plants (see Reviews by Cooke et al., 2012; Rios et al., 2014; Wisniewski et al., 2014). Research by Wisniewski et al. (2011) has documented that overexpression of a peach CBF gene (PpCBF1) in a rootstock variety (M.26) of apple resulted in an increase in both non-acclimated and acclimated cold hardiness, early induction of budset and dormancy stimulated by exposure to short day length, and growth inhibition. These observations were further confirmed by 3 years of field observations of trees of the transgenic line (T166) and the non-transformed parent line (M.26) and also revealed that, in addition to early leaf senescence and dormancy, T166 also exhibited delayed bud break in the spring (Artlip et al., 2014). These findings were further confirmed by a year of extra data presented in the current study (Figures 1–4). Thus, these plants may serve as a useful model for investigating how the regulation of these various life-cycle parameters are integrated. The objective of the present study was to document the expression of all of the apple CBF genes in response to cold acclimating conditions and to compare the expression of various genes that have been reported to play a major role in dormancy, bud break, and growth in the T166 and M.26 genotypes. This was done in an attempt to better understand how the regulation of these various life-cycle parameters are integrated.
CBF gene expression
Five CBF genes have been identified in the genome of apple (Zhuang et al., 2011; Wisniewski et al., 2014). The expression of only three (MdCBF1, 2, and 4) of the five CBF genes could be discerned in leaf tissues collected from M.26 trees subjected to cold acclimating conditions (4°C, 8/16 h light/dark photoperiod) suggesting that not all the CBF genes in apple are responsive to cold acclimating conditions or that expression of individual CBF genes may be tissue specific (Figure 5). The strongest induction occurred in MdCBF4 which was induced after 2 h and peaked at 24 h. The next highest level of induction was observed in MdCBF4. An earlier report (Wisniewski et al., 2011) indicated that the normal expression pattern of MdCBF1 and 2 were not altered by the ectopic expression of PpCBF1, so this aspect was not further investigated in the present study.
Differential expression in different tissues and differential levels of transcript accumulation of CBF genes has been commonly observed (Wisniewski et al., 2014). In detailed studies of grape, Xiao et al. (2008) reported that VrCBF4 exhibited induced expression for 0.5 h to 2 days in both younger and older leaves, while VrCBF1, 2, and 3 accumulated only in young tissues with maximum expression occurring at 30 min, 8 h, and 5 days, respectively (Xiao et al., 2006). CBF genes have also been demonstrated to respond to varying degrees to different abiotic stresses. Overexpression of AtCBF1, 2 or 3 enhanced cold tolerance and also drought and salt tolerance (Mizoi et al., 2012). Mantri et al. (2012) have noted a significant overlap between drought- and cold-stress induced transcriptomes. Epigenetic regulation may also play a role in the expression of specific transcription factors (Rios et al., 2014). Therefore, it would not be unexpected to find the different levels of expression and perhaps tissue-specific expression noted in the present study.
The impact of CBF expression on other plant developmental processes has also been noted, reduced growth and late flowering being two notable effects (Gilmour et al., 1998; Lazaro et al., 2012). The involvement of CBF genes in seed dormancy, via their induction of genes that lower GA expression has also been reported (Kendall et al., 2011). Many of these same features have been observed in the T166 transgenic apple line investigated in the present study. In this regard, it is interesting to note that all of the MdCBF genes, except MdCBF4, and other transcription factors, except MdRGL3b, examined in the present study contain C-repeat elements in the promoter regions of their genes (Figure 10). Although the integrated regulation of cold acclimation, dormancy, and growth will inevitably be shown to be complex, this is one way in which self-regulation of CBF gene expression, and cross-regulation amongst CBF genes and between CBF genes and other key regulatory transcription factors could occur and be integrated. Evidence for this level of cross-talk and regulation has yet to be conclusively demonstrated in our transgenic apple but the current study lays the foundation for future experiments.
Dormancy and bud break-related gene expression
Dormancy Associated MADS-box (DAM) genes have been reported to be directly associated with the regulation of dormancy onset and release in peach (Bielenberg et al., 2008; Li et al., 2009; Jimenez et al., 2010), pear (Saito et al., 2013), and apricot (Sasaki et al., 2011) trees, as well the herbaceous plant, leafy spurge (Euphorbia esula) (Horvath et al., 2010), and indirectly in apple (Falavigna et al., 2014). T166 trees exhibit early induction of dormancy and leaf senescence in the summer\autumn and delayed bud break in the spring. This altered phenology was initially reported in greenhouse-grown plants (Wisniewski et al., 2011), then in field-grown plants (Artlip et al., 2014), and further confirmed in the current study (Figures 1, 2). Both previous studies postulated a role for DAM genes in the altered phenotype and so they were further investigated in the current study.
The putative apple DAM genes, MdDAM1, MdDAM2, and MdDAM3, defined in the present study, have high similarity to the Pyrus pyrifolia MADS13 genes (Saito et al., 2013), and other DAM genes reported in apricot and peach, however, phylogenetic relationships and analogous functions still need to be determined. While a complete set (a full year) of data is available on the expression of three of the putative apple genes (Figure 6) in bark tissues only a partial set (late winter through spring) of data are available for DAM gene expression in vegetative apple buds (Figure 7). In bark tissues, the pattern of seasonal expression of all three apple DAM genes (MdDAM1, MdDAM2, MdDAM3) were similar in both genotypes, however, a sharp spike in expression was noted in the expression of MdDAM2 and MdDAM3 in T166 trees in April/May. This suggests that high levels of expression of these genes may inhibit bud break as trees would have fulfilled their chilling requirement by this time and presumably would have been ecodormant rather than endodormant. In this regard, expression of MdDAM2 was significantly greater in T166 trees in January through March and may have played a more direct role in dormancy release (transition from endodormancy to ecodormancy). Expression of only two (MdDAM1 and MdDAM3) could be detected in apple vegetative buds (Figure 7). While the overall pattern of expression of these two DAM genes was again similar in both genotypes, a single sharp spike was observed in the T166 buds in early April and several spikes in expression were observed in MdDAM1 during January through April in T166 buds. Interestingly, expression levels for MdDAM1 did not differ at all in bark tissues. This suggests that this DAM gene may play a more important role in the regulation of bud dormancy/bud break while the other DAM genes may play a more important role in the regulation of cambial dormancy (collection of bark tissues from current year shoots would have included sampling of phloem and cambial tissues).
A variety of seasonal patterns of DAM gene expression have been noted in other woody plant species (Li et al., 2009; Sasaki et al., 2011; Saito et al., 2013) as was observed in the present study. In peach, Li et al. (2009) reported that DAM1, 2, and 4 were the most likely candidates for control of seasonal elongation cessation and bud formation, while Jimenez et al. (2010) reported that DAM5 and DAM6 were negatively correlated with chill hour accumulation and the rate of bud break. In apricot, PmDAM4 and PmDAM6 have been suggested as the primary candidates regulating endodormancy and chilling requirement (Sasaki et al., 2011), and in pear all three reported DAM genes (PpMADS13-1, 13-2, and 13-3) were reported to be associated with endodormancy establishment and release and were impacted by the application of dormancy-releasing agents (Saito et al., 2013). In the latter study PpMADS13-1 was reported to be specifically associated with dormancy release. In apple, the seasonal pattern of expression of MdDAM1 has been reported to exhibit a dormancy-related expression in three different cultivars (Falavigna et al., 2014), and to differ in the level of expression in low and high chill varieties. In the current study in apple, three DAM genes (MdDAM1, MdDAM2, MdDAM3) exhibited patterns of expression that could be associated with dormancy onset and release. However, only the expression pattern of MdDAM2 and MdDAM3 differed between the two genotypes (T166 and M.26) making them the most likely candidates for fine tuning the regulation of dormancy in apple, and perhaps more specifically cambial dormancy. In buds, only the expression of two DAM genes (MdDAM1 and MdDAM3) could be detected, with both genes showing a different pattern of expression in T166 vs. M.26 plants that may have impacted bud break. Collectively, our results in apple buds confirm the results obtained by Falavigna et al. (2014) and provide new information on the potential role of DAM genes in cambial dormancy. Importantly, as noted with the CBF genes, all the DAM genes identified in the current study possess C-repeat elements in their promoters, although the C-repeat element in MdDAM3 is somewhat removed (1380 bp) from the coding region of the gene (Figure 10).
It is important to note that release from dormancy and time to bud break are two different phenological events and may have different modes of genetic regulation. While release from dormancy is associated with the accumulation of chilling hours, time to bud break after dormancy release is associated with the accumulation of heat units. Yordanov et al. (2014) recently reported that overexpression of a putative poplar APETALA2/Ethylene responsive factor transcription factor in poplar caused early bud-flush, whereas down-regulation delayed budbreak. The gene was highly expressed in actively growing apices, and was undetectable in dormant buds. Two EBB like genes were identified in the current study, however, expression of only one of them (MdEBB1), could be detected in vegetative apple buds. No expression was observed in bark tissues (data not shown). Expression of MdEBB1, as in poplar, was low to non-detectable in dormant buds and began to rapidly increase just prior to the onset of budbreak (Figure 8). Notably, levels of MdEBB1 began to increase earlier in the non-transformed M.26 genotype than in the transgenic T166 genotype. The M.26 trees also exhibited earlier bud break than T166 trees (Figures 1, 2), hence MdEBB1 should be considered a strong candidate, in addition to DAM genes, for regulating bud break in apple trees. As with other genes investigated in this study, MdEBB1, also possesses a C-repeat element in its promoter region. This is the first association of this putative class of transcription factors with the regulation of dormancy and/or bud break in apple.
RGL (DELLA protein) gene expression
T166 trees exhibit reduced overall growth, as well as a reduced number of lateral branches compared to non-transformed M.26 trees (Figures 3, 4). Growth inhibition due to over-expression of native and foreign CBF genes has been reported in Arabidopsis (Jaglo-Ottosen et al., 1998; Kasuga et al., 1999; Gilmour et al., 2000; Welling and Palva, 2008). Over-expression of two different native Eucalyptus CBF genes in Eucalyptus and the native VvCBF4 gene in grape (Vitus vinifera) also resulted in reduced growth (Navarro et al., 2011; Tillett et al., 2012).
The reduced-growth phenotype in Line T166 and other systems is most likely caused directly or indirectly by a reduction in the level of bio-active giberellic acid (GA). Achard et al. (2008), Suo et al. (2012) and Niu et al. (2014) have all reported changes in the expression levels of GA-biosynthetic and GA-deactivating genes in plants over-expressing CBF genes (Arabidopsis thaliana, soybean (Glycine max) and tobacco (Nicotiana tabacum), respectively), with Suo et al. (2012) and Niu et al. (2014) noting decreased GA levels in such plants. The reduction in bioactive GA levels have been attributed to increases in GA2ox (GA deactivating) enzymes (Achard et al., 2008; Suo et al., 2012) or to a decrease in the geranylgeranyl diphosphate precursor to GAs (Niu et al., 2014). Application of exogenous bio-active GA was shown to overcome the dwarfism associated with CBF gene over-expression (Achard et al., 2008; Suo et al., 2012). Yang et al. (2013) indicate that GA3 is primary bioactive GA in apple vegetative tissues. Examination of GA3 levels along with expression data on GA-biosynthetic and GA-deactivating genes in Line T166 and non-transformed M.26, however, were not conducted in the present study.
An additional cause of reduced growth levels in Line T166 may stem from an up-regulation of DELLA proteins which normally repress GA responses. Achard et al. (2008) demonstrated that over-expression of AtCBF1in Arabidopsis thaliana up-regulates RGL3 which leads to enhanced levels of DELLA protein(s). GAs normally mediate the turnover of DELLAs by direct and indirect means (Achard and Genschik, 2009), hence a reduction in bio-active GAs also leads to enhanced levels of DELLA proteins.
There are six apple DELLA genes, MdRGL1a, 1b, 2a, 2b, 3a, and 3b, with each a-b pair highly related to each other and having nearly identical expression patterns (Foster et al., 2007). The seasonal expression of four MdRGL genes (1a, 1b, 3a, and 3b) was examined in bark tissues (Figure 9). The overall pattern of seasonal expression for these genes was similar in T166 and non-transformed M.26 trees. All four genes exhibited increased expression during the summer, with peaks occurring in July. Artlip et al. (2014) reported that current year shoot growth in both these genotypes dramatically slows in July–August. This slowing down in growth coincides with a peak in MdRGL expression in July (Figure 7). Foster et al. (2007) noted that shoots with arrested growth had higher MdRGL expression levels than actively growing shoots. In general, higher fold increases were observed for these genes in T166 bark tissues than in M.26 bark tissues. This was especially apparent for MdRGL1a during the period of late summer into early winter, MdRGL1b during summer and early autumn, MdRGL3a during summer and early autumn, and MdRGL3b during summer into early winter. These data indicate that MdRGL gene products may play a role in the reduced growth in T166 trees, relative to non-transformed M.26 trees.
Notably, expression of the RGL genes in the scion cultivar “CrimsonCrisp” was much greater than in the transgenic and non-transgenic M.26 trees, except for MdRGL1b (Figure 7). Achard and Genschik (2009) have suggested that levels of DELLA proteins are coupled with GA levels in order to maintain GA homeostasis. It is possible that scion cultivars such as “CrimsonCrisp” synthesize higher levels of GA compared to root-stock trees, and require greater levels of DELLA proteins to maintain GA homeostasis. This premise is supported by the observation that own-rooted, “Royal Gala” trees (“very vigorous” growth habit) have greater levels of root-sourced GA than “Royal Gala” on root stocks varying in their vigor (van Hooijdonk et al., 2011).
Potential regulation of the examined MdRGL genes by CBF is possible, as MdRGL1a has two canonical dicot C-repeats, while MdRGL1b and MdRGL3a have monocot-related C-repeats. (Figure 10; Table S2). These genes all show greater fold changes in Line T166 compared to non-transformed M.26. MdRGL1b, however, also displayed similar pattern of expression, despite having no C-repeat (Figure 10; Table S2). This suggests that CBF genes represent only one method of regulating RGL gene expression. Indeed, various promoter elements related to light and dehydration are present in all of the MdRGL genes (Table S2). The similar pattern of expression for all the examined MdRGL genes in apple (Figure 9) also supports the premise stated by Foster et al. (2007), of overlapping or perhaps redundant functions between MdRGL gene products.
Promoter analyses
The upstream 1000 bp prior to the translational start sites of the investigated genes were examined for the presence of various promoter elements (Table S2). In addition to the Low Temperature Response Element (LTRE or C-repeat), other promoter elements were identified, including ABA and dehydration responsive (ABRE, MYB1A, and MYB2), light or circadian rhythm (GATA, PIF, Evening Element, CircadianLHC) and bud or floral development (RAV or AGAMOUS). All of the promoter regions of the investigated genes had these elements, suggesting multiple regulatory possibilities. The presence of dehydration- or ABA-responsive elements in growth repressing DAM or DELLA genes is consistent with a need to repress growth during periods of water-limitation. Regulatory elements associated with light or circadian rhythm are also consistent with the need to integrate appropriate light cues for growth. An analysis to determine the presence of regulatory elements frequently found in CBF genes such as MYC (core of the ICEr1 binding site) and TGGAGGC (ICEr2 binding site) (Zarka et al., 2003), CAMTA (Calmodulin binding Transcription Activator) and Common Motifs (CMs) (Doherty et al., 2009) was also conducted. In contrast to Arabidopsis thaliana, few of these motifs were consistently found in the promoter region of the MdCBF genes. A CAMTA motif was found in MdCBF1, implying regulation by Ca2+ (Doherty et al., 2009). MdCBF4 had an ICEr1-like motif but no ICEr2 motif, while MdCBF5 had an ICEr2 motif but no recognizable ICEr1 motif.
Conclusions
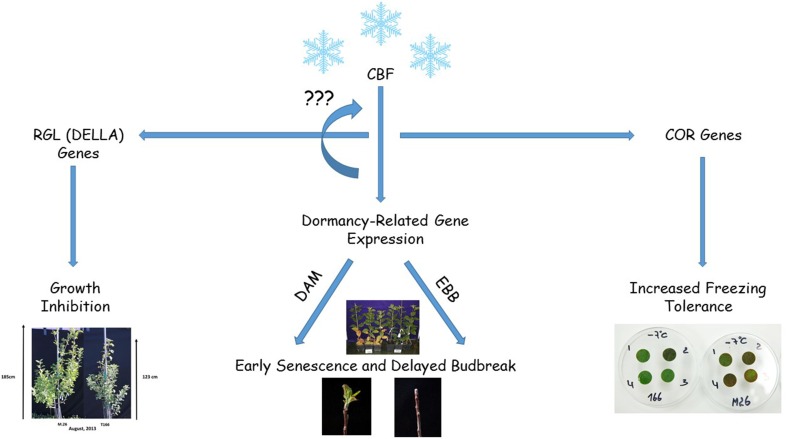
The timing of cold acclimation and deacclimation, the onset and release from dormancy, and the timing of bud break and onset of growth are overlapping, integrated processes that play an essential role in the life cycle of woody plants and their adaptation to the external environment. Overexpression of a peach CBF (PpCBF1) in a rootstock variety of apple (M.26) alters many of these parameters, exhibiting increased cold hardiness, early cessation of growth and leaf senescence, delayed bud break in the spring, growth inhibition, and increased sensitivity to short photoperiod with respect to the onset of dormancy. In the current study, several transcription factor genes that have been reported to regulate one or more of these processes was examined. Results indicated that expression of several of these key genes, including MdDAM, MdRGL, and MdEBB was altered in transgenic T166 trees relative to non-transformed M.26 trees. In particular, several MdDAM genes, associated with the dormancy-cycle in other species of woody plants in the Rosaceae, exhibited different patterns of expression in the T166 vs. M.26 trees. Additionally, for the first time a putative APETALA2/Ethylene responsive factor transcription factor, originally described in poplar and shown to regulate the timing of bud break, was shown to be associated with the timing of bud break in apple. Since the overexpression of PpCBF1 in apple results in a dramatic alteration in cold acclimation, dormancy, and growth, this transgenic line (T166 and others) may represent a useful model for studying the integration of these seasonal life-cycle parameters (Figure 11).
Figure 11.
Diagram summarizing the results obtained in the current study where a peach CBF (PpCBF1) was overexpressed in an apple (Malus × domestica) rootstock variety, M.26. Overxpression of the CBF gene (PpCBF1) results in altered expression of apple dormancy-related genes (MdDAMx and MdEBBx) resulting in early senescence in the fall and delayed budbreak in the spring. Overexpression of the CBF gene also leads to altered expression of RGL genes (MdRGLx) which results in growth inhibition. CBF overexpression also induces the expression of COR genes which results in increased freezing tolerance (Wisniewski et al., 2011). The regulation of the various genes and other CBF genes may be due to the presence of C-repeat motifs present in the promoter region of the studied genes.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The technical assistance of Erik Burchard and Phil Welser in conducting the RT-qPCR and generation and propagation of the transgenic plants are greatly appreciated.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2015.00085/abstract
Alignment of conceptual MdCBF1-5 and PpCBF1 amino acid sequences. Light red residues indicate different residues between the sequences. The alignment was performed with CLUSTALW (Thompson et al., 1994).
Alignment of conceptual MdDAM, peach DAM5 and DAM6, and pear PpMADS13-1, 13-2 and 13-3 amino acid sequences. Light red residues indicate different residues between the sequences. The alignment was performed with CLUSTALW (Thompson et al., 1994).
Alignment of conceptual MdEBB and poplar EBB1 amino acid sequences. Light red residues indicate different residues between the sequences. The alignment was performed with CLUSTALW (Thompson et al., 1994).
Alignment of conceptual MdRGL (DELLA) amino acid sequences. Light red residues indicate different residues between the sequences. The alignment was performed with CLUSTALW (Thompson et al., 1994).
References
- Achard P., Genschik P. (2009). Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J. Exp. Bot. 60, 1085–1092. 10.1093/jxb/ern301 [DOI] [PubMed] [Google Scholar]
- Achard P., Gong F., Cheminant S., Alioua M., Hedden P., Genschik P. (2008). The cold-inducible CBF1 factor–dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20, 2117–2129. 10.1105/tpc.108.058941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C. L., Jensen J. L., Ømtoft T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. 10.1158/0008-5472.CAN-04-0496 [DOI] [PubMed] [Google Scholar]
- Artlip T. S., Wisniewski M. E., Norelli J. L. (2014). Field evaluation of apple overexpressing a peach CBF gene confirms its effect on cold hardiness, dormancy, and growth. Environ. Exp. Bot. 106, 79–86 10.1016/j.envexpbot.2013.12.008 [DOI] [Google Scholar]
- Belknap W. R., Rockhold D. R., McCue K. F. (2008). pBINPLUS/ARS: an improved plant transformation vector based on pBINPLUS. Biotechniques 44, 753–756. 10.2144/000112731 [DOI] [PubMed] [Google Scholar]
- Benedict C., Skinner J. S., Meng R., Chang Y., Bhalerao R., Huner N. P. A., et al. (2006). The CBF1-dependent low temperature signalling pathway, regulon and increase in freeze tolerance are conserved in Populus spp. Plant Cell Environ. 29, 1259–1272. 10.1111/j.1365-3040.2006.01505.x [DOI] [PubMed] [Google Scholar]
- Bielenberg D. G., Wang Y., Li Z., Zhebentyayeva T., Fan S., Reighard G. L., et al. (2008). Sequencing and annotation of the evergrowing locus in peach (Prunus persica [L.] Batsch) reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet. Genomes 4, 495–507 10.1007/s11295-007-0126-9 [DOI] [Google Scholar]
- Chang W. C., Lee T. Y., Huang H. D., Pan R. L. (2008). PlantPAN: plant promoter analysis navigator for identifying combinatorial cis-regulatory elements with distance constrain in plant gene group. BMC Genomics 9:561. 10.1186/1471-2164-9-561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys H., De Bodt S., Inze D. (2014). Giberellins and DELLAs: central nodes in growth regulatory networks. Trends Plant Sci. 19, 231–239. 10.1016/j.tplants.2013.10.001 [DOI] [PubMed] [Google Scholar]
- Cooke J. E. K., Eriksson M. E., Junttila O. (2012). The dynamic nature of bud dormancy in trees: environmental control and molecular mechanisms. Plant Cell Environ. 35, 1707–1728. 10.1111/j.1365-3040.2012.02552.x [DOI] [PubMed] [Google Scholar]
- Doherty C. J., Van Buskirk H. A., Myers S. J., Thomashow M. F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21, 972–984. 10.1105/tpc.108.063958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falavigna V., da S., Porto D. D., Buffon V., Margis-Pinheiro M., Pasquali G., Revers L. F. (2014). Differential transcriptional profiles of dormancy-related genes in apple buds. Plant Mol. Biol. Rep. 32, 796–813 10.1007/s11105-013-0690-0 [DOI] [Google Scholar]
- Foster T., Kirk C., Jones W. T., Allan A. C., Espley R., Karunairetnam S., et al. (2007). Characterization of the DELLA subfamily in apple (Malus x domestica Borkh.). Tree Genet. Genomes 3, 187–197 10.1007/s11295-006-0047-z [DOI] [Google Scholar]
- Gilmour S. J., Sebolt A. M., Salazar M. P., Everard J. D., Thomashow M. F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124, 1854–1865. 10.1104/pp.124.4.1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour S. J., Zarka D. G., Stockinger E. J., Salazar M. P., Houghton J. M., Thomashow M. F. (1998). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16, 443–442. 10.1046/j.1365-313x.1998.00310.x [DOI] [PubMed] [Google Scholar]
- Hänninen H., Tanino K. (2011). Tree seasonality in a warming climate. Trends Plant Sci. 16, 412–416. 10.1016/j.tplants.2011.05.001 [DOI] [PubMed] [Google Scholar]
- Heide O. M., Prestrud A. K. (2005). Low temperature, but not photoperiod, controls growth cessation and dormancy induction and release in apple and pear. Tree Physiol. 25, 109–114. 10.1093/treephys/25.1.109 [DOI] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. 10.1093/nar/27.1.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath D. P., Sung S., Kim D., Chao W., Anderson J. (2010). Characterization, expression and function of DORMANCY ASSOCIATED MADS-BOX genes from leafy spurge. Plant Mol. Biol. 73, 169–179. 10.1007/s11103-009-9596-5 [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen K. R., Gilmour S. J., Zarka D. G., Schabenberger O., Thomashow M. F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280, 104–106. 10.1126/science.280.5360.104 [DOI] [PubMed] [Google Scholar]
- Jimenez S., Reighard G. L., Bielenberg D. G. (2010). Gene expression of DAM5 and DAM6 is suppressed by chilling temperatures and inversely correlated with bud break rate. Plant Mol. Biol. 73, 157–167. 10.1007/s11103-010-9608-5 [DOI] [PubMed] [Google Scholar]
- Kasuga M., Liu Q., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. 10.1038/7036 [DOI] [PubMed] [Google Scholar]
- Kendall S. L., Hellwege A., Marriot P., Whalley C., Graham I. A., Penfield S. (2011). Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell 23, 2568–2580. 10.1105/tpc.111.087643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A., Valverde F., Pineiro M., Jarillo J. A. (2012). The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 24, 982–999. 10.1105/tpc.110.081885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leida C., Conesa A., Llácer G., Badenes M. L., Ríos G. (2012). Histone modifications and expression of DAM6 gene in peach are modulated during bud dormancy release in a cultivar-dependent manner. New Phytol. 193, 67–80. 10.1111/j.1469-8137.2011.03863.x [DOI] [PubMed] [Google Scholar]
- Lescot M., Dehais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. 10.1093/nar/30.1.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Reighard G. L., Abbott A. G., Bielenberg D. G. (2009). Dormancy- associated MADS genes from the EVG locus of peach (Prunus persica [L.] Batsch) have distinct seasonal and photoperiodic expression patterns. J. Exp. Bot. 60, 3521–3530. 10.1093/jxb/erp195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantri N., Patade V., Penna S., Ford R., Pang E. (2012). Abiotic stress responses in plants: present and future, in Abiotic Stress Responses in Plants: Metabolism, Productivity and Sustainability, eds Ahmad P., Prasad M. N. V. (New York, NY: Springer; ), 1–19. [Google Scholar]
- Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 86–96. 10.1016/j.bbagrm.2011.08.004 [DOI] [PubMed] [Google Scholar]
- Navarro M., Ayax C., Martinez Y., Laur J., El Kayal W., Marque C., et al. (2011). Two EguCBF1 genes overexpressed in Eucalyptus display a different impact on stress tolerance and plant development. Plant Biotech. J. 9, 50–63. 10.1111/j.1467-7652.2010.00530.x [DOI] [PubMed] [Google Scholar]
- Niu S., Gao Q., Li Z., Chen X., Li W. (2014). The role of gibberellin in the CBF1-mediated stress-response pathway. Plant Mol. Biol. Rep. 32, 852–863 10.1007/s11105-013-0693-x [DOI] [Google Scholar]
- Restrepo M. A., Freed D. D., Carrington J. C. (1990). Nuclear transport of plant potyviral proteins. Plant Cell 2, 987–998. 10.1105/tpc.2.10.987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios G., Leida C., Conejero A., Badenes M. L. (2014). Epigenetic regulation of bud dormancy events in perennial plants. Front. Plant Sci. 5:247. 10.3389/fpls.2014.00247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J., Sherman W. B., Scorza R., Wisniewski M., Okie W. R. (1994). Evergreen peach, its inheritance and dormant behavior. J. Amer. Soc. Hort. Sci. 119, 789–792. [Google Scholar]
- Rohde A., Bhalerao R. P. (2007). Plant dormancy in the perennial context. Trends Plant Sci. 12, 217–223. 10.1016/j.tplants.2007.03.012 [DOI] [PubMed] [Google Scholar]
- Rohde A., Ruttink T., Hostyn V., Sterck L., Driessche K. V., Boerjan W. (2007). Gene expression during the induction, maintenance, and release of dormancy in apical buds of poplar. J. Exp. Bot. 58, 4047–4060. 10.1093/jxb/erm261 [DOI] [PubMed] [Google Scholar]
- Rohde A., Storme V., Jorge V., Gaudet M., Vitacolonna N., Fabbrini E., et al. (2011). Bud set in poplar—genetic dissection of a complex trait in natural and hybrid populations. New Phytologist 189, 106–121. 10.1111/j.1469-8137.2010.03469.x [DOI] [PubMed] [Google Scholar]
- Ruttink T., Arend M., Morreel K., Storme V., Rombauts S., Fromm J., et al. (2007). A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell 19, 2370–2390. 10.1105/tpc.107.052811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Bai S., Ito A., Sakamoto D., Saito T., Ewa Ubi B., et al. (2013). Expression and genomic structure of the dormancy-associated MADS box genes MADS13 in Japanese pears (Pyrus pyrifolia Nakia) that differ in their chilling requirement for endodormancy release. Tree Physiol. 33, 654–667. 10.1093/treephys/tpt037 [DOI] [PubMed] [Google Scholar]
- Sasaki R., Yamane H., Ooka T., Jotatsu H., Kitamura Y., Akagi T., et al. (2011). Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot. Plant Physiol. 157, 485–497. 10.1104/pp.111.181982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo H., Ma Q., Ye K., Yang C., Tang Y., Hao J., et al. (2012). Overexpression of AtDREB1A causes a severe dwarf phenotype by decreasing endogenous gibberellin levels in soybean [Glycine max (L.) Merr.]. PLoS ONE 7:e45568. 10.1371/journal.pone.0045568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow M. F., Gilmour S. J., Stockinger E. J., Jaglo-Ottosen K., Zarka D. G. (2001). Role of the Arabidopsis CBF transcriptional activatorsin cold acclimation. Physiol. Plantarum 112, 171–175 10.1034/j.1399-3054.2001.1120204.x [DOI] [Google Scholar]
- Thompson J. D., Higgins D. G, Gibson T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillett R. L., Wheatley M. D., Tattersall E. A. R., Schlauch K. A., Cramer G. R., Cushman J. C. (2012). The Vitis vinifera C-repeat binding protein 4 (VvCBF4) transcriptional factor enhances freezing tolerance in wine grape. Plant Biotech. J. 10, 105–124 10.1111/j.1467-7652.2011.00648.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hooijdonk B., Woolley D., Warrington I. (2011). Rootstocks modify scion architecture, endogenous hormones, and root growth of newly grafted ‘Royal Gala’ apple trees. J. Amer. Soc. Hort. Sci. 136, 93–102. [Google Scholar]
- Welling A., Palva E. T. (2006). Molecular control of cold acclimation in trees. Physiol. Plantarum 127, 167–181 10.1111/j.1399-3054.2006.00672.x [DOI] [Google Scholar]
- Welling A., Palva E. T. (2008). Involvement of CBF transcription factors in winter hardiness in birch. Plant Physiol. 147, 1199–1211. 10.1104/pp.108.117812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski M., Nassuth A., Teulieres C., Marque C., Rowland J., Cao P-B., et al. (2014). Genomics of cold hardiness in woody plants. Crit. Rev. Plant Sci. 33, 92–124 10.1080/07352689.2014.870408 [DOI] [Google Scholar]
- Wisniewski M., Norelli J., Bassett C., Artlip T., Macarisin D. (2011). Ectopic expression of a novel peach (Prunus persica) CBF transcription factor in apple (Malus x domestica) results in short-day induced dormancy and increased cold hardiness. Planta 233, 971–983. 10.1007/s00425-011-1358-3 [DOI] [PubMed] [Google Scholar]
- Xiao H., Siddiqua M., Braybrook S., Nassuth A. (2006). Three grape CBF/DREB1 genes respond to low temperature, drought, and abscisic acid. Plant Cell Environ. 29, 1410–1421. 10.1111/j.1365-3040.2006.01524.x [DOI] [PubMed] [Google Scholar]
- Xiao H., Tattersall E. A. R., Siddiqua M., Cramer G. R., Nassuth A. (2008). CBF4 is a unique member of the CBF transcription factor family of Vitis vinifera and Vitis riparia. Plant Cell Environ. 31, 1–10. 10.1111/j.1365-3040.2007.01741.x [DOI] [PubMed] [Google Scholar]
- Yamane H., Ooka T., Jotatsu H., Hosaka Y., Sasaki R., Tao R. (2011). Expressional regulation of PpDAM5 and PpDAM6, peach (Prunuspersica) dormancy-associated MADS-box genes, by lowtemperature and dormancy-breaking reagent treatment. J. Exp. Bot. 62, 3481–3488. 10.1093/jxb/err028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Brown S. K., Davies P. J. (2013). The content and in vivo metabolism of gibberellin in apple vegetative tissues. J. Amer. Soc. Hort. Sci. 138, 173–183. [Google Scholar]
- Yordanov Y. S., Ma C., Strauss S. H., Buscov V. B. (2014). EARLY BUD-BREAK 1 (EBB1) is a regulator of release from seasonal dormancy in poplar trees. Proc. Natl. Acad. Sci. U.S.A. 111, 10001–10006. 10.1073/pnas.1405621111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarka D. G., Vogel J. T., Cook D., Thomashow M. F. (2003). Cold induction of Arabidopsis CBF genes involved multiple ICE (inducer of CBF expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiol. 133, 910–918. 10.1104/pp.103.027169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang J., Yao Q-H., Xiong A-S., Zhang J. (2011). Isolation, phylogeny and expression pattersn of AP2-like genes in apple (Malus x domestica Borkh.). Plant Mol. Biol. Rep. 29, 209–216 10.1007/s11105-010-0227-8 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of conceptual MdCBF1-5 and PpCBF1 amino acid sequences. Light red residues indicate different residues between the sequences. The alignment was performed with CLUSTALW (Thompson et al., 1994).
Alignment of conceptual MdDAM, peach DAM5 and DAM6, and pear PpMADS13-1, 13-2 and 13-3 amino acid sequences. Light red residues indicate different residues between the sequences. The alignment was performed with CLUSTALW (Thompson et al., 1994).
Alignment of conceptual MdEBB and poplar EBB1 amino acid sequences. Light red residues indicate different residues between the sequences. The alignment was performed with CLUSTALW (Thompson et al., 1994).
Alignment of conceptual MdRGL (DELLA) amino acid sequences. Light red residues indicate different residues between the sequences. The alignment was performed with CLUSTALW (Thompson et al., 1994).