Abstract
Mesoderm induction as a result of the interaction between endoderm and ectoderm is one of the most crucial events in vertebrate development. We identified activin as a strong mesoderm-inducing factor in an animal cap assay, an in vitro assay system using amphibian pluripotential cell mass. Activin induces mesodermal tisswes including most dorsal mesodermal tissue, notochord (which has important roles in neural induction, somite segmentation, and endodermal organogenesis), and its effects are concentration-dependent. Animal cap cells treated with high concentrations of activin differentiate into anterior endoderm, which can act as an organizer, or center of body patterning. We have established an in vitro induction system for 22 different organs and tissues using animal cap cells, and have isolated many organ-specific genes. With these useful methods, and analysis of newly isolated tissue- and organ-specific genes, the molecular biological “road map” for organogenesis is being established.
Keywords: Activin, organogenesis, animal cap, mesoderm induction, organizer
Introduction
Embryonic development is a dynamic event, consisting of sequential cell division from a single cell to a population of diverse cell types with complex interactions. Identification of the mechanisms and factors that underlie this process remains a fundamental aim of developmental biology. Fertilization is the first step in the morphogenesis of early embryonic development. In amphibia, the fertilized egg develops from a single cell into the blastula by the process of cleavage. After formation of three germ layers (endoderm, mesoderm, and ectoderm), gastrulation and neural induction proceed simultaneously. The tissues become differentiated during axial body patterning, and the organs are formed via interactions between the various cells and tissues.
The ability of mesoderm to induce neural tissue formation in ectoderm was established by Spemann and Mangold in a landmark study in 1924.1) They found that transplantation of blastoporal lip (organizer) into the blastocoel resulted in ectopic neural axis formation from host ectoderm. Nieuwkoop later showed the induction of mesoderm directly, using microsurgical combination of endodermal cells with ectodermal cells.2) Mesoderm induction came to be considered a key event in early development, and the characterization of mesoderm-inducing factor(s) became an important focus of study in this field. A mass of pluripotential ectodermal cells within the amphibian blastula, termed the animal cap, has been used to evaluate the activity of many putative mesodermal inducers.
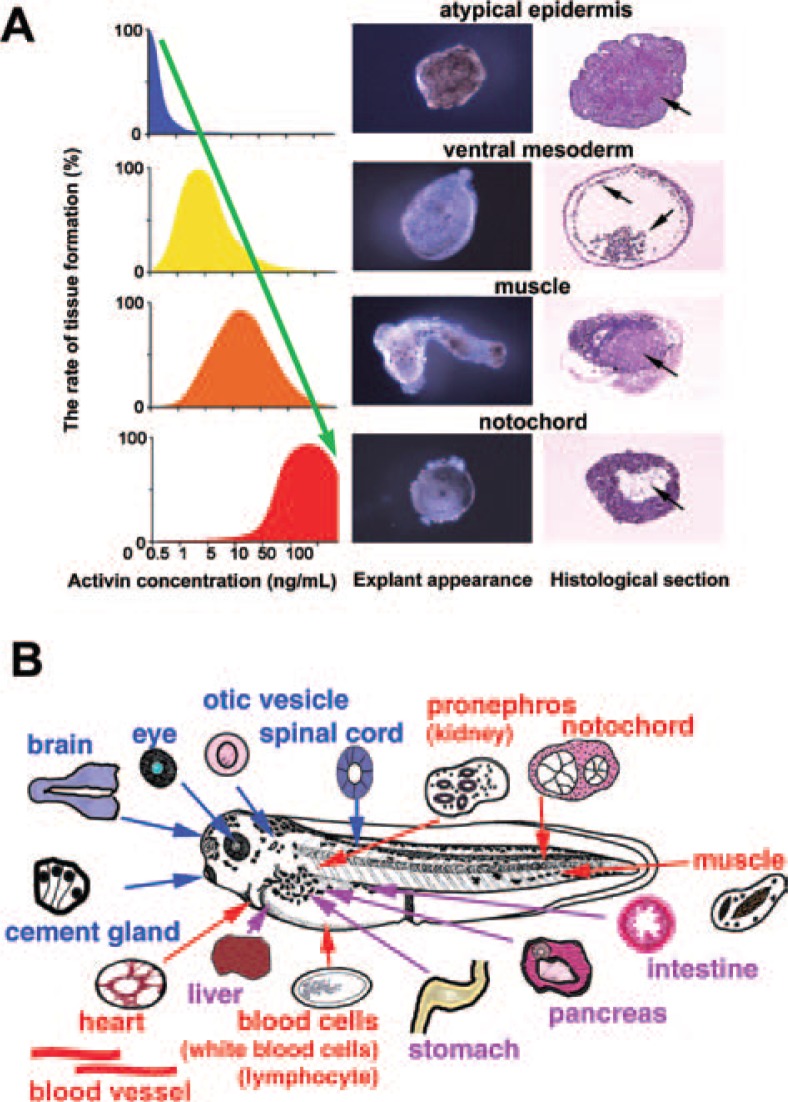
At the end of the 1980s, several growth factors were identified as candidate mesoderm-inducing factors. We reported first that activin3), 4) could induce all types of typical mesodermal tissues, including notochord (the most dorsal of mesodermal tissues), in the animal cap cell mass. The effects of activin were found to be concentration-dependent.5),6) We have since established in vitro induction methods and appropriate culture conditions using activin and other inducing factors for the differentiation of animal cap cells into 22 different organs and tissues, including heart, pronephros, pancreas, cartilage, eye, neural tissues, blood cells and other mesodermal and endodermal tissues (Fig. 1, Fig. 2B). These methods are stable and reproducible, so that specific tissues can be reliably induced by simple manipulation of animal cap cells. In this report, we describe our recent work on the mechanisms of body patterning and organogenesis in amphibian embryos, using techniques that are based on these experimental systems in combination with molecular biological methods.
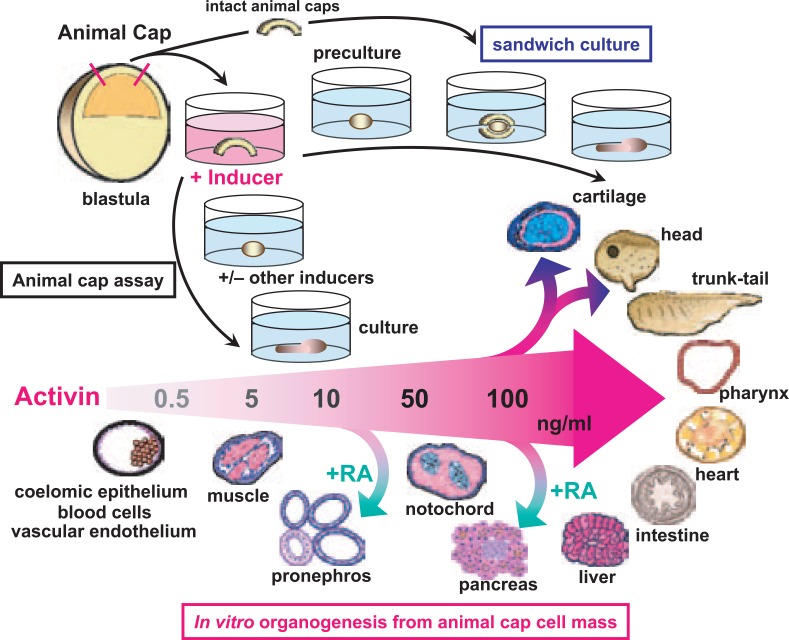
Fig. 1.
In vitro tissue and organ induction using animal caps. The animal cap, which forms the blastocoelic roof of the amphibian blastula, is excised and cultured in saline containing inducers such as activin and retinoic acid (RA). Differentiated explants are examined by histological and immunohistological techniques, and analyzed for expression of molecular markers.
Fig. 2.
Induction of tissues and organs by activin treatment of animal caps. (A) Activin treatment induces mesodermal tissue in the animal cap cell mass in a concentration-dependent manner. Induced tissue was dorsalized by treatment with higher concentrations of activin. (B) The organs and tissues successfully induced using animal cap cell mass in our laboratory.
Induction of mesodermal and endodermal tissues by animal cap assay
Amphibia have been widely used as experimental animals in the field of developmental biology because of their rapid embryonic development and because they are easy to observe. They are also readily cultivated in vitro with simple saline solutions, and they can be manipulated microsurgically. Fertilized newt (Urodela) eggs, which are about 2 mm in diameter, have been used for many of the classical microsurgical experiments with amphibian embryos. At present, Xenopus laevis tends to be used as the representative amphibian, because it has been extensively investigated in molecular biological studies. A fertilized egg of Xenopus is spherical, with a diameter of approximately 1.2 mm, and develops into a blastula by sequential cleavage, about 7 hours after fertilization at 20 °C. The blastula of Xenopus consists of about 10,000 cells, and has large cavity (the blastocoel) in the animal hemisphere, which is the upper hemisphere of the spherical blastula. In Xenopus, the animal cap region forms the blastocoelic roof. It consists of a few layers of presumptive ectodermal cells, which can be easily excised using watchmakers’ forceps and sharp needles under stereoscopic microscope. During normal development of the amphibian, animal cap cells differentiate into ectodermal structures such as neural tissues or epidermis. When the animal cap cell mass is excised from the blastula and cultivated in a saline solution containing the appropriate induction factors, it can be directed to differentiate into various types of tissues including those normally derived from mesoderm and endoderm, as well as ectoderm7) (Fig. 1, “Animal cap assay”). The explants formed by animal caps in vitro can be maintained in stable cultivation for a week or longer, a time that normally sufficient for organogenesis in the Xenopus tadpole. Highly differentiated tissues and organs with three-dimensional structures can be formed in explants, which indicates the effectiveness of the animal cap assay as a tool for investigation of embryonic development and organ generation in vitro.
Using the animal cap assay, we isolated a mesoderm-inducing factor from conditioned medium of the human K-562 cell line,5) and identified it as activin (EDF).3), 8) Activin treatment causes differentiation of animal cap cells into various mesodermal tissues in a concentration-dependent manner (Fig. 2A). Untreated animal caps form atypical epidermis when cultured for a few days. In culture medium with 0.3–1 ng/mL activin, animal caps differentiate into ventral mesoderm-derived cells and tissues such as blood cells, coelomic epithelium and mesenchymal cells. Animal caps cultured in 5–10 ng/mL of activin show elongation movement and muscle differentiation.
Elongation movement is regarded as a reproduction of the gastrulation movement of mesodermal cells (known as convergent extension), and it provides a useful model for investigating the mechanisms of gastrulation and body patterning. We examined the effects of cytochalasin B (an inhibitor of actin polymerization) on this cell movement, and found that inhibition of the elongation causes a decrease in expression of muscle specific genes in the activin-treated explants.9)
Treatment with 50–100 ng/mL of activin induces animal caps to differentiate into notochord, the most dorsal mesoderm, and into endodermal tissue containing abundant yolk. These results suggest that the gradient concentration of activin regulates the differentiation of animal cap cells into mesodermal tissues along the dorsoventral axis.4), 6), 10), 11)
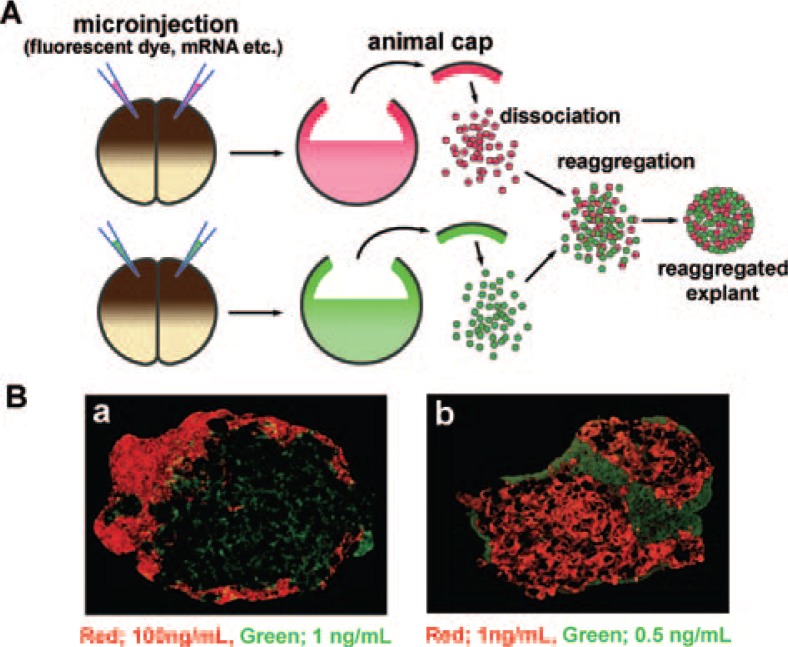
The animal cap can be easily dissociated to single cells by incubation in Ca2+/Mg2+-free culture medium. Treatment with activin at concentrations of 0–0.5 ng/mL, 1–5 ng/mL and 2.5–100 ng/mL causes dissociated cells to differentiate into atypical epidermis, notochord and yolk-rich tissue respectively, when they reaggregate after the activin exposure12) (dissociation and reaggregation method, Fig. 3A). When two groups of animal cap cells that have been treated with different concentrations of activin are cultured as a mixed aggregate, the cells migrate in the aggregate and within 5–10 hours after treatment form separate clusters made up of cells treated with the same concentrations of activin (Fig. 3B). This autonomous cell-sorting in the mixed aggregate indicates that activin treatment causes a change in the adhesive properties of animal cap cells. This result suggests that there is an important relationship between the function of cell adhesion molecules and the induction of various tissues by activin treatment in animal cap cells. We identified axial protocadherin (AXPC) as a candidate gene for changing the adhesion properties of animal cap cells treated with activin, and showed it to be essential in prenotochord cell sorting.13) The dissociation-reaggregation method used in this study is valuable for interrogating cell-cell interactions and the differentiation of various organs in vitro.
Fig. 3.
Dissociation-reaggregation method using animal cap cells. (A) Schematic diagram of the dissociation-reaggregation method. Fluorescent dyes (tracer, red or green) were microinjected into the animal pole of 2-cell-stage embryos. Animal cap cells were excised and dissociated in a Ca2+/Mg2+-free solution, treated in a solution containing inducers, and then mixed and reaggregated. This method was used to evaluate cell-cell interactions (secondary induction) between the two groups of cells, treated with different conditions. (B) Histological sections of the reaggregated explants. The cells treated with activin under the same conditions for notochord induction (1 ng/mL of activin) migrated and formed a cluster autonomously inside the explant. a) Green cells (treated with 1 ng/mL of activin) were observed on the inner side of the explant in contrast to the red cells (treated with 100 ng/mL of activin). b) Red cells (treated with 1 ng/mL of activin) were observed on the inner side of the explant in contrast to the green cells (treated with 0.5 ng/mL of activin).
A cDNA screen of mesodermal tissues using these induction methods revealed novel genes that are required for the differentiation of mesodermal tissues and for body patterning. One of these genes, XBtg2, is essential for notochord differentiation14) and anterior neural development.15) We also found a novel gene group, comprising Bowline16) and Ledger-line,17) with important roles in somite segmentation.
Animal caps acquire organizer-like activity by treatment with activin
To evaluate the secondary induction between two animal cap cell-masses where one has been treated with an inducer and the other has not, a cultivation method that combines intact animal caps with a treated animal cap can be used18) (Fig. 1, “sandwich culture”). When a newt animal cap is treated with 100 ng/mL activin for 1 hour and cultured for an additional 6–12 hours (“preculture”), then sandwiched between intact animal caps for culturing, the combined animal cap cell mass (explant) differentiates into a head structure containing hindbrain with otic vesicle. If the preculture duration is extended to 18–24 hours, the explant instead differentiates into a head structure containing a forebrain with eyes. When the preculture step is omitted from this procedure, the explant differentiates into a trunk-and-tail structure containing axial structures such as spinal cord, notochord and somites19) (Fig. 1). The tissues of endodermal organs such as liver, pancreas and intestine can also be induced in newt animal caps in sandwich culture by an animal cap treated with 100 ng/mL activin.20) Transplantation of a newt animal cap treated with activin into the ventral side of a newt gastrula (host embryo) causes the formation of a well-organized secondary axis, as shown in a classical experiment of organizer transplantation.21), 22) The tracing of cell lineage by labeling animal cap cells with fluorescein-dextran-amine (FDA) has revealed that animal caps treated with high concentrations (100 ng/mL) of activin differentiate into anterior endoderm when transplanted into the host embryo.20) These results show that a newt animal cap treated with high concentrations of activin acquires organizer-like activity. We have found that one of the organizer-related genes, newt homolog of chordin (Cychd), is expressed in both the organizer region of embryos and in the head-inducible animal caps treated with activin.23)
In Xenopus, animal caps treated with high concentrations of activin usually differentiate into notochord (dorsal mesodermal tissue) and yolk-rich cells (endodermal tissue). The Xenopus animal cap (blastocoelic roof) has a few layers of ectodermal cells, in contrast to the single cell layer in the animal cap of newt (Cynops). On the assumption that this structural difference causes the different response to activin treatment in Xenopus and Cynops animal caps, we separated the Xenopus animal cap into outer-layer ectoderm (OLE) and inner-layer ectoderm (ILE), so that the layers could be treated uniformly by activin. When treated separately with 100 ng/mL activin, the OLE and ILE usually differentiated into yolk-rich endodermal tissue at a high rate. RT-PCR analysis showed that after activin treatment, ILE exclusively expressed endodermin (an endodermal marker), in contrast to the combined expression of endodermin, ms-actin (mesodermal markers) and N-CAM (a neural marker) in OLE. The simultaneous transplantation of activin-treated (100 ng/ml) ILE and OLE into the ventral marginal zone of a blastula (host embryo) resulted in secondary axis formation with an evident head-like structure including eyes. In these embryos, the grafted ILE and OLE differentiated into endodermal tissue.24) From these results, we concluded that Xenopus animal caps consist of outer and inner layers that have different respective responses to activin, but that both layers can acquire organizer-like activity after uniform treatment with high concentrations of activin. The activin treatment of animal caps is therefore a useful method for investigating the organizer formation, head induction and determination of axial patterning in Xenopus, as well as in Cynops.
These results suggested that animal caps treated with high concentrations of activin express genes related to neural induction. Based on this, we performed a screen for genes upregulated by activin and identified XSIP-1 (Xenopus Smad-interacting protein-1),25) which was found to be essential for neural differentiation.26)
Eye formation in vitro and transplantation in the embryo
As mentioned above, we induced a head-like structure in vitro by the sandwich culture method. Further improvement of this method also revealed the optimal conditions for eye induction in vitro. The sandwich culture of intact animal caps in combination with excised dorsal blastopore lip and the lateral marginal zone of early gastrula induced differentiation of the explant into eye at a high frequency.27) When the eye region of the explant was excised and implanted into normal tadpole embryos (stage 33), the grafted eye was integrated into the host embryo. The optic nerve extended and connected normally to the optic tectum, and functional eye was formed. This eye remained functional even after metamorphosis to the frog. This result indicated that we could reproduce the induction of eye in vitro.
Maxillofacial cartilage formation in vitro
Based on our method that uses activin treatment to induce formation of a head-like structure from animal caps, we aimed to establish a procedure that would induce maxillofacial cartilage with a high success rate. When Xenopus animal caps are treated with 100 ng/mL activin for 1 hour, precultured for 1 hour, sandwiched between untreated animal caps, and maintained in culture for 7 days, formation of chondrocytes in the explant is the most frequently observed outcome.28) For maxillofacial cartilage induction, a method using dissociated animal cap cells was also developed, as follows: dissociated animal cap cells are treated with 25 ng/mL activin for 1 hour, mixed with untreated cells in a ratio of 1:5 to form an aggregate, then cultured for 7 days as an explant.29) Col2 and Cart-1, marker genes expressed in the process of cartilage differentiation, are expressed in these explants in a similar way to the normal development of maxillofacial cartilage. Goosecoid and X-dll4, markers of cephalic ventral mesenchyme and anterior ectoderm, are also expressed in these explants. Ectopic tooth germ-like structures are formed after transplantation of an explant made by the dissociation-reaggregation method into the abdominal region of a host embryo.29) These findings strongly suggest that maxillofacial cartilage is induced in Xenopus animal cap explants by activin treatment.
Induction of blood cells and vascular cells in vitro
Animal caps differentiate into ventral mesodermal tissues including blood cells following treatment with 0.3–1 ng/ml of activin. We established additional conditions involving the simultaneous treatment of animal caps with 0.5 ng/ml activin and 30 ng/ml SCF (stem cell factor) or 10 ng/ml IL-11 (interleukin-11) that could induce erythrocyte or leukocyte differentiation in explants at a high frequency.30) We also found that cotreatment with 10 ng/mL VEGF (vascular endothelial growth factor) and 10 ng/mL activin induced animal caps to differentiate into a duct-like structure composed of Flk-1-positive cells.31) Flk-1 is a VEGF receptor, and a marker for vascular endothelium and hematopoietic cells. This result indicated that animal cap cells differentiated into hemangioblasts.
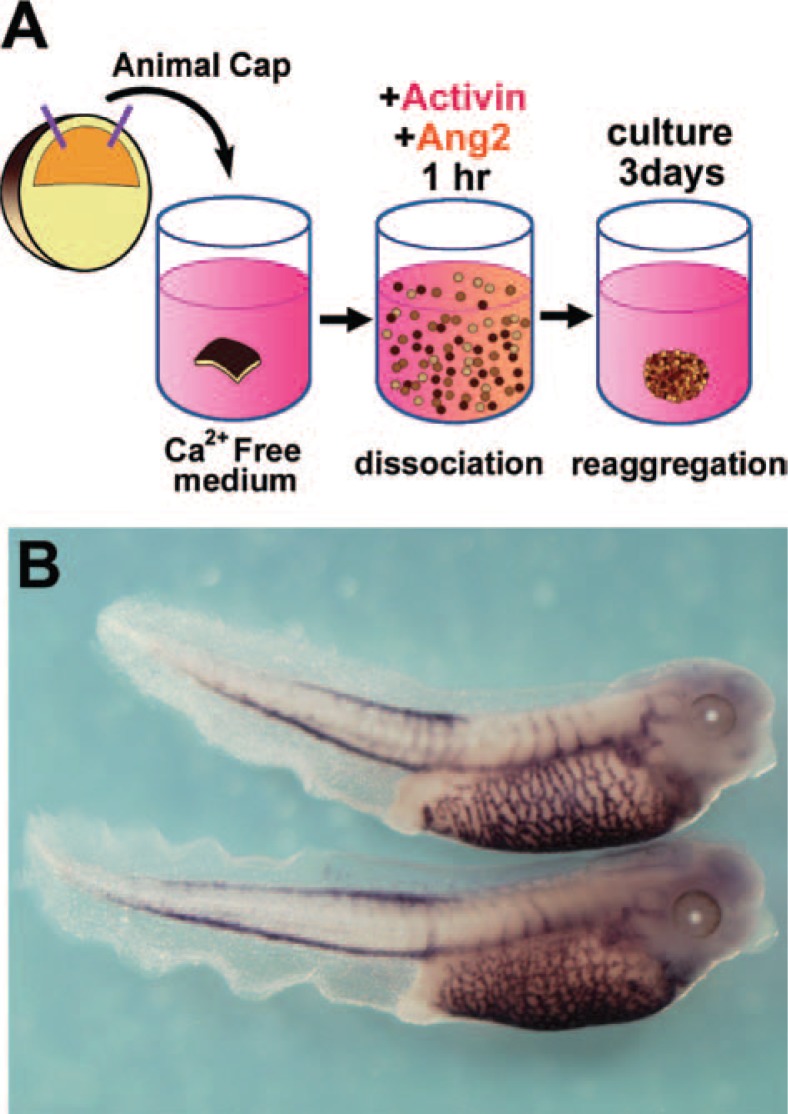
Next we attempted to establish the optimal conditions for inducing vascular endothelial cells in animal cap explants, because hematopoietic cells and vascular endothelial cells share a common precursor. We treated dissociated animal cap cells with activin and another inducer, angiopoietin, which is important in the later stages of vessel differentiation (Fig. 4A). Cotreatment of these dissociated animal cap cells with 0.4 ng/mL activin and 100 ng/mL angiopoietin-2 caused the evident expression of vascular endothelial markers (X-msr and Xtie2), but no expression of hematopoietic markers in the reaggregated explants. Microarray analysis was used to compare gene expression in the activin-treated and activin/angiopoietin-2-treated reaggregates, and revealed some vascular-specific genes.32) We also recently isolated a novel vascular-related gene, named Ami (“mesh” in Japanese).33) Ami mRNA expression was detected in the endothelium of the forming vasculature (Fig. 4B). This induction method for vascular endothelium and novel marker genes provides a useful tool for future studies of angiogenesis.
Fig. 4.
In vitro induction of vascular endothelium in animal caps. (A) Method for the induction of vascular endothelium in the reaggregated animal cap cells. Animal caps were dissociated, treated with 0.4 ng/mL activin and 100 ng/mL angiopoietin-2 for one hour, then reaggregated and cultured for 3 days. (Ang2; angiopoietin-2) (B) The expression pattern of Ami mRNA. Ami was isolated in a screen for cardiovascular-related genes.
Heart formation in vitro and transplantation in the embryo
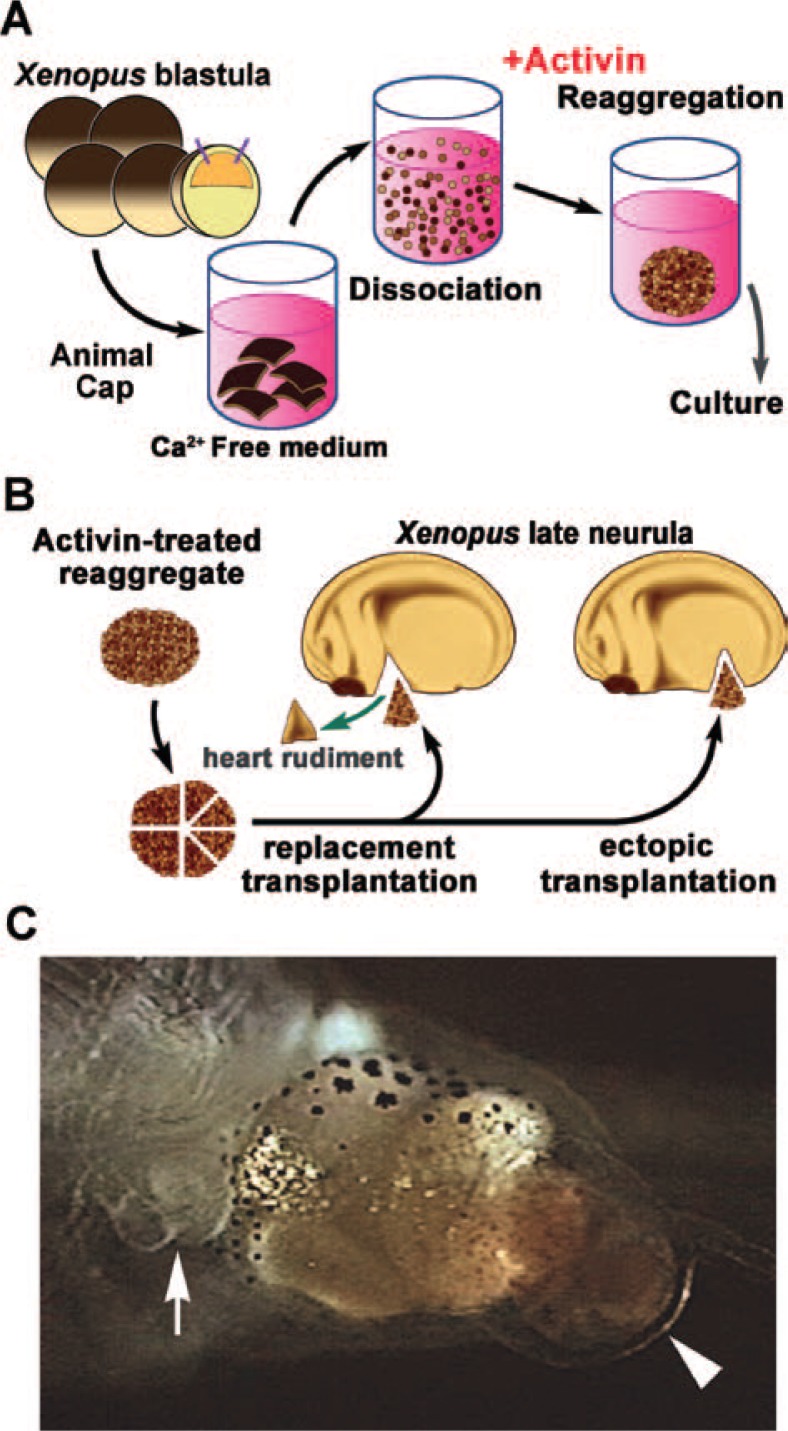
We found that the in vitro induction method can lead to formation of a heart-like structure in the newt animal cap. When newt animal caps are treated with 100 ng/mL activin, the explants differentiate into anterior endoderm, but about 20%–30% of explants differentiate into beating heart-like structures.34) These explants are positive for anti-α-sarcomeric actin, and the characteristic microstructures of myocardium, Z band, and intercalated discs are apparent on electron microscopy. Recently, we established an optimal method for inducing beating hearts using Xenopus animal cap cells, as follows; 5 animal caps (approximately 1000 cells in total) are dissociated and treated with 100 ng/mL of activin for 5 hours, then reaggregated and cultured for 3 days as an explant (Fig. 5A). The expression of XGATA-4, an early marker for heart and anterior endoderm, is prolonged in this dissociation-reaggregation explant as compared to intact animal caps treated with the same concentration of activin. The expression of XNkx2.5, an early cardiomyocyte marker, is also increased in this explant. When the original heart primordium of the host embryo is replaced with this explant (“replacement transplantation”), the host embryo develops with the heart derived from the donor explant, and the heart functions normally (Fig. 5B). When the explant is transplanted into the abdominal region of the host embryo (“ectopic transplantation”), the host embryo develops into a frog with two hearts; one derived from the original heart primordium and one from the donor explant35) (Fig. 5C). These results suggest that the dissociation-reaggregation method used to induce the beating heart-like structures may reproduce the induction of heart primordium that occurs in the normal development of Xenopus.
Fig. 5.
In vitro induction of heart rudiment in animal caps. (A) The method used to induce cardiac tissues in animal cap cells of Xenopus. Five animal caps were dissociated and treated with 100 ng/mL activin for 5 h, then reaggregated and cultured for 3 days. (B) The transplantation procedure. Explants were implanted as a replacement of the heart rudiment or ectopically. (C) A 10-day-old tad-pole with a well developed ectopic heart that formed after transplantation of activin-treated reaggregates into the abdominal region at the late neurula stage. (arrow; original heart, arrow head; ectopic heart).
Kidney formation in vitro and transplantation in the embryo
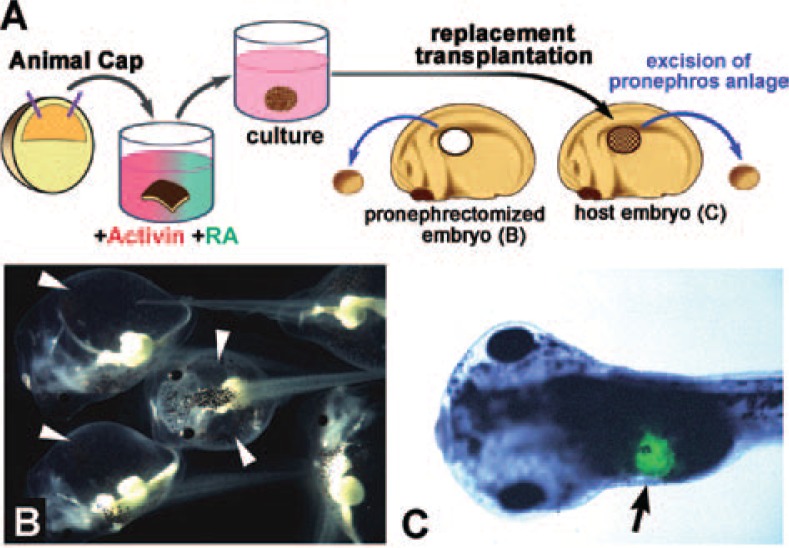
Retinoic acid (RA) is distributed endogenously in a concentration gradient along the antero-posterior axis in the Xenopus embryo, and is considered a candidate for determining body patterning in embryonic development. In the Xenopus animal cap assay, RA affects mesoderm induction and modifies its fate to lateral or posterior property. We found an effective method for inducing pronephros formation in vitro in animal caps, using a combination of RA and activin. By simultaneous treatment with 10 ng/mL activin and 10−4 M RA, pronephric tubules were formed at a high frequency in animal cap explants36) (Fig. 6A). Pronephros-specific marker genes were expressed in these explants, as they are in the normal embryo.37) Immunohistological staining and histological examination by electron microscopy revealed that these explants contained all three components of a nephron: glomeruli, pronephric tubules, and the pronephric duct.38) When Xlim-1, the essential gene for the differentiation of pronephros in normal development, was inhibited in animal cap cells, pronephros formation was suppressed in the animal caps treated with activin and RA.39) These results suggest that the method for pronephric induction in animal caps is a good replication of normal kidney development in Xenopus. By replacement transplantation of the presumptive pronephros region of the Xenopus embryo with the explant treated with activin and RA, we showed that the donor explant was integrated into the host embryo, and a normally functioning pronephros was formed40) (Fig. 6A–C). We investigated the detailed expression of gene markers for pronephric development in the pronephric structure formed in the explant, and found that the timing of their expression was the same as that in normal development.41) Screening of cDNA libraries constructed from the explants induced by activin and RA identified some pronephros-specific genes42)–49) (Fig. 7), of which at least one was essential for normal kidney development in mice as well as in Xenopus.50), 51)
Fig. 6.
In vitro induction of pronephric tubules in animal caps. (A) The method for pronephros induction in animal caps and transplantation into the late neurula of Xenopus. Animal caps were treated with 10 ng/mL activin and 10−4 M of RA. The explant was implanted to replace the pronephros anlage. (B) Embryos from which the pronephric primordium has been excised are unable to eliminate water and clearly show edema (arrowheads). (C) An embryo transplanted with a pronephric explant shows normal development. Green fluorescent dye previously introduced into the explant indicates that it has been incorporated into the pronephric area (arrow).
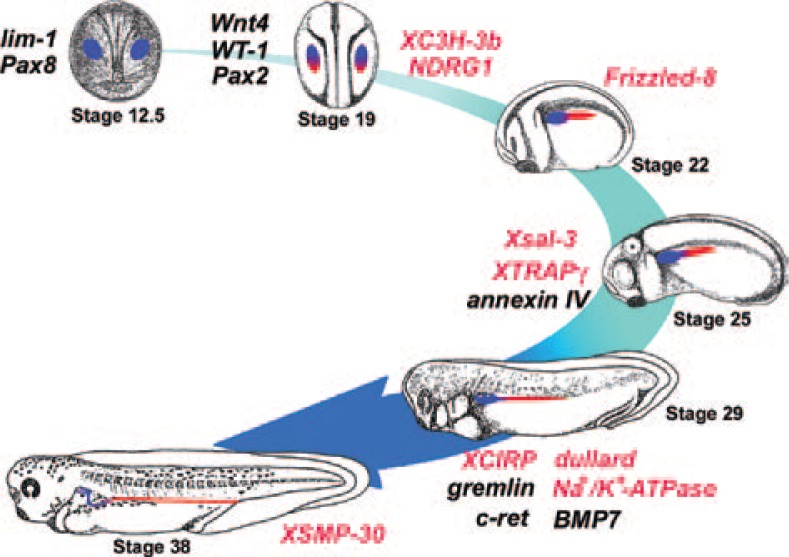
Fig. 7.
Sequential expression of genes involved in kidney development in Xenopus. By analyzing the function of genes expressed in developing kidney, a “road map” of the genes essential for normal kidney development will be established. The genes written in red have been newly identified as pronephros-related genes as a result of our studies.
Pancreas formation in vitro
By modifying the timing and length of the treatment with activin and RA, we established a method to differentiate animal cap cells into pancreatic tissue, as follows; animal caps are treated with 100 ng/mL activin, cultured for 5 hours, and then treated with 10−4 MRA.52) The induction of pancreatic tissue by this method can be explained by the anterior endodermal tissue (which is induced by high concentrations of activin) being posteriorized by RA, where it differentiates into pancreas.53) Explants formed using this protocol contain normal pancreatic structure, including both exocrine regions and endocrine regions, and glucagon and insulin can be detected by immunohistological staining. These results indicated that we could induce functional pancreatic tissue in vitro using Xenopus pluripotent cells. By the above–mentioned method, pronephric tissue was also formed infrequently in the explant as a mixture with pancreas tissue. By modifying this method, we successfully induced pancreatic tissue containing no apparent pronephric tissue by increasing the concentration of activin to 400 ng/mL. Subtractive hybridization screening with the cDNA clones obtained by this induction method identified many pancreas-specific genes including four novel genes.54)
Analysis of activin signaling-related genes in embryogenesis
Activin plays a key role in our methods in regulating the induction of various organs and tissues in animal cap cells. We have extensively investigated the function of activin and activin signaling in the embryonic development of Xenopus in many previous studies. To initially confirm the distribution of maternal activin protein in vivo, we purified activin from Xenopus early embryos.55), 56) We also detected activin and activin receptor in vitellogenic oocytes.57), 58) These results showed that Xenopus embryos express activin protein maternally, and that activin has some role in early development. A subsequent screen for the earliest zygotic factors that cause dorsalization in embryos isolated novel nodal-related genes, Xnr5 and Xnr6, which share the same receptors with activin.59) Microinjection of Xnr5 and Xnr6 mRNAs into the ventral vegetal sides of blastula induced formation of the secondary axis. Further, endogenous Xnr5 and Xnr6 were detected in the dorsal-vegetal region of the blastula including the Nieuwkoop center, which induces the Spemann’s organizer in the adjacent dorsal marginal zone. These results implicated activin/nodal signaling as having a crucial role in organizer formation and early body patterning in Xenopus. We subsequently investigated the functions of nodal-related genes to elucidate the mechanism of activin signaling in embryonic development and organizer formation.60)–65) By screening for novel genes related to endoderm specification, we isolated the organizer-related gene, Mig30 (Mixer inducible gene 30),66) which could be induced by activin. Mig30 is expressed at the Spemann’s organizer region, and affects gastrulation movement. We aim to further clarify the mechanism of body patterning and organizer formation in future studies using these approaches.
We have also investigated the unknown factors and mechanisms that regulate activin/nodal signaling. Activin is a secreted protein that belongs to the transforming growth factor-beta (TGF-beta) superfamily. Activin signal is transduced by the formation of heterotetrameric complexes comprising activin receptor serine kinases (type I and type II receptors), and sequential phosphorylation of intracellular mediators, the receptor-regulated Smads (RSmad).67) EGF-CFC proteins act together with activin receptors as essential coreceptors in the regulation of activin/nodal signaling.68), 69) We isolated the EGF-CFC gene, FRL-1, and demonstrated its important role in early neural differentiation.70) Screening for novel genes upregulated by activin directly identified Xantivin as a feedback inhibitor of activin signaling during mesoderm induction.71) Xantivin antagonized activin/nodal signaling to regulate anteroposterior axis formation by inhibiting EGFCFC genes.72) Studies into the mechanism underlying the loss of mesodermal competence in animal cap assays following activin treatment demonstrated that Notch signaling modulates the nuclear localization of Smad2 (R-Smad in Xenopus), while activin signaling activates Notch signaling.73), 74) Notch signaling is also involved in regulating the multipotency of undifferentiated cells during embryonic development.75) The interaction between activin/nodal signaling and Notch signaling may therefore have an essential role in organogenesis. With further analysis based on these studies, we aim to define the regulatory mechanisms of activin/nodal signaling.
To investigate the mechanisms of organogenesis and embryogenesis using a genetic approach, we recently studied Xenopus (Silurana) tropicalis, which has a diploid genome76) in contrast to the allotetraploid genome in Xenopus laevis. Our comparison of Xenopus laevis and Xenopus tropicalis animal caps revealed a similar competence in response to activin.77) We have also isolated Xtdazl (Xenopus tropicalis DAZ -like gene) as a specific marker of germ plasm, and pPGC (presumptive primordial germ cell) for the investigation of germ cells.78) By genetic analysis of Xenopus tropicalis in combination with our methods for organogenesis in vitro, we aim to analyze effectively the functions of a variety of organogenesis-specific genes and the genes related to activin/nodal signaling.
Concluding remarks
We have established techniques for the artificial induction of various tissues and organs using amphibian animal cap cells. These are simple, stable and reproducible methods for in vitro organogenesis, and they are also useful tools for investigating the mechanisms of organ formation during normal embryonic development in vertebrates. We have shown that the tissues and organs induced in vitro are highly similar to the ones produced in the normal embryo, when compared by histology and molecular biology. By these induction methods, we have isolated many genes related to organogenesis and body patterning. With comprehensive analysis of the sequential expressions of these genes, we aim to construct a temporal “road map” of gene expressions for each organ. Building on this “road map” for each organ will help to clarify the mechanisms of organogenesis.
Our strategy is effective and unique in that we take a synergistic approach in combining our in vitro organogenesis methods and detailed molecular biological analyses. Through further improvements in these methods and approaches, and a detailed analysis of induction mechanisms in the explants, we aim to study mammalian pluripotent cell lines such as mouse ES cells and human stem cells. Such work also promises to assist, by the elucidation of basic mechanisms, the therapeutic application of such cells in regenerative medicine.
Acknowledgments
The authors would like to acknowledge the financial assistance provided by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, Culture and Technology of Japan, and by ICORP (International Cooperative Research Project) of the Japan Science and Technology Agency.
Profile
Born in 1944, Makoto Asashima embarked on a research career in 1967 studying the developmental biology of regenerating rat liver and sea urchin embryos.
After graduating from the Faculty of Science (Doctor of Science, March 1972) at the University of Tokyo, he took up a position with Prof. Dr. (Prof. Dr. Dr.) Heinz Tiedemann at the Institute for Molecular Biology at the Free University of Berlin in Germany. It was here that Asashima fuelled his life-long interest in embryonic development by isolating inducing factors from chick embryos known as mesoderm or vegetalizing factors. Prof. Dr. (Prof. Dr. Dr.) Heinz Tiedemann and his wife, Dr. Hildegard Tiedemann, were important career mentors and taught him everything from classical embryology to a host of molecular biological techniques.
In October 1974, Asashima was appointed as an Associate Professor at the Yokohama City University in Japan. Despite a lack of funding, laboratory equipment, and help in the form of graduate students or staff, he continued to look for organizing factors. Finally, he succeeded in partially isolating an important inducing factor and was promoted to professor at the same University in 1985. In 1989 he presented his work as an invited speaker at the International Developmental Congress in The Netherlands, with a paper titled, “Activin A has mesodermal inducing activity”. The work was subsequently published in February 1990. Following these key findings of Asashima, many scientists reported similar results about activin A and mesodermal development, and renewed interest in this field was generated among developmental biologists, molecular biologists, and geneticists. Professor Asashima went on to establish several in vitro organ and tissue models using animal caps (undifferentiated cells mass) of newt or Xenopus embryos.
Asashima was awarded widely for this pioneering and original work in embryonic development, including prizes from the Zoological Society of Japan in 1991, the Inoue Academic Prize in 1991, the Kihara Memorial Academic Prize in 1994, and the Philip von Siebold Prize in 1994 in Germany.
In 1993, he moved to the Department of Biology in the Faculty of Arts and Sciences at the University of Tokyo, where he encouraged and mentored many students in the field of developmental biology through his teaching. He was subsequently also awarded the Torey Science Technology Prize in 1999, the Naito Memorial Academic Prize in 1999, the Uehara Prize in 2000, Imperial Prize and Japan Academy Prize in 2001, and the Purple Award from the Japanese Government in 2001.
Between 2003 and 2005, Professor Asashima was Dean of the Graduate School of Arts and Sciences and became the Vice President of the Science Council of Japan in 2005. He is currently President of the Zoological Society of Japan and the Japanese Society of Developmental Biologists.

References
- 1).Spemann, H., and Mangold, H. (1924) Wilhelm Roux’s Arch. Entwicklungsmech. Org. 100, 599–638. [Google Scholar]
- 2).Nieuwkoop, P. D. (1969) Wilhelm Roux’s Arch. Entwicklungsmech. Org. 162, 341–373. [DOI] [PubMed] [Google Scholar]
- 3).Asashima, M., Shimada, K., Nakano, H., Kinoshita, K., and Ueno, N. (1989) Cell Differ. Dev. 27 (suppl.), 53. [Google Scholar]
- 4).Asashima, M., Nakano, H., Shimada, K., Kinoshita, K., Ishii, K., Shibai, H., and Ueno, N. (1990) Roux’s Arch. Dev. Biol. 198, 330–335. [DOI] [PubMed] [Google Scholar]
- 5).Nakano, H., Kinoshita, K., Ishii, K., Shibai, H., and Asashima, M. (1990) Dev. Growth Differ. 32, 165–170. [DOI] [PubMed] [Google Scholar]
- 6).Ariizumi, T., Sawamura, K., Uchiyama, H., and Asashima, M. (1991) Int. J. Dev. Biol. 35, 407–414. [PubMed] [Google Scholar]
- 7).Yamada, T., and Takata, T. (1961) Dev. Biol. 3, 411–423. [DOI] [PubMed] [Google Scholar]
- 8).Asashima, M. (1994) Dev. Growth Differ. 36, 343–355. [DOI] [PubMed] [Google Scholar]
- 9).Tamai, K., Yokota, C., Ariizumi, T., and Asashima, M. (1999) Dev. Growth Differ. 41, 41–49. [DOI] [PubMed] [Google Scholar]
- 10).Okabayashi, K., Asashima, M. (2003) Curr. Opin. Genet. Dev. 13, 502–507. [DOI] [PubMed] [Google Scholar]
- 11).Furue, M., and Asashima, M. (2004) InHandbook of Stem Cells Volume 1 (ed. Lanza R.). Academic Press, San Diego, pp. 483–492. [Google Scholar]
- 12).Kuroda, H., Sakumoto, H., Kinoshita, K., and Asashima, M. (1999) Dev. Growth Differ. 41, 283–291. [DOI] [PubMed] [Google Scholar]
- 13).Kuroda, H., Inui, M., Sugimoto, K., Hayata, T., and Asashima, M. (2002) Dev. Biol. 244, 267–277. [DOI] [PubMed] [Google Scholar]
- 14).Sugimoto, K., Hayata, T., and Asashima, M. (2005) Dev. Growth Differ. 47, 435–443. [DOI] [PubMed] [Google Scholar]
- 15).Sugimoto, K., Okabayashi, K., Sedohara, A., Hayata, T., and Asashima, M. (2006) Dev. Neurosci. (in press). [DOI] [PubMed] [Google Scholar]
- 16).Kondow, A., Hitachi, K., Ikegame, T., and Asashima, M. (2006) Int. J. Dev. Biol. 50, 473–479. [DOI] [PubMed] [Google Scholar]
- 17).Chan, T. C., Satow, R., Kitagawa, H., Kato, S., and Asashima, M. (2006) Zool. Sci. 23, 689–697. [DOI] [PubMed] [Google Scholar]
- 18).Holtfreter, J. (1933) Wilhelm Roux’s Arch. Entwicklungsmech. Org. 128, 584–633. [DOI] [PubMed] [Google Scholar]
- 19).Ariizumi, T., and Asashima, M. (1995) Rouxs Arch. Dev. Biol. 204, 427–435. [DOI] [PubMed] [Google Scholar]
- 20).Ariizumi, T., Komazaki, S., and Asashima, M. (1999) Zool. Sci. 16, 115–124. [Google Scholar]
- 21).Ninomiya, H., Ariizumi, T., and Asashima, M. (1998) Dev. Growth Differ. 40, 199–208. [DOI] [PubMed] [Google Scholar]
- 22).Ariizumi, T., and Asashima, M. (2001) Int. J. Dev. Biol. 45, 273–279. [PubMed] [Google Scholar]
- 23).Yokota, C., Ariizumi, T., and Asashima, M. (1998) Dev. Growth Differ. 40, 335–341. [DOI] [PubMed] [Google Scholar]
- 24).Ninomiya, H., Takahashi, S., Tanegashima, K., Yokota, C., and Asashima, M. (1999) Dev. Growth Differ. 41, 391–400. [DOI] [PubMed] [Google Scholar]
- 25).Eisaki, A., Kuroda, H., Fukui, A., and Asashima, M. (2000) Biochem. Biophys. Res. Commun. 271, 151–157. [DOI] [PubMed] [Google Scholar]
- 26).Nitta, K. R., Tanegashima, K., Takahashi, S., and Asashima, M. (2004) Dev. Biol. 275, 258–267. [DOI] [PubMed] [Google Scholar]
- 27).Sedohara, A., Komazaki, S., and Asashima, M. (2003). Dev. Growth Differ. 45, 463–471. [DOI] [PubMed] [Google Scholar]
- 28).Furue, M., Myoishi, Y., Fukui, Y., Ariizumi, T., Okamoto, T., and Asashima, M. (2002) Proc. Natl. Acad. Sci. USA 99, 15474–15479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Myoishi, Y., Furue, M., Fukui, Y., Okamoto, T., and Asashima, M. (2004) Int. J. Dev. Biol. 48, 1105–1112. [DOI] [PubMed] [Google Scholar]
- 30).Miyanaga, Y., Shiurba, R., Nagata, S., Pfeiffer, J., and Asashima, M. (1998) Dev. Genes Evol. 207, 417–426. [DOI] [PubMed] [Google Scholar]
- 31).Yoshida, S., Furue, M., Nagamine, K., Abe, T., Fukui, Y., Myoishi, Y., Fujii, T., Okamoto, T., Taketani, Y., and Asashima, M. (2005). In Vitro Cell. Dev. Biol. Anim. 41, 104–110. [DOI] [PubMed] [Google Scholar]
- 32).Nagamine, K., Furue, M., Fukui, A., and Asashima, M. (2005) Zool. Sci. 22, 755–761. [DOI] [PubMed] [Google Scholar]
- 33).Inui, M., and Asashima, M. (2006) Gene Expr. Patterns 6, 613–619. [DOI] [PubMed] [Google Scholar]
- 34).Ariizumi, T., Komazaki, S., Asashima, M., and Malacinski, G. M. (1996) Int. J. Dev. Biol. 40, 715–718. [PubMed] [Google Scholar]
- 35).Ariizumi, T., Kinoshita, M., Yokota, C., Takano, K., Fukuda, K., Moriyama, N., Malacinski, G. M., and Asashima, M. (2003) Int. J. Dev. Biol. 47, 405–410. [PubMed] [Google Scholar]
- 36).Moriya, N., Uchiyama, H., and Aasashima, M. (1993) Dev. Growth Differ. 35, 123–128. [DOI] [PubMed] [Google Scholar]
- 37).Uochi, T., and Asashima, M. (1996) Dev. Growth Differ. 38, 625–634. [DOI] [PubMed] [Google Scholar]
- 38).Osafune, K., Nishinakamura, R., Komazaki, S., and Asashima, M. (2002) Dev. Growth Differ. 44, 161–167. [DOI] [PubMed] [Google Scholar]
- 39).Chan, T. C., Takahashi, S., and Asashima, M. (2000) Dev. Biol. 228, 256–269. [DOI] [PubMed] [Google Scholar]
- 40).Chan, T. C., Ariizumi, T., and Asashima, M. (1999) Naturwissenschaften 86, 224–227. [DOI] [PubMed] [Google Scholar]
- 41).Chan, T. C., and Asashima, M. (2006) Nephron Exp. Nephrol. 103, e81–85. [DOI] [PubMed] [Google Scholar]
- 42).Uochi, T., Takahashi, S., Ninomiya, H., Fukui, A., and Asashima, M. (1997) Dev. Growth Differ. 39, 571–580. [DOI] [PubMed] [Google Scholar]
- 43).Uochi, T., and Asashima, M. (1998) Gene 211, 245–250. [DOI] [PubMed] [Google Scholar]
- 44).Sato, A., Asashima, M., Yokota, T., and Nishinakamura, R. (2000) Mech. Dev. 92, 273–275. [DOI] [PubMed] [Google Scholar]
- 45).Satow, R., Chan, T. C., and Asashima, M. (2002) Biochem. Biophys. Res. Commun. 295, 85–91. [DOI] [PubMed] [Google Scholar]
- 46).Kyuno, J., Fukui, A., Michiue, T., and Asashima, M. (2003) Biochem. Biophys. Res. Commun. 309, 52–57. [DOI] [PubMed] [Google Scholar]
- 47).Kaneko, T., Chan, T., Satow, R., Fujita, T., and Asashima, M. (2003) Biochem. Biophys. Res. Commun. 308, 566–572. [DOI] [PubMed] [Google Scholar]
- 48).Satow, R., Chan, T. C., and Asashima, M. (2004) Biochem. Biophys. Res. Commun. 321, 487–494. [DOI] [PubMed] [Google Scholar]
- 49).Li, D. H., Chan, T., Satow, R., Komazaki, S., Hashizume, K., and Asashima, M. (2005) Int. J. Dev. Biol. 49, 401–408. [DOI] [PubMed] [Google Scholar]
- 50).Nishinakamura, R., Matsumoto, Y., Nakao, K., Nakamura, K., Sato, A., Copeland, N. G., Gilbert, D. J., Jenkins, N. A., Scully, S., Lacey, D. L.et al. (2001) Development 128, 3105–3115. [DOI] [PubMed] [Google Scholar]
- 51).Onuma, Y., Nishinakamura, R., Takahashi, S., Yokota, T., and Asashima, M. (1999) Biochem. Biophys. Res. Commun. 264, 151–156. [DOI] [PubMed] [Google Scholar]
- 52).Moriya, N., Komazaki, S., Takahashi, S., Yokota, C., and Asashima, M (2000) Dev. Growth Differ. 42, 593–602. [DOI] [PubMed] [Google Scholar]
- 53).Moriya, N., Komazaki, S., and Asashima, M. (2000) Dev. Growth Differ. 42, 175–185. [DOI] [PubMed] [Google Scholar]
- 54).Sogame, A., Hayata, T., and Asashima, M. (2003) Dev. Growth Differ. 45, 143–152. [DOI] [PubMed] [Google Scholar]
- 55).Asashima, M., Nakano, H., Uchiyama, H., Sugino, H., Nakamura, T., Eto, Y., Ejima, D., Nishimatsu, S., Ueno, N., and Kinoshita, K. (1991) Proc. Natl. Acad. Sci. USA 88, 6511–6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56).Fukui, A., Nakamura, T., Uchiyama, H., Sugino, K., Sugino, H., and Asashima, M. (1994) Dev. Biol. 163, 279–281. [DOI] [PubMed] [Google Scholar]
- 57).Fukui, A., Shiurba, R., and Asashima, M. (1999) Cell. Mol. Biol. 45, 545–554. [PubMed] [Google Scholar]
- 58).Fukui, A., Komazaki, S., Miyoshi, O., and Asashima, M. (2003) Dev. Growth Differ. 45, 113–119. [DOI] [PubMed] [Google Scholar]
- 59).Takahashi, S., Yokota, C., Takano, K., Tanegashima, K., Onuma, Y., Goto, J., and Asashima, M. (2000) Development 127, 5319–5329. [DOI] [PubMed] [Google Scholar]
- 60).Onuma, Y., Takahashi, S., Yokota, C., and Asashima, M. (2002) Dev. Biol. 241, 94–105. [DOI] [PubMed] [Google Scholar]
- 61).Haramoto, Y., Tanegashima, K., Onuma, Y., Takahashi, S., Sekizaki, H., and Asashima, M. (2004) Dev. Biol. 265, 155–168. [DOI] [PubMed] [Google Scholar]
- 62).Onuma, Y., Takahashi, S., Haramoto, Y., Tanegashima, K., Yokota, C., Whitman, M., and Asashima, M. (2005) Dev. Dyn. 234, 900–910. [DOI] [PubMed] [Google Scholar]
- 63).Haramoto, Y., Takahashi, S., and Asashima, M. (2006) Biochem. Biophys. Res. Commun. 346, 470–478. [DOI] [PubMed] [Google Scholar]
- 64).Takahashi, S., Onuma, Y., Yokota, C., Westmoreland, J. J., Asashima, M., and Wright, C. V. (2006) Genesis 44, 309–321. [DOI] [PubMed] [Google Scholar]
- 65).Ito, Y., Oinuma, T., Takano, K., Komazaki, S., Obata, S., and Asashima, M. (2006) Gene. Expr. Patterns 6, 294–298. [DOI] [PubMed] [Google Scholar]
- 66).Hayata, T., Tanegashima, K., Takahashi, S., Sogame, A., and Asashima, M. (2002) Mech. Dev. 112, 37–51. [DOI] [PubMed] [Google Scholar]
- 67).Massague, J., Seoane, J., and Wotton, D. (2005) Genes Dev. 19, 2783–2810. [DOI] [PubMed] [Google Scholar]
- 68).Shen, M. M., and Schier, A. F. (2000) Trends Genet. 16, 303–309. [DOI] [PubMed] [Google Scholar]
- 69).Whitman, M. (2001) Dev. Cell. 1, 605–617. [DOI] [PubMed] [Google Scholar]
- 70).Yabe, S., Tanegashima, K., Haramoto, Y., Takahashi, S., Fujii, T., Kozuma, S., Taketani, Y., and Asashima, M. (2003) Development 130, 2071–2081. [DOI] [PubMed] [Google Scholar]
- 71).Tanegashima, K., Yokota, C., Takahashi, S., and Asashima, M. (2000) Mech. Dev. 99, 3–14. [DOI] [PubMed] [Google Scholar]
- 72).Tanegashima, K., Haramoto, Y., Yokota, C., Takahashi, S., and Asashima, M. (2004) Int. J. Dev. Biol. 48, 275–283. [DOI] [PubMed] [Google Scholar]
- 73).Abe, T., Furue, M., Myoishi, Y., Okamoto, T., Kondow, A., and Asashima, M. (2004) Int. J. Dev. Biol. 48, 327–332. [DOI] [PubMed] [Google Scholar]
- 74).Abe, T., Furue, M., Kondow, A., Matsuzaki, K., and Asashima, M. (2005) Mech. Dev. 122, 671–680. [DOI] [PubMed] [Google Scholar]
- 75).Hitoshi, S., Alexson, T., Tropepe, V., Donoviel, D., Elia, A. J., Nye, J. S., Conlon, R. A., Mak, T. W., Bernstein, A., van der Kooy, D. (2002) Genes Dev. 16, 846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76).Uehara, M., Haramoto, Y., Sekizaki, H., Takahashi, S., Asashima, M. (2002) Dev. Growth Differ. 44, 427–436. [DOI] [PubMed] [Google Scholar]
- 77).Sedohara, A., Suzawa, K., and Asashima, M. (2006) Int. J. Dev. Biol. 50, 385–392. [DOI] [PubMed] [Google Scholar]
- 78).Sekizaki, H., Takahashi, S., Tanegashima, K., Onuma, Y., Haramoto, Y., and Asashima, M. (2004) Dev. Dyn. 229, 367–372. [DOI] [PubMed] [Google Scholar]