Significance
Germinal center (GC) response is central for generation of memory B cells and plasma cells that produce high-affinity antibodies, which are crucial for protective immunity against various foreign antigens. Even though key genetic factors for the GC formation are known, it is largely unknown how this process is controlled by epigenetic factors. Here we have demonstrated that the activity of SWI/SNF chromatin remodeling complex is required for the development of both GC B cells and follicular helper T cells. The SWI/SNF complex modulates Bcl-6–mediated Blimp-1 repression, and thus plays as a key mediator in the GC response. Our results provide fundamental insights into the epigenetic regulation in the GC reaction.
Keywords: SWI/SNF chromatin remodeling complex, follicular helper T cell, germinal center reaction, Blimp-1
Abstract
Germinal center (GC) reaction is crucial in adaptive immune responses. The formation of GC is coordinated by the expression of specific genes including Blimp-1 and Bcl-6. Although gene expression is critically influenced by the status of chromatin structure, little is known about the role of chromatin remodeling factors for regulation of GC formation. Here, we show that the SWI/SNF chromatin remodeling complex is required for GC reactions. Mice lacking Srg3/mBaf155, a core component of the SWI/SNF complex, showed impaired differentiation of GC B and follicular helper T cells in response to T cell-dependent antigen challenge. The SWI/SNF complex regulates chromatin structure at the Blimp-1 locus and represses its expression by interacting cooperatively with Bcl-6 and corepressors. The defect in GC reactions in mice lacking Srg3 was due to the derepression of Blimp-1 as supported by genetic studies with Blimp-1–ablated mice. Hence, our study identifies the SWI/SNF complex as a key mediator in GC reactions by modulating Bcl-6–dependent Blimp-1 repression.
The germinal centers (GCs) represent sites of humoral immunity for immune protection against foreign antigens. The primary function of GCs is to generate memory B cells as well as plasma B cells (PCs) producing high-affinity antibodies (1). For GC reaction, interaction between B and T cells is crucial. Follicular helper CD4+ T (TFH) cells are distinct subsets of CD4+ effector T cells that migrate into the boundary between the T-cell zone and B-cell follicles and provide cognate help for GC formation (2). Defective function of TFH results in impaired GC reactions in the context of a T cell-dependent antibody response and is also associated with autoimmune diseases such as systemic lupus erythematosus (3, 4).
The differentiation of GC B and TFH cells is genetically controlled by specific transcription factors, such as B lymphocyte-induced maturation protein-1 (Blimp-1) and B-cell lymphoma 6 (Bcl-6). Blimp-1 is a key regulator in the differentiation of PCs. Blimp-1 represses a large number of genes that are required for GC formation (5). In addition, Blimp-1 blocks the generation of TFH cells. Constitutive expression of Blimp-1 in CD4+ T cells impairs TFH differentiation and conversely, deletion of Blimp-1 in CD4+ T cells augments TFH differentiation (3). Blimp-1 is antagonistically repressed by Bcl-6. Bcl-6−/− mice show defects in T cell-dependent antibody response due to the impairment of GC formation and ectopic Bcl-6 expression inhibits plasmacytic differentiation of B cells (6). Bcl-6−/− CD4+ T cells are defective in TFH differentiation, and ectopic expression of Bcl-6 drives CD4+ T cells into TFH cells.
As chromatin structure organized in higher-order nucleosomes impedes the transcriptional process in eukaryotes, epigenetic regulation of chromatin structure is required for proper gene expression (7). The SWI/SNF chromatin remodeling complex is an ATP-dependent remodeler that drives conformational changes of nucleosomes by using the energy from ATP hydrolysis. The SWI/SNF complex is a multisubunit complex composed of at least 12 different subunits including Brg1, a core subunit containing the ATPase activity, and Srg3/mBaf155 (hereafter referred to as Srg3), a core subunit that controls the stability of this complex (8–11). Whereas the SWI/SNF complex has been known to be essential for gene activation, increasing evidence also indicates its involvement in gene repression (12–14).
In this study, we show that the SWI/SNF complex is required for the differentiation of GC B and TFH cells. The SWI/SNF complex modulates the chromatin conformation and the transcriptional repression of Blimp-1 through cooperative interaction with Bcl-6 and histone modifiers. Our data suggest that the SWI/SNF complex is an essential mediator for GC reactions.
Results
GC Formation Is Defective in Srg3fl/flCD19Cre+ Mice.
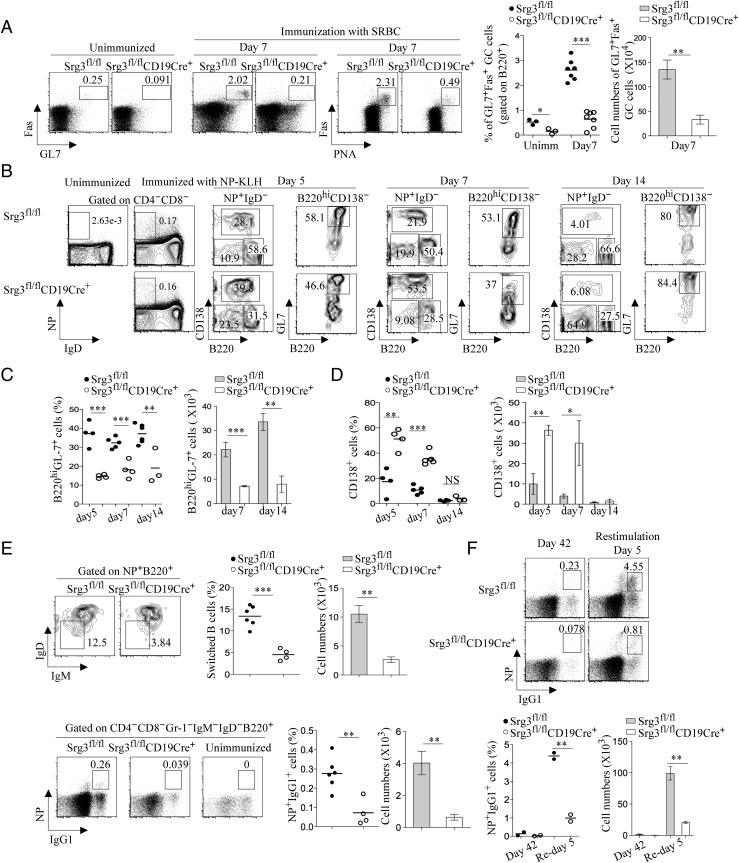
We specifically ablated Srg3 in B lineage cells by crossing Srg3fl/fl mice with CD19-Cre mice (15). Srg3fl/flCD19Cre+ mice showed normal B-cell development in the bone marrow (SI Appendix, Fig. S1 A and B). The V(D)J rearrangement of the Ig heavy chain (Igh) and κ-light chain genes of Srg3-deficient B cells was similar to that of control B cells (SI Appendix, Fig. S1C). No significant changes in architecture, such as the size and the distribution of red and white pulp, were observed in the spleen of Srg3-deficient mice (SI Appendix, Fig. S2A). B-cell development including follicular (FO) and marginal zone (MZ) B cells in the periphery was not significantly altered by Srg3 loss (SI Appendix, Fig. S2 B and C). In addition, the proliferation of Srg3-deficient B cells in response to cross-linking of the B-cell receptor (BCR) with anti-IgM or anti-IgM+IL-4 was also similar to that of control B cells (SI Appendix, Fig. S2D). However, spontaneous generation of GC B cells was defective in Peyer's patches of unimmunized Srg3fl/flCD19Cre+ mice (SI Appendix, Fig. S3). To assess the formation of GC B cells, we immunized mice with T cell-dependent antigen, sheep red blood cells (SRBCs). Srg3fl/flCD19Cre+ mice showed severe defects in the generation of GL7+Fas+ or peanut agglutinin (PNA)-positive GC B cells (Fig. 1A). When we immunized mice with nitrophenol coupled to keyhole limpet hemocyanin (NP-KLH), another T cell-dependent antigen, Srg3fl/flCD19Cre+ mice failed to develop GC B cells (SI Appendix, Fig. S4). In addition, H&E staining of splenic sections showed a substantial deficiency in the GC formation (SI Appendix, Fig. S5). Specifically, there was a significant decrease in the frequency and cell numbers of NP-specific GC B cells in Srg3fl/flCD19Cre+ mice (Fig. 1 B and C). In contrast to the depletion of GC B cells, Srg3fl/flCD19Cre+ mice showed an increase in the frequency and cell numbers of NP+B220low/−CD138+ short-lived PCs after immunization (Fig. 1 B and D). Srg3fl/flCD19Cre+ mice also had a defect in the generation of IgD−IgM− switched B cells and there were considerably fewer NP-specific IgG1+ B cells on day 14 after immunization (Fig. 1E). When we assess the memory B-cell response on day 5 after renewed antigen encounter, Srg3fl/flCD19Cre+ mice had an impaired secondary antibody response (Fig. 1F). We also found that Srg3 is expressed in high levels in GC B cells (SI Appendix, Fig. S6). Together, these results suggest that the SWI/SNF complex is required for GC formation.
Fig. 1.
The generation of GC B cells is impaired in Srg3fl/flCD19Cre+ mice. (A) Flow cytometry of GC B cells obtained from the spleen of unimmunized or SRBC-immunized mice at day 7. Cells gated on B220+ were analyzed for the expression of surface molecules, Fas, GL-7, and PNA. Frequency and cell numbers of Fas+GL-7+ GC B cells were calculated (Right). Each individual dot represents one mouse; the lines show the mean percentage. Error bars represent a SEM; *P < 0.05, **P < 0.01, ***P < 0.001. (B) Flow cytometry of NP-specific PCs and GC B cells immunized with NP-KLH after immunization. NP and IgD levels on CD4−CD8− cells obtained from unimmunized or immunized mice with NP-KLH. Surface expression of CD138 and B220 in cells gated on NP+IgD− and expression of GL-7 on CD138−B220+ cells were analyzed. (C) Frequency (Left) and cell numbers (Right) of NP-specific B220hiCD138−GL-7+ GC B cells. (D) Frequency (Left) and cell numbers (Right) of NP-specific B220−CD138+ PCs. (E) Flow cytometry of the expression of IgM and IgD in cells gated on NP+B220+ at day 14 after immunization (Upper Left). Frequency and cell numbers (Upper Right) of NP-specific IgM−IgD− switched B cells 14 d after immunization. Flow cytometry of NP+IgG1+ cells gated on CD4−CD8−IgM−IgD−Gr-1−B220+ isotype-switched B cells was analyzed (Lower Left). Frequency and cell numbers (Lower Right) of NP+IgG1+ B cells were calculated. (F) Flow cytometry of splenocytes at day 42 primary and 5 d after antigen recall (Upper). Frequency and cell numbers of NP-specific memory cells for each group at day 42 and 5 d after rechallenge of antigen were calculated (Lower).
Blimp-1 Expression Is Derepressed in the Absence of the SWI/SNF Complex.
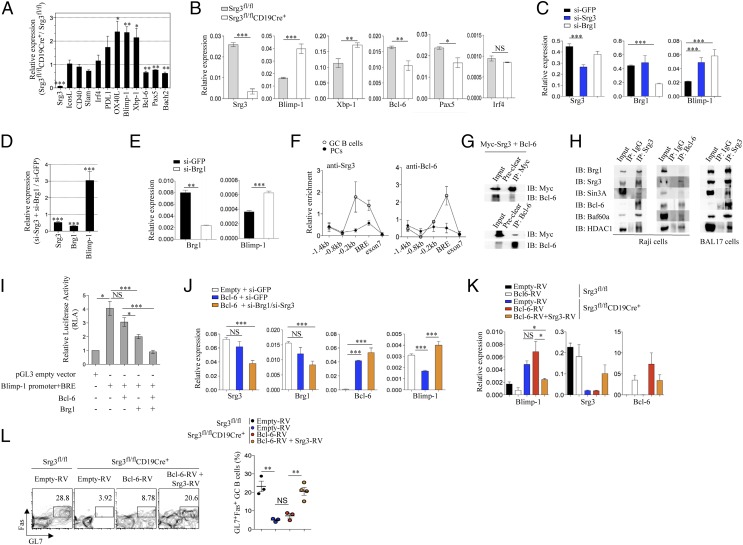
When we analyzed gene expression profiles in FO B cells, there was much higher expression of Blimp-1 in Srg3-deficient B cells than in control cells (Fig. 2A). Xbp-1, which is induced by Blimp-1, was also highly expressed and the expression of Pax5, Bcl-6, each of which is repressed by Blimp-1, was down-regulated in Srg3-deficient FO B cells (5, 16, 17). The amounts of Blimp-1 transcript were much higher in Srg3-deficient B cells after activation with LPS+IL-4 (Fig. 2B). In addition, Blimp-1 expression was substantially elevated in Srg3 knockdown BAL17 cells (Fig. 2C). The knockdown of Brg1 also expressed Blimp-1 highly and the depletion of both Srg3 and Brg1 together also induced up-regulation of the Blimp-1 expression (Fig. 2D). We also confirmed the derepression of Blimp-1 by Brg1 depletion in another B-cell line, Raji cells (Fig. 2E). These data collectively suggest that the SWI/SNF complex mediates Blimp-1 repression.
Fig. 2.
The SWI/SNF complex is required for Bcl-6–mediated Blimp-1 repression. (A) Quantitative RT-PCR analysis of the expression of genes required for the differentiation of PCs and GC B cells in FO B cells purified from Srg3fl/flCD19Cre+ mice; results are presented relative to those of FO B cells purified from Srg3fl/fl mice. *P < 0.05, **P < 0.01, ***P < 0.001. (B) FO B cells of Srg3fl/fl or Srg3fl/flCD19Cre+ mice were stimulated for 3 d in the presence of LPS (20 μg/mL) plus IL-4 (10 ng/mL) and the expression of Srg3, Blimp-1, Xbp-1, Bcl-6, Pax5, and Irf4 was analyzed using quantitative RT-PCR. NS, not significant. (C) Transcription levels for the indicated genes were determined by quantitative RT-PCR analysis from BAL17 cells transfected with siRNA targeting GFP, Brg1, or Srg3. (D) Quantitative RT-PCR analysis of Srg3, Brg1, and Blimp-1 in BAL17 B cells transfected together with si-Brg1 and si-Srg3; results are presented relative to those of si-GFP–treated cells. (E) Transcription levels for the indicated genes in Brg1-depleted Raji B cells. (F) ChIP analysis of DNA precipitated with anti-Srg3 or anti–Bcl-6 from GC B cells and PCs obtained from mice immunized with NP-KLH, followed by amplification with indicated primers specific to regions at the Blimp-1 locus. The value of bound/input was normalized to the value of nonspecific binding to control region (GAPDH intron). (G) Bcl-6 and Myc-tagged Srg3 were ectopically expressed in 293T cells and lysates were immunoprecipitated with anti-Myc or anti-Srg3 followed by immunoblotting with the indicated antibodies. (H) Immunoprecipitation of Srg3 or Bcl-6 using nuclear extracts purified from Raji or BAL17 B cells was performed. The subunits of the SWI/SNF complex and other proteins associated with Srg3 or Bcl-6 were analyzed by immunoblotting using the indicated antibodies. (I) SW-13 cells lacking Brg1 were transiently cotransfected with BRE-containing Blimp-1 luciferase reporters and Bcl-6 or Brg1. The results are shown as fold activation compared with reporter activity of cells transfected with pGL3 vector only. Data represent the average ± SEM (n = 5). (J) Bcl-6 was transiently transfected to Brg1/Srg3-deficient or -sufficient BAL17 cells, and Blimp-1 expression was analyzed using quantitative RT-PCR. (K) FO B cells of Srg3fl/fl or Srg3fl/flCD19Cre+ mice were activated with LPS and IL-4 during 18–20 h. These cells were transduced with empty retroviral vector expressing GFP only (Empty-RV), Bcl-6 (Bcl-6–RV), or Bcl-6–RV together with Srg3-expressing retroviral vector (Srg3-RV) and further cultured with LPS and IL-4. After 48 h, GFP+ cells were purified by flow cytometry and expression of Blimp-1, Srg3, and Bcl-6 was analyzed. (L) FO B cells of Srg3fl/flCD19Cre+ mice infected with Empty-RV or Bcl-6–RV or Bcl-6–RV together with Srg3-RV were mixed with WT naïve CD4+ T cells and adoptively transferred into Rag2−/− mice subsequently immunized with NP-KLH. After 7 d, the formation of GC B cells was analyzed by flow cytometric detection.
The SWI/SNF Complex Cooperatively Interacts with Bcl-6 for Blimp-1 Repression.
To investigate whether the SWI/SNF complex directly controls Blimp-1 expression, we analyzed the potential binding of the SWI/SNF complex to regulatory regions of the Blimp-1 gene. ChIP analysis revealed a direct and specific association of BRG1 and SRG3 with the proximal promoter and BRE (Bcl-6 responsive element) regions at the Blimp-1 locus and the occupancies of BRG1 and SRG3 were severely reduced in Brg1/Srg3 knockdown cells, similarly to isotype control (SI Appendix, Fig. S7). Furthermore, we found the specific binding of SRG3 to the proximal promoter and BRE regions in GC B cells, in contrast to PCs (Fig. 2F). As Bcl-6 is known to bind to BRE to inhibit Blimp-1 expression, it is likely that Srg3 might be involved in Bcl-6–mediated Blimp-1 regulation. Thus, we attempted to examine the physical interaction between Bcl-6 and Srg3. For this, two plasmids expressing BCL-6 or Myc-tagged SRG3 were cotransfected into 293T cells. Reciprocal coimmunoprecipitation analysis with anti-Myc and anti–Bcl-6 antibodies showed the interaction between SRG3 and BCL-6 (Fig. 2G). To confirm the endogenous interactions, whole-cell extracts of Raji B cells were immunoprecipitated with antibodies to SRG3 or BCL-6. BCL-6 was recovered by immunoprecipitation with antibody to SRG3 and, conversely, immunoprecipitation of BCL-6 revealed subunits of the SWI/SNF complex including SRG3 (Fig. 2H, Left). We also confirmed this association in BAL17 cells (Fig. 2H, Right). Notably, corepressors including mSin3A and HDAC1 were also found to be associated with components of the SWI/SNF complex and BCL-6, indicating that the mSin3A-containing SWI/SNF complex interacts with BCL-6. To test whether repressional activity of Bcl-6 on Blimp-1 expression requires the SWI/SNF complex, we generated luciferase reporters containing the Blimp-1 promoter and BRE as previously described (18). When Bcl-6 was transiently transfected into SW-13 cells, which is deficient in Brg1, reporter activity was not altered (Fig. 2I). When Bcl-6 was cotransfected with Brg1, however, reporter activity was repressed. Ectopic expression of Bcl-6 failed to repress Blimp-1 in Brg1/Srg3-depleted cells, whereas Blimp-1 transcripts were down-regulated by Bcl-6 in control cells (Fig. 2J). Moreover, when we transduced Bcl-6 into Srg3-deficient B cells followed by the activation with LPS+IL-4, repression of Blimp-1 was not detected (Fig. 2K). However, coexpresion of Bcl-6 with Srg3 in Srg3-deficient B cells augmented the effects of Bcl-6 on the inhibition of Blimp-1 expression. When Srg3-deficient B cells transduced with empty, Bcl-6, or Bcl-6 and Srg3 together were adoptively transferred with WT naïve CD4+ T cells into Rag2−/− mice and immunized with NP-KLH, cells transduced with Bcl-6 alone failed to develop GC B cells (Fig. 2L). These results indicate that the SWI/SNF complex is required for Bcl-6–mediated Blimp-1 repression.
The SWI/SNF Complex Regulates Chromatin Structure and Histone Modifications at the Blimp-1 Locus.
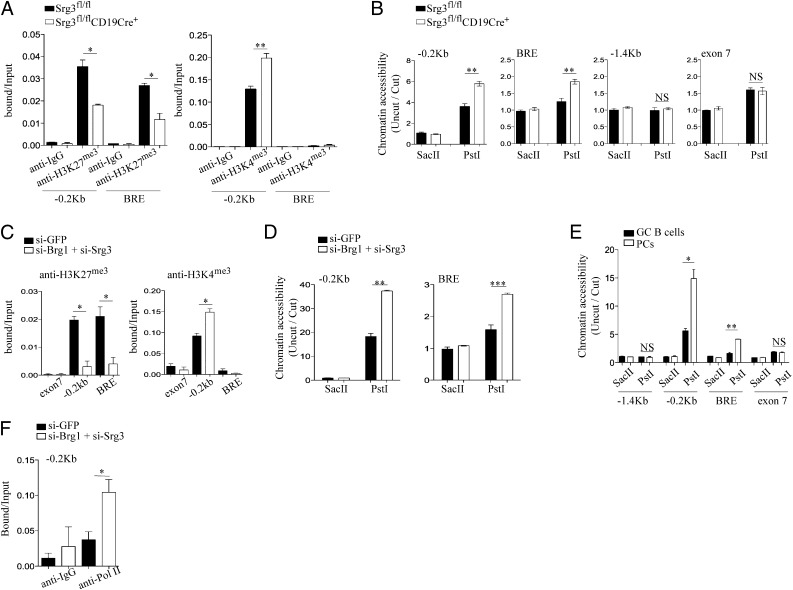
We tested whether the loss of the SWI/SNF complex affects histone modifications at the Blimp-1 locus. The levels of repressive trimethylated histone H3 at lysine 27 (H3K27me3) at the proximal promoter and BRE region were significantly reduced in FO B cells obtained from Srg3fl/flCD19Cre+ mice, whereas significant increase in the permissive methylation at lysine 4 of H3 (H3K4me3) levels at the proximal promoter locus was observed (Fig. 3A). To test whether the loss of the SWI/SNF complex affects chromatin structure, we performed a restriction endonuclease accessibility assay within the Blimp-1 locus (19, 20). BRE and the proximal promoter regions contain a PstI recognition site, but no SacII restriction site. The PstI cleavage efficiencies were increased in BRE and the proximal promoter in Srg3-deficient B cells, whereas the chromatin status of regions including −1.4 kb and exon 7, which showed no bindings of the SWI/SNF complex, was not affected by Srg3 loss, indicating that the lack of the SWI/SNF complex makes the chromatin status of both BRE and the proximal promoter regions accessible (Fig. 3B). Consistently, knockdown of Brg1 and Srg3 in BAL17 cells reduced H3K27me3 levels at the proximal promoter and BRE regions and significantly elevated H3K4me3 levels at the proximal promoter region (Fig. 3C). The loss of Brg1 and Srg3 also affected the efficiencies of PstI cleavage at BRE and the proximal promoter regions (Fig. 3D). We also found that the chromatin status of BRE and the proximal promoter regions at the Blimp-1 locus were more accessible in PCs compared with GC B cells (Fig. 3E). Moreover, recruitment of Pol II to the proximal promoter region was significantly increased in Brg1/Srg3 knockdown BAL17 cells (Fig. 3F). Collectively, these data indicate that the SWI/SNF complex modulates repressive chromatin structures of Blimp-1 by facilitating histone modification.
Fig. 3.
Srg3 loss impacts on the histone modification and chromatin structures at the Blimp-1 locus. (A) ChIP analysis of FO B cells obtained from Srg3fl/fl or Srg3fl/flCD19Cre+ mice with anti-isotype IgG, anti-H3K4me3, or anti-H3K27me3. The precipitated DNA was analyzed by quantitative RT-PCR using the indicated primers amplifying the promoter region (−0.2 kb) or BRE at the Blimp-1 locus. (B) Restriction endonuclease accessibility assay was performed on nuclei isolated from FO B cells of Srg3fl/fl or Srg3fl/flCD19Cre+ mice. Nuclei were digested with SacII or PstI and then subjected to quantitative RT-PCR analysis for the Blimp-1 locus with the indicated primers; −0.2 kb, BRE and exon 7 contain PstI recognition site, but not SacII site. The −1.4 kb does not contain both PstI and SacII recognition sites. (C) ChIP analysis of Brg1/Srg3-depleted or -intact BAL17 cells with anti-H3K27me3 or anti-H3K4me3. The precipitated DNA was analyzed by quantitative RT-PCR using the indicated primers amplifying the promoter region (−0.2 kb), BRE, or exon 7. (D) Restriction endonuclease accessibility with nuclei isolated from Brg1/Srg3-depleted or intact BAL17 cells. Nuclei digested with SacII or PstI were subjected to quantitative RT-PCR analysis for the promoter region and BRE. (E) Restriction endonuclease accessibility with nuclei isolated from PCs and GC B cells obtained from mice immunization with NP-KLH. Nuclei digested with SacII or PstI were subjected to quantitative RT-PCR analysis for the Blimp-1 locus with the indicated primers. (F) ChIP analysis of Brg1/Srg3-depleted or intact BAL17 cells with anti-Pol II. The precipitated DNA was analyzed by quantitative RT-PCR using the indicated primers amplifying the proximal promoter region (−0.2 kb). *P < 0.05, **P < 0.01, ***P < 0.001.
Defect in the Generation of GC B Cells in Srg3fl/flCD19Cre+ Mice Is Dependent on the Derepression of Blimp-1 Expression.
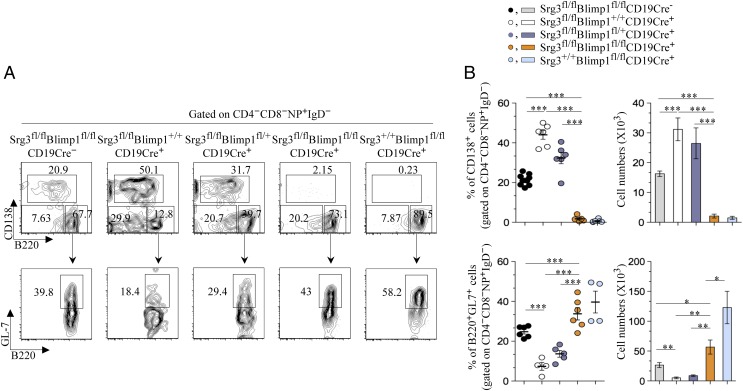
As the numbers of short-lived PCs were increased with augmented expression of Blimp-1 and reciprocally GC B-cell formation was compromised in Srg3fl/flCD19Cre+ mice, we hypothesized that increased expression of Blimp-1 in mice lacking Srg3 resulted in the defect in the generation of GC B cells. To directly confirm this hypothesis, we mated Blimp-1fl/fl mice with Srg3fl/fl mice to genetically ablate Blimp-1 in B cells using CD19-Cre transgenic mice and analyzed GC B-cell generation in these mice. Consistent with previous results, Blimp-1 deficiency (Srg3+/+Blimp-1fl/flCD19Cre+ mice) caused a dramatic increase in GC B cells (Fig. 4 and SI Appendix, Fig. S8). Whereas there was severe reduction in GC B cells in Srg3fl/flBlimp-1+/+CD19Cre+ mice, deletion of Blimp-1 in mice lacking Srg3 (Srg3fl/flBlimp-1fl/flCD19Cre+ mice) was sufficient to rescue the defect in GC B-cell development (Fig. 4 and SI Appendix, Fig. S8). These findings were further corroborated by H&E staining of splenic section (SI Appendix, Fig. S9A). In addition, heterozygous deletion of Blimp-1 in mice lacking Srg3 (Srg3fl/flBlimp-1fl/+CD19Cre+ mice) also marginally increased the generation of GC B cells compared with that in Srg3fl/flBlimp-1+/+CD19Cre+ mice, indicating that impaired GC B-cell development and reciprocal increase in short-lived PCs in mice lacking Srg3 were dependent on the depression of Blimp-1. The severely decreased production of NP-specific IgG1+ B cells in mice lacking Srg3 was also restored by Blimp-1 ablation on day 14 (SI Appendix, Fig. S9B). Together, these data demonstrate that the SWI/SNF complex-mediated repression of Blimp-1 is essential for GC B-cell formation.
Fig. 4.
The impaired differentiation of GC B-cells in Srg3fl/flCD19Cre+ mice is rescued by the genetic ablation of Blimp-1. (A) Flow cytometry of NP-specific PCs and GC B cells in the spleen of NP-KLH–immunized mice on day 7. Surface expression levels of CD138 and B220 of cells gated on CD4+CD8−NP+IgD− and GL-7 levels on CD138−B220+ cells were analyzed. Numbers adjacent to the outlined area indicate the percentage of populations. (B) Frequency and cell numbers of NP-specific B220−CD138+ PCs (Upper) and CD138−B220+GL7+ GC B cells in mice as described. Error bars represent a SEM; *P < 0.05, **P < 0.01, ***P < 0.001.
TFH Cell Differentiation Is Impaired in Srg3fl/flCD4Cre+ Mice.
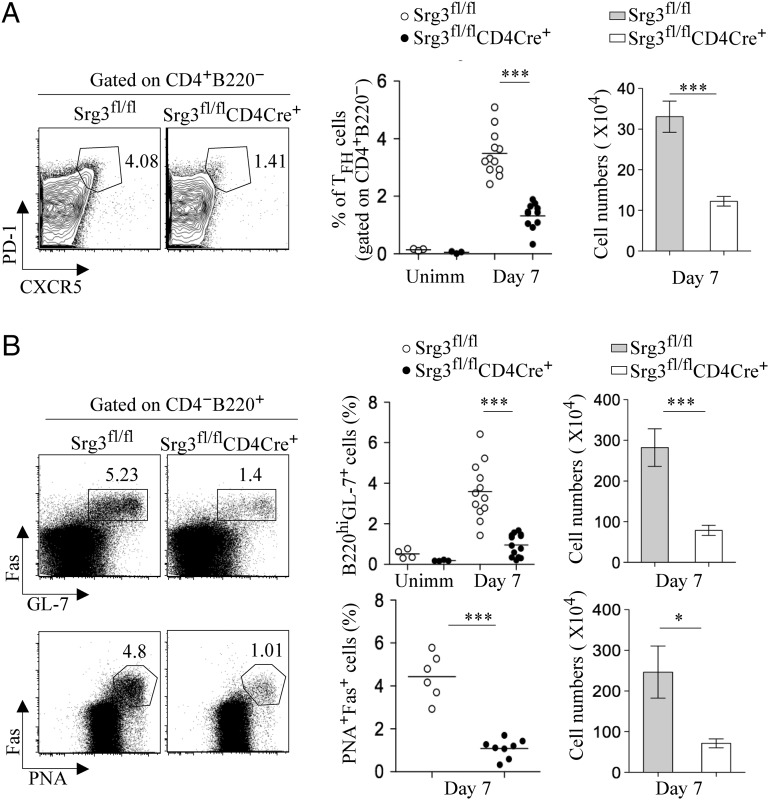
In addition to GC B cells, Bcl-6–mediated Blimp-1 repression is essential for TFH cell differentiation. To investigate whether the SWI/SNF complex is required for TFH cell differentiation, we specifically ablated Srg3 by crossing Srg3fl/fl mice with CD4-Cre transgenic mice (21). Srg3fl/flCD4Cre+ mice showed grossly normal CD4/CD8 T-cell development and the proliferation of Srg3-deficient CD4+ T cells was similar to that of control CD4+ T cells upon in vitro T-cell receptor (TCR) stimulation (SI Appendix, Fig. S10). Interestingly, we found that spontaneous generation of GC B cells was impaired in Peyer's patches of unimmunized Srg3fl/flCD4Cre+ mice (SI Appendix, Fig. S11). As T-cell help in follicles is known to be crucial in GC B-cell formation, we assessed TFH differentiation by immunizing mice with SRBCs. Srg3fl/flCD4Cre+ mice had a much lower frequency and cell numbers of PD-1hiCXCR5hi TFH cells on day 7 after immunization (Fig. 5A). GC B cells were also substantially decreased in Srg3fl/flCD4Cre+ mice (Fig. 5B). We also confirmed the expression of Srg3 in TFH cells (SI Appendix, Fig. S12). These data indicate that the SWI/SNF complex is essential for TFH cell differentiation.
Fig. 5.
The differentiation of TFH cells is defective in Srg3fl/flCD4Cre+ mice. (A) Flow cytometry of splenocytes of Srg3fl/fl or Srg3fl/flCD4Cre+ mice immunized with SRBCs on day 7. Cells gated on B220−CD4+ were analyzed for expression of PD-1 and CXCR5 (Left). Statistical quantitation of CXCR5+PD-1hi TFH cells as frequency gated on CD4+ T cells (B220−CD4+) and cell numbers (Right) was analyzed. Each individual dot represents one mouse and data are presented as mean and SEM. ***P < 0.001. (B) Flow cytometry of GC B cells (Fas+GL-7+ or Fas+PNA+ gated on B220+CD4−cells) obtained from the spleen of SRBC-immunized mice on day 7. Frequency and cell numbers of GC B cells were statistically analyzed. *P < 0.05, ***P < 0.001.
Impaired TFH Cell Differentiation in Srg3fl/flCD4Cre+ Mice Is Rescued by the Genetic Ablation of Blimp-1.
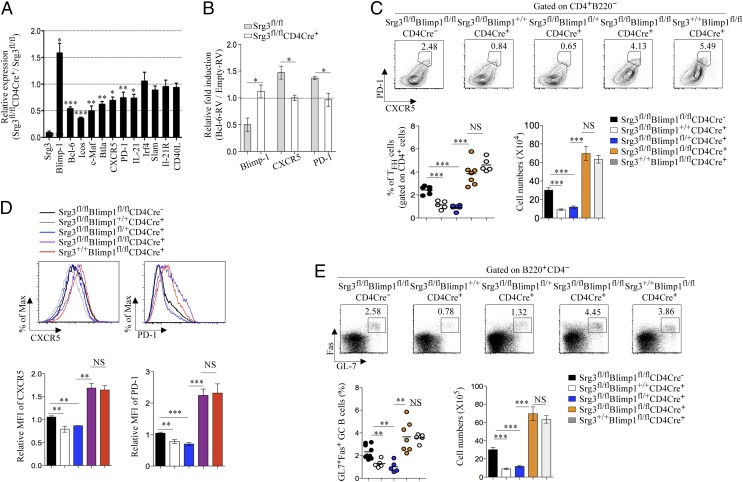
We surveyed mRNA expression profiles in CD4+ T cells purified from Srg3fl/fl and Srg3fl/flCD4Cre+ mice on day 1 after in vitro activation under TFH-like cell differentiation conditions (4, 22). There was much lower expression of genes that are required for TFH cell differentiation, such as Bcl-6, c-Maf, ICOS, CXCR5, and PD-1 in Srg3-deficient CD4+ T cells than in control cells (Fig. 6A). Notably, the expression of Blimp-1, which inhibits TFH differentiation (3), was significantly higher in Srg3-deficient CD4+ T cells. In addition, Blimp-1 repression by Bcl-6 expression was compromised in Srg3-deficient CD4+ T cells, whereas Blimp-1 expression was down-regulated in control CD4+ T cells transduced with retroviral vector expressing Bcl-6 (Bcl-6-RV) compared with those transduced with empty retroviral vector expressing GFP only (Empty-RV) (Fig. 6B). We hypothesized that impaired TFH cell differentiation in Srg3fl/flCD4Cre+ mice might result from increased expression of Blimp-1. To address this possibility, we genetically ablated Blimp-1 in Srg3fl/flCD4Cre+ mice and analyzed TFH cell differentiation. Srg3fl/flBlimp-1fl/+CD4Cre+ or Srg3fl/flBlimp-1+/+CD4Cre+ mice showed a severe defect in TFH cell development (Fig. 6 C and D). However, the genetic ablation of Blimp-1 in mice lacking Srg3 (Srg3fl/flBlimp-1fl/flCD4Cre+ mice) restored the differentiation of TFH cells with up-regulation of CXCR5 and PD-1 (Fig. 6 C and D). Moreover, blockade of GC B-cell generation in mice lacking Srg3 was also sufficiently restored by the ablation of Blimp-1 (Fig. 6E). However, consistent with previous results (23, 24), Blimp-1–deficient CD4+ T cells in Srg3fl/flBlimp-1fl/flCD4Cre+ had an abnormally activated phenotype displaying CD44hiCD25Llow on the surface (SI Appendix, Fig. S13). In addition, Blimp-1 deficiency in T cells is known to be associated with autoimmunity (23, 24). To circumvent these complications, we deleted Blimp-1 or Srg3 genes in vitro in naïve CD4+ T cells purified from Srg3fl/flBlimp-1fl/+ or Srg3fl/flBlimp-1fl/+ mice by using a Cre-expressing retroviral vector (Cre-RV) (3). We adoptively transferred the Srg3fl/flBlimp-1fl/+ CD4+ T cells transduced with Empty-RV or Cre-RV and Srg3fl/flBlimp-1fl/fl CD4+ T cells transduced with Cre-RV with WT B cells into Rag2−/− mice subsequently immunized with NP-KLH. All CD4+GFP+ T cells in Rag2−/− mice after immunization expended equivalently without altering proliferation (SI Appendix, Fig. S14A). Deletion of Blimp-1 together with Srg3 sufficiently restored TFH differentiation, whereas there was substantial decrease in TFH cell in the absence of Srg3 (SI Appendix, Fig. S14B). GC B cells were also restored by ablation of Blimp-1 (SI Appendix, Fig. S14C). Together, these data indicate that impaired differentiation of TFH cells in mice lacking Srg3 was mediated by derepression of Blimp-1. These data indicate that impaired differentiation of TFH cells in mice lacking Srg3 was mediated by derepression of Blimp-1.
Fig. 6.
The deletion of Blimp-1 rescues the perturbed differentiations of TFH and GC B cells in Srg3fl/flCD4Cre+ mice. (A) CD4+ T cells of Srg3fl/fl CD4Cre+ mice were stimulated for 24 h with anti-CD3 and anti-CD28 in the presence of IL-6, anti–IL-4, anti–IFN-γ, and anti–TGF-β, followed by quantitative RT-PCR analysis for the expression of genes involved in differentiation or function in TFH cells; results are presented relative to those of CD4+ T cells of Srg3fl/fl mice. Data are presented by mean and SEM; *P < 0.05, **P < 0.01, ***P < 0.001. n = 4–8. (B) CD4+ T cells of Srg3fl/fl or Srg3fl/flCD4Cre+ mice were transduced with Empty-RV or Bcl-6–RV after 18-h activation with anti-CD3 and anti-CD28 and further cultured in condition as described in A. After 48 h, GFP+ cells were purified by flow cytometry and expression of Blimp-1, CXCR5, and PD-1 was analyzed by quantitative RT-PCR; results in CD4+ T cells transduced with Bcl-6–RV are presented as fold induction relative to those in CD4+ T cells transduced with Empty-RV. Data are presented as mean and SEM; *P < 0.05. n = 3–4. (C) Flow cytometry of CXCR5+PD-1hi TFH cells obtained from the spleen of NP-KLH–immunized mice on day 7. The statistical quantitation of frequency in CD4+ T cells and cell numbers for CXCR5+PD-1hi TFH cells was analyzed. Each individual dot represents one mouse and data are presented as mean and SEM. (D) CXCR5 and PD-1 expressions in CD4+ cells. Bar graphs show mean fluorescent intensity of CXCR5 (Left) and PD-1 (Right). (E) Flow cytometry of GC B cells obtained from the spleen of NP-KLH–immunized mice on day 7. Cells gated on CD4−B220+ were analyzed for the expression of Fas and GL-7 (Upper). Numbers adjacent to outlined area indicate the percentage of populations. The statistical quantitation of frequency in CD4−B220+ B cells and cell numbers for CXCR5+PD-1hi TFH cells (Lower) was analyzed.
Discussion
Recently, the antagonistic functions of two major transcriptional repressors, Bcl-6 and Blimp-1, in the differentiation of TFH cells and GC B cells have been revealed (2). We found that the SWI/SNF complex plays a critical role in the formation of GC B cells and TFH differentiation. We have shown here that the SWI/SNF complex directly interacts with Bcl-6 and regulates the transcriptional repression of Blimp-1 by modulating chromatin structure and histone modification. Our findings indicate that the SWI/SNF complex coordinates the fates of activated B cells, as well as CD4+ T cells by cooperatively interacting with transcription factors and epigenetic modifiers.
We found that the genetic ablation of Srg3 in B cells severely impaired GC formation upon immunization with T cell-dependent antigen. In contrast, short-lived PCs were significantly increased in Srg3fl/flCD19Cre+ mice. Our data have also shown that GC reaction was compromised by deleting Srg3 in T cells due to the defect in TFH cell differentiation. These overt characteristics of impairment in GC B and TFH cell generation closely resemble those observed in sustained expression of Blimp-1. It has been reported that Blimp-1 favors differentiation of PCs and inhibits the genetic program of GC B-cell development (25, 26). On the contrary, loss of Blimp-1 resulted in the enhanced differentiation of GC B cells (27). Ectopic expression of Blimp-1 in CD4+ T cells suppresses TFH cell differentiation (3). Indeed, Srg3-deficient B cells showed higher expression of Blimp-1 and its target gene, Xbp-1. Consistently, when CD4+ T cells were activated in TFH-like polarizing conditions, Blimp-1 expression was significantly higher in Srg3-deficient CD4+ T cells. Furthermore, we found that defective differentiation of GC B and TFH cells observed in mice lacking Srg3 was restored by genetic ablation of Blimp-1. Therefore, derepressed expression of Blimp-1 itself and/or downstream target genes of Blimp-1 is a likely explanation for the impairment of GC formation and TFH cell differentiation in Srg3-deficient mice. However, the cell numbers of GC B cells in Srg3fl/flBlimp-1fl/flCD19Cre+ mice were not completely rescued by Blimp-1 deletion compared with Srg3+/+Blimp-1fl/flCD4Cre+ mice, even though there was not a substantial difference in the frequency of GC B cells between two mice. Blimp-1–deficient B cells are known to be hyperproliferative and this characteristic is associated with the increased expression of genes that are necessary for cell division and cell cycle, such as c-Myc, E2F-1, and p21 (5, 27, 28). Notably, the SWI/SNF complex is required for the transcriptional activities of c-Myc, E2F-1, and for the up-regulation of p21 (29, 30). Thus, it is likely that the functional activity of genes described above for proliferation of GC B cells may be impaired in doubly mutant mice lacking both Blimp-1 and Srg3, compared with mice lacking Blimp-1 but having intact Srg3.
Our results provided direct evidences that the SWI/SNF complex is required for Bcl-6–mediated transcriptional repression of Blimp-1. It has been shown that Bcl-6 antagonistically inhibits Blimp-1 expression and this is a key genetic program for the differentiation of GC and TFH cells (3, 18). We found that subunits of the SWI/SNF complex including Brg1 and Srg3 bound to BRE, which is a functional response element mediating Blimp-1 repression by associating with Bcl-6. Ablation of Srg3 compromised the Bcl-6 activity of Blimp-1 repression and, as a result, the differentiation of GC B cells and TFH cells. Thus, our data reflect the indispensable requirement of the SWI/SNF complex for the functional activity of Bcl-6 in GC reactions. Previous studies have shown that the SWI/SNF complex physically remodels nucleosomal structure of chromatin and mediates transcriptional regulation through cooperative interaction with chromatin modifying enzymes (7, 12, 31–33). We have also found that the SWI/SNF complex interacts with subunits of histone deacetylases complex, such as HDAC1 and Sin3A. In accordance with this result, the SWI/SNF complex seems to regulate chromatin structures by facilitating histone modification at BRE region of Blimp-1 locus. In intact condition, BRE region was marked by the repressive histone modification of H3K27me3 and formed an inaccessible chromatin structure. However, in the absence of Srg3 or Brg1, the level of H3K27me3 modification was substantially reduced and chromatin structure was altered into accessible conformations. Thus, our data indicate that the recruitment of the SWI/SNF complex by Bcl-6 mediates the repressive conformation at the BRE region through facilitation of histone modification. In addition, we found that the SWI/SNF complex also binds to the proximal promoter of the Blimp-1 gene and that the level of active histone marker H3K4me3 was significantly increased in the absence of Srg3. Chromatin conformation of the proximal promoter region was altered to become accessible and Pol II binding was enhanced in the absence of Srg3. Thus, it is likely that binding of the SWI/SNF complex to both the proximal promoter and BRE regions may promote an inaccessible chromatin structure probably by forming a looping structure and this conformational change mediates Blimp-1 repression.
The decrease of H3K27me3 by loss of the SWI/SNF complex at the Blimp-1 locus is interesting because this repressive histone marker is modulated by the Ezh2–PRC2 complex (34, 35). Previous studies have shown that Ezh2 is required for GC formation and the SWI/SNF complex synergistically regulates several genes together with the Ezh2–PRC2 complex (36–39). MTA3, a subunit of the NuRD complex, is also known to interact with Bcl-6 and to inhibit Blimp-1 expression (40). It has been shown that the NuRD complex mediates the recruitment of the Ezh2–PRC2 complex and that the SWI/SNF complex physically interacts with subunits of the NuRD complex including Mi-2β, HDAC1/2, MTA2, and MBD3 (31, 33, 41–44). In addition, it has been reported that the association of the NuRD complex in regulatory regions that are co-occupied with the SWI/SNF complex is severely reduced in the absence of the SWI/SNF complex in various biological contexts, whereas the binding of the SWI/SNF complex is not affected by the NuRD complex (42, 45). Thus, it appears that the cooperative interplay between the SWI/SNF and NuRD complexes is required for the maintenance of the H3K27me3 histone marker mediated by the PRC2 complex at the Blimp-1 locus for Blimp-1 repression.
Blimp-1 is known to be highly expressed in follicular regulatory T (TFR) cells. These Foxp3-expresssing CXCR5hiPD-1hi TFR cells control TFH cells and limit germinal center reactions (46–48). It may be possible that the function or development of Foxp3+ TFR, which express high levels of Blimp-1, is impaired in the absence of Blimp-1. Thus, a lack of TFR cells leads to an increase in the TFH cell numbers and enhances the GC reactions, so the rescued development of TFH and GC B cells observed in Srg3fl/flBlimp-1fl/flCD4Cre+ may be due to the defect in Foxp3+ TFR cells. Our results described in SI Appendix, Fig. S14 have shown that deletion of Blimp-1 together with Srg3 in adoptively transferred CD4+ T cells sufficiently restored GC reactions. As TFR cells originate from thymic-derived natural Treg cells, these results may reduce the possible effect of TFR cells in rescued TFH cells in this system. However, we cannot completely rule out the possibility that defective Foxp3+ TFR cells in the absence of Blimp-1 may contribute to the increase in TFH cell differentiation and GC response observed in mice lacking Srg3 and Blimp-1.
Blimp-1 plays a critical role for GC reactions by regulating the differentiations of GC B and TFH cells. Thus, the misregulation of Blimp-1 impairs GC reactions and finally results in uncontrolled immune responses upon variable antigens. Whereas the target genes regulated by Blimp-1 is different between B and T cells, the repression of Blimp-1 by transcriptional repressor Bcl-6 is indispensable for differentiation of both GC B and TFH cells. Our study provides evidences that the SWI/SNF complex mediates the repressive chromatin structures in the Blimp-1 gene by facilitating the histone modifications. Thus, the SWI/SNF complex appears to be a key regulator for GC reactions.
Materials and Methods
Mice.
Targeting constructs and strategy for the generation of conditional Srg3/mBaf155 knockout mice have been described (49). LoxP-flanked Blimp-1 conditional KO mice (27), Rag2−/−, TCRβ−/−, CD19-Cre, and CD4-Cre transgenic mice were purchased from The Jackson Laboratories. All mice were bred and maintained in specific pathogen-free barrier facilities at Seoul National University and Institute of Molecular Biology and Genetics and were used according to protocols approved by the Institutional Animal Care and Use Committees of Seoul National University.
Immunization.
For evaluation of T cell-dependent antibody responses, 8- to 10-wk-old mice were immunized intraperitoneally with 100 μg NP-KLH (Bioresearch Technologies) precipitated in aluminum hydroxide (Imject Alum; Pierce) or with 200 μL of 10% (vol/vol) SRBCs (Innovative Research) in PBS. Secondary challenge was done 6 wk after primary immunization.
Immunoprecipitation and Western Blotting.
The immunoblot assay was performed with indicated antibodies: anti-Myc (Sigma), Bcl-6 (Santa Cruz Biotechnology), mSin3A (Santa Cruz Biotechnology), HDAC1 (Santa Cruz Biotechnology), and Baf60a (BD Biosciences). Antisera for Brg1 and Srg3 were raised from rabbits in our laboratories. Detailed methods are presented in SI Appendix, SI Materials and Methods.
Chromatin Immunoprecipitation Assay.
Sample preparation and immunoprecipitation were performed as previously described (49). DNA was purified with a QIAquick Spin kit (Qiagen) and eluted DNA was analyzed by PCR with indicated primers (SI Appendix, Table S1). Detailed methods are presented in SI Appendix, SI Materials and Methods.
Flow Cytometry.
Cell sorting was performed using FACSAria II (BD Biosciences) at the National Center for Inter-University Research Facilities at Seoul National University. Data were collected using FACSCanto II (BD Biosciences) and were analyzed with FlowJo software (Tree Star). Sample preparation and cell suspension were performed as described (49). Detailed methods are presented in SI Appendix, SI Materials and Methods.
Restriction Endonuclease Accessibility Assay.
Chromatin accessibility assay was performed as described (19, 50) with minor modifications. Detailed methods are presented in SI Appendix, SI Materials and Methods.
Statistical Analysis.
All statistical analyses were performed using Prism 5 (GraphPad Prism) software. Two-tailed Student's t tests were used to assess the statistical significance of differences (P values) between groups. P values less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank members of our laboratories for useful advice and discussions and Y. Kim for critical feedback on the manuscript. This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT, and Future Planning (2014073789 and 2012R1A2A2A01046164 and Research Center for Functional Cellulomics). J.C., K.P., S.C., and S.J. are supported by the BK21 Plus Program (F14SN01D1305) from the Ministry of Education and the NRF. J.C. is supported by a Seoul Science Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418592112/-/DCSupplemental.
References
- 1.MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117–139. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 2.Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 3.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325(5943):1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nurieva RI, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29(1):138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaffer AL, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17(1):51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 6.Reljic R, Wagner SD, Peakman LJ, Fearon DT. Suppression of signal transducer and activator of transcription 3-dependent B lymphocyte terminal differentiation by BCL-6. J Exp Med. 2000;192(12):1841–1848. doi: 10.1084/jem.192.12.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108(4):475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 8.Sohn DH, et al. SRG3 interacts directly with the major components of the SWI/SNF chromatin remodeling complex and protects them from proteasomal degradation. J Biol Chem. 2007;282(14):10614–10624. doi: 10.1074/jbc.M610563200. [DOI] [PubMed] [Google Scholar]
- 9.Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3(2):247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, et al. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10(17):2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 11.Jeon SH, et al. A new mouse gene, SRG3, related to the SWI3 of Saccharomyces cerevisiae, is required for apoptosis induced by glucocorticoids in a thymoma cell line. J Exp Med. 1997;185(10):1827–1836. doi: 10.1084/jem.185.10.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sif S, Saurin AJ, Imbalzano AN, Kingston RE. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 2001;15(5):603–618. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barker N, et al. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J. 2001;20(17):4935–4943. doi: 10.1093/emboj/20.17.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martens JA, Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr Opin Genet Dev. 2003;13(2):136–142. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 15.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25(6):1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin KI, Angelin-Duclos C, Kuo TC, Calame K. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol Cell Biol. 2002;22(13):4771–4780. doi: 10.1128/MCB.22.13.4771-4780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaffer AL, et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21(1):81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Tunyaplin C, et al. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol. 2004;173(2):1158–1165. doi: 10.4049/jimmunol.173.2.1158. [DOI] [PubMed] [Google Scholar]
- 19.Rao S, Procko E, Shannon MF. Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J Immunol. 2001;167(8):4494–4503. doi: 10.4049/jimmunol.167.8.4494. [DOI] [PubMed] [Google Scholar]
- 20.Hagège H, et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat Protoc. 2007;2(7):1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 21.Lee PP, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15(5):763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 22.Ise W, et al. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat Immunol. 2011;12(6):536–543. doi: 10.1038/ni.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kallies A, et al. Transcriptional repressor Blimp-1 is essential for T cell homeostasis and self-tolerance. Nat Immunol. 2006;7(5):466–474. doi: 10.1038/ni1321. [DOI] [PubMed] [Google Scholar]
- 24.Martins GA, et al. Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat Immunol. 2006;7(5):457–465. doi: 10.1038/ni1320. [DOI] [PubMed] [Google Scholar]
- 25.Martins G, Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol. 2008;26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- 26.Shapiro-Shelef M, Calame K. Regulation of plasma-cell development. Nat Rev Immunol. 2005;5(3):230–242. doi: 10.1038/nri1572. [DOI] [PubMed] [Google Scholar]
- 27.Shapiro-Shelef M, et al. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19(4):607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 28.Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276(5312):596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- 29.Cheng SW, et al. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet. 1999;22(1):102–105. doi: 10.1038/8811. [DOI] [PubMed] [Google Scholar]
- 30.Giraud S, Hurlstone A, Avril S, Coqueret O. Implication of BRG1 and cdk9 in the STAT3-mediated activation of the p21waf1 gene. Oncogene. 2004;23(44):7391–7398. doi: 10.1038/sj.onc.1207972. [DOI] [PubMed] [Google Scholar]
- 31.Datta J, et al. Physical and functional interaction of DNA methyltransferase 3A with Mbd3 and Brg1 in mouse lymphosarcoma cells. Cancer Res. 2005;65(23):10891–10900. doi: 10.1158/0008-5472.CAN-05-1455. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Pal S, et al. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol Cell Biol. 2003;23(21):7475–7487. doi: 10.1128/MCB.23.21.7475-7487.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trotter KW, Archer TK. The BRG1 transcriptional coregulator. Nucl Recept Signal. 2008;6:e004. doi: 10.1621/nrs.06004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reynolds N, et al. NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. EMBO J. 2012;31(3):593–605. doi: 10.1038/emboj.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32(4):491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Béguelin W, et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 2013;23(5):677–692. doi: 10.1016/j.ccr.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caganova M, et al. Germinal center dysregulation by histone methyltransferase EZH2 promotes lymphomagenesis. J Clin Invest. 2013;123(12):5009–5022. doi: 10.1172/JCI70626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho L, et al. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol. 2011;13(8):903–913. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2009;27(2):317–328. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- 40.Fujita N, et al. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004;119(1):75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 41.Liang J, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10(6):731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 42.Yildirim O, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147(7):1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brand M, et al. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat Struct Mol Biol. 2004;11(1):73–80. doi: 10.1038/nsmb713. [DOI] [PubMed] [Google Scholar]
- 44.Mahajan MC, Narlikar GJ, Boyapaty G, Kingston RE, Weissman SM. Heterogeneous nuclear ribonucleoprotein C1/C2, MeCP1, and SWI/SNF form a chromatin remodeling complex at the beta-globin locus control region. Proc Natl Acad Sci USA. 2005;102(42):15012–15017. doi: 10.1073/pnas.0507596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramirez-Carrozzi VR, et al. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20(3):282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chung Y, et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med. 2011;17(8):983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Linterman MA, et al. Foxp3+ follicular regulatory T cells control the germinal center response. Nat Med. 2011;17(8):975–982. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sage PT, Francisco LM, Carman CV, Sharpe AH. The receptor PD-1 controls follicular regulatory T cells in the lymph nodes and blood. Nat Immunol. 2013;14(2):152–161. doi: 10.1038/ni.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi J, et al. The SWI/SNF-like BAF complex is essential for early B cell development. J Immunol. 2012;188(8):3791–3803. doi: 10.4049/jimmunol.1103390. [DOI] [PubMed] [Google Scholar]
- 50.Weinmann AS, Plevy SE, Smale ST. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity. 1999;11(6):665–675. doi: 10.1016/s1074-7613(00)80141-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.