Significance
The origin of hepatitis C virus (HCV) has long remained a mystery. Unexpectedly, a plethora of HCV-related hepaciviruses was recently discovered in horses, monkeys, rodents, and bats. These discoveries are of particular interest and may aid in understanding HCV evolution, molecular biology, and natural history. Currently, immunocompetent HCV animal models are lacking, impeding vaccine development; novel hepaciviruses and their natural hosts could provide such models. Here, we demonstrate that the closest HCV homolog, nonprimate hepacivirus (NPHV), is a hepatotropic equine virus with many similarities to HCV, including the capacity to establish persistent infection, delayed-onset seroconversion, and liver pathology. We identify the complete NPHV genome and establish a functional clone infectious in horses, a key advance providing a direct link between virus infection and clinical outcome.
Keywords: hepatitis C virus, infectious cDNA clone, equine liver disease, animal model, 3′-untranslated region
Abstract
Nonprimate hepacivirus (NPHV) is the closest known relative of hepatitis C virus (HCV) and its study could enrich our understanding of HCV evolution, immunity, and pathogenesis. High seropositivity is found in horses worldwide with ∼3% viremic. NPHV natural history and molecular virology remain largely unexplored, however. Here, we show that NPHV, like HCV, can cause persistent infection for over a decade, with high titers and negative strand RNA in the liver. NPHV is a near-universal contaminant of commercial horse sera for cell culture. The complete NPHV 3′-UTR was determined and consists of interspersed homopolymer tracts and an HCV-like 3′-terminal poly(U)-X-tail. NPHV translation is stimulated by miR-122 and the 3′-UTR and, similar to HCV, the NPHV NS3-4A protease can cleave mitochondrial antiviral-signaling protein to inactivate the retinoic acid-inducible gene I pathway. Using an NPHV consensus cDNA clone, replication was not observed in primary equine fetal liver cultures or after electroporation of selectable replicons. However, intrahepatic RNA inoculation of a horse initiated infection, yielding high RNA titers in the serum and liver. Delayed seroconversion, slightly elevated circulating liver enzymes and mild hepatitis was observed, followed by viral clearance. This establishes the molecular components of a functional NPHV genome. Thus, NPHV appears to resemble HCV not only in genome structure but also in its ability to establish chronic infection with delayed seroconversion and hepatitis. This NPHV infectious clone and resulting acute phase sera will facilitate more detailed studies on the natural history, pathogenesis, and immunity of this novel hepacivirus in its natural host.
Until recently, hepatitis C virus (HCV) and the enigmatic GB-virus (GBV) B were the only known members of the Hepacivirus genus in the Flaviviridae family. Searches for related viruses in primates were unsuccessful, leaving the history and origin of HCV unknown. Recently, developments in deep sequencing methods accelerated identification of a number of related viruses (1). Nonprimate hepacivirus (NPHV) was the first to be discovered, initially identified in samples from dogs and termed canine hepacivirus (2). However, with the exception of a single seropositive farm dog (3), subsequent studies did not detect viral RNA or antibodies in dogs (4–6). Instead, horses appear to be the natural host for NPHV, with 30–40% antibody positive (4, 7) and 2–7% RNA positive among US and European horses (4–7) and an even higher RNA prevalence (35%) in a small Japanese herd (8). Tissue tropism and disease association remain largely uncharacterized. Subsequently, hepaciviruses have also been discovered in rodents (6, 9), bats (10), and Old World primates (11). Pegiviruses are closely related to hepaciviruses and include human (HPgV/GBV-C) and simian (SPgV/GBV-A) pegiviruses, none of which are associated with disease (12). More recently, rodent (RPgV) (9), bat (BPgV) (10), and equine (EPgV and Theiler’s disease associated virus, TDAV) (13, 14) pegiviruses were discovered. Interestingly, strong epidemiological evidence links serum hepatitis of horses (Theiler’s disease) to infection with TDAV (13).
The NPHV genome resembles that of HCV, with a long ORF of around 8,826 nt encoding 2,942 aa predicted to yield the structural proteins Core, E1, and E2 and the nonstructural proteins p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B. The 5′-UTR of around 384 nt resembles that of HCV but with a longer stem-loop I and only a single miR-122 seed site (4). The internal ribosome entry site (IRES) is similar in sequence and predicted structure to that of HCV (4) and is capable of driving translation of the downstream ORF (15). Whether NPHV engages and requires the liver-specific miR-122 in a manner similar to that of HCV remains to be established.
Here, we characterize NPHV in horses and find that NPHV can cause persistent hepatotropic infection. We further determine the complete NPHV 3′-UTR, the longest among hepaciviruses, encompassing several interspersed homopolymer tracts. This allowed the construction of a full-length cDNA clone that was infectious in a horse after intrahepatic inoculation of in vitro transcribed RNA. Taken together, our findings demonstrate exciting parallels between HCV in humans and NPHV in horses, offering an attractive model for hepacivirus studies.
Results
NPHV Is Associated with Persistent Hepatotropic Infection in Horses.
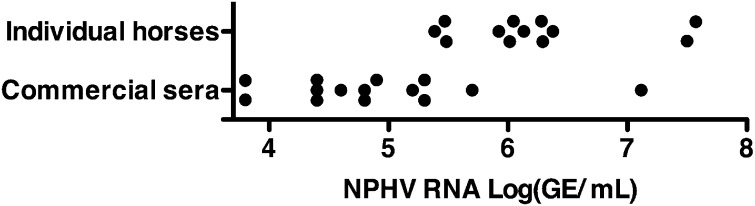
To determine the prevalence of NPHV infection, we tested sera from two groups of healthy US horses for viral RNA. In one group, we found one NPHV RNA-positive horse among serum samples drawn from 18 animals in 2008–2010. In consecutive samples from 2011 and 2012, the originally infected horse had cleared the infection, whereas a newly introduced horse (M303) was positive. Sequencing of the 5′-UTR revealed the presence of the same isolate for both horses and in consecutive samples from the second animal taken 1 y apart. We previously reported an EPgV prevalence of 15–32% in this herd (14). In another group, we found unusually high NPHV prevalence, with 7/17 NPHV positive (41%) and one coinfected with EPgV. No history was available to explain this high prevalence. An unrelated NPHV-positive horse was identified with retrospective samples, revealing chronic infection for more than 12 y. From NPHV-infected horses in the two groups and a previously described group (4), we determined viral RNA levels in serum ranging from 5.4 log10 genome equivalents (GE)/mL to 7.5 log10(GE)/mL, with horse M303 among the highest titers (Fig. 1).
Fig. 1.
NPHV RNA titers in individual animal and commercial horse sera. NPHV RNA genome equivalents (GE) per milliliter were measured by qRT-PCR.
To investigate NPHV tissue tropism, we collected serum, peripheral blood mononuclear cells (PBMCs), liver and lymph node biopsies, and a transtracheal wash sample from horse M303. NPHV RNA levels were undetectable in the tracheal sample and in PBMCs, whereas low levels were found in the lymph node and high levels in serum and in the liver (Table 1). We further found negative-strand RNA in the liver of M303, a hallmark of viral replication. Similar findings were made in two other horses. Although our analysis does not exclude replication in other tissues, it provides strong evidence that NPHV is an equine hepatotropic virus with the ability to establish persistent infection.
Table 1.
Tissue tropism and persistence of NPHV
| Infection parameter | Individual horses | ||
| Time infected | |||
| Persistence | >1 y | 4.5 mo | >12 y |
| Viral RNA titer, Log10(GE/mL) | |||
| Serum | 7.5 | 5.2–7.7 | 5.0 |
| Viral RNA titer, Log10(GE/ng total RNA) | |||
| Liver biopsy | 3.7 | 2.5–3.3 | 2.5 |
| Lymph node biopsy | 1.1 | 1.1 | N/A |
| PBMCs | <LLOD | <LLOD | N/A |
| Tracheal wash | <LLOD | N/A | N/A |
| Presence of (−)RNA | |||
| Liver biopsy | Yes | Yes | N/A |
N/A, not available; LLOD, lower level of detection.
Commercial Horse Serum Is Contaminated with NPHV.
Horse serum is a common additive to cell culture media and is typically pooled from a number of animals. Analyzing 15 sera from different companies, we found that 14 were positive for NPHV RNA with titers ranging from 3.8 log10(GE)/mL to 6.7 log10(GE)/mL (Fig. 1). Even one lot of FBS was positive, whereas donkey, goat, sheep, and rabbit sera tested were negative. Sequence analysis of the NPHV structural region revealed a broad diversity of virus isolates in selected serum lots, some of which were vastly different, suggesting pooling from several infected animals (Fig. S1). In general, the range of viral diversity fell within that previously reported (4).
Identification of the Complete NPHV 3′-UTR.
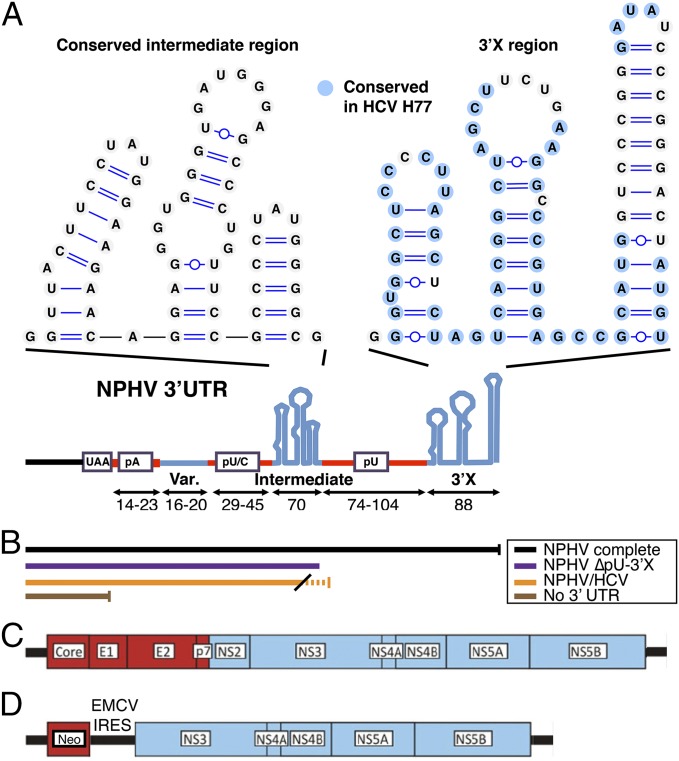
Evidence that the complete NPHV 3′-UTR has been identified is lacking. We therefore used NZP1 serum (4) and serum and liver samples from M303 to determine the complete 3′-UTR of NPHV. Our results revealed to our knowledge the longest hepacivirus 3′-UTR of around 328 nt, including a short poly(A) tract, a variable region, a poly(U/C) tract, a conserved intermediate region, a long poly(U) tract, and a conserved 3′X region (Fig. 2A and Fig. S2). The conserved intermediate and 3′X regions were each predicted to fold into three stem loops. Whereas the upstream part of the 3′-UTR had no resemblance to known sequences, the poly(U) and 3′X regions were structurally similar to HCV but with sequence differences, in particular within the terminal stem loop. The fact that the extreme terminus of the 3′-UTR sequence was predicted to fold into a stem loop resembling that of HCV with identical terminal nucleotides suggested that the complete NPHV 3′-UTR had been determined.
Fig. 2.
Structure of the NPHV 3′-UTR and constructed consensus clones. (A) Elements and predicted structure of the complete NPHV 3′-UTR. Homopolymer tracts are shown in red. Lengths in nucleotides are indicated. (B) The 3′-UTR regions according to A included in versions of translation reporters. The dotted line indicates the HCV 3′-terminal stem loop. (C) Schematic of the full-length NPHV sequence, as inserted downstream of a T7 promoter in the NZP1 consensus clone. (D) Schematic of the NZP1 subgenomic replicon.
Due to the high content of homopolymer stretches, determination of the 3′-UTR was achieved only through a series of linker ligation and homopolymer tailing reactions on (+)RNA and 5′-RACE on (−)RNA. Curiously, using linker ligation on serum-derived RNA, we identified genomes terminating in much longer poly(A) tails immediately downstream of the stop codon (A28–128 for NZP1 and A9–11GA5GA32–40 for B10), compared with the ∼20 nt of the internal poly(A) tract of the complete 3′-UTR. Having identified the complete NPHV 3′-UTR sequence, we next examined its conservation across isolates. We found the overall structure was conserved among isolates with only minor variation in the intermediate and 3′X regions, whereas the region downstream of the poly(A) tract and the length of homopolymer tracts were highly variable (Fig. 2A and Fig. S2).
Construction of an NPHV Full-Length Consensus Clone.
To enable reverse genetic studies of NPHV, we constructed a full-length consensus clone. We used the NZP1 isolate due to (i) its relatedness to the original canine hepacivirus isolate (2, 4), indicating possible cross-species transmission, and (ii) the relatively high RNA titers found in this sample. We confirmed the previously determined 5′-UTR sequence (4) and engineered this downstream of a T7 promoter and a single G residue. The consensus sequence of the complete NZP1 ORF was determined and a sequence with five noncoding differences from consensus, all residues with variation among published isolates, was assembled (Table S1). Compared with the consensus sequence, the published NZP1 bulk sequence (JQ434001) differed in five noncoding and two coding residues (Table S1). The 3′-UTR sequence of the consensus clone is shown in Fig. S2. To allow linearization of the clone before RNA in vitro transcription, a BspEI site was placed immediately downstream of the terminal nucleotide. Thus, the final NZP1 consensus clone consisted of 9,538 nt, including a 5′-UTR of 384 nt, an ORF of 8,826 nt, and a 3′-UTR of 328 nt that carried a poly(U) tract of 96 nt (Fig. 2C and Fig. S1). In addition, residues known to be of critical importance to HCV replication in the NS3 protease, helicase, NS4A, NS5A, and NS5B were conserved in the final NPHV clone (details in Table S1).
NPHV in Vitro Translation Mediates NS3-4A Cleavage of Mitochondrial Antiviral-Signaling Protein.
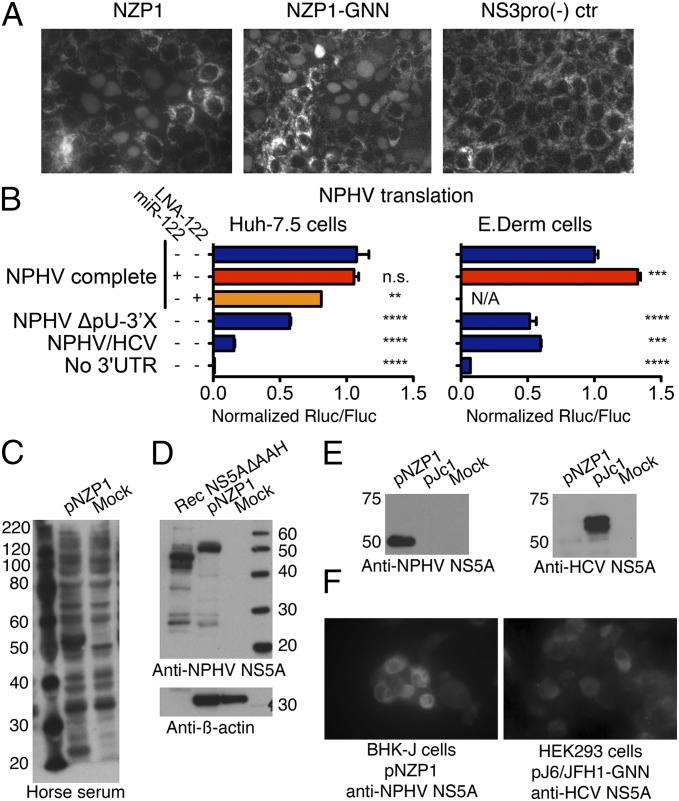
An important strategy for HCV antagonizing induction of IFN-mediated innate immunity is cleavage of the retinoic acid-inducible gene I pathway adaptor mitochondrial antiviral-signaling protein (MAVS) by the viral NS3-4A protease (16). Human hepatoma cell (Huh-7)-derived Clone8 cells carry a highly sensitive MAVS cleavage reporter that translocates RFP to the nucleus upon HCV NS3-4A cleavage (17). Upon transfection of Clone8 cells with RNA transcribed from the NZP1 full-length consensus clone, we observed nuclear translocation (Fig. 3A). This confirmed that NPHV translation occurs in human hepatoma cells and that the NPHV NS3-4A protease is capable of cleaving human MAVS. NPHV did not replicate in Clone8 cells, however, because the percentage of cells with nuclear translocation decreased with similar kinetics for NZP1 and a polymerase-defective mutant (NZP1-GNN).
Fig. 3.
Translation of NPHV and detection of viral proteins. (A) Representative images of Clone8 cells 1 d posttransfection with RNA from NZP1, NZP1-GNN, or the protease-deficient control, NS3pro(−). (B) Translation from monocistronic translation reporters in Huh-7.5 or E.Derm cells. Relative Renilla (Rluc) to firefly (Fluc) luciferase values are shown after normalization for RNA amounts. Mean and SD are shown. Differences were evaluated by ANOVA. For P values, **P < 0.01, ***P < 0.001, and ****P < 0.0001. N/A, not applicable. (C) Western blot (WB) of lysates from T7-expressing HEK293 cells. Crude horse serum (lot 8211574) was used as a primary antibody. (D) WB of recombinant NS5A(ΔAAH) and the HEK293 lysates from B, using purified polyclonal NPHV NS5A8211574 antibody. The size difference for NS5A is due to absence of the amphipathic α-helix (AAH) in the recombinant protein. (E) NPHV and HCV NS5A antibodies do not cross-react. Lysates from T7-expressing HEK293 cells transfected with pNZP1 (NPHV) or pJc1 (HCV) were used for WB, using NPHV NS5A8211574 antibody or HCV NS5A9E10 antibody. (F) Immunostaining of T7-expressing HEK293 or BHK-J cells with and without NPHV (pNZP1) or HCV (pJ6/JFH1-GNN) detected with NPHV NS5A8211574 or HCV NS5A9E10 antibody.
The NPHV 3′-UTR and miR-122 Stimulate Translation.
For HCV, the 3′-UTR (18, 19) and miR-122 (20) have been shown to enhance IRES-dependent translation. To determine whether similar functions were evident for NPHV, we constructed monocistronic reporters in which Renilla luciferase (Rluc) was flanked by the complete NPHV UTRs. For comparison, we constructed a second version truncated before the poly(U) tract (ΔpU-3′X), a third version in which the last stem loop of the intermediate region was replaced by the terminal stem loop of HCV (NPHV/HCV), and a fourth version with no 3′-UTR (Fig. 2B). The effect of the 3′-UTR on translation was evaluated in human hepatoma (Huh-7.5) and equine fibroblasts (E.Derm) by measuring the relative Rluc signal compared with a cotransfected capped firefly luciferase mRNA, after normalization for relative RNA levels. In both cell lines, the presence of the complete NPHV 3′-UTR was critical for efficient translation (Fig. 3B), with the least translation occurring after complete deletion of the 3′-UTR. miR-122 supplementation of Huh-7.5 cells already expressing this miRNA did not change the level of translation; however, sequestrating miR-122 using locked nucleic acid (LNA) decreased translation. Conversely, when E.Derm cells that do not endogenously express miR-122 were engineered to do so, the level of translation was increased (Fig. 3B). Thus, both the NPHV 3′-UTR and miR-122 appeared to stimulate IRES-dependent translation. Despite controlling for RNA levels by quantitative RT (qRT)-PCR, it is possible, however, that differences in RNA stability contribute in part to these results, e.g., through biases introduced by detection of residual extracellular or partially degraded RNA.
Purification of NPHV Antibodies from Horse Serum.
Given the high prevalence of NPHV in commercial horse serum and access to large quantities, we next examined commercial serum as a source of NPHV polyclonal antibodies. To produce NPHV proteins for screening of serum lots, the NZP1 DNA clone was transfected into T7 polymerase expressing HEK293 or BHK-J cells. We then screened eight commercial donor sera, using cell lysates and Western blotting. Two prominent NPHV-specific bands of 53 kDa and 23 kDa were identified by six of eight sera (Fig. 3C). Based on predicted molecular weights, we hypothesized the 53-kDa species to be NS5A and the 23-kDa species to be Core, NS2, or NS4B. Using recombinant NS5A, we successfully affinity purified polyclonal antibodies recognizing both recombinant NS5A and NS5A from transiently transfected cells with minimal background (Fig. 3D). There was no cross-reactivity between NPHV and HCV NS5A antibodies (Fig. 3E). The affinity-purified NS5A antibody was also useful for immunostaining transiently transfected HEK293 and BHK-J cells (Fig. 3F).
NPHV Replication Is Not Readily Established in Vitro.
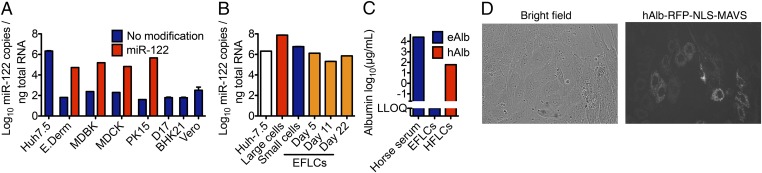
To determine whether RNA transcripts from the NZP1 clone were replication competent in vitro, we transfected RNA from NZP1, NZP1-GNN, and NZP1-Ypet, having the fluorescent protein Ypet inserted between duplicated NS5A and NS5B cleavage sites, into Huh-7.5 (human hepatoma), E.Derm (equine fibroblast), MDBK (bovine kidney), MDCK (canine kidney), D-17 (canine pulmonary osteosarcoma metastasis), PK-15 (porcine kidney), Vero (green monkey kidney), and BHK-21 (hamster kidney) cells. No indication of viral replication was evident in any of the cell lines by NS5A immunostaining, Ypet expression, or intracellular RNA levels (Fig. S3). Although NPHV dependence on miR-122 remains to be established, we tested NPHV replication in E.Derm, MDBK, MDCK, and PK-15 cells transduced with lentivirus for stable miR-122 expression (Fig. 4A). However, ectopic expression of miR-122 was unable to mediate NPHV replication in these cells.
Fig. 4.
Characterization of cell lines and primary EFLCs. (A) miR-122 levels in unmodified cell lines and in cell lines transduced with lentiviruses to express miR-122. (B) miR-122 levels in processed equine fetal liver tissue. Large cell (EFLCs, red) and small cell (blue) preparations before plating and EFLC cultures postplating (days 5–22, orange) were compared with Huh-7.5 cells (white). (C) Equine albumin (eAlb) concentration in EFLCs compared with horse serum and typical human albumin (hAlb) for HFLCs (23). (D) Representative phase-contrast and fluorescence images of EFLC cultures on day 6 postplating. EFLCs were transduced with lentiviruses expressing RFP-NLS-MAVS driven from the human albumin promoter. Error bars in A and B represent SD.
To allow positive selection of replicating genomes, we constructed subgenomic replicons for NPHV (pNZP1-SGR), similar to those described for HCV (21, 22) (Fig. 2D). In parallel with a polymerase-defective negative control, pNZP1-SGR was electroporated into Huh-7.5, E.Derm, E.Derm/miR-122, MDBK, MDBK/miR-122, MDCK/miR-122, PK-15, and PK15/miR-122 cells. No colonies were observed after 2–3 wk of G418 selection, indicating absence of NPHV replication or replication at levels too low to confer resistance.
Given our finding that NPHV is a hepatotropic equine virus, the narrow host and tissue tropism of HCV, and the lack of equine liver cell lines, we attempted to establish equine fetal liver cultures (EFLCs) similar to human fetal liver cultures (HFLCs) known to support HCV replication (23). Primary EFLCs were obtained from an equine fetus at 80 d of gestation, which is an earlier stage of development than that used to establish HCV permissive HFLCs (16–24 wk). EFLCs of hepatocytes, fibroblasts, and presumably other cells were established, and the presence of hepatocytes was confirmed by morphology (23) and high levels of miR-122 (Fig. 4 B and D). In addition, a fraction of cells in the culture became positive for the lentivirus transduced MAVS-RFP-NLS reporter driven by a liver-specific albumin promoter (Fig. 4D). Secreted equine albumin levels, however, were low at all times (Fig. 4C). Despite relatively high, sustained levels of miR-122 and MAVS-RFP-NLS reporter expression in EFLCs, we were unable to confirm infection by NPHV as measured by RFP nuclear translocation after inoculation with serum from three different NPHV-positive horses or transfection of in vitro transcribed RNA from pNZP1.
NPHV RNA Transcripts Are Infectious After in Vivo Intrahepatic Inoculation.
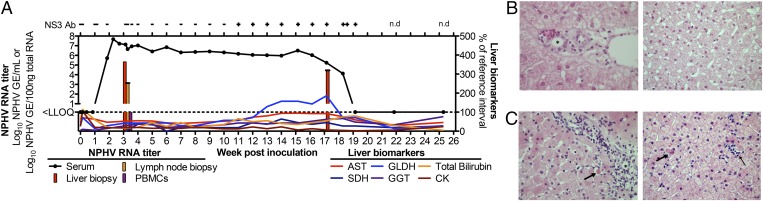
Given the unsuccessful attempts to establish NPHV replication in culture, we next tested the infectivity of the NPHV full-length consensus clone in vivo. We identified a horse negative for NPHV, TDAV, and EPgV RNA and seronegative for NPHV antibodies. A total of ∼350 µg in vitro transcribed NZP1 consensus clone RNA was delivered at seven sites in the liver by video-guided laparoscopy (Movie S1). The horse was negative for circulating NPHV RNA on days 1, 3, and 6 after inoculation, but became positive at week 2, peaking with a viral load of 7.7 log GE/mL at 2.5 wk (Fig. 5A). High viral titers were observed in the liver at weeks 3 and 17 with comparably low or absent titers in the lymph node and PBMCs. Negative strand RNA was also found in the liver at week 3. Late seroconversion was observed at week 11, when NS3-specific antibodies were first detected. This was followed by mild elevations in circulating liver enzymes from week 13 to week 17 (Fig. 5A). Mild but demonstrable changes were observed by liver histology during the early acute phase of infection and at the time of peak glutamate dehydrogenase (GLDH) elevation (weeks 3 and 17) compared with the preinoculation histology. At week 17, lymphocytic infiltration of portal tracts was observed with breaching of the limiting plate accompanied by piecemeal hepatocyte necrosis. Small foci of inflammation were dispersed in the lobules with mild increases in mitotic cells, suggesting compensation for loss of hepatocytes (Fig. 5 B and C). The observed immune response was followed by apparent clearance of the virus, with NPHV RNA titers falling below the limit of quantification from week 19 onward and liver histology reverted to normal at week 22. No changes to the consensus ORF were observed when sequenced at weeks 2, 6, and 17. The complete UTRs of serum-derived virus also had no consensus differences. Among sequenced clones, the poly(U) tract ranged from 55 nt to 92 nt, compared with the 96 nt of the consensus clone and 81–86 nt of the original NZP1 serum. Thus, the NZP1 full-length consensus clone was infectious in vivo, confirming the presence of all essential NPHV genetic elements.
Fig. 5.
Course of infection after intrahepatic inoculation of NPHV RNA. (A) In vitro transcribed RNA from the NZP1 consensus clone was inoculated directly into the liver of a horse (Movie S1), and viral RNA, NS3 antibodies, and liver biomarkers were quantified over time. Liver biomarkers are plotted as percentage of reference interval (AST, 199–374 units/L; SDH, 0–11 units/L; GLDH, 1–8 units/L; GGT, 8–29 units/L; bilirubin, 0.5–2.5 mg/dL (total) and 0.1–0.3 mg/dL (direct); creatine kinase, 142–548 units/L). n.d., not done. (B and C) Representative sections of liver portal tract regions (Left) and lobules (Right) before (B) and 17 wk post (C)-NPHV RNA inoculation. Portal lymphocytic infiltrates (blue nuclei) breach the limiting plate (C, Left). Individual necrotic hepatocytes in the limiting plate indicative of piecemeal necrosis are indicated by the arrow. Individual necrotic hepatocytes are dispersed in the lobules (C, Right, thick arrow). Small foci of lymphocytic inflammation are highlighted (C, Right, thin arrow). The asterisk denotes a bile duct.
Discussion
NPHV is the closest relative of HCV and a thorough understanding of this virus could not only further our understanding of HCV evolution, molecular biology, immune responses, and pathogenesis, but also provide a useful model to study hepacivirus infection in a natural host. Here we report an initial characterization of NPHV infection in horses, the determination of the complete genome sequence, and the successful development of a reverse genetic system. To establish in vitro and in vivo systems, verification of the complete set of genetic elements of the virus is of crucial importance. Only after successful reverse genetic studies launching HCV infection from cDNA-transcribed synthetic RNA in chimpanzees was the complete genetic makeup of the virus known and the direct cause–effect relationship of HCV and liver disease confirmed (24, 25). For HCV, efforts to determine the complete genome sequence were pivotal given that the 3′-terminal highly conserved X tail remained undiscovered for 6 y after the discovery of HCV (26, 27). This enabled the establishment of in vivo and in vitro assays for fundamental virologic studies, target identification and validation, and drug development (21, 22, 28).
In this report, we determined the complete NPHV 3′-UTR, which contains several internal homopolymer tracts, a variable and a conserved region, and a long poly(U) tract and 3′X region highly similar to HCV in predicted structure. Given the finding of genomes terminating with long poly(A) tails immediately downstream of the ORF stop codon, it is possible that in some instances, the internal poly(A) tract causes the viral polymerase to dissociate from its (−)RNA template or acts as a polyadenylation site. Such genomes lacking the complete 3′-UTR could play a role in the viral life cycle, e.g., as mRNAs. It remains to be determined whether putative kissing-loop interactions, as can be predicted for the NPHV 3′-UTR (Fig. S4), are important for replication, as have been described for HCV (29).
Importantly, intrahepatic inoculation of RNA transcripts of the NPHV consensus clone resulted in productive equine infection. Thus, all of the essential elements of the NPHV genome are now defined and reverse genetic studies can be conducted in vivo. Interestingly, although only a single animal has been studied to date, the course of NPHV infection appears remarkably similar to that of HCV, with a delay in onset of seroconversion followed by modest elevations in liver enzymes and histopathological evidence of hepatic inflammation (30). Whereas clearance of HCV infection is often associated with robust elevations in circulating liver enzymes, NPHV infection was cleared following only mild elevations, with only GLDH activity exceeding the reference range. This was consistent with recent observations of mild elevations in liver enzymes in two horses during the clearance of naturally acquired NPHV infection (7). In that study, two chronically infected animals followed for the same period did not show such elevations. In our study, during viremia and particularly during clearance, we observed mild hepatitis with infiltrating lymphocytes breaching the limiting plate, piecemeal necrosis, and foci of hepatic necrosis in the parenchyma. It is of particular interest that this picture mirrors HCV-induced pathogenesis (31), although the extent and penetrance of NPHV-induced clinical liver disease will require additional study. The more distantly related pegivirus, TDAV, appears to be associated with a more severe clinical form of acute liver injury (13).
Similar to previous NPHV prevalence studies (4–6), we found 3–5% of horses with NPHV viremia except for one herd with an unexplained high prevalence of ∼40%. Viral RNA titers in serum were comparable to those found for HCV (31). The prevalent contamination of pooled commercial donor horse serum is therefore unsurprising and resembles contamination of commercial FBS with bovine viral diarrhea virus (BVDV) of the related Pestivirus genus (32). Whether the hepacivirus sequence found in FBS originates from infected cows is interesting but will need to be examined further by sampling individual animals (4). Given the high prevalence of NPHV RNA and antibodies in commercial horse sera, this source should be considered and ruled out as a potential contaminant in hepacivirus discovery studies.
Although sample availability was too limited to quantitatively study persistence, we observed spontaneous clearance, as well as persistent infection for up to 12 y. Another recent study over a period of only 4 mo reported an NPHV chronicity rate of less than 40%, and likely as low as 20% (7), which is lower than that observed for HCV (∼70%) (30). High viral RNA titers and the presence of (−)RNA documented that NPHV is an equine hepatotropic virus. Titers in lymph nodes and PBMCs were low or absent, and although a postmortem organ-wide screen was not possible, our data are consistent with primary or exclusive replication in the liver. Similar results were recently reported in another individual horse (7). These findings are in contrast to what we previously reported for EPgV, where no major differences were observed between levels of viral RNA in liver, lymph node, and PBMCs (14). Thus, the natural history of NPHV infection in horses resembles HCV in many respects: (i) it is a hepatotropic infection affecting ∼3% of the population; (ii) it is capable of establishing persistent infection, although the chronicity rate seems lower than for HCV; (iii) similar levels of viral RNA are found in serum; and (iv) the host response to infection includes delayed seroconversion with concurrent elevations in circulating liver enzymes and liver-infiltrating lymphocytes and hepatocyte necrosis. NPHV could therefore present a useful hepacivirus animal model with marked parallels to HCV. This is particularly important given that the best immunocompetent experimental model for HCV, the chimpanzee, is no longer available for most National Institutes of Health-sponsored research. The human liver-chimeric mouse models and genetically engineered mice are useful for some studies, but lack functional immune systems or allow only limited viral replication (31). GBV-B infection of New World monkeys has been used as a surrogate model, but persistent infection is rare (12, 31). Despite recent breakthroughs in HCV therapy (28), vaccine efforts lag far behind and would be aided by a better understanding of hepacivirus immunity and a preclinical model for testing vaccine candidate concepts (30, 31). Although the large size and associated animal care costs are potential disadvantages of the equine NPHV model, these may be offset by the availability of reagents to follow immune responses as well as extensive experience with equine vaccination and passive immunoprophylaxis.
Our efforts to coax NPHV replication in vitro have thus far proved unsuccessful. This should come as no surprise given the enormous difficulties encountered in attempts to establish robust culture systems for hepatitis viruses, including HBV and HCV (21, 22). One major obstacle to establishing NPHV replication in culture is likely the cellular environment, in this case exacerbated by the lack of equine liver cell lines. For HCV, miR-122 supplementation was sufficient to promote replication in hepatic cells, with lower replication in nonhepatic cells (21, 22). For NPHV, this strategy has been unsuccessful for the nonhepatic cell lines tested thus far. Our attempt to establish NPHV infection in EFLCs was unsuccessful but it is possible that fetal cells at this early gestational age lack the proper hepatic phenotype. In future work, attempts should be made to evaluate later stage EFLCs, micropatterned cocultures of adult equine hepatocytes, hepatocyte-differentiated equine stem cells, or immortalized equine liver cells, similar to successful approaches for HCV (21, 22).
Despite the absence of replication, the NPHV consensus clone was efficiently translated in culture. Using luciferase reporters, we demonstrated the importance of the 3′-UTR and miR-122 for efficient translation from the NPHV IRES, although differences in RNA stability could be a confounding factor. These results mirrored previous reports for HCV (18–20). Future studies are warranted to understand the miR-122 requirement for NPHV replication. In contrast to the HCV 5′-UTR, which has two miR-122 sites, the NPHV 5′-UTR has only seed site 2, with an extended stem-loop I instead of site 1. This suggests that this genome may be a useful tool to further study the role of miR-122 in the hepacivirus life cycle. Our study confirmed the previous finding that the NPHV NS3-4A protease is capable of cleaving MAVS (33, 34). It remains unclear, however, whether NPHV is capable of cleaving equine MAVS, given the significant sequence differences at the cleavage site (ref. 34 and XM_001496561).
In conclusion, this study defines a complete functional NPHV genome and describes key reverse genetic experiments in vivo that fulfill the modern version of Koch’s postulates. Our data demonstrate that NPHV is an equine hepatotropic virus capable of establishing long-term persistent infection with a number of similarities to HCV in its molecular biology and natural history. Further epidemiological studies are needed, however, to determine whether NPHV may be linked to serum hepatitis in horses. Future studies of NPHV also could prove valuable in understanding the immunopathogenesis of hepacivirus infection and determining whether NPHV in horses can be a surrogate model for HCV vaccine development.
Materials and Methods
All animal husbandry and sampling adhered to the Institutional Animal Care and Use Committee protocols at the institutions involved. Detection, quantification, and sequencing of viral RNA and miRNA; determination of the NPHV UTRs; construction of NPHV plasmids; cell culture and preparation of EFLCs; lentivirus transduction for miR-122 expression; NPHV protein expression; purification of NS5A antibodies and Western blotting; translation luciferase assays; in vitro transcription, transfection, and electroporation; and in vivo RNA inoculation and monitoring of infection were done as described in SI Materials and Methods and Tables S2 and S3.
Supplementary Material
Acknowledgments
We are grateful to Drs. William Schneider, Mohsan Saeed, and Margaret MacDonald for important reagents and methodological advice. This study was supported by grants from the Public Health Service; the National Institutes of Health (NIH) (AI072613 and AI107631); the National Institute for Food and Agriculture (230554); the United States Department of Agriculture, Office of the Director through the NIH Roadmap for Medical Research (DK085713); the National Cancer Institute (CA057973); The Rockefeller University Center for Clinical and Translational Science (UL1RR024143); the Center for Basic and Translational Research on Disorders of the Digestive System through the generosity of the Leona M. and Harry B. Helmsley Charitable Trust; the Greenberg Medical Research Institute; the Starr Foundation; the Jack Lowe Cornell University Equine Research Fund; the Harry M. Zweig Memorial Fund for Equine Research; and in part from the National Center for Advancing Translational Sciences, National Institutes of Health Clinical and Translational Science Award program (UL1 TR000043). T.K.H.S. is supported by a Postdoctoral Fellowship and a Sapere Aude Research Talent Award from The Danish Council for Independent Research.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KP325401–KP325403).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1500265112/-/DCSupplemental.
References
- 1.Scheel TK, Simmonds P, Kapoor A. Surveying the global virome: Identification and characterization of HCV-related animal hepaciviruses. Antiviral Res. 2014 doi: 10.1016/j.antiviral.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapoor A, et al. Characterization of a canine homolog of hepatitis C virus. Proc Natl Acad Sci USA. 2011;108(28):11608–11613. doi: 10.1073/pnas.1101794108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lyons S, et al. Viraemic frequencies and seroprevalence of non-primate hepacivirus and equine pegiviruses in horses and other mammalian species. J Gen Virol. 2014;95(Pt 8):1701–1711. doi: 10.1099/vir.0.065094-0. [DOI] [PubMed] [Google Scholar]
- 4.Burbelo PD, et al. Serology-enabled discovery of genetically diverse hepaciviruses in a new host. J Virol. 2012;86(11):6171–6178. doi: 10.1128/JVI.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyons S, et al. Nonprimate hepaciviruses in domestic horses, United Kingdom. Emerg Infect Dis. 2012;18(12):1976–1982. doi: 10.3201/eid1812.120498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drexler JF, et al. Evidence for novel hepaciviruses in rodents. PLoS Pathog. 2013;9(6):e1003438. doi: 10.1371/journal.ppat.1003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfaender S, et al. Clinical course of infection and viral tissue tropism of hepatitis C virus-like nonprimate hepaciviruses in horses. Hepatology. 2015;61:448–459. doi: 10.1002/hep.27440. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka T, et al. Hallmarks of hepatitis C virus in equine hepacivirus. J Virol. 2014;88(22):13352–13366. doi: 10.1128/JVI.02280-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kapoor A, et al. Identification of rodent homologs of hepatitis C virus and pegiviruses. MBio. 2013;4(2):e00216-13. doi: 10.1128/mBio.00216-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quan PL, et al. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc Natl Acad Sci USA. 2013;110(20):8194–8199. doi: 10.1073/pnas.1303037110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lauck M, et al. A novel hepacivirus with an unusually long and intrinsically disordered NS5A protein in a wild Old World primate. J Virol. 2013;87(16):8971–8981. doi: 10.1128/JVI.00888-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stapleton JT, Foung S, Muerhoff AS, Bukh J, Simmonds P. The GB viruses: A review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J Gen Virol. 2011;92(Pt 2):233–246. doi: 10.1099/vir.0.027490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandriani S, et al. Identification of a previously undescribed divergent virus from the Flaviviridae family in an outbreak of equine serum hepatitis. Proc Natl Acad Sci USA. 2013;110(15):E1407–E1415. doi: 10.1073/pnas.1219217110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapoor A, et al. Identification of a pegivirus (GB virus-like virus) that infects horses. J Virol. 2013;87(12):7185–7190. doi: 10.1128/JVI.00324-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stewart H, et al. The non-primate hepacivirus 5′ untranslated region possesses internal ribosomal entry site activity. J Gen Virol. 2013;94(Pt 12):2657–2663. doi: 10.1099/vir.0.055764-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horner SM, Gale M., Jr Regulation of hepatic innate immunity by hepatitis C virus. Nat Med. 2013;19(7):879–888. doi: 10.1038/nm.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones CT, et al. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol. 2010;28(2):167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song Y, et al. The hepatitis C virus RNA 3′-untranslated region strongly enhances translation directed by the internal ribosome entry site. J Virol. 2006;80(23):11579–11588. doi: 10.1128/JVI.00675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradrick SS, Walters RW, Gromeier M. The hepatitis C virus 3′-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acids Res. 2006;34(4):1293–1303. doi: 10.1093/nar/gkl019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jangra RK, Yi M, Lemon SM. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J Virol. 2010;84(13):6615–6625. doi: 10.1128/JVI.00417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinmann E, Pietschmann T. Cell culture systems for hepatitis C virus. Curr Top Microbiol Immunol. 2013;369:17–48. doi: 10.1007/978-3-642-27340-7_2. [DOI] [PubMed] [Google Scholar]
- 22.Lohmann V, Bartenschlager R. On the history of hepatitis C virus cell culture systems. J Med Chem. 2014;57(5):1627–1642. doi: 10.1021/jm401401n. [DOI] [PubMed] [Google Scholar]
- 23.Andrus L, et al. Expression of paramyxovirus V proteins promotes replication and spread of hepatitis C virus in cultures of primary human fetal liver cells. Hepatology. 2011;54(6):1901–1912. doi: 10.1002/hep.24557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolykhalov AA, et al. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277(5325):570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 25.Yanagi M, Purcell RH, Emerson SU, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci USA. 1997;94(16):8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka T, Kato N, Cho MJ, Shimotohno K. A novel sequence found at the 3′ terminus of hepatitis C virus genome. Biochem Biophys Res Commun. 1995;215(2):744–749. doi: 10.1006/bbrc.1995.2526. [DOI] [PubMed] [Google Scholar]
- 27.Kolykhalov AA, Feinstone SM, Rice CM. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1996;70(6):3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013;19(7):837–849. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friebe P, Boudet J, Simorre JP, Bartenschlager R. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J Virol. 2005;79(1):380–392. doi: 10.1128/JVI.79.1.380-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park SH, Rehermann B. Immune responses to HCV and other hepatitis viruses. Immunity. 2014;40(1):13–24. doi: 10.1016/j.immuni.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bukh J. Animal models for the study of hepatitis C virus infection and related liver disease. Gastroenterology. 2012;142(6):1279–1287. doi: 10.1053/j.gastro.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 32.Yanagi M, Bukh J, Emerson SU, Purcell RH. Contamination of commercially available fetal bovine sera with bovine viral diarrhea virus genomes: Implications for the study of hepatitis C virus in cell cultures. J Infect Dis. 1996;174(6):1324–1327. doi: 10.1093/infdis/174.6.1324. [DOI] [PubMed] [Google Scholar]
- 33.Parera M, Martrus G, Franco S, Clotet B, Martinez MA. Canine hepacivirus NS3 serine protease can cleave the human adaptor proteins MAVS and TRIF. PLoS ONE. 2012;7(8):e42481. doi: 10.1371/journal.pone.0042481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel MR, Loo YM, Horner SM, Gale M, Jr, Malik HS. Convergent evolution of escape from hepaciviral antagonism in primates. PLoS Biol. 2012;10(3):e1001282. doi: 10.1371/journal.pbio.1001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.