Significance
The small nucleotide cyclic di-3′,5′-adenosine monophosphate (c-di-AMP) recently emerged as a ubiquitous signaling molecule among bacteria, with essential roles in both bacterial physiology and host–pathogen interactions. Bacterial mutants with abnormal c-di-AMP levels exhibit growth and virulence defects, reflecting the importance of regulating c-di-AMP synthesis and degradation for normal signal transduction and adaptation to changing environments. Previously documented phosphodiesterases hydrolyze c-di-AMP via the DHH-DHHA1 domain, but they are not present in all c-di-AMP synthesizing species. We identified a previously unrecognized class of His-Asp -domain phosphodiesterases that are widespread across several taxonomic groups. Furthermore, for the bacterial pathogen Listeria monocytogenes, phosphodiesterase mutants exhibit enhanced host inflammation, growth defects inside host cells, and significantly attenuated virulence in a murine model of infection.
Keywords: c-di-AMP, Listeria monocytogenes, phosphodiesterase, HD domain, bacterial signal transduction
Abstract
The nucleotide cyclic di-3′,5′- adenosine monophosphate (c-di-AMP) was recently identified as an essential and widespread second messenger in bacterial signaling. Among c-di-AMP–producing bacteria, altered nucleotide levels result in several physiological defects and attenuated virulence. Thus, a detailed molecular understanding of c-di-AMP metabolism is of both fundamental and practical interest. Currently, c-di-AMP degradation is recognized solely among DHH-DHHA1 domain-containing phosphodiesterases. Using chemical proteomics, we identified the Listeria monocytogenes protein PgpH as a molecular target of c-di-AMP. Biochemical and structural studies revealed that the PgpH His-Asp (HD) domain bound c-di-AMP with high affinity and specifically hydrolyzed this nucleotide to 5′-pApA. PgpH hydrolysis activity was inhibited by ppGpp, indicating a cross-talk between c-di-AMP signaling and the stringent response. Genetic analyses supported coordinated regulation of c-di-AMP levels in and out of the host. Intriguingly, a L. monocytogenes mutant that lacks c-di-AMP phosphodiesterases exhibited elevated c-di-AMP levels, hyperinduced a host type-I IFN response, and was significantly attenuated for infection. Furthermore, PgpH homologs, which belong to the 7TMR-HD family, are widespread among hundreds of c-di-AMP synthesizing microorganisms. Thus, PgpH represents a broadly conserved class of c-di-AMP phosphodiesterase with possibly other physiological functions in this crucial signaling network.
Nucleotide-based signal transduction governs many aspects of bacterial physiology, including sporulation, bacterial cell stability, antibiotic sensitivity, metabolic activity, virulence, and immune activation during infection (1–3). Among such molecules, cyclic di-3′,5′- adenosine monophosphate (c-di-AMP) is a widespread bacterial second messenger, synthesized by more than 2,000 species of diverse taxonomic phyla (4, 5). Furthermore, a balanced level of this nucleotide appears essential for normal bacterial physiology and virulence, as demonstrated in several species, including Staphylococcus aureus, Bacillus subtilis, Streptococcus pneumoniae, and Listeria monocytogenes (6–10).
In bacteria, c-di-AMP synthesis is catalyzed by the diadenylate cyclase activity of the DisA_N domain (Pfam PF02457), whereas c-di-AMP hydrolysis has been shown to be catalyzed by the phosphodiesterase activity of the DHH-DHHA1 domain (4, 5). Previous studies reported two families of DHH-DHHA1 domain-containing proteins for c-di-AMP degradation. The first family comprises homologs of membrane-bound GdpP proteins, which contain a PAS domain, a degenerate GGDEF domain, and a catalytic DHH-DHHA1 domain (11). The second family comprises soluble, stand-alone DHH-DHHA1 domains (9, 12, 13).
The intracellular pathogen L. monocytogenes synthesizes and secretes c-di-AMP into broth growth medium and the host cytosol during infection (14). This bacterium harbors a single homolog of GdpP, named PdeA, for c-di-AMP degradation (10). PdeA shares substantial sequence and structural similarities with GdpP and exhibits c-di-AMP hydrolysis activity in vitro. However, a pdeA null allele does not confer a considerable effect on secreted c-di-AMP levels during L. monocytogenes growth in broth (10). This observation suggests the presence of another protein for c-di-AMP degradation in L. monocytogenes.
In this study, we identified and characterized PgpH as representing a previously unidentified class of c-di-AMP phosphodiesterases with a catalytic His-Asp (HD) domain. The PgpH HD domain interacted with c-di-AMP at high affinity and specificity. We also obtained a crystal structure of the PgpH HD domain in complex with metal ions and the c-di-AMP substrate. Consistent with structure-based predictions, the HD domain degraded c-di-AMP into 5′-pApA in a reaction that requires Mn2+ ions and the His-Asp active site. Deletion of the pdeA and pgpH alleles conferred elevated c-di-AMP accumulation during broth growth and infection, with differential effects by each allele in these environments. Furthermore, the absence of both phosphodiesterases greatly diminished bacterial growth in and out of the host. Finally, our phylogenetic survey revealed that PgpH homologs are broadly distributed among c-di-AMP synthesizing species, suggesting that hydrolysis by the HD domain is a conserved and widespread mechanism.
Results
The PgpH HD Domain Interacts Specifically with c-di-AMP.
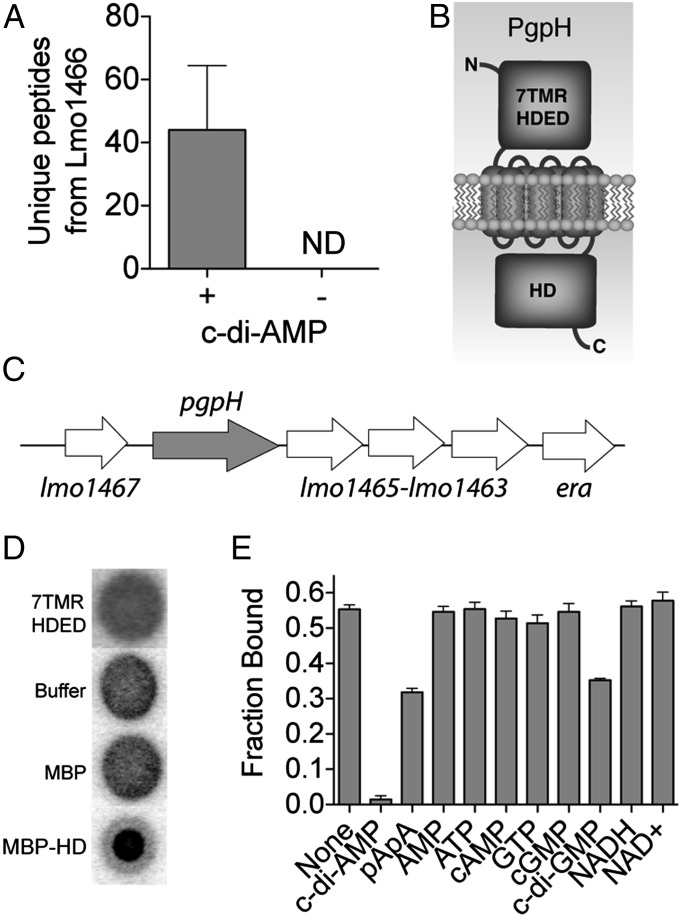
To identify proteins that bind c-di-AMP, we used an affinity pull-down strategy in which c-di-AMP–conjugated Sepharose beads were incubated with L. monocytogenes cell lysate (15). Ethanolamine-conjugated beads were used as a control to evaluate nonspecific binding to the Sepharose scaffold. Among protein candidates found to interact with c-di-AMP Sepharose beads, Lmo1466 (numbered according to L. monocytogenes EGD-e strain) exhibited the highest level of enrichment, and it showed no interaction with control Sepharose (Fig. 1A), consistent with a specific, high-affinity interaction.
Fig. 1.
Identification and characterization of PgpH and c-di-AMP interactions. (A) Spectral counting of Lmo1466 signature peptides interacting with c-di-AMP Sepharose beads (+) and control (-) beads. ND, none detected. Error bars represent the SEM of spectral counts from three independent experiments. (B) Schematic diagram of Lmo1466 domain organization: extracellular domain (7TMR-HDED), seven transmembrane helices, and cytoplasmic HD domain (HD). (C) Operon organization surrounding pgpH. (D) Nucleotide binding assays (DRACALA) for 32P-c-di-AMP with PgpH extracellular domain (7TMR HDED; 220 μM), MBP-HD domain (7 μM), and maltose binding protein (MBP, 21 μM) as a control. (E) Binding of PgpH HD domain with 32P-c-di-AMP in the presence of competing unlabeled nucleotides, each at 200 μM concentration. For binding assays, error bars represent the SEM of duplicate measures and are representative of two independent experiments.
Lmo1466 was previously named PgpH and was linked to L. monocytogenes growth at low temperatures (16). The protein contains an amino-terminal extracellular domain, seven transmembrane helices, and a carboxyl-terminal HD domain, belonging to a superfamily of phosphohydrolases with a characteristic His-Asp motif (17). PgpH also has a similar domain architecture to members of the 7TMR-HD protein family (7TMR; 7TM receptor) (18). Additionally, pgpH is found in a six-gene operon (19) that is highly conserved among Firmicutes (Fig. 1 B and C). Homologous proteins encoded in the operon have several biological functions spanning phosphate starvation response (lmo1467) (20), ribosome maturation and assembly (lmo1465 and era) (21, 22), lipid biogenesis (lmo1464) (23), and nucleotide metabolism (lmo1463) (24).
Because L. monocytogenes synthesizes c-di-AMP in the cytoplasm and secretes the nucleotide into the extracellular space, c-di-AMP may interact with the extracellular (ED) domain and/or the cytoplasmic HD domain of PgpH. To interrogate these possibilities, we expressed the PgpH ED domain as a His-tagged recombinant protein (ED-His construct) and the HD domain as a fusion with an amino-terminal maltose-binding protein (MBP-HD construct). To directly evaluate the interaction between the domains of PgpH and c-di-AMP, we used a differential radial action of ligand assay (DRACALA), which tests for protein binding with radiolabeled nucleotides (25). These binding experiments indicated that the ED domain and MBP alone did not interact with 32P-c-di-AMP, whereas the MBP-HD domain construct exhibited clear binding (Fig. 1D).
To evaluate binding specificity, we conducted competition assays, in which unlabeled nucleotides were added in excess to disrupt 32P-c-di-AMP interaction with MBP-HD protein. Among the nucleotides tested, only 5′-pApA and c-di-GMP diminished radiolabeled c-di-AMP binding, although much less than unlabeled c-di-AMP (Fig. 1E). These results indicated that the HD domain of PgpH specifically interacted with c-di-AMP.
The PgpH HD Domain Binds c-di-AMP with High Affinity and Exhibits c-di-AMP Hydrolysis Activity.
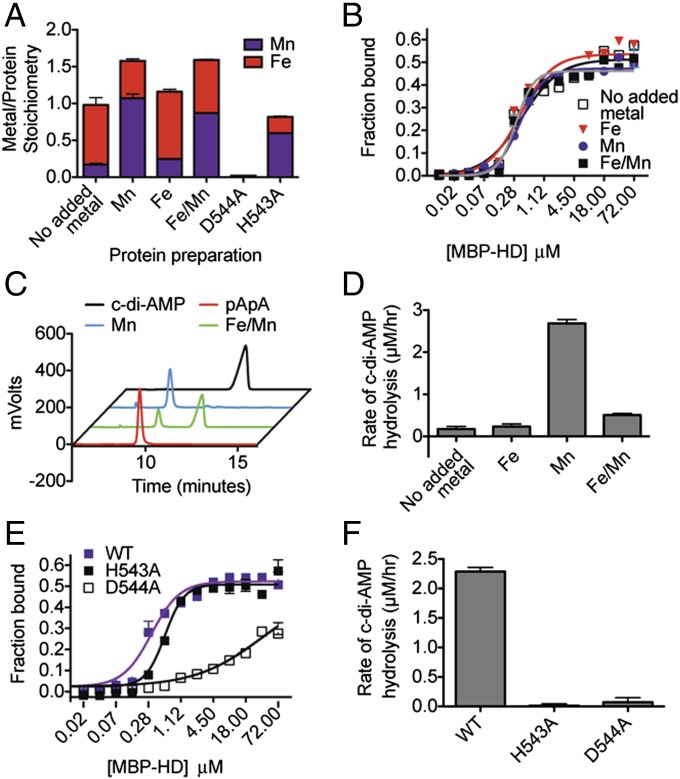
The structurally related nucleotide c-di-GMP is hydrolyzed by HD-GYP phosphodiesterases, which are also HD superfamily members (26). Thus, we hypothesized that PgpH was a c-di-AMP phosphodiesterase. Because HD family proteins require two metal ions for phosphohydrolase activity (17), we investigated the effects of different metals on protein function. The MBP-HD protein was purified using an amylose affinity column and inductively coupled plasma–mass spectromotry (ICP-MS) was used to analyze metal incorporation. This protein preparation contained mostly Fe, Zn, and Mn (61%, 20%, and 13% of total metal content, respectively) (Table S1). Other metals (Mg, Co, Ni, and Cu) were found at very low levels. Accordingly, we analyzed different preparations of MBP-HD protein, each with metal salts added to the expression cultures at the time of induction as follows: (i) zinc salt (ZnCl2), (ii) ferrous salt (FeSO4), (iii) manganous salt (MnCl2), and (iv) an equimolar mixture of both ferrous and manganous salts (FeSO4/MnCl2). The Zn preparation was inactive and the stoichiometry of this ion did not correlate with the active site HD motif, as described below. Together with the observation that the activities of GdpP (11) and HD-GYP proteins that hydrolyze c-di-GMP (27–29) are Mn- or Fe-dependent hydrolases, we focused further characterization on these ions. As expected, metal incorporation patterns corresponded to the metal supplements. Protein preparations purified with FeSO4 and MnCl2 had a higher ratio of incorporated Fe and Mn ions, respectively, whereas the preparation with FeSO4/MnCl2 had similar amounts of both ions (Fig. 2A).
Fig. 2.
Metal cations and the HD motif are required for c-di-AMP binding and hydrolysis by PgpH HD domain. PgpH HD domain was purified as an MBP-HD construct with different metal salts (0.35 mM in total when added to expression cultures) and analyzed for stoichiometry of bound metal (error bars represent average of two measurements) (A); binding affinity with 32P-c-di-AMP (error bars represent two independent experiments, each performed in duplicates) (B); and c-di-AMP hydrolysis activity by HPLC (C) and LC-MS/MS (D). The H543A and D544A mutants were purified with added MnCl2 and analyzed for stoichiometry of bound metal (A), binding affinity with 32P -c-di-AMP (E), and c-di-AMP hydrolysis (F) by LC-MS/MS. LC-MS/MS data are representative of two independent experiments, and error bars represent the SE derived from fitting reaction rates by linear regression.
Despite different metal content, all preparations bound 32P-c-di-AMP with similar dissociation constants (Kd = 0.3–0.4 μM) (Fig. 2B). We then tested the activity of MBP-HD preparations toward bis(p-nitrophenyl) phosphate (bis-pNPP), a widely used colorimetric reporter substrate for phosphodiesterase activity (Fig. S1A). The Zn preparation was inactive. Among those protein preparations that exhibited bis-pNPP hydrolysis, activity was highest with Mn, intermediate with Mn/Fe, and lowest with Fe or without added metal. C-di-AMP inhibited p-nitrophenol product formation in a concentration-dependent manner (Fig. S1B), suggesting competitive binding between c-di-AMP and bis-pNPP at the enzyme active site. To directly examine MBP-HD activity toward c-di-AMP, we incubated the protein preparations with c-di-AMP and analyzed the reactions by HPLC and LC-MS/MS (Fig. 2 C and D). The protein preparation with added Mn displayed nearly complete turnover of c-di-AMP into 5′-pApA. Preparations without metal, or with added Fe, exhibited very weak activities. The preparation with added Fe/Mn displayed an intermediate activity. Thus, Mn2+ was optimal for phosphodiesterase activity toward c-di-AMP, and the formation of 5′-pApA was consistent with structural predictions (see below).
The MBP-HD protein also hydrolyzed c-di-GMP into 5′-pGpG (Fig. S2A), albeit at a slower rate than c-di-AMP hydrolysis. For similar incubation periods, ∼70% of c-di-AMP was degraded, whereas only 25% of c-di-GMP was degraded. Additionally, c-di-GMP interacted with MBP-HD protein at a much lower affinity than c-di-AMP. In competition nucleotide binding assays, micromolar levels of unlabeled c-di-AMP readily competed with bound 32P-c-di-AMP, whereas millimolar levels of unlabeled c-di-GMP exhibited only partial competition (Fig. S2B). Together, the results indicated that c-di-GMP is unlikely to be a physiological substrate for PgpH activity. These observations are reminiscent of GdpP, which hydrolyzes both c-di-AMP and c-di-GMP, but its diminished affinity for c-di-GMP results in specificity toward the physiological substrate c-di-AMP (11).
Among HD-family hydrolases, the metals required for catalytic activity are coordinated by the conserved active site His-Asp motif, corresponding to PgpH residues His543 and Asp544. To examine the requirement of the HD active site, we generated the MBP-HD H543A and MBP-HD D544A mutant proteins, and purified them with added Mn. Relative to wild-type (WT) protein, both mutants were impaired for Fe and Mn incorporation (Fig. 2A). Zn incorporation remained unchanged, consistent with binding to another region of the protein. The H543A mutant incorporated significantly less total metal and the D544A mutant exhibited nearly undetectable metal incorporation. In DRACALA assays, the H543A mutant was slightly defective for c-di-AMP binding (Kd = 0.6 µM), whereas the D544A mutant was severely diminished (Kd ∼36 µM; Fig. 2E). Finally, both the H543A and D544A mutants were inactive for substrate turnover (Fig. 2F). Thus, the HD active site was required for metal incorporation, nucleotide binding, and c-di-AMP hydrolysis. In particular, the Asp544 residue of this motif was critical for protein function.
PgpH HD Activity Is Inhibited by ppGpp.
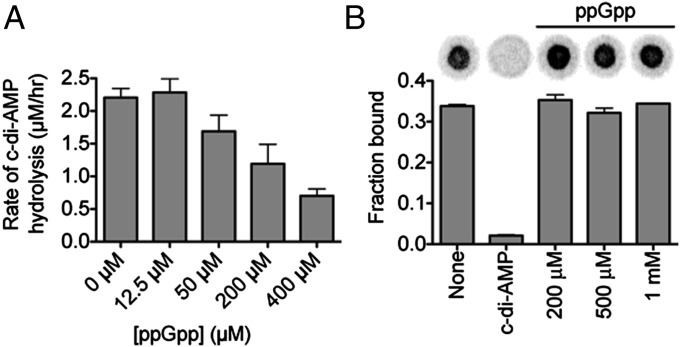
Previous studies suggested a cross-talk between the stringent response and c-di-AMP signaling. In the L. monocytogenes cld-14 strain, a transposon insertion in the pgpH gene resulted in an elevated accumulation of ppGpp (16). Furthermore, ppGpp is a competitive inhibitor of c-di-AMP hydrolysis by GdpP (11). Here, we investigated the effects of ppGpp on PgpH HD domain activity. C-di-AMP hydrolysis was inhibited by ppGpp in a concentration-dependent manner with an approximate IC50 of 200–400 µM (Fig. 3A). However, the PgpH HD domain did not hydrolyze ppGpp. Finally, in a competitive nucleotide-binding assay, increasing concentrations of ppGpp, from 200 μM to 1 mM, did not inhibit 32P-c-di-AMP binding by the MBP-HD construct (Fig. 3B). These results suggested that ppGpp regulates PgpH activity through a distinct site, separate from the c-di-AMP binding site.
Fig. 3.
PgpH hydrolysis of c-di-AMP is inhibited by ppGpp. (A) The rate of c-di-AMP hydrolysis in the presence of ppGpp monitored by LC-MS/MS. Error bars are the SE derived from fitting reaction rates by linear regression. (B) Binding of PgpH HD domain with 32P-c-di-AMP in the presence of competing unlabeled c-di-AMP (200 μM) or ppGpp (200 μM–1 mM). Error bars represent the SEM of two measurements.
A Crystal Structure of the PgpH HD Domain Reveals c-di-AMP Binding Mode.
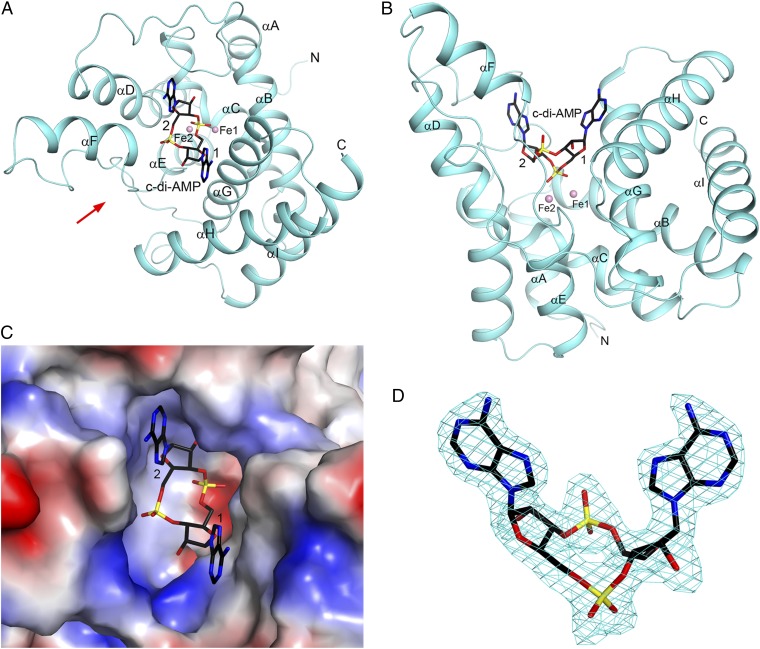
The Fe-containing HD protein retained nucleotide binding but exhibited limited substrate turnover. Thus, we used this protein for structural characterization of the substrate–protein complex. We obtained a crystal structure of the PgpH HD domain in complex with c-di-AMP at 2.1 Å resolution. The atomic model has good agreement with the crystallographic data and the expected bond lengths, bond angles, and other geometric parameters (Table S2). One hundred percent of the residues in the structure are in the favored region of the Ramachandran plot, and no residues are in the disallowed region. There are two molecules of the c-di-AMP complex in the asymmetric unit. The overall structures of the HD domain and the binding modes of the compound are essentially the same in the two complexes, with rms distance of 0.5 Å for equivalent Cα atoms of the two HD domains. The structure is missing ∼15 residues at the N terminus of the loop that connects to the transmembrane domain of PgpH and ∼10 residues at the C terminus of PgpH.
We also tried to define the binding site of ppGpp and obtained crystals of the HD domain grown in the presence of 5 mM of the compound. The structure was determined at 2.4-Å resolution (Table S2), in a different crystal form, but we were not able to locate any ppGpp based on the diffraction data. There are only conformational differences in a few of the surface loops of the HD domain between this “free enzyme” structure and the c-di-AMP complex, with rms distance of 0.5 Å for their equivalent Cα atoms. The lack of ppGpp binding was probably due to its weaker affinity for the protein.
The PgpH HD domain has an all-helical structure, with nine mostly antiparallel helices (αA through αI; Fig. 4 A and B). Helix αC has a two-turn 310 helix extension at the C-terminal end, and the HD motif is located in this 310 segment. Helix αD has a kink near the middle, and one of the ligands to the metal ions (His580) is located at this position. The two metal ions bound in the active site are likely predominantly Fe3+, based on our analysis of the anomalous diffraction data, although we cannot exclude the possibility that a small amount of Mn2+ may be present as well.
Fig. 4.
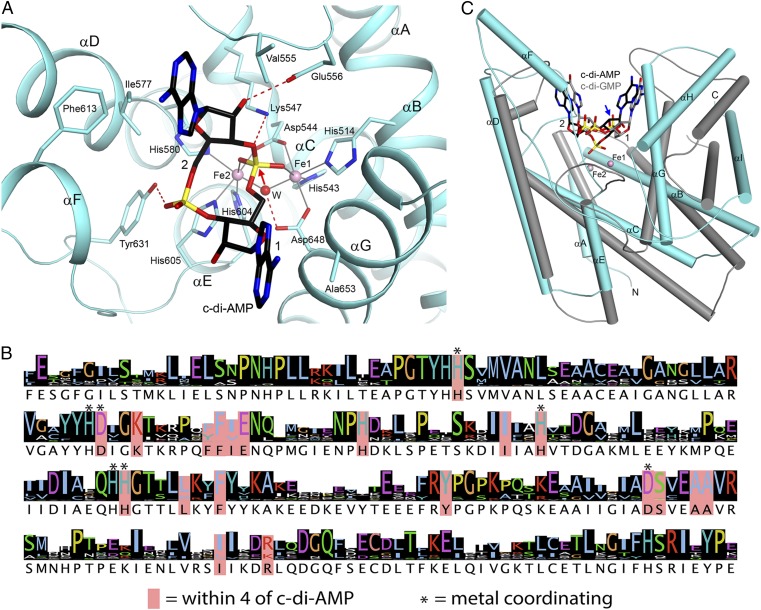
Crystal structure of the HD domain of PgpH in complex with c-di-AMP. (A) Schematic drawing of the structure of PgpH HD domain (in cyan) in complex with c-di-AMP (stick model with carbon atoms in black). The two metal ions in the active site are shown as pink spheres and labeled. The two nucleotides of c-di-AMP are labeled 1 and 2. (B) Structure of the PgpH HD domain in complex with c-di-AMP, viewed along the red arrow in A. (C) Molecular surface of the PgpH HD domain near the c-di-AMP binding site, colored by the electrostatic potential (red, negative; blue, positive). (D) Omit Fo–Fc electron density map for c-di-AMP at 2.1 Å resolution, contoured at 2.5σ.
The helices of the HD domain are arranged such that there is a large depression on one face of the structure, where the metal ions and c-di-AMP are bound (Fig. 4 A–C). Clear electron density was observed for c-di-AMP (Fig. 4D), and the compound has extensive interactions with the HD domain (Fig. 5A). The two terminal oxygen atoms of the 5′ phosphate group of nucleotide 1 are coordinated to the two metal ions in the active site. Each metal ion is octahedrally coordinated. Within the HD motif, the His residue coordinates one metal ion (Fe1), whereas the Asp residue has bidentate coordination to both metal ions, consistent with our observation that mutation of this residue results in complete loss of metal incorporation (Fig. 2A). In addition, a water molecule is a bridging ligand to both metal ions. Other ligands of Fe1 include His514 (helix αB) and Asp648 (αG), whereas other ligands of Fe2 include His580 (αD), His604, and His605 (near the C-terminal end of αE). In comparison, the 5′ phosphate group of nucleotide 2 has limited interaction with the HD domain, making one hydrogen bond with the side chain of Tyr631 (αF-αG loop).
Fig. 5.
Binding mode of c-di-AMP in PgpH HD domain. (A) Detailed interactions between c-di-AMP and the PgpH HD domain. Hydrogen-bonding interactions are indicated with dashed lines (in red) and liganding interactions as thin lines (in gray). The bridging water between the two metal ions is shown as a red sphere and labeled W. (B) Sequence conservation of the PgpH HD domain among the top 250 closest homologs identified by BLAST. Sequence logo was generated by using Jalview. (C) Overlay of the structure of the PgpH HD domain in complex with c-di-AMP (in color) with that of the HD-GYP domain in complex with c-di-GMP (in gray) (29). A large difference in the position of the scissile phosphate group in the c-di-GMP structure is indicated with the blue arrow. All structure figures were produced with PyMOL (www.pymol.org).
Both adenine bases of c-di-AMP are in the “anti” configuration. The adenine base of nucleotide 1 lies against helix αG, especially the peptide bond between Ala652 and Ala653 and the side chain of Ala653. The other face of this adenine is exposed to the solvent. The 2′ hydroxyl of this nucleotide is hydrogen-bonded to Tyr631. The adenine base of nucleotide 2 is positioned in a pocket, with Ile577 (αD) and Phe613 (αF) on one face and Val555 (αC-αD loop) on the other face. A guanine base at this position could have steric clashes between its 2-amino group and the protein (Fig. 4C), possibly explaining why the HD domain has lower affinity for c-di-GMP. The 2′ hydroxyl of this nucleotide is hydrogen-bonded to Glu556 (αC-αD loop), which may help distinguish between ribose and deoxyribose. Finally, Lys547 is located near the 3′ hydroxyl of the scissile phosphate. All of the residues in the active site region of the HD domain are highly conserved among the PgpH homologs (Fig. 5B). The binding mode of c-di-AMP provides a clear mechanism for the hydrolytic activity of the HD domain (see below), confirming that the PgpH HD domain is a c-di-AMP hydrolase.
The structure of the HD domain of PgpH has homology to that of other HD domain metal-dependent phosphohydrolases, although the sequence conservation is low (<20% identity). The homologs include the HD-GYP domain c-di-GMP phosphodiesterase (27, 29), with a Z score of 13.0 from DaliLite (4), the Escherichia coli 5′ deoxyribonucleotidase YfbR (Z score 9.7) (30), and other HD family proteins (PDB ID codes including 3HC1, 3M1T, 3CCG, and 3P3Q). Although the positions of several helices in the core of these structures are generally similar, there are large variations in the positions of the other helices and especially the loops (Fig. 5C). Moreover, in the structure of the HD-GYP domain in complex with c-di-GMP (29), only one metal ion (equivalent to Fe2 in PgpH) is bound in the active site, although this site can bind three metal ions. The scissile phosphate is located far from the metal ion and is not in a productive binding mode.
Listeria Phosphodiesterases Exhibit Differential Activities in Distinct Environments.
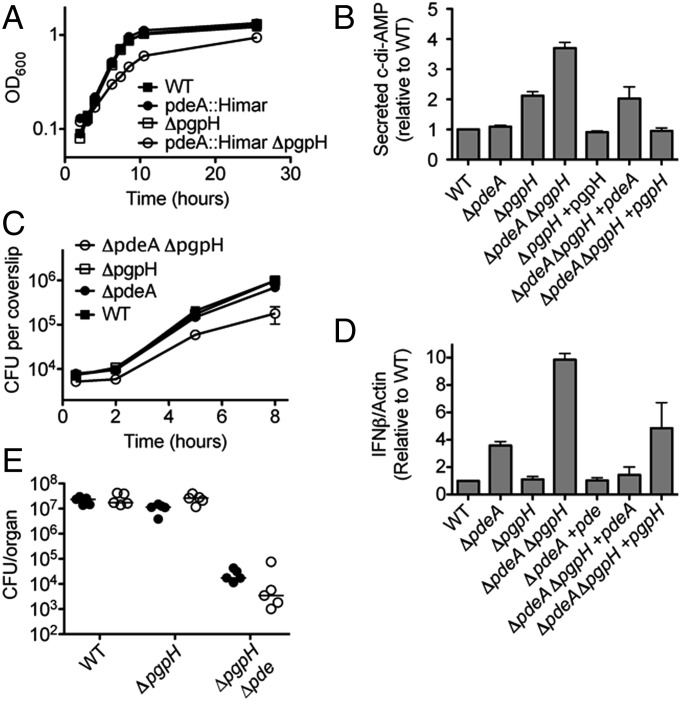
L. monocytogenes encodes a DHH-DHHA1 phosphodiesterase, PdeA, which hydrolyzes c-di-AMP in vitro. Our biochemical and structural analyses suggested that PgpH functioned as another c-di-AMP–specific phosphodiesterase, acting via a catalytic HD domain. We conducted a genetic analysis to investigate the physiological relevance of PdeA and PgpH. During broth growth, L. monocytogenes secretes c-di-AMP via the multidrug efflux pumps (31, 32). The ΔpdeA and ΔpgpH mutants were indistinguishable from the WT strain for both growth rate and yield, and the ΔpdeA ΔpgpH mutant was slightly defective (Fig. 6A). In addition, all mutants exhibited similarly low levels of bacterial cell lysis in broth cultures as assessed by a β-gal release assay (Fig. S3A). To evaluate secreted c-di-AMP, we analyzed the supernatants of stationary phase cultures grown in minimal medium, using liquid chromatography-mass spectrometry. As previously reported, the ΔpdeA mutant secreted a similar amount of c-di-AMP as the WT strain (Fig. 6B). By contrast, the ΔpgpH and ΔpdeA ΔpgpH mutants secreted considerably more c-di-AMP, approximately two- and fourfold compared with the WT, respectively. Consistent with our biochemical data, these results suggested that PgpH was a c-di-AMP phosphodiesterase in broth growth. Furthermore, compared with PdeA, PgpH appeared to be the primary phosphodiesterase in broth cultures. Because the ΔpdeA and ΔpgpH alleles conferred additive phenotypes, PdeA and PgpH must be involved in independent pathways for c-di-AMP degradation.
Fig. 6.
Characterization of L. monocytogenes c-di-AMP phosphodiesterase mutants. (A) Growth curves of L. monocytogenes strain in chemically defined minimal medium. (B) c-di-AMP secretion by various L. monocytogenes strains in chemically defined minimal medium. Secreted c-di-AMP is normalized to WT. (C) Growth curves in bone marrow-derived macrophages. (D) IFN-β expression by qPCR at 4 h after infection, normalized to WT-infected macrophages. (E) Female C57BL/6 mice (6–8 wk old) were infected with 1 × 105 cfu of the indicated strains. Organs were harvested at 48 hpi, and bacterial burden per spleen (filled circles) and liver (open circles) was enumerated. Median values are presented as horizontal lines. Error bars represent the SEM and are representative of at least two independent experiments.
We complemented the mutant phenotypes in trans by chromosomal integration of the pgpH+ and pdeA+ alleles at the tRNAArg locus (33). In both the ΔpgpH and ΔpdeA ΔpgpH backgrounds, the pgpH+ allele reduced c-di-AMP secretion to the WT level (Fig. 6B). In the ΔpdeA ΔpgpH background, the pdeA+ allele reduced c-di-AMP secretion to an intermediate level comparable to the ∆pgpH single mutant. Thus, PgpH regulated the levels of c-di-AMP secreted by L. monocytogenes in broth cultures. Furthermore, PdeA appeared to be a functional phosphodiesterase in this condition, but its effects were only measurable in the absence of PgpH.
During infection, L. monocytogenes secretes c-di-AMP into the host cytosol, triggering a robust type I IFN response that leads to the production and secretion of IFN-β (IFN-β) by the infected host cell (31, 34, 35). To evaluate secreted c-di-AMP levels during intracellular growth, we infected bone marrow-derived macrophages with L. monocytogenes, and measured IFN-β expression by quantitative RT-PCR (qRT-PCR). The ΔpdeA and ΔpgpH mutants were virtually identical to the WT strain for growth rate and yield, and the ΔpdeA ΔpgpH mutant was slightly defective (Fig. 6C). Compared with the WT, the ΔpdeA mutant induced an elevated level of IFN-β expression, whereas the ΔpgpH mutant was indistinguishable from the WT (Fig. 6D). The ΔpdeA ΔpgpH mutant induced the highest IFN-β expression level, at least fourfold higher than the WT strain. In addition to c-di-AMP secretion, L. monocytogenes can also activate IFN-β through DNA release, which stimulates cell death and lactate dehydrogenase release (36, 37). However, for all mutants, altered type I IFN responses were not due to increased DNA released during infection, because similar levels of luciferase reporter plasmid were released within host cells by the different strains (Fig. S3B).
Similar to observations for broth culture, the mutant phenotypes were complemented in trans (Fig. 6D). In both the ΔpdeA and ΔpdeA ΔpgpH mutant backgrounds, the pdeA+ allele reduced IFN-β expression to the WT level. In the ΔpdeA ΔpgpH background, the pgpH+ allele reduced IFN-β expression to an intermediate level, approximately that conferred by the ΔpdeA strain (Fig. 6D). Thus, PgpH was also involved in c-di-AMP hydrolysis in the host environment, but its effects were only observable in the absence of PdeA. Compared with PgpH, PdeA appeared to be the primary c-di-AMP phosphodiesterase during intracellular growth.
Elevated c-di-AMP Levels Diminish L. monocytogenes Virulence.
Relative to the WT and individual mutants, the ΔpdeA ΔpgpH strain exhibited the highest levels of c-di-AMP and IFN-β from infected host cells. This mutant was also defective for growth in macrophages (Fig. 6C). To evaluate virulence, we i.v. infected mice with L. monocytogenes, and recovered bacterial loads from the livers and spleens 48 h after infection (Fig. 6E). The ΔpgpH mutant was indistinguishable from WT, and we reported a similar observation for the ∆pdeA mutant (10). However, the ΔpdeA ΔpgpH strain exhibited a 3-log reduction for bacterial growth. Thus, elevated c-di-AMP levels diminished L. monocytogenes virulence.
HD-Domain c-di-AMP Phosphodiesterases Are Widespread Among Bacteria.
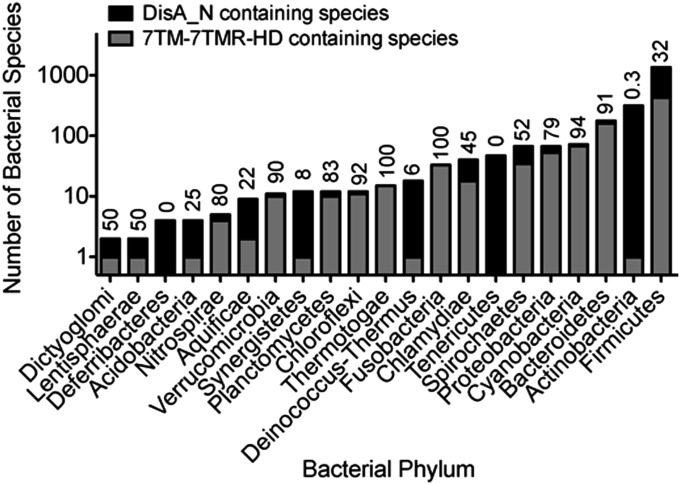
We defined PgpH homologs as those with a seven transmembrane domain and a cytoplasmic HD domain belonging to the 7TM-7TMR_HD family of bacterial receptors (Pfam PF07698). Most proteins within this family, including PgpH, contain an amino-terminal extracellular domain, likely involved in signal detection and transmission (18). PgpH homologs are present in ∼36% of nearly 2,400 DisA_N containing organisms recognized by the Pfam database. These proteins span across different taxonomic groups, and include most DisA_N phyla, except for Deferribacteres, Tenericutes, and Actinobacteria (Fig. 7). The majority of PgpH-harboring species are Firmicutes. However, within this phylum, PgpH is absent in the Staphylococcaceae and the Streptococcaceae families. Interestingly, PgpH is also present in more than 90% of DisA_N containing Cyanobacteria, Bacteroidetes, Fusobacteria, and δ-Proteobacteria. Together with our experimental evidence of PgpH phosphodiesterase activity, the widespread presence of its homologs suggests that these 7TMR-HD hydrolases constitute a major mechanism for c-di-AMP degradation.
Fig. 7.
Taxonomic distribution of di-adenylate cyclase and PgpH proteins. The number of DisA_N and 7TM_7TMR_HD containing organisms are reported based on bacterial phylum. Numbers above each bar represent the percentage of DisA_N-containing organisms within a phylum that also harbor a PgpH homolog.
Discussion
Many second messenger signaling systems have dedicated mechanisms to control both synthesis and degradation of the signal molecules. To date, studies characterizing c-di-AMP degradation have focused on DHH-DHHA1 domain-containing phosphodiesterases. Here, we present biochemical, structural, and genetic evidence that PgpH represents a widespread class of 7TMR-HD hydrolases that degrade c-di-AMP via a catalytic HD domain.
An HD Domain-Based Mechanism for c-di-AMP Hydrolysis.
The PgpH HD domain bound c-di-AMP with a high affinity and hydrolyzed it into 5′-pApA. Our structural characterization allowed us to elucidate a mechanism for c-di-AMP hydrolysis by the HD domain. In the HD active site with bound Fe, the bridging water between the two metal ions is positioned directly below the 5′ phosphate group of nucleotide 1 (Fig. 5A), the scissile phosphate group of the hydrolysis. This water is activated by the two metal ions, as well as by a hydrogen bond with Asp648, which is also a ligand to Fe1. The water/hydroxide acts as a nucleophile to attack the phosphorus atom, and the 3′ hydroxyl of nucleotide 2 is then the leaving group. The oxyanion is stabilized by the side chain ammonium ion of Lys547. This side chain, or a solvent water molecule, protonates the oxyanion to complete the reaction, and the product is pA1pA2, consistent with in vitro hydrolysis activity of the MBP-HD protein. Considering the conserved structure of the HD domain, this mechanism is likely common among other HD-domain phosphodiesterases.
Although DHH-DHHA1 domain c-di-AMP phosphodiesterases have not been structurally characterized, they are also binuclear Mn-dependent proteins (5). Thus, they may exhibit a similar mechanism of nucleotide hydrolysis to HD-domain phosphodiesterases. If this prediction is true, mechanism-based inhibitors can be designed to target both enzyme families simultaneously. Such compounds would exhibit therapeutic or industrial potential for preventing microbial growth.
PgpH and HD-GYP c-di-GMP phosphodiesterases both belong to the HD superfamily and both share an overall similar fold, but they exhibit different active sites and nucleotide binding modes. Although the PgpH HD domain interacted weakly with c-di-GMP and hydrolyzed it into 5′-pGpG, our nucleotide binding data indicated that this activity was not physiologically relevant. Furthermore, whereas HD-GYP proteins hydrolyze c-di-GMP and 5′-pGpG sequentially into GMP (27, 29), PgpH showed activity only toward c-di-AMP with 5′-pApA as the final product. Thus, the structurally related nucleotides c-di-AMP and c-di-GMP are hydrolyzed by distinct enzyme families.
Coordinated Activities of c-di-AMP Phosphodiesterases.
Our results indicated that L. monocytogenes encodes at least two c-di-AMP phoshodiesterases, PdeA with a catalytic DHH-DHHA1 domain and PgpH with a catalytic HD domain. We also found that these two proteins exhibited cooperative activities in regulating c-di-AMP levels. Whereas PgpH appeared more prominent in broth growth, PdeA appeared more prominent in intracellular growth inside host cells (Fig. 6 B and D). Thus, they may be regulated by distinct signals in each environment. Previous reports linked PdeA activity to heme and nitric oxide binding, consistent with a specific function in the host compartment (38). By contrast, PgpH may respond to environmental signals via the extracellular domain and/or the seven-transmembrane helices (18). In addition, both PdeA and PgpH activities were inhibited by ppGpp, suggesting that nutrient limitation was another regulatory factor for c-di-AMP hydrolysis. Comparative analyses of their functions will be necessary to separate their physiological roles in c-di-AMP signaling across the diverse range of conditions encountered by L. monocytogenes and other bacteria.
In addition to PgpH and PdeA, other proteins may be involved in c-di-AMP hydrolysis in L. monocytogenes. Listeria species encode a stand-alone DHH-DHHA1 protein, Lmo1575, homologous to other DHH-DHHA1 proteins that are reported to degrade c-di-AMP (9, 12, 13). Although PgpH is the only 7TMR_HD protein found in L. monocytogenes, seven other proteins of the HD superfamily (Pfam PF01966) are also present within the genome. Thus, it is possible that other DHH-DHHA1 and HD domain containing hydrolases contribute to c-di-AMP hydrolysis in Listeria. It should be noted, however, that none of these other proteins was isolated in our affinity pull-down with c-di-AMP beads from exponential phase cultures, indicating either low interaction affinity with the nucleotide or low abundance under the probed conditions.
Beyond Listeria, it is not clear whether PgpH and DHH-DHHA1 domain-containing phosphodiesterases represent the only protein families responsible for c-di-AMP hydrolysis. Organisms within the archaea domain and bacteria in the Deferribacteres, Tenericutes, Spirochetes, and Actinobacteria phyla have no GdpP or PgpH homologs, suggesting the presence of alternative c-di-AMP phosphodiesterases. Stand-alone DHH-DHHA1 domain proteins in the Spirochete Borrelia burgdorferi and the Actinobacterium Mycobacterium tuberculosis have been reported to modulate c-di-AMP nucleotide levels within these species, illustrating that GdpP homologs are not the only c-di-AMP phosphodiestereases that act via the DHH-DHHA1 domain. However, stand-alone DHH-DHHA1 proteins also hydrolyze substrates other than c-di-AMP, such as nanoRNA and pAp (39, 40). Furthermore, they are widely conserved in organisms that do not synthesize c-di-AMP, including all classes of Proteobacteria and several Eukaryotes. As such, the presence of a DHH-DHHA1 protein cannot definitively predict c-di-AMP hydrolysis activity. Future studies will be required to define the molecular determinants of substrate recognition by the c-di-AMP–specific DHH-DHHA1 phosphodiesterases, and to examine the distribution of this subfamily and the presence of alternative c-di-AMP hydrolases throughout bacteria and archaea.
Attenuated Virulence by Phosphodiesterase Mutants.
Regulation of c-di-AMP levels is critical to bacterial growth and metabolism. Several studies have established that c-di-AMP depletion increases bacterial susceptibility to cell wall antibiotics, cell lysis, and is lethal for many species (5). Conversely, c-di-AMP elevation also appears to be similarly detrimental, affecting growth, division, peptidoglycan synthesis, and virulence (6, 8, 9, 13). Here, we observed that the ΔpdeA ΔpgpH mutant was considerably attenuated in a murine infection model. Because this strain also exhibited moderate broth and intracellular growth defects, this observation was not unexpected. However, in comparison with other slow-growing L. monocytogenes strains related to c-di-AMP production, the level of attenuation that we observed was particularly striking. For instance, both the ΔpdeA ΔpgpH and the previously characterized ΔdacA ΔcitZ mutants were similarly impaired for growth in vitro, but the ΔpdeA ΔpgpH mutant exhibited a three-log reduction in bacterial load, whereas the ΔdacA ΔcitZ mutant exhibited a one-log defect (15). Thus, c-di-AMP accumulation was specifically detrimental to bacteria inside the host, a phenotype that is not solely attributable to altered bacterial growth rate.
The exact mechanism by which increased c-di-AMP levels affect bacterial growth and infection is not yet understood, but could be attributed to host cell or bacterial factors. In host cells, c-di-AMP stimulates robust IFN-β production in a STING-dependent pathway (34, 41). Similar to the L. monocytogenes ΔpdeA ΔpgpH mutant, the tetR mutant secretes high levels of c-di-AMP and is also attenuated in infection (31). However, this defect is IFN-β independent, indicating STING activation is not responsible for virulence defects by c-di-AMP elevation. In a different immune signaling pathway, c-di-AMP also activates NLRP3 and Caspase-1, which induces pyroptosis and limits bacterial burden in the host (41, 42). However, the ΔpdeA ΔpgpH strain did not increase cell death, suggesting that Caspase-1 is not involved. Thus, the virulence defect of the ΔpdeA ΔpgpH mutant may reflect aberrant bacterial physiology in excess of c-di-AMP. In our study, c-di-AMP accumulation did not cause bacterial lysis and must impair other cellular aspects important for infection. Examples of affected bacterial processes may include potassium uptake and central metabolism (43, 44). Future studies to characterize the role of individual c-di-AMP protein signaling partners should be proposed to detail the consequences of elevated nucleotide levels on virulence factor expression and bacterial adaptation to the dynamic environments encountered in the host.
Methods
Bacterial Strains, Culture Conditions, and Tissue Culture Infections.
L. monocytogenes strains used in this study are listed in Table S3. Deletion of pdeA and pgpH alleles was achieved by allelic exchange as described (45). The ORF for PdeA or PgpH was introduced into pPL2 with a Pspac promoter. Plasmids were subsequently introduced into the ∆pdeA, ∆pgpH, or ∆pdeA ∆pgpH strains to complement each gene in trans. For in vitro growth curves, cultures were grown in brain heart infusion (BHI) broth overnight at 37 °C, and transferred to a defined minimal medium at 37 °C with shaking. For assessment of bacterial intracellular growth, standing cultures were grown in BHI broth overnight at 30 °C and used to infect J2 immortalized bone-derived macrophages as described (10). For broth lysis assessment, a β-galactosidase–encoding transposon insertion was transduced to constitutively promote β-galactosidase expression within the appropriate strains. Broth lysis and interacellular lysis assays were performed as described (10, 36).
Protein Cloning, Expression, and Purification.
The DNA fragment encoding the PgpH HD domain (amino acids 494–718) was PCR-amplified and cloned into the pMAL-c2x vector with the BamHI and HindIII restriction sites, generating an amino-terminal fusion of maltose binding protein. The resulting plasmid was transformed into E. coli BL21 (DE3) strain for expression.
For protein expression, overnight E. coli cultures were used to inoculate 1.5 L of LB medium containing 0.2% glucose and 100 μg/mL ampicillin, and grown at 37 °C, 200 rpm (MaxQ 6000, Thermo Scientific) until late-exponential phase (OD600 ∼ 0.8). At this stage, cultures were supplemented with 0.8 mM IPTG, metal salts (146 mg of FeSO4⋅7H2O, or 104 mg of MnCl2⋅4H2O, or 73 mg of FeSO4⋅7H2O + 57 mg of MnCl2⋅4H2O) and shaken at 16 °C, 200 rpm (MaxQ 6000, Thermo Scientific) for ∼14 h. Cell pellets were collected by centrifugation and lyzed by sonication in lysis buffer [20 mM Tris, pH 7.5, 200 mM NaCl, 5% (vol/vol) glycerol, 1 mM phenylmethylsulfonyl fluoride, 0.1% β-mercaptoethanol]. The following purification steps were performed at 4 °C by using a Biologic system (Bio-Rad) at a flow rate of 1 mL/min. Cell lysate containing soluble proteins were run through 2.5 mL of amylose resin (New England Biolabs), washed with column buffer [20 mM Tris, pH 7.5, 200 mM NaCl, 5% (vol/vol) glycerol] until the A280 of the flow through was less than 0.01 AU, and proteins were eluted with elution buffer (column buffer supplemented with 10 mM maltose monohydrate). After SDS/PAGE analysis, protein fractions were pooled, concentrated with a 20-mL, 5-kDa MWCO spin column concentrator (GE Healthcare) to a volume of 1 mL. The concentrated protein was diluted with protein storage buffer (20 mM Tris, pH 7.5, 50 mM NaCl) and reconcentrated for a total of three times to remove maltose and salt. Protein concentration was determined by Bradford assay. Final protein preparations were stored at −80 °C in storage buffer containing 25% (vol/vol) glycerol.
Synthesis of 32P-c-di-AMP.
Radio-labeled c-di-AMP was synthesized by incubating 1.2 μM α-[32P]ATP (Perkin-Elmer) with 1 μM of the diadenylate cyclase protein DisA in binding buffer (40 mM Tris pH 7.5, 100 mM NaCl, 20 mM MgCl2) at room temperature overnight. The sample was incubated for 5 min at 95 °C, and the DisA protein was removed by centrifugation. The mouse STING protein was used for further purification of synthesized 32P-c-di-AMP. Thirty-micromolar STING was bound to HisPur Ni-NTA resin (Thermo Scientific) and incubated with the remaining c-di-AMP synthesis reaction mix for 10 min at room temperature. The Ni-NTA resin was washed two times with cold binding buffer, incubated with 50 μL of binding buffer for 5 min at 95 °C, and transferred to a minispin column to elute 32P-c-di-AMP. The purity of this preparation, determined to be ∼96%, was evaluated by TLC, separating 1 μL on Polygram CEL300 PEI TLC plates (Machery-Nagel) in buffer containing 1:1.5 (vol/vol) saturated (NH4)2SO4 and 1.5 M NaH2PO4 pH 3.6. The yield was 2 nM 32P-c-di-AMP on average.
Nucleotide Binding Assay.
Interactions between MBP-HD proteins and nucleotides were evaluated by the DRACALA method (differential radial capillary action of ligand assay) as described (25). Binding reactions of 10 μL were performed in protein storage buffer. For the evaluation of binding affinities, varying concentrations of protein were incubated with ∼0.2 nM 32P-c-di-AMP. For the evaluation of binding specificity, 7 μM protein was incubated with ∼0.2 nM 32P-c-di-AMP, and 200 μM unlabeled nucleotides. After 5 min of incubation, nitrocellulose membrane was blotted with 4 μL of the reaction mixtures in duplicates, air-dried, and exposed to a PhosphorImager system for visualization. The fraction of ligand bound and Kd values were calculated as described (25).
Phosphodiesterase Activity Assay.
Phosphodiesterase activity was screened with bis-p-nitrophenol (bis-pNPP) substrate in 96-well reactions of 200 μL containing 20 μM protein and 1 mM bis-pNPP in storage buffer, with or without varying concentrations of c-di-AMP or ppGpp. The reactions were incubated at 37 °C, and the formation of p-nitrophenol was monitored by absorbance at 410 nm.
Phosphodiesterase activity toward c-di-AMP was determined in 300-μL reactions containing 20 μM protein and 100 μM c-di-AMP in storage buffer. The reactions were incubated at room temperature. Time point samples were heated at 95 °C for 5 min, centrifuged at maximum speed to remove precipitate, and analyzed by HPLC or LC/MS for c-di-AMP degradation. For HPLC, 20 μL of the clear supernatant was injected into a reverse phase column equilibrated with 20 mM potassium phosphate, pH 6.0 and 8% ethanol. Nucleotides were eluted in a mobile phase containing 20 mM potassium phosphate, pH 6.0 and 8% ethanol. For LC/MS, the supernatant was diluted 50 times in storage buffer.
Protein Expression and Purification for Structural Characterization.
The HD domain of PgpH (residues 494–718) was cloned into a modified pET-28a vector with a 6-His and SMT3 protein tag at the N terminus. Protein was overexpressed in E. coli BL21 Star (DE3) strain (Novagen). The cells were induced with 0.4 mM isopropyl β-d-1-thiogalactopyranoside for 18 h at 20 °C. The harvested cells were resuspended in a buffer containing 50 mM phosphate (pH 7.6), 500 mM NaCl, 10 mM imidazole, and lysed by sonication. Cell lysates were centrifuged for 30 min at 4 °C before incubating with nickel beads (Qiagen). After 1 h, beads were transferred to a gravity flow column (Bio-Rad) and washed extensively with a buffer containing 50 mM phosphate (pH 7.6), 500 mM NaCl, and 20 mM imidazole. Protein was eluted with a buffer containing 50 mM phosphate (pH 7.6), 250 mM NaCl, and 250 mM imidazole. Protein eluate was diluted by the wash buffer and incubated with ULP1 protease to cleave the His-SMT3 fusion tag, which was then retained on anion and cation exchange columns placed in tandem. The flow-through from the columns was further purified by gel filtration using Sephacryl S300 column (GE Healthcare) equilibrated in a buffer containing 5 mM Hepes (pH 7.6) and 150 mM NaCl. The protein sample were concentrated to 20 mg/mL and stored at –80 °C.
Protein Crystallization.
Protein solution (5 mg/mL) with 2 mM cyclic-di-AMP was used to set up crystallization. High-quality crystals were grown by mixing 1 μL of protein solution with 1 μL of well solution [0.2 M MgCl2, 0.1 M Mes (pH 5.6), 30% (wt/vol) polyethylene glycol 3350] using sitting drop method at 20 °C. Crystals appeared after 5 d and were transferred to the well solution before being flash-frozen in liquid nitrogen. The crystals belong to space group P61, and there are two complexes in the asymmetric unit.
Data Collection, Structure Determination, and Refinement.
A single-wavelength anomalous diffraction dataset at 2.7 Å resolution was collected at the peak absorption wavelength of iron (1.74 Å) at the X25 beamline of the National Synchrotron Light Source (NSLS), using a Pilatus 6M detector. A native dataset at 2.1 Å resolution for refinement was collected at wavelength of 1.10 Å at the same beamline. All data were processed with the HKL package (46). The structure was solved with the program autoSHARP (47). The structure model was manually built with Coot (48) and refined with Phenix (49). The crystallographic information is summarized in Table S2.
Quantification of Secreted and Intracellular c-di-AMP.
For measurement of intracellular levels of c-di-AMP, L. monocytogenes strains from overnight BHI broth cultures were grown again in fresh BHI broth to midexponential phase (OD600 ∼ 0.7). Cell pellets from 0.5- to 1-mL cultures were resuspended in 50 μL of 0.5 μM heavy-labeled (C13 N15) c-di-AMP, then mixed with 500 μL of methanol and sonicated. Methanol-extracted c-di-AMP was collected in two fractions. The first fraction of ∼500 μL was collected by centrifugation of lysed cells. The remaining pellets were resuspended in 50 μL of H2O, mixed with 500 μL of methanol, and centrifuged again to collect the second 500-μL fraction. Both fractions were pooled and evaporated to dryness in a speed vacuum concentrator. The resulting pellets were resuspended in 50 μL of ddH2O for mass spectrometry analysis.
For measurement of secreted c-di-AMP, L. monocytogenes strains were first grown in BHI broth at 37 °C overnight, and these cultures were used to inoculate minimal medium (50) cultures, which were then grown for 24 h at 37 °C. Aliquots were centrifuged to collect the supernatants. Mass spectrometry analysis was performed on supernatants mixed with heavy-labeled c-di-AMP in 1:1 volume ratio.
Quantification of c-di-AMP by LC-MS.
The chromatographic separation was performed with an analytical Synergi 4u Hydro-RP 80A column (50 × 2 mm, 4 μM particle size; Phenomenex), using a G6460 triple quadrupole LC-MS System (Agilent Technologies) equipped with a binary pump system. The injection volume was 10 μL. Each chromatographic run was 6.9 min. Eluent A was 10 mM formic acid in water, and eluent B was 10 mM formic acid in methanol. Ninety-eight percent A was used from 0 to 2 min, 70% A was used from 2 to 2.5 min, 20% A was used from 2.5 to 3 min, 100% B was used from 3 to 5 min, and 98% A was used until 6.9 min. Flow rates were 0.25 mL/min from 0 to 3 min, 0.35 mL/min from 3 to 6.5 min, and 0.25 mL/min from 6.5 to 6.9 min. The internal standard 13C 15N c-di-AMP and c-di-AMP were eluted at 2.7 min. The mass spectrometer parameters were as follows: capillary voltage: 3,500 V; charging voltage: 500 V; gas temperature: 350 °C; nebulizer gas: 45 psi.
The analyte detection was performed in the same LC-MS system with an electron spray ionization source, using multiple-reaction monitoring (MRM) analysis in positive ionization mode. The following MRM transitions were detected: c-di-AMP: +659.1/524.0 (qualifier), +659.1/330.0 (qualifier), +659.1/312.0 (qualifier), +659.1/136.0 (quantifier), and 13C 15N c-di-AMP: +689.0/146.0 (quantifier). The MRM transitions labeled as “quantifier” were used to quantify the compound of interest whereas “qualifier” transitions were monitored as confirmatory signals. A standard curve was constructed using standard 1 (250 nM 13C 15N c-di-AMP, 10 nM c-di-AMP) and standard 2 (250 nM 13C 15N c-di-AMP, 1000 nM c-di-AMP).
qRT-PCR.
Immortalized bone marrow derived macrophages were infected with L. monocytogenes for 4 h. RNA was harvested with an Ambion RNA extraction kit (Life Technologies) and IFN-β expression relative to actin was measured by qRT-PCR, as described (10).
Supplementary Material
Acknowledgments
We thank Dale Whittington for assistance in developing mass spectrometry methods and Samuel Miller for HPLC access. We thank Neil Whalen, Rick Jackimowicz, and Howard Robinson for access to the X29A beamline at the NSLS. The in-house instrument for X-ray diffraction was purchased with NIH Grant S10OD012018 (to L.T.). This work was supported by NIH Grants R56AI108698 (to J.J.W.) and R01DK067238 (to L.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 1921.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416485112/-/DCSupplemental.
References
- 1.Gomelsky M. cAMP, c-di-GMP, c-di-AMP and now cGMP: Bacteria use them all! Mol Microbiol. 2011;79(3):562–565. doi: 10.1111/j.1365-2958.2010.07514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Danilchanka O, Mekalanos JJ. Cyclic dinucleotides and the innate immune response. Cell. 2013;154(5):962–970. doi: 10.1016/j.cell.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalia D, et al. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem Soc Rev. 2013;42(1):305–341. doi: 10.1039/c2cs35206k. [DOI] [PubMed] [Google Scholar]
- 4.Römling U. Great times for small molecules: c-di-AMP, a second messenger candidate in Bacteria and Archaea. Sci Signal. 2008;1(33):pe39. doi: 10.1126/scisignal.133pe39. [DOI] [PubMed] [Google Scholar]
- 5.Corrigan RM, Gründling A. Cyclic di-AMP: Another second messenger enters the fray. Nat Rev Microbiol. 2013;11(8):513–524. doi: 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- 6.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Gründling A. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011;7(9):e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo Y, Helmann JD. Analysis of the role of Bacillus subtilis σ(M) in β-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol Microbiol. 2012;83(3):623–639. doi: 10.1111/j.1365-2958.2011.07953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehne FM, et al. Cyclic di-AMP homeostasis in bacillus subtilis: Both lack and high level accumulation of the nucleotide are detrimental for cell growth. J Biol Chem. 2013;288(3):2004–2017. doi: 10.1074/jbc.M112.395491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai Y, et al. Two DHH subfamily 1 proteins in Streptococcus pneumoniae possess cyclic di-AMP phosphodiesterase activity and affect bacterial growth and virulence. J Bacteriol. 2013;195(22):5123–5132. doi: 10.1128/JB.00769-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Witte CE, et al. Cyclic di-AMP is critical for Listeria monocytogenes growth, cell wall homeostasis, and establishment of infection. MBio. 2013;4(3):e00282–e13. doi: 10.1128/mBio.00282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao F, et al. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J Biol Chem. 2010;285(1):473–482. doi: 10.1074/jbc.M109.040238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manikandan K, et al. Two-step synthesis and hydrolysis of cyclic di-AMP in Mycobacterium tuberculosis. PLoS ONE. 2014;9(1):e86096. doi: 10.1371/journal.pone.0086096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye M, et al. DhhP, a c-di-AMP phosphodiesterase of Borrelia burgdorferi, is essential for cell growth and virulence. Infect Immun. 2014;82(5):1840–1849. doi: 10.1128/IAI.00030-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328(5986):1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sureka K, et al. The cyclic dinucleotide c-di-AMP is an allosteric regulator of metabolic enzyme function. Cell. 2014;158(6):1389–1401. doi: 10.1016/j.cell.2014.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S, Bayles DO, Mason TM, Wilkinson BJ. A cold-sensitive Listeria monocytogenes mutant has a transposon insertion in a gene encoding a putative membrane protein and shows altered (p)ppGpp levels. Appl Environ Microbiol. 2006;72(6):3955–3959. doi: 10.1128/AEM.02607-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aravind L, Koonin EV. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci. 1998;23(12):469–472. doi: 10.1016/s0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- 18.Anantharaman V, Aravind L. Application of comparative genomics in the identification and analysis of novel families of membrane-associated receptors in bacteria. BMC Genomics. 2003;4(1):34. doi: 10.1186/1471-2164-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wurtzel O, et al. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol Syst Biol. 2012;8:583. doi: 10.1038/msb.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim SK, Makino K, Amemura M, Shinagawa H, Nakata A. Molecular analysis of the phoH gene, belonging to the phosphate regulon in Escherichia coli. J Bacteriol. 1993;175(5):1316–1324. doi: 10.1128/jb.175.5.1316-1324.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Britton RA. Role of GTPases in bacterial ribosome assembly. Annu Rev Microbiol. 2009;63:155–176. doi: 10.1146/annurev.micro.091208.073225. [DOI] [PubMed] [Google Scholar]
- 22.Davies BW, et al. Role of Escherichia coli YbeY, a highly conserved protein, in rRNA processing. Mol Microbiol. 2010;78(2):506–518. doi: 10.1111/j.1365-2958.2010.07351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Horn WD, Sanders CR. Prokaryotic diacylglycerol kinase and undecaprenol kinase. Ann Rev Biophys. 2012;41:81–101. doi: 10.1146/annurev-biophys-050511-102330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song BH, Neuhard J. Chromosomal location, cloning and nucleotide sequence of the Bacillus subtilis cdd gene encoding cytidine/deoxycytidine deaminase. Mol Gen Genet. 1989;216(2-3):462–468. doi: 10.1007/BF00334391. [DOI] [PubMed] [Google Scholar]
- 25.Roelofs KG, Wang J, Sintim HO, Lee VT. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc Natl Acad Sci USA. 2011;108(37):15528–15533. doi: 10.1073/pnas.1018949108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galperin MY, Natale DA, Aravind L, Koonin EV. A specialized version of the HD hydrolase domain implicated in signal transduction. J Mol Microbiol Biotechnol. 1999;1(2):303–305. [PMC free article] [PubMed] [Google Scholar]
- 27.Lovering AL, Capeness MJ, Lambert C, Hobley L, Sockett RE. The structure of an unconventional HD-GYP protein from Bdellovibrio reveals the roles of conserved residues in this class of cyclic-di-GMP phosphodiesterases. MBio. 2011;2(5):e00163–11. doi: 10.1128/mBio.00163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miner KD, Klose KE, Kurtz DM., Jr An HD-GYP cyclic di-guanosine monophosphate phosphodiesterase with a non-heme diiron-carboxylate active site. Biochemistry. 2013;52(32):5329–5331. doi: 10.1021/bi4009215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellini D, et al. Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre. Mol Microbiol. 2014;91(1):26–38. doi: 10.1111/mmi.12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zimmerman MD, Proudfoot M, Yakunin A, Minor W. Structural insight into the mechanism of substrate specificity and catalytic activity of an HD-domain phosphohydrolase: The 5′-deoxyribonucleotidase YfbR from Escherichia coli. J Mol Biol. 2008;378(1):215–226. doi: 10.1016/j.jmb.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartz KT, et al. Hyperinduction of host beta interferon by a Listeria monocytogenes strain naturally overexpressing the multidrug efflux pump MdrT. Infect Immun. 2012;80(4):1537–1545. doi: 10.1128/IAI.06286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaplan Zeevi M, et al. Listeria monocytogenes multidrug resistance transporters and cyclic di-AMP, which contribute to type I interferon induction, play a role in cell wall stress. J Bacteriol. 2013;195(23):5250–5261. doi: 10.1128/JB.00794-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lauer P, Chow MY, Loessner MJ, Portnoy DA, Calendar R. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J Bacteriol. 2002;184(15):4177–4186. doi: 10.1128/JB.184.15.4177-4186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sauer JD, et al. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79(2):688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tadmor K, et al. Listeria monocytogenes MDR transporters are involved in LTA synthesis and triggering of innate immunity during infection. Front Cell Infect Microbiol. 2014;4:16. doi: 10.3389/fcimb.2014.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sauer JD, et al. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe. 2010;7(5):412–419. doi: 10.1016/j.chom.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Witte CE, et al. Innate immune pathways triggered by Listeria monocytogenes and their role in the induction of cell-mediated immunity. Adv Immunol. 2012;113:135–156. doi: 10.1016/B978-0-12-394590-7.00002-6. [DOI] [PubMed] [Google Scholar]
- 38.Rao F, Ji Q, Soehano I, Liang ZX. Unusual heme-binding PAS domain from YybT family proteins. J Bacteriol. 2011;193(7):1543–1551. doi: 10.1128/JB.01364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mechold U, Fang G, Ngo S, Ogryzko V, Danchin A. YtqI from Bacillus subtilis has both oligoribonuclease and pAp-phosphatase activity. Nucleic Acids Res. 2007;35(13):4552–4561. doi: 10.1093/nar/gkm462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Postic G, Danchin A, Mechold U. Characterization of NrnA homologs from Mycobacterium tuberculosis and Mycoplasma pneumoniae. RNA. 2012;18(1):155–165. doi: 10.1261/rna.029132.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abdul-Sater AA, et al. Cyclic-di-GMP and cyclic-di-AMP activate the NLRP3 inflammasome. EMBO Rep. 2013;14(10):900–906. doi: 10.1038/embor.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miao EA, Rajan JV, Aderem A. Caspase-1-induced pyroptotic cell death. Immunol Rev. 2011;243(1):206–214. doi: 10.1111/j.1600-065X.2011.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corrigan RM, et al. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci USA. 2013;110(22):9084–9089. doi: 10.1073/pnas.1300595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bai Y, et al. Cyclic di-AMP impairs potassium uptake mediated by a cyclic di-AMP binding protein in Streptococcus pneumoniae. J Bacteriol. 2014;196(3):614–623. doi: 10.1128/JB.01041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Camilli A, Tilney LG, Portnoy DA. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol Microbiol. 1993;8(1):143–157. doi: 10.1111/j.1365-2958.1993.tb01211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otwinowski ZaM, W Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 47.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 50.Phan-Thanh L, Gormon T. A chemically defined minimal medium for the optimal culture of Listeria. Int J Food Microbiol. 1997;35(1):91–95. doi: 10.1016/s0168-1605(96)01205-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.