Significance
Alphaviruses infect human cells through endocytosis and low pH-triggered membrane fusion. Because of the recent epidemics and lack of specific treatment for alphaviral infections, numerous investigations have aimed to deduce the mechanism of the alphaviral infection pathway. Building on previous structural models of the viral envelope proteins, we identify critical histidine protonation-state changes responsible for pH-triggered conformational transitions between key fusion intermediates through quantitative calculation of pKa and free energies and further show that the key contributions arise from the His residues having conserved sequences and chemical environments. Our study provides insight into the mechanistic roles of these residues and suggests hotspots for targeting inhibitors that could shift the pH profile of membrane fusion and thus, interfere with infectivity.
Keywords: alphavirus, envelope protein, pH, membrane fusion, constant pH molecular dynamics
Abstract
Alphavirus envelope proteins, organized as trimers of E2–E1 heterodimers on the surface of the pathogenic alphavirus, mediate the low pH-triggered fusion of viral and endosomal membranes in human cells. The lack of specific treatment for alphaviral infections motivates our exploration of potential antiviral approaches by inhibiting one or more fusion steps in the common endocytic viral entry pathway. In this work, we performed constant pH molecular dynamics based on an atomic model of the alphavirus envelope with icosahedral symmetry. We have identified pH-sensitive residues that cause the largest shifts in thermodynamic driving forces under neutral and acidic pH conditions for various fusion steps. A series of conserved interdomain His residues is identified to be responsible for the pH-dependent conformational changes in the fusion process, and ligand binding sites in their vicinity are anticipated to be potential drug targets aimed at inhibiting viral infections.
Alphaviruses, mosquito-borne human pathogens causing severe inflammations and fatal fevers, have infected many millions of people in recent outbreaks worldwide since 2005 (1–3). The lack of a vaccine or specific treatment prompts investigations of the fundamental mechanisms of the alphaviral lifecycle to facilitate the development of effective antiviral therapies (4). Alphaviruses have been reported to enter the cell through receptor-mediated endocytosis. Here, alphaviruses are ferried toward the perinuclear space of the host cell inside vesicles towed by molecular motors and delivered to specific locations for productive replication (5–11). Even when direct entry into the cytoplasm is possible (11–15), the endocytic entry pathway facilitates the transportation of viruses across the crowded cytoplasmic space and delays detection by the immune system without leaving empty capsid or envelope as obvious evidence of the viral infection exposed outside the host cell (10, 11). Before the delivery of its viral genome into the cytoplasm of a host cell, the alphavirus must undergo a critical step of low pH-triggered membrane fusion, which is a common mechanism in the endocytic viral entry pathway among many different viruses. Understanding the mechanism of the low pH-triggered alphaviral membrane fusion is essential for the development of therapies against alphavirus as well as other viruses using similar endocytic entry mechanisms.
Recent studies of the lifecycle of alphavirus reveal that a precursor, p62, is first synthesized as a chaperon forming a heterodimer with E1, which is essential for viral budding (16); p62 protects the E1 protein in the low-pH environment of the secretory pathway before being cleaved by cellular furin to produce mature E2-E1 and a smaller fragment, E3 (17–21). After the virus buds from the cytoplasmic membrane, E3 is released from the virus particle under neutral pH conditions outside the host cell (13, 22–24).
On the surface of a mature alphavirus, 80 (E2–E1)3 viral spikes, organized in T = 4 icosahedral symmetry on the viral lipid membrane, enclose the viral capsid and genome (25–43). On internalization of the mature virus in the endosome of the host cell in a new round of infection cycle, the increasingly acidified endosomal environment triggers a series of conformational changes in the alphaviral spike (E2–E1)3 (38), including the dissociation of E2 (42, 44, 45), release of a fusion loop on E1 (46, 47), and trimerization of E1 (48). The fusion loop, roughly residues 83–100 on the cd loop of each E1 protein (13, 49, 50), in the newly formed E1 homotrimer (HT), inserts into the endosomal membrane. Then, the E1 proteins fold back, pulling the viral and endosomal membranes together and thus, promoting membrane fusion (13, 24).
Recently solved high-resolution structures of the alphavirus envelope proteins E2–E1 fitted into cryo-EM data representing the intact virus under both acidic and neutral pH conditions (43, 51, 52) provide excellent atomic models for studies of the low pH-triggered fusion process. The structure of Chikungunya virus (CHIKV) obtained at pH 8.0 represents the initial mature state (M state) of the (E2–E1)3 viral spike before the fusion process (51). Under pH 5.6, domain B (DB) of E2, which protects the E1 fusion loop, is observed to be disordered in Sindbis virus (52). The rest of the domains of the (E2–E1)3 spike show moderate conformational differences with an rmsd = 4.0 Å among Cα atoms compared with the structures obtained at pH 8.0 for CHIKV (43, 51). The structure of the envelope proteins in acidic conditions most likely depicts a fusion intermediate (FI) state (52) before E2 dissociation during the low pH-triggered fusion process. In addition, the crystal structure of the folded-back E1 HT (53) is a good model to describe the postfusion state.
Based on these atomic models of the E2 and E1 envelope proteins and our previously developed constant pH molecular dynamics (CPHMD) method (54–58), we simulated the envelope proteins with icosahedral symmetry under various pH conditions covering pH 2.0–9.0. We used pH replica exchange in CPHMD and calculated pKa values using pH titration fitting, which has been shown as a reliable and accurate approach to capture pKa values of protein residues in various systems (59–64). Through the CPHMD modeling, we calculated the pKa of the possible pH-sensitive residues (Asp, Glu, and His) in the M, FI, dissociated E2 (Dis), and HT states. We, therefore, derive the shifts in the thermodynamic stabilities originating from each titrating residue for the steps from the M to the FI state (M→FI) of (E2–E1)3, from the FI to the Dis state (FI→Dis) of E2 proteins, and from the FI to the HT state (FI→HT) of E1 proteins as shown in Fig. 1D. For these processes, we assume that the virus is in the endosomal environment, and we do not consider possible receptor-induced conformational changes. Our residue-level resolution simulations and analyses allow us to identify the critical functional residues with significant pKa shifts and changes in thermodynamic stability in the low pH-triggered fusion activation. Our results suggest that the most pH-sensitive residues are highly conserved among different alphaviral species and that these critical residues control the pH threshold of fusion activities, provide guidance to further mutagenesis experiments, and lead to more fundamental understanding of low pH-triggered alphaviral membrane fusion.
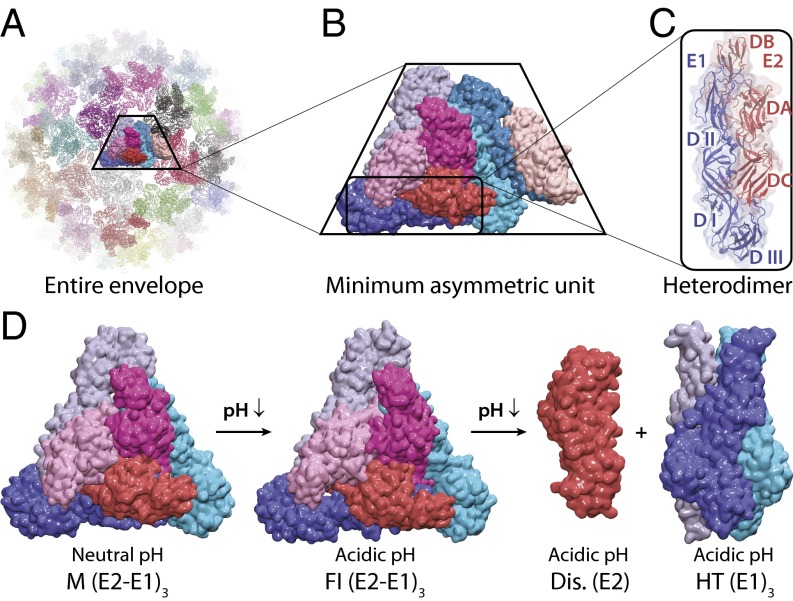
Fig. 1.
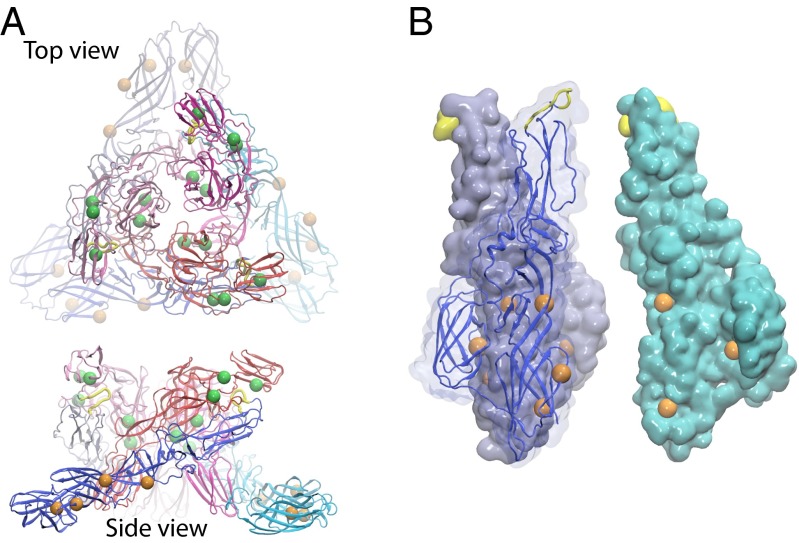
Structure and organization of alphaviral envelope proteins. (A) The alphaviral envelope modeled in our simulations. (B) The alphaviral envelope proteins in an MAU. (C) The heterodimer of E2 (DA–DB–DC) and E1 (DI–DII–DIII). (D) Structures of a viral spike in different conformational states simulated for shifts in pKa values and thermodynamic stabilities. E1 proteins are shown in blue, cyan, and light blue. E2 proteins are shown in red, magenta, and pink.
Results and Discussion
pKa Shifts and pH-Dependent Thermodynamic Stabilities at a Residue-Level Resolution.
The organization of the alphaviral envelope proteins is shown in Fig. 1. The envelope proteins form 80 spikes on the surface of the alphavirus. Each spike is a complex of the (E2–E1)3 composed of a trimer of heterodimers of proteins E2 and E1. The alphaviral envelope is observed to adopt T = 4 icosahedral symmetry, with a minimum asymmetric unit (MAU) containing four of the E2–E1 dimers (Fig. 1 B and C) for both M and FI states (30, 35, 37, 49). In this work, the ectodomains of the envelope proteins in an MAU (E2: DA, DB, and DC; E1: DI, DII, and DIII) were used as our simulation system, and the icosahedral symmetric neighbors were generated to construct the entire viral envelope (Fig. 1A) using rotational symmetry boundary conditions in CHARMM (65, 66), which provide a significant advantage to efficiently handle the interspike interactions in the viral envelope. The icosahedral symmetry constrains the residues from other rotationally related image units to adopt the same conformations and pKa values; this constraint may result in inherent error to microscopic pKa values because of the possible coupling between symmetry-related neighbors, which is discussed in more detail in SI Appendix; however, calculations without the use of such symmetry constraints are beyond the scope of this work and practical calculations of this sort.
In our simulations, the pKa values of the titrating residues were computed for the states of the alphavirus envelope proteins representing the M and FI states of (E2–E1)3, E2 Dis, and E1 HT. The relative difference in free energy cost associated with changing the pH from neutral (pHref 7.5) to acidic (pH 5.5) conditions was calculated according to Eq. 2 to model the change from the extracellular to the endosomal environment for all fusion steps (67, 68). The pKa values of the same residues in the viral spike (E2–E1)3 of the MAU were averaged.
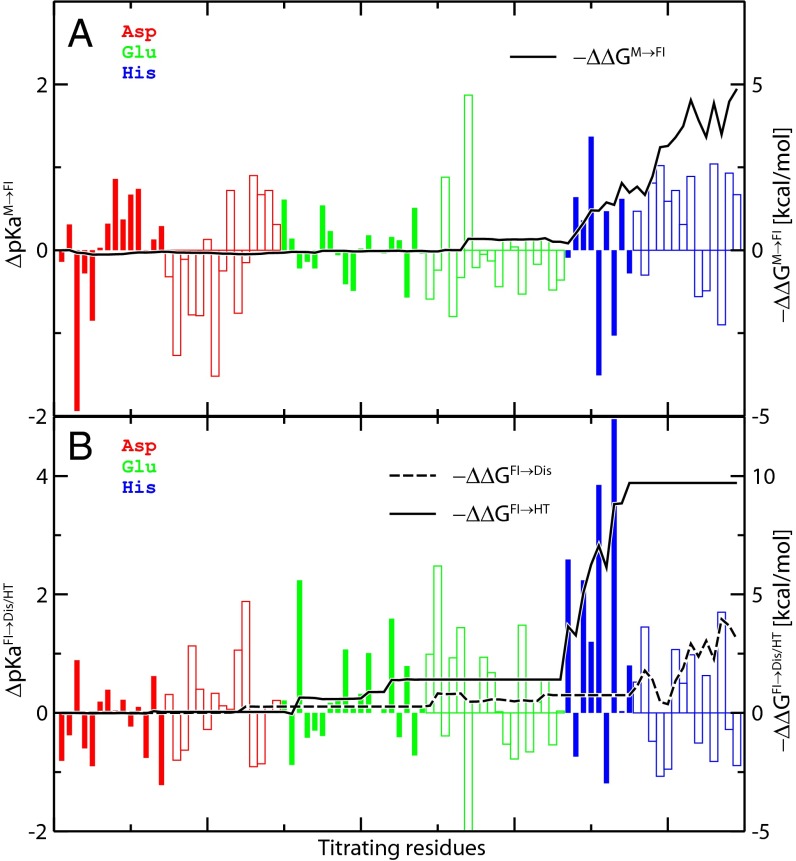
In Fig. 2, we show the pKa shifts and corresponding relative free energy components for each titrating residue involved in the M→FI activation (Fig. 2A), the Dis, and the E1 HT formation steps (Fig. 2B). The majority of the Asp and Glu has pKa values within the range of pH 2–5, and most His are between pH 5 and pH 8, with estimated simulation errors of about 0.2–0.3 pH unit (SI Appendix, Fig. S2). The significant upshift in pKa is mainly a consequence of the conformational changes and the corresponding reorganizations of H-bond interactions (Table 1 and SI Appendix, Table S1). The change in the nearby chemical environment from proton donors or positively charged groups to strong proton acceptors or negatively charged groups leads to the significant upshifts in the titrating residues.
Fig. 2.
ΔpKa values and the corresponding shifts in thermodynamic stability (ΔΔG) at residue-level resolution for (A) the M→FI activation step and (B) the E2 dissociation (FI→Dis) and HT formation (FI→HT) steps. ΔpKaA→B values of individual residues are represented by vertical bars for Asp (red), Glu (green), and His (blue) residues in E1 (solid bars) and E2 (outlined bars) proteins, with scales on the left y axes. Integration of −ΔΔGM→FI, −ΔΔGFI→HT, and −ΔΔGFI→Dis per monomer (with pH 5.5 and pHref 7.5) is shown in black lines, with scales on the right y axes.
Table 1.
Summary of strictly conserved His residues with corresponding pKa shifts, ΔΔG, and interaction partners in different alphaviruses
| H-bond interaction partners | M→FI | FI→Dis/HT | ||||||
| Residue | M | FI | HT/Dis | ΔpKa | −ΔΔG | ΔpKa | −ΔΔG | Domain interface* |
| H3(E1) | PCP(E1)† | PCP (E1) | E284/F287(E1/E1′)‡ | −0.1 | 0.0 | 2.6 | 2.2 | HT: DI/DIII′ |
| H125(E1) | T126(E1′) | T126 (E1′) | D174(E1′) | 0.4 | 0.3 | 2.2 | 1.8 | HT: DII/DII′ |
| H230(E1) | R267(E2′) | R267 (E2′) | E67(E1′) | −1.5 | 0.0 | 3.9 | 0.8 | HT: DII/DII′ |
| H331(E1) | PCP(E1) | PCP (E1) | N149(E1′) | −1.0 | 0.1 | 5.0 | 2.7 | HT: DIII/DI′ |
| H386(E1) | K280(E2) | K280(E2) | F192(E1′) | 0.1 | 0.1 | 1.1 | 1.0 | E2–E1: DC/stem |
| H170(E2) | S57(E1)/R244(E2) | — | — | 1.1 | 1.0 | −0.8 | −0.7 | E2–E1: β/DII |
| H256(E2) | K254(E2) | K254(E2) | K254/E166(E2) | −0.1 | −0.1 | 0.4 | 0.4 | — |
Domain interface of the relative stronger interaction is listed.
PCP, positively charged pocket composed of Y15(E1), K16(E1), and T17(E1).
E1′ refers to another E1 protein belonging to the nearest monomeric or dimeric neighbor interacting with the protein of focus.
According to Eq. 2, ΔΔG has negative correlation with ΔpKa. Thus, in Fig. 2, values of −ΔΔG instead of ΔΔG are displayed (solid and dashed curves in Fig. 2) to maintain a consistent sign with the ΔpKa (solid and outlined vertical bars in Fig. 2). We can see that most −ΔΔGM→FI contributions originate from His residues, despite the fact that many Asp and Glu residues possess relatively large ΔpKaM→FI values (vertical bars), because those residues with pKa values in the relevant range for the pH-induced transition (pH 5.5–7.5) contribute more than the rest. Therefore, even if both D250(E2) (pKaM = 3.2, pKaFI = 4.2) and H170(E2) (pKaM = 6.5, pKaFI = 7.5) show similar pKa upshifts of about 1.0 unit, the free energy component of D250(E2), |ΔΔGM→FI [D250(E2)]|, is less than 0.1 kcal/mol per monomer compared with ΔΔGM→FI[H170(E2)] = −1.0 kcal/mol per monomer. Thus, a large pKa upshift in the relevant pH range leads to a significant negative ΔΔG component, providing the effect of favoring the corresponding conformational change under acidic conditions.
In the first fusion activation step, M→FI, the His residues from the E2 proteins were found to be the main source (∼70% contribution) of the thermodynamic shift (ΔΔGM→FI(E2):ΔΔGM→FI(total) ≈ −9.8:−14.5 kcal/mol for each spike). After the viral spike is activated and reaches the FI state under acidic pH conditions, the E2 residues cause an additional shift of ΔΔGFI→Dis ≈ −10.6 kcal/mol per spike to the dissociation step, showing dominant control of the early fusion activation processes. However, residues on the E1 proteins were found to be relatively passive during the first step (M→FI), with ΔΔGM→FI(E1) ≈ −4.7 kcal/mol per spike. However, they become significantly pH-sensitive in the HT formation and fold back after E2 dissociation, with ΔΔGHT(E1) ≈ −32.1 kcal/mol per spike.
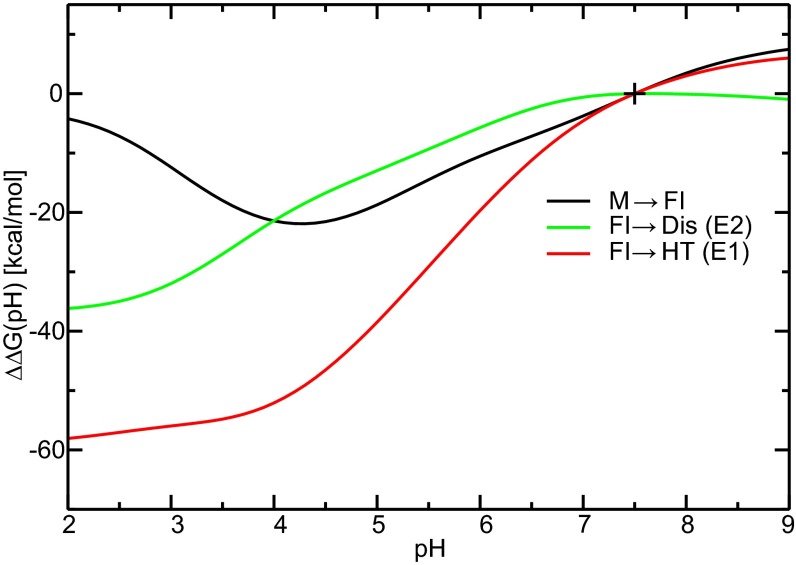
According to Eq. 2, we construct the free energy shifts of the fusion activation steps over the pH spectrum using the pKa shifts and characterize the pH transitions of these conformational changes. Using pH 2.0–9.0 and pHref 7.5, the free energy shift as a function of pH conditions, ΔΔG(pH), was calculated and is displayed in Fig. 3 for each fusion step. All three fusion steps show a pH transition at pH ≈ 5–6 and reach the most favorable free energy conditions at pH ≈ 3–4. The M→FI step then becomes much less favorable at lower pH, mainly because of the opposing contributions from Asp residues. The ΔΔG(pH) profiles of the FI→Dis/HT steps become flat at lower pH conditions, which is in qualitative agreement with the biochemical experiments showing the threshold of pH ≈ 6 for E2 dissociation and E1 HT formation and reaching plateaus at pH < 5 (69).
Fig. 3.
pH dependence of the shifts of thermodynamic stabilities per viral spike, ΔΔG(pH), for the M→FI, FI→Dis (E2), and FI→HT (E1) processes. ΔΔG at the reference pH, pHref 7.5, is set to 0 kcal/mol, indicated by +.
Correlation Between Residue Sequence Conservation and pH Sensitivity.
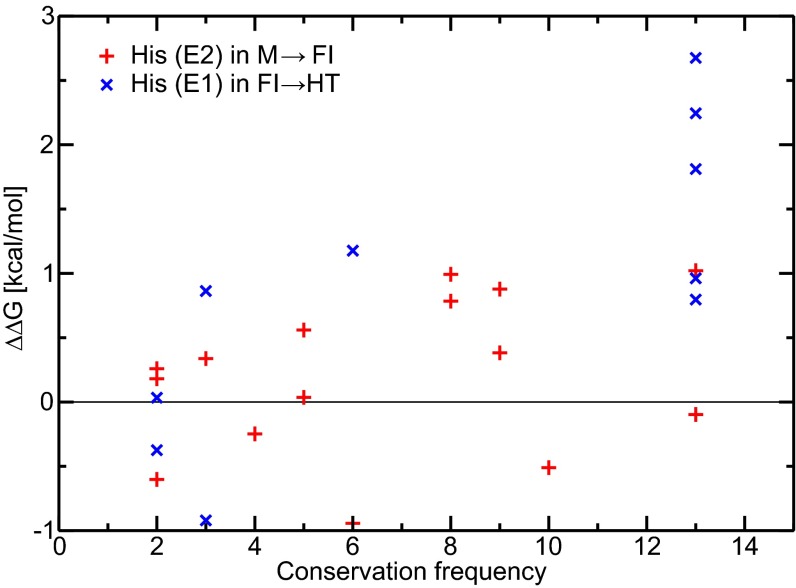
To explore the common features in the membrane fusion process of alphaviruses, it is critical to identify the conservation of pH-sensitive residues and examine the roles of conserved residues. We performed a survey on all His residues found in the CHIKV E2–E1 proteins and obtained the conservation frequency through alignment and comparison of 13 alphaviruses (51). The conservation frequency of each His residue and the corresponding pH-dependent ΔΔG components are shown in Fig. 4. As reported in Table 1, we listed all of the strictly conserved His residues with the corresponding shifts in pKa and thermodynamic stability. The conserved H170(E2) is determined to contribute most to the first M→FI activation step. All five conserved residues on the E1 protein show relatively large ΔΔG contributions to the FI→HT step. In summary, six of seven strictly conserved His residues were found to contribute to the pH-dependent fusion steps, consistent with Fig. 2. H256(E2) is the only strictly conserved His residue that shows no significant contribution to the two steps investigated in this work. However, previous experimental results suggest that the H-bond network around H256(E2) is important in the pH-dependent regulation of the binding and dissociation of the peripheral glycoprotein E3 (21, 23).
Fig. 4.
Correlation of sequence conservation and pH sensitivity. Sequence conservation frequencies of all His residues on E2 and E1 of CHIKV with the corresponding ΔΔG contributions to the M→FI and FI→HT steps are shown by red + and blue ×, respectively. A conservation frequency of 13 represents a strictly conserved sequence among the 13 alphaviral species surveyed here.
From another perspective, we ranked the residues with largest free energy contributions (with |ΔΔG| > 1.0 kcal/mol per monomer) in SI Appendix, Table S1 with the corresponding conservation frequency. Among the most pH-sensitive residues, specifically for CHIKV virus, we found that the residues on the E2 proteins in the FI→Dis are relatively the least conserved compared with other steps. In the M→FI step, other than the strictly conserved H170(E2), H232(E2) shows a relatively high conservation frequency of 8 of 13, indicating that the activation from M state to FI state is a more generic step to regulate the dissociation of E2 among different alphaviruses. In addition, the pH-sensitive His residues in the FI→HT step are also highly conserved (all on E1 proteins), with H152(E1) as the only residue not strictly conserved (conservation frequency of 6 of 13), suggesting the common mechanism of the HT formation among different alphaviruses.
Hydrogen Bond Interactions and Origins of the pH-Dependent Fusion States.
To illustrate the origins of the shifts in pKa and pH-dependent thermodynamic stabilities, we examined the changes of the chemical environment and reorganization of the H-bond networking in the conformational changes among the different fusion states. The interaction partners of each conserved His residue are reported in Table 1. It is encouraging to observe that not only the pH-sensitive His residues themselves but also, their interaction partners show high levels of conservation among different alphaviral species in terms of the residue sequence or the chemical environment that they provide (SI Appendix, Table S2).
As shown in SI Appendix, Fig. S3–S7, all five strictly conserved His residues on the E1 protein were found to interact with positively charged or neutral polar residues in both the M and FI states. In the final HT state, however, these His residues all form relatively strong salt bridges with acidic residues or H bonds with electron-rich groups. The chemical environments of these pH-sensitive His residues are highly conserved in all fusion states (SI Appendix, Table S2). The dramatic reorganization in the interaction network caused by the conformational change leads to the large upshifts in pKa and the significant negative shifts in ΔΔG, which strongly favor the FI→HT step.
In a similar scenario shown in SI Appendix, Fig. S8, H170(E2) forms a relatively stable H bond in the M state with S57(E1), a moderate proton donor, as previously suggested in experimental mutagenesis (18). In addition, the nearby E2 R244 is also likely to interact with the deprotonated H170(E2) and reinforce the H170(E2)–S57(E1) interaction, further stabilizing the E2–E1 interface in the M state. After H170(E2) is protonated under acidic pH and E2–E1 transforms to the FI state, R244(E2) is most likely repelled by the positively charged H170(E2), and the H170(E2)–S57(E1) interaction is destabilized, resulting in the upshift in the pKa of H170(E2).
Functional Roles of the Conserved pH-Sensitive Residues and Mutation Effects.
According to our simulation results, all conserved His residues, except for H256(E2), actively contribute to the low pH-triggered fusion process by shifting thermodynamic stabilities to favor the conformational changes of M→FI or FI→HT under acidic pH conditions. Depending on the different locations and types of H bonds that they form, we have analyzed the mechanisms, functional roles, and possible mutation effects of these conserved His residues underlying the fusion steps.
H3(E1), H125(E1), and H230(E1) are all found to form relatively strong salt-bridge interactions, stabilizing the trimeric interfaces on the E1s in the HT state, whereas they have relatively weak H-bond interactions in the M and FI states. Eliminating any of these interactions through mutations to nonpolar residues is expected to directly affect the HT formation. According to Fig. 3, the pH threshold of the HT formation will be lowered if the stabilizing interactions on these sites are weakened. According to experimental mutagenesis, H3A(E1) and H125A(E1) mutants show fusion activities at lower pH conditions with shifts of about 1.5 and 0.5 pH units, respectively (69). Particularly, the pH threshold for HT formation is lowered from pH 6 to pH 5 in the H3A(E1) mutant (69). In addition, it is reported that HT formation in the H230A(E1) mutant requires pH conditions at about 0.5 pH units lower (70), although the major effect of H230A(E1) is more likely to interfere with the late fusion stage (71).
During the FI→HT step, H331(E1) breaks the interaction H331(E1)–K16(E1) (FI) and forms a new H331(E1)–N149(E1′) H bond (HT). Both interactions are highly conserved and near the DI/DIII linker region. It is likely that breaking of H331(E1)–K16(E1) is involved in the opening of DIII and the formation of the extended HT state. More specific contribution to this step remains less clear, because the extended form of HT is not simulated in this work because of the lack of structural information. The newly formed H331(E1)–N149(E1) interaction, however, is likely to contribute to the DIII fold-back process subsequent to the E1s trimer core formation. However, the DIII fold back is found to be a highly spontaneous process with a thermodynamic driving force of about 7–10 kcal/mol (72). Thus, the ΔΔG contribution from H331(E1) seems to be less critical for the fold-back process, which suggests that the mutation of H331A(E1) is likely to show no apparent effect on the overall fusion activity in a bioassay (69). Based on this model, mutations of H331D/E(E1) are expected to further stabilize the D/E331(E1)–K16(E1) interaction, preventing opening of DI/DIII and inhibiting the proper refolding of DIII; thus, these mutations should introduce notable interference into fusion activity.
H386(E1) and H170(E2) both form relatively stable H bonds on the E2–E1 interface. Thus, we suggest that H386(E1) and H170(E2) mainly contribute to the regulation of the pH-dependent E2 dissociation. Elimination of these interactions is expected to destabilize the E2–E1 interface in the M state and trigger E2 dissociation at a higher pH threshold. This conjecture qualitatively agrees with experimental mutagenesis results showing increased fusion activity at pH 6.25 as a result of mutations including H386A(E1) (73) as well as the observation of E2–E1 dissociation in the S57A(E1)–H170A(E2) double mutant under neutral conditions with pH up to 8.0 (18).
In addition to the pH-sensitive His residues, with protonation states that are directly influenced by the decreasing endosomal pH conditions, experimental mutagenesis suggests that D188(E1), G91(E1), and the stem region 384–413(E1) may play essential structural roles during the early stages of endocytic membrane fusion (73–75). It is unlikely for the structural residues to change their protonation states under endosomal pH conditions; however, mutations of these residues may cause structural disruptions (e.g., D188K may lead to extra repulsion at the trimeric interface) (75). The structural disruption from these mutations must be compensated for by the pH-sensitive residues to reach the desired conformational states, thus indirectly lowering the pH thresholds of the fusion stages.
Potential Hotspots to Inhibit Fusion Activity.
The correlation between the pH sensitivity (ΔΔG) and the conserved residual interactions points to possible common features in the fusion pathway and universal critical sites on the envelope proteins among different species of alphavirus, suggesting the possibility to develop fusion-interfering antiviral therapies effective against multiple species of alphaviruses.
Investigation of the locations of the conserved pH-sensitive residues indicates several hotspots with conserved interfacial interactions that drive the reorganization of the domains during the conformational changes of the fusion process. In Fig. 5, the pH-sensitive residues identified to contribute to the M→FI and FI→HT steps are shown in green and orange beads, respectively. These residues mainly concentrate in two sites: (i) the E2/E1–DII region [H170(E2), H232(E2), and H230(E1)] adjacent to the fusion loop and (ii) the E1–DI region, including DI/DII hinge [H125(E1)], DI/DIII linker [H331(E1)], and DI/DI′ core trimeric [H3(E1)] interfaces. Multimutants of the pH-sensitive residues and their interaction partners in these regions are expected to introduce significant inhibition of alphaviral membrane fusion. In addition, the highly localized distribution of critical residues suggests that ligands binding to the domain interfaces in the two hotspots may dramatically hinder the fusion process.
Fig. 5.
Locations of pH-sensitive residues on (A) the (E2–E1)3 viral spike complex in the M state and (B) the (E1)3 in the HT state. Strictly conserved pH-sensitive residues that contribute to the M→FI and FI→HT steps are mapped in green and orange beads, respectively, on the alphaviral spike. Two nonstrictly conserved residues with ΔΔG > 1 kcal/mol in the M→FI and FI→HT steps are shown in smaller beads. In B, one of the E1 monomers (cyan) is displaced and rotated to show the residues on the trimeric interface inside the HT.
Conclusion
In this work, we simulated the alphaviral E2 and E1 proteins in different conformational states under varying pH conditions. Through the calculation of pKa values and pH-dependent shifts in the thermodynamic stability of each titrating residue among the conformational states, we provide residue-level mechanistic insights into the low pH-triggered alphaviral membrane fusion steps. We identified a series of pH-sensitive His residues found to be highly conserved among 13 species that hold primary responsibility for fusion activation and HT formation. The correlation in the high level of conservation and pH sensitivity suggests universal interaction networks and a possible common pathway of the fusion mechanism among different alphaviral species. Our simulation results qualitatively agree with previous mutagenesis observations in terms of the varying pH conditions for fusion activities caused by the mutation of several conserved His residues. Additional mutagenesis and corresponding biochemical assays on the pH-sensitive residues as well as their H-bond partners should be extremely helpful in verifying the functional roles of these residues and enriching our understanding of the low pH-triggered fusion process. Moreover, the localized distribution of the critical residues identified here suggests possible pH-sensitive hotspots in the fusion process that may be used as potential drug targets against the alphaviral infections.
This work bridges the fundamental H-bond interactions at the residue level and the macroscopic pH dependence of the endocytic membrane fusion process. The agreement between the computational and experimental results from mutagenesis points to the exciting potential application of our approach and paves the road to additional studies of other low pH-triggered endocytic viral entries of numerous different viruses.
Theory and Methods
The M and FI states of the (E2–E1)3 complex and the E1 HT state were built from Protein Data Bank ID codes 3J2W (43, 51), 3MUW (52), and 1RER (53), respectively. The structure of the E2 monomer has not been reported previously; thus, the isolated E2 was adapted from the FI state with some equilibration through molecular dynamics to serve as a reference accounting for the possible metastable postdissociation state. The pH replica exchange MD (molecular dynamics) simulations were carried out to determine the pKa values of Asp, Glu, and His residues (54–57). During the pH replica exchange MD simulations, the alchemical variable-λ is propagated according to the extended Hamiltonian equations of motions (54, 55), characterizing varying protonation from fully deprotonated (λ = 1) to protonated (λ = 0) residues. The pKa of a titrating residue was determined according to the generalized Henderson–Hasselbach equation:
| [1] |
where Su is the fraction of the deprotonated states. Np and Nu were counted for snapshots with λ < 0.1 and λ > 0.9, respectively. The error bars of the pKa values reported in SI Appendix, Fig. S2 were estimated based on the rms residual in the nonlinear fitting of Su to Eq. 1. Based on the pKa values in different conformational states, we can estimate the shifts of the relative thermodynamic stabilities of a given conformational change step under different pH conditions using the Wyman–Tanford linkage equation (60, 76, 77):
| [2] |
where i represents the ith titrating residue. pHref is the reference pH, which is selected to be 7.5 to represent the extracellular conditions (67). The quantity of ΔΔGA→B(pH) represents the relative difference in the thermodynamic free energy cost of a given conformational change step, A→B, under a lower pH with respect to pHref. A residue with pKa upshifts (i.e., ΔpKaA→B > 0 during the A→B step) leads to a component of ΔΔGA→B(pH) < 0, corresponding to a contribution favoring the A→B step under the given acidic pH relative to the neutral conditions at pHref. A full description of simulation details is included in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Bin Zhang, Prof. Eric May, and Dr. Karunesh Arora for discussions. Financial support through NIH Grants GM107233, GM057513, and GM037554 and NSF XSEDE Allocation MCB140005 is greatly appreciated.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1414190112/-/DCSupplemental.
References
- 1.Enserink M. Infectious diseases. Chikungunya: No longer a third world disease. Science. 2007;318(5858):1860–1861. doi: 10.1126/science.318.5858.1860. [DOI] [PubMed] [Google Scholar]
- 2.Sourisseau M, et al. Characterization of reemerging chikungunya virus. PLoS Pathog. 2007;3(6):e89. doi: 10.1371/journal.ppat.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz O, Albert ML. Biology and pathogenesis of chikungunya virus. Nat Rev Microbiol. 2010;8(7):491–500. doi: 10.1038/nrmicro2368. [DOI] [PubMed] [Google Scholar]
- 4.Kaur P, Chu JJH. Chikungunya virus: An update on antiviral development and challenges. Drug Discov Today. 2013;18(19-20):969–983. doi: 10.1016/j.drudis.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sieczkarski SB, Whittaker GR. Dissecting virus entry via endocytosis. J Gen Virol. 2002;83(Pt 7):1535–1545. doi: 10.1099/0022-1317-83-7-1535. [DOI] [PubMed] [Google Scholar]
- 6.Sieczkarski SB, Whittaker GR. Viral entry. Curr Top Microbiol Immunol. 2005;285:1–23. doi: 10.1007/3-540-26764-6_1. [DOI] [PubMed] [Google Scholar]
- 7.Earp LJ, Delos SE, Park HE, White JM. The many mechanisms of viral membrane fusion proteins. Curr Top Microbiol Immunol. 2005;285:25–66. doi: 10.1007/3-540-26764-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruenberg J. Viruses and endosome membrane dynamics. Curr Opin Cell Biol. 2009;21(4):582–588. doi: 10.1016/j.ceb.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15(7):690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marsh M, Helenius A. Virus entry: Open sesame. Cell. 2006;124(4):729–740. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mercer J, Schelhaas M, Helenius A. Virus entry by endocytosis. Annu Rev Biochem. 2010;79(1):803–833. doi: 10.1146/annurev-biochem-060208-104626. [DOI] [PubMed] [Google Scholar]
- 12.Hernandez R, Luo T, Brown DT. Exposure to low pH is not required for penetration of mosquito cells by Sindbis virus. J Virol. 2001;75(4):2010–2013. doi: 10.1128/JVI.75.4.2010-2013.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kielian M, Chanel-Vos C, Liao M. Alphavirus entry and membrane fusion. Viruses. 2010;2(4):796–825. doi: 10.3390/v2040796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kononchik JP, Jr, Hernandez R, Brown DT. An alternative pathway for alphavirus entry. Virol J. 2011;8(1):304. doi: 10.1186/1743-422X-8-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vancini R, Wang G, Ferreira D, Hernandez R, Brown DT. Alphavirus genome delivery occurs directly at the plasma membrane in a time- and temperature-dependent process. J Virol. 2013;87(8):4352–4359. doi: 10.1128/JVI.03412-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim KH, Strauss EG, Strauss JH. Adaptive mutations in Sindbis virus E2 and Ross River virus E1 that allow efficient budding of chimeric viruses. J Virol. 2000;74(6):2663–2670. doi: 10.1128/jvi.74.6.2663-2670.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, et al. Dengue structure differs at the temperatures of its human and mosquito hosts. Proc Natl Acad Sci USA. 2013;110(17):6795–6799. doi: 10.1073/pnas.1304300110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fields W, Kielian M. A key interaction between the alphavirus envelope proteins responsible for initial dimer dissociation during fusion. J Virol. 2013;87(7):3774–3781. doi: 10.1128/JVI.03310-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Fugère M, Day R, Kielian M. Furin processing and proteolytic activation of Semliki Forest virus. J Virol. 2003;77(5):2981–2989. doi: 10.1128/JVI.77.5.2981-2989.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sjöberg M, Lindqvist B, Garoff H. Activation of the alphavirus spike protein is suppressed by bound E3. J Virol. 2011;85(11):5644–5650. doi: 10.1128/JVI.00130-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snyder AJ, Mukhopadhyay S. The alphavirus E3 glycoprotein functions in a clade-specific manner. J Virol. 2012;86(24):13609–13620. doi: 10.1128/JVI.01805-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jose J, Snyder JE, Kuhn RJ. A structural and functional perspective of alphavirus replication and assembly. Future Microbiol. 2009;4(7):837–856. doi: 10.2217/fmb.09.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uchime O, Fields W, Kielian M. The role of E3 in pH protection during alphavirus assembly and exit. J Virol. 2013;87(18):10255–10262. doi: 10.1128/JVI.01507-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaney M-C, Duquerroy S, Rey FA. Alphavirus structure: Activation for entry at the target cell surface. Curr Opin Virol. 2013;3(2):151–158. doi: 10.1016/j.coviro.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Cheng RH, et al. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80(4):621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith TJ, et al. Putative receptor binding sites on alphaviruses as visualized by cryoelectron microscopy. Proc Natl Acad Sci USA. 1995;92(23):10648–10652. doi: 10.1073/pnas.92.23.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferlenghi I, et al. The first step: Activation of the Semliki Forest virus spike protein precursor causes a localized conformational change in the trimeric spike. J Mol Biol. 1998;283(1):71–81. doi: 10.1006/jmbi.1998.2066. [DOI] [PubMed] [Google Scholar]
- 28.Paredes AM, et al. Structural localization of the E3 glycoprotein in attenuated Sindbis virus mutants. J Virol. 1998;72(2):1534–1541. doi: 10.1128/jvi.72.2.1534-1541.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forsell K, Xing L, Kozlovska T, Cheng RH, Garoff H. Membrane proteins organize a symmetrical virus. EMBO J. 2000;19(19):5081–5091. doi: 10.1093/emboj/19.19.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mancini EJ, Clarke M, Gowen BE, Rutten T, Fuller SD. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol Cell. 2000;5(2):255–266. doi: 10.1016/s1097-2765(00)80421-9. [DOI] [PubMed] [Google Scholar]
- 31.Paredes A, Alwell-Warda K, Weaver SC, Chiu W, Watowich SJ. Venezuelan equine encephalomyelitis virus structure and its divergence from old world alphaviruses. J Virol. 2001;75(19):9532–9537. doi: 10.1128/JVI.75.19.9532-9537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pletnev SV, et al. Locations of carbohydrate sites on alphavirus glycoproteins show that E1 forms an icosahedral scaffold. Cell. 2001;105(1):127–136. doi: 10.1016/s0092-8674(01)00302-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haag L, et al. Acid-induced movements in the glycoprotein shell of an alphavirus turn the spikes into membrane fusion mode. EMBO J. 2002;21(17):4402–4410. doi: 10.1093/emboj/cdf442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W, et al. Aura virus structure suggests that the T=4 organization is a fundamental property of viral structural proteins. J Virol. 2002;76(14):7239–7246. doi: 10.1128/JVI.76.14.7239-7246.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang W, et al. Placement of the structural proteins in Sindbis virus. J Virol. 2002;76(22):11645–11658. doi: 10.1128/JVI.76.22.11645-11658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang W, Heil M, Kuhn RJ, Baker TS. Heparin binding sites on Ross River virus revealed by electron cryo-microscopy. Virology. 2005;332(2):511–518. doi: 10.1016/j.virol.2004.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukhopadhyay S, et al. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure. 2006;14(1):63–73. doi: 10.1016/j.str.2005.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu S-R, et al. The dynamic envelope of a fusion class II virus. Prefusion stages of semliki forest virus revealed by electron cryomicroscopy. J Biol Chem. 2007;282(9):6752–6762. doi: 10.1074/jbc.M609125200. [DOI] [PubMed] [Google Scholar]
- 39.Kostyuchenko VA, et al. The structure of barmah forest virus as revealed by cryo-electron microscopy at a 6-angstrom resolution has detailed transmembrane protein architecture and interactions. J Virol. 2011;85(18):9327–9333. doi: 10.1128/JVI.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang J, et al. Molecular links between the E2 envelope glycoprotein and nucleocapsid core in Sindbis virus. J Mol Biol. 2011;414(3):442–459. doi: 10.1016/j.jmb.2011.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang R, et al. 4.4 Å cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. EMBO J. 2011;30(18):3854–3863. doi: 10.1038/emboj.2011.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao S, Zhang W. Characterization of an early-stage fusion intermediate of Sindbis virus using cryoelectron microscopy. Proc Natl Acad Sci USA. 2013;110(33):13362–13367. doi: 10.1073/pnas.1301911110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun S, et al. Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization. eLife. 2013;2:e00435. doi: 10.7554/eLife.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wahlberg JM, Garoff H. Membrane fusion process of Semliki Forest virus. I: Low pH-induced rearrangement in spike protein quaternary structure precedes virus penetration into cells. J Cell Biol. 1992;116(2):339–348. doi: 10.1083/jcb.116.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Justman J, Klimjack MR, Kielian M. Role of spike protein conformational changes in fusion of Semliki Forest virus. J Virol. 1993;67(12):7597–7607. doi: 10.1128/jvi.67.12.7597-7607.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hammar L, et al. Prefusion rearrangements resulting in fusion Peptide exposure in Semliki forest virus. J Biol Chem. 2003;278(9):7189–7198. doi: 10.1074/jbc.M206015200. [DOI] [PubMed] [Google Scholar]
- 47.Gibbons DL, et al. Multistep regulation of membrane insertion of the fusion peptide of Semliki Forest virus. J Virol. 2004;78(7):3312–3318. doi: 10.1128/JVI.78.7.3312-3318.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wahlberg JM, Bron R, Wilschut J, Garoff H. Membrane fusion of Semliki Forest virus involves homotrimers of the fusion protein. J Virol. 1992;66(12):7309–7318. doi: 10.1128/jvi.66.12.7309-7318.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roussel A, et al. Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus. Structure. 2006;14(1):75–86. doi: 10.1016/j.str.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 50.Lescar J, et al. The Fusion glycoprotein shell of Semliki Forest virus: An icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105(1):137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- 51.Voss JE, et al. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature. 2010;468(7324):709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- 52.Li L, Jose J, Xiang Y, Kuhn RJ, Rossmann MG. Structural changes of envelope proteins during alphavirus fusion. Nature. 2010;468(7324):705–708. doi: 10.1038/nature09546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gibbons DL, et al. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature. 2004;427(6972):320–325. doi: 10.1038/nature02239. [DOI] [PubMed] [Google Scholar]
- 54.Lee MS, Salsbury FR, Jr, Brooks CL., III Constant-pH molecular dynamics using continuous titration coordinates. Proteins. 2004;56(4):738–752. doi: 10.1002/prot.20128. [DOI] [PubMed] [Google Scholar]
- 55.Khandogin J, Brooks CL., III Constant pH molecular dynamics with proton tautomerism. Biophys J. 2005;89(1):141–157. doi: 10.1529/biophysj.105.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wallace JA, Shen JK. Continuous constant pH molecular dynamics in explicit solvent with pH-based replica exchange. J Chem Theory Comput. 2011;7(8):2617–2629. doi: 10.1021/ct200146j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sabri Dashti D, Meng Y, Roitberg AE. pH-replica exchange molecular dynamics in proteins using a discrete protonation method. J Phys Chem B. 2012;116(30):8805–8811. doi: 10.1021/jp303385x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.May ER, Arora K, Brooks CL., III pH-induced stability switching of the bacteriophage HK97 maturation pathway. J Am Chem Soc. 2014;136(8):3097–3107. doi: 10.1021/ja410860n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khandogin J, Brooks CL., III Molecular simulations of pH-mediated biological processes. Annu Rep Comput Chem. 2007;3:3–13. [Google Scholar]
- 60.Shen JK. A method to determine residue-specific unfolded-state pKa values from analysis of stability changes in single mutant cycles. J Am Chem Soc. 2010;132(21):7258–7259. doi: 10.1021/ja101761m. [DOI] [PubMed] [Google Scholar]
- 61.Arthur EJ, Yesselman JD, Brooks CL., III Predicting extreme pKa shifts in staphylococcal nuclease mutants with constant pH molecular dynamics. Proteins. 2011;79(12):3276–3286. doi: 10.1002/prot.23195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang BW, Brunetti L, Brooks CL., III Probing pH-dependent dissociation of HdeA dimers. J Am Chem Soc. 2011;133(48):19393–19398. doi: 10.1021/ja2060066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Law SM, Zhang BW, Brooks CL., 3rd pH-sensitive residues in the p19 RNA silencing suppressor protein from carnation Italian ringspot virus affect siRNA binding stability. Protein Sci. 2013;22(5):595–604. doi: 10.1002/pro.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goh GB, Hulbert BS, Zhou H, Brooks CL., III Constant pH molecular dynamics of proteins in explicit solvent with proton tautomerism. Proteins. 2014;82(7):1319–1331. doi: 10.1002/prot.24499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Çağin T, Holder M, Pettitt BM. A method for modeling icosahedral virions: Rotational symmetry boundary conditions. J Comput Chem. 1991;12(5):627–634. [Google Scholar]
- 67.Roos A, Boron WF. Intracellular pH. Physiol Rev. 1981;61(2):296–434. doi: 10.1152/physrev.1981.61.2.296. [DOI] [PubMed] [Google Scholar]
- 68.Geisow MJ, Evans WH. pH in the endosome. Measurements during pinocytosis and receptor-mediated endocytosis. Exp Cell Res. 1984;150(1):36–46. doi: 10.1016/0014-4827(84)90699-2. [DOI] [PubMed] [Google Scholar]
- 69.Qin Z-L, Zheng Y, Kielian M. Role of conserved histidine residues in the low-pH dependence of the Semliki Forest virus fusion protein. J Virol. 2009;83(9):4670–4677. doi: 10.1128/JVI.02646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chanel-Vos C, Kielian M. A conserved histidine in the ij loop of the Semliki Forest virus E1 protein plays an important role in membrane fusion. J Virol. 2004;78(24):13543–13552. doi: 10.1128/JVI.78.24.13543-13552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roman-Sosa G, Kielian M. The interaction of alphavirus E1 protein with exogenous domain III defines stages in virus-membrane fusion. J Virol. 2011;85(23):12271–12279. doi: 10.1128/JVI.05902-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sánchez-San Martín C, Nanda S, Zheng Y, Fields W, Kielian M. Cross-inhibition of chikungunya virus fusion and infection by alphavirus E1 domain III proteins. J Virol. 2013;87(13):7680–7687. doi: 10.1128/JVI.00814-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liao M, Kielian M. Functions of the stem region of the Semliki Forest virus fusion protein during virus fusion and assembly. J Virol. 2006;80(22):11362–11369. doi: 10.1128/JVI.01679-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kielian M, Klimjack MR, Ghosh S, Duffus WA. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J Cell Biol. 1996;134(4):863–872. doi: 10.1083/jcb.134.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu CY, Kielian M. E1 mutants identify a critical region in the trimer interface of the Semliki forest virus fusion protein. J Virol. 2009;83(21):11298–11306. doi: 10.1128/JVI.01147-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wyman J., Jr Linked functions and reciprocal effects in hemoglobin: A second look. Adv Protein Chem. 1964;19:223–286. doi: 10.1016/s0065-3233(08)60190-4. [DOI] [PubMed] [Google Scholar]
- 77.Tanford C. Protein denaturation. C. Theoretical models for the mechanism of denaturation. Adv Protein Chem. 1970;24:1–95. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.