Significance
The germline, which produces sperm or oocyte, is separated from other cells that generate the rest of the body, the soma, during early development in most animals. Somatic cells experience and respond to the environment in each generation, and it is unknown whether they can transmit information to the germline for inheritance into subsequent generations. We found that neurons of the worm Caenorhabditis elegans can transmit double-stranded RNA to the germline to initiate transgenerational silencing of a gene of matching sequence. To our knowledge, these results demonstrate for the first time that a somatic tissue of an animal can have transgenerational effects on a gene through the transport of double-stranded RNA to the germline.
Keywords: epigenetics, mobile RNAs, soma to germline, Weismann barrier
Abstract
An animal that can transfer gene-regulatory information from somatic cells to germ cells may be able to communicate changes in the soma from one generation to the next. In the worm Caenorhabditis elegans, expression of double-stranded RNA (dsRNA) in neurons can result in the export of dsRNA-derived mobile RNAs to other distant cells. Here, we show that neuronal mobile RNAs can cause transgenerational silencing of a gene of matching sequence in germ cells. Consistent with neuronal mobile RNAs being forms of dsRNA, silencing of target genes that are expressed either in somatic cells or in the germline requires the dsRNA-selective importer SID-1. In contrast to silencing in somatic cells, which requires dsRNA expression in each generation, silencing in the germline is heritable after a single generation of exposure to neuronal mobile RNAs. Although initiation of inherited silencing within the germline requires SID-1, a primary Argonaute RDE-1, a secondary Argonaute HRDE-1, and an RNase D homolog MUT-7, maintenance of inherited silencing is independent of SID-1 and RDE-1, but requires HRDE-1 and MUT-7. Inherited silencing can persist for >25 generations in the absence of the ancestral source of neuronal dsRNA. Therefore, our results suggest that sequence-specific regulatory information in the form of dsRNA can be transferred from neurons to the germline to cause transgenerational silencing.
The germline is separated from the rest of the body, or soma, during early development in most animals, consistent with the suggestion that environmental effects on soma throughout the lifetime of an animal cannot influence inheritance through the germline (1). However, some environmental changes can cause effects that last for three or more generations, even in the apparent absence of changes in the genotype (reviewed in ref. 2). These transgenerational epigenetic effects are presumably initiated either by direct changes within the ancestral germline or by the transfer of information from ancestral somatic cells to the ancestral germline. It is difficult to distinguish between these possibilities because complex ancestral changes that affect subsequent generations, such as diet (3–5) or endocrine disruption (6), perturb many genes in many tissues in ways that are as yet unclear. Manipulating the activity of a single gene in specific tissues and across generations can help distinguish between these possibilities. Such specific inactivation of a single gene can be achieved by using double-stranded RNA (dsRNA) to trigger RNA interference (RNAi) in the worm Caenorhabditis elegans (7).
As in most animals, the C. elegans germline is set aside early in development—after four cell divisions (8). Gene silencing initiated through RNAi-related mechanisms within the C. elegans germline can last for many generations (9–13). Such transgenerational silencing can be triggered by both injected dsRNA (14–16) and ingested dsRNA (16–19). However, both injection and ingestion can deliver dsRNA directly into the fluid-filled body cavity that surrounds the germline, without entry into the cytosol of any somatic cell (20, 21). Thus, it remains unknown whether somatic cells in C. elegans can export signals for delivery into the germline to cause transgenerational gene silencing.
The transfer of gene-specific information from one somatic tissue to another somatic tissue during RNAi has been observed in C. elegans (22). Such intertissue transfer of gene-regulatory information appears to occur through the transport of forms of dsRNA called mobile RNAs (23). Entry of these mobile RNAs into the cytosol requires the dsRNA-selective importer SID-1 (22, 24, 25). Consequently, when dsRNA is expressed in a variety of somatic tissues such as the gut, muscles, or neurons, SID-1–dependent silencing of genes of matching sequence is observed in other somatic tissues (20). Because gene silencing by mobile RNAs from neurons appears to be stronger than that by mobile RNAs from other somatic tissues (20), we examined whether neurons export mobile RNAs that can enter the germline to cause transgenerational gene silencing.
Here, we show that neuronal mobile RNAs can enter both somatic and germ cells to trigger gene silencing. Although silencing in somatic tissues is not detectably inherited despite multigenerational exposure to neuronal mobile RNAs, silencing in the germline is inherited for many generations after a single generation of exposure to neuronal mobile RNAs.
Results
Neuronal Mobile RNAs Can Enter Most Somatic Tissues and the Germline.
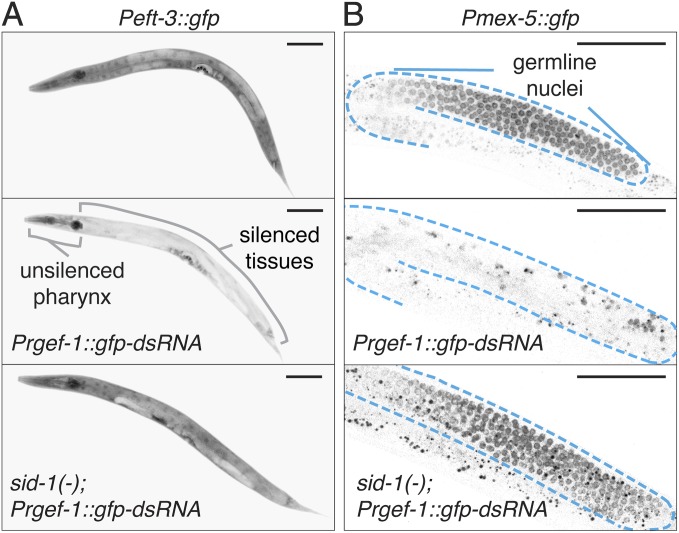
Genetic analyses suggest that neuronal mobile RNAs are forms of dsRNA (23). Mobile RNAs generated from dsRNA expressed in neurons against the muscle gene unc-22 can enter muscle cells through the dsRNA importer SID-1 and cause unc-22 silencing (23). To examine silencing of a gene expressed in multiple tissues by a single source of dsRNA in neurons, we used animals that expressed cytosolic gfp (Peft-3::gfp) in all somatic tissues and gfp–dsRNA in all neurons (Prgef-1::gfp–dsRNA) (Fig. 1). GFP expression was detectably reduced in most somatic tissues (with the notable exception of the pharynx) in the presence of Prgef-1::gfp–dsRNA (Fig. 1A, Top vs. Fig. 1A, Middle), and this silencing was enhanced in the absence of the exonuclease ERI-1 (Fig. S1), consistent with ERI-1 acting to inhibit silencing by imported neuronal mobile RNAs (23). Silencing in all somatic tissues, even in the eri-1(−) background, was lost upon removal of the mobile RNA importer SID-1 (Fig. 1A, Bottom, and Fig. S1), suggesting that all observed silencing was due to mobile RNAs made in neurons.
Fig. 1.
Neuronal mobile RNAs can cause gene silencing in most somatic tissues and in the germline. (A) Representative fourth larval (L4)-staged animals that express GFP (black) in somatic tissues (Peft-3::gfp) in a wild-type (Top) background and animals that in addition express dsRNA in neurons against gfp (Prgef-1::gfp–dsRNA) in wild-type (Middle) or sid-1(−) (Bottom) backgrounds are shown. Silenced tissues and unsilenced pharynx are indicated (Middle). Detectable silencing was observed in 100% of wild-type animals (n = 135) and 0% of sid-1(−) animals (n = 115). (Scale bars, 50 µm.) Also see Fig. S1. (B) Representative L4-staged animals that express GFP (black) in the germline (Pmex-5::gfp; outlined in cyan) in a wild-type (Top) background and animals that in addition express Prgef-1::gfp–dsRNA in wild-type (Middle) or sid-1(−) (Bottom) backgrounds are shown. Because of the long exposure time required to acquire these images, variable and irregular autofluorescence due to gut granules was also detected. Detectable silencing was observed in 87% of wild-type animals (n = 54) and 27% of sid-1(−) animals (n = 59). (Scale bars, 50 µm.)
To test whether the germline is susceptible to silencing by mobile RNAs, we examined silencing of GFP expression in animals that express gfp in the germline (Pmex-5::gfp) and neuronal mobile RNAs from a Prgef-1::gfp–dsRNA transgene. Like most somatic cells, the germline was susceptible to silencing by neuronal mobile RNAs, and the silencing was predominantly dependent on SID-1 (Fig. 1B). The silencing was sequence-specific and did not occur in animals with transgenic expression of a co-injection marker (Fig. S2A) or in animals with transgenic expression of unc-22 dsRNA in neurons (Fig. S2B). Furthermore, silencing, as detected by the loss of GFP fluorescence within the germline (Fig. S3A), was correlated with a reduction in gfp mRNA levels (Fig. S3B). Consistent with mobile RNAs that are imported into the germline being forms of gfp–dsRNA, silencing was strongly dependent on the dsRNA importer SID-1 and the primary Argonaute RDE-1 that acts on short dsRNA (26), but independent of the RNA-dependent RNA polymerase RRF-1 that generates single-stranded secondary small RNAs in somatic cells (27) (Fig. S3C). The residual silencing observed in sid-1(−) and rde-1(−) animals may reflect additional sid-1– and rde-1–independent gene silencing mechanisms that can act in the germline (9–12). Because silencing of a germline target due to dsRNA expression in neurons is greatly reduced in the absence of SID-1 (Fig. 1B, Bottom, and Fig. S3C), we conclude that SID-1–dependent neuronal mobile RNAs can enter the germline.
Together, our results suggest that neuronal mobile RNAs can enter most somatic tissues as well as the germline to silence genes of matching sequence. Because injection of in vitro-synthesized dsRNA can generate signals that are inherited in C. elegans (7, 14–16), our observations raise the possibility that neuronal mobile RNAs may also generate such inherited signals upon silencing a gene within the germline or upon silencing a gene in other somatic cells.
Silencing in the Germline by Neuronal Mobile RNAs Is Inherited for Many Generations.
Injected or ingested dsRNA can cause transgenerational gene silencing of germline genes in C. elegans (10, 14–18). However, both forms of dsRNA delivery could result in the direct entry of dsRNA into the germline without entry into the cytoplasm of somatic cells. Ingested dsRNA is transcytosed across the gut into the body cavity that surrounds the germline (20, 21), and it is difficult to avoid spillage of injected dsRNA into the body cavity. These experimental considerations suggest that to test the possibility of somatic tissues initiating transgenerational gene silencing, it is necessary to express silencing triggers within somatic tissues and examine gene silencing within the germline. Although induction using heat shock of a transgene that encodes a viral genome in somatic tissues caused transgenerational silencing in C. elegans (13), such heat-shock induction also leads to expression within the germline (figure S5 in ref. 28). Therefore, because of the inherent difficulty in ensuring lack of expression within the germline from transgenes, only germline silencing that is reduced in the absence of the dsRNA importer SID-1 (Fig. 1B, Bottom, and Fig. S3C) can be interpreted as being caused by mobile RNAs.
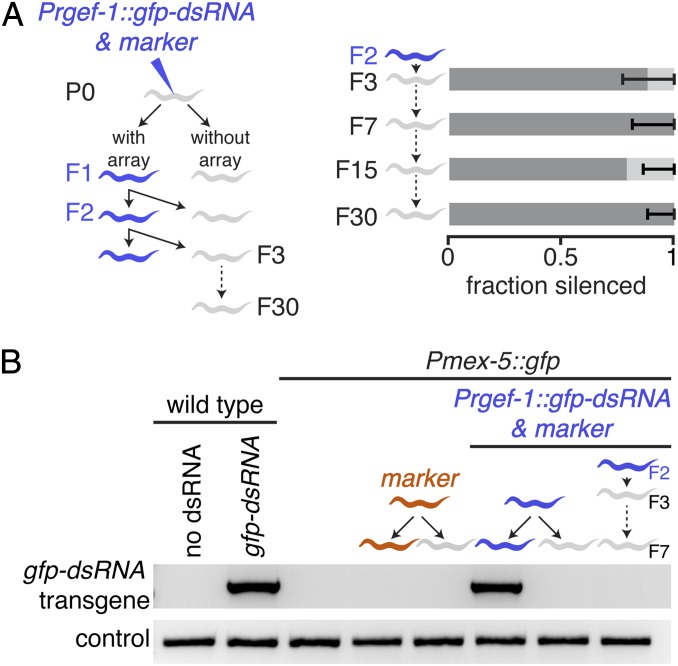
To determine whether neuronal mobile RNAs that are imported into the germline can cause transgenerational silencing, we examined animals that lack the DNA for gfp–dsRNA but whose ancestors expressed neuronal dsRNAs. Because stable transgenic lines of extrachromosomal arrays are generated in C. elegans two generations after an animal [parental generation (P0)] is transformed with DNA (i.e., in the F2 generation) (29), we examined the silencing of GFP expression in wild-type animals of the F3 generation that lacked the gfp–dsRNA transgene and in their descendents (Fig. 2A, Left). Animals that lack the gfp–dsRNA transgene can be identified by the loss of a red fluorescent co-injection marker, the DNA for which is expected to be incorporated along with the DNA for gfp–dsRNA into a single extrachromosomal array upon cotransformation. All F3 animals without the extrachromosomal array showed silencing of GFP expression in the germline (Fig. 2A, Right). Inherited silencing due to the ancestral production of neuronal mobile RNAs persisted for >25 subsequent generations, despite unbiased passaging of worms from one generation to the next (Fig. 2A, Right, and Fig. S4). Consistent with the loss of the gfp–dsRNA transgene in animals that lack fluorescence from the co-injection marker, we failed to detect the gfp–dsRNA transgene in the DNA of worms that lacked the co-injection marker after 35 cycles of PCR amplification (Fig. 2B). These results suggest that neuronal mobile RNAs imported into the germline can initiate gene silencing that lasts for many generations in the absence of the ancestral source of neuronal dsRNA.
Fig. 2.
Neuronal mobile RNAs can cause transgenerational silencing of a germline gene. (A) Inherited silencing in the germline lasts for >25 generations after the source of neuronal mobile RNAs is lost. (A, Left) Pmex-5::gfp animals (P0) were injected with constructs to express neuronal mobile RNAs (Prgef-1::gfp–dsRNA) along with a co-injection marker (Pmyo-2::DsRed) to generate F2 transgenic lines (blue worm). (A, Right) The proportions of animals that all lack fluorescence from the co-injection marker (gray worm) but that show either strong (dark gray bars) or weak (light gray bars) silencing in the F3 generation and in successive generations (F4–F30) were determined. Error bars indicate 95% CI and n > 14 L4-staged animals for each generation. Also see Fig. S4. Dark gray bars and light gray bars are as in Fig. S3C. (B) Animals that lack the co-injection marker also lack the gfp–dsRNA transgene. Genomic DNA from wild-type animals (no dsRNA), from wild-type animals that express Prgef-1::gfp–dsRNA, from Pmex-5::gfp animals, and from Pmex-5::gfp animals that either have or whose ancestors had extrachromosomal transgenes [i.e., Pmex-5::gfp animals that in addition express the co-injection marker alone (marker; orange worm) or along with Prgef-1::gfp–dsRNA (Prgef-1::gfp–dsRNA & marker; blue worm) or apparently lack these extrachromosomal transgenes (gray worm) but that were derived from ancestors that expressed these transgenes] were analyzed. Although the control gene was detected in all cases, a PCR product for the gfp–dsRNA transgene was detected only in wild-type animals with gfp–dsRNA and in Pmex5::gfp animals with gfp–dsRNA as evidenced by fluorescence from the co-injection marker.
Transgenerational Silencing by Neuronal Mobile RNAs Has Distinct Genetic Requirements for Initiation and Maintenance.
Although transgenerational silencing is reliably observed by using multiple transgenic sources of neuronal mobile RNAs (Fig. S4), the number of generations that show silencing varied from one transgenic line to another, possibly due to differences in the levels of expression of dsRNA in different transgenic lines. To facilitate comparison of transgenerational silencing across multiple genetic backgrounds and to expose animals to mobile RNAs in defined generations, we chose a single extrachromosomal transgenic line that expresses neuronal mobile RNAs against gfp in wild-type animals and crossed it into animals that express gfp in the germline. This experimental scheme was then used to determine the genetic requirements for the initiation and maintenance of transgenerational gene silencing.
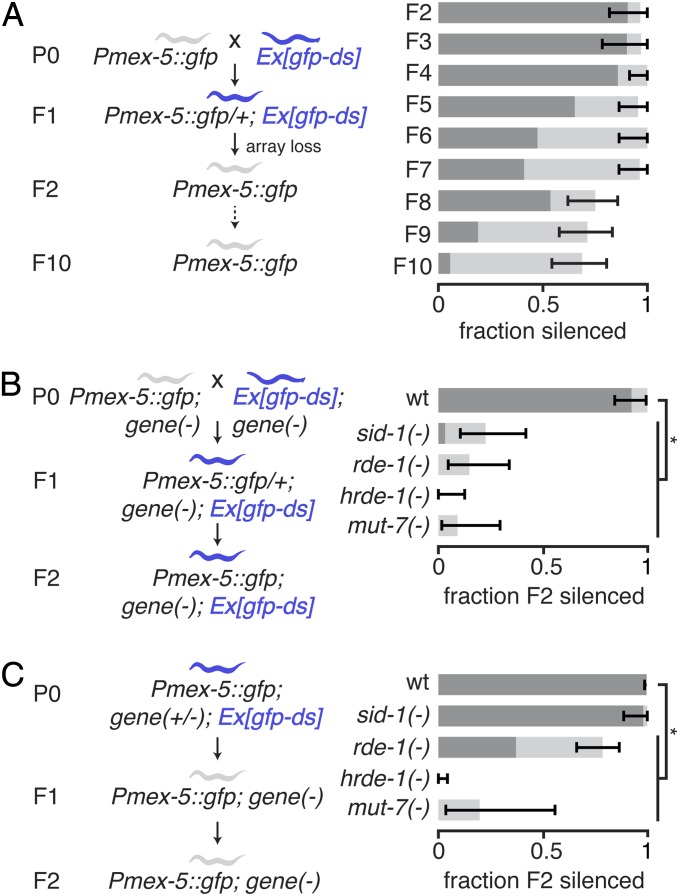
Using this experimental scheme, we found that exposure of a germline target gene to neuronal mobile RNAs for a single generation was sufficient to cause transgenerational silencing (Fig. 3A). Specifically, when animals with Pmex-5::gfp and animals with Prgef-1::gfp–dsRNA were mated, the F1 cross progeny that inherited the Prgef-1::gfp–dsRNA transgene could initiate transgenerational silencing. This silencing persisted for many generations, despite the loss of the source of neuronal mobile RNAs in the F2 generation (Fig. 3A).
Fig. 3.
Neuronal mobile RNAs have distinct requirements for the initiation and maintenance of transgenerational silencing. (A) Expression of neuronal mobile RNAs for one generation is sufficient to initiate multigenerational silencing. Pmex-5::gfp animals were crossed with animals that express neuronal dsRNA from an extrachromosomal array (Ex[gfp–ds]) and the proportions of animals that lack the extrachromosomal array (gray worm) but that show either strong (dark gray bars) or weak (light gray bars) silencing in the F2 generation and in successive generations (F3–F10) were determined. The loss of Ex[gfp–ds] was determined by the loss of the fluorescent co-injection marker. (B) Initiation of silencing by neuronal mobile RNAs requires sid-1, rde-1, hrde-1, and mut-7. Wild-type (wt), sid-1(−), rde-1(−), hrde-1(−), or mut-7(−) animals that all express Pmex-5::gfp were mated with animals of identical genetic backgrounds that all express neuronal dsRNA (Ex[gfp–ds]), and the silencing in descendants that had both Pmex-5::gfp and Ex[gfp–ds] was measured as in A. (C) Maintenance of germline gene silencing by neuronal mobile RNAs requires HRDE-1 and MUT-7, but not SID-1 or RDE-1. Wild-type (wt), sid-1(+/−), rde-1(+/−), hrde-1(+/−), or mut-7(+/−) animals that all had both Pmex-5::gfp and Ex[gfp–ds] were allowed to have progeny, and the silencing in wild-type (wt), sid-1(−), rde-1(−), hrde-1(−), or mut-7(−) grand progeny animals that all had Pmex-5::gfp but that all lacked Ex[gfp–ds] was measured as in A. The analyzed grand progeny were progeny of animals that also lacked Ex[gfp–ds]. Error bars indicate 95% CI. *P < 0.05. n > 19 L4-staged animals, except for mut-7(−) animals in C, where n = 10 L4-staged animals. Dark gray bars and light gray bars are as in Fig. S3C.
To test whether a gene is required for germline silencing by neuronal mobile RNAs, we used the same experimental scheme as above, but with animals that also had a mutation in the gene being tested (Fig. 3B). For example, to test the requirement for sid-1, we mated sid-1 null mutants [sid-1(−)] that express Pmex-5::gfp with sid-1(−) animals that express Prgef-1::gfp–dsRNA and examined silencing in sid-1(−) animals of a later generation that express both Pmex-5::gfp and Prgef-1::gfp–dsRNA. Germline silencing using this experimental scheme also required SID-1 and RDE-1, in agreement with the results obtained for silencing by transgenic lines that were independently generated in mutant backgrounds (Fig. S3C). Thus, germline silencing due to neuronal mobile RNAs likely relies on the import of forms of dsRNA through SID-1 and subsequent processing by the primary Argonaute RDE-1 within the germline. Further processing within the germline leads to the production of secondary single-stranded small RNAs. These secondary small RNAs eventually cause gene silencing through mechanisms that require many proteins (reviewed in ref. 30), including the secondary nuclear Argonaute HRDE-1 (18) and the RNase D homolog MUT-7 (31). We found that both HRDE-1 and MUT-7 were required for silencing by neuronal mobile RNAs, suggesting that silencing within the germline is executed by secondary small RNAs and downstream genes. For all genes tested above, the source of neuronal mobile RNAs was present in the animals that were tested. Therefore, the lack of silencing in sid-1(−), rde-1(−), hrde-1(−), and mut-7(−) animals reflects a requirement for the corresponding genes in the initiation of germline silencing by neuronal mobile RNAs.
The observed genetic requirements for silencing by neuronal mobile RNAs are distinct from those observed for silencing by ingested or injected dsRNA. Whereas the requirement for SID-1 and RDE-1 is in agreement with the requirement for these genes when silencing is triggered using ingested and injected dsRNA, the requirement for HRDE-1 and MUT-7 is in contrast to the HRDE-1–independent silencing observed in response to ingested dsRNA (ref. 18 and Fig. S5) and the MUT-7–independent silencing observed in response to injected dsRNA (14). These differences might reflect differences in the dosage of dsRNA delivered into the germline using the different methods or the differential engagement of silencing machinery by the different sources of dsRNA used to trigger gene silencing.
To test whether a gene is required for the maintenance of transgenerational silencing by neuronal mobile RNAs, we examined silencing in animals that had mutations in the gene but were descendants of ancestors that had a wild-type copy of the gene during exposure to neuronal mobile RNAs (Fig. 3C). For example, to test the requirement for sid-1 in the maintenance of transgenerational silencing, we examined silencing in sid-1 null mutants [sid-1(−)] that were grand progeny of sid-1(+/−) heterozygous animals that were exposed to Prgef-1::gfp–dsRNA. Grand progeny were examined for silencing instead of progeny because maternal deposition of mRNA or protein from heterozygous parents can complicate interpretation of results in sid-1(−) progeny. We observed silencing in the sid-1(−) grand progeny of sid-1(+/−) heterozygous animals that were exposed to Prgef-1::gfp–dsRNA, which suggests that SID-1 is not required for the maintenance of transgenerational silencing. Similar experiments with null mutants of rde-1, hrde-1, and mut-7 revealed that RDE-1, like SID-1, is dispensable for the maintenance of transgenerational silencing, but HRDE-1 and MUT-7 are required for the maintenance of transgenerational silencing.
In summary, our results suggest a model where mobile RNAs exported from neurons enter the germline through SID-1 to cause RDE-1–, MUT-7–, and HRDE-1–dependent silencing in the parent, which is subsequently maintained through a MUT-7– and HRDE-1–dependent, but SID-1– and RDE-1–independent mechanism. Because HRDE-1 has been shown to use secondary small RNAs to guide trimethylation of the histone H3 on lysine 9 (H3K9me3) at genes of matching sequence (17, 18), our results suggest that the initiation and maintenance of transgenerational silencing by neuronal mobile RNAs is associated with the deposition of H3K9me3 marks on genes of matching sequence. Although the response to ingested or injected dsRNA strongly suggests that secondary small RNAs are inherited (14, 17, 19), it is possible that in response to neuronal mobile RNAs chromatin marks are inherited across generations. Furthermore, although silencing of somatic genes has been reported to be inherited for a few generations when the silencing is triggered by using ingested dsRNA (17, 19), it is unclear whether silencing of a somatic gene by neuronal mobile RNAs is inherited and whether transgenerational silencing by neuronal mobile RNAs within the germline can spread to somatic cells.
Silencing in Somatic Cells by Neuronal Mobile RNAs Is Not Detectably Inherited.
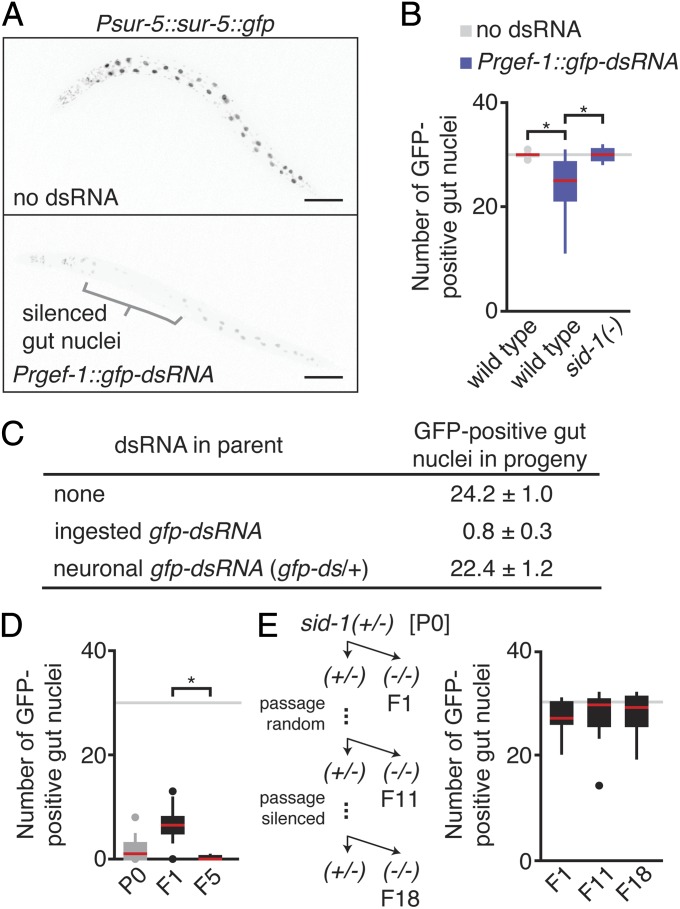
To measure silencing by neuronal mobile RNAs in somatic cells, we used animals that have two different integrated transgenes—one that expresses nuclear-localized GFP (sur-5::gfp) under the control of a promoter that drives expression in all somatic cells (Psur-5) and one that expresses gfp–dsRNA under the control of a promoter that drives expression in all neurons (Prgef-1). Silencing due to neuronal mobile RNAs made from the Prgef-1::gfp–dsRNA transgene results in a reduction in fluorescence of nuclear-localized GFP made from the Psur-5::sur-5::gfp transgene (Fig. 4A). This silencing can be most easily observed in the large intestinal cell nuclei, and counting the number of GFP-positive gut nuclei provides a reliable measure of silencing that correlates with reduction in gfp mRNA levels (Fig. S6 A and B). Wild-type animals with neuronal mobile RNAs had, on average, fewer GFP-positive gut nuclei than did animals without neuronal mobile RNAs (Fig. 4B; 24.2 vs. 29.9 GFP-positive gut nuclei; P < 0.05). Consistent with silencing by neuronal mobile RNAs, this silencing was abolished in sid-1(−) animals (Fig. 4B) and not observed in wild-type animals that were merely cocultured with animals that express neuronal mobile RNAs (Fig. S6C).
Fig. 4.
Silencing of a somatic gene by neuronal mobile RNAs in parents is not detectably inherited by progeny. (A) Neuronal mobile RNAs can silence GFP expression in gut cells. Representative L4-staged animals that express GFP (black) in all somatic cells (Psur-5::sur-5::gfp) (Upper) or that in addition express dsRNA in all neurons (Prgef-1::gfp–dsRNA) (Lower) are shown. Brackets indicate strongly silenced gut nuclei. (Scale bars, 50 µm.) Also see Fig. S6. (B) Double-stranded RNAs expressed in neurons against gfp require SID-1 to silence GFP expression in gut cells. The numbers of GFP-expressing gut nuclei were counted in wild-type animals that do not express dsRNA against gfp (no dsRNA; gray) and in wild-type, or sid-1(−) animals that express Prgef-1::gfp–dsRNA (blue). Gray line indicates average number of gut nuclei in L4-staged animals, n > 19 L4-staged animals, and red bar in box plots indicates median. *P < 0.05 (Student’s t test). (C–E) An enhanced RNAi background [eri-1(−)] was used to maximize the ability to detect inherited silencing. (C) Unlike silencing by ingested dsRNA, silencing by neuronal mobile RNAs is not detectably inherited by progeny. Numbers of GFP-positive gut nuclei in genetically identical progeny of animals that were not exposed to gfp–dsRNA (none) or that were exposed to ingested gfp–dsRNA or that had one copy of an integrated transgene that expresses Prgef-1::gfp–dsRNA (gfp–ds/+) were counted. Errors indicate SEM. (D) Unbiased passaging of worms for multiple generations can lead to small differences in gene silencing. Worms that express Prgef-1::gfp–dsRNA (P0) were passaged for five generations (F1–F5) by picking a random worm at each generation, and the numbers of GFP-positive gut nuclei in animals of each generation were determined (see Fig. S8 for additional data). Gray line, n, red bar, and asterisks are as in B, except for F5, which had n = 8 L4-staged animals. (E) SID-1 is required for silencing by neuronal mobile RNAs even after 17 generations of ancestral silencing by neuronal mobile RNAs. (E, Left) Schematic of experimental design to test the requirement for SID-1 in each generation for silencing by neuronal mobile RNAs. At each generation, the numbers of GFP-positive gut nuclei in sid-1(−/−) animals were counted, and heterozygous [sid-1(+/−)] siblings of any sid-1(−/−) animal (F1–F11) or heterozygous siblings of the most silenced sid-1(−/−) animal (F12–F18) were passaged. (E, Right) The extent of silencing in F1, F11, and F18 are shown (see Fig. S9 for additional data). Gray line, red bar, and n are as in B.
Because the initiation of inherited silencing occurs more frequently in animals that lack the exonuclease ERI-1 (16), we examined the ability of neuronal mobile RNAs to trigger inherited silencing in an eri-1(−) background, where trace amounts of dsRNA (32) and additional mobile RNAs (20) made from the multicopy Psur-5::sur-5::gfp transgene could also contribute to silencing. Using this sensitive genetic background, we did not detect inherited silencing by neuronal mobile RNAs (Fig. 4C), but did detect inherited silencing by ingested dsRNAs as reported earlier (Fig. 4C and ref. 19). We noticed a correlation between an increase in silencing by neuronal mobile RNAs and an increase in parental or ancestral exposure to mobile RNAs (Fig. S7). However, the increases in silencing were small and comparable to the small variations in silencing observed in successive generations when worms with Prgef-1::gfp–dsRNA of identical genotype were simply passaged (Fig. 4D). Furthermore, selection of the most silenced or most desilenced animal for four generations introduced marginal differences in silencing between the first and fifth generations (Fig. S8). Nevertheless, if marginal increases in inherited silencing accrued over many generations due to the presence of parental neuronal mobile RNAs, such inherited silencing might become independent of neuronal mobile RNAs and thus independent of SID-1 in later generations. However, we did not detect such SID-1–independent silencing, even after exposure to 17 generations of silencing by neuronal mobile RNAs (Fig. 4E and Fig. S9). The requirement for sid-1 in every generation for silencing by neuronal mobile RNAs suggests that transport of neuronal mobile RNAs must occur in every generation to observe silencing in somatic cells.
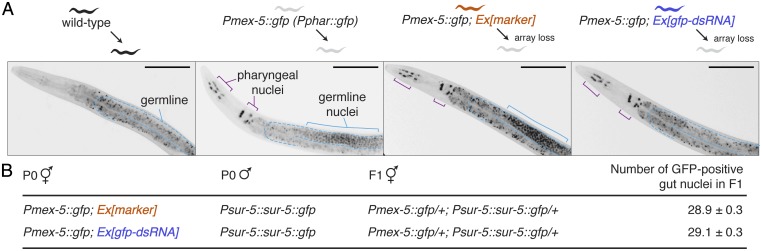
The absence of robust inherited silencing by neuronal mobile RNAs of genes expressed in somatic cells could be either because somatic silencing does not generate signals for transmission to the next generation or because such signals require a template of matching sequence in the germline for stability. To test this latter possibility, we examined inherited somatic silencing by neuronal mobile RNAs in animals that express the target gene (gfp) in somatic cells as well as in the germline either from a single transgene (germline expression due to Pmex-5::gfp and pharyngeal expression due to an additional promoter in the Pmex-5::gfp transgene) (Fig. 5A) or from two separate transgenes (germline expression due to Pmex-5::gfp and gut expression due to sur-5::gfp) (Fig. 5B). In both cases, no inherited silencing was detected in somatic cells.
Fig. 5.
Inherited silencing of a germline gene by neuronal mobile RNAs in the parent does not spread to the soma of the progeny. (A) Silencing of GFP expression within the germline by neuronal mobile RNAs does not cause detectable inherited silencing of GFP expressed from the same locus in the pharynx of progeny. Pmex-5::gfp animals and Pmex-5::gfp animals that in addition express an extrachromosomal source of either the co-injection marker (Ex[marker]) or neuronal mobile RNAs (Ex[gfp–dsRNA]) were passaged and L4-staged progeny that lack the extrachromosomal arrays were imaged under identical conditions. The pharyngeal expression of GFP (black) is from an additional uncharacterized promoter (Pphar::gfp) within the Pmex-5::gfp transgene and is absent in wild-type worms. Germline (outlined in cyan), GFP expression in the germline nuclei (cyan brackets), and GFP expression in pharyngeal nuclei (purple brackets) are indicated. (Scale bars, 50 µm.) (B) Silencing of GFP expression within the germline by neuronal mobile RNAs in the parent does not cause detectable inherited silencing of GFP expressed from a different locus in gut cells of progeny. Pmex-5::gfp animals (P0 hermaphrodite) that in addition expressed Ex[marker] or Ex[gfp–dsRNA] were crossed with Psur-5::sur-5::gfp animals (P0 male), and the numbers of GFP-positive gut nuclei were counted in the resulting F1 progeny that lack extrachromosomal arrays. Errors indicate SEM, and n > 18 L4-staged animals.
Together, our results suggest that neuronal mobile RNAs generate transgenerational silencing signals that have a strong effect on gene expression in the germline and a minimal effect, if any, on gene expression in somatic tissues.
Discussion
We found that neurons can transport forms of dsRNA into the germline to cause silencing that can last for many generations and that such transgenerational silencing is restricted to the germline with distinct genetic requirements for initiation and maintenance.
Mobile RNAs that enter the germline can provide an organism with the ability to transfer gene-specific regulatory information from somatic cells across generations and could be one mechanism by which the environment elicits transgenerational effects in animals. Although restricted to the germline, transgenerational silencing by mobile RNAs could underlie effects of the environment across generations in some cases. For example, expression of some genes within the germline can affect longevity (33), and transgenerational silencing of such genes might underlie the longevity that results from ancestral starvation in C. elegans (5). Thus, additional experiments are needed to determine the role of mobile RNAs, if any, in the transport of such experience-dependent information from somatic cells to subsequent generations in C. elegans.
The presence of a mammalian homolog of the dsRNA importer SID-1 that is also required for the uptake of dsRNAs into cells (34) raises the possibility that dsRNA generated from distant somatic cells—potentially in response to environmental influences—may be imported through SID-1 into the mammalian germline to trigger transgenerational epigenetic changes. Consistent with this possibility, small RNAs have been found in circulation in mammals (35); dsRNAs have been detected in mammalian germ cells (36–38); and injection of RNAs into the early mouse embryo can trigger epigenetic silencing (39). However, even if RNAs from somatic cells are transported to the germline in mammals, they may not always initiate transgenerational inherited effects because they have to escape mechanisms that reprogram epigenetic information in each generation (40). Additional studies are required to determine whether specific mechanisms prevent environmental influences from triggering transmission of information in the form of mobile RNAs from somatic cells to the germline.
Materials and Methods
All C. elegans strains were generated and maintained by using standard methods (41). Transgenic animals were generated by injecting PCR fragments or plasmids into the germline (29) of wild-type or mutant animals, and transgenes were also introduced into different genetic backgrounds through genetic crosses. Visible markers were used to balance sid-1(−) and to mark Pmex-5::gfp. Silencing of Pmex-5::gfp and Peft-3::gfp were measured by imaging under identical nonsaturating conditions using a Nikon AZ100 microscope. Silencing of sur-5::gfp was quantified by counting the number of gut nuclei that showed GFP expression above a fixed threshold of brightness. DNA of Prgef-1::gfp–dsRNA and Pmex-5::gfp transgenes were detected by using PCR in crosses and in inheritance experiments. Semiquantitative RT-PCR was used to determine relative mRNA levels by carrying out reverse transcription with gene-specific primers for gfp and tbb-2 followed by <31 cycles of PCR. Inherited silencing by feeding RNAi (19) and statistical analyses (20) were performed as described earlier. Detailed procedures are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Tom Kocher, Geraldine Seydoux, Norma Andrews, Steve Mount, Steve Wolniak, Karen Carleton, and members of the A.M.J. laboratory for critical reading of the manuscript; the Caenorhabditis elegans Genetic stock Center and the Hunter laboratory (Harvard University) for some worm strains; the Hamza laboratory (University of Maryland) for bacteria that express gfp–dsRNA; and Amy Beaven from the Department of Cell Biology and Molecular Genetics imaging core for microscopy advice. This work was supported by National Institutes of Health Grants R00GM085200 and R01GM111457 (to A.M.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423333112/-/DCSupplemental.
References
- 1.Weismann A. On heredity. In: Poulton EB, Schonland S, Shipley AE, editors. Essays upon Heredity and Kindred Biological Problems. Clarendon; Oxford: 1883. pp. 67–106. [Google Scholar]
- 2.Heard E, Martienssen RA. Transgenerational epigenetic inheritance: Myths and mechanisms. Cell. 2014;157(1):95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carone BR, et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143(7):1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng SF, et al. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature. 2010;467(7318):963–966. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- 5.Rechavi O, et al. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell. 2014;158(2):277–287. doi: 10.1016/j.cell.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308(5727):1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 8.Strome S. 2005 Specification of the germ line. WormBook, ed The C. elegans Research Community, 10.1895/wormbook.1.9.1, www.wormbook.org.
- 9.Shirayama M, et al. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150(1):65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashe A, et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150(1):88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee HC, et al. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell. 2012;150(1):78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luteijn MJ, et al. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 2012;31(16):3422–3430. doi: 10.1038/emboj.2012.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rechavi O, Minevich G, Hobert O. Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell. 2011;147(6):1248–1256. doi: 10.1016/j.cell.2011.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans. Science. 2000;287(5462):2494–2497. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- 15.Alcazar RM, Lin R, Fire AZ. Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis elegans. Genetics. 2008;180(3):1275–1288. doi: 10.1534/genetics.108.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vastenhouw NL, et al. Gene expression: Long-term gene silencing by RNAi. Nature. 2006;442(7105):882. doi: 10.1038/442882a. [DOI] [PubMed] [Google Scholar]
- 17.Gu SG, et al. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat Genet. 2012;44(2):157–164. doi: 10.1038/ng.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buckley BA, et al. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489(7416):447–451. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burton NO, Burkhart KB, Kennedy S. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2011;108(49):19683–19688. doi: 10.1073/pnas.1113310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jose AM, Smith JJ, Hunter CP. Export of RNA silencing from C. elegans tissues does not require the RNA channel SID-1. Proc Natl Acad Sci USA. 2009;106(7):2283–2288. doi: 10.1073/pnas.0809760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calixto A, Chelur D, Topalidou I, Chen X, Chalfie M. Enhanced neuronal RNAi in C. elegans using SID-1. Nat Methods. 2010;7(7):554–559. doi: 10.1038/nmeth.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295(5564):2456–2459. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- 23.Jose AM, Garcia GA, Hunter CP. Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Nat Struct Mol Biol. 2011;18(11):1184–1188. doi: 10.1038/nsmb.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feinberg EH, Hunter CP. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 2003;301(5639):1545–1547. doi: 10.1126/science.1087117. [DOI] [PubMed] [Google Scholar]
- 25.Shih JD, Hunter CP. SID-1 is a dsRNA-selective dsRNA-gated channel. RNA. 2011;17(6):1057–1065. doi: 10.1261/rna.2596511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tabara H, et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99(2):123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 27.Sijen T, et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107(4):465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 28.Sheth U, Pitt J, Dennis S, Priess JR. Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development. 2010;137(8):1305–1314. doi: 10.1242/dev.044255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10(12):3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grishok A. Biology and mechanisms of short RNAs in Caenorhabditis elegans. Adv Genet. 2013;83:1–69. doi: 10.1016/B978-0-12-407675-4.00001-8. [DOI] [PubMed] [Google Scholar]
- 31.Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RH. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99(2):133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 32.Hellwig S, Bass BL. A starvation-induced noncoding RNA modulates expression of Dicer-regulated genes. Proc Natl Acad Sci USA. 2008;105(35):12897–12902. doi: 10.1073/pnas.0805118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greer EL, et al. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479(7373):365–371. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolfrum C, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol. 2007;25(10):1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 35.Arroyo JD, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song R, et al. Male germ cells express abundant endogenous siRNAs. Proc Natl Acad Sci USA. 2011;108(32):13159–13164. doi: 10.1073/pnas.1108567108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tam OH, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453(7194):534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453(7194):539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 39.Rassoulzadegan M, et al. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441(7092):469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 40.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330(6004):622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.