Significance
CRYPTOCHROMES (CRYs) are blue light photoreceptors that mediate phototransduction in brain arousal neurons, as well as circadian light entrainment in Drosophila fruit flies. We describe how light-activated Drosophila CRY couples to membrane depolarization and increased action potential firing rate in large ventral lateral arousal neurons. Pharmacological treatments that specifically disrupt the CRY redox-sensitive flavin chromophore or block voltage-gated K+ channels abolish the light response. Correspondingly, we find that the Kvβ channel subunit Hyperkinetic with a well conserved redox sensor domain links light-evoked redox changes in CRY to rapid changes in membrane electrical potential.
Keywords: phototransduction, potassium channel, redox, cryptochrome
Abstract
Blue light activation of the photoreceptor CRYPTOCHROME (CRY) evokes rapid depolarization and increased action potential firing in a subset of circadian and arousal neurons in Drosophila melanogaster. Here we show that acute arousal behavioral responses to blue light significantly differ in mutants lacking CRY, as well as mutants with disrupted opsin-based phototransduction. Light-activated CRY couples to membrane depolarization via a well conserved redox sensor of the voltage-gated potassium (K+) channel β-subunit (Kvβ) Hyperkinetic (Hk). The neuronal light response is almost completely absent in hk−/− mutants, but is functionally rescued by genetically targeted neuronal expression of WT Hk, but not by Hk point mutations that disable Hk redox sensor function. Multiple K+ channel α-subunits that coassemble with Hk, including Shaker, Ether-a-go-go, and Ether-a-go-go–related gene, are ion conducting channels for CRY/Hk-coupled light response. Light activation of CRY is transduced to membrane depolarization, increased firing rate, and acute behavioral responses by the Kvβ subunit redox sensor.
CRYPTOCHROME (CRY) is a photoreceptor that mediates rapid membrane depolarization and increased spontaneous action potential firing rate in response to blue light in arousal and circadian neurons in Drosophila melanogaster (1, 2). CRY regulates circadian entrainment by targeting circadian clock proteins to proteasomal degradation in response to light (3–6). CRY is expressed in a small subset of central brain circadian, arousal, and photoreceptor neurons in D. melanogaster and other insects, including the large lateral ventral neuron (LNv; l-LNv) subset (1, 2, 7, 8). The l-LNvs are light-activated arousal neurons (1, 2, 9–11), whereas the small lateral ventral neurons (s-LNvs) are critical for circadian function (5, 12). Previous results suggest that light activated arousal is likely attenuated in cry-null mutants. In addition to modulating light-activated firing rate, membrane excitability in the LNv neurons helps maintain circadian rhythms (9, 13, 14), and LNv firing rate is circadian regulated (2, 16).
Based on our previous work suggesting that l-LNv electrophysiological light response requires a flavin-specific redox reaction and modulation of membrane K+ channels, we investigated the molecular mechanism for CRY phototransduction to determine how light-activated CRY is coupled to rapid membrane electrical changes. Sequence and structural data suggest that the cytoplasmic Kvβs are redox sensors based on a highly conserved aldo-keto-reductase domain (AKR) (17–21). Although no functional role for redox sensing by Kvβ subunits has been established yet in vivo, studies with heterologously expressed WT and mutant Kvβ subunits show that they confer modulatory sensitivity to coexpressed K+ channels in response to oxidizing and reducing chemical agents (22–24). Mammals express six Kvβ genes, whereas Drosophila expresses a single Kvβ designated HYPERKINETIC (Hk) (18). We find that the light-activated redox reaction of the flavin adenine dinucleotide (FAD) chromophore in CRY has a distinct phototransduction mechanism that evokes membrane electrical responses via the Kvβ subunit Hk, which we show is a functional redox sensor in vivo.
Results
Acute Behavioral Responses to Blue Light Are Altered in CRY Mutants.
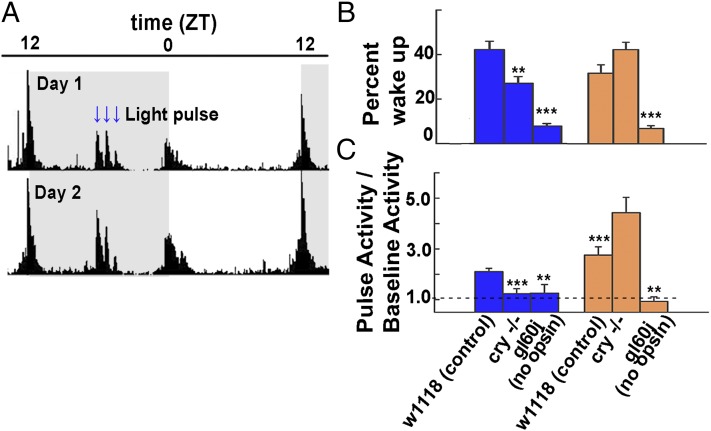
Mutants lacking CRY or opsin-based phototransduction exhibit defects in light entrainment (5). Subsequent work on the LNv indicates that these neurons also mediate light-driven arousal behavior (9–11). The CRY-dependent rapid electrophysiological response to blue light recorded in the LNv suggests that the loss of CRY attenuates acute behavioral arousal responses to blue light, but not to wavelengths beyond its absorbance cutoff above 530 nm (1). The behavioral locomotor responses were compared with 460-nm blue vs. 595-nm orange 5-min LED light pulses given in the middle of the night at ZT18, ZT19, and ZT20 for three successive nights to control, cry−/−, and gl60j mutant flies (which lack all opsin-based external photoreceptors). The averaged behavioral actograms of control flies to blue light for two successive nights are shown (Fig. 1A). We examined flies which were asleep, defined as inactive for at least 5 min, immediately before the light pulse for the percentage that awakened during the pulse. For control flies, a 5-min pulse of blue light woke 41 ± 3.7% of the sleeping flies; the numbers of cry−/− and gl60j mutant flies awakened were significantly lower (Fig. 1B). We also scored the behavioral response of awake flies to light pulses. For awake control flies, blue and orange light pulses in the middle of the night evoked twofold and threefold increases in locomotor activity relative to baseline activity in the dark (Fig. 1C). In contrast, both cry−/− and gl60j mutant awake flies show significantly attenuated behavioral responses to nighttime blue light pulses (Fig. 1C). CRY- and opsin-mediated phototransduction pathways contribute to the arousal behavioral response to nighttime blue light and mediate weaker responses to nighttime blue light when either component is absent. As expected, gl60j mutant awake flies do not behaviorally respond to nighttime orange light pulses (Fig. 1C), whereas cry−/− mutant awake flies show significantly greater behavioral response to nighttime orange light pulses (Fig. 1C).
Fig. 1.
Blue light activation of CRY contributes to rapid acute behavioral arousal responses. (A) Two days of averaged locomotor activity of control flies maintained in 12-h:12-h light:dark cycles. Entrainment shifts following the blue or orange nighttime light pulse protocol do not occur for any genotypes tested. (B) The averaged values of sleeping flies that wake in arousal response to blue and orange nighttime light pulses are shown for control (41 ± 4%, n = 40 blue; 32.6 ± 4%, n = 32 orange), cry−/− (26.3 ± 3%, n = 26 blue; 43.6 ± 3%, n = 17 orange), and gl60j (7.6 ± 1%, n = 14 blue; 7.1 ± 1%, n = 18 orange) mutant flies. Significantly fewer sleeping cry−/− flies wake in response to blue light pulses (P = 0.007), whereas the percentage of sleeping cry−/− flies that wake in response to orange light pulses does not differ from control (P = 0.09). Sleeping flies lacking all external opsin-based photoreceptors (gl60j mutants) show similar severe defects in their wake arousal responses to blue (P < 0.001) and orange (P = 0.001) light pulses. (C) The averaged normalized (pulse activity/baseline activity) values of behavioral arousal locomotor responses of awake flies are shown for control (2.1 ± 0.1, n = 509 blue; 2.8 ± 0.3, n = 286 orange), cry−/− (1.2 ± 0.2, n = 173 blue; 4.4 ± 0.6, n = 79 orange), and gl60j (1.3 ± 0.3, n = 86 blue; 0.9 ± 0.2, n = 112 orange) flies. The arousal response to blue light pulses is significantly attenuated in awake cry−/− flies (P = 0.002), but these flies show a significantly higher arousal response to orange light pulses (P = 0.02). In contrast, the arousal response to blue (P = 0.034) and orange (P = 0.003) light is significantly decreased in awake gl60j mutant flies. All data values and statistics are presented in detail in Dataset S1.
The CRY-Mediated Electrophysiological Light Response Is Attenuated in Mutants Lacking the Kvβ Subunit Hyperkinetic.
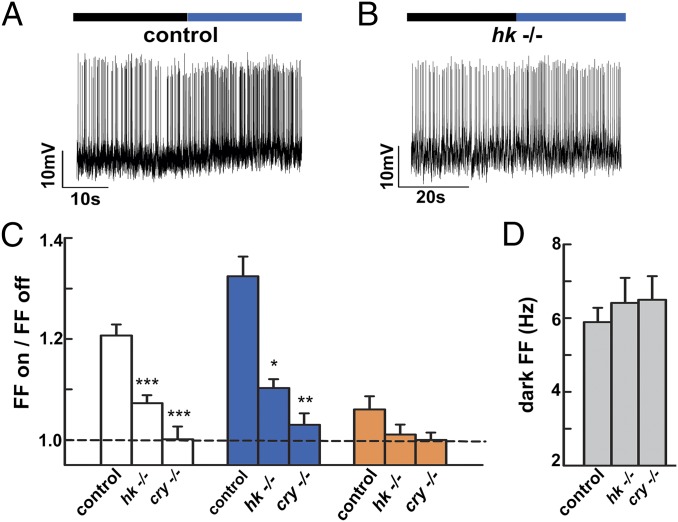
Acute pharmacological block of K+ channel currents eliminates the CRY-mediated l-LNv light response (1). Acute block of the l-LNv light response by the flavin-specific redox inhibitor diphenyleneiodonium (DPI) requires light-activated reduction of CRY flavin (Fig. S1). As redox signaling appears to be important for the CRY light response, we tested light responses in l-LNv recordings of flies null for hyperkinetic (hk−/−) Kvβ subunit (18). Kvβ proteins are highly conserved members of the NADP+-dependent AKR superfamily, but as yet are not linked to any known in vivo physiological function (17–21, 23, 24). The l-LNv blue light response (firing frequency light on/light off) in hk−/− flies is significantly attenuated (Fig. 2 B and C) relative to control (Fig. 2 A and C). This electrophysiological property is CRY light response-specific, as the l-LNv dark spontaneous firing rate in hk−/− vs. control does not differ from control or cry−/− (P = 0.769, ANOVA; Fig. 2D and Dataset S1). The l-LNv light response to white and blue wavelengths is significantly decreased in hk−/− flies relative to control (ANOVA; Fig. 2C). Control, hk−/−, and cry−/− all show no response to orange light and do not differ.
Fig. 2.
Mutant flies lacking the redox-sensor Kvβ subunit hyperkinetic have a significantly reduced l-LNv light response that is indistinguishable from cry−/−. (A) Representative traces for control blue l-LNv light response (0.6 mW/cm2, 375–450 nm, purple bar; black bar indicates no light) vs. (B) genetic null hk−/−. (C) Bar graph quantifies the l-LNv light response for control, hk−/−, and cry−/− flies. In white light (4 mW/cm2), control flies (1.21 ± 0.02, n = 26) are significantly different from hk−/− (1.07 ± 0.02, n = 16, P < 0.001) and cry−/− (1.00 ± 0.03, n = 14, P < 0.001 vs. control) flies. In blue light, control flies (1.32 ± 0.04, n = 30) are also significantly different from hk−/− (1.10 ± 0.02, n = 23, P = 0.001) and cry−/− flies (1.03 ± 0.02, n = 17, P = 0.0001). The severely attenuated light responses of hk−/− and cry−/− flies do not differ from each other (P = 0.14 in white; P = 0.36 in violet). In orange light (4 mW/cm2, >550 nm), control (1.04 ± 0.11, n = 23), hk−/− (1.01 ± 0.08, n = 17), and cry−/− (1.00 ± 0.05, n = 13) flies, responses do not differ (P > 0.35 in all pairwise comparisons). (D) Basal firing frequencies under dark conditions. The values for control (5.89 Hz ± 0.39, n = 77), hk−/− (6.41 Hz ± 0.68, n = 37), and cry−/− (6.5 Hz ± 0.64, n = 38) do not differ (P = 0.77 and P = 0.69 vs. control, respectively). All data values and statistics are presented in detail in Dataset S1.
LNv-Directed Expression of Hyperkinetic RNAi Attenuates the CRY-Mediated Light Response.
K+ channel subunits are widely expressed in the Drosophila nervous system and photoreceptors (25). To test the contribution of Hk to the CRY-mediated light response in the 8–10 pair of LNv neurons, the electrophysiological light response was measured in neurons that express Hk RNAi and Dicer directed by the pdf-GAL4 promoter. Normal electrophysiological responses to blue, white, and orange light (firing frequency light on/light off) were measured in l-LNv whole-cell patch-clamp recordings by using the RNAi genetic background control genotype (Fig. S2 A and C), but blue and white (but not orange) light responses are significantly reduced relative to controls in transgenic flies that express HkRNAi and DICER in the LNv (Fig. S2 B and C). As an additional control, we tested the effects of LNv-directed ShabRNAi knockdown of the Kv2-family voltage-gated K+ channel Shab subunit and found no effect on the light response recorded in l-LNv to blue, white, or orange light, which are all indistinguishable vs. RNAi controls (Fig. S2C). The baseline l-LNv action potential firing rates recorded in the dark were calculated for each genotype, and none significantly differed from control (Fig. S2C and Dataset S1). Thus, RNAi knockdown restricted to the LNv closely resembles the electrophysiological phenotype of loss of l-LNv response to blue and white light seen in hk−/−.
The l-LNv Light Response Is Occluded by Genetic or Acute Disruption of the Cellular Redox Environment and Is Dependent on the Hk Redox Sensor.
To test whether the l-LNv light response is modulated by the cellular redox environment, we measured the l-LNv light response under conditions that genetically or chemically disrupt the cellular redox environment. Superoxide dismutase (SOD) enzymes (SOD1, expressed in cytoplasm; and SOD2, expressed in mitochondria) regulate the redox environment by limiting cellular superoxide radicals (26). Blue and white, but not orange, light responses in the l-LNv are significantly lower in sod1−/− (but not sod2−/−) relative to genetic WT control (Fig. S3 A and B). Acute treatment with the oxidizer H2O2 abolishes response to blue light relative to vehicle control. Conditions that disrupt the cellular redox environment lead to increased spontaneous action potential firing in the absence of light thus occlude the l-LNv light response. Dark spontaneous firing frequency of l-LNv is significantly increased in sod1−/− and sod2−/− flies relative to genetic controls (Dataset S1). Dark spontaneous firing rate of l-LNv is significantly increased following acute H2O2 treatment relative to vehicle controls in genetic WT control flies (Fig. S3 C, D, and G and Dataset S1). Acute H2O2-induced increases in dark spontaneous firing rate of l-LNv are Hk dependent, as no significant increase in dark spontaneous firing rate of l-LNv is observed following acute H2O2 treatment in hk−/− flies (which resemble vehicle controls for the hk−/− genotype and the genetic controls), but the dark spontaneous firing rate of l-LNv is significantly increased in flies with LNv-directed WT Hk expression in the hk−/− genetic background, which in turn resemble the H2O2 response of WT controls (Fig. S3 E–G). Thus, acute H2O2-induced increases in dark spontaneous firing rate of l-LNv are Hk dependent, demonstrating an in vivo role for Hk connecting cellular redox environment and membrane firing properties.
WT Hk Expression Functionally Rescues the Cry-Mediated Light Response in hk−/− Flies, Whereas Mutants That Disrupt the Hk Redox Sensor Fail to Rescue.
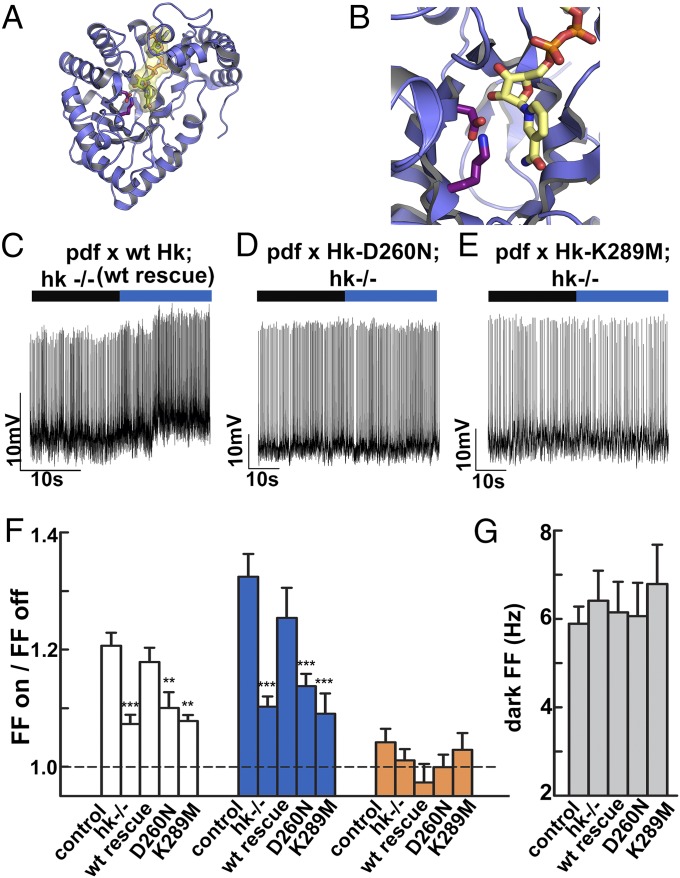
Ion channels express motifs that confer modulation to signaling pathways (27, 28) or metabolic cues (29). Structural analysis of fly and mammalian Kvβ proteins reveal that they are functional AKRs (17–21), measured by using heterologous expression systems (22–24). To determine whether the loss of the CRY-mediated l-LNv light response in hk−/− is a result of the loss of the redox sensor in this Kvβ subunit, we tested a matrix of genetic rescue experiments in the hk−/− genetic background by LNv-targeted expression of functional WT Hk or point mutants that disable the enzymatic activity of Kvβ without affecting protein expression levels (23, 24, 30). Fig. 3A shows a structural model of the Kvβ subunit in blue, with the NADP cofactor depicted in yellow. Two highly conserved residues (31) near the NADP+ cofactor, D260 and K289, depicted in magenta, are critical for enzymatic activity of Kvβ (Fig. 3B) (23, 24). We focused on these two sites because extensive previous work shows that mutations at these residues impair redox sensing without influencing protein expression or trafficking (23, 24, 30). To confirm that the Drosophila Hk D260 and K289 mutations do not affect protein expression or trafficking, they were tested in a GFP-tagged K isoform of Hk, and they express at equivalent or higher levels relative to WT Hk with no discernible changes in cellular protein distribution (Fig. S4). The l-LNv light response to blue light is cell-autonomously restored to levels indistinguishable from controls by WT Hk expression in the hk−/− genetic background (Fig. 3C). In contrast, the l-LNv light response to blue light is not functionally rescued by expression of the D260N-Hk mutant (Fig. 3D) or the K289M-Hk mutant in the hk−/− genetic background (Fig. 3E). The blue and white light response recorded in l-LNv neurons with LNv-targeted expression of WT Hk in the hk−/− genetic background is indistinguishable from the control light response, whereas similar LNv targeted expression of D260N- and K289M-Hk are significantly lower than the control light response and are indistinguishable from the severely attenuated light response seen in hk−/− (Fig. 3F, middle bars). Control, hk−/−, and WT Hk, D260N-Hk, and K289M-Hk expressed in the LNv in the hk−/− genetic background all show no response to orange light (Fig. 3F). The baseline l-LNv spontaneous firing rate recorded in the absence of light for control, hk−/−, and WT Hk, D260N-Hk, and K289M-Hk expressed in the hk−/− genetic background show no significant differences, indicating that the electrophysiological effects noted previously are specific for Cry-mediated light response (Fig. 3G and Dataset S1).
Fig. 3.
LNv-directed expression of WT Hk in hk−/− flies functionally rescues the Cry-mediated light response, whereas expression of Hk redox sensor-disabling mutants fail to rescue. (A) Structural model of the Kvβ subunit (blue) bound to the NADP+ cofactor (yellow). (B) Positional model of two key residues for Kvβ redox sensing (D260 and K289) depicted in stick form in magenta relative to the NADP+ cofactor (yellow). (C) WT Hk expressed in LNv neurons in the hk−/− genetic background rescues the l-LNv light response (black bar indicates no light; purple bar indicates 375–450 nm blue light, 0.6 mW/cm2). In contrast, LNv-directed expression of redox-disabled point mutants D260N-Hk (D) and K289M-Hk (E) in hk−/− fail to rescue the l-LNv blue light response. (F) The white light response (firing frequency lights on/lights off, 4 mW/cm2) recorded in l-LNv expressing WT Hk in hk−/− (WT rescue, 1.18 ± 0.02, n = 22) is indistinguishable from the control response (1.21 ± 0.02, n = 26, P = 0.79) but is significantly different from hk−/− (1.07 ± 0.02, n = 16, P = 0.004), showing functional rescue. In contrast, LNv-directed expression of the redox-disabled D260N-Hk mutant in hk−/− (1.12 ± 0.03, n = 13) is significantly different from control (P = 0.036) but not hk−/− (P = 0.58) recordings, indicating failure to rescue. Similarly, the white light response recorded in l-LNv expressing the redox disabled K289M-Hk mutant in hk−/− (1.08 ± 0.01, n = 12) is significantly different from control (P < 0.001) but not null (P = 0.99) recordings. The response to blue light in WT rescue l-LNv recordings (1.25 ± 0.05, n = 16) is not different from control (P = 0.61), but differs from hk−/− (1.10 ± 0.02, n = 23, P = 0.038); the light response in recordings of l-LNv expressing D260N-Hk is significantly less than from control (1.15 ± 0.02, n = 15, P = 0.01) and not different from null (P = 0.88); the light response in recordings of l-LNv expressing K289M-Hk in hk−/− (1.06 ± 0.02, n = 13) is significantly less than control (P < 0.001) and not different from null (P = 0.95). In orange light (4 mW/cm2, >550 nm), WT rescue (0.96 ± 0.1, n = 11), D260N-Hk (1.00 ± 0.06, n = 8), and K289M-Hk (1.03 ± 0.09, n = 10) are not significantly different from each other, control (1.04 ± 0.11, n = 23), or hk−/− (1.01 ± 0.08, n = 17) recordings (P > 0.1 in all comparisons). (G) The basal firing frequencies in the dark for control (5.89 Hz ± 0.39, n = 77), hk−/− (6.41 Hz ± 0.68, n = 37), Hk WT rescue (6.15 Hz ± 0.69, n = 31), Hk D260N rescue (6.06 Hz ± 0.76, n = 23), and Hk K289M rescue (6.79 Hz ± 0.89, n = 18) do not differ (P = 0.77, P = 0.941, P = 0.999, and P = 0.874 vs. control, respectively). All data values and statistics are presented in detail in Dataset S1.
The Ether-a-Go-Go Family K+ Channels Underlie CRY-Mediated Membrane Depolarization and Increased Neuronal Firing Rate in Response to Light.
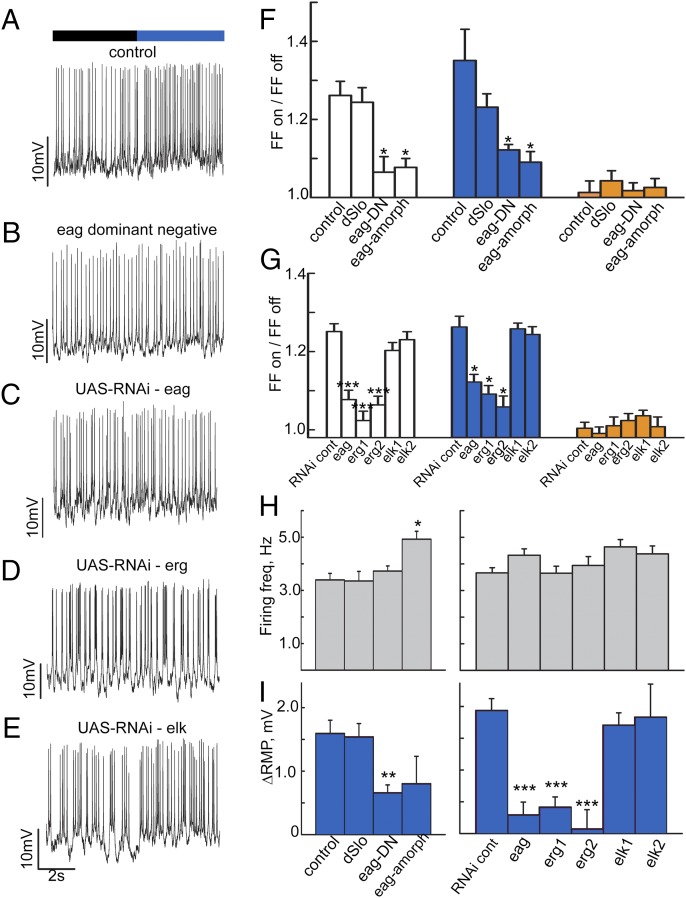
Hk also coassembles with D. melanogaster EAG-family Kvα subunits (32, 33). To determine whether EAG-family Kvα subunits contribute to the l-LNv CRY-mediated light response, including membrane depolarization, we expressed the dominant-negative eag∆932 transgene (eag-DN) (34) in the LNv driven by the pdf-GAL4 promoter. LNv-targeted expression of eag-DN eliminates blue and white light responses seen in controls (Fig. 4 A, B, and F). Similar to the LNv targeted expression of eag-DN, no blue or white light responses are seen in l-LNv recordings prepared from eag genetic null mutants (eag amorphic; Fig. 4F). In contrast, blue and white light responses recorded from the l-LNv of dslo-null mutant flies, a Ca-sensitive K channel that is not reported to interact with Hk but has been reported to be modulated by redox state (35, 36), are indistinguishable from control (Fig. 4F). No orange light responses were observed for all genotypes tested (Fig. 4F).
Fig. 4.
The Ether-a-go-go family K+ channels underlie light evoked membrane depolarization and increased neuronal firing rate. Representative traces for control (A), eag-dominant negative (B), pdfGAL4-driven UAS-RNAi against eag (C), pdfGAL4-driven UAS-RNAi against erg (D), and pdfGAL4-driven UAS-RNAi against elk (E). Bar graphs (F) quantify firing frequency change for control flies in white light (1.26 ± 0.04, n = 19), which is not different from the response of dSlo flies (1.23 ± 0.03, n = 24, P > 0.05). Eag-dominant negative (1.06 ± 0.04, n = 18) and eag amorphic flies (1.08 ± 0.02, n = 13) are both different from control and dSlo (P < 0.05 in each case). In blue light, control (1.35 ± 0.08, n = 19) and dSlo (1.23 ± 0.03, n = 26) do not differ (P > 0.05). However, eag-DN (1.12 ± 0.01, n = 19) and eag amorphic (1.09 ± 0.03, n = 13) both differ from control and dSlo (P > 0.05 in each case). The responses to orange light do not differ. (G) The white light response for RNAi genotype-control flies is 1.25 ± 0.02 (n = 19). This is significantly different from the response of flies with pdfGAL4-driven RNAi against eag (1.08 ± 0.02, n = 16, P < 0.001), against erg line 1 (1.02 ± 0.02, n = 16, P < 0.001), and against erg line 2 (1.06 ± 0.02, n = 17). It is not different from the responses of the two elk RNAi lines (1.20 ± 0.02, n = 24, P = 0.50; and 1.23 ± 0.02, n = 17, P = 0.98, respectively). In blue light, the control response (1.26 ± 0.03, n = 23) is different from that of the eag RNAi line (1.12 ± 0.02, n = 19, P < 0.05) and both erg RNAi lines (1.09 ± 0.02, n = 18, P < 0.05; and 1.06 ± 0.03, n = 16, P < 0.05), but not the elk lines (1.26 ± 0.02, n = 19, P > 0.05; and 1.24 ± 0.02, n = 19, P > 0.05). The responses to orange light again do not differ. (H) The basal dark firing frequencies for control (3.39 Hz ± 0.24, n = 58), dSlo (3.35 Hz ± 0.36, n = 42), and eag-dominant negative (3.72 Hz ± 0.19, n = 47) do not differ (P > 0.05), but that of eag amorphic (eagsc29; 4.92 Hz ± 0.30, n = 25) is different from control (P < 0.05). The firing frequencies for RNAi control (3.66 Hz ± 0.20, n = 43), eag-RNAi (4.32 Hz ± 0.24, n = 39), erg RNAi-1 (3.65 Hz ± 0.26, n = 33), erg RNAi-2 (3.94 Hz ± 0.33, n = 30), elk RNAi line 1 (4.64 Hz ± 0.28, n = 41), and elk RNAi line 2 (4.37 Hz ± 0.30, n = 30) do not differ (P > 0.5 in each case.) (I) Bar graphs quantify the change in resting membrane potential during periods of blue light vs. darkness. Control (1.60 mV ± 0.21, n = 22) and dSlo (1.54 mV ± 0.21, n = 17) do not differ (P > 0.05). eag-DN, however, is different from control and dSlo (0.66 mV ± 0.12, n = 30, P < 0.05). eag amorphic (0.80 mV ± 0.43, n = 13) trended lower, but its difference does not reach significance (P > 0.05). The change in RMP for RNAi control flies is 1.94 mV ± 0.19 (n = 27). This is significantly different from eag RNAi-expressing flies (0.30 mV ± 0.20, n = 15), erg RNAi 1 flies (0.42 mV ± 0.16, n = 16), and erg RNAi 2 flies (0.07 mV ± 0.30, n = 10; P < 0.001 in each case). Elk RNAi-expressing flies lines 1 and 2 (1.71 mV ± 0.19, n = 18; and 1.83 mV ± 0.52, n = 13) do not differ from control (P = 0.978 and P = 1.00, respectively). All data values and statistics are presented in detail in Dataset S1.
As eag-DN expression could potentially disrupt the function of all three closely related members of the Drosophila melanogaster EAG family [EAG, EAG-related gene (ERG), and EAG-like K+ channel gene (ELK)], we measured the blue and white light response following LNv-targeted expression of a single RNAi line for EAG and two independent RNAi lines each for ERG and ELK. Compared with the normal blue and white light responses seen in l-LNv recordings prepared from an RNAi control line (Fig. 4G), significantly lower blue and white light responses are recorded following the LNv targeted expression of eag RNAi and both lines for erg RNAi (Fig. 4 C, D, and G). In contrast, the blue and white light responses are indistinguishable from controls in l-LNv recordings following LNv targeted expression of both elk RNAi lines (Fig. 4 E and G). No orange light responses were observed for all RNAi line genotypes tested (Fig. 4G). Baseline l-LNv firing rate in the dark was measured for all genotypes tested and all were indistinguishable from control, except for the eag amorphic-null mutant, which exhibits a significantly higher baseline firing rate (Fig. 4H and Dataset S1). Membrane depolarization response to blue light in the dslo-null mutant is indistinguishable from control recordings (Fig. 4I), but is significantly lower in neurons recorded from the eag amorphic-null mutant or following LNv-targeted expression of eag-DN (Fig. 4I). Similarly, blue light-evoked l-LNv depolarization is significantly lower relative to control following LNv targeted expression of eag RNAi and two independent erg RNAi lines, but not two independent elk RNAi lines (Fig. 4I). EAG-family channels regulate membrane resting potential and the action potential repolarization (37), whereas Shaker-type K+ channels, which also coassemble with Hk, have very little effect on membrane resting potential, but regulate firing rate subject to rapid inactivation that contributes to cumulative inactivation (38). Taken together, the results indicate that Hk-coassembled EAG and ERG (but not ELK) K+ channels underlie CRY-mediated light-evoked increased firing rate and depolarization.
Discussion
Acute behavioral arousal to blue light is significantly attenuated in CRY mutants. We identify a redox signaling couple between blue light-activated CRY and rapid membrane depolarization via the redox sensor of Kvβ channel subunits coassembled with Kvα channel subunits. Additional unknown factors may act as intermediates between CRY and Hk. This finding provides in vivo validation of a very longstanding hypothesis that the highly conserved redox sensor of Kvβ subunit functionally senses cellular redox events to physiological changes in membrane electrical potential. Genetic loss of any single component functionally disrupts the CRY-mediated blue light response, which is functionally rescued by LNv restricted expression of their WT genes in the null backgrounds. Although little is known about the structural contacts between Kvβ and EAG subunits, Kvβ subunits make extensive physical contacts in a fourfold symmetric fashion in 1:1 stoichiometry with the T1 assembly domain of other coassembled tetrameric Kvα subunits that form the complete functional channel (19–21). Application of Kvβ redox chemical substrate modulates voltage-evoked channel peak current, steady-state current, and inactivation in heterologously expressed α-β channels, which are reversed by fresh NADPH (23, 24). These results indicate that measurements of channel biophysical properties can reflect the redox enzymatic cycle of Kvβ as these channel modulatory effects are absent in preparations that lack the expression of WT Kvβ subunits or express redox sensor mutant Kvβ subunits (23, 24). Whether direct chemical redox reactions occur between CRY and Hk is unclear. For CRY, light or chemical reduction induces one-electron reduction of the FAD cofactor of CRY (39–41), whereas the reductive catalytic mechanism of AKRs (such as Hk) requires a hydride ion transferred from NADPH to a substrate carbonyl, then a solvent-donated proton reduces the substrate carbonyl to an alcohol (42). These differences in redox chemistry between CRY and Hk suggest that other intermediates, such as oxygen, are possibly required for redox coupling.
Spectroscopic analysis of animal and plant CRYs suggest that light activation causes reduction of the FAD oxidized base state (39–41, 43). Light activation of Drosophila CRY also evokes conformational changes in the C terminus of CRY that clearly promotes CRY C-terminal access to proteolytic degradation and subsequent interactions with the TIMELESS clock protein, thus signaling degradation and circadian entrainment (44–47). However, all existing evidence suggests that light activated CRY-mediated circadian entrainment and membrane electrical phototransduction operate under different mechanisms, including their different activation thresholds and relative dependence on the C terminus of CRY (1). Further distinguishing the distinct mechanisms of the downstream effects of light-activated CRY, the light-induced conformational changes that couple CRY to ubiquitin ligase binding (thus causing circadian entrainment) occur in oxidized and reduced states of CRY and are unaffected in CRY tryptophan mutants that presumably are responsible for intraprotein electron transfer reactions following light-evoked reduction of the FAD cofactor (47). Another recent study shows that light- or chemical-evoked reduction of Drosophila CRY FAD is coupled to conformational changes of the CRY C terminus (41), along with reporting a surprising negative result that DPI has no effect on the reoxidation of the reduced anionic semiquinone of purified Drosophila CRY. DPI could hypothetically influence the electrophysiological light response by blocking the pentose phosphate pathway (48) which produces the Hk redox cofactor NADPH, but this does not explain the light dependence for DPI blocking the electrophysiological light response herein. The available evidence indicates that CRY-mediated light evoked membrane depolarization occurs independently of conformational changes in the CRY C-terminal domain but depends on redox changes in CRY, whereas CRY-mediated light evoked circadian entrainment depends on conformational changes in the CRY C-terminal domain and may or may not depend on CRY redox state.
Light-activated CRY evokes rapid membrane depolarization through the redox sensor of the Kvβ subunit Hk. A general role for circadian regulation of redox state coupled to membrane excitability has been described recently in mammalian suprachiasmatic neurons (49). Redox modulation of circadian neural excitability may be a well-conserved feature.
Materials and Methods
Locomotor activity was recorded using TriKinetics Drosophila Activity Monitor system (9). l-LNv recordings were performed on acutely dissected adult fly brains in whole-cell current-clamp mode (1, 2). DPI and H2O2 were obtained from Sigma and prepared in H2O or standard external recording solution (1). Extended information on materials and methods is provided in SI Materials and Methods, including protocols for electrophysiology, optics, genetics, behavioral testing, and statistical analysis.
Supplementary Material
Acknowledgments
The authors thank Jeff Hall (cry01) and the Bloomington Stock Center for reagents, Sheeba Vasu for helpful discussion, Vinh Nguy and Dana Mumford for technical assistance, Luette Forrest for HEK293 cells, and Rongsheng Jin for additional Hk 3D modeling. This work was funded by National Institutes of Health Grants NS046750 (to T.C.H.), GM102965 (to T.C.H.), GM107405 (to T.C.H.), and HL086392 (to M.Z.), and benefited from access to the Optical Biology Shared Resource of Cancer Center Support Grant CA-62203 at the University of California, Irvine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416586112/-/DCSupplemental.
References
- 1.Fogle KJ, Parson KG, Dahm NA, Holmes TC. CRYPTOCHROME is a blue-light sensor that regulates neuronal firing rate. Science. 2011;331(6023):1409–1413. doi: 10.1126/science.1199702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sheeba V, Gu H, Sharma VK, O’Dowd DK, Holmes TC. Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J Neurophysiol. 2008;99(2):976–988. doi: 10.1152/jn.00930.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95(5):669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 4.Stanewsky R, et al. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95(5):681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 5.Helfrich-Förster C, Winter C, Hofbauer A, Hall JC, Stanewsky R. The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron. 2001;30(1):249–261. doi: 10.1016/s0896-6273(01)00277-x. [DOI] [PubMed] [Google Scholar]
- 6.Koh K, Zheng X, Sehgal A. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science. 2006;312(5781):1809–1812. doi: 10.1126/science.1124951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshii T, Todo T, Wülbeck C, Stanewsky R, Helfrich-Förster C. Cryptochrome is present in the compound eyes and a subset of Drosophila’s clock neurons. J Comp Neurol. 2008;508(6):952–966. doi: 10.1002/cne.21702. [DOI] [PubMed] [Google Scholar]
- 8.Benito J, Houl JH, Roman GW, Hardin PE. The blue-light photoreceptor CRYPTOCHROME is expressed in a subset of circadian oscillator neurons in the Drosophila CNS. J Biol Rhythms. 2008;23(4):296–307. doi: 10.1177/0748730408318588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheeba V, et al. Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol. 2008;18(20):1537–1545. doi: 10.1016/j.cub.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parisky KM, et al. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron. 2008;60(4):672–682. doi: 10.1016/j.neuron.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shang Y, Griffith LC, Rosbash M. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci USA. 2008;105(50):19587–19594. doi: 10.1073/pnas.0809577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helfrich-Förster C. Robust circadian rhythmicity of Drosophila melanogaster requires the presence of lateral neurons: A brain-behavioral study of disconnected mutants. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1998;182(4):435–453. doi: 10.1007/s003590050192. [DOI] [PubMed] [Google Scholar]
- 13.Nitabach MN, Blau J, Holmes TC. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell. 2002;109(4):485–495. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- 14.Nitabach MN, et al. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J Neurosci. 2006;26(2):479–489. doi: 10.1523/JNEUROSCI.3915-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu MN, Koh K, Yue Z, Joiner WJ, Sehgal A. A genetic screen for sleep and circadian mutants reveals mechanisms underlying regulation of sleep in Drosophila. Sleep. 2008;31(4):465–472. doi: 10.1093/sleep/31.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao G, Nitabach MN. Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J Neurosci. 2008;28(25):6493–6501. doi: 10.1523/JNEUROSCI.1503-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCormack T, McCormack K. Shaker K+ channel beta subunits belong to an NAD(P)H-dependent oxidoreductase superfamily. Cell. 1994;79(7):1133–1135. doi: 10.1016/0092-8674(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 18.Chouinard SW, Wilson GF, Schlimgen AK, Ganetzky B. A potassium channel beta subunit related to the aldo-keto reductase superfamily is encoded by the Drosophila hyperkinetic locus. Proc Natl Acad Sci USA. 1995;92(15):6763–6767. doi: 10.1073/pnas.92.15.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulbis JM, Mann S, MacKinnon R. Structure of a voltage-dependent K+ channel beta subunit. Cell. 1999;97(7):943–952. doi: 10.1016/s0092-8674(00)80805-3. [DOI] [PubMed] [Google Scholar]
- 20.Gulbis JM, Zhou M, Mann S, MacKinnon R. Structure of the cytoplasmic beta subunit-T1 assembly of voltage-dependent K+ channels. Science. 2000;289(5476):123–127. doi: 10.1126/science.289.5476.123. [DOI] [PubMed] [Google Scholar]
- 21.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309(5736):897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 22.Pérez-García MT, López-López JR, González C. Kvbeta1.2 subunit coexpression in HEK293 cells confers O2 sensitivity to kv4.2 but not to Shaker channels. J Gen Physiol. 1999;113(6):897–907. doi: 10.1085/jgp.113.6.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weng J, Cao Y, Moss N, Zhou M. Modulation of voltage-dependent Shaker family potassium channels by an aldo-keto reductase. J Biol Chem. 2006;281(22):15194–15200. doi: 10.1074/jbc.M513809200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan Y, Weng J, Cao Y, Bhosle RC, Zhou M. Functional coupling between the Kv1.1 channel and aldoketoreductase Kvbeta1. J Biol Chem. 2008;283(13):8634–8642. doi: 10.1074/jbc.M709304200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardie RC. Voltage-sensitive potassium channels in Drosophila photoreceptors. J Neurosci. 1991;11(10):3079–3095. doi: 10.1523/JNEUROSCI.11-10-03079.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Godenschwege T, et al. Mitochondrial superoxide radicals differentially affect muscle activity and neural function. Genetics. 2009;183(1):175–184. doi: 10.1534/genetics.109.103515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmes TC, Fadool DA, Ren R, Levitan IB. Association of Src tyrosine kinase with a human potassium channel mediated by SH3 domain. Science. 1996;274(5295):2089–2091. doi: 10.1126/science.274.5295.2089. [DOI] [PubMed] [Google Scholar]
- 28.Nitabach MN, et al. A mechanism for combinatorial regulation of electrical activity: Potassium channel subunits capable of functioning as Src homology 3-dependent adaptors. Proc Natl Acad Sci USA. 2001;98(2):705–710. doi: 10.1073/pnas.031446198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noma A. ATP-regulated K+ channels in cardiac muscle. Nature. 1983;305(5930):147–148. doi: 10.1038/305147a0. [DOI] [PubMed] [Google Scholar]
- 30.Campomanes CR, et al. Kv beta subunit oxidoreductase activity and Kv1 potassium channel trafficking. J Biol Chem. 2002;277(10):8298–8305. doi: 10.1074/jbc.M110276200. [DOI] [PubMed] [Google Scholar]
- 31.Scott VE, et al. Primary structure of a beta subunit of alpha-dendrotoxin-sensitive K+ channels from bovine brain. Proc Natl Acad Sci USA. 1994;91(5):1637–1641. doi: 10.1073/pnas.91.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson GF, Wang Z, Chouinard SW, Griffith LC, Ganetzky B. Interaction of the K channel beta subunit, Hyperkinetic, with eag family members. J Biol Chem. 1998;273(11):6389–6394. doi: 10.1074/jbc.273.11.6389. [DOI] [PubMed] [Google Scholar]
- 33.Petersen CI, et al. In vivo identification of genes that modify ether-a-go-go-related gene activity in Caenorhabditis elegans may also affect human cardiac arrhythmia. Proc Natl Acad Sci USA. 2004;101(32):11773–11778. doi: 10.1073/pnas.0306005101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Broughton SJ, Kitamoto T, Greenspan RJ. Excitatory and inhibitory switches for courtship in the brain of Drosophila melanogaster. Curr Biol. 2004;14(7):538–547. doi: 10.1016/j.cub.2004.03.037. [DOI] [PubMed] [Google Scholar]
- 35.Tang XD, Garcia ML, Heinemann SH, Hoshi T. Reactive oxygen species impair Slo1 BK channel function by altering cysteine-mediated calcium sensing. Nat Struct Mol Biol. 2004;11(2):171–178. doi: 10.1038/nsmb725. [DOI] [PubMed] [Google Scholar]
- 36.Santarelli LC, Wassef R, Heinemann SH, Hoshi T. Three methionine residues located within the regulator of conductance for K+ (RCK) domains confer oxidative sensitivity to large-conductance Ca2+-activated K+ channels. J Physiol. 2006;571(pt 2):329–348. doi: 10.1113/jphysiol.2005.101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ganetzky B, Robertson GA, Wilson GF, Trudeau MC, Titus SA. The eag family of K+ channels in Drosophila and mammals. Ann N Y Acad Sci. 1999;868:356–369. doi: 10.1111/j.1749-6632.1999.tb11297.x. [DOI] [PubMed] [Google Scholar]
- 38.Hoffman DA, Magee JC, Colbert CM, Johnston D. K+ channel regulation of signal propagation in dendrites of hippocampal pyramidal neurons. Nature. 1997;387(6636):869–875. doi: 10.1038/43119. [DOI] [PubMed] [Google Scholar]
- 39.Berndt A, et al. A novel photoreaction mechanism for the circadian blue light photoreceptor Drosophila cryptochrome. J Biol Chem. 2007;282(17):13011–13021. doi: 10.1074/jbc.M608872200. [DOI] [PubMed] [Google Scholar]
- 40.Kao YT, et al. Ultrafast dynamics and anionic active states of the flavin cofactor in cryptochrome and photolyase. J Am Chem Soc. 2008;130(24):7695–7701. doi: 10.1021/ja801152h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaidya AT, et al. Flavin reduction activates Drosophila cryptochrome. Proc Natl Acad Sci USA. 2013;110(51):20455–20460. doi: 10.1073/pnas.1313336110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barski OA, Tipparaju SM, Bhatnagar A. The aldo-keto reductase superfamily and its role in drug metabolism and detoxification. Drug Metab Rev. 2008;40(4):553–624. doi: 10.1080/03602530802431439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin C, et al. Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science. 1995;269(5226):968–970. doi: 10.1126/science.7638620. [DOI] [PubMed] [Google Scholar]
- 44.Dissel S, et al. A constitutively active cryptochrome in Drosophila melanogaster. Nat Neurosci. 2004;7(8):834–840. doi: 10.1038/nn1285. [DOI] [PubMed] [Google Scholar]
- 45.Peschel N, Chen KF, Szabo G, Stanewsky R. Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Curr Biol. 2009;19(3):241–247. doi: 10.1016/j.cub.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 46.Ozturk N, Selby CP, Annayev Y, Zhong D, Sancar A. Reaction mechanism of Drosophila cryptochrome. Proc Natl Acad Sci USA. 2011;108(2):516–521. doi: 10.1073/pnas.1017093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ozturk N, Selby CP, Zhong D, Sancar A. Mechanism of photosignaling by Drosophila cryptochrome: Role of the redox status of the flavin chromophore. J Biol Chem. 2014;289(8):4634–4642. doi: 10.1074/jbc.M113.542498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riganti C, et al. Diphenyleneiodonium inhibits the cell redox metabolism and induces oxidative stress. J Biol Chem. 2004;279(46):47726–47731. doi: 10.1074/jbc.M406314200. [DOI] [PubMed] [Google Scholar]
- 49.Wang TA, et al. Circadian rhythm of redox state regulates excitability in suprachiasmatic nucleus neurons. Science. 2012;337(6096):839–842. doi: 10.1126/science.1222826. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.