Significance
We present experimental evidence showing that meiosis is an important source of germline mutations. Because sites of meiotic recombination experience recurrent double-strand breaks at hotspots, recombination has been previously suspected to be mutagenic. Yet inferences made from sequence comparisons have not found strong evidence for a mutagenic effect of recombination. Here, we directly sequenced a large number of single sperm DNA molecules and found more new mutations in molecules with a crossover than in molecules without a recombination event. We also observed that GC alleles are transmitted more often than AT alleles at polymorphic sites. Our data demonstrate that both mutagenesis and biased transmission occur during crossing over in meiosis and are important modifiers of the sequence content at recombination hotspots.
Keywords: meiotic recombination, crossover, sequence evolution, biased gene conversion, mutation
Abstract
Meiosis is a potentially important source of germline mutations, as sites of meiotic recombination experience recurrent double-strand breaks (DSBs). However, evidence for a local mutagenic effect of recombination from population sequence data has been equivocal, likely because mutation is only one of several forces shaping sequence variation. By sequencing large numbers of single crossover molecules obtained from human sperm for two recombination hotspots, we find direct evidence that recombination is mutagenic: Crossovers carry more de novo mutations than nonrecombinant DNA molecules analyzed for the same donors and hotspots. The observed mutations were primarily CG to TA transitions, with a higher frequency of transitions at CpG than non-CpGs sites. This enrichment of mutations at CpG sites at hotspots could predominate in methylated regions involving frequent single-stranded DNA processing as part of DSB repair. In addition, our data set provides evidence that GC alleles are preferentially transmitted during crossing over, opposing mutation, and shows that GC-biased gene conversion (gBGC) predominates over mutation in the sequence evolution of hotspots. These findings are consistent with the idea that gBGC could be an adaptation to counteract the mutational load of recombination.
Meiotic recombination, localized in recombination hotspots, not only increases genetic diversity via the formation of new haplotypes but is also an important driver of sequence evolution. The binding sites used by the human recombination machinery involving PRDM9 (PR domain containing 9) are more eroded in humans than the same sequences in chimps, given that PRDM9 in chimps uses different binding sites (1). Moreover, regions in close vicinity to these PRDM9 binding sites also showed a significant enrichment of polymorphisms in humans (2). In addition, within- and between-species sequence diversity positively correlates with regions of high recombination activity in humans (3–7) and other eukaryotes (reviewed in refs. 8–10).
One process recognized as a major evolutionary force reshaping the genomic nucleotide landscape at recombination hotspots, as shown in humans (6), chimpanzees (6, 11), mice (12), yeast (13), and metazoans (14), is GC-biased gene conversion (gBGC). In gBGC, the repair of heteroduplex tracts formed during meiotic recombination leads to the non-Mendelian segregation of alleles favoring GC over AT variants. The precise molecular mechanisms leading to gBGC have yet to be unraveled, but experimental evidence has shown that in crossovers (COs) of fission yeast, GC alleles can be overtransmitted within ∼1–2 kb in length of the double-strand break (DSB) region (13), implicating mismatch repair (15).
However, it is also plausible that the higher sequence variation observed at recombination hotspots is a result of a mutagenic effect of recombination: meiotic recombination is initiated by DSBs, which are associated with an increased mutation frequency. In the nonreducing division, mitosis, genetic experiments have shown that the repair of DSBs in homologous recombination involves error-prone translesion polymerases, increasing the mutation frequency at DSB sites in Drosophila (16) and yeast (17–19). In humans as well, an error-prone polymerase (DNA polymerase θ) carries out translesion synthesis in the repair of DSBs (20). In addition to mitotic DSB repair, it was recently shown that error-prone translesion synthesis polymerases (Rev1, PolZeta, and Rad30) are also involved in the repair of DSBs in meiosis in yeast (21) and could potentially contribute to a higher mutation rate in the germ cells at recombination hotspots, which are recurrent sites for DSBs.
Although high mutation rates might be an important driver of the high genetic diversity and elevated divergence in regions of high recombination (refs. 5–7, 22, and 23 and reviewed by refs. 23 and 24), the mutagenic signature in population data may be obscured by a complex interplay of other factors, including selection, demographic history, and gBGC (8–10). Therefore, to detect and quantify any elevation in mutation rates arising during meiosis, we measured the frequency of de novo mutations in a large number of single COs. We provide, for the first time to our knowledge, experimental data showing that recombination associated with COs is mutagenic in humans. In our large survey of recombination products, we also find evidence that the transmission of GC alleles is favored during crossing over and that associated gBGC is acting in opposition to the introduced mutational bias.
Results and Discussion
Mutation Frequencies Are Increased in COs.
We amplified single CO products from a pool of sperm at two previously identified recombination hotspots (HSI and HSII) (25, 26), using allele-specific PCR (26). In total, we sequenced 5,796 COs, including both reciprocal recombination products, from six Caucasian donors. As a control, we screened single nonrecombinants (NRs) in the same region and subset of donors, using the same experimental conditions (SI Appendix, Table S1). COs had a mutation frequency ∼3.6 times higher than NR controls (Fisher’s exact test, P = 0.037), suggesting that the observed mutations are associated with CO formation and are independent of other site-specific factors such as base composition (Table 1). As mutations in NRs probably reflect a combination of rare de novo mutations and PCR artifacts, we conservatively used the mutation frequency of NRs to adjust CO mutation frequencies (Table 1). The enrichment of mutations in COs was similar for both hotspots, and we did not observe significant heterogeneity between hotspots and among donors (SI Appendix, Fig. S1), although the small sample size provides little power for detecting potential differences. Because most of our donors came from HSI, we focused our main analysis on data from this hotspot.
Table 1.
Observed mutations in hotspots
| Hotspot | CO | NR | µCO/µNR | µCO* | µHS | c | µHS/µhAve | ||||
| n | Effective sites | µCO | n | Effective sites | µNR | ||||||
| HSI | 14 | 12,068,100 | 1.16 × 10−6 | 2 | 7,143,800 | 2.80 × 10−7 | 4.1*1 | 8.80 × 10−7 | 2.07 × 10−8 | 1.00 × 10−2 | 1.72 |
| HSII | 3 | 1,152,900 | 2.60 × 10−6 | 1 | 1,188,600 | 8.41 × 10−7 | 3.1 | 1.76 × 10−6 | 1.46 × 10−8 | 1.50 × 10−3 | 1.22 |
| Total | 17 | 13,221,000 | 1.29 × 10−6 | 3 | 8,332,400 | 3.60 × 10−7 | 3.6*2 | 9.26 × 10−7 | 1.79 × 10−8 | 6.47 × 10−3 | 1.49 |
µCO and µNR are the mutation counts (n) measured in COs or NRs per effective number of sequenced sites; *1 and *2 indicate a statistically significant difference between the mutation fractions for CO versus NR molecules (Fisher’s exact test; P = 0.041 and 0.037, respectively); µCO* is the corrected mutation fraction of CO (µCO) by the mutation fraction of NRs (µNR); and µHS is the mutation rate at hotspots estimated as the sum of the mutations in the fraction of gametes with a CO and the fraction of the gametes without a CO with a human average mutational load, expressed as c(µCO*)+(1-c)µhAve, where µhAve is the human mutation average rate of 1.2 × 10−8 (27) and c is the CO frequency estimated from the data (SI Appendix, Table S5).
On average, we observed 0.29% [95% Poisson confidence interval (CI), 0.17–0.47%] new mutations per CO (SI Appendix, Table S1). Approximately half occurred between the DSB and the CO breakpoint, with an average distance to the hotspot center of 348 nucleotides (Fig. 1 and SI Appendix, Fig. S2 and Table S1). Despite the rarity of COs, we estimated that the overall mutation rate at hotspots driven by COs is increased compared with genome-wide average mutation rates (Table 1). Given the dependency of the mutation rate at hotspots (µHS) with the CO frequency (shown in Table 1), the increase of mutations at hotspots is associated with the hotspot strength, with more active hotspots exerting a stronger mutagenic effect than weaker hotspots. This also assumes that the number of mutations per CO is similar among hotspots, which remains to be verified with a larger data set.
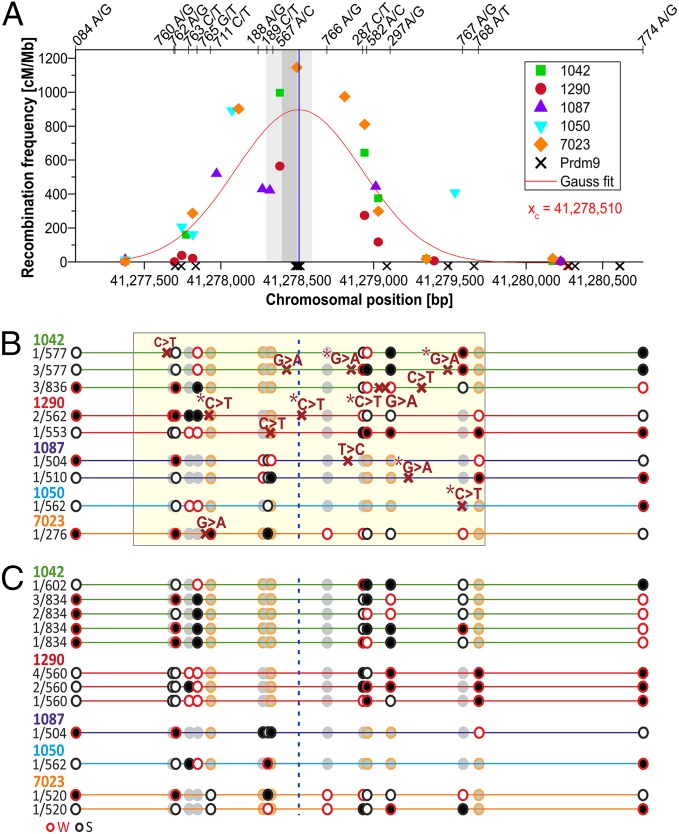
Fig. 1.
COs, mutations, and CCOs in HSI. (A) Distribution of both reciprocal COs in HSI (marks represent different donors). A best-fit normal distribution (Gaussian function) of the CO breakpoints represents the hotspot center at xc (vertical line), verified by data representing the DSB region (33), shaded in gray, and the Myers motif (allowing one mismatch or less) (1) represented as crosses on the x-axis. (B) Distribution of mutations. The sequenced region (yellow shaded area) shows the new mutations with red crosses (asterisk denotes a CpG) on different haplotypes (mutations per haplotype and donor identification shown on the left). Black and white circles denote heterozygous SNPs (red rim = AT-Weak alleles; black rim = GC-Strong alleles), and gray circles are homozygous SNPs. The vertical dotted line shows the hotspot center. (C) Distribution of CCOs. Different CCOs identified in the same donors as above (frequency of each CCO per haplotype to the left). CCOs are within 60 bp of another heterozygous site in 56% of the cases, suggesting conversion tracks in CCOs involved a single SNP, although the SNP involved cannot be determined unequivocally (SI Appendix, Table S4).
All the observed de novo mutations were transitions, occurring mainly at CpG sites, predominantly from strong (S) to weak (W) base pairs (S > W or CG > TA; Table 2). A detailed analysis of the mutational bias associated with COs for HSI reveals a significantly elevated S > W transition rate compared with the reciprocal rate W > S (Table 2; Fisher’s exact test, P = 5.5 × 10−4), which is also reflected in a significant bias of mutations at CpG versus non-CpG sites in COs by ∼37-fold (Fisher’s exact test, P = 2.5 × 10−8), an observation made also for genome-wide mutation averages [9.5–18.2-fold (27–29)].
Table 2.
Types of mutations in HSI CO products
| Mutation type | µCO | µNR | µCO* | µHS | µhAve | |
| S > W/W > S*1 | ||||||
| S > W | 2.31 × 10−6 | 6.00 × 10−7 | 1.71 × 10−6 | 2.91 × 10−8 | 1.21 × 10−8 | |
| 11 | ||||||
| W > S | 1.55 × 10−7 | 0.00 | 1.55 × 10−7 | 6.92 × 10−9 | 5.42 × 10−9 | |
| CpG/non-CpG*2 | ||||||
| CpG | 2.21 × 10−5 | 5.35 × 10−6 | 1.68 × 10−5 | 2.79 × 10−7 | 1.12 × 10−7 | |
| 37 | ||||||
| non-CpG | 5.96 × 10−7 | 1.44 × 10−7 | 4.52 × 10−7 | 1.06 × 10−8 | 6.18 × 10−9 |
CO mutation fractions (µCO) are mutation counts (n) per effective number of sequenced sites corrected by the NR fraction of the equivalent mutation type µNR resulting in µCO* and are estimated for strong (S: GC) to weak (W: AT) or vice versa transitions, CpG and non-CpG dinucleotides. The asterisk for *1 and *2 denotes a significant difference between the CO mutation fractions (Fisher’s exact test; P = 5.5 × 10−4 and 2.5 × 10−8, respectively).
Although the strong mutational bias at CpGs observed in our data may not be exclusive to COs, it could be explained by single-stranded DNA processing. CpG dinucleotides generally have high mutation rates resulting from spontaneous oxidative deamination of methylated cytosines (5-meC), and biochemical experiments have shown that they are ∼1,000 times more susceptible to deamination in single-stranded DNA than double-stranded DNA (ref. 30 and references within). Moreover, repair of deaminated 5-meC (equal to thymine) is initiated by the recognition of mismatched base pairs (G:T) in double-stranded DNA, which is not possible in single-stranded DNA (31). Thus, the higher mutation rate at CpGs in COs could be linked to the formation of single-stranded resected 3′-ends introduced after the DSB (Fig. 2A), extending as far as 2 kb from the DSB, as described in mice (32), and our hotspots (33). Methylation levels at CpG sites are very high in our hotspots (83–88%) in both testis and sperm, which represent the cellular states before and after meiosis, respectively (SI Appendix, Table S2). Because methylation levels stay constant throughout male spermatogenesis until shortly before fertilization, at least in mice (34), we assume that CpG sites in our hotspots were methylated throughout meiosis. Alternatively, the mutational bias could also be explained by the effect of translesion polymerases active during the repair of the DSB (21).
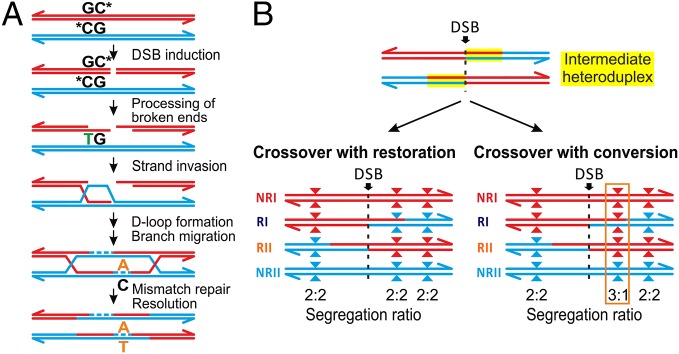
Fig. 2.
Model of CO-driven evolution. (A) Mutagenic model. Mutagenic activity of recombination could be associated to the deamination of methyl-C at a CpG site during 3′-end resection and single-stranded DNA formation, which introduces a thymine that remains unrepaired. In addition to deamination of CpG sites, translesion polymerases may also introduce mutations at hotspots (21) if the repair of heteroduplexes by the mismatch repair machinery active during meiosis is biased towards the newly synthesized strand. (B) gBGC. During the repair of DSBs, mismatches in intermediate heteroduplex tracts at polymorphic sites (triangles) can be either resolved restoring the original allele or can lead to gene conversion (gBGC) favoring GC alleles (red) versus AT alleles (blue). In the case of gBGC, more COs will have breakpoints with GC alleles distal to the DSB than proximal, distorting the segregation ratio of alleles between reciprocals.
Gene Conversion in CO Products Favor GC Alleles.
We also analyzed our large survey of COs for indications of allelic transmission bias. In principle, non-Mendelian segregation of alleles at hotspots arising during DSB repair could be either a result of an initiation bias, in which DSB-suppressing alleles are used to repair the broken homolog (refs. 35 and 36 and references in ref. 37), or gene conversion favoring GC-alleles, leading to gBGC (15). Patterns seen in our data (Fig. 3 and SI Appendix, Table S3) indicate a biased transmission in favor of GC alleles representative of gBGC, rather than of an initiation bias: (i) all of the donors are homozygous at the DSB site [determined by inferring the CO center fitting a Gaussian distribution on all of the CO breakpoints of the hotspot (Fig. 1) and by DSB break genome-wide maps (33)], making an initiation bias unlikely, and (ii) sites with the strongest evidence of unequal transmission favor strong (GC) versus weak (AT) alleles, with the exception of one case in which an insertion was favored at an InDel polymorphism. Further, initiation biases do not appear to favor strong over weak alleles in humans (35, 36) or in yeast (15), whereas gBGC does: mismatches in heteroduplexes produced during DSB repair have been shown to involve preferential repair favoring the GC allele up to 2 kb or up to 0.5 kb away from the DSB in yeast (13, 15) and mice (12), respectively. Our data further suggest a contribution of a repair system that produces short conversion tracks (e.g., short patch repair by base excision repair), rather than long conversion tracks (e.g., mismatch repair). The transmission bias we observe affects single SNPs independent of nearby informative SNPs (in most cases within ∼150 bp or less), with SNPs with preferential transmission sometimes closer to and sometimes further away from the DSB than nearby SNPs with equal transmission (Fig. 3). Moreover, base excision repair intrinsically favors GC alleles as a result of glycosylases excising thymine at DNA mismatches (8, 38), consistent with the direction of the observed gBGC.
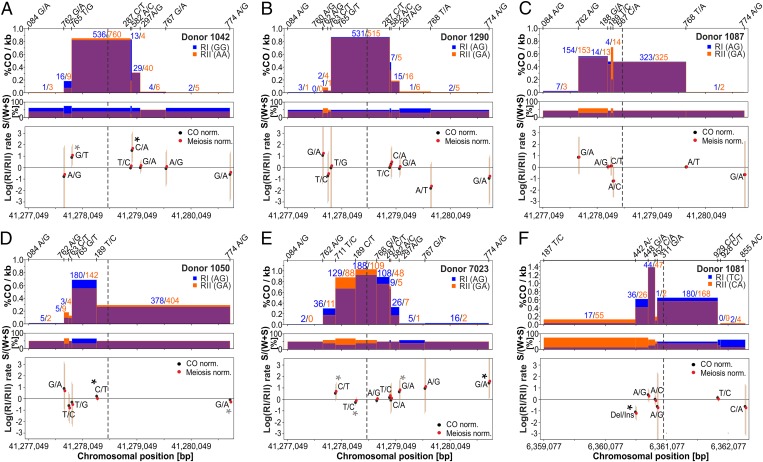
Fig. 3.
Transmission distortion between reciprocal COs. (Top) Frequencies of CO breakpoints are compared between reciprocals (blue and orange) in HSI (donors 1042, 1290, 1087, 1050, and 7023) and HSII (donor 1081), A–F, respectively, with numbers representing CO breakpoint counts (nRI vs. nRII) and the position of phased alleles of heterozygous SNPs of NRs shown on top. (Middle) Proportion of CG (S) alleles per heterozygous sites of the donor. (Lower) Log of the rate ratios of the different recombinant haplotypes, calculated as log[(nRI/totalRI)/(nRII/totalRII)], where the denominator is the total number of either COs (black) or meiosis (red) surveyed per reciprocal. Asterisks denote significant transmission distortion, based on the standardized Pearson residual (black asterisks denote the haplotype with the strongest evidence of heterogeneity; SI Appendix, Table S3). Note that for HSII, the largest skew occurred at an indel polymorphism of a homopolymeric run of six or seven consecutive As in donor 1081.
Other Sources of gBGC: Complex COs Are Also Biased for S over W Conversions.
Another potential source of gBGC in the 6,085 mapped COs is the formation of complex COs (CCOs), observed as COs with rare, discontinuous conversion tracks containing two CO breakpoints (Fig. 1C and SI Appendix, Fig. S2C). We measured 0.41% (95% Poisson CI, 0.26–0.60%) CCOs out of all COs and 0.35% for HSI alone (95% Poisson CI, 0.21–0.54), consistent with the previous estimate of 0.33% obtained for human sperm (39) (SI Appendix, Table S4). The observed CCOs were located on average ∼505 bp from the hotspot center (range, 169–1,073 bp). The exact length of the conversion tracts is difficult to estimate, but it is likely that the CCOs are short, as evidenced by those cases in which neighboring SNPs, as close as 60 or 26 bp apart, are differentially affected (56% or 40% of the cases, respectively). Although the converted SNP involved cannot be unambiguously determined, we can estimate the fraction involving conversion to strong versus weak alleles. If we assume the CCOs occurred in the more frequent CO type, then all CCOs involved conversion of a weak to a strong allele; if we consider all possibilities, 87% of the CCOs favored the strong allele in HSI (SI Appendix, Table S4). Thus, CCOs could also be another source of gBGC in humans. In HSII, the only CCOs that occurred involved either the InDel site (A7/A6) or the adjacent SNP, 207 bp away (SI Appendix, Fig. S2C and Table S4). Intriguingly, more CCOs seemed to accumulate in one reciprocal CO product than in the other for two of our donors (SI Appendix, Fig. S1C); although a larger data set is needed to validate this trend, inspection of previously published data also reveals an uneven distribution of CCOs between reciprocals (39).
Opposing Effects of gBGC and Mutation.
The tendency of recombination to increase GC content via gBGC opposes the AT-biased mutagenic activity of meiosis. To examine the relative contribution of these factors, we first quantitatively estimated the effect of gBGC in our survey of COs (excluding CCOs). We assumed the following model for gBGC occurring in COs: in the absence of gBGC, both recombination products, formed by the exchange of flanking regions of the double Holliday junction, are produced with equal probability. The number of CO breakpoints will decrease as the distance from the hotspot center increases, but in the absence of gBGC, this reduction should occur equally for breakpoints on either side of both strong and weak alleles. During the processing of the DSB, heteroduplex tracts form in polymorphic regions, and if there is gBGC acting on these heteroduplex tracts, COs will tend to include strong alleles and exclude weak alleles, thus appearing as COs with a more distal breakpoint to the DSB for strong alleles (or with a more proximal breakpoint for weak alleles), distorting transmission ratios (Fig. 2B and SI Appendix, Fig. S5). By comparing CO breakpoints proximal and distal to strong versus weak alleles in a contingency table analysis (Materials and Methods), we find that an excess of recovered COs include strong alleles, as assessed by the Cochrane Mantel Haenszel test for strong versus weak alleles, with a weighted odds ratio for HSI of 1.203 (95% CI, 1.035–1.398; χ2 = 5.67; df = 1; P = 0.017). We calculated that the GC alleles at polymorphic sites in HSI would be favored by 52.3% in sperm with a CO, instead of the expected 50%, or ∼50.023% including NRs, assuming both males and females show the same CO rate and bias (SI Appendix, Materials and Methods). The transmission advantage resulting from gBGC estimated for the ∼2-kb sequenced region of HSI is 4.62 × 10−4. Note that this estimate depends on the CO frequency at this hotspot, and thus may be unusually strong (SI Appendix, Materials and Methods). However, it is similar to that measured for gene conversions in noncrossovers (NCOs) in humans [3.34 × 10−4 (37)], but lower than that obtained for yeast [1.3 × 10−2 (15)].
Although the gBGC effect appears small, it could have a profound effect on equilibrium GC content, despite the elevated rate of AT-biased mutations at hotspots. To investigate this, we evaluated the contribution of mutagenesis and gBGC on the sequence composition at hotspots. Considering our mutation and gBGC estimates for HSI and assuming that hotspots exist indefinitely, we predict a neutral GC content at equilibrium close to 100% (SI Appendix, Materials and Methods), demonstrating that gBGC is the dominant factor. The strength of the hotspot plays an important role here: gBGC and mutagenesis have little influence on equilibrium GC at low CO frequencies. When CO frequencies are ∼0.1 cM/Mb or lower, equilibrium GC content is determined only by genome-wide average mutational biases (SI Appendix, Fig. S3A). The equilibrium GC content under gBGC is likely never reached, as the recombination initiation machinery involves PRDM9 acting in a sequence-specific manner (1, 32, 35), and sequence erosion caused by mutagenesis and gBGC likely affects PRDM9 binding, and thus hotspot intensity over time, consistent with the short lifespan of hotspots (40). How fast crossing over drives the decimation of a hotspot via mutagenesis and gBGC depends on how sequence changes affect CO frequencies, which is still a mystery.
Conclusions
We demonstrate that crossing over is an important source of new mutations and gBGC at recombination hotspots associated with DSB repair. If, as we speculate here, the formation of single-stranded DNA at methylated CpG sites is the main driver for de novo mutations, then DSBs resolved alternatively as NCOs might also experience a higher mutation frequency. Because NCO are also subject to gBGC (37), the overall effect of NCOs is expected to be similar to the one observed for COs. Our results thus contribute to the understanding of the long-term evolutionary dynamics of sequence composition at recombination hotspots. In particular, they suggest that gBGC is the dominant force shaping the nucleotide composition at hotspots during crossing over, and potentially also in other recombination products, which might explain the high GC content associated with recombination (8). Finally, given the opposing effects of mutation and gBGC on base composition, it is possible that gBGC is an adaptation to reduce the mutational load of recombination (or DSB), as has been previously suggested (8, 41).
Materials and Methods
Sample Collection and Preparation.
Human samples were collected from anonymous donors by informed consent approved by the ethics commission of Upper Austria (Approval: F1-11). Sperm DNA was prepared as described previously (42) and was measured for quality and quantity with a spectrophotometer. In brief, DNA was extracted from ∼106 sperm cells (or 35 mg testis biopsy), using the Gentra Puregene Cell Kit (Qiagen), with the addition of 24 µM DTT (Sigma-Aldrich) and 60 µg/mL proteinase K (Qiagen) during the cell lysis step, followed by an overnight incubation at 37 °C; 1 µL glycogen solution (Qiagen) was added during DNA precipitation. For mixing, all vortexing steps were replaced by repeated inversion of tubes to avoid DNA damage from shear forces.
Identification of Informative Donors.
To collect CO products of a selected hotspot region, informative donors (heterozygotes) were identified by genotyping SNPs selected from the SNP database in the National Center for Biotechnology Information (NCBI) with a high heterozygosity. Informative donors were identified by genotyping all donors for at least 4 SNPs flanking the recombination hotspot. Additional genotyping of up to 8 SNPs lying within the recombination hotspot was performed. Genotyping was carried out in-house by real-time PCR (CFX384 System, Bio-Rad), as described previously (26). In brief, allele-specific primers were used, with the last three phosphodiester bonds at the 3′-end substituted by phosphorothioate bonds to increase allele specificity. PCRs for genotyping were carried out in a volume of 10 µL, using OneTaq DNA Polymerase (NEB) or Phusion Hot Start II High-Fidelity DNA Polymerase (ThermoFisher Scientific). Reactions contained 5 µL total genomic DNA (blood or sperm) (2 ng/µL), 0.2 µM allele-specific primer, 0.2 µM outer primer, 1× SYBR Green I (Invitrogen), and either 1× OneTaq Reaction Buffer (NEB), 0.125 U OneTaq Hot Start DNA polymerase, 0.2 mM dNTPs, and 2.5 mM MgCl2 or 1× Phusion HF Buffer (ThermoFisher Scientific), 0.1 U Phusion Hot Start II High-Fidelity DNA Polymerase, and 0.16 mM dNTPs. The reactions were carried out with an initial heating step of 95 °C for 2 min, followed by 45 cycles at 95 °C for 30 s, annealing temperature for 30 s, and 68 °C for 15 s when using OneTaq Hot Start DNA polymerase or with 98 °C for 30 s, followed by 45 cycles at 98 °C for 5 s, annealing temperature for 15 s, and 72 °C for 5 s when using Phusion Hot Start II High-Fidelity DNA Polymerase. Two reactions were amplified for each sample, one for each allele, differing only by the allele-specific primer. Primer sequences and specific annealing temperatures are shown in SI Appendix, Table S6.
The different haplotypes (phase of alleles) of the four flanking SNPs were determined using long-range allele-specific PCR (26). Sixteen reactions were set up covering all possible combinations of the allele-specific primers (for primer sequences, see SI Appendix, Materials and Methods). Reactions contained 50 ng genomic DNA (blood or sperm), 0.5 µM each allele-specific primer, 1× SYBR Green I (Invitrogen), 1× Expand Long Range Buffer with MgCl2, and 0.35 U Expand Long Range Enzyme Mix (Roche). The reactions were carried out with an initial heating step of 92 °C for 2 min, followed by 55 cycles at 92 °C for 10 s, 60 °C (for HSI)/57 °C (for HSII) for 15 s, and 68 °C for 270 s.
Collecting Single COs and NRs.
CO products were collected for six donors (five from HSI on chromosome 21 and one from HSII on chromosome 16). All donors were of European descent ranging in age from 27 to 40 years and were carriers of PRDM9 allele A. Allele-specific primers were selected on the basis of SNPs flanking the hotspot informative for the largest number of donors. The collection of CO products included two rounds of nested PCR with allele-specific primers that preferentially amplified the single recombinant over the excess of NR sperm genomes, as described previously (26). Genomic sperm DNA molecules were prepared to a dilution of 0.2 CO products on average, based on single-molecule Poisson distribution, to minimize the number of reactions with more than one CO per aliquot to less than 2%. For all donors (except 1042 and 1050), 1 µg sperm DNA was treated with 1.6 U Fpg (NEB) in a reaction volume of 10 µL at 37 °C for 1 h 30 min, followed by a 1:10 dilution and a treatment with 0.5 U USER Enzyme (NEB) at 37 °C for 30 min before amplification to reduce oxidated and deaminated bases in the sperm DNA. Both enzyme mixes produce abasic sites and single-stranded breaks at 8-oxoguanine or uracil sites, respectively, rendering the template unamplifiable.
The first round of PCR was carried out in a volume of 10 µL. Reactions contained genomic DNA from 100 to 600 total sperm heads (quantified via a spectrophotometer), 0.25 µM of the appropriate forward and reverse allele-specific primer, 0.16 mM dNTPs, 1× Phusion HF Buffer (ThermoFisher Scientific), and 0.1 U Phusion Hot Start II High-Fidelity DNA Polymerase, a thermostable polymerase with the lowest reported error rate (43); primer sequences; and cycling conditions are shown in SI Appendix, Materials and Methods. The second round of PCR was carried out in a volume of 20 µL with the same components as above, but with an aliquot of 1 or 2 µL of the first PCR instead of genomic DNA and 1× EvaGreen (Jena Bioscience) in a real-time PCR system (CFX384 System, Bio-Rad). Reactions for the first and second round of PCR were set up in different laminar flow hoods located in separate rooms to avoid carry-over contamination.
NRs were collected using the same PCR conditions as for COs. NR sperm aliquots were prepared at single-molecule dilution such that ∼20% of the reactions contained, on average, one NR genome in a pool of 100–600 sperm or blood genomes of another donor with the recombinant haplotype. Given that for each donor we collected NRs at a single-molecule dilution, we used the number of positive reactions from these experiments (containing initially a single amplifiable NR) to estimate based on the Poisson distribution the number of amplifiable NRs of several independent experiments for each chromosome per donor. We then calculated the deviation from the input molecules (quantified via a spectrophotometer) from the estimated number of amplifiable NR and used a correction factor to adjust the number of meiosis in the CO frequency estimates (SI Appendix, Table S5). For donor 7023, we used an average correction factor derived from the other four donors of HSI.
In most cases, either both reciprocal COs or one reciprocal and NR control were amplified in the same experiment prepared with the same mastermix. Experiments included, on average, 180 reactions per CO or NR type, two to four no-template controls, and ∼10 negative controls for each collected sample type of only blood DNA from the same donor (or a donor with the same haplotype) to monitor the specificity of the amplification.
Sequencing and Mutation Analysis.
We focused our search for mutations around the center of the hotspot because this region harbors the DSBs and most of the recombination exchanges (36, 44). Amplicons were sequenced using standard capillary Sanger sequencing in a 96-well format (by LGC Genomics GmbH), using three overlapping sequencing reactions covering 2,300 and 2,100 bp of the HSI and HSII regions, respectively; sequence read lengths were ∼800 bp (primer sequences are shown in SI Appendix, Table S7). All of the 20 µL of the second-round amplification reaction was cleaned with PEG before sequencing, and an aliquot of ∼5 µL was sequenced with BigDye 3.1.
Chromatograms were analyzed for new mutations using the Mutation Surveyor package (45). The NCBI sequence of the selected hotspot region and a consensus chromatogram of all of the sequencing reads of one experiment were used as a reference sequence to identify a mutation. A chromatogram peak with a different base from the reference was called a mutation if it exceeded a certain threshold in both forward and reverse sequencing reads. The threshold was determined on the basis of the overlap, signal-to-noise ratio, quality score (0–100), and drop (fraction of alternate nucleotides in a peak) of the chromatograms and was used by the Mutation Surveyor package to categorize alternate chromatogram peaks as homogenous or heterogenous (SI Appendix, Fig. S6). We only used the homogenous peaks as bona fide mutations (drop averaged among two to four measurements of 0.98) because heterogeneous peaks (drop, <0.85; average, ∼0.55) likely represented sequencing or PCR artifacts, given that the sequenced templates were derived from an initial single CO. In total, we observed nine heterogeneous peaks in 5,796 COs, and six in 3,672 NRs that do not significantly differ between COs and NRs (Fisher’s exact test, P = 1). Identified mutations were verified by both sequencing in both directions and repeating two to four times the second PCR for CO collection and sequencing again.
CpG Methylation Analysis.
CpG methylation levels of 11 CpG sites lying within HSI were analyzed using bisulfite sequencing. Sperm DNA of donors 1042 and 1050 and testis DNA from an additional donor were converted using the EZ DNA Methylation-Lightning Kit (Zymo Research) according to manufacturer’s instructions. Five hundred nanograms genomic DNA were used for bisulfite treatment and further amplified either with TaKaRa Ex Taq Hot Start DNA polymerase (Takara) (Region 1 → CpG #1+2) or Platinum Taq DNA Polymerase (Region 2 → CpG #3+4 and Region 3 → CpG #5–11). Reactions contained 0.5 µL converted DNA, 0.2 µM of the appropriate forward and reverse primer (SI Appendix, Table S8), 0.2 mM dNTPs, 1× EvaGreen Fluorescent DNA Stain, 1× Ex Taq Buffer or 1× PCR buffer + 1.5 mM MgCl2, and 0.25 U TaKaRa Ex Taq HS or 0.2 U Platinum Taq DNA Polymerase. The reactions were carried out with an initial heating step of 94 °C for 2 min, followed by 45 cycles at 94 °C for 30 s, 60 °C (region 1)/55 °C (region 2+3) for 30 s, and 68 °C for 40 s. Sequences of the PCR products were obtained by Sanger sequencing, and methylation levels were estimated by assessing the sequence with the Mutation Surveyor software (45).
Estimation of gBGC in Simple COs.
BGC associated with COs can occur on either side of the DSB during the repair of heteroduplex tracts. In contrast, GC-biased repair will result in more CO breakpoints occurring distal to GC alleles from the hotspot center and proximal to AT alleles. The reason is that at a site segregating for GC and AT alleles, repair of a heteroduplex to the GC allele would yield a CO breakpoint distal from the DSB for the GC allele (or proximal for the AT allele; Fig. 2B and SI Appendix, Fig. S5). To test for such an effect, we assumed that COs start with a DSB at the center of the hotspot, estimated according to the maximum of the Gaussian distribution of all of the CO breakpoints of the analyzed donors for that hotspot (for HSI, near chr21: 41278510; for HSII, near chr16: 6361054), verified by DSBs measurements (33). We analyzed sites segregating for weak versus strong alleles, focusing on COs ending immediately proximal and distal of each SNP, which best represent the effect of alleles segregating at that site. We compared the ratio of COs occurring proximal and distal from the strong allele with those ending proximal and distal from the weak allele (SI Appendix, Table S9) in a contingency table analysis, using the Cochrane Mantel Haenszel test (46) implemented in the rma.mh function in the metafor package for R. This test is an extension of the χ2 test, for which an overall odds ratio is calculated for multiple 2 × 2 contingency tables, weighted by the amount of data in each table. The odds ratio was calculated so that a value over 1 indicates a preference for the strong allele; that is, odds ratio equals strong(distal/proximal)/weak(distal/proximal). Qualitatively similar results were obtained using Tarone’s estimator (47). The calculations for estimating gBGC from this analysis and simulations testing its validity are detailed in SI Appendix, Materials and Methods and Fig. S3, respectively.
Supplementary Material
Acknowledgments
We are very thankful to B. Charlesworth, P. Keightley, C. Vogl, C. Huber, and N. Arnheim for helpful discussions and A. Futschik for his input in the biostatistical analysis. This work was supported by the Austrian Science Fund (Grant P23811-B12, to I.T-B.), the doctoral (DOC) Fellowship of the Austrian Academy of Sciences at the Institute of Biophysics Johannes Kepler University (to B.A.), and a Career Track Fellowship from the Vetmeduni Vienna (to A.J.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. L.D.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416622112/-/DCSupplemental.
References
- 1.Myers S, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327(5967):876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montgomery SB, et al. 1000 Genomes Project Consortium The origin, evolution, and functional impact of short insertion-deletion variants identified in 179 human genomes. Genome Res. 2013;23(5):749–761. doi: 10.1101/gr.148718.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lercher MJ, Hurst LD. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 2002;18(7):337–340. doi: 10.1016/s0168-9525(02)02669-0. [DOI] [PubMed] [Google Scholar]
- 4.Nachman MW. Single nucleotide polymorphisms and recombination rate in humans. Trends Genet. 2001;17(9):481–485. doi: 10.1016/s0168-9525(01)02409-x. [DOI] [PubMed] [Google Scholar]
- 5.Spencer CC, et al. The influence of recombination on human genetic diversity. PLoS Genet. 2006;2(9):e148. doi: 10.1371/journal.pgen.0020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duret L, Arndt PF. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008;4(5):e1000071. doi: 10.1371/journal.pgen.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hellmann I, et al. Why do human diversity levels vary at a megabase scale? Genome Res. 2005;15(9):1222–1231. doi: 10.1101/gr.3461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- 9.Cutter AD, Payseur BA. Genomic signatures of selection at linked sites: Unifying the disparity among species. Nat Rev Genet. 2013;14(4):262–274. doi: 10.1038/nrg3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Webster MT, Hurst LD. Direct and indirect consequences of meiotic recombination: Implications for genome evolution. Trends Genet. 2012;28(3):101–109. doi: 10.1016/j.tig.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Auton A, et al. A fine-scale chimpanzee genetic map from population sequencing. Science. 2012;336(6078):193–198. doi: 10.1126/science.1216872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clément Y, Arndt PF. Meiotic recombination strongly influences GC-content evolution in short regions in the mouse genome. Mol Biol Evol. 2013;30(12):2612–2618. doi: 10.1093/molbev/mst154. [DOI] [PubMed] [Google Scholar]
- 13.Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454(7203):479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capra JA, Pollard KS. Substitution patterns are GC-biased in divergent sequences across the metazoans. Genome Biol Evol. 2011;3:516–527. doi: 10.1093/gbe/evr051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lesecque Y, Mouchiroud D, Duret L. GC-biased gene conversion in yeast is specifically associated with crossovers: Molecular mechanisms and evolutionary significance. Mol Biol Evol. 2013;30(6):1409–1419. doi: 10.1093/molbev/mst056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kane DP, Shusterman M, Rong Y, McVey M. Competition between replicative and translesion polymerases during homologous recombination repair in Drosophila. PLoS Genet. 2012;8(4):e1002659. doi: 10.1371/journal.pgen.1002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strathern JN, Shafer BK, McGill CB. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140(3):965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deem A, et al. Break-induced replication is highly inaccurate. PLoS Biol. 2011;9(2):e1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rattray AJ, McGill CB, Shafer BK, Strathern JN. Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: A role for SAE2/COM1. Genetics. 2001;158(1):109–122. doi: 10.1093/genetics/158.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hogg M, Sauer-Eriksson AE, Johansson E. Promiscuous DNA synthesis by human DNA polymerase θ. Nucleic Acids Res. 2012;40(6):2611–2622. doi: 10.1093/nar/gkr1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arbel-Eden A, et al. March 11, 2013. Trans-lesion DNA Polymerases may be Involved in Yeast Meiosis. G3 (Bethesda), 10.1534/g3.113.005603.
- 22.Schaibley VM, et al. The influence of genomic context on mutation patterns in the human genome inferred from rare variants. Genome Res. 2013;23(12):1974–1984. doi: 10.1101/gr.154971.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berglund J, Pollard KS, Webster MT. Hotspots of biased nucleotide substitutions in human genes. PLoS Biol. 2009;7(1):e26. doi: 10.1371/journal.pbio.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodgkinson A, Eyre-Walker A. Variation in the mutation rate across mammalian genomes. Nat Rev Genet. 2011;12(11):756–766. doi: 10.1038/nrg3098. [DOI] [PubMed] [Google Scholar]
- 25.Frazer KA, et al. International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tiemann-Boege I, Calabrese P, Cochran DM, Sokol R, Arnheim N. High-resolution recombination patterns in a region of human chromosome 21 measured by sperm typing. PLoS Genet. 2006;2(5):e70. doi: 10.1371/journal.pgen.0020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campbell CD, et al. Estimating the human mutation rate using autozygosity in a founder population. Nat Genet. 2012;44(11):1277–1281. doi: 10.1038/ng.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong A, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488(7412):471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156(1):297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen JC, Rideout WM, 3rd, Jones PA. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res. 1994;22(6):972–976. doi: 10.1093/nar/22.6.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neddermann P, Jiricny J. Efficient removal of uracil from G.U mispairs by the mismatch-specific thymine DNA glycosylase from HeLa cells. Proc Natl Acad Sci USA. 1994;91(5):1642–1646. doi: 10.1073/pnas.91.5.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smagulova F, et al. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011;472(7343):375–378. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pratto F, et al. DNA recombination. Recombination initiation maps of individual human genomes. Science. 2014;346(6211):1256442. doi: 10.1126/science.1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lees-Murdock DJ, Walsh CP. DNA methylation reprogramming in the germ line. Epigenetics. 2008;3(1):5–13. doi: 10.4161/epi.3.1.5553. [DOI] [PubMed] [Google Scholar]
- 35.Berg IL, et al. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 2010;42(10):859–863. doi: 10.1038/ng.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeffreys AJ, Neumann R. Reciprocal crossover asymmetry and meiotic drive in a human recombination hot spot. Nat Genet. 2002;31(3):267–271. doi: 10.1038/ng910. [DOI] [PubMed] [Google Scholar]
- 37.Odenthal-Hesse L, Berg IL, Veselis A, Jeffreys AJ, May CA. Transmission distortion affecting human noncrossover but not crossover recombination: A hidden source of meiotic drive. PLoS Genet. 2014;10(2):e1004106. doi: 10.1371/journal.pgen.1004106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hardeland U, Bentele M, Jiricny J, Schär P. The versatile thymine DNA-glycosylase: A comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res. 2003;31(9):2261–2271. doi: 10.1093/nar/gkg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Webb AJ, Berg IL, Jeffreys A. Sperm cross-over activity in regions of the human genome showing extreme breakdown of marker association. Proc Natl Acad Sci USA. 2008;105(30):10471–10476. doi: 10.1073/pnas.0804933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeffreys AJ, Neumann R. The rise and fall of a human recombination hot spot. Nat Genet. 2009;41(5):625–629. doi: 10.1038/ng.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Birdsell JA. Integrating genomics, bioinformatics, and classical genetics to study the effects of recombination on genome evolution. Mol Biol Evol. 2002;19(7):1181–1197. doi: 10.1093/oxfordjournals.molbev.a004176. [DOI] [PubMed] [Google Scholar]
- 42.Meyer WK, et al. Evaluating the evidence for transmission distortion in human pedigrees. Genetics. 2012;191(1):215–232. doi: 10.1534/genetics.112.139576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boulanger J, Muresan L, Tiemann-Boege I. Massively parallel haplotyping on microscopic beads for the high-throughput phase analysis of single molecules. PLoS ONE. 2012;7(4):e36064. doi: 10.1371/journal.pone.0036064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485(7400):642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minton JA, Flanagan SE, Ellard S. Mutation surveyor: Software for DNA sequence analysis. Methods Mol Biol. 2011;688:143–153. doi: 10.1007/978-1-60761-947-5_10. [DOI] [PubMed] [Google Scholar]
- 46.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 47.Tarone RE. On summary estimators of relative risk. J Chronic Dis. 1981;34(9-10):463–468. doi: 10.1016/0021-9681(81)90006-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.