Significance
Influenza A viruses (IAVs) are a cause of major morbidity in the human population. Being RNA viruses, replication is error prone, and proteins such as viral envelope hemagglutinin rapidly mutate. Current vaccines stimulate antibodies targeting exposed virion proteins but require annual reformation due to constant sequence variation. In contrast, vaccines that stimulate CD8 T cells directed at conserved peptides from internal proteins would offer stable immunity if these peptides are displayed by HLA proteins on infected cells. Currently, functional readouts infer the IAV peptides displayed. Using new MS technology, epitopes on infected human HLA-A2+ lung epithelium are identified and abundances characterized. The data show interconnections between viral evasion, immunodominance, and stealth responses that will aid in developing cellular vaccines against influenza.
Keywords: mass spectrometry, T-cell epitopes, influenza A viruses, antigen presentation, T-cell avidity
Abstract
Vaccines eliciting immunity against influenza A viruses (IAVs) are currently antibody-based with hemagglutinin-directed antibody titer the only universally accepted immune correlate of protection. To investigate the disconnection between observed CD8 T-cell responses and immunity to IAV, we used a Poisson liquid chromatography data-independent acquisition MS method to physically detect PR8/34 (H1N1), X31 (H3N2), and Victoria/75 (H3N2) epitopes bound to HLA-A*02:01 on human epithelial cells following in vitro infection. Among 32 PR8 peptides (8–10mers) with predicted IC50 < 60 nM, 9 were present, whereas 23 were absent. At 18 h postinfection, epitope copies per cell varied from a low of 0.5 for M13–11 to a high of >500 for M158–66 with PA, HA, PB1, PB2, and NA epitopes also detected. However, aside from M158–66, natural CD8 memory responses against conserved presented epitopes were either absent or only weakly observed by blood Elispot. Moreover, the functional avidities of the immunodominant M158–66/HLA-A*02:01-specific T cells were so poor as to be unable to effectively recognize infected human epithelium. Analysis of T-cell responses to primary PR8 infection in HLA-A*02:01 transgenic B6 mice underscores the poor avidity of T cells recognizing M158–66. By maintaining high levels of surface expression of this epitope on epithelial and dendritic cells, the virus exploits the combination of immunodominance and functional inadequacy to evade HLA-A*02:01-restricted T-cell immunity. A rational approach to CD8 vaccines must characterize processing and presentation of pathogen-derived epitopes as well as resultant immune responses. Correspondingly, vaccines may be directed against “stealth” epitopes, overriding viral chicanery.
Human influenza is an acute respiratory infection caused by Orthomyxoviridae, a family of single-stranded, negative sense RNA viruses containing a segmented genome. The influenza virion is enveloped by a lipid bilayer derived from the host cell with hemagglutinin (HA), neuraminidase (NA), and matrix 2 (M2) transmembrane proteins of the virus exposed and antibody (Ab) accessible. HA is the most abundant protein on the surface and is the principle antigen recognized by humoral immunity (1). RNA replication is error prone, with influenza A virus (IAV) averaging roughly one error for each replicated genome (2). Notably, the HA protein manifests a high functional tolerance to sequence variation not evident in some of the internal proteins (3). Current vaccines protect largely by inducing Abs against the HA and NA proteins but require continual reformulation based on inferring the dominant strains that will circulate in the upcoming flu season, an imperfect science at best. Recognizing the challenges for universal protection against influenza via antibodies, there has been extensive discussion about vaccines harnessing cellular immunity (4–6). Cytotoxic T lymphocytes (CTLs), primarily CD8+ T cells, can recognize and kill infected cells when their T-cell receptors (TCRs) recognize fragments of viral proteins that are in turn bound to major histocompatibility complex (MHC in general; for humans, HLA) proteins on the surface of infected cells. These peptides can derive from segments of internal proteins conserved among IAV strains so that CTLs targeting these sequence-constrained peptides would remain effective as surface HA and NA antigens, in contrast, change by genetic drift or shift. There is substantial evidence for T-cell–mediated protection in mice (7, 8), but qualified evidence in humans (9, 10). The limited CTL protection has been ascribed to slow cellular responses against IAV, a latency reflecting the expansion and activation of central memory T cells in lymph nodes and the subsequent recruitment of CTLs to infected lung (11). The recent identification of CD8+ effector resident memory T cells (TRM) within barrier tissues postinfection (12, 13) could challenge this picture. However, vaccines for cellular immunity are at an early stage of development and what formulations and delivery methods are needed to harness TRM immunity is not well understood. In particular, the pathogen-derived peptides bound to HLA molecules on the surface of IAV-infected lung epithelial cells are unknown. As a T cell monitors a single MHC-restricted antigen, surveillance is constrained by the numbers of antigen-specific cells and their motility in lung parenchyma under homeostatic conditions. Sentinel TRM cell populations lodged in barrier tissues are limited, so that inducing irrelevant specificities will necessarily displace those with useful specificities. In contrast, the fluid volume surrounding a cell at the site of infection can contain a large set of antibodies recognizing diverse antigens. Rational T-cell vaccine development must focus on the peptides directly presented by infected lung epithelium and identify those peptides that can be recognized by high avidity TRM cells.
IAV peptides displayed by infected cells are conventionally identified by T-cell functional assays. In principle this “reverse immunology” only identifies what epitopes the antigen-experienced host has recognized, not what can or should be recognized. As such, reverse immunology is a poor guide for vaccine development. Here new acquisition and analysis methods in mass spectrometry (MS) are applied to directly identify these peptides. New methodology is necessary as conventional data-dependent acquisition (DDA) MS, identifying ions largely in order of signal intensity, cannot plow deep enough into the sample to uncover the IAV peptides underneath an ocean of “self” peptides. Reflecting the prevalence of the HLA-A2 allele in the population and the large number of studies of IAV infection and CD8+ T-cell responses restricted by this allele, this study focuses on antigen presentation by HLA-A*02:01 (A2). Our results identify previously unrecognized IAV epitopes that may be useful in vaccine formulations, question the veracity of functionally ascribed epitope display, and show via stable isotope labeling by amino acids in culture (SILAC) that positive strand RNA-derived translation and not protein cross-presentation is the basis of A2 IAV peptidome array on dendritic cells (DCs) that phagocytose non-A2 IAV-infected UV-irradiated cells. In addition, our data rationalize why an immunodominant CD8 T-cell response to the highly conserved M158–66 peptide GILGFVFTL does not provide sterilizing immunity to IAV.
Results
IAV Peptides Bound to A2 Can Be Detected from a Few Million Influenza A Infected Cells by MS.
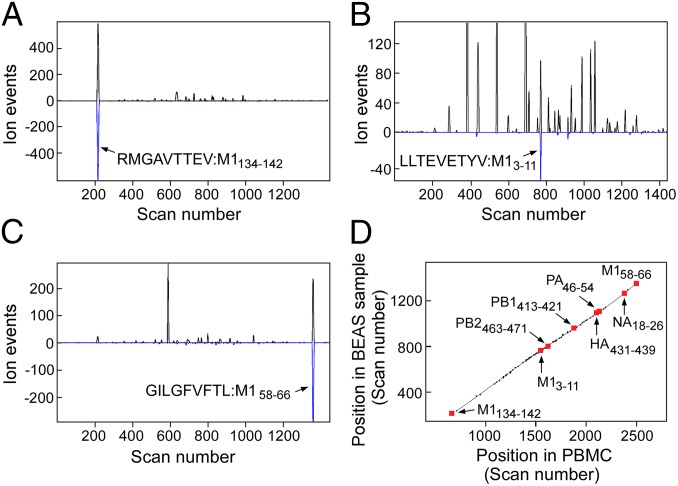
Two orthogonal MS methods, Poisson nanospray MS3 (14) and a Poisson liquid chromatography data-independent acquisition (Poisson LC-DIAMS) are applied. In nanospray MS3, the sample is not separated by chromatography but directly loaded into the nanospray emitter. For each target to be detected, the instrument generates a fragment spectrum of a selected ion fragment of a selected precursor ion (an MS3 spectrum). LC-DIAMS uses online LC and transmits a sequence of overlapping bands of ions into a collision cell at high energy (to collect a set of MS2 spectra) and a single wide band at low energy (to collect an MS spectrum). Spectra are collected in cycles throughout the LC elution. Both methods use a Poisson metric to detect a reference fragmentation pattern embedded in a background of ion fragments. In MS3 detection the target's pattern is embedded in an MS3 spectrum; in LC-DIAMS, the pattern is embedded in a series of band MS2 spectra covering the target's elution. Broadly, targeted detection using MS3 or MS2 signatures enhances detection sensitivity. For MS3, the pair of selected m/z values is specific to a target so that acquired data can only be used for different targets in the rare event they share the same pair of selection values. In contrast, LC-DIAMS data acquisition is independent of targeting, as the overlapping MS2 bands are set to cover the entire m/z range. Collected data can be analyzed at future times for new targets, pending acquisition of the target's reference pattern. Detecting IAV peptides from a few million infected cells requires the highest MS sensitivity. As electrospray MS sensitivity is largely determined by analyte concentration, ultralow flow LC (10 nL/min) is implemented using 20-μm monolithic columns (Materials and Methods) with direct pressure-bomb loading of the column. Poisson detection enhances discrimination of an arrival rate pattern when ion counts are low, signatures are dominated by sampling noise, and background chemical noise obscures individual fragment ion chromatograms.
For Poisson MS3 and LC-DIAMS detection, 31 calculated high A2 binding peptides (15) from the proteome of H1N1 viral strain A/Puerto Rico/8/1934 (PR8) were synthesized (SI Appendix, Table S1, excluding AIMDKNIIL and ILGFVFTL). Reference patterns were acquired from MS3 and LC-DIAMS runs of the synthetic peptides. For elution mapping, the synthetic peptides were added to an extract of A2 peptides from uninfected cells and this sample was run by LC-DIAMS. Elution positions of the synthetic IAV peptides along with the positions of a reference set of abundant endogenous peptides in this run were recorded. To detect IAV peptides, A2 complexes were affinity purified from lysates of 1–5 million PR8-infected bronchial epithelial airway BEAS-2B-A2 (BEAS) cells. Peptides eluted from the complexes with acid (Material and Methods) were analyzed by MS3 or LC-DIAMS. Seven IAV peptides were identified using reference patterns from the set of 31 synthesized peptides by Poisson nanospray MS3 (SI Appendix, Fig. S9) and the same peptides plus M1134–142 (RMGAVTTEV) were detected by Poisson segmented LC-DIAMS (SI Appendix, Fig. S10). MS3 of synthetic RMGAVTTEV had shown detection would be heavily compromised by dominating neutral losses in ion fragmentation and the experiment was not attempted. Poisson detection for three peptides from the IAV M1 protein using segmented LC-DIAMS data of 5 million infected BEAS cells is shown in Fig. 1 A–C. The combination of a dominant Poisson peak (fragmentation pattern), coincident XIC peak (precursor m/z), and placement of the coincidence on an elution curve with a low degree of scatter provides high confidence detection. Eight of the 31 synthesized IAV peptides are detected by LC-DIAMS (SI Appendix, Fig. S10). Negative Poisson detection for the 23 remaining synthesized peptides in the PR8-infected BEAS sample are shown in SI Appendix, Fig. S11. SI Appendix, Fig. S12 shows a negative control (naïve or uninfected BEAS cells) for the 8 IAV peptides that were detected in infected cells. IAV strain A/X-31(H3N2) contains the same core proteins as PR8 but different HA and NA surface proteins. Poisson detection chromatograms for a sample of 18-h A/X-31 infection of 1.67 million BEAS cells and the corresponding elution map for the detected peptides identifies the expected epitopes from the common core proteins, whereas the NA and HA epitopes are absent (SI Appendix, Fig. S13). SI Appendix, Fig. S14 shows the PR8 M1, PB1, and PA epitopes are detected from 18 h A/Victoria/3/1975 H3N2 infection but now the PB2463–471 epitope is also absent. The M158–66, M13–11, PB1413–421, PB2463–471, NA18–26, HA431–439, and PA46–54 epitopes were also identified in primary human lung epithelial cells obtained from Lonza and infected for 18 h with IAV strain PR8 (SI Appendix, Fig. S15 A and B).
Fig. 1.
(A–C) Poisson detection plots of three M1 peptides extracted from affinity-purified HLA-A*02:01 complexes isolated from 5 million bronchial epithelial airway (BEAS-2B) cells infected with influenza strain A/Puerto Rico/8/1934. Black traces are extracted precursor ion chromatograms with the y scale being ion arrival events at the detector (counts) and the x scale, scan number (time). The blue trace is a Poisson chromatogram reflecting a likelihood of the peptide’s fragmentation pattern with units of scaled ion counts (SI Appendix, Outline of Poisson Segmented LC-DIAMS Method). Arrows mark the elution of the indicated peptide. (D) Elution mapping. Synthetic peptides of 31 targeted influenza epitopes were added to an extract of HLA-A2:01 bound peptides from 2.5 million A2+ peripheral blood mononuclear cells (PBMCs). Elution positions of major endogenous and synthetic peptides were recorded. When infected BEAS cells were analyzed, the elution positions of shared endogenous peptides and detected influenza epitopes were again recorded. Paired elution positions are plotted with the y axis as the scan position in the BEAS sample and the x axis, the position in PBMCs. Detected epitopes fall near the elution line.
Segmented LC-DIAMS Data also Can Be Searched Using Numerical Models.
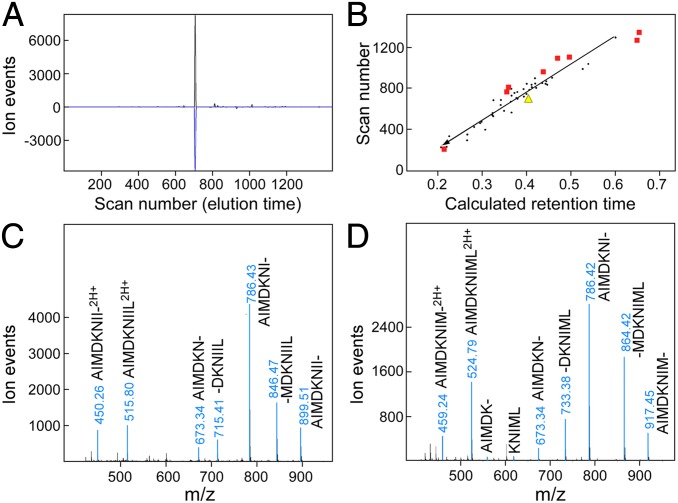
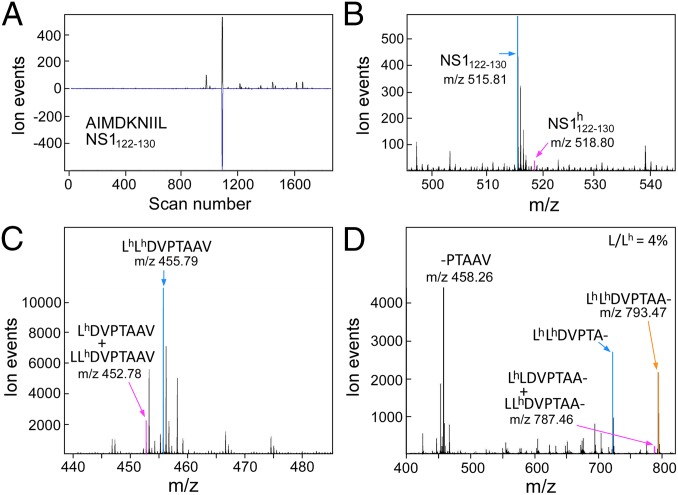
The LC-DIAMS format allows the detection of targets using elution and fragmentation patterns measured with synthetic peptides after the sample data are collected. Because peptide synthesis is a substantial bottleneck, we have found it profitable to use numerical predictors in a search for more intense ion signatures. Using triplets of b or y ions as a simple model of ion fragmentation and the Kangas/Petritis predictor (16, 17) for elution position, a peptide that was not in the target set, NS1122–130 (AIMDKNIIL), gave a strong detection signature (Fig. 2A). Given the substantial scatter of the Kangas/Petritis predictor (Fig. 2B), the confidence from the predicted elution position is only moderate. However, this is an intense ion signal and the corresponding band MS2 spectrum, dominated by AIMDKNIIL fragments, can be easily assigned (Fig. 2C). The feature is also observed in A/X-31 infections but not in naïve BEAS cells or A/Victoria/3/75 infection (SI Appendix, Fig. S16). Interestingly, the same exercise for the corresponding NS1122–130 peptide AIMDKNIML in the Victoria strain infection produces another intense detection signature with unambiguous assignment of its MS2 spectrum (Fig. 2D). Examining M1 peptides identified as A2 restricted by reverse immunology, (18) the highly conserved M159–66 peptide ILGFVFTL was also indicated by prediction and verified by fragment assignment (SI Appendix, Fig. S17). Computer predicted ion signatures and retention times can be used to identify intense signatures as well as to rank target candidates for synthesis and more definitive identification.
Fig. 2.
Detection without synthetic peptides is possible with strong ion signals. NS1122–130 peptides AIMDKNIIL from IAV strain A/Puerto Rico/8/1934 and AIMDKNIML from IAV strain A/Victoria/3/1975 are detected using numerical models. (A) Poisson detection chromatogram for AIMDKNIIL in sample of Fig. 1 uses a simple model of fragmentation (three b-type ions). (B) Elution mapping using normalized elution time (NET) Prediction Utility. The identified elution positions of some endogenous peptides (black dots) and detected IAV peptides (red squares) are plotted as a function of calculated retention time. Yellow triangle is scan position of XIC/Poisson coincidence in A plotted against predicted retention time. (C and D) MS2 spectra at elution peak for NS1122–130 peptides in PR8 and Victoria strain infections identifies each peptide by sequence ions.
Quantitative Analyses of IAV Peptides Shows the Immunodominant M158–66 Epitope Is Presented at Much Higher Copies per Infected Cell than Other IAV Peptides.
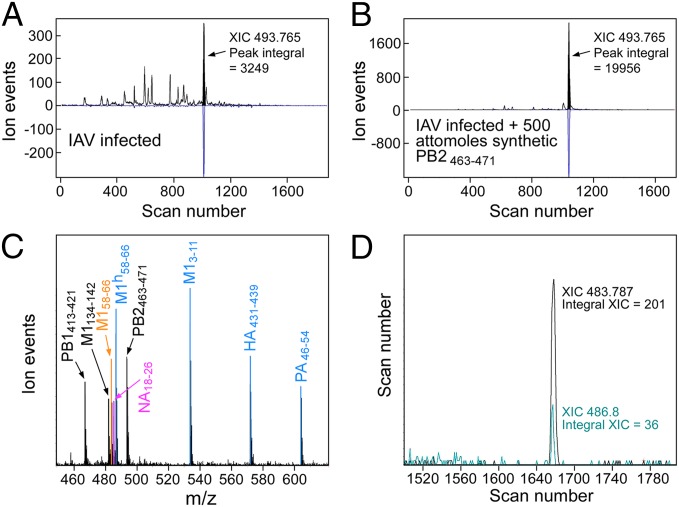
High purity synthetic analogs were obtained for eight detected IAV peptides (Fig. 3C). Although relative ion amplitudes by static nanospray MS ranged over factors of 2–3 when the peptides were directly diluted, similar relative amplitudes are not observed when these peptides are spiked into affinity-purified complexes at the acid elution step and analyzed by LC-DIAMS. In particular, surface exposure and poorly characterized electrospray factors strongly suppress the hydrophobic NA18–26 and M158–66 peptides. Optimal quantitation would use isotopically labeled analogs as internal standards for all peptides but considering expense and anticipating the discussion to follow, this was restricted to the M158–66 peptide. Quantitation using unlabeled peptides is described first. An infected sample was split into equal aliquots and 500 attomoles (amols) of the synthetic mixture added to one aliquot. Both samples were analyzed by LC-DIAMS and precursor ion amplitudes for both the IAV peptides and a set of endogenous peptides were determined. As the endogenous peptides are equally abundant in the split sample, measuring amplitude ratios provide a scan-dependent intensity normalization factor for the paired runs. Ion amplitudes for IAV peptides in both halves combined with the known 500 amols of synthetic peptide added to the second half can be translated into the moles present in infection. Divided by the number of infected cells in the sample recovers the copy number per cell and this is illustrated for PB2463–471 (Fig. 3 A and B). The method was extended to other IAV peptides (SI Appendix, Quantitation of IAV Peptides as Copies/Infected Cell).
Fig. 3.
Measuring copy numbers per cell after 18 h infection of BEAS cells by IAV strain A/X-31. (A and B) Sample of affinity beads with immobilized A2 complexes is split into equal portions of 1.67 million cells and 500 attomoles of PB2463–471 peptide added to one part. Both portions are run, with the intensity of major endogenous peptides normalizing the ion flux (e.g., variation due to position of the emitter tip). (A) The PB2 peptide in the first run has an area of 3,249 ion events. (B) Adding 500 attomoles PB2 peptide increases peak area to 19,956 events. From measuring endogenous peptide intensities, the A run is roughly twice as sensitive as B at this elution position. Hence, 500 attomoles in B gives a peak area of 18,332 (19,956–3,249/2), or infection produces 500 × 1,624/18,332 = 44 attomoles from 1.67 million cells. This corresponds to 16 copies per cell. (C) Nanospray MS of purified synthetic peptides used in the quantitation experiment. (D) Isotope-labeled analogs. Adding 500 attomoles of isotope-labeled M158–66 and measuring XIC peak areas (now distinguished by m/z) gives the amount of unlabeled peptide from infection as 500 × 201/36 = 2,792 attomoles or roughly 1,000 copies per cell.
The hydrophobic M158–66 peptide's ion signal is strongly suppressed, as adding 500 amols increased the observed ion signal by only a small fraction. Given the immunological significance of this peptide, an isotopic analog, 13C-labeled at the leucine in the third position (GILhGFVFTL), was synthesized and 500 amols added at the early acid elution step. Because isotope labeling does not impact ion suppression factors, but shifts the molecular mass by 6 Da, XIC peaks at m/z 483.8 and 486.8 (doubly charged ions) coelute (Fig. 3D) at a position coincident with their Poisson detection. The ratio of XIC peak areas × 500 amols gives 2,800 amols M158–66 in 1.7 million cells or a copy number of roughly 1,000 per cell. Quantitation of M158–66 using isotope labels is also possible by MS3 and we found 860 copies per cell for A/Victoria/3/75 infection of BEAS cells. Quantitation results for IAV A/X-31 infection are shown in SI Appendix, Table S1 with data and details provided in SI Appendix, Quantitation of IAV Peptides as Copies/Infected Cell).
Human Monocyte-Derived DCs Do Not Cross-Present Influenza Proteins.
DCs can endocytose exogenous proteins, which are cleaved into peptide fragments and loaded on MHC class I molecules (MHCI) for cross-presentation to CD8 T cells. Cross-presentation by migratory CD103+ DCs is thought to play a critical role in T-cell priming during IAV infection (19). In vitro culture of blood monocytes with GM-CSF/IL-4 generates phagocytic cells (moDCs) sharing morphological and functional properties of conventional CDllc+ DCs, including the capacity to cross-present antigens to T cells. Although blood monocytes are not thought to be precursors of lung resident DCs, cross-presentation by moDCs has been extensively studied and they are readily accessible on sample scales of a million cells for MS analyses. Infecting A2− BEAS cells for 18 h induces a fraction of the cells to become apoptotic and lose adherence; these “floaters” can be collected and cultured with A2+ moDCs. Examining the IAV peptides displayed by the moDCs gave the same set of peptides as direct IAV infection on A2+ BEAS cells. To minimize direct viral infectivity of the floaters, they were UV irradiated before culture with moDCs. Comparable UV irradiation of virus blocked infection of A2+ BEAS cells as assayed by M1 staining (SI Appendix, Fig. S18A) and MS (SI Appendix, Fig. S18B). However, coculture of irradiated virus with moDCs induced their development and gave both positive surface HA staining with mAbs (SI Appendix, Fig. S19 A and B) and the same set of IAV peptides found on infected BEAS cells (SI Appendix, Fig. S18B, data for NS1122–130 shown). To distinguish the fraction of presented IAV peptides that reflected protein synthesis in the moDC from the fraction that reflected processing exogenous protein, A2− LAZ 468 cells were grown in SILAC media with isotope-labeled leucine and arginine. LAZ cells were used in this experiment as BEAS cells did not grow well in the media. Fig. 4 C and D shows the high degree of heavy isotope incorporation by MS and MS2 signatures, respectively, of the prominent endogenous GILT peptide from SILAC-labeled A2+ LAZ 509 cells. Eighteen-hour coculture in normal media of SILAC-labeled, 18 h IAV A/X-31 infected, UV irradiated, A2− LAZ 468 cells with A2+ moDCs produced dominantly unlabeled IAV peptides (Fig. 4 A and B and SI Appendix, Fig. S20), consistent with new protein translation. The very minor fraction of isotope-labeled peptide in Fig. 4B may also arise from residual SILAC media and/or metabolism of endocytosed heavy protein and not necessarily cross-presentation.
Fig. 4.
Human monocyte-derived DCs generate primarily light NS1122–130 peptide after culture with UV irradiated, IAV A/X-31 infected, SILAC-labeled, A2− BEAS cells. (A) Poisson detection chromatogram for light NS1122–130 identifies elution at scan 1,088. (B) MS at scan 1,088. The heavy form at m/z 518.8 is about 5% of the light form at m/z 515.8. (C) SILAC labeling of an A2+ LAZ cell line produces dominantly heavy-labeled peptides as shown by the low energy (C) precursor spectrum and high energy (D) fragment spectrum of the gamma-IFN–inducible lysosomal thiol reductase peptide, an almost universally observed and abundant endogenous peptide with HLA-A2+ cells.
Poor Functional Avidity of Polyclonal M158–66-Specific CD8 T Cells.
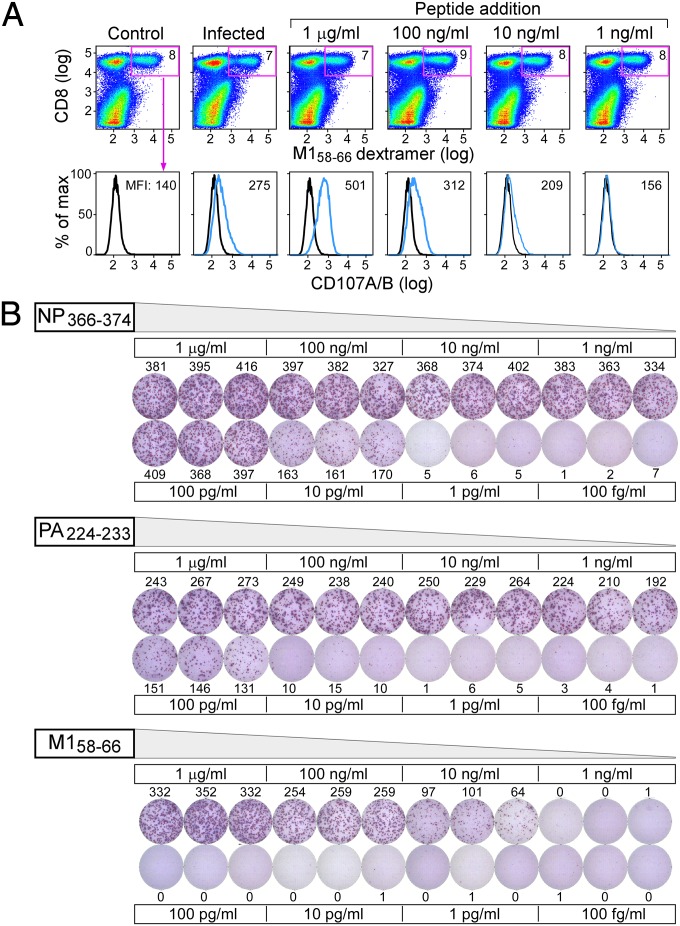
Given the abundant M158–66 expression on infected lung epithelial cells and the conserved nature of this epitope across IAV strains, this peptide might represent an ideal focus for CTL-mediated protection in A2 individuals. However, whereas CD8 T cells with this specificity are plentiful and persistent in peripheral blood mononuclear cells (PBMCs) of IAV-experienced individuals, they do not provide absolute protective heterosubtypic immunity. To assess whether this disparity might be a consequence of poor functional avidity to this epitope, two types of experiments were performed. In the first, polyclonal short-term in vitro M158–66–stimulated CD8 T cells were expanded and assessed for their ability to undergo granular exocytosis when exposed to A2+ BEAS epithelial cells infected with PR8, or in comparison, when exposed to uninfected A2+ BEAS cells incubated with different concentrations of M158–66 peptide. The CD8+ M158–66 dextramer subset was gated and CD107A/B (LAMP1/2) surface expression associated with lytic granule exocytosis determined by FACS analysis. As shown in Fig. 5A, none of the antigen-specific T cells (7–8% of total) exposed to uninfected cells expressed CD107A/B, whereas a low level staining of a fraction of the T cells was observed after exposure to IAV-infected cells. By contrast, virtually all dextramer+ CD8 T cells were CD107A/B+ at 1 μg/mL M158–66 peptide loading, less at lower concentrations, and none detected at 1 ng/mL. Connecting IAV infection with loading concentration, MS3 quantitation (SI Appendix, Fig. S21) determined 3.6 copies of M158–66/cell at 1 ng/mL and 28 copies at 10 ng/mL loading. By extrapolation, this scale is consistent with the other quantifications of infection: postinfection, many hundreds of M158–66 copies per cell are observed, whereas the copy number required for the majority of CD8 M158–66-specific T cells to degranulate (1 μg/mL loading) is several thousand. These data imply that the functional avidity of the bulk polyclonal human peripheral blood T cells to M158–66 is poor.
Fig. 5.
Poor functional avidity of M158–66-specific T cells. (A) An M158–66-specific T-cell line was generated from an HLA-A02 positive donor previously identified to have high frequency influenza A M158–66 memory responses. After 2 wk of in vitro peptide stimulations, the resulting T-cell line was plated with uninfected (control), IAV A/PR8/34 infected, or uninfected but peptide pulsed (concentrations indicated) BEAS-2B HLA-A02 positive cells to assess cytotoxic T-cell responses. CD8 and M158–66 dextramer positive populations were gated (percentage given in Right Upper quadrant of each graph in the Top row) and CD107A/B staining on the M158–66-specific T cells were determined as shown in the second row. (B) HLA-A02 transgenic mice were infected with a sublethal dose of influenza A/PR8/34 virus to determine IAV peptide responses. Splenocytes were stimulated 3 wk postinfection with various concentrations of H2-Db restricted NP366–374 and PA224–236 and HLA-A2 restricted M158–66 peptides to test T-cell avidity in an IFN gamma Elispot assay.
To exclude the possibility that this poor responsiveness was a consequence of multiple IAV exposures, a second type of experiment was performed. We examined the functional avidity of M158–66-specific CD8 T cells from A2 transgenic mice (C57BL/6-Tg HLA-A2.1) (Jackson Labs) stimulated with various doses of M158–66 3 wk after a single PR8 infection using IFN-γ Elispot assay. For comparison, the immunodominant H2-Db-restricted CD8 T-cell responses against the NP366–374 9-mer and PA224–233 10-mer were also assessed (Fig. 5B). As with the granular exocytosis assay in the human, the CD8 response in HLA-A2.1 transgenic mouse was no longer detected at 1 ng/mL of M158–66. In contrast, the Db-restricted responses titered to 100- to 1,000-fold lower concentrations. These data show that the human and mouse T-cell repertoires responding to M158–66 are dominated by low-avidity T cells (20). In line with previous reports, we also found robust responses to M158–66 among 7 of the 10 HLA-A02 donors studied, whereas 2 and 1 individuals responded to PB1413–421 and PB1501–509, and none responded significantly to M13–11 or PA46–54 (SI Appendix, Fig. S22). Hence epitopes with both full conservation as well as limited variability among IAV strains (SI Appendix, Fig. S23) are expressed on lung epithelium but engender little or no T-cell response.
Discussion
Here we identify IAV-derived T-cell epitopes on lung epithelium associated with HLA-A*02:01 using ultralow flow chromatography, DIAMS and a Poisson metric (14) for target detection. In contrast to previous MS efforts to identify IAV peptides in association with A2 (21, 22), the highly conserved M158–66 peptide GILGFVFTL was not only detected, but shown to be abundantly presented by infected lung epithelium. Previously unrecognized IAV epitopes were identified and characterized by their copy number on infected cells. In comparing reverse immunology to MS for identifying IAV epitopes, the overall concordance is poor with the exception of M158–66. The main problem is inconsistent identification by reverse immunology. The M159–66, PA46–54, PB1413–421, and NS1122–130 peptides identified by MS are only sporadically identified in the T-cell responses of A2 donors. None of the other MS-identified epitopes in SI Appendix, Table S1 were functionally detected (18, 20, 23). In a few examples, peptides with positive functional responses in blood were also in the set examined by MS but were not detected. Whether these discordances represent manifestations of MS detection limits or cross-reactivities of T cells to peptides presented via the same or other alleles is yet to be determined. In contrast, MS identification is robust: over replicates, across moDCs, primary human lung tissue, epithelial (BEAS), and B-cell lines (LAZ) and, where viral strains permitted, the same epitopes were identified or excluded in a predictable manner.
Of the nine detected peptides, PA46–54, M13–11, PB1413–421, and M158–66 sequences are ≥99% conserved from analysis of more than 10,000 human influenza A virus strains from 1902 to 2014 globally (SI Appendix, Table S1 and Fig. S23). Whereas the other six peptides are not fully conserved, variability is still finite, with three variants each covering the HA431–439 stem region segment and PB2463–471 and eight peptide variants covering NA18–26 and NS1122–130. M1134–142 global strain variability is accounted for by three peptides but the RMGAVTTES variant found in 13% is predicted to be a very weak A2 binder, potentially permitting viral escape. HLA-A*02:01 accounts for up to 40% of the Caucasian population and if HLA binding can be extended to other alleles in the A2 supertype (24), these epitopes would be excellent candidates for vaccine inclusion. Aside from M158–66, little is known about the TCR repertoire that could target these pMHC. Immunodominance of the M158–66 response to IAV has been observed in both HLA-A2 humans and HLA-A2.1 transgenic mice (20, 25). If the immunodominant response is of low functional avidity, effective targeting and destruction of IAV-infected epithelium is not possible despite the large number of responding T cells, as is the case with M158–66. The utility of this epitope is ill-advised, unless vaccination can expand a rare, high-avidity M158–66 T-cell specificity. By contrast, because T cells with high functional avidity can recognize a single or several copies of a given pMHC (26), the other detected pMHC complexes listed in SI Appendix, Table S1 are potential vaccine candidates, with the possible exception of the low-copy M13–11. Both mouse and nonhuman primate models of IAV infection as well as immune correlates of human protection against naturally acquired 2009 H1N1 pandemic infection argue that cross-reactive CD8 T cells are important (10). These data showing reduction in symptoms of infection and viral shedding are also consistent with earlier experimental IAV challenge in human subjects lacking preexisting antibodies (9).
An in vivo study demonstrated that migratory CD103+ DCs phagocytose cellular material containing GFP-labeled viral gene products without themselves becoming overtly infected, a response dependent on type I IFN signaling (19). Migratory DCs are assumed to prime T cells in lymph nodes through protein cross-presentation. However, CD8 T-cell priming through proteasome dependent cross-presentation of cell-associated viral antigens seems to face an impossible stoichiometry in several respects: proteomic analyses of IAV infection cannot detect any influenza proteins against the overwhelming load of endogenous proteins in an infected cell (27); an immune focus on recent translation by defective ribosomal product presentation (28) is inoperative if the infected and ingested cell proteome is uniformly subjected to processing/presentation; and all cross-presented peptides would be diluted by the DC's endogenous MHCI display. Perhaps chaperones might bind recently translated proteins in the stressed and dying cell and provide targets for specific endosomal translocation into the cytosol and/or the DC may somehow limit self-presentation in favor of cross-presentation as mechanisms to explain cross-presentation. Using in vitro GM-CSF/IL-4 differentiated moDCs and isotope-labeled apoptotic cells we examined the evidence for DC cross-presentation as isotope labeling unambiguously identifies the source of an IAV epitope as exogenous or endogenous. Both CD103+ DCs isolated from näive lung (19) and immature moDCs can be infected in vitro. As phagocytosis of IAV-infected cells induced the immature moDCs to mature, a level of direct presentation in our experiment was expected. To maximize cross-presentation over direct presentation, moDCs were cultured with a 10-fold excess of UV-irradiated apoptotic cells at a UV dose that rendered virus and virally infected apoptotic cells noninfectious to epithelial cells, as assayed by FACS and MS (SI Appendix, Fig. S18). After 18 h coculture of moDCs with infected and SILAC-labeled apoptotic cells we found by MS very little evidence for protein cross-presentation of IAV peptides. There is no doubt that DCs can take up protein and cross-present MHCI peptides if the endocytosed protein is abundant (e.g., protein-coated micro beads) but we did not find evidence in our experimental system for processes that circumvent the stoichiometric paradox and make cross-presentation conceivable for cell-associated viral antigens. Although we cannot exclude the existence of cross-presentation in other DC subsets or the requirement of T-cell–DC interactions to induce cross-presentation, the current data suggest that rather than protein cross-presentation, migratory DCs permit infection (perhaps limited in nature) or RNA translation from phagocytosed material as a significant means to provide epitopes for CD8 T-cell priming.
Our data imply that the immunodominant CD8 T-cell response to the highly conserved M158–66 peptide GILGFVFTL does not provide sterilizing protective immunity to IAV in HLA-A*02:01 individuals because of generic low functional avidity. Conventional wisdom argues that vaccine-induced immune responses should focus on viral segments that are conserved and where mutations therein would impose high fitness costs (29, 30). Conservation of potential T-cell epitopes in a viral protein sequence otherwise usually implies that the virus is under no immune selection pressure, as would be the case if a potential epitope were not processed and presented. In contrast, the invariant M158–66 is displayed on the cell surface. Most strikingly, alanine mutations at eight of the nine positions of this viral peptide are well tolerated with respect to viral fitness when recombinant viruses are generated, eliminating that basis for conservation (31). Notwithstanding, introducing single amino acid changes at most positions in the M158–66 peptide diminished M158–66-specific CTL recognition. These paradoxical data collectively argue that in the case of A2 individuals, a significant human subpopulation, high M158–66 pMHCI copy number may be maintained by IAV to induce nonprotective low-avidity M158–66-specific CTLs, precluding deployment of effective high-avidity sentinel CD8 TRM in barrier tissues and/or CD8 T effector memory (TEM) in the lung parenchyma more generally. Although myriad CTL evasion mechanisms have been described for viruses including IAV (32, 33), this mechanism involving an immunodominant epitope has not been previously uncovered. One could assume that equivalent forms of viral chicanery will involve other conserved IAV peptides bound to distinct HLA alleles.
Materials and Methods
MS3 Experiments.
MS3 analysis was done as previously described (14) using an ABSciex QTrap 4000 MS.
LC-DIAMS Experiments.
The LC was an Acuity UPLC using an in-house, 15-cm long, 20 μm inner diameter (ID) C18-modified polystyrene-divinylbenzene (PS-DVB) monolith with a 1.5-cm long, 50 μm inner diameter (ID) C10-modified PS-DVB monolith as a precolumn. Flow rate through the column was measured at 10 nL/min. The LC-DIAMS acquisition was done with an ABSciex Elite quadrupole-TOF.
Additional detailed information is provides in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant UO1 AI90043 as well as Dana-Farber Cancer Institute institutional funds provided to the Cancer Vaccine Center.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: MS data has been deposited to ImmPort, https://immport.niaid.nih.gov, as part of HIPC effort.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423482112/-/DCSupplemental.
References
- 1.Dormitzer PR, et al. Influenza vaccine immunology. Immunol Rev. 2011;239(1):167–177. doi: 10.1111/j.1600-065X.2010.00974.x. [DOI] [PubMed] [Google Scholar]
- 2.Drake JW. Rates of spontaneous mutation among RNA viruses. Proc Natl Acad Sci USA. 1993;90(9):4171–4175. doi: 10.1073/pnas.90.9.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heaton NS, Sachs D, Chen CJ, Hai R, Palese P. Genome-wide mutagenesis of influenza virus reveals unique plasticity of the hemagglutinin and NS1 proteins. Proc Natl Acad Sci USA. 2013;110(50):20248–20253. doi: 10.1073/pnas.1320524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doherty PC, Turner SJ, Webby RG, Thomas PG. Influenza and the challenge for immunology. Nat Immunol. 2006;7(5):449–455. doi: 10.1038/ni1343. [DOI] [PubMed] [Google Scholar]
- 5.Krammer F, Palese P, Steel J. Advances in universal influenza virus vaccine design and antibody mediated therapies based on conserved regions of the hemagglutinin. Curr Top Microbiol Immunol. 2015;386:301–321. doi: 10.1007/82_2014_408. [DOI] [PubMed] [Google Scholar]
- 6.Miller MS, Palese P. Peering into the crystal ball: Influenza pandemics and vaccine efficacy. Cell. 2014;157(2):294–299. doi: 10.1016/j.cell.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 7.Kumar P, Khanna M, Kumar B, Rajput R, Banerjea AC. A conserved matrix epitope based DNA vaccine protects mice against influenza A virus challenge. Antiviral Res. 2012;93(1):78–85. doi: 10.1016/j.antiviral.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 8.Tan AC, et al. The design and proof of concept for a CD8(+) T cell-based vaccine inducing cross-subtype protection against influenza A virus. Immunol Cell Biol. 2013;91(1):96–104. doi: 10.1038/icb.2012.54. [DOI] [PubMed] [Google Scholar]
- 9.McMichael AJ, Gotch FM, Noble GR, Beare PA. Cytotoxic T-cell immunity to influenza. N Engl J Med. 1983;309(1):13–17. doi: 10.1056/NEJM198307073090103. [DOI] [PubMed] [Google Scholar]
- 10.Sridhar S, et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med. 2013;19(10):1305–1312. doi: 10.1038/nm.3350. [DOI] [PubMed] [Google Scholar]
- 11.Doherty PC, Kelso A. Toward a broadly protective influenza vaccine. J Clin Invest. 2008;118(10):3273–3275. doi: 10.1172/JCI37232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slütter B, Pewe LL, Kaech SM, Harty JT. Lung airway-surveilling CXCR3(hi) memory CD8(+) T cells are critical for protection against influenza A virus. Immunity. 2013;39(5):939–948. doi: 10.1016/j.immuni.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu T, et al. Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J Leukoc Biol. 2014;95(2):215–224. doi: 10.1189/jlb.0313180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reinhold B, Keskin DB, Reinherz EL. Molecular detection of targeted major histocompatibility complex I-bound peptides using a probabilistic measure and nanospray MS3 on a hybrid quadrupole-linear ion trap. Anal Chem. 2010;82(21):9090–9099. doi: 10.1021/ac102387t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundegaard C, Lund O, Nielsen M. Accurate approximation method for prediction of class I MHC affinities for peptides of length 8, 10 and 11 using prediction tools trained on 9mers. Bioinformatics. 2008;24(11):1397–1398. doi: 10.1093/bioinformatics/btn128. [DOI] [PubMed] [Google Scholar]
- 16.Monroe M. 2013. Normalized Elution Time (NET) Prediction Utility. Available at panomics.pnnl.gov. Accessed January 23, 2015.
- 17.Petritis K, et al. Use of artificial neural networks for the accurate prediction of peptide liquid chromatography elution times in proteome analyses. Anal Chem. 2003;75(5):1039–1048. doi: 10.1021/ac0205154. [DOI] [PubMed] [Google Scholar]
- 18.Assarsson E, et al. Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. J Virol. 2008;82(24):12241–12251. doi: 10.1128/JVI.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helft J, et al. Cross-presenting CD103+ dendritic cells are protected from influenza virus infection. J Clin Invest. 2012;122(11):4037–4047. doi: 10.1172/JCI60659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan AC, La Gruta NL, Zeng W, Jackson DC. Precursor frequency and competition dictate the HLA-A2-restricted CD8+ T cell responses to influenza A infection and vaccination in HLA-A2.1 transgenic mice. J Immunol. 2011;187(4):1895–1902. doi: 10.4049/jimmunol.1100664. [DOI] [PubMed] [Google Scholar]
- 21.Testa JS, et al. MHC class I-presented T cell epitopes identified by immunoproteomics analysis are targets for a cross reactive influenza-specific T cell response. PLoS ONE. 2012;7(11):e48484. doi: 10.1371/journal.pone.0048484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wahl A, Schafer F, Bardet W, Hildebrand WH. HLA class I molecules reflect an altered host proteome after influenza virus infection. Hum Immunol. 2010;71(1):14–22. doi: 10.1016/j.humimm.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gianfrani C, Oseroff C, Sidney J, Chesnut RW, Sette A. Human memory CTL response specific for influenza A virus is broad and multispecific. Hum Immunol. 2000;61(5):438–452. doi: 10.1016/s0198-8859(00)00105-1. [DOI] [PubMed] [Google Scholar]
- 24.Zhang GL, et al. MULTIPRED2: A computational system for large-scale identification of peptides predicted to bind to HLA supertypes and alleles. J Immunol Methods. 2011;374(1-2):53–61. doi: 10.1016/j.jim.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishizuka J, et al. The structural dynamics and energetics of an immunodominant T cell receptor are programmed by its Vbeta domain. Immunity. 2008;28(2):171–182. doi: 10.1016/j.immuni.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 26.Sykulev Y, Joo M, Vturina I, Tsomides TJ, Eisen HN. Evidence that a single peptide-MHC complex on a target cell can elicit a cytolytic T cell response. Immunity. 1996;4(6):565–571. doi: 10.1016/s1074-7613(00)80483-5. [DOI] [PubMed] [Google Scholar]
- 27.Kroeker AL, Ezzati P, Coombs KM, Halayko AJ. Influenza A infection of primary human airway epithelial cells up-regulates proteins related to purine metabolism and ubiquitin-related signaling. J Proteome Res. 2013;12(7):3139–3151. doi: 10.1021/pr400464p. [DOI] [PubMed] [Google Scholar]
- 28.Antón LC, Yewdell JW. Translating DRiPs: MHC class I immunosurveillance of pathogens and tumors. J Leukoc Biol. 2014;95(4):551–562. doi: 10.1189/jlb.1113599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferguson AL, et al. Translating HIV sequences into quantitative fitness landscapes predicts viral vulnerabilities for rational immunogen design. Immunity. 2013;38(3):606–617. doi: 10.1016/j.immuni.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goulder PJ, Watkins DI. HIV and SIV CTL escape: Implications for vaccine design. Nat Rev Immunol. 2004;4(8):630–640. doi: 10.1038/nri1417. [DOI] [PubMed] [Google Scholar]
- 31.Berkhoff EG, et al. Functional constraints of influenza A virus epitopes limit escape from cytotoxic T lymphocytes. J Virol. 2005;79(17):11239–11246. doi: 10.1128/JVI.79.17.11239-11246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horst D, Verweij MC, Davison AJ, Ressing ME, Wiertz EJ. Viral evasion of T cell immunity: Ancient mechanisms offering new applications. Curr Opin Immunol. 2011;23(1):96–103. doi: 10.1016/j.coi.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 33.Schmolke M, García-Sastre A. Evasion of innate and adaptive immune responses by influenza A virus. Cell Microbiol. 2010;12(7):873–880. doi: 10.1111/j.1462-5822.2010.01475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.